Abstract
In recent years there have been major advances in our understanding of the role of free fatty acids (FAs) and their metabolism in shaping the functional properties of macrophages and DCs. This review presents the most recent insights into how cell intrinsic FA metabolism controls DC and macrophage function, as well as the current evidence of the importance of various exogenous FAs (such as polyunsaturated FAs and their oxidation products—prostaglandins, leukotrienes, and proresolving lipid mediators) in affecting DC and macrophage biology, by modulating their metabolic properties. Finally, we explore whether targeted modulation of FA metabolism of myeloid cells to steer their function could hold promise in therapeutic settings.
Keywords: Dendritic cells, Fatty Acids, Macrophages, Metabolism, Proresolving mediators
Fatty acids play key roles in shaping the functional properties of macrophages and DCs. We here discuss the latest insights into how intracellular as well as exogenous fatty acids, such as proresolving mediators, regulate macrophage and DC metabolism and function, and how this may be exploited for therapeutic gain.

Introduction
Macrophages are innate immune cells of myeloid origin with a diverse range of functions. They not only play a key role in maintaining tissue homeostasis under noninflammatory conditions, but also in phagocytosis and killing of microbes during an infection and as well as in driving wound healing and tissue regeneration during the resolution phase of an inflammatory response [1, 2]. To be able to acquire these distinct functional traits, macrophages can adopt various polarization states, which are imprinted by the local cues they are exposed to in the tissues where they reside [3, 4, 5]. The most well‐studied differentiation states are classically activated macrophages (also called M1 macrophages) and alternatively activated macrophages (also referred to as M2 macrophages), and both can be modelled in in vitro culture systems. Although these two polarization states fail to capture the full diversity of functional states that macrophages can adopt in vivo, they are commonly used as models for studying the proinflammatory and anti‐inflammatory properties of these cells, respectively [6].
Like macrophages, DCs also belong to the group of myeloid innate immune cells. DCs sit at the crossroads between the innate and the adaptative immune system, working as specialized APCs that are capable of initiating T‐cell responses [7]. During steady state, DCs reside in peripheral tissues in a quiescent state. Upon sensing of pathogens or tissue‐derived danger signals, DCs undergo a phenotypic and functional change involving enhanced internalization and processing of Ags [8, 9, 10] and migration towards tissue draining lymph nodes, where they can induce an adaptive immune response by priming and activating Ag‐specific T cells [9, 11]. Furthermore, DCs also play a role in the induction of tolerance during steady state due to exposure to tolerizing signals. These tolerogenic DCs can mediate tolerance by promoting peripheral T‐cell anergy and apoptosis, decreasing effector and memory T‐cell responses, and inducing the differentiation and activation of Tregs [12, 13, 14, 15].
Due to the central role played by DCs and macrophages in the immune response, it is important to understand how their function is regulated and what kind of stimuli are needed to initiate/sustain their activation and polarization in specific scenarios. For instance, there has been a longstanding interest in defining the signaling pathways regulating macrophage and DC function in the context of “classical” immunological cues such as cytokines, chemokines, and PAMPs [16, 17]. More recently, there has been a growing appreciation that metabolic signals and alterations in cellular metabolism can also dictate immune cell function (see [18] for a detailed introduction to immune cell metabolism). Recent research about immunometabolism has contributed to the realization that stimuli and changes in the environment macrophages and DCs are exposed to, eventually converge into alterations in their metabolic properties. It has become clear that reprogramming of metabolic pathways, such as glycolysis, oxidative phosphorylation (OXPHOS), fatty acid (FA) synthesis and oxidation (FAO) are not only associated with, but are also crucial for shaping functional responses of DCs and polarization of macrophages to environmental cues [18].
The notion that FAs (both intracellular as well as extracellular) and their metabolism play a central role in shaping DC and macrophage biology has gained significant traction in recent years [19]. Extracellular FAs can be synthesized de novo from carbons derived from other core metabolic pathways such as the TCA cycle, glycolysis and glutaminolysis, hydrolyzed from intracellular lipid stores, or directly obtained from extracellular space (Fig. 1). These FAs play a pivotal structural role when used for incorporation into cellular membranes. Moreover, through their oxidation in mitochondria they serve an important role in generating energy via OXPHOS as well as in generating various TCA cycle intermediates that can act as signaling metabolites or that can be used for the synthesis of other macromolecules. As a result, metabolism of intracellular FAs is a central regulator of DC and macrophage function. In addition, various FAs present in the extracellular environment, released by other cells, including adipocytes, tumor cells and other immune cells, or obtained through diet [20], have also been shown to have the potential to alter the functional properties of DCs and macrophages [20, 21, 22].
Figure 1.
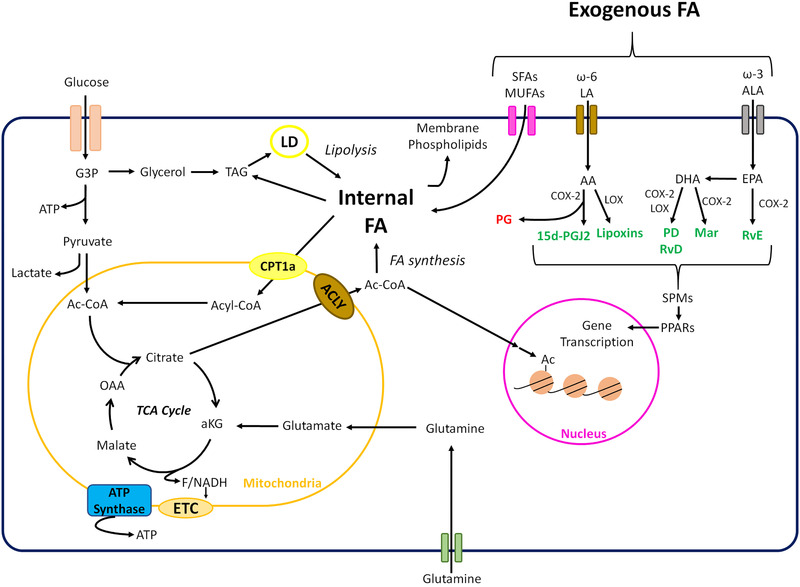
Cellular metabolism of intra‐ and extracellular fatty acids. A schematic overview of key processes involved in cellular FA uptake and metabolism. Core metabolic pathways connected to FA metabolism are indicated as well as the main processes involved in SPM synthesis from PUFAs such as ω‐3 and ω‐6 FAs. Specifically, PUFAs, many of which are essential FAs obtained from food, can be metabolized by cyclooxygenases (COX) and lipoxygenases (LOX) to give rise to SPMs. Main PUFAs that serve as substrate for these enzymes are linoleic acid (LA) and arachidonic acid (AA), which are ω‐6 PUFAs, and eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA), both ω‐3 PUFAs. Several enzymatic reactions lead to synthesis of SPMs (highlighted in green). The metabolism of EPA by COX‐2 eventually gives rise to E‐series Resolvins (RvE), while with DHA the products are more diverse. The actions of LOX on DHA lead to products from the Maresin (MaR) family, while both LOX and COX‐2 can give products from the Protectin family (PD) or D‐series Resolvins (RvD). In the case of ω‐6 PUFAs, such as AA, COX and LOX, give products.
The most well‐studied exogenous FAs in the context of myeloid cell biology are short‐chain fatty acids (SCFAs), saturated fatty acids (SFAs), and unsaturated fatty acids (UFAs), which could be monounsaturated (MUFAs) and polyunsaturated (PUFAs). In addition, specialized proresolving mediators (SPMs) which are products from PUFA metabolism (mainly ω‐3 and ω‐6 PUFAs; see Fig. 1) [23], can also have strong modulatory effects on myeloid cells including macrophages and DCs [23, 24]. Various mechanisms have been proposed through which these exogenous FAs can affect DC and macrophage function. These include acting as signaling molecules engaging receptors, serving as structural components, and interestingly, altering the metabolism of these cells.
In this review, we will discuss the most recent insights into how intrinsic FA metabolism controls DC and macrophage function, as well as the current evidence showing how various exogenous FAs (such as PUFAs and their oxidation products—prostaglandins, leukotrienes, and SPMs) affect DC and macrophage function, by modulating their metabolic properties.
Role of intrinsic fatty acid metabolism in myeloid cell function
Fatty acid oxidation
Macrophages
Free FAs used for FAO can be acquired either by uptake of dietary fats and subsequent hydrolysis, by lipolysis of stored acylglycerols, or by de novo FA synthesis [20]. These FAs can be oxidized either in peroxisomes, and/or be transported into the mitochondria by Carnitine Palmitoyltransferase (CPT) where they will undergo FAO. FAO results in the formation of multiple units of acetyl‐CoA, which can serve as substrate in the TCA cycle to fuel OXPHOS. It is a tightly regulated pathway, with the rate limiting step being the transport of acyl‐CoA into the mitochondrial matrix by CPT1. The control of CPT1 activity is, therefore, a key checkpoint for regulating mitochondrial FAO (Fig. 1) [25].
Murine macrophages stimulated with IL‐4 in vitro are characterized by increased FAO activity [26]. Likewise, tumor‐associated macrophages (TAMs) which share phenotypic and anti‐inflammatory characteristics with in vitro‐generated M2 macrophages [27] were shown to also have high levels of FAO [28]. Moreover, increased FAO is also correlated with efferocytosis, a process used by M2‐like macrophages to remove apoptotic cells to maintain tissue homeostasis [29]. Initial studies have suggested that FAO itself is crucial for M2 polarization, as pharmacological inhibition of CPT1 using etomoxir prevented M2 differentiation [26, 30–32]. However, more recent work has questioned these findings [33]. Using genetic approaches, it was shown that conditional deletion of CPT1a had no effect on acquisition of an M2 phenotype. Moreover, etomoxir was found to have substantial off‐target effects at concentrations used in these earlier studies [33]. However, it should be noted that long‐term genetic deletion of crucial FAO enzymes, such as CPT1a, may result in metabolic adaptation by usage of compensatory pathways to support cellular processes normally dependent on long‐chain FAO, that cells may not be able to resort to upon acute pharmacological inhibition [34]. These issues will need to be resolved to fully understand the importance of FAO in alternative activation of macrophages.
In vitro‐generated murine M1 macrophages are characterized by high expression of iNOS leading to the synthesis of NO [35], which is known to impair OXPHOS by inhibiting the electron transport chain in an auto‐ or paracrine manner (Fig. 2A) [36], and promote the synthesis of ROS that help in the microbicidal activity of M1 macrophages. As a consequence, FAO is severely compromised in these cells [30]. However, LPS‐stimulated peritoneal macrophages (pMacrophages) are characterized by increased OXPHOS [37], which may stem from a lower potential to produce NO by pMacrophages compared to their in vitro counterparts [30]. Under certain conditions, FAO can also be important for specific proinflammatory properties of macrophages. NLRP3 inflammasome activation, and synthesis of IL‐1β was impaired following pharmacological inhibition of CPT1a with etomoxir suggesting dependency on FAO [38, 39, 40]. However, the exact mechanism by which FAO boosts NLRP3 activations is still not clear, although increased FAO‐fuelled mitochondrial ROS production has been implicated [40, 41].
Figure 2.
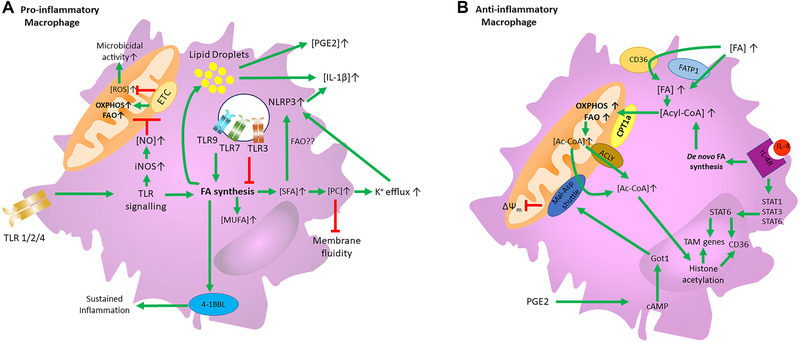
FA metabolism of pro‐ and anti‐inflammatory macrophage Schematic depiction of how FA metabolism and uptake control the function of (A) pro‐ and (B) anti‐inflammatory macrophages. Green lines indicate positive signaling, while red lines indicate an inhibitory effect. (A) Proinflammatory macrophages increase FA synthesis upon TLR signaling. This increase in FA synthesis drives 4‐1BBL activity which helps to sustain inflammation. TLR‐driven FA synthesis also leads to an increase in lipid droplets which is associated with increased PGE2 and IL‐1β synthesis. Moreover, TLR signaling induces iNOS expression and subsequent NO synthesis which inhibits the ETC, thereby reducing FAO, and promoting ROS formation which helps with microbicidal functions. Increased FA synthesis upon TLR stimulation also leads to SFA synthesis, which increases PC levels in the cell membrane, leading to less fluidity and K+ efflux, thereby activating NLPR3 and IL‐1β synthesis. TLR1/2, TLR7, and TLR9 activation increased FA synthesis and de novo SFA and MUFA synthesis. TLR3 activation leads to the opposite effect, inhibiting FA synthesis, along with MUFA and SFA synthesis. (B) IL‐4R signaling activates STAT1, STAT3, and STAT6 which promote the transcription of TAM genes and CD36. IL‐4R signaling also promotes de novo FA synthesis which increases Acyl‐CoA levels in the cell. Extrinsic FAs can also increase Acyl‐CoA levels by being transported intracellularly by CD36 or FATP‐1. Increased Acyl‐CoA promotes OXPHOS and FAO in a CPT1a‐dependent manner. The increased flux in OXPHOS and FAO results in elevated levels of mitochondrial Acetyl‐CoA which can be transported to the cytosol in an ACLY‐dependent manner or through the Malate‐Aspartate shuttle. Cytosolic Acetyl‐CoA can participate in histone acetylation of M2‐like and TAM genes. PGE2 impairs mitochondrial membrane potential in M2‐like macrophages by dysregulation of the Malate‐Aspartate shuttle by increasing cAMP‐induced Got1 expression.
One of the mechanisms by which FA metabolism regulates macrophage polarization involves changes in histone acetylation of IL‐4‐inducible genes fuelled by an increase in cellular concentrations of acetyl‐CoA (Ac‐CoA) [31]. It was found that IL‐4 receptor (IL‐4R) signaling increases ATP citrate lyase (ACLY) expression and activity (see Fig. 2B), resulting in an accumulation of nuclear and cytosolic Ac‐CoA to enable efficient histone acetylation. Tracing experiments revealed that of glutamine, glucose and palmitate (PA), the latter was the largest source of carbon for Ac‐CoA [31]. This may indicate that FAO in M2 macrophages fuels the TCA cycle to increase ACLY‐driven Ac‐CoA output to support histone acetylation required for expression of M2 macrophage‐associated genes.
Thus, the relationship between macrophage phenotype and FAO does not appear to be as black and white, as initially thought. A picture is emerging that FAO can—depending on the context—support both pro‐ or anti‐inflammatory properties by appropriating specific functions. For instance, in proinflammatory macrophages, FAO may be used to produce ROS to support activation of the NLRP3 inflammasome, as well as potentially fuel the TCA cycle to compensate for cataplerosis of intermediates that are being extracted from the TCA cycle for synthesis of amino acids and other macromolecules needed for proinflammatory activation [38, 39, 40, 41]. On the other hand, in M2 macrophages there is evidence that FAO, in addition to being involved in epigenetic remodeling by serving as a source of Ac‐CoA required for acquisition of an M2 phenotype, also contributes to maintenance of anti‐inflammatory activities, such as efferocytosis [29], by burning through FAs that otherwise accumulate in these cells as a consequence of this process. Moreover, β‐oxidation of FAs from apoptotic cells enhanced IL‐10 transcription and synthesis, thereby reinforcing their anti‐inflammatory phenotype [42].
Dendritic cells
Several studies have highlighted the importance of FAO in regulating the functional properties of DCs in a stimulus‐ and subset‐specific manner [43, 44, 45]. Different TLR stimuli engage FAO to a different extent in human moDCs [43]. While TLR4 stimulation was found to induce glycolysis, TLR7/8 stimulation with pRNA increased FAO and OXPHOS in human moDCs (Fig. 3A). This increase in FAO and OXPHOS was due to branched‐chain alpha‐keto acid dehydrogenase complex E1‐alpha subunit (BCKDE1α) phosphorylation in a PTEN‐induced putative kinase 1 (PINK1)‐dependent manner. Interestingly, inducing PINK1 activity in tolerogenic DCs stimulated FAO and rendered these DCs immunostimulatory [43], suggesting FAO can promote proinflammatory functions in DCs. Other studies have also implicated FAO in supporting proinflammatory DC activation by showing that its pharmacological inhibition by etomoxir suppressed both murine cDC and pDC activation stimulated with a TLR9 agonist, as evidenced by decreased expression of costimulatory molecules and decreased synthesis of proinflammatory cytokines [44]. Similarly, murine pDCs increase FAO upon stimulation of TLR9, which was found to be required for efficient type 1 interferon (I‐IFN) production [45]. Tumor cells may appropriate this mechanism by secreting α‐Fetoprotein, which inhibits FAO and OXPHOS in DCs, leading to impaired stimulation of Ag‐specific effector functions [46]. This points towards a link between FAO and triggering of endosomal TLRs, which may be explained by fact that engagement of endosomal TLRs generally lead to strong type 1‐IFN production, which can promote FAO in an autocrine manner [41]. Possibly, in this context, FAO may take over the role from glycolysis as main carbon source to fuel the synthesis of TCA cycle intermediates, which in DCs stimulated with cell membrane‐associated TLRs is required to support the anabolic demands of immunogenic DC activation [47].
Figure 3.
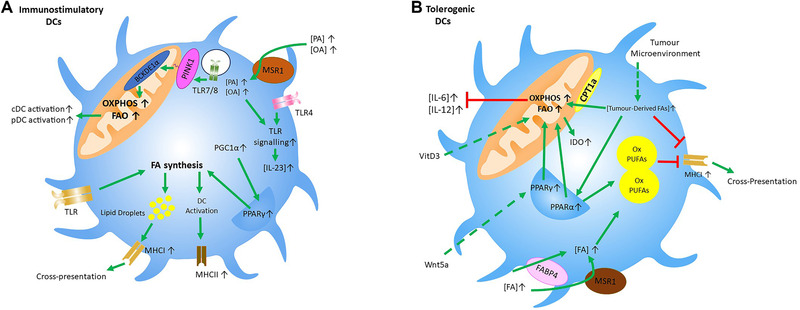
FA metabolism of immunostimulatory and tolerogenic DCs: Schematic depiction of how FA metabolism and uptake control the function of (A) immunostimulatory and (B) tolerogenic dendritic cells. Green lines indicate positive signaling, while red lines indicate an inhibitory effect. Dashed lines represent effects from exogenous stimuli. (A) In immunostimulatory DCs, TLR signaling increases FA synthesis to promote ER expansion and LD formation, which contributes to upregulation of MHCII and MHCI. PGC1α and PPARγ activation is also associated with increased FA synthesis in immunostimulatory DCs. TLR7/8 stimulation can lead to the phosphorylation of BCKDE1α in a PINK‐mediated manner. This results in increased OXPHOS and FAO which in turn is associated with increased activation of cDCs and pDCs. Additionally, extrinsic FAs, such as OA and PA, which are transported intracellularly by MSR1, can boost TLR4 signaling and increase IL‐23 synthesis. (B) In tolerogenic DCs, increased OXPHOS and FAO are associated with reduced IL‐6 and IL‐12 synthesis and increased IDO expression. Tumor‐derived FAs can increase OXPHOS and FAO by either being transported by CPT1a or by activating PPARα. This PPARα activation can also increase LD formation, which have a high content of oxidized PUFAs. Extrinsic FAs transported by MSR1 and FABP4 can feed these LDs and increase their oxidized PUFA content. These LDs, and tumor‐derived FAs themselves, impair cross‐presentation by suppressing MHCI surface expression. External stimuli, such as VitD3 and Wnt5a, can also increase FAO and OXPHOS, with Wnt5a doing so in a PPARγ‐dependent manner.
However, increased FAO has also been implicated in supporting tolerogenic properties of DCs (Fig. 2B). DCs rendered tolerogenic in vitro using Vitamin‐D3 or in vivo through Wnt5a signaling in the tumor microenvironment, display increased FAO [48, 49, 50] on which they in part rely for induction of Tregs [50, 51]. In the latter study, FAO was mechanistically shown to enhance indolamine 2,3‐dioxygenase‐1 (IDO) activity and suppress IL‐6 and IL‐12 cytokine expression by DCs, culminating in Treg generation. Additionally, failure of TLR‐stimulated murine CD11c+ DCs or BM‐DCs to switch from OXPHOS to glycolysis, due to deficiency in miRNA‐142, which normally suppresses CPT1a activity, locked these cells in a tolerogenic state with reduced synthesis of proinflammatory cytokines and reduced ability to activate T cells both in vitro and in vivo [52].
Taken together, FAO may support, depending on the studied DC subset and/or nature of activating signal, either proinflammatory or anti‐inflammatory properties of these cells. How exactly the same metabolic pathway can underpin these divergent immunological properties in DCs remains to be determined.
Fatty acid synthesis
De novo FA synthesis is a process in the cytoplasm whereby acyl chains are generated from Ac‐CoA through the action of FA synthases. Most of the Ac‐CoA which is converted into FAs is derived from carbohydrates originating from the glycolytic pathway and TCA cycle [15, 46].
Macrophages
There are several studies that have linked de novo FA synthesis to supporting proinflammatory function of macrophages (Fig. 2A). For instance, genetic models to block FA synthesis have shown that NO production and proinflammatory signaling are reduced in both human and murine macrophages upon TLR stimulation [53, 54, 55, 56]. Additionally, FA synthesis also appears to support TLR‐driven 4‐1BBL activity, a member of the TNF superfamily which regulates the sustained production of proinflammatory cytokines in TLR‐activated macrophages [57]. Consistent with this latter finding, inhibition of FA synthesis in a murine model of psoriasis, in which prolonged proinflammatory activity of M1 macrophages is a driving factor behind the disease, alleviated the symptoms [57]. However, conditional deletion of ACC1, the enzyme that converts Acetyl‐CoA into Malonyl‐CoA used for FA synthesis, did not compromise proinflammatory macrophage responses [58]. These discrepancies in outcome might, as alluded to above, arise from the difference in effects of instant pharmacological targeting with potential off‐target effects versus long‐term deletion. In addition, different TLR ligands appear to have different effects on FA synthesis and the lipidome of macrophages [59]. Stimulation of MyD88‐dependent TLRs (i.e. TLR1/2, TLR7, or TLR9) increased de novo SFA and MUFA synthesis, while TLR3 stimulation reduced both SFA and MUFA synthesis. Moreover, inhibition of MUFA synthesis, without affecting SFA synthesis, disturbed the TLR‐driven reprogramming of the lipidome, resulting in an increased inflammatory response [59].
Together, this links SFA synthesis to proinflammatory properties of macrophages. Several mechanisms have been proposed through which FA synthesis can support proinflammatory properties of macrophages. First, it may help in changing membrane lipid composition to alter fluidity to facilitate membrane‐associated effector functions, such as phagocytosis [60, 61, 62], as has been shown following TLR4 stimulation [63]. Here, sterol regulatory element binding protein‐1a (SREBP‐1a), a key transcriptional regulatory protein of FA metabolism, was activated downstream of TLR4 and increased FA synthesis. Inhibiting this pathway led to defective phagocytosis, resulting from a reduction in the interaction between lipid rafts and the cytoskeleton, presumably due to reduced accumulation of newly synthesized fatty acyl chains within membrane phospholipids [63]. Secondly, synthesized FAs can be used to form lipid droplets (LDs) that have a role in the first line of defense against pathogens, by serving as anchors for immune proteins and as docking sites for phagocytic membranes. This facilitates the encounter between immune proteins and phagocytized pathogens, while also protecting the cell from possible unwanted damage due to the cytotoxic properties of these proteins [64]. Additionally, LDs can change cellular metabolism by uncoupling themselves from mitochondria upon infection, to lower mitochondrial FAO. Finally, it was recently shown that LD development, as a consequence of commitment to triacylglycerol synthesis following TLR stimulation, was needed for increased synthesis of M1‐associated inflammatory mediators such as IL‐1β and PGE2 [65].
Dendritic cells
Studies with murine BMDCs have shown that LPS stimulation promotes de novo FA synthesis to support expression of cytokines and costimulatory markers required for potent T‐cell activation [47]. Moreover, DC differentiation and subsequent upregulation of MHCII requires FA synthesis [66, 67, 68] (Fig. 3A). However, in contrast to pharmacological targeting of FA synthesis as used in the aforementioned study to interrogate the role of de novo FA synthesis, genetic targeting of ACC1 did not appear to affect DC cytokine expression [58].
Mechanistically, FA synthesis‐driven DC activation was linked to increased ER and Golgi expansion to allow for efficient translation of these proteins [47]. It is interesting to note that in this study increased LD formation dependent on FA synthesis was also observed. This increase in LD formation has been reported by others as well following LPS or IL‐4 stimulation, both in in vitro cultured BM‐DCs as well as primary CD11c+ murine splenic, and lymph node DCs [69], and was associated with enhanced T‐cell activation [47, 69]. This suggests that these LDs serve not just as passive lipid storage organelles but may also be linked to key processes fundamental to DC biology, such as Ag processing and presentation. Indeed inactivation of genes which regulate the assembly of lipid bodies abrogated cross‐presentation by DCs [70]. The exact mechanism by which LDs participate in Ag cross‐presentation is not yet known, but previous work showed that lipid composition of LDs affected MHC‐I expression in DCs [71].
Role of exogenous fatty acids on myeloid cell function
Apart from intracellular FA metabolism, free FAs present in the extracellular space, which can come from the diet or released by other cells, can also affect myeloid cell function [20, 72, 73]. This can be either by serving as structural components, by being used as nutrients to fuel cellular metabolic pathways following uptake, or by acting as signaling molecules through engagement of surface or intracellular receptors. Here, we will specifically focus on their effect on macrophage and DC metabolism and thereby function.
Fatty acid uptake
Macrophages
For many of their biological effects, extracellular FAs first need to be imported into the cell. Macrophages express various receptors and transporters that mediate this process (Fig. 2B). Studies have shown that FA transport protein 1 (FATP1), an important transporter for FA uptake, plays a role in the metabolic reprogramming of macrophages during inflammation [74]. FATP1 overexpression in murine BM‐macrophages induced FAO and, in turn reduced an LPS‐induced inflammatory response. Additionally, by inhibiting FATP1, instead a proinflammatory macrophage phenotype could be promoted. CD36, a scavenger receptor, is also involved in the transport of exogenous FAs into the cytosol [32], where they can fuel FAO as shown in IL‐4‐driven M2 differentiation [32]. This was functionally important as CD36‐deficient macrophages were impaired in their M2 polarization. Likewise, human macrophages displayed a reduction in LPS‐induced IL‐12 and TNF‐α synthesis following exposure to FAs [75]. Taken together, this suggests that uptake of exogenous FAs is generally linked to promotion of anti‐inflammatory macrophage function.
Dendritic cells
The role for FA uptake in regulating DC function seems to be more complex than in macrophages. On the one hand, activation of both in vitro‐cultured BMDCs and primary CD11c+ murine DCs (isolated from spleen and lymph nodes) resulted in increased FA accumulation and LD formation (Fig. 3A), which was correlated with increased expression of scavenger receptors, such as Macrophage scavenger receptor 1 (MSR1) [69], suggesting FA uptake may support DC immunogenicity. On the other hand, there is also evidence for a tolerising effect of FA uptake by DCs, especially in the tumor microenvironment [21] (Fig. 3B). Tumor‐associated DCs (TADCs) upregulate scavenger receptors, including MSR1, FA binding protein 4 (FABP4), and lipoprotein lipase, which promote exogenous FA uptake, [76, 77, 78], and correspondingly display high lipid content and LD accumulation. Functionally, these TADCs showed impaired Ag presentation and subsequent T‐cell stimulating ability. Interestingly, inhibiting FA synthesis or MSR1 activity restored their lipid content to normal levels and as well as their T‐cell priming abilities [77]. Recently, it was shown that FAs taken up by TADCs can serve as ligands for peroxisome proliferator‐activated receptor alpha (PPARα) which is a member of ligand‐activated nuclear transcription factors regulating lipid metabolism. PPARα binding promoted LD synthesis, as well as increased FAO, which resulted in reduced DC immunogenicity [79]. Interestingly, correspondingly, inhibition of PPARα activation in this context, restored DCs function and enhanced antitumor immune responses in a therapeutic setting. Even though the anti‐inflammatory properties of PPARα and PPARδ are well documented (as reviewed in [80]), in some contexts their activity is associated with inflammatory responses. For instance, a recent publication showed that deletion of PPARδ in CD11c+ cells in mice dampened palmitic acid‐induced IL‐12 and TNF synthesis, and upregulation of costimulatory molecules, resulting in attenuated development of atherosclerosis [81].
An explanation for why FA uptake and LD formation can, depending on the context, either support or interfere with DC immunogenicity, may come from the nature of the FAs these cells accumulate. LDs in TADCs were shown to contain high levels of oxidized PUFAs [82, 83] compared to non‐TADCs, which has been linked to tumor‐derived molecules that prompt lipid peroxidation in TADCs [84]. This was found to drive accumulation of MHC‐I‐peptide complexes in lysosomes and late endosomes, limiting cross‐presentation and, subsequently, cytotoxic T‐cell priming. This would be consistent with recent work by Ugolini et al. [85] showing that uptake of oxidized truncated FAs impaired DC Ag cross‐presentation in cancer, without affecting direct presentation.
Fatty acids as signaling molecules
Saturated fatty acids
It is well described that in general signaling by exogenous SFAs exert proinflammatory effects on DCs and macrophages [86]. Interestingly, several recent studies have now also revealed an important role for metabolic rewiring in this process. Activation of exogenous SFAs into Acyl‐CoA, was shown to activate the NLPR3 inflammasome, driving an M1 type while UFAs prevented this [38] (Fig. 2A). The authors showed that these SFAs promoted the synthesis of phosphatidylcholine, leading to loss of membrane fluidity and K+ efflux, enabling subsequent NLRP3 activation. UFAs were able to inhibit this effect by instead redirecting SFAs to triacylglycerol synthesis. Furthermore, exposure of macrophages to palmitic acid (PA), a SFA, was associated with impaired wound healing, a state of low‐grade chronic inflammation and increased IL‐1β and IL‐23 synthesis [87, 88, 89]. In an environment rich in FAs, DCs are also stimulated toward a proinflammatory phenotype. Specifically, accumulation of PA and oleic acid (OA) amplified TLR signaling and led to an increase in IL‐23 expression, which in a model of psoriasis worsened disease progression [88] (Fig. 3A). This was linked to PA inhibiting hexokinase activity and perturbing TCA metabolism in TLR‐activated cells, leading to an increase in mtROS and proinflammatory cytokines. Nevertheless, the exact mechanisms or receptors by which SFAs can modulate macrophage or DC function are still not fully elucidated. It was previously thought that SFAs could bind TLRs, thus, activating a proinflammatory phenotype in macrophages. However, recent data show that, while TLR4 signaling is needed for SFA‐induced inflammation, SFAs do not bind directly to TLR4 [90].
Polyunsaturated fatty acids
One of the most well‐studied bioactive FAs known to modulate myeloid cell function is Prostaglandin E2 (PGE2), an oxidation product of AA that can bind specific receptors (Fig. 1). While PGE2 was already reported to inhibit murine BM‐macrophage activation and polarization both in vitro and in vivo [91], more recent work elucidated metabolic effects of PGE2 on M2 macrophages. The authors observed that PGE2, alongside a drop in expression of a subset of M2 markers, caused a dissipation of the mitochondrial membrane potential in IL‐4‐stimulated M2 macrophages [92] (Fig. 2B). This was due to PGE2 affecting the transcription of several genes related to maintenance of mitochondrial membrane potential in a cAMP‐mediated manner. PGE2 initiated the transcription of genes that regulate the malate‐aspartate shuttle, including Got1. Another PUFA, Leukotriene‐B4 (LTB4) has also recently been linked to regulating macrophage metabolism [93]. Type 1 diabetic mice have increased levels of circulating LTB4. Macrophages from these mice displayed increased FAO and CPT2 expression when compared to macrophages from control mice. This was associated with an increased proinflammatory signature. These effects were reduced upon blocking LTB4 signaling using a receptor antagonist.
Specialized proresolving mediators
Recently, SPMs have received considerable attention given the growing evidence for their key role in active resolution of inflammation. There are already some strong correlations between deregulation of SPM metabolism, and certain chronic inflammatory diseases such as Alzheimer's disease, atherosclerosis, arthritis, and type‐2 diabetes [23] (Table 1). The proresolving properties of SPMs stem in a large part from their ability to suppress inflammatory properties of macrophages and DCs. SPMs promote the shift from M1‐like to M2‐like macrophages, increase phagocytic and efferocytotic activities of macrophages, and reduce IL‐12 synthesis by DCs [94, 95]. However, until now, little is known about whether SPMs may affect metabolism of these cells or how cellular metabolism of those cells affects SPM synthesis.
Table 1.
SPMs and their association with protection against inflammatory diseases
Given the known dampening effects of SPMs on proinflammatory DC and macrophage activation and the clear functional link between engagement of certain metabolic pathways and anti‐inflammatory properties of these cells, it is tempting to speculate that a mechanism by which SPMs mediate these effects is by modulating DC and macrophage metabolism. One way SPMs might achieve this is by binding to PPARs. PPARs can be activated by many different ligands, including long‐chain SFAs and UFAs, eicosanoids or other products of PUFA oxidation such as SPM 15‐deoxy‐δ‐12,14‐prostaglandin J2 and Marensin‐1 [96, 97, 98, 99, 100]. Interestingly, PPAR signaling is known to play a role in attenuating the inflammatory function of macrophages as well as DCs by regulating their metabolism [79, 101]. Additionally, SPMs can bind surface receptors within the family of G‐protein‐coupled receptors (GPRs) [102]. Although it remains to be established whether signaling through GPRs that bind SPMs (e.g. GPR32, ALX/FPR2, ChemR23 [103, 104]) could drive metabolic reprogramming, the fact that signaling through other GPRs, such as GPR120 and GPR40, has already been described to affect FA synthesis in adipocytes and hepatocytes, makes it tempting to speculate that SPMs may also alter macrophage and DC metabolism and thereby their function via GPR signaling [105, 106].
Perspectives and outlook
There is a growing body of evidence for a key role of FAs in the regulation of myeloid cell function and the inflammatory response by serving as nutrients, structural components of cells, signaling molecules, and/or epigenetic regulators. Many studies point to the increasing importance of FA metabolism in the function of DCs, such as T‐cell priming and Ag‐presentation, and that of macrophages such as microbicidal activity, phagocytosis, and efferocytosis. While pro‐ and anti‐inflammatory macrophage and DC phenotypes are often characterized by, and dependent on, engagement of FA synthesis and FAO, respectively, it is becoming increasingly clear that, depending on the context, particularly FAO can support both pro‐ and anti‐inflammatory properties of these cells. How exactly a single metabolic pathway can appropriate these different functions that allow it to support such diverse immunological responses is still poorly understood and it is one of the key outstanding questions that awaits to be addressed. However, one could hypothesize that the activity of metabolic pathways directly connected to FA metabolism, controlled by specific environmental cues and/or the nature of cell subset intrinsic metabolic imprinting, can play a decisive role in how products derived from FA metabolism are being redirected and used, and thereby what the final functional output is of such a pathway.
Many important insights in the effects of FAs on myeloid cell metabolism and function have been gained from in vitro studies. However, more in‐depth in vivo studies will be crucial to fully capture the complexity of this interaction and will be needed further in this field. First, this pertains to the complexity of the FA composition and metabolic changes in the local microenvironment that are not easily reproduced in vitro. Metabolic conditions in general, and FA composition in particular—such as those found at sites of inflammation or the tumor microenvironment—are highly complex and are likely fluctuating over time. Second, the diversity in macrophage phenotypes and DC subsets and associated metabolic properties found in tissues cannot be fully modelled in vitro. For instance, this is well illustrated by tissue‐resident macrophages which have a very different metabolic profile than BM‐macrophages generated in vitro [107]. Likewise, evidence is accumulating that specific DC subsets found in vivo, have distinct metabolic properties and requirements for FA metabolism for their function [108]. To what extent most aforementioned studies, often using in vitro‐generated BMDCs, faithfully recapitulate the metabolic programs that are engaged and needed for primary DCs in situ is questionable. Moreover, what role FA metabolism plays in the biology of the recently discovered new DC subsets is yet to be addressed [109, 110].
An important hurdle in studying FA metabolism of macrophages and DCs in situ is that they are generally present at very low frequencies. However, recent advances in single‐cell technologies, such single‐cell RNA sequencing and high dimensional flow cytometry [111], are now making it possible to characterize in unprecedented depth phenotypes and metabolic characteristics of rare cell populations at single‐cell level in clinical and tissue samples [112]. Moreover, imaging mass cytometry combined with imaging mass spectrometry could provide crucial additional spatial information about local metabolite and FA abundance and myeloid cell phenotype in tissues. These are some of the promising technological advances that will no doubt spur new discoveries in this exciting field of immunometabolism.
As exemplified in this review, there is a growing number of studies that show that extracellular FAs and SPMs can modulate DC and macrophage function by altering their metabolic properties. An aspect that has thus far received less attention is, if and how cellular metabolism shapes the production and release of these lipid mediators. There are first indications that show indeed synthesis of certain PUFAs is dependent on LD formation fuelled by de novo FA synthesis [65]. Whether production of anti‐inflammatory FAs, such as SPMs, is supported by different metabolic programs than proinflammatory FAs warrants further investigation.
Finally, there is increasing evidence that suggests that deregulated FA metabolism in macrophages and DCs can contribute to development of several inflammatory diseases. This link has been particularly well established in atherosclerosis and type 2 diabetes where dysfunctional lipid handling and metabolism by macrophages have been shown to be an important driver of pathology (i.e. plaque formation and tissue‐specific insulin resistance, respectively) [74, 113, 114]. Moreover, altered lipid handling by macrophages and DCs due to hyperlipidemia as in human studies are associated with, and in murine models causally linked to increased chances of developing autoimmune diseases, such as psoriasis, rheumatoid arthritis (RA), and systemic lupus erythematosus (SLE) [57, 81, 115]. Therefore, therapies aimed at targeting FA metabolism have proven their value in treatment of such disorders. For example, PPARγ agonists are successfully used in the clinic to counteract hyperlipidemia and hyperglycemia. However, the beneficial effects of these drugs have been primarily attributed to alterations in FA metabolism of metabolic tissues rather than myeloid cells [116]. Nonetheless, given the important role of FA metabolism in the regulation of macrophage and DC biology, now also efforts are undertaken to evaluate whether direct manipulation of FA metabolism of those cells could be used to shape their functional properties for therapeutic purposes. In this respect, there are several preclinical mouse studies that have given promising results. For instance, pharmacological inhibition of FAO with etomoxir in myeloid‐derived suppressor cells enhanced the effectiveness of cancer therapies in mice [117]. Additionally, etomoxir improved antitumor response following checkpoint blockade treatment in vivo [50], which was associated with a switch from a tolerogenic to an immunogenic TADC phenotype. Apart from targeting core FA metabolism in DCs or macrophages to shape their function for therapeutic purposes, there are also interesting developments aimed at promoting the synthesis of SPMs for treatment of inflammatory disorders. In this respect it is interesting to note that it is already known that aspirin, a common anti‐inflammatory drug, triggers the synthesis of several SPMs by modifying COX‐2 activity and inhibiting COX‐1 [118]. These data suggest the anti‐inflammatory properties which have been attributed to aspirin lie, in part, in its ability to promote SPM synthesis. The therapeutic potential of SPMs has also been evaluated more directly. It was shown that RvD1 leads to cartilage protection and better disease outcome in a murine arthritis model [119]. Additionally, through lipidomic analysis of human samples, 17‐HDHA was shown to be associated with lower pain in arthritis patients, pointing toward a possible therapeutic application of these compounds in treating inflammatory diseases [120]. Indeed, in a human skin blister model, it was shown that administration of SPMs into the inflamed site promoted resolution [121]. These findings have paved the way for a phase I clinical trial (NCT04308889) that is currently ongoing, in which the effects of dietary supplementation with ω‐3 FAs in a human inflammation blister model are assessed, to determine whether SPMs may promote resolution of inflammation. Additional work will be needed to establish to what extent the potential beneficial effects of these treatments are dependent on functional modulation of myeloid cells.
In conclusion, the intricate connection between FA metabolism and myeloid cell function, makes it a highly interesting target for therapeutic intervention to modulate immune responses and to potentially treat diseases marked by a compromised inflammatory response, such as cancer, or by a failure to resolve inflammation, as occurs in various chronic inflammatory disorders.
Conflict of interest
The authors declare no financial or commercial conflict of interest.
Abbreviations
- Ac‐CoA
acetyl‐CoA
- ACLY
ATP citrate lyase
- CPT
Carnitine Palmitoyltransferase
- FA
fatty acid
- FAO
fatty acid oxidation
- FATP1
FA transport protein 1
- GPRs
G‐protein‐coupled receptors
- IDO
indolamine 2,3‐dioxygenase‐1
- IL‐4R
IL‐4 receptor
- LDs
lipid droplets
- LTB4
leukotriene‐B4
- MSR1
macrophage scavenger receptor 1
- MUFAs
monounsaturated
- OA
oleic acid
- OXPHOS
oxidative phosphorylation
- PINK1
PTEN‐induced putative kinase 1
- PGE2
prostaglandin E2
- pMacrophages
peritoneal macrophages
- PPARα
peroxisome proliferator‐activated receptor alpha
- PUFAs
polyunsaturated
- RA
rheumatoid arthritis
- SCFAs
short‐chain fatty acids
- SREBP‐1a
sterol regulatory element binding protein‐1a
- SPMs
specialized proresolving mediators
- SFAs
saturated fatty acids
- TADCs
tumour‐associated DCs
- TAMs
tumour‐associated macrophages
- UFAs
unsaturated fatty acids
Acknowledgments
This work was supported by funding from the European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska‐Curie Grant agreement No. 812890.
References
- 1.Wynn, T. and Barron, L., Master regulators of inflammation and fibrosis. Semin. Liver Dis. 2010. 30: 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gordon, S., Alternative activation of macrophages. Nat. Rev. Immunol. 2003. 3: 23–35. [DOI] [PubMed] [Google Scholar]
- 3.Gordon, S. and Taylor, P. R., Monocyte and macrophage heterogeneity. Nat. Rev. Immunol. 2005. 5: 953–964. [DOI] [PubMed] [Google Scholar]
- 4.Murray, P. J. and Wynn, T. A., Protective and pathogenic functions of macrophage subsets. Nat. Rev. Immunol. 2011. 11: 723–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mosser, D. M. and Edwards, J. P., Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 2008. 8: 958–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murray, P. J., Allen, J. E., Biswas, S. K., Fisher, E. A., Gilroy, D. W., Goerdt, S., Gordon, S., et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 2014. 41: 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sallusto, F. and Lanzavecchia, A., Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony‐stimulating factor plus Interleukin 4 and downregulated by tumor necrosis factor a. J. Exp. Med. 1994. 179: 1109–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reis, E. and Sousa, C., Toll‐like receptors and dendritic cells: for whom the bug tolls. Semin. Immunol. 2004. 16: 27–34. [DOI] [PubMed] [Google Scholar]
- 9.Henri, S., Guilliams, M., Poulin, L. F., Tamoutounour, S., Ardouin, L., Dalod, M. and Malissen, B., Disentangling the complexity of the skin dendritic cell network. Immunol. Cell Biol. 2010. 88: 366–375. [DOI] [PubMed] [Google Scholar]
- 10.Platt, C. D., Ma, J. K., Chalouni, C., Ebersold, M., Bou‐Reslan, H., Carano, R. A. D., Mellman, I., et al., Mature dendritic cells use endocytic receptors to capture and present antigens. Proc. Natl. Acad. Sci. USA 2010. 107: 4287–4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martín‐Fontecha, A., Sebastiani, S., HöPken, U. E., Uguccioni, M., Lipp, M., Lanzavecchia, A. and Sallusto, F., Regulation of dendritic cell migration to the draining lymph node: impact on T lymphocyte traffic and priming. J. Exp. Med. 2003. 198: 615–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kryczanowsky, F., Raker, V., Graulich, E., Domogalla, M. P. and Steinbrink, K., IL‐10–modulated human dendritic cells for clinical use: identification of a stable and migratory subset with improved tolerogenic activity. J. Immunol. 2016. 197: 3607–3617. [DOI] [PubMed] [Google Scholar]
- 13.Raker, V. K., Domogalla, M. P. and Steinbrink, K., Tolerogenic dendritic cells for regulatory T cell induction in man. Front. Immunol. 2015. 6: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boks, M. A., Kager‐Groenland, J. R., Haasjes, M. S. P., Zwaginga, J. J., van Ham, S. M. and ten Brinke, A., IL‐10‐generated tolerogenic dendritic cells are optimal for functional regulatory T cell induction: a comparative study of human clinical‐applicable DC. Clin. Immunol. 2012. 142: 332–342. [DOI] [PubMed] [Google Scholar]
- 15.Li, H. and Shi, B., Tolerogenic dendritic cells and their applications in transplantation. Cell Mol. Immunol. 2015. 12: 24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murray, P. J., Macrophage polarization. Annu. Rev. Physiol. 2017. 79: 541–566. [DOI] [PubMed] [Google Scholar]
- 17.Kapsenberg, M. L., Dendritic‐cell control of pathogen‐driven T‐cell polarization. Nat. Rev. Immunol. 2003. 3: 984–993. [DOI] [PubMed] [Google Scholar]
- 18.O'neill, L. A. J., Kishton, R. J. and Rathmell, J., A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 2016. 16: 553–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'neill, L. A. J. and Pearce, E. J., Immunometabolism governs dendritic cell and macrophage function. J. Exp. Med. 2016. 213: 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galli, C. and Risé, P., Origin of fatty acids in the body: endogenous synthesis versus dietary intakes. Eur. J. Lipid Sci. Technol. 2006. 108: 521–525. [Google Scholar]
- 21.Koundouros, N. and Poulogiannis, G., Reprogramming of fatty acid metabolism in cancer. Br J. Cancer 2020. 122: 4–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu, H., Han, Y., Rodriguez Sillke, Y., Deng, H., Siddiqui, S., Treese, C., Schmidt, F., et al., Lipid droplet‐dependent fatty acid metabolism controls the immune suppressive phenotype of tumor‐associated macrophages. EMBO Mol. Med. 2019. 11: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Serhan, C. N. and Levy, B. D., Resolvins in inflammation: emergence of the pro‐resolving superfamily of mediators. J. Clin. Invest. 2018. 128: 2657–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Serhan, C. N., Chiang, N. and Dalli, J., The resolution code of acute inflammation: novel pro‐resolving lipidmediators in resolution. Semin. Immunol. 2015. 27: 200–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lehner, R. and Quiroga, A. D., Fatty acid handling in mammalian cells [Internet]. Biochemistry of lipids, lipoproteins and membranes: Sixth edition. Elsevier, Amsterdam, The Netherlands, 2016; pp. 149–184. 10.1016/B978-0-444-63438-2.00005-5 [DOI] [Google Scholar]
- 26.Huang, S. C. C., Smith, A. M., Everts, B., Colonna, M., Pearce, E., Schilling, J. D. and Pearce, E. J., Metabolic reprogramming mediated by the mTORC2‐IRF4 signaling axis is essential for macrophage alternative activation. Immunity 2016. 45: 817–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vitale, I., Manic, G., Coussens, L. M., Kroemer, G. and Galluzzi, L., Macrophages and metabolism in the tumor microenvironment. Cell Metab. 2019. 30: 36–50. [DOI] [PubMed] [Google Scholar]
- 28.Su, P., Wang, Q., Bi, E., Ma, X., Liu, L., Yang, M., Qian, J. and Yi, Q., Enhanced lipid accumulation and metabolism are required for the differentiation and activation of tumor‐associated macrophages. Cancer Res. 2020. 80: 1438–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park, D., Han, C. Z., Elliott, M. R., Kinchen, J. M., Trampont, P. C., Das, S., Collins, S., et al. Continued clearance of apoptotic cells critically depends on the phagocyte Ucp2 protein. Nature 2011. 477: 220–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Den, B. J. V., Lao, N. and Menon, D., Macrophage immunometabolism : where are we (going)? Trends Immunol. 2017. 38: 395–406. [DOI] [PubMed] [Google Scholar]
- 31.Covarrubias, A. J., Aksoylar, H. I., Yu, J., Snyder, N. W., Worth, A. J., Iyer, S. S., Wang, J., et al. Akt‐mTORC1 signaling regulates ACLY to integrate metabolic input to control of macrophage activation. Elife 2016. 5: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang, S. C.‐.C., Everts, B., Ivanova, Y., O'sullivan, D., Nascimento, M., Smith, A. M., Beatty, W., et al., Cell‐intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat. Immunol. 2014. 15: 846–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Divakaruni, A. S., Hsieh, W. Y., Minarrieta, L., Duong, T. N., Kim, K. K.O., Desousa, B. R., Andreyev, A. Y., et al., Etomoxir inhibits macrophage polarization by disrupting COA homeostasis. Cell Metab. 2018. 28: 490–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Den Bossche, J. and Van Der Windt, G. J. W., Fatty acid oxidation in macrophages and T cells: time for reassessment? Cell Metab. 2018. 28: 538–540. [DOI] [PubMed] [Google Scholar]
- 35.Macmicking, J. D., North, R. J., Lacourse, R., Mudgett, J. S., Shah, S. K. and Nathan, C. F., Identification of nitric oxide synthase as a protective locus. Proc. Natl Acad. Sci. 1997. 94: 5243–5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tengan, C. H. and Moraes, C. T., NO control of mitochondrial function in normal and transformed cells. Biochim. Biophys. Acta‐Bioenerg. 2017. 1858: 573–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Artyomov, M. N., Sergushichev, A. and Schilling, J. D., Integrating immunometabolism and macrophage diversity. Semin. Immunol. [Internet]. 2016. 28: 417–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gianfrancesco, M. A., Dehairs, J., L'homme, L., Herinckx, G., Esser, N., Jansen, O., Habraken, Y., et al., Saturated fatty acids induce NLRP3 activation in human macrophages through K + efflux resulting from phospholipid saturation and Na, K‐ATPase disruption. Biochim. Biophys. Acta—Mol. Cell Biol. Lipids 2019. 1864: 1017–1030. [DOI] [PubMed] [Google Scholar]
- 39.Yang, Q., Liu, R., Yu, Q., Bi, Y. and Liu, G., Metabolic regulation of inflammasomes in inflammation. Immunology 2019. 157: 95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moon, J. ‐ S., Nakahira, K., Chung, K. ‐ P., Denicola, G. M., Koo, M. J., Pabón, M. A., Rooney, K. T., et al., NOX4‐dependent fatty acid oxidation promotes NLRP3 inflammasome activation in macrophages. Nat. Med. 2016. 22: 1002–1012. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Martinon, F., Signaling by ROS drives inflammasome activation. Eur. J. Immunol. 2010. 40: 616–619. [DOI] [PubMed] [Google Scholar]
- 42.Zhang, S., Weinberg, S., DeBerge, M., Gainullina, A., Schipma, M., Kinchen, J. M., Ben‐Sahra, I., et al. Efferocytosis fuels requirements of fatty acid oxidation and the electron transport chain to polarize macrophages for tissue repair. Cell Metab. 2019. 29: 443–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Basit, F. and De Vries, I. J. M., Dendritic cells require PINK1‐mediated phosphorylation of BCKDE1 α to promote fatty acid oxidation for immune function. Front. Immunol. 2019. 10: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qiu, C. C., Atencio, A. E. and Gallucci, S., Inhibition of fatty acid metabolism by etomoxir or TOFA suppresses murine dendritic cell activation without affecting viability. Immunopharmacol. Immunotoxicol. 2019. 41: 361–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu, D., Sanin, D. E., Everts, B., Chen, Q., Qiu, J., Buck, M. D., Patterson, A., et al. Type 1 interferons induce changes in core metabolism that are critical for immune function. Immunity 2016. 44: 1325–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Santos, P. M., Menk, A. V., Shi, J., Tsung, A., Delgoffe, G. M. and Butterfield, L. H., Tumor‐derived a‐fetoprotein suppresses fatty acid metabolism and oxidative phosphorylation in dendritic cells. Cancer Immunol. Res. 2019. 7: 1001–1013. [DOI] [PubMed] [Google Scholar]
- 47.Everts, B., Amiel, E., Huang, S. C.‐C., Smith, A. M., Chang, C. ‐ H., Lam, W. Y., Redmann, V., et al., TLR‐driven early glycolytic reprogramming via the kinases TBK1‐IKKε supports the anabolic demands of dendritic cell activation. Nat. Immunol. 2014. 15: 323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferreira, G. B., Van Etten, E., Lage, K., Hansen, D. A., Moreau, Y., Workman, C. T., Waer, M., et al., Proteome analysis demonstrates profound alterations in human dendritic cell nature by TX527, an analogue of vitamin D. Proteomics 2009. 9: 3752–3764. [DOI] [PubMed] [Google Scholar]
- 49.Ferreira, G. B., Kleijwegt, F. S., Waelkens, E., Lage, K., Nikolic, T., Hansen, D. A., Workman, C. T., et al., Differential protein pathways in 1, 25‐dihydroxyvitamin D3 and dexamethasone modulated tolerogenic human dendritic cells. J. Proteome Res. 2012. 11: 941–971. [DOI] [PubMed] [Google Scholar]
- 50.Zhao, F., Xiao, C., Evans, K. S., Theivanthiran, T., Devito, N., Holtzhausen, A., Liu, J., et al., Paracrine Wnt5a‐β‐catenin signaling triggers a metabolic program that drives dendritic cell tolerization. Immunity 2018. 48: 147–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Malinarich, F., Duan, K., Hamid, R. A., Bijin, A., Lin, W. X., Poidinger, M., Fairhurst, A. ‐ M., et al., High mitochondrial respiration and glycolytic capacity represent a metabolic phenotype of human tolerogenic dendritic cells. J. Immunol. 2015. 194: 5174–5186. [DOI] [PubMed] [Google Scholar]
- 52.Sun, Y., Oravecz‐Wilson, K., Bridges, S., Mceachin, R., Wu, J., Kim, S. H., Taylor, A., et al., miR‐142 controls metabolic reprogramming that regulates dendritic cell activation. J. Clin. Invest. 2019. 129: 2029–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Infantino, V., Iacobazzi, V., Palmieri, F. and Menga, A., ATP‐citrate lyase is essential for macrophage inflammatory response. Biochem. Biophys. Res. Commun. 2013. 440: 105–111. [DOI] [PubMed] [Google Scholar]
- 54.Infantino, V., Convertini, P., Cucci, L., Panaro, M. A., Di Noia, M. A., Calvello, R., Palmieri, F., et al., The mitochondrial citrate carrier: a new player in inflammation. Biochem. J. 2011. 438: 433–436. [DOI] [PubMed] [Google Scholar]
- 55.Wei, X., Song, H., Yin, L., Rizzo, M. G., Sidhu, R., Covey, D. F., Ory, D. S., et al., Fatty acid synthesis configures the plasma membrane for inflammation in diabetes. Nature 2016. 539: 294–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moon, J.‐S., Lee, S., Park, Mi‐Ae, Siempos, I. I., Haslip, M., Lee, P. J., Yun, M., et al., UCP2‐induced fatty acid synthase promotes NLRP3 inflammasome activation during sepsis. J. Clin. Invest. 2015. 125: 665–680. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57.Miki, H., Han, K. H., Scott, D., Croft, M. and Kang, Y. J., 4‐1BBL regulates the polarization of macrophages, and inhibition of 4‐1BBL signaling alleviates imiquimod‐induced psoriasis. J. Immunol. 2020. 204: 1892–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stüve, P., Minarrieta, L., Erdmann, H., Arnold‐Schrauf, C., Swallow, M., Guderian, M., Krull, F., et al. De novo fatty acid synthesis during mycobacterial infection is a prerequisite for the function of highly proliferative T cells, but not for dendritic cells or macrophages. Front. Immunol. 2018. 9: 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hsieh, W.‐Y., Zhou, Q. D., York, A. G., Williams, K. J., Scumpia, P. O., Kronenberger, E. B., Hoi, X. P., et al., Toll‐like receptors induce signal‐specific reprogramming of the macrophage lipidome. Cell Metab. 2020. 32: 128–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Araldi, E., Fernández‐Fuertes, M., Canfrán‐Duque, A., Tang, W., Cline, G. W., Madrigal‐Matute, J., Pober, J. S., et al., Lanosterol modulates TLR4‐mediated innate immune responses in macrophages. Cell Rep. 2017. 19: 2743–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blanc, M., Hsieh, W. Y., Robertson, K. A., Kropp, K. A., Forster, T., Shui, G., Lacaze, P., et al., The transcription factor STAT‐1 couples macrophage synthesis of 25‐hydroxycholesterol to the interferon antiviral response. Immunity 2013. 38: 106–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schumann, J., It is all about fluidity: fatty acids and macrophage phagocytosis. Eur. J. Pharmacol. 2016. 785: 18–23. [DOI] [PubMed] [Google Scholar]
- 63.Oishi, Y., Spann, N. J., Link, V. M., Muse, E. D., Strid, T., Edillor, C., Kolar, M. J., et al. SREBP1 contributes to resolution of pro‐inflammatory TLR4 signaling by reprogramming fatty acid metabolism. Cell Metab. 2017. 25: 412–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bosch, M., Sánchez‐Álvarez, M., Fajardo, A., Kapetanovic, R., Steiner, B., Dutra, F., Moreira, L., et al. Mammalian lipid droplets are innate immune hubs integrating cell metabolism and host defense. Science 2020. 370: eaay8085. [DOI] [PubMed] [Google Scholar]
- 65.Castoldi, A., Monteiro, L. B., van Teijlingen Bakker, N., Sanin, D. E., Rana, N., Corrado, M., Cameron, A. M., et al. Triacylglycerol synthesis enhances macrophage inflammatory function. Nat. Commun. 2020. 11: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ishikawa, F., Niiro, H., Iino, T., Yoshida, S., Saito, N., Onohara, S., Miyamoto, T., et al., The developmental program of human dendritic cells is operated independently of conventional myeloid and lymphoid pathways. Blood 2007. 110: 3591–3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Le Naour, F., Hohenkirk, L., Grolleau, A., Misek, D. E., Lescure, P., Geiger, J. D., Hanash, S. and Beretta, L., Profiling changes in gene expression during differentiation and maturation of monocyte‐derived dendritic cells using both oligonucleotide microarrays and proteomics. J. Biol. Chem. 2001. 276: 17920–17931. [DOI] [PubMed] [Google Scholar]
- 68.Rehman, A., Hemmert, K. C., Ochi, A., Jamal, M., Henning, J. R., Barilla, R., Quesada, J. P., et al., Role of fatty‐acid synthesis in dendritic cell generation and function. J. Immunol. 2013. 190: 4640–4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maroof, A., English, N. R., Bedford, P. A., Gabrilovich, D. I. and Knight, S. C., Developing dendritic cells become “lacy” cells packed with fat and glycogen. Immunology 2005. 115: 473–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bougnères, L., Helft, J., Tiwari, S., Vargas, P., Chang, B. H.‐.J., Chan, L., Campisi, L., et al., A role for lipid bodies in the cross‐presentation of phagocytosed antigens by MHC class I in dendritic cells. Immunity 2009. 31: 232–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shaikh, S. R., Mitchell, D., Carroll, E., Li, M., Schneck, J. and Edidin, M., Differential effects of a saturated and a monounsaturated fatty acid on MHC class I antigen presentation. Scand. J. Immunol. 2008. 68: 30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nordestgaard, B. G., Chapman, M. J., Ray, K., Borén, J., Andreotti, F., Watts, G. F., Ginsberg, H., et al., Lipoprotein(a) as a cardiovascular risk factor: current status. Eur. Heart J. 2010. 31: 2844–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Willnow, T. E., Sheng, Z., Ishibashi, S., Herz, J. Inhibition of hepatic chylomicron remnant uptake by gene transfer of a receptor antagonist. Science 1994. 264: 1471–1474. [DOI] [PubMed] [Google Scholar]
- 74.Johnson, A. R., Qin, Y., Cozzo, A. J., Freemerman, A. J., Huang, M. J., Zhao, L., Sampey, B. P., et al. Metabolic reprogramming through fatty acid transport protein 1 (FATP1) regulates macrophage inflammatory potential and adipose inflammation. Mol. Metab. 2016. 5: 506–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Klein‐Wieringa, I. R., Andersen, S. N., Kwekkeboom, J. C., Giera, M., De Lange‐Brokaar, B. J. E., Van Osch, G. J. V. M., Zuurmond, A.‐M., et al., Adipocytes modulate the phenotype of human macrophages through secreted lipids. J. Immunol. 2013. 191: 1356–1363. [DOI] [PubMed] [Google Scholar]
- 76.Gao, F., Liu, C., Guo, J., Sun, W., Xian, L., Bai, D., Liu, H., et al. Radiation‐driven lipid accumulation and dendritic cell dysfunction in cancer. Sci. Rep. 2015. 5: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Herber, D. L., Cao, W., Nefedova, Y., Novitskiy, S. V., Nagaraj, S., Tyurin, V. A., Corzo, A., et al. Lipid accumulation and dendritic cell dysfunction in cancer. Nat. Med. 2010. 16: 880–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gardner, J. K., Mamotte, C. D. S., Patel, P., Yeoh, T. L., Jackaman, C. and Nelson, D. J., Mesothelioma tumor cells modulate dendritic cell lipid content, phenotype and function. PLoS One 2015. 10: 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yin, X., Zeng, W., Wu, B., Wang, L., Wang, Z., Tian, H., Wang, L., et al., PPARα inhibition overcomes tumor‐derived exosomal lipid‐induced dendritic cell dysfunction. Cell Rep. 2020. 33: 108278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Varga, T., Czimmerer, Z. and Nagy, L., PPARs are a unique set of fatty acid regulated transcription factors controlling both lipid metabolism and inflammation. Biochim. Biophys. Acta—Mol. Basis Dis. 2011. 1812: 1007–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tian, D., Hong, H., Shang, W., Ho, C. C., Dong, J. and Tian, X. Y., Deletion of Ppard in CD11c+ cells attenuates atherosclerosis in ApoE knockout mice. FASEB J. 2020. 34: 3367–3378. [DOI] [PubMed] [Google Scholar]
- 82.Cao, W., Ramakrishnan, R., Tuyrin, V. A., Veglia, F., Condamine, T., Amoscato, A., Mohammadyani, D., et al. Oxidized lipids block antigen cross‐presentation by dendritic cells in cancer. J. Immunol. 2014. 192: 2920–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Veglia, F., Tyurin, V. A., Mohammadyani, D., Blasi, M., Duperret, E. K., Donthireddy, L., Hashimoto, A., et al. Lipid bodies containing oxidatively truncated lipids block antigen cross‐presentation by dendritic cells in cancer. Nat. Commun. 2017. 8: 2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Garris, C. S. and Pittet, M. J., ER stress in dendritic cells promotes cancer. Cell 2015. 161: 1492–1493. [DOI] [PubMed] [Google Scholar]
- 85.Ugolini, A., Tyurin, V., Tyurina, Y., Tsyganov, E., Donthireddy, L., Kagan, V. E., et al. Polymorphonuclear myeloid‐derived suppressor cells limit antigen cross‐presentation by dendritic cells in cancer. JCI Insight. 2020. 5: e138581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fritsche, K. L., The science of fatty acids and inflammation. Adv. Nutr. 2015. 6: 293S–301S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Snodgrass, R. G., Boß, M., Zezina, E., Weigert, A., Dehne, N., Fleming, I., Brüne, B. and Namgaladze, D., Hypoxia potentiates palmitate‐induced pro‐inflammatory activation of primary human macrophages. J. Biol. Chem. 2016. 291: 413–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mogilenko, D. A., Haas, J. T., L'homme, L., Fleury, S., Quemener, S., Levavasseur, M., Becquart, C., et al., Metabolic and innate immune cues merge into a specific inflammatory response via the UPR. Cell 2019. 177: 1201–1216. [DOI] [PubMed] [Google Scholar]
- 89.Davis, F. M., Dendekker, A., Joshi, A. D., Wolf, S. J., Audu, C., Melvin, W. J., Mangum, K., et al., Palmitate‐TLR4 signaling regulates the histone demethylase, JMJD3, in macrophages and impairs diabetic wound healing. Eur. J. Immunol. 2020. 50: 1929–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lancaster, G. I., Langley, K. G., Berglund, N. A., Kammoun, H. L., Reibe, S., Estevez, E., Weir, J., et al., Evidence that TLR4 is not a receptor for saturated fatty acids but mediates lipid‐induced inflammation by reprogramming macrophage metabolism article evidence that TLR4 is not a receptor for saturated fatty acids but mediates lipid‐induced inflammation by. Cell Metab. 2018. 1–15. [DOI] [PubMed] [Google Scholar]
- 91.Zasłona, Z., Serezani, C. H., Okunishi, K., Aronoff, D. M. and Peters‐Golden, M., Prostaglandin E2 restrains macrophage maturation via E prostanoid receptor 2/protein kinase A signaling. Blood. 2012. 119: 2358–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sanin, D. E., Matsushita, M., Klein Geltink, R. I., Grzes, K. M., Van Teijlingen Bakker, N., Corrado, M., Kabat, A. M., et al., Mitochondrial membrane potential regulates nuclear gene expression in macrophages exposed to prostaglandin E2. Immunity 2018. 49: 1021–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ramalho, T., Ramalingam, L., Filgueiras, L., Festuccia, W., Jancar, S. and Moustaid‐Moussa, N., Leukotriene‐B4 modulates macrophage metabolism and fat loss in type 1 diabetic mice. J. Leukoc. Biol. 2019. 106: 665–675. [DOI] [PubMed] [Google Scholar]
- 94.Chiu, C. ‐ Y., Gomolka, B., Dierkes, C., Huang, N. R., Schroeder, M., Purschke, M., Manstein, D., et al., Omega‐6 docosapentaenoic acid‐derived resolvins and 17‐hydroxydocosahexaenoic acid modulate macrophage function and alleviate experimental colitis. Inflamm. Res. 2012. 61: 967–976. [DOI] [PubMed] [Google Scholar]
- 95.Duffney, P. F., Falsetta, M. L., Rackow, A. R., Thatcher, T. H., Phipps, R. P. and Sime, P. J., Key roles for lipid mediators in the adaptive immune response. J. Clin. Invest. 2018. 128: 2724–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhao, X., Zhang, Y., Strong, R., Grotta, J. C. and Aronowski, J., 15d‐Prostaglandin J2 activates peroxisome proliferator‐activated receptor‐γ, promotes expression of catalase, and reduces inflammation, behavioral dysfunction, and neuronal loss after intracerebral hemorrhage in rats. J. Cereb. Blood Flow Metab. 2006. 26: 811–820. [DOI] [PubMed] [Google Scholar]
- 97.Desvergne, B. and Wahli, W., Peroxisome proliferator‐activated receptors: nuclear control of metabolism. Endocr. Rev. 1999. 20: 649–688. [DOI] [PubMed] [Google Scholar]
- 98.Surh, Y. J., Na, H. K., Park, J. M., Lee, H. N., Kim, W., Yoon, I. S. and Kim, D. ‐ D., 15‐Deoxy‐Δ 12,14‐prostaglandin J 2, an electrophilic lipid mediator of anti‐inflammatory and pro‐resolving signaling. Biochem. Pharmacol. 2011. 82: 1335–1351. [DOI] [PubMed] [Google Scholar]
- 99.Bernardo, A., Levi, G. and Minghetti, L., Role of the peroxisome proliferator‐activated receptor‐γ (PPAR‐γ) and its natural ligand 15‐deoxy‐Δ(12,14)‐prostaglandin J2 in the regulation of microglial functions. Eur. J. Neurosci. 2000. 12: 2215–2223. [DOI] [PubMed] [Google Scholar]
- 100.Jung, T. W., Park, H. S., Choi, G. H., Kim, D., Ahn, S. H., Kim, D. S., Lee, T. et al. Maresin 1 attenuates pro‐inflammatory reactions and ER stress in HUVECs via PPARα‐mediated pathway. Mol. Cell Biochem. 2018. 448: 335–347. [DOI] [PubMed] [Google Scholar]
- 101.Vats, D., Mukundan, L., Odegaard, J. I., Zhang, L., Smith, K. L., Morel, C. R., Greaves, D. R., et al., Oxidative metabolism and PGC‐1β attenuate macrophage‐mediated inflammation. Cell Metab. 2006. 4: 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Spite, M., Clària, J. and Serhan, C. N., Resolvins, specialized proresolving lipid mediators, and their potential roles in metabolic diseases. Cell Metab. 2014. 19: 21–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Arita, M., Bianchini, F., Aliberti, J., Sher, A., Chiang, N., Hong, S., Yang, R., et al., Stereochemical assignment, antiinflammatory properties, and receptor for the omega‐3 lipid mediator resolvin E1. J. Exp. Med. 2005. 201: 713–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Krishnamoorthy, S., Recchiuti, A., Chiang, N., Yacoubian, S., Lee, C. ‐ H., Yang, R., Petasis, N. A., et al., Resolvin D1 binds human phagocytes with evidence for proresolving receptors. Proc. Natl. Acad Sci. USA 2010. 107: 1660–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Satapati, S., Qian, Y., Wu, M. S., Petrov, A., Dai, Ge, Wang, S. ‐ P., Zhu, Y., et al., GPR120 suppresses adipose tissue lipolysis and synergizes with GPR40 in antidiabetic efficacy. J. Lipid Res. 2017. 58: 1561–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ichimura, A., Hirasawa, A., Poulain‐Godefroy, O., Bonnefond, A., Hara, T., Yengo, L., Kimura, I., et al., Dysfunction of lipid sensor GPR120 leads to obesity in both mouse and human. Nature 2012. 483: 350–354. [DOI] [PubMed] [Google Scholar]
- 107.Davies, L. C., Rice, C. M., Palmieri, E. M., Taylor, P. R., Kuhns, D. B. and McVicar, D. W., Peritoneal tissue‐resident macrophages are metabolically poised to engage microbes using tissue‐niche fuels. Nat. Commun. 2017. 8: 2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Basit, F., Mathan, T., Sancho, D. and De Vries, J. M., Human dendritic cell subsets undergo distinct metabolic reprogramming for immune response. Front. Immunol. 2018. 9: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bourdely, P., Anselmi, G., Vaivode, K., Ramos, R. N., Missolo‐Koussou, Y., Hidalgo, S., Tosselo, J., et al., Transcriptional and functional analysis of CD1c+ Human dendritic cells identifies a CD163+ subset priming CD8+ CD103+ T cells. Immunity 2020. 53: 335–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Villani, A. ‐ C., Satija, R., Reynolds, G., Sarkizova, S., Shekhar, K., Fletcher, J., Griesbeck, M., et al., Single‐cell RNA‐seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science (80‐) 2017. 356: eaah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Galli, E., Friebel, E., Ingelfinger, F., Unger, S., Núñez, N. G. and Becher, B., The end of omics? High dimensional single cell analysis in precision medicine. Eur. J. Immunol. 2019. 49: 212–220. [DOI] [PubMed] [Google Scholar]
- 112.Artyomov, M. N. and Van Den Bossche, J., Immunometabolism in the single‐cell era. Cell Metab. 2020. 32: 710–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Furuhashi, M., Fucho, R., Görgün, C. Z., Tuncman, G., Cao, H. and Hotamisligil, G. S., Adipocyte/macrophage fatty acid‐binding proteins contribute to metabolic deterioration through actions in both macrophages and adipocytes in mice. J. Clin. Invest. 2008. 118: 2640–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ménégaut, L., Jalil, A., Thomas, C. and Masson, D., Macrophage fatty acid metabolism and atherosclerosis: the rise of PUFAs. Atherosclerosis 2019. 291: 52–61. [DOI] [PubMed] [Google Scholar]
- 115.Ryu, H., Kim, J., Kim, D., Lee, J. ‐ E. and Chung, Y., Cellular and molecular links between autoimmunity and lipid metabolism. Mol. Cells 2019. 42: 747–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cheng, H. S., Tan, W. R., Low, Z. S., Marvalim, C., Lee, J. Y. H. and Tan, N. S., Exploration and development of PPAR modulators in health and disease: an update of clinical evidence. Int. J. Mol. Sci. 2019. 20:1–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hossain, F., Al‐Khami, A. A., Wyczechowska, D., Hernandez, C., Zheng, L., Reiss, K., Valle, L. D., et al., Inhibition of fatty acid oxidation modulates immunosuppressive functions of myeloid‐derived suppressor cells and enhances cancer therapies. Cancer Immunol. Res. 2015. 3: 1236–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Serhan, C. N., Clish, C. B., Brannon, J., Colgan, S. P., Chiang, N. and Gronert, K., Novel functional sets of lipid‐derived mediators with antiinflammatory actions generated from omega‐3 fatty acids via cyclooxygenase 2‐nonsteroidal antiinflammatory drugs and transcellular processing. J. Exp. Med. 2000. 192: 1197–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Norling, L. V., Headland, S. E., Dalli, J., Arnardottir, H. H., Haworth, O., Jones, H. R., Irimia, D., et al., Proresolving and cartilage‐protective actions of resolvin D1 in inflammatory arthritis. JCI Insight 2016. 1: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Valdes, A. M., Ravipati, S., Menni, C., Abhishek, A., Metrustry, S., Harris, J., Nessa, A., et al. Association of the resolvin precursor 17‐HDHA, but not D‐or E‐series resolvins, with heat pain sensitivity and osteoarthritis pain in humans. Sci. Rep. 2017. 7: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Motwani, M. P., Colas, R. A., George, M. J., Flint, J. D., Dalli, J., Richard‐Loendt, A., De Maeyer, R. P. H., et al., Pro‐resolving mediators promote resolution in a human skin model of UV‐killed Escherichia coli‐driven acute inflammation. JCI Insight 2018. 3: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Emre, C., Hjorth, E., Bharani, K., Carroll, S., Granholm, A. ‐ C., and Schultzberg, M., Receptors for pro‐resolving mediators are increased in Alzheimer's disease brain. Brain Pathol. 2020. 30: 614–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mizwicki, M. T., Liu, G., Fiala, M., Magpantay, L., Sayre, J., Siani, A., Mahanian, M., et al., 1α,25‐dihydroxyvitamin D3 and resolvin D1 retune the balance between amyloid‐β phagocytosis and inflammation in Alzheimer's disease patients. J. Alzheimer's Dis. 2013. 34: 155–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Arnardottir, H. H., Dalli, J., Norling, L. V., Colas, R. A., Perretti, M. and Serhan, C. N., Resolvin D3 is dysregulated in arthritis and reduces arthritic inflammation. J. Immunol. 2016. 197: 2362–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hersberger, M., Potential role of the lipoxygenase derived lipid mediators in atherosclerosis: leukotrienes, lipoxins and resolvins. Clin. Chem. Lab. Med. 2010. 48: 1063–1073. [DOI] [PubMed] [Google Scholar]
- 126.De Gaetano, M., McEvoy, C., Andrews, D., Cacace, A., Hunter, J., Brennan, E. and Godson, C., Specialized pro‐resolving lipid mediators: modulation of diabetes‐associated cardio‐, reno‐, and retino‐vascular complications. Front. Pharmacol. 2018. 9: 1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Krashia, P., Cordella, A., Nobili, A., La Barbera, L., Federici, M., Leuti, A., Campanelli, F., et al. Blunting neuroinflammation with resolvin D1 prevents early pathology in a rat model of Parkinson's disease. Nat. Commun. 2019. 10: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]


