Abstract
Background
Early detection/prediction of flare‐ups in asthma, commonly triggered by viruses, would enable timely treatment. Previous studies on exhaled breath analysis by electronic nose (eNose) technology could discriminate between stable and unstable episodes of asthma, using single/few time‐points. To investigate its monitoring properties during these episodes, we examined day‐to‐day fluctuations in exhaled breath profiles, before and after a rhinovirus‐16 (RV16) challenge, in healthy and asthmatic adults.
Methods
In this proof‐of‐concept study, 12 atopic asthmatic and 12 non‐atopic healthy adults were prospectively followed thrice weekly, 60 days before, and 30 days after a RV16 challenge. Exhaled breath profiles were detected using an eNose, consisting of 7 different sensors. Per sensor, individual means were calculated using pre‐challenge visits. Absolute deviations (|%|) from this baseline were derived for all visits. Within‐group comparisons were tested with Mann‐Whitney U tests and receiver operating characteristic (ROC) analysis. Finally, Spearman's correlations between the total change in eNose deviations and fractional exhaled nitric oxide (FeNO), cold‐like symptoms, and pro‐inflammatory cytokines were examined.
Results
Both groups had significantly increased eNose fluctuations post‐challenge, which in asthma started 1 day post‐challenge, before the onset of symptoms. Discrimination between pre‐ and post‐challenge reached an area under the ROC curve of 0.82 (95% CI = 0.65–0.99) in healthy and 0.97 (95% CI = 0.91–1.00) in asthmatic adults. The total change in eNose deviations moderately correlated with IL‐8 and TNFα (ρ ≈ .50–0.60) in asthmatics.
Conclusion
Electronic nose fluctuations rapidly increase after a RV16 challenge, with distinct differences between healthy and asthmatic adults, suggesting that this technology could be useful in monitoring virus‐driven unstable episodes in asthma.
Keywords: asthma, biomarkers, omics and systems biology, virus
Electronic nose sensor fluctuations increased after a RV‐16 challenge in asthmatic and healthy adults, more evidently in asthma. In asthma, the increase in eNose fluctuations occurred before the onset of cold‐like symptoms. The magnitude of the change in eNose fluctuations was not correlated with FeNO and cold‐like symptoms, but moderately correlated with pre‐ and post‐challenge cytokine levels in asthma.

Abbreviations
- FeNO
fractional exhaled nitric oxide
- RV‐16
rhinovirus‐16
1. INTRODUCTION
One of the major burdens in asthma is episodes of loss of control and exacerbations,1, 2 characterized by acute flare‐ups of respiratory symptoms, such as shortness of breath, cough, wheezing, and chest tightness. Commonly, these episodes are triggered by respiratory viral infections,3 especially rhinoviruses,4, 5, 6 and can potentially lead to emergency visits and hospitalizations, possibly requiring urgent medical interventions1, 2 and incurring high healthcare expenses.
Unfortunately, episodes of loss of control are hard to detect due to the poor correlation between clinical symptoms and the underlying disease activity.7 Furthermore, predictors of upcoming exacerbations in an individual patient are currently lacking, except that recent (severe) asthma exacerbations are predictive of future (severe) exacerbations.8, 9 Moreover, treatment is modestly effective during exacerbations,10 which emphasizes the importance of strategies that limit the development of exacerbations, preferably in the pre‐symptomatic phase. Robust biomarkers indicating disease severity and control over time could help predict (the severity of) episodic flare‐ups in asthma. A metabolomic approach might be able to sufficiently capture subtle changes in asthma control, possibly before symptoms occur.
Analysis of the exhaled breath's metabolic content (ie, breathomics) is relatively new and of interest in asthma, because of its non‐invasive character and its potential to detect changes of inflammatory profiles in asthmatic patients.11 Exhaled breath consists of volatile organic compounds (VOCs), which are gaseous organic molecules that originate either from the body itself (endogenous) or from the environment (exogenous). Endogenous VOCs can reflect the metabolic processes occurring in the lungs and beyond.12 The studies by Brinkman et al.13 and Fens et al.14 showed that exhaled breath could distinguish clinically stable from unstable episodes in asthmatic patients; however, only three time‐points (ie, baseline, loss of control, and recovery) were compared with weeks to months between visits. Moreover, the studies by Robroeks et al.15 and van Vliet et al.16 showed that exacerbations could be predicted based on two‐monthly exhaled VOC measurements, using an offline breath analysis technique. Furthermore, our group has previously shown that breath profiles change after an experimental rhinovirus‐16 (RV16) infection in healthy and asthmatic adults.17 However, only single time‐point comparisons and group averages were examined. Now, studies investigating the potential of real‐time exhaled breath analysis to monitor and predict such episodes on a day‐to‐day basis should follow.
It is believed that biological and thereby metabolic processes fluctuate over time to maintain homeokinesis and that external triggers can influence such fluctuations.18 These metabolic fluctuations may be reflected in exhaled breath metabolites and possibly be of value in detection and prediction of loss of control/exacerbations in asthma. Since most exhaled breath studies are cross‐sectional or longitudinal with low temporal resolution, knowledge about the day‐to‐day fluctuations in exhaled breath profiles, with and without external triggers, in patients and healthy controls, is lacking.
We hypothesized that the day‐to‐day fluctuations of exhaled breath would change after an external trigger and that this response would differ between healthy controls and asthmatic patients, due to differences in biological processes. Our first objective was to compare the day‐to‐day fluctuations from personal baselines in exhaled breath profiles between asthmatic and healthy controls, before and after a rhinovirus (RV) challenge. Our second objective was to investigate whether the magnitude of the altered eNose fluctuations was linked to pre‐challenge inflammatory markers and was reflected in post‐challenge symptoms and inflammation, to identify possible differences in biological processes between and within groups.
2. METHODS
2.1. Study design
In this prospective, intervention study, asthmatic patients and healthy controls were followed for 60 days before and 30 days after a RV16 challenge (Figure 1). The study was conducted at the Amsterdam UMC, location AMC (Amsterdam, The Netherlands), from February 2016 till June 2017. Before inclusion, participants were screened and provided informed consent. Prior to the start of the study period, participants went through a run‐in phase to familiarize themselves with all the different measurements performed in the study. Exhaled breath analysis was performed 3 times per week using an electronic nose (eNose).
FIGURE 1.

Overview of the study design: all participants were screened (1 visit) to test eligibility and had a run‐in phase (2 visits) to make participants familiarized with all measurements. During the study period of 90 days, participants had around 2–3 visits per week, with ~20 visits in the pre‐challenge phase (60 days) and ~10 visits in the post‐challenge phase (30 days)
This study, along with the safety of the virus, was approved by the medical ethical committee of the Amsterdam UMC, location AMC (Amsterdam, the Netherlands), and registered at the Netherlands Trial Register (NTR5426).
2.2. Study population
In this study, 12 intermittent or mild‐to‐moderate atopic asthmatics (based on the Global Initiative for Asthma criteria 2014, www.ginasthma.org) and 12 non‐atopic healthy controls were included, aged 18–35 years, all non‐smokers. Atopy was based on a skin prick test with common aeroallergens (See Appendix S1). Asthmatics had to be clinically stable at inclusion, defined by no use of corticosteroids and no exacerbations, 6 weeks prior to inclusion.
Subjects were excluded when they had a cold (4 weeks prior to screening), a RV16 titer ≥1:8 in serum (at the time of screening and before the RV16 challenge visit), and a positive PCR for any respiratory virus in nasal lavage (at the day before the RV16 challenge) or when they were pregnant. Details about the inclusion and exclusion criteria can be found in the published work by Sinha et al.18
All participants were recruited via advertisements at the outpatient clinics of various hospitals in Amsterdam and via social media. Furthermore, subjects from previous study cohorts of our hospital were contacted, only when they had provided informed consent to be invited for future research.
2.3. Rhinovirus challenge
After the 60‐day pre‐challenge phase, participants were exposed to RV16 using a standardized and validated challenge approach, as previously described.18, 19 An experimental RV16 infection was used with a nasal dose of inoculum of 100 TCID50, tissue culture infective dose of the virus required to cause cytopathy in 50% of the cells. This has been considered safe for in vivo testing in human volunteers, during a scientific advice meeting at BfArM (Bonn, Germany, April 30, 2013). The RV16 was prepared under good manufacturing practice (GMP), as part of the U‐BIOPRED study, and tested in a dose‐dependent manner in healthy individuals and mild asthma patients (manuscript in preparation), which revealed that 100TCID50 was the lowest dose that effectively infected those exposed and caused expected symptomology.
2.4. Exhaled breath analysis
2.4.1. Measurement setup
Real‐time exhaled breath analysis was performed using an eNose, the SpiroNose, connected in series with a spirometer (SpiroPerfectTM, Welch Allyn) (Figure S1). The SpiroNose consists of seven different cross‐reactive metal oxide sensors (Table S1), present in fourfold, twice on the inside and twice on the outside of the device. The mixture of VOCs in exhaled breath is detected by the inner sensors during the exhalation, while ambient VOCs are detected by the outer sensors.
2.4.2. eNose measurement
The eNose measurement was performed as described previously.20 In short, subjects were asked to rinse their mouth three times thoroughly with water. Subsequently, exhaled breath analysis was performed in duplicate with a 2‐min interval. All participants were instructed to perform five tidal breaths, followed by a single inspiratory capacity maneuver up to total lung capacity, a 5‐s breath‐hold, and slow (<0.4 L/s) maximal expiration toward residual volume, with their nose clipped. A new mouthpiece, bacterial filter, and nose clamp were used for each subject.
2.4.3. Data processing
Processing of the eNose sensor data was performed using MATLAB® as described in De Vries et al.20 and included filtering, detrending, ambient correction, and automated peak detection. The highest sensor peak of each sensor signal was selected as the variable for further analysis. All sensor peaks were normalized to the most stable sensor, sensor 2, to minimize the inter‐array differences. Therefore, data from sensor 2 are not included in the fluctuation analysis.
2.5. Other outcomes
2.5.1. Home monitoring
During the study, several clinical parameters were monitored at home: spirometry, the Wisconsin Upper Respiratory Symptom Survey (WURSS‐21), and Asthma Control Questionnaire (ACQ). Spirometry and the ACQ‐6 (six questions, with a score range of 0–6)21 were monitored daily, in the morning at home, by the volunteers themselves, using a hand‐held spirometry device (MicroDiary, CareFusion). Lung function parameters of interest were the forced vital capacity (FVC), forced expiratory volume in 1 s (FEV1), and peak expiratory flow (PEF), expressed as percentages of predicted. The WURSS‐21 questionnaire (see Appendix S1) monitoring started at the day of challenge.
2.5.2. FeNO
Double fractional exhaled nitric oxide (FeNO) measurements were performed using the NIOX MINO (Aerocrine AB, Sweden) during the study visits (thrice weekly) at the Amsterdam UMC, location AMC (Amsterdam, the Netherlands), according to American Thoracic Society recommendations.22 We used the average of the two measurements in our analysis.
2.5.3. Nasal lavages
Nasal lavages were collected once weekly pre‐challenge and thrice weekly post‐challenge during the study visits, as described previously.18 Standardized washings collected from the nose were used for cytokine analyses by luminex: IFN‐γ, IL‐1β, IL‐6, IL‐8, IL‐10, IL‐13, IL‐17A, IL‐33, IP‐10, and TNF‐α (after the cells were removed by centrifugation).
2.6. Sample size
This proof‐of‐concept study had an explorative nature and was based on initial estimates of fluctuating inflammatory biomarkers. Our sample size of 12 individuals per group was based on previous studies by Turner et al.,23, 24 in which detection of temporal variability in exhaled VOCs was possible with fewer data points (once weekly, for 6 months). Moreover, our sample size provides adequate power for multi‐omics analysis according to the study by Li et al.25
2.7. Statistical analysis
2.7.1. Baseline characteristics and clinical presentation
Baseline characteristics are presented as mean and standard deviation (SD) for normally distributed variables, median and interquartile range (IQR) for skewed data, and n (%) for categorical variables. The distribution of the data was visually examined using histograms and Q–Q (quantile‐quantile) plots. Differences between groups were compared using the Mann‐Whitney U or Kruskal‐Wallis tests for continuous variables and chi‐square test for categorical variables. Lung function, FeNO, WURSS‐21, and ACQ scores were summarized by calculating means of the pre‐ and post‐challenge and minima/maxima of the post‐challenge at an individual level, followed by medians at group level, to compare baseline (pre‐challenge) with the (maximal) response to the RV challenge. For the WURSS‐21 score, data from the day of the RV16 challenge and the first day post‐challenge were used for the “pre‐challenge” phase, as daily home monitoring started on the day of challenge.
2.7.2. eNose deviations
Fluctuations in the eNose signals were examined for the pre‐ and post‐challenge phase, separately, in both asthmatic and healthy volunteers. For each sensor and subject, means were calculated based on all pre‐challenge visits, serving as an individual baseline () (Figure 2A, left graph). Per subject, deviations from this personal baseline were derived for all study visits, expressed as absolute percentages: with x being an observation and the sample mean (Figure 2B, right graph). Next, the mean of these absolute deviation percentages was calculated for the pre‐ () and post‐challenge () phase, at individual and group level, consecutively. Within‐ and between‐group comparisons were made using Wilcoxon signed‐rank and Mann‐Whitney U tests, respectively. Because of multiple testing, false discovery rate (FDR)–adjusted p‐values (q‐values) were also calculated.26 Finally, receiver operating characteristic (ROC) analysis was performed to calculate the discriminative power of the individual sensor fluctuations to distinguish between pre‐ and post‐challenge within and between groups.
FIGURE 2.
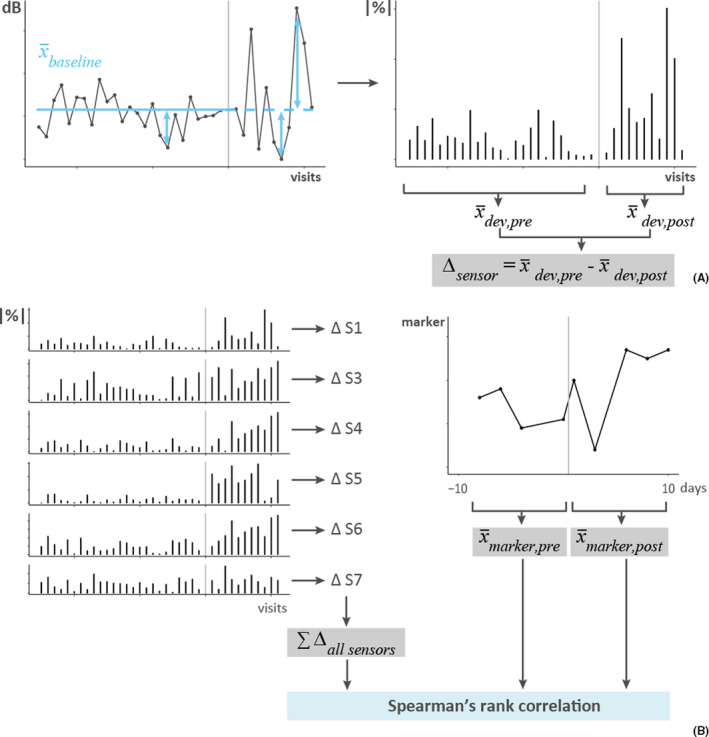
Calculation of eNose deviations. Data from one subject with the vertical gray lines representing the RV challenge. (A) The left graph shows the sensor values (dB) of one sensor, for all visits pre‐ and post‐challenge. The mean of the sensor values pre‐challenge (blue solid line, ) served as the baseline, also for the post‐challenge phase (blue dashed line). Next, deviations from this personal baseline were calculated (blue double arrows) and expressed as absolute percentages (|%|), as shown on the right. Finally, the average deviation pre‐challenge () and post‐challenge () was calculated and the difference () between these, per subject and sensor. (B) The left graph shows the deviations from the personal baseline for all sensors (S1, S3–S7), expressed as absolute percentages (|%|). The difference in deviations () between the pre‐ and post‐challenge phase was calculated for each sensor and summed to determine the total change in deviations (). On the right, data from one of the markers of interest (FeNO, cytokines, and WURSS‐21) are shown, from which 10‐day averages were calculated for the pre‐challenge () and post‐challenge () phase (note that 10 days ≠10 visits). Finally, Spearman's rank correlations were calculated between the total change in deviation and the pre‐ and post‐averages of the marker of interest, separately
2.7.3. Linking change in eNose deviations to inflammatory markers and symptoms
First, we calculated the overall change in eNose fluctuations by summing up all the differences between pre‐ and post‐challenge mean deviation percentages of all sensors (i), at an individual level () (Figure 2B, left graph). Next, we explored the link between the magnitude of the change in eNose fluctuations and post‐challenge cold‐like symptoms (WURSS‐21), as well as, pre‐ and post‐challenge inflammatory marker levels (FeNO and cytokines). For this, the FeNO, WURSS‐21, and cytokine data were log10‐transformed and averaged over the last 10 days pre‐challenge () and the first 10 days post‐challenge (), separately, for each subject (Figure 2B, right graph). Finally, Spearman's rank correlations between the total deviation difference and the mean FeNO, cytokine levels, or WURSS‐21 scores, pre‐ and post‐challenge separately, were determined.
Statistics were performed in R (version 3.6.1) combined with R packages “pROC” and “RVAideMemoire”. p‐values and q‐values <0.05 were considered significant.
3. RESULTS
3.1. Study cohort
In total, 24 participants were included in this study: 12 healthy and 12 asthmatic participants (inclusion chart, Figure S2). At baseline, there were no major differences between groups regarding sex, age, ethnicity, body mass index (BMI), pack years, and lung function (Table 1). Only FeNO was significantly different between groups (p < .01), with a median of 14 ppb (IQR: 12–21) in healthy controls and 45 ppb (IQR: 30–63) in asthmatics. The number of visits was similar between and within groups, with on average ~23 visits (range 20–29) before and ~11 visits (range 10–16) after the RV16 challenge.
TABLE 1.
Baseline characteristics and the number of visits per group
| Healthy (n = 12) | Asthma (n = 12) | |
|---|---|---|
| Sex (female) | 7 (58%) | 8 (67%) |
| Age (years) | 21 (±1.5) | 22.2 (±2.2) |
| Ethnicity (Caucasian) | 11 (92%) | 9 (75%) |
| BMI (kg/m2) | 22.2 (±1.6) | 22.8 (±3.1) |
| Smoking (pack years) | 1 (± 0.17) | – |
| Baseline spirometry | ||
| FEV1% of predicted | 106 (±12) | 101 (±10) |
| FVC % of predicted | 104 (±11) | 104 (±10) |
| PEF % of predicted | 108 (±14) | 105 (±12) |
| FeNO (ppb) | 14 (12–21)* | 45 (30–63)* |
| Number of visits (mean; range) | ||
| Total | 34 (30–40) | 35 (33–38) |
| Before challenge | 23 (20–29) | 23 (21–26) |
| After challenge | 11 (10–12) | 12 (10–16) |
Data are presented as mean (±SD), median (IQR), n (%) or else when stated, and have partly been published previously.18 BMI, body mass index; FEV1, forced expiratory volume in 1 s; FVC, forced vital capacity; PEF, peak expiratory flow; FeNO, fractional exhaled nitric oxide. Differences in the number of visits were due to personal reasons (ie, missing visits) or when there was a delay in the RV16 challenge due to logistical reasons (ie, extra visits). * Significant difference (p < .01).
3.2. Clinical presentation (pre‐ and post‐challenge)
Either or a combination of serum antibody tests, along with clinical symptoms and RV polymerase chain reaction (PCR) conducted on nasal lavage samples, confirmed that all the study participants were successfully inoculated with the RV16, as published previously.18 During the whole study period, no exacerbations/loss of control, as defined by Reddel et al.,1 occurred in the participants. The strongest clinical effect was seen in the WURSS‐21 score, which increased after the RV16 challenge in both groups. The maximal WURSS‐21 score occurred 3 days post‐challenge, with an average score of 35 ± 33 in asthmatics and 19 ± 9 in healthy participants (Figure S3). Using descriptive statistics, we did not find major changes in lung function, ACQ score, or FeNO (Table S2). However, in‐depth analysis on the development of the response to the challenge was carefully studied before using time series analysis, as previously published.18
3.3. eNose deviations
The following comparisons were made regarding the eNose deviations: 1) pre‐viral challenge with post‐viral challenge states and 2) diseased (asthma) cohort with healthy (control) cohort.
3.3.1. Pre‐ vs. post‐challenge
For both groups, the mean deviation of the eNose signals increased after the rhinovirus challenge in the majority of the sensors (Figure 3; Table S3). For healthy subjects, mean deviations increased significantly in sensor 5 (Δ = 2% ±2; p < .05) and 7 (Δ = 8% ±8; p < .01) and for asthmatics in sensor 1 (Δ = 6% ±7; p < .05), 4 (Δ = 3% ±4; p < .05), 5 (Δ = 9% ±7; p < .001), and 6 (Δ = 3% ±3; p < .001) (Figure 3). The change in mean deviations per subject is shown in Figure 4 and S4. The highest area under the ROC curve (AUROCC) for discrimination between pre‐ and post‐challenge was found in sensor 5, for both healthy 0.82 (95% CI: 0.65–0.99) and asthmatic subjects 0.97 (95% CI: 0.91–1.00) (Table 2). The eNose fluctuations of sensor 5 increased (on average) directly 1 day after the RV challenge in asthmatics (Figure S5). This was less evident in healthy subjects.
FIGURE 3.
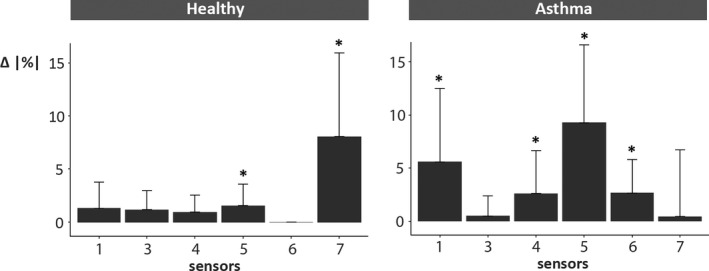
Total change in absolute eNose deviations (per sensor). The change (Δ = post ‐ pre) in eNose fluctuations after the RV16 challenge, expressed as absolute mean deviation percentages (|%|). First, the difference between the personal mean deviations (pre‐ and post‐challenge) was determined (). Next, at group level, the average differences were calculated and are depicted in the graph for each sensor separately. In healthy controls, sensor 5 (p < .05) and sensor 7 (p < .01) showed significant increases in fluctuations post‐challenge, while in asthmatics sensor 1, 4 (both p < .05), 5, and 6 (both p < .01) were significantly different. *significant difference (p < .05)
FIGURE 4.
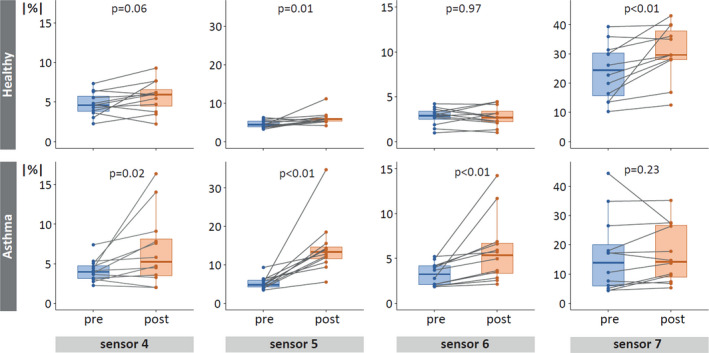
Absolute eNose deviations pre‐ and post‐challenge. Absolute mean deviation percentages (|%|) of all healthy (top) and asthmatic (bottom) participants. Only sensors with significant differences (also after FDR adjustment) between the pre‐ (blue) and post‐challenge (orange) phase, for either one or both groups, are shown. Each dot represents the personal mean deviation pre‐ or post‐challenge, connected by a line, to visualize individual differences between the two study phases. Note that scales differ between sensors, but not between groups.
TABLE 2.
Area under the ROC curves for discrimination between study phase and groups
| Sensor |
Healthy Pre vs. post |
Asthma Pre vs. post |
Pre Healthy vs. asthma |
Post Healthy vs. asthma |
|---|---|---|---|---|
| 1 | 0.67 (CI: 0.44–0.90) | 0.76 (CI: 0.55–0.96) | 0.82 (CI: 0.65–0.99) | 0.54 (CI: 0.27–0.81) |
| 3 | 0.69 (CI: 0.46–0.92) | 0.51 (CI: 0.26–0.76) | 0.63 (CI: 0.39–0.87) | 0.74 (CI: 0.52–0.95) |
| 4 | 0.64 (CI: 0.40–0.88) | 0.67 (CI: 0.43–0.91) | 0.61 (CI: 0.37–0.85) | 0.52 (CI: 0.27–0.77) |
| 5 | 0.82 (CI: 0.65–0.99) | 0.97 (CI: 0.91–1.00) | 0.62 (CI: 0.39–0.86) | 0.94 (CI: 0.83–1.00) |
| 6 | 0.53 (CI: 0.28–0.78) | 0.77 (CI: 0.57–0.97) | 0.59 (CI: 0.34–0.84) | 0.81 (CI: 0.64–0.99) |
| 7 | 0.71 (CI: 0.49–0.92) | 0.56 (CI: 0.32–0.81) | 0.71 (CI: 0.49–0.93) | 0.88 (CI: 0.74–1.00) |
The area under receiver operating characteristic curve (AUROCC) for discrimination between pre‐ and post‐challenge, as well as healthy and asthma, using mean sensor deviations. Numbers in bold are AUROCC >0.80; CI, 95% confidence interval.
3.3.2. Healthy vs. asthma
When comparing the two groups, healthy controls had significantly larger deviations (8% ±6) than asthmatics (6% ±5) before the RV16 challenge in sensor 1 (p < .01) (Table S3), with an AUROCC of 0.82 (95% CI: 0.65–0.99) (Table 2). Post‐challenge, mean deviations were larger in asthmatics compared to healthy controls in sensor 5 (15% ±13 and 6% ±5, respectively; p < .001) and 6 (6% ±5 and 3% ±3, respectively; p < .01), and vice versa for sensor 7 (17% ±15 and 32% ±26, respectively; p < .01) (Table S3). The highest AUROCC for discrimination in post‐challenge eNose fluctuations between asthma and healthy was 0.94 (95% CI: 0.83–1.00) based on sensor 5 (Table 2).
All differences remained (almost) statistically significant (q ≤ 0.051) after FDR adjustment; all p‐values and q‐values are shown in Table S4. Individual absolute deviations for all visits and each sensor are depicted in Figures S6–S8. Pearson correlations between eNose sensor peak values are listed in Table S5 and show a strong correlation between sensor 5 and 6 (R = 0.91) due to the cross‐reactive nature of the sensors.
3.4. Linking change in eNose deviations to inflammatory markers and symptoms
The total change in absolute eNose sensor deviations (ie, the change in mean deviations summed up for all sensors, per subject) was slightly larger in asthma (20.4%, IQR 7.3–23.5) than in healthy (11.7%, IQR 7.3–21.7), although not statistically significant (p = .48) (Figure S9). Only for asthma, this total change in deviations moderately correlated (R>≈0.50) with four cytokines (Table 3); an increase in eNose fluctuations was inversely correlated with pre‐ and post‐challenge IL‐8 levels (pre: ρ = −.50, p = .10; post: ρ = −.60, p < .05) and post‐challenge IL‐1β (ρ = −.49, p = .11), IL‐17A (ρ = −.49, p = .11), and TNF‐α (ρ = −.55; p = .07) levels. For all outcomes (ie, FeNO, WURSS‐21, and cytokines) in healthy and asthma (except for the four previously mentioned cytokines), the absolute correlation coefficients were <.49 and p‐values ≥.14 (Table 3).
TABLE 3.
Correlation between total change in eNose fluctuations and symptoms or inflammation
| Healthy | Asthma | |||||||
|---|---|---|---|---|---|---|---|---|
| Pre | Post | Pre | Post | |||||
| ρ (95% CI) | p‐value | ρ (95% CI) | p‐value | ρ (95% CI) | p‐value | ρ (95% CI) | p‐value | |
| FeNO | −.16 (−0.64–0.47) | .62 | .20 (−0.52–0.73) | .53 | .00 (−0.73–0.61) | 1.00 | .03 (−0.72–0.61) | .94 |
| WURSS−21 | ‐ | ‐ | .41 (−0.23–0.79) | .18 | ‐ | ‐ | −.14 (−0.72–0.55) | .67 |
| IFN‐γ | −.15 (−0.71–0.52) | .65 | .19 (−0.45–0.76) | .56 | .12 (−0.59–0.68) | .70 | −.03 (−0.66–0.56) | .92 |
| IL−1β | .17 (−0.54–0.75) | .60 | .41 (−0.27–0.87) | .18 | −.36 (−0.87–0.37) | .26 | −.49 (−0.89–0.21) | .11 |
| IL−10 | −.13 (−0.77–0.63) | .70 | .30 (−0.41–0.83) | .34 | −.28 (−0.73–0.30) | .38 | −.22 (−0.77–0.42) | .48 |
| IL−13 | .26 (−0.40–0.73) | .42 | .34 (−0.41–0.87) | .28 | −.20 (−0.77–0.48) | .54 | −.49 (−0.82–0.12) | .11 |
| IL−17A | .06 (−0.55–0.62) | .85 | .14 (−0.53–0.72) | .67 | −.37 (−0.89–0.33) | .24 | −.45 (−0.87–0.18) | .14 |
| IL−33 | −.11 (−0.69–0.57) | .73 | −.05 (−0.61–0.51) | .89 | .00 (−0.70–0.66) | 1.00 | .01 (−0.67–0.80) | .99 |
| IL−6 | .33 (−0.32–0.82) | .30 | .05 (−0.57–0.68) | .89 | −.26 (−0.81–0.44) | .42 | −.41 (−0.76–0.24) | .18 |
| IL−8 | .29 (−0.40–0.80) | .37 | .29 (−0.33–0.81) | .37 | −.50 (−0.98–0.10) | .10 | −.60 (−0.89–−0.03) | .04 |
| IP−10 | −.38 (−0.93–0.41) | .22 | −.01 (−0.60–0.58) | .97 | −.19 (−0.80–0.43) | .56 | −.36 (−0.89–0.38) | .26 |
| TNF‐α | .22 (−0.45–0.73) | .50 | .25 (−0.41–0.83) | .43 | −.45 (−0.90–0.23) | .14 | −.55 (−0.92–0.01) | .07 |
For FeNO, WURSS‐21, and the cytokine levels, individual means were calculated based on 10 days pre‐ or post‐challenge; WURSS‐21 scores were only available post‐challenge. Next, Spearman's rank correlation coefficients (ρ) and bootstrapped (n = 1000) 95% confidence intervals (CI) were calculated between the total change in eNose fluctuations and FeNO, WURSS‐21, or cytokines in nasal lavage. Results with p‐values <.05 and/or ρ ≥ .50 are marked in bold.
Abbreviations: FeNO, fractional exhaled nitric oxide; WURSS, Wisconsin Upper Respiratory Symptom Survey.
4. DISCUSSION
In this prospective 90‐day follow‐up study, day‐to‐day fluctuations in exhaled breath signals rapidly increased after a RV16 challenge, in both non‐atopic healthy and atopic asthmatic volunteers. We could distinguish between a stable and relatively unstable period with high accuracy, reaching a maximal AUROCC of 0.97 in asthmatics and 0.82 in healthy controls. Asthmatics with a relatively larger increase in eNose fluctuations had (toward) significantly lower IL‐1β and IL‐17A levels (pre‐ and post‐challenge) and a trend toward lower IL‐8 and TNF‐α levels (pre‐challenge), possibly due to differences in biological processes between groups. In both groups, the change in eNose fluctuations due to RV16 challenge did not correlate with FeNO and cold‐like symptoms (WURSS‐21).
To our knowledge, we are the first to examine fluctuations in exhaled breath profiles in such an extensive follow‐up study, additionally with a highly controlled exposure, in a well‐defined study cohort with cases and controls. Our study complements the study of Brinkman et al,13 in which unstable (loss of control induced by steroid withdrawal) and stable (baseline and recovery) asthma periods were correctly classified with 86–95% accuracy, using breath analysis detected by an eNose platform. However, in our study, we did not provoke loss of control, so a direct comparison cannot be made. Our study extends the study of Brinkman et al, as we had a controlled exposure and included more (frequent) time‐points (days vs. weeks/months) designed exactly around the viral inoculation event. Moreover, we have improved the accuracy for discriminating pre‐ and post‐challenge phases evidently, when compared to our previous study by Abdel‐Aziz et al.17 In that study, although not the main focus, the discrimination between pre‐ and post‐challenge only reached an AUROCC of 0.79 in asthma and 0.76 in healthy, using single time‐point comparisons and “raw” sensor peak values. The improved discriminative accuracy of our current analysis shows the added value of considering all time‐points and personal baseline sensor values.
As we had hypothesized, fluctuations in the eNose signals existed regardless of the exposure27, 28, 29 and increased after the RV exposure. Fluctuations play an important role in the adaptive capacity of physiological systems to respond to a changing environment and can be too rigid or overly unstable in asthma.30, 31 Analysis of lung function, nasal eosinophils and neutrophils, and FeNO from the same study as ours, showed that the adaptive capacity was lower in asthma,18 possibly explaining the larger increase in eNose fluctuations after the RV16 challenge in asthmatics compared to healthy controls.
Secondly, the increase in eNose fluctuations after the RV16 challenge was detected by different sensors between the two groups. One explanation could be the cross‐reactivity of the sensors, meaning a compound can be detected by several sensors and vice versa, which makes it possible that the same or similar compounds were involved between groups. Another explanation could be differences in (patho)physiological mechanisms between healthy and asthmatic participants that become more prominent when local cells are activated (eg, by viral infection). There are several indications that metabolic activity in local cell asthma differs from that in healthy individuals.32 These metabolites are often of low molecular weight that can be detected by eNose technology. It is intriguing that we did find moderate inverse correlations between the magnitude of the increase in eNose fluctuations and the cytokine levels of IL‐1β, IL‐17A, IL‐8, and TNF‐α, and not for any other mediators. As macrophages are abundantly present in the nasal compartment,33 and M1‐like macrophages are activated by RVs34at an early stage, it is likely that M1‐like macrophages gave rise to these cytokine levels, which can lead to neutrophilic inflammation.35 Allergic asthmatics have reduced numbers of M1‐like macrophages during RV‐induced exacerbations,36 which may explain the differences in eNose fluctuations. In addition, airway epithelial cells are activated at an early stage by RVs and give rise to mediators like IL‐1β and IL‐8.37, 38 Together, this suggests that the eNose may detect RV‐induced metabolic changes.
Finally, we showed that the change in eNose fluctuations was not correlated with FeNO or WURSS‐21 scores. It has been shown before that exhaled breath profiles or VOCs minimally correlate with FeNO.13, 39, 40 This could possibly be explained by eNose signals representing a composite interplay of multiple molecular constituents, making it multidimensional as compared to FeNO. Regarding symptoms, a study by van der Schee et al.41 showed differences in VOC profiles between wheezing and asymptomatic children, regardless of the presence (AUROCC 0.77) or absence (AUROCC 0.81) of a RV. This distinction remained accurate even after symptoms recovered in the RV‐positive group (AUROCC 0.84), but not as accurate in the RV‐negative group (AUROCC 0.67), illustrating how breath profiles may reflect complex inflammatory processes and/or (pre‐existing) biological host‐response differences regardless of symptoms.
The first strength of this carefully designed study was the controlled RV exposure and the long follow‐up period of 3 months, with multiple measurements per week, throughout the study. On top of that, we included both healthy and asthmatic volunteers, allowing for investigation of differences in response to the RV16 challenge between a diseased and control group. Secondly, we examined fluctuations at individual level first, using personal baselines, before looking at group averages. This reduced the issue of averaging out individual effects and enabled us to compare normal day‐to‐day fluctuations with that when triggered with a perturbation. Finally, we made steps toward clinical applicability by using a non‐invasive and real‐time method, namely eNose technology, for detection of the exhaled breath profiles.
This study also has a few limitations. First of all, the RV16 exposure induced cold‐like symptoms, but no loss of control or exacerbation in asthma. This was likely due to our choice to investigate mild asthmatics and a relatively mild RV strain, a choice driven by ethical concerns. Nevertheless, the eNose was capable to detect differences in breath profile fluctuations between the pre‐ and post‐challenge phases, as well as, between groups. Secondly, exhaled breath profiles detected by eNose technology cannot be directly linked to (patho)physiological pathways, as eNose sensors cannot detect and identify single VOCs, due to their cross‐reactivity. However, the eNose possibly did reflect macrophage activity or downstream interacting processes. On top of that, it is not easy to delineate differences between groups that arise from disease or atopy, as we included non‐atopic healthy and atopic asthmatic volunteers. Nevertheless, the majority of asthmatic subjects in reality also suffer from atopy and hence our study resembled a real‐life scenario,42 in which unknown allergic exposures may have influenced the eNose fluctuations. Even so, it would still be of interest to study eNose fluctuations in atopic healthy and non‐atopic asthmatics as well, also with respect to the generalizability of our results. Our method of fluctuation analysis (ie, summarizing all deviations per study phase, at individual and group level) may have caused a loss of temporal and quantitative information on day‐to‐day fluctuations. Furthermore, we did not perform internal or external validation due to the limited sample size and the unavailability of comparable data matching the sampling frequency to this intensive study. However, adding multi‐omics’ analyses offsets the requirement for a large sample size,25 although we admit that it does not exclude the possible risk of overfitting (ie, unrepresentative AUROCCs).43, 44 Consequently, we believe that our data merit a prospective longitudinal study with a larger cohort, preferably in a real‐life setting, including daily home monitoring of exhaled breath during stable and unstable periods of asthma. Finally, a sample size estimation was not possible due to the unknown effect sizes of the novel VOC markers, which was compensated by unprecedented high sampling frequency in individuals.
Although these results cannot be directly applied in clinical practice in the current form, they show that exhaled breath analysis could potentially be a useful and additional tool in monitoring disease instability, as it captures a more comprehensive biomarker signal than that merely captured by FeNO and clinical symptoms (WURSS‐21). In addition, the increase in eNose fluctuations appeared to start before the onset of cold‐like symptoms in asthma, supporting its potential for patient management at the point‐of‐care. A better understanding of normal and protective homeokinesis vs. diseased and damaging fluctuations is required, to discover how treatment could be guided and the development of exacerbations could be limited, in asthmatic patients. Non‐invasive monitoring of exhaled markers using eNose technology can play an important role in this.
5. CONCLUSION
Day‐to‐day fluctuations in exhaled breath profiles rapidly increased after a RV16 challenge with distinct differences between non‐atopic healthy and atopic asthmatic volunteers. The increase in fluctuations did not seem to correlate with cold‐like symptoms and FeNO, but slightly with some of the investigated pro‐inflammatory biomarkers, making it a complementary tool for investigation of disease stability monitoring and possibly treatment adjustment purposes, in a non‐invasive manner. Our data justify the design of a longitudinal home monitoring study in a real‐life setting and larger population.
CONFLICT OF INTEREST
Ariana Lammers has nothing to disclose; Paul Brinkman has nothing to disclose; Louwrina H. te Nijenhuis has nothing to disclose; Rianne de Vries reports personal fees and other from Breathomix BV, outside the submitted work; Yennece W.F. Dagelet has nothing to disclose; Erik Duijvelaar has nothing to disclose; Binbin Xu has nothing to disclose; Mahmoud I. Abdel‐Aziz has nothing to disclose; Susanne J. Vijverberg has nothing to disclose; Anne H. Neerincx has nothing to disclose; Urs Frey has nothing to disclose; Rene Lutter reports grants from European Respiratory Society/Marie Curie (EU), during the conduct of the study; Anke H. Maitland ‐ van der Zee reports grants from GSK, grants from Boehringer Ingelheim, grants from Chiesi, personal fees from Astra Zeneca, personal fees from Boehringer Ingelheim, grants from Vertex, outside the submitted work; Peter J. Sterk reports other from being Scientific Advisor and having an officially non‐substantial interest in the start‐up company Breathomix BV that produces eNoses and accompanying cloud algorithms, during the conduct of the study; and Anirban Sinha has nothing to disclose.
Supporting information
App S1
ACKNOWLEDGEMENTS
The salary of Anirban Sinha was sponsored by the European Respiratory Society‐Marie Sklodowska Curieactions COFUND RESPIRE two fellowships (MCF‐7077–2014) and also from grants supported by Swiss Lung Association(2017_14) and Swiss Lung Foundation. The work was supported by an unrestricted grant from Chiesi Pharmaceuticals, institutional funding from the Amsterdam UMC location AMC, Amsterdam UMC, University of Amsterdam (IA601011). The authors would like to thank all the participants for their time and dedication to the study.
REFERENCES
- 1.Reddel HK, Taylor DR, Bateman ED, et al. An official American Thoracic Society/European Respiratory Society statement: asthma control and exacerbations ‐ Standardizing endpoints for clinical asthma trials and clinical practice. Am J Respir Crit Care Med. 2009;180(1):59‐99. 10.1164/rccm.200801-060ST [DOI] [PubMed] [Google Scholar]
- 2.Suruki RY, Daugherty JB, Boudiaf N, Albers FC. The frequency of asthma exacerbations and healthcare utilization in patients with asthma from the UK and USA. BMC Pulm Med. 2017;17:74. 10.1186/s12890-017-0409-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnston NW, Johnston SL, Duncan JM, et al. The September epidemic of asthma exacerbations in children: a search for etiology. J Allergy Clin Immunol. 2005;115(1):132‐138. 10.1016/j.jaci.2004.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnston SL, Pattemore PK, Sanderson G, et al. Community study of role of viral infections in exacerbations of asthma in 9–11 year old children. BMJ. 1995;310(6989):1225‐1229. 10.1136/bmj.310.6989.1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnston NW, Sears MR. Asthma exacerbations · 1: epidemiology. Thorax. 2006;(61):722‐728. 10.1136/thx.2005.045161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nicholson KG, Kent J, Ireland DC. Respiratory viruses and exacerbations of asthma in adults. Br Med J. 1993;307(6910):982‐986. 10.1136/bmj.307.6910.982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haldar P, Pavord ID, Shaw DE, et al. Cluster analysis and clinical asthma phenotypes. Am J Respir Crit Care Med. 2008;178(3):218‐224. 10.1164/rccm.200711-1754OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haselkorn T, Fish JE, Zeiger RS, et al. Consistently very poorly controlled asthma, as defined by the impairment domain of the expert panel report 3 guidelines, increases risk for future severe asthma exacerbations in the epidemiology and natural history of asthma: outcomes and treatment regime. J Allergy Clin Immunol. 2009; 124(5):895‐902. 10.1016/j.jaci.2009.07.035 [DOI] [PubMed] [Google Scholar]
- 9.Haselkorn T, Zeiger RS, Chipps BE, et al. Recent asthma exacerbations predict future exacerbations in children with severe or difficult‐to‐treat asthma. J Allergy Clin Immunol. 2009;124(5):921‐927. 10.1016/j.jaci.2009.09.006 [DOI] [PubMed] [Google Scholar]
- 10.Loymans RJB, Gemperli A, Cohen J, et al. Comparative effectiveness of long term drug treatment strategies to prevent asthma exacerbations: network meta‐analysis. BMJ. 2014;348(may13 3):g3009. 10.1136/bmj.g3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brinkman P, Wagener AH, Hekking PP, et al. Identification and prospective stability of electronic nose (eNose)–derived inflammatory phenotypes in patients with severe asthma. J Allergy Clin Immunol. 2019;143(5):1811. 10.1016/j.jaci.2018.10.058 [DOI] [PubMed] [Google Scholar]
- 12.Van Der Schee MP, Paff T, Brinkman P, Van Aalderen WMC, Haarman EG, Sterk PJ. Breathomics in lung disease. Chest. 2015;147(1):224‐231. 10.1378/chest.14-0781 [DOI] [PubMed] [Google Scholar]
- 13.Brinkman P, van de Pol MA, Gerritsen MG, et al. Exhaled breath profiles in the monitoring of loss of control and clinical recovery in asthma. Clin Exp Allergy. 2017;47(9):1159‐1169. 10.1111/cea.12965 [DOI] [PubMed] [Google Scholar]
- 14.Fens N, Van De Pol MA, Brinkman P, et al. Repeated exhaled breath profiling by electronic noses identifies asthma exacerbations. Am J Respir Crit Care Med. 2014;A4247. [Google Scholar]
- 15.Robroeks CM, van Berkel JJ, Jöbsis Q, et al. Exhaled volatile organic compounds predict exacerbations of childhood asthma in a 1‐year prospective study. Eur Respir J. 2013;42(1):98‐106. 10.1183/09031936.00010712 [DOI] [PubMed] [Google Scholar]
- 16.van Vliet D, Smolinska A, Jöbsis Q, et al. Can exhaled volatile organic compounds predict asthma exacerbations in children? J Breath Res. 2017;11(1):16016. 10.1088/1752-7163/aa5a8b [DOI] [PubMed] [Google Scholar]
- 17.Abdel‐Aziz MI, de Vries R, Lammers A, et al. Cross‐sectional biomarker comparisons in asthma monitoring using a longitudinal design: the eNose premise. Allergy. 2020;75(10):2690‐2693. 10.1111/all.14354 [DOI] [PubMed] [Google Scholar]
- 18.Sinha A, Lutter R, Xu B, et al. Loss of adaptive capacity in asthmatic patients revealed by biomarker fluctuation dynamics after rhinovirus challenge. Elife. 2019;8:e47969. 10.7554/eLife.47969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Der SKFV, De PMAV, Kulik W, et al. Systemic tryptophan and kynurenine catabolite levels relate to severity of rhinovirus‐induced asthma exacerbation: a prospective study with a parallel‐group design. Thorax. 2013;68(12):1122‐1130. 10.1136/thoraxjnl-2013-203728 [DOI] [PubMed] [Google Scholar]
- 20.De Vries R, Dagelet YWF, Spoor P, et al. Clinical and inflammatory phenotyping by breathomics in chronic airway diseases irrespective of the diagnostic label. Eur Respir J. 2018;51(1):1701817. 10.1183/13993003.01817-2017 [DOI] [PubMed] [Google Scholar]
- 21.Juniper EF, O’Byrne PM, Ferrie PJ, King DR, Roberts JN. Measuring asthma control: clinic questionnaire or daily diary? Am J Respir Crit Care Med. 2000;162(4):1330‐1334. 10.1164/ajrccm.162.4.9912138 [DOI] [PubMed] [Google Scholar]
- 22.Dweik RA, Boggs PB, Erzurum SC, et al. An official ATS clinical practice guideline: interpretation of exhaled nitric oxide levels (FENO) for clinical applications. Am J Respir Crit Care Med. 2011;184(5):602‐615. 10.1164/rccm.9120-11ST [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turner C, Španěl P, Smith D. A longitudinal study of ethanol and acetaldehyde in the exhaled breath of healthy volunteers using selected‐ion flow‐tube mass spectrometry. Rapid Commun Mass Spectrom. 2006;20(1):61‐68. 10.1002/rcm.2275 [DOI] [PubMed] [Google Scholar]
- 24.Turner C, Španěl P, Smith D. A longitudinal study of breath isoprene in healthy volunteers using selected ion flow tube mass spectrometry (SIFT‐MS). Physiol Meas. 2006;27(1):13‐22. 10.1088/0967-3334/27/1/002 [DOI] [PubMed] [Google Scholar]
- 25.Li C‐X, Wheelock CE, Sköld CM, Wheelock ÅM. Integration of multi‐omics datasets enables molecular classification of COPD. Eur Respir J. 2018;51(5):1701930. 10.1183/13993003.01930-2017 [DOI] [PubMed] [Google Scholar]
- 26.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57(1):289‐300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- 27.Que CL, Kenyon CM, Olivenstein R, Macklem PT, Maksym GN. Homeokinesis and short‐term variability of human airway caliber. J Appl Physiol. 2001;91(3):1131‐1141. 10.1152/jappl.2001.91.3.1131 [DOI] [PubMed] [Google Scholar]
- 28.Tirone TA, Brunicardi FC. Overview of glucose regulation. World J Surg. 2001;25(4):461‐467. 10.1007/s002680020338 [DOI] [PubMed] [Google Scholar]
- 29.Palmer BF, Clegg DJ. Physiology and pathophysiology of potassium homeostasis. Adv Physiol Educ. 2016;40(4):480‐490. 10.1152/advan.00121.2016 [DOI] [PubMed] [Google Scholar]
- 30.Frey U, Maksym G, Suki B. Temporal complexity in clinical manifestations of lung disease. J Appl Physiol. 2011;110(6):1723‐1731. 10.1152/japplphysiol.01297.2010 [DOI] [PubMed] [Google Scholar]
- 31.Goldberger AL, Amaral LAN, Hausdorff JM, Ivanov PC, Peng CK, Stanley HE. Fractal dynamics in physiology: alterations with disease and aging. Proc Natl Acad Sci USA. 2002;99(Supplement 1):2466‐2472. 10.1073/pnas.012579499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michaeloudes C, Bhavsar PK, Mumby S, et al. Role of metabolic reprogramming in pulmonary innate immunity and its impact on lung diseases. J Innate Immun. 2020;12(1):31‐46. 10.1159/000504344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryu G, Bae JS, Kim JH, et al. Role of IL‐17A in chronic rhinosinusitis with nasal polyp. Allergy Asthma Immunol Res. 2020;12(3):507‐522. 10.4168/aair.2020.12.3.507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rajput C, Walsh MP, Eder BN, Metitiri EE, Popova AP, Hershenson MB. Rhinovirus infection induces distinct transcriptome profiles in polarized human macrophages. Physiol Genomics. 2018;50(5):299‐312. 10.1152/physiolgenomics.00122.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bullone M, Carriero V, Bertolini F, et al. Elevated serum IgE, oral corticosteroid dependence and IL‐17/22 expression in highly neutrophilic asthma. Eur Respir J. 2019;54(5):1900068. 10.1183/13993003.00068-2019 [DOI] [PubMed] [Google Scholar]
- 36.Nikonova A, Khaitov M, Jackson DJ, et al. M1‐like macrophages are potent producers of anti‐viral interferons and M1‐associated marker‐positive lung macrophages are decreased during rhinovirus‐induced asthma exacerbations. EBioMedicine. 2020;54:102734. 10.1016/j.ebiom.2020.102734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piper SC, Ferguson J, Kay L, et al. The role of interleukin‐1 and interleukin‐18 in pro‐inflammatory and anti‐viral responses to rhinovirus in primary bronchial epithelial cells. PLoS One. 2013;8(5):e63365. 10.1371/journal.pone.0063365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamaya M, Nomura K, Arakawa K, et al. Clarithromycin decreases rhinovirus replication and cytokine production in nasal epithelial cells from subjects with bronchial asthma: effects on IL‐6, IL‐8 and IL‐33. Arch Pharm Res. 2020;43(5):526‐539. 10.1007/s12272-017-0950-x [DOI] [PubMed] [Google Scholar]
- 39.Van Vliet D, Smolinska A, Jöbsis Q, et al. Association between exhaled inflammatory markers and asthma control in children. J Breath Res. 2016;10(1):16014. 10.1088/1752-7155/10/1/016014 [DOI] [PubMed] [Google Scholar]
- 40.Van der Schee MP, Palmay R, Cowan JO, Taylor DR. Predicting steroid responsiveness in patients with asthma using exhaled breath profiling. Clin Exp Allergy. 2013;43(11):1217‐1225. 10.1111/cea.12147 [DOI] [PubMed] [Google Scholar]
- 41.Van Der Schee MP, Hashimoto S, Schuurman AC, et al. Altered exhaled biomarker profiles in children during and after rhinovirusinduced wheeze. Eur Respir J. 2015;45(2):440‐448. 10.1183/09031936.00044414 [DOI] [PubMed] [Google Scholar]
- 42.Bourdin A, Gras D, Vachier I, Chanez P. Upper airway · 1: Allergic rhinitis and asthma: United disease through epithelial cells. Thorax. 2009;64:999‐1004. 10.1136/thx.2008.112862 [DOI] [PubMed] [Google Scholar]
- 43.Azim A, Barber C, Dennison P, Riley J, Howarth P. Exhaled volatile organic compounds in adult asthma: a systematic review. Eur Respir J. 2019;54(3):1900056. 10.1183/13993003.00056-2019 [DOI] [PubMed] [Google Scholar]
- 44.Sola Martínez RA, Pastor Hernández JM, Yanes Torrado Ó, Cánovas Díaz M, de Diego PT, Vinaixa CM. Exhaled volatile organic compounds analysis in clinical pediatrics: a systematic review. Pediatr Res. 2021;89(6):1352‐1363. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
App S1


