Abstract
Early case detection and isolation of infected individuals are critical to controlling coronavirus disease 2019 (COVID-19). Reverse transcription polymerase chain reaction (RT-PCR) is considered the gold standard for the diagnosis of severe acute respiratory syndrome coronavirus 2 infection, but false negatives do occur. We built a user-friendly online tool to estimate the probability of having COVID-19 with negative RT-PCR results and thus avoid preventable transmission.
Keywords: Bayesian, COVID, 19, online tool, RT-PCR test, SARS-CoV-2
The coronavirus disease 2019 (COVID-19) pandemic has imposed a catastrophic toll worldwide, with about 130 million reported cases and 2.8 million reported deaths as of March 30, 2021 [1]. Despite the historical development and approval of several effective COVID-19 vaccines [2], epidemic control in most countries still critically depends on nonpharmaceutical interventions, such as social distancing and early detection and effective quarantine of infected individuals [3]. Community- and travel-related severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) transmission risk will remain important until a large proportion of the population is immunized.
Compared with other viruses, SARS-CoV-2, the virus that causes COVID-19 disease, has 2 characteristics that make early case detection and quarantine of infected individuals particularly critical for epidemic control. First, the virus is an efficient spreader, with an average of about 2.5 secondary infections caused by a single infected individual in a susceptible population [4]. Second, the highest risk of transmission occurs very early in the disease, before or within the first days of symptom onset [5, 6]. These characteristics hinder transmission control because early case detection and effective quarantine are challenging. Thus far, COVID-19 surveillance has been primarily based on reverse transcription polymerase chain reaction (RT-PCR) assays, which are considered the most reliable tests for diagnosing SARS-CoV-2 infection [7, 8]. However, RT-PCR positivity varies among infected patients depending on, for example, the timing of sample collection in relation to symptom onset or the sampling technique used (eg, nasopharyngeal swabs, sputum, bronchoalveolar lavage) [9, 10].
This lack of a clear-cut result presents a non-negligible challenge in the workplace and health care settings because a negative RT-PCR should be interpreted as a person being less likely to be infected. Training and experience in clinical epidemiological reasoning may help health care workers make safer decisions and recommendations. A negative RT-PCR result may be complemented in such settings, for example, with chest radiographs, repeat swabs, tomography scans, and or a patient’s contact history [10]. However, RT-PCR testing strategies are often used in community settings where false-negative results may be interpreted at face value—for example, to clear individuals for work in industry, attend college, or in pretravel evaluations [11], increasing the risk of preventable transmission of the virus.
We addressed this problem by designing a readily available, easy-to-use online tool to estimate the probability that an individual is infected with SARS-CoV-2 conditional on having 1 or more negative RT-PCR test results. The tool helps individuals interpret test results by incorporating information about the person and the test. Specifically, we use the pretest probability (anchor) of the person being infected and adjust this probability with the information provided by the test and the time elapsed since the onset of symptoms. Our tool, based mainly on the work by Kucirka et al. [12], requires users to first choose between 1, 2, or 3 consecutive negative RT-PCR tests. Second, users need to provide (i) the estimated probability of being positive before taking the RT-PCR diagnostic test (range, 0 to 1), and (ii) the user provides the number of days elapsed between the first symptoms and the first, second, or third RT-PCR test (range, –4 to 16 days).
METHODS
We built upon Kucirka et al.’s work [12] and adjusted a hierarchical logistic model to estimate the rate of false negatives for different moments in time from the onset of symptoms. We implemented the model using JAGS and the RJAGS library in R [13]. We generated a Markov chain of 420 000 samples; the first 20 000 samples were discarded, and the rest were resampled to generate a subchain of size 20 000. In contrast to Kucirka et al. [12], we estimated a non-study-specific marginal rate of false-negative RT-PCR tests. Our online tool assumes a specificity of 1 of the RT-PCR tests for detecting SARS-CoV-2 and independence of the results of different tests when considering >1 test (technical details are shown in Section 1 of the Supplementary Data and Supplementary Figure 1).
To help potential users, our tool also provides an empirical estimate of the pretest probability of SARS-CoV-2 infection. This estimate, including uncertainty, is based on a real-world sample of 926 individuals with suspected SARS-CoV-2 infection or close contact with a laboratory-confirmed COVID-19 patient from 5 medical centers in Santiago, Chile [14]. Participants responded to a brief questionnaire of COVID-19-related signs and symptoms (eg, fever, cough, rhinitis, breathing difficulty, muscular pain) and then got tested for Covid-19 using an RT-PCR assay. Based on these data, we used a Bayesian logistic regression model, along with a classification tree, to estimate the pretest probability based on the available covariates (Supplementary Data, Supplementary Figure 2). We generated a Markov chain of 420 000 samples; the first 20 000 samples were discarded, and the rest were resampled to generate a subchain of size 20 000. Users of our tool can consider this estimate their pretest probability or adjust it based on other criteria (eg, results from a chest radiograph). We provide this data set as an example, but the pretest probability can be easily estimated for a different population (Supplementary Data, Section 2).
RESULTS AND DISCUSSION
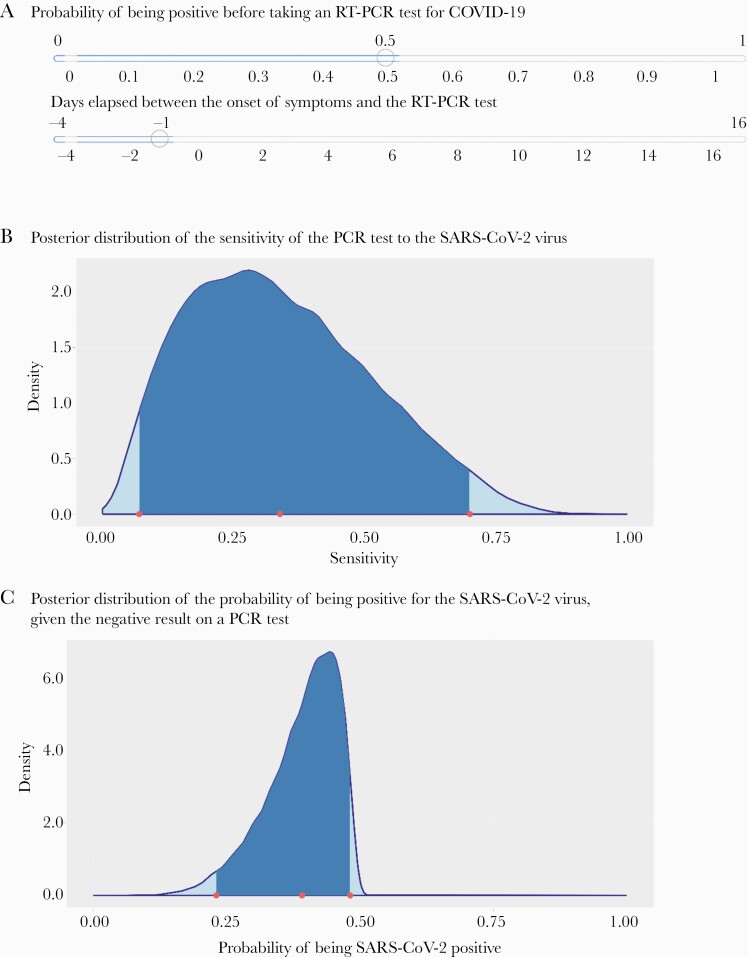
Our online tool enables users to estimate the probability of having COVID-19 with 1 or more negative RT-PCR results. The tool is available in English and Spanish at https://midas-uc.shinyapps.io/Calculadora-COVID19/. To show the tool’s utility, we input data from a health care worker in a hospital setting in Chile who was not working with COVID-19 patients. The worker was inadvertently exposed to SARS-CoV-2 by an asymptomatic patient who later developed symptoms and had confirmed COVID-19. The worker had a negative nasopharyngeal swab RT-PCR result 3 days after exposure. However, she reported symptoms the day after the first negative RT-PCR and had a second RT-PCR test that showed positive results (Supplementary Data). Because she was exposed for a relatively long period, we used a 50% pretest probability of infection (Figure 1).
Figure 1.
Illustrative results from the tool to estimate the probability of having COVID-19 with 1 negative RT-PCR tool. A, The worker reported symptoms the day after the first negative RT-PCR; because she was exposed for a relatively long period of time, we used a 50% pretest probability of infection. B, The posterior distribution of the sensitivity of the PCR test to the SARS-CoV-2 virus. C, The posterior distribution of the probability of being positive for the SARS-CoV-2 virus, given the negative result on a PCR test. Abbreviations: COVID-19, coronavirus disease 2019; PCR, polymerase chain reaction; RT-PCR, reverse transcription polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
One of the main strengths of our tool is its relative simplicity. However, the tool does not include all factors that may affect the RT-PCR-negative predictive value. For instance, clinicians often estimate the pretest probability of disease (anchor) based on several criteria such as local disease prevalence, occupational risk, alternative diagnoses, contact history, setting (primary or secondary health care), and other diagnostic tools such as a chest radiograph, then adjust this probability based on RT-PCR test results (post-test probability). Therefore, the negative predictive value of the RT-PCR test result may increase based on a clinician’s experience, good judgment, and available resources, which are not included in our model. Our tool is intended for use beyond clinical settings, such as in a mining operation where all workers are tested or in combination with other diagnostic strategies to reopen society [15]. To illustrate, we chose a random patient from our sample. This person had a negative RT-PCR result. We assumed that she and the lab would not have the skills or experience to estimate her pretest probability of SARS-CoV-2 infection. She reported having fever and cough and no other symptoms and had taken an RT-PCR 2 days after the onset of fever. Using the data-based pretest probability module in our tool, she would obtain a probability distribution with an estimated 56% pretest probability of infection (Supplementary Data, Section 3, and Supplementary Figure 3). The tool also shows the posterior distribution for the sensitivity of the RT-PCR test to the SARS-CoV-2 virus and the posterior distribution for the probability of being positive for the SARS-CoV-2 virus, given the negative result on 1 RT-PCR test (Supplementary Data).
One remaining question is how these results should be used. Specific recommendations for patients depend on the pretest probability of disease and the potential consequences of a false-negative diagnosis. In principle, we recommend that anyone with a clinical COVID-19 diagnosis isolate, despite having a negative RT-PCR result. This is more important for people working with vulnerable populations or who have frequent face-to-face interactions with others.
Accurate test results are critical to prevent onward SARS-CoV-2 transmission in community and hospital settings. As the subsequent waves of infection continue, straining health system capacity even further, essential workers and travelers with known exposures to the virus will continue to use RT-PCR tests to rule out SARS-CoV-2 infection. False-negative RT-PCR results are particularly problematic among health care and other essential workers such as firefighters and police, as well as travelers, who may inadvertently become superspreaders [16]. As social distancing measures are relaxed, RT-PCR false negatives also affect community control measures if individuals with suspected infection are cleared to return to work or travel. If false-negative RT-PCR results are treated as evidence of no infection, there is a non-negligible risk of preventable transmission of the virus.
We hope this publicly available online tool will help workers, travelers, and decision-makers avoid the preventable transmission of SARS-CoV-2 by making safer decisions.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
We thank Eduardo Agosín (Pontificia Universidad Católica de Chile), Pedro Saa (Pontificia Universidad Católica de Chile), the Millenium Institute Foundational Research on Data, and the KOR team for primary data collection and management.
Financial support. This work was supported by the Agencia Nacional de Investigación y Desarrollo (ANID) Millennium Science Initiative grants MIDAS NCN17_059 to A.J. and MICROB-R NCN17_081 to E.U., R.A.
Potential conflicts of interest. All authors: no conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Author contributions. A.J. and R.A. conceived the idea and analysis strategy. A.J. performed the computations and created the tool. E.U. wrote the first draft. A.J., E.U., and R.A. contributed to the final version of the manuscript and figures.
Data availability. The tool is available in English and Spanish at https://midas-uc.shinyapps.io/Calculadora-COVID19/. Data used to estimate the pretest probability are available at 10.17632/z4ktvcwfp6.2#file-c8232f24–5acf-4d93–84ef-ff82b0cc4310.
Patient consent. This study used anonymized secondary data from a sample of patients in 5 medical centers in Santiago, Chile. Primary data collection and study procedures, including obtaining the patients’ written consent, were approved by the Comité Científico de Ciencias de la Salud UC, Pontificia Universidad Catolica de Chile’ ethics committee.
References
- 1.Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis 2020; 20:533–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zimmer C, Corum J, Wee S-L. Coronavirus Vaccine Tracker. 2021. Available at: https://nyti.ms/3nvAEc4. Accessed 30 April 2021.
- 3.Li Y, Campbell H, Kulkarni D, et al. ; Usher Network for COVID-19 Evidence Reviews (UNCOVER) group. The temporal association of introducing and lifting non-pharmaceutical interventions with the time-varying reproduction number (R) of SARS-CoV-2: a modelling study across 131 countries. Lancet Infect Dis 2021; 21:193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu JT, Leung K, Leung GM. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet 2020; 395:689–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cevik M, Tate M, Lloyd O, et al. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe 2021; 2: e13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hauser A, Counotte MJ, Margossian CC, et al. Estimation of SARS-CoV-2 mortality during the early stages of an epidemic: a modeling study in Hubei, China, and six regions in Europe. PLoS Med 2020; 17:e1003189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiersinga WJ, Rhodes A, Cheng AC, et al. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA 2020; 324:782–93. [DOI] [PubMed] [Google Scholar]
- 8.Böger B, Fachi MM, Vilhena RO, de Fátima Cobre A, Tonin FS, Pontarolo R. Systematic review with meta-analysis of the accuracy of diagnostic tests for COVID-19. Am J Infect Control 2020; 49:21–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sethuraman N, Jeremiah SS, Ryo A. Interpreting diagnostic tests for SARS-CoV-2. JAMA 2020; 323:2249–51. [DOI] [PubMed] [Google Scholar]
- 10.Watson J, Whiting PF, Brush JE. Interpreting a COVID-19 test result. BMJ 2020; 369:m1808. [DOI] [PubMed] [Google Scholar]
- 11.Centers for Disease Control & Prevention. COVID-19. Testing and international air travel. 2021. Available at: https://www.cdc.gov/coronavirus/2019-ncov/travelers/testing-air-travel.html. Accessed 25 March 2021.
- 12.Kucirka LM, Lauer SA, Laeyendecker O, et al. Variation in false-negative rate of reverse transcriptase polymerase chain reaction-based SARS-CoV-2 tests by time since exposure. Ann Intern Med 2020; 173:262–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.R: A Language and Environment for Statistical Computing [Computer Program]. Vienna: R Foundation for Statistical Computing; 2019. [Google Scholar]
- 14.Eyheramendy S, Saa PA, Undurraga EA, et al. Improved screening of COVID-19 cases through a Bayesian network symptoms model and psychophysical olfactory test. medRxiv 2021.2001.2018.21249821 [Preprint]. 31 May 2021. Available at: 10.1101/2021.01.18.21249821. Accessed 31 July 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Service RF. Fast, cheap tests could enable safer reopening. Science 2020; 369:608–9. [DOI] [PubMed] [Google Scholar]
- 16.Chang S, Pierson E, Koh PW, et al. Mobility network models of COVID-19 explain inequities and inform reopening. Nature 2021; 589:82–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.