Summary
During primary growth, plant tissues increase their length, and as these tissues mature, they initiate secondary growth to increase thickness.1 It is not known what activates this transition to secondary growth. Cytokinins are key plant hormones regulating vascular development during both primary and secondary growth. During primary growth of Arabidopsis roots, cytokinins promote procambial cell proliferation2,3 and vascular patterning together with the hormone auxin.4, 5, 6, 7 In the absence of cytokinins, secondary growth fails to initiate.8 Enhanced cytokinin levels, in turn, promote secondary growth.8,9 Despite the importance of cytokinins, little is known about the downstream signaling events in this process. Here, we show that cytokinins and a few downstream LATERAL ORGAN BOUNDARIES DOMAIN (LBD) family of transcription factors are rate-limiting components in activating and further promoting secondary growth in Arabidopsis roots. Cytokinins directly activate transcription of two homologous LBD genes, LBD3 and LBD4. Two other homologous LBDs, LBD1 and LBD11, are induced only after prolonged cytokinin treatment. Our genetic studies revealed a two-stage mechanism downstream of cytokinin signaling: while LBD3 and LBD4 regulate activation of secondary growth, LBD1, LBD3, LBD4, and LBD11 together promote further radial growth and maintenance of cambial stem cells. LBD overexpression promoted rapid cell growth followed by accelerated cell divisions, thus leading to enhanced secondary growth. Finally, we show that LBDs rapidly inhibit cytokinin signaling. Together, our data suggest that the cambium-promoting LBDs negatively feed back into cytokinin signaling to keep root secondary growth in balance.
Keywords: cytokinin, transcription factor, secondary growth, primary growth, cell growth, procambium, cambium, vascular tissue, pericycle
Graphical abstract
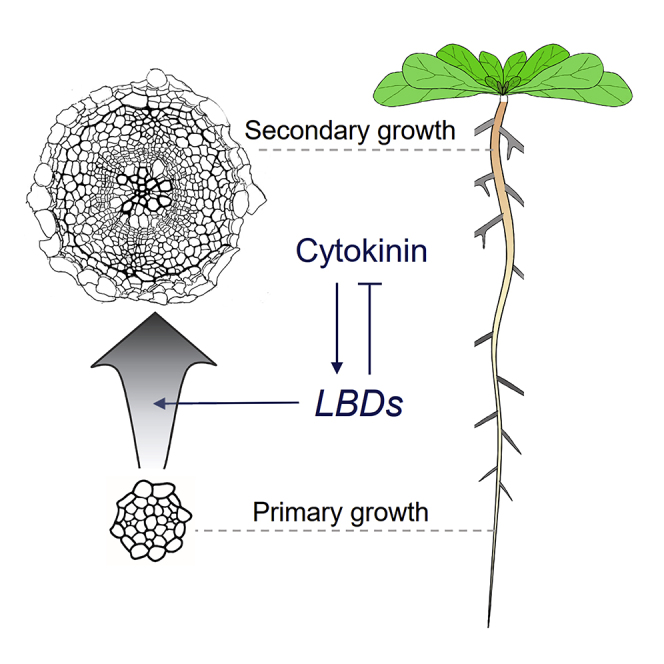
Highlights
-
•
Transition from primary to secondary growth occurs gradually in Arabidopsis root
-
•
Cytokinins activate secondary growth through a set of LBD genes
-
•
LBDs are required for both cell division and cell growth during secondary growth
-
•
LBDs rapidly inhibit cytokinin signaling
Ye et al. demonstrate that phytohormone cytokinin and four downstream LATERAL ORGAN BOUNDARIES DOMAIN (LBD) family of transcription factors promote the transition from primary to secondary growth in Arabidopsis root. LBDs negatively feed back to cytokinin signaling to keep secondary growth in balance.
Results
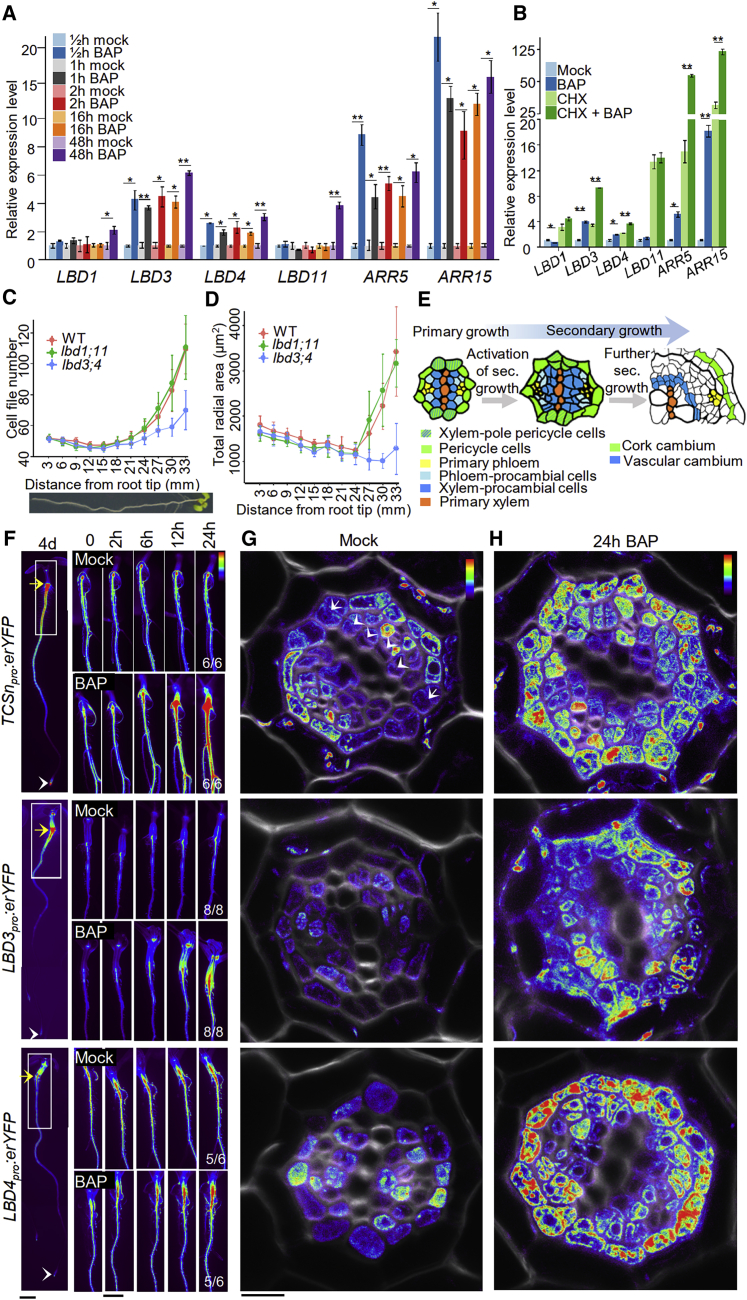
Recently, we used genome-wide analysis to identify a large set of transcription factors required for normal cambium development.10 In order to identify which of these factors operate downstream of cytokinin signaling during cambium development, we investigated whether any of their transcriptional regulatory regions are bound by type-B ARABIDOPSIS RESPONSE REGULATORS (ARRs),11, 12, 13 the key transcription factors conveying cytokinin signaling (Figure S1A). To further shorten the candidate list, we only considered genes that are also rapidly induced by cytokinins.14 Using these three criteria, we identified three candidate genes (SHORT HYPOCOTYL 2, LBD3, and LBD4), two of which, LBD3 and LBD4, are close homologs (Figure S1B).15 We therefore focused on these two genes. First, we tested the response of LBD3 and LBD4 to cytokinin by carrying out a qRT-PCR analysis using RNA isolated from Arabidopsis roots. LBD3 and LBD4 were rapidly induced by cytokinin after just 30 min, a timescale similar to the cytokinin primary response genes ARR5 and ARR15 (Figure 1A), confirming previous findings obtained from whole plants.16 This transcriptional induction also occurred in the presence of the protein synthesis inhibitor cycloheximide (Figure 1B). Furthermore, the basal expression level and cytokinin responsiveness of LBD3 and LBD4 were reduced in type-B ARR double mutants (Figure S1C). These data together suggest that LBD3 and LBD4 are direct targets of type-B ARRs.
Figure 1.
LBD3 and LBD4 are cytokinin primary response genes
(A) qRT-PCR analysis of gene transcription after a time course of BAP treatment in 5-day-old roots.
(B) qRT-PCR analysis of gene transcription in 5-day-old plants (whole plants) after mock or BAP treatment in the absence or presence of cycloheximide (CHX).
(C and D) Cell file number (C) and total radial area (D) of pericycle and procambium lineage of 7-day-old roots were quantified (data are presented as mean ± SD, n = 7–31). See Figure S2A for details. Roots were cross-sectioned in 3 mm intervals. x axis indicates the distance of cross-sections from root tip.
(E) Schematic illustration of the developmental progression of root primary vascular tissue into secondary vascular tissue. Adopted from Smetana et al.17
(F) Stereo microscopy of fluorescent reporter lines of 4-day-old (left panel) and 6-day-old (right panels) roots. Time course visualization after BAP or mock treatment (right panels). Numbers represent the frequency of the observed expression in independent roots. Yellow arrows indicate the root-hypocotyl junction. White arrowheads mark root tips. White boxes approximately represent the corresponding region visualized in the right panels.
(G and H) Confocal microscopy (heatmap) of TCSnpro:erYFP, LBD3pro:erYFP and LBD4pro:erYFP root cross-sections. 6-day-old plants were treated for 24 h with mock (G) or 1 μM BAP (H). Sections were collected from the region undergoing activation of secondary growth (~1.5 cm below the root-hypocotyl junction). Arrowheads and arrows indicate cell divisions in the procambium and pericycle, respectively (G).
Data are presented as mean ± SE from three biological replicates in (A) and (B). Two-tailed t test. ∗p < 0.05; ∗∗p < 0.01. Scale bars, 1 mm (F) and 10 μm (G and H). See also Figures S1 and S2A.
Next, we compared the spatial expression of LBD3pro:erYFP and LBD4pro:erYFP with the cytokinin response marker TCSnpro:erYFP18,19 along the primary root. Each reporter had a graded expression pattern along the root with a maximum in the upper, mature region of root or in the hypocotyl (Figure 1F). There was also a local maximum in the root tip (Figure 1F). Cytokinin treatment rapidly induced the expression of TCSn, LBD3, and LBD4 (Figure 1F), and reduction of cytokinin levels by inducible overexpression of the cytokinin degradation enzyme CYTOKININ OXIDASE 7 (CKX7)20 led to reduced fluorescence of all three reporters (Figure S1D). These data demonstrate that LBD3 and LBD4 expression in the root is tightly controlled by cytokinins.
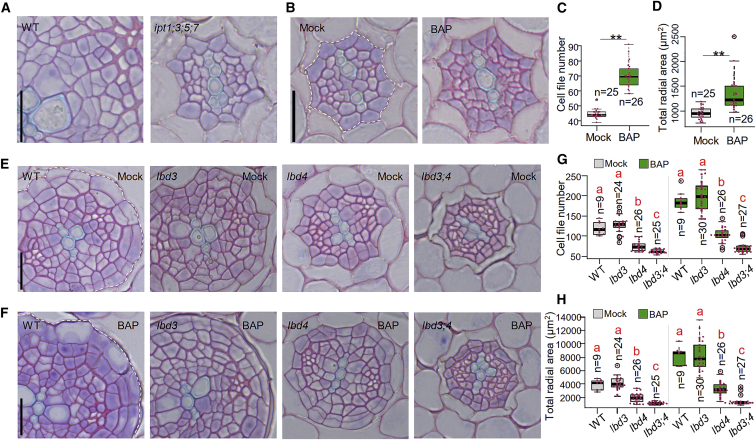
During primary growth, procambial cells within the root apical meristem undergo periclinal cell division (i.e., division along the plane parallel to the root surface) controlled by cytokinin and auxin and a set of downstream transcription factors.2, 3, 4, 5, 6, 7,21,22 These cell divisions terminate as the cells exit the meristem, and thus, the vascular cell file number remains stable until the activation of secondary growth (i.e., radial growth; Figures 1C–1E). During activation, the first divisions in procambium and pericycle cells are sporadic (15∼18 mm from root tip), followed by frequent divisions and radial cell growth along the maturing root (Figures 1C, 1D, and S2A), thus leading to secondary growth (∼24–27 mm from the root tip; Figure 1D). Pericycle cells and a subset of procambial cells (xylem procambial cells) give rise to two secondary meristems, cork cambium and vascular cambium, respectively. Xylem-pole pericycle cells produce both vascular and cork cambial cells from their circumferential position (Figure 1E).17 LBD3 and LBD4 are expressed in procambial and pericycle cell lineages during the activation of secondary growth (Figure 1G), and this expression is enhanced in the same cell lineages after cytokinin treatment (Figure 1H). The elevated LBD and TCS expression in the mature, upper region of the root (Figure 1F) coincides with the activation of procambial and pericycle cell divisions (Figure 1G). Mutant lacking four genes encoding ATP/ADP ISOPENTENYLTRANSFERASES (IPTs),23 key enzymes for cytokinin biosynthesis, fails to initiate secondary growth in the root (Figure 2A).8 Thus, our results raised the hypothesis that the gradual increase of cytokinin signaling as a root matures triggers secondary growth. To test this hypothesis, we transferred 3-day-old seedlings (i.e., before cambium activation) onto cytokinin-containing medium for 2 days. While procambium and pericycle lineages in control seedlings underwent no or only a few cambial cells divisions, cytokinin-treated roots had a number of cell divisions in both lineages, resulting in an increased number of cell files in pericycle and procambial cell lineages (Figures 2B and 2C) and increased radial growth (Figure 2D). These results demonstrate that cytokinins are rate-limiting factors in the activation of secondary growth.
Figure 2.
Cytokinin is sufficient to activate cambium prematurely, and this requires LBD3 and LBD4
(A) Cross-sections of 10-day-old wild-type (WT) and ipt1;3;5;7 roots.
(B) Cross-sections of 5-day-old WT roots. 3-day-old roots were treated for 2 days with mock or 1 μM BAP. Cells and area inside of dotted line were considered in cell file number (C) and total radial area quantifications (D), respectively.
(C and D) Quantification of cell file number (C) and total radial area (D) in the experiment presented in (B). Two-tailed t test. ∗∗p < 0.01.
(E and F) Cross-sections of WT, lbd3, lbd4, and lbd3;4 in 8-day-old roots. Six-day-old roots were treated for 2 days with mock (E) or 1 μM BAP (F). Cells and area inside of dotted line were considered in cell file number (G) and total radial area quantifications (H), respectively.
(G and H) Quantification of cell file number (G) and total radial area (H) in the experiment presented in (E) and (F).
Sections were collected from the main roots 5 mm below the hypocotyl-root junction. Scale bars, 20 μm. Red dots indicate cell numbers or radial area in individual roots (C, D, G, and H). n, number of independent roots analyzed. A separate ANOVA test was performed for mock and BAP treatment. Different red letters indicate significant differences at level alpha = 0.05, as determined by a one-way ANOVA with Tamhane’s post-test. The exact p values for each comparison can be found in Data S2A–S2D. See also Figures S3A–S3E and Data S2A–S2D.
Next, we asked whether LBD3 and LBD4 have a role during and after the activation of secondary growth. We generated knockout mutants of both genes via CRISPR-Cas9-mediated genome editing (Figure S2B). Before the onset of secondary growth, both single mutants and the lbd3;lbd4 double mutant had a normal number of pericycle and vascular cell files (Figure S3A). At the initial stages of secondary development (5-day-old roots; 5 mm below the hypocotyl-root junction), the lbd3;lbd4 double mutant already had significantly fewer cell files than wild type (Figures S3B and S3D). 3 days later (8-day-old roots), lbd4, but not lbd3, also displayed a reduced number of cell files (Figures 2E and 2G). While 8-day-old wild-type roots had produced several layers of cells from both the vascular and cork cambia, lbd3;lbd4 roots of the same age had undergone just a few cell divisions. To investigate the cell division and radial growth dynamics during the transition to secondary growth, lbd3;lbd4 roots were serial sectioned from root tip to hypocotyl. Even though the mutant shows progressively less cell files than wild type, the activation of the first cell divisions appeared not to be delayed (Figures 1C and S2A). However, an increase in cross-sectional area was observed later in lbd3;lbd4 (33 mm from root tip) than in wild type (24 mm; Figure 1D), indicating that LBD3 and LBD4 are required for activation of secondary growth. These two LBDs seem to mainly operate during secondary growth, because the primary root length and shoot size of the lbd3;lbd4 double mutant appeared similar to wild type (Figures S3G and S3H). We also tested whether LBD3 and LBD4 mediate cytokinin-induced secondary growth. After a 2-day cytokinin treatment, lbd4 had far fewer additional cell files than wild type, and even fewer were seen in lbd3;lbd4 (Figures 2E–2G and S3B–S3D). Similar resistance for cytokinin was observed also in radial growth (Figures 2H and S3E). Together, these results indicate that LBD3 and LBD4 are major factors acting downstream of cytokinin signaling to activate secondary growth.
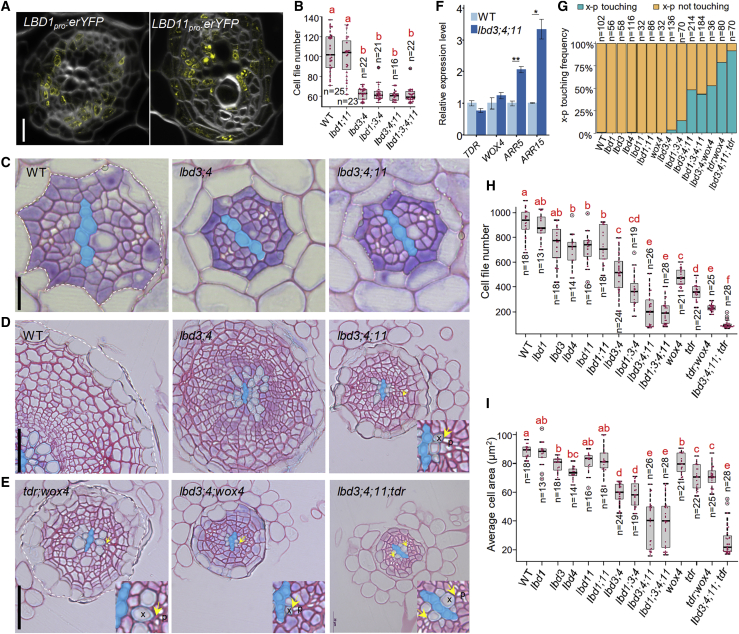
To dig deeper into potential redundancy for LBD3 and LBD4, we turned our attention to the closest homologs, LBD1 and LBD11 (Figure S1B).15 Unlike LBD3 and LBD4, LBD1 and LBD11 were induced only after prolonged cytokinin treatment, suggesting that they are not primary response genes (Figures 1A, 1B, S1E, and S1F). Similar to LBD3pro:erYFP and LBD4pro:erYFP, both LBD1pro:erYFP and LBD11pro:erYFP are expressed in the secondary tissue, albeit more weakly (Figure 3A). While LBD1pro:erYFP is expressed predominantly in the secondary phloem and vascular cambium, expression of LBD11pro:erYFP is somewhat variable in the secondary xylem, vascular cambium, and periderm (Figure S1G). To investigate whether LBD1 and LBD11 have a role in cambium development, we generated knockout mutants through genome editing (Figure S2B). Because the lbd1;lbd11 double mutant did not have defects during activation of secondary growth (Figures 1C, 1D, 3B, and S2A) and had only a slightly reduced number of cell files in 14-day-old roots (Figure 3H), we combined lbd3;lbd4 with lbd1 and lbd11. 7-day old lbd1;lbd3;lbd4, lbd1;lbd3;lbd4;lbd11, and lbd3;lbd4;lbd11 roots showed a reduction in cell file number similar to lbd3;lbd4 (Figures 3B and 3C); however, in 14-day-old roots, the triple and quadruple mutants showed a further reduction in cell file number (Figures 3D and 3H). This phenotype was associated with the frequent differentiation of vascular cambium cells located between the primary xylem and the phloem (Figures 3D and 3G), indicating that the LBDs are required for stem cell maintenance. Despite the strong cambial phenotype, shoots of lbd1;lbd3;lbd4;lbd11 appeared similar to wild type (Figure S3H). Taken together, these results indicate that cytokinin signaling initiates a two-stage process at the onset of secondary growth: LBD3 and LBD4 mediate the transition from primary to secondary growth, and this is followed by co-operation between LBD1, LBD3, LBD4, and LBD11 to promote further secondary growth and cambium stem cell maintenance.
Figure 3.
LBDs redundantly promote cambium stem cell maintenance together with TDR and WOX4
(A) Confocal microscopy of LBD1pro:erYFP and LBD11pro:erYFP root cross-sections. Sections were collected from 5 mm below the root-hypocotyl junction of 7-day-old roots.
(B) Quantification of cell file number in pericycle and procambium lineage. Seven-day-old roots are shown.
(C) Root cross-sections in 7-day-old roots. Cells inside of dotted line were considered in cell file number quantifications (B).
(D and E) Root cross-sections in 14-day-old roots. p, phloem cell (i.e., white sieve element cell); x, xylem vessel. Arrows mark xylem vessels adjacent to phloem cells, indicating that vascular cambium cells have been differentiated in this position, which is not observed in WT. Cells inside of dotted line were considered in cell file number quantifications (H) and in average cell area calculations (I).
(F) qRT-PCR analysis of gene transcription in WT and the lbd3;4;11 triple mutant. RNA was extracted from the upper 1 cm part of the main root just below the root-hypocotyl junction in 14-day-old plants. Data are presented as mean ± SE from three biological replicates. Two-tailed t test. ∗p < 0.05; ∗∗p < 0.01.
(G) Frequency of observed phloem adjacent to the xylem (x-p touching) phenotype in WT and in mutants. n, total number of events analyzed.
(H and I) Quantification of root cell file number (H) and average radial cell area (I) in 14-day-old WT and mutants.
Primary xylem cells (i.e., xylem axis) are false colored in blue (C–E). Red dots indicate cell file number or average cell area in individual roots (B, H, and I). n, number of independent roots analyzed. Different red letters indicate significant differences at level alpha = 0.05, as determined by one-way ANOVA with Tamhane’s post-test. The exact p values for each comparison can be found in Data S2E–S2I. Scale bars, 10 μm (A), 25 μm (C), and 50 μm (D and E). See also Figures S3G and S3H and Data S2E–S2I.
Differentiation of the cambial cells between the primary xylem and the phloem is reminiscent of the phenotype of a double mutant lacking both PHLOEM INTERCALATED WITH XYLEM/TDIF RECEPTOR (PXY/TDR) and WUSCHEL-RELATED HOMEOBOX4 (WOX4).24 We therefore wondered whether TDR and WOX4 operate in the same pathway as the LBDs. To test this, we first analyzed the transcription levels of TDR and WOX4 in 14-day-old lbd3;lbd4;lbd11 mutant. The transcript level of both appeared unchanged compared with wild type (Figure 3F). Additionally, the lbd3;lbd4;wox4 mutant displayed frequent cambium cell differentiation adjacent to primary phloem, a phenotype that is not seen in either lbd3;lbd4 or wox4 alone (Figures 3E and 3G). The cambium cell differentiation phenotype was more frequent in the lbd3;lbd4;lbd11;tdr mutant than in any of the parental mutant combinations and was accompanied by a further reduction of secondary growth (Figures 3E, 3G, and 3H). The additive phenotype indicates redundant roles for the LBDs and TDR-WOX4 in cambium stem cell maintenance; however, it is unclear whether they operate in the same or parallel pathways.
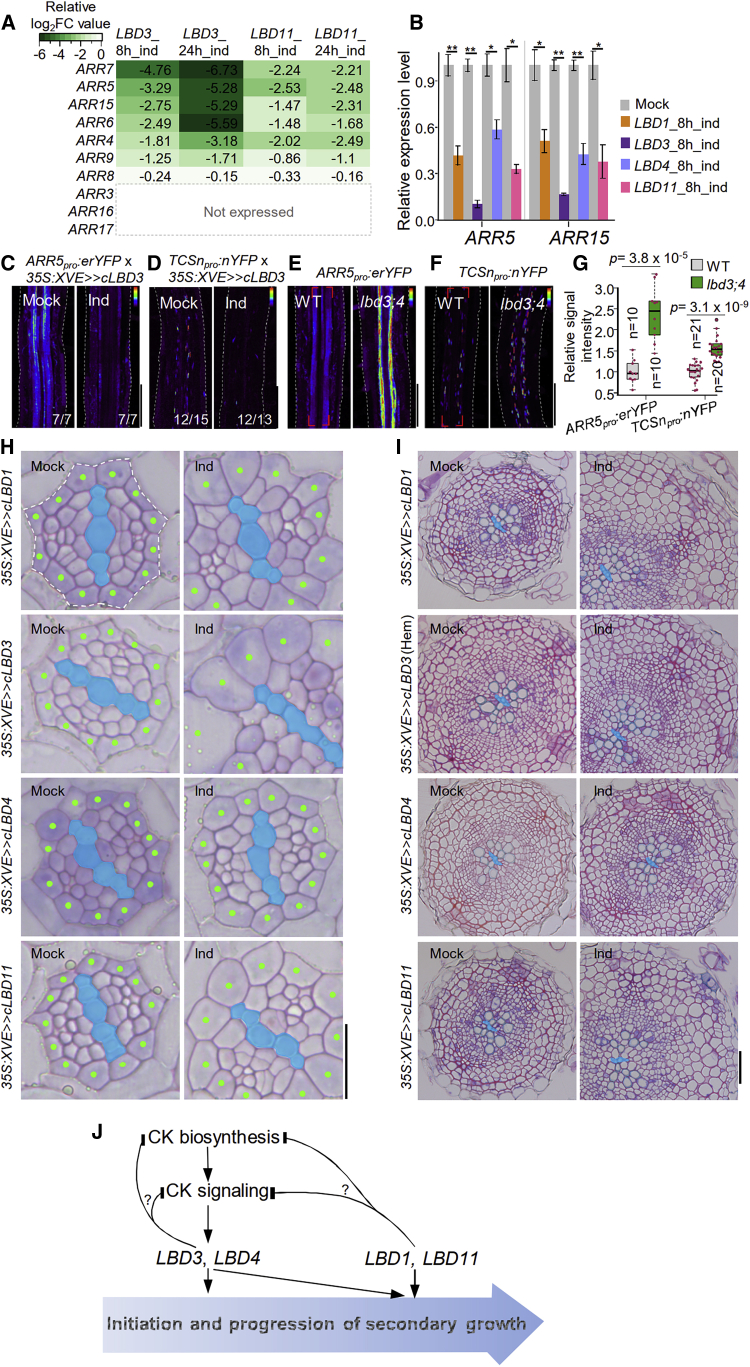
Because the LBDs encode transcription factors,25 we examined signaling events downstream of them by generating inducible overexpression lines of each of the four LBDs. We carried out transcript profiling of LBD3 and LBD11 lines, thus covering one representative from each subclade (Figure S1B). RNA was isolated from the mature part of the root after an 8-h and 24-h induction, and this was followed by RNA sequencing (RNA-seq) analysis (Data S1A–S1H). Examination of hormone-related genes revealed differential regulation of cytokinin, auxin, and abscisic acid biosynthesis and signaling (Table S2). This is in accordance with the reduced cytokinin, auxin, and abscisic acid levels in Arabidopsis shoots when LBD3 was constitutively overexpressed.16
The most striking result was that all of the type-A ARRs expressed in the mature root were already downregulated after 8 h of LBD3 or LBD11 induction (Figure 4A). Type-A ARRs are rapidly transcriptionally upregulated by cytokinins and are thus considered to be the primary cytokinin response genes.26 qRT-PCR analysis after induction of any of the four LBDs also showed a rapid reduction of ARR5 and ARR15 transcript levels, confirming our RNA-seq findings (Figure 4B). In addition, induction of LBD3 led to reduced fluorescence of ARR5pro:erYFP and TCSnpro:nYFP (Figures 4C and 4D). It has been previously shown that constitutive overexpression of LBD3 reduces cytokinin levels in aerial parts of seedlings,16 suggesting that LBD3 inhibits cytokinin signaling by inhibiting cytokinin biosynthesis or promoting its degradation. However, our RNA-seq data revealed that LBD3 or LBD11 induction led to downregulation of genes encoding both cytokinin biosynthesis and degradation enzymes as well as signaling components (Table S2). Additionally, TCSnpro:nYFP downregulation by LBD3 induction occurred equally rapidly (8 h) with induction of CKX7 (Figures S4A and S4B). Therefore, the mechanism by which the LBDs inhibit cytokinin signaling appears to be complex and thus remains unresolved. Finally, TCSnpro:nYFP and ARR5pro:erYFP fluorescence was enhanced in lbd3;lbd4 (Figures 4E–4G), and ARR5 and ARR15 were significantly elevated in lbd3;lbd4;lbd11 (Figure 3F). Together, these findings demonstrate that LBD1, LBD3, LBD4, and LBD11 inhibit cytokinin signaling by regulating cytokinin levels and/or signaling.
Figure 4.
LBDs negatively regulate cytokinin signaling and promote cell growth during secondary growth
(A) Heatmap showing normalized log2FoldChange (FC) of A-type ARRs mRNA in LBD3 or LBD11 inducible overexpression RNA-seq data. We considered a gene “not expressed” in mature root if read counts were less than 10.
(B) qRT-PCR analysis of A-type ARR (ARR5 and ARR15) transcription in 9-day-old roots (0.5–2 cm below the root-hypocotyl junction, undergoing secondary growth) with 8 h mock or induction. Data are presented as mean ± SE from three biological replicates. Two-tailed t test. ∗p < 0.05; ∗∗p < 0.01.
(C and D) Confocal microscopy of ARR5pro:erYFP (C) and TCSnpro:nYFP (D) after 1-day LBD3 induction in 6-day-old roots. ARR5pro:erYFP was analyzed in F1 generation.
(E and F) Confocal microscopy of ARR5pro:erYFP (E) and TCSnpro:nYFP (F) in 7-day-old WT and lbd3;4 roots.
(G) Quantification of average fluorescent signal intensity in (E) and (F). Area marked with brackets (E and F) was quantified. Red dots indicate average fluorescent signal intensity in individual roots. n, number of independent roots analyzed.
(H) Cross-sections of 5-day-old roots of LBD inducible overexpression lines. Three-day-old roots were treated for 2 days with mock or 5 μM 17-β, except in the case of LBD3, which was treated with 0.5 μM 17-β. Green dots represent pericycle cells. Cells inside of dotted line were used for cell file quantification in Figure S4G.
(I) Cross-sections of 14-day-old roots of LBD inducible overexpression lines. Eight-day-old roots were treated for 6 days with mock or 5 μM 17-β. Note that 0.5 μM 17-β was used for the LBD3 hemizygous (hem) line due to dose-dependent effect (Figure S4M).
(J) A model presenting the roles of LBDs during the progression of root secondary growth (large blue arrow) downstream of cytokinin (CK).
Dashed lines represent root boundaries (C–F). Primary xylem cells (i.e., xylem axis) are false colored in blue (H and I). Scale bars, 100 μm (C–F), 20 μm (H), and 50 μm (I). See also Figures S1G and S4, Table S2, and Data S1 and S3.
Because the LBDs regulate cambium development, we compared the list of differentially expressed genes after LBD induction with a list of cambium-enriched genes.10 Approximately 25% of cambial-enriched genes were already differentially regulated after an 8-h induction of LBD3 or LBD11, and the proportion reached 47% after a 24-h induction of LBD3, indicating an intimate link between LBDs and cambium development (Figure S4D; Data S1I). We also conducted a gene ontology (GO) enrichment analysis of the LBD3 and LBD11 transcript profiling data. Several categories were over-represented, but processes associated with primary cell wall modification, such as “pectin catabolic process” and “plant-type cell wall modification,” were the most notable (Figures S4E and S4F). These data suggest that the LBDs regulate primary cell wall composition and thus may regulate cell wall extensibility.27 Indeed, 2 days after LBD1, LBD3, or LBD11 induction, we observed rapid procambial and pericycle cell growth (Figures 4H and S4G–S4I). When LBDs were induced after cambium activation, enhanced cell growth was associated with more cell divisions and thus accelerated secondary growth (Figures 4I and S4J–S4L). Furthermore, in addition to having fewer cell files, lbd mutants also have a reduced average cell size in radial dimension (Figure 3I). While also tdr;wox4 show reduced number of cell files, this phenotype is not associated with markedly reduced average cell size as observed in lbd3;lbd4;lbd11 (Figure 3I). These data suggest that the LBD proteins promote secondary growth at least in part through promoting controlled cell growth.
Discussion
Previous studies have shown that LBD3 and LBD4 are positive regulators of secondary growth in Arabidopsis.10,28 Here, we show that LBD3 and LBD4 act redundantly directly downstream of cytokinin signaling to activate secondary growth in the Arabidopsis root. Later in development, LBD1 and LBD11 also act to promote further secondary growth. These four LBDs are also required to keep cambial stem cells undifferentiated, and they are able to rapidly inhibit cytokinin signaling. These findings allowed us to generate a model (Figure 4J) in which elevated cytokinin signaling in the mature root leads to two stages of LBD expression to first initiate and then maintain secondary growth. The exact role of the negative feedback of LBD expression on cytokinin levels or signaling remains unclear, as does the mechanism by which it is mediated; however, it may serve to maintain stable and responsive signaling, as has been suggested for other genetic networks with negative feedback.29
Tissue growth is a result of delicate coordination of cell growth and cell division. Loss of LBDs leads to a reduced number of cell files and reduced radial cell size, thus resulting in diminished secondary growth. However, induction of LBDs promotes rapid cell growth, and accelerated cell divisions appear only after a longer induction. It is possible that LBDs require co-operation with other factors to promote cell division. Alternatively, there could be other factors downstream of cytokinin that promote cell divisions, while LBDs are in charge of promoting cell growth. Such factors could be the PEAR genes21 or Dof2.1,22 which has been shown to promote periclinal cell divisions downstream of cytokinin in root primary vascular tissue.
Loss of LBD1, LBD3, LBD4, and LBD11 led to differentiation of cambium cells adjacent to phloem, a phenotype similar to loss of both TDR and WOX4.24 Our genetic analysis did not determine whether the LBDs and TDR-WOX4 operate in the same pathway or parallel pathways to maintain cambial stem cells. In support of the idea that they function in the same pathway, it has also been shown that LBD4 operates downstream of auxin and TDR signaling,28 and both WOX4 and TDR are regulated by auxin.17,30 LBDs seem to broadly regulate the transcription of genes encoding signaling components or metabolic enzymes for several plant hormones (Table S2). Thus, the interaction of the LBDs with TDR-WOX4 and hormonal pathways appears complex and warrants further investigation.
Cytokinins are also critical players in secondary growth in poplar trees.31,32 Additionally, overexpression of the poplar homolog of LBD1 or its dominant-negative version has been shown to lead to enhanced or decreased secondary growth in poplar stems, respectively.33 It remains to be determined whether this and the other related LBDs regulate cambial development in tree stems by promoting cell growth and whether there is a negative feedback mechanism with cytokinin signaling similar to in Arabidopsis.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Agrobacterium tumefaciens c58 GV3101 pMP90 | Koncz and Schell34 | N/A |
| Escherichia coli DH5α | N/A | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| 17-β-oestradiol (17-β) | Sigma-Aldrich | Cat# 3301 |
| 6-benzylaminopurine (BAP) | Sigma-Aldrich | Cat# B3408 |
| Cycloheximide (CHX) | Sigma-Aldrich | Cat#C7698 |
| DdeI (HpyF3I) | ThermoFisher | Cat# FD1884 |
| Dimethyl Sulfoxide (DMSO) | Sigma-Aldrich | Cat# D8418 |
| DL-Phosphinothricin (PPT) | Duchefa Biochemie | Cat# P0159.1000 |
| Dream Taq DNA Polymerase | ThermoFisher | Cat# EP0713 |
| DNase I | ThermoFisher | Cat# EN0521 |
| FTA Classic Card | Sigma-Aldrich | Cat# WHAWB120206 |
| Gateway LR Clonase II Plus enzyme | ThermoFisher | Cat# 12538120 |
| Hygromycin B Gold | InvivoGen | Cat# ant-hg-1 |
| Maxima H Minus Reverse Transcriptase | ThermoFisher | Cat# EP0752 |
| Murashige & Skoog medium | Murashige and Skoog35 and Duchefa Biochemie | Cat# M 0222.0050 |
| NotI | ThermoFisher | Cat# FD0595 |
| Oligo(dT)18 Primer | ThermoFisher | Cat# SO132 |
| Phusion High-Fidelity DNA Polymerase | ThermoFisher | Cat# F530L |
| RiboLock RNase Inhibitor | ThermoFisher | Cat# EO0382 |
| Rifampicin | Duchefa Biochemie | Cat# R0146 |
| Ruthenium red | Fluka | Cat# 84071 |
| ScaI | ThermoFisher | Cat# FD0434 |
| SCRI Renaissance 2200 Stain | Renaissance Chemicals | N/A |
| Sodium deoxycholate | Sigma-Aldrich | Cat# 30970 |
| Solis Biodyne 5x HOT FIREPol EvaGreen qPCR Mix Plus | Solis Biodyne | Cat# 08-25-00001 |
| Spectinomycin | Duchefa Biochemie | Cat# S0188 |
| Toluidine blue O | Sigma-Aldrich | Cat# T3260 |
| Urea | Sigma-Aldrich | Cat# U0631 |
| Xylitol | Sigma-Aldrich | Cat# X3375 |
| Critical commercial assays | ||
| GeneJET GEL Extraction Kit | ThermoFisher | Cat# K0692 |
| GeneJET Plant RNA purification kit | ThermoFisher | Cat# K0802 |
| GeneJET Plasmid Miniprep Kit | ThermoFisher | Cat# K0503 |
| Heat & Run gDNA removal kit | ArticZymes | SKU# 80200-50 |
| Plant Ribo-Zero rRNA Removal Kit | Illumina | Cat# MRZSR116 |
| TruSeq Stranded Total RNA HT Sample Prep Kit (with Ribo-Zero Plant) | Illumina | Cat# RS-122-2403 |
| Deposited data | ||
| RNA-seq data files | BioProject | PRJNA684618 |
| Experimental models: Organisms/strains | ||
| Arabidopsis: Col-0 | Nottingham Arabidopsis Stock Centre | N/A |
| Arabidopsis: TCSnpro:nYFP | Vatén et al.18 | N/A |
| Arabidopsis: TCSnpro:erYFP | This manuscript | N/A |
| Arabidopsis: ARR5pro:erYFP | Siligato et al.36 | N/A |
| Arabidopsis: LBD1pro:erYFP | This manuscript | N/A |
| Arabidopsis: LBD3pro:erYFP | This manuscript | N/A |
| Arabidopsis: LBD4pro:erYFP | This manuscript | N/A |
| Arabidopsis: LBD11pro:erYFP | This manuscript | N/A |
| Arabidopsis: ipt1;3;5;7 | Miyawaki et al.23 | N/A |
| Arabidopsis: arr1-3;arr10-5 | Argyros et al.37 | N/A |
| Arabidopsis: arr1-3;arr12-1 | Argyros et al.37 | N/A |
| Arabidopsis: lbd1c | This manuscript | N/A |
| Arabidopsis: lbd3c | This manuscript | N/A |
| Arabidopsis: lbd4c | This manuscript | N/A |
| Arabidopsis: lbd11c | This manuscript | N/A |
| Arabidopsis: lbd3c;lbd4c | This manuscript | N/A |
| Arabidopsis: lbd1c;lbd11c | This manuscript | N/A |
| Arabidopsis: lbd1c;lbd3c;lbd4c | This manuscript | N/A |
| Arabidopsis: lbd3c;lbd4c;lbd11c | This manuscript | N/A |
| Arabidopsis: lbd1c;lbd3c;lbd4c;lbd11c | This manuscript | N/A |
| Arabidopsis: tdr | Hirakawa et al.24 | N/A |
| Arabidopsis: wox4 | Hirakawa et al.24 | N/A |
| Arabidopsis: tdr;wox4 | Hirakawa et al.24 | N/A |
| Arabidopsis: lbd3c;lbd4c;wox4 | This manuscript | N/A |
| Arabidopsis: lbd3c;lbd4c;lbd11c;tdr | This manuscript | N/A |
| Arabidopsis: 35S:XVE≫cLBD1 | This manuscript | N/A |
| Arabidopsis: 35S:XVE≫cLBD3 | This manuscript | N/A |
| Arabidopsis: 35S:XVE≫cLBD4 | This manuscript | N/A |
| Arabidopsis: 35S:XVE≫cLBD11 | This manuscript | N/A |
| Arabidopsis: 35S:XVE≫CKX7-RFP | This manuscript | N/A |
| Arabidopsis: TCSnpro:nYFP;35S:XVE≫CKX7-RFP | This manuscript | N/A |
| Arabidopsis: LBD3pro:erYFP;35S:XVE≫CKX7-RFP | This manuscript | N/A |
| Arabidopsis: LBD4pro:erYFP;35S:XVE≫CKX7-RFP | This manuscript | N/A |
| Arabidopsis: LBD3pro:gLBD3-YFP;lbd3c;lbd4c | This manuscript | N/A |
| Arabidopsis: LBD4pro:gLBD4-YFP;lbd3c;lbd4c | This manuscript | N/A |
| Arabidopsis: LBD11pro:gLBD11-YFP;lbd1c;lbd3c;lbd4c;lbd11c | This manuscript | N/A |
| Arabidopsis: ARR5pro:erYFP;35S:XVE≫cLBD3 | This manuscript | N/A |
| Arabidopsis: TCSnpro:nYFP;35S:XVE≫cLBD3 | This manuscript | N/A |
| Oligonucleotides | ||
| See Table S1 | N/A | N/A |
| Recombinant DNA | ||
| pDONRP41R | ThermoFisher | N/A |
| p1R4z-LBD1pro | This manuscript | N/A |
| p1R4z-LBD3pro | This manuscript | N/A |
| p1R4z-LBD4pro | This manuscript | N/A |
| p1R4z-LBD11pro | This manuscript | N/A |
| p221z-erYFP | Siligato et al.36 | N/A |
| p2R3a-nosT | Siligato et al.36 | N/A |
| pBm43GW | Siligato et al.36 | N/A |
| pHm43GW | Siligato et al.36 | N/A |
| pDONR221 | ThermoFisher | N/A |
| p1R4-35S:XVE | Siligato et al.36 | N/A |
| p1R4z-TCSnpro | This manuscript | N/A |
| pHm43GW-pTCSn:erYFP | This manuscript | N/A |
| p221z-cLBD1 | This manuscript | N/A |
| p221z-cLBD11 | This manuscript | N/A |
| pBm43GW-35S:XVE≫cLBD3 | Zhang et al.10 | N/A |
| pBm43GW-35S:XVE≫cLBD4 | Zhang et al.10 | N/A |
| pBm43GW-35S:XVE≫cLBD1 | This manuscript | N/A |
| pBm43GW-35S:XVE≫cLBD11 | This manuscript | N/A |
| p221z-CKX7 | This manuscript | N/A |
| 2R3z-tagRFP | Siligato et al.36 | N/A |
| pCAM-kan-R4R3 | Siligato et al.36 | N/A |
| pCAM-kan-R4R3-35S:XVE≫CKX7-RFP | This manuscript | N/A |
| pHEE2E-TRI | Wang et al.38 | N/A |
| 2R3z-Bsa I-ccdB-Bsa I | Wang et al.39 | N/A |
| p221z-zCas9- rbcS-E9t | This manuscript | N/A |
| p1R4- pEC1.2en EC1.1p | This manuscript | N/A |
| p2R3z-AtU3b-sgRNALBD1+AtU3d-sgRNALBD3+ AtU6-1-sgRNALBD4+AtU6-29-sgRNALBD4 | This manuscript | N/A |
| pEC1.2en EC1.1p:zCas9- rbcS-E9t-AtU3b-sgRNALBD1+AtU3d-sgRNALBD3+ AtU6-1-sgRNALBD4+AtU6-29-sgRNALBD4 | This manuscript | N/A |
| pHEE2E-TRI-LBD4 | This manuscript | N/A |
| p221z-gLBD3 | This manuscript | N/A |
| p221z-gLBD4 | This manuscript | N/A |
| p221z-gLBD11 | This manuscript | N/A |
| pBm43GW-LBD3pro:gLBD3-YFP | This manuscript | N/A |
| pBm43GW-LBD4pro:gLBD4-YFP | This manuscript | N/A |
| pBm43GW-LBD11pro:gLBD11-YFP | This manuscript | N/A |
| Software and algorithms | ||
| Chipster v3.11- v3.16 | Kallio et al.40 | https://chipster.csc.fi/; RRID: SCR_010939 |
| Clustal X 2.1 | Larkin et al.41 | http://www.clustal.org/clustal2/; RRID: SCR_017055 |
| clusterProfiler v3.16.1 | Yu et al.42 | http://bioconductor.org/packages/release/bioc/html/clusterProfiler.html; RRID: SCR_016884 |
| CorelDRAW Graphics Suite 2020 | CorelDRAW | http://www.coreldraw.com/en/product/graphic-design-software/; RRID: SCR_014235 |
| dCAPs Finder v2.0 | Neff et al.43 | http://helix.wustl.edu/dcaps/ |
| DESeq2 | Love et al.44 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html; RRID: SCR_015687 |
| FastQC v0.11.3 | Andrews45 | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/; RRID: SCR_014583 |
| FIJI ImageJ v1.52 | Schindelin et al.46 | https://fiji.sc/; RRID: SCR_002285 |
| ggplot2 v3.3.2 | Wickham47 | https://cran.r-project.org/web/packages/ggplot2/index.html; RRID: SCR_014601 |
| gplots v3.1.0 | Warnes et al.48 | https://cran.r-project.org/web/packages/gplots/index.html |
| HTSeq | Anders et al.49 | https://htseq.readthedocs.io/en/release_0.9.1/; RRID: SCR_005514 |
| Leica Application Suite X | Leica Microsystems | https://www.leica-microsystems.com/products/microscope-software/p/leica-las-x-ls/; RRID: SCR_013673 |
| Leica LAS AF Lite 2.6.x | Leica Microsystems | https://leica-las-af-lite.software.informer.com/2.6/ |
| MEGA7 | Kumar et al.50 | https://megasoftware.net/; RRID: SCR_000667 |
| R v4.0.2 | The R Development Core Team51 | https://www.r-project.org/ |
| RStudio v1.4.1106 | Racine52 | https://www.rstudio.com/; RRID: SCR_000432 |
| SPSS Statistics 26 | IBM | https://www.ibm.com/products/spss-statistics; RRID: SCR_019096 |
| TopHat2 | Kim et al.53 | http://ccb.jhu.edu/software/tophat/index.shtml; RRID: SCR_013035 |
| Venny v2.1.0 | Oliveros54 | https://bioinfogp.cnb.csic.es/tools/venny/; RRID: SCR_016561 |
| Other | ||
| GENEWIZ | GENEWIZ | https://www.genewiz.com/ |
| Bioanalyzer 2100 | Agilent Technologies | https://www.agilent.com/cs/library/posters/Public/BioAnalyzer.PDF; RRID: SCR_019715 |
| Leica DM2500 microscope | Leica Microsystems | https://www.leica-microsystems.com/products/light-microscopes/p/leica-dm2500/; RRID: SCR_020224 |
| Leica M165 FC fluorescent stereo microscope | Leica Microsystems | https://www.leica-microsystems.com/products/stereo-microscopes-macroscopes/p/leica-m165-fc/ |
| Leica Stellaris 8 confocal microscope | Leica Microsystems | https://www.leica-microsystems.com/products/confocal-microscopes/p/stellaris-8/ |
| Leica TCS SP5 II confocal microscope | Leica Microsystems | https://downloads.leica-microsystems.com/Leica%20TCS%20SP5%20II/Brochures/Leica%20TCS_SP5_II-Brochure_Technical_Data.EN.pdf; RRID: SCR_018714 |
| NanoDrop 1000 Spectrophotometer | ThermoFisher | http://tools.thermofisher.com/content/sfs/manuals/nd-1000-v3.8-users-manual-8%205x11.pdf; RRID: SCR_016517 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by Ari Pekka Mähönen (AriPekka.Mahonen@helsinki.fi).
Materials availability
Transgenic plant seeds and mutants generated in this study will be available on request.
Data and code availability
The RNA-seq data files are deposited under BioProject of NCBI (https://www.ncbi.nlm.nih.gov/bioproject) with accession number (BioProject ID: PRJNA684618).
Experimental model and subject details
All Arabidopsis lines used in this study were in Col-0 background as detailed in the Key resources table. Arabidopsis seedlings were cultivated in a 22°C growth chamber under long day condition (16 h light and 8 h dark).
Method details
Plant material and growth conditions
Seeds were surface sterilized by sequentially incubating them in 20% chlorine and 70% ethanol for 1 min with vortexing, followed by washing twice in sterile water. The sterilized seeds were stratified in darkness at 4°C for two days. Seeds were plated on half-strength Germination Medium (½GM) containing 0.5x Murashige and Skoog salts,35 0.8% plant agar, 1% sucrose and 0,5g/l MES pH 5.8. The plates were placed vertically in a 22°C growth chamber with long day settings (16 h light and 8 h dark).
For cytokinin treatments, a 10 mM 6-benzylaminopurine (BAP, Sigma) stock solution dissolved in dimethyl sulfoxide (DMSO, Sigma) was prepared and a 1 μM working concentration was used. BAP or mock treatment was performed by transferring plants to ½GM plates supplied with an equal volume of BAP or DMSO and continuing growth for the indicated time. 17-β-oestradiol (17-β, Sigma) treatment was conducted in a similar way. The working concentration of 17-β for XVE-based gene induction was 5 μM unless stated otherwise. For cycloheximide treatment, 5-day-old plants were first immersed in liquid ½GM (without agar) supplemented with/without 50 μM cycloheximide (CHX) for 30 min with gentle shaking. BAP was added to a final concentration of 1 μM and plants were incubated for 1h. An equal volume of DMSO was used as a mock treatment.
The following transgenic lines and mutant alleles were published previously: ipt1;3;5;723, ARR5pro:erYFP,36 TCSnpro:nYFP18 (a modified version of Zürcher et al.19), tdr,24wox4,24 tdr;wox4,24 arr1-3;arr10-537 and arr1-3;arr12-1.37
Molecular cloning and transformation
LBD1 (AT1G07900), LBD4 (AT1G31320) and LBD11 (AT2G28500) promoters were cloned by amplifying a length of 3152 bp, 4434 bp and 4943 bp upstream of the transcription start site, respectively. A 2932 bp promoter sequence of LBD3 (AT1G16530) was synthesized in GENEWIZ and the TCS promoter sequence was amplified from a published TCSnpro:nYFP reporter line18 (a modified version of Zürcher et al.19). All these promoter sequences were cloned into the pDONRP41R entry vector. The resulting entry vectors harboring different promoters, together with p221z-erYFP36 and p2R3a-nosT,36 were recombined into the destination vector pBm43GW55 for LBDs or pHm43GW55 for TCS by a MultiSite Gateway LR reaction. The coding sequence of LBD1 and LBD11 (with a stop codon) and the genomic sequence of CKX7 (without a stop codon) were cloned into the pDONR221 entry vector. The inducible overexpression constructs of LBD1 and LBD11 were generated by combining p1R4-35S:XVE,36 p221z-cLBD1/11 and p2R3a-nosT36 with the destination vector pBm43GW.55 The constructs 35S:XVE≫cLBD3 and 35S:XVE≫cLBD4 have been published before.10 The inducible construct overexpressing CKX7-tagRFP fusion was made by integrating p1R4-35S:XVE,36 p221z-CKX7 and p2R3z-tagRFP36 into pCAM-kan-R4R3.36 All newly generated entry vectors were verified by Sanger sequencing and all binary constructs were transformed into Col-0 ecotype by floral dipping.56 Transgenic seedlings were screened (15-25 transgenic individuals per each construct) on selective ½GM plates supplied with 20 μg/ml DL-phosphinothricin (PPT, Duchefa Biochemie) or 20 μg/ml Hygromycin B Gold (InvivoGen). Homozygous single insertion lines were selected according to Mendelian segregation of a selection marker. For each construct, at least two transgenic lines with similar phenotype were identified for further analysis, and one was used as the representative in our analysis. The primers used in this study are listed in Table S1.
Mutant generation by CRISPR/Cas9
Since we did not find potential knock-out alleles for all four LBDs, we decided to generate new knock-out mutants for all of them by gene editing. The egg-cell specific promoter pEC1.2en EC1.1p and a codon optimized zCas9 coding sequence with rbcS-E9t terminator were amplified from pHEE2E-TRI38 and separately cloned into pDONRP41R and pDONR221. Four sgRNA expression cassettes targeting LBD1, LBD3, LBD4 and LBD11, transcribed under promoters AtU3b, AtU3d, AtU6-1 and AtU6-29,57 respectively, were combined into 2R3z-Bsa I-ccdB-Bsa I39 by Golden Gate cloning as described previously.39 All three entry vectors were assembled into pBm43GW by an LR reaction to make the CRISPR/Cas9 binary construct.
The mutant screen was done in the T1 generation by sequencing PCR products of all four LBDs from 51 individual samples. T2 seeds were harvested from plants with heterozygous or homozygous mutations in the T1 generation for a construct-free mutant screen. To achieve this, seeds were geminated on ½GM plates containing 20 μg/ml PPT for 5 days, followed by transferal of PPT sensitive plants to PPT-free ½GM plates for another week to rescue them. The LBD4 mutation was not obtained in T1 generation, likely because the sgRNA was driven by the AtU6-1 promoter, which is less active in genome editing.39 Therefore, another independent CRISPR/Cas9 construct targeting only LBD4 was generated and transformed into the lbd3c background to make the lbd3c;4c double mutant. After segregating out the construct, the obtained lbd3c;4c knock-out mutant was crossed with the lbd1c;11c knock-out mutant, and the different mutant combinations used in this study were acquired in the F2 population. All lbd mutants used in this study contain frameshift mutations which give rise to a premature termination of translation and thus to truncated proteins. For clarity, elsewhere in this article lbd1c, lbd3c, lbd4c and lbd11c were written as lbd1, lbd3, lbd4 and lbd11, respectively. More details can be found in Figure S2B.
To confirm that the observed phenotype was a consequence of the CRISPR/Cas9 caused LBD mutation, a complementation assay was performed. We first cloned the genome sequence (without a stop codon) of LBD3, LBD4 and LBD11 into pDONR221. The resulting entry vectors p221z-gLBD3/4/11 were recombined into the pBm43GW55 destination vector together with entry vectors of the respective promoter and p2R3a-YFP36 to generate the translational reporters LBD3pro:gLBD3-YFP,LBD4pro:gLBD4-YFP and LBD11pro:gLBD11-YFP, respectively. Both LBD3pro:gLBD3-YFP and LBD4pro:gLBD4-YFP reporters complemented the lbd3;lbd4 double mutant, and LBD11pro:gLBD11-YFP reporter complemented the lbd1;lbd3;lbd4;lbd11 quadruple mutant (Figures S2C–S2F).
Once the mutation type was determined, in the following cross experiments, we also performed dCAPs43 (derived cleaved amplified polymorphic sequence, http://helix.wustl.edu/dcaps/) assays for genotyping analysis by digesting the PCR products of LBD1, LBD3, LBD11 containing the target sites with ScaI, DdeI and NotI, respectively. The homozygous mutations identified by dCAPs were further confirmed by Sanger sequencing. The LBD4 mutation was always genotyped by Sanger sequencing. All related primers are listed in Table S1.
Microscopy and image processing
To observe the anatomy of vascular tissue, plastic cross-sections were made from sections 5 mm below the root–hypocotyl junction unless otherwise stated. The sections were sequentially stained in 0.05% (w/v) toluidine blue solution and 0.05% (w/v) ruthenium red solution before imaging. The detailed sectioning methodology has been described before.58 Plastic cross-section images were captured using a Leica 2500 microscope.
The live imaging of fluorescence reporters was conducted on a Leica M165 FC fluorescent stereo microscope equipped with the Leica Application Suite X package. In other cases, fluorescence observations were performed on a Leica TCS SP5 II and Stellaris 8 confocal microscopes. The samples were first fixed by incubating roots in 4% paraformaldehyde (dissolved in 1 × PBS, pH 7.2) for 1 h with vacuum, and then incubated at 4°C for several hours or overnight. Lateral view observations used fixed samples which were first washed twice in 1xPBS and then cleared with ClearSee solution59 at room temperature for at least 1 day. Transverse observations used section cut with a vibratome as described elsewhere.17 Cell walls were stained with 0.1% (v/v) SCRI Renaissance 2200 Stain. Confocal images were obtained with the Leica LAS AF Software.
To make the results comparable, microscopy settings were kept unchanged throughout each experiment. Heatmaps were generated using Leica AF Lite 2.6.x. Microsoft PowerPoint was used to crop and organize images. Images were sometimes rotated in CorelDRAW Graphics Suite 2020. The brightness of the fluorescence signal was sometimes manually adjusted for better visualization. In cases where images from mock and treatment were compared, the adjustments were always equal.
qRT-PCR
Total RNA was extracted with a GeneJET Plant RNA purification kit (ThermoFisher) according to the manufacturer’s instructions and treated with DNase I (ThermoFisher). The cDNA was synthesized from 500 ng total RNA by using Oligo(dT)18 primer (ThermoFisher) and the Maxima H Minus Reverse Transcriptase (ThermoFisher). Synthesized cDNA was diluted by adding equal volume of nuclease-free water. qPCR were performed in a 10 μL reaction volume consisting of 1 μL diluted cDNA, 2 μL 5 x EvaGreen qPCR mix (Solis Biodyne), 0.5 μL 5 μM forward and reverse primers and 6.5 μL nuclease-free water. qPCR was run on a Bio-Rad CFX384 cycler with following program: 95°C for 12 min, 45 cycles (95°C for 15 s, 60°C for 20 s, 72°C for 20 s). Amplification of target genes was analyzed by melting curve. Three biological repeats were conducted for each experiment and three technical repeats were included for each biological repeat. The relative expression levels were calculated by the 2−ΔΔCt method60 and normalized against three reference genes UBQ10, ACT2 and TIP41. All qRT-PCR primers can be found in Table S1.
RNA-Seq
35S:XVE≫cLBD3 and 35S:XVE≫cLBD11 seeds were first germinated on ½GM plates for nine days, and then the 9-day-old seedlings were transferred to 5 μM 17-β induction or mock-plates for 8h or 24h. For each sample, root segments 0.5 cm-2 cm below the root-hypocotyl junction were collected from about 15 individuals. Visible lateral roots were removed. Three biological repeats were conducted for each time point. Total RNA was isolated using a GeneJET Plant RNA purification kit (ThermoFisher) following the manufacturer’s instructions. RNA concentration and integrity were monitored using a NanoDrop 1000 Spectrophotometer (ThermoFisher) and a Bioanalyzer 2100 (Agilent Technologies). Genomic DNA and ribosomal RNA were removed using a Heat & Run gDNA removal kit (ArticZymes) and a Plant Ribo-Zero rRNA Removal Kit (Illumina), respectively. RNA-seq libraries were constructed using a TruSeq Stranded Total RNA Library prep kit (Illumina). The pooled samples were single-end sequenced on a NextSeq 500 sequencer with an HT 75 cycle kit v2.5 in 2 runs. Library construction and sequencing was done by the Sequencing and Genomics Laboratory, Institute of Biotechnology, University of Helsinki. Raw data can be accessed at BioProject of NCBI (https://www.ncbi.nlm.nih.gov/bioproject) with accession number (BioProject ID: PRJNA684618).
RNA-seq data analysis was performed using packages available in Chipster40 (https://chipster.csc.fi/) and RStudio52 (https://www.rstudio.com/) with the program R v4.0.251 (https://www.r-project.org/). FastQC45 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used to check the quality of the raw reads merged from 2 separate runs, and single-end reads (76 bp) were then mapped to the Arabidopsis reference genome (TAIR 10.36) using TopHat253 (http://ccb.jhu.edu/software/tophat/index.shtml). The reads aligning to exons of each gene were counted with HTSeq49 (https://htseq.readthedocs.io/en/release_0.9.1/). Differential expression analysis between the mock and inductions was determined using the DESeq2 package44 (https://bioconductor.org/packages/release/bioc/html/DESeq2.html). Subsequently, |log2FoldChange| > 1 and padjusted < 0.05 were applied to identify differentially expressed genes (DEGs). DEGs are listed in Data S1. For the hormone-related genes analysis in Table S2, the gene sets of Arabidopsis hormone synthetic pathway and signaling transduction were obtained from the Plant Hormone Research Network (RIKEN) database (http://hormones.psc.riken.jp/pathway_hormones.html) and Kyoto Encyclopedia of Genes and Genomes (KEGG) database61 (https://www.genome.jp/kegg-bin/show_pathway?ath04075), respectively. Gene Ontology (GO) enrichment of biological processes in Data S3 and Figure S4 was performed using the clusterProfiler package42 (http://bioconductor.org/packages/release/bioc/html/clusterProfiler.html) in R with p < 0.05. A Venn diagram (Figure S4C) was generated using Venny v2.1.054 (https://bioinfogp.cnb.csic.es/tools/venny/). The heatmap of A-type ARRs based on log2FC was done with the gplots48 package in RStudio52 (https://www.rstudio.com/) with the program R v4.0.251 (https://www.r-project.org/).
Quantification and statistical analysis
Cell file number, diameter and total area were measured manually in FIJI ImageJ v1.52.46 The average cell area was calculated by dividing total area by cell file number. Plots were created with boxplot or the ggplot247 package in RStudio52 (https://www.rstudio.com/) with the program R v4.0.251 (https://www.r-project.org/). Boxplots show the first quartile (lower limit of boxes), median (center line) and third quartile (upper limit of boxes). The red dots and empty circles in the boxplots represent individual samples and outliers, respectively. All experiments were repeated at least twice.
Statistical analyses were carried out using SPSS Statistics 26 and RStudio52 (https://www.rstudio.com/) with the program R v4.0.251 (https://www.r-project.org/). Student’s t test was used for comparing two groups. For multiple comparisons, one-way ANOVA analysis was done in SPSS Statistics 26. Levene’s Test was used to test for homogeneity of variances. The significant differences between each dataset was calculated using a Tukey post hoc test (for equal homogeneous variance) or Tamhane’s T2 test (for equal variance not assumed) at significance level alpha = 0.05. All ANOVA results can be found in Data S2.
Acknowledgments
We thank A. Vatén and H. Fukuda for sharing published materials; lab members, Y. Helariutta, and A. Bishopp for helpful comments on the manuscript; M. Herpola and M. Iida for technical assistance; B. Wybouw for help on statistical analysis; Biodata Analytics Unit, Light Microscopy Unit, and Sequencing and Genomics Laboratory at Institute of Biotechnology; CSC–IT Center for Science, Finland, for computational resources; and S. el-Showk for proofreading of this manuscript. This work was supported by the Academy of Finland (grants no. 316544, no. 266431, no. 304806, and no. 307335), European Research Council (ERC-CoG CORKtheCAMBIA, agreement 819422), and University of Helsinki HiLIFE fellowship. X.W. is also supported by a grant from the Chinese Scholarship Council (CSC). The ORCID IDs for this article are as follows: 0000-0003-3834-862X (L.Y.); 0000-0002-8982-9848 (X.W.); 0000-0003-1645-3488 (M.L.); 0000-0002-1310-2757 (R.S.); 0000-0002-3439-3696 (G.E.); 0000-0001-7113-1604 (T.B.); 0000-0003-4463-7516 (J.Z.); and 0000-0001-6051-866X (A.P.M.).
Author contributions
L.Y., X.W., and A.P.M. designed the experiments. L.Y. conducted most experiments together with X.W. M.L. performed qRT-PCR. R.S. made the initial discovery on cytokinin response gradient and cytokinin activating cambium prematurely. R.S., T.B., and M.L. generated and analyzed CKX7 inducible overexpression lines. L.Y., X.W., G.E., and L.V. quantified cell file number in cross-sections. Earlier findings by J.Z. and T.B. directed our interest toward the LBD genes. T.B. and L.Y. performed RNA-seq data analysis. L.Y., X.W., and A.P.M. analyzed the results and wrote the manuscript, with input from all co-authors.
Declaration of interests
The authors declare no competing interests.
Published: June 14, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cub.2021.05.036.
Contributor Information
Xin Wang, Email: xin.wang@helsinki.fi.
Ari Pekka Mähönen, Email: aripekka.mahonen@helsinki.fi.
Supplemental information
(A, B) Differentially upregulated genes (A) and downregulated genes (B) in LBD3 overexpression line with 8 h induction. (C, D) Differentially upregulated genes (C) and downregulated genes (D) in LBD3 overexpression line with 24 h induction. (E, F) Differentially upregulated genes (E) and downregulated genes (F) in LBD11 overexpression line with 8 h induction. (G, H) Differentially upregulated genes (G) and downregulated genes (H) in LBD11 overexpression line with 24 h induction. (I) Cambium enriched genes overlapped with DEGs in LBD inducible overexpression lines.
ANOVA output data relative to the experiment shown in Figure 2G left panel (A), Figure 2G right panel (B), Figure 2H left panel (C), Figure 2H right panel (D), Figure 3B (E), Figure 3H (F), Figure 3I (G), Figure S3A (H), Figure S3D left panel (I), Figure S3D right panel (J), Figure S3E left panel (K) and Figure S3E right panel (L).
(A, B) GO enrichment analysis of differentially upregulated genes in LBD3 line (A) and LBD11 line (B) with 8h induction.
References
- 1.Evert R.F. Third Edition. John Wiley & Sons; 2006. Esau’s Plant Anatomy: Meristems, Cells, and Tissues of the Plant Body: Their Structure, Function, and Development. [Google Scholar]
- 2.Mähönen A.P., Bishopp A., Higuchi M., Nieminen K.M., Kinoshita K., Törmäkangas K., Ikeda Y., Oka A., Kakimoto T., Helariutta Y. Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science. 2006;311:94–98. doi: 10.1126/science.1118875. [DOI] [PubMed] [Google Scholar]
- 3.Mähönen A.P., Bonke M., Kauppinen L., Riikonen M., Benfey P.N., Helariutta Y. A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev. 2000;14:2938–2943. doi: 10.1101/gad.189200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bishopp A., Lehesranta S., Vatén A., Help H., El-Showk S., Scheres B., Helariutta K., Mähönen A.P., Sakakibara H., Helariutta Y. Phloem-transported cytokinin regulates polar auxin transport and maintains vascular pattern in the root meristem. Curr. Biol. 2011;21:927–932. doi: 10.1016/j.cub.2011.04.049. [DOI] [PubMed] [Google Scholar]
- 5.De Rybel B., Adibi M., Breda A.S., Wendrich J.R., Smit M.E., Novák O., Yamaguchi N., Yoshida S., Van Isterdael G., Palovaara J. Plant development. Integration of growth and patterning during vascular tissue formation in Arabidopsis. Science. 2014;345:1255215. doi: 10.1126/science.1255215. [DOI] [PubMed] [Google Scholar]
- 6.el-Showk S., Help-Rinta-Rahko H., Blomster T., Siligato R., Marée A.F., Mähönen A.P., Grieneisen V.A. Parsimonious model of vascular patterning links transverse hormone fluxes to lateral root initiation: auxin leads the way, while cytokinin levels out. PLoS Comput. Biol. 2015;11:e1004450. doi: 10.1371/journal.pcbi.1004450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muraro D., Mellor N., Pound M.P., Help H., Lucas M., Chopard J., Byrne H.M., Godin C., Hodgman T.C., King J.R. Integration of hormonal signaling networks and mobile microRNAs is required for vascular patterning in Arabidopsis roots. Proc. Natl. Acad. Sci. USA. 2014;111:857–862. doi: 10.1073/pnas.1221766111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsumoto-Kitano M., Kusumoto T., Tarkowski P., Kinoshita-Tsujimura K., Václavíková K., Miyawaki K., Kakimoto T. Cytokinins are central regulators of cambial activity. Proc. Natl. Acad. Sci. USA. 2008;105:20027–20031. doi: 10.1073/pnas.0805619105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Randall R.S., Miyashima S., Blomster T., Zhang J., Elo A., Karlberg A., Immanen J., Nieminen K., Lee J.Y., Kakimoto T. AINTEGUMENTA and the D-type cyclin CYCD3;1 regulate root secondary growth and respond to cytokinins. Biol. Open. 2015;4:1229–1236. doi: 10.1242/bio.013128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J., Eswaran G., Alonso-Serra J., Kucukoglu M., Xiang J., Yang W., Elo A., Nieminen K., Damén T., Joung J.G. Transcriptional regulatory framework for vascular cambium development in Arabidopsis roots. Nat. Plants. 2019;5:1033–1042. doi: 10.1038/s41477-019-0522-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Potter K.C., Wang J., Schaller G.E., Kieber J.J. Cytokinin modulates context-dependent chromatin accessibility through the type-B response regulators. Nat. Plants. 2018;4:1102–1111. doi: 10.1038/s41477-018-0290-y. [DOI] [PubMed] [Google Scholar]
- 12.Xie M., Chen H., Huang L., O’Neil R.C., Shokhirev M.N., Ecker J.R. A B-ARR-mediated cytokinin transcriptional network directs hormone cross-regulation and shoot development. Nat. Commun. 2018;9:1604. doi: 10.1038/s41467-018-03921-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zubo Y.O., Blakley I.C., Yamburenko M.V., Worthen J.M., Street I.H., Franco-Zorrilla J.M., Zhang W., Hill K., Raines T., Solano R. Cytokinin induces genome-wide binding of the type-B response regulator ARR10 to regulate growth and development in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2017;114:E5995–E6004. doi: 10.1073/pnas.1620749114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhargava A., Clabaugh I., To J.P., Maxwell B.B., Chiang Y.H., Schaller G.E., Loraine A., Kieber J.J. Identification of cytokinin-responsive genes using microarray meta-analysis and RNA-Seq in Arabidopsis. Plant Physiol. 2013;162:272–294. doi: 10.1104/pp.113.217026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iwakawa H., Ueno Y., Semiarti E., Onouchi H., Kojima S., Tsukaya H., Hasebe M., Soma T., Ikezaki M., Machida C., Machida Y. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 2002;43:467–478. doi: 10.1093/pcp/pcf077. [DOI] [PubMed] [Google Scholar]
- 16.Naito T., Yamashino T., Kiba T., Koizumi N., Kojima M., Sakakibara H., Mizuno T. A link between cytokinin and ASL9 (ASYMMETRIC LEAVES 2 LIKE 9) that belongs to the AS2/LOB (LATERAL ORGAN BOUNDARIES) family genes in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2007;71:1269–1278. doi: 10.1271/bbb.60681. [DOI] [PubMed] [Google Scholar]
- 17.Smetana O., Mäkilä R., Lyu M., Amiryousefi A., Sánchez Rodríguez F., Wu M.F., Solé-Gil A., Leal Gavarrón M., Siligato R., Miyashima S. High levels of auxin signalling define the stem-cell organizer of the vascular cambium. Nature. 2019;565:485–489. doi: 10.1038/s41586-018-0837-0. [DOI] [PubMed] [Google Scholar]
- 18.Vatén A., Soyars C.L., Tarr P.T., Nimchuk Z.L., Bergmann D.C. Modulation of asymmetric division diversity through cytokinin and SPEECHLESS regulatory interactions in the Arabidopsis stomatal lineage. Dev. Cell. 2018;47:53–66.e5. doi: 10.1016/j.devcel.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zürcher E., Tavor-Deslex D., Lituiev D., Enkerli K., Tarr P.T., Müller B. A robust and sensitive synthetic sensor to monitor the transcriptional output of the cytokinin signaling network in planta. Plant Physiol. 2013;161:1066–1075. doi: 10.1104/pp.112.211763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Köllmer I., Novák O., Strnad M., Schmülling T., Werner T. Overexpression of the cytosolic cytokinin oxidase/dehydrogenase (CKX7) from Arabidopsis causes specific changes in root growth and xylem differentiation. Plant J. 2014;78:359–371. doi: 10.1111/tpj.12477. [DOI] [PubMed] [Google Scholar]
- 21.Miyashima S., Roszak P., Sevilem I., Toyokura K., Blob B., Heo J.O., Mellor N., Help-Rinta-Rahko H., Otero S., Smet W. Mobile PEAR transcription factors integrate positional cues to prime cambial growth. Nature. 2019;565:490–494. doi: 10.1038/s41586-018-0839-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smet W., Sevilem I., de Luis Balaguer M.A., Wybouw B., Mor E., Miyashima S., Blob B., Roszak P., Jacobs T.B., Boekschoten M. DOF2.1 controls cytokinin-dependent vascular cell proliferation downstream of TMO5/LHW. Curr. Biol. 2019;29:520–529.e6. doi: 10.1016/j.cub.2018.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyawaki K., Tarkowski P., Matsumoto-Kitano M., Kato T., Sato S., Tarkowska D., Tabata S., Sandberg G., Kakimoto T. Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc. Natl. Acad. Sci. USA. 2006;103:16598–16603. doi: 10.1073/pnas.0603522103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirakawa Y., Kondo Y., Fukuda H. TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. Plant Cell. 2010;22:2618–2629. doi: 10.1105/tpc.110.076083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Husbands A., Bell E.M., Shuai B., Smith H.M.S., Springer P.S. LATERAL ORGAN BOUNDARIES defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res. 2007;35:6663–6671. doi: 10.1093/nar/gkm775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.To J.P., Kieber J.J. Cytokinin signaling: two-components and more. Trends Plant Sci. 2008;13:85–92. doi: 10.1016/j.tplants.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 27.Cosgrove D.J. Plant cell wall extensibility: connecting plant cell growth with cell wall structure, mechanics, and the action of wall-modifying enzymes. J. Exp. Bot. 2016;67:463–476. doi: 10.1093/jxb/erv511. [DOI] [PubMed] [Google Scholar]
- 28.Smit M.E., McGregor S.R., Sun H., Gough C., Bågman A.M., Soyars C.L., Kroon J.T., Gaudinier A., Williams C.J., Yang X. A PXY-mediated transcriptional network integrates signaling mechanisms to control vascular development in Arabidopsis. Plant Cell. 2020;32:319–335. doi: 10.1105/tpc.19.00562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alon U. CRC; 2006. An Introduction to Systems Biology: Design Principles of Biological Circuits. [Google Scholar]
- 30.Brackmann K., Qi J., Gebert M., Jouannet V., Schlamp T., Grünwald K., Wallner E.S., Novikova D.D., Levitsky V.G., Agustí J. Spatial specificity of auxin responses coordinates wood formation. Nat. Commun. 2018;9:875. doi: 10.1038/s41467-018-03256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Immanen J., Nieminen K., Smolander O.P., Kojima M., Alonso Serra J., Koskinen P., Zhang J., Elo A., Mähönen A.P., Street N. Cytokinin and auxin display distinct but interconnected distribution and signaling profiles to stimulate cambial activity. Curr. Biol. 2016;26:1990–1997. doi: 10.1016/j.cub.2016.05.053. [DOI] [PubMed] [Google Scholar]
- 32.Nieminen K., Immanen J., Laxell M., Kauppinen L., Tarkowski P., Dolezal K., Tähtiharju S., Elo A., Decourteix M., Ljung K. Cytokinin signaling regulates cambial development in poplar. Proc. Natl. Acad. Sci. USA. 2008;105:20032–20037. doi: 10.1073/pnas.0805617106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yordanov Y.S., Regan S., Busov V. Members of the LATERAL ORGAN BOUNDARIES DOMAIN transcription factor family are involved in the regulation of secondary growth in Populus. Plant Cell. 2010;22:3662–3677. doi: 10.1105/tpc.110.078634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koncz C., Schell J. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 1986;204:383–396. [Google Scholar]
- 35.Murashige T., Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962;15:473–497. [Google Scholar]
- 36.Siligato R., Wang X., Yadav S.R., Lehesranta S., Ma G., Ursache R., Sevilem I., Zhang J., Gorte M., Prasad K. MultiSite gateway-compatible cell type-specific gene-inducible system for plants. Plant Physiol. 2016;170:627–641. doi: 10.1104/pp.15.01246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Argyros R.D., Mathews D.E., Chiang Y.H., Palmer C.M., Thibault D.M., Etheridge N., Argyros D.A., Mason M.G., Kieber J.J., Schaller G.E. Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell. 2008;20:2102–2116. doi: 10.1105/tpc.108.059584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Z.P., Xing H.L., Dong L., Zhang H.Y., Han C.Y., Wang X.C., Chen Q.J. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 2015;16:144. doi: 10.1186/s13059-015-0715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X., Ye L., Lyu M., Ursache R., Löytynoja A., Mähönen A.P. An inducible genome editing system for plants. Nat. Plants. 2020;6:766–772. doi: 10.1038/s41477-020-0695-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kallio M.A., Tuimala J.T., Hupponen T., Klemelä P., Gentile M., Scheinin I., Koski M., Käki J., Korpelainen E.I. Chipster: user-friendly analysis software for microarray and other high-throughput data. BMC Genomics. 2011;12:507. doi: 10.1186/1471-2164-12-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 42.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neff M.M., Turk E., Kalishman M. Web-based primer design for single nucleotide polymorphism analysis. Trends Genet. 2002;18:613–615. doi: 10.1016/s0168-9525(02)02820-2. [DOI] [PubMed] [Google Scholar]
- 44.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data.https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Google Scholar]
- 46.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wickham H. Springer-Verlag; 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- 48.Warnes G.R., Bolker B., Bonebakker L., Gentleman R., Huber W., Liaw A., Lumley T., Maechler M., Magnusson A., Moeller S. 2016. Package ‘gplots’. Various R programming tools for plotting data.https://cran.r-project.org/web/packages/gplots/gplots.pdf [Google Scholar]
- 49.Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.The R Development Core Team . R Foundation for Statistical Computing; 2013. R: A language and environment for statistical computing. [Google Scholar]
- 52.Racine J.S. RStudio: a platform-independent IDE for R and Sweave. J. Appl. Econ. 2012;27:167–172. [Google Scholar]
- 53.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oliveros J. 2016. Venny. An interactive tool for comparing lists with Venn’s diagrams. 2007–2015.https://bioinfogp.cnb.csic.es/tools/venny/index.html [Google Scholar]
- 55.Karimi M., Inzé D., Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- 56.Clough S.J., Bent A.F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 57.Ma X., Zhang Q., Zhu Q., Liu W., Chen Y., Qiu R., Wang B., Yang Z., Li H., Lin Y. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant. 2015;8:1274–1284. doi: 10.1016/j.molp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 58.Kareem A., Radhakrishnan D., Wang X., Bagavathiappan S., Trivedi Z.B., Sugimoto K., Xu J., Mähönen A.P., Prasad K. Protocol: a method to study the direct reprogramming of lateral root primordia to fertile shoots. Plant Methods. 2016;12:27. doi: 10.1186/s13007-016-0127-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ursache R., Andersen T.G., Marhavý P., Geldner N. A protocol for combining fluorescent proteins with histological stains for diverse cell wall components. Plant J. 2018;93:399–412. doi: 10.1111/tpj.13784. [DOI] [PubMed] [Google Scholar]
- 60.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 61.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A, B) Differentially upregulated genes (A) and downregulated genes (B) in LBD3 overexpression line with 8 h induction. (C, D) Differentially upregulated genes (C) and downregulated genes (D) in LBD3 overexpression line with 24 h induction. (E, F) Differentially upregulated genes (E) and downregulated genes (F) in LBD11 overexpression line with 8 h induction. (G, H) Differentially upregulated genes (G) and downregulated genes (H) in LBD11 overexpression line with 24 h induction. (I) Cambium enriched genes overlapped with DEGs in LBD inducible overexpression lines.
ANOVA output data relative to the experiment shown in Figure 2G left panel (A), Figure 2G right panel (B), Figure 2H left panel (C), Figure 2H right panel (D), Figure 3B (E), Figure 3H (F), Figure 3I (G), Figure S3A (H), Figure S3D left panel (I), Figure S3D right panel (J), Figure S3E left panel (K) and Figure S3E right panel (L).
(A, B) GO enrichment analysis of differentially upregulated genes in LBD3 line (A) and LBD11 line (B) with 8h induction.
Data Availability Statement
The RNA-seq data files are deposited under BioProject of NCBI (https://www.ncbi.nlm.nih.gov/bioproject) with accession number (BioProject ID: PRJNA684618).