Abstract
Objectives
With the ongoing emergence of SARS‐CoV‐2 variants and potential to evade vaccine‐induced neutralisation, understanding the magnitude and breadth of vaccine‐induced T‐cell immunity will be critical for the ongoing optimisation of vaccine approaches. Strategies that provide a rapid and easily translatable means of assessing virus‐specific T‐cell responses provide an opportunity to monitor the impact of vaccine rollouts in the community. In this study, we assessed whether our recently developed SARS‐CoV‐2 whole‐blood assay could be used effectively to analyse T‐cell responses following vaccination.
Methods
Following a median of 15 days after the first dose of the ChAdOx1‐S (AstraZeneca®) vaccine, peripheral blood was isolated from 58 participants. Blood was incubated overnight with an overlapping set of spike protein peptides and assessed for cytokine production using a cytometric bead array.
Results
The majority of vaccine recipients (51/58) generated a T helper 1 response (IFN‐γ and/or IL‐2) following a single dose of ChAdOx1‐S. The magnitude of the IFN‐γ and IL‐2 response strongly correlated in vaccine recipients. While the production of other cytokines was evident in individuals who did not generate IFN‐γ and IL‐2, they showed no correlation in magnitude, nor did we see a correlation between sex or age and the magnitude of the response.
Conclusions
The whole‐blood cytokine assay provides a rapid approach to assessing T‐cell immunity against SARS‐CoV‐2 in vaccine recipients. While the majority of participants generated a robust SARS‐CoV‐2‐specific T‐cell response following their first dose, some did not, demonstrating the likely importance of the booster dose in improving T‐cell immunity.
Keywords: COVID‐19, SARS‐CoV‐2, T cells, vaccine
Strategies that provide a rapid and easily translatable means of assessing SARS‐CoV‐2‐specific T‐cell responses provide an opportunity to monitor the impact of vaccine rollouts in the community. In this study, we assessed whether our recently developed SARS‐CoV‐2 whole‐blood assay could be used effectively to analyse T‐cell responses following vaccination. We demonstrated that the whole‐blood cytokine assay provides a rapid approach to assessing T‐cell immunity against SARS‐CoV‐2 in vaccine recipients.

Introduction
The ongoing emergence of novel SARS‐CoV‐2 variants highlights the likely benefit of generating broad cellular and humoral immunity in response to vaccination.1, 2, 3 Memory T cells play a critical role in protection against all viral pathogens.4 They act directly by killing virally infected cells and also provide help for the maturation of B cells and their subsequent production of neutralising antibodies. Recent observations have shown that vaccine‐induced memory T cells retain the capacity to recognise novel SARS‐CoV‐2 variants.2 Thus, the coordinated action of memory T‐cell immunity will likely provide the most effective mechanism for controlling emerging variants with the capacity to evade neutralising antibody responses. While currently approved vaccines are known to induce T cells that specifically recognise the SARS‐CoV‐2 spike antigen, qualitative aspects of these T‐cell responses are not well defined, particularly in large cohort studies.5, 6 Understanding the magnitude and breadth of the vaccine‐induced memory T‐cell response will be critical for defining which subsets of the population display poor immunological memory and, as such, may be more susceptible upon exposure to viral variants.
Assessment of T‐cell immunity in humans typically relies on the isolation of peripheral blood mononuclear cells (PBMC) in order to assess antigen specificity and T‐cell function via intracellular cytokine analysis, ELISA or ELISPOT assays.7 While these assays have proven robust in the measurement of T‐cell immunity, the need to isolate PBMC limits the feasibility of these approaches for large‐scale, high‐throughput analysis. We recently demonstrated that a whole‐blood approach, eliminating the need for PBMC isolation, could be used to assess T‐cell responses in convalescent individuals previously exposed to SARS‐CoV‐2.8 This response showed a T helper (Th)‐1 bias, characterised by the production of interferon (IFN)‐γ and interleukin (IL)‐2 following stimulation with SARS‐CoV‐2 antigens. We have now used this assay to study SARS‐CoV‐2‐specific T‐cell responses following vaccination. In a cohort of 58 participants recruited a median of 15 days after they received the first dose of ChAdOx1‐S COVID‐19 vaccine, we demonstrate a similar Th1 bias, with significant increase in IFN‐γ and IL‐2 production in comparison with a cohort of 26 unvaccinated controls. While the majority of participants demonstrated a T‐cell response following this single dose of ChAdOx1‐S, a small percentage failed to generate either IFN‐γ or IL‐2.
Results
We previously reported the application of a whole‐blood assay to assess T‐cell responses in individuals who had recovered from SARS‐CoV‐2.8 To determine whether a similar approach could be used for the high‐throughput screening of vaccine recipients, we recruited a cohort of 58 recipients of the ChAdOx1‐S COVID‐19 vaccine 12–23 days after their first dose (PostVax). To assess vaccine‐induced T‐cell responses, peripheral blood was incubated overnight with overlapping peptides covering the SARS‐CoV‐2 spike antigen. Because of the large size of the spike protein, these overlapping peptides were divided into two separate pools herein referred to as spike pool 1 and spike pool 2. Plasma was harvested and cytokine production was determined using a cytometric bead array (CBA). To accommodate the potential impact of pre‐existing or cross‐reactive immune responses in the detection of spike‐specific immunity, we used a cohort of 26 non‐vaccinated and SARS‐CoV‐2 unexposed donors as a control (PreVax). Participant demographics are provided in Supplementary table 1. Similar to the observations in SARS‐CoV‐2‐recovered individuals, we saw a significant increase in the quantity of the two Th1 cytokines, IFN‐γ and IL‐2, produced in response to both spike peptide pools in the PostVax cohort (Figure 1a, b). The number of participants with a cytokine response < 1 pg mL−1 is provided in Supplementary table 2. We also noted a significant increase in the production of other cytokines measured in response to at least one of the peptide pools following vaccination. Pairwise analysis from five participants for whom we had both PreVax and PostVax samples confirmed an increase in cytokine production in 4 of 5 participants (Figure 1c, d). However, we also noted that the increase in cytokine production was not evident in all vaccine recipients, with some demonstrating no detectable change in their cytokine response.
Figure 1.
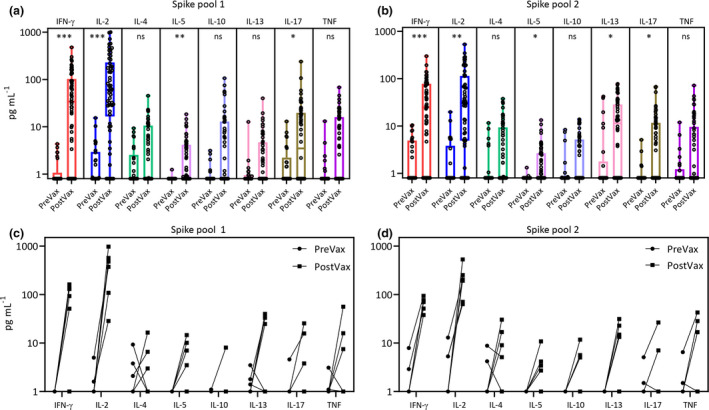
Spike‐specific cytokine detection following the first dose of ChAdOx1‐S. Box and whisker plots displaying cytokine responses to spike pool 1 (a) and spike pool 2 (b) comparing vaccinated (PostVax; n = 58 and unvaccinated individuals (PreVax, n = 26). Data represent values after subtraction of background cytokine levels following incubation of blood with no antigen. Pairwise cytokine analysis from seven participants with PreVax and PostVax samples is shown in response to spike pool 1 (c) and spike pool 2 (d). Statistical analysis using multiple t‐tests (n = 8) and corrected for multiple comparisons using the Holm–Sidak method was used to assess significant differences in cytokine production. Adjusted P‐values: *** < 0.005; ** 0.005–0.01; * 0.01–0.05.
To set a stringent cut‐off for the detection of a cytokine response, the maximum value detected in the PreVax cohort was used to determine a baseline for each cytokine. As such, PostVax participants were only considered positive if their cytokine response was greater than the established PreVax cut‐off. We then collated the data from both peptide pools to determine the number of participants with a positive cytokine response (Figure 2a). As anticipated, IFN‐γ and IL‐2 responses were the most frequently produced cytokines, detected in samples from 43 participants. Assessment of these cytokine profiles revealed that IFN‐γ and IL‐2 were produced by participants with strong SARS‐CoV‐2 immunity, while the production of other cytokines was variable (Figure 2b). Positive IFN‐γ or IL‐2 responses were detected in 51/58 (87.9%) samples, and both cytokines were detected in 37/58 (63.8%) samples. Of the remaining seven (12.1%) participants who did not express detectable IFN‐γ or IL‐2 following spike peptide stimulation, six showed detectable responses through other cytokines (IL‐4, IL‐5, IL‐10, IL‐13, IL‐17 or TNF), while one had no detectable cytokine responses after the first vaccine dose. To determine whether there was a correlation between the quantities of different cytokines, we compared the quantity of IFN‐γ with all other cytokines. Consistent with our published analysis in SARS‐CoV‐2‐recovered individuals, a strong correlation was evident between IFN‐γ and IL‐2 (Figure 2c). We also noted a weak correlation between IFN‐γ and IL‐5, and IFN‐γ and TNF (Supplementary figure 1). No correlation was evident between IFN‐γ and any of the remaining cytokines. The heat map in Figure 2d further emphasises the strong association between IFN‐γ and IL‐2 production, and no correlation between the levels of other cytokines.
Figure 2.
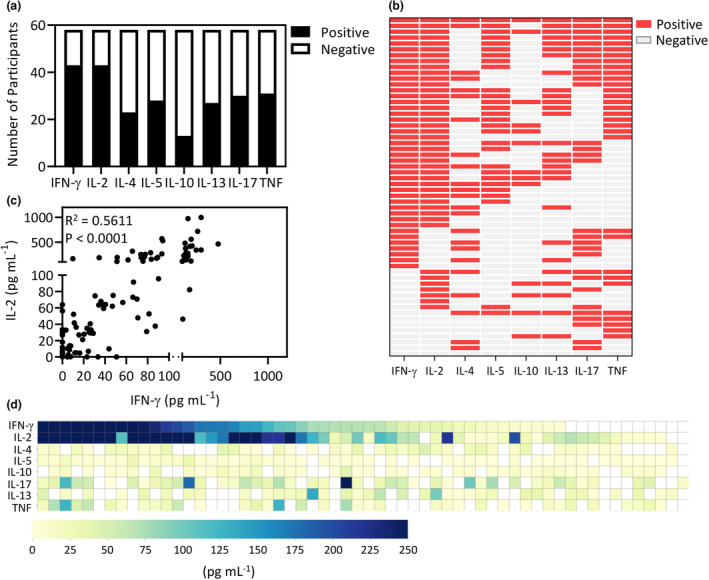
Proportional analysis of participants responding to either spike pool 1 or 2. (a) PostVax samples were considered cytokine positive if the response exceeded the maximum cytokine concentration from the PreVax cohort. Data represent the number of vaccinated participants generating a cytokine response to spike pool 1 and/or spike pool 2. (b) The heat map represents the pattern of cytokine responses from each vaccinated participant. Detected cytokines are represented by red boxes, while cytokines that were not detected are represented by grey boxes. (c) Data represent the concentration of IFN‐γ and IL‐2 from PostVax samples stimulated with either spike pool 1 or spike pool 2. Correlation analysis was performed using a Pearson correlation. (d) The heat map displays spike pool 1 and spike pool 2 aggregate cytokine response levels.
Age and sex have both been shown to impact risk of COVID‐19.9 Age is also well known to have an impact on vaccine responsiveness.10 To assess the impact of these factors on T‐cell responses following the first dose of the ChAdOx1‐S vaccine, we compared cumulative spike‐specific cytokine responses in females and males and in younger (< 50 years of age) and older vaccine recipients (> 50 years of age) from 53 participants for whom we had year of birth available. We noted no significant differences in the cytokine responses in females and males, nor in younger or older vaccine recipients (Figure 3a, b). Participants who demonstrated no detectable IFN‐γ and/or IL‐2 response following the first dose were seen in all subgroups. Although only one donor was aged greater than 70, they generated both IFN‐γ and IL‐2 post‐vaccination. To assess the impact of time post‐vaccination on cytokine titres, we correlated cytokine responses with days post‐vaccination. There was no correlation between days post‐vaccination and titres of IFN‐γ (Figure 3c), IL‐2 (Figure 3d) or IL‐13 (Figure 3e). High cytokine responses were evident within 12 days and up to 23 days following vaccination. These observations indicate that differences in the magnitude of the immune response against SARS‐CoV‐2 are attributable to factors independent of sex, age and time post‐vaccination in our cohort.
Figure 3.
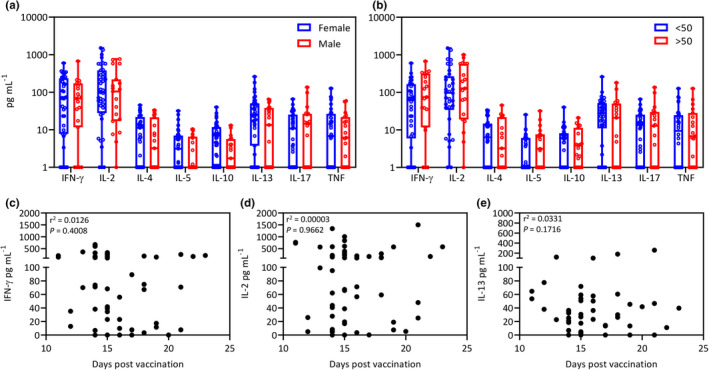
Impact of sex, age and days post‐vaccination on cytokine responses. (a) Box and whisker plots displaying post‐vaccination aggregate cytokine responses in females (n = 41) and males (n = 19). Data represent values after subtraction of background cytokine levels following incubation of blood with no antigen. Statistical significance was determined using multiple t‐tests. No significant differences were observed. (b) Box and whisker plots displaying post‐vaccination aggregate cytokine responses in participants < 50 years of age (n = 34) and > 50 years of age (n = 19). Data represent values after subtraction of background cytokine levels following incubation of blood with no antigen. Statistical significance was determined using multiple t‐tests. (c–e) No significant differences were observed. Data represent correlation of the production of IFN‐γ (c), IL‐2 (d) and IL‐13 (c) with days post‐vaccination. No significant correlations were observed.
Discussion
The worldwide implementation of a number of different COVID‐19 vaccines has provided hope that the COVID‐19 pandemic will soon be under control.11, 12, 13 However, the emergence of multiple SARS‐CoV‐2 variants that may limit the effectiveness of vaccine‐induced neutralising antibodies14, 15 highlights the need to understand all aspects of vaccine‐induced immunity. In this study, using a whole‐blood T‐cell assay, we demonstrate that the majority of ChAdOx1‐S vaccinees generate a strong SARS‐CoV‐2 spike‐specific T‐cell response within 2–3 weeks of their first dose. However, we also noted that a Th1 cytokine (IFN‐γ and/or IL‐2) response was not detectable in 7 of 58 (12.1%) participants. It remains to be determined whether the response in these individuals will be boosted following administration of the second dose or whether they represent a proportion of the population who do not generate significant T‐cell immunity against the spike antigen.
The observations in our current study are consistent with those reported in the analysis of T‐cell responses in a phase 1/2 study of the ChAdOx1‐S vaccine.16 Similarly, an increase in the spike‐specific production of IFN‐γ and IL‐2 by PBMC was evident in vaccine recipients after a single dose. It was also evident in this study that some recipients did not generate responses above those detected in non‐vaccinated individuals. While the threshold for effective T‐cell immunity has not been established, these observations suggest that some individuals may not generate efficient T‐cell responses following a single dose. Observations from the initial phase 1/2 study indicate that these low responses may be boosted following the second dose.5 Most recently, studies have also suggested that these responses could be further amplified using a heterologous boost with the BNT162b2 vaccines.17 In this study, the ChAdOx1‐S vaccine boost did not appear to significantly augment spike‐specific T‐cell immunity.
To provide primary assessment of parameters that may impact the T‐cell response generated post‐vaccination, we assessed the magnitude of the response in the context of sex and age. As previously reported, the participant’s sex did not impact the magnitude of the T‐cell response.16 We also noted no impact of age on the magnitude of the T‐cell response, although our cohort was primarily limited to participants between the ages of 27 and 61, with only a single participant over the age of 70. However, given the variability in responsiveness identified in our cohort, it is likely that additional drivers of differential T‐cell immunity post‐vaccination are involved, including comorbidities (which we did not collect in our cohort). Another potentially influential variable lays with patient‐specific human leukocyte antigens, which is known to influence vaccine responsiveness.18, 19 It also remains to be determined whether differential T‐cell immunity correlates with antibody‐mediated immunity.
Taken together, our observations provide evidence that whole‐blood cytokine assays can provide an effective and rapid strategy for the high‐throughput assessment of T‐cell immunity following vaccination for COVID‐19, providing a platform for expansion into larger cohorts, particularly those known to be at an increased risk of developing severe COVID‐19.
Methods
Study participants
This study was performed according to the principles of the Declaration of Helsinki. Ethics approval to undertake the research was obtained from QIMR Berghofer Medical Research Institute Human Research Ethics Committee. Informed consent was obtained from all participants. Peripheral blood samples were collected from 58 participants, composed of 39 females and 19 males with a median age of 40, who had received the first dose of vaccine between 12 and 23 days previously. A cohort of 26 healthy, non‐vaccinated and non‐COVID‐exposed individuals, composed of 13 females and 13 males with a median age of 39, was also recruited to determine positive cytokine responses.
Whole‐blood measurement of SARS‐CoV‐2‐specific T‐cell immunity
The small‐scale whole‐blood assay used to assess T‐cell responses following vaccination has recently been described. The protocol was modified to only assess spike‐specific T‐cell immunity. The peptide pools using in the study were supplied by JPT Technologies (Berlin, Germany). A T‐cell cytometric bead array (CBA) was performed using the BD Biosciences Flex‐sets (Franklin Lakes, New Jersey). Samples were acquired using a BD LSRFortessa with FACSDiva software (BD Biosciences) and analysed using FCAP array (BD Biosciences) software.
Statistical analysis
GraphPad Prism 8.2.1 (San Diego, CA) was used to perform statistical analysis. Statistical comparisons between participant groups (post‐Vax and unvaccinated) were made using multiple unpaired t‐tests using the Holm–Sidak method. Correlative analysis was performed using the Pearson correlation coefficient. Box plots were used to represent median (horizontal line), 25th and 75th percentiles (boxes) and minimum and maximum values (whiskers). P < 0.05 was considered statistically significant.
Conflicts of interest
The authors report no conflict of interest.
Author contribution
Katie Lineburg: Conceptualization; Data curation; Formal analysis; Funding acquisition; Investigation; Methodology; Project administration; Supervision; Visualization; Writing‐review & editing. Michelle Anne Neller: Conceptualization; Project administration; Writing‐review & editing. George Ambalathingal: Investigation; Methodology; Writing‐review & editing. Jyothy Raju: Investigation; Methodology; Writing‐review & editing. Laetitia LeTexier: Investigation; Methodology; Writing‐review & editing. Srividhya Swaminathan: Formal analysis; Investigation; Writing‐review & editing. Lea Lekieffre: Investigation; Writing‐review & editing. Caitlyn Smith: Investigation; Writing‐review & editing. Sweera Rehan: Investigation; Writing‐review & editing. Pauline Crooks: Investigation; Writing‐review & editing. Archana Panikkar: Investigation; Writing‐review & editing. Sriganesh Srihari: Data curation; Formal analysis; Writing‐review & editing. Rajiv Khanna: Conceptualization; Supervision; Writing‐review & editing. Corey Smith: Conceptualization; Data curation; Formal analysis; Funding acquisition; Investigation; Methodology; Supervision; Writing‐original draft.
Supporting information
Supplementary table 1
Supplementary table 2
Supplementary table 3
Supplementary figure 1
Acknowledgments
We thank all of the participants who generously donated their blood for this study. This work was supported by generous donations to the QIMR Berghofer COVID‐19 appeal and the Medical Research Future Fund (MRFF, APP2005654). SSw is supported by an Australian Government Research Training Program Scholarship, and RK is supported by a National Health and Medical Research Council Fellowship.
References
- 1.Altmann DM, Reynolds CJ, Boyton RJ. SARS‐CoV‐2 variants: Subversion of antibody response and predicted impact on T cell recognition. Cell Rep Med 2021; 2: 100286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geers D, Shamier MC, Bogers Set al. SARS‐CoV‐2 variants of concern partially escape humoral but not T‐cell responses in COVID‐19 convalescent donors and vaccinees. Sci Immunol 2021; 6: eabj1750. 10.1126/sciimmunol.abj1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore PL, Moyo‐Gwete T, Hermanus Tet al. Neutralizing antibodies elicited by the Ad26.COV2.S COVID‐19 vaccine show reduced activity against 501Y.V2 (B.1.351), despite protection against severe disease by this variant. bioRxiv 2021: 2021.2006.2009.447722.
- 4.Swain SL, McKinstry KK, Strutt TM. Expanding roles for CD4+ T cells in immunity to viruses. Nat Rev Immunol 2012; 12: 136–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Folegatti PM, Ewer KJ, Aley PKet al. Safety and immunogenicity of the ChAdOx1 nCoV‐19 vaccine against SARS‐CoV‐2: a preliminary report of a phase 1/2, single‐blind, randomised controlled trial. Lancet 2020; 396: 467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sadoff J, Le Gars M, Shukarev Get al. Interim results of a phase 1–2a trial of Ad26.COV2.S Covid‐19 vaccine. N Engl J Med 2021; 384: 1824–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tischer S, Dieks D, Sukdolak Cet al. Evaluation of suitable target antigens and immunoassays for high‐accuracy immune monitoring of cytomegalovirus and Epstein‐Barr virus‐specific T cells as targets of interest in immunotherapeutic approaches. J Immunol Methods 2014; 408: 101–113. [DOI] [PubMed] [Google Scholar]
- 8.Lineburg KE, Srihari S, Altaf Met al. Rapid detection of SARS‐CoV‐2‐specific memory T‐cell immunity in recovered COVID‐19 cases. Clin Transl Immunology 2020; 9: e1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gebhard C, Regitz‐Zagrosek V, Neuhauser HK, Morgan R, Klein SL. Impact of sex and gender on COVID‐19 outcomes in Europe. Biol Sex Differ 2020; 11: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grijalva CG, Feldstein LR, Talbot HKet al. Influenza vaccine effectiveness for prevention of severe influenza‐associated illness among adults in the United States, 2019–2020: a test‐negative study. Clin Infect Dis 2021. 10.1093/cid/ciab462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Polack FP, Thomas SJ, Kitchin Net al. Safety and Efficacy of the BNT162b2 mRNA Covid‐19 Vaccine. N Engl J Med 2020; 383: 2603–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voysey M, Clemens SAC, Madhi SAet al. Safety and efficacy of the ChAdOx1 nCoV‐19 vaccine (AZD1222) against SARS‐CoV‐2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 2021; 397: 99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baden LR, El Sahly HM, Essink Bet al. Efficacy and safety of the mRNA‐1273 SARS‐CoV‐2 vaccine. N Engl J Med 2021; 384: 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia‐Beltran WF, Lam EC, St Denis Ket al. Multiple SARS‐CoV‐2 variants escape neutralization by vaccine‐induced humoral immunity. Cell 2021; 184: 2372‐2383 e2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jangra S, Ye C, Rathnasinghe Ret al. SARS‐CoV‐2 spike E484K mutation reduces antibody neutralisation. Lancet Microbe 2021; 2: e283–e284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ewer KJ, Barrett JR, Belij‐Rammerstorfer Set al. T cell and antibody responses induced by a single dose of ChAdOx1 nCoV‐19 (AZD1222) vaccine in a phase 1/2 clinical trial. Nat Med 2021; 27: 270–278. [DOI] [PubMed] [Google Scholar]
- 17.Georg B, Joana B‐M & Swantje H et al. Humoral and cellular immune response against SARS‐CoV‐2 variants following heterologous and homologous ChAdOx1 nCoV‐19/BNT162b2 vaccination. Nature Portfolio 2021. 10.21203/rs.3.rs-580444/v1. [DOI]
- 18.Wang C, Tang J, Song W, Lobashevsky E, Wilson CM, Kaslow RA. HLA and cytokine gene polymorphisms are independently associated with responses to hepatitis B vaccination. Hepatology 2004; 39: 978–988. [DOI] [PubMed] [Google Scholar]
- 19.Gelder CM, Lambkin R, Hart KWet al. Associations between human leukocyte antigens and nonresponsiveness to influenza vaccine. J Infect Dis 2002; 185: 114–117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary table 1
Supplementary table 2
Supplementary table 3
Supplementary figure 1


