Abstract
Background: Peptidyl arginine deiminase 4 (PAD4) is an enzyme that converts arginine into citrulline. PAD4 is expressed in neutrophils that, when activated, can drive the formation of neutrophil extracellular traps (NETs). Uncontrolled activation of PAD4 and subsequent citrullination of proteins is increasingly recognized as a driver of (auto)immune diseases. Currently, our understanding of PAD4 structure–function relationships and activity control in vivo is incomplete.
Aims: To provide the current state‐of‐the‐art on PAD4 structure‐activity relationships and involvement of PAD4 in autoimmune disorders as well as in thrombo‐inflammatory disease.
Materials & Methods: Literature review and molecular modelling
Results: In this review, we used molecular modelling to generate a three‐dimensional structure of the complete PAD4 molecule. Using our model, we discuss the catalytic conversion of the arginine substrate to citrulline. Besides mechanistic insight into PAD4 function, we give an overview of biological functions of PAD4 and mechanisms that influence its activation. In addition, we discuss the crucial role of PAD4‐mediated citrullination of histones during the formation of NETs. Subsequently, we focus on the role of PAD4‐mediated NET formation and its role in pathogenesis of rheumatoid arthritis, sepsis and (immune‐)thrombosis. Finally, we summarize current efforts to design different classes of PAD4 inhibitors that are being developed for improved treatment of autoimmune disorders as well as thrombo‐inflammatory disease.
Discussion: Advances in PAD4 structure‐function are still necessary to gain a complete insight in mechanisms that control PAD4 activity in vivo. The involvement of PAD4 in several diseases signifies the need for a PAD4 inhibitor. Although progress has been made to produce an isotype specific and potent PAD4 inhibitor, currently no PAD4 inhibitor is ready for clinical use.
Conclusion: More research into PAD4 structure and function and into the regulation of its activity is required for the development of PAD4 specific inhibitors that may prove vital to combat and prevent autoimmune disorders and (thrombo)inflammatory disease.
Keywords: cancer, citrullination, immune disease, NETosis, neutrophil extracellular traps (NETs), PAD4, PAD4 inhibitors, protein arginine deiminase, rheumatoid arthritis (RA), sepsis, thrombosis
1. INTRODUCTION
Posttranslational modification (PTM) of amino acids through the addition or removal of chemical groups greatly increases the level of regulation and diversity of protein function.1 Over the past 2 decades, citrullination, or deimination, has been increasingly recognized as a modifier of immune‐mediated disorders. Citrullination is the irreversible conversion of an arginine to citrulline, which is catalyzed by a group of enzymes that belong to the peptidyl arginine deiminase (PAD) family. The proposed chemical modification of arginine into a citrulline by PAD42 is described in Figure 1.
FIGURE 1.
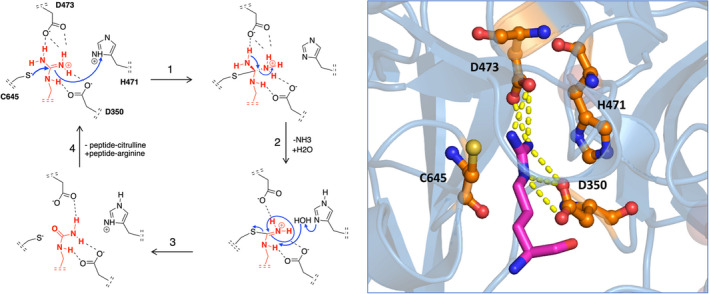
Chemical modification of arginine to citrulline by PAD4. The mechanism of citrullination by PAD4, as adapted from Knuckley et al.2 The PAD4 residues Asp350 and Asp473 function as “anchor points” and sit at the bottom of the tunnel, forming strong interactions via two salt bridges with the guanidinium group of the arginine substrate. The actual catalytic process of deimination is initiated by nucleophilic attack of thiolate‐form Cys645 on the guanidinium carbon of arginine, followed by stabilization of the tetrahedral intermediate through protonation by His471 (1). Subsequent collapse of the intermediate (2) yields free ammonia and an S‐alkylthiouronium intermediate. This intermediate is then hydrolyzed by a water molecule yielding the original Cys645 thiolate and citrulline (3). Release of the peptide‐citrulline product finishes the catalytic cycle (4). On the right, the crystal structure of PAD4 together with the substrate arginine is shown. The PAD4 active site resides are shown in orange, arginine is shown in magenta. Yellow dotted lines represent hydrogen bonds between arginine and aspartic acids D350 and D473. PAD, peptidyl arginine deiminase
Citrullination can greatly affect proteins, mainly because the positive charge of arginine is lost when it is converted to a citrulline.3 This way, citrullination can introduce novel protein structures, functions, and interactions, as summarized in Figure 2. When citrullination alters the surface electrostatic potential of a protein, its inter‐/intra‐protein interactions can be affected.4 Likewise, protein dysfunction or unfolding may occur.5 Importantly, citrullination of proteins has been linked to the formation of autoimmune antibodies in several diseases, including rheumatoid arthritis (RA).6
FIGURE 2.
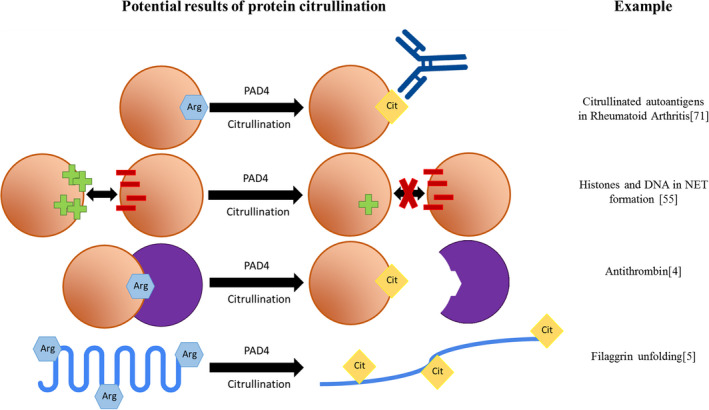
Consequences of citrullination. Overview of the possible consequences of citrullination. An orange circle represents a protein, a purple circle represents a protein interactor. For each mechanism, an example is given on the right. Arginine residues are shown as blue hexagons. Citrulline residues are shown as yellow diamonds
In humans, five PAD isotypes are expressed: PAD1‐4 and PAD6 (PAD5 is identical to PAD4). PADs are highly conserved among different isotypes as well as among different species.7 For example, PADs share approximately 50% to 55% sequence identity within a single species, and each PAD isotype exhibits 70% to 95% identity among different species. The high percentage of sequence similarity and identity of human PAD isoforms suggests that all human PAD isotypes have a similar tertiary structure. Moreover, the high conservation of amino acids at the active site indicates that these PADs catalyze the conversion of arginine to citrulline via the same mechanism.
The focus of this article, human PAD4, was first described in HL‐60 cells and was found to participate in differentiation of HL‐60 cells into granulocytes and monocytes/macrophages.8 PAD4 is the only PAD isotype that carries a nuclear localization signal9 and is highly expressed in immune cells including granulocytes, monocytes, macrophages,8 CD34(+) bone marrow cells, and multipotent progenitor cells.10 PAD4 mainly targets histones in the nucleus,11 but other proteins, such as glycogen synthase kinase‐3β (GSK3β),12 p300,13 inhibitor of growth 4 (ING4),14 40S ribosomal protein S2,15 and a disintegrin and metalloproteinase with a thrombospondin type 1 motif, member 13 (ADAMTS13)16 have been reported to be citrullinated by PAD4. Moreover, more than 100 different PAD4 substrates were identified in HEK293 T cells via mass spectrometric analysis, revealing a rather promiscuous substrate specificity for PAD4 with a modest preference for an RGG motif.17 Generally, for PAD4 the citrullination turnover rate is inversely correlated with the structural order of the substrate.18 The only substrate‐specific limitation for citrullination seems to be the requirement for a small side chain at the R‐2 position, to prevent steric inhibition.19
The aforementioned citrullination of nuclear histones by PAD4 is recognized as a trigger in the formation of neutrophil extracellular traps (NETs). NETs have recently been discovered as an integral part of the innate immune system.20 NETs have the capacity to enhance immune responses by trapping and killing bacteria. However, at the same time, NETs are indiscriminate with respect to their cytotoxicity, and uncontrolled NET formation contributes to damage of healthy tissue, which is why PAD4 has been described as a “double‐edged sword.”21, 22 In neutrophils, different stimuli can result in the formation of NETs, which are produced in distinct cellular pathways.23, 24 Unlike the classic NETosis pathway, PAD4 activation by calcium results in reactive oxygen species (ROS)‐independent NET formation, in a process that has been named leukotoxic hypercitrullination.23 More and more studies have connected PAD4 or leukotoxic hypercitrullination to human diseases, including RA, sepsis, thrombosis, and cancer.25, 26, 27, 28, 29
Although the role of PAD4 in the development of disease is being increasingly uncovered, a complete mechanistic insight into factors that are required for the expression and regulation of PAD4 activity is lacking. In this review, we summarize the available PAD4 structure–function data, discuss physiologic functions, and provide an insight into mechanisms of PAD4 activation and regulation. Moreover, we provide an overview of the role of PAD4 in the pathogenesis of thrombo‐inflammatory and autoimmune disorders. We provide a complete structural overview of PAD4 as a scaffold that may be useful for the development of novel classes of inhibitors to more efficiently combat PAD4‐mediated diseases.
2. THE STRUCTURE OF PAD4
The human PADI4 gene of 55,806 bases encodes for a 663‐amino acid PAD4 monomer of 74 kDa. A PAD4 monomer consists of two domains: an N‐terminal domain (residues Met1‐Pro300) and a C‐terminal catalytic domain (residues Asn301‐Pro663). The N‐terminal domain can be divided into two immunoglobulin‐like subdomains (subdomain I: residues Met1‐Cys118; and subdomain II: residues Ala119‐Pro300)30 (Figure 3A). The immunoglobulin‐like domains are involved in different functions including cell–cell recognition, cell‐surface receptors, and have been reported to serve functions in the immune system.31 The catalytic domain contains the active site and forms an α/β propeller structure.30 Human PAD4 contains five Ca2+ binding sites. Ca1 and Ca2 are located in the C‐terminal domain and are structurally involved in the calcium‐induced conformational changes that occur when the functional active site cleft is generated.30 Ca3, Ca4, and Ca5 are noncatalytic calcium ions situated in the N‐terminal domain and are thought to be required for stabilization of the active conformation of PAD4.32 At the N‐terminus, there is a single nuclear localization signal (NLS) (Figure 3A), which is a (56‐PPAKKKST‐63) motif in which three continuous lysine residues 59 through 61 follow two proline residues.9 We have used the Homology Model suite implemented in YASARA program33 to build the regions that are missing from the reported incomplete X‐ray structures of PAD4 in order to obtain a complete three‐dimensional structure of human PAD4. From our homology model, it shows that the NLS adopts a loop conformation (Figure 3A), which is responsible for interaction with nuclear transport receptors to guide the passage of PAD4 across the nuclear membrane.9 Under physiological conditions, PAD4 has a head to tail homodimeric conformation, and it was demonstrated that dimeric PAD4 expresses full catalytic activity and calcium binding.30 The dimerization of PAD4 is mediated by several electrostatic interactions of which those between Arg8 from one subunit and Asp547 from another subunit is most important, as was concluded from mutagenesis studies.34 Dissociation of dimer formation could reduce PAD4 enzymatic activity and calcium‐binding cooperativity. Monomeric PAD4 has a residual 25% to 50% enzyme activity compared with the dimeric enzyme.34
FIGURE 3.
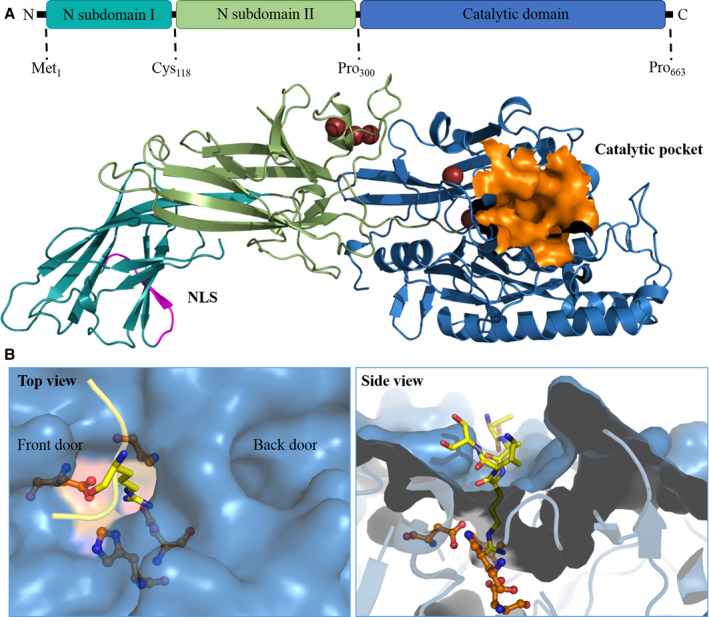
The three‐dimensional structure of PAD4. A, Full‐length PAD4 monomer. The homology model of PAD4 is made using YASARA software based on the 3B1U PDB template. The NLS is shown in magenta, and the catalytic pocket is shown in orange. B, The catalytic site and substrate‐binding pocket of PAD4. The arginine substrate containing histone tail (TARKS) is shown in yellow; active site residues are orange. The left panel shows the top view, where the TAKS residues of the histone tails are not shown. The front door and back door of the active site are indicated in the image. The right panel contains the side view of the binding pocket, where it is clear how only the substrate arginine protrudes into the active site. PAD, peptidyl arginine deiminase
The PAD4 active site contains a negatively charged U‐shaped tunnel that allows the arginine‐containing substrates to bind through a so‐called “front door” (Figure 3B).35 After catalysis, reaction products are released from the active site and leave the active site region through a so‐called “back door” (Figure 3B).35 Enzymes with substrate tunnels generally have high substrate selectivity, influenced by the depth and shape of the tunnel, with which they may select particular substrates of an appropriate chain length.36 The catalytic site is mainly formed by four active site residues (Asp350, His471, Asp473, Cys645) that are deeply buried in the tunnel (Figure 3B).30 The conformation of arginine in the active site, flanked by Cys645 and His471, is displayed in Figure 3B.
3. ACTIVATION AND REGULATION OF PAD4 ACTIVITY
Our understanding of the mechanisms that govern PAD4 activation and regulation is incomplete. Here, we provide an overview of factors or conditions that are known to contribute to the up‐ or down‐regulation of PAD4 enzyme activity, which have been graphically summarized in Figure [Link], [Link].
As previously introduced, PAD4 is activated intracellularly following a Ca2+ wave. Calcium ionophores, such as Ionomycin and A23187 are therefore frequently used to activate PAD4 in vitro.37 Calcium is known as a key regulator of PAD4 catalytic activity by its capacity to induce structural changes in PAD4. These conformational changes accompany the transition of an inactive to an active PAD4 conformation.30, 32 To achieve maximal activation in vitro, PAD4 requires typically 5 to 10 mM calcium to occupy its five calcium sites, which can be recognized from the PAD4 structure as described previously.30, 32 However, intracellular calcium concentration range from 10 nM to 100 μM and the calcium concentration in blood ranges from 2.2 to 2.6 mM.38 This suggests that in vivo, other factors than calcium play a role in PAD4 activation. It has been suggested that cofactors may lower the requirement of PAD4 for calcium and assist in activation of PAD4, although this is, as of yet, a poorly understood mechanism. One example where the threshold for PAD4 activation by calcium is lowered is present in some RA patients who produce antibodies that react with both PAD3 and PAD4.39 Compared with patients who only have anti‐PAD4 antibodies, patients with combined anti‐PAD3/PAD4 cross‐reactive antibodies have the most erosive joint disease. Darrah et al. showed that the anti‐PAD3/PAD4 cross‐reactive antibodies can activate and increase the catalytic efficiency of PAD4 at low calcium concentrations (0.2 mM).39 It was hypothesized that stabilization of the active conformation of PAD4 was achieved by binding of cross‐reactive antibodies in a calcium‐independent fashion.39 However, conclusive data supporting this mechanism are currently lacking.
Furthermore, independent of calcium concentration, it was shown that bicarbonate increases PAD4 activity, without altering the affinity of PAD4 for calcium.40 In the presence of bicarbonate, an increase of PAD4 catalyzed citrullination of histone H3 and fibrinogen was described. However, the in vitro determined optimal bicarbonate concentration for the modulation of PAD4 activity was around 22 mM. This concentration favors the activity of PAD4 in the extracellular environment, given that the bicarbonate concentration in human serum is 17 to 29 mM and inside cells it is <10 mM.41 How exactly bicarbonate can modulate PAD4 catalytic activity is still unknown, and further experimental studies are needed to understand its mode of action.
3.1. Redox regulation
As early as in 1991, it was shown that three mouse PAD isoforms (PAD1‐3) require a reducing environment for their enzymatic activity.42 PADs can be rapidly oxidized and become inactive after release into the extracellular space, and both H2O2 and ROS formation can inhibit PAD activity in a dose‐dependent manner.43, 44 Most likely, this requirement for a reducing environment for PAD4 activity is due to the Cys645 residue requiring reduction to the thiolate form to allow nucleophilic attack of the guanidinium carbon of arginine (Figure 1). Reducing agents, such as dithiothreitol, Tris(2‐carboxyethyl) phosphine, or beta‐mercaptoethanol, are used in vitro to preserve PAD4 activity. The intracellular environment is also regarded as a reducing environment. Damgaard et al. showed that lower millimolar range glutathione (GSH) as present in the cytoplasm was capable of activating PAD4 in an in vitro assay.44 However, the extracellular level of GSH (<1 mM) is too low to prevent the oxidation of PAD4. Additionally, elevation of the oxidoreductase enzyme thioredoxin was observed in RA synovial fluid and plasma, and Nagar et al. proved the PAD4‐activating capacity of thioredoxin at physiological concentration.45 Taken together, the redox state of the PAD4 environment may contribute to the regulation of PAD4 activity under physiological conditions.
3.2. PAD4 auto‐citrullination
A further candidate for control of PAD4 activation is the process of auto‐citrullination. PAD4 has been shown to be capable of citrullinating itself on several positions, which was shown to affect PAD4 protein–protein interactions.46 Regarding the effect of auto‐citrullination on PAD4 activity, we showed in a recent publication that PAD4 auto‐citrullination has no effect on its activity.47
4. PAD4 IN NET FORMATION
Neutrophils function as a first line of defense of the human innate immune system.48 They migrate to sites of infection from the blood to engulf and kill invasive pathogens like bacteria, fungi, and viruses.49 In response to an inflammatory environment, neutrophils can respond in several ways. More established are the NETs that neutrophils form as part of the innate immune response. NETs were first characterized in 2004 as a new mechanism of neutrophil activation in response to pathogens.50 Decondensed chromatin is regarded as the scaffold of NETs, which is decorated with granular and cytoplasmic proteins, including neutrophil elastase, myeloperoxidase, cathepsin G, as well as histones.20 Recently, Thiama et al. used high‐resolution microscopy to unravel the sequence of events during neutrophil extracellular trap formation (NETosis). They report that, following stimulation, NET‐forming neutrophils quickly disassemble their actin cytoskeleton and start shedding plasma membrane microvesicles. Subsequent remodeling of the vimentin and microtubule cytoskeletons precedes endoplasmic reticulum (ER) vesiculation. This is followed by chromatin decondensation and loss of nuclear lobularity, after which permeabilization of the nuclear envelope and plasma membrane occurs. Last, chromatin exits through the ruptured nuclear envelope into the cytoplasm and enters the extracellular matrix after rupture of the plasma membrane.51 Importantly, Thiama et al. additionally report that, chromatin decondensation, lamin meshwork destabilization, nuclear envelope rupture, and extracellular DNA release are PAD4‐dependent processes during NET formation.51 Interestingly, NETs can be formed as a result of several pathways. Initially, NETosis was reported as a mechanism that depends on ROS generated by neutrophil NADPH oxidase.52 In this mechanism, ROS production leads to destabilization of granule membranes and the nuclear envelope, which allows mixing of cytoplasmic and nuclear components. Cytoplastic neutrophil elastase (NE) can subsequently cleave the nuclear histones, which results in a decondensation of chromatin. However, it was discovered that inhibition of NADPH oxidase cannot prevent NET formation in response to the bacteria,53 hinting at an alternative NET formation pathway.50, 54
Recent studies have revealed that PAD4‐catalyzed histone citrullination is a crucial step for NET formation in a ROS‐independent pathway. During PAD4‐mediated NET formation, or leukotoxic hypercitrullination,23 PAD4 citrullinates histones in neutrophils. Citrullinated histones lose positive surface charges that interact with the negatively charged DNA backbone, which results in decondensation of chromatin.55 Non‐DNA‐bound histones can subsequently break the nuclear envelope and plasma membranes.56 The inability for PAD4‐deficient mice to form NETs in response to ionomycin shows the critical role PAD4 plays in this process.50, 57 In line with this, inhibition of PAD4 by the known PAD4 inhibitor Cl‐amidine significantly reduces histone citrullination and prevents the formation of NET‐like structures after stimulation of neutrophils with ionomycin.58
As introduced previously, NETs themselves are viewed as a double‐edged sword.21, 22 NETs are known to activate the Complement system through binding of C1q to extracellular DNA.59 Additionally, NET's prime CD4+ T cells in a process that engages the T‐cell receptor, and in doing so lower their activation threshold.60 Additionally, NETs exacerbate inflammation through danger‐associated molecular pattern signaling.61 Especially extracellular histones resulting from NETs have been shown to damage tissues through activation of Toll‐like receptors 2 and 461 and have a pro‐thrombotic effect through their capacity to activate platelets.62 Furthermore, NETs can act as a scaffold for thrombus formation through binding of platelets, red blood cells and procoagulant molecules.63 In addition to this, extracellular activity of PAD4 has been postulated to be a driver of diseases through uncontrolled citrullination of proteins. As such, NET formation is seen as a link between inflammation and onset of several (autoimmune) diseases,64 some of which will be discussed in the next paragraph.
5. ROLE OF PAD4 IN PATHOGENESIS OF DISEASES
Citrullination of proteins by PAD4 is associated with a multitude of disease. Here, we provide a detailed overview of the role of PAD4 in rheumatoid arthritis, sepsis, and thrombosis.
5.1. Rheumatoid arthritis
RA is one of the most common inflammatory autoimmune diseases and affects around 0.5% to 1.0% of the population worldwide.65 RA patients suffer from systemic inflammation, which leads to damaging of synovial tissues resulting in movement restriction and disability.66 In past decades, a growing number of studies have pointed to dysregulation of PAD4‐mediated citrullination of extracellular proteins as a driver of the autoimmune response in RA, as 75% of patients produce anti‐citrullinated protein antibodies (ACPA). This leads to the working hypothesis that ongoing citrullination in RA synovial fluid provides autoantigens that lead to continuation of the inflammatory response.67 Associations between genetic variants of PAD4 and increased risk in RA development have been made. Recently, a recent meta‐analysis of several studies that focused on the single nucleotide PAD4 polymorphism 104C/T indicated that the polymorphism associates with an increased risk for developing RA.68, 69 The haplotype of the PADI4 gene that increases RA risk is linked to increased production of citrullinated peptides, further strengthening the connection between citrullination and RA.68 In 2018, Tivawala et al. published the RA citrullinome, and identified many different citrullinated proteins in synovial fluid of patients with RA.70 Citrullinated proteins can be considered autoantigens that recognized as non‐self by the immune system and induce autoantibody generation against citrulline‐containing proteins.71 In short, uncontrolled citrullination of proteins can result in the immune system losing tolerance‐to‐self. This principle was illustrated in a study with rats, which reports that the presence of citrullinated protein and PAD4 levels correlate with the severity of collagen‐induced arthritis.6 PAD4 knockout mice present with a significantly reduced arthritis severity in a glucose‐6‐phosphate isomerase‐induced arthritis model.72 In a collagen‐induced arthritis mouse model it was shown that anti‐collagen antibodies and inflammatory cytokine levels were remarkably decreased in PAD4 deficient mice compared with wild‐type mice.27
Importantly, ACPAs appear in an early stage of the disease and some are even regarded as a hallmark of RA diagnosis and prediction. Because immunoglobulin G anti‐filaggrin autoantibodies can recognize a citrulline epitope of various forms of filaggrin, they are regarded a very specific marker of RA.73 Importantly, higher ACPA titers correlate with disease severity.74 Anti‐cyclic citrullinated peptide autoantibodies have ~98% specificity and 68% to 75% sensitivity and can be used in early stage RA diagnosis.75
5.2. PAD4 in sepsis
Sepsis‐related deaths account for approximately 20% of all global deaths, with approximately 50 million reported cases in 2017.76 A growing number of studies have demonstrated that NETs are one of the major contributors to disease severity in sepsis.77, 78, 79 As mentioned, citrullination of nuclear histones by PAD4 leading to decondensation of chromatin is a key step in the onset of NET formation. NETs are formed within the vasculature during sepsis, in response to circulating pathogens.80 In the early stages of sepsis, depletion of NETs is not beneficial for the prevention or containment of a systemic infection and it can even exacerbate the pathology, as was observed in a mouse model of polymicrobial sepsis.81 In severe sepsis, NETs and platelets can synergistically participate in pathogen containment. Histones were reported as major mediators of death in sepsis by contributing to endothelial damage and organ failure.77 Increased histone levels correlated with mortality in sepsis patients and inversely correlated with platelet counts.28, 82 Direct targeting of extracellular histones with histone antibodies, activated protein C, or heparin can significantly reduce mortality in different mouse models of sepsis.77 Recently, it was shown that citrullinated extracellular histone H3 levels, a significant marker of NET formation,83 are elevated in wild‐type mice compared with PAD4 knockout mice in a pneumonia‐derived sepsis model.84 In a dual‐insult mouse model of hemorrhagic shock and sepsis, deficiency of PAD4 resulted in decreased organ dysfunction and improved survival compared with wild‐type animals.29 Therefore, prevention of NET formation by inhibition of PAD4 to block histone release is regarded as an attractive strategy to prevent NET‐induced tissue damage in septic patients.
5.3. PAD4 and thrombosis
Thromboembolic conditions are currently the leading global cause of mortality.85 NETs, and by extension PAD4, are known mediators of thrombosis.86 This was established originally because cleavage of tissue factor pathway inhibitor by NET‐bound serine proteases (NE, cathepsin G) has a pro‐thrombotic effect.87 Interestingly NETs have been found in both venous and arterial thrombi, where they serve as a scaffold for platelets and red blood cells, facilitating their adhesion and aggregation.88, 89, 90 Additionally NETs bind plasma proteins including fibronectin and von Willebrand factor (VWF),25 further contributing to thrombus formation. Constituents of NETs, primarily extracellular DNA and histones, also profoundly influence coagulation.25 Levels of these NET biomarkers correlate with disease severity in thrombotic disorders including thrombotic microangiopathies and deep vein thrombosis. Extracellular DNA leads to thrombin generation in patients with sepsis,88 and in an experimental deep vein thrombosis model, was able to promote thrombosis through induction of platelet aggregation.25 Extracellular histones released from NETs increase plasma thrombin generation and thereby promote coagulation through TLR2 and TLR4 mediated activation of platelets.91 Importantly, because NETs are primarily discussed in the context of inflammation, NETs can be formed in noninflammatory conditions, such as during exhaustive exercise.92 This finding, combined with the previously mentioned pro‐coagulant properties of NETs has led researchers to view NET formation as a homeostatic process that must be tightly controlled.86 Inflammation driven activation of neutrophils, platelets, and vascular endothelial cells may promote a state of immune‐thrombosis which is most likely driving to disease severity in for example COVID‐19 patients.93
Because PAD4 is essential for NET formation, an increasing number of studies focus on the involvement of PAD4 in thrombosis. It was found that PAD4 knockout mice generate much less thrombi than wild‐type mice after induction of an inferior vena cava stenosis.94 Depletion of PAD4 significantly reduced neutrophil and platelet accumulation at the areas of vascular injury, thereby preventing venous thrombosis in a murine model of heparin‐induced thrombocytopenia.26, 95 PAD4 is a key player in the pathogenesis of thrombosis not only by triggering NET formation but also by citrullinating proteins that are involved in maintaining hemostasis. Evidence has been obtained for PAD4‐mediated citrullination of serine protease inhibitors such as antithrombin, as well as fibrinogen and the VWF‐cleaving protease ADAMTS13.70 Citrullination of serine protease inhibitors can result in activity loss, which in turn could result in dysregulation of blood coagulation.96 Citrullination of antithrombin abolished its ability to inhibit thrombin activation; however, increased levels of citrullinated antithrombin found in the plasma of RA and colorectal adenocarcinoma patients did not show association with thrombosis.97 Additionally, injection of PAD4 in vivo can induce citrullination of plasma proteins like ADAMTS13, thereby reducing the ability of ADAMTS13 to cleave VWF, resulting in the formation of prothrombotic VWF‐platelet strings.16 Based on these findings, we hypothesize that PAD4‐dependent citrullination of hemostatic proteins further modulates thrombus formation on the highly prothrombotic three‐dimensional scaffold that results from NETosis. Therefore, PAD4‐directed inhibitors provide a highly attractive strategy to combat or prevent (immune‐)thrombosis.
6. PAD4 INHIBITION
The contribution of PAD4 in the pathologies described above is a promising premise for therapeutic use of PAD4 inhibitors. Unfortunately, no physiological PAD4 inhibitors are known that could help guide inhibitor design. Furthermore, design of a PAD4 isotype‐specific inhibitor is complicated by the high structural conservation of the PAD active site among all isotypes. Inhibiting several PAD isotypes simultaneously is undesirable because each isotype is involved in separate biological pathways.7 An overview of the expression location and targets of each PAD isotype is given in Table [Link], [Link]. Nonetheless, recent years have seen substantial efforts made toward the design of therapeutically usable PAD4 inhibitors, especially by Thompson and colleagues. Both irreversible and reversible PAD4 inhibitors have been explored. Irreversible inhibitors contain a halo (commonly F‐, Cl‐) acetamidine group which covalently binds to Cys645 in the PAD4 active site (Figure 4B). The first described PAD inhibitor of this class was 2‐chloroacetamidine.98 However, since then, multiple generations that improve upon the 2‐chloroacetamine characteristics have been developed. Reversible PAD4 inhibitors are chemically more diverse, and rely on different mechanisms of PAD4 inhibition, such as guanidine‐containing compounds that interact with PAD4 active site residues (Figure 4C) and compounds that occupy both the “front door” and “back door” of the U‐shaped tunnel. This topic is extensively covered in a recent article by Nemmara and Thompson.99 The different modes of action of irreversible and reversible PAD4 inhibitors are described in Figure 4. In conclusion, it appears very difficult to design PAD inhibitors to have a high selectivity for the PAD4 isotype in combination with achieving high potency. Currently, there is no PAD4 inhibitor with the required characteristics to be used therapeutically. We anticipate that high‐throughput screening of small compounds in conjunction with in silico biomolecular modelling will have the potential to design novel classes of high‐affinity, highly specific PAD4 inhibitors that can be successfully used for treatment of patients suffering from thrombosis and/or inflammatory disorders.
FIGURE 4.
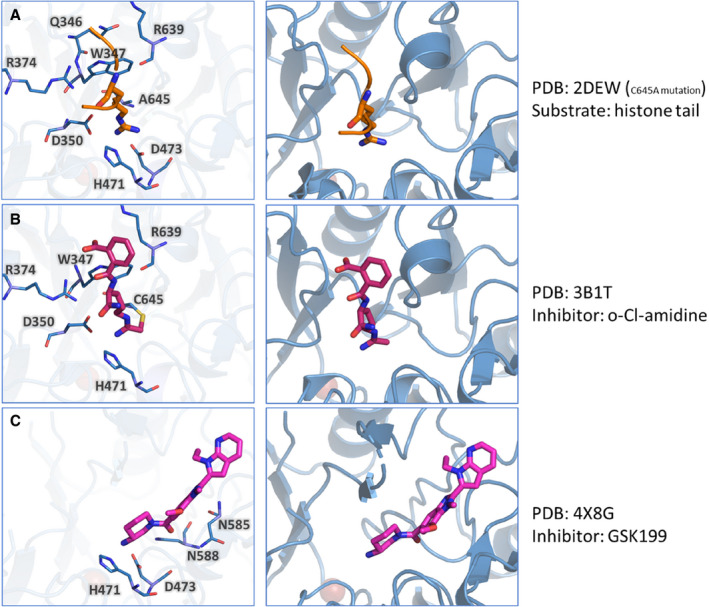
Irreversible and reversible inhibitors of PAD4. Here the crystal structure of PAD4 is shown together with a histone arginine (natural substrate, orange), the irreversible inhibitor o‐Cl‐amidine (magenta) and the reversible inhibitor GSK199 (magenta). In the left panel, the PAD4 residues that interact with the substrate or inhibitor are highlighted. In the right panel, the position of the substrate/inhibitor within the crystal structure is shown. A, PAD4 interactions with arginine from a histone tail (sequence 6‐TARKS‐10). B, PAD4 interactions with irreversible inhibitor o‐Cl‐amidine, where the covalent interaction with C645 is present. C, PAD4 interactions with reversible inhibitor GSK199. PAD, peptidyl arginine deiminase; TARKS, arginine substrate containing histone tail, represent 3‐letter abbreviation for amino acids Threonine, Alanine, Arginine, Lysine, and Serine
7. CONCLUSION AND FUTURE PERSPECTIVE
In recent years, PAD4, through its crucial role in NET formation, has emerged as a key driver of immune mediated diseases, thrombosis, and inflammatory disorders. In this review, we have summarized the state‐of‐the‐art of PAD4 structure, activation, cellular functions, and its role in disease. Our knowledge on the structure–function relationship of PAD4 is currently incomplete. A complete experimental structure of PAD4 is not yet available, and we have to partially rely on homology models as presented in this review for the design of novel classes of PAD4 inhibitors.
There are also still many unknowns in our understanding of the mechanisms that control PAD4 activity. Often, nonphysiological reducing agents like dithiothreitol are used to activate PAD4 in vitro, in combination with Ca2+ concentrations that far exceed physiological levels.32, 45 Although hypothesized physiological coactivators of PAD4 are being investigated, there is currently no consensus on how PAD4 reaches full activity within the neutrophil nuclei. Additionally, the observation that PAD4 citrullinates plasma proteins like antithrombin and ADAMTS13seems to be in direct contradiction to the fact that PAD4 requires a reducing environment to remain active in vitro. Clearly, additional research into the control of PAD4 activity in vivo is crucial to improve our understanding of this enigmatic and promiscuous enzyme.
The double‐edged sword nature of PAD4‐facilitated NET formation, however, is uncontested. The contribution of PAD4 to NET formation currently appears to be the major biological function of PAD4. NETs are capable of neutralizing pathogens, but also exacerbate inflammation and have multiple pro‐thrombotic effects. Taken together, this indicates that NET formation is a balanced process, the full inhibition of which may not be desirable, and a rebalancing of this process could be a better approach to optimally combat disease.100 Nevertheless, PAD4 qualifies as a target for pharmacological intervention. In this review we presented multiple generations of PAD4 inhibitors. However, there currently is no inhibitor that combines PAD4 isotype specificity with high potency. Especially the high conservation in active site structure between the different PAD isoforms makes specificity hard to achieve. Therefore, to design novel and specific PAD4 inhibitors, we propose that it is essential to focus on residues at the substrate binding pocket which could form specific interactions with PAD4‐selective inhibitors. Several residues qualify for such condition, such as Arg374, which exists in PAD1 and PAD4 but not in PAD2 and PAD3; Arg639 is only present in PAD4; His640 only exists in PAD4 and PAD3. Disrupting PAD4 dimerization, or blocking of the previously mentioned “back door” rather than “front door” could also be used to derive novel and selective PAD4 inhibitors.
In conclusion, although the structures, functions, and inhibitions of PAD4 have been investigated and studied during the past few decades, there are still many questions surrounding PAD4 structure and function. Improving our understanding of this remarkable enzyme will pave the way for the development of potent and specific inhibitors, with exciting potential for therapeutic use.
CONFLICT OF INTEREST
The authors declare to have no conflict of interest.
AUTHOR CONTRIBUTIONS
Xiaosong Liu and Tom Arfman designed and performed experiments, analyzed data, interpreted data, and wrote the manuscript. Kanin Wichapong contributed to data acquisition and analysis. Chris P. M. Reutelingsperger and Jan Voorberg contributed to data analyses. Gerry A. F. Nicolaes conceived and supervised the study, provided funding, and wrote the paper. All authors were involved in the proofreading of the manuscript.
Supporting information
Fig S1
Tab S1
App S1
ACKNOWLEDGMENTS
We thank China Scholarship Council for providing a scholarship to author X. L.. T. A. is supported by a grant from the Dutch Thrombosis Society (TSN2019.3)
Xiaosong Liu and Tom Arfman contributed equally.
Manuscript handled by: James Morrissey
Final decision: James Morrissey, 22‐Mar‐2021
REFERENCES
- 1.Mann M, Jensen ON. Proteomic analysis of post‐translational modifications. Nat Biotechnol. 2003;21:255‐261. [DOI] [PubMed] [Google Scholar]
- 2.Knuckley B, Bhatia M, Thompson PR. Protein arginine deiminase 4: evidence for a reverse protonation mechanism. Biochemistry. 2007;46:6578‐6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Venrooij WJ, Pruijn GJ. Citrullination: a small change for a protein with great consequences for rheumatoid arthritis. Arthritis Res. 2000;2:249‐251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ordóñez A, Martínez‐Martínez I, Corrales FJ, et al. Effect of citrullination on the function and conformation of antithrombin. FEBS J. 2009;276:6763‐6772. [DOI] [PubMed] [Google Scholar]
- 5.Tarcsa E, Marekov LN, Mei G, Melino G, Lee S‐C, Steinert PM. Protein unfolding by peptidylarginine deiminase. J Biol Chem. 1996;271:30709‐30716. [DOI] [PubMed] [Google Scholar]
- 6.Lundberg K, Nijenhuis S, Vossenaar ER, et al. Citrullinated proteins have increased immunogenicity and arthritogenicity and their presence in arthritic joints correlates with disease severity. Arthritis Res Ther. 2005;7:R458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vossenaar ER, Zendman AJ, van Venrooij WJ, Pruijn GJ. PAD, a growing family of citrullinating enzymes: genes, features and involvement in disease. BioEssays. 2003;25:1106‐1118. [DOI] [PubMed] [Google Scholar]
- 8.Nakashima K, Hagiwara T, Ishigami A, et al. Molecular characterization of peptidylarginine deiminase in HL‐60 cells induced by retinoic acid and 1alpha,25‐dihydroxyvitamin D(3). J Biol Chem. 1999;274:27786‐27792. [DOI] [PubMed] [Google Scholar]
- 9.Nakashima K, Hagiwara T, Yamada M. Nuclear localization of peptidylarginine deiminase V and histone deimination in granulocytes. J Biol Chem. 2002;277:49562‐49568. [DOI] [PubMed] [Google Scholar]
- 10.Chang X, Han J. Expression of peptidylarginine deiminase type 4 (PAD4) in various tumors. Mol Carcinog. 2006;45:183‐196. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Li M, Stadler S, et al. Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation. J Cell Biol. 2009;184:205‐213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stadler SC, Vincent CT, Fedorov VD, et al. Dysregulation of PAD4‐mediated citrullination of nuclear GSK3β activates TGF‐β signaling and induces epithelialto‐mesenchymal transition in breast cancer cells. Proc Natl Acad Sci USA. 2013;110:11851‐11856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee YH, Coonrod SA, Kraus WL, Jelinek MA, Stallcup MR. Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proc Natl Acad Sci USA. 2005;102:3611‐3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo Q, Fast W. Citrullination of Inhibitor of Growth 4 (ING4) by Peptidylarginine Deminase 4 (PAD4) disrupts the interaction between ING4 and p53. J Biol Chem. 2011;286:17069‐17078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo Q, Bedford MT, Fast W. Discovery of peptidylarginine deiminase‐4 substrates by protein array: antagonistic citrullination and methylation of human ribosomal protein S2. Mol BioSyst. 2011;7:2286‐2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sorvillo N, Mizurini DM, Coxon C, et al. Plasma peptidylarginine deiminase IV promotes VWF‐platelet string formation and accelerates thrombosis after vessel injury. Circ Res. 2019;125:507‐519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanikawa C, Ueda K, Suzuki A, et al. Citrullination of RGG motifs in FET proteins by PAD4 regulates protein aggregation and ALS susceptibility. Cell Rep. 2018;22:1473‐1483. [DOI] [PubMed] [Google Scholar]
- 18.Rohrbach AS, Slade DJ, Thompson PR, Mowen KA. Activation of PAD4 in NET formation. Front Immunol. 2012;3:360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arita K, Shimizu T, Hashimoto H, Hidaka Y, Yamada M, Sato M. Structural basis for histone N‐terminal recognition by human peptidylarginine deiminase 4. Proc Natl Acad Sci USA. 2006;103(14):5291‐5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brinkmann V. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532‐1535. [DOI] [PubMed] [Google Scholar]
- 21.Euler M, Hoffmann MH. The double‐edged role of neutrophil extracellular traps in inflammation. Biochem Soc Trans. 2019;47:1921‐1930. [DOI] [PubMed] [Google Scholar]
- 22.Kaplan MJ, Radic M. Neutrophil extracellular traps: double‐edged swords of innate immunity. J Immunol. 2012;189:2689‐2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konig MF, Andrade F. A critical reappraisal of neutrophil extracellular traps and NETosis mimics based on differential requirements for protein citrullination. Front Immunol. 2016;7:461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kenny EF, Herzig A, Kruger R, et al. Diverse stimuli engage different neutrophil extracellular trap pathways. Elife. 2017;6:e24437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fuchs TA, Brill A, Duerschmied D, et al. Extracellular DNA traps promote thrombosis. Proc Natl Acad Sci USA. 2010;107:15880‐15885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perdomo J, Leung HHL, Ahmadi Z, et al. Neutrophil activation and NETosis are the major drivers of thrombosis in heparin‐induced thrombocytopenia. Nat Commun. 2019;10:1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki A, Kochi Y, Shoda H, et al. Decreased severity of experimental autoimmune arthritis in peptidylarginine deiminase type 4 knockout mice. BMC Musculoskelet Disord. 2016;17:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yokoyama Y, Ito T, Yasuda T, et al. Circulating histone H3 levels in septic patients are associated with coagulopathy, multiple organ failure, and death: a single‐center observational study. Thromb J. 2019;17:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biron BM, Chung CS, Chen Y, et al. PAD4 deficiency leads to decreased organ dysfunction and improved survival in a dual insult model of hemorrhagic shock and sepsis. J Immunol. 2018;200:1817‐1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arita K, Hashimoto H, Shimizu T, Nakashima K, Yamada M, Sato M. Structural basis for Ca2+‐induced activation of human PAD4. Nat Struct Mol Biol. 2004;11:777‐783. [DOI] [PubMed] [Google Scholar]
- 31.Barclay AN. Ig‐like domains: evolution from simple interaction molecules to sophisticated antigen recognition. Proc Natl Acad Sci USA. 1999;96:14672‐14674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y‐L, Lee C‐Y, Huang Y‐N, Chen H‐Y, Liu G‐Y, Hung H‐C. Probing the roles of calcium‐binding sites during the folding of human peptidylarginine deiminase 4. Sci Rep. 2017;7:2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krieger E, Vriend G. YASARA view ‐ molecular graphics for all devices ‐ from smartphones to workstations. Bioinformatics (Oxford, England). 2014;30:2981‐2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu YL, Chiang YH, Liu GY, Hung HC. Functional role of dimerization of human peptidylarginine deiminase 4 (PAD4). PLoS ONE. 2011;6:e21314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teo CY, Shave S, Chor AL, et al. Discovery of a new class of inhibitors for the protein arginine deiminase type 4 (PAD4) by structure‐based virtual screening. BMC Bioinformatics. 2012;13(Suppl 17):S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kingsley LJ, Lill MA. Substrate tunnels in enzymes: structure‐function relationships and computational methodology. Proteins. 2015;83:599‐611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holmes CL, Shim D, Kernien J, Johnson CJ, Nett JE, Shelef MA. Insight into neutrophil extracellular traps through systematic evaluation of citrullination and peptidylarginine deiminases. J Immunol Res. 2019;2019:1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robertson WG, Marshall RW, Bowers GN. Ionized calcium in body fluids. Crit Rev Clin Lab Sci. 1981;15:85‐125. [DOI] [PubMed] [Google Scholar]
- 39.Darrah E, Giles JT, Ols ML, Bull HG, Andrade F, Rosen A. Erosive rheumatoid arthritis is associated with antibodies that activate PAD4 by increasing calcium sensitivity. Sci Transl Med. 2013;5:186ra65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou Y, Mittereder N, Sims GP. Perspective on protein arginine deiminase activity—Bicarbonate is a pH‐independent regulator of citrullination. Front Immunol. 2018;9:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Connell E. Tietz textbook of clinical chemistry and molecular diagnostics (5th edn). Ann Clin Biochem Int J Lab Med. 2012;49(6):615. [Google Scholar]
- 42.Terakawa H, Takahara H, Sugawara K. Three types of mouse peptidylarginine deiminase: characterization and tissue distribution. J Biochem. 1991;110:661‐666. [DOI] [PubMed] [Google Scholar]
- 43.Damgaard D, Bjørn ME, Jensen PØ, Nielsen CH. Reactive oxygen species inhibit catalytic activity of peptidylarginine deiminase. J Enzyme Inhib Med Chem. 2017;32:1203‐1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Damgaard D, Bjorn ME, Steffensen MA, Pruijn GJ, Nielsen CH. Reduced glutathione as a physiological co‐activator in the activation of peptidylarginine deiminase. Arthritis Res Ther. 2016;18:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagar M, Tilvawala R, Thompson PR. Thioredoxin modulates protein arginine deiminase 4 (PAD4)‐catalyzed citrullination. Front Immunol. 2019;10:244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Slack JL, Jones LE, Bhatia MM, Thompson PR. Autodeimination of protein arginine deiminase 4 alters protein‐protein interactions but not activity. Biochemistry. 2011;50:3997‐4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu X, Wichapong K, Lamers B, Reutelingsperger CPM, Nicolaes GAF. Autocitrullination of PAD4 does not alter its enzymatic activity: in vitro and in silico studies. Int J Biochem Cell Biol. 2021;134:105938. [DOI] [PubMed] [Google Scholar]
- 48.Nathan C. Neutrophils and immunity: challenges and opportunities. Nat Rev Immunol. 2006;6:173‐182. [DOI] [PubMed] [Google Scholar]
- 49.Segal AW. How neutrophils kill microbes. Annu Rev Immunol. 2005;23:197‐223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li P, Li M, Lindberg MR, Kennett MJ, Xiong N, Wang Y. PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. J Exp Med. 2010;207:1853‐1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thiama HR, Wong SL, Qiu R, et al. NETosis proceeds by cytoskeleton and endomembrane disassembly and PAD4‐mediated chromatin decondensation and nuclear envelope rupture. Proc Natl Acad Sci USA. 2020;117:7326‐7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fuchs TA, Abed U, Goosmann C, et al. Novel cell death program leads to neutrophil extracellular traps. J Cell Biol. 2007;176:231‐241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hirschfeld J, White PC, Milward MR, Cooper PR, Chapple ILC. Modulation of neutrophil extracellular trap and reactive oxygen species release by periodontal bacteria. Infect Immun. 2017;85:e00297‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tatsiy O, McDonald PP. Physiological Stimuli induce PAD4‐dependent, ROS‐independent NETosis, with early and late events controlled by discrete signaling pathways. Front Immunol. 2018;9:2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leshner M, Wang S, Lewis C, et al. PAD4 mediated histone hypercitrullination induces heterochromatin decondensation and chromatin unfolding to form neutrophil extracellular trap‐like structures. Front Immunol. 2012;3:307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neeli I, Khan SN, Radic M. Histone deimination as a response to inflammatory stimuli in neutrophils. J Immunol. 2008;180:1895‐1902. [DOI] [PubMed] [Google Scholar]
- 57.Saha P, Yeoh BS, Xiao X, et al. PAD4‐dependent NETs generation are indispensable for intestinal clearance of Citrobacter rodentium. Mucosal Immunol. 2019;12:761‐771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lewis HD, Liddle J, Coote JE, et al. Inhibition of PAD4 activity is sufficient to disrupt mouse and human NET formation. Nat Chem Biol. 2015;11:189‐191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leffler J, Martin M, Gullstrand B, et al. Neutrophil extracellular traps that are not degraded in systemic lupus erythematosus activate complement exacerbating the disease. J Immunol. 2012;188:3522‐3531. [DOI] [PubMed] [Google Scholar]
- 60.Tillack K, Breiden P, Martin R, Sospedra M. T lymphocyte priming by neutrophil extracellular traps links innate and adaptive immune responses. J Immunol. 2012;188:3150‐3159. [DOI] [PubMed] [Google Scholar]
- 61.Kumar SVR, Kulkarni OP, Mulay SR, et al. Neutrophil extracellular trap‐related extracellular histones cause vascular necrosis in severe GN. J Am Soc Nephrol. 2015;26:2399‐2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fuchs TA, Bhandari AA, Wagner DD. Histones induce rapid and profound thrombocytopenia in mice. Blood. 2011;118:3708‐3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thalin C, Hisada Y, Lundstrom S, Mackman N, Wallen H. Neutrophil extracellular traps: villains and targets in arterial, venous, and cancer‐associated thrombosis. Arterioscler Thromb Vasc Biol. 2019;39:1724‐1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fousert E, Toes R, Desai J. Neutrophil extracellular traps (NETs) take the central stage in driving autoimmune responses. Cells. 2020;9:915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Silman AJ, Pearson JE. Epidemiology and genetics of rheumatoid arthritis. Arthritis Res. 2002;4:S265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fujii K, Tsuji M, Tajima M. Rheumatoid arthritis: a synovial disease? Ann Rheum Dis. 1999;58:727‐730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fisher BA, Venables PJ. Inhibiting citrullination in rheumatoid arthritis: taking fuel from the fire. Arthritis Res Ther. 2012;14:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Suzuki A, Yamada R, Chang X, et al. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat Genet. 2003;34:395‐402. [DOI] [PubMed] [Google Scholar]
- 69.Hua J, Huang W. Peptidylarginine deiminase 4–104C/T polymorphism and risk of rheumatoid arthritis: A pooled analysis based on different populations. PLoS ONE. 2018;13:e0193674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tilvawala R, Nguyen SH, Maurais AJ, et al. The rheumatoid arthritis‐associated citrullinome. Cell Chem Biol. 2018;25:691.e6‐704.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kuhn KA. Antibodies against citrullinated proteins enhance tissue injury in experimental autoimmune arthritis. J Clin Invest. 2006;116:961‐973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Seri Y, Shoda H, Suzuki A, et al. Peptidylarginine deiminase type 4 deficiency reduced arthritis severity in a glucose‐6‐phosphate isomerase‐induced arthritis model. Sci Rep. 2015;5:13041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vincent C, Simon M, Sebbag M, et al. Immunoblotting detection of autoantibodies to human epidermis filaggrin: a new diagnostic test for rheumatoid arthritis. J Rheumatol. 1998;25:838‐846. [PubMed] [Google Scholar]
- 74.Mewar D, Coote A, Moore DJ, et al. Independent associations of anti‐cyclic citrullinated peptide antibodies and rheumatoid factor with radiographic severity of rheumatoid arthritis. Arthritis Res Ther. 2006;8:R128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van Venrooij WJ, Zendman AJ, Pruijn GJ. Autoantibodies to citrullinated antigens in (early) rheumatoid arthritis. Autoimmun Rev. 2006;6:37‐41. [DOI] [PubMed] [Google Scholar]
- 76.Rudd KE, Johnson SC, Agesa KM, et al. Global, regional, and national sepsis incidence and mortality, 1990‐2017: analysis for the global burden of disease study. Lancet. 2020;395:200‐211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu J, Zhang X, Pelayo R, et al. Extracellular histones are major mediators of death in sepsis. Nat Med. 2009;15:1318‐1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dwivedi DJ, Toltl LJ, Swystun LL, et al. Prognostic utility and characterization of cell‐free DNA in patients with severe sepsis. Crit Care. 2012;16:R151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rhodes A, Wort SJ, Thomas H, Collinson P, Bennett ED. Plasma DNA concentration as a predictor of mortality and sepsis in critically ill patients. Crit Care. 2006;10:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Clark SR, Ma AC, Tavener SA, et al. Platelet TLR4 activates neutrophil extracellular traps to ensnare bacteria in septic blood. Nat Med. 2007;13:463‐469. [DOI] [PubMed] [Google Scholar]
- 81.Meng W, Paunel‐Görgülü A, Flohé S, et al. Depletion of neutrophil extracellular traps in vivo results in hypersusceptibility to polymicrobial sepsis in mice. Crit Care. 2012;16:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cheng L, Yan J, Han S, et al. Comparative efficacy of vasoactive medications in patients with septic shock: a network meta‐analysis of randomized controlled trials. Crit Care. 2019;23:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wildhagen KCAA, Wiewel MA, Schultz MJ, et al. Extracellular histone H3 levels are inversely correlated with antithrombin levels and platelet counts and are associated with mortality in sepsis patients. Thromb Res. 2015;136:542‐547. [DOI] [PubMed] [Google Scholar]
- 84.Claushuis TAM, van der Donk LEH, Luitse AL, et al. Role of peptidylarginine deiminase 4 in neutrophil extracellular trap formation and host defense during Klebsiella pneumoniae‐induced pneumonia‐derived sepsis. J Immunol. 2018;201:1241‐1252. [DOI] [PubMed] [Google Scholar]
- 85.Wendelboe AM, Raskob GE. Global burden of thrombosis: epidemiologic aspects. Circ Res. 2016;118:1340‐1347. [DOI] [PubMed] [Google Scholar]
- 86.Kimball AS, Obi AT, Diaz JA, Henke PK. The emerging role of NETs in venous thrombosis and immunothrombosis. Front Immunol. 2016;7:236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Massberg S, Grahl L, Von Bruehl ML, et al. Reciprocal coupling of coagulation and innate immunity via neutrophil serine proteases. Nat Med. 2010;16:887‐896. [DOI] [PubMed] [Google Scholar]
- 88.Gould TJ, Vu TT, Swystun LL, et al. Neutrophil extracellular traps promote thrombin generation through platelet‐dependent and platelet‐independent mechanisms. Arterioscler Thromb Vasc Biol. 2014;34:1977‐1984. [DOI] [PubMed] [Google Scholar]
- 89.Laridan E, Martinod K, De Meyer S. Neutrophil extracellular traps in arterial and venous thrombosis. Semin Thromb Hemost. 2019;45:86‐93. [DOI] [PubMed] [Google Scholar]
- 90.McDonald B, Davis RP, Kim SJ, et al. Platelets and neutrophil extracellular traps collaborate to promote intravascular coagulation during sepsis in mice. Blood. 2017;129:1357‐1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Semeraro F, Ammollo CT, Morrissey JH, et al. Extracellular histones promote thrombin generation through platelet‐dependent mechanisms: involvement of platelet TLR2 and TLR4. Blood. 2011;118:1952‐1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Beiter T, Fragasso A, Hudemann J, et al. Neutrophils release extracellular DNA traps in response to exercise. J Appl Physiol. 2014;117:325‐333. [DOI] [PubMed] [Google Scholar]
- 93.Morris G, Bortolasci CC, Puri BK, et al. Preventing the development of severe COVID‐19 by modifying immunothrombosis. Life Sci. 2021;264:118617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Martinod K, Demers M, Fuchs TA, et al. Neutrophil histone modification by peptidylarginine deiminase 4 is critical for deep vein thrombosis in mice. Proc Natl Acad Sci USA. 2013;110:8674‐8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gollomp K, Hayes V, Johnston I, et al. Inhibition of neutrophil extracellular trap release through the disruption of PAD4 leads to decreased neutrophil and platelet accumulation at sites of venular vascular injury in a murine model of heparin‐induced thrombocytopenia. Blood. 2017;130:2268. [Google Scholar]
- 96.Thompson PR. Citrullination inhibits SERPIN activity. FASEB J. 2018;32:104.3‐3. [Google Scholar]
- 97.Ordóñez A, Yélamos J, Pedersen S, et al. Increased levels of citrullinated antithrombin in plasma of patients with rheumatoid arthritis and colorectal adenocarcinoma determined by a newly developed ELISA using a specific monoclonal antibody. Thromb Haemost. 2010;104:1143‐1149. [DOI] [PubMed] [Google Scholar]
- 98.Stone EM, Schaller TH, Bianchi H, Person MD, Fast W. Inactivation of two diverse enzymes in the amidinotransferase superfamily by 2‐chloroacetamidine: Dimethylargininase and peptidylarginine deiminase. Biochemistry. 2005;44:13744‐13752. [DOI] [PubMed] [Google Scholar]
- 99.Nemmara VV, Thompson PR. Development of activity‐based proteomic probes for protein citrullination. Curr Top Microbiol Immunol. 2019;420:233‐251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cheng OZ, Palaniyar N. NET balancing: a problem in inflammatory lung diseases. Front Immunol. 2013;4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Tab S1
App S1


