Abstract
Purpose:
To develop T2-weighted (T2w) time-resolved (TR) 4DMRI reconstruction technique with higher soft-tissue contrast for multiple breathing-cycle motion assessment by building a super-resolution (SR) framework using the T1w TR-4DMRI reconstruction as guidance.
Methods:
The multi-breath T1w TR-4DMRI was reconstructed by deforming a high-resolution (HR:2x2x2mm3) volumetric breath-hold (BH, 20s) 3DMRI image to a series of low-resolution (LR:5x5x5mm3) 3D cine images at a 2Hz frame rate in free-breathing (FB, 40s) using an enhanced Demons algorithm, namely [T1BH→FB] reconstruction. Within the same imaging session, respiratory-correlated (RC) T2w 4DMRI (2x2x2mm3) was acquired based on an internal navigator to gain HR T2w (T2HR) in three states (full exhalation and mid and full inhalation) in ~5 minutes. Minor binning artifacts in the RC-4DMRI were automatically identified based on voxel intensity correlation (VIC) between consecutive slices as outliers (VIC<VICmean-σ) and corrected by deforming the artifact slices to interpolated slices from the adjacent slices iteratively until no outliers were identified. A T2HR image with minimal deformation (<1cm at the diaphragm) from the T1BH image was selected for multi-modal B-Spline DIR to establish the T2HR-T1BH voxel correspondence. Two approaches to reconstruct T2w TR-4DMRI were investigated: (A) T2HR→[T1BH→FB]: to deform T2w HR to T1w BH only as T1w TR-4DMRI was reconstructed, and combine the two displacement vector fields (DVFs) to reconstruct T2w TR-4DMRI, and (B) [T2HR←T1BH]→FB: to deform T1w BH to T2w HR first and apply the deformed T1w BH to reconstruct T2w TR-4DMRI. The reconstruction times were similar, 8-12 minutes per volume. To validate the two methods, T2w- and T1w-mapped 4D XCAT digital phantoms were utilized with three synthetic spherical tumors (ϕ=2.0, 3.0 and 4.0cm) in the lower or mid lobes as the ground truth to evaluate the tumor location (the center of mass, COM), size (volume ratio, %V), and shape (Dice index). Six lung cancer patients were scanned under an IRB-approved protocol and the T2w TR-4DMRI images reconstructed from the two methods were compared based on the preservation of the three tumor characteristics. The local tumor-contained image quality was characterized using the VIC and structure similarity (SSIM).
Results:
In the 4D digital phantom, excellent tumor alignment after T2HR-T1HR DIR is achieved: ΔCOM=0.8±0.5 mm, %V=1.06±0.02, and Dice=0.91±0.03, in both deformation directions using the DIR-target image as the reference. In patients, binning artifacts are corrected with improved image quality: average VIC increases from 0.92±0.03 to 0.95±0.01. Both T2w TR-4DMRI reconstruction methods produce similar tumor alignment errors ΔCOM=2.9±0.6 mm. However, method B ([T2HR←T1BH]→FB) produces superior preservation in preserving more T2w tumor features with a higher %V=0.99±0.03, Dice=0.81±0.06, VIC=0.85±0.06, and SSIM=0.65±0.10 in the T2w TR-4DMRI images.
Conclusion:
This study has demonstrated the feasibility of T2w TR-4DMRI reconstruction with high soft-tissue contrast and adequately-preserved tumor position, size, and shape in multiple breathing cycles. The T2w-centric DIR (method B) produces a superior solution for the SR-based framework of T2w TR-4DMRI reconstruction with highly preserved tumor characteristics, including position, size, and shape, which is useful for tumor delineation and motion management in radiation therapy.
Keywords: T2w Time-resolved 4DMRI, Super-resolution image reconstruction, Deformable image registration (DIR), Radiotherapy motion simulation, Radiotherapy planning
INTRODUCTION
Time-resolved four-dimensional magnetic resonance imaging (TR-4DMRI) provides multi-breathing cycles, which can better characterize respiratory-induced tumor motion that may be irreproducible due to breathing irregularities1–3. Substantial tumor motion variation has recently been evidenced under clinical conditions in lung and liver patients during radiotherapy treatment delivery, deviating the motion assessment in treatment simulation and planning4,5. Tumor motion variation may have a greater impact on hypo-fractional stereotactic body radiotherapy (SBRT) than conventional treatments, due to loss of the statistical averaging power in 1-5 treatment fractions as well as the sharp dose fall-off outside of the planning tumor volume (PTV). Existing T2-weighted (T2w) respiratory-correlated (RC) 4DMRI, although it provides higher soft-tissue contrast than T1w 4DMDI for target delineation and organ segmentation6–11, only produces a motion snapshot with one composite breathing cycle with little indication of tumor motion variation over a longer time period during treatment delivery. Therefore, T2w RC-4DMRI is incapable of providing multi-breath tumor motion, which has become a clinical concern in managing patient motion variation due to breathing irregularities.
Direct acquisition of dynamic volumetric 4DMRI via 3D cine scan, although preferable, has reached its fundamental limit to achieve high resolution in both space and time, due to slow MR relaxation and large clinical field of view, even using the state-of-the-art technologies, such as parallel imaging and compressed sensing12–14. To overcome this clinical problem, different approaches have been developed with a priori knowledge of a patient anatomy/motion model and fast pulse sequence and acquisition with short TE/TR, such as T1w or balanced steady-state free precession. Harris et al. have developed a 2D-cine-guided method to reconstruct TR-4DMRI through motion modeling based on RC-4DMRI using deformable image registration (DIR) and principal component analysis (PCA)15. Stemkens et al have also developed a similar technique for abdominal cancer but using radial acquisition and internal navigator in RC-4DMRI acquisition for DIR/PCA modeling16. These methods are 2D-cine-guided reconstruction and the quality depends on the motion model derived from RC-4DMRI. Li et al reported a 3D-cine-guided, super-resolution (SR) method to reconstruct T1w TR-4DMRI using DIR to combine two complementary image sets: adequate temporal-resolution free-breathing (FB: 5x5x5mm3 at 2Hz) 3D cine and high spatial-resolution breath-hold (BH: 2x2x2mm3) 3D static images17–19. Recently, this SR approach has also been implemented using cascade deep learning technique with limited training data for MR-guided adaptive radiotherapy20. The SR approach does not assume motion periodicity, and therefore can handle both regular and irregular motions, including breathing irregularities and non-respiratory motions. However, clinically-preferred MR contrasts, such as T2w, may be needed to better visualize soft-tissue for tumor and OAR delineation6,8,21–25, demanding a generic SR reconstruction framework for the TR-4DMRI modality.
In this study, we investigate the feasibility of a new SR approach to reconstruct T2w TR-4DMRI using T1w TR-4DMRI as the guidance. This novel SR approach combines three sets of MR images, rather than two in previous studies17–19. First, high-resolution (HR) T2w MRI was acquired using navigator-triggered 3-bin RC-4DMRI scan within the same imaging session of T1w HR BH and low-resolution (LR) FB 3D cine acquisitions. Second, mild binning artifacts in T2w MRI were automatically identified and corrected based on voxel intensity correlation (VIC) among adjacent slices using 2D DIR. Third, the voxel correspondence between T2wHR and T1wBH was established by DIR in either T2w→T1w or T2w←T1w directions. Fourth, either original or deformed T1w HR was used to deform to LR 3D cine. Last, the displacement vector fields (DVFs) in the above processes were combined and applied to the T2w HR image for T2w TR-4DMRI reconstruction. T2w and T1w-mapped 4D digital phantoms with a 3.0cm motion range at the diaphragm and three spherical tumors (2.0, 3.0, and 4.0 cm in diameter) were created as the ground truth to validate this new approach, focusing on the preservation of tumor position, volume, and shape. Six lung cancer patients were scanned under an IRB-approved protocol to test these tumor characteristics in the SR-reconstructed T2w multi-breath TR-4DMRI images. As many acronyms are used in the manuscript, a list of abbreviation is provided in the Appendix.
METHODS AND MATERIALS
General description of the T2w TR-4DMRI reconstruction framework
The reconstruction of T2w TR-4DMRI is based on the SR approach to combine three image sets, namely T2w HR, T1w BH and FB 3D cine images of a patient that were acquired in the same imaging session. Five steps were involved from acquisition to reconstruction of T2w TR-4DMRI (1) to acquire T2w HR images using the navigator-triggered RC-4DMRI with 3 bins and voxel size of 2x2x2 mm3, together with T1w BH and FB 3D cine images, (2) to select one T2w image closest to the T1w BH image (<1cm displacement at the diaphragm) for an iterative correction of binning artifacts using the outliers of the voxel intensity correlation (VIC < - σ, σ ≈ 0.02) for identification and 2D DIR to interpolated slices for correction, (3) to establish the T2-T1 voxel correspondence by deforming the corrected T2w HR image to T1w BH image (3a) or vice versa (3b), using multi-modal, B-Spline 3D DIR, (4) to establish the static-cine voxel correspondence using the original or deformed T1w BH image to map HR texture to 3D cine, and (5) to combine all DVFs in step (3) to (4) and apply it to T2w HR image to build the T2w TR-4DMRI. The workflow is shown in Fig. 1.
Figure 1.
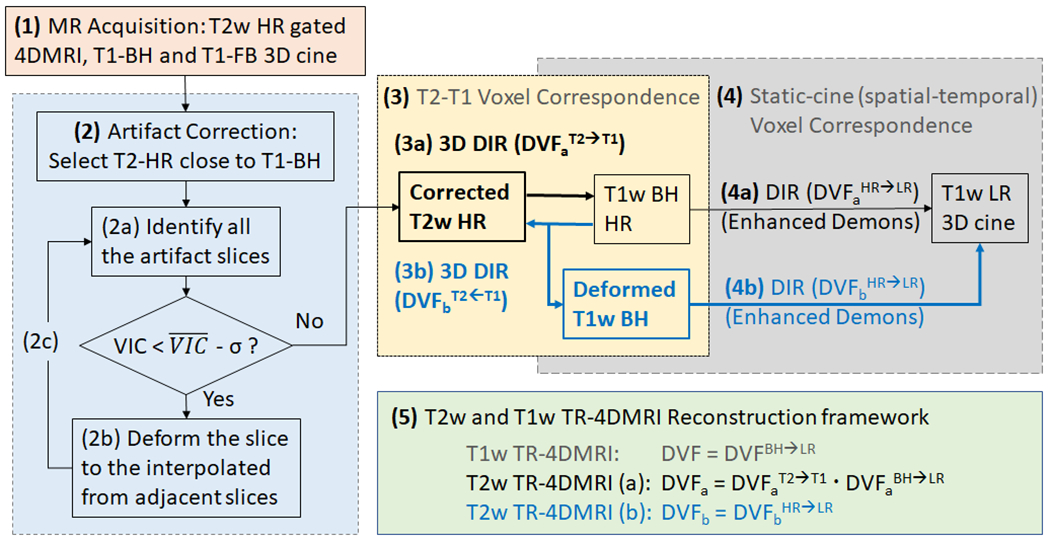
The workflow of T2w TR-4DMRI image reconstruction using two DIR directions [(3a) and (3b)]. (1) Acquisition of T2w high-resolution (HR), T1w breath-hold (BH), and T1w free-breathing (FB) images, (2) Correction of binning artifacts in T2w image using voxel intensity correlation (VIC) as criteria, (3) Two approaches (a and b) to establish T2-T1 voxel correspondence, (4) Deformation (a and b) to map T1w HR to T1w 3D cine images, and (5) Use of combined displacement vector fields (DVFs) for T2w TR-4DMRI reconstruction.
Acquisition of T2w and T1w MRI patient images
Both T2w and T1w MRI images were acquired on a 3-Tesla MRI scanner (3T Ingenia, Philips Healthcare, Amsterdam, the Netherlands) under an IRB-approved protocol. Six lung cancer patients were scanned in a body mold with both arms up for all MR acquisitions in the coronal direction. In brief, high-resolution (HR: 2x2x2 mm3) T2w RC-4DMRI was acquired using a pulse sequence of a single-shot, turbo spin-echo with echo/repetition time 80/5000-7000 ms and flip angle 90°, together with SENSE (2.0) and partial Fourier (0.7) for acceleration. An MR navigator box was placed at the right diaphragm dome to serve as an internal respiratory surrogate to trigger a prospective RC-4DMRI acquisition. Three respiratory bins were utilized with a narrow acquisition amplitude level (window) and the scanning time was about 5 minutes. More T2w RC-4DMRI acquisition details were described before8.
T1w 3D cine images in BH (HR: 2x2x2 mm3) and FB (LR: 5x5x5 mm3, 2Hz) were acquired using multi-shot, fast turbo field echo with echo/repetition time 1.9/4.2 ms and flip angle 15°, together with SENSE (4 for HR and 6 for LR) and partial Fourier (0.8) approximation. A BH scan took 20 s and the FB scan lasted 40 s at a 2Hz frame rate. The field of view was set the same for T1w and T2w scans, covering the full lungs and liver. More details of BH and FB 3D cine MR acquisition can be found elsewhere17.
Identification and correction of minor binning artifacts in T2w RC-4DMRI
A binning artifact correction method was developed in MatLab (MathWorks, MA) to remove minor binning artifacts in the navigator-triggered RC-4DMRI. A small, graduate tissue change across adjacent slices (spacing = 2.0 mm) was assumed and quantified by the VIC index. Negative peaks (sudden drop from the mean value) in the VIC plot as a function of slice number suggested high local tissue discontinuity of images, likely caused by binning artifacts26. In other words, a missorted slice would cause large anatomic discontinuity, leading a sudden VIC drop outside the 68% distribution range (). The VIC was defined as:
| (1) |
where the Im and Is denote the voxel intensity of the moving and static images, respectively. The cov is the covariance and σ is the standard deviation (roughly ΔVIC ≈ 0.02). To correct it, the low-VIC slices were deformed to linearly-interpolated slices from the adjacent slices using 2D DIR. The correction was performed iteratively until no more VIC outliers were identified. The VIC values before and after the 2D DIR correction were compared to quantify the effectiveness of the iterative correction method. One of the three T2w HR images with the smallest diaphragm displacement from the T1w BH (<1.0 cm) was selected for binning-artifact correction by deforming it to the T1w BH image.
Multi-modal B-Spline DIR and uni-modal enhanced-Demons DIR
The T2HR↔T1BH image deformation was performed using a multi-modal DIR (PlastiMatch27 and ITK[Insight ToolKits]28). The B-spline algorithm was used to define a continuous DVF based on a finite number of control points with a cost function of mutual information. The DIR was first performed within the body contour, followed by two separated fine-tuning DIR in the lung and non-lung regions based on automatic segmentation of the lung, establishing the T2-T1 voxel correspondence. The binning artifact corrected T2w HR image was used to deform to T1w BH image or vice versa, so two different voxel correspondences were obtained for comparison and evaluation, leading to two different reconstructions of T2w TR-4DMRI.
The T1w image deformation from the original or deformed BH to 3D cine (or BH/HR→FB) was performed using an enhanced demons DIR, developed for T1w TR-4DMRI reconstruction18. This enhanced demons () DIR algorithm introduced a pseudo demons force () into the demons force () to accelerate and enlarge coarse deformation in a multi-resolution approach.
| (2) |
where voxel i is surrounded by neighbor voxels j within a Gaussian Cube (gCube: 5x5x5 voxels) and is a normalization factor. The demons and pseudo demons forces can be expressed as:
| (3) |
| (4) |
Where and are the voxel intensity gradient at voxel i in moving (m) and static (s) images, is the voxel intensity difference, and α is a normalization factor (0.2 to 0.7). In the pseudo demons force, is a Gaussian probability density function (σ=1 voxel) serving as a weighting factor and is accumulated demons force from a neighbor voxel j within the gCube. Therefore, the pseudo force allows the demons force from its neighbor voxels (j) to contribute its own deformation, enhancing the deformation range. Adding the pseudo force in a coarse alignment was equivalent to move the regulatory Gaussian filter upfront to have a “forward” impact on voxel clusters in the deformation process, as described in a previous work18.
Assessment of the T2w TR-4DMRI reconstruction using a 4D digital phantom29
T1w- and T2w-mapped 4DMRI (10 respiratory states) HR (2x2x2 mm3) images of a digital phantom (4D XCAT30) were built with 3.0-cm superior-inferior and 0.5-cm anterior-posterior motion at the diaphragm. Three synthesized spherical tumors were placed in the left inferior posterior and left and right mid posterior of the lungs. Because the gross tumor volume (GTV) observed in lung cancer patients was slightly larger on average in T2w than in T1w31, the T1w diameters of the 3 spherical tumors were 2.0, 3.0 and 4.0 cm, while their T2w diameters were set at 2.2, 3.3 and 4.4 cm in diameter, respectively, leading to a diameter increase by 10% and tumor volume increase by 31-33%. The T1w MRI images were down-sampled to create low-resolution (LR: 5x5x5 mm3) with the addition of 2% Rayleigh noise, simulating the LR 3D cine images.
Two directional DIR (T2w→T1w and T2w←T1w) were performed to align the T2w and T1w HR images to set up the T2-T1 correspondence. The feature presentation of the T2w HR image in the T2w TR-4DMRI was evaluated based on the GTV alignment in position (center of mass, ΔCOM), size (volume ratio, %V), and shape (Dice index). The GTV in deformed images was automatically segmented using the region-growing algorithm.
Evaluation of the T2w TR-4DMRI reconstruction in lung cancer patients
In the T2w TR-4DMRI reconstruction workflow (see Fig. 1), both DIR (T2HR→T1BH and T2HR←T1BH) were performed and compared, similar to the 4D digital phantom experiment. The GTV COM in both HR and LR was calculated by automatical or semi-automatical delineation using a region grown algorithm. The tumor position (ΔCOM), size (%V), and shape (Dice) were evaluated for the two different DIR directions. In the tumor delineation when it is attached to the chest wall, a region of interest was manually drawn first (30x30x30 voxels centered at the GTV), followed by automatic region-growing segmentation to complete tumor delineation. Because of DIR uncertainty (~2 mm)17,18, when calculating the Dice index, rigid alignment on the tumor was performed first based on the ΔCOM. The quality of a local tumor-centered ROI of reconstructed T2w TR-4DMRI was compared with the original T2w HR MRI using the structure similarity (SSIM) and VIC indexes. Prior to comparison, the closest diaphragm match (left and right) between the reconstructed images (80 during 40s acquisition) and the original image was used to pair the images. The COM of GTV was then rigidly aligned and a common ROI of 30x30x30 voxels with the GTV at the center was created for comparison. It was worthwhile to mention that, unlike the digital phantom, a tumor in LR patient images was much more blurred with low tumor-lung contrast. Therefore, the tumor size and shape were less determined by the low-resolution GTV because of a fuzzy tumor edge in the 3D cine images.
RESULTS
Evaluation of T2w TR-4DMRI reconstructions based on a 4D digital phantom
Two reconstruction methods (A: T2HR→[T1BH→FB] and B: [T2HR←T1BH]→FB) of T2w TR-4DMRI are assessed using the 4D digital phantom and the results are shown in Table 1. Because of the well-defined GTV edges in 4DMRI phantoms, including the down-sampled LR images (with 2% Rayleigh noise mimicking the 3D LR cine), the size and shape of the deformed GTV follows those in final target images, as shown in Table 1. Fig. 2 shows that two well-defined tumors in the reconstructed images of two extreme T2w TR-4DMRI at full exhalation and full inhalation using methods A and B, together with T1w TR-4DMRI reconstruction as a control. The overall image quality of the super-resolution T2w TR-4DMRI images is acceptable. Regardless of using method A or B, the tumor position (ΔCOM = 1.0±0.6 mm), size (%V = 1.00±0.03) and shape (Dice = 0.91±0.04) in the simulated LR images are well preserved in the reconstruction process. However, method A produces a smaller GTV (%V = 0.93±0.04), while method B produces a larger GTV (%V = 1.06±0.02).
Table 1.
Quantitative evaluation of two T2w TR-4DMRI reconstruction methods in 4D digital phantom using T1w TR-4DMRI as the control. Three quantitative measures are used, including the difference in the center of mass (ΔCOM), tumor volume ratio, and Dice similarity index. The position difference (Δd) at the diaphragm domes is also provided.
| Measure | Tumor diameter (mm) | Motion (SI, AP) (mm) | T1w TR-4DMRI | T2w TR-4DMRI by Method A# | T2w TR-4DMRI by Method B # | ||
|---|---|---|---|---|---|---|---|
|
| |||||||
| T1→FB | T2→T1 | T2→[T1→FB] | T2←T1 | [T2←T1]→FB | |||
| ΔCOM (mm) | 20 | 12, 6 | 0.8 | 1.2 | 1.8 | 0.8 | 1.5 |
| 30 | 20, 1 | 0.6 | 0.8 | 0.7 | 0.4 | 0.4 | |
| 40 | 12, 1 | 0.4 | 0.8 | 1.4 | 0.2 | 0.5 | |
|
| |||||||
| Mean | 0.6 | 0.8 | 1.2 | 0.4 | 0.8 | ||
| STD | 0.2 | 0.4 | 0.5 | 0.3 | 0.5 | ||
|
| |||||||
| Δd (mm) | Dome* | 30, 4 | 0.4 | 0.3 | 0.7 | 0.2 | 0.6 |
|
| |||||||
| Volume Ratio (%V) | 20 | 12, 6 | 0.95 | 0.98 | 0.90 | 1.00 | 1.08 |
| 30 | 20, 1 | 0.98 | 1.00 | 0.93 | 1.01 | 1.05 | |
| 40 | 12, 1 | 0.99 | 0.99 | 0.97 | 1.01 | 1.05 | |
|
| |||||||
| Mean | 0.97 | 0.99 | 0.93 | 1.01 | 1.06 | ||
| STD | 0.02 | 0.01 | 0.04 | 0.01 | 0.02 | ||
|
| |||||||
| Dice Index | 20 | 12, 6 | 0.89 | 0.89 | 0.85 | 0.93 | 0.87 |
| 30 | 20, 1 | 0.93 | 0.94 | 0.94 | 0.96 | 0.93 | |
| 40 | 12, 1 | 0.94 | 0.94 | 0.93 | 0.96 | 0.93 | |
|
| |||||||
| Mean | 0.92 | 0.92 | 0.91 | 0.95 | 0.91 | ||
| STD | 0.03 | 0.03 | 0.05 | 0.02 | 0.03 | ||
The alignment of the left and right diaphragm domes is evaluated against the ground truth in the digital motion phantom.
The reconstruction follows the order of T2w→[T1w→FB] in method A and [T2w←T1w]→FB in method B.
Figure 2.
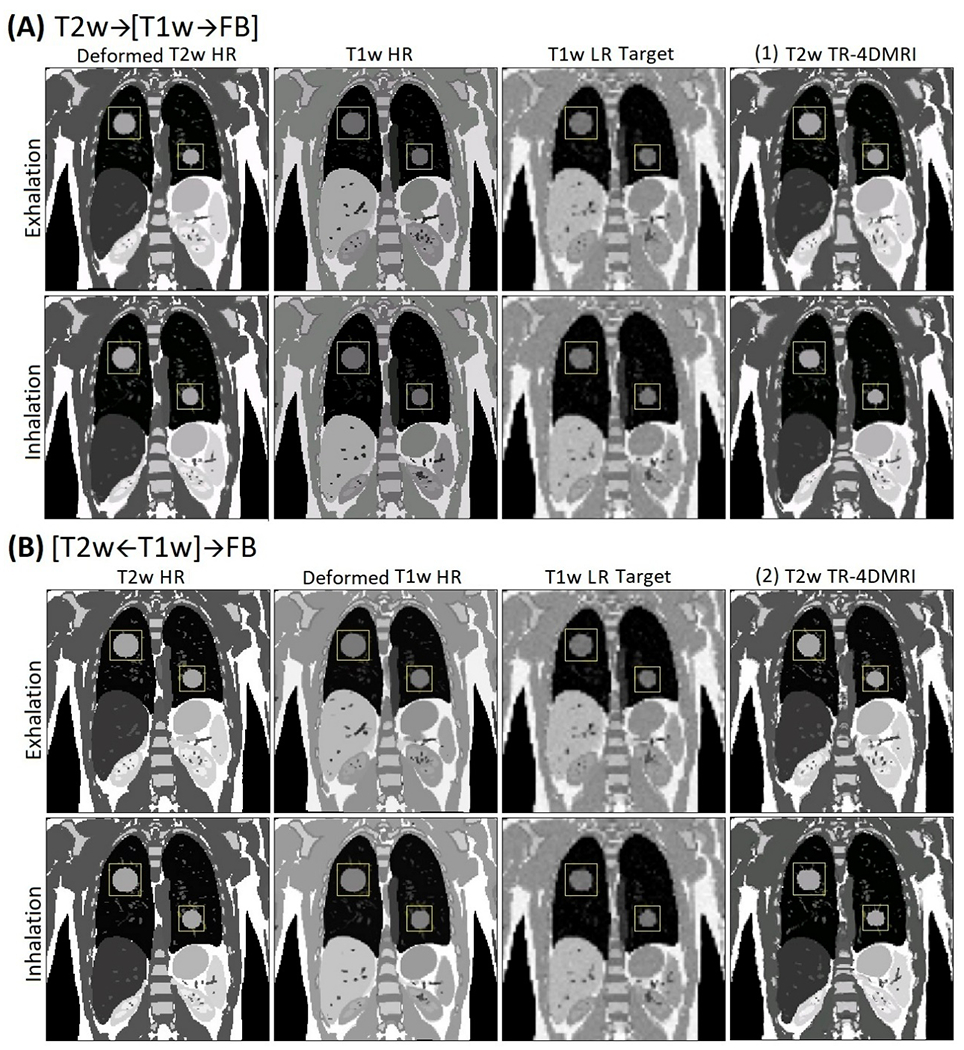
Demonstration of two reconstruction methods (A and B) of T2w time-resolved (TR) 4DMRI via multi-modal deformable image registration (DIR) between high-resolution (HR) T2w and HR T1w BH. Low-resolution (LR) T1w images are downsampled and noise-added from HR, simulating 3D cine. The final tumor size in T2w TR-4DMRI is similar to T1w tumor using method A and to T2w tumor using method B in both the exhalation and inhalation cases.
Correction of binning artifacts of patient RC-4DMRI images
Fig. 3 illustrates a case of identification and iterative correction of artifact slices that are mis-binned using the VIC plot (negative peaks) and 2D DIR to the interpolated slice from adjacent slices. The workflow is shown in Fig. 1. This image contains the most severe artifacts among 6 patients, taking 5-15 iterations for correction. Artifacts in the T2w RC-4DMRI image are generally mild because of using an internal navigator for reconstruction8. After correction, the image quality is visually improved (Fig. 3), and quantitatively, the VIC curve becomes much smoother and VIC value increases from 0.92±0.03 to 0.95±0.01 with a large reduction of the standard deviation, suggesting improved tissue continuity/similarity between consecutive slices.
Figure 3.
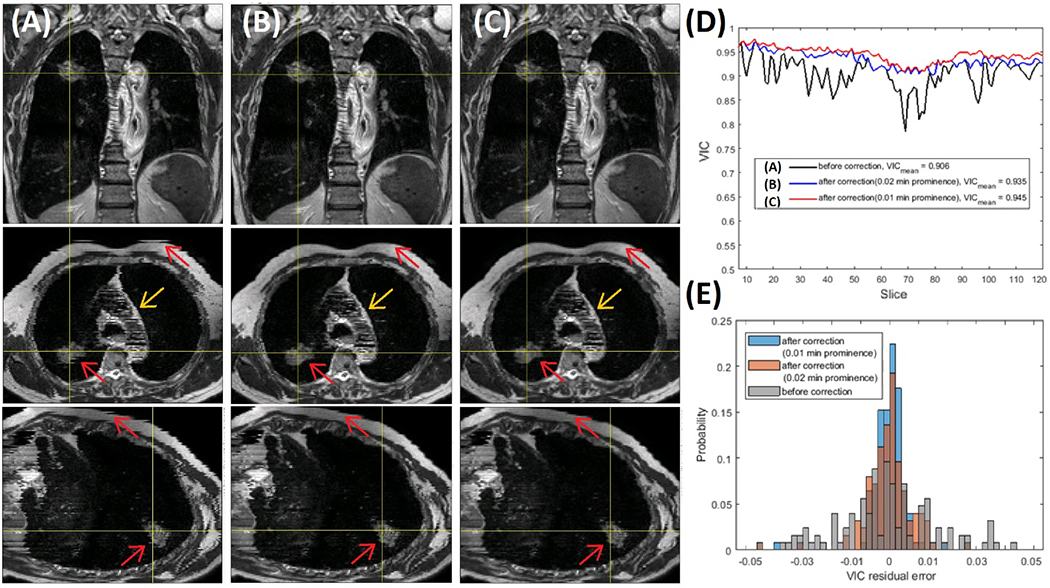
Illustration of minor binning artifact identification and correction of T2w RC-4DMRI with relatively “large” binning artifacts. (A) and (B & C) are images before and after artifact correction using 2D DIR to the interpolated slice from the adjacent slices with improvements (red arrows). (D) and (E) are the curve and histogram of the voxel intensity correlation (VIC), respectively, before (Gray) and after (B: Blue with VIC>0.02 and C: orange with VIC>0.01) the iterative correction. Note: the striking artifact within the heart (yellow arrows) is caused by the variation of blood flow at different acquisition times and cannot be corrected.
Evaluation of T2w-T1w alignment in patients
In patients, the alignment of T2w and T1w HR image is a key step to establish the voxel-to-voxel correspondence via the multi-modal DIR. Table 2 tabulates the GTV characteristics, including the ΔCOM, %V, and Dice using the GTV in the DIR-target image as the reference. Because the deformation between T2w HR and T1w BH is within 10.0 mm (6.7±2.1mm), the uncertainty of the COM alignment is small, about 1 voxel (2.2-2.5 mm) on average. Since the GTVs are irregular and small, the Dice index is more sensitive to the segmentation uncertainty than the volume ratio. Because of slightly larger GTV in T2w HR than T1w BH image31, the reconstructed tumor size varies between methods A and B, depending on T2↔T1 DIR direction.
Table 2.
Preservation of patient gross tumor volume (GTV) characters between T2w and T1w high-resolution (HR) images using multi-modal DIR. The largest T2w-T1w displacement at the diaphragm is within 1.0 cm. The original GTV in HR images is used as a reference.
| Patient | ΔCOM (mm) | %V of GTV | Dice Index of GTV | |||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Before DIR | After DIR | Before DIR | After DIR vs. Before | After DIR vs. Before | ||||
|
| ||||||||
| T2-T1 | T2→T1 | T2←T1 | T1/T2 | [T2→T1]/T1 | [T2←T1]/T2 | [T2→T1] vs. T1 | [T2←T1] vs. T2 | |
| 1 | 4.5 | 2.3 | 2.0 | 0.85 | 0.99 | 0.95 | 0.79 | 0.83 |
| 2 | 8.5 | 2.1 | 2.5 | 0.74 | 1.03 | 0.94 | 0.81 | 0.83 |
| 3 | 5.6 | 1.9 | 2.3 | 0.51 | 0.99 | 1.00 | 0.74 | 0.82 |
| 4 | 5.4 | 2.8 | 3.9 | 0.75 | 0.92 | 0.96 | 0.81 | 0.79 |
| 5 | 6.2 | 2.9 | 1.3 | 0.86 | 1.04 | 0.98 | 0.85 | 0.84 |
| 6 | 10.0 | 2.0 | 2.4 | 0.76 | 1.01 | 1.00 | 0.82 | 0.85 |
|
| ||||||||
| Mean | 6.7 | 2.2 | 2.4 | 0.75 | 1.00 | 0.97 | 0.80 | 0.83 |
| St Dev | 2.1 | 0.5 | 0.9 | 0.13 | 0.05 | 0.02 | 0.04 | 0.02 |
Evaluation of T2w TR-4DMRI reconstructions in patients
In the six patients, the assessment of the reconstruction results at two extreme states (full inhalation and full exhalation) is tabulated in Table 3, representing the largest uncertainties in the reconstruction. The LR 3D cine images of a patient are much more blurring than the down-sampled, noise-added phantom image, so that patient 3D cine images provide the guidance mostly for tumor position alignment, but not enough to make a substantial change the volume and shape. Therefore, the reference of comparison in ΔCOM is set to LR FB, whereas those of %V and Dice (after COM alignment) are set to the original HR images. The tumor volume (%V = 0.99±0.03) and shape (Dice = 0.81±0.06) are preserved well using reconstruction method B ([T2w←T1w]→FB), while its position is accurately updated (ΔCOM = 2.8±0.8 mm), similar to the results in T1w TR-4DMRI (ΔCOM = 3.0±0.9 mm). Method A, however, produces high %V and Dice values when using the T1w HR image as the reference.
Table 3.
Preservation of patient gross tumor volume (GTV) characters in T2w TR-4DMRI using two reconstruction methods, T2wA: T2HR→[T1BH→FB] and T2wB: [T2HR←T1BH]→FB, together with T1w TR-4DMRI. Results at the two extreme respiratory states are shown, full exhalation (Ex) and full inhalation (In), carrying the upper limit of uncertainty. The GTV in T2w high-resolution (HR) or T1w breath-hold (BH) images are used as the reference.
| Patient | State | ΔCOM (mm) | %V | Dice Index | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| (Ref to T1 FB) | (Ref to T1-BH) | (Ref to T2-HR) | (Ref to T2-HR) | |||||||
|
| ||||||||||
| T1w | T2wA | T2wB | T1w | T2wA | T2wA | T2wB | T2wA | T2wB | ||
| 1 | Ex | 2.4 | 2.5 | 3.2 | 1.01 | 1.06 | 0.90 | 1.03 | 0.85 | 0.87 |
| In | 3.1 | 2.7 | 3.1 | 1.02 | 1.12 | 0.95 | 1.00 | 0.83 | 0.90 | |
| 2 | Ex | 3.5 | 3.2 | 2.1 | 1.03 | 1.01 | 0.75 | 0.97 | 0.83 | 0.86 |
| In | 3.9 | 3.9 | 3.7 | 1.03 | 1.04 | 0.77 | 0.94 | 0.81 | 0.84 | |
| 3 | Ex | 4.0 | 2.1 | 2.6 | 1.07 | 1.07 | 0.47 | 0.95 | 0.62 | 0.87 |
| In | 3.1 | 2.3 | 2.3 | 1.10 | 1.13 | 0.50 | 1.00 | 0.64 | 0.85 | |
| 4 | Ex | 3.1 | 3.8 | 3.0 | 0.99 | 1.03 | 0.78 | 1.02 | 0.80 | 0.79 |
| In | 3.3 | 1.6 | 2.3 | 0.96 | 1.06 | 0.80 | 0.99 | 0.78 | 0.80 | |
| 5 | Ex | 3.3 | 1.0 | 3.8 | 1.00 | 1.01 | 0.89 | 1.01 | 0.68 | 0.71 |
| In | 0.6 | 3.6 | 3.2 | 1.00 | 1.05 | 0.92 | 0.98 | 0.63 | 0.74 | |
| 6 | Ex | 2.0 | 1.7 | 2.3 | 1.03 | 1.10 | 0.84 | 1.04 | 0.69 | 0.80 |
| In | 3.2 | 3.4 | 3.1 | 1.01 | 1.10 | 0.84 | 1.00 | 0.65 | 0.72 | |
|
| ||||||||||
| Mean | 3.0 | 2.7 | 2.9 | 1.02 | 1.06 | 0.78 | 0.99 | 0.73 | 0.81 | |
|
| ||||||||||
| St Dev | 0.9 | 0.9 | 0.6 | 0.04 | 0.04 | 0.15 | 0.03 | 0.09 | 0.06 | |
The image qualities of the T2w TR-4DMRI reconstruction is quantified by SSIM and VIC based on the local, tumor-centered ROI (30x30x30 voxels) of the same respiratory stage 4DMRI comparing with the original HR T2w MRI, as shown in Table 4. Because a respiratory image was found from the RC-4DMRI to match to the HR T2w MRI based on the tumor, right and left diaphragm positions among 80 volumetric images during 40s scan, as well as small residual tumor mismatch was corrected by rigid alignment, the comparison between the initial HR T2w MRI and reconstructed MRI at the same respiratory stage is valid, providing the local image similarity between them and image quality of the reconstructed T2w TR-4DMRI. Method B provides consistently superior similarity results than method A, providing acceptable SSIM and VIC values and confirming the excellent tumor COM, V% and DICE results above.
Table 4.
Local image quality around the tumor in reconstructed T2w time-resolved (TR) 4DMRI in comparison with the initial T2w high-resolution (HR) MRI image using the structure similarity (SSIM) and voxel intensity correlation (VIC) indexes. The two images of comparison were on the same respiratory phase (left and right diaphragms matches are about 1 voxel) and local tumor-centered anatomy (30x30x30 voxels) was rigidly aligned to the center of mass (COM) of the tumor, prior to index calculation. The two reconstruction methods are noted as A and B.
| Respiratory stage consistency measure | Image similarity measure | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Patient | Diaphragm match (mm) | Tumor match (mm) | SSIM | VIC | ||||
| Left | Right | before | after | T2wA | T2wB | T2wA | T2wB | |
|
| ||||||||
| 1 | 0.3 | 2.0 | 1.0 | 0.0 | 0.61 | 0.75 | 0.79 | 0.92 |
| 2 | 2.7 | 2.7 | 2.0 | 0.0 | 0.47 | 0.56 | 0.80 | 0.86 |
| 3 | 1.3 | −0.3 | 0.7 | 0.0 | 0.57 | 0.65 | 0.78 | 0.84 |
| 4 | 1.3 | 2.7 | −2.3 | 0.0 | 0.44 | 0.59 | 0.77 | 0.87 |
| 5 | −2.0 | −3.3 | −2.0 | 0.0 | 0.52 | 0.54 | 0.65 | 0.73 |
| 6 | −2.7 | −2.7 | −0.3 | 0.0 | 0.61 | 0.77 | 0.81 | 0.86 |
|
| ||||||||
| Mean | 0.2 | 0.2 | −0.2 | 0.00 | 0.54 | 0.65 | 0.77 | 0.85 |
| St dev | 2.1 | 2.7 | 1.7 | 0.00 | 0.07 | 0.10 | 0.06 | 0.06 |
DISCUSSION
The quality of the T2w TR-4DMRI image reconstruction
The image quality of T2w TR-4DMRI depends on the T2w-T1w mapping that establishes the voxel-to-voxel correspondence. The ability to correct minor binning artifacts and select a T2w HR MR with a similar motion to the T1w BH image minimizes the uncertainty in the T2w-T1w DIR. Therefore, only minor uncertainty is added beyond the T1w TR-4DMRI reconstruction. Generally, the smaller the deformation between the moving and target images, the more accurate the DIR would be for a DIR algorithm. In this study, the diaphragm motion difference between T2w and T1w HR images are within 1.0 cm, leading to an overall misalignment of 1.9±0.9 mm at the diaphragm dome in T2w TR-4DMRI, slightly higher than −0.9±0.5mm in T1w TR-4DMRI reconstruction17,18. In reference to the FB 3D cine images, the uncertainties of the diaphragm dome position are similar between T2w TR-4DMRI (1.8±0.7 mm) and T1w TR-4DMRI (1.4±0.7 mm). Therefore, T2w TR-4DMRI reconstruction is expected to be accurate.
In the phantom, the tumor position (ΔCOM=1.0±0.5 mm), size (%V=1.06±0.02), and shape (Dice=0.91±0.02) are pertained during the T2w TR-4DMRI reconstruction using method B (similar to method A), as shown in Table 1. In the patient data, only the tumor position is assessed in reference to the tumor image in LR 3D cine that is much more blurred than that in the digital phantom image. As shown in Table 2, the uncertainty of T2w-T1w alignment at the tumor is at the level of 1.0 voxel, the overall uncertainty of T2w TR-4DMRI is about 1.5 voxel (ΔCOM=2.9±0.6 mm). The blurred GTV does not regulate the tumor size and shape well, so the GTV in the moving image dominant the final results. Therefore, methods B ([T2HR←T1BH]→FB]) depicts a distinguished difference from method A (T2HR→[T1BH→FB]) and provides a superior result. Based on this study of both 4D phantom and patients, it demonstrates that the reconstruction accuracy of the T2w TR-4DMRI and T1w TR-4DMRI is similar. Further improvements of acquired image resolution and image quality used for the TR-4DMRI image reconstruction are desirable and helpful to reduce the uncertainty.
In addition to the quantification of the GTV in the reconstructed T2w TR-4DMRI, the local image qualities around the tumor are quantified using the SSIM and VIC indexes, which illustrates high image similarity around lung tumors between anatomically-matched image sets, namely between the original HR T2w MRI and the reconstructed T2w TR-4DMRI among the six patients. It is worthwhile to emphasize that the ability to find an anatomical matched 4DMRI is owing to the continuous multi-breath image acquisition, which provides about 80 volumetric images during the 40s acquisition for the selection. In addition, the final rigid tumor alignment is important, similar to the Dice index calculation.
Preservation of tumor size and shape in the reconstruction of T2w TR-4DMRI
It is interesting that the two reconstruction methods are not equivalent, and in fact, method B consistently outperforms method A with better preserved T2w tumor characteristics, including size and shape, although tumor positions are similar by both methods. Because the authenticity of GTV is of foremost importance in target delineation for radiotherapy applications, we consider method B is a superior approach in T2w TR-4DMRI reconstruction.
As T2w and T1w HR MRI images have somewhat different tumor appearance31, the direction of DIR affects the T1w-T2w voxel correspondence, and usually, the resulting tumor characteristics are determined by the target MR image. In other words, a T1w-like tumor will be produced by DIR with T2HR→T1BH in method A, whereas a T2w-like tumor will be produced by DIR with T2HR←T1BH in method B. Therefore, different tumor characteristics are applied in T1HR→FBLR DIR for the reconstruction, depending on which method is used. However, the tumor in the FB images is not only of low resolution but also motion-blurred (as shown in Fig. 4), as the result, the LR cine image DIR behaves differently from HR image DIR. In fact, the LR image provides sufficient voxel intensity gradience (VIG) and difference (VID) in the Demons force to move the tumor position, but not enough to shape up the tumor size and shape, as shown in Table 3. Therefore, method B that has the T2w-like GTV produces a superior result in keeping the T2w tumor characteristics.
Figure 4.
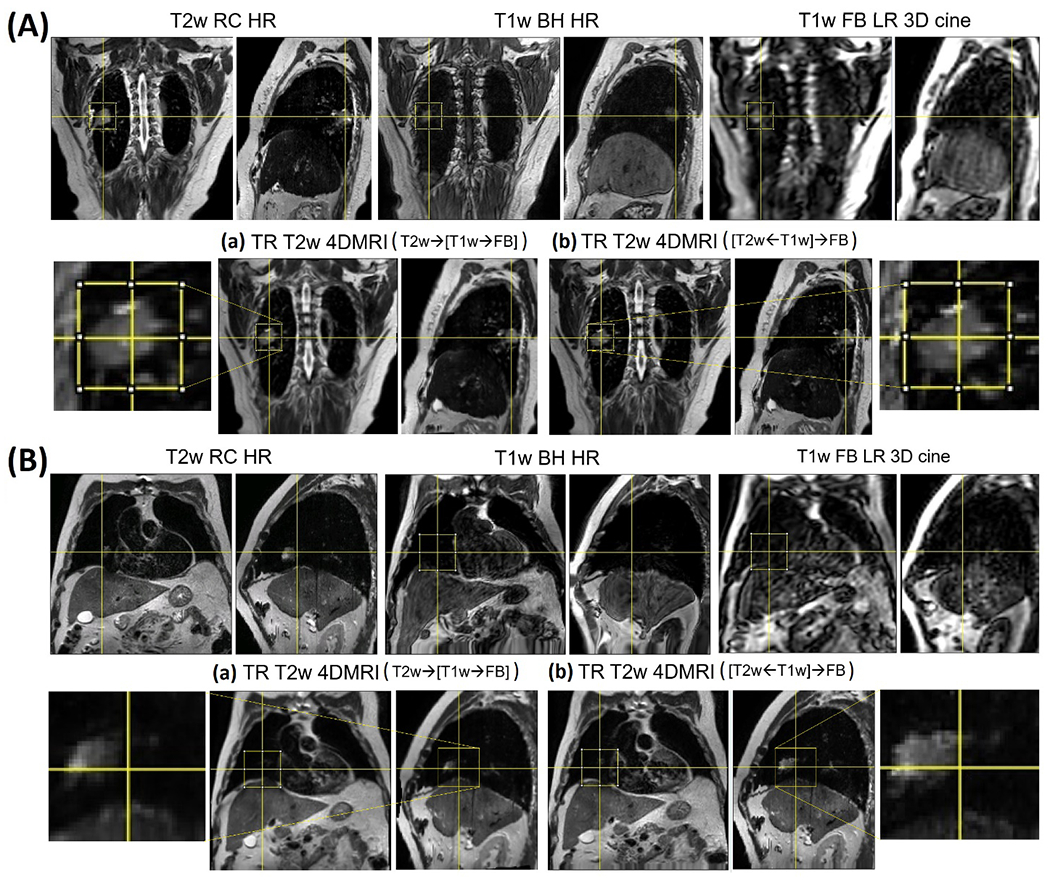
T2w TR-4DMRI of two patients (A and B). For each patient, the results from the method (a) and (b) are shown, together with inserts of enlarged gross tumor volume (GTV). Method (b) preserves the T2w GTV well while method (a) keeps the T1w-like GTV.
In patients, there is no ground truth on whether T1w or T2w tumor is more accurate, but T2w tumor size is more consistent with that delineated from 4DCT31. The GTV ratio is 0.75±0.13 between T1w BH MR and T2w RC HR images, similar to the previous report of a 24% difference. An almost identical volume ratio (%V=0.99±0.02) is found between T2w TR-4DMRI and T2w RC-4DMRI from method B, suggesting that the tumor volume is preserved. Similarly, the tumor volume ratio is %V=1.02±0.04 between T1w TR-4DMRI and T1w BH MR image. In contrast, method A retains the tumor volume from T1w BH image (p<0.0001), serving as a control for comparison. Therefore, the tumor volume in the final T2w TR-4DMRI depends on the DIR direction in the reconstruction and method B is superior to method A. The Dice index for tumor shape comparison between T2w TR-4DMRI and the artifact-corrected HR MRI is calculated after the COM alignment. The Dice indexes differ significantly (p = 0.006) between method A (0.73±0.09) and method B (0.81±0.06). Therefore, method B is superior to method A in preserving original tumor characters, which are important for accurate tumor delineation.
A generalized TR-4DMRI reconstruction framework using any MR contrasts
The significance of MR contrast extension from T1w to T2w in TR-4DMRI reconstruction is that it provides a generalized framework to reconstruct TR-4DMRI with an MR contrast of interest for a clinical application. In this study, we have demonstrated the feasibility of T2w TR-4DMRI reconstruction, while the same procedure can apply to other MR contrasts. For instance, the Dixon scanning sequence32, which has been developed for an optimal liver scan with and without fat suppression via multi-parameter scans, can be incorporated as long as an HR Dixon image can be acquired33. Therefore, the methodology developed here can be applied to achieve TR-4DMRI with any MR contrast.
In practice, the HR T2w or other MR contrast images can be acquired by respiratory-gated acquisition using a navigator echo set on the patient’s diaphragm, similar to RC-4DMRI. In the radial acquisition, self-navigation can be applied using heavily under-sampled images to extract diaphragm motion for surrogating. Additionally, a golden-angle radial acquisition method could be applied with compressed sensing reconstruction to provide a high-resolution 4DMRI image with little motion artifacts34. Furthermore, if the same gating criterion is applied to acquisition for both T1w and T2w, then the T2-T1 voxel correspondence can be achieved at the scanning level, minimizing the need for T2-T1 DIR. Therefore, a high-resolution image with the desired MR contrast can be acquired to reconstruct the new TR-4DMRI.
Because TR-4MDRI provides multi-breath motion information without restricting to periodic motion, it is a desirable method to imaging not only respiratory motion but also motion variation and irregular motions, such as breathing irregularities, digestive or even voluntary motion of a patient. Such motion assessment should be beneficial to radiotherapy treatment planning as well as treatment delivery evaluation. With the ability to reconstruct the TR-4DMRI with any MR contrast, it promises to facilitate the versatile and diversified clinical applications.
Advantages, limitations, and future directions
Advantages of the super-resolution TR-4DMRI technique include that (1) it provides multi-breath motion image data, unlike RC-4DMRI that produces one breathing cycle, (2) it can image irregular motion because it does not assume motion periodicity, unlike RC-4DMRI, and (3) it offers a generalized TR-4DMRI reconstruction framework using any MR contrasts that could be optimal for certain clinical applications. For a modest tumor size (>2cm), we have demonstrated that the estimation of tumor position and preservation of tumor characteristics are in a clinically acceptable range for radiotherapy and there are rooms for improvements.
A major limitation of the super-resolution approach for T1w/T2w TR-4DMRI reconstruction is associated with DIR uncertainty. This uncertainty can be reduced by improving the 3D cine image quality and minimizing the need for deformation. First, the reconstruction quality of TR-4DMRI image depends on the quality of FB 3D cine images, which provide the template for BH image to align. With recently-added compressed sensing capability in the 3T MR scanner, the 3D cine image quality has been improved, and therefore, the T1w TR-4DMRI quality is expected to be further improved as well. Second, multiple T1w HR images can be acquired using golden-angle radial acquisition with compressed sensing reconstruction for T1w RC-4DMRI with little binning artifacts35. Unlike Cartesian MR acquisition, the radial acquisition is insensitive to the motion. Therefore, the HR MR image quality can be improved and the DIR range to LR FB images is substantially reduced, leading to an improved T1w TR-4DMRI. Third, a navigator-gated acquisition at a specified state of respiration can be applied to acquire T2w 3DMRI image (or with another MR contrast), so that the deformation for T2w-T1w is also minimized. Fourth, the registration images can be finetuned with optimal visualization and the DIR algorithm can be further improved18,19,36,37, aiming to reduce the uncertainties related to the DIR reconstruction. In addition to the 5 similarity indexes, other parameters could also be applied to quantify image quality38. Overall, these approaches can be readily applied to achieve more accurate and more reliable T1w/T2w TR-4DMRI, facilitating the clinical implementation of TR-4DMRI.
Last, it is also worthwhile to mention that deep learning techniques have been applied in super-resolution reconstruction of dynamic 4DMRI20,39 and higher-resolution MRI image40,41 using limited training image sets. These studies focused on a single MR contrast, unlike this study that develops a method to extend TR-4DMRI from one MR contrast to another, focusing on the generalization of the SR approach. Recently, machine learning technique has been increasingly applied to radiation therapy, including cancer image segmentation42, deformable image registration43,44, and MR image reconstruction38,45, and can be explored for further improvement of T2w TR-4DMRI reconstruction in the future.
CONCLUSION
We have demonstrated a novel SR-based framework to reconstruct TR-4DMRI with T2w, T2w, or any other MR contrasts. The feasibility, accuracy, and performance are evaluated, based on the study of a 4D digital phantom and 6 lung cancer patients. The T2w-centric approach, namely reconstruction method B: [T2wHR←T1wBH]→FBLR, is preferable to preserve the T2w image features of the GTV with higher VIC = 0.85±0.06 and SSIM = 0.65±0.10 index values. Good tumor alignment and similarity are observed: ΔCOM = 0.8±0.5 mm, %V = 1.06±0.02, and Dice = 0.91±0.03 in phantom and ΔCOM = 2.9±0.6 mm, %V = 0.99±0.03, and Dice = 0.81±0.06 in patients. These quantitative evaluation results are similar to T1w TR-4DMRI, suggesting that minimal uncertainty is added in the T2w TR-4DMRI reconstruction. The generalized SR-based reconstruction framework of TR-4DMRI promises to have clinical application in multi-breath tumor motion assessment and motion monitoring.
Acknowledgments:
This research is supported in part by the Breast Cancer Research Foundation (BCRF-17–193) and by the MSK Cancer Center Support Grant (P30 CA008748). We would like to thank Christian Czmielewski for assistance in consenting patients and MR technologists/therapists in assisting the patient scans for the IRB-approved 4DMRI protocol study.
APPENDIX
The acronyms used in this paper
| MRI: | Magnetic Resonance Imaging |
| TR-4DMRI: | Time-resolved four-dimensional MRI |
| RC-4DMRI | Respiratory-correlated 4DMRI |
| T1w (T1): | T1-weighted |
| T2w (T2): | T2-weighted |
| BH: | Breath Hold |
| FB: | Free Breathing |
| HR: | High Resolution |
| LR: | Low Resolution |
| IDR: | Deformable Image Registration |
| DVF: | Displacement Vector Field |
| GTV: | Gross Tumor Volume |
| COM: | Center of Mass |
| ΔCOM: | COM difference |
| %V: | Volume Ratio |
| VID: | Voxel Intensity Difference |
| VIG: | Voxel Intensity Gradience |
| VIC: | Voxel Intensity Correlation |
| SSIM: | Structure Similarity index |
Footnotes
Conflict of Interest: Memorial Sloan Kettering Cancer Center has a master research agreement (MRA) with Philips Healthcare.
REFERENCES
- 1.Keall PJ, Mageras GS, Balter JM, et al. The management of respiratory motion in radiation oncology report of AAPM Task Group 76 [published online ahead of print 2006/11/09]. Med Phys. 2006;33(10):3874–3900. [DOI] [PubMed] [Google Scholar]
- 2.Sonke JJ, Lebesque J, van Herk M. Variability of four-dimensional computed tomography patient models [published online ahead of print 2007/11/27]. Int J Radiat Oncol Biol Phys. 2008;70(2):590–598. [DOI] [PubMed] [Google Scholar]
- 3.Cai J, Read PW, Larner JM, Jones DR, Benedict SH, Sheng K. Reproducibility of interfraction lung motion probability distribution function using dynamic MRI: statistical analysis. Int J Radiat Oncol Biol Phys. 2008;72(4):1228–1235. [DOI] [PubMed] [Google Scholar]
- 4.Thomas DH, Santhanam A, Kishan AU, et al. Initial observations of intra- and inter- fractional motion variation in MR guided lung SBRT. The British journal of radiology. 2017;91(1083):20170522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dhont J, Vandemeulebroucke J, Burghelea M, et al. The long- and short-term variability of breathing induced tumor motion in lung and liver over the course of a radiotherapy treatment. Radiother Oncol. 2018;126(2):339–346. [DOI] [PubMed] [Google Scholar]
- 6.Du D, Caruthers SD, Glide-Hurst C, et al. High-quality t2-weighted 4-dimensional magnetic resonance imaging for radiation therapy applications. Int J Radiat Oncol Biol Phys. 2015;92(2):430–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y, Yin FF, Czito BG, Bashir MR, Cai J. T2-weighted four dimensional magnetic resonance imaging with result-driven phase sorting. Med Phys. 2015;42(8):4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li G, Wei J, Olek D, et al. Direct Comparison of Respiration-Correlated Four-Dimensional Magnetic Resonance Imaging Reconstructed Using Concurrent Internal Navigator and External Bellows. Int J Radiat Oncol Biol Phys. 2017;97(3):596–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freedman JN, Collins DJ, Gurney-Champion OJ, et al. Super-resolution T2-weighted 4D MRI for image guided radiotherapy [published online ahead of print 2018/06/07]. Radiother Oncol. 2018;129(3):486–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J, Markova S, Garcia A, et al. Evaluation of automatic contour propagation in T2-weighted 4DMRI for normal-tissue motion assessment using internal organ-at-risk volume (IRV). Journal of applied clinical medical physics / American College of Medical Physics. 2018;19(5):598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J, Srivastava S, Wang C, et al. Clinical evaluation of 4D MRI in the delineation of gross and internal tumor volumes in comparison with 4DCT [published online ahead of print 2019/09/21]. Journal of applied clinical medical physics / American College of Medical Physics. 2019;20(9):51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plathow C, Klopp M, Schoebinger M, et al. Monitoring of lung motion in patients with malignant pleural mesothelioma using two-dimensional and three-dimensional dynamic magnetic resonance imaging: comparison with spirometry. Investigative radiology. 2006;41(5):443–448. [DOI] [PubMed] [Google Scholar]
- 13.Li G, Citrin D, Camphausen K, et al. Advances in 4D medical imaging and 4D radiation therapy [published online ahead of print 2008/01/18]. Technol cancer research treatment. 2008;7(1):67–81. [DOI] [PubMed] [Google Scholar]
- 14.Miquel ME, Blackall JM, Uribe S, Hawkes DJ, Schaeffter T. Patient-specific respiratory models using dynamic 3D MRI: preliminary volunteer results. Phys Med. 2013;29(2):214–220. [DOI] [PubMed] [Google Scholar]
- 15.Harris W, Ren L, Cai J, Zhang Y, Chang Z, Yin FF. A Technique for Generating Volumetric Cine-Magnetic Resonance Imaging. Int J Radiat Oncol Biol Phys. 2016;95(2):844–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stemkens B, Tijssen RH, de Senneville BD, Lagendijk JJ, van den Berg CA. Image-driven, model-based 3D abdominal motion estimation for MR-guided radiotherapy. Physics in medicine and biology. 2016;61(14):5335–5355. [DOI] [PubMed] [Google Scholar]
- 17.Li G, Wei J, Kadbi M, et al. Novel Super-Resolution Approach to Time-Resolved Volumetric 4-Dimensional Magnetic Resonance Imaging With High Spatiotemporal Resolution for Multi-Breathing Cycle Motion Assessment. Int J Radiat Oncol Biol Phys. 2017;98(2):454–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li G, Sun A, Nie X, et al. Introduction of a pseudo demons force to enhance deformation range for robust reconstruction of super-resolution time-resolved 4DMRI. Med Phys. 2018;45(11):5197–5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li G, Liu Y, Nie X. Respiratory-correlated (RC) versus time-resolved (TR) four-dimensional magnetic resonance imaging (4DMRI) for radiotherapy of thoracic and abdominal cancer. Frontiers in oncology. 2019;Special issue on MRI in Radiaiton Therapy(9):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chun J, Zhang H, Gach HM, et al. MRI super-resolution reconstruction for MRI-guided adaptive radiotherapy using cascaded deep learning: In the presence of limited training data and unknown translation model [published online ahead of print 2019/07/17]. Med Phys. 2019;46(9):4148–4164. [DOI] [PubMed] [Google Scholar]
- 21.Liu YL, Yin FF, Czito BG, Bashir MR, Cai J. T2-weighted four dimensional magnetic resonance imaging with result-driven phase sorting. Med Phys. 2015;42(8):4460–4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freedman JN, Collins DJ, Bainbridge H, et al. T2-Weighted 4D Magnetic Resonance Imaging for Application in Magnetic Resonance-Guided Radiotherapy Treatment Planning. Invest Radiol. 2017;52(10):563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freedman JN CD, Gurney-Champion OJ, McClelland JR, Nill S, Oelfke U, Leach MO, Wetscherek A. Super-resolution T2-weighted 4D MRI for image guided radiotherapy. Radiother Oncol. 2018. 10.1016/j.radonc.2018.05.015 Accessed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lichy MP, Wietek BM, Mugler JP, et al. Magnetic resonance imaging of the body trunk using a single-slab, 3-dimensional, T2-weighted turbo-spin-echo sequence with high sampling efficiency (SPACE) for high spatial resolution imaging - Initial clinical experiences. Invest Radiol. 2005;40(12):754–760. [DOI] [PubMed] [Google Scholar]
- 25.Clarke G PN. MRI for Diagnosis and Treatment of Cancer - AAPM. In.: https://www.aapm.org/meetings/amos2/pdf/34-8205-79886-720.pdf.
- 26.Hertanto A, Zhang Q, Hu YC, Dzyubak O, Rimner A, Mageras GS. Reduction of irregular breathing artifacts in respiration-correlated CT images using a respiratory motion model. Med Phys. 2012;39(6):3070–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shackleford JA, Kandasamy N, Sharp GC. On developing B-spline registration algorithms for multi-core processors. Physics in medicine and biology. 2010;55(21):6329–6351. [DOI] [PubMed] [Google Scholar]
- 28.McCormick M, Liu X, Jomier J, Marion C, Ibanez L. ITK: enabling reproducible research and open science. Front Neuroinform. 2014;8:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panta RK, Segars P, Yin FF, Cai J. Establishing a framework to implement 4D XCAT phantom for 4D radiotherapy research. Journal of cancer research and therapeutics. 2012;8(4):565–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Segars WP, Sturgeon G, Mendonca S, Grimes J, Tsui BM. 4D XCAT phantom for multimodality imaging research [published online ahead of print 2010/10/23]. Med Phys. 2010;37(9):4902–4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J, Srivastava S, Wang C, et al. Clinical evaluation of 4D MRI in the delineation of gross and internal tumor volumes in comparison with 4DCT. Journal of applied clinical medical physics / American College of Medical Physics. 2019;in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma J Dixon techniques for water and fat imaging [published online ahead of print 2008/09/09]. Journal of magnetic resonance imaging : JMRI. 2008;28(3):543–558. [DOI] [PubMed] [Google Scholar]
- 33.Eiber M, Martinez-Moller A, Souvatzoglou M, et al. Value of a Dixon-based MR/PET attenuation correction sequence for the localization and evaluation of PET-positive lesions [published online ahead of print 2011/06/21]. European journal of nuclear medicine and molecular imaging. 2011;38(9):1691–1701. [DOI] [PubMed] [Google Scholar]
- 34.Feng L, Axel L, Chandarana H, Block KT, Sodickson DK, Otazo R. XD-GRASP: Golden-angle radial MRI with reconstruction of extra motion-state dimensions using compressed sensing. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2015. doi: 10.1002/mrm.25665:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nie X, Feng L, Zakian K, Otazo R, Li G. A novel library-matching approach to reconstruct time-resolved 4DMRI via full and sparse sampling using golden-angle radial acquisition and compressed sensing reconstruction: A simulation study. Paper presented at: AAPM Annual Meeting; 7/14–18, 2019; San Antonio, TX. [Google Scholar]
- 36.Nie X, Deasy J, Li G. Novel hybrid deformable image registration for robust reconstruction of time-resolved 4DMRI with sliding motion via automatic body-cavity segmentation Paper presented at: AAPM Annual Meeting; 7/14–18, 2019; San Antonio, TX. [Google Scholar]
- 37.Liu Y, Nie X, Kadbi M, Deasy JO, Li G. (ORAL) Investigation on fast reconstruction of time-resolved 4DMRI with dynamic keyhole method using respiratory-correlated image library. Paper presented at: AAPM Annual Meeting; 7/14–18, 2019; San Antonio, TX. [Google Scholar]
- 38.Seitzer M, Yang G, Schlemper J, et al. Adversarial and Perceptual Refinement for Compressed Sensing MRI Reconstruction. The 21st International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), . 2018.pp. 232–240. [Google Scholar]
- 39.Dutta J, Huang C, Li Q, El Fakhri G. Pulmonary imaging using respiratory motion compensated simultaneous PET/MR. Med Phys. 2015;42(7):4227–4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu J, Yang G, Lio P. How Can We Make Gan Perform Better in Single Medical Image Super-Resolution? A Lesion Focused Multi-Scale Approach. 2019 Ieee 16th International Symposium on Biomedical Imaging (Isbi 2019). 2019.1669–1673. [Google Scholar]
- 41.Zhao X, Zhang Y, Zhang T, Zou X. Channel Splitting Network for Single MR Image Super-Resolution [published online ahead of print 2019/06/21]. IEEE transactions on image processing : a publication of the IEEE Signal Processing Society. 2019;28(11):5649–5662. [DOI] [PubMed] [Google Scholar]
- 42.Jiang J, Hu YC, Tyagi N, et al. Cross-modality (CT-MRI) prior augmented deep learning for robust lung tumor segmentation from small MR datasets [published online ahead of print 2019/07/06]. Med Phys. 2019;46(10):4392–4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Balakrishnan G, Zhao A, Sabuncu MR, Guttag J, Dalca AV. An Unsupervised Learning Model for Deformable Medical Image Registration. Proc Cvpr Ieee. 2018. doi: 10.1109/Cvpr.2018.00964:9252-9260. [DOI] [Google Scholar]
- 44.Liao R, Miao S, de Tournemire P, et al. An Artificial Agent for Robust Image Registration. Thirty-First Aaai Conference on Artificial Intelligence. 2017.4168–4175. [Google Scholar]
- 45.Yang G, Yu S, Dong H, et al. DAGAN: Deep De-Aliasing Generative Adversarial Networks for Fast Compressed Sensing MRI Reconstruction [published online ahead of print 2018/06/06]. IEEE Trans Med Imaging. 2018;37(6):1310–1321. [DOI] [PubMed] [Google Scholar]


