ABSTRACT
The complete genome sequence of Lactobacillus crispatus type strain ATCC 33820 was obtained by combining Nanopore and Illumina sequencing technologies. The genome consists of a 2.2-Mb circular chromosome with 2,194 open reading frames and an average GC content of 37.0%.
ANNOUNCEMENT
Lactobacillus crispatus is the most frequently isolated species among the vaginal lactobacilli of the human microbiota of healthy women; its presence is associated with reduced risk of preterm delivery, viral sexually transmitted infections, and bacterial vaginosis (1). To date (June 2021), only eight L. crispatus complete genomes are available in the NCBI database (https://www.ncbi.nlm.nih.gov/genome/browse#!/prokaryotes/1815/). Here, we contribute to the genomic characterization of this species by publicly releasing the genome of strain ATCC 33820, the type strain of Lactobacillus crispatus (Fig. 1). The strain was purchased from the American Type Culture Collection in October 2020 and grown in 250 ml of DeMan-Rogosa-Sharpe (MRS) broth at 37°C to an optical density at 590 nm (OD590) of 1.9. Bacterial cells were harvested by centrifugation (5,000 × g for 30 min at 4°C), and the cell pellet was dry-vortexed and incubated for 1 h at 37°C in protoplasting buffer (20% raffinose, 50 mM Tris-HCl [pH 8.0], 5 mM EDTA) containing 4 mg/ml lysozyme. Protoplasts were centrifuged (5,000 × g for 5 min), resuspended in 15 ml of deionized H2O with 100 μg/ml proteinase K (Merck KGaA, Darmstadt, Germany), and incubated for 30 min at 37°C to obtain osmotic lysis, with 0.5% SDS added after 15 min. Then, 0.55 M NaCl was added, and the mixture was incubated for 10 min at room temperature. High-molecular-weight DNA was purified by three extractions with 1 volume of Sevag (chloroform-isoamyl alcohol, 24:1 [vol:vol]), precipitated in 0.6 volume of cold isopropanol, and spooled on a glass rod. DNA was resuspended in saline-sodium citrate (SSC)/10 buffer and then adjusted to 600 μl SSC 1×. The DNA solution was homogenized using a rotator mixer and stored at 4°C. DNA sequencing was performed with both Oxford Nanopore GridION and Illumina NovaSeq 6000 instruments. The Nanopore sequencing library was prepared using the Nanopore sequencing kit SQK-LSK 109 (Oxford Nanopore Technologies, Oxford, UK), and the sample was sequenced using an R9.4 flow cell (FLO-MIN106). Real-time high-accuracy base calling (quality cutoff, >Q7) of Nanopore reads was performed using Guppy v4.0.11 (https://github.com/nanoporetech/pyguppyclient), and base-called reads were analyzed with NanoPlot v1.18.2 (2). Illumina sequencing was performed at MicrobesNG (University of Birmingham, UK) using a Nextera XT library preparation kit (Illumina Inc., San Diego, CA, USA), followed by paired-end sequencing. Illumina reads were trimmed using Trimmomatic v0.30 (3) and analyzed with FastQC v0.11.5 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Nanopore and Illumina sequencing generated 136,000 long reads (630,559,194 bp; N50, 8.7 kb) and 762,936 read pairs (2 × 250 bp), respectively. Nanopore reads were filtered using Filtlong v0.2.0 with the parameter -target_bases to retain a total of 230 Mbp (https://github.com/rrwick/Filtlong) (N50, 19,822 bp) and assembled using Unicycler v0.4.7 (4). The resulting circular contig was polished using Medaka v0.7.1 (https://github.com/nanoporetech/medaka) with all Nanopore reads, followed by two polishing rounds with Pilon v1.22 using the Illumina reads (5). Assembly quality was evaluated using Ideel (https://github.com/mw55309/ideel). Annotation was performed with the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) v5.1 (6). Default parameters were used for all software unless otherwise specified. The genome of L. crispatus ATCC 33820 consists of a single circular chromosome (2,239,089 bp) with an overall GC content of 37.0%. The assembly contains 2,194 open reading frames, 78.8% with putative biological function, 64 tRNA genes, 3 rRNA operons, and 3 structural RNAs.
FIG 1.
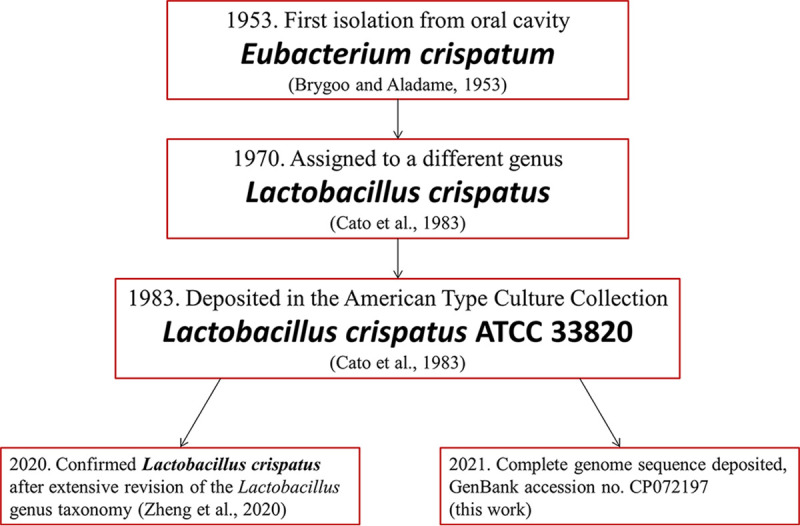
History of Lactobacillus crispatus type strain ATCC 33820. L. crispatus type strain ATCC 33820 was isolated at the Institut Pasteur in 1953 by E. R. Brygoo and N. Aladame from an oral sample of a European individual in Saigon and was considered a new species of the genus Eubacterium (Collection of the Institut Pasteur, Paris, strain II) (7). Later, it was deposited in the Virginia Polytechnic Institute and State University as VPI 3199 and identified as Lactobacillus (8). Further characterization upon American Type Culture Collection deposition indicated that ATCC 33820 DNA was 100% homologous to the previously defined L. acidophilus group A2 (8). Over the years, the L. crispatus type strain has been distributed among different collections and also designated DSM 20584 = CCUG 30722 = CIP 102990 = CIPP II = JCM 1185 = LMG 9479. Recently, Zheng and colleagues (9) reclassified the genus Lactobacillus into 25 genera through a polyphasic approach; however, the nomenclature of Lactobacillus crispatus remained unchanged. Strain ATCC 33820 was acquired by our laboratory in October 2020. Arrows indicate sequential steps in the history of the L. crispatus type strain. Red boxes contain the year, followed by a brief description of the event, the strain name (in bold), and the reference (in parentheses).
Data availability.
Sample information and sequence and genomic assembly/annotation are accessible under the NCBI BioProject, BioSample, and whole-genome sequence accession numbers PRJNA716945, SAMN18472633, and CP072197, respectively. Raw Nanopore and Illumina sequencing reads are accessible under Sequence Read Archive accession numbers SRR14509463 and SRR14509462, respectively.
ACKNOWLEDGMENTS
This work was funded by the Italian Ministry of Education, University and Research (MIUR-Italy), under grant number 20177J5Y3P (“Progetti di Ricerca di Rilevante Interesse Nazionale–Bando 2017”). The GridION x-5 was acquired through the “Excellence Departments” grant from MIUR to the Department of Medical Biotechnologies, University of Siena.
Illumina genome sequencing was provided by MicrobesNG.
Contributor Information
Francesco Santoro, Email: santorof@unisi.it.
David A. Baltrus, University of Arizona
REFERENCES
- 1.Petrova MI, Lievens E, Malik S, Imholz N, Lebeer S. 2015. Lactobacillus species as biomarkers and agents that can promote various aspects of vaginal health. Front Physiol 6:81. doi: 10.3389/fphys.2015.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Coster W, D’Hert S, Schultz DT, Cruts M, Van Broeckhoven C. 2018. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics 34:2666–2669. doi: 10.1093/bioinformatics/bty149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI Prokaryotic Genome Annotation Pipeline. Nucleic Acids Res 44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brygoo ER, Aladame N. 1953. Study of a new strictly anaerobic species of the genus Eubacterium: Eubacterium crispatum n. sp. Ann Inst Pasteur (Paris) 84:640–641. [PubMed] [Google Scholar]
- 8.Cato EP, Moore WEC, Johnson JL. 1983. Synonymy of strains of “Lactobacillus acidophilus” group A2 (Johnson et al. 1980) with the type strain of Lactobacillus crispatus (Brygoo and Aladame 1953) Moore and Holdeman 1970. Int J Syst Evol Microbiol 33:426–428. [Google Scholar]
- 9.Zheng J, Wittouck S, Salvetti E, Franz CMAP, Harris HMB, Mattarelli P, O’Toole PW, Pot B, Vandamme P, Walter J, Watanabe K, Wuyts S, Felis GE, Gänzle MG, Lebeer S. 2020. A taxonomic note on the genus Lactobacillus: description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int J Syst Evol Microbiol 70:2782–2858. doi: 10.1099/ijsem.0.004107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sample information and sequence and genomic assembly/annotation are accessible under the NCBI BioProject, BioSample, and whole-genome sequence accession numbers PRJNA716945, SAMN18472633, and CP072197, respectively. Raw Nanopore and Illumina sequencing reads are accessible under Sequence Read Archive accession numbers SRR14509463 and SRR14509462, respectively.


