Abstract
HIV represents a persistent infection which negatively alters the immune system. New tools to reinvigorate different immune cell populations to impact HIV are needed. Herein, a novel nanotool for the specific enhancement of the natural killer (NK) immune response towards HIV-infected T-cells has been developed. Bispecific Au nanoparticles (BiAb-AuNPs), dually conjugated with IgG anti-HIVgp120 and IgG anti-human CD16 antibodies, were generated by a new controlled, linker-free and cooperative conjugation method promoting the ordered distribution and segregation of antibodies in domains. The cooperatively-adsorbed antibodies fully retained the capabilities to recognize their cognate antigen and were able to significantly enhance cell-to-cell contact between HIV-expressing cells and NK cells. As a consequence, the BiAb-AuNPs triggered a potent cytotoxic response against HIV-infected cells in blood and human tonsil explants. Remarkably, the BiAb-AuNPs were able to significantly reduce latent HIV infection after viral reactivation in a primary cell model of HIV latency. This novel molecularly-targeted strategy using a bispecific nanotool to enhance the immune system represents a new approximation with potential applications beyond HIV.
Keywords: HIV, Bispecific nanoparticles, NK cells, Polarization
Introduction
The augmentation of the effector immune responses towards specific targets, such as virally-infected or cancerous cells, is positioning itself as a new promising therapeutic intervention [1]. Indeed, an efficient immunological synapse between immune effectors and aberrant cells is a key factor to promote an effective immune-mediated killing of such undesired cells. In this context, cell-to-cell contact is required for natural killer (NK) cells, lymphocytes of the innate immune system that eliminate abnormal cells through several sophisticated biological mechanisms, including antibody-dependent cellular cytotoxicity (ADCC), to accomplish their mission [2–4]. Unfortunately, in the context of a persistent viral infection, like the one established by the human immunodeficiency virus (HIV), the virus integrates into the genome of host cells and is able to escape from the immune response, therefore perpetuating the infection [5]. In turn, chronic infection leads to a persistent immune activation that promotes the appearance of dysfunctional NK cells with diminished capacity to kill infected cells [6], further impairing immune defences. Importantly, this functionality is not completely restored with the current antiretroviral therapy, and discontinuation of the treatment leads to a rapid viral rebound [7]. Thus, new targeted interventions are needed to help the immune system of HIV-infected patients to control the virus. In this regard, novel therapeutic strategies focused on the re-direction, potentiation and/or promotion of an efficient immunological synapse between NK and infected T-cells are highly desirable to efficiently combat such persistent viral infections.
Nano-based approaches with a dual capacity of recognition are emerging as promising bispecific formats to engage cell-to-cell contacts and potentiate antiviral and antitumor immunity [8]. Such nanotools based on nanoparticles (NPs) displaying mixtures of antibodies showed significant advantages over the more conventional bispecific antibodies, such as the possibility to regulate biological processes in a size-dependent manner, their capacity to penetrate into some tissues and cells and their enhanced potency mediated by their multivalent properties [9–11].
The interaction of proteins, including antibodies, onto inorganic surfaces, such as gold, has been a subject of intense studies in the field of biochemistry and medicine since implants were developed. These studies on the adsorption of proteins at surfaces, a phenomenon ubiquitous in nature, have shown two models: Random Sequential (disordered) vs Clustering (cooperative) adsorption [12]. The first indications on the mode in which IgGs adsorb onto AuNPs is the kinetics of the adsorption process that already points towards a cooperative mode: proteins at the NPs surface evolve from forming a transient corona to become permanent with time, known as soft and hard NP-Protein Corona (PC) respectively [13]. As it has been well described, when NPs are dispersed in protein solutions, these rapidly form a soft protein corona around the NP, formed of loosely bound proteins, in equilibrium with free proteins in solution. With time, this PC evolves, due to rearrangements and crowding effects, towards a permanent corona since protein-dense clusters are more stable at the NP surface than single proteins. Indeed, cooperative adsorption implies that a protein adsorbed to a surface binds more strongly to it if it is surrounded by similar proteins thanks to the possibility of build compact arrangements with identical building blocks, increasing coating stability (cooperative effect) as the protein domain grows. This evolves towards the formation of a complete hard corona of proteins permanently attached to the NP surface that remains associated to the NP even after NP precipitation and resuspension into protein-free media [13,14], thus conferring biological identity to the NP [15]. In these conditions, similar proteins pack better than those which are different, favoring thus the formation of homogeneous coating domains from a mix of coating molecules [16]. This evolution towards a more stable state, an expression of the Vroman effect on the surface of NPs [17], is what allows to explore enthalpy-driven, rather than entropy-driven states, which implies macromolecular ordering and domain segregation [18]. Thus, by controlling the principal parameters affecting the NP-protein interactions, such as relative concentrations and affinity, incubation time, pH, temperature, media ionic composition and solubility, it is possible to find the optimal conditions to promote the cooperative adsorption of proteins onto the NP [14] and consequently the formation of highly polarized NP conjugates [12,19,20].
Here, with the purpose to enhance the NK immune response towards HIV-expressing cells we have constructed bispecific nanoparticles following a novel cooperative adsorption conjugation protocol of a mixture of antibodies, inducing their domain segregation at the NP surface and resulting in a certain degree of NP functionality polarization. This novel molecularly-targeted approach uses the antibodies A32 (IgG anti-HIVgp120), which recognizes the protein gp120 expressed on the membrane of HIV-infected cells, and the 3G8 (IgG anti-human CD16, clone 3G8) that recognizes the CD16 receptor expressed on NK cells, conjugated to a 40 nm AuNP. This novel nanotool appears very promising for the targeting and immune-mediated elimination of HIV-infected cells.
Results
Generation of functionalized bispecific nanoparticles
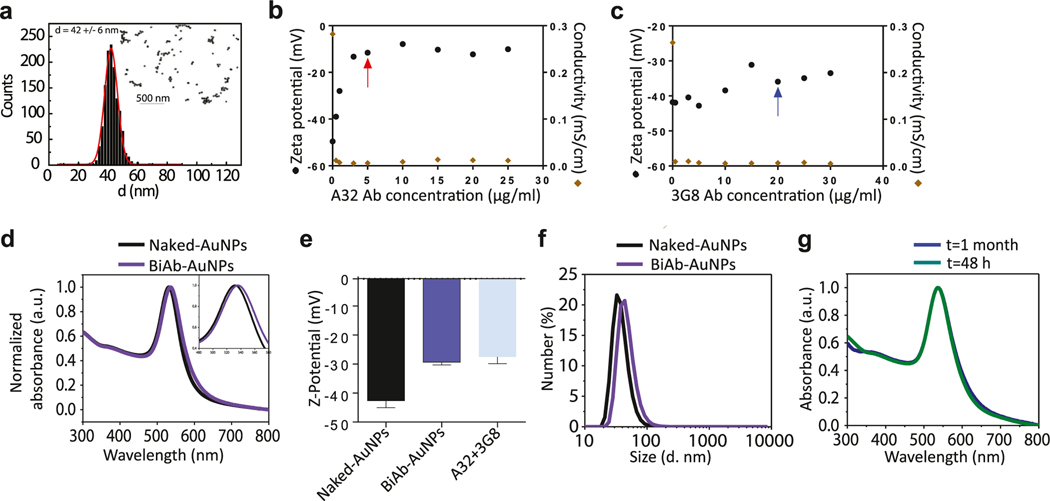
Our goal was to develop multivalent bispecific NPs after a cooperative adsorption of the antibodies at their surface. For that, we carefully studied the conjugation medium and the conditions to achieve a cooperative adsorption protocol and we validated the approach. First, we generated monoconjugates with the antibodies A32 and 3G8 separately and studied the impact of the conjugation process on the capacity of these antibodies to still recognizing their cognate antigens. In order to assess the affinity of the different antibodies for the AuNP surface, we performed Langmuir-type isothermal adsorption curves to identify the concentration needed to obtain surface-saturated monoconjugates. AuNPs of 42 ± 6 nm (Fig. 1a) with a surface charge of −44 mV, colloidally stable in a 2.2 mM sodium citrate (SC) at 5 × 1010 NPs/ml concentration and with an UV-Vis Spectroscopy absorption peak at 532 nm, were exposed to the different antibodies dispersed in a 2.2–4 mM SC-Borate selected conjugating medium at different concentrations. From the adsorption curves, 5 μg/ml and 20 μg/ml were identified as the antibody saturation concentrations needed for A32 and 3G8, respectively (Fig. 1b and c). The specific cell-surface binding abilities of both monoconjugates were evaluated by flow cytometry. We found that A32-AuNPs retained the same binding capability than the free antibody and recognized the entire population of HIV+ cells, indicating that the conjugation process did not alter its targeting capacity (median 98.60% for the conjugate and 99.60% for free A32) (Supplementary Fig. 1a). Similarly, 3G8-AuNPs were able to recognize the CD16 molecules expressed on NK cells in a similar manner as the non-conjugated free antibody (median 58.60% for the conjugate vs. 51.30% for free 3G8) (Supplementary Fig. 1b), indicating that antibody adsorption did not result in any decrease in its ability to recognize the cognate antigen.
Fig. 1.
Characterization of the bispecific gold nanoparticles (BiAb-AuNPs). (a) Representative TEM image of the ~40 nm AuNPs. (b) A32-AuNPs saturation curve. Zeta potential (mV) versus the antibody concentration is plotted. The red arrow indicates the saturating concentration for the A32 antibody. (c) 3G8-AuNPs saturation curve. The blue arrow indicates the saturating concentration for the 3G8 antibody. (d) Representative UV-Vis spectra of the BiAb-AuNPs compared to naked AuNPs. (e) Z-potential measurements of BiAb-AuNPs, naked AuNPs, and the single antibodies A32 and 3G8 (n = 3, mean with SD). (f) Representative Dynamic light scattering (DLS) characterization of the BiAb-AuNPs compared to naked nanoparticles. (g) Representative UV-Vis spectra of the BiAb-AuNPs at 48 h and 1 month after antibody conjugation.
Next, we generated the polarized bispecific nanoparticles (pBiAb-AuNPs) by mixing equal volumes of the AuNPs solution with the mix of antibodies in solution for 48 h at 4 °C carefully controlling the conjugating medium. By mildly destabilizing ~40 nm AuNPs in a mix of A32 and 3G8 antibodies, we observed how the system slowly evolved towards a new equilibrium state where NPs got fully coated by the two antibodies (vide infra). In a close-up, A32 and 3G8 present differences in their fine structure and physicochemical properties, that enabled its segregation when adsorbed at the NP surface. For instance, A32 has a reported isoelectric point (pI) of 8.8 [21] and a surface charge of ≃−15 mV, meanwhile the 3G8 antibody has an estimated pI of 8.2 (according to its sequence) and a more negative measured surface charge. The aim was to work under conditions where antibodies and NPs were rather stable and therefore slowly conjugated, allowing them to explore their minimum energy positions which correspond with clustering and crystallization.
The ratio of each antibody in the solution was chosen according to the saturation curves and their different relative affinities for the AuNP surface. In order to obtain an approximate 50:50 proportion of each antibody at the NP surface, a ratio of 100:60 for 3G8 and A32 respectively was finally chosen. After conjugation, polarized bispecific nanoparticles (pBiAb-AuNPs) presented a redshift in the UV–Vis spectra of 4 nm (Fig. 1d), consistent with a full coating of the NP surface with proteins [22]. The surface charge of the pBiAb-AuNPs was also characterized (Fig. 1e), reporting a lower negative value (−31.4 mV), corresponding to half way between the values of the two homoconjugates (Supplementary Table 1) indicating the presence of a 50% mixture of antibodies in the fully coated surface of AuNPs [23]. The DLS (Dynamic Light Scattering) hydrodynamic radius signal also increased upon conjugation (about 10 nm), correlating well with the UV-Vis spectroscopy measurements and attributed to the formation of a dense antibody monolayer on top of the AuNP (Fig. 1f). No further shifts of the UV–Vis post conjugation were observed after a month (Fig. 1g), indicating the high stability of the formed conjugates. Note that if antibodies were denaturalized as a consequence of the interaction with the AuNP surface, they would inevitably lead to antibody aggregation and detectable NP cross-linking [24]. Importantly, the characteristic width of the measured peaks after conjugation remained similar to the measurements before conjugation, further confirming the formation of homogeneous antibody coronas [25]. Otherwise, if a dispersion in the conjugation process would occur leading to NPs with different degrees or types of antibody coatings, UV–Vis, DLS and Z-potential peaks would become broader than in the case of non-conjugated NPs, integrating the dispersion of both, the NP core and the NP coating.
Using a theoretical approximation of 150 nm2 per antibody on dense antibody coatings [26], we estimated that 32 antibodies were displayed on the surface of our pBiAb-AuNPs (Supplementary Note 1). Moreover, thermogravimetric analysis (TGA) of the conjugates, considering a density of 1.35 and 19.30 g/ml for an antibody and the Au, respectively, led to similar results. In this case, the evaporation at 450 °C of a 10 nm corona of antibodies in a ~40 nm diameter of solid AuNP accounted for about 10% of the total weight loss (Supplementary Fig. 2).
Polarization assessment after following the cooperative adsorption conjugation protocol
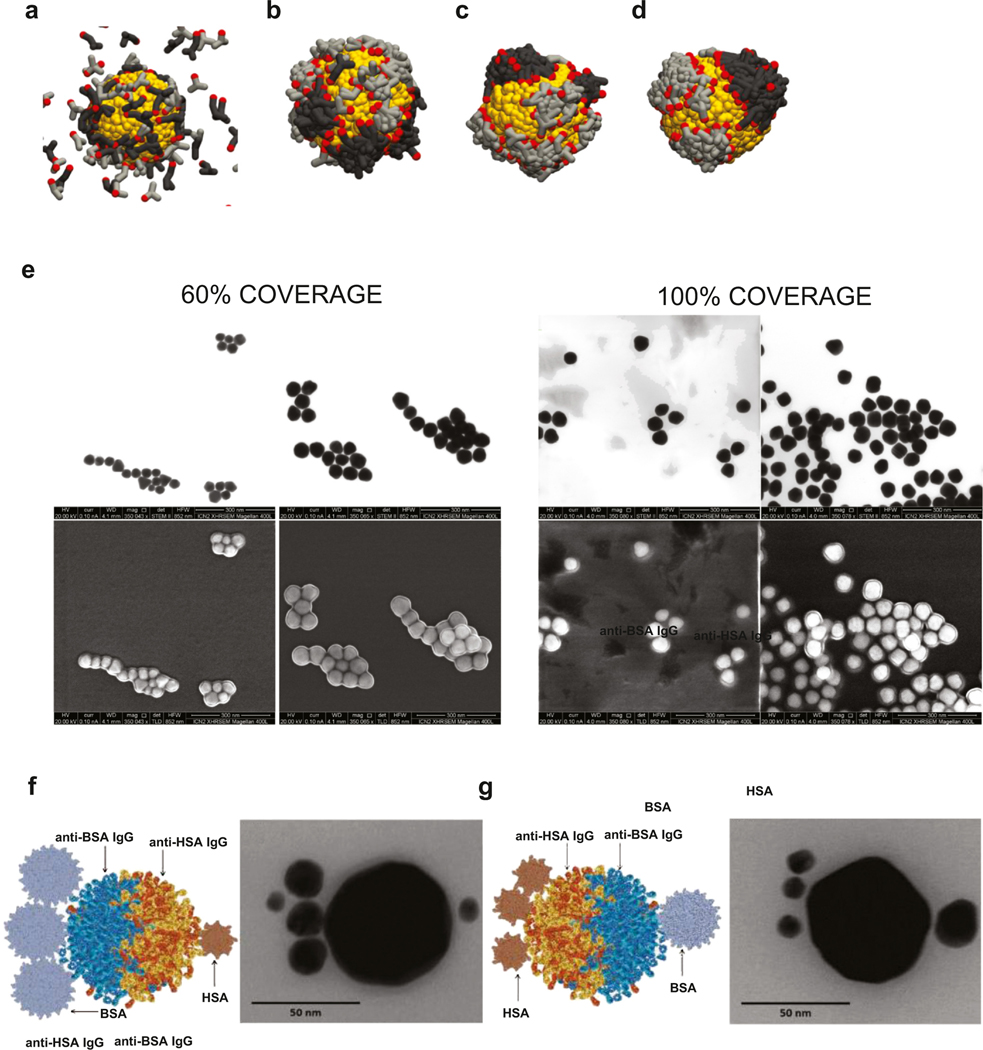
Molecular mixtures on a NP surface are difficult to characterize [27], especially in their working environment. In the early 1990s, studies suggested that two alkanethiols of different chain length segregate on a gold surface, forming microscopic islands, despite the strong bond between thiols groups and Au [16]. Since then, other mixtures of alkanethiols [19], organic molecules as MUA and PEG [28] and also peptides did show a tendency to segregate onto the NP surface, and this effect was discussed not without controversy [19,29–31]. This tendency to grow domains was described to occur during the self-assembly process onto the NP surface, and not when the molecules were free in solution [32]. Simulations performed mimicking our experimental conditions showed the tendency of cooperative adsorption of antibodies onto the NP surface when enough time was allowed to the antibodies to organize themselves onto the AuNP surface. Similarly, once few clusters had nucleated onto the NP surface, these grew until the whole surface was densely covered (Fig. 1a–d).
Describing the organization of molecules at the NP surface is challenging. Despite the literature is full with examples of functionalized nanoparticles generally illustrated with inspiring schemes, the actual distribution and conformation of molecules at the NP surface is still unknown and often remain poorly characterized. We obtained the first evidence for the formation of antibody domains on top of the AuNPs from the antibody loading curves and geometrical considerations. The progressive coating of the antibodies on the NPs can be observed in the UV–Vis, DLS and Z-potential measurements after purification as a function of incubation time. If the adsorbed proteins were distributed purely at random, those NPs with just above the 50% of coverage should be resistant to aggregation during the purification process (that forces NPs together). In a perfect random distribution, after loading more than 50%, the remaining grooves would not be large enough to efficiently fit another protein (carrying an attached NP), nor the two AuNP surfaces could be in contact (see schema in Supplementary Fig. 3). In our experiments, NPs aggregated during purification up to surface coverages close to the 80%, well away from the theoretical 50% suggesting cooperative growth of the antibody coating (Supplementary Fig. 4a and b). This time-dependent hardening of the protein corona was also confirmed by the analysis of the evolution of the surface charges. A minimum of 24 h of antibody exposure was needed for the formation of a stable antibody layer (Supplementary Fig. 4c). This time point for conjugation (24 h) was associated to an 80% of surface coverage (Supplementary Fig. 4d). Note that in a Langmuir-like isothermal adsorption behavior, the observed shift of the characteristic plasmonic peak position between zero and saturation is directly proportional to the degree of coating [28]. We used this information to prepare partially and fully antibody-coated AuNPs, and gain further insight into the growth regimen of the antibody layer under our protocol conditions by visualizing the prepared conjugates through transmission electron microscopy (TEM) and scanning electron microscopy (SEM). We cannot distinguish by TEM different antibodies on top of the NP, but we can observe if before saturation, the distribution of proteins is random or if the coating grows cooperatively onto the AuNP. In Fig. 1e, representative TEM and SEM images of partially (at the 60%) and fully antibody coated AuNPs are shown. In the TEM images (Fig. 1e upper panel) we observe how in case of saturation, AuNPs do not touch each other, while for the unsaturated samples the AuNP-AuNP direct contact is more evident. In the SEM images (Fig. 1e lower panel), the charging effects of the organic layer on the edge of the NPs can be clearly seen, and it can be observed how the coating is complete in the case of antibody saturated samples while only one side of the partially coated AuNPs appears to wear it.
In addition, to directly analyze the distribution of antibodies per NP following our conjugation protocol, we used doubly functionalized AuNPs with anti-HSA and anti-BSA IgGs. We then used 25 nm AuNPs functionalized with BSA and 15 nm AuNPs functionalized with HSA, as labels to directly observe the presence and distribution of the two antibodies at the AuNP surface. This experiment was not performed with the A32 and 3G8 antibodies because their cognate antigens are embedded at the cell surface, and therefore, it will require the use of recombinant gp120 and CD16 to functionalize the label nanoparticles. Moreover, anti-human secondary antibodies could not be used because it does recognize not only the A32 but also the 3G8 antibody, providing a high unspecific background signal and preventing the real discrimination between the A32 and 3G8 domains. After conjugation, the three functionalized AuNPs were incubated together and analyzed using TEM. The cooperative adsorption approach was confirmed by the observation of NPs with a hemispherical distribution of the labels (Fig. 1f and g). After fully characterization, 50% of the sample presented non- or mono-conjugated NPs (identified by the absence of labels or by only one type of the secondary labels), 26% of the multiconjugated NPs showed some polarity, while 24% of them seemed to present stochastic orientation of the antibodies. All in all showing the tendency of proteins to grow forming domains in these conditions. A gallery of TEM images representing different structures indicating the cooperative adsorption of the antibodies is shown in Supplementary Information 2.
BiAb-AuNPs promote specific cell-to-cell contact between CD4-expressing HIV and NK cells
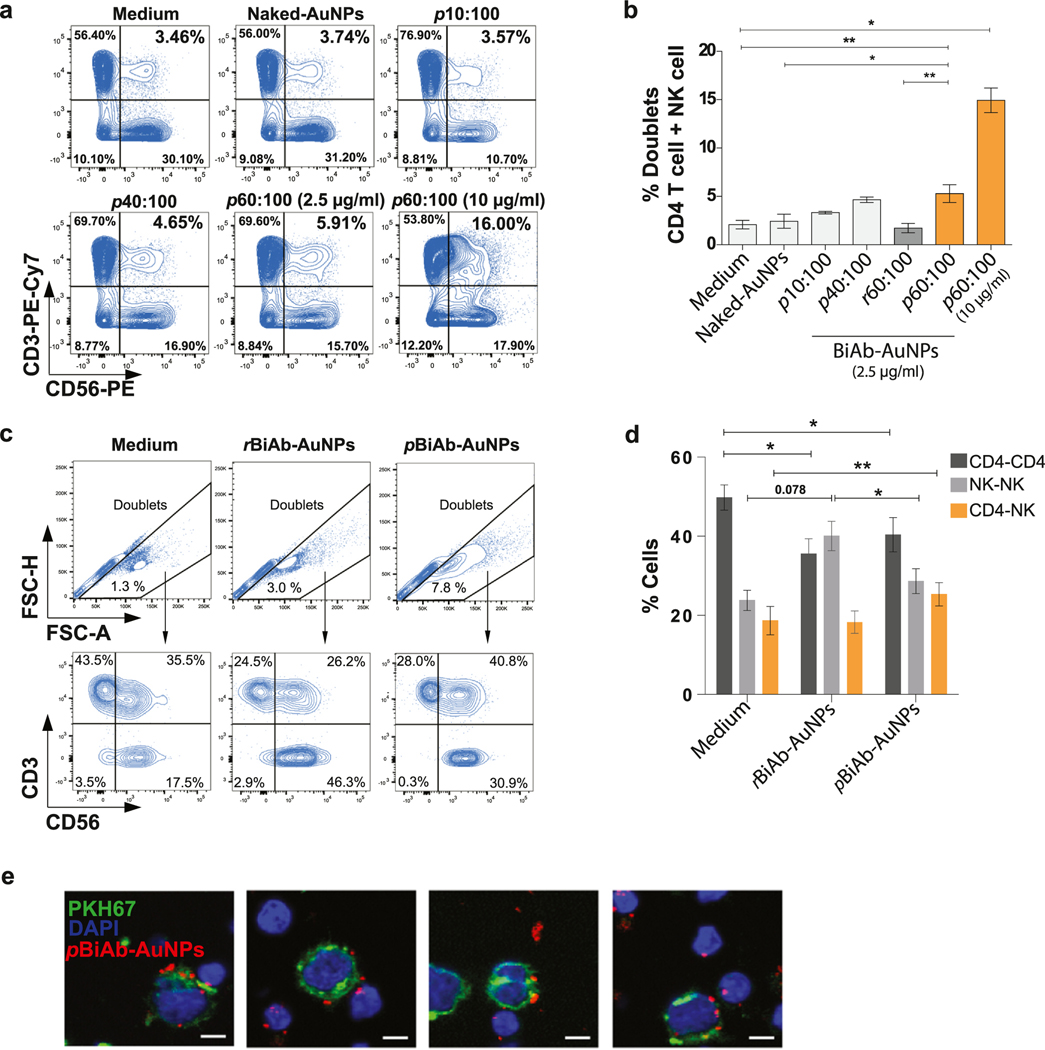
Once the pBiAb-AuNPs presenting A32 and 3G8 antibodies were characterized, we investigated their potential to bring closer cytotoxic NK cells (characterized by dim expression of the marker CD56 and high levels of the CD16 molecule) and primary CD4 T cells coated with the viral protein gp120. We performed cell-to-cell contact experiments by culturing both cell types in the presence of different bioconjugates. As control, irrelevant bispecific AuNPs (Irre-AuNPs) coated with polyclonal IgG mouse and polyclonal IgG rat antibodies, were similarly prepared. In addition, we prepared 10:100 and 40:100 A32:3G8 ratio conjugates. The conjugation ratio 100:10 A32:3G8 was also prepared but it rapidly led to the irreversible aggregation of the AuNPs precluding its further use. Besides, we also compared our pBiAb-AuNPs generated using the cooperative approach, with an already well described conjugation protocol expected to yield randomly antibody-coated NPs (rBiAb-AuNPs) [33]. Results for the employed conjugates are shown in Supplementary Table 1. No direct toxicity in ex-vivo HIV-infected CD4+ T cells was associated with the presence of any pBiAb-AuNPs or Irre-AuNPs in culture after 4 h incubation (mean percentage 95.83 ± 6.56 and 90.10 ± 7.78 for pBiAb-AuNPs and Irre-AuNPs, respectively). Longer incubation times (48 h) did not significantly affect cell viability either (91.2% and 86.67% cell viability, for pBiAb-AuNPs and Irre-AuNPs, respectively). We observed that 60% and 100% of the saturating concentration for each antibody (A32 and 3G8, respectively) produced the highest number of NK-HIV+ CD4 T doublets, with a 2.5-fold increase compared with controls (median % doublets of 5.66 compared to 2.23%, 3.30% and 4.65% for control medium, 10:100 and 40:100 ratios, respectively) (Fig. 2a and b). Thus, 60% and 100% of the saturating concentration for A32 and 3G8 were selected as the optimized concentrations for the generation of the pBiAb-AuNPs. Importantly, a dose-dependent response was observed, and the addition of the pBiAb-AuNPs coated with 10 μg/ml of antibodies significantly increased the mean of cell doublets by over 7-fold (p = 0.01) (Fig. 2a and b). Of note, the rBiAb-AuNPs with a random (non-polarized) disposition of the A32 and 3G8 antibodies at their surface, were not efficient at performing cell-to-cell engagement (Fig. 2b). These results position our pBiAb-AuNPs as better candidates to achieve such biological effects. In addition, we performed a different analysis and assessed the composition of the cell doublets in the absence and presence of pBiAb-AuNPs and rBiAb-AuNPs. First, we observed that our pBiAb-NPs were better nanoconjugates at inducing the formation of cell-doublets (Fig. 2c upper panel), and within these cell doublets, we observed an increase in the specific CD4-NK doublets and formed less homodoublets CD4-CD4 compared to the medium control (Fig. 2c lower panel and 3d), further supporting their polarized nature. Besides, rBiAb-AuNPs generated significantly less cell doublets, with increased percentages of NK-NK doublets (Fig. 2d). Note that in the absence of AuNP polarization, one would expect also an increase of homodoublets (NK-NK or CD4-CD4) due to the random distribution of both antibodies on the surface of the NPs. This is consistent with results observed for rBiAb-AuNPs which increased NK-NK doublets (Fig. 2d). After the precise quantification by flow cytometry, we imaged the preparation by confocal microscopy, since the very low dielectric constant of AuNPs scatters light very efficiently [34] and therefore the ~40 nm AuNPs can be observed in the confocal microscope, especially if they form any kind of aggregates. Direct contact of NK and HIV+ cells zipped by the pBiAb-AuNPs, which appear in red, can be observed (Fig. 2e).
Fig. 2.
Simulations and Transmission electron microscopy (TEM) and Scanning electron microscopy (SEM) analysis of conjugates following the cooperative adsorption protocol. Four snapshots of the resultant simulations are shown (a–d). From the left to the right, the water-antibodies bead interacting parameters are increased from 24 to 50. (a) The adsorption of these proteins starts to be observed but without any preferences. (b–d) All the antibodies are adsorbed onto AuNP for minimizing their interaction with water. Note that, the higher is the repulsion with water, the closer will be the antibodies. Polarized AuNPs are observed when water-antibodies repulsion is bigger than its own interaction. (e) In the upper panel TEM images of partially-coated (60% surface cover) and fully-coated Au conjugates can be seen. In the lower panel SEM images are shown. (f) AuNPs functionalized following cooperative adsorption with anti-BSA IgG and anti-HSA IgG, and exposed to smaller conjugates as labels (AuNPs of 25 nm functionalized with BSA and of 15 nm with HSA), are shown. It can be observed a hemispherical distribution of the labels, with three BSA-AuNPs distributed in one side of the bispecific AuNP and one HSA-AuNP in the opposite side. (g) The same can be observed in this panel but with an inverted proportion of the labels, further confirming polarity.
pBiAb-AuNPs induce a potent cytotoxic response against primary CD4 T cells from HIV-infected patients and against ex vivo human infected tonsillar explants
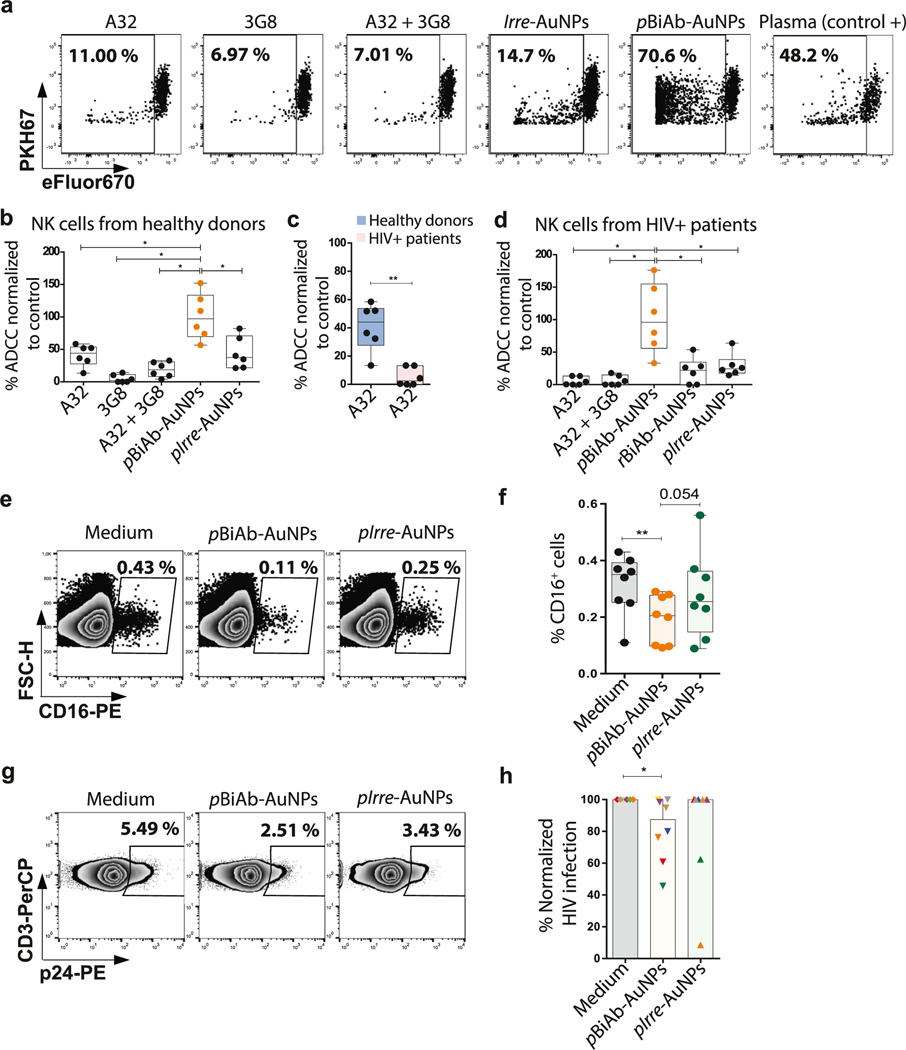
Next, in order to evaluate the ADCC-like activity of NK cells towards HIV-expressing cells in the presence of the pBiAb-AuNPs, we performed the corresponding functional assays. CEM NKR CCR5 cells coated with gp120 were co-cultured with NK cells during 4 h in the presence of different nanoconjugates. Cells non-coated with gp120 were included as a control. Cell killing was measured by the loss of the marker eFluor670 by flow cytometry. A representative example and gating strategy are shown in Fig. 3a and Supplementary Fig. 5, respectively. As a positive control we used plasma from a viremic HIV+ patient containing a pool of antibodies targeting the gp120 viral protein. Values were normalized to this positive control which is considered the maximum ADCC response that could be achieved for the individual samples. First, we tested NK cells from healthy donors and we observed that pBiAb-AuNPs were able to promote a highly potent and specific ADCC-like response (Fig. 3b). The effect of the pBiAb-AuNPs was significantly better than the cytotoxicity reached by the addition of IrreAuNPs or the free A32 antibody, which is described in the literature as one of the most potent non-neutralizing antibodies [35]. Value for the pBiAb-AuNPs was close to 100% of the maximum NK immune effector response (median 97.04% vs. 44.09%, 37.37% and 18.63% for A32, Irre-AuNPs and the combination of A32 and 3G8 unconjugated antibodies, respectively) (Fig. 3b). Note that the addition of both 3G8 and A32 antibodies to cell culture decreased the ADCC levels compared to A32 alone. 3G8 is commonly used to block Fc receptors (through the interaction with the variable fraction) at concentrations near 5 μg/ml, thus when presented in a non-conjugated form it may block part of the ADCC response promoted by A32. Next, we assessed the capacity of the pBiAb-AuNPs to restore the functional activity of NK cells from HIV+ patients, since it is well known that HIV infection impairs their effector immune responses [6]. These patients presented a median CD4 T cell count in blood of 590 cells/μl and a median time under suppressive antiretroviral treatment of 69 months (plasma viral load < 50 copies/ml). Using the A32 antibody we observed, as expected, that NK cells from HIV-infected patients had a diminished capacity to kill by ADCC in comparison to healthy donors (Fig. 3c). Importantly, after the addition of the polarized pBiAb-AuNPs, the functionality of NK cells was significantly restored (Fig. 3d). Of note, polarization of the antibodies at the AuNPs surface was a requisite for the enhancement of the NK cell immune response, since we found that non-polarized rBiAb-AuNPs could not strongly potentiate the NK cytotoxic response (Fig. 3d).
Fig. 3.
The pBiAb-AuNPs induce cell-to-cell contact between HIV-expressing cells and NK cells. (a) Representative flow cytometry plots showing cell doublets corresponding to primary CD4+ T cells coated with gp120 (CD3+) and primary NK cells (CD56+) after 20 min co-culture in the presence of the different nanoconjugates. Double positive cells for the markers CD3 and CD56 denote pairs of cells formed by CD4-HIV+ and NK cells. Controls corresponding to medium alone and non-conjugated (naked) nanoparticles are shown. Different ratios for the generation of BiAb-AuNPs and different total antibody concentrations on the nanoparticles (2.5 μg/ml for all conjugates and an additional dose of 10 μg/ml for 60:100 ratio) are shown. (b) Summary graph for the induction of cell-to-cell contacts in the presence of different nanoconjugates. Mean with SEM for the experimental replicates are shown. Statistical analysis consisted on a paired t-test. *p < 0.05. (c) Representative example of the gating strategy to identify cell doublets using the FSC-H and FSH-A signals, and the subsequent analysis of the frequency of different types of cell pairs using the cell markers CD3 and CD56. (d) Distribution of the cell doublets. Cell doublets were identified by flow cytometry using the FSH-H and FSH-A plot. The total cell doublets were distributed in T-T cells (CD3+CD3+), NK-NK (CD56+CD56+) or T-NK (CD3+CD56+) doublets and the percentages are plotted. Data shows mean with SEM of n = 6 independent experiments. Statistical comparisons were performed using Wilcoxon matched-pairs signed rank test. (e) Representative confocal micrographs of HIV-expressing cells (CEM.NKR CCR5+ coated with HIV gp120 recombinant protein and stained with PKH67 in green, and DAPI nuclei staining in blue) and primary NK cells (DAPI nuclei staining). Both cell types were co-cultured at 1:1 ratio for 20 min in the presence of the pBiAb-AuNPs. pBiAb-AuNPs are visualized in red. Scale bar represents 5 μm.
As the triggering through the CD16 receptor can be a potent activating signal, we addressed the possibility of direct NK activation by our pBiAb-AuNPs. Human peripheral blood mononuclear cells (PBMCs) were cultured with the pBiAb-AuNPs for 4.5 h and the different activation and cytotoxicity markers were measured by flow cytometry. We observed that after incubation with the pBiAb-AuNPs, primary NK cells (CD3− CD56+) produced IFN-γ (Supplementary Fig. 6a). Accordingly, 3G8 free antibody and IrreAuNPs also increased IFN-γ production in NK cells, showing that in general, targeting Fc receptors could also stimulate NK cells to produce IFN-γ, as previously reported [36]. By contrast, the pBiAb-AuNPs not always induced the expression of CD107a (a degranulation marker) (Supplementary Fig. 6b), nor significantly affected cell activation, measured by the expression of HLA-DR (Supplementary Fig. 6c).
We also used a human tissue histoculture model [37] to assess the penetrability of the pBiAb-AuNPs in blocks of lymphoid tissue and the NK immune effector function during ex-vivo HIV infection (Supplementary Fig. 7). Note that the only valuable animal model to study the NK biology in tissues during HIV disease are primarily non-human primates. Tissue blocks were infected with HIV, and after 15 min pBiAb-AuNPs were added to the culture. After 24 h incubation, we checked the penetrability of the pBiAb-AuNPs by detection of the CD16 marker in NK cells. 6 days later, the infection was measured by intracellular staining of the HIV p24 viral protein. We observed that pBiAb-AuNPs were able to penetrate these cultured tonsil blocks, as demonstrated by the specific loss in the detection of the CD16 marker by flow cytometry in the presence of the pBiAb-AuNPs (Fig. 3e and f). This reduction of CD16 expression is attributed to the blockage of CD16 by the 3G8 antibody present in the pBiAb-AuNPs after tissue penetration. In addition, the pBiAb-AuNPs significantly decreased HIV infection in this lymphoid tissue, reaching up to 50% reduction in some cases (Fig. 3g and h). This is noteworthy, since the majority of the persistent HIV infection in antiretroviral treated individuals reside in secondary lymphoid tissue such as lymph nodes and the gut-associated lymphoid tissue (GALT), being the main barrier to reach a cure [38]. Interestingly, NPs are being extensively studied as drug carriers and targeting vehicles for their ability to target some of these tissue compartments [39].
pBiAb-AuNPs strongly enhance the NK-mediated killing of latent HIV-infected cells after viral reactivation
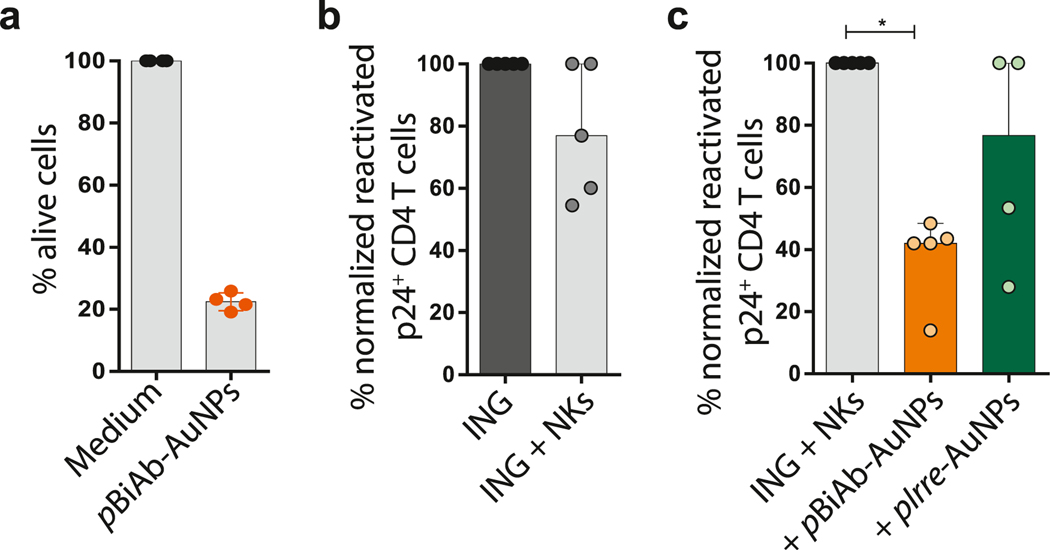
Reducing or eliminating the persistent reservoir is the main challenge in HIV research field. Recently, therapeutic strategies named “shock and kill” are being designed and tested to clear the HIV reservoir. This approach tries to induce the expression of dormant HIV viruses with latency reversal agents (LRAs), relying on achieving also a potent immune response able to eliminate the HIV-infected cells. For assessing the suitability of the pBiAb-AuNPs to be used in the context of a shock and kill strategy, we tested their capacity to induce a cytotoxic response on viral reactivated HIV-infected cells with LRAs, first on the latently infected cell line ACH-2 and then on a primary cell model of HIV latency. When pBiAb-AuNPs were added to ACH-2 cells and subjected to viral reactivation, we consistently observed a highly potent response against HIV+ cells, inducing a median of 80.0% of cell death in 4 h (Fig. 4a). This potent response was also reproduced with NK cells when pBiAb-AuNPs were added to the primary cell model of HIV latency after viral reactivation; in the absence of nanoparticles, NK cells were able to reduce the pool of reactivated cells by a median of 23.08% (Fig. 4b). However, this killing was highly enhanced when cells were cultured in the presence of the pBiAb-AuNPs, further reducing the pool of viral reactivated CD4 T cells by a median of 58.0% (in comparison to median of 23.20% achieved by the control Irre-AuNPs) (Fig. 4c). Therefore, the properties of our pBiAb-AuNPs make them also highly valuable in the context of emerging shock and kill clinical strategies.
Fig. 4.
NK-mediated cytotoxic response against HIV-expressing cells promoted by the pBiAb-AuNPs. (a) Representative flow cytometry plots for NK-mediated cytotoxicity assays. CEM-NKR.CCR5+ cells were double-stained with PKH67 and eF670, and coated with the HIVBal gp120 recombinant protein. Cells were co-cultured with primary NK for 4 h at 1:10 target/effector ratio and in the presence of different nanoconjugates at the following antibody doses: 10 μg/ml for pIrre-AuNPs, 10 μg/ml for pBiAb-AuNPs and 5 μg/ml for each free antibody. Loss of the eF670 marker was used to determine the percentage of dead cells in an eF670 versus PKH67 plot. (b) Summary graph (n = 6) of the ADCC activity mediated by NK cells from healthy donors in the presence of different nanoconjugates at the following antibody doses: 10 μg/ml for irrelevant AuNPs, 10 μg/ml for pBiAb-AuNPs and 5 μg/ml for each free antibody. Represented data is normalized to the positive control condition (plasma from a viremic HIV+ patient). Statistical comparisons were performed using Wilcoxon matched-pairs signed rank test. Median with range (min-max) are shown. *p < 0.05. (c) Graph comparing the ADCC NK-mediated response between healthy donors and treated HIV-infected patients in the presence of the anti-gp120 antibody A32. Statistical comparisons were performed using Mann-Whitney test. Median with range (min-max) are shown. *p < 0.05; **p < 0.01. (d) Summary graph (n = 6) of the ADCC activity mediated by NK cells from treated HIV-infected patients in the presence of different nanoconjugates at the following antibody doses: 10 μg/ml for irrelevant AuNPs, 10 μg/ml for pBiAb-AuNPs and non-polarized rBiAb-AuNPs, and 5 μg/ml for each free antibody. Represented data is normalized to the positive control condition (plasma from a viremic HIV+ patient). Statistical comparisons were performed using Wilcoxon matched-pairs signed rank test. Median with range (min-max) are shown. *p < 0.05. (e) Representative flow cytometry plots showing CD16-expressing cells in digested cells after 24 h of tonsil histoculture in the presence of the pBiAb-AuNPs or Irre-AuNPs (5 μg/ml of antibody burden dose). (f) Summary graph showing CD16-expressing cells after different tissue culture conditions (basal condition, 24 h incubation with 5 μl of pBiAb-AuNPs at 5 μg/ml dose or 5 μl of Irre-AuNPs at 5 μg/ml dose per block) in n = 8 histocultures. Median and ranges (min-max) are shown. Statistical comparisons were performed using Wilcoxon matched-pairs one tailed signed rank test. *p < 0.05; **p < 0.01. (g) Representative flow cytometry plots showing reduction in HIV infection (by detection of the viral protein p24) after 6 days of infection of the tonsil histocultures, in the presence of the pBiAb-AuNPs or Irre-AuNPs (5 μl of the 5 μg/ml dose per tonsil block). (h) Graph summarizing the results of the HIV inhibition assays (measured at day 6 of HIV infection by detecting the p24 antigen expression by flow cytometry) in tonsil histocultures in the presence of the pBiAb-AuNPs or Irre-AuNPs (5 μl of the 5 μg/ml dose per tonsil block) (n = 8). Median with ranges are represented. Statistical comparisons were performed using Wilcoxon matched-pairs signed rank test. *p < 0.05.
Discussion
The development of novel therapeutic strategies to target persistent HIV in patients treated with antiretroviral therapy represents a current priority in the search for an HIV cure [40,41]. Here, with the aim to generate a potent immune-based approach to specifically target and deplete productively HIV-infected and reactivated latently-infected cells, we have constructed novel targeted and polarized bispecific nanoparticles. For that, we have optimized and developed a linker-free method for promoting cooperative antibody conjugation, generating AuNPs with 2 specificities; on the one hand the A32 antibody recognizes the HIV protein gp120 displayed in the cell membrane of productively or viral-reactivated HIV-infected cells, and on the other hand, the anti-CD16 monoclonal antibody (3G8) recognizes the molecule CD16 mainly expressed on the cytotoxic subset of NK cells (CD56dim NK cells). Using this novel conjugation strategy, antibodies were attached to the AuNPs surface forming ordered domains and fully retaining their antigen specificity. Thus, pBiAb-AuNPs were able to significantly enhance the generation of desired cell doublets, and activate NK cells which induced a potent cytotoxic response against both, cultured HIV-expressing cells and infected lymphoid tissue cells. Importantly, and supporting its application in “shock-and-kill” approaches, the pBiAb-AuNPs efficiently reduced the pool of latent HIV-infected cells after pharmacological reactivation.
The results presented here support not only the beneficial effects of the pBiAb-AuNPs bispecific platform to eliminate HIV-infected cells, but also the importance of developing polarizing antibody conjugation methods to construct bispecific nanotools in order to achieve better biological results. Herein, we carefully studied the conjugation medium and the process to obtain bispecific hard protein coronas, driven by a cooperative adsorption of the mix of antibodies, leading to their organization in domains at the AuNP surface. In the case of non-conjugated non-neutralizing anti-HIV antibodies, such as free A32, indirectly recruit NK cells through their Fc portion and promote ADCC activity. The potency of the ADCC immune response depends on many factors, such as the valence of the antibody-epitope binding, the antibody orientation on the bounded antigen and the resulting size of the immune complex [42–44]. Interestingly, pBiAb-AuNPs multivalency (they held about 32 antibodies at their surface) favored all these conditions to take place and triggered a potent anti-HIV NK-mediated cytotoxic response, which was also facilitated by their specific cell engaging capacity.
NK cells express a broad variety of inhibitory and activating cell receptors, and only a specific balance between the expressions of these receptors triggers a cytotoxic immune response [45]. However, CD16, the cognate antigen of 3G8 antibody, has been reported as the only receptor able to activate cytotoxicity and cytokine secretion without the need of additional signals [46]. We proved that direct CD16 engagement by pBiAb-AuNPs helped to achieve a potent anti-HIV cytotoxic response mediated by the stimulation of NK cells and the enhanced contact with cell targets. Free 3G8 antibody and IrreAuNPs slightly increased the percentage of NK cells producing IFN-γ, showing that in general, targeting Fc receptors could stimulate NK cells to produce this cytokine, as previously reported [36]. These irrelevant nanoparticles (IrreAuNPs), used as an unspecific control, are composed of polyclonal IgG mouse and polyclonal IgG rat antibodies, which can bind Fc receptors expressed on NK cells, such as CD16, and provide unspecific cell activation. Of note, it is uncertain the exact orientation of the antibodies on top of the AuNPs. The cell targeting results indicate that the variable chain of the antibodies remains available for antigen recognition. However, it is also possible that the Fc portion stills accesible and might stimulate NK cells through Fc receptors. Both scenarios are likely possible if antibodies are adsorbed laterally on AuNPs. This would explain the results obtained in the cytotoxic assays, both against cultured gp120-coated cells and HIV-infected tonsillar tissue cells, in which control Irre-AuNPs showed certain effect. In the mentioned cytotoxicity assays, pBiAb-AuNPs efficiently eliminated gp120-coated cells, providing better response than Irre-AuNPs and the A32 free antibody, a non-neutralizing antibody highly potent at performing ADCC [35]. Besides, the combination of unconjugated A32 and 3G8 resulted in a weak cytotoxic response, further supporting that their combination and confinement in a polarized and multivalent AuNP platform is essential to mediate potent anti-HIV effect.
Furthermore, pBiAb-AuNPs were able to rescue NK cells from HIV-infected patients, triggering a cytotoxic response similar to the one provided by the positive control. This is noteworthy, since HIV persistent infection leads to the appearance of dysfunctional NK cells with diminished capacity to kill cell targets [6], and this NK cell exhaustion persists despite the antiretroviral treatment [47–49]. Thus, all these properties place pBiAb-AuNPs as an attractive molecularly-targeted nanotool to redirect and potentiate the NK cell response to combat persistent HIV infection. Other nano-based approaches trying to boost anti-viral immunity have been previously developed [50,51].
To our knowledge, the pBiAb-AuNPs are the first nanotool described engaging NK cells with HIV-infected cells. Bispecific AuNPs have been previously developed to target malignant B cells [52]. However, such dual-affinity AuNPs were generated attaching both antibodies via a linker in a randomly-distributed manner, which consequently also attached cells of a similar type. In a similar manner, several bispecific antibody derivatives targeting NK cells through CD16 have been explored for cancer therapy [53,54], showing CD16 engagement as a powerful way for tilting the balance in favor of NK activation.
Importantly, the pBiAb-AuNPs were able to penetrate and greatly reduce the HIV infection in tonsillar lymphoid human tissue. This is of relevance since the majority of the persistent HIV infection in antiretroviral treated individuals, which hinders the eradication of the HIV [55], reside in secondary lymphoid tissue such as the lymph nodes and the GALT [38]. The histocultures assays performed here are highly valuable as evidence for in vivo performance, since the only animal models that fully recapitulate NK cell function are non-human primates [56].
Finally, we tested the potential of the pBiAb-AuNPs to promote a NK-mediated clearance of the HIV-latently infected cells after viral reactivation, mimicking “shock and kill” assays. Latently HIV-infected cells are supposed to be efficiently killed by the immune system after the pharmacologically reactivation of dormant viruses. However, reactivation of viral expression alone has appeared not sufficient to eradicate cell reservoirs [57–59]. Thus, the appropriate potentiation of the immune system is essential to fully eliminate the latent reservoir. NK cells could help to completely clear viral-reactivated cells [4,60,61]. The A32 antibody is a good candidate for targeting latently infected cells in the setting of viral reactivation for several reasons; it broadly reacts against envelopes from all HIV-1 clades [62], A32-like epitope residues are highly conserved [63], there is no evidence of epitope escape mutations [64], and it targets the earliest known epitope expressed post-infection [35]. Importantly, pBiAb-AuNPs extensively reduced the pool of reactivated latently infected cells by triggering a potent NK immune response. This is very relevant since viral-reactivated cells have been reported to be partially resistant to the well-studied CD8 cell-mediated immune responses, and therefore novel strategies are urgently needed [65].
Materials and methods
Ethics statement
PBMCs from healthy donors were obtained from the Blood and Tissue Bank, Barcelona, Spain. The study protocol was approved by the Comitè d’Ètica d’Investigació Clínica (Institutional Review Board number PR(AG)350/2017) of the Hospital Universitari Vall d’Hebron, Barcelona, Spain. Samples were obtained from adults, who all provided written informed consent, and were prospectively collected and cryopreserved in the Biobanc (register number C.0003590). All samples received were totally anonymous and untraceable. PBMCs from HIV-1-infected patients were also obtained from the HIV unit of the Hospital Universitari Vall d’Hebron (Institutional Review Board number PR(AG)350/2017). In addition, tonsil tissue was obtained from eight individuals who were undergoing routine tonsillectomy from the Otolaryngology Department of the Hospital Universitari Vall d´Hebron, Barcelona, Spain (Institutional Review Board number PR(AG)116/2018). All participants gave written informed consent for their participation in these studies.
Cells, antibodies and reagents
We used the cell lines CEM.NKR CCR5+ and ACH-2 (obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH, from Dr. Alexandra Trkola [66,67] and Dr. Thomas Folks [68,69], respectively). PBMCs were isolated by Ficoll-Paque density gradient centrifugation and cryopreserved in liquid nitrogen. Cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS; Gibco, Life Technologies, Inc.), 100 U/ml penicillin, and 100 μg/ml streptomycin (Life Technologies, Inc.) (R10; control medium), and maintained at 37 °C in a 5% CO2 incubator. The A32 antibody was obtained through the AIDS Research and Reference Program, NIAID, NIH (Cat#11438) from Dr. James E. Robinson [70,71]. The anti-CD16 clone 3G8 was purchased from Stemcell. BaL gp120 recombinant protein was also obtained through the NIH AIDS Reagent Program. Polyclonal IgG mouse and IgG rat were purchased from Sigma-Aldrich. IL-2 was obtained from the Vall d´Hebron Hospital pharmacy.
Synthesis of gold nanoparticles and conjugation with antibodies Chemicals
Sodium citrate tribasic dihydrate (≥99%), gold (III) chloride trihydrate HAuCl4•3H2O (99.9% purity) and sodium tetraborate decahydrate were purchased from Sigma-Aldrich. All reagents were used as received without further purification and all glass material was sterilized and dehydrogenated in an oven prior to use. Mili-Q water was used in the preparation of all solutions.
Nanoparticle synthesis
Aqueous solutions of sterile endotoxin free ~40 nm citrate-stabilized AuNPs were synthetized according to previously developed seeded-growth method following a citrate reduction protocol as previously described [72]. AuNPs were obtained after different sequential steps of growing, yielding 5 × 1010 NPs/ml (33 μg/ml). After purification by centrifugation, to discard by-products and contaminants, NPs were resuspended in a solution of 2.2 mM sodium citrate (SC) and stored at 4 °C in the dark. All the particles were used within 20 days after their synthesis.
Nanoparticle conjugation
Following our protocol, antibodies were dispersed in the conjugating medium, consisting in a 4 mM borate 2.2 mM SC solution. The initial antibody concentrations were 3 μg/ml of the anti-HIVgp120 (A32), and 20 μg/ml of the anti-CD16 (clone 3G8). Then, the solution of AuNPs 2.2 mM was added drop-wise (volume ratio antibodies solution:NP solution of 1:1). The mixture was incubated for 48 h at 4 °C under mild agitation conditions (without magnet). Finally, we removed the conjugates from the unbound antibodies by centrifugation at 5000 g× for 40 min. The difference in the antibody concentration used for each antibody is directly related to their different affinity for the AuNP surface, as demonstrated in the isothermal adsorption curves (Fig. 1b and c). We optimized our protocol of conjugation by exploring different mixtures of ions to increase the stability of the process and the conjugates. We found that 3–10 mM range of borate and 2.2 mM SC solution conferred high stability to both antibodies and the nanoparticles (data not shown), allowing a soft interaction between them. In addition, it is well known that a slow and controlled addition of nanoparticles to an ion-optimized solution of diluted antibody (protein), and long-term incubation of the mixture at 4 °C under mild agitation conditions, lead to an ordered disposition of the different antibodies into segregated domains, forming polarized nanoparticles [13]. This cooperative adsorption phenomenon, facilitated by a weak perturbation of the conjugation system, translates the mixture to a new equilibrium, leading to polarized NPs [12,19].
In addition, we reproduced a previously reported antibody-adsorption conjugation protocol to generate bispecific gold nanoparticles [33]. This protocol leads to a random disposition of the antibodies on the surface of gold nanoparticles (non-polarized). Briefly, 133 μl of 15 mM borate buffer pH 8.7 was added to 1 ml of AuNPs. 3 μg/ml of A32 antibody and 20 μg/ml of 3G8 were added and allowed to react under agitation for 30 min. Afterwards, sucrose was incorporated to a final concentration of 5% and incubated for 30 min. Then, 160 μl of 3% bovine serum albumin (BSA) were added and shaken for 10 min. Thereafter, we centrifuged the sample (7500 g× for 30 min) to remove unbound antibodies and AuNPs were re-suspended in 1 ml of 2 mM borate buffer pH 8.7 containing 5% sucrose, 2% glycerol, 0.5% BSA, and 0.01% Tween. The washing step was repeated and the conjugate was re-suspended in 100 μl of the mentioned borate buffer.
Physicochemical characterization of BiAb-AuNPs
Physicochemical characteristics of the synthesized nanoparticles were assessed by different methodologies, such as electron microscopy. To characterize the different conjugates, we evaluated samples by UV-Vis spectrophotometry (Cary 60 UV-Vis Spectrophotometer, Agilent), Dynamic Light Scattering (DLS) and Zeta potential experiments. DLS and Zeta potential experiments were conducted in a Zetasizer Nano series instrument (Malvern). The particles size and Zeta potential were measured simultaneously three times. Previously to the characterization experiments, samples were centrifuged to remove the unbound antibodies.
TEM - SEM analysis
Transmission Electron Microscopy. Gold nanoparticles and Gold nanoparticles – antibody complexes were visualized using FEI MAGELLAN 400 L XHR SEM. This microscope allows for images acquisition in the high energy (15–30 kV) SEM and STEM mode. The ultrathin Formvar-coated 200-mesh copper grid (Ted-pella, Inc.) were directly dipped in the sample solution and left to dry in air overnight. TEM images of the prepared colloidal AuNP were used for the size distribution measurements. For each sample, the size of at least 100 particles was measured and the average size and the standard distribution were obtained. TEM and SEM images of the prepared AuNP – antibody complexes were used for the analysis of surface contact between AuNPs in the case of total antibody surface passivation or partial antibody surface passivation. The charging effect observable on the edge of the NPs visualized by SEM is associated to the presence of the organic layer. This effect provides information on the degree of NP surface passivation by the antibody.
UV-Visible absorption spectroscopy
UV-Visible absorption (UV-Vis) spectra were acquired on an Agilent Cary 60 UV-Vis Spectrophotometer. A total volume of 1 ml of the analysed sample was placed in a plastic cell and the spectral analysis was performed between 300 nm and 800 nm at room temperature. Cary WinUV software by Agilent has been used to collect the spectrophotometer output.
DLS and Zeta potential measurements
The hydrodynamic diameter and the surface charge of the colloids were measured by Dynamic Light Scattering (DLS) and Zeta Potential on a Malvern Zetasizer Nano ZS90 which incorporates a Z-potential analyser (Malvern Instruments Ltd, Worcestershire, UK).
Bioconjugates analysis by TEM-SEM
For conjugation, 50 nm AuNPs were used at final concentration 4 × 1010 NP/ml (total surface area approximately 2 ×1014 nm2/ml). AuNPs were exposed to IgG molecules at two different final protein concentration, corresponding to (i) a situation in which the number of IgG molecules theoretically cover the 60% of the total NPs surface, and (ii) a situation in which IgG molecules theoretically cover the total NP surface with a 100x excess of antibody molecules. The total NP surface was calculated knowing the surface area of the 50 nm AuNPs (7.85 × 103 nm2 per particle). The IgG surface area used to estimate the theoretical number of IgG molecules in solutions, and consequently, the total NP surface coverage was calculated according to the relation with molecular weight specific for globular proteins [73] and approximating the IgG molecules to a sphere. IgG was adsorbed onto AuNPs by incubating the AuNPs with IgG in 10 mM Borate Buffer pH 8.5 for a total time of 48 h at 4 °C under mild agitation (650 RPM). After 48 h of incubation time, IgG-conjugated NPs were analyzed without further purification.
Bioconjugation to Nivolumab
50 nm AuNPs were conjugated to Nivolumab antibody. To perform bioconjugation, we calculated the theoretical number of biomolecules required to cover the whole NP surface knowing the surface area of the 50 nm AuNPs (7.85 × 103 nm2 per particle) and the surface coverage of Nivolumab molecule. For Nivolumab, we estimated the number of antibodies molecules needed to cover the NP surface according to the relation with molecular weight specific for globular proteins [73]. Incubation was performed in 10 mM Phosphate Buffer, pH 7.4 at Room Temperature, by directly diluting the AuNP in the protein solution in the presence of a 100x Nivolumab excess, in order to ensure complete surface coverage.
Distribution of antibodies on cooperatively-conjugated AuNPs by TEM
With the aim to directly observe if the optimized conjugation method leads to the segregation of a mix of antibodies in two different domains (polarization), we performed a straight forward experiment. On the one hand, AuNPs of ~60 nm were doubly conjugated with rabbit anti-HSA IgG (18 μg/ml) and rabbit anti-BSA IgG (18 μg/ml). On the other hand, we conjugated smaller AuNPs as labels: AuNPs of 25 nm were functionalized with BSA (5 μM) and of 15 nm functionalized with HSA (5 μM). The bispecific conjugates were incubated with the AuNPs functionalized with BSA and the AuNPs functionalized with HSA for 24 h. Then, the sample containing the three types of AuNPs was purified from the excess of smaller AuNPs by decantation and after that, the TEM grids were prepared by dipping, by directly immersing the grids in the AuNPs solution and letting them dry in an open atmosphere overnight. Finally, the sample was analyzed by TEM, using an 80-keV TEM JEOL 1010 equipped with an Orius (Gatan) CCD Camera. TEM grids consisted on an ultrathin-formvar coated 200 mesh copper grid covered with a layer of carbon (Ted-Pella). The TEM images of 232 AuNPs of 60 nm were randomly obtained and classified depending on the apparent polarization formed by the specific recognition between 25 nm and 15 nm label AuNPs.
Thermogravimetric assay
Thermogravimetric analyses (TGA) were carried out on a Pyris TGA 8000 under N2 flow with a temperature range from 30 °C to 600 °C at a heating rate of 5 °C min−1. Considering that the limit of detection is a loss in mass of 5 μg of antibody, a total volume of 417 μl of AuNPs was conjugated with mouse IgG producing 0.269 mg of bioconjugates. The purified bioconjugates were deposited in a ceramic crucible, allowing the excess of water to evaporate during 24 h in a laminar flow cabinet.
Simulation model
Coarse-grained (CG) models were employed to extract the relevant characteristics for the studied processes [74]. In our model, one bead comprises three heavy atoms. The bead interaction parameters, topology and composition are chosen to mimic the inherent chemistry for the atoms. The employed model is Dissipative Particle Dynamics (DPD) [75], a stochastic CG simulation model that has been used to study soft matter [76], complex fluids and biomolecular systems, such as self-assembling amphiphilic micelles and bilayers, polymers, proteins, nanoparticles [18,77] and colloidal systems [78].
The cooperative adsorption of immunoglobulins onto AuNPs is studied by DPD-MC hybrid model. All the simulations are performed in a NPT ensemble (constant number of particles N, pressure P and temperature, T) using a dimensionless units system. For our parameterization, ρ = 3, m = 3 and V w = 30 Å and the friction constant (λ) necessary for evaluating the dissipative force is set equal to 4.5. For each system, the length of the simulation is normally 300,000 cycles, equivalent to 1350 ns if using 0.03τ as time step. The working temperature use throughout the simulations is KbT = 0.42 [79,80].
All experiments are performed in a cubic box water of Lx × Ly × Lz = 30 × 30 × 30d 3. Simulations contain 80,000 beads of water (equivalent to 240,000 molecules of water), 3000 beads of gold (equivalent to 9000 gold atoms) that form a unique AuNP and 100 molecules of each biological ligand. All compounds are randomly distributed along the box.
Cell targeting assays
We assessed the binding capabilities of conjugated A32 (anti-HIVgp120) and 3G8 (anti-CD16) antibodies to their targets by flow cytometry. Briefly, for evaluation of A32-monoconjugates we used the CEM.NKR CCR5+ cell line. Cells were coated with 1 μg of the HIV-1 BaL gp120 recombinant protein during 1 h at room temperature (RT). After incubation, cells were extensively washed in ice-cold R10 medium and staining buffer (PBS containing 3% FBS). Thereafter, cells were incubated for 20 min at RT in R10 medium containing naked AuNPs, AuNPs conjugated with A32 or free A32, and stained with an anti-human FITC-labelled secondary antibody (dil 1:100) (Thermo Fisher) for A32 detection (20 min at RT). To assess the targeting capacity of 3G8 monoconjugates, NK cells were obtained from PBMCs of healthy donors by negative isolation using magnetic beads (MagniSort™ Human NK cell Enrichment Kit, eBioscience).
Then, NK cells were incubated in medium containing naked AuNPs, AuNPs conjugated with 3G8 or free 3G8. For detection of 3G8 by flow cytometry, we used an anti-mouse AF647-labelled secondary antibody (dil 1:200) (Invitrogen). R10 media controls were included for both conjugates. Cells were then fixed with paraformaldehyde solution (PFA; Affymetrix) (2%) and acquired in an LSR Fortessa flow cytometer (Becton Dickinson). Results were analyzed with FlowJo v10 software (Tree Star).
Cell-to-cell contact assays
CD4+ T cells and NK cells were isolated from cryopreserved PBMCs of healthy donors by commercial kits (MagniSort Human CD4+ T Cell Enrichment; Affymetrix, and MagniSort™ Human NK cell Enrichment; eBioscience). CD4+ T cells were coated with 1 μg of recombinant gp120 protein during 1 h at RT. After extensive washes, CD4+ T cells were mixed with NK cells in a 1:1 ratio and stained with anti-CD56 (PE; Becton Dickinson) and anti-CD3 (PE-Cy7; Becton Dickinson) antibodies for detection of NK cells and CD4 T cells, respectively. After washing, we added naked AuNPs or BiAb-AuNPs at two different doses (2.5 μg/ml and 10 μg/ml of total antibody). After 20 min incubation at RT, cells were washed and fixed with PFA (2%). Samples were then acquired on a LSR Fortessa flow cytometer (Becton Dickinson) and data were analyzed using FlowJo V10 software.
Confocal microscopy
Attachment of BiAb-AuNPs to their targets and subsequently promotion of dual cell conjugates were assessed by confocal microscopy. First, CEM.NKR CCR5+ cells were stained with 2 μM of the membrane lipid marker PKH67 (Sigma-Aldrich) according to the manufacturer instructions. These cells were also coated with 1 μg of HIV-1 BaL gp120 recombinant protein for 1 h at RT. Then, NK cells isolated from PBMCs of healthy donors were obtained as described above. NKs and PKH67-labelled CEM.NKR CCR5+ cells were mixed at 1:1 ratio and incubated for 20 min at RT with pBiAb-AuNPs (2.5 μg/ml of total antibody burden), then attached to coverslips (by centrifugation at 800 ×g for 5 min) previously coated with 0.1 mg/ml of poly-L-lysine (Sigma-Aldrich) and fixed with PFA (4%) for 15 min at RT. Cells were finally stained with DAPI (1:5000 dilution) (Thermo Fisher) and mounted with Fluoromount G (eBioscience). Preparations were imaged with an Olympus Spectral Confocal Microscope FV1000. ImageJ software was used for image compositions. A total of 20 images were counted, containing 1650 cells. Note that AuNPs of 40 nm do not reflect or diffract light because they are below the visible light resolution limit, but they are good at dispersing light, especially if aggregated. Thus, the light scattering/dispersion can be observed with a confocal microscope [25].
NK cell activation and functional activity assays
The activation and cytotoxicity phenotype of NK cells after stimulation with pBiAb-AuNPs was evaluated. Cytotoxicity was assessed by the measurement of CD107a and IFN-ɤ. Briefly, PBMCs were cultured in R10 medium with 10 μg/ml pBiAb-AuNPs, 10 μg/ml Irre-AuNPs, 5 μg/ml of A32, 5 μg/ml of 3G8 or 10 ng/ml PMA plus 1 μM ionomycin (positive control) for 4.5 h in a 96-well plate at 37°C and 5% CO2. CD107a-PE-Cy5 (H4A3; Becton Dickinson), BD GolgiPlug Protein Transport Inhibitor (Beckton Dickinson) and BD GolgiStop Protein Transport Inhibitor containing monensin (Becton Dickinson) were also added to each well at the recommended concentrations. Cells were then washed and stained with a viability dye (LIVE/DEAD Fixable Violet dead cell stain; Thermo Fisher). To measure the activation marker HLA-DR, cells were stained with anti-CD56-FITC (B159; Becton Dickinson), anti-CD3-PE-Cy7 (SK7; Becton Dickinson) and anti-HLA-DR-SB600 (LN3; eBioscience) antibodies for 20 min at RT. Cells were then fixed and permeabilized with Fixation/Permeabilization Solution (Becton Dickinson) for 20 min at 4 °C, washed with BD Perm/Wash buffer and stained with anti-IFN-ɤ AF700 (Life technologies) for 30 min at 4 °C. After washing, cells were fixed with PFA (2%) and acquired on an LSR Fortessa flow cytometer (Becton Dickinson). Data were analyzed using FlowJo V10 software.
Antibody-dependent cell-mediated cytotoxicity (ADCC) assay
The ADCC assay was performed as described by others with some modifications [81]. Briefly, CEM-NKR. CCR5 cells were used as target cells after being double-stained with PKH67 (Sigma-Aldrich) and eF670 (Labclinics) dyes following manufacturer’s instructions. Then, cells were coated with 1 μg of the HIV-1 BaL gp120 recombinant protein for 1 h at RT and extensively washed in ice-cold R10 medium. Target cells were dispensed in U-bottom 96-well plates (5000 cells/well) and incubated for 15 min with 5 μg/ml of free A32, 5 μg/ml of free 3G8, Irre-AuNPs or BiAb-AuNPs (10 μg/ml antibody burden), and plasma (1:1000 dilution) from a viremic (high viral load in blood) HIV+ patient as a positive control. After incubation, NK effector cells isolated from PBMCs of healthy donors or antiretroviral therapy (ART) suppressed HIV+ patients were added at 1:10 target/effector ratio. Plates were centrifuged and incubated for 4 h at 37 °C and 5% CO2. Finally, cells were washed, fixed with PFA (2%), acquired on a LSR Fortessa flow cytometer (Becton Dickinson) and analyzed using FlowJo software. Flow cytometry count beads (AccuCount Blank Particles, Cytognos) are used to normalize cell collection to a constant number of particles (1000 events). Target cells were identified in a PKH67-versus side scatter (SSC) plot. Loss of the eF670 marker was used to determine the percentage of killing in an eF670 versus PKH67 plot. ADCC on virally-reactivated cells was performed as follows; the latently-infected cell line ACH-2 was stimulated to produce HIV by the addition of 10 μg/ml PHA (Phytohemagglutinin; Fisher Scientific) and 10 nM PMA (phorbol myristate acetate; Abcam) during 17 h at 37 °C. Then, cells were subjected to the ADCC assay as described above. ADCC of viral-reactivated cells was calculated as the fraction of cells that disappeared within the target population after the addition of pBiAb-AuNPs in comparison to the control condition with targets and NK cells, but lacking the nanoparticles. To assess the absolute number of cells killed by ADCC, we added to each well flow cytometry particles for absolute cell counting (5 × 104/ml) (AccuCount Blank 5.0–5.9 μm, Cytognos).
Tonsillar tissue processing and infection
Tonsils from healthy children were obtained and processed as previously described [82]. Briefly, the tissue was processed within the first 2 h following surgery. We dissected the tissue into uniform blocks of approximately 2 mm × 2 mm x 1 mm, after discarding all cauterized, bloody or necrotic areas. Ten blocks per condition were placed on top of a piece of sponge (Absorbable gelatin Surgispon; AEGIS LIFESCIENCES) suspended in RPMI1640 medium supplemented with 20% FBS (R20) and 100 U/ml penicillin and 100 μg/ml streptomycin in a 6-well plate. Then, for viral infection experiments, 5 μl of HIVBal virus (457 TCID50) were added on top of each tissue block. Some pieces were not infected as a control. After 15 min incubation, we added 5 μl of pBiAb-AuNPs or Irre-AuNPs at 5 μg/ml of total antibody burden on each block and placed the plate in the incubator (37 °C 5% CO2). We performed two different assays with the tonsillar blocks. First, after one day incubation with the nanoparticles, we digested the blocks as previously described [82] and performed the staining with LIVE/DEAD Far Red viability (Thermo Fisher) to identify dead cells for 30 min at RT. After washing with PBS, we stained the digested cells with the following labelled antibodies: anti-CD45-FITC (HI30; Biolegend), anti-CD3-PerCP (SK7; Becton Dickinson), anti-CD8-APC (RPA-T8; Becton Dickinson) and anti-CD16-PE (3G8; Biolegend), for 20 min at RT. Then, cells were washed and fixed with PFA (2%). Second, we assessed HIV infection at day 6 of tonsil culture by tissue digestion and staining with LIVE/DEAD Far Red viability, anti-CD45-FITC, anti-CD3-PerCP and anti-CD8-APC as described. After the surface staining, cells were fixed and permeabilized with Fixation/Permeabilization Solution (Becton Dickinson) for 20 min at 4 °C, washed with BD Perm/Wash buffer and stained with anti-p24-PE (Beckman Coulter) for 20 min on ice and 20 min at RT. Finally, cells were washed and fixed with PFA (2%). Samples were acquired in a FACSCalibur flow cytometer (Becton Dickinson) and analyzed with FlowJo V10 software.
Generation of latency model, viral reactivation and cytotoxicity assays
PBMCs from healthy donors or HIV-infected patients were thawed. Then, CD4 T cells were isolated using a commercial kit (MagniSort Human CD4+ T Cell Enrichment; Affymetrix) and infected with HIV (350,000 TDCI50/million cells) with NL4.3 viral strain for 4 h at 37 °C 5% CO2 or by spinoculation at 1200 ×g and 37 °C for 2 h. Cells were washed twice with PBS and cultured in R10 with IL-2 (10 U/ml) and IL-7 (1 nM) (Bio-Techne R&D Systems). After 2 days of infection, Raltegravir (1 μM), Darunavir (1 μM) and Nevirapin (1 μM) were added to the cell culture to prevent new rounds of viral infection. Next day, cells were incubated with Q-VD (10 μM) for 2 h at 37 °C and 5% CO2, in R10 medium containing IL-2, IL-7, Raltegravir, Darunavir and Nevirapin at the indicated concentrations, and then latent viral infection was reactivated during 22 h with Ingenol (100 nM) and subjected to the cytotoxic NK assay. NK cells were added at ratio 1:1 in the presence or absence of pBiAb-AuNPs or irrelevant AuNPs (2.5 ug/ml total antibody burden), for 22 h at 37 °C and 5% CO2. In some cases, we included as a negative control AuNPs conjugated with the antibody Palivizumab. The cytotoxic assays were performed in round bottom 96 wells plates, at a cellular concentration of 2.5 M cells/ml. Finally, cells were stained with LIVE/DEAD violet viability (Thermo Fisher) for 20 min at RT. After washing once with staining buffer, cells were stained with anti-CD3-AF700 (SK7; Biolegend) and anti-CD56-FITC (B159; Becton Dickinson) antibodies for 20 min at RT. Cells were then fixed and permeabilized with Fixation/Permeabilization Solution (Becton Dickinson) for 20 min at 4 °C, washed with BD Perm/Wash buffer and stained with anti-p24-PE (Beckman Coulter) for 20 min on ice and 20 min at RT. After washing, cells were fixed with PFA (2%). Samples were acquired on an LSR Fortessa flow cytometer (Becton Dickinson) and data analyzed using FlowJo V10 software.
Cell toxicity of pBiAb-AuNPs and Irre-AuNPs was measured using the primary cell line CEM-NKR and ex-vivo infected primary CD4+ T cells. Cells were incubated with the different nanoconjugates for 4 and 48 h. Then, cell viability was determined by staining with a LIVE/DEAD viability dye and measured by flow cytometry.
Statistical analysis
Analyses were performed with GraphPad Prism v6 and statistical tests are reported within each figure legend. p values < 0.05 were considered statistically significant.
Supplementary Material
Fig. 5.
The pBiAb-AuNPs enhance the killing of the latently HIV-infected cells after viral reactivation. (a) Cytotoxic response against latently-infected ACH-2 cells after viral reactivation. PHA and PMA-reactivated ACH-2 cells were cultured with primary NK cells at ratio 1:10 target/effector for 4 h in the presence or absence of the pBiAb-AuNPs at 10 μg/ml. Cytotoxicity was calculated as the disappearance of ACH-2 target cells using flow cytometry count beads. Data is normalized to the control condition in absence of the pBiAb-AuNPs. Mean with SD of 4 independent experiments is shown. (b) Graph representing the cytotoxic response mediated by NK cells in a primary cell model of HIV latency and after viral reactivation with Ingenol (ING) (100 nM) (ratio reactivated cells:NK cells 1:1) in the absence of pBiAb-AuNPs. Median with range of n = 5 experiments is represented. (c) Summary graph of the enhancement of the NK-mediated cytotoxic response against viral reactivated CD4+ T cells induced by the presence of the pBiAb-AuNPs or Irre-AuNPs (2.5 μg/ml total antibody burden) (n = 5). Median with range is represented. Statistical comparisons were performed using the one sample t-test. *p < 0.05.
Acknowledgements
The authors thank Jana Oliveras and Javy Patarroyo for helping in the acquisition of TEM and SEM images of NP antibody complexes.
Funding
This study was supported by the Spanish Secretariat of Science and Innovation and FEDER funds (grants SAF2015-67334-R and RTI2018-101082-B-I00 [MINECO/FEDER]), American National Institutes of Health (grant R21AI118411 to M.B), an unrestricted research grant from Bristol-Myers Squibb S.A.U (PfC-2015-AI424-564) to M.B, the Spanish “Ministerio de Economía y Competitividad, Instituto de Salud Carlos III” (ISCIII, PI17/01470) to M.G and the Spanish “Ministerio de Economía y Competitividad, Instituto de Salud Carlos III” (ISCIII, PI14/01058) to J.G.P, a research grant from Gilead Sciences (GLD17-00204 and GLD19-00084) to M.B, GeSIDA and the Spanish AIDS network “Red Temática Cooperativa de Investigación en SIDA” (RD16/0025/0007). The Miguel Servet program funded by the Spanish Health Institute Carlos III (CP17/00179) to M.B and J.G.P (CPII15/00014). The “Pla estratègic de recerca i innovació en salut” (PERIS), from the Catalan Government to M.G. The Spanish Secretariat of Science and Innovation Ph.D. fellowship to A.A-G (BES-2016-076382), AGAUR-FI-B-00582 Ph.D. fellowship from the Catalan Government to O.BL, and PIF-UAB Ph.D. fellowship from Universitat Autònoma de Barcelona to R.SL. The funders had no role in study design, data collection and analysis, the decision to publish or preparation of the manuscript.
Footnotes
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.nantod.2020.101056.
References
- [1].Zhukovsky EA, Morse RJ, Maus MV, Bispecific antibodies and CARs: generalized immunotherapeutics harnessing T cell redirection, Curr. Opin. Immunol 40 (2016) 24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Huot N, Jacquelin B, Garcia-Tellez T, Rascle P, Ploquin MJ, Madec Y, Reeves RK, Derreudre-Bosquet N, Müller-Trutwin M, Natural killer cells migrate into and control simian immunodeficiency virus replication in lymph node follicles in African green monkeys, Nat. Med 23 (11) (2017) 1277–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Alter G, Martin MP, Teigen N, Carr WH, Suscovich TJ, Schneidewind A, Streeck H, Waring M, Meier A, Brander C, Lifson JD, Allen TM, Carrington M, Altfeld M, Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes, J. Exp. Med 204 (12) (2007) 3027–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Marras F, Casabianca A, Bozzano F, Ascierto ML, Orlandi C, Di Biagio A, Pontali E, Dentone C, Orofino G, Nicolini L, Taramasso L, Magnani M, Marincola FM, Wang E, Moretta L, De Maria A, Control of the HIV-1 DNA reservoir is associated in vivo and in vitro with NKp46/NKp30 (CD335 CD337) inducibility and interferon gamma production by transcriptionally unique NK cells, J. Virol 91 (23) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Chun TW, Stuyver L, Mizell SB, Ehler LA, Mican JAM, Baseler M, Lloyd AL, Nowak MA, Fauci AS, Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy, Proc. Natl. Acad. Sci. U. S. A 94 (24) (1997) 13193–13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bi J, Tian Z, NK cell exhaustion, Front. Immunol 8 (2017) 760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zhang L, Chung C, Hu BS, He T, Guo Y, Kim AJ, Skulsky E, Jin X, Hurley A, Ramratnam B, Markowitz M, Ho DD, Genetic characterization of rebounding HIV-1 after cessation of highly active antiretroviral therapy, J. Clin. Investig 106 (7) (2000) 839–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Yuan H, Jiang W, von Roemeling CA, Qie Y, Liu X, Chen Y, Wang Y, Wharen RE, Yun K, Bu G, Knutson KL, Kim BYS, Multivalent bi-specific nanobioconjugate engager for targeted cancer immunotherapy, Nat. Nanotechnol 12 (8) (2017) 763–769. [DOI] [PubMed] [Google Scholar]
- [9].Jiang W, Kim BYS, Rutka JT, Chan WCW, Nanoparticle-mediated cellular response is size-dependent, Nat. Nanotechnol 3 (3) (2008) 145–150. [DOI] [PubMed] [Google Scholar]
- [10].Bhattacharyya S, Bhattacharya R, Curley S, McNiven MA, Mukherjee P, Nanoconjugation modulates the trafficking and mechanism of antibody induced receptor endocytosis, Proc. Natl. Acad. Sci. U. S. A 107 (33) (2010) 14541–14546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Garrido C, Simpson CA, Dahl NP, Bresee J, Whitehead DC, Lindsey EA, Harris TL, Smith CA, Carter CJ, Feldheim DL, Melander C, Margolis DM, Gold nanoparticles to improve HIV drug delivery, Future Med. Chem 7 (9) (2015) 1097–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ramsden JJ, Bachmanova GI, Archakov AI, Kinetic evidence for protein clustering at a surface, Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Top 50 (6) (1994) 5072–5076. [DOI] [PubMed] [Google Scholar]
- [13].Casals E, Pfaller T, Duschl A, Oostingh GJ, Puntes V, Time evolution of the nanoparticle protein corona, ACS Nano 4 (7) (2010) 3623–3632. [DOI] [PubMed] [Google Scholar]
- [14].Hlady VV, Buijs J, Protein adsorption on solid surfaces, Curr. Opin. Biotechnol 7 (1) (1996) 72–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Monopoli MP, Åberg C, Salvati A, Dawson KA, Biomolecular coronas provide the biological identity of nanosized materials, Nat. Nanotechnol 7 (12) (2012) 779–786. [DOI] [PubMed] [Google Scholar]
- [16].Colangelo E, Comenge J, Paramelle D, Volk M, Chen Q, Lévy R, Characterizing self-assembled monolayers on gold nanoparticles, Bioconjugate Chem. 28 (1) (2017) 11–22. [DOI] [PubMed] [Google Scholar]
- [17].Vroman L, Effect of absorbed proteins on the wettability of hydrophilic and hydrophobic solids, Nature 196 (1962) 476–477. [DOI] [PubMed] [Google Scholar]
- [18].Raman A, Jaime C, Puntes VF, Domain formation and conformational changes in gold nanoparticle conjugates studied using DPD simulations, Langmuir 33 (50) (2017) 14502–14512. [DOI] [PubMed] [Google Scholar]
- [19].Jackson AM, Myerson JW, Stellacci F, Spontaneous assembly of subnanometre-ordered domains in the ligand shell of monolayer-protected nanoparticles, Nat. Mater 3 (5) (2004) 330–336. [DOI] [PubMed] [Google Scholar]
- [20].Singh C, Ghorai PK, Horsch MA, Jackson AM, Larson RG, Stellacci F, Glotzer SC, Entropy-mediated patterning of surfactant-coated nanoparticles and surfaces, Phys. Rev. Lett 99 (22) (2007) 226106. [DOI] [PubMed] [Google Scholar]
- [21].Sajadi MM, Lewis GK, Seaman MS, Guan Y, Redfield RR, DeVico AL, Signature biochemical properties of broadly cross-reactive HIV-1 neutralizing antibodies in human plasma, J. Virol 86 (9) (2012) 5014–5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Piella J, Bastus NG, Puntes V, Size-dependent protein-nanoparticle interactions in citrate-stabilized gold nanoparticles: the emergence of the protein corona, Bioconjugate Chem. 28 (1) (2017) 88–97. [DOI] [PubMed] [Google Scholar]
- [23].Pillai PP, Kowalczyk B, Pudlo WJ, Grzybowski BA, Electrostatic titrations reveal surface compositions of mixed, on-nanoparticle monolayers comprising positively and negatively charged ligands, J. Phys. Chem C 120 (2016) 4139–4144. [Google Scholar]
- [24].Zhang D, Neumann O, Wang H, Yuwono VM, Barhoumi A, Perham M, Hartgerink JD, Wittung-Stafshede P, Halas NJ, Gold nanoparticles can induce the formation of protein-based aggregates at physiological pH, Nano Lett. 9 (2) (2009) 666–671. [DOI] [PubMed] [Google Scholar]
- [25].García-Fernández L, Garcia-Pardo J, Tort O, Prior I, Brust M, Casals E, Lorenzo J, Puntes VF, Conserved effects and altered trafficking of Cetuximab antibodies conjugated to gold nanoparticles with precise control of their number and orientation, Nanoscale 9 (18) (2017) 6111–6121. [DOI] [PubMed] [Google Scholar]
- [26].Saha B, Evers TH, Prins MW, How antibody surface coverage on nanoparticles determines the activity and kinetics of antigen capturing for biosensing, Anal. Chem 86 (16) (2014) 8158–8166. [DOI] [PubMed] [Google Scholar]
- [27].Ong Q, Luo Z, Stellacci F, Characterization of ligand shell for mixed-ligand coated gold nanoparticles, Acc. Chem. Res 50 (8) (2017) 1911–1919. [DOI] [PubMed] [Google Scholar]
- [28].Comenge J, The role of PEG conformation in mixed layers: from protein corona substrate to steric stabilization avoiding protein adsorption, Sci. Open Res (2015). [Google Scholar]
- [29].Cesbron Y, Shaw CP, Birchall JP, Free P, Lévy R, Stripy nanoparticles revisited, Small 8 (24) (2012) 3714–3719 author reply 3720–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ong QK, Stellacci F, Response to “Critical assessment of the evidence for striped nanoparticles”, PLoS One 10 (11) (2015) e0135594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Duchesne L, Wells G, Fernig DG, Harris SA, Lévy R, Supramolecular domains in mixed peptide self-assembled monolayers on gold nanoparticles, Chembiochem 9 (13) (2008) 2127–2134. [DOI] [PubMed] [Google Scholar]
- [32].Bonomi R, Cazzolaro A, Prins LJ, Assessment of the morphology of mixed SAMs on Au nanoparticles using a fluorescent probe, Chem. Commun 47 (1) (2011) 445–447. [DOI] [PubMed] [Google Scholar]
- [33].Ciaurriz P, Fernández F, Tellechea E, Moran JF, Asensio AC, Comparison of four functionalization methods of gold nanoparticles for enhancing the enzyme-linked immunosorbent assay (ELISA), Beilstein J. Nanotechnol 8 (2017) 244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Jain PK, Lee KS, El-Sayed IH, El-Sayed MA, Calculated absorption and scattering properties of gold nanoparticles of different size, shape, and composition: applications in biological imaging and biomedicine, J. Phys. Chem. B 110 (14) (2006) 7238–7248. [DOI] [PubMed] [Google Scholar]
- [35].Ferrari G, Pollara J, Kozink D, Harms T, Drinker M, Freel S, Moody MA, Alam SM, Tomaras GD, Ochsenbauer C, Kappes JC, Shaw GM, Hoxie JA,Robinson JE, Haynes BF, An HIV-1 gp120 envelope human monoclonal antibody that recognizes a C1 conformational epitope mediates potent antibody-dependent cellular cytotoxicity (ADCC) activity and defines a common ADCC epitope in human HIV-1 serum, J. Virol 85 (14) (2011) 7029–7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Jacobi C, Claus M, Wildemann B, Wingert S, Korporal M, Römisch J, Meuer S, Watzl C, Giese T, Exposure of NK cells to intravenous immunoglobulin induces IFN gamma release and degranulation but inhibits their cytotoxic activity, Clin. Immunol 133 (3) (2009) 393–401. [DOI] [PubMed] [Google Scholar]
- [37].Cantero J, Genesca M, Maximizing the immunological output of the cervicovaginal explant model, J. Immunol. Methods 460 (2018) 26–35. [DOI] [PubMed] [Google Scholar]
- [38].Estes JD, Kityo C, Ssali F, Swainson L, Makamdop KN, Del Prete GQ, Deeks SG, Luciw PA, Chipman JG, Beilman GJ, Hoskuldsson T, Khoruts A, Anderson J, Deleage C, Jasurda J, Schmidt TE, Hafertepe M, Callisto SP, Pearson H, Reimann T, Schuster J, Schoephoerster J, Southern P, Perkey K, Shang L, Wietgrefe SW, Fletcher CV, Lifson JD, Douek DC, McCune JM, Haase AT, Schacker TW, Defining total-body AIDS-virus burden with implications for curative strategies, Nat. Med 23 (11) (2017) 1271–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kennedy J, Larrañeta E, McCrudden MTC, McCrudden CM, Brady AJ, Fallows SJ, McCarthy HO, Kissenpfennig A, Donnelly RF, In vivo studies investigating biodistribution of nanoparticle-encapsulated rhodamine B delivered via dissolving microneedles, J. Control. Release 265 (2017) 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Cao S, Woodrow KA, Nanotechnology approaches to eradicating HIV reservoirs, Eur. J. Pharm. Biopharm 138 (2019) 48–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Bowen A, Sweeney EE, Fernandes R, Nanoparticle-based immunoengineered approaches for combating HIV, Front. Immunol 11 (2020) 789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Boesch AW, Brown EP, Ackerman ME, The role of Fc receptors in HIV prevention and therapy, Immunol. Rev 268 (1) (2015) 296–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lux A, Yu X, Scanlan CN, Nimmerjahn F, Impact of immune complex size and glycosylation on IgG binding to human FcgammaRs, J. Immunol 190 (8) (2013) 4315–4323. [DOI] [PubMed] [Google Scholar]
- [44].Acharya P, Tolbert WD, Gohain N, Wu X, Yu L, Liu T, Huang W, Huang C, Kwon YD, Louder RK, Luongo TS, McLellan JS, Pancera M, Yang Y, Zhang B, Flinko R, Foulke JS, Sajadi MM, Kamin-Lewis R, Robinson JE, Martin L, Kwong PD, Guan Y, DeVico AL, Lewis GK, Pazgier M, Structural definition of an antibody-dependent cellular cytotoxicity response implicated in reduced risk for HIV-1 infection, J. Virol 88 (21) (2014) 12895–12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lanier LL, Up on the tightrope: natural killer cell activation and inhibition, Nat. Immunol 9 (5) (2008) 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Bryceson YT, March ME, Ljunggren HG, Long EO, Synergy among receptors on resting NK cells for the activation of natural cytotoxicity and cytokine secretion, Blood 107 (1) (2006) 159–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Azzoni L, Papasavvas E, Chehimi J, Kostman JR, Mounzer K, Ondercin J, Perussia B, Montaner LJ, Sustained impairment of IFN-gamma secretion in suppressed HIV-infected patients despite mature NK cell recovery: evidence for a defective reconstitution of innate immunity, J. Immunol 168 (11) (2002) 5764–5770. [DOI] [PubMed] [Google Scholar]
- [48].Chehimi J, Azzoni L, Farabaugh M, Creer SA, Tomescu C, Hancock A, Mackiewicz A, D’Alessandro L, Ghanekar S, Foulkes AS, Mounzer K, Kostman J, Montaner LJ, Baseline viral load and immune activation determine the extent of reconstitution of innate immune effectors in HIV-1-infected subjects undergoing antiretroviral treatment, J. Immunol 179 (4) (2007) 2642–2650. [DOI] [PubMed] [Google Scholar]
- [49].Lichtfuss GF, Cheng WJ, Farsakoglu Y, Paukovics G, Rajasuriar R, Velayudham P, Kramski M, Hearps AC, Cameron PU, Lewin SR, Crowe SM, Jaworowski A, Virologically suppressed HIV patients show activation of NK cells and persistent innate immune activation, J. Immunol 189 (3) (2012) 1491–1499. [DOI] [PubMed] [Google Scholar]
- [50].Sweeney EE, Balakrishnan PB, Powell AB, Bowen A, Sarabia I, Burga RA, Jones RB, Bosque A, Cruz CRY, Fernandes R, PLGA nanodepots co-encapsulating prostratin and anti-CD25 enhance primary natural killer cell antiviral and antitumor function, Nano Res. 13 (3) (2020) 736–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Jones RB, Mueller S, Kumari S, Vrbanac V, Genel S, Tager AM, Allen TM, Walker BD, Irvine DJ, Antigen recognition-triggered drug delivery mediated by nanocapsule-functionalized cytotoxic T-cells, Biomaterials 117 (2017) 44–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Yeheskely-Hayon D, Minai L, Golan L, Dann EJ, Yelin D, Optically induced cell fusion using bispecific nanoparticles, Small 9 (22) (2013) 3771–3777. [DOI] [PubMed] [Google Scholar]
- [53].Wiernik A, Foley B, Zhang B, Verneris MR, Warlick E, Gleason MK, Ross JA, Luo X, Weisdorf DJ, Walcheck B, Vallera DA, Miller JS, Targeting natural killer cells to acute myeloid leukemia in vitro with a CD16 × 33 bispecific killer cell engager and ADAM17 inhibition, Clin. Cancer Res 19 (14) (2013) 3844–3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Gleason MK, Verneris MR, Todhunter DA, Zhang B, McCullar V, Zhou SX, Panoskaltsis-Mortari A, Weiner LM, Vallera DA, Miller JS, Bispecific and trispecific killer cell engagers directly activate human NK cells through CD16 signaling and induce cytotoxicity and cytokine production, Mol. Cancer Ther 11 (12) (2012) 2674–2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Joos B, Fischer M, Kuster H, Pillai SK, Wong JK, Boni J, Hirschel B, Weber R, Trkola A, Gunthard HF, HIV rebounds from latently infected cells, rather than from continuing low-level replication, Proc. Natl. Acad. Sci. U. S. A 105 (43) (2008) 16725–16730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Manickam C, Shah SV, Nohara J, Ferrari G, Reeves RK, Monkeying around: using non-human primate models to study NK cell biology in HIV infections, Front. Immunol 10 (2019) 1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Rasmussen TA, Tolstrup M, Brinkmann CR, Olesen R, Erikstrup C, Solomon A, Winckelmann A, Palmer S, Dinarello C, Buzon M, Lichterfeld M, Lewin SR, Østergaard L, Søgaard OS, Panobinostat, a histone deacetylase inhibitor, for latent-virus reactivation in HIV-infected patients on suppressive antiretroviral therapy: a phase 1/2, single group, clinical trial, Lancet HIV 1 (1) (2014) e13–e21. [DOI] [PubMed] [Google Scholar]
- [58].Archin NM, Liberty AL, Kashuba AD, Choudhary SK, Kuruc JD, Crooks AM, Parker DC, Anderson EM, Kearney MF, Strain MC, Richman DD, Hudgens MG, Bosch RJ, Coffin JM, Eron JJ, Hazuda DJ, Margolis DM, Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy, Nature 487 (7408) (2012) 482–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Elliott JH, Wightman F, Solomon A, Ghneim K, Ahlers J, Cameron MJ, Smith MZ, Spelman T, McMahon J, Velayudham P, Brown G, Roney J, Watson J, Prince MH, Hoy JF, Chomont N, Fromentin R, Procopio FA, Zeidan J, Palmer S, Odevall L, Johnstone RW, Martin BP, Sinclair E, Deeks SG, Hazuda DJ, Cameron PU, Sékaly RP, Lewin SR, Activation of HIV transcription with short-course vorinostat in HIV-infected patients on suppressive antiretroviral therapy, PLoS Pathog. 10 (10) (2014) e1004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, Liao HX, DeVico AL, Lewis GK, Williams C, Pinter A, Fong Y, Janes H, DeCamp A, Huang Y, Rao M, Billings E, Karasavvas N, Robb ML, Ngauy V, de Souza MS, Paris R, Ferrari G, Bailer RT, Soderberg KA, Andrews C, Berman PW, Frahm N, De Rosa SC, Alpert MD, Yates NL, Shen X, Koup RA, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Michael NL, Kim JH, Immune-correlates analysis of an HIV-1 vaccine efficacy trial, N. Engl. J. Med 366 (14) (2012) 1275–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Olesen R, Vigano S, Rasmussen TA, Søgaard OS, Ouyang Z, Buzon M, Bashirova A, Carrington M, Palmer S, Brinkmann CR, Yu XG, Østergaard L, Tolstrup M, Lichterfeld M, Innate immune activity correlates with CD4 T cell-associated HIV-1 DNA decline during latency-reversing treatment with panobinostat, J. Virol 89 (20) (2015) 10176–10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Moore JP, McCutchan FE, Poon SW, Mascola J, Liu J, Cao Y, Ho DD, Exploration of antigenic variation in gp120 from clades A through F of human immunodeficiency virus type 1 by using monoclonal antibodies, J. Virol 68 (12) (1994) 8350–8364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Tolbert WD, Gohain N, Veillette M, Chapleau JP, Orlandi C, Visciano ML, Ebadi M, DeVico AL, Fouts TR, Finzi A, Lewis GK, Pazgier M, Paring down HIV Env: design and crystal structure of a stabilized inner domain of HIV-1 gp120 displaying a major ADCC target of the A32 region, Structure 24 (5) (2016) 697–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Lewis GK, Pazgier M, DeVico AL, Survivors remorse: antibody-mediated protection against HIV-1, Immunol. Rev 275 (1) (2017) 271–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Deng K, Pertea M, Rongvaux A, Wang L, Durand CM, Ghiaur G, Lai J, McHugh HL, Hao H, Zhang H, Margolick JB, Gurer C, Murphy AJ, Valenzuela DM, Yancopoulos GD, Deeks SG, Strowig T, Kumar P, Siliciano JD, Salzberg SL, Flavell RA, Shan L, Siliciano RF, Broad CTL response is required to clear latent HIV-1 due to dominance of escape mutations, Nature 517 (7534) (2015) 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Howell DN, et al. , Natural killing target antigens as inducers of interferon: studies with an immunoselected, natural killing-resistant human T lymphoblastoid cell line, J. Immunol 134 (2) (1985) 971–976. [PubMed] [Google Scholar]
- [67].Trkola A, Matthews J, Gordon C, Ketas T, Moore JP, A cell line-based neutralization assay for primary human immunodeficiency virus type 1 isolates that use either the CCR5 or the CXCR4 coreceptor, J. Virol 73 (11) (1999) 8966–8974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Clouse KA, et al. , Monokine regulation of human immunodeficiency virus-1 expression in a chronically infected human T cell clone, J. Immunol 142 (2) (1989) 431–438. [PubMed] [Google Scholar]
- [69].Folks TM, Clouse KA, Justement J, Rabson A, Duh E, Kehrl JH, Fauci AS, Tumor necrosis factor alpha induces expression of human immunodeficiency virus in a chronically infected T-cell clone, Proc. Natl. Acad. Sci. U. S. A 86 (7) (1989) 2365–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Moore JP, Thali M, Jameson BA, Vignaux F, Lewis GK, Poon SW, Charles M, Fung MS, Sun B, Durda PJ, Immunochemical analysis of the gp120 surface glycoprotein of human immunodeficiency virus type 1: probing the structure of the C4 and V4 domains and the interaction of the C4 domain with the V3 loop, J. Virol 67 (8) (1993) 4785–4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Wyatt R, Moore J, Accola M, Desjardin E, Robinson J, Sodroski J, Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding, J. Virol 69 (9) (1995) 5723–5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Bastus NG, Comenge J, Puntes V, Kinetically controlled seeded growth synthesis of citrate-stabilized gold nanoparticles of up to 200 nm: size focusing versus Ostwald ripening, Langmuir 27 (17) (2011) 11098–11105. [DOI] [PubMed] [Google Scholar]
- [73].Erickson HP, Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy, Biol. Proced. Online 11 (2009) 32–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Monticelli L, Kandasamy SK, Periole X, Larson RG, Tieleman DP, Marrink SJ, The MARTINI coarse-grained force field: extension to proteins, J Chem Theory Comput 4 (5) (2008) 819–834. [DOI] [PubMed] [Google Scholar]
- [75].Afshar Y SF, Pishevar A, Worley S, Exploiting seeding of random number generators for efficient domain decomposition parallelization of dissipative particle dynamics, Comput. Phys. Commun 184 (4) (2013) 1119–1128. [Google Scholar]
- [76].Aa MAJ, Dissipative particle dynamics: effects of parameterization and thermostating schemes on rheology, Soft Mater. 13 (2) (2015) 106–117. [Google Scholar]
- [77].Franconetti A, Carnerero JM, Prado-Gotor R, Cabrera-Escribano F, Jaime C, Chitosan as a capping agent: Insights on the stabilization of gold nanoparticles, Carbohydr. Polym 207 (2019) 806–814. [DOI] [PubMed] [Google Scholar]
- [78].Moreno A, A J, Grillo R, Fraceto LF, Jaime C, A study on the molecular existing interactions in nanoherbicides: a chitooligosaccharide/tripolyphosphate kloaded with paraquat case, Colloids Surf. A 562 (2018) 220–228. [Google Scholar]
- [79].De Meyer FJM, M RJ, Willems TF, Smit B, Molecular simulation of the effect of cholesterol on lipid-mediated protein-protein interactions, Biophys. J 99 (11) (2010) 3629–3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Venturoli M, a.S SB, M M, Simulation studies of protein-induced bilayer deformations, and lipid-induced protein tilting, on a mesoscopic model for lipid bilayers with embedded proteins, Biophys. J 88 (3) (2005) 1778–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Gómez-Román VR, Florese RH, Patterson LJ, Peng B, Venzon D, Aldrich K, Robert-Guroff M, A simplified method for the rapid fluorometric assessment of antibody-dependent cell-mediated cytotoxicity, J. Immunol. Methods 308 (1–2) (2006) 53–67. [DOI] [PubMed] [Google Scholar]
- [82].Grivel JC, Margolis L, Use of human tissue explants to study human infectious agents, Nat. Protoc 4 (2) (2009) 256–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.