Abstract
Background
A definitive diagnosis of prostate cancer requires a biopsy to obtain tissue for pathologic analysis, but this is an invasive procedure and is associated with complications.
Purpose
To develop an artificial intelligence (AI)‐based model (named AI‐biopsy) for the early diagnosis of prostate cancer using magnetic resonance (MR) images labeled with histopathology information.
Study Type
Retrospective.
Population
Magnetic resonance imaging (MRI) data sets from 400 patients with suspected prostate cancer and with histological data (228 acquired in‐house and 172 from external publicly available databases).
Field Strength/Sequence
1.5 to 3.0 Tesla, T2‐weighted image pulse sequences.
Assessment
MR images reviewed and selected by two radiologists (with 6 and 17 years of experience). The patient images were labeled with prostate biopsy including Gleason Score (6 to 10) or Grade Group (1 to 5) and reviewed by one pathologist (with 15 years of experience). Deep learning models were developed to distinguish 1) benign from cancerous tumor and 2) high‐risk tumor from low‐risk tumor.
Statistical Tests
To evaluate our models, we calculated negative predictive value, positive predictive value, specificity, sensitivity, and accuracy. We also calculated areas under the receiver operating characteristic (ROC) curves (AUCs) and Cohen's kappa.
Results
Our computational method (https://github.com/ih-lab/AI-biopsy) achieved AUCs of 0.89 (95% confidence interval [CI]: [0.86–0.92]) and 0.78 (95% CI: [0.74–0.82]) to classify cancer vs. benign and high‐ vs. low‐risk of prostate disease, respectively.
Data Conclusion
AI‐biopsy provided a data‐driven and reproducible way to assess cancer risk from MR images and a personalized strategy to potentially reduce the number of unnecessary biopsies. AI‐biopsy highlighted the regions of MR images that contained the predictive features the algorithm used for diagnosis using the class activation map method. It is a fully automatic method with a drag‐and‐drop web interface (https://ai-biopsy.eipm-research.org) that allows radiologists to review AI‐assessed MR images in real time.
Level of Evidence
1
Technical Efficacy Stage
2
Keywords: artificial intelligence, biopsy, deep neural networks, MRI images, PI‐RADS, prostate cancer
Prostate cancer is the most commonly diagnosed cancer in adult men.1 Distinguishing patients with high‐risk (tumor tissue growing faster) and low‐risk (tumor tissue growing slowly) forms of prostate cancer is important because early detection of high‐risk prostate cancer improves survival rate2 and accurate diagnosis prevents overtreatment.3
The European Society of Urogenital Radiology established the Prostate Imaging Reporting and Data System (PI‐RADS), a standardized guideline for interpretation and reporting prostate magnetic resonance imaging (MRI).4 PI‐RADS is designed to improve and standardize detection, localization, characterization, and risk stratification in patients with suspected cancer.5 Radiologists apply PI‐RADS' subjective features such as lesion shape and margins for categorization of prostate cancer6 and assignment of a score ranging from 1 to 5.7 Although PI‐RADS has been found to be effective in evaluating the clinical risk associated with prostate cancer,8 it requires experts' visual assessment, which introduces an element of subjectivity.9
There are currently two main scoring systems used to assess histology slides for prostate cancer aggressiveness. The Gleason Score (GS) is the most commonly used prognostic score to predict the clinical status of prostate cancer based on biopsy material. The GS describes how much the tissue from a biopsy looks like healthy tissue (lower score) or abnormal tissue (higher score).10 GS is the sum of primary and secondary scores, each with a range of 3 to 5. Thus, GSs range from 6 (3 + 3) to 10 (5 + 5) (Table 1). Grade Group (GG) is an alternative scoring system that subdivides prostate cancer into five categories using pathological characteristics.11 Pathologists use either of these scores in routine clinical practice.
TABLE 1.
Grade Group and Gleason Score and Their Association With the Risk Level of Prostate Cancer
| Grade Group | Gleason Score | Combined Gleason Score | Aggressiveness degree |
|---|---|---|---|
| Grade Group 1 | 3 + 3 | 6 | Low risk |
| Grade Group 2 | 3 + 4 | 7 | Intermediate risk but closer to low risk |
| Grade Group 3 | 4 + 3 | 7 | Intermediate risk but closer to high risk |
| Grade Group 4 | 4 + 4, 3 + 5, 5 + 3 | 8 | High risk |
| Grade Group 5 | 4 + 5, 5 + 4, 5 + 5 | 9 and 10 | High risk |
These two different systems are mapped together using the table that was provided and simplified based on the NCCN guidelines version 4.2018 prostate cancer.27
Although a biopsy provides a definitive diagnosis of prostate cancer, patients undergoing prostate biopsy may experience incorrect staging and complications such as infection; 2% to 3% of patients will develop sepsis that is associated with life‐threatening organ dysfunction and death.12
We hypothesized that prostate cancer aggressiveness can be predicted directly from MR images using machine learning (ML) techniques, perhaps reducing the need for a tissue biopsy by optimizing PI‐RADS assessment and increasing diagnosis accuracy. In recent years, ML and especially deep learning (DL) approaches have been applied to a variety of medical conditions,13, 14, 15 such as lung cancer subtype diagnosis using pathology images,16 assessing human blastocyst quality after in vitro fertilization,17 and prostate cancer classification by MR images.18 In the latter study,18 the authors used DL and non‐DL algorithms to differentiate benign prostate from prostate cancer using axial T2‐weighted (T2w) MR images of 172 patients. They were able to distinguish benign from malignant lesions with areas under the receiver operating characteristic (ROC) curves (AUCs) of 0.84 and 0.70 using DL and non‐DL methods, respectively.18 In another related study, Kwon et al described a radiomics‐based approach to classify clinically important lesions in multiparametric MRI (mp‐MRI) using feature‐based methods such as regression trees and random forests. Random forest achieved the highest performance with an AUC of 0.82.19
Recent research indicates that multimodal diagnosis using DL methods has exhibited notable improvement over conventional unimodal approaches in classifying radiology and pathology images.20 Moreover, when MR images are limited, using convolutional neural networks (CNNs) for feature extraction across data concatenation can yield better CNN‐based classification performance.
The aim of this study is to develop a CNN‐based method that uses MR imaging data as input and recognizes benign from cancerous tumor and high‐risk prostate cancer from low‐risk forms, as defined by pathology assessments such as GS and GG. While the training combines MRI data with pathology assessment, our objective was to develop predictive models that could provide assessments from MR images alone.
Materials and Methods
Ethics Statement
All experiments and methods were performed in accordance with the Institutional Review Board at Weill Cornell Medicine. The study used fully de‐identified data and was approved by the ethics committee of our institution (IRB number: 1601016896).
Combined Database
This study included 228 patients from our own urology center (imaged between 2015/02 and 2019/03). We refer to this data set as in‐house throughout this manuscript. All images were acquired on GE and Siemens platforms confirming to PI‐RADS v2.1 specification (T2w—slice thickness 3 mm, no gap; field of view: generally, 12–20 cm; in‐plane dimension: ≤0.7 mm [phase] × ≤0.4 mm [frequency]). The MR images were labeled by cancer GS and GG information obtained from corresponding fusion‐guided biopsy (transrectal and transperineal). We also used four external public data sets obtained from The Cancer Imaging Archive (TCIA)21 that de‐identifies and hosts a large archive of medical images from cancer patients that are accessible for public download. We used data from the PROSTATEx Challenge (n = 99 patients),22, 23 PROSTATE‐MRI (n = 26),24 PROSTATE‐DIAGNOSIS (n = 38 patients),25 and TCGA‐PRAD (n = 9 patients).26 The collection of public data therefore comprised a total of 172 patients with T2w prostate MR images along with histopathology information from corresponding prostate core needle biopsy or prostatectomy specimens. All public and in‐house data were converted from DICOM to PNG format and regularized for intensity inhomogeneity using a Python script. For each patient, consistent sequences of seven axial image slices containing the prostate gland (=2800 images interpolated to 512 × 512 pixels) were reviewed and selected by two radiologists (one advanced imaging technologist with 6 years of experience and one uroradiologist with 17 years of experience) (Fig. 1a). Also, for in‐house data, the assigned PI‐RADS1, 2, 3, 4, 5 scores (if it is available) were reviewed by these two radiologists. Then, we categorized all MR images based on their corresponding pathology reports (Fig. 1b). This means that all the patients' MR images were labeled with a benign, GS,6, 7, 8, 9, 10 or GG1, 2, 3, 4, 5 pathology evaluation performed by experienced pathologists (from different clinics) and then reviewed by one pathologist from our institution (with 15 years of experience). Table 1 shows how we mapped the cancer GG with GS that were obtained from different data sets to use for determining the risk level of prostate cancer. This table was provided and simplified based on the National Comprehensive Cancer Network guidelines version 4.2018 prostate cancer.27 We have follow‐up biopsy for all in‐house cases (benign and malignant) but not for publicly available cases. When multiple biopsies were available, we considered the maximum GS or GG as the final label. In other words, only cases where all biopsies were benign were labeled as benign. Characteristics of all five data sets and their images are summarized in Table 2.
FIGURE 1.
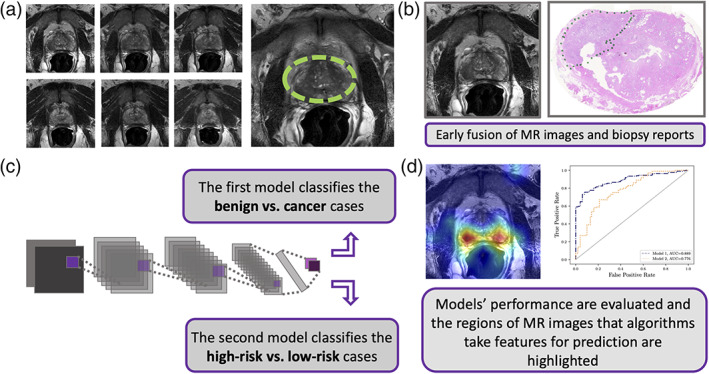
Method flow chart. (a) Unsegmented consistent sequences of seven axial T2w magnetic resonance (MR) image slices for each patient were selected that represent the prostate glands. (b) Each patient's MRI slice labeled by their corresponding biopsy result based on its Grade Group (GG) and Gleason Score (GS). (c) A convolutional neural network (CNN)‐based model (Model 1) classifies the cancer vs. benign and subsequently, and the second CNN‐based model (Model 2) predicts the risk level for each patient. (d) We highlighted the regions of MR images that algorithms focus on for prediction and compared the output of Model 2 with Prostate Imaging Reporting and Data System (PI‐RADS) using pathology labels as ground truth for a subset of test set. Receiver operating characteristic curves (ROCs) were used to assess the performance of different models based on individual patient.
TABLE 2.
Characteristics of All Five Cohorts and the Comprised Biopsy Reports and T2w Images Obtained from TCIA21 and In‐House
| Databases and references | Selected cases and MRI types | Annotation method (biopsy types) | Cancer patients | Benign cases | |||
|---|---|---|---|---|---|---|---|
| High‐risk | Low‐risk | Intermediate‐risk | Intermediate‐risk | Benign | |||
| (GS ≥ 8) | (GS = 6) | (GS = 7) | (GS = 7) | ||||
| (GG = 4 & GG = 5) | (GG = 1) | (GG = 2) | (GG = 3) | ||||
| Weill Cornell Medicine | 228, age (52–85), 3.0 T | GS and GG (fusion guided biopsy), PI‐RADS | 11 | 48 | 37 | 15 | 117 |
| PROSTATEx22, 23 | 99, 3.0 T | GG (core needle biopsy) | 13 | 29 | 38 | 19 | 0 |
| PROSTATE‐DIAGNOSIS25 | 38, 1.5 T | GS (core needle biopsy) | 9 | 5 | 15 | 9 | 0 |
| PROSTATE‐MRI24 | 26, 3.0 T | GS (prostatectomy) | 11 | 0 | 13 | 2 | 0 |
| TCGA‐PRAD26 | 9, 3.0 T | GS and GG (core needle biopsy) | 4 | 0 | 3 | 2 | 0 |
| Total | 400, 1.5 T to 3.0 T | GG and GS (reviewed pathology report) | 48 | 82 | 106 | 47 | 117 |
T = Tesla; GS = Gleason Score; GG = Grade Group; T2w = T2‐weighted; TCIA = The Cancer Imaging Archive; MRI = magnetic resonance imaging.
Models' Architecture and Implementation
We used Google's Inception‐V128 (GoogLeNet) as the main architecture of our models, which offers an effective run‐time and computational cost. To train this architecture, we used transfer learning and pretrained the network on the ImageNet data set.29 We then fine‐tuned all outer layers using the training set of MRI and evaluated the trained model by validation and test sets of MR images obtained from our in‐house and public resources.
To implement our framework (AI‐biopsy), we used TensorFlow, version 1.7, and the TF‐Slim Python library for defining, training, and evaluating models. Training of our deep neural network (DNN) models were performed on a server running the SMP Linux operating system. This server was powered by four NVIDIA GeForce GTX 1080 GPUs with 8 GB of memory for each GPU and 12 1.7‐GHz Intel Xeon CPUs.30 We used Python open‐source libraries such as Pydicom, scikit‐learn, NumPy, SciPy, and Matplotlib for all the statistical analyses.
Training Method
The prostate cancer group (n = 283 patients) included high‐risk patients (n = 48, with GG = 4 and 5), intermediate‐risk patients (n = 153, with GG = 2 and 3), and low‐risk patients (n = 82, with GG = 1). The benign group contains 117 patients (Table 2).
For training the first model (cancer vs. benign), we grouped GG = 3, 4, and 5 together in one class (n = 95 patients) and trained the algorithm vs. the benign class (n = 117). We did not use GG = 1 and 2 patients for training this model so as to allow the algorithm to learn the two ends of the spectrum and take more associated features for classifying cancer vs. benign. However, we tested the trained model on all GGs (GG = 1, 2, 3, 4, 5) as well as benign subjects (Table 3).
TABLE 3.
Characteristics of Both Trained Models and the Comprised Patients
| Model | Data resources | Number of patients with cancerous tumor in training and validation sets | Number of patients with benign tumor in training and validation sets | Number of patients in test set |
|---|---|---|---|---|
|
Model 1: Benign vs. cancer |
In‐house and public | 75 patients (37 GG = 3, 38 GG = 4 and GG = 5) | 107 patients (benign) |
10 benign 10 GG = 1 10 GG = 2 10 GG = 3 10 GG = 4&5 |
| Model | Data resources | Number of patients with high‐risk tumor in training and validation sets | Number of patients with low‐risk tumor in training and validation sets | Number of patients in test set |
|
Model 2: High‐risk vs. low‐risk |
In‐house and public | 75 patients (37 GG = 3, 38 GG = 4 and GG = 5) | 168 patients (72 GG = 1 and 96 GG = 2) |
10 GG = 1 10 GG = 2 10 GG = 3 10 GG = 4&5 |
GG = Grade Group.
For training the second model (high‐risk vs. low‐risk), we grouped GG = 3, 4, and 5 together in one class as high‐risk (n = 95 patients) and trained the algorithm vs. the low‐risk class that combined GG = 1 and 2 (n = 188). We used one of the oversampling techniques, adding Gaussian noise to the images, to address the class imbalance problem. Noise injection consists of injecting a matrix of random values usually drawn from a Gaussian distribution.31 Then, we tested the trained model for all the GG groups (GG = 1, 2, 3, 4, 5) (Table 3).
The cancer/benign images (GG = 3, 4, 5/benign) from 212 patients include a total of 1484 images. One thousand two hundred and seventy‐four images (= 182 patients) were randomly selected for training and validation, and 210 remaining images (= 30 patients for GG = 3, 4, 5, benign) with the addition of 140 images (= 20 patients for GG = 1, 2) were selected for test set (Table 3). Also, the high‐risk/low‐risk images (GG = 3, 4, 5/GG = 1, 2) for 283 patients include a total of 1981 images. Out of these, 1701 images (= 243 patients for GG = 1, 2, 3, 4, 5) were randomly selected for training and validation, and the 280 remaining images (= 40 patients for GG = 1, 2, 3, 4, 5) were selected for test set (Table 3).
Deep Feature Analysis
We applied class activation map (CAM)32 using global average pooling (GAP) in CNNs. Before the final output layer (softmax) of the AI‐biopsy, we performed GAP on the convolutional feature maps and used those as features for a fully connected layer. Given this connectivity structure, we could identify the importance of the image regions by projecting back the weights of the output layer onto the convolutional feature maps.
Evaluation and Statistical Analysis of the Developed Method
We divided the images into training, validation, and test groups. The images and the patients in training, validation, and test sets did not overlap. For each model (Fig. 1c), we performed 5‐fold cross‐validation (resampling procedure) and measured the performance of the algorithm for the test set using AUCs with 95% confidence interval (CI) (Fig. 1d). Characteristics of training, validation, and test set images of each model are summarized in Table 3.
To measure the accuracy of the trained algorithm for individual patients (with sequence of seven axial image slices), we used a simple voting system. For Model 1 (differentiating between malignant and benign tumor), if the number of image slices with cancer (with P ≥ 0.5) from a patient was ≥1, the final label of that patient was “cancer.” Otherwise, the final label of the patient was “benign.” For Model 2 (differentiating between high‐risk and low‐risk tumor), if the number of image slices with high‐risk (with P ≥ 0.5) from a patient was ≥2, the final label of the patient was “high‐risk.” Otherwise, the final label of the patient was “low‐risk.” We employed these threshold conditions on the outputs of the algorithms to reduce false‐negative prediction by giving more weight to the cancer and high‐risk classes.
To evaluate our method, we used negative predictive value, positive predictive value, specificity, sensitivity, and accuracy. We also calculated the AUCs and Cohen's kappa.33
Code Availability
The source code and the guidelines for using the source code are publicly available at https://github.com/ih-lab/AI-biopsy. In addition, AI‐biopsy is available through a web‐based user interface at https://ai-biopsy.eipm-research.org.
Results
Classification of MR Images Based on their Pathology Labels
Our trained Model 1 was able to distinguish cancer patients from benign patients with an AUC of 0.89 (95% CI: [0.86–0.92]), negative predictive value (= 81.6), positive predictive value (= 81.9), specificity (= 82), sensitivity (= 81.5), and accuracy (= 81.8) (Fig. 2a,b). Also, Model 2 was able to classify high‐risk vs. low‐risk (GS = 5 + 5, 5 + 4, 4 + 5, 4 + 4, 4 + 3 vs. GS = 3 + 3, 3 + 4) cancer with an AUC of 0.78 (95% CI: [0.74–0.82]), negative predictive value (= 73), positive predictive value (= 67), specificity (= 68.9), sensitivity (= 71.3), and accuracy (= 70) (Fig. 2c,d). While the performance of Model 2 in classifying GS ≥ 8 vs. GS = 3 + 3 was high (AUC = 0.86), the ability of Model 2 to classify intermediate‐risk cases (GS = 3 + 4 vs. GS = 4 + 3) of prostate cancer was lower (AUC = 0.71).
FIGURE 2.
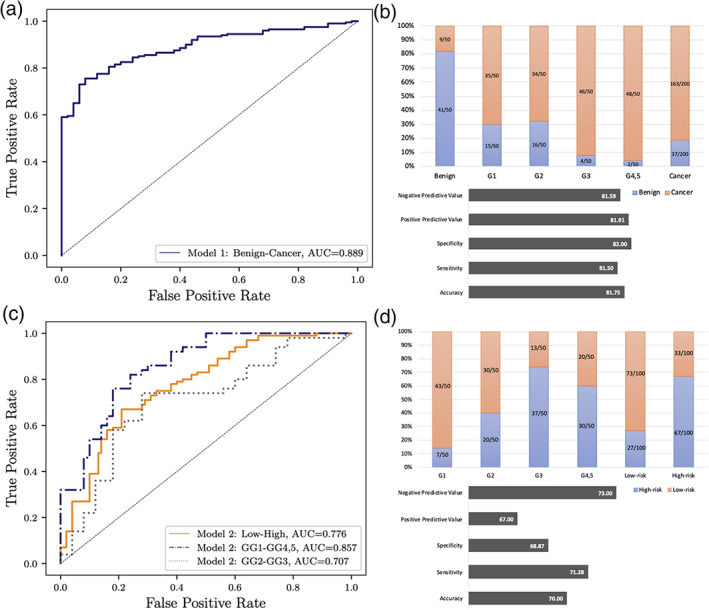
Performance of two trained models for individual patient in the test set. (a) Model 1 performance for classifying cancer vs. benign. (b) The number of patients that were identified correct or incorrect by Model 1, negative predictive value, positive predictive value, specificity, sensitivity, and accuracy for cancer vs. benign. (c) Model 2 performance for classifying high risk vs. low risk. (d) The number of patients that were identified correct or incorrect by Model 2, negative predictive value, positive predictive value, specificity, sensitivity, and accuracy for high risk vs. low risk.
Deep Learning Algorithm Outperforms PI‐RADS for Classification
To further evaluate our model (AI‐biopsy), we were able to compare the PI‐RADS scores with the output of the trained model applied to images that have not been used in the training set. Specifically, we tested the trained model with 28 patients (11 high‐risk and 17 low‐risk, with available PI‐RADS scores), which were not used for training the algorithm (as a blind test set) obtained from the in‐house database. Our model correctly identified 75% (= 21/28 patients) of the labels for high‐risk (= 7/11 patients) and low‐risk (= 14/17 patients). Of the 28 patients, the PI‐RADS score identified 11 patients as high‐risk (PI‐RADS score = 4 and 5) and four patients as low‐risk (PI‐RADS score = 2 and 3). When compared to ground truth (pathology labels), the PI‐RADS score predicted 53.6% (= 15/28 patients) of the patients correctly.
Cohen's kappa values for AI‐biopsy and PI‐RADS in comparison to pathology results as a reference standard were 0.467 (moderate) and 0.195 (slight), respectively.
Discriminative Localization Using Deep Feature Detection
We reviewed the AI‐biopsy results for the above test set (n = 28 patients) to determine whether the disagreement between AI‐biopsy and pathology (reference standard) was due to incorrect feature selection by the AI‐biopsy. A comparison of the CAM result with the radiologists' results demonstrated that AI‐biopsy algorithm is able to detect the prostate gland when it predicts the pathology label correctly (Fig. 3a), while the AI biopsy prediction is incorrect, when the algorithm does not detect the prostate gland (Fig. 3b).
FIGURE 3.
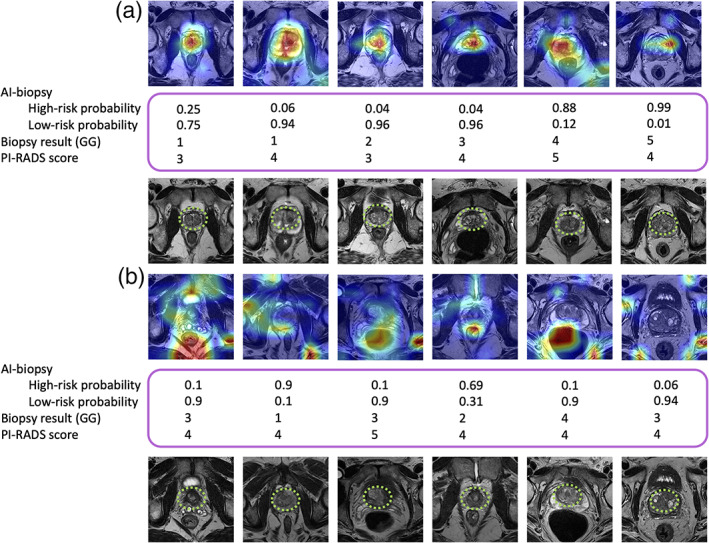
The highlighted prostate glands using class activation map (CAM) and radiologists. Model 2 classifies each image as high risk or low risk, and the deep feature analysis highlights the discriminative regions of the images. A radiologist marked the prostate gland of the images using green square dots. Biopsy results (based on Grade Groups [GGs]) as ground truth and Prostate Imaging Reporting and Data System (PI‐RADS) also are indicated in the figure. (a) Artificial intelligence (AI)‐biopsy predicts the risk level of cases (with a probability score for each class) and highlighted the prostate gland correctly. (b) AI‐biopsy is not able to predict the correct risk level of cases in which the prostate glands are not correctly detected. Red color illustrates features with higher weight.
Discussion
The early and precise diagnosis of prostate cancer is important for proper management of patients. Integrating multimodal clinical data using DL methods has induced useful perceptions and denoted harmonious implementations of this approach to promise next‐generation diagnosis.
The aim of this study was to determine whether a DL method using MRI data that were labeled according to histology results could improve accuracy of prostate cancer diagnosis. We trained a multimodal model to integrate the MR image and pathology score as predictors and further encode the interdependency between these diagnosis sets.
There are two main levels of data integration: early fusion (data are integrated before feeding to the model) and late fusing (different trained model will be integrated using various ML techniques).20 We used the early fusion technique and proposed CNN‐based system, AI‐biopsy, to fully utilize MR images and biopsy results to detect prostate cancer. We trained and validated AI‐biopsy using MR images of 400 patients that were labeled with histopathology information. In addition, compared to the PI‐RADS score, our model indicated higher agreement with biopsy results.
Several groups have attempted to use DL‐based approaches for assessment of prostate cancer aggressiveness with varying degrees of success.34, 35, 36 Cao and colleagues proposed a multiclass CNN (named FocalNet) to detect prostate lesions.36 They used the MR images of 417 patients to predict GS using a homogeneous cohort and showed that their method outperformed U‐Net37 and DeepLab,38 both of which are CNN‐based methods.36 They trained their model to predict four GSs: GS ≥ 7 vs. GS < 7; GS ≥ 4 + 3 vs. GS ≤ 3 + 4; GS ≥ 8 vs. GS < 8; and GS ≥ 9 vs. GS < 9. Their result showed that FocalNet achieved AUCs of 0.81, 0.79, 0.67, and 0.57, respectively.36 A recent study34 has used apparent diffusion coefficient, a metric that is correlated with GS and an important component of mp‐MRI for determining aggressiveness of prostate cancer. They used MR images of 165 patients and predicted high‐risk (GS ≥ 7) from low‐risk (GS = 6) prostate cancer with an AUC of 0.79.34 In addition, Yuan et al presented a DL‐based method to classify 123 patients with high‐risk cancer (GS = 4 + 3, and 8) and 98 patients with low‐risk cancer (GS = 3 + 4, and 3 + 3)35 based on cropped mp‐MRI images. The best performance was obtained using a patch size of 28 × 28 pixels, which led to classifying the two groups with an AUC of 0.896.35
Although these methods achieved good accuracy in assessing prostate cancer aggressiveness, they required several time‐consuming preprocessing steps. Also, they were based on limited homogeneous data sets that did not cover all GSs. The advantage of our method is that instead of only focusing on predetermined, segmented features to analyze, the unsegmented image of the prostate (without bounding box) is assessed, allowing for quantification of all the available data. Our study used a large heterogeneous data set compared to those used in previous studies and included all GS lesion ranges (GS = 3 + 3, 3 + 4, 4 + 3, 4 + 4, 3 + 5, 5 + 3, 4 + 5, 5 + 4, and 5 + 5) as well as benign cases. Previous studies revealed that despite the heterogeneity between data, which is likely due to a combination of technical differences during data acquisition and the biological differences between study cohorts, the deep CNN models are able to accurately extract related signals from noises.16 These studies found that the heterogeneity is gradually mitigated across the layers of the deep CNN model. The heterogeneity is strongest at the input layer but became insignificant at the output layer that makes a CNN model robust and generalizable to data outside the training data set.39
Limitations
Our data were obtained from five different data sets, and they were provided by different techniques (eg, imaging parameters and biopsy types) and annotated by various pathologists and radiologists who may use slightly different methods to assign the scores to each case. To address the heterogeneity among cases as well as lack of details about the clinical information of all cases (eg, patient's age and PSA level), we evaluated the algorithms through 5‐fold cross‐validation to indicate the generalizability of our models to various data sets. In addition, we only used axial T2w MR images in this study because we had more data in this category for both public and in‐house data sets. T2w MRI is routinely used for diagnosis and staging of prostate cancer; however, there is no limitation for using other types of images such as T1w by provided codes. Finally, our MR images were labeled using pathology labels that may include inaccurate histologic findings. Further studies are needed to consolidate the connection between MRI and prostate cancer diagnosis, particularly with available molecular subtypes of prostate cancer.
Conclusion
AI‐biopsy is an automated DNN method (Fig. 4) that increases the accuracy of PI‐RADS scoring for prostate cancer. The trained model integrates complementary information from biopsy report and improves prediction beyond what is possible with MR images alone. It does not require any manual segmentation for testing new images and can be implemented in clinical practice by providing a straightforward platform to use without requiring sophisticated computational knowledge (Fig. 4).
FIGURE 4.
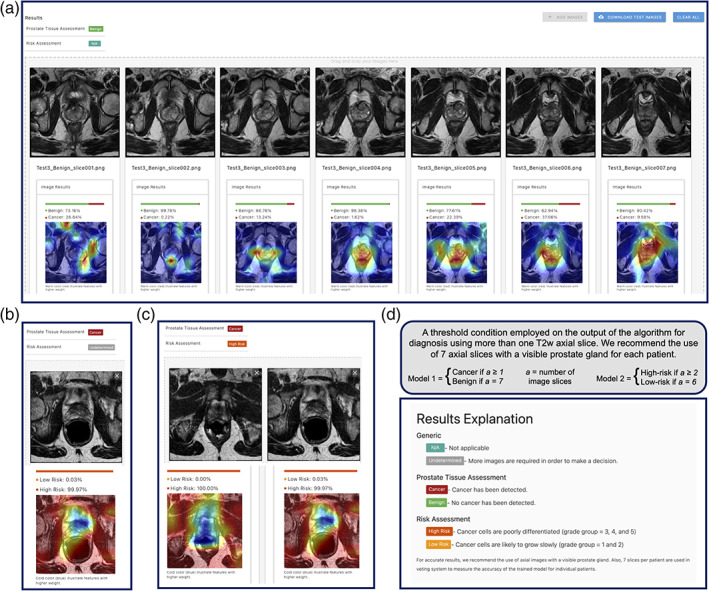
AI‐biopsy is a fully automated framework to use in clinics for evaluation of the prostate cancer risk level. We employed a threshold condition on the output of both models for diagnosis using minimum seven T2w axial image slices. (a) While for prediction of benign diagnosis, all seven image slices should get P ≥ 0.5 for the benign class; (b) one image slice (out of seven imported image slices) with P ≥ 0.5 is enough for Model 1 to result in cancer prediction; (c) Model 2 needs at least two image slices (out of seven imported image slices) with high‐risk P ≥ 0.5 for a patient to result in high‐risk diagnosis; and (d) the result explanation could be seen by clicking on “N/A” option in the web interface (https://ai‐biopsy.eipm‐research.org).
Conflict of Interest
The authors declare no competing interests.
Author Contributions
P.K., A.Sb., O.E., B.C., and I.H. conceived the study. P.K., M.B., Q.L., E.K., and J.B. conceived the method and designed the algorithmic techniques. P.K., M.L., and M.E. generated the data sets and prepared and labeled the images for various Grade groups and Gleason scores. P.K., Q.L., E.K., C.R., and D.M. wrote the codes. P.K. performed computational analysis with input from A.Sb., O.E., B.C., and I.H. P.Z. and A.Si. developed the web interface. T.D.M. and A.Y. reviewed the MR images and PI‐RADS scores. B.D.R. reviewed pathological images, Gleason scores, and Grade groups. P.K. wrote the paper, and all authors read, edited, and approved the final manuscript.
Acknowledgments
We thank Dr. Sohrab P. Shah and Dr. Nicole Rusk for helpful comments on the manuscript. This work was supported by start‐up funds (Weill Cornell Medicine) to I.H. This work used the Extreme Science and Engineering Discovery Environment (XSEDE) GPU servers through allocation IRI200018 to P.K. The authors thank Richard Kneppe, Tom Maiden, John Ruffing, and Hanif Khalak for their assistance with porting and optimization, which was made possible through the XSEDE Extended Collaborative Support Service (ECSS) program. P.K. also thanks Sinan Ramazanoglu and Hamid Mohamadi for their help with the source code and debugging. I.H. also acknowledges NVIDIA for the donation of Titan Xp to support this research through a GPU gift grant as well as XSEDE GPU servers through allocation ASC180052.
[Correction added on 04 May 2021, after first online publication: The affiliation for Ehsan Kazemi has been updated.]
Contributor Information
Bilal Chughtai, Email: bic9008@med.cornell.edu.
Iman Hajirasouliha, Email: imh2003@med.cornell.edu.
REFERENCES
- 1.Pilleron S, Sarfati D, Janssen‐Heijnen M, et al. Global cancer incidence in older adults, 2012 and 2035: A population‐based study. Int J Cancer 2019;144(1):49‐58. [DOI] [PubMed] [Google Scholar]
- 2.Hricak H, Choyke PL, Eberhardt SC, Leibel SA, Scardino PT. Imaging prostate cancer: A multidisciplinary perspective. Radiology 2007;243(1):28‐53. [DOI] [PubMed] [Google Scholar]
- 3.Barrett T, Haider MA. The emerging role of MRI in prostate cancer active surveillance and ongoing challenges. AJR Am J Roentgenol 2017;208(1):131‐139. [DOI] [PubMed] [Google Scholar]
- 4.Hamdy FC, Donovan JL, Lane JA, et al. 10‐year outcomes after monitoring, surgery, or radiotherapy for localized prostate cancer. N Engl J Med 2016;375(15):1415‐1424. [DOI] [PubMed] [Google Scholar]
- 5.Padhani AR, Weinreb J, Rosenkrantz AB, Villeirs G, Turkbey B, Barentsz J. Prostate Imaging‐Reporting and Data System Steering Committee: PI‐RADS v2 status update and future directions. Eur Urol 2019;75(3):385‐396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krishna S, Schieda N, McInnes MD, Flood TA, Thornhill RE. Diagnosis of transition zone prostate cancer using T2‐weighted (T2W) MRI: Comparison of subjective features and quantitative shape analysis. Eur Radiol 2019;29(3):1133‐1143. [DOI] [PubMed] [Google Scholar]
- 7.Vargas HA, Hotker AM, Goldman DA, et al. Updated prostate imaging reporting and data system (PIRADS v2) recommendations for the detection of clinically significant prostate cancer using multiparametric MRI: Critical evaluation using whole‐mount pathology as standard of reference. Eur Radiol 2016;26(6):1606‐1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Portalez D, Mozer P, Cornud F, et al. Validation of the European Society of Urogenital Radiology Scoring System for prostate cancer diagnosis on multiparametric magnetic resonance imaging in a cohort of repeat biopsy patients. Eur Urol 2012;62(6):986‐996. [DOI] [PubMed] [Google Scholar]
- 9.Khalvati F, Zhang Y, Le PHU, Gujrathi I, Haider MA. PI‐RADS guided discovery radiomics for characterization of prostate lesions with diffusion‐weighted MRI. SPIE Medical Imaging: Computer‐Aided Diagnosis, Vol. 10950. San Diego, California, United States: International Society for Optics and Photonics; 2019. 1095042 p. 10.1117/12.2512550. [DOI] [Google Scholar]
- 10.Donovan MJ, Fernandez G, Scott R, et al. Development and validation of a novel automated Gleason grade and molecular profile that define a highly predictive prostate cancer progression algorithm‐based test. Prostate Cancer Prostatic Dis 2018;21(4):594‐603. [DOI] [PubMed] [Google Scholar]
- 11.Epstein JI, Egevad L, Amin MB, Delahunt B, Srigley JR, Humphrey PA. The 2014 International Society of Urological Pathology (ISUP) consensus conference on Gleason grading of prostatic carcinoma: Definition of grading patterns and proposal for a new grading system. Am J Surg Pathol 2016;40(2):244‐252. [DOI] [PubMed] [Google Scholar]
- 12.Jones TA, Radtke JP, Hadaschik B, Marks LS. Optimizing safety and accuracy of prostate biopsy. Curr Opin Urol 2016;26(5):472‐480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal 2017;42:60‐88. [DOI] [PubMed] [Google Scholar]
- 14.Shen D, Wu G, Suk HI. Deep learning in medical image analysis. Annu Rev Biomed Eng 2017;19:221‐248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suzuki K. Overview of deep learning in medical imaging. Radiol Phys Technol 2017;10(3):257‐273. [DOI] [PubMed] [Google Scholar]
- 16.Khosravi P, Kazemi E, Imielinski M, Elemento O, Hajirasouliha I. Deep convolutional neural networks enable discrimination of heterogeneous digital pathology images. EBioMedicine 2018;27:317‐328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khosravi P, Kazemi E, Zhan Q, et al. Deep learning enables robust assessment and selection of human blastocysts after in vitro fertilization. NPJ Digit Med 2019;2(1):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Yang W, Weinreb J, et al. Searching for prostate cancer by fully automated magnetic resonance imaging classification: Deep learning versus non‐deep learning. Sci Rep 2017;7(1):15415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwon D, Reis IM, Breto AL, et al. Classification of suspicious lesions on prostate multiparametric MRI using machine learning. J Med Imaging (Bellingham) 2018;5(3):034502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez K, Fodeh SJ, Allam A, Brandt CA, Krauthammer M. Reducing annotation burden through multimodal learning. Front Big Data 2020;3:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clark K, Vendt B, Smith K, et al. The cancer imaging archive (TCIA): Maintaining and operating a public information repository. J Digit Imaging 2013;26(6):1045‐1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Litjens G, Debats O, Barentsz J, Karssemeijer N, Huisman H. Computer‐aided detection of prostate cancer in MRI. IEEE Trans Med Imaging 2014;33(5):1083‐1092. [DOI] [PubMed] [Google Scholar]
- 23.Geert Litjens OD, Barentsz J, Karssemeijer N, Huisman H. ProstateX Challenge data. Cancer Imaging Arch; 2017;10:K9TCIA.
- 24.Choyke PTB, Pinto P, Merino M, Wood B. Data from PROSTATE‐MRI. Cancer Imaging Arch; 2016;9. http://doi.org/10.7937K.
- 25.Bloch BN, Jain A, Jaffe CC. Data from PROSTATE‐DIAGNOSIS. Cancer Imaging Arch; 2015;9:10.7937.
- 26.Zuley ML, Jarosz R, Drake BF, et al. Radiology Data from The Cancer Genome Atlas Prostate Adenocarcinoma [TCGA‐PRAD] collection. Cancer Imaging Arch; 2016;9. http://doi.org/10.7937K.
- 27.(NCCN) NCCN . Prostate Cancer (Version 4.2018). 2018.
- 28.Szegedy C, Liu W, Jia Y, et al. Going deeper with convolutions. Computer vision and pattern recognition (CVPR). Boston, MA: IEEE; 2015. p 1‐9. [Google Scholar]
- 29.Deng J, Dong W, Socher R, Li L, Kai L, Li F‐F. ImageNet: A large‐scale hierarchical image database. 2009 IEEE Conference on Computer Vision and Pattern Recognition. Miami, FL: IEEE; 2009. p 248‐255. [Google Scholar]
- 30.Towns J, Cockerill T, Dahan M, et al. XSEDE: Accelerating scientific discovery. Comput Sci Eng 2014;16(5):62‐74. [Google Scholar]
- 31.Shorten C, Taghi M. Khoshgoftaar. A survey on image data augmentation for deep learning. J Big Data 2019;6(1):1‐48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A. Learning deep features for discriminative localization. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) ; 2016. p 2921–2929.
- 33.Cohen JA. Coefficient of agreement for nominal scales. Educ Psychol Meas 1960;20(1):37‐46. [Google Scholar]
- 34.Woo S, Kim SY, Cho JY, Kim SH. Preoperative evaluation of prostate cancer aggressiveness: Using ADC and ADC ratio in determining Gleason score. Am J Roentgenol 2016;207(1):114‐120. [DOI] [PubMed] [Google Scholar]
- 35.Yuan YX, Qin WJ, Buyyounouski M, et al. Prostate cancer classification with multiparametric MRI transfer learning model. Med Phys 2019;46(2):756‐765. [DOI] [PubMed] [Google Scholar]
- 36.Cao R, Bajgiran AM, Mirak SA, et al. Joint prostate cancer detection and Gleason score prediction in mp‐MRI via FocalNet. IEEE Trans Med Imaging 2019;38:2496‐2506. [DOI] [PubMed] [Google Scholar]
- 37.Ronneberger O, Fischer P, Brox T. U‐Net: Convolutional networks for biomedical image segmentation. Medical image computing and computer‐assisted intervention, Pt III, Vol 9351; Switzerland: Springer; 2015. p 234‐241. 10.1007/978-3-319-24574-4_28. [DOI] [Google Scholar]
- 38.Chen LC, Papandreou G, Kokkinos I, Murphy K, Yuille AL. DeepLab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. IEEE Trans Pattern Anal Mach Intell 2018;40(4):834‐848. [DOI] [PubMed] [Google Scholar]
- 39.Hu Z, Tang A, Singh J, Bhattacharya S, Butte AJ. A robust and interpretable end‐to‐end deep learning model for cytometry data. Proc Natl Acad Sci U S A 2020;117(35):21373‐21380. [DOI] [PMC free article] [PubMed] [Google Scholar]


