Abstract
Mesenchymal stem cells (MSCs) are the likely precursors of multiple lines of mesenchymal cells. The existence of bona fide MSCs with self‐renewal capacity and differentiation potential into all mesenchymal lineages, however, has been unclear because of the lack of MSC‐specific marker(s) that are not expressed by the terminally differentiated progeny. Meflin, a glycosylphosphatidylinositol‐anchored protein, is an MSC marker candidate that is specifically expressed in rare stromal cells in all tissues. Our previous report showed that Meflin expression becomes down‐regulated in bone marrow‐derived MSCs cultured on plastic, making it difficult to examine the self‐renewal and differentiation of Meflin‐positive cells at the single‐cell level. Here, we traced the lineage of Meflin‐positive cells in postnatal and adult mice, showing that those cells differentiated into white and brown adipocytes, osteocytes, chondrocytes and skeletal myocytes. Interestingly, cells derived from Meflin‐positive cells formed clusters of differentiated cells, implying the in situ proliferation of Meflin‐positive cells or their lineage‐committed progenitors. These results, taken together with previous findings that Meflin expression in cultured MSCs was lost upon their multilineage differentiation, suggest that Meflin is a useful potential marker to localize MSCs and/or their immature progenitors in multiple tissues.
Keywords: immunoglobulin superfamily containing leucine‐rich repeat, Islr, Meflin, Mesenchymal stem cells, Mesenchymal stromal cells, satellite cell
Contribution of Meflin‐positive cells to the development of multiple mesenchymal tissues. Meflin is a marker of MSCs/SSCs in the cartilage and bone, ADSCs in the adipose tissue, Pax7‐positive satellite cells and PDGFRα‐positive FAPs in the skeletal muscle.
![]()
1. INTRODUCTION
Mesenchymal stem cells (MSCs) are thought to be tissue stem cells with the capacity for both self‐renewal and multilineage differentiation. They are found in many tissues such as bone marrow (BM), adipose tissues, umbilical cord and virtually all tissues and organs of the body (Andrzejewska et al., 2019; Caplan, 1991; Pittenger, 1999). The nature and precise in vivo localization of MSCs, however, have not been explicitly determined due to the lack of specific marker(s). Therefore, the definition of MSCs has differed, depending on research contexts. In research fields that are focused on the medical use of cultured and ex vivo‐expanded MSCs, they are defined by their ability to adhere to plastic in culture and to express CD105, CD73 and CD90 cell surface markers and to lack the expression of CD14, CD31, CD34 and CD45 (Dominici et al., 2006). They can be induced to differentiate into osteoblasts, chondroblasts, adipose cells, fibroblasts and skeletal muscle cells in vitro (Beresford et al., 1992; Pittenger, 1999; Wakitani et al., 1995; Woodbury et al., 2000). Although this definition has been used for many years by the International Society for Cellular Therapy (ISCT), it has been clear that this population contains not only bona fide MSCs but also heterogeneous populations of progenitors committed to each of the mesenchymal lineages (Dominici et al., 2006; Horwitz et al., 2005). Another concern in this field has been that the expressed markers are not specific to MSCs and are expressed by many types of terminally differentiated cells derived from multiple germ layers.
Not surprisingly, there have been debates regarding the best method for MSC isolation from multiple types of tissues, and many markers for prospective isolation of MSCs have been identified (Lv et al., 2014; Nicodemou & Danisovic, 2017). Evidence showed that platelet‐derived growth factor receptor‐α (PDGFRα)/Sca‐1 double‐positive cells (PαS cells) in mouse bone marrow (BM) represent the most primitive BM‐MSCs with the highest clonogenic potential (Morikawa et al., 2009). In contrast, sorting CD271+/CD90+ (Thy‐1) cells is an effective method to isolate human MSCs from BM and other tissues (Bühring et al., 2007; Mabuchi et al., 2013). There are more isolation methods using other MSC markers such as Stro‐1, CD106 and CD146 (Mabuchi et al., 2013; Nicodemou & Danisovic, 2017; Sacchetti et al., 2007; Simmons & Torok‐Storb, 1991). However, none of these markers are specific to MSCs, and there is no single marker that is available to isolate MSCs or localize them across multiple organs.
The in vivo differentiation capacity of MSCs has been shown by fate mapping using transgenic or knock‐in mice expressing Cre recombinase or its tamoxifen‐inducible form (Cre‐ERT2) under the control of promoters of genes that are preferentially expressed in MSCs or their progenitors. Most studies have focused on lineage tracing of MSCs in the bone and BM. Those analyses showed that the Leptin receptor (LepR), GLI family zinc finger 1 (Gli1), paired related homeobox 1 (Prrx1, also termed Prx1), CXC motif ligand 12 (CXCL12) and Gremlin 1 (Grem1)‐expressing cells give rise to terminally differentiated osteocytes, chondrocytes, adipocytes and fibroblasts (Greenbaum et al., 2013; Kramann et al., 2015; Omatsu et al., 2010; Worthley et al., 2015; Zhou et al., 2014). The differentiation capacities of these cells and their directions of differentiation vary by marker, suggesting that these marker genes do not uniformly label bona fide MSCs but instead progenitors that are precommitted to specific lineages. However, the number of studies that investigated the differentiation of cells positive for the above markers that exist outside of the bone has been limited. Other studies have shown that several marker genes, such as PDGFRα, Wilms tumor 1 homologue (WT1), paired box 7 (Pax7) and myogenic factor 5 (Myf5), are useful for fate mapping of tissue‐resident mesenchymal progenitors that give rise to mature adipocytes, fibroblasts and skeletal myocytes in adipose tissues and muscles (Chau et al., 2014; Halder et al., 2008; Relaix et al., 2005). However, information on the contribution of progenitors positive for those markers to osteochondrogenic and other mesenchymal cells in the bone and BM has not been provided, leaving the question unanswered whether there exists a single marker that labels MSCs that undergo multilineage differentiation across multiple tissues and organs.
Meflin is a glycosylphosphatidylinositol (GPI)‐anchored membrane protein. It is encoded by the immunoglobulin superfamily containing leucine‐rich repeat (Islr) gene and is expressed by cultured BM‐MSCs but not epithelial, endothelial, smooth muscle, peripheral neuronal or blood cells (Maeda et al., 2016). Notably, our analysis showed that Meflin expression was most enriched in PαS cells in the BM, suggesting that Meflin might be a marker of the most primitive BM‐MSCs (Maeda et al., 2016). Interestingly, Meflin expression was significantly down‐regulated or became negative when cultured BM‐MSCs were induced to differentiate into osteoblasts, chondroblasts, adipocytes or myofibroblasts in vitro (Hara et al., 2019; Maeda et al., 2016). Meflin depletion in BM‐MSCs resulted in decreased colony‐forming unit‐fibroblasts and their spontaneous commitment toward osteochondrocyte or myofibroblast lineages, which suggested a role for Meflin in maintaining BM‐MSCs in their undifferentiated state (Hara et al., 2019; Maeda et al., 2016). Histological analyses showed that Meflin was expressed in rare fibroblastoid stromal cells that were preferentially localized in perivascular areas in the BM as well as in almost all tissues and organs including the skin, adipose tissue, skeletal muscle, brain, heart, intestine and pancreas (Hara et al., 2019; Kobayashi et al., 2021; Maeda et al., 2016; Mizutani et al., 2019; Xu et al., 2020). These data implied that Meflin is a marker of MSCs that are distributed throughout the body.
A recent study from another group showed the differentiation of Meflin‐positive cells into mature white adipocytes and beige‐like adipocytes in physiological and cold stress conditions, respectively, using a transgenic mouse line that expresses inducible Cre recombinase under the Meflin promoter (Kuwano et al., 2021). Here, we conducted lineage tracing experiments using knock‐in mouse lines that express inducible or constitutive Cre recombinase under the control of the endogenous Meflin promoter and a reporter mouse that expresses tdTomato only after Cre‐mediated genomic recombination. We showed that Meflin‐positive cells, which were sparse immediately after the labeling, differentiate into clusters of mature white and brown adipocytes, chondrocytes, osteocytes and myocytes in both postnatal and adult periods. These data suggest that Meflin may be a useful potential marker to localize MSCs and/or their early progeny throughout the body in the postnatal and adult stages.
2. RESULTS
2.1. Meflin‐positive cells gave rise to white adipocytes of the inguinal fat pad in postnatal development
To perform genetic labeling of Meflin‐expressing cells and tracing their lineage in postnatal development, we generated a knock‐in mouse line expressing tamoxifen‐inducible Cre recombinase driven by the Meflin promoter (Meflin‐CreERT2) (Mizutani et al., 2019) (Figure 1A). We crossed it with a mouse line harboring a Rosa26‐LoxP‐stop‐LoxP (LSL)‐tdTomato allele to specifically label Meflin‐expressing cells by tdTomato (Meflin‐CreERT2; Rosa26‐LSL‐tdTomato, hereafter termed Meflin‐CreERT2; LSL‐tdTomato, Figure 1A). For the induction of Cre recombinase and tdTomato expression in Meflin‐expressing cells, we administered 50 µg tamoxifen (TAM) by intragastric injection (i.g.) at postnatal days 1 (P1), 2 and 3 (Figure 1B). We first prepared tissues from Meflin‐CreERT2; LSL‐tdTomato mice 1 day after the last administration of TAM (P4) to localize cells expressing Meflin at the time of the TAM treatment (hereafter termed Meflin+ cells) by tdTomato immunohistochemical (IHC) staining. We subsequently harvested tissues from the mice at P21 and P49 to trace the fate of cells derived from Meflin‐expressing cells. Thus, we examined tdTomato‐positive (tdTomato+) cells, that is, cells in the Meflin lineage that included cells that were no longer Meflin+ (Figure 1B). Throughout the study, we visualized tdTomato+ cells by IHC using anti‐tdTomato antibody on formalin‐fixed paraffin‐embedded (FFPE) sections or by fluorescence analysis of frozen tissues. The specificity of IHC for tdTomato was validated by control experiments in which we stained for tdTomato on tissue sections prepared from Meflin‐CreERT2; LSL‐tdTomato mice that did not receive TAM (Figure S1). No signals were observed in any of the tissues tested, showing the specificity of the staining. Moreover, there was no leaky Cre activity in the absence of TAM. In situ hybridization (ISH) for detection of Meflin mRNA showed that approximately 25%–30% of Meflin mRNA+ cells were labeled with tdTomato at P4 and at the age of 9 weeks, suggesting that Cre recombination efficiency was modest at least in the current experimental setup (Figure S2A,B). All the tdTomato cells were positive for Meflin mRNA, indicating that tdTomato expression faithfully marked the Meflin lineage.
FIGURE 1.
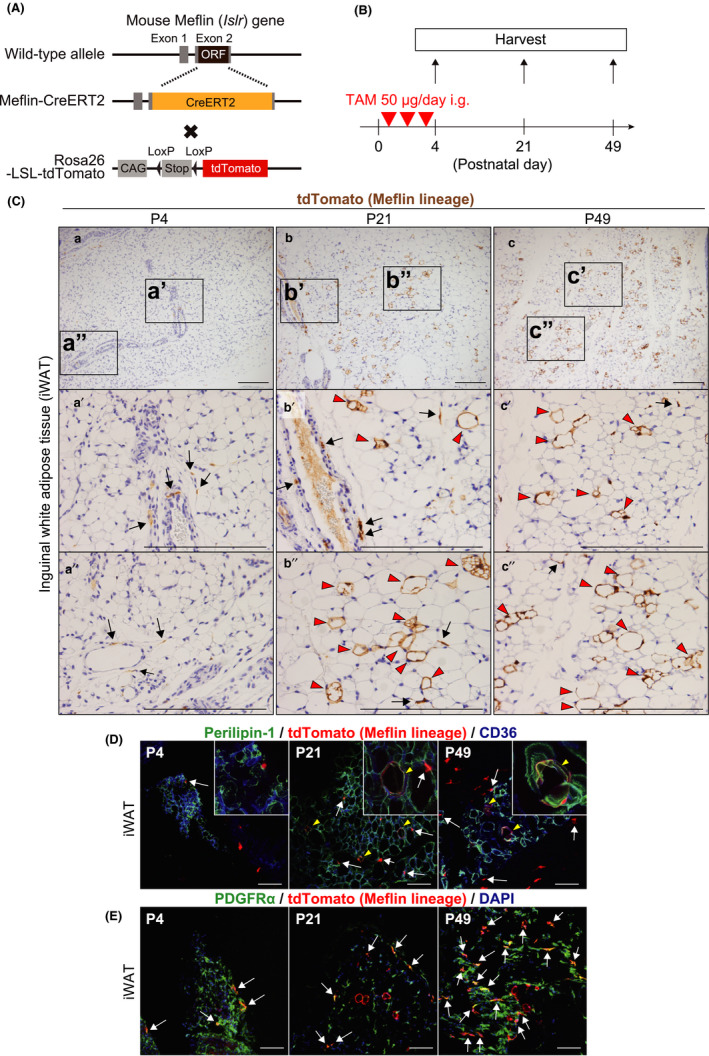
Meflin+ cells are fibroblastoid cells that differentiate into white adipocytes in the postnatal period. (A) A diagram of the generation of Meflin‐CreERT2; LSL‐tdTomato mice. ORF; open reading frame, CAG; chicken ß‐actin promoter with cytomegalovirus enhancer and Stop; stop element. (B) The experimental design for lineage tracing of Meflin+ cells in the postnatal period. The Meflin‐CreERT2; LSL‐tdTomato pups were administered 50 µg tamoxifen (TAM) at P1, P2 and P3 to label Meflin+ cells with tdTomato. (C) iWAT harvested from Meflin‐CreERT2; LSL‐tdTomato mice was stained for tdTomato by IHC. tdTomato+ cells (brown) represent Meflin+ cells at P4 (a) and their descendants (Meflin lineage cells) at P21 (b) and P49 (c). Boxed regions in a–c are magnified in lower panels (a′–c′ and a″–c″). Arrows denote tdTomato+ stromal cells with a fibroblastoid morphology. Red arrowheads indicate tdTomato+ terminally differentiated mature white adipocytes (Meflin lineage cells). Scale bars, 200 µm. (D, E) iWAT harvested from Meflin‐CreERT2; LSL‐tdTomato mice was stained for the adipocyte marker Perilipin‐1 (D) and the fibroblast marker PDGFRα (E) by IF (green). CD36 is a marker of adipocytes and endothelial cells (blue). Magnified regions are indicated in insets. Arrows indicate tdTomato+ stromal cells with a fibroblastoid morphology, whereas arrowheads show tdTomato+ mature adipocytes. Note that tdTomato+/PDGFRα+ cells were observed throughout P4, P21 and P49 in (E). Scale bars, 100 µm
We first found that Meflin+ cells were present in inguinal white adipose tissue (iWAT) at P4, the majority of which resided in the perivascular area in the interstitium (Figure 1Ca‐a″). This finding was consistent with the results of ISH analyses in the previous report (Maeda et al., 2016) and the prevailing notion that most MSCs reside in the perivascular areas of multiple organs (Crisan et al., 2008). Consistent with the previous report (Maeda et al., 2016), Meflin was not expressed by mature adipocytes filled with large unilocular lipid droplets throughout iWAT.
We next examined iWATs prepared from P21 and P49 Meflin‐CreERT2; LSL‐tdTomato mice that had been administered TAM. Our data showed that there were tdTomato+ Meflin lineage cells that resembled mature adipocytes based on their morphology (Figure 1Cb‐b″, c‐c″). Immunofluorescent (IF) staining showed that these tdTomato+ cells were positive for both Perilipin‐1, a major lipid droplet coat protein and a marker of terminally differentiated adipocytes, and CD36, a marker of adipocyte progenitors (Figure 1D). These data suggested that Meflin+ cells differentiated into cells of the adipocyte lineage during postnatal development.
Interestingly, some tdTomato+ Meflin lineage adipocytes were found in groups or clusters at P21 and P49, implying that they might originate from the same stem cells or progenitors (Figure 1Cb‐b″, c‐c″). Moreover, there were rare tdTomato+ cells with a spindle‐shaped fibroblastoid morphology in or around vessel walls or the interstitium of iWAT at P21 and P49 (Figure 1C,D, arrows). IF staining showed that these spindle cells were positive for PDGFRα, a marker of fibroblasts and MSCs, including adipose tissue‐derived stem cells (ADSCs) (Berry & Rodeheffer, 2013; Zuk et al., 2001) (Figure 1E), an intriguing finding that we will address below.
We also found that Meflin+ cells did not give rise to platelet endothelial cell adhesion molecule‐1 (PECAM1)‐positive or endomucin‐positive endothelial cells (Figure S3). MSC‐specific expression of Meflin was confirmed by the analysis of a recent dataset of single‐cell RNA sequencing (scRNA‐seq) of murine adipose tissue (Tabula Muris Consortium, 2018), where Meflin was specifically detected in MSC subsets that were enriched for PDGFRα expression (Figure S4). Taken together, these findings indicate that Meflin+ cells represent subset(s) of MSCs in WAT that have an adipogenic differentiation capacity in vivo.
2.2. Meflin+ cells gave rise to brown adipocytes during postnatal development
We next examined the localization of Meflin+ cells and their subsequent offspring in brown adipose tissue (BAT) using Meflin‐CreERT2; LSL‐tdTomato mice. BAT is composed of mature brown adipocytes that store fat in multilocular droplets and express uncoupling protein 1 (UCP‐1), a key determinant of mitochondrial thermogenesis, and adipose stem cells (Fedorenko et al., 2012; Zuk et al., 2001). Similar to the findings obtained from iWAT, Meflin+ cells were found in the interstitium of the BAT and had a spindle‐shaped fibroblastoid morphology at P4 (immediately after TAM administration) (Figure 2Aa‐a″). Meflin was not expressed by mature brown adipocytes at P4. At P21 and P49, we found that Meflin+ cells had differentiated into mature multilocular brown adipocytes (Figure 2Ab‐b″, c‐c″). This was corroborated by IF staining that showed that tdTomato+ Meflin lineage cells expressed the lipid droplet coat protein Perilipin‐1 as well as the mature brown adipocyte marker UCP‐1 (Figure 2B,C). Similar to the findings on iWAT, we found that there were some tdTomato+ Meflin lineage cells at P21 and P49 that had the same morphology as Meflin+ cells at P4 and were positive for PDGFRα (Figure 2Ab′,b″,c′,c″,D).
FIGURE 2.
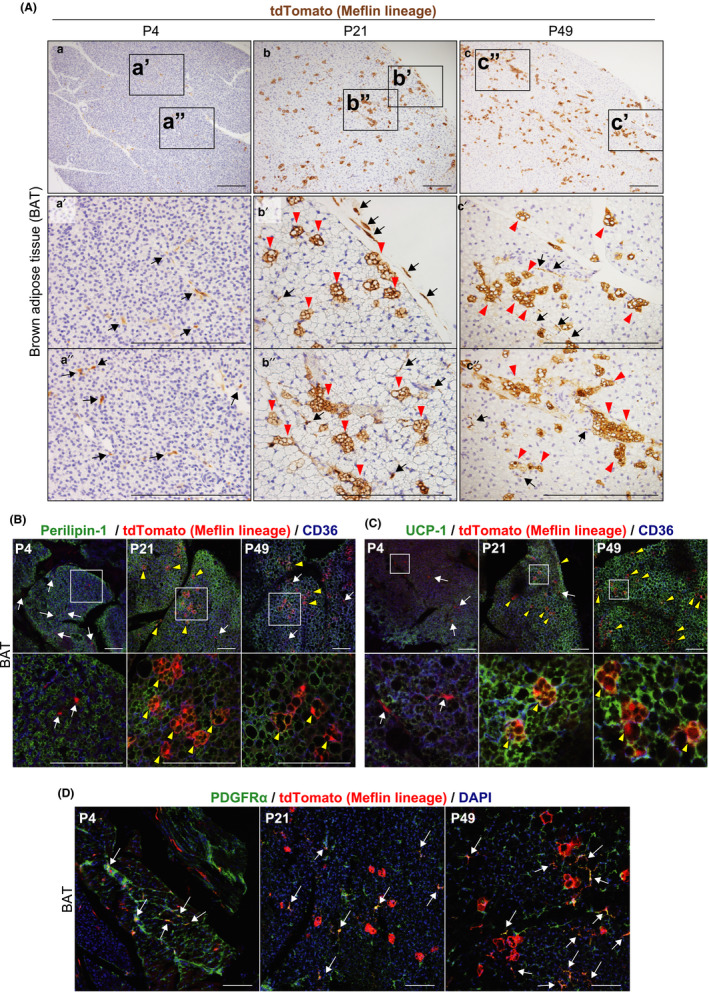
Meflin+ cells are fibroblastoid cells that differentiate into brown adipocytes in the postnatal period. (A) BAT harvested from Meflin‐CreERT2; LSL‐tdTomato mice was stained for tdTomato. tdTomato+ cells (brown) represent Meflin+ cells at P4 (a) and Meflin lineage cells at P21 (b) and P49 (c). Boxed regions in a – c are magnified in lower panels (a′–c′ and a″–c″). Arrows denote tdTomato+ stromal cells with a fibroblastoid morphology that reside in the interstitium, and arrowheads indicate tdTomato+ terminally differentiated multilocular brown adipocytes (Meflin lineage cells). Scale bars, 200 µm. (B, C) BAT harvested from Meflin‐CreERT2; LSL‐tdTomato mice was stained for the adipocyte marker Perilipin‐1 (B) and the brown adipocyte marker UCP‐1 (C) by IF (green). Boxed regions are magnified in lower panels. Arrows indicate tdTomato+ stromal cells with a fibroblastoid morphology, whereas arrowheads show tdTomato+ mature brown adipocytes. Scale bars, 100 µm. (D) PDGFRα expression (green) was found in Meflin+ cells at P4 and tdTomato+ cells at P21 and P49 with a fibroblastoid morphology (red) in postnatal BAT (arrows), suggesting that some Meflin+ cells retain their undifferentiated state even at P21 and P49. Scale bars, 100 µm
2.3. Meflin+ cells gave rise to osteocytes and chondrocytes of the bone in postnatal development
One of the most established features of MSCs is that they have the capacity to differentiate into both chondrogenic and osteogenic lineages (Caplan, 1991; Prockop, 1997). MSCs are heterogeneous and comprise bona fide MSCs and progenitors that are committed to restricted pathways of differentiation in varying degrees, examples of which are Grem1‐ or homeobox A11 (Hoxa11)‐positive skeletal stem cells (SSCs) (Rux et al., 2016; Worthley et al., 2015). Using ISH analyses, we previously showed that Meflin mRNA was present in immature chondroblasts in the growth plate and immature osteoblasts condensing in and around the periosteum, but not in mature osteocytes and chondrocytes of the long bones in adult (P56) mice (Maeda et al., 2016). These data suggest that Meflin is a marker of MSCs or SSCs in the bone and/or their downstream progenitors but not their terminally differentiated progeny.
In the present study, we examined the fate of Meflin+ cells in the femoral epiphysis, where osteogenesis and chondrogenesis are active in the postnatal period. We found that Meflin+ cells displayed a rounded or polygonal chondrocyte‐like morphology in the proliferating zone at P4, 1 day after TAM administration (Figure 3Aa,a′). They are small sized and positive for SRY‐box transcription factor 9 (Sox9), a marker of chondrocyte differentiation, suggesting that they are immature chondroblasts (Figure 3B). Meflin lineage chondrocytes formed clusters, some of which aligned in a single‐filed linear pattern at P21 and P49 (Figure 3Ab′,B). Those data are consistent with the established notion that resting chondroprogenitors in the growth plate undergo sequential differentiation to form the proliferating and hypertrophic zones (Bianco & Robey, 2015; Kronenberg, 2003). The tdTomato+ Meflin lineage cells were either Sox9‐positive or negative at P21 and P49 (Figure 3B), suggesting that these cells include chondroblasts in different stages of maturation.
FIGURE 3.
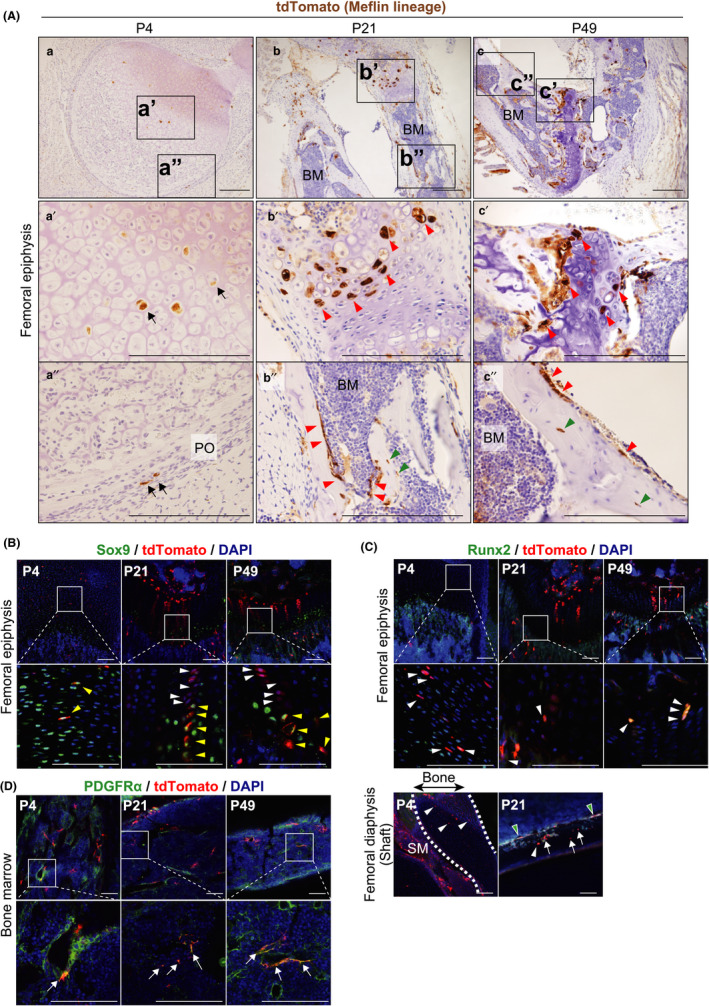
In postnatal development, Meflin+ cells differentiate into bone chondrocytes and osteocytes and PDGFRα‐positive stromal cells in BM. (A) tdTomato IHC showed Meflin+ immature chondroblast‐like cells in the articular cartilage (a′) and fibroblastoid cells in the periosteum (PO) (a″) of the femoral epiphysis at P4 (arrows). Those cells give rise to chondrocytes and osteoblasts that line the surfaces of the trabecular bone at P21 and P49 (red arrowheads). tdTomato+ Meflin lineage cells include mature osteocytes (green arrowheads). Scale bars, 200 µm. (B) Meflin+ cells (red) are positive for Sox9 (green) at P4 (yellow arrowheads), whereas Meflin lineage cells are either positive (yellow arrowheads) or negative (white arrowheads) for Sox9 at P21 and P49, suggesting the tdTomato+ cells include cells in different stages of chondroblast maturation. Boxed regions are magnified in lower panels. Scale bars, 100 µm. (C) Meflin+ cells at P4 and Meflin lineage cells at P21 and P49 (red) are positive for Runx2 (green) in the femoral epiphysis and diaphysis (arrowheads). Note that most Meflin lineage cells that line the surface of the bone are positive for Runx2 (green arrowheads), whereas there are some Runx2‐negative Meflin lineage cells in the periosteum (lower panels, arrows) at P21 and P49. Dashed lines indicate the area of the femoral diaphysis at P4. SM, skeletal muscle. Scale bars, 100 µm. (D) Meflin+ cells at P4 and Meflin lineage cells at P21 and P49 (red) were detected in the perisinusoidal region. They were positive for PDGFRα (green) in the BM in the postnatal development (arrows)
Meflin+ cells were also found in the periosteum known to contain osteoprogenitors at P4 (Bianco & Robey, 2015) (Figure 3Aa″). These cells were positive for runt‐related transcription factor 2 (Runx2), a key regulator for osteogenic differentiation (Figure 3C). The fate mapping of Meflin+ cells showed that they differentiated into osteoblasts that are located adjacent to the growth plate or line bone surfaces at P21 and P49; those cells are either positive or negative for Runx2 (Figure 3Ab″,c″,C). tdTomato expression was observed in some mature osteocytes embedded within the bone mineral, indicating terminal osteogenic differentiation of Meflin+ cells (Figure 3Ab″,c″, green arrowheads). Taken together, these data showed that Meflin is a marker of MSCs, SSCs and/or their downstream progenitors in the postnatal development of the skeletal system.
2.4. Meflin+ cells represent PDGFRα+ stromal cells in the BM
We previously showed that Meflin mRNA was expressed in stromal cells that are distributed sparsely within the BM. Most of those cells are localized adjacent to sinusoidal vessels or small arteries (Maeda et al., 2016; Mizutani et al., 2019). In the present study, we showed that most Meflin+ cells found within the BM at P4 are positive for PDGFRα, a marker of the most primitive MSCs in the BM (Morikawa et al., 2009; Omatsu et al., 2010) (Figure 3D). Intriguingly, most tdTomato+ Meflin lineage cells are also positive for PDGFRα at P21 and P49. In addition, the density of tdTomato+ Meflin lineage cells in the BM at P21 and P49 seemed not to be much different from that of Meflin+ cells at P4. These findings suggest the possibilities that Meflin+ cells in the BM are undifferentiated and slow cycling or that they remain positive for PDGFRα even after cell division or self‐renewal (Figure 3D). Given the view that MSCs provide niches for hematopoietic stem cells (HSCs) and are localized in the perivascular area in the BM (Wei & Frenette, 2018), it seems possible that Meflin+ cells are involved in the maintenance of HSCs in the BM. Supporting this hypothesis, the analysis of a published dataset of scRNA‐seq of murine BM showed that Meflin mRNA was detected in not only PDGFRα+ MSC clusters but also in those that were positive for LepR and CXCL12, which are known to be niche cells for HSCs in the BM (Baccin et al., 2020; Wei & Frenette, 2018) (Figure S5).
2.5. Meflin+ cells gave rise to skeletal myocytes in postnatal development
In skeletal muscle, there are tissue stem cells called satellite cells. They differentiate into skeletal myocytes in embryonic development and regenerate upon injury (Relaix et al., 2005; Seale et al., 2000; Zhang et al., 2018). In the present study, we found that there were spindle‐shaped Meflin+ cells interspersed among muscle fibers in the rectus femoris muscle at P4 (Figure 4Aa‐a″). Some of these cells seemed to be attached to the surface of muscle fibers and were positive for paired box 7 (Pax7), a marker of satellite cells (Figure 4A,B). Lineage tracing of Meflin+ cells showed that they developed into skeletal myocytes and myotubes at P21 and P49, respectively, and they expressed myogenic differentiation 1 (MyoD1) (Figure 4A,B). Limited numbers of tdTomato+ Meflin lineage cells were double‐positive for Pax7 and MyoD1 at P21, suggesting that these cells represented developing myoblasts (Figure 4B).
FIGURE 4.
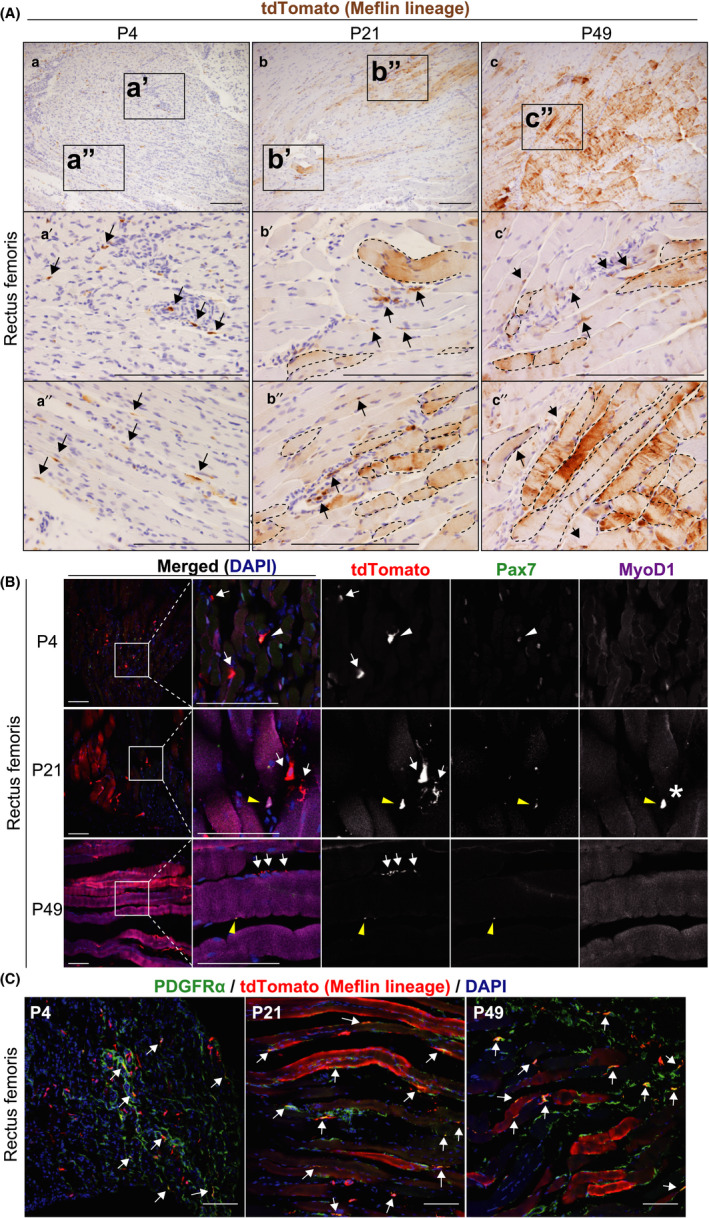
Meflin is a marker of Pax7+ satellite cells and PDGFRα+ FAPs in postnatal skeletal muscle tissue. (A) Skeletal muscle tissues harvested from the rectus femoris of Meflin‐CreERT2; LSL‐tdTomato mice were stained for tdTomato. tdTomato+ cells (brown) represent Meflin+ cells at P4 (a) and their descendants (Meflin lineage cells) at P21 (b) and P49 (c). Arrows denote tdTomato+ cells that were adjacent to muscle fibers or resided in the interstitium. Areas indicated by dashed lines show tdTomato+ skeletal myocytes (Meflin lineage cells). Scale bars, 200 µm. (B) Some Meflin+ cells were positive for the satellite cell marker Pax7 (white arrowhead), whereas the others are negative for Pax7 at P4 (white arrows). Some Meflin lineage cells are also positive for Pax7 at P21 and P49 (yellow arrowheads). A few Meflin lineage cells are double‐positive for both Pax7 and MyoD1, which may represent developing myoblasts (asterisk). Note that the magenta signals found in myotubes are due to the autofluorescence of skeletal fibers (647 nm). Scale bars, 100 µm. (C) Some Meflin+ cells at P4 and Meflin lineage cells at P21 and P49 are positive for PDGFRα (arrows), suggesting that these cells constitute FAPs in the stroma of the skeletal muscle. Scale bars, 100 µm
Our previous study showed that Meflin mRNA was also expressed by PDGFRα‐positive and PECAM1‐negative stromal cells, which are known as fibro‐adipogenic progenitors (FAPs), in the skeletal muscle tissue of P56 mice (Maeda et al., 2016; Uezumi et al., 2010). Consistent with this, our present study detected Meflin+ cells that exhibited a fibroblastoid appearance and were positive for PDGFRα at P4 (Figure 4C). Some of these Meflin+/PDGFRα+ cells remained positive for PDGFRα and did not differentiate into myoblasts even at P21 and P49 (Figure 4C). These data suggest that Meflin is expressed by both Pax7+ satellite cells and PDGFRα+ FAPs in the skeletal muscle. Supporting this view, an analysis of a published dataset of scRNA‐seq of murine skeletal muscle supported this finding. Specifically, Meflin mRNA was detected in PDGFRα+ FAPs as well as some subsets of satellite cells (Tabula Muris Consortium, 2018) (Figure S6).
2.6. Meflin marked MSCs and/or their early progenitors across multiple tissues in adult mice
Next, to investigate the fate of Meflin+ cells in adult mice, 8‐week‐old Meflin‐CreERT2; LSL‐tdTomato mice were administered TAM intraperitoneally (i.p.) 3 times, which was immediately followed by the observation of tdTomato+ cells at 9 weeks of age (referred to as Meflin+ cells) and tdTomato+ Meflin lineage cells at 12 and 40 weeks of age (Figure 5A). The Cre recombination efficiency was approximately 30% in adipose, bone and skeletal muscle tissues (Figure S2B). As observed in the postnatal lineage tracing experiments, Meflin+ cells showed a fibroblastoid appearance and were localized in the interstitium of white and brown adipose, bone and skeletal muscle tissues at 9 weeks of age (Figure 5B). At the ages of 12 and 40 weeks, these cells gave rise to differentiated white and brown adipose cells filled with lipid droplets, chondroblasts/chondrocytes, osteocytes and skeletal myocytes (Figure 5B). Compared to the postnatal lineage tracing experiments, the number of differentiated cells was low, reflecting the low frequency of differentiation and turnover of these mesenchymal cells in the adult stage. Indeed, we observed some fibroblastic or satellite cell‐like tdTomato+ cells even at 12 and 40 weeks of age, implying that these cells remained stem cells or progenitors (Figure 5Bb,e,f,k). tdTomato+ cells with an undifferentiated morphology were also present in the BM at 40 weeks of age (Figure 5Bi). These results indicate that Meflin+ cells may retain their status as tissue stem cells or progenitors as adults.
FIGURE 5.
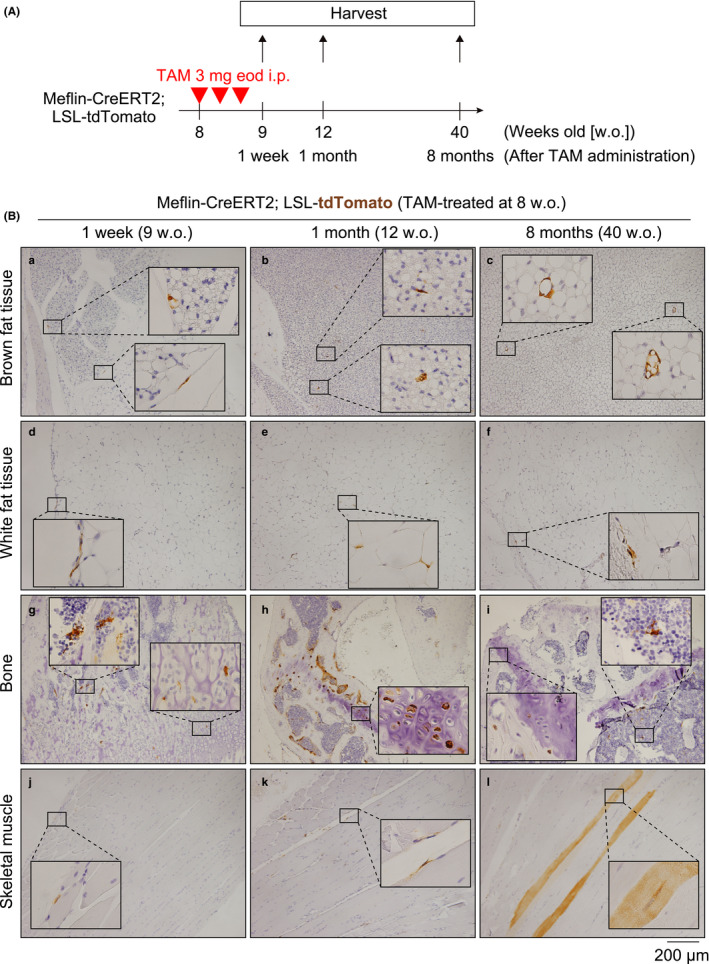
Meflin+ cells differentiate into multilineage cells in the adult period. (A) The experimental design for lineage tracing of Meflin+ cells into adulthood. Meflin‐CreERT2; LSL‐tdTomato mice were intraperitoneally administered 3 mg TAM 3 times every other day (eod) at the age of 8 weeks. Tissues were harvested at 9, 12 and 40 weeks of age. (B) Lineage tracing of Meflin+ cells labeled with tdTomato in brown (a–c) and white (d–f) adipose tissues, bone (g–i) and skeletal muscle (j–l) in the adult period. tdTomato+ cells appear to be fibroblastoid cells in the adipose tissues and skeletal muscle, immature chondroblasts in the bone, and perisinusoidal stromal cells in the BM one week after TAM administration (a, d, g, j). Those cells gave rise to mature brown adipocytes (c), mature chondrocytes (h), osteocytes (i) and skeletal myocytes (l). Some Meflin lineage cells retained their fibroblastoid morphology even 12 weeks and 8 months after TAM administration (b, e, f, i, k). Scale bars, 200 µm
2.7. Contribution of Meflin+ cells to the development of mesenchymal tissues in embryonic, postnatal and adult mice
Finally, we asked to what extent mesenchymal lineage cells were derived from Meflin+ cells across multiple tissues in the embryonic, postnatal and adulthood periods. Thus, we conducted a lineage tracing experiment by crossing Meflin‐Cre mice, which constitutively express Cre recombinase under the control of the Meflin promoter (Hara et al., 2019) with LSL‐tdTomato mice (Meflin‐Cre; LSL‐tdTomato). We examined various tissues of 8‐week‐old Meflin‐Cre; LSL‐tdTomato mice by tdTomato staining (Figure 6A). We found that Meflin+ cells gave rise to terminally differentiated mature white and brown adipocytes that were distributed in a mosaic pattern at 8 weeks of age (Figure 6Aa,b). Notably, there were tdTomato‐negative terminally differentiated adipocytes, implying the possibility that there exist Meflin‐negative MSCs or progenitors in the early stage of embryonic development (Figure 6Aa,b). This was also the case with chondrocytes and osteocytes. That is, the majority of chondrocytes and osteoblasts were positive for tdTomato, but some were negative (Figure 6Ac). Conversely, almost all of the skeletal myotubes of the rectus femoris muscle were positive for tdTomato at the age of 8 weeks. Those data indicate that most of the skeletal muscles originated from Meflin+ MSCs or progenitors, including satellite cells, during embryonic development (Figure 6Ad).
FIGURE 6.
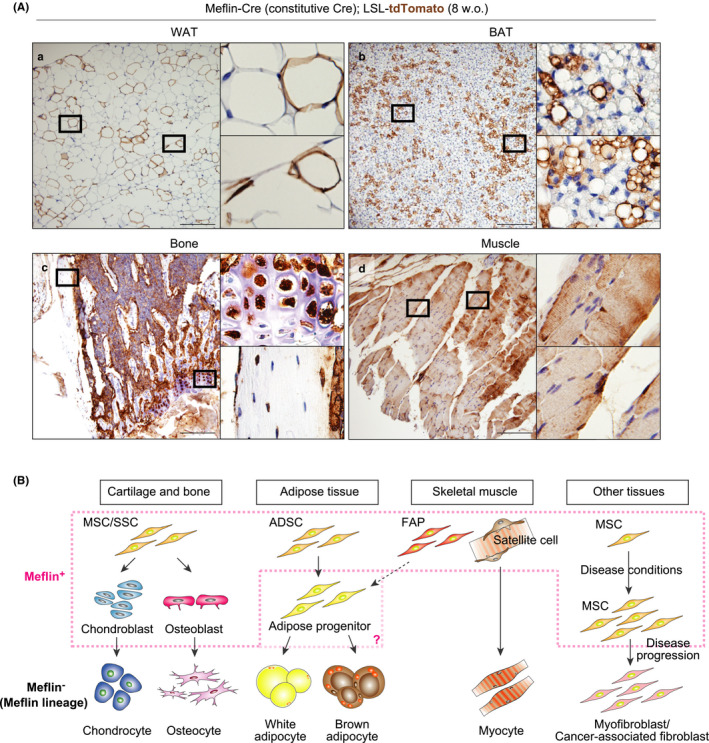
Contribution of Meflin+ cells to multiple mesenchymal lineage cells throughout life. (A) Meflin‐Cre mice, which constitutively express Cre under the control of the Meflin promoter, were crossed with Rosa26‐LSL‐tdTomato mice, followed by tissue harvest at 8 weeks of age and tdTomato staining. Boxed regions are magnified in adjacent panels. Note that almost all myotubes are Meflin lineage cells, whereas chondrocytes, adipocytes and osteocytes exhibited a mosaic pattern with some terminally differentiated cells expressing tdTomato. Scale bars, 200 µm. (B) A schematic illustration of our working hypothesis regarding the contribution of Meflin+ cells to the development of multiple mesenchymal tissues. Based on the findings in the present study and our previous studies (Hara et al., 2019; Maeda et al., 2016; Mizutani et al., 2019), we speculate that Meflin is a marker of MSCs/SSCs in the cartilage and bone, ADSCs in the adipose tissue, Pax7+ satellite cells and PDGFRα+ FAPs in the skeletal muscle (dashed pink line). Meflin is also expressed by a subset(s) of early progenitors of MSCs/SSCs. It is possible that early progenitors of ADSCs are also positive for Meflin. The present study did not address the roles of Meflin+ cells in regeneration or diseases of the mesenchymal tissues. Previous studies showed that Meflin+ MSCs significantly proliferate in fibrotic diseases and cancer to repair tissue injury and restrain disease progression. In contrast, those cells can differentiate into α‐SMA+ myofibroblasts or cancer‐associated fibroblasts that are weakly positive or negative for Meflin and promote disease (Hara et al., 2019; Mizutani et al., 2019)
3. DISCUSSION
In the present study, we showed that Meflin+ cells with an immature morphology reside in various mesenchymal tissues and that these cells give rise to mature adipocytes, chondrocytes, osteocytes and skeletal myocytes in postnatal and adult periods. Given that Meflin is expressed by Sox9+ chondroblasts, Runx2+ stromal cells in the periosteum and Pax7+ satellite cells, one cannot conclude that Meflin expression is restricted to bona fide MSCs. However, previous studies showed that Meflin expression is sharply down‐regulated or lost upon the induction of cell differentiation into chondroblasts, osteoblasts, adipocytes and myofibroblasts in cultured BM‐MSCs (Hara et al., 2019; Maeda et al., 2016; Mizutani et al., 2019). Moreover, Meflin knockout (KO) or knock‐down led to the loss of stemness and deregulated differentiation (e.g., Runx2 over‐expression in osteoblasts and their increased number in the bone) (Hara et al., 2019; Maeda et al., 2016). Thus, it appears that Meflin is a functional marker of MSCs and/or their immature lineage‐committed progenitors (Figure 6B). Meflin is not expressed by other types of cells, including epithelial, smooth muscle, endothelial, peripheral nerve and blood and immune cells (Maeda et al., 2016). That finding makes Meflin a useful marker to localize MSCs and/or their immature progenitors in situ and to monitor the undifferentiated status of cultured MSCs. Meflin expression significantly decreases during aging in mice (Hara et al., 2019). Such down‐regulation is compatible with the established fact that the number of MSCs decreases during aging in both mice and humans (Stenderup et al., 2003), further supporting the validity of Meflin as a marker of MSCs.
An appealing feature of Meflin as a marker of MSCs and their immature progenitors is that Meflin+ cells are present in almost all tissues, although it is unknown whether Meflin+ cells in various tissues are identical. Our previous report and the present studies showed that Meflin is preferentially expressed by fibroblastoid stromal cells that are perivascular, independent of tissue type, which is consistent with the view that MSCs reside in all tissues and organs as “pericytes” (Crisan et al., 2008; Maeda et al., 2016). In our previous study, ISH analysis of various tissues showed that the number of Meflin+ cells is much lower than the number of conventional pericytes that line the abluminal side of the vessels (Maeda et al., 2016). We suppose that a small subset of all pericytes or a distinct population of perivascular fibroblasts with a multilineage differentiation capacity expresses Meflin across multiple tissues. This interpretation is consistent with a previous study that showed that CD34+/PDGFRα+/Sca‐1+ fibroblasts but not conventional pericytes or vascular smooth muscle cells represent adipose stem cells that give rise to adipocytes (Hepler & Gupta, 2017).
The Cre recombination efficiency of Meflin‐CreERT2; LSL‐tdTomato mice was modest, which enabled us to observe the fate of single Meflin+ or Meflin lineage cells. Interestingly, Meflin lineage cells with terminally differentiated features tended to form clusters at P21 and P49. In addition, some Meflin lineage cells with an undifferentiated morphology that expressed PDGFRα were also present in these tissues (Figures 1E, 2D, 3D, 4C). These results suggest 3 distinct but not mutually exclusive possibilities. That is, (a) some Meflin+ cells gave rise to both mature lineage cells and PDGFRα‐positive fibroblasts, (b) some Meflin+ cells at P4 remain quiescent until P21 and P49, maintaining their undifferentiated state, or (c) some Meflin+ cells underwent asymmetric division such that each Meflin+ cell divided to generate one daughter cell with a stem cell fate and one daughter that differentiated into a mature lineage cell. Future identification of bona fide MSC marker(s) will help resolve which of these possibilities is most plausible.
The present study did not address the roles of Meflin and Meflin+ cells in disease. We previously showed that Meflin+ cells significantly proliferate in the contexts of acute tissue injury and fibrosis of the heart and cancer development (Hara et al., 2019; Kobayashi et al., 2019; Miyai et al., 2020; Mizutani et al., 2019) (Figure 6B). Most of these cells were positive for PDGFRα, leading to the speculation that Meflin+ MSCs themselves proliferate or they differentiate into PDGFRα+ fibroblasts in response to acute tissue injury and carcinogenesis. Notably, our data from mouse models and pathological human tissues showed that Meflin expression in these MSCs/fibroblasts was essential for tissue repair after injury. However, further studies showed that Meflin function seemed incompatible with the well‐known functions of fibroblasts to promote fibrosis and cancer. For example, Meflin KO mice exhibited accelerated fibrosis in a model of chronic heart failure as well as cancer progression with an increased number of α‐smooth muscle actin (SMA)‐positive cancer‐associated fibroblasts in a pancreatic cancer model (Hara et al., 2019; Kobayashi et al., 2021; Mizutani et al., 2019). Lineage tracing experiments showed that Meflin+ MSCs/fibroblasts differentiate into Meflin‐negative and α‐SMA‐positive fibroblasts that promote disease progression (Hara et al., 2019; Mizutani et al., 2019) (Figure 6B). Based on these findings, we propose a hypothesis that Meflin expression confers MSCs/fibroblasts disease‐restraining functions both in the context of fibrosis and cancer. The role of Meflin in disease and tissue regeneration should be further investigated in the future.
In conclusion, we presented the result of lineage tracing of Meflin+ cells in the postnatal and adult periods under physiological conditions in mice. Meflin may be a useful single marker to localize MSCs and/or their progenitors in situ, the significance of which will be evaluated by comparing it with other MSC markers in the future.
4. EXPERIMENTAL PROCEDURES
4.1. Animal experiments and generation of Meflin‐CreERT2 knock‐in mice
All animal experiments were carried out according to the protocols approved by the Animal Care and Use Committee of Nagoya University Graduate School of Medicine, Japan. To generate the Meflin‐CreERT2 knock‐in mice, conventional gene‐targeting techniques were used as described previously (Mizutani et al., 2019). Briefly, the 5′ homology arm (2.8 kb) and 3′ homology arm (5.6 kb) were isolated by PCR with PrimeStar Max DNA polymerase (Takara) using genomic DNA from the 129S6 mouse as a template. A targeting vector that contained the 5′‐homology arm, the CreERT2 gene, the Flippase Recognition Target (FRT)‐flanked PGK‐neo resistance gene and the 3′‐homology arm was generated to insert the CreERT2 9 bp downstream from the start codon of the Meflin gene (Islr). The linearized targeting vector was electroporated into CSL3 embryonic stem cells (a gift from Chyan‐Sheng Lin, Columbia University), followed by G418 selection. Targeted embryonic stem cell clones identified by Southern blot analysis were injected into C57BL/6 blastocysts at the Institute of Immunology Co., Ltd. (Tokyo, Japan) for the generation of chimeric mice. Chimeras were mated with Rosa26‐FLP mice (JAX Stock Number: 003946) to excise the neo resistance cassette. The knock‐in mice were mated with C57BL/6 mice to generate mice with a C57BL/6 genetic background. Genomic DNAs extracted from mouse tails were used for PCR genotyping. Sequences of the primers were as follows: WT forward, ACACACGACCTTGGCAAGTCCCAGC; WT reverse, GTCTGCAATCTGGAAGCCATACTTCTCC; Meflin‐CreERT2 forward, ACACACGACCTTGGCAAGTCCCAGC; and Meflin‐ERT2 reverse, CGATCCCTGAACATGTCCATCAGG. The PCR product sizes from WT and meflin‐CreERT2 alleles were 291 and 359 bp, respectively.
To conduct lineage tracing experiments, Meflin‐CreERT2 mice were crossed with Rosa26‐LoxP‐stop‐LoxP (LSL)‐tdTomato mice (JAX Stock Number: 007909) to generate Meflin‐CreERT2; Rosa26‐LSL‐tdTomato mice. For postnatal lineage tracing, pups were administered 50 µg tamoxifen through an intragastric route at postnatal days 1, 2 and 3. Tissues were harvested at ages 4, 21 and 49 days. For lineage tracing in adulthood, 8‐week‐old Meflin‐CreERT2; Rosa26‐LSL‐tdTomato mice were administrated 3 mg tamoxifen by intraperitoneal injection 3 times every other day. Tissues were harvested 2 days and 1 and 8 months after the third injection.
Meflin‐Cre mice, which constitutively express the fluorescent protein ZsGreen, diphtheria toxin receptor (DTR) and the Cre recombinase under the control of the Meflin promoter, were generated as previously described, followed by crossing them with Rosa26‐LSL‐tdTomato mice (Hara et al., 2019). Sequences of the primers were as follows: Meflin‐Cre forward, TAGGTGGTATTGGATTCTGGCTGGG; Meflin‐Cre reverse, TTGAAGTAGTCGACGATGTCCTGG.
4.2. IHC
For IHC on FFPE tissues, sections were deparaffinized with xylene and rehydrated with PBS, followed by immersion in an antigen retrieval buffer (HistoVT One, pH 7, Nacalai Tesque) for 30 min at 98°C. The sections were washed 3 times in phosphate‐buffered saline (PBS) and incubated in blocking buffer (X0909, Dako) for 30 min, followed by incubation with anti‐RFP rabbit polyclonal antibody (1:1,000, 600‐401‐379, Rockland Immunochemicals) (which also detects the tdTomato fluorescent protein), diluted with antibody diluent buffer (1% BSA, 0.1% NaN3 in PBS) overnight at 4°C. The sections were washed 3 times with PBS and immersed in 0.5% H2O2 in methanol for 15 min for the inactivation of endogenous peroxidase. The sections were washed 3 times in PBS, followed by incubation in EnVision + System‐ HRP Labelled Polymer Anti‐Rabbit (K4003, Dako) for 30 min at RT. The sections were washed in PBS, incubated with Liquid DAB + Substrate Chromogen System (K3468, Dako) for 5 min at RT, dehydrated in ethanol and cleared in xylene, followed by mounting with xylene‐based mounting media Entellan new (100869, Merck Millipore).
4.3. Immunofluorescent staining
Harvested adipose tissues were fixed in ice‐cold 4% paraformaldehyde (PFA, 163‐18435, Wako) in PBS at 4°C overnight, followed by transfer through increasing concentrations of sucrose (5%, 10%, 15% and 20% in PBS) for 6–12 hr each. Tissues were placed in an embedding solution (8% gelatin, 1.5% polyvinylpyrrolinone, 15% sucrose in PBS) and frozen using liquid nitrogen for sectioning. Bones were dissected and placed in ice‐cold 4% PFA in PBS overnight at 4°C. Samples were then decalcified in 0.5 M EDTA (pH 8.0) for 24 hr, followed by 20% sucrose in PBS overnight and stored at −80°C as previously described (Kusumbe et al., 2015). For samples of skeletal muscles, the rectus femoris was dissected and placed in 4% PFA in PBS overnight, followed by submersion in 20% sucrose in PBS and embedding.
Frozen samples were sectioned to 100, 20 and 10 µm thickness for bones, adipose tissues and skeletal muscles, respectively, using a cryostat (CM3050S, Leica). Sections rehydrated in PBS were permeabilized with 0.3% Triton X‐100 in PBS, except those prepared from adipose tissues. The sections were incubated with blocking solutions (5% normal donkey serum, 0.1% Triton X‐100 in PBS for bones; 10% normal donkey serum, 0.1% Triton X‐100 in PBS for adipose tissues; 5% normal donkey serum, 0.5% Triton X‐100 in PBS for skeletal muscles) for 30 min at room temperature (RT), followed by incubation with primary antibodies diluted in the blocking solutions overnight at 4°C. Following 3 washes with buffer (PBS for bones and muscles; 0.1% Triton X‐100 in PBS for adipose tissues), the sections were treated with secondary antibodies in blocking solution overnight at 4°C. Sections were washed 3 times with wash buffer and counterstained with 4′,6‐diamidino‐2‐phenylindole (DAPI, 10‐236‐276‐001, Roche) for nuclear staining, followed by mounting in PermaFluor Aqueous Mounting Medium (TA‐030‐FM, Thermo Fisher Scientific).
For IF staining, the following primary antibodies were used: rat monoclonal anti‐PECAM1/CD31 (MEC13.3, 553370, BD Biosciences, 1:100); goat polyclonal anti‐CD36 (AF2519, R&D Systems, 1:200); rat monoclonal anti‐Endomucin (V.7C7, eBioscience, 1:50); goat polyclonal anti‐mouse PDGFRα (AF1062, R&D Systems, 1:200); rat monoclonal anti‐MyoD (5F11, MABE132, Merck Millipore, 1:200); goat polyclonal anti‐Osteopontin (AF808, R&D Systems, 1:200); rabbit polyclonal anti‐Pax7 (PA1‐117, Invitrogen, 1:200); rabbit monoclonal anti‐Perilipin‐1 (D1D8, 9349S, Cell Signaling Technology, 1:300); goat polyclonal anti‐RFP (200‐101‐379, Rockland Immunochemicals, 1:500); rabbit polyclonal anti‐RFP (600‐401‐379, Rockland Immunochemicals, 1:500); chicken polyclonal anti‐Red Fluorescent Protein (RFP) (600‐901‐379, Rockland Immunochemicals, 1:500); rat monoclonal anti‐Runx2 (232902, MAB2006, R&D Systems, 1:100); goat polyclonal anti‐Sox9 (AF3075, R&D Systems, 1:200); and rabbit polyclonal anti‐UCP1 (ab10983, Abcam, 1:300) antibodies.
For the detection of the primary antibodies, the following fluorophore‐conjugated secondary antibodies were used: Alexa Fluor 488‐conjugated donkey anti‐rat IgG (A21208, Invitrogen, 1:500); Alexa Fluor 488‐conjugated donkey anti‐rabbit IgG (A21206, Invitrogen, 1:500); Cy3‐conjugated donkey anti‐chicken IgY (703‐165‐155, Jackson ImmunoResearch, 1:500); Cy3‐conjugated donkey anti‐rat IgG (112‐165‐003, Jackson ImmunoResearch, 1:500); Alexa Fluor 555‐conjugated donkey anti‐rabbit IgG (A31572, Invitrogen, 1:500); Alexa Fluor 555‐conjugated donkey anti‐goat IgG (A21432, Invitrogen, 1:500); CF647‐conjugated donkey anti‐rabbit IgG (20047, Biotium, 1:500); Alexa Fluor 647‐conjugated donkey anti‐rat IgG (ab15015, Abcam, 1:500); and Alexa Fluor 647‐conjugated donkey anti‐goat IgG (A21447, Invitrogen, 1:500) antibodies.
4.4. In situ hybridization assay combined with IF
For the combined detection of mRNAs by ISH and proteins by IF, we first performed IF using Opal fluorophores (NEL810001KT, Akoya Biosciences) according to the manufacturer's instruction. Briefly, FFPE sections were deparaffinized, followed by antigen retrieval in AR9 (AR900250ML, Akoya Biosciences) for 15 min at 98°C. After blocking with Antibody Diluent/Blocking (ARD1001EA, Akoya Biosciences), the sections were incubated with primary antibodies for 2 hr at RT, washed 3 times in Tris‐buffered saline (150 mM NaCl, 25 mM Tris‐HCl) with 0.05% polyoxyethylene sorbitan monolaurate (Tween 20) (TBST) and incubated in Opal Polymer HRP Secondary Antibody for 10 min at RT. After 3 washes in TBST, Opal 570 in 1X Plus Amplification Diluent (1:125) was added and reacted for 10 min at RT. Next, we performed ISH using RNAscope Multiplex Fluorescent Reagent Kit v2 (323100, ACD Bio) per the manufacturer's instructions. For mRNA retrieval and the removal of antibody–HRP complexes, the slides were immersed in 1X target retrieval buffer using a steam cooker (VC100571, T‐fal) for 15 min, followed by treatment with Proteinase Plus (322331, ACD Bio) for 30 min at 40°C. The sections were twice washed in deionized water, incubated with target probe against mouse Islr for 2 hr at 40°C, twice washed in 1X wash buffer and then incubated in amplification reagents (AMP1‐3), followed by HRP‐C1 Reagent (323104, ACD Bio). The signals were amplified with TSA‐Plus Cyanine 5 System (NEL745001KT, PerkinElmer), followed by mounting the sections with PermaFluor Aqueous Mounting Medium (TA‐030‐FM, Thermo Fisher Scientific).
4.5. Image acquisition and analysis
The fluorescence images were acquired using a confocal laser scanning microscope (LSM700, Carl Zeiss) and ZEN black software (ver. 7.1, Carl Zeiss). The bright field images were acquired using an upright microscope (BX53, OLYMPUS) equipped with a digital camera (DP73, OLYMPUS) and CellSens Standard software (ver. 1.13, OLYMPUS). Cre recombination efficiency was calculated manually using ImageJ software (ver. 1.59, National Institute of Health) as follows: recombination efficiency (%) = 100 * number of tdTomato+ cells/(number of Meflin mRNA+ cells + number of tdTomato+ cells). We examined 5 or 6 randomly selected areas for each tissue under high magnification view for the determination of the Cre recombination efficiency.
4.6. Analysis of publicly available single‐cell RNA‐seq datasets
Sequencing count data from single‐cell murine brown, gonadal, mesenteric and subcutaneous fat tissues [accession number GSM2967049], murine limb muscle [accession number GSM2967056] at the age of 3 months and murine bone marrow [accession number GSE122465] at the age of 2 months were obtained from Gene Expression Omnibus (GEO) (Baccin et al., 2020; Tabula Muris Consortium, 2018). The data were preprocessed using the functions that normalize the feature expression measurements for each cell by the total expression, multiplies this by a scale factor and log‐transforms the result. Next, principal components analysis was performed and significant PCs were used as input for graph‐based clustering. Dimensionality reduction was performed with t‐Distributed Stochastic Neighbor Embedding (t‐SNE). We next applied modularity optimization techniques such as the Leiden algorithm to iteratively group cells together. Each cluster was annotated according to the cell metadata obtained from the database. We also visualized marker expression of several genes by violin plots. All these procedures were performed with Monocle 3, an R toolkit for single‐cell genomics (Trapnell et al., 2014).
4.7. Statistical analysis
Statistical analyses for the calculation and the visualization were performed using the base package or Tidyverse in R version 4.0.
CONFLICT OF INTERESTS
The authors declare no competing interests.
Supporting information
Supplementary Material
ACKNOWLEDGMENTS
We gratefully thank Kaori Ushida and Kozo Uchiyama (Nagoya University) for support in immunostaining and tissue preparation. This work was supported by a Grant‐in‐Aid for Scientific Research (B) (18H02638 to A.E., 20H03467 to M.T.) commissioned by the Ministry of Education, Culture, Sports, Science and Technology of Japan; AMED‐CREST (Japan Agency for Medical Research and Development, Core Research for Evolutional Science and Technology; 19gm0810007h0104 and 19gm1210008s0101 to A.E.); and the Project for Cancer Research and Therapeutic Evolution (P‐CREATE) from AMED (19cm0106332h0002 to A.E.).
Hara A, Kato K, Ishihara T, et al. Meflin defines mesenchymal stem cells and/or their early progenitors with multilineage differentiation capacity. Genes Cells.2021;26:495–512. 10.1111/gtc.12855
Communicated by: Hideyuki Saya
REFERENCES
- Andrzejewska, A., Lukomska, B., & Janowski, M. (2019). Concise review: Mesenchymal stem cells: From roots to boost. Stem Cells, 37, 855–864. 10.1002/stem.3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccin, C., Al‐Sabah, J., Velten, L., Helbling, P. M., Grünschläger, F., Hernández‐Malmierca, P., Nombela‐Arrieta, C., Steinmetz, L. M., Trumpp, A., & Haas, S. (2020). Combined single‐cell and spatial transcriptomics reveal the molecular, cellular and spatial bone marrow niche organization. Nature Cell Biology, 22, 38–48. 10.1038/s41556-019-0439-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beresford, J. N., Bennett, J. H., Devlin, C., Leboy, P. S., & Owen, M. E. (1992). Evidence for an inverse relationship between the differentiation of adipocytic and osteogenic cells in rat marrow stromal cell cultures. Journal of Cell Science, 102, 341–351. 10.1242/jcs.102.2.341 [DOI] [PubMed] [Google Scholar]
- Berry, R., & Rodeheffer, M. S. (2013). Characterization of the adipocyte cellular lineage in vivo. Nature Cell Biology, 15, 302–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco, P., & Robey, P. G. (2015). Skeletal stem cells. Development, 142, 1023–1027. 10.1242/dev.102210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bühring, H. J., Battula, V. L., Treml, S., Schewe, B., Kanz, L., & Vogel, W. (2007). Novel markers for the prospective isolation of human MSC. Annals of the New York Academy of Sciences, 1106, 262–271. 10.1196/annals.1392.000 [DOI] [PubMed] [Google Scholar]
- Caplan, A. I. (1991). Mesenchymal stem cells. Journal of Orthopaedic Research, 9, 641–650. 10.1002/jor.1100090504 [DOI] [PubMed] [Google Scholar]
- Chau, Y.‐Y., Bandiera, R., Serrels, A., Martínez‐Estrada, O. M., Qing, W., Lee, M., Slight, J., Thornburn, A., Berry, R., McHaffie, S., Stimson, R. H., Walker, B. R., Chapuli, R. M., Schedl, A., & Hastie, N. (2014). Visceral and subcutaneous fat have different origins and evidence supports a mesothelial source. Nature Cell Biology, 16, 367–375. 10.1038/ncb2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisan, M., Yap, S., Casteilla, L., Chen, C.‐W., Corselli, M., Park, T. S., Andriolo, G., Sun, B., Zheng, B., Zhang, L., Norotte, C., Teng, P.‐N., Traas, J., Schugar, R., Deasy, B. M., Badylak, S., Buhring, H.‐J., Giacobino, J.‐P., Lazzari, L., … Péault, B. (2008). A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell, 3, 301–313. [DOI] [PubMed] [Google Scholar]
- Dominici, M., Le Blanc, K., Mueller, I., Slaper‐Cortenbach, I., Marini, F. C., Krause, D. S., Deans, R. J., Keating, A., Prockop, D. J., & Horwitz, E. M. (2006). Minimal criteria for defining multipotent mesenchymal stromal cells. The international society for cellular therapy position statement. Cytotherapy, 8, 315–317. [DOI] [PubMed] [Google Scholar]
- Fedorenko, A., Lishko, P. V., & Kirichok, Y. (2012). Mechanism of fatty‐acid‐dependent UCP1 uncoupling in brown fat mitochondria. Cell, 151, 400–413. 10.1016/j.cell.2012.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum, A., Hsu, Y.‐M.‐S., Day, R. B., Schuettpelz, L. G., Christopher, M. J., Borgerding, J. N., Nagasawa, T., & Link, D. C. (2013). CXCL12 in early mesenchymal progenitors is required for haematopoietic stem‐cell maintenance. Nature, 495, 227–230. 10.1038/nature11926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar, M., Karan, G., Tvrdik, P., & Capecchi, M. R. (2008). Two cell lineages, myf5 and myf5‐independent, participate in mouse skeletal myogenesis. Developmental Cell, 14, 437–445. 10.1016/j.devcel.2008.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara, A., Kobayashi, H., Asai, N., Saito, S., Higuchi, T., Kato, K., Okumura, T., Bando, Y. K., Takefuji, M., Mizutani, Y., Miyai, Y., Saito, S., Maruyama, S., Maeda, K., Ouchi, N., Nagasaka, A., Miyata, T., Mii, S., Kioka, N., … Enomoto, A. (2019). Roles of the mesenchymal stromal/stem cell marker meflin in cardiac tissue repair and the development of diastolic dysfunction. Circulation Research, 125, 414–430. 10.1161/CIRCRESAHA.119.314806 [DOI] [PubMed] [Google Scholar]
- Hepler, C., & Gupta, R. K. (2017). The expanding problem of adipose depot remodeling and postnatal adipocyte progenitor recruitment. Molecular and Cellular Endocrinology, 445, 95–108. 10.1016/j.mce.2016.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz, E. M., Le Blanc, K., Dominici, M., Mueller, I., Slaper‐Cortenbach, I., Marini, F. C., Deans, R. J., Krause, D. S., & Keating, A., & International Society for Cellular Therapy . (2005). Clarification of the nomenclature for MSC: The International Society for Cellular Therapy position statement. Cytotherapy, 7, 393–395. 10.1080/14653240500319234 [DOI] [PubMed] [Google Scholar]
- Kobayashi, H., Enomoto, A., Woods, S. L., Burt, A. D., Takahashi, M., & Worthley, D. L. (2019). Cancer‐associated fibroblasts in gastrointestinal cancer. Nature Reviews Gastroenterology & Hepatology, 16, 282–295. 10.1038/s41575-019-0115-0 [DOI] [PubMed] [Google Scholar]
- Kobayashi, H., Gieniec, K. A., Wright, J. A., Wang, T., Asai, N., Mizutani, Y., Iida, T., Ando, R., Suzuki, N., Lannagan, T. R. M., Ng, J. Q., Hara, A., Shiraki, Y., Mii, S., Ichinose, M., Vrbanac, L., Lawrence, M. J., Sammour, T., Uehara, K., … Wood, S. L. (2021). The balance of stromal BMP signaling mediated by GREM1 and ISLR drives colorectal carcinogenesis. Gastroenterology, 160, 1224–1239. 10.1053/j.gastro.2020.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramann, R., Schneider, R. K., DiRocco, D. P., Machado, F., Fleig, S., Bondzie, P. A., Henderson, J. M., Ebert, B. L., & Humphreys, B. D. (2015). Perivascular Gli1+ progenitors are key contributors to injury‐induced organ fibrosis. Cell Stem Cell, 16, 51–66. 10.1016/j.stem.2014.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberg, H. M. (2003). Developmental regulation of the growth plate. Nature, 423, 332–336. 10.1038/nature01657 [DOI] [PubMed] [Google Scholar]
- Kusumbe, A. P., Ramasamy, S. K., Starsichova, A., & Adams, R. H. (2015). Sample preparation for high‐resolution 3D confocal imaging of mouse skeletal tissue. Nature Protocols, 10, 1904–1914. 10.1038/nprot.2015.125 [DOI] [PubMed] [Google Scholar]
- Kuwano, T., Izumi, H., Aslam, M. R., Igarashi, Y., Bilal, M., Nishimura, A., Watanabe, Y., Nawaz, A., Kado, T., Ikuta, K., Yamamoto, S., Sasahara, M., Fujisaka, S., Yagi, K., Mori, H., & Tobe, K. (2021). Generation and characterization of a Meflin‐CreERT2 transgenic line for lineage tracing in white adipose tissue. PLoS One, 16, e0248267. 10.1371/journal.pone.0248267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv, F.‐J., Tuan, R. S., Cheung, K. M. C., & Leung, V. Y. L. (2014). Concise review: The surface markers and identity of human mesenchymal stem cells. Stem Cells, 32, 1408–1419. 10.1002/stem.1681 [DOI] [PubMed] [Google Scholar]
- Mabuchi, Y., Morikawa, S., Harada, S., Niibe, K., Suzuki, S., Renault‐Mihara, F., Houlihan, D. D., Akazawa, C., Okano, H., & Matsuzaki, Y. (2013). LNGFR+THY‐1+VCAM‐1hi+ cells reveal functionally distinct subpopulations in mesenchymal stem cells. Stem Cell Reports, 1, 152–165. 10.1016/j.stemcr.2013.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, K., Enomoto, A., Hara, A., Asai, N., Kobayashi, T., Horinouchi, A., Maruyama, S., Ishikawa, Y., Nishiyama, T., Kiyoi, H., Kato, T., Ando, K., Weng, L., Mii, S., Asai, M., Mizutani, Y., Watanabe, O., Hirooka, Y., Goto, H., & Takahashi, M. (2016). Identification of meflin as a potential marker for mesenchymal stromal cells. Scientific Reports, 6, 22288. 10.1038/srep22288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyai, Y., Esaki, N., Takahashi, M., & Enomoto, A. (2020). Cancer‐associated fibroblasts that restrain cancer progression: Hypotheses and perspectives. Cancer Science, 111, 1047–1057. 10.1111/cas.14346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani, Y., Kobayashi, H., Iida, T., Asai, N., Masamune, A., Hara, A., Esaki, N., Ushida, K., Mii, S., Shiraki, Y., Ando, K., Weng, L., Ishihara, S., Ponik, S. M., Conklin, M. W., Haga, H., Nagasaka, A., Miyata, T., Matsuyama, M., … Takahashi, M. (2019). Meflin‐positive cancer‐associated fibroblasts inhibit pancreatic carcinogenesis. Cancer Research, 79, 5367–5381. 10.1158/0008-5472.CAN-19-0454 [DOI] [PubMed] [Google Scholar]
- Morikawa, S., Mabuchi, Y. O., Kubota, Y., Nagai, Y., Niibe, K., Hiratsu, E., Suzuki, S., Miyauchi‐Hara, C., Nagoshi, N., Sunabori, T., Shimmura, S., Miyawaki, A., Nakagawa, T., Suda, T., Okano, H., & Matsuzaki, Y. (2009). Prospective identification, isolation, and systemic transplantation of multipotent mesenchymal stem cells in murine bone marrow. Journal of Experimental Medicine, 206, 2483–2496. 10.1084/jem.20091046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicodemou, A., & Danisovic, L. (2017). Mesenchymal stromal/stem cell separation methods: Concise review. Cell Tissue Bank, 18, 443–460. 10.1007/s10561-017-9658-x [DOI] [PubMed] [Google Scholar]
- Omatsu, Y., Sugiyama, T., Kohara, H., Kondoh, G., Fujii, N., Kohno, K., & Nagasawa, T. (2010). The essential functions of adipo‐osteogenic progenitors as the hematopoietic stem and progenitor cell niche. Immunity, 33, 387–399. 10.1016/j.immuni.2010.08.017 [DOI] [PubMed] [Google Scholar]
- Pittenger, M. F. (1999). Multilineage potential of adult human mesenchymal stem cells. Science, 284, 143–147. 10.1126/science.284.5411.143 [DOI] [PubMed] [Google Scholar]
- Prockop, D. J. (1997). Marrow stromal cells as stem cells for nonhematopoietic tissues. Science, 276, 71–74. 10.1126/science.276.5309.71 [DOI] [PubMed] [Google Scholar]
- Relaix, F., Rocancourt, D., Mansouri, A., & Buckingham, M. (2005). A Pax3/Pax7‐dependent population of skeletal muscle progenitor cells. Nature, 435, 948–953. 10.1038/nature03594 [DOI] [PubMed] [Google Scholar]
- Rux, D. R., Song, J. Y., Swinehart, I. T., Pineault, K. M., Schlientz, A. J., Trulik, K. G., Goldstein, S. A., Kozloff, K. M., Lucas, D., & Wellik, D. M. (2016). Regionally restricted hox function in adult bone marrow multipotent mesenchymal stem/stromal cells. Developmental Cell, 39, 653–666. 10.1016/j.devcel.2016.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacchetti, B., Funari, A., Michienzi, S., Di Cesare, S., Piersanti, S., Saggio, I., Tagliafico, E., Ferrari, S., Robey, P. G., Riminucci, M., & Bianco, P. (2007). Self‐renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell, 131, 324–336. [DOI] [PubMed] [Google Scholar]
- Seale, P., Sabourin, L. A., Girgis‐Gabardo, A., Mansouri, A., Gruss, P., & Rudnicki, M. A. (2000). Pax7 is required for the specification of myogenic satellite cells. Cell, 102, 777–786. 10.1016/S0092-8674(00)00066-0 [DOI] [PubMed] [Google Scholar]
- Simmons, P. J., & Torok‐Storb, B. (1991). Identification of stromal cell precursors in human bone marrow by a novel monoclonal antibody, STRO‐1. Blood, 78(1), 55–62. [PubMed] [Google Scholar]
- Stenderup, K., Justesen, J., Clausen, C., & Kassem, M. (2003). Aging is associated with decreased maximal life span and accelerated senescence of bone marrow stromal cells. Bone, 33, 919–926. 10.1016/j.bone.2003.07.005 [DOI] [PubMed] [Google Scholar]
- Tabula Muris Consortium . (2018). Single‐cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature, 562, 367–372. 10.1038/s41586-018-0590-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell, C., Cacchiarelli, D., Grimsby, J., Pokharel, P., Li, S., Morse, M., Lennon, N. J., Livak, K. J., Mikkelsen, T. S., & Rinn, J. L. (2014). The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nature Biotechnology, 32, 381–386. 10.1038/nbt.2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uezumi, A., Fukada, S.‐I., Yamamoto, N., Takeda, S., & Tsuchida, K. (2010). Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nature Cell Biology, 12, 143–152. 10.1038/ncb2014 [DOI] [PubMed] [Google Scholar]
- Wakitani, S., Saito, T., & Caplan, A. I. (1995). Myogenic cells derived from rat bone marrow mesenchymal stem cells exposed to 5‐azacytidine. Muscle and Nerve, 18, 1417–1426. 10.1002/mus.880181212 [DOI] [PubMed] [Google Scholar]
- Wei, Q., & Frenette, P. S. (2018). Niches for hematopoietic stem cells and their progeny. Immunity, 48, 632–648. 10.1016/j.immuni.2018.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodbury, D., Schwarz, E. J., Prockop, D. J., & Black, I. B. (2000). Adult rat and human bone marrow stromal cells differentiate into neurons. Journal of Neuroscience Research, 61, 364–370. 10.1002/1097-4547 [DOI] [PubMed] [Google Scholar]
- Worthley, D. L., Churchill, M., Compton, J. T., Tailor, Y., Rao, M., Si, Y., Levin, D., Schwartz, M. G., Uygur, A., Hayakawa, Y., Gross, S., Renz, B. W., Setlik, W., Martinez, A. N., Chen, X., Nizami, S., Lee, H. G., Kang, H. P., Caldwell, J.‐M., … Wang, T. C. (2015). Gremlin 1 identifies a skeletal stem cell with bone, cartilage, and reticular stromal potential. Cell, 160, 269–284. 10.1016/j.cell.2014.11.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J., Tang, Y., Sheng, X., Tian, Y., Deng, M., Du, S., Lv, C., Li, G., Pan, Y., Song, Y., Lou, P., Luo, Y., Li, Y., Zhang, B., Chen, Y., Liu, Z., Cong, Y., Plikus, M. V., Meng, Q., … Yu, Z. (2020). Secreted stromal protein ISLR promotes intestinal regeneration by suppressing epithelial Hippo signaling. EMBO Journal, 39, e103255. 10.15252/embj.2019103255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, K., Zhang, Y., Gu, L., Lan, M., Liu, C., Wang, M., Su, Y., Ge, M., Wang, T., Yu, Y., Liu, C., Li, L., Li, Q., Zhao, Y., Yu, Z., Wang, F., Li, N., & Meng, Q. (2018). Islr regulates canonical Wnt signaling‐mediated skeletal muscle regeneration by stabilizing Dishevelled‐2 and preventing autophagy. Nature Communications, 9, 1–16. 10.1038/s41467-018-07638-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, B. O., Yue, R., Murphy, M. M., Peyer, J. G., & Morrison, S. J. (2014). Leptin‐receptor‐expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell, 15, 154–168. 10.1016/j.stem.2014.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk, P. A., Zhu, M., Mizuno, H., Huang, J., Futrell, J. W., Katz, A. J., Benhaim, P., Lorenz, H. P., & Hedrick, M. H. (2001). Multilineage cells from human adipose tissue: Implications for cell‐based therapies. Tissue Engineering, 7, 211–228. 10.1089/107632701300062859 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


