To the Editor,
Anaphylaxis is a life‐threatening allergic disorder1 for which there is a need for improved diagnostic techniques and a deeper understanding of the molecular mechanisms involved.2 Extracellular vesicles (EVs) play a key role in cellular communication, offering new possibilities with which to analyze patient particularities.3
We hypothesized that anaphylaxis‐derived EVs could provide potential markers of anaphylaxis due to their participation in the underlying molecular mechanisms. Vesicles were purified from 86 plasma samples (collected from 43 patients) during the acute phase of anaphylaxis (AnEVs) and baseline (BEVs). Clinical description, demographic characteristics, and their experimental utilities are displayed in Supplementary Table S1.
AnEVs and BEVs were isolated and characterized. Electron microscopy and nanoparticle tracking analysis (NTA) showed particles that were consistent with the reported size range for EVs (100–250 nm) (Figure 1A–B). These vesicles exhibited markers previously related to EVs (Figure 1C). Their proteome is increasingly considered a source of biomarkers for various disease states.3
FIGURE 1.
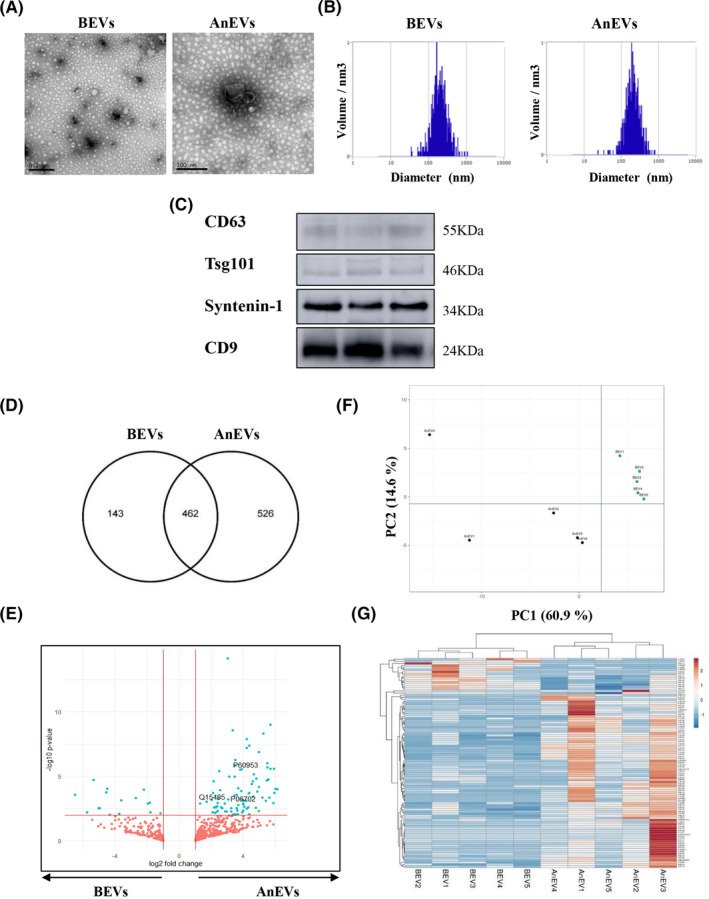
Characterization, proteomic profiling, and differential analysis of AnEVs and BEVs. (A) Transmission electron microscopy images from representative circulating BEVs and AnEVs. Scale bar: 200 nm. Images depict representative EVs from both samples. (B) Seven different EV preparations from each experimental condition were analyzed by NTA. Representative NTA histograms showed average particle sizes of 200 nm for BEVs and AnEvs. (C) AnEVs were characterized by Western blot. Panels show immunoblots from three representative patients. Bona fide EV markers, such as CD63, TSG101, Syntenin‐1, and CD9 were detected. (D) Venn diagram of brute data obtained by mass spectrometry‐based quantitative proteomics representing the intersection between detected proteins in BEVs and AnEVs. (E) Volcano plot showing all proteins identified. Statistically significant differences (p > .05) in the AnEVs (right side) and in the BEVs (left side) appear in blue. Access number (UniProt) for proteins of interest (CDC42, Ficolin‐2, S100A9) are shown. Principal component analysis (F) and unsupervised hierarchical clustering (G) of plasma‐derived EVs
To investigate anaphylaxis‐derived EVs, we analyzed the protein pattern profiles in 10 acute and baseline paired samples using mass spectrometry. The label‐free quantification method identified 1206 proteins, and by comparing brute values of AnEVs and BEVs, 526 proteins were exclusively detected in AnEVs (Figure 1D). The whole amount of proteins identified was depicted through a volcano plot (Figure 1E). Principal component analysis (PCA) was used to gather biological replicates, and separation of BEV and AnEVs groups was performed by PC1 (Figure 1F). Statistical analysis revealed 99 differentially expressed EV proteins, of which 83 were increased in the acute phase, thus suggesting their potential as candidate biomarkers (Supplementary Table S2). The differential clustering of these proteins is shown in Figure 1G. Only a few proteins found in this panel have been previously related to human anaphylaxis, and 2% had not been included in comprehensive EV resources (Vesiclepedia).
Three proteins were selected to confirm our results in a larger cohort. Specific analysis of CDC42, Ficolin‐2, and S100A9 demonstrated an increased abundance in AnEVs, thus supporting an anaphylactic EV‐protein signature (Figure 2A). CDC42 is one of the most enriched proteins in AnEVs and its role in cytoskeletal reorganization is essential to EV secretion. Ficolin‐2 is closely related to immune processes participating in signaling pathways which release inflammatory mediators such as IFNγ, IL‐6, TNFα, and nitric oxide. S100‐family proteins are activated under cell stress and contribute regulating inflammatory processes and endothelium activation. From the studies performed here, S100A7, S100A8, and S100A9 were found to be increased in AnEVs, supporting a possible role for alarmins in this event.4, 5
FIGURE 2.
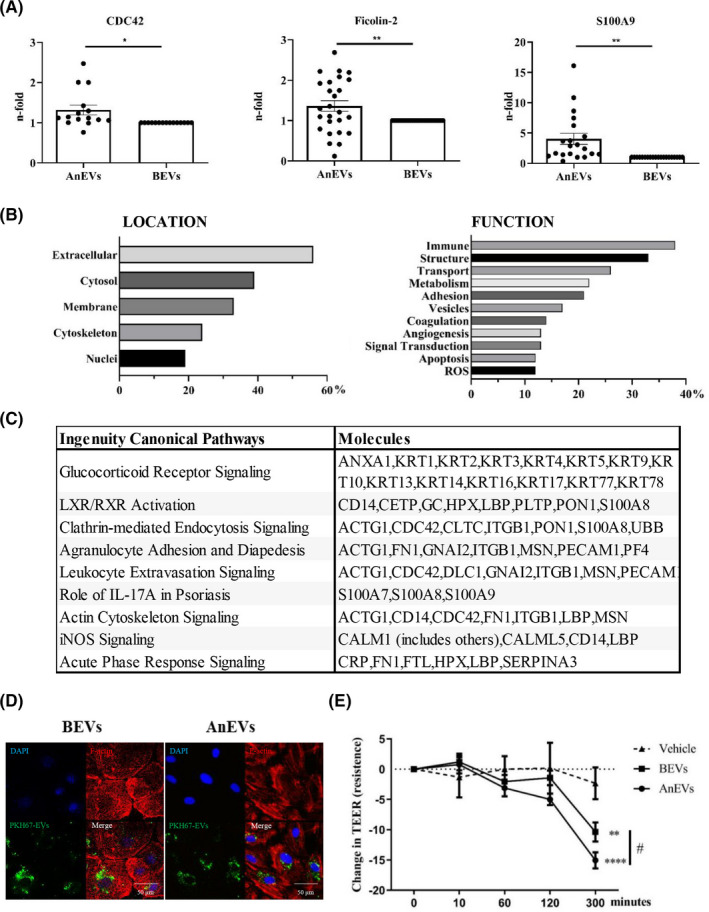
Functional protein association networks in AnEVs. (A) CDC42, S100A9, and Ficolin‐2 are increased in AnEVs. The graphics represent the abundance protein levels expressed as the quantified ratio (AnEVs/BEVs and BEVs/BEVs) of a larger group of anaphylactic plasma paired samples. CDC42 (*p = .0137, n = 15, MW=21 KDa), S100A9 (**p = .0017, n = 20, MW = 14 KDa), Ficolin‐2 (**p = .0092, n = 26, MW = 34 KDa). (B) Classification of the main cellular localization and function of the protein panel based on the UniProt database. (C) Top Canonical Pathways obtained by IPA and symbols of the molecules. (D–E) HMVEC‐Ls were incubated with 100 µg/ml of purified BEVs and AnEVs. The images reveal PKH67‐EVs around the nuclei (DAPI) and cytoskeletal fibers (F‐actin) from a representative paired EVs patient sample from 4 performed (D). (E) The graphic shows the change in TEER measurements after the addition of BEVs and AnEVs (n = 16 patients). Two‐way ANOVA followed by the Bonferroni test was performed: **p = .0033, ****p ≤ .0001 versus Vehicle (EBM + PBS); #p = .0285 versus BEVs)
A coordinated function displayed in the contents of anaphylaxis‐derived EVs may provide information about the molecular bases of the reaction. Therefore, a comprehensive analysis based on the Uniprot database revealed a group of major immune proteins participating in the cellular structure (Figure 2B). In addition, in silico Ingenuity pathway analysis (IPA) showed around 25% of the protein signature participating in leukocyte trans‐endothelial migration and cell degranulation (Figure 2C and Supplementary Table S3).
Another process closely associated with anaphylactic reactions is vascular permeability (v.p.).6 To evaluate the impact of EVs in v.p., we incubated them together with human microvascular endothelial cells‐lung (HMVEC‐Ls), revealing no differences in cellular uptake and showing a perinuclear localization (Figure 2D). However, AnEVs uptaked‐ECs exhibited morphological contractile changes. In agreement, v.p. assays in 16 patients demonstrated that AnEVs induced a greater increase in the cellular permeability compared to BEVs (Figure 2E and Supplementary Figure S1).
Use of EVs as disease biomarkers is a matter of intense research3 that could improve knowledge and management of anaphylaxis. Using a challenging sample dataset, we identified an anaphylaxis EV signature for the first time and performing pilot biomarker studies. Functional involvement of EVs in anaphylaxis and v.p. needs further clarification. Future studies are necessary to determine their possible diagnostic utility. Though exploratory, our findings suggest the clinical potential of EVs, possibly leading to new therapeutic directions. Proteomic profiling of these plasma‐derived AnEVs is a great resource for the allergy community.
CONFLICT OF INTEREST
There are no conflicts of interest.
Supporting information
Supplementary Material
ACKNOWLEDGMENTS
We thank Maria del Mar Gonzalez García‐Parreño for technical support and Oliver Shaw for editing the manuscript and matters concerning English use.
Benito‐Martin Alberto and Esteban Vanesa equal contribution.
Funding information
This research was supported by the Spanish Council Ministry of Science and Innovation (Ramón y Cajal Program RyC‐12880‐2013), grants from the Instituto de Salud Carlos III (PI16/00888, PI16/01334 and PI18/00348) and FEDER Thematic Networks and Cooperative Research Centers RETICS ARADyAL (RD16/0006/0003, RD16/0006/0013, RD16/0006/0033). This work was also supported by the SEAIC (19_A08) and Alfonso X el Sabio University Foundations. ENB is the beneficiary of grant funding from the Community of Madrid included in the project FOOD‐AL (CM_P2018/BAAA‐4574). Proteomic analysis was performed in the Proteomics Unit of UCM, a member of ProteoRed and is supported by Grant PRB3 (IPT17/0019‐ISCIII‐SGEFI/ERDF).
Contributor Information
Alberto Benito‐Martin, Email: albertobenitomartin@gmail.com.
Vanesa Esteban, Email: vesteban@fjd.es.
REFERENCES
- 1.Reber LL, Hernandez JD, Galli SJ. The pathophysiology of anaphylaxis. J Allergy Clin Immunol 2017;140:335‐348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castells M. Diagnosis and management of anaphylaxis in precision medicine. J Allergy Clin. 2017;140(2):321‐333. [DOI] [PubMed] [Google Scholar]
- 3.Hoshino A, Kim HS, Bojmar L, et al. Extracellular vesicle and particle biomarkers define multiple human cancers. Cell 2020;182(4):1044‐1061.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jönsson F, de Chaisemartin L, Granger V, et al. An IgG‐induced neutrophil activation pathway contributes to human drug‐induced anaphylaxis. Sci Transl Med 2019;11(500):eaat1479. [DOI] [PubMed] [Google Scholar]
- 5.Francis A, Bosio E, Stone SF, et al. Markers involved in innate immunity and neutrophil activation are elevated during acute human anaphylaxis: validation of a microarray study. J Innate Immun 2019;11:63‐73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakamura T, Murata T. Regulation of vascular permeability in anaphylaxis. Br J Pharmacol 2018;175:2538‐2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


