Abstract
Mucin 1 (MUC1) is a membrane-bound, highly glycosylated protein that is overexpressed in all stages of malignant transformation. Overexpression of MUC1 together with loss of polarization and hypoglycosylation are associated with resistance to apoptosis, which is the process that results in efficient removal of damaged cells. Inhibition of the apoptotic process is responsible for tumor development, tumor progression and drug resistance. MUC1 is considered as an oncogenic molecule that is involved in various signaling pathways responsible for the regulation of apoptosis. Based on this, the aim of the present study was to discuss the involvement of MUC1 in the divergent mechanisms regulating programmed cell death.
Keywords: anoikis, apoptosis, cancer, MUC1, glycosylation
1. Introduction
The epithelium is a layer of cells that separates an organism from the external environment; it is coated by mucous gel, which is mainly composed of mucins (MUCs). MUCs are high-molecular-weight O-linked glycoproteins synthesized by goblet cells. The human MUC family contains 22 characterized members (MUC1 to MUC22), which are divided into secreted and transmembrane glycoproteins (1,2). Secreted gel-forming MUCs, without a transmembrane domain, include MUC2, MUC5AC, MUC5B, MUC6, MUC7, MUC8 and MUC9. Membrane-bound MUCs form rod-like structures that extend from 200 to 500 nm beyond the glycocalyx, and members include MUC3A, MUC3B, MUC4, MUC12, MUC13, MUC15, MUC16, MUC17, MUC20, MUC21 and MUC22 (3-6). To date, MUC1 is the best-characterized MUC. Cancer-associated MUC1 notably differs from that of normal cells with regards to its function, biochemical features and cellular distribution (7,8). It has been reported that MUC1, which is present in epithelial cancer cells, is implicated in cell invasion, migration, adhesion, proliferation, and resistance to apoptosis and chemoradiotherapy (9). Therefore, MUC1 has been attracting considerable attention as an oncogenic molecule due to its crucial role in cancer progression (10). Apoptosis is a fundamental biological process that allows organisms to remove unwanted cells. One of the features of malignant transformation is the alteration of the cell death pathways, which is associated with increased cell survival. MUC1-dependent suppression of apoptosis causes an imbalance between cell proliferation and cell death, which in turn influences cancer progression (9). Therefore, the aim of the present study was to summarize the divergent mechanisms of action of MUC1 with regard to the alteration of programmed cell death.
2. Apoptosis
Among all types of cell death, apoptosis (also known as programmed cell death) is the most common; it is a genetically controlled process, which eliminates unnecessary or damaged individual cells (11). The process is crucial in the regulation of cell death and survival during the development of multicellular organisms and during normal homeostasis. Apoptosis provides the conditions necessary for appropriate regulation of numerous physiological processes, such as embryonic development, immune system function and maintenance of body homeostasis (12). In cancerous diseases, an imbalance between cell division and death occurs due to a lack of appropriate signaling. For example, downregulation of the tumor suppressor gene p53 may result in continuous cancer cell proliferation. Therefore, changes in the ability of the cells to undergo apoptosis may cause malignant transformation. In this context, the development of novel drugs that target numerous steps of the apoptotic process is beneficial in cancer treatment (11,13,14).
The induction of apoptosis is mediated via two main pathways, namely the receptor- and mitochondria-mediated pathways of apoptosis. Both pathways are regulated by the B-cell lymphoma (Bcl)-2 family of proteins, which includes pro- and anti-apoptotic members. Pro-apoptotic proteins include Bcl-2-associated X protein (Bax), Bcl-2 antagonist/killer 1 (Bak), BH3 interacting domain death agonist (Bid), Bad, Bax inhibitor motif (Bim), Bcl-2 interacting killer, Bcl-10, B-lymphocyte kinase, NADPH oxidase activator and p53 upregulated modulator of apoptosis (PUMA), while anti-apoptotic proteins mainly include Bcl-2, apoptosis regulator Bcl-x, B-cell lymphoma-extra large (Bcl-xL), Bcl-extra small, Bcl-w, induced myeloid leukemia cell differentiation protein (Mcl-1) and Bcl-2-associated athanogene (12,15,16).
The mitochondrial pathway is also known as the intrinsic pathway of apoptosis; it is initiated by various non-receptor-mediated stimuli, such as increased reactive oxygen species (ROS) or calcium ion levels, release of selected cytokines [e.g., interferon (IFN)] and immune cells (e.g., T cells), hormone deficiency, the presence of pathogens (e.g., viruses, bacteria and their products) or induction of DNA damage (12). Pro-apoptotic signals induce the translocation of Bax and Bak to the mitochondrial outer membrane (MOM), where they form pores, thereby altering cell membrane permeability. The assembly of Bax/Bak oligomers within MOM is promoted by Bid and Bim (17). Subsequently, mitochondrial cytochrome c is released into the cytosol where it forms the apoptosome. The latter is a complex including cytochrome c, apoptotic protease activating factor-1 (18,19), pro-caspase-9 and deoxyadenosine triphosphate (20). Other molecules released from the mitochondria include second mitochondria-derived activator of caspase/direct inhibitor of apoptosis-binding protein with low pI (Smac/DIABLO) and the mitochondrial serine protease high temperature requirement factor A2, which contribute to activation of apoptosis inducing factor (AIF), endonuclease G and caspases. The activation of inactive caspase-9 pro-enzyme results in cleavage of effector pro-caspases-3, -6 and -7 (21).
Caspases are cysteine proteases, which are divided into inflammatory (caspase-1, -4 and -5) and apoptotic proteins. The latter include initiator enzymes, such as caspase-2, -8 and -9, and executioner enzymes, such as caspase-3, -6 and -7 (17,20,22). Caspases are synthesized as pro-enzymes. Following apoptotic stimuli, they are converted into mature enzymes. Caspase substrates include both enzymatic and structural proteins. Specific examples include cytokeratins, the plasma membrane cytoskeletal protein α-fodrin (spectrin-like protein found in most cells) and microtubule-binding proteins, which are involved in the formation of the spindle poles and the segregation of chromosomes during mitosis (nuclear mitotic apparatus protein). These ultimately lead to the morphological and biochemical changes of the apoptotic cells. Proteolytic cleavage of caspase substrates, such as poly (ADP-ribose) polymerase, renders DNA repair impossible (22-24). Eventually, activation of caspases results in cell death (14,21).
The initiation of the receptor-associated pathway of apoptosis, which is also known as the extrinsic pathway of apoptosis, involves receptor-ligand interactions. These ligands belong to cytokines of the tumor necrosis factor (TNF) superfamily, which are termed cognate ligands. Approximately 40 ligand-receptor pairs have been characterized to date. The most widely known ligands are fatty acid synthetase ligand (FasL), which binds to the fatty acid synthetase receptor, TNF-α and TNF-related apoptosis-inducing ligand or Apo2 ligand (TRAIL/Apo2L). TNF-α can bind to TNF-receptor 1 (TNF-R1) and TNF-R2, and TRAIL/Apo2L interacts with four cell surface receptors, namely TRAIL receptor-1 (TRAIL-R1), TRAIL-R2, TRAIL-R3 and TRAIL-R4 (25,26). These receptors are also termed death receptor DR4, DR5/TRICK2/KILLER, decoy receptor (DcR)1/TRID/LIT and DcR2/TRUNDD, respectively. The majority of these receptors contain a C-terminal region, which includes an 80-amino acid (aa) death domain, an extracellular N-terminal region and a transmembrane domain. The signal transduction machinery has been described in detail using FasL/Fas and TNF-α/TNF-R1 models (26). Receptor-ligand binding results in receptor trimerization and the recruitment of adapter proteins, which contain similar death domains. FasL activation occurs following binding of this ligand with Fas, which leads to the interaction of the receptor with the adapter protein FAS-associated with death domain (FADD/MORT1). TNF-α binding to TNF-R allows the recruitment of the adapter protein TNF receptor type 1-associated death domain (TRADD), FADD and receptor-interacting protein (20,26). TRADD participates in signal transduction mediated by TNF-R, but not by TRAIL and Fas (25). Subsequently, FADD aggregates with pro-caspase-8 via the death effector domain (DED) sequence, leading to formation of the death-inducing signaling complex (DISC). Following proteolytic cleavage of its precursor, active caspase-8 is released and triggers activation of executive caspases, primarily caspase-3 (12). In addition, caspase-8 cleaves and activates Bid to t-Bid, which is its truncated form. The latter induces the release of mitochondrial cytochrome c and pro-apoptotic factors, and links the extrinsic with the intrinsic pathway of apoptosis (27-29). Cellular FADD-like IL-1β-converting enzyme-inhibitory protein (FLIP) can bind to FADD and caspase-8, causing their inactivation and, consequently, the inhibition of the apoptotic response (12).
Anoikis is a type of programmed cell death that prevents epithelial cells from seeding to abnormal sites and takes place when cells lose adhesion to the extracellular matrix (30,31). During anoikis, both apoptotic pathways are activated (32,33). Detachment from the surrounding matrix induces translocation of the pro-apoptotic protein Bim to the mitochondria, where it binds to Bcl-xL, neutralizing its anti-apoptotic functions. Moreover, decreased proteosomal degradation of Bim promotes Bax/Bak oligomerization, whereas matrix detachment results in upregulation of FasL and Fas expression, FLIP downregulation and consequent activation of caspase-8 (34-36).
The apoptotic response of the cells may be deregulated by various factors. One of the features of human cancer is the high level of MUC1 expression. Accumulating evidence has shown that MUC1 can modulate the apoptotic response in different ways as a result of its interaction with multiple proteins that participate in the regulation of this process (37,38).
3. MUC1 as a cancer-associated membrane-bound molecule
MUC1 localization and structure
MUCs are highly glycosylated proteins, and are the most abundant components of mucus on the epithelial surface. Among transmembrane MUCs, MUC1 has been widely investigated due to its role in oncogenesis (39-43). MUC1 is also known as episialin, polymorphic epithelial MUC, H23 antigen, mucin-like carcinoma-associated antigen, epithelial membrane antigen, cluster of differentiation 227, Krebs von den Lungen-6, peanut-reactive urinary mucin, carcinoma antigen CA15.3 and CA27.29 (the commonly used serum markers for breast cancer), human milk fat globule antigen, carcinoma-associated MUC and peanut-reactive urinary MUC (44). MUC1 (for humans) and Muc1 (for other species) are the most commonly used names assigned at the 1st International Workshop on Carcinoma-Associated Mucins (San Francisco, USA; 1990) and are in accordance with the Human Genome Project mapping conventions (45).
MUC1 is a heterodimeric transmembrane protein that normally resides at the apical borders of epithelial cells of the respiratory and gastrointestinal tracts, as well as in the ducts of organs such as the liver, kidney, pancreas and mammary gland (46,47). MUC1 is also expressed by non-epithelial cells, such as hematopoietic cells, T cells and male germ cells (48-51).
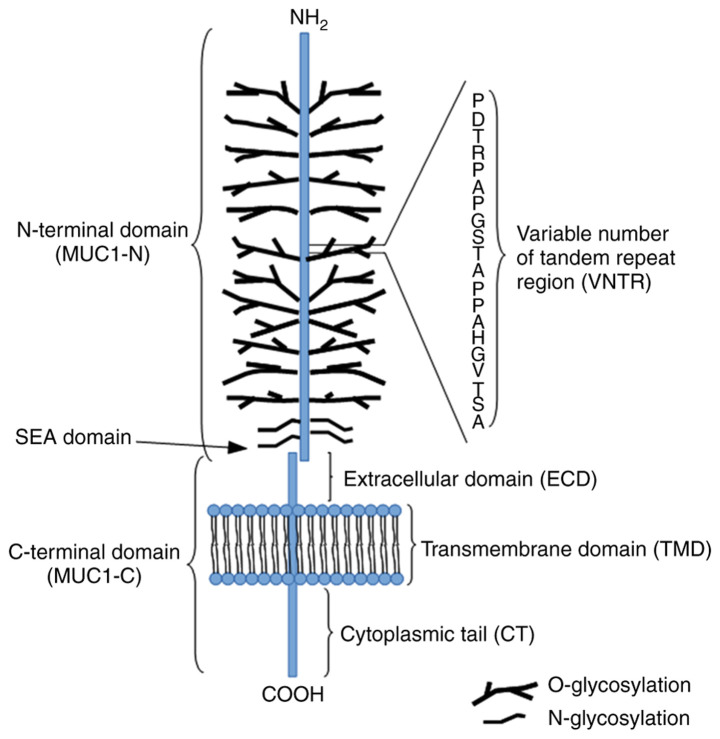
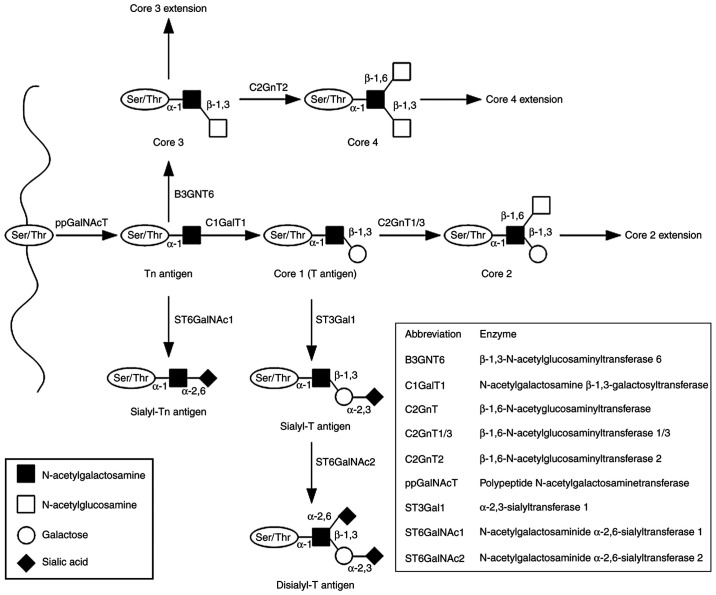
The MUC1 structure contains two distinctive subunits, one large N-terminal subunit (MUC1-N; subunit α) and one short C-terminal subunit (MUC1-C; subunit β) derived from autoproteolytic cleavage of a single polypeptide chain at the sea-urchin sperm protein enterokinase and agrin domain in the endoplasmic reticulum. These subunits are bound by non-covalent interactions (stable hydrogen bonds; Fig. 1) (5,52). The MUC1-N domain contains an N-terminus (104 aa) and a variable number of tandem repeat (VNTR) segment (20 aa) sequence (PDTRPAPGSTAPPAHGVTSA), which is repeated 20-200 times. The domain is also rich in serine and threonine residues that constitute potential sites of O-glycosylation, and contains a large C-terminus 170 aa in length (5,52). The molecular mass range of MUC1 is estimated to 1-40×106 Da. The majority of the protein modifications include sugar moieties, which constitute 60-80% of the total weight of the protein and are principally O-glycans. The complex synthesis of these polysaccharides is based on enzymatic attachment of monosaccharides to a polypeptide chain by ≥30 or more glycosyltransferase enzymes (53,54). The detailed synthesis of the O-glycan basic core structure is presented in Fig. 2. The VNTR region is followed by a segment containing five N-linked glycosylation sites (5). The MUC1-C is composed of three regions, including the extracellular domain (ECD), composed of 58 aa, which is responsible for anchoring MUC1 to the cell membrane, the transmembrane domain, composed of 28 aa, and a cytoplasmic tail (MUC1-CT), composed of 72 aa. Moreover, the MUC1-C ECD contains asparagine residues that are used as N-glycosylation sites (41,55). The MUC1-C cysteine-glutamine-cysteine (CQC) motif is located below the transmembrane region and is crucial for the formation of homodimers (42).
Figure 1.
Structure of MUC1. MUC1, mucin 1.
Figure 2.
Mucin-type O-glycan synthesis. O-glycosylation is initiated by attachment of GalNAc to the hydroxyl groups of the Ser/Thr of the protein chain. The reaction is catalyzed by ppGalNAcT enzymes and results in the formation of the Tn antigen (GalNAcα1-O- Ser/Thr). The four basic core structures are generated in the Golgi apparatus by specific glycosyltransferases. The Tn antigen can be elongated through galactose addition catalyzed by C1GalT1 or core 1 synthase, which results in the synthesis of the T antigen or core 1. The antigen can also be elongated by B3GNT6, which transfers GlcNAc to the GalNAc-Ser/Thr structure to form core 3. Subsequent GlcNAc addition to core 3 forms core 4. Core structures can be further elongated or terminated by attachment of fucose or sialic acid. GalNAc, N-acetylgalactosamine; Ser, serine; Thr, threonine; ppGalNAcT, N-acetylgalactosaminyltransferase; C1GalT1, N-acetylgalactosamine β-1,3-galactosyltransferase; Galβ1, galactosamine β1; T, Galβ1-3GalNAcα1-O-Ser/Thr; Tn, GalNAcα1-O-Ser/Thr.
Functions of MUC1 in physiology
In the past years, a considerable amount of data has been gathered regarding MUC1 functions. It has been shown that this MUC participates in complex interactions and is involved in the regulation of a wide variety of cellular pathways, affecting both physiological and pathological processes. Substantial glycosylation of the MUC1 extracellular subunit in normal tissues aims to protect them from the entry of harmful substances and provide lubrication to the underlying epithelia (55). Therefore, the N-terminal subunit can be shed from the cell surface and released into the extracellular space as a result of proteolytic cleavage in order to provide a barrier to invading pathogens. Such events may be induced by a number of inflammatory stimuli, such as IFN-γ and TNF-α (56,57). This process is also mediated by specific enzymes, such as TNF-α-converting enzyme/a disintegrin and metalloproteinase 17 (56,57). Therefore, MUC1-N can act as a receptor when transmitting stress signals to the interior of the cell (41). Furthermore, glycans can participate in cell-cell and cell-matrix interactions and are involved in the recognition of normal cells by the immune system (58). Moreover, glycans participate in the appropriate distribution of proteins, which are newly synthesized in the endoplasmic reticulum. By affecting proteolysis, they protect proteins from intra- or extracellular degradation (58). In addition, MUC1-CT may be phosphorylated by multiple kinases, including the epidermal growth factor receptor (EGFR), glycogen synthase kinase-3β, tyrosine-protein kinase MET, non-receptor tyrosine kinase (Src), protein kinase C (PKC), glycogen synthase kinase 3 (GSK3) and tyrosine-protein kinase Abl. Previous studies have shown that MUC1 interacts with diverse molecules and modulates their activity (41,55).
MUC1 in cancer
Tumor progression is associated with a notable increase in MUC1 expression, which has been reported in diverse cancer types, such as colon, breast, lung, pancreatic and prostate cancer, as well as in hematological malignancies (59-62).
The changes in the oligosaccharide structure of glycoproteins have received considerable attention due to the key role of glycans in processes such as cell proliferation, differentiation, invasion, metastasis and immune surveillance (63-66). It has been shown that, during the process of neoplasia, the MUC1 ECD undergoes significant modifications, including changes in the glycosylation profile via the following main mechanisms: Incomplete synthesis and synthesis of atypical forms of glycans (67). The attachment of the truncated oligosaccharides to the VNTR region changes the MUC1 spatial structure and increases the availability of the peptide backbone to other proteins, which in turn affects potential protein-protein interactions and intracellular signaling (52,68,69).
The majority of human carcinomas, including gastric and colorectal cancer, contain truncated forms of glycans, such as N-acetylgalactosamine (GalNAcα)1-O-serine (Ser)/threonine (Thr) (Tn) and galactose (Gal) β1-3GalNAcα1-O-Ser/Thr (T) antigens and their sialylated forms Neu5Acα2-6GalNAcα1-O-Ser/Thr (sTn) and Neu5Acα2-3Galβ1-3GalNAcα1-O-Ser/Thr (sT). These forms are combined with decreased levels of core 3 and core 4 structures (6,69,70) (Fig. 2); their presence is predominantly associated with changes in the expression levels of different glycosyltransferases (71,72). For example, the activity of the core 1 synthase, also known as N-acetylgalactosamine β-1,3-galactosyltransferase (C1GalT1), depends on the co-expression of the specific chaperone core 1 β3GalT-specific molecular chaperone (COSMC). C1GalT1 is an enzyme that catalyzes galactose addition to the Tn antigen. In the absence of COSMC, the enzyme loses its function, resulting in higher levels of Tn antigen than those noted in normal cells (52). Moreover, abnormal expression of sialyltransferases, including N-acetylgalactosaminide α-2,6-sialyltransferase 1 and β-galactoside α-2,3-sialyltransferase 1, may be associated with increased MUC1 sialylation and the formation of sTn and sT antigens (52). The transfer of fucose residues to oligosaccharides and proteins is catalyzed by fucosyltransferases. Aberrant fucosylation is associated with neoplasia and may lead to EGFR stimulation, which in turn affects the functions of integrins and selectins, and the induction of apoptosis or oncogenesis. Large amounts of sialylated and fucosylated core 1 and 2 structures have been detected in gastric, ovarian, renal, colon and prostate cancer (73-78). Sialylated and fucosylated glycans (Lewis-type antigens) are composed of free monosaccharides, namely N-acetylglucosamine, Gal and fucose, which differ in terms of their corresponding glycosidic bonds. Core 1 structures contain the Galβ1-3GlcNAc bond (Lewis a, Lewis b), whereas core 2 structures contain the Galβ1-4GlcNAc bond (Lewis x, Lewis y). Further addition of sialic acid leads to formation of their sialylated forms (sLea and sLex); their expression may be associated with intensification of the neoplastic transformation of cells (69,74). The sLex and sLea carbohydrate ligands have been shown to adhere to E-selectin in vascular endothelial cells. The expression of this protein may be induced by proinflammatory agents, such as cytokines (69).
MUC1 function in cancer is associated with its cellular localization. Loss of cell polarity during epithelial transformation causes the translocation of the MUC1-N/MUC1-C complex from the apical to the entire surface of the cell membrane (5). Therefore, MUC1 may interact with molecules normally expressed at the basolateral membrane, such as receptor tyrosine kinases (RTKs), including EGFR. These RTKs, in turn, activate several signaling pathways, including the PI3K/protein kinase B/Akt, the p38 mitogen-activated protein (MAP) kinase, the c-Jun N-terminal kinases (JNK), the Janus kinase/signal transducers and activators of transcription (JAK/STAT) and the Src pathways, which are involved in cell proliferation, survival and differentiation under normal and pathological conditions (79). Formation of extracellular connections between MUC1 and EGFR requires the MUC1-C/galectin-3 interaction (80). Galectin-3 is member of a family of β-galactoside-binding lectins, which influence biological processes, such as cell adhesion, proliferation, differentiation, inflammation, angiogenesis and oncogenesis (81,82). In addition, altered glycosylation may also affect appropriate oligomerization of cell surface receptors and, therefore, their sensitivity to stimulation (55,83).
In vitro studies have shown that MUC1-C can translocate from the cell membrane to the mitochondria, where it most likely localizes to the MOM (84). The process may be stimulated by phosphorylation of the MUC1-CT tyrosine induced by fibroblast growth factor-1 (FGF-1), which results in MUC1 binding to the heat shock protein HSP90 chaperone and its consequent translocation to the mitochondria (85). It is still not fully understood how these proteins are targeted and anchored to the MOM. However, it has been shown that Bcl-xL and Bcl-2 integrate with MOM by their C-termini (84). In cancer cells, MUC1-C is also imported into the nucleus, where it directly interacts with specific transcription factors, such as nuclear factor-κB (NF-κB), and stimulates their transcription (86-89). In addition, MUC1 hypoglycosylation affects its subcellular localization via increased intracellular MUC endocytic trafficking by clathrin-coated pits (10).
Kufe (83) and Li et al (90) demonstrated that MUC1-C participated in signal transmission from the Wnt/β-catenin pathway to the nucleus. This was facilitated following its interactions with p53, STAT3 and estrogen receptor α (ERα) (91). The aforementioned pathways are associated with oncogenesis (41). In addition, MUC1 can contribute to constitutive stimulation of various processes. For example, Ahmad et al (92) demonstrated that MUC1-C interacted with TNF-R1 in mammary epithelial MCF-10A cells and participated in TAK1-mediated phosphorylation of IκB kinase β (IKKβ), formation of the IKKβ-inhibitor of NF-κB kinase subunit γ (IKKγ) complex and autophosphorylation of IKKβ. Furthermore, MUC1 can be regulated by hormone receptors, such as ERα, nuclear retinitis pigmentosa GTPase regulator and the androgen receptor. Estrogen treatment combined with MUC1 knockdown results in increased cell death in aromatase inhibitor-resistant cells (93).
4. Role of MUC1 in apoptosis
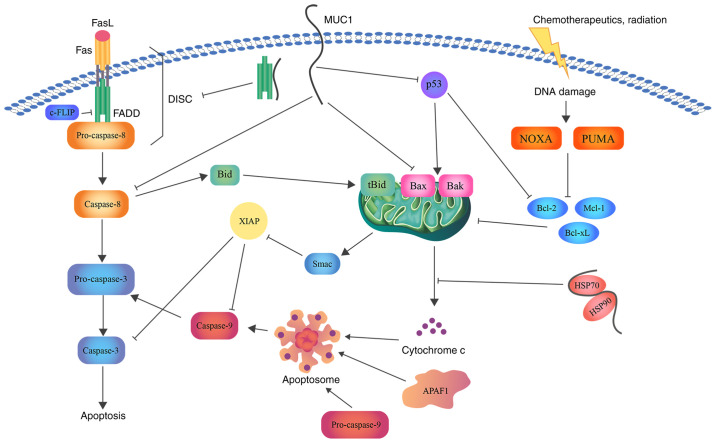
Several articles have been published regarding the effects of MUC1 on apoptosis. In the present study, the diverse interactions of MUC1 with different factors that lead to the inhibition of this process were assessed (Fig. 3).
Figure 3.
Schematic representation of the role of MUC1 in apoptosis. MUC1 interacts with FADD DED, blocking the formation of DISC and suppressing the induction of the extrinsic apoptotic pathway. Direct association with caspase-8 inhibits its activation. In addition, MUC1-C suppresses Bax translocation to the MOM and cytochrome c release. Binding of MUC1 to the HSP90/HSP70 complex weakens the activation of the mitochondrial pathway. Moreover, direct binding of MUC1 to the p53 regulatory domain is associated with stimulation of growth-arresting gene transcription, thereby inhibiting apoptosis. Bcl, B-cell lymphoma; Bax, Bcl-2-associated X protein; Bcl-xL, Bcl-extra large; MUC1, mucin 1; FADD, FAS-associated with death domain; DED, death effector domain; DISC, death inducing signaling complex; MUC1-C, MUC1 C-terminal subunit; MOM, mitochondrial outer membrane; HSP, heat shock protein; FasL, fatty acid synthetase ligand; FLIP, FLICE (FADD-like IL-1β-converting enzyme)-inhibitory protein; Bid, BH3 interacting domain death agonist; Mcl-1, induced myeloid leukemia cell differentiation protein; NOXA, NADPH oxidase activator; PUMA, p53 upregulated modulator of apoptosis; Smac, second mitochondria-derived activator of caspase; XIAP, X-linked inhibitor of apoptosis protein.
Role of MUC1 in the intrinsic apoptotic pathway
The intrinsic (mitochondrial) pathway of apoptosis is activated in response to various stimuli. It has been previously mentioned that the treatment of cells with genotoxic anticancer agents leads to the release of cytochrome c from the mitochondria and the activation of the intrinsic pathway of apoptosis (12,94). Ren et al (84) revealed that MUC1-C expression was associated with decreased release of apoptogenic proteins, including cytochrome c, Smac/DIABLO and AIF, as well as attenuated activation of caspase-3 and PKC δ type (PKCδ) following cisplatin treatment. It was shown that PKCδ may induce apoptosis in lipopolysaccharide-activated macrophages via regulation of TNF production (95). Similar results were obtained when HCT116 colon carcinoma cancer cells were treated with etoposide (84). Moreover, the release of the aforementioned molecules was associated with loss of the mitochondrial membrane potential. Therefore, MUC1-C-dependent impairment of mitochondrial pro-apoptotic factor release attenuates the apoptotic response, notably the intrinsic pathway of apoptosis, in response to DNA damage. In contrast to this finding, cisplatin treatment of A549 and ZR-75-1 cells with transiently downregulated MUC1 expression increased the apoptotic response (84).
The mitochondrial localization of MUC1 impacts signal transduction from the cell membrane to the mitochondria, leading to decreased activation of the intrinsic apoptotic pathway (96,97). Bax plays a crucial role among other members of the Bcl-2 protein family in the activation of the mitochondrial pathway of apoptosis (98). During the apoptotic process, Bax localizes from the cytosol to MOM, where it undergoes conformational changes followed by oligomerization to form a pore necessary for cytochrome c release (99). Bax is composed of nine α-helices. The α2 helix contains the Bcl-2 homology BH3 domain, which is essential for Bax homo- or hetero-dimerization with other cysteine-containing proteins, such as the Bcl-2, Bcl-xL and Mcl proteins (99,100). Ahmad et al (101) showed that MUC1 interacted directly with Bax in breast and colon cancer cells. Two cysteines present in the MUC1-C CQC motif participated in the formation of MUC1-C heterodimers. Studies have demonstrated that the MUC1-C CQC motif binds to Cys-62 in the BH3 domain of Bax. This interaction may block the ability of Bax to dimerize. It is not currently known whether MUC1-C binds to a secondary Bax BH3 cysteine residue at position 126. It has been shown that oxidative stress may promote the association between MUC1-C and Bax (101-103). In conclusion, the current data indicate that MUC1-C attenuates Bax dimerization, translocation to MOM and, in turn, the release of cytochrome c, which suppresses the activation of the intrinsic apoptotic pathway. Mutations in the MUC1-C CQC motif impair MUC1-C transport to the nucleus and MOM. This mutation abolishes the oncogenic function of MUC1-C and can be used for the development of molecules that may block homodimerization and, in turn, MUC1-C signal transduction (55).
Nickel acetate (Ni2+) is an agent with genotoxic abilities (104,105). The study by Castorina and Giunta (106) revealed that cell death resistance of human bronchial epithelial cells (Beas-2B) exposed to Ni2+ was associated with direct stimulation of the EGFR by MUC1. This glycoprotein had the ability to activate EGFR/ERK1/2 signaling. Decreased levels of cleaved caspase-3 were also noted (106). These results are consistent with those reported in the study by Schroeder et al (107), which examined the ability of MUC1 to enhance EGF binding to the EGFR in breast tumors. In non-small lung cancer, MUC1 promoted activation of the PI3K/Akt pathway (108), which was necessary for NF-κB signaling activation (109). Previous studies have shown that the interaction of MUC1-C with IKKβ/IKKγ complex activates NF-κB signaling (92,110). Moreover, Akt has demonstrated an anti-apoptotic effect by inhibiting the functions of specific Bcl-2 proteins, such as Bad, which interact with chaperone protein 14-3-3, resulting in the release of specific anti-apoptotic proteins (111,112). Therefore, it has been hypothesized that chronic exposure to nickel compounds in combination with increased MUC1 expression enhances epithelial cell resistance to apoptosis and promotes the development of carcinogenesis (106).
An additional study has revealed that MUC1 overexpression may protect cells from oxidative stress-induced apoptosis (85). The abundance of ROS, which are derived from the mitochondria, leads to the activation of cell death pathways (113,114). Yin et al (115) observed an increase in MUC1 transcription and translation in vitro following exposure to H2O2. MUC1-positive cells further demonstrated higher levels of antioxidant enzymes. Since H2O2 easily diffuses across the cell membrane, it was hypothesized that its transmembrane subunit may be associated with a decrease in ROS levels in the cells. These results indicated that MUC1 inhibited the apoptotic response to oxidative stress by decreasing the concentration levels of oxidative molecules (85). The increase in ROS levels following MUC1-C inhibition was recently confirmed in mouse embryonic stem cells (116).
Moreover, MUC1 can interact with forkhead box class O (FOXO)3a, which is also known as forkhead in rhabdomyo-sarcoma-like 1. FOXO3a is a member of the FOXO family of transcription factors that mediate gene transcription following dephosphorylation in the nucleus (115,117). MUC1 decreases its phosphorylation by attenuating activation of the PI3K/Akt pathway. Downregulation of MUC1 in breast cancer cells causes inactivation of FOXO3a, which increases the necrotic cell response to oxidative stress (115). Therefore, MUC1 can play a role in protecting the epithelium from apoptosis following injury (84).
The tumor suppressor p53 protein is a product of the TP53 gene. This gene is located in humans at the short arm of chromosome 17 (17p13.1) (118). p53 plays a role in induction of the cell apoptotic response by stimulation of Bax/Bak oligomerization or inhibition of anti-apoptotic Bcl-2 family member function. Loss of p53 activity prevents cells from forming a normal response to DNA damage or stress (119). Wei et al (120) reported that MUC1 inhibited the cellular response to DNA damage mediated by p53. MUC1 direct binding to the regulatory domain of p53 was associated with enhanced transcription of growth arrest genes and in turn depletion of apoptosis.
Additional evidence has confirmed that MUC1 interacts with small non-coding RNA molecules and microRNAs (miR/miRNAs). It is known that miRNAs, such as miR-136, are implicated in specific biological processes, such as the cell cycle, proliferation, migration and apoptosis (121,122). For example, miR-136 may function as a suppressor in the development of multiple cancer types and its decreased levels in human glioma cells can stimulate apoptosis via inhibition of astrocyte elevated gene-1 and Bcl-2 proteins. It was demonstrated that miR-136 upregulation contributed to the induction of apoptosis in esophageal squamous cell carcinoma cells via MUC1 inhibition (121). An additional study indicated that miR-145 decreased ovarian cancer cell proliferation and invasion by suppressing MUC1 (123). Moreover, breast cancer cells with silenced MUC1 expression and overexpression of miR-485-5p demonstrated inhibition of cell proliferation, invasiveness and migration (124).
Role of MUC1 in the extrinsic apoptotic pathway
Cleavage of the pro-apoptotic protein Bid is mediated by caspase-8 and results in the formation of tBid, which may induce the mitochondrial release of cytochrome c. This allows the interaction between the receptor-mediated and mitochondrial pathways (27,28,125). Therefore, the modulation of the extrinsic pathway response by MUC1 is important. Ren et al (84) demonstrated that in HCT116 cells MUC1 attenuated TRAIL-induced apoptosis. This effect was reversed by the addition of cycloheximide. It has also been shown that MUC1-C may inhibit caspase-8 activation induced by TRAIL, TNF-α and FasL, and consequently block death-receptor signaling (91). Caspase-8 is composed of an N-terminal region containing two DEDs (1-183 aa). The p18 (217-374 aa) and p10 (385-480 aa) fragments are derived following cleavage of each region. The association of the adaptor protein FADD with caspase-8 via DEDs leads to caspase-8 dimerization and its cleavage to the p18/p10 fragments (91,126). It has been shown that MUC1-C can bind directly to caspase-8 p18 via its specific regions, which contain the aa residues 270-322 and 1-20, respectively. This binding occurs by interactions other than disulfide bonds. MUC1 binding to the p18 region may interrupt interdimer processing and block caspase activation (91).
MUC1 competition with caspase-8 for binding to the FADD DED may interrupt formation of DISC in vitro. The same region of MUC1-C can bind to other protein partners, such as β-catenin, IKKγ or the HSP90/HSP70 complex (40,92,127). Based on this evidence, Agata et al (91) suggested that MUC1-C may have both transmembrane receptor and chaperone-like functions. Moreover, MCF-10A cells with downregulated MUC1 expression exhibited an increase in caspase-8 activity following TNF-α, FasL and TRAIL stimulation in comparison to non-transformed MCF-10A cells. Therefore, the ability of MUC1-C to attenuate caspase-8-mediated activation of apoptosis could be used by normal epithelial or malignant cells to protect them from cell death under inflammatory conditions or to enable their survival in an adverse environment (91).
Certain enzymes, such as c-Jun N-terminal kinase 1 (JNK1), are involved in the regulation of the apoptotic process. Chen et al (128) reported that MUC1-C (1-45 aa) directly binds to JNK1. JNK1 belongs to the superfamily of MAP kinases (129). The three following isoforms of JNK have been identified: JNK1, JNK2 and JNK3. The first two are expressed in a variety of tissues, whereas JNK3 is mainly limited to the neurons and heart (130). The JNK signaling pathway can be activated by different stimuli, including genotoxic agents, TNF-α, MAP kinase 4 (MKK4) or MKK7. Following its activation, JNK1 localizes to the nucleus and phosphorylates the effector protein c-Jun, which in turn influences transcription of multiple target genes, including activator protein 1. MUC1 overexpression contributes to increased activation of JNK1 and its target c-Jun following treatment of the cells with genotoxic anticancer agents, such as cisplatin or doxorubicin (128). This process decreases the cellular response to apoptotic stimuli (128).
Several other studies have also confirmed the significant role of MUC1 in apoptosis. Zhang et al (131) demonstrated that cell proliferation, invasion, migration, epithelial-mesenchymal transition and apoptosis in oral squamous cell carcinoma (OSCC) may be affected by the activity of specific transcription factors, such as Snail and Slug. Slug is an invasion-promoting factor that plays a major role in the inhibition of E-cadherin transcription and the repression of the function of the pro-apoptotic protein PUMA, which in turn leads to the induction of cell survival (132,133). It has been shown that MUC1 expression in OSCC is positively correlated with the expression of Slug, whereas MUC1 gene silencing is correlated with a decrease in Slug levels. Therefore, is has been suggested that MUC1 silencing is associated with the induction of apoptosis and the inhibition of cell proliferation, invasion and migration via downregulation of Slug expression (131).
The JAK/STAT signaling pathway plays an important role in transferring signals from cell membrane receptors to the nucleus (134). Overexpression of MUC1 was associated with decreased caspase-3 activation, resulting in a decreased apoptotic response in the irradiated hepatocellular carcinoma (HCC) SMMC-7721 cell line. An increase in the expression levels of the anti-apoptotic proteins Mcl-1 and Bcl-xL was also observed. Yi and Lu (135) reported that resistance to irradiation-induced apoptosis was associated with JAK2/STAT3 signaling pathway activation by MUC1. Therefore, this protein contributed to the radioresistance of the HCC cells. Escher et al (93) demonstrated the role of MUC1 in enhancing the activity of JAK/STAT, which stimulated IFN-induced transmembrane protein 1 expression and an aggressive phenotype in breast cancer cells resistant to aromatase inhibitors.
Kato et al (136) demonstrated that cells treated with polyinosinic:polycytidilic acid exhibited increased activation of caspase-3 and -8, IFN regulatory factor 3, NF-κB and IFN-β following MUC1 silencing in comparison to MUC1-expressing cells. This study further showed that MUC1-CT attenuated Toll-like receptor 3 (TRL3)-induced apoptosis in lung epithelial cells by blocking the interaction between TRL3 and TIR-domain-containing adapter-inducing IFN-β (136). Moreover, inhibition of MUC1 significantly increased the sensitivity of lung and pancreatic cancer cells to the induction of apoptosis by anticancer drugs (137,138).
Role of MUC1 in anoikis
Loss of cell adhesion to the surrounding matrix or its inappropriate adherence results in the activation of a specific type of apoptosis, termed anoikis (139). Cell resistance to anoikis is a biological process that precedes metastasis. Despite the current understanding of the apoptotic process, the mechanism by which metastatic cancer cells evade anoikis remains poorly defined. It has been previously shown that MUC1 overexpression, which occurs mainly in the ECD, blocks anoikis activation. This effect may be associated with extensive glycosylation, which forms a specific microenvironment on the cell surface and protects from activation of anoikis-initiating factors and death receptors, such as integrins and Fas, respectively (32,33). One of the key enzymes in the O-glycosylation process is C1GalT1; its deficiency leads to the formation of abnormal shortened forms of O-linked sugar chains and is associated with increased availability of cell surface receptors for integrin1β, E-cadherin or FasL. These molecular events result in activation of the extracellular pathway of apoptosis (31,52). Piyush et al (140) demonstrated that suppression of C1GalT1 expression resulted in increased expression of the Tn antigen in MUC1-negative HCT116 cells (human colon cancer). However, activation of anoikis with a concomitant increase in caspase-8 activity due to binding of FasL to Fas was only noted in MUC1-positive cells (SW620). Therefore, an evident association between excessive O-glycosylation of MUC1 and anoikis resistance was observed, primarily due to inhibition of anoikis-initiating molecule activation.
5. Conclusions
MUC1 is a component of mucus that plays a protective role in normal epithelial cells. However, during malignant transformation, the changes in the expression and glycosylation pattern of MUC1 modulate its interactions with other proteins, which in turn regulate signal transmission. High levels of MUC1 are correlated with a poor prognosis and shorter survival time in patients with cancer. In addition, aberrant expression of this protein may block drug diffusion through the cell membrane and promote survival of cancerous cells, since it has been shown to impact both extrinsic and intrinsic apoptotic pathways. Therefore, the upregulation of MUC1-dependent attenuation of apoptotic response indicates the potential role of this protein in cancer therapy. Despite extensive evidence reported on the mechanism of action of MUC1 with regard to cell death, a number of aspects remain unresolved. Therefore, additional studies are necessary to further elucidate such interactions.
Acknowledgments
Not applicable.
Abbreviations
- AIF
apoptosis-inducing factor
- C1GalT1
N-acetylgalactosamine β-1,3-galactosyltransferase (core 1 synthase)
- CQC
cysteine-glutamine-cysteine motif
- DED
death effector domain
- DISC
death inducing signaling complex
- ERα
estrogen receptor α
- FADD/MORT1
FAS-associated with death domain
- FasL
fatty acid synthase ligand
- FLIP
FLICE (FADD-like IL-1β-converting enzyme)-inhibitory protein
- FOXO
forkhead box class O family of transcription factors
- FOXO3a
forkhead box class O3a
- HCC
hepatocellular carcinoma
- Gal
galactose
- HSP90
heat shock protein 90
- IKKβ
IκB kinase β
- JAK/STAT
Janus kinase/signal transducers and activators of transcription
- MKK
mitogen-activated protein kinase kinase
- MOM
mitochondrial outer membrane
- MUC1
mucin 1
- OSCC
oral squamous cell carcinoma
- PKC
protein kinase C
- PKCδ
PKC δ type
- ROS
reactive oxygen species
- RTKs
receptor tyrosine kinases
- Smac/DIABLO
second mitochondria-derived activator of caspase/direct inhibitor of apoptosis-binding protein with low pI
- Src
non-receptor tyrosine kinase
- TRADD
TNF receptor type 1-associated death domain protein
- TRAIL/Apo2L
tumor necrosis factor-related apoptosis-inducing ligand
- VNTR
variable number of tandem repeats region
Funding Statement
This study was supported by the Medical University of Bialystok (Bialystok, Poland; grant no. SUB/2/DN/19/004/2203).
Availability of data and materials
Not applicable.
Authors' contributions
KS developed the concept for the study and drafted the manuscript. IR reviewed and edited the manuscript. All authors read and approved the final manuscript. Data authentication is not applicable.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Behera SK, Praharaj AB, Dehury B, Negi S. Exploring the role and diversity of mucins in health and disease with special insight into non-communicable diseases. Glyconconj J. 2015;32:575–613. doi: 10.1007/s10719-015-9606-6. [DOI] [PubMed] [Google Scholar]
- 2.Dhanisha SS, Guruvayoorappan C, Drishya S, Abeesh P. Mucins: Structural diversity, biosynthesis, its role in pathogenesis and as possible therapeutic targets. Crit Rev Oncol Hematol. 2018;122:98–122. doi: 10.1016/j.critrevonc.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 3.Cornick S, Tawiah A, Chadee K. Roles and regulation of the mucus barrier in the gut. Tissue Barriers. 2015;3:e982426. doi: 10.4161/21688370.2014.982426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moniaux N, Escande F, Porchet N, Aubert JP, Batra SK. Structural organization and classification of the human mucin genes. Front Biosci. 2001;6:D1192–D1206. doi: 10.2741/Moniaux. [DOI] [PubMed] [Google Scholar]
- 5.Nath S, Mukherjee P. MUC1: A multifaceted oncoprotein with a key role in cancer progression. Trends Mol Med. 2014;20:332–342. doi: 10.1016/j.molmed.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanson RL, Hollingsworth MA. Functional consequences of differential O-glycosylation of MUC1, MUC4, and MUC16 (downstream effects on signaling) Biomolecules. 2016;6:34. doi: 10.3390/biom6030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jonckheere N, Van Seuningen I. The membrane-bound mucins: How large O-glycoproteins play key roles in epithelial cancers and hold promise as biological tools for gene-based and immunotherapies. Crit Rev Oncog. 2008;14:177–196. doi: 10.1615/CritRevOncog.v14.i2-3.30. [DOI] [PubMed] [Google Scholar]
- 8.Lau SK, Weiss LM, Chu PG. Differential expression of MUC1, MUC2, and MUC5AC in carcinomas of various sites: An immunohistochemical study. Am J Clin Pathol. 2004;122:61–69. doi: 10.1309/9R6673QEC06D86Y4. [DOI] [PubMed] [Google Scholar]
- 9.Reynolds IS, Fichtner M, McNamara DA, Kay EW, Prehn JHM, Burke JP. Mucin glycoproteins block apoptosis; promote invasion, proliferation, and migration; and cause chemoresistance through diverse pathways in epithelial cancers. Cancer Metastasis Rev. 2019;38:237–257. doi: 10.1007/s10555-019-09781-w. [DOI] [PubMed] [Google Scholar]
- 10.Altschuler Y, Kinlough CL, Poland PA, Bruns JB, Apodaca G, Weisz OA, Hughey RP. Clathrin-mediated endocytosis of MUC1 is modulated by its glycosylation state. Mol Biol Cell. 2000;11:819–831. doi: 10.1091/mbc.11.3.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katoch B, Sebastian S, Sahdev S, Padh H, Hasnain SE, Begum R. Programmed cell death and its clinical implications. Indian J Exp Biol. 2002;40:513–524. [PubMed] [Google Scholar]
- 12.Elmore S. Apoptosis: A review of programmed cell death. Toxicol Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D'Arcy MS. Cell death: A review of the major forms of apoptosis, necrosis and autophagy. Cell Biol Int. 2019;43:582–592. doi: 10.1002/cbin.11137. [DOI] [PubMed] [Google Scholar]
- 14.Wong RS. Apoptosis in cancer: From pathogenesis to treatment. J Exp Clin Cancer Res. 2011;30:87. doi: 10.1186/1756-9966-30-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jan R, Chaudhry GE. Understanding apoptosis and apoptotic pathways targeted cancer therapeutics. Adv Pharm Bull. 2019;9:205–218. doi: 10.15171/apb.2019.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papaliagkas V, Anogianaki A, Anogianakis G, Ilonidis G. The proteins and the mechanisms of apoptosis: A mini-review of the fundamentals. Hippokratia. 2007;11:108–113. [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor R, Cullen S, Martin S. Apoptosis: Controlled demolition at the cellular level. Nat Rev Mol Cell Biol. 2008;9:231–241. doi: 10.1038/nrm2312. [DOI] [PubMed] [Google Scholar]
- 18.Shakeri R, Kheirollahi A, Davoodi J. Apaf-1: Regulation and function in cell death. Biochimie. 2017;135:111–125. doi: 10.1016/j.biochi.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 19.Shimizu S, Narita M, Tsujimoto Y. Bcl-2 family proteins regulate the release of apoptogenic cytochrome c by the mitochondrial channel VDAC. Nature. 1999;399:483–487. doi: 10.1038/20959. [DOI] [PubMed] [Google Scholar]
- 20.Savitskaya MA, Onishchenko GE. Mechanisms of apoptosis. Biochemistry (Mosc) 2015;80:1393–1405. doi: 10.1134/S0006297915110012. [DOI] [PubMed] [Google Scholar]
- 21.O'Brien MA, Kirby R. Apoptosis: A review of pro-apoptotic and anti-apoptotic pathways and dysregulation in disease. J Vet Emerg Crit Care (San Antonio) 2008;18:572–585. doi: 10.1111/j.1476-4431.2008.00363.x. [DOI] [Google Scholar]
- 22.Julien O, Wells JA. Caspases and their substrates. Cell Death Differ. 2017;24:1380–1389. doi: 10.1038/cdd.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Porter AG, Jänicke RU. Emerging roles of caspase-3 in apoptosis. Cell Death Differ. 1999;6:99–104. doi: 10.1038/sj.cdd.4400476. [DOI] [PubMed] [Google Scholar]
- 24.Van Ba H, Hwang I. Role of caspase-9 in the effector caspases and genome expressions, and growth of bovine skeletal myoblasts. Dev Growth Differ. 2014;56:131–142. doi: 10.1111/dgd.12098. [DOI] [PubMed] [Google Scholar]
- 25.Dempsey PW, Doyle SE, He JQ, Cheng G. The signaling adaptors and pathways activated by TNF superfamily. Cytokine Growth Factor Rev. 2003;14:193–209. doi: 10.1016/S1359-6101(03)00021-2. [DOI] [PubMed] [Google Scholar]
- 26.Srivastava RK. TRAIL/Apo-2L: Mechanisms and clinical applications in cancer. Neoplasia. 2001;3:535–546. doi: 10.1038/sj.neo.7900203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Zhu H, Xu CJ, Yuan J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell. 1998;94:491–501. doi: 10.1016/S0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- 28.Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell. 1998;94:481–490. doi: 10.1016/S0092-8674(00)81589-5. [DOI] [PubMed] [Google Scholar]
- 29.Valentijn AJ, Gilmore AP. Translocation of full-length Bid to mitochondria during anoikis. J Biol Chem. 2004;279:32848–32857. doi: 10.1074/jbc.M313375200. [DOI] [PubMed] [Google Scholar]
- 30.Woods NT, Yamaguchi H, Lee FY, Bhalla KN, Wang HG. Anoikis, initiated by Mcl-1 degradation and Bim induction, is deregulated during oncogenesis. Cancer Res. 2007;67:10744–10752. doi: 10.1158/0008-5472.CAN-07-3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao Q, Piyush T, Chen C, Hollingsworth MA, Hilkens J, Rhodes JM, Yu LG. MUC1 extracellular domain confers resistance of epithelial cancer cells to anoikis. Cell Death Dis. 2014;5:e1438. doi: 10.1038/cddis.2014.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim YN, Koo KH, Sung JY, Yun UJ, Kim H. Anoikis resistance: An essential prerequisite for tumor metastasis. Int J Cell Biol. 2012;2012:306879. doi: 10.1155/2012/306879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paoli P, Giannoni E, Chiarugi P. Anoikis molecular pathways and its role in cancer progression. Biochim Biophys Acta. 2013;1833:3481–3498. doi: 10.1016/j.bbamcr.2013.06.026. [DOI] [PubMed] [Google Scholar]
- 34.Yang MC, Lin RW, Huang SB, Huang SY, Chen WJ, Wang S, Hong YR, Wang C. Bim directly antagonizes Bcl-xl in doxorubicin-induced prostate cancer cell apoptosis independently of p53. Cell Cycle. 2016;15:394–402. doi: 10.1080/15384101.2015.1127470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aoudjit F, Vuori K. Matrix attachment regulates Fas-induced apoptosis in endothelial cells: A role for c-flip and implications for anoikis. J Cell Biol. 2001;152:633–643. doi: 10.1083/jcb.152.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marconi A, Atzei P, Panza C, Fila C, Tiberio R, Truzzi F, Wachter T, Leverkus M, Pincelli C. FLICE/caspase-8 activation triggers anoikis induced by beta1-integrin blockade in human keratinocytes. J Cell Sci. 2004;117:5815–5823. doi: 10.1242/jcs.01490. [DOI] [PubMed] [Google Scholar]
- 37.Hattrup CL, Gendler SJ. Structure and function of the cell surface (tethered) mucins. Annu Rev Physiol. 2008;70:431–457. doi: 10.1146/annurev.physiol.70.113006.100659. [DOI] [PubMed] [Google Scholar]
- 38.Singh PK, Hollingsworth MA. Cell surface-associated mucins in signal transduction. Trends Cell Biol. 2006;16:467–476. doi: 10.1016/j.tcb.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 39.Hagiwara M, Yasumizu Y, Yamashita N, Rajabi H, Fushimi A, Long MD, Li W, Bhattacharya A, Ahmad R, Oya M, et al. MUC1-C Activates the BAF (mSWI/SNF) complex in prostate cancer stem cells. Cancer Res. 2021;81:1111–1122. doi: 10.1158/0008-5472.CAN-20-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanson JM, BroweIl DA, Cunliffe WJ, Varma J, Allen A, Hemming D, Shenton BK, Young JR, Higgs MJ, Brotherick I, Pearson JP. MUC1 expression in primary breast cancer: The effect of tamoxifen treatment. Breast Cancer Res Treat. 2001;67:215–222. doi: 10.1023/A:1017955726902. [DOI] [PubMed] [Google Scholar]
- 41.Kufe DW. Mucins in cancer: Function, prognosis and therapy. Nat Rev Cancer. 2009;9:874–885. doi: 10.1038/nrc2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raina D, Ahmad R, Rajabi H, Panchamoorthy G, Kharbanda S, Kufe D. Targeting cysteine-mediated dimerization of the MUC1-C oncoprotein in human cancer cells. Int J Oncol. 2012;40:1643–1649. doi: 10.3892/ijo.2011.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang J. Identification of novel biomarkers, MUC5AC, MUC1, KRT7, GAPDH, CD44 for gastric cancer. Med Oncol. 2020;37:34. doi: 10.1007/s12032-020-01362-0. [DOI] [PubMed] [Google Scholar]
- 44.Apostolopoulos V, Stojanovska L, Gargosky SE. MUC1 (CD227): A multi-tasked molecule. Cell Mol Life Sci. 2015;72:4475–4500. doi: 10.1007/s00018-015-2014-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor-Papadimitriou J. Report on the first international workshop on carcinoma-associated mucins. Int J Cancer. 1991;49:1–5. doi: 10.1002/ijc.2910490102. [DOI] [PubMed] [Google Scholar]
- 46.Gendler SJ, Spicer AP. Epithelial mucin genes. Annu Rev Physiol. 1995;57:607–634. doi: 10.1146/annurev.ph.57.030195.003135. [DOI] [PubMed] [Google Scholar]
- 47.Hanisch FG, Müller S. MUC1: The polymorphic appearance of a human mucin. Glycobiology. 2000;10:439–449. doi: 10.1093/glycob/10.5.439. [DOI] [PubMed] [Google Scholar]
- 48.Agrawal B, Krantz MJ, Parker J, Longenecker BM. Expression of MUC1 mucin on activated human T cells: Implications for a role of MUC1 in normal immune regulation. Cancer Res. 1998;58:4079–4081. [PubMed] [Google Scholar]
- 49.Dent GA, Civalier CJ, Brecher ME, Bentley SA. MUC1 expression in hematopoietic tissues. Am J Clin Pathol. 1999;111:741–747. doi: 10.1093/ajcp/111.6.741. [DOI] [PubMed] [Google Scholar]
- 50.Franke FE, Kraus S, Eiermann C, Pauls K, Lalani EN, Bergmann M. MUC1 in normal and impaired spermatogenesis. Mol Hum Reprod. 2001;7:505–512. doi: 10.1093/molehr/7.6.505. [DOI] [PubMed] [Google Scholar]
- 51.Seo JT, Lee JS, Jun JH, Yang MH. Expression of mucin genes in the human testis and its relationship to spermatogenesis. Yonsei Med J. 2005;46:667–672. doi: 10.3349/ymj.2005.46.5.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cascio S, Finn OJ. Intra- and extra-cellular events related to altered glycosylation of MUC1 promote chronic inflammation, tumor progression, invasion, and metastasis. Biomolecules. 2016;6:39. doi: 10.3390/biom6040039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bennett EP, Mandel U, Clausen H, Gerken TA, Fritz TA, Tabak LA. Control of mucin-type O-glycosylation: A classification of the polypeptide GalNAc-transferase gene family. Glycobiology. 2012;22:736–756. doi: 10.1093/glycob/cwr182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taherali F, Varum F, Basit AW. A slippery slope: On the origin, role and physiology of mucus. Adv Drug Deliv Rev. 2018;124:16–33. doi: 10.1016/j.addr.2017.10.014. [DOI] [PubMed] [Google Scholar]
- 55.Raina D, Agarwal P, Lee J, Bharti A, McKnight CJ, Sharma P, Kharbanda S, Kufe D. Characterization of the MUC1-C cytoplasmic domain as a cancer target. PLoS One. 2015;10:e0135156. doi: 10.1371/journal.pone.0135156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McAuley JL, Corcilius L, Tan HX, Payne RJ, McGuckin MA, Brown LE. The cell surface mucin MUC1 limits the severity of influenza A virus infection. Mucosal Immunol. 2017;10:1581–1593. doi: 10.1038/mi.2017.16. [DOI] [PubMed] [Google Scholar]
- 57.Thathiah A, Blobel CP, Carson DD. Tumor necrosis factor-alpha converting enzyme/ADAM 17 mediates MUC1 shedding. J Biol Chem. 2003;278:3386–3394. doi: 10.1074/jbc.M208326200. [DOI] [PubMed] [Google Scholar]
- 58.Tarp MA, Clausen H. Mucin-type O-glycosylation and its potential use in drug and vaccine development. Biochim Biophys Acta. 2008;1780:546–563. doi: 10.1016/j.bbagen.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 59.Awaya H, Takeshima Y, Yamasaki M, Inai K. Expression of MUC1, MUC2, MUC5AC, and MUC6 in atypical adenomatous hyperplasia, bronchioloalveolar carcinoma, adenocarcinoma with mixed subtypes, and mucinous bronchioloalveolar carcinoma of the lung. Am J Clin Pathol. 2004;121:644–653. doi: 10.1309/U4WGE9EBFJN6CM8R. [DOI] [PubMed] [Google Scholar]
- 60.Horm TM, Schroeder JA. MUC1 and metastatic cancer: Expression, function and therapeutic targeting. Cell Adh Migr. 2013;7:187–198. doi: 10.4161/cam.23131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krishn SR, Kaur S, Smith LM, Johansson SL, Jain M, Patel A, Gautam SK, Hollingsworth MA, Mandel U, Clausen H, et al. Mucins and associated glycan signatures in colon adenoma-carcinoma sequence: Prospective pathological implication(s) for early diagnosis of colon cancer. Cancer Lett. 2016;374:304–314. doi: 10.1016/j.canlet.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Singh AP, Chauhan SC, Bafna S, Johansson SL, Smith LM, Moniaux N, Lin MF, Batra SK. Aberrant expression of transmembrane mucins, MUC1 and MUC4, in human prostate carcinomas. Prostate. 2006;66:421–429. doi: 10.1002/pros.20372. [DOI] [PubMed] [Google Scholar]
- 63.Gao Y, Liu Z, Feng J, Sun Q, Zhang B, Zheng W, Ma W. Expression pattern of polypeptide N-acetylgalactosaminyltransferase-10 in gastric carcinoma. Oncol Lett. 2013;5:113–116. doi: 10.3892/ol.2012.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guda K, Moinova H, He J, Jamison O, Ravi L, Natale L, Lutterbaugh J, Lawrence E, Lewis S, Willson JK, et al. Inactivating germ-line and somatic mutations in polypeptide N-acetylgalactosaminyltransferase 12 in human colon cancers. Proc Natl Acad Sci USA. 2009;106:12921–12925. doi: 10.1073/pnas.0901454106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stowell SR, Ju T, Cummings RD. Protein glycosylation in cancer. Annu Rev Pathol. 2015;10:473–510. doi: 10.1146/annurev-pathol-012414-040438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liesche F, Kölbl AC, Ilmer M, Hutter S, Jeschke U, Andergassen U. Role of N-acetylgalactosaminyltransferase 6 in early tumorigenesis and formation of metastasis. Mol Med Rep. 2016;13:4309–4314. doi: 10.3892/mmr.2016.5044. [DOI] [PubMed] [Google Scholar]
- 67.Hakomori S. Aberrant glycosylation in cancer cell membranes as focused on glycolipids: Overview and perspectives. Cancer Res. 1985;45:2405–2414. [PubMed] [Google Scholar]
- 68.Radziejewska I, Supruniuk K, Nazaruk J, Karna E, Popławska B, Bielawska A, Galicka A. Rosmarinic acid influences collagen, MMPs, TIMPs, glycosylation and MUC1 in CRL-1739 gastric cancer cell line. Biomed Pharmacother. 2018;107:397–407. doi: 10.1016/j.biopha.2018.07.123. [DOI] [PubMed] [Google Scholar]
- 69.Syrkina MS, Maslakova AA, Potashnikova DM, Veiko VP, Vassetzky YS, Rubtsov MA. Dual role of the extracellular domain of human mucin MUC1 in metastasis. J Cell Biochem. 2017;118:4002–4011. doi: 10.1002/jcb.26056. [DOI] [PubMed] [Google Scholar]
- 70.Ho WL, Hsu WM, Huang MC, Kadomatsu K, Nakagawara A. Protein glycosylation in cancers and its potential therapeutic applications in neuroblastoma. J Hematol Oncol. 2016;9:100. doi: 10.1186/s13045-016-0334-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu B, Pan S, Xiao Y, Liu Q, Xu J, Jia L. LINC01296/miR-26a/GALNT3 axis contributes to colorectal cancer progression by regulating O-glycosylated MUC1 via PI3K/AKT pathway. J Exp Clin Cancer Res. 2018;37:316. doi: 10.1186/s13046-018-0994-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mao Y, Zhang Y, Fan S, Chen L, Tang L, Chen X, Lyu J. GALNT6 promotes tumorigenicity and metastasis of breast cancer cell via β-catenin/MUC1-C signaling pathway. Int J Biol Sci. 2019;15:169–182. doi: 10.7150/ijbs.29048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bäckström M, Thomsson KA, Karlsson H, Hansson GC. Sensitive liquid chromatography-electrospray mass spectrometry allows for the analysis of the O-glycosylation of immunoprecipitated proteins from cells or tissues: Application to MUC1 glycosylation in cancer. J Proteome Res. 2009;8:538–545. doi: 10.1021/pr800713h. [DOI] [PubMed] [Google Scholar]
- 74.Blanas A, Sahasrabudhe NM, Rodríguez E, van Kooyk Y, van Vliet SJ. Fucosylated antigens in cancer: An alliance toward tumor progression, metastasis, and resistance to chemotherapy. Front Oncol. 2018;8:39. doi: 10.3389/fonc.2018.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jia L, Zhang J, Ma T, Guo Y, Yu Y, Cui J. The Function of Fucosylation in Progression of Lung Cancer. Front Oncol. 2018;8:565. doi: 10.3389/fonc.2018.00565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen Z, Gulzar ZG, St Hill CA, Walcheck B, Brooks JD. Increased expression of GCNT1 is associated with altered O-glycosylation of PSA, PAP, and MUC1 in human prostate cancers. Prostate. 2014;74:1059–1067. doi: 10.1002/pros.22826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nakamori S, Kameyama M, Imaoka S, Furukawa H, Ishikawa O, Sasaki Y, Kabuto T, Iwanaga T, Matsushita Y, Irimura T. Increased expression of sialyl Lewisx antigen correlates with poor survival in patients with colorectal carcinoma: Clinicopathological and immunohistochemical study. Cancer Res. 1993;53:3632–3637. [PubMed] [Google Scholar]
- 78.Ricardo S, Marcos-Silva L, Valente C, Coelho R, Gomes R, David L. Mucins MUC16 and MUC1 are major carriers of SLe(a) and SLe(x) in borderline and malignant serous ovarian tumors. Virchows Arch. 2016;468:715–722. doi: 10.1007/s00428-016-1929-6. [DOI] [PubMed] [Google Scholar]
- 79.Mori Y, Akita K, Yashiro M, Sawada T, Hirakawa K, Murata T, Nakada H. Binding of galectin-3, a β-galactoside-binding lectin, to MUC1 protein enhances phosphorylation of extracellular signal-regulated kinase 1/2 (ERK1/2) and Akt, promoting tumor cell malignancy. J Biol Chem. 2015;290:26125–26140. doi: 10.1074/jbc.M115.651489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ramasamy S, Duraisamy S, Barbashov S, Kawano T, Kharbanda S, Kufe D. The MUC1 and galectin-3 oncoproteins function in a microRNA-dependent regulatory loop. Mol Cell. 2007;27:992–1004. doi: 10.1016/j.molcel.2007.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sciacchitano S, Lavra L, Morgante A, Ulivieri A, Magi F, De Francesco GP, Bellotti C, Salehi LB, Ricci A. Galectin-3: One molecule for an alphabet of diseases, from A to Z. Int J Mol Sci. 2018;19:379. doi: 10.3390/ijms19020379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhao Q, Guo X, Nash GB, Stone PC, Hilkens J, Rhodes JM, Yu LG. Circulating galectin-3 promotes metastasis by modifying MUC1 localization on cancer cell surface. Cancer Res. 2009;69:6799–6806. doi: 10.1158/0008-5472.CAN-09-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kufe DW. MUC1-C oncoprotein as a target in breast cancer: Activation of signaling pathways and therapeutic approaches. Oncogene. 2013;32:1073–1081. doi: 10.1038/onc.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ren J, Agata N, Chen D, Li Y, Yu WH, Huang L, Raina D, Chen W, Kharbanda S, Kufe D. Human MUC1 carcinoma-associated protein confers resistance to genotoxic anticancer agents. Cancer Cell. 2004;5:163–175. doi: 10.1016/S1535-6108(04)00020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yin L, Kufe D. Human MUC1 carcinoma antigen regulates intracellular oxidant levels and the apoptotic response to oxidative stress. J Biol Chem. 2003;278:35458–35464. doi: 10.1074/jbc.M301987200. [DOI] [PubMed] [Google Scholar]
- 86.Baldwin AS. Control of oncogenesis and cancer therapy resistance by the transcription factor NF-κB. J Clin Invest. 2001;107:241–246. doi: 10.1172/JCI11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dyomin VG, Palanisamy N, Lloyd KO, Dyomina K, Jhanwar SC, Houldsworth J, Chaganti RS. MUC1 is activated in a B-cell lymphoma by the t(1;14)(q21;q32) translocation and is rearranged and amplified in B-cell lymphoma subsets. Blood. 2000;95:2666–2671. doi: 10.1182/blood.V95.8.2666. [DOI] [PubMed] [Google Scholar]
- 88.Nakshatri H, Bhat-Nakshatri P, Martin DA, Goulet RJ, Jr, Sledge GW., Jr Constitutive activation of NF-κB during progression of breast cancer to hormone-independent growth. Mol Cell Biol. 1997;17:3629–3639. doi: 10.1128/MCB.17.7.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stroopinsky D, Rosenblatt J, Ito K, Mills H, Yin L, Rajabi H, Vasir B, Kufe T, Luptakova K, Arnason J, et al. MUC1 is a potential target for the treatment of acute myeloid leukemia stem cells. Cancer Res. 2013;73:5569–5579. doi: 10.1158/0008-5472.CAN-13-0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li Y, Yu WH, Ren J, Huang L, Kharbanda S, Loda M, Kufe D. Heregulin targets γ-catenin to the nucleolus by a mechanism dependent on the DF3/MUC1 protein. Mol Cancer Res. 2003;1:765–775. [PubMed] [Google Scholar]
- 91.Agata N, Ahmad R, Kawano T, Raina D, Kharbanda S, Kufe D. MUC1 oncoprotein blocks death receptor-mediated apoptosis by inhibiting recruitment of caspase-8. Cancer Res. 2008;68:6136–6144. doi: 10.1158/0008-5472.CAN-08-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ahmad R, Raina D, Trivedi V, Ren J, Rajabi H, Kharbanda S, Kufe D. MUC1 oncoprotein activates the IkappaB kinase beta complex and constitutive NF-kappaBsignalling. Nat Cell Biol. 2007;9:1419–1427. doi: 10.1038/ncb1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Escher TE, Lui AJ, Geanes ES, Walter KR, Tawfik O, Hagan CR, Lewis-Wambi J. Interaction between MUC1 and STAT1 drives IFITM1 overexpression in aromatase inhibitor-resistant breast cancer cells and mediates estrogen-induced apoptosis. Mol Cancer Res. 2019;17:1180–1194. doi: 10.1158/1541-7786.MCR-18-0916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pistritto G, Trisciuoglio D, Ceci C, Garufi A, D'Orazi G. Apoptosis as anticancer mechanism: Function and dysfunction of its modulators and targeted therapeutic strategies. Aging (Albany NY) 2016;8:603–619. doi: 10.18632/aging.100934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Comalada M, Xaus J, Valledor AF, López-López C, Pennington DJ, Celada A. PKC epsilon is involved in JNK activation that mediates LPS-induced TNF-alpha, which induces apoptosis in macrophages. Am J Physiol Cell Physiol. 2003;285:C1235–1245. doi: 10.1152/ajpcell.00228.2003. [DOI] [PubMed] [Google Scholar]
- 96.Rajabi H, Kufe D. MUC1-C oncoprotein integrates a program of EMT, epigenetic reprogramming and immune evasion in human carcinomas. Biochim Biophys Acta Rev Cancer. 2017;1868:117–122. doi: 10.1016/j.bbcan.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rajabi H, Hiraki M, Kufe D. MUC1-C activates polycomb complexes and downregulates tumor suppressor genes in human cancer cells. Oncogene. 2018;37:2079–2088. doi: 10.1038/s41388-017-0096-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, Ross AJ, Roth KA, MacGregor GR, Thompson CB, Korsmeyer SJ. Proapoptotic BAX and BAK: A requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Westphal D, Dewson G, Czabotar PE, Kluck RM. Molecular biology of Bax and Bak activation and action. Biochim Biophys Acta. 2011;1813:521–531. doi: 10.1016/j.bbamcr.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 100.Peña-Blanco A, García-Sáez AJ. Bax, Bak and beyond - mitochondrial performance in apoptosis. FEBS J. 2018;285:416–431. doi: 10.1111/febs.14186. [DOI] [PubMed] [Google Scholar]
- 101.Ahmad R, Alam M, Rajabi H, Kufe D. The MUC1-C oncoprotein binds to the BH3 domain of the pro-apoptotic BAX protein and blocks BAX function. J Biol Chem. 2012;287:20866–20875. doi: 10.1074/jbc.M112.357293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.D'Alessio M, De Nicola M, Coppola S, Gualandi G, Pugliese L, Cerella C, Cristofanon S, Civitareale P, Ciriolo MR, Bergamaschi A, et al. Oxidative Bax dimerization promotes its translocation to mitochondria independently of apoptosis. FASEB J. 2005;19:1504–1506. doi: 10.1096/fj.04-3329fje. [DOI] [PubMed] [Google Scholar]
- 103.Leng Y, Cao C, Ren J, Huang L, Chen D, Ito M, Kufe D. Nuclear import of the MUC1-C oncoprotein is mediated by nucleoporin Nup62. J Biol Chem. 2007;282:19321–19330. doi: 10.1074/jbc.M703222200. [DOI] [PubMed] [Google Scholar]
- 104.Cangul H, Broday L, Salnikow K, Sutherland J, Peng W, Zhang Q, Poltaratsky V, Yee H, Zoroddu MA, Costa M. Molecular mechanisms of nickel carcinogenesis. Toxicol Lett. 2002;127:69–75. doi: 10.1016/S0378-4274(01)00485-4. [DOI] [PubMed] [Google Scholar]
- 105.Lu H, Shi X, Costa M, Huang C. Carcinogenic effect of nickel compounds. Mol Cell Biochem. 2005;279:45–67. doi: 10.1007/s11010-005-8215-2. [DOI] [PubMed] [Google Scholar]
- 106.Castorina A, Giunta S. Mucin 1 (MUC1) signalling contributes to increase the resistance to cell death in human bronchial epithelial cells exposed to nickel acetate. Biometals. 2014;27:1149–1158. doi: 10.1007/s10534-014-9776-x. [DOI] [PubMed] [Google Scholar]
- 107.Schroeder JA, Masri AA, Adriance MC, Tessier JC, Kotlarczyk KL, Thompson MC, Gendler SJ. MUC1 overexpression results in mammary gland tumorigenesis and prolonged alveolar differentiation. Oncogene. 2004;23:5739–5747. doi: 10.1038/sj.onc.1207713. [DOI] [PubMed] [Google Scholar]
- 108.Raina D, Kosugi M, Ahmad R, Panchamoorthy G, Rajabi H, Alam M, Shimamura T, Shapiro GI, Supko J, Kharbanda S, Kufe D. Dependence on the MUC1-C oncoprotein in non-small cell lung cancer cells. Mol Cancer Ther. 2011;10:806–816. doi: 10.1158/1535-7163.MCT-10-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Andjelic S, Hsia C, Suzuki H, Kadowaki T, Koyasu S, Liou HC. Phosphatidylinositol 3-kinase and NF-kappa B/Rel are at the divergence of CD40-mediated proliferation and survival pathways. J Immunol. 2000;165:3860–3867. doi: 10.4049/jimmunol.165.7.3860. [DOI] [PubMed] [Google Scholar]
- 110.Ahmad R, Raina D, Joshi MD, Kawano T, Ren J, Kharbanda S, Kufe D. MUC1-C oncoprotein functions as a direct activator of the nuclear factor-kappaB p65 transcription factor. Cancer Res. 2009;69:7013–7021. doi: 10.1158/0008-5472.CAN-09-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg ME. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–241. doi: 10.1016/S0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 112.Datta SR, Katsov A, Hu L, Petros A, Fesik SW, Yaffe MB, Greenberg ME. 143-3 proteins and survival kinases cooperate to inactivate BAD by BH3 domain phosphorylation. Mol Cell. 2000;6:41–51. doi: 10.1016/S1097-2765(05)00012-2. [DOI] [PubMed] [Google Scholar]
- 113.Fleury C, Mignotte B, Vayssière JL. Mitochondrial reactive oxygen species in cell death signaling. Biochimie. 2002;84:131–141. doi: 10.1016/S0300-9084(02)01369-X. [DOI] [PubMed] [Google Scholar]
- 114.Redza-Dutordoir M, Averill-Bates DA. Activation of apoptosis signalling pathways by reactive oxygen species. Biochim Biophys Acta. 2016;1863:2977–2992. doi: 10.1016/j.bbamcr.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 115.Yin L, Huang L, Kufe D. MUC1 oncoprotein activates the FOXO3a transcription factor in a survival response to oxidative stress. J Biol Chem. 2004;279:45721–45727. doi: 10.1074/jbc.M408027200. [DOI] [PubMed] [Google Scholar]
- 116.Park JA, Park S, Choi JK, Han MK, Lee Y. Inhibition of MUC1-C Increases ROS and Cell Death in Mouse Embryonic Stem Cells. Int J Stem Cells. 2021;14:180–190. doi: 10.15283/ijsc20089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu Y, Ao X, Ding W, Ponnusamy M, Wu W, Hao X, Yu W, Wang Y, Li P, Wang J. Critical role of FOXO3a in carcinogenesis. Mol Cancer. 2018;17:104. doi: 10.1186/s12943-018-0856-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Levine AJ, Momand J, Finlay CA. The p53 tumour suppressor gene. Nature. 1991;351:453–456. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- 119.Amaral JD, Xavier JM, Steer CJ, Rodrigues CM. The role of p53 in apoptosis. Discov Med. 2010;9:145–152. [PubMed] [Google Scholar]
- 120.Wei X, Xu H, Kufe D. Human MUC1 oncoprotein regulates p53 responsive gene transcription in the genotoxic stress response. Cancer Cell. 2005;7:167–178. doi: 10.1016/j.ccr.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 121.Huang HZ, Yin YF, Wan WJ, Xia D, Wang R, Shen XM. Up-regulation of microRNA-136 induces apoptosis and radiosensitivity of esophageal squamous cell carcinoma cells by inhibiting the expression of MUC1. Exp Mol Pathol. 2019;110:104278. doi: 10.1016/j.yexmp.2019.104278. [DOI] [PubMed] [Google Scholar]
- 122.Wang JJ, Li ZF, Li XJ, Han Z, Zhang L, Liu ZJ. Effects of microRNA-136 on melanoma cell proliferation, apoptosis, and epithelial-mesenchymal transition by targetting PMEL through the Wnt signaling pathway. Biosci Rep. 2017;37:BSR20170743. doi: 10.1042/BSR20170743. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 123.Wang L, Wu X, Wang B, Wang Q, Han L. Mechanisms of miR-145 regulating invasion and metastasis of ovarian carcinoma. Am J Transl Res. 2017;9:3443–3451. [PMC free article] [PubMed] [Google Scholar]
- 124.Wang X, Zhou X, Zeng F, Wu X, Li H. miR-485-5p inhibits the progression of breast cancer cells by negatively regulating MUC1. Breast Cancer. 2020;27:765–775. doi: 10.1007/s12282-020-01075-2. [DOI] [PubMed] [Google Scholar]
- 125.Du C, Fang M, Li Y, Li L, Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33–42. doi: 10.1016/S0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- 126.Tummers B, Green DR. Caspase-8: Regulating life and death. Immunol Rev. 2017;277:76–89. doi: 10.1111/imr.12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ren J, Raina D, Chen W, Li G, Huang L, Kufe D. MUC1 oncoprotein functions in activation of fibroblast growth factor receptor signaling. Mol Cancer Res. 2006;4:873–883. doi: 10.1158/1541-7786.MCR-06-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chen Q, Li D, Ren J, Li C, Xiao ZX. MUC1 activates JNK1 and inhibits apoptosis under genotoxic stress. Biochem Biophys Res Commun. 2013;440:179–183. doi: 10.1016/j.bbrc.2013.09.055. [DOI] [PubMed] [Google Scholar]
- 129.Dunn C, Wiltshire C, MacLaren A, Gillespie DA. Molecular mechanism and biological functions of c-Jun N-terminal kinase signalling via the c-Jun transcription factor. Cell Signal. 2002;14:585–593. doi: 10.1016/S0898-6568(01)00275-3. [DOI] [PubMed] [Google Scholar]
- 130.Yarza R, Vela S, Solas M, Ramirez MJ. c-Jun N-terminal Kinase (JNK) signaling as a therapeutic target for Alzheimer's disease. Front Pharmacol. 2016;6:321. doi: 10.3389/fphar.2015.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhang AM, Chi XH, Bo ZQ, Huang XF, Zhang J. MUC1 gene silencing inhibits proliferation, invasion, and migration while promoting apoptosis of oral squamous cell carcinoma cells. Biosci Rep. 2019;39:BSR20182193. doi: 10.1042/BSR20182193. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 132.Tanaka Y, Terai Y, Kawaguchi H, Fujiwara S, Yoo S, Tsunetoh S, Takai M, Kanemura M, Tanabe A, Ohmichi M. Prognostic impact of EMT (epithelial-mesenchymal-transition)-related protein expression in endometrial cancer. Cancer Biol Ther. 2013;14:13–19. doi: 10.4161/cbt.22625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Vega S, Morales AV, Ocaña OH, Valdés F, Fabregat I, Nieto MA. Snail blocks the cell cycle and confers resistance to cell death. Genes Dev. 2004;18:1131–1143. doi: 10.1101/gad.294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Renauld JC. Class II cytokine receptors and their ligands: Key antiviral and inflammatory modulators. Nat Rev Immunol. 2003;3:667–676. doi: 10.1038/nri1153. [DOI] [PubMed] [Google Scholar]
- 135.Yi FT, Lu QP. Mucin 1 promotes radioresistance in hepatocellular carcinoma cells through activation of JAK2/STAT3 signaling. Oncol Lett. 2017;14:7571–7576. doi: 10.3892/ol.2017.7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kato K, Lillehoj EP, Kim KC. MUC1 regulates epithelial inflammation and apoptosis by PolyI:C through inhibition of Toll/IL-1 receptor-domain-containing adapter-inducing IFN-β (TRIF) recruitment to Toll-like receptor 3. Am J Respir Cell Mol Biol. 2014;51:446–454. doi: 10.1165/rcmb.2014-0018OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tréhoux S, Duchêne B, Jonckheere N, Van Seuningen I. The MUC1 oncomucin regulates pancreatic cancer cell biological properties and chemoresistance. Implication of p42-44 MAPK, Akt, Bcl-2 and MMP13 pathways. Biochem Biophys Res Commun. 2015;456:757–762. doi: 10.1016/j.bbrc.2014.12.025. [DOI] [PubMed] [Google Scholar]
- 138.Xu X, Wells A, Padilla MT, Kato K, Kim KC, Lin Y. A signaling pathway consisting of miR-551b, catalase and MUC1 contributes to acquired apoptosis resistance and chemoresistance. Carcinogenesis. 2014;35:2457–2466. doi: 10.1093/carcin/bgu159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Frisch SM, Ruoslahti E. Integrins and anoikis. Curr Opin Cell Biol. 1997;9:701–706. doi: 10.1016/S0955-0674(97)80124-X. [DOI] [PubMed] [Google Scholar]
- 140.Piyush T, Rhodes JM, Yu LG. MUC1 O-glycosylation contributes to anoikis resistance in epithelial cancer cells. Cell Death Discov. 2017;3:17044. doi: 10.1038/cddiscovery.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.