Abstract
Background:
Colorectal cancer has a high incidence rate worldwide, with over 1.8 million new cases and 880,792 deaths in 2018. Fortunately, its early detection significantly increases the survival rate, reaching a cure rate of 90% when diagnosed at a localized stage. Colonoscopy is the gold standard technique for detection and removal of colorectal lesions with potential to evolve into cancer. When polyps are found in a patient, the current procedure is their complete removal. However, in this process, gastroenterologists cannot assure complete resection and clean margins which are given by the histopathology analysis of the removed tissue, which is performed at laboratory.
Aims:
In this paper, we demonstrate the capabilities of multiphoton microscopy (MPM) technology to provide imaging biomarkers that can be extracted by deep learning techniques to identify malignant neoplastic colon lesions and distinguish them from healthy, hyperplastic, or benign neoplastic tissue, without the need for histopathological staining.
Materials and Methods:
To this end, we present a novel MPM public dataset containing 14,712 images obtained from 42 patients and grouped into 2 classes. A convolutional neural network is trained on this dataset and a spatially coherent predictions scheme is applied for performance improvement.
Results:
We obtained a sensitivity of 0.8228 ± 0.1575 and a specificity of 0.9114 ± 0.0814 on detecting malignant neoplastic lesions. We also validated this approach to estimate the self-confidence of the network on its own predictions, obtaining a mean sensitivity of 0.8697 and a mean specificity of 0.9524 with the 18.67% of the images classified as uncertain.
Conclusions:
This work lays the foundations for performing in vivo optical colon biopsies by combining this novel imaging technology together with deep learning algorithms, hence avoiding unnecessary polyp resection and allowing in situ diagnosis assessment.
Keywords: Colorectal polyps, convolutional neural network, dataset, multiphoton microscopy, optical biopsy
INTRODUCTION
Colorectal cancer ranks as one of the predominant cancers, being the third most commonly occurring cancer in men and the second most commonly occurring cancer in women. There were over 1.8 million new cases and 880,792 deaths in 2018.[1] Fortunately, its early detection significantly increases the survival rate, reaching a cure rate of 90% when diagnosed at a localized stage.[2] Furthermore, colorectal cancer can be prevented by detection of colonic adenomatous polyps, premalignant lesions which may progress toward colorectal cancer. It is estimated that 20%–40%[3] of patients undergoing colonoscopy present polyps. Of these detected polyps, 29%–42% are hyperplastic and entail little or no malignant risk, whereas the rest are neoplastic and could progress to colorectal cancer if not removed.[3,4,5] When multiple polyps (i.e., 5–10) are detected in a patient, the current gold standard procedure is the complete removal of all the polyps, using a technique appropriate to each polyp size and shape, followed by subsequent histopathological analysis. Resection of hyperplastic polyps, which carry no malignant potential, and the subsequent costly analysis together with patient trauma and bleeding risks could be avoided if they were accurately and objectively identified at the time of endoscopy.
Besides this, during a conventional colonoscopic polypectomy using endoscopic mucosal resection, residual adenomatous tissue rates of 46% and postprocedure recurrence rates of 12%–21.9% have been reported.[6,7] This makes follow-up and reintervention necessary, which negatively affects the prognosis of the patient and increases the risk of complications (bleeding or perforation). This fact reinforces the necessity of new in situ and in vivo diagnostic technologies to assist decision making on polyp resection by measuring the presence and degree of malignancy for the identified tissue and thus, allowing safer resection with clean margins, as the polyp margins could be analyzed prior and after resection.
The latest advances in optical imaging technologies enable structural and functional tissue microscopy in real time. Of relevance, recent studies[8,9] conclude that images of human colon tissue obtained with multiphoton excitation microscopy at high resolution (×40 objective with 1.3 NA, ×25 objective with 1.1 NA, respectively) contain morphological and functional information for discriminating between cancer, adenoma, and normal tissue. Moreover, this imaging technology takes advantage of the endogenous tissue fluorescence avoiding the use of fluorescent dyes which are time-consuming and may present the risk of toxicity. A systematic review by Tatjana et al. published in 2019[10] discusses the diagnostic value, advantages, and challenges in the practical use of multiphoton microscopy (MPM) in surgical oncology. They identified specific tumor characteristics in MPM imaging and compared this type of images to gold standard histopathology images.
While deep learning has been previously applied to the analysis of colorectal cancer (Xu et al.[11]), to the best of the authors' knowledge, there is no published work about the use of deep learning to characterize MPM images of colon tissue. In this paper, we propose a deep learning algorithm to demonstrate the capabilities of multiphoton excitation to distinguish among healthy, hyperplastic, adenomatous, and adenocarcinomatous tissue by extracting imaging biomarkers present in human colon tissue images acquired with a multiphoton microscope and histopathologically confirmed. The implemented algorithm consists of a convolutional neural network (CNN) that classifies MPM images into two classes or categories: benign (which includes images of healthy, hyperplastic, and benign neoplastic tissues) or malignant (which includes images of malignant neoplastic tissues). Adenomatous polyps are considered as benign neoplastic, whereas adenocarcinoma polyps are considered as malignant neoplastic.
MATERIALS AND METHODS
Dataset definition
The dataset consists of a set of 44 samples of lesions obtained during colonoscopies and colectomies carried out between 2012 and 2017 at the Basurto University Hospital (Spain). These are 23 malignant neoplasms (adenocarcinomas), 19 preneoplastic lesions (adenomas), and 2 hyperplastic polyps, obtained from 24 men and 18 women. The samples were diagnosed by the Pathological Anatomy Department and formalin-fixed paraffin-embedded (FFPE) blocks were stored in the Basque Biobank (structure accredited by the Health Department of Spain and inscribed in the register of the Instituto de Salud Carlos III). All the samples were processed after patients signing informed consent and following standard operation procedures. Samples obtained by cutting the FFPE blocks at 30 μm were scanned using a multiphoton microscope and later stained with hematoxylin and eosin (H and E) as explained in the next subsection (acquisition procedure). The histopathological analysis of the samples was done by the pathology department. The results are shown in Table 1 according to the nomenclature specified in Nagtegaal et al.,[12] In the case of adenocarcinomas, the terms “low grade” and “high grade” refer to the tumor grading. Low includes the well and moderately differentiated grades, whereas high refers to the poorly differentiated grade.
Table 1.
Dataset histopathological description
| Sample identified | Slide content description | Histological analysis | Scanned tissue sections |
|---|---|---|---|
| 56 | 2.2 cm part of a 7 cm size polyp obtained from the descending colon | Villous adenoma with high grade dysplasia | 1 |
| 57 | 1 cm part of a 3.7 cm size polyp obtained from the ascending colon | Tubulovillous adenoma with high grade dysplasia | 1 |
| 58 | 2.3 cm part of a 4 cm size polyp obtained from the descending colon | Villous adenoma with high grade dysplasia | 1 |
| 59 | 0.4 cm size polyp obtained from the ascending colon | Tubular adenoma | 2 |
| 60 | 3.3 cm size polyp obtained from the ascending colon | Tubulovillous adenoma with high grade dysplasia | 2 |
| 61 | 2.1 cm part of a 9 cm size polyp obtained from the descending colon | Villous adenoma with high grade dysplasia | 1 |
| 62 | 0.5 cm size polyp obtained from the ascending colon | Tubular adenoma | 1 |
| 63-1 | 1.1 cm part of a 2.8 cm size polyp obtained from the descending colon | Tubular adenoma with low grade dysplasia | 1 |
| 63-2 | 1.65 cm part of a 2.8 cm size polyp obtained from the descending colon | Adenocarcinoma over tubulovillous adenoma with high grade dysplasia | 1 |
| 64 | 0.9 cm part of a 1.2 cm size polyp obtained from the ascending colon | Tubular adenoma with low grade dysplasia | 1 |
| 65 | 6 polyps with sizes between 0.32 and 0.54 cm, belonging to a case of 118 polyps with sizes between 0.6 and 6 cm, obtained from the ascending colon | Tubular adenoma with low grade dysplasia | 1 |
| 66 | 3.1 cm part of a 9 cm size polyp obtained from the ascending colon | Tubulovillous adenoma with high grade dysplasia | 1 |
| 67 | 1.4 cm size polyp obtained from the ascending colon | Sessile tubular adenoma, low grade | 1 |
| 68 | 0.2 cm part of a 0.3 cm size polyp obtained from the descending colon | Tubular adenoma with low grade dysplasia | 1 |
| 69-1 | 2 polyps with sizes of 0.2 and 0.3 cm, belonging to a case of 5 polyps, obtained from the descending colon | Hyperplastic polyp | 1 |
| 69-2 | 0.36 cm part of a 0.4 cm size polyp obtained from the descending colon | Tubular adenoma with low grade dysplasia | 1 |
| 70 | 0.8 cm part of a 1 cm size polyp obtained from the ascending colon | Tubular adenoma with low grade dysplasia | 1 |
| 71 | 2.2 cm part of a 2.5 cm size polyp obtained from the ascending colon | Tubulovillous adenoma with high grade dysplasia | 1 |
| 72 | 3.2 cm part of a 4 cm size polyp obtained from the ascending colon | Tubular adenoma with low grade dysplasia | 1 |
| 73 | 0.2 cm size polyp obtained from the descending colon | Hyperplastic polyp | 1 |
| 74 | 1.2 cm part of a 1.8 cm size polyp obtained from the descending colon | Tubulovillous adenoma | 1 |
| 75 | No polyp from a case with a 3 cm size polyp obtained from the descending colon | Invasive colloid adenocarcinoma | 1 |
| 76 | 0.4 cm part of a 0.6 cm size polyp obtained from the transverse colon | Tubular adenoma | 6 |
| 77 | No polyp obtained from the ascending colon | Low grade adenocarcinoma, NOS | 1 |
| 78 | 2.2 cm part of a 3 cm size polyp obtained from the transverse colon | Low grade adenocarcinoma, NOS over high grade tubulovillous adenoma | 1 |
| 79 | No polyp obtained from the ascending colon | Low grade adenocarcinoma, NOS | 1 |
| 80 | No polyp obtained from the descending colon | Low grade adenocarcinoma, NOS | 1 |
| 82 | 1.4 cm part of a 4 cm size polyp obtained from the descending colon | Low grade adenocarcinoma, NOS | 1 |
| 83 | 2 cm part of a 2.3 cm size polyp obtained from the descending colon | Low grade adenocarcinoma, NOS | 1 |
| 84 | 2.6 cm part of a 4 cm size polyp obtained from the descending colon | Low grade adenocarcinoma, NOS | 1 |
| 85 | 1.5 cm part of a 2.5 cm size polyp obtained from the ascending colon | Low grade adenocarcinoma, NOS | 1 |
| 86 | 1.2 cm part of a 1.5 cm size polyp obtained from the descending colon | Low grade adenocarcinoma, NOS | 1 |
| 87 | 1.6 cm part of a 2.6 cm size polyp obtained from the ascending colon | Low grade adenocarcinoma, NOS | 1 |
| 88 | 1.9 cm part of a 4.5 cm size polyp obtained from the ascending colon | Low grade adenocarcinoma, NOS | 1 |
| 89 | 1.9 cm part of an 8.7 cm size polyp obtained from the descending colon | Low grade adenocarcinoma, NOS | 1 |
| 90 | 1.6 cm part of a 3.5 cm size polyp obtained from the descending colon | Low grade adenocarcinoma, NOS | 1 |
| 91 | 1.8 cm part of a 6.5 cm size polyp obtained from the descending colon | Low grade adenocarcinoma, NOS | 1 |
| 92 | 2.7 cm part of an 8 cm size polyp obtained from the transverse colon | High grade, adenocarcinoma NOS | 1 |
| 93 | 1 cm part of an 8 cm size polyp obtained from the descending colon | High grade adenocarcinoma, NOS | 1 |
| 94 | 2.4 cm part of a 6 cm size polyp obtained from the ascending colon | High grade adenocarcinoma, NOS | 1 |
| 95 | 1.3 cm part of a 4 cm size polyp obtained from the descending colon | Low grade adenocarcinoma, NOS | 1 |
| 96 | 1.7 cm part of a 5 cm size polyp obtained from the descending colon | Low grade adenocarcinoma, NOS | 1 |
| 97 | 1.3 cm part of a 5 cm size polyp obtained from the descending colon | High grade adenocarcinoma, NOS | 1 |
| 98 | 2 cm part of a 5 cm size polyp obtained from the descending colon | High grade adenocarcinoma, NOS | 1 |
NOS: Not otherwise specified
Note: These samples are a subset of a larger set of samples scanned with other technologies. For traceability purposes, the identifiers assigned by the histology lab are kept. Samples scanned with the multiphoton microscope for the study presented in this paper have identifiers from 56 to 98.
Acquisition procedure
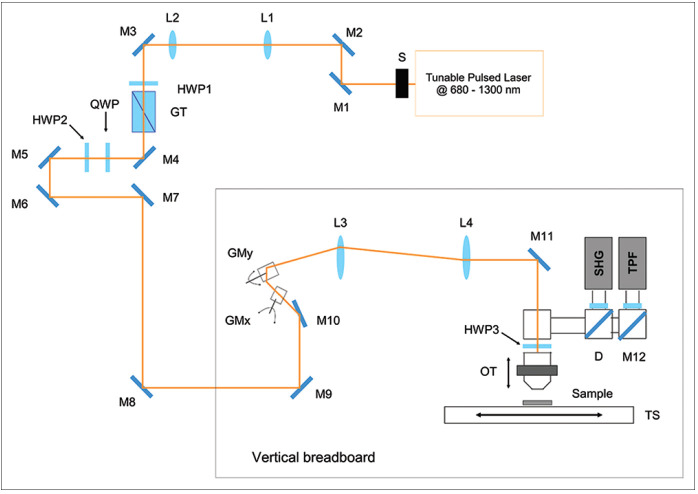
The experimental setup used for the acquisition of the maps consists of a custom-made multimodal multiphoton microscope, having the optical scheme shown in Figure 1. The excitation source is a chameleon discovery (Coherent, Santa Clara, CA, USA), an Yb-based femtosecond pulsed laser at 80 MHz rate with two synchronous outputs. The beam used in the experiments here described is tunable in the 680–1300 nm range. Immediately after the laser output, a mechanical shutter allows minimizing the sample exposure when acquiring images, a telescope collimates the beam and adjusts its size, and a motorized half-waveplate, mounted in tandem with a Glan-Taylor polarizer, is used for power dimming. Laser light is then directed onto a vertical stainless-steel optical breadboard, mounted vertically onto an antivibration optical table (Thorlabs Inc., Newton, NJ, USA), where two galvanometric mirrors (Cambridge Technology, Bedford, MA, USA) provide beam scanning. A telescope optically relays the beam to the objective lens, which is mounted on an optomechanical support equipped with both mechanical and piezoelectric (P-725KHDS PIFOC, Physik Instrumente, Karlsruhe, Germany) actuators for gross and fine movements, respectively. The samples are placed onto a xy-translator (M-687 PIline, Physik Instrumente, Karlsruhe, Germany) that allows moving the sample over a broad range with sub-micrometric resolution for mapping large areas. Fluorescence light emitted by the sample is then collected in Epi-detection by the same objective lens used for excitation, reflected by a dichroic mirror (FF665-Di02-25 × 36, Semrock Inc. New York, NY, USA) and focused on the active area of a photomultiplier tube H7422-40 (Hamamatsu, Hamamatsu City, Japan). The photocurrent is integrated using custom electronics and acquired on a PC through an acquisition board PCI-MIO (National Instruments, Austin, TX, USA) that allows synchronous signal sampling and scanner driving. System control and data acquisition are controlled using a custom software developed using LabView 2015 (National Instruments, Austin, TX, USA) development module. A more detailed description of the experimental setup can be found in literature.[13,14]
Figure 1.
Schematic of the custom-made multimodal multiphoton microscope: tunable source; shutter (S); mirrors (M); telescope lenses (L1-L2); half wave plate; quarter wave plate; Glan-Taylor polarizer; galvanometric mirrors (GMx, GMy); scan lens (L3); tube lens (L4); objective translator; XY-translation stage (TS); dichroic mirror (D)
Multiphoton fluorescence images were acquired using an excitation wavelength of 785 nm, focused on the sample by means of a Plan-Apochromat ×10 objective lens (NA 0.45, WD 2.1 mm, Carl Zeiss Microscopy, Jena, Germany). Image tiles were acquired using a field of view of 511 × 511 μm2, with a resolution of 1024 × 1024 pixel2, a pixel dwell time of 5 μs, and an average laser power of about 20 mW on the sample.
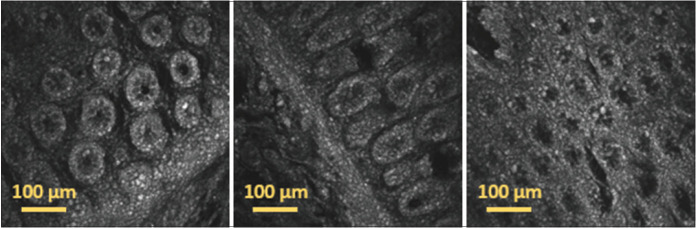
As example of the captured images, Figure 2 shows several 511 × 511 μm2 image tiles acquired with the multiphoton microscope using two-photon fluorescence (TPF) in different positions of the same tissue slide.
Figure 2.
Individual image tiles acquired using two-photon fluorescence in different positions of a 30 μm thick paraffin-embedded tissue slide with sample 73 diagnosed as hyperplastic polyp. The images show cells with different shape and morphology acquired in different regions of the sample, demonstrating the capability of two-photon fluorescence for the label-free morphological assessment of tissues
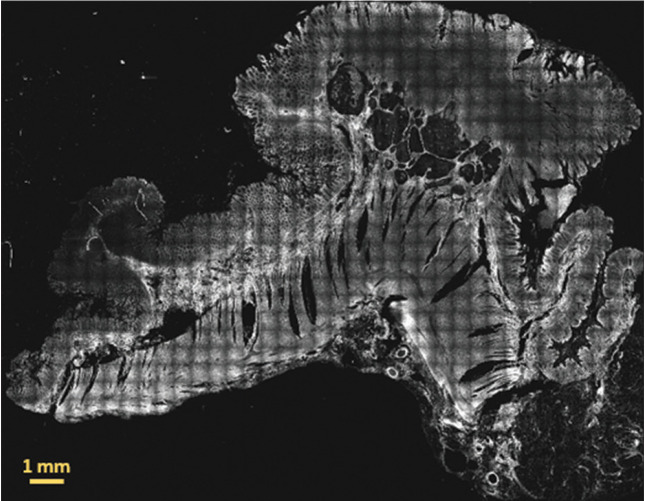
Figure 3 shows an example of a whole 30 μm thick paraffin-embedded tissue slide scanned with the multiphoton microscope. The image has been generated by concatenating all the individual TPF image tiles.
Figure 3.
Two-photon fluorescence image of a whole 30 μm thick paraffin-embedded tissue slide with sample 86 diagnosed as low grade adenocarcinoma. The signal originates mainly from mitochondrial NADH in the cell cytoplasm and from elastic fibers and other fluorescent molecules in the extracellular matrix. This image has been obtained by merging 37 by 29 image tiles, resulting in an overall field of view: 18.907 mm by 14.819 mm
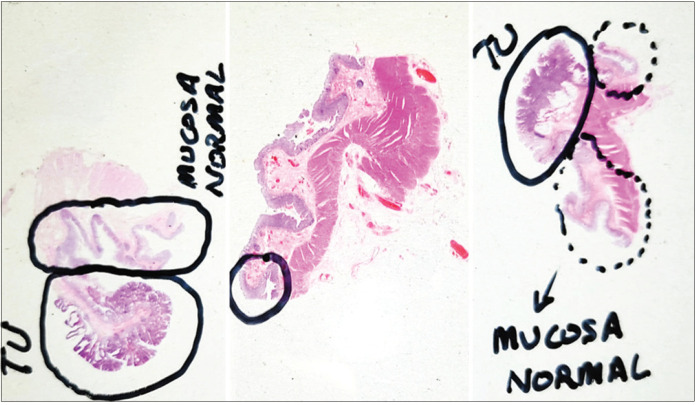
Samples analyzed for this study were cut with a rotary microtome (RM2255, Leica Biosystems, Wetzlar, Germany) at 3 and 30 μm from FFPE blocks of human tissue. Then, superfrost slides (LineaLAB, Badalona (Barcelona), Spain) were H and E stained in an automated slide stainer (SIMPHONY system, F. Hoffmann-La Roche Ltd, Basel, Switzerland). The histopathologists analyzed the stained slides under a microscope and annotated them as shown in the examples of Figure 4. Finally, all the slides were scanned with a fully motorized microscope (BX 61, Olympus Corporation, Tokyo, Japan) equipped with Ariol software platform, where all the images had ×1.25 and ×20s magnification.
Figure 4.
Examples of tissue slides annotated by the histopathologists. Left: specimen 57 with tumoral area diagnosed as tubulovillous adenoma with high grade dysplasia; centre: specimen 73 with marked area diagnosed as hyperplastic polyp; right: specimen 86 with tumoral area diagnosed as low grade adenocarcinoma. TU stands for “tumoral”
Clinical capabilities of multiphoton fluorescence images
To gain some understanding of the potential uses for the MPM images, we aimed to establish what clinical information can be obtained from them. A representative panel of 18 tissue samples acquired with the multiphoton microscope were presented to four immunoglobulins pathologists, as well as the equivalent images from the H and E slides. The images were viewed at a low-power view (the complete sample image including all tiles) and at a higher-power view by examining individual tiles within a region of interest, in much the same manner as examining a conventional H and E slide. Figure 5 shows an example of one of the analyzed MPM images and its corresponding co-registered H and E image.
Figure 5.
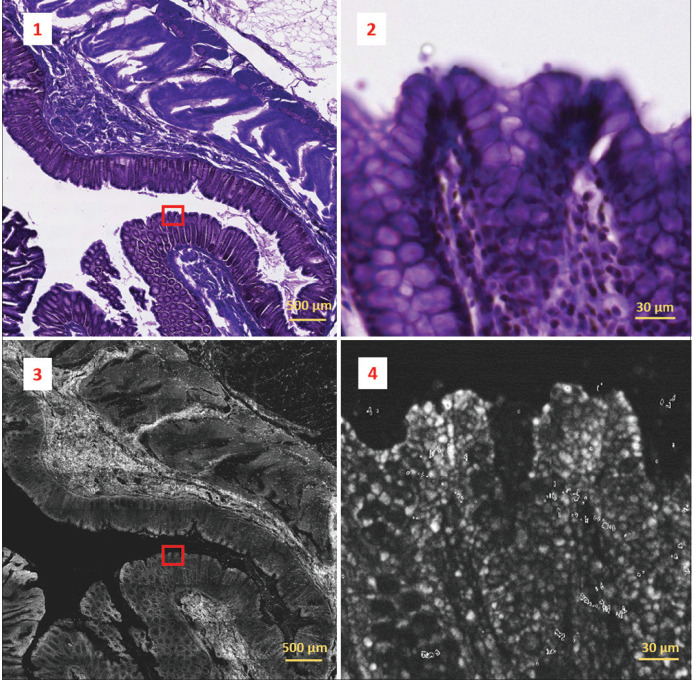
Two-photon fluorescence image (bottom-left) acquired from a 30 μm thick paraffin-embedded tissue slide with sample 69-2 diagnosed as tubular adenoma with low grade dysplasia and its co-registered corresponding H&E image (top-left). Two-photon fluorescence image was obtained by concatenating 9 by 12 image tiles. The overall field of view results in 4.599 mm by 6.132 mm. Detail marked by the red box in the images on the left is represented on a magnified scale on the right. Colonic crypts and goblet cells can be identified in the H&E crop (top-right), but these features are not appreciable on the corresponding two-photon fluorescence crop (bottom-right)
The findings of the MPM image analysis showed promise, in that the tiled images viewed as a whole, showed good fidelity with the H and E samples and allowed for the assessment of gross cellular architectural structures. On closer examination of the individual image tiles, the level of detail appreciated on the MPM images was felt to be less than that found in traditional H and E slides. Part of the reason for this was the reduced visual contrast as the MPM images are grayscale as opposed to the colored H and E staining. It was also felt that the intracellular features such as cell nuclei were less apparent on the MPM images. Overall, the MPM images do show tissue architecture, but not at the level of detail shown in traditional H and E images as presented in Figure 5 which shows representative microscopic images of human colon, obtained using traditional H and E staining (1,2) and MPM (3,4) and viewed at low-power (1,3) and high-power (2,4) magnification. High-power magnification of the H and E stained slide identifies the colonic crypts and goblet cells, although these features are not appreciable on the high-power MPM image.
From a diagnostic point of view, it was felt to be difficult to make a confident tissue diagnosis using the MPM images. The reasons for this are complex, but relate in part to the reduced level of detail within the images, and partly to the unfamiliarity with MPM images when compared with H and E slides; it is difficult for a human operator to confidently interpret a new image modality until this is fully understood and more widely studied. From the point of view of machine-learning, however, the observed similarities between H and E and MPM slides suggest that there is potential for a deep learning approach to analysis of the MPM images.
Dataset partitioning
In order to feed the proposed CNN, the dataset consists of the TPF images acquired with the multimodal multiphoton microscope and converted to PNG file format. To generate these image files, the values returned by the multiphoton microscope have been normalized using the maximum and minimum values of all the tiles belonging to the same sample and then rescaled between 0 and 255. Each image corresponds to an area of 511 × 511 μm2, so the scanning of each sample results in several images that are consecutive tiles of the whole slide. The images are grouped into two categories or classes: benign for images of tissue samples with benign lesions, that is, hyperplastic polyps or adenomas; and malignant for images of tissue samples with adenocarcinomas.
Initial image tiles have been filtered discarding those images without tissue (only the microscope slide appears in the image), with little tissue or with strong artifacts. The rest of the image tiles of the benign samples are included in the dataset including tiles containing normal (that is, healthy) tissue. In the case of the malignant samples, the image tiles that are outside the tissue area marked as adenocarcinoma by the pathologists are considered as tiles with uncertain diagnosis and not included in the dataset.
The resulting dataset is composed of 14,712 images of 1024 × 1024 pixels and it is well balanced with 6985 images from benign lesions and 7727 images from malignant lesions. Then, for algorithm development purposes, the dataset has been partitioned into training (70%), validation (15%), and test (15%) sequences as described in Table 2. Six different partitions have been generated for statistics purpose. The dataset is openly available at https://www.biobancovasco.org/en/Sample-and-data-e-catalog/PD177-Databases-EN.html after download request form completion (https://forms.office.com/r/ycdFxGaMEE).
Table 2.
Dataset partition summary
| Partition | Class | Training | Validation | Test | |||
|---|---|---|---|---|---|---|---|
|
|
|
|
|||||
| Image number | Sample identified | Image number | Sample identified | Image number | Sample identified | ||
| K1 | Benign | 4843 | 56, 58, 60, 631, 66, 67, 691, 692, 70, 71, 73, 76 | 1028 | 57, 62, 64, 68 | 1114 | 59, 61, 65, 72, 74 |
| Malignant | 5401 | 632, 77, 82, 83, 84, 85, 87, 88, 90, 91, 94, 95, 96, 97, 98 | 1173 | 75, 78, 80, 86, 93 | 1153 | 79, 89, 92 | |
| K2 | Benign | 4931 | 56, 57, 59, 60, 61, 631, 65, 66, 67, 68, 691, 692, 73, 76 | 991 | 71, 72, 74 | 1063 | 58, 62, 64, 70 |
| Malignant | 5428 | 632, 75, 78, 80, 82, 83, 84, 86, 87, 88, 91, 93, 95, 96, 97, 98 | 1153 | 79, 89, 92 | 1146 | 77, 85, 90, 94 | |
| K3 | Benign | 4914 | 57, 58, 59, 61, 62, 631, 64, 66, 67, 68, 70, 71, 72, 73, 74, 76 | 1044 | 60, 65, 691 | 1027 | 56, 692 |
| Malignant | 5392 | 632, 75, 77, 78, 79, 84, 85, 87, 89, 90, 91, 92, 94, 95, 96, 98 | 1168 | 83, 88, 97 | 1167 | 80, 82, 86, 93 | |
| K4 | Benign | 4902 | 57, 58, 60, 61, 62, 64, 65, 67, 68, 70, 71, 72, 73, 74, 76 | 1052 | 56, 59, 692 | 1031 | 631, 66, 691 |
| Malignant | 5431 | 78, 79, 80, 82, 84, 86, 87, 88, 89, 91, 92, 93, 95, 96, 98 | 1146 | 77, 85, 90, 94 | 1150 | 632, 75, 83, 97 | |
| K5 | Benign | 4852 | 56, 59, 60, 61, 62, 64, 65, 66, 691, 692, 71, 72, 73, 74, 76 | 1064 | 58, 631, 70 | 1069 | 57, 67, 68 |
| Malignant | 5485 | 75, 77, 78, 79, 80, 82, 83, 85, 86, 87, 88, 89, 90, 92, 93, 94, 97 | 1109 | 632, 84, 96 | 1133 | 91, 95, 98 | |
| K6 | Benign | 4899 | 56, 57, 58, 59, 60, 61, 62, 631, 64, 65, 68, 691, 692, 70, 72, 74 | 1025 | 66, 67 | 1061 | 71, 73, 76 |
| Malignant | 5467 | 632, 75, 77, 79, 80, 82, 83, 85, 86, 89, 90, 91, 92, 93, 94, 96, 97 | 1190 | 87, 95, 98 | 1070 | 84, 88, 78 | |
Later, during the network training phase, data augmentation has been applied on the training set in order to have more training images. Specifically, the left-right flip and 90°, 180° and 270° rotation transformations have been randomly performed with a probability of 0.5.
Deep learning architecture
Proposed architecture
The purpose of the present analysis is to study if the human colon tissue images acquired with a multiphoton microscope include enough information, so that a deep learning-based algorithm can recognize images of malignant neoplastic lesions and distinguish them from images of healthy, hyperplastic, or benign neoplastic tissues. To this end, we have built a two-class classifier based on the Xception model architecture.[15]
The network architecture is modified following the methodology explained in.[16,17] In this case, a CNN with Xception architecture has been instantiated removing the top fully connected layer. After the last convolutional block, a global average pooling layer is added followed by a fully connected layer with two outputs and softmax activation. The network is initialized with the weights pretrained on image net dataset.[18] Details on training are given in next section.
Dataset images are grouped into training, validation, and test sequences as described in the previous section. These images need to be preprocessed. Since pretrained image net weights are used for network initialization, the images in our dataset were resized from (1024, 1024) to the expected input shape of (299, 299, 3). In order to perform image net based fine tuning, the same gray-scale image has been repeated into the three channels and the preprocessing method for the Xception model is applied.
Baseline multiphoton microscopy classifier
The described network model has been trained in three phases: first, only the top Dense layer (which weights are randomly initialized) and the Batch Normalization layers are trained, while the weights of all convolutional layers are frozen; in the second phase, the weights of the convolutional layers of the last three Xception blocks are unfrozen and trained together with the Batch Normalization and top Dense layers; and finally, in the third phase, all the layers are unfrozen and trained. The momentum parameter of the Batch Normalization layers is set to 0.9. The training tries to minimize a categorical cross-entropy loss function. Each training phase runs for 30 epochs and data is divided into batches of 4 images. The optimization configuration is the same in the three training phases: The Adam optimizer is used with an initial learning rate equal to 1e-4 and the learning rate is reduced by a factor of 10 if the validation loss does not improve for 10 training epochs.
Once the model has been trained and tested following the described strategy, the last step has been to look for the threshold value between the two classes (by default this threshold is set to 0.5) that maximizes the balanced accuracy (BAC) for the validation set. The results are very promising as shown later in the results section.
Multiphoton microscopy classifier with spatially coherent predictions
Two strategies have been followed to improve the performance of the baseline MPM classifier: test-time augmentation (TTA) and prediction based on adjacent tiles or spatially coherent predictions (SCP).
TTA consists in creating several copies of each image in the test set by performing different image transformations. Then, predictions are made for the original image and each copy, and the final prediction for the image to test has been generated as the mean of all the predictions. In our experiment, the transformations have consisted in 90°, 180°, and 270° rotations and left-right flips, generating seven augmented copies per test image. This strategy does not improve the performance of the baseline model, leading to similar metrics values.
The second strategy relies on the fact that a lesion extends over several tiles and therefore, the image tiles that belong to a certain lesion area have the same diagnosis. In the practice, the algorithm identifies all the image tiles that are adjacent to each image to test, and it is analogous to a human pathologist examining a region of tissue and interpreting their findings in the context of surrounding tissue structure. We can find between 0 and 8 adjacent tiles per image in our test set. Then, the final prediction for each image is calculated as the mean of the predictions for the image and its adjacent tiles. This mechanism improves the mean BAC of the baseline model in almost 4 points, as it is described in the next section.
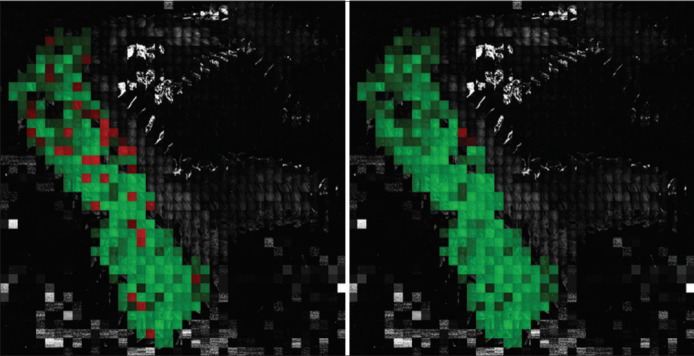
An example of the classification improvement with the SCP method over the baseline model is shown in Figure 6. Two images of a full slide created by concatenating its image tiles after resizing the original 1024 × 1024 pixels tiles into 128 × 128 pixels tiles are shown in the figure. Concretely, the slide corresponds to a malignant tissue sample. The tiles that are correctly classified by the algorithm are highlighted in green, while those tiles that are misclassified are highlighted in red color. The gray tiles are the sample tiles that have not been included in the dataset, because they are tiles without tissue, with little tissue, with strong artifacts, or tiles outside the area marked as adenocarcinoma by the pathologist. In this example, 37 tiles out of the 268 tiles are misclassified with the baseline model. However, with the SCP scheme, only 3 tiles result in wrong classification.
Figure 6.
Classification improvement with the spatially coherent predictions method versus the baseline model: correctly classified tiles are shown in green, while misclassified tiles are highlighted in red; tiles in gray are the tiles not included in the dataset. 37 tiles out of 268 were misclassified with the baseline model (left). Only 3 tiles resulted misclassified with the spatially coherent predictions method (right)
RESULTS
The proposed classifier architecture and training mechanism results in a BAC of 0.8293 ± 0.0895 for the baseline model, calculating the mean and standard deviation of the results obtained with the six dataset partitions detailed in Table 2. When the adjacent tiles are considered in the prediction (SCP scheme), the BAC increases up to 0.8671 ± 0.0966. These results confirm that the combination of this novel imaging technology together with deep learning algorithms leads the way to perform real-time optical biopsies for in vivo diagnosis.
The confusion matrixes over the test sequence in partition K1 for both, baseline and SCP models, are presented in Tables 3 and 4. The performance metrics of the implemented network model for all dataset partitions are shown in Table 5.
Table 3.
Confusion matrix over the testing subset for the baseline model
| Actual | Prediction | |
|---|---|---|
|
| ||
| Benign | Malignant | |
| Benign | 859 | 255 |
| Malignant | 142 | 1008 |
Table 4.
Confusion matrix over the testing subset for the spatially coherent predictions model
| Actual | Predict | |
|---|---|---|
|
| ||
| Benign | Malignant | |
| Benign | 948 | 166 |
| Malignant | 106 | 1047 |
Table 5.
Multiphoton microscopy classifier performance metrics
| Model | Partition | BAC | Sensitivity | Specificity | PPV | NPV | Threshold |
|---|---|---|---|---|---|---|---|
| Baseline | K1 | 0.8238 | 0.8765 | 0.7711 | 0.7981 | 0.8581 | 0.10 |
| K2 | 0.9419 | 0.9083 | 0.9755 | 0.9756 | 0.9081 | 0.48 | |
| K3 | 0.6654 | 0.6249 | 0.7059 | 0.7068 | 0.6239 | 0.59 | |
| K4 | 0.8871 | 0.8460 | 0.9282 | 0.9293 | 0.8439 | 0.71 | |
| K5 | 0.7783 | 0.6295 | 0.9270 | 0.9013 | 0.7028 | 0.14 | |
| K6 | 0.8796 | 0.9213 | 0.8379 | 0.8511 | 0.9137 | 0.13 | |
| Mean±SD | 0.8293±0.0895 | 0.8011±0.1252 | 0.8576±0.0954 | 0.8604±0.0887 | 0.8084±0.1080 | N/A | |
| SCP | K1 | 0.8795 | 0.9081 | 0.8510 | 0.8632 | 0.8994 | 0.38 |
| K2 | 0.9932 | 0.9939 | 0.9925 | 0.9930 | 0.9934 | 0.34 | |
| K3 | 0.7003 | 0.6255 | 0.7751 | 0.7596 | 0.6456 | 0.51 | |
| K4 | 0.9095 | 0.8296 | 0.9893 | 0.9886 | 0.8388 | 0.72 | |
| K5 | 0.7884 | 0.6011 | 0.9757 | 0.9632 | 0.6977 | 0.42 | |
| K6 | 0.9318 | 0.9785 | 0.8850 | 0.8956 | 0.9761 | 0.39 | |
| Mean±SD | 0.8671±0.0966 | 0.8228±0.1575 | 0.9114±0.0814 | 0.9105±0.0826 | 0.8418±0.1314 | N/A |
BAC: Balanced accuracy, PPV: Positive predictive values, NPV: Negative predictive values, SCP: Spatially coherent predictions, SD: Standard deviation, N/A: Not applicable
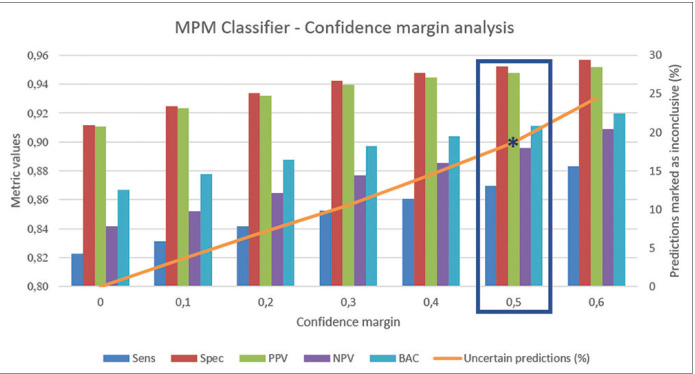
We have additionally analyzed the effect of the self-confidence estimation. For this, we have taken the SCP model (the one that takes adjacent tiles into account) and the threshold value that maximizes the BAC, and we have defined a confidence margin around this threshold value that makes network predictions falling in that range (threshold ± margin) to be marked as uncertain. Then, we apply this threshold and uncertainty margin to the predictions for the test set of the six partitions. The effect of the confidence margin on the mean performance metrics is shown in Figure 7. We can appreciate the increase on the performance as we increase the uncertainty margin, getting a BAC of 0.9111 (0.8697 sensitivity and 0.9524 specificity) when we classify as uncertain the 18.67% of the test images.
Figure 7.
Effect of the confidence margin on the performance metrics: as the uncertainty margin increases (orange line) and more predictions are marked as uncertain, the performance for the rest of the predictions increases resulting in better metrics. A confidence margin of 0, 5 around the classification threshold results in a balanced accuracy of 0.9111 (0.8697 sensitivity and 0.9524 specificity) and 18.67% of the test images marked as uncertain
DISCUSSIONS
The use of deep learning methods to efficiently characterize MPM datasets is still in an early stage. However, some recently published studies illustrate the potential derived from using MPM imaging together with deep learning. For example, the work presented by Huttunen et al.,[19] shows a successful classification of MPM images of murine ovarian tissue as healthy or high-grade serous carcinoma achieving 95% sensitivity and 97% specificity; Lin et al. demonstrate[20] the fusion of MPM and deep learning to classify different stages of hepatocellular carcinoma with an accuracy over 90%; Huttunen et al. show[21] that MPM images collected on the dermo-epidermal junction can be automatically classified into healthy and dysplastic classes with high precision (sensitivity, specificity and accuracy all exceeding 90%) using a deep learning method and existing pretrained CNNs; in the work presented by Guimarães et al.,[22] they train and tune a CNN to detect the presence of living cells, and if so, to diagnose atopic dermatitis from MPM images; finally, Yu et al. explain[23] the use of a pretrained deep learning network for automated MPM image classification and scoring of liver fibrosis stages in a Wistar Rat model. These previous works have encouraged our study about the use of deep learning to characterize MPM images of human colon tissue.
The results presented in Table 5 are good, but other published deep learning studies using images of human colon or gastrointestinal cancer tissues can be found presenting better metric results. For example, the work from Xu et al.[11] achieves an accuracy of 99.9% for normal slides and 94.8% for cancer slides on H and E stained histology slides, or the work from Wei et al.[24] obtains a mean accuracy of 93.5% (86.8% sensitivity and 95.7and specificity) when classifying histology slides into four colorectal polyp types. However, it is not straightforward to compare the results of our work with this type of studies, because the nature of the images is different. As described in the “Clinical capabilities of multiphoton fluorescence images” section, the level of detail of the features presented in MPM images is not the same as in images of H and E stained tissue. There are some imaging biomarkers present in H and E images that are not present in MPM images, such as colonic crypts and goblet cells. In addition, most works, such as the mentioned references, report classification results for whole slides, while our metrics refer to image tiles or patches which is a more complicated approach.
Regarding studies using MPM images, the works presented by Li et al.[25,26] related to colorectal and gastric tissue respectively, or the study shown by He et al.[27] about colorectal tissue, obtain sensitivity and specificity values higher than those obtained in our study. However, the data have been obtained on fresh colon tissue and on frozen stomach tissue. Hence, it is a bit difficult to compare the results as we used paraffine fixed tissue specimens. It can be expected that the diagnostic accuracy is higher when using fresh tissue since the molecules contributing to the auto fluorescence signal are less degraded and the signal is stronger and hence, more functional information available. As a confirmation of this hypothesis, in the studies examined in the review paper,[10] the diagnostic accuracy is almost always higher when using fresh or frozen tissue than using fixed tissues, even if it is difficult to compare data obtained on different tissue types.
CONCLUSIONS
At present, colonoscopy is the gold standard technique for the detection and removal of colorectal lesions with potential to evolve into cancer. However, in this process, gastroenterologists cannot assure complete resection and clean margins which are given by the posterior histopathology analysis of the removed tissue that is performed at laboratory, leading to potential recurrence of the lesions. Besides, the current clinical procedure involves the complete removal of all the polyps found in the colonoscopy, including hyperplastic polyps, which carry no malignant potential. Therefore, clinicians demand new in situ and in vivo diagnostic technologies that help them in the detection and discrimination of hyperplastic and neoplastic polyps and assist them during the polyp resection process assuring clean margin detection.
The work presented in this paper demonstrates that a deep learning algorithm can recognize images of malignant neoplastic lesions and distinguish them from images of healthy, hyperplastic, or benign neoplastic tissues, by autonomously extracting imaging biomarkers present in human colon tissue images acquired with a multiphoton microscope. The dataset generated to carry out this analysis has also been presented and it is openly available. The implemented classifier gets a BAC of 0.8671 ± 0.0966 (0.8228 ± 0.1575 sensitivity and 0.9114 ± 0.0814 specificity).
Current work presents a first attempt of applying deep learning methods over a dataset of human colon MPM images acquired from tissue slides. Although obtained results are promising in this direction, the deep learning strategy can be further improved. The relevance of our research is that it shows the potential of both technologies (multiphoton imaging and deep learning) for colon cancer detection. Some published studies about multiphoton imaging and deep learning with other types of tissue show classification accuracies over 90%[20,21] or even higher, such as in Huttunen et al.,[19] where they have achieved 95% sensitivity and 97% specificity. Considering this, the following future steps could be implemented with the aim of reducing the false negative rate:
Implement multiple instance learning approaches, since it could reduce the problem of the weakly supervised tiles
Add contextual information on close tiles or use multiscale networks, which will help integrate fine details and contextual information
Study whether combining TPF images with second harmonic generation images could lead to better results. The study presented by Huttunen et al.,[21] reports sensitivity and specificity improvement when combining both types of multiphoton images, although the results published by Huttunen et al.,[19] show only a slight improvement.
As mentioned before, the dataset is public and available for download after request form completion which can lead to further research of other groups working on this subject.
Apart from being used to implement the deep learning model, the acquired MPM images have been reviewed by an external panel of pathologists who have concluded that although cellular structure is apparent, the level of diagnostic information is not adequate for making a confident informed diagnosis of tissue type. This is due in part to the lower level of detail in comparison with H and E images, but it is also related to the novel nature of the images and consequent unfamiliarity with them. This gives us insight that a method for comparison of MPM images with an imaging modality well-known by clinicians, such as H and E stained microscopy images, is necessary to break down the barriers for incorporating new imaging technologies into real clinical practices. In fact, in parallel to this study, we have worked on an algorithm to transform multiphoton images into virtually H and E stained images which can be interpreted by clinicians, since it is their current gold standard image modality. This work has been presented by Picon et al.,[28] and it also includes the results of classifying these virtually stained images with an algorithm trained with native H and E images. Further steps in this direction are to be included in a new publication soon.
Clinicians also distrust black-box methods (as deep learning) where further assessment of the reasoning process cannot be done, preventing their adoption in the daily clinical practice. The application of explainability methods could therefore be useful to understand and demonstrate what imaging features a neural network looks for. This is another interesting work line complementary to our work presented here, to facilitate the adoption of new optical imaging modalities for clinical diagnosis. In this regard, the adequacy of the currently available methods, designed to detect features at macroscopic level, to explain deep learning models that identify features at microscopic level should be investigated.
Finally, the SCP scheme has been used to perform diagnosis prediction for a tile taking into account the predictions for its adjacent tiles, increasing, in this way, the algorithm performance. This scheme could also be used to perform diagnosis of the whole lesion considering the mean score for all its tiles. This opens the possibility to perform digital pathology for automatic slide classification and diagnosis without H and E staining.
Financial support and sponsorship
This work was supported by the PICCOLO project. This project has received funding from the European Union's Horizon2020 Research and Innovation Programme under grant agreement No. 732111. The sole responsibility of this publication lies with the authors. The European Union is not responsible for any use that may be made of the information contained therein. This research has also been supported by the project ONKOTOOLS (KK-2020/00069) funded by the Basque Government Industry Department under the ELKARTEK program.
Conflicts of interest
There are no conflicts of interest.
Acknowledgments
The authors would like to thank Roberto Bilbao, director of the Basque Biobank, Ainara Egia Bizkarralegorra and biobank technicians from Basurto University Hospital (Spain) and pathologist Prof. Rob Goldin from Imperial College London (UK).
Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2021/12/1/27/320051
REFERENCES
- 1.GLOBOCAN 2018 Database, Accessible Online at the Global Cancer Observatory. [Last accessed 2020 Oct 30]. Available from: https://gco.iarc.fr .
- 2.SEER 18, Relative Survival by Stage (2010-2016) Database, Accessible Online At. [Last accessed 2020 Oct 30]. Available from: https://seer.cancer.gov/canques/survival.html .
- 3.Rajasekhar PT, Mason J, Wilson A, Close H, Rutter M, Saunders B, et al. Detect inspect characterise resect and discard 2: Are we ready to dispense with histology? Gut. 2015;64:A13. [Google Scholar]
- 4.Kaltenbach T, Rastogi A, Rouse RV, McQuaid KR, Sato T, Bansal A, et al. Real-time optical diagnosis for diminutive colorectal polyps using narrow-band imaging: The VALID randomised clinical trial. Gut. 2015;64:1569–77. doi: 10.1136/gutjnl-2014-307742. [DOI] [PubMed] [Google Scholar]
- 5.Hale MF, Kurien M, Basumani P, Slater R, Sanders DS, Hopper AD. Endoscopy II: PTU-233 In vivo polyp size and histology assessment at colonoscopy: Are we ready to resect and discard.A multi-centre analysis of 1212 polypectomies? Gut. 2012;61:A280–1. [Google Scholar]
- 6.Kedia P, Waye JD. Colon polypectomy: A review of routine and advanced techniques. J Clin Gastroenterol. 2013;47:657–65. doi: 10.1097/MCG.0b013e31829ebda7. [DOI] [PubMed] [Google Scholar]
- 7.Sánchez-Peralta LF, Bote-Curiel L, Picón A, Sánchez-Margallo FM, Pagador JB. Deep learning to find colorectal polyps in colonoscopy: A systematic literature review. Artif Intell Med. 2020;108:101923. doi: 10.1016/j.artmed.2020.101923. [DOI] [PubMed] [Google Scholar]
- 8.Cicchi R, Sturiale A, Nesi G, Kapsokalyvas D, Alemanno G, Tonelli F, et al. Multiphoton morpho-functional imaging of healthy colon mucosa, adenomatous polyp and adenocarcinoma. Biomed Opt Express. 2013;4:1204–13. doi: 10.1364/BOE.4.001204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsui T, Mizuno H, Sudo T, Kikuta J, Haraguchi N, Ikeda JI, et al. Non-labeling multiphoton excitation microscopy as a novel diagnostic tool for discriminating normal tissue and colorectal cancer lesions. Sci Rep. 2017;7:6959. doi: 10.1038/s41598-017-07244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tatjana TK, Jan G, Oliver JM. Multiphoton microscopy in surgical oncology – A systematic review and guide for clinical translatability. Surg Oncol. 2019;31:119–31. doi: 10.1016/j.suronc.2019.10.011. [DOI] [PubMed] [Google Scholar]
- 11.Xu L, Walker B, Liang PI, Tong Y, Xu C, Su YC, et al. Colorectal cancer detection based on deep learning. J Pathol Inform. 2020;11:28. doi: 10.4103/jpi.jpi_68_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagtegaal I, Odze R, Klimstra D, Paradis V, Rugge M, Schirmacher P, et al. 5th ed. Vol. 1. Lyon (France): IARC; 2019. Digestive System Tumours: WHO Classification of Tumours. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marchetti M, Baria E, Cicchi R, Pavone FS. Custom multiphoton/raman microscopy setup for imaging and characterization of biological samples. Methods Protoc. 2019;2:51. doi: 10.3390/mps2020051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dal Fovo A, Sanz M, Mattana S, Oujja M, Marchetti M, Pavone FS, et al. Safe limits for the application of nonlinear optical microscopies to cultural heritage: A new method for in-situ assessment. Microchem J. 2020;154:104568. [Google Scholar]
- 15.Chollet F. Honolulu, HI: IEEE; 2017. Deep learning with depthwise separable convolutions. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) pp. 1800–7. [Google Scholar]
- 16.Medela A, Picon A, Saratxaga CL, Belar O, Cabezón V, Cicchi R, et al. Few shot learning in histopathological images: Reducing the need of labelled data on biological datasets. IEEE Int Symp Biomed Imaging. 2019:1860–4. [Google Scholar]
- 17.Medela A, Picon A. Constellation loss: Improving the efficiency of deep metric learning loss functions for the optimal embedding of histopathological images. J Pathol Inform. 2020;11:38. doi: 10.4103/jpi.jpi_41_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, et al. Image net large scale visual recognition challenge. Int J Comput Vis. 2015;115:211–52. [Google Scholar]
- 19.Huttunen MJ, Hassan A, McCloskey CW, Fasih S, Upham J, Vanderhyden BC, et al. Automated classification of multiphoton microscopy images of ovarian tissue using deep learning. J Biomed Opt. 2018;23:1–7. doi: 10.1117/1.JBO.23.6.066002. [DOI] [PubMed] [Google Scholar]
- 20.Lin H, Wei C, Wang G, Chen H, Lin L, Ni M, et al. Automated classification of hepatocellular carcinoma differentiation using multiphoton microscopy and deep learning. J Biophotonics. 2019;12:e201800435. doi: 10.1002/jbio.201800435. [DOI] [PubMed] [Google Scholar]
- 21.Huttunen MJ, Hristu R, Dumitru A, Floroiu I, Costache M, Stanciu SG. Multiphoton microscopy of the dermoepidermal junction and automated identification of dysplastic tissues with deep learning. Biomed Opt Express. 2020;11:186–99. doi: 10.1364/BOE.11.000186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guimarães P, Batista A, Zieger M, Kaatz M, Koenig K. Artificial intelligence in multiphoton tomography: Atopic dermatitis diagnosis. Sci Rep. 2020;10:7968. doi: 10.1038/s41598-020-64937-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu Y, Wang J, Ng CW, Ma Y, Mo S, Fong ELS, et al. Deep learning enables automated scoring of liver fibrosis stages. Sci Rep. 2018;8:16016. doi: 10.1038/s41598-018-34300-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei JW, Suriawinata AA, Vaickus LJ, Ren B, Liu X, Lisovsky M, et al. Evaluation of a deep neural network for automated classification of colorectal polyps on histopathologic slides. JAMA Netw Open. 2020;3:e203398. doi: 10.1001/jamanetworkopen.2020.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L, Chen Z, Wang X, Li H, Jiang W, Zhuo S, et al. Detection of morphologic alterations in rectal carcinoma following preoperative radiochemotherapy based on multiphoton microscopy imaging. BMC Cancer. 2015;15:142. doi: 10.1186/s12885-015-1157-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li L, Kang D, Huang Z, Zhan Z, Feng C, Zhou Y, et al. Multimodal multiphoton imaging for label-free monitoring of early gastric cancer. BMC Cancer. 2019;19:295. doi: 10.1186/s12885-019-5497-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He K, Zhao L, Chen Y, Huang X, Ding Y, Hua H, et al. Label-free multiphoton microscopic imaging as a novel real-time approach for discriminating colorectal lesions: A preliminary study. J Gastroenterol Hepatol. 2019;34:2144–51. doi: 10.1111/jgh.14772. [DOI] [PubMed] [Google Scholar]
- 28.Picon A, Medela A, Sanchez-Peralta LF, Cicchi R, Bilbao R, Alfieri D, et al. Autofluorescence image reconstruction and virtual staining for in-vivo optical biopsying. IEEE Access. 2021;9:32081–93.s. [Google Scholar]