SUMMARY
CD8+ T cell exhaustion is a major barrier to current anti-cancer immunotherapies. Despite this, the developmental biology of exhausted CD8+ T cells (Tex) remains poorly defined, restraining improvement of strategies aimed at “re-invigorating” Tex cells. Here, we defined a four-cell-stage developmental framework for Tex cells. Two TCF1+ progenitor subsets were identified, one tissue restricted and quiescent and one more blood accessible, that gradually lost TCF1 as it divided and converted to a third intermediate Tex subset. This intermediate subset re-engaged some effector biology and increased upon PD-L1 blockade but ultimately converted into a fourth, terminally exhausted subset. By using transcriptional and epigenetic analyses, we identified the control mechanisms underlying subset transitions and defined a key interplay between TCF1, T-bet, and Tox in the process. These data reveal a four-stage developmental hierarchy for Tex cells and define the molecular, transcriptional, and epigenetic mechanisms that could provide opportunities to improve cancer immunotherapy.
Graphical Abstract

In Brief
Beltra et al. define a hierarchical developmental pathway for CD8+ T cell exhaustion, revealing four stages and multistep transcriptional and epigenetic dynamics underlying subset transitions and subset-associated biological changes.
INTRODUCTION
Chronic viral infections (HIV, HCV, HBV) and cancers subvert CD8+ T cell differentiation toward exhaustion. CD8+ T cell exhaustion has a central role in these settings because blockade of the PD-1 inhibitory receptor (IR) pathway reinvigorates exhausted CD8+ T cells (Tex) and improves immune control of chronic infections and cancer (McLane et al., 2019). Conversely, as exhaustion gets more severe, clinical symptoms of autoimmunity decrease (McKinney et al., 2015). Indeed, Tex cells are a major cell type responding to PD-1 blockade (Huang et al., 2017; Miller et al., 2019; Siddiqui et al., 2019; Thommen et al., 2018), and trials are underway to reinvigorate Tex cells in human chronic viral infections (Wykes and Lewin, 2018). A hallmark of Tex populations is heterogeneity (Blackburn et al., 2008; He et al., 2016; Im et al., 2016; Paley et al., 2012; Utzschneider et al., 2016; Wu et al., 2016). This heterogeneity is relevant for disease because the distribution of these Tex subsets in tumors could relate to response to checkpoint blockade (Daud et al., 2016; Huang et al., 2019; Huang et al., 2017; Kurtulus et al., 2019; Sade-Feldman et al., 2019). Despite the clinical importance of Tex cells, our understanding of their developmental biology and underlying cellular and molecular mechanisms regulating their formation, maintenance, and response to immunotherapies remains incomplete.
PD-1-intermediate Tex progenitors selectively respond to PD-1 pathway blockade, whereas PD-1-high Tex cells are terminally exhausted and do not respond (Blackburn et al., 2008). A developmental hierarchy was defined in which the progenitor Tex subset self-renews and also gives rise to the more terminal Tex subset through proliferation (Paley et al., 2012). T cell factor family member TCF1 (Tcf7) is a key transcription factor (TF) for the progenitor Tex population (Chen et al., 2019; Im et al., 2016; Utzschneider et al., 2016; Wu et al., 2016). However, different studies used distinct markers to define Tex subsets (e.g., PD-1, the T-box TFs T-bet and Eomes, or CXCR5 and Tim-3), but inconsistencies between the proposed models suggest a more heterogeneous Tex subset landscape and developmental biology (Im et al., 2016; Paley et al., 2012; Utzschneider et al., 2016). Indeed, recent studies of CX3CR1+ Tex populations (Hudson et al., 2019; Zander et al., 2019) and high-dimensional single-cell profiling of Tex cells in humans and mice (Kurtulus et al., 2019; Li et al., 2019; Miller et al., 2019; Sade-Feldman et al., 2019; Zander et al., 2019) indicate a more complex Tex subset biology.
The transcriptional and epigenetic landscape of Tex cells defines exhaustion as a distinct branch of CD8+ T cell differentiation (Pauken et al., 2016; Philip et al., 2017; Scott-Browne et al., 2016; Sen et al., 2016). The high-mobility group (HMG)-protein Tox drives epigenetic programming of Tex cells and establishes the exhaustion-specific epigenetic landscape (Alfei et al., 2019; Khan et al., 2019; Scott et al., 2019; Seo et al., 2019; Yao et al., 2019). Tex cells maintain high Tox at late time points of chronic viral infections and cancer. Nevertheless, the role of Tox has only been defined during initiation of Tex cell development, and the impact of this TF once Tex cells have been established remains unclear. Moreover, one consequence of a stable epigenetic identity of Tex cells is that interventions such as PD-1 pathway blockade only provide temporary benefit by reactivating existing transcriptional circuits (Pauken et al., 2016). These reinvigorated Tex cells, however, are not epigenetically reprogrammed and reacquire their exhausted features over time (Pauken et al., 2016). A major question is whether Tex cell heterogeneity can be exploited to achieve better efficacy of immunotherapies. However, the key steps in Tex cell differentiation, underlying TFs, and epigenetic landscape changes involved remain to be defined.
To address these questions, we used chronic lymphocytic choriomeningitis virus (LCMV) infection in mice and performed complementary analysis in mouse and human tumors. We defined four distinct Tex subsets based on Ly108 (Slamf6) and CD69 expression and linked these subsets in a hierarchical developmental scheme. Two interconverting TCF1+ progenitor cell-states were identified: one quiescent and blood inaccessible and a second that initiated robust cell cycling and gained access to circulation. This second TCF1+ Tex subset gave rise to a TCF1− T-bethi intermediate Tex subset that (re)acquired some effector-like features. These intermediate Tex cells ultimately became terminally differentiated, losing T-bet (and gaining Eomes) and permanently exiting cell cycle. This final transition was coordinated by Tox-mediated antagonism of T-bet. PD-1 pathway blockade preferentially expanded the second progenitor and the T-bethi intermediate Tex subsets. These data help clarify previous Tex subset models by defining additional heterogeneity and connecting this heterogeneity to differentiation and underlying transcriptional and epigenetic control.
RESULTS
Ly108 (Slamf6) and CD69 Define Four Tex Subsets during Chronic Viral Infection and Cancer
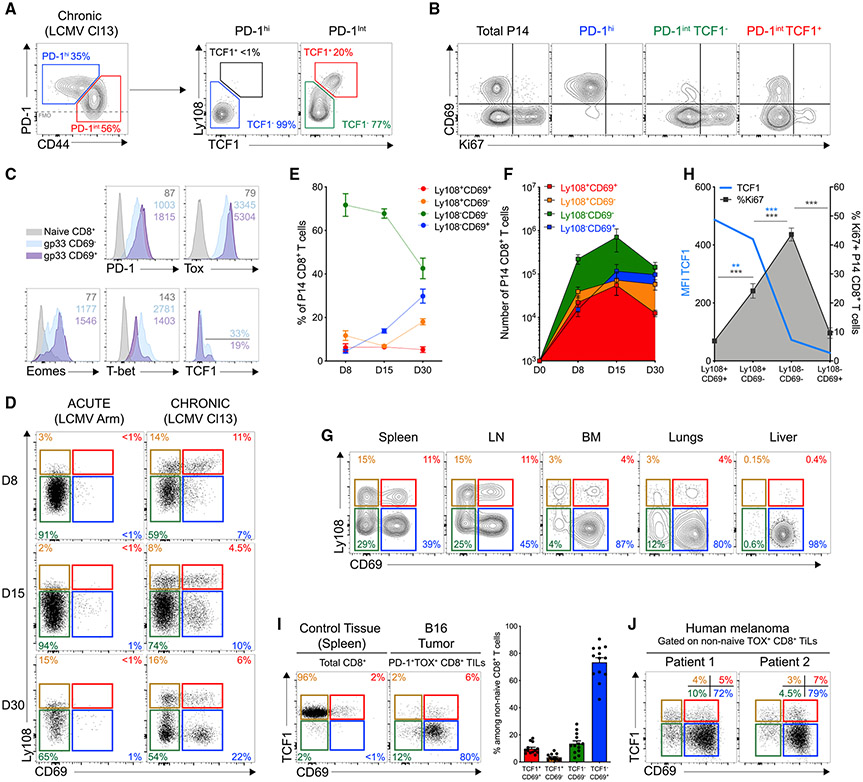
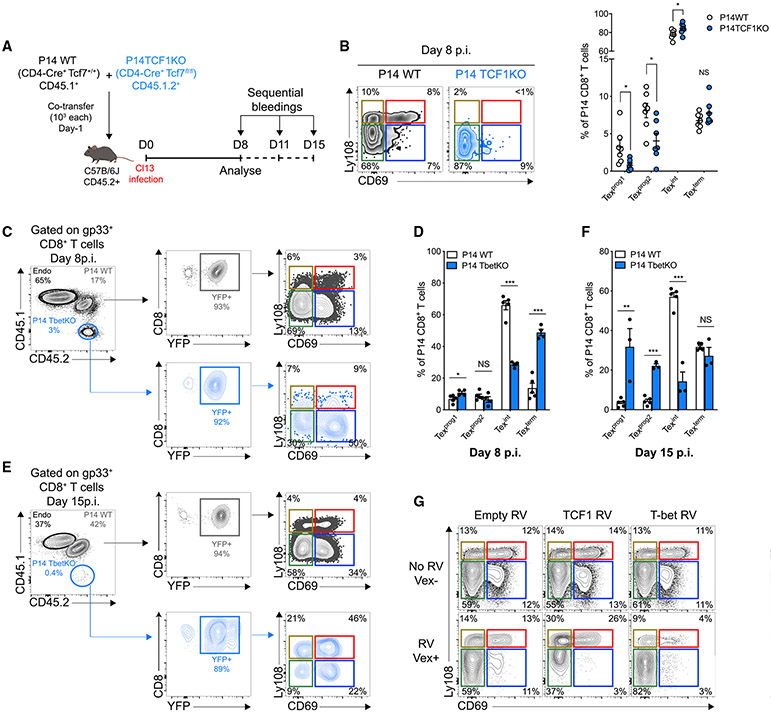
To interrogate Tex cell heterogeneity, we first analyzed adoptively transferred LCMV DbGP33-41-specific P14 CD8+ T cells in LCMV-clone-13-infected mice. At day 30 post-infection (d30pi), P14 cells could be separated into PD-1 intermediate (PD-1int) and PD-1 high (PD-1hi) subsets (Figure 1A) (Blackburn et al., 2008). PD-1int Tex cells could be further subdivided by TCF1 expression (Figure 1A) identifying PD-1intTCF1+, PD-1int TCF1−, and PD-1hiTCF1− cells. PD-1intTCF1− cells were more abundant than either PD-1hi or PD-1intTCF1+ populations and displayed evidence of recent proliferation in vivo, reminiscent of an active developmental transition (Figures S1A and S1B) (Paley et al., 2012). Proliferating cells were also observed in the PD-1intTCF1+ population, whereas few PD-1hi cells were Ki67+ (Figures 1B and S1B). CD69 distinguished PD-1int TCF1− cells (CD69−) from the PD-1hi Tex subset (CD69+), and expression of CD69 was anti-correlated with proliferation (Figures 1B and S1C). CD69 expression also correlated with Tox and Eomes but was negatively associated with T-bet (Figures 1C and S1D). CD69 also demarcated a fraction of TCF1+ cells that preferentially lacked Ki67 expression (Figures 1B, 1C, and S1E). Thus, combining CD69 with Ly108 (Slamf6), a surrogate of TCF1 expression (Figure S1F) identified four Tex subpopulations: Ly108+CD69+, Ly108+CD69−, Ly108−CD69− and Ly108−CD69+ in chronic, but not acute, infection (Figure 1D). This pattern was present at all time points, was also observed for non-T cell receptor (TCR) transgenic responses, and was seen in multiple organs, although different subsets were enriched in different tissues (Figures 1D-1G and S1G). TCF1 expression was lost as Ki67 increased, peaking in the Ly108−CD69− population before declining in the Ly108−CD69+ subset. This latter subset was consistent with PD-1hi terminal Tex cells (Figure 1H). The Ly108+CD69+ subset had higher TCF1 but low ex vivo Ki67 in comparison with Ly108+CD69− cells (Figure 1H). Hence, differential CD69 expression distinguished quiescent (Ly108+CD69+) and cycling (Ly108+CD69−) TCF1+ Tex subsets. A similar pattern of four Tex subsets was found in mouse B16 tumors and also for tumor-infiltrating lymphocytes (TILs) from human melanoma (Figures 1I,1J, and S1H-S1K). Thus, we identified four Tex subsets based on Ly108 (or TCF1) and CD69 expression in mice and humans.
Figure 1. Four Tex Subsets Develop during Chronic Viral Infections and Cancer.
Naive CD45.1+ P14 CD8+ T (103) were adoptively transferred into C57BL/6J mice (CD45.2+) after infection with LCMV clone 13 and analyzed at d30pi.
(A) Ly108 and TCF1 expression by PD-1int and PD-1hi P14 CD8+ T cells. Numbers are frequencies.
(B) Representative Ki67 and CD69 co-expression in indicated populations of P14 CD8+ T cells.
(C) Representative histograms for indicated markers on subpopulations among endogenous Dbgp33 tetramer+ CD8+ T cells. Numbers are mean fluorescence intensity (MFI).
(D) Representative Ly108 and CD69 co-expression on splenic P14 CD8+ T cells at indicated time post-infection (pi) with LCMV Armstrong (left) or clone 13 (right). Numbers are frequencies.
(E) Frequencies of different P14 populations at indicated time after clone-13 infection.
(F) Absolute number of different P14 populations at indicated time after clone-13 infection.
(G) Representative Ly108 and CD69 co-expression in tissues. Numbers are frequencies.
(H) Frequencies of Ki67+ cells among indicated P14 populations (gray line; right axis) and MFI for TCF1 (blue line; left axis).
(I) Representative TCF1 and CD69 co-expression on PD-1+Tox+ TILs from B16 tumors (see Figure S1I for gating). Right graph shows cumulative data.
(J) Representative TCF1 and CD69 co-expression on non-naive Tox+ TILs (see Figure S1J for gating) analyzed from human melanoma tumors. n = 7 patients.
(A)–(H), n = 5 with 6 to 16 mice per group/time point.
(I), n = 2 with 11 mice/group.
Developmental Relationships between the Four Ly108- and CD69-Defined Tex Subsets
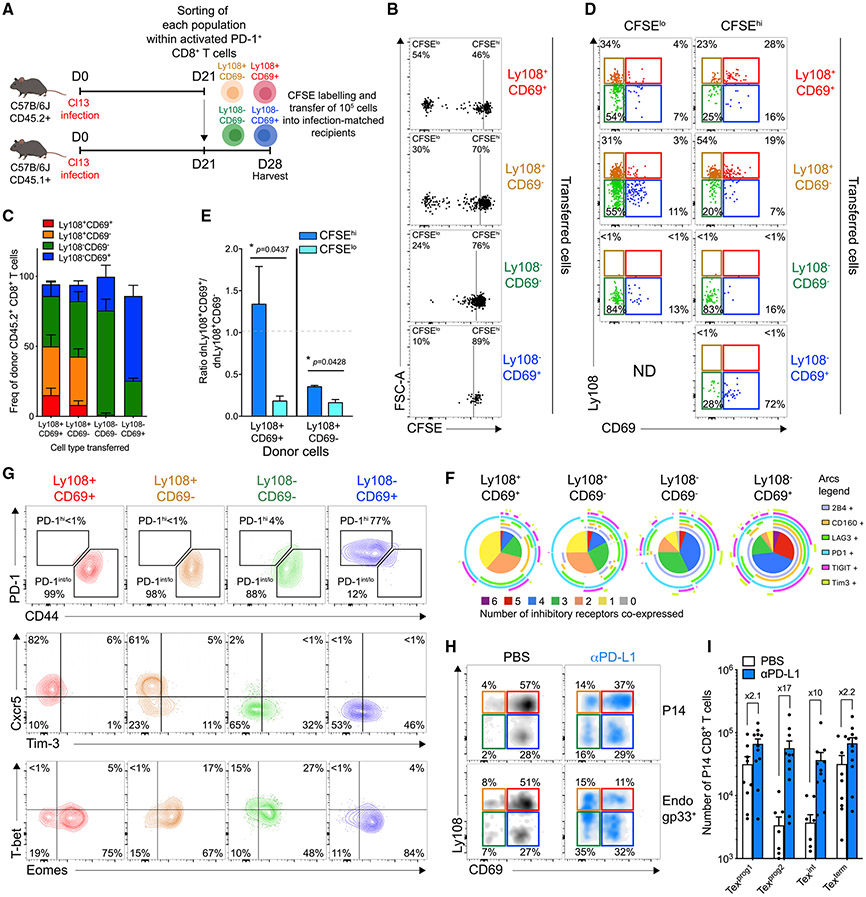
To next investigate lineage relationships, we adoptively transferred sorted and CFSE-labeled Tex subsets into infection-matched recipients (Figures 2A and S2A). After seven days, the Ly108+CD69+ donor population gave rise to the most divided cells followed by the Ly108+CD69− subset. Both populations persisted better in vivo than did the Ly108− subsets and regenerated all four Tex populations (Figures 2B, 2C, S2B, and S2C). Notably, the Ly108+CD69− Tex subset also gave rise to Ly108+CD69+ cells (Figure 2C). Ly108−CD69− and Ly108−CD69+ transferred cells divided poorly, had limited persistence, and restrained developmental plasticity (Figures 2B, 2C, S2B, and S2C). The Ly108−CD69− cells also mainly retained their identity and gave rise to Ly108−CD69+ cells. Donor Ly108−CD69+ cells gave rise to essentially no proliferated progeny and mainly retained their original state (Figures 2B and 2C). The few Ly108−CD69− cells recovered from the Ly108−CD69+ donor population likely reflected few contaminating Ly108−CD69− cells in the donor pool (though some inter-conversion cannot be excluded). Neither of the Ly108− subsets gave rise to Ly108+ cells, consistent with irreversible commitment when TCF1 was lost. Based on the undivided (CFSEhi) transferred cells, Ly108+CD69+ and Ly108+CD69− donor Tex cells were capable of generating each other (Figures 2D, S2D, and S2E). Ly108+CD69− cells accumulated more than the Ly108+CD69+ in the divided (CFSElo) fraction independently of the donor population (Figures 2D, 2E, S2D, and S2E). Accordingly, Ly108+CD69+ donor cells that retained their identity after seven days had undergone few cell divisions, whereas the Ly108+CD69− progeny from these donor cells had proliferated extensively (Figure S2F). Hence, Ly108+CD69+ cells had features of quiescent progenitors and restrained in vivo cell cycle (Figure 1H). Thus, Ly108+CD69+ cells likely first passively transitioned to a secondary Ly108+CD69− stage. Extensive proliferation then occurred with the formation of Ly108−CD69− cells (Figures 1H and 2D). Although a low frequency of Ly108−CD69− cells could be detected without proliferation from Ly108+CD69+ (13.9%) or Ly108+CD69− (16.2%) donor cells, the proportion of the Ly108−CD69− subset increased substantially with proliferation, outnumbering Ly108+CD69− cells among the dividing cells (Figures 2D, S2D, S2E, and S2G). The high Ki67 expression found at steady state in Ly108−CD69− cells (Figure 1H) contrasted their lack of proliferative potential in this setting (Figure 2B), suggesting that this population arises from dividing Ly108+CD69− progenitors and loses further proliferative potential. Lastly, the few cells recovered from the donor Ly108−CD69+ cells remained undivided and largely conserved their original identity (Figures 2B-2D and S2B-S2E). Ly108−CD69+ cells could also be generated from all other subsets (Figures 2C, 2D, S2D, and S2E). The minor Ki67 expression found in the Ly108−CD69+ subset coupled with the lack of proliferative potential of Ly108−CD69− cells suggested that the former cells differentiate from the post-mitotic fraction of Ly108−CD69− cells. Co-expression of IRs and phenotypic changes consistent with terminal exhaustion accompanied the progression from Ly108+CD69+ → Ly108+CD69− → Ly108−CD69− → Ly108−CD69+ (Figures 2F and 2G). For clarity, these Tex subsets will be referred to here as progenitor 1 (Ly108+CD69+; Texprog1), progenitor 2 (Ly108+CD69−; Texprog2), intermediate (Ly108−CD69−; Texint), and terminal (Ly108−CD69+; Texterm).
Figure 2. Developmental Lineage Relationships between Four Tex Subsets and Changes upon PD-L1 Blockade.
(A) Experimental design. Tex subsets were sorted from endogenous PD-1+ CD8+ T cells at d21pi, labeled with CFSE, transferred back (1 × 105 each) into infection-matched recipients and analyzed 7 days post-transfer (d28pi).
(B) CFSE profile of indicated transferred populations. Numbers are frequencies.
(C) Frequencies of P14 populations from indicated donor cells.
(D) Ly108 and CD69 co-expression on divided (CFSElo) and undivided (CFSEhi) cells that developed from indicated transferred cells.
(E) Ratio of Ly108+CD69+/Ly108+CD69− within CFSElo (light blue) and CFSEhi (dark blue) cells from indicated donor origin.
(F) IRs co-expression on indicated P14 populations.
(G) Representative PD-1/CD44, Cxcr5/Tim-3, and T-bet/Eomes co-expression in indicated P14 populations.
(H) Representative Ly108 and CD69 co-expression on P14 (CD45.1+) and endogenous Dbgp33+ (CD45.2+) CD8+ T cells 1 day after treatment (d35pi) with either PBS (CTR) or anti-PD-L1.
(I) Absolute number of indicated P14 populations 1 day post-treatment (d35pi) with either PBS (CTR) or anti-PD-L1.
(B)–(E), n = 2 with 6–8 mice/group; (F), n = 3 with 10 mice/group; (G), n = 2 with 9 mice/group; (H) and (I), n = 2 with 7–11 mice/group.
PD-L1 Blockade Selectively Expands Transitioning Tex Subsets
PD-1/PD-L1 pathway blockade likely impacts Tex subset distribution. Thus, we interrogated changes in the four Tex subsets after PD-L1 blockade (Figure S2H). In the absence of CD4+ T cell help, the Texprog2 and Texint subsets were reduced in comparison with the CD4-sufficient settings at d30pi (Figures S2I and S2J) (Zander et al., 2019). PD-L1 blockade preferentially promoted accumulation of the Texprog2 and Texint subsets. One day after treatment, the absolute number of Texprog1 and Texterm cells increased 2.1- and 2.2-fold, respectively, whereas the number of Texprog2 and Texint cells increased 17- and 10-fold, respectively (Figures 2H and 2I). These data indicate that Tex cell reinvigoration by PD-1 pathway blockade partially rebalanced Tex subset distribution by fostering the formation of the Texprog2 and Texint subsets.
Dynamic Changes in T-bet and Eomes in Four Subsets of Tex Cells
We previously identified differences in T-bet and Eomes expression in Tex subsets (Figure S3A) (Paley et al., 2012). More recent studies that defined Tex subsets based on CXCR5 and Tim-3 found a different expression pattern for T-bet and Eomes (Figure S3A) (Im et al., 2016; Utzschneider et al., 2016). We therefore asked whether the four-stage developmental hierarchy defined above could better resolve the dynamics of T-bet and Eomes in Tex cells. First, gating on Tex subsets based on PD-1 versus CXCR5 and Tim-3 revealed different patterns of the four subsets defined by Ly108 and CD69. CXCR5+ Tex captured Texprog1 and Texprog2 cells whereas the PD-1int population also contained the Texint subset (Figure S3B). This Ly108−CD69− Texint subset was nearly completely absent from the PD-1hi population but was contained in the subset defined as CXCR5− (Figure S3B). Eomes was highest in Texprog1 cells, declining slightly in Texprog2 and even more in the Texint subset (Figures S3C-S3E). However, Eomes expression rebounded at the Texterm stage (Figures S3C-S3H). T-bet expression, in contrast, gradually increased from Texprog1 and was highest in Texint cells from both LCMV-infected mice and human tumors before dropping at the Texterm stage (Figures S3C-S3H). Consistent with T-bet-repressing PD-1 (Kao et al., 2011), we observed a slight decrease in PD-1 at the Texprog2 and Texint stages, whereas other IRs gradually increased from Texprog1 to Texterm (Figures 2F and S3I). Thus, the initial stages of Tex subset differentiation (i.e., from Texprog1 to Texint) were accompanied by an Eomeshi to T-bethi conversion concomitant with loss of TCF1 and decreased PD-1 expression, whereas the later transition to the Texterm subset was accompanied by a T-bethi to Eomeshi conversion and higher PD-1 expression.
Location and Function of Ly108- and CD69-Defined Tex Subsets
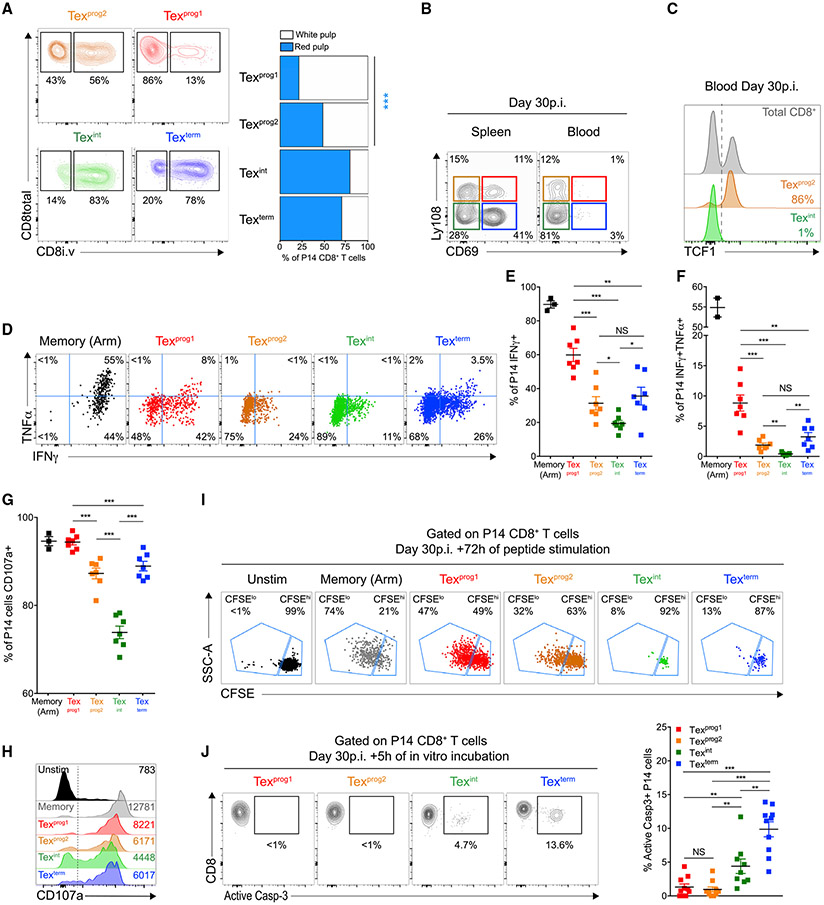
CXCR5-defined Tex progenitors (CXCR5+) are resident of lymphoid tissues whereas CXCR5− Tex cells are found in circulation and peripheral organs (He et al., 2016; Im et al., 2016). Because the Tex subsets described above were defined by CD69 that can regulate lymphocyte egress, we next performed intravascular antibody (i.v. Ab) staining to assess blood accessibility (Anderson et al., 2014). Consistent with previous work (Beura et al., 2015), 73% (±2% SEM) of splenic P14 CD8+ T cells were labeled by i.v. anti-CD8 Ab (Figures S4A and S4B). Texprog1 cells, however, were almost exclusively (~80%) in the i.v. Ab− fraction, consistent with localization to the white pulp of the spleen (Figure 3A). In contrast, 47% (±2% SEM) of the Texprog2 population was marked by i.v. Ab staining, indicating blood accessibility likely in the red pulp (Figure 3A). Moreover, a majority of Texint and Texterm were i.v. Ab labeled (Figure 3A). The two CD69− subsets, Texprog2 and Texint, were present in blood, whereas Texprog1 and Texterm cells were not (Figure 3B). Circulating Texprog2 cells retained expression of TCF1 in the blood (Figure 3C). The absence of Texterm cells in blood despite robust i.v. Ab staining suggested that these cells were resident of the blood-accessible splenic red pulp. Thus, the Ly108+ (TCF-1+) Texprog1 and Texprog2 subsets differed in anatomical location, with the latter gaining access to blood. The Texint subset was also highly blood accessible whereas the re-acquisition of CD69 expression by Texterm cells was associated with tissue-residency but in distinct environments in comparison with Texprog1 cells.
Figure 3. Developmental Transitions Alter Tex Subset Localization and Effector Functions.
(A) LCMV-clone-13-infected mice were injected with 3μg/mouse of anti-CD8 i.v. at d30pi and sacrificed 3 min later. Representative dot plots (left) and frequencies (right) of indicated P14 populations located either in the white pulp (CD8 i.v. antibody negative) or the red pulp (CD8 i.v. antibody positive) of the spleen. Numbers are frequencies.
(B) Representative Ly108 and CD69 co-expression on P14 CD8+ T cells isolated from the spleen (left plot) or blood (right plot). Numbers are frequencies.
(C) TCF1 expression in indicated P14 populations isolated from the blood.
(D) Representative IFNγ and TNF co-production by indicated P14 populations at d30pi. Numbers are frequencies.
(E–G) Frequencies of IFNγ+ (E), IFNγ+TNFα+ (F), and CD107a+ (G) cells among indicated P14 populations.
(H) Representative CD107a expression by indicated P14 populations. Numbers are MFI.
(I) Indicated P14 populations were sorted at d30pi, labeled with CFSE, and incubated with gp33-peptide coated dendritic cells (ratio 1/1). Dot plots display CFSE dilution profile after 3 days of co-culture. Numbers are frequencies.
(J) Representative dot plots and frequencies of active caspase-3-positive cells within indicated P14 populations. Numbers are frequencies.
(A)–(C), 3 independent experiments with 12–18 mice/group; (D)–(H), n = 2 with 7 mice/group; (I), n = 2 with 4–10 mice/group; (J), n = 3 with 10 mice/group.
We next evaluated effector functions. Among the Tex subsets, Texprog1 contained the highest frequency of IFNγ-producers, IFNγ and TNF coproducers (Figures 3D-3F) and also made the most IFNγ per cell (Figure S4C). Cytokine coproduction declined in Texprog2 and Texint subsets (Figures 3D-3F and S4C) but then rebounded slightly at the Texterm stage, although these cells were inferior to Texprog1 on a per-cell basis (Figures 3D-3F and S4C). However, Texint and Texterm accounted for a respective 49% and 34% of the absolute number of IFNγ-producing P14 cells, and Texterm contributed the largest number of IFNγ and TNF co-producers (Figures S4D and S4E). Note, expression of Ly108 and CD69 on Tex cells was unaffected by peptide re-stimulation (Figures S4F and S4G), and memory T cells (Tmem) outperformed all Tex subsets (Figures 3D-3F and S4C). CD107a staining followed a similar trend to cytokine secretion profiles (Figures 3G and 3H). However, Texint and Texterm cells mediated the most target killing (Figure S4H). Ex vivo proliferative potential was restricted to Texprog1 and to a lesser extent Texprog2 cells, although this proliferation was substantially worse than Tmem cells (Figure 3I). The robust proliferation of Texprog1 cells upon antigen stimulation but evidence of low in vivo proliferation and relative lack of Ki67 (Figure 1H) pointed to a high proliferative potential but low ongoing cell cycle by this quiescent subset in the steady state. In contrast, Texint and Texterm cells were almost incapable of undergoing cell division upon re-stimulation in vitro despite, at least for Texint cells, ex vivo Ki67 expression as evidence of recent in vivo cell cycling (Figure 3I). Texprog1 and Texprog2 cells also had little evidence of ex vivo cell death, whereas apoptotic cells were readily detectable for Texint and Texterm subsets in both the LCMV system and human melanoma tumors (Figures 3J and S4I). Collectively, these data identify a specific set of functional properties for each Tex subset.
Transcriptional Programs of Tex Subsets
We next performed RNA sequencing (RNA-seq) to further define the biology of Tex subsets. Principal-component analysis (PCA) highlighted a distinct transcriptional program for each Tex subset defined by Ly108 and CD69, and each population also differed from conventional effector (Teff) or Tmem cells (Figures 4A and S5A). Pairwise comparisons defined differentially expressed genes (DEGs) and subset-specific transcriptional signatures (Figures 4B and S5B; Table S1). Tex subsets shared upregulated genes (DEGsUP) with their direct neighbors from the developmental relationships above, but fewer with more distant subsets. For example, ~40% of Texprog1 DEGsup were also up in Texprog2 whereas few of these genes were shared by Texint and Texterm cells (Figure 4C). We then asked how the Tex subset transcriptional signatures compared with previous Tex cell scRNA-seq data (Figure S5C) (Miller et al., 2019). Bi-clustering analysis classified DEGs from Ly108- and CD69-defined subsets into seven modules (Figure S5D, top row; Table S2). These DEGs modules were then mapped onto the scRNA-seq data (Figure S5D, bottom row). This analysis revealed similarity of Texprog1 and Texprog2 cells with a Tcf7+ progenitor-like single cell cluster (Figures S5C and S5D; module 1), with broader mapping of the Texprog2 signature in scRNA-seq space (Figures S5C and S5D; modules 3 and 4). DEGs from the Texint subset mapped mainly to a scRNA-seq cluster enriched for effector-like biology exemplified by genes like Cx3cr1 (Figures S5C and S5D; module 5). DEGs biased to the Texterm subset mapped mainly to the cluster identified as terminal Tex cells (Figures S5C and S5D; module 7). Thus, each Tex population had a distinct transcriptional program, and the relationship between these transcriptional signatures was consistent with the developmental scheme above.
Figure 4. Subset Transitions Induce Major Transcriptional Changes and Drive Acquisition of Population-Specific Transcriptomic Signatures.
(A) PCA of RNA-seq profiles.
(B) Venn diagram displaying overlaps between DEGs from indicated pairwise comparisons (lfc ≥ 1, p = 0.01).
(C) DEGsUP overlaps between indicated P14 populations. Bubble size represents the proportion of DEGsUP from each individual Tex population (y axis) also found to be upregulated in indicated Tex subsets (x axis) in at least one pairwise comparison.
(D) Heatmap displaying all DEGs (lfc ≥ 1, p = 0.01) clustered by using Pearson correlation as distance measure. Color legend indicates row z scores.
(E) Pathway enrichment analysis. Bubble graph displays the five most significantly enriched pathways by log(q-value) for each cluster established in Figure 4D.
(F) Representative histograms showing the expression of indicated markers by different P14 populations. Numbers are MFIs.
(G) GSEA displaying enrichment of each individual Tex subset for signatures of indicated cell types. NES, Normalized Enrichment Score.
n = 3 biological replicates.
To identify subset-specific biology, we clustered DEGs and examined gene ontology. Genes upregulated in Texprog1 cells were involved in progenitor biology (Tcf7, Myb, Il7r, Self), Tfh biology (Cxcr5, Icos), and positive co-stimulation (Cd28) (Figure 4D, clusters 5 and 6; Table S3). Many of these genes were also expressed in Texprog2, but often at lower amounts (e.g., Tcf7, Cxcr5, CD28), an observation confirmed for protein (Figures 4D, 4F, S5E, and S5F). Cluster 5, biased to the Texprog1 subset, also contained interferon-stimulated genes (ISGs) including Irf7, Oas1, and Stat1 contributing to an enrichment for antiviral defense signatures (Figures 4D and 4E). Cluster 1 contained genes preferentially upregulated in Texprog2 and was enriched for genes related to cell-cycle (e.g., cyclins and mki67) and cell motility (Anxa2, Itgb7, Alcam) (Figures 4D, 4E, and S5E). The transcriptional signature of Texint cells was enriched in clusters 3, 4, and 8, which contained effector-related genes (e.g., Grzma, Grzmb, Prf1 [cluster 4]; Klrg1 [cluster 3]; Cx3cr1 [cluster 8]) and TFs (Tbx21, Zeb2, Id2, Prdm1 [cluster 4]), with corresponding protein expression and gene ontology pathways (cluster 4) (Figures 4D-4F, S5E, and S5F). Cluster 3 also captured biology related to natural killer (NK) cells for Texint (e.g., Klr family genes). Lastly, Texterm cells upregulated mRNA and protein for IRs (Pdcd1, Lag3, Tigit, Cd244 [clusters 2 and 7]) and molecules associated with terminal exhaustion (Entpd1, Cd101, Cd38 [clusters 2 and 7]). Cluster 2 was highly biased to Texterm and enriched for pathways including negative regulation of cell activation (Figure 4E). Texterm cells also showed signs of recent TCR signaling including high expression of Zap70, Nfatc1, and calcium influx pathway enrichment (cluster 7) (Figures 4D, 4E, and S5E, and S5F). Although Tox was expressed by all Tex subsets (Figure 1C), Texterm had the highest expression (Figures S5E and S5F). Gene-set enrichment analysis (GSEA) revealed that Texprog1 and Texprog2 shared similarity with CXCR5+Tim-3− Tex cells and memory precursor effector cells (MPEC), although Texprog1 had stronger enrichment (Figure 4G). Texprog2 and Texint were enriched for signatures from circulating effectors (Tcirc) (Figure 4G). Texint and Texterm enriched for the signature of CXCR5−Tim-3+ Tex cells, but Texint also showed similarity to short-lived effector cells (SLECs) (Figure 4G). Texterm selectively enriched for exhaustion and resident-memory (Trm) signatures that coincided with elevated Runx3, a TF associated with tissue-residency (Figures 4G, S5E, and S5F) (Milner et al., 2017). Finally, each Tex subset enriched for transcriptional signatures of different populations of mouse and human TILs (Figure S5G). These transcriptional programs are consistent not only with a stepwise developmental program but also with acquisition of Tex subset-specific biological functions.
Distinct Epigenetic Remodeling and Associated TF Control of Tex Subsets
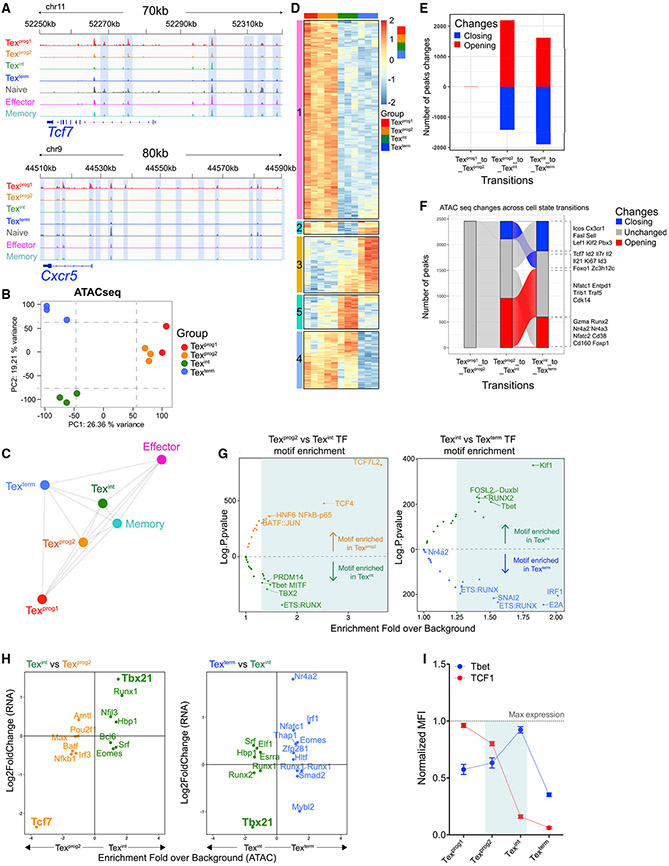
To define the epigenetic relationships between these Tex subsets, we next compared the open-chromatin landscape of the four Tex subsets, as well as Teff and Tmem cells, by assay for transposase-accessible chromatin followed by deep sequencing (ATAC-seq) (Buenrostro et al., 2013). Chromatin accessibility changes mostly occurred at intronic and intergenic regions and correlated with transcriptional changes of the nearest genes (Figures S6A and S6B). We identified specific patterns of chromatin accessibility in each Tex subset. For example, Tcf7 and Cxcr5 loci were more accessible in Texprog1 and Texprog2, whereas Cx3cr1 or Cd38 had more accessibility in Texint or Texterm, respectively (Figures 5A and S6C). By PCA, Texprog1 and Texprog2 were similar, whereas Texint and Texterm were distinct from each other and also from either Texprog subset (Figure 5B). Each Tex subset also differed epigenetically from Teff and Tmem cells (Figures 5C, S6D, and S6E). Although Texint cells mapped closer to Teff and Tmem cells, this Tex subset remained distinct from Teff and Tmem by ≈ 5,000 open chromatin regions (OCRs) (including OCRs near Tox and Pdcd1). Many of these Texint OCRs or specific modules were also shared by other Tex subsets (Figures S6F and S6G; Table S4). Unsupervised clustering revealed five epigenetically distinct clusters between Tex subsets (Figure 5D; Table S5). Cluster 1 identified a set of OCRs uniquely shared between Texprog1 and Texprog2. Many OCRs in this cluster mapped near genes related to progenitor biology (i.e., Il2, Lef1, Wnt2) (Table S5). Texprog1 and Texprog2 also enriched for cluster 2 that overlapped with Texterm and cluster 4 associated with Texint. Texprog2 showed more accessibility than Texprog1 in cluster 5, a cluster also associated with Texint cells that contained OCRs associated with mKi67 (Figure 5D; Table S5). Cluster 3, absent from either Texprog subset, was highly biased to Texterm with some features moderately enriched in Texint. Overall, these open-chromatin landscapes highlighted an epigenetic similarity between Texprog1 and Texprog2 but also reveal aspects of Texprog2 that were intermediate between Texprog1 and Texint. However, robust epigenetic remodeling occurred during the Texprog2 to Texint and Texint to Texterm transitions (Figure 5E; Table S6). Most of the chromatin regions either opening or closing during the Texint to Texterm transition occurred at sites that were unaffected during Texprog2 to Texint differentiation and vice versa. Genes related to the progenitor biology (Tcf7, Il7r) were epigenetically silenced during the Texprog2 to Texint transition and remained closed thereafter (Figure 5F; Table S6). Conversely, OCRs in genes encoding effector- (Cx3cr1) and exhaustion-related genes (Cd160, Nr4a family members) became, respectively, less or more accessible during the Texint to Texterm transition. Thus, a clear set of epigenetic landscape changes accompanied transitions between Tex subsets.
Figure 5. Epigenetic Remodeling during Tex Subset Transitions Identifies Subset-Specific TF Accessibility.
(A) ATAC-seq tracks at indicated loci.
(B) PCA of normalized ATAC-seq counts (top 25% peaks by variance are displayed).
(C) Spearman correlation network analysis showing similarities between indicated populations based on all ATAC-seq peaks.
(D) Heatmap of peak intensity displaying the top 500 peaks by variance and clustered by using K-means method (k = 5). Color legend indicates row z scores.
(E) Number of statistically significant peaks (lfc ≥ 1, p < 0.05) either opening (red) or closing (blue) upon indicated subset transitions.
(F) Alluvial plot displaying the dynamics of transition-specific peaks (E) and refined as lfc ≥ 1, p < 0.01.
(G) TF motifs enrichment analysis (HOMER) of subset-transition specific peaks (E). Volcano plots show TFs with the highest predicted binding activity to the identified DNA sequence.
(H) Comparing TF motif enrichment with differential mRNA expression (Log2FoldChange) upon the indicated subset-transition. Plotted are TFs with a motif enrichment fold over background >1.25 in (G).
(I) Protein expression dynamics of TCF1 and T-bet in the indicated P14 populations at d30pi.
(A)–(H), n = 3 biological replicates; (I), n = 2 with 7mice/group.
We next examined enrichment for TF binding sequences in chromatin accessible regions altered during Tex subset transitions using HOMER. These analyses identified TFs with potential subset-specific roles (Figure 5G; Table S7). For example, the most significantly enriched DNA binding sequence in chromatin differentially accessible in Texprog2 versus Texint cells was for T cell factor family of TFs (e.g., Tcf7; TCF4 and TCF7L2 motifs) (Figure 5G left plot; Table S7). TF binding motifs enriched in Texint compared to Texprog2 included ETS:RUNX (Pu.1, Etv1, Etv2, Etv6) and PRDM14 (Znf264 and Rfx7). T-box motifs (e.g., Tbet, TBX2) were highly enriched in Texint compared to in Texprog2 or Texterm (Figure 5G; Table S7). Texterm-specific OCRs were enriched for interferon-related TF motifs including the IRF1 sequence that can be bound by IRF1, IRF2, and IRF7 as well as sequences bound by STAT1 and Blimp-1. Combining motif enrichment analysis with RNA expression refined the list of TFs predicted to have Tex subset-specific roles. TCF1 (Tcf7) was the top-enriched TF in Texprog2 (Figure 5H, left), whereas T-bet (Tbx21) was the most enriched for Texint (Figure 5H). Nr4a2 was the most-enriched predicted TF for the Texterm subset, followed by Irf1, Nfatc1, and Eomes (Figure 5H). In the comparison of Texprog2 to Texint, the enrichment for T-bet versus TCF1 was strongly anti-correlated, and these TFs displayed opposing protein-expression kinetics during the Texprog2 to Texint transition (Figures 5H and 5I). Together, these data demonstrate de novo epigenetic changes occurring during the Texprog2 to Texint and Texint to Texterm transitions that coincided with gain or loss of accessibility for key TFs including potential opposing functions for TCF1 and T-bet in the Texprog2 to Texint transition.
T-bet and TCF1 Control the Developmental Transition from Texprog2 to Texint Cells
We next directly tested the impact of TCF1 and T-bet on Tex subset development. Co-adoptive transfer of naive P14 WT and P14 TCF1 cKO cells into LCMV-clone-13-infected mice revealed an essential role for TCF1 in early development of Texprog1 and Texprog2 cells (Figures 6A, 6B, and S7A-S7C) (Chen et al., 2019; Im et al., 2016; Utzschneider et al., 2016; Wu et al., 2016). TCF1-deficient P14 cells had almost exclusively a Texint phenotype at d8pi, although these cells were not maintained over time (Figures 6B and S7B). In the absence of T-bet (Figures S7D and S7E), there was a modest increase in the Texprog1 subset at d8pi but marked reduction of the Texint population and accumulation of Texterm cells (Figures 6C and 6D). At d15pi the loss of Texint cells was even more prominent, but the accumulation of Texterm was lost (Figures 6E, 6F, and S7H). Consistent with previous reports (Kao et al., 2011; Paley et al., 2012), the number of all Tex cells was decreased in the absence of T-bet (Figures S7F, S7G and S7I), although there was a bias of T-bet cKO P14 cells toward the Texprog1 and Texprog2 compartments at d15pi (Figures 6E and 6F). These data identified a crucial role for T-bet specifically in the Texint subset. The higher frequency of Texterm at d8pi in P14 T-bet cKO suggested that T-bet could stabilize the Texint stage, restraining conversion into Texterm cells (Figures 6C and 6D), although there is likely also a role for T-bet in the non-exhausted KLRG1+ Teff lineage at these early time points of infection (Chen et al., 2019). The inability to sustain the development of the Texterm population at d15pi in the T-bet cKO likely reflected a lack of replenishment caused by defective generation of Texint cells from Texprog2 (Figures 6C-6F and S7H). Thus, the defect in Texint cell development in the absence of T-bet disrupted Tex subset dynamics, causing attrition of the overall CD8+ T cell response during chronic infection.
Figure 6. TCF1 and T-bet Mediate Opposing Regulation of Tex Subset Differentiation.
(A) Experimental design.
(B) Representative flow cytometry plots (left) and frequencies (right) of indicated populations among splenic WT (gray; open circles) or TCF1KO (blue) P14 CD8+ T cells at d8pi. Numbers are frequencies.
(C–F) Gating strategy, representative flow cytometry plots and frequencies of indicated populations of WT P14 (gray; open bars) or T-bet KO P14 (blue) at d8 ([C] and [D]) and 15 ([E] and [F]) pi. Numbers are frequencies.
(G) Representative Ly108 and CD69 co-expression by P14 CD8+ T cells transduced (VEX+; lower line) or not (VEX−; upper line) with either empty (Empty RV), TCF1 (short isoform; TCF1 RV), or T-bet (Tbet RV) encoding RVs at d15pi. Numbers are frequencies.
(B), n = 2 with 7 mice/group; (C)–(F), n = 3–5 with 11–17 mice/group; (G), n = 1 with 4 mice/group.
To confirm these results, we enforced expression of either TCF1 (short isoform) or T-bet in P14 CD8+ T cells with retroviral (RV) transduction (Kurachi et al., 2017) (Figure S7J). TCF1 overexpression (VEX+) favored accumulation of Texprog1 and Texprog2 at d15pi, reducing the proportion of Texint and Texterm cells in comparison with the non-transduced (VEX–) or empty RV controls (Figures 6G and S7K). Conversely, enforcing T-bet expression led to a substantial accumulation of Texint cells, whereas the proportion of Texprog1, Texprog2, and also Texterm cells was reduced (Figures 6G and S7K). These data further illustrated the role of T-bet in fostering and stabilizing the Texint subset. Taken together, we defined the TCF1-T-bet axis as a major transcriptional control mechanism in the conversion of progenitor cells into Texint cells.
The High-Mobility Group (HMG) Box Protein TOX Antagonizes T-bet Expression to Favor Terminal Exhaustion
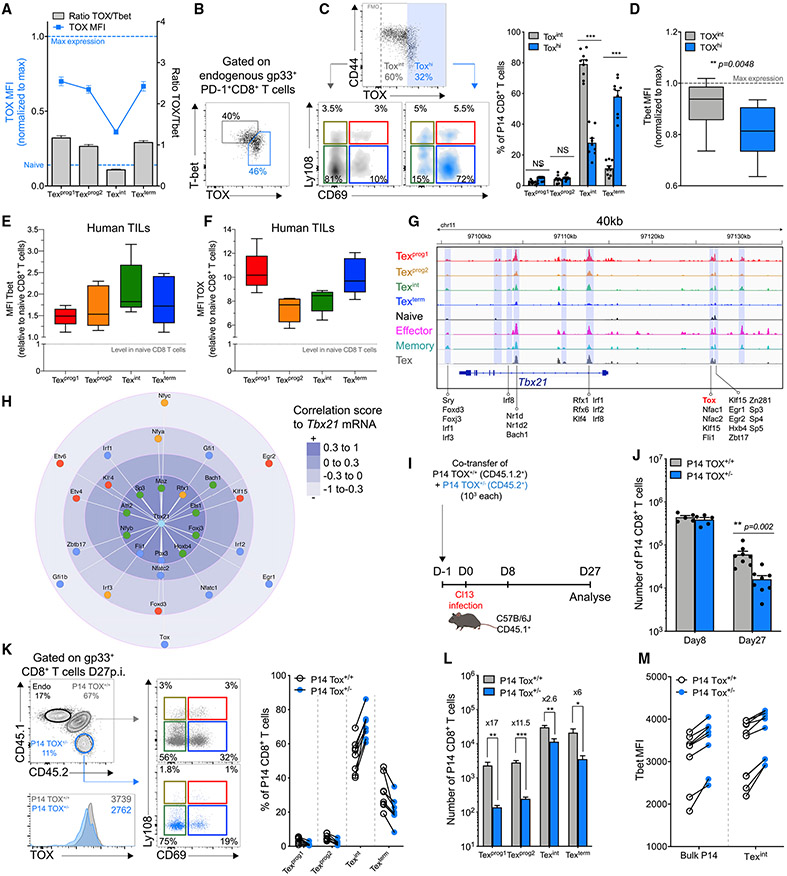
We next aimed to investigate the molecular events promoting final conversion of Texint cells to terminal Texterm cells. We and others recently reported a role for Tox in early Tex versus Teff cell-fate commitment (Alfei et al., 2019; Khan et al., 2019; Scott et al., 2019; Seo et al., 2019; Yao et al., 2019). We then asked whether Tox might antagonize the T-bet-driven Teff-like Texint subset to favor terminal exhaustion. Indeed, Tox expression was lowest in Texint cells but increased robustly at the Texterm stage (Figures 7A, S5E, and S5F). Texint cells also had the lowest Tox/T-bet ratio. Moreover, when examined directly, the Tox and T-bet co-expression pattern revealed distinct ToxhiT-betlo and ToxintT-bethi populations at d27pi (Figure 7B). Accordingly, in chronic infection and cancer, lower Tox was characteristic of T-bethi Texint cells whereas higher Tox was associated with Texterm and/or Texprog subsets that were T-betlo (Figures 7C-7F). Tox also had a predictive binding site at an OCR distal to the Tbx21 TSS, and among all TFs predicted to bind near Tbx21 in Tex cells, Tox was among the most anti-correlated with Tbx21 mRNA (Figures 7G and 7H). These data suggested potential direct negative regulation of T-bet by Tox.
Figure 7. Tox Modulates Tex Subset Dynamics through Regulation of T-bet.
(A) Tox MFI (blue line; left axis) and Tox/T-bet ratio (gray bars; right axis) in indicated populations among Dbgp33 tetramer+CD8+ T cells at d27pi.
(B) Representative T-bet and Tox co-expression in PD-1+ Dbgp33 tetramer+ CD8+ T cells at d27pi.
(C) Representative plots (left) and frequency (right) of indicated populations among Toxint (gray) and Toxhi (blue) endogenous Dbgp33 tetramer+ CD8+ T cells.
(D) T-bet MFI in Toxint and Toxhi endogenous Dbgp33 tetramer+ CD8+ T cells.
(E and F) T-bet (E) and Tox (F) MFI in indicated populations (gated on TCF1 and CD69; Figure 1J) within non-naive Tox+ TILs isolated from human melanoma tumors.
(G) ATACseq tracks at the Tbx21 locus highlighting TFs with predictive binding sites at the indicated OCRs.
(H) Correlation score by mRNA between T-bet and TFs predicted to bind near the Tbx21 locus (filtering out TFs without detectable mRNA in Tex subsets). Each TF is colored by the cell-type in which mRNA expression is highest (red-Texprog1, orange Texprog2, green-Texint, blue-Texterm).
(I) Experimental design.
(J) Numbers of Tox+/+ P14 (gray) and Tox+/− P14 (blue) cells at the indicated time points.
(K) Representative dot plots (left) and frequencies (right) of the indicated populations among Tox+/+ (gray; open circles) and Tox+/− (blue) P14 cells. Numbers indicate frequencies. Numbers in the histogram are MFI.
(L) Number of indicated populations within Tox+/+ P14 (gray) and Tox+/− P14 (blue) at d27pi.
(M) T-bet MFI in indicated populations among Tox+/+ P14 (open circles) and Tox+/− P14 (blue circles). Linked circles represent individual mice.
(A)–(D), 3 experiments with 22 mice/group; (E) and (F), n = 7 patients; (J)–(M), 2 experiments with 15 mice/group.
To test this relationship, we performed co-transfer of Tox haplo-insufficient P14 cells (Tox+/−) with WT littermate (Tox+/+) in congenic mice followed by clone-13 infection (Figure 7I). Tox+/− P14 cells had ~20%–30% reduction in Tox expression (Figure 7K, bottom) in ~60% of the recipient mice by d27pi. Whereas a complete abrogation of Tox precludes Tex cell development (Alfei et al., 2019; Khan et al., 2019; Scott et al., 2019; Seo et al., 2019; Yao et al., 2019), Tox+/− P14 cells expanded normally, were maintained up to d27pi (although at reduced frequencies and numbers compared to Tox+/+ P14 cells), and sustained high expression of PD-1 consistent with exhaustion (Figures 7J, 7K left, and S7L). However, tempering Tox expression reduced the Texprog1, Texprog2, and Texterm compartments and favored accumulation of Texint cells (Figure 7K). Although the absolute number of all Tex subsets was reduced in the Tox+/− setting, the Texint cells were affected the least (Figure 7L). These data confirmed a developmental advantage for Texint cells in Tox+/− P14 that corresponded to an overall increase in T-bet expression in comparison with Tox+/+ P14 controls, a difference also apparent in the Texint subset (Figure 7M). These data indicated that Tox control over T-bet expression in Tex cells represents a key switch point regulating the balance between Texint and Texterm cell accumulation.
DISCUSSION
We and others have previously identified Tex subsets. However, the number of subsets, their underlying transcriptional and epigenetic control, and their developmental relationships have remained poorly understood. Here, we identified a four-stage developmental trajectory of Tex cell subsets defined by Ly108 (a surrogate of TCF1) and CD69. Our data revealed two interchangeable progenitor states (Texprog1 and Texprog2) with transcriptional, phenotypic, functional, and anatomical differences but a related open-chromatin landscape. Texprog1 are quiescent and resident and convert to Texprog2. This conversion was associated with relocalization from blood-inaccessible lymphoid residence to blood-accessible locations and a proliferation-driven transition to a downstream Texint developmental stage. This Texint subset had similarities to circulating “effector-like” cells, although Texint cells were epigenetically distinct from bona fide Teff cells. Texint cells ultimately converted to Texterm cells that were absent from the blood but accumulated in tissues and the blood-accessible locations in the spleen. We also discovered a TF cascade as Tex cells transitioned from TCF1hiToxhi to TCF1 intToxhi to TCF1negT-bethiToxint and finally to TCF1negT-betloToxhiEomeshi during Tex subset conversion. Moreover, we defined key roles for these TFs in this developmental trajectory, although other TFs likely also have a role. These findings also reconcile previous differences between descriptions of Tex subsets that could have related to whether the T-bethi Texint subset was included in a progenitor or terminal population in earlier two subset models (Im et al., 2016; Paley et al., 2012; Utzschneider et al., 2016). Moreover, although there will likely be disease-specific features, the general biology identified here appears to be preserved across diseases and species, suggesting a conserved core developmental biology.
These studies also provided additional insights about the biological role of T-bet in Tex cells. First, T-bet drove conversion of TCF1+ progenitor Tex cells to TCF1− Texint cells. Second, the Texint subset re-engaged some effector-like activity and migration reminiscent of T-bet functions in other CD8+ T cells (Berrien-Elliott et al., 2015; Intlekofer et al., 2008; Intlekofer et al., 2005; Pearce et al., 2003; Taqueti et al., 2006). Third, T-bet stabilized the Texint stage, preventing terminal exhaustion, whereas this activity was counterbalanced by Tox. Moreover, two recent studies highlight a CX3CR1+ Tex subset that possesses an effector-like transcriptional program, expresses T-bet, and is similar to the Texint subset defined here (Hudson et al., 2019; Zander et al., 2019). Together, these data highlight additional functions for T-bet as a central coordinator of an intermediate Tex subset and identify the TCF1, T-bet, and Tox axes as essential components regulating the development and balance between Tex subsets.
PD-1 pathway blockade preferentially amplifies the Tex subsets with circulatory potential (i.e., Texprog2 and Texint cells) consistent with detection of responses in the blood after PD-1 blockade in cancer patients (Huang et al., 2017). The observed Increase in effector-like transcriptional features in the Texint subset despite a Tex cell epigenetic landscape Is consistent with reengagement of effector gene transcription but lack of epigenetic changes associated with PD-1 pathway blockade (Pauken et al., 2016). Because Texint cells also convert to Texterm, these data also predict a potential replacement of Texterm in tumors with new Texint converting to Texterm cells after PD-1 blockade consistent with accumulation of Eomeshi Tex cells after blockade or loss of PD-1 (Huang et al., 2017; Odorizzi et al., 2015; Pauken et al., 2016). Moreover, these dynamics could explain the clonal replacement observed in humans after PD-1 blockade (Yost et al., 2019). T-bet directly represses PD-1 in Tex cells (Kao et al., 2011), and we consistently observed a modest decrease in PD-1 expression in the Texprog1 → Texprog2 and Texprog2 → Texint transitions. Thus, it is possible that by restraining antigen-dependent TCR signals, PD-1 protects Tex progenitors. A subtle transient downregulation of PD-1 by T-bet, and therefore increase In TCR and/or costimulatory signaling, could provoke loss of TCF1, re-entry to cell cycle, and differentiation to Texint, although whether loss of TCF1 instigates T-bet expression or vice versa remains currently unclear. Blocking PD-1 signals could provoke a similar effect, causing a synchronous and robust conversion of Texprog2 into Texint.
These observations also have relevance for understanding Tex biology in humans with chronic viral infections and cancers. For example, CyTOF analysis in patients with HIV or non-small-cell lung cancer identified 9 clusters of Tex cells that expressed IRs and Tox (Bengsch et al., 2018). One rare cluster in the blood was TCF1+, PD-1+, and CXCR5+, which was associated with less severe disease and might represent Texprog1-like cells (Bengsch et al., 2018). Several other clusters expressed lower TCF1 and/or lacked CXCR5 and were Ki67+ bearing resemblance to Texprog2. Three additional clusters resembled Texint, expressing T-bet, Ki67, and/or cytotoxicity molecules. Finally, two clusters were more similar to Texterm or Texint just transitioning to Texterm and had high Tox, IRs, and Eomes and lacked Ki67. Notably, some of these latter clusters correlated with more severe disease and/or were enriched in lung tumors in comparison with blood (Bengsch et al., 2018). In addition, other studies in humans identified cells similar to Texprog1 in lymphoid aggregates in tumors (Jansen et al., 2019), proliferating in the peripheral blood after checkpoint blockade similar to Texprog2 and/or Texint (Huang et al., 2019; Huang et al., 2017; Kamphorst et al., 2017) or clonally related populations in tumors with features of Texprog cells and Texint or Texterm (Li et al., 2019; Wu et al., 2020; Zhang et al., 2018) consistent with the lineage relationships defined here.
Most tissues are organized into proliferative hierarchies, and these hierarchies are important in settings of proliferative stress (Grompe, 2012). Often, these regenerative hierarchies involve a quiescent tissue stem cell or progenitor cell that could receive local tissue signals, for example the crypt base columnar cell or hematopoietic stem cell (Gehart and Clevers, 2019; McCracken et al., 2016). There is often also a second progenitor population that can display more steady-state cell cycle (Grompe, 2012; McCracken et al., 2016). In some settings these two progenitor populations could interconvert, especially if proliferative stress is increased (Grompe, 2012). In these tissues there is often also a transient amplifying population that is more numerically abundant and occupies that part of the developmental hierarchy where most proliferation has occurred, but also where cells acquire major biological features of the function of the tissue. Finally, terminally differentiated tissue cells are formed. Although these cells, such as post-mitotic intestinal enterocytes (Gehart and Clevers, 2019; Grompe, 2012), are terminally differentiated, they are critical for the function of the tissue. Each step in these tissue proliferative hierarchies, which allow cells to manage proliferative stress and regeneration, has possible analogies to Tex cell biology including two distinct but interconvertible progenitor populations: a transient amplifying population that acquires key functions of the tissue and a terminal population that is short-lived, but likely has key biological functions. In some of these settings, the proliferative hierarchy has branches allowing different types of more differentiated descendant cells to be formed (Gehart and Clevers, 2019; McCracken et al., 2016). Whether the Tex cells proliferative hierarchy contains more complexity than revealed here remains to be determined.
Together, these studies help clarify our understanding of Tex subset heterogeneity as well as define the biological properties and associated underlying transcriptional and epigenetic control of Tex cell heterogeneity. These “maps” of Tex cell biology could allow more detailed understanding of data from cancer patients to determine which aspects of Tex cells have been altered by therapeutic interventions. Finally, these data also illustrate a developmental biology structure for Tex cell differentiation and highlight similarities to other biological settings where a hierarchy of progenitor cell maintenance and differentiation balances proliferative stress and homeostatic functions of the “tissue.”
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information, requests for resources and reagents should be directed to the lead contact, E. John Wherry (wherry@pennmedicine.upenn.edu).
Materials Availability
Materials generated by this study are available upon request.
Data and Code Availability
The accession number for the RNAseq and ATACseq reported in this paper is GEO:GSE149879.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Patients samples
Results presented in this study were obtained by re-analyzing tumor resection data from clinical stage III or stage IV melanoma patients involved in a previously reported clinical trial at the University of Pennsylvania (NCT02434354) (Huang et al., 2019) or collected from patients under IRB protocol 08607 at the University of Pennsylvania. Flow cytometry and staining were performed as previously described. (Huang et al., 2019) Samples were collected either before or 3 weeks after treatment with a single flat dose of pembrolizumab 200mg intravenously. The protocol and its amendments were approved by the Institutional Review Board at the University of Pennsylvania, and all patients provided written informed consent.
Mice
Six-week old female C57BL/6 mice (CD45.2, Charles River, NCI) were used for most experiments. Alternatively, six-week old female NCI B6-Ly5.1/Cr (NCI Charles River) or male B6.SJL-Ptprc Pepc/BoyJ mice (The Jackson laboratory) expressing the congenic marker CD45.1 were used as recipients for some P14 knockout adoptive transfers and CFSE tracing experiments. P14 TCR transgenic mice expressing a TCR specific for the LCMV Dbgp33-41 peptide were bred in house and backcrossed onto the C57BL/6 background. P14 Tcf7flox/flox CD4-cre (P14TCF1KO) mice were bred in house by crossing CD4-cre mice (The Jackson laboratory) to Tcf7flox/flox mice on the P14 background. Tbx21flox/flox from the Jackson Laboratory (B6.128-Tbx21tm2Srnr/J) were bred in house with P14 ERT2-Cre+/− Rosa26-YFP+/− to generate P14 Tbx21flox/flox ERT2-CRE+/− Rosa-YFP+/− mice (P14TbetKO). ToxKO mice were kindly provided by Jonathan Kaye, PhD (Cedars-Sinai Medical Center, Los Angeles, CA, USA) and bred in house onto the P14 background to generate P14 Tox+/− mice. All experiments and breeding conditions were in accordance with Institutional Animal Care and Use Committee (IACUC) guidelines for the University of Pennsylvania.
Viruses and Infections
LCMV Armstrong and clone 13 were grown in BHK cells and titrated using plaque assay on VERO cells. Recipient mice were infected either intraperitoneally (i.p.) with LCMV Armstrong (2x105 plaque forming units [PFU]) or intravenously (i.v.) with LCMV clone 13 (4x106 PFU) one day post adoptive transfer of P14 cells where noted.
Cell line and tumor transplant
B16 and MC-38 tumor cells were maintained in DMEM supplemented with 10% FCS, 1% L-glut and 1% Pen/Strep. Tumor cells (5x105) cultured for less than two weeks were implanted subcutaneously in the flank of recipient mice using 29G1/2 syringes. Tumor size was monitored using a manual caliper and tumors were excised before exceeding the volume permitted by the IACUC guidelines for the University of Pennsylvania.
Retroviral vectors
Tbx21 and Tcf7 short isoform (p33; Origene) cDNAs were cloned into a MSCV-IRES-VEX plasmid. Retroviral particles were produced by transfection of 293T cells. Briefly, 293T cells were pre-incubated with warmed cDMEM supplemented with chloroquine (25μM; Sigma). Cells were transduced with a pCL-Eco plasmid (15μg) and MSCV-IRES-VEX expression plasmid (15μg) using Lipofectamine 3000 (ThermoFisher Scientific) for 6 h at 37°C 5%CO2. After incubation, transduction medium was replaced with fresh cDMEM. RV supernatant were collected at days 3 and 4 of culture and titrated on NIH 3T3 cells.
METHODS DETAILS
Adoptive T cell transfer
Recipient mice were adoptively transferred with PBMCs containing 1x103 P14 CD8+ T cells 24 h prior infection with either LCMV Armstrong or LCMV clone 13. P14TCF1KO (CD45.1.2+) or P14 TOX+/− (CD45.2+) were co-transferred along with their relative WT P14 littermate controls (CD4-Cre+ Tcf7+/+ [CD45.1+] or Tox+/+ [CD45.1.2+] respectively), 1x103 each. For T-bet KO experiments, P14WT (ERT2-Cre+/− Rosa26-YFP+/− Tbx21+/+; CD45.1.2+) and P14TbetKO (ERT2-Cre+/− Rosa26 YFP+/− Tbx21flox/flox, CD45.2+) were treated in vitro with TAT-cre (50μg/mL) in serum free RPMI medium for 45min at 37°C, 5%CO2 and 1x103 of each were co-transferred into naive CD45.1 recipients 24 h before infection. (Gordon et al., 2012) Markers typically associated with early T cell activation (i.e., CD69, Ly6C, PD-1, CD25, CD62L, CD127) were checked in each P14 populations before infusion into recipient mice to ensure transfer of naive cells.
Cell preparation, flow cytometry and cell sorting
Spleens and inguinal lymph nodes (LN) were mechanically disrupted onto a 70μM cell strainer using the plunger of a 3mL syringe and resuspended in 1mL of ACK red blood cell lysing buffer (GIBCO) for 3 min at room temperature (RT). Cell suspensions were washed in cRPMI supplemented with 10% FBS, 1% penn/strep, 1% L-glut, HEPES 10mM (Cell Center, UPenn), MEM non-essential amino acids 1% (GIBCO), Sodium Pyruvate 1mM (Cell Center Upenn), β-mercaptoethanol (0.05mM) and resuspended in FACs buffer (PBS1%, 1%FBS, EDTA [2mM]). Bone marrow suspensions were harvested by flushing cells out of the femur and tibia of infected mice with a 29G syringe and cRPMI. Cells were then treated as above. For lungs and livers, mice were perfused with cold PBS to maximize removal of blood stream cells from organs. Lungs were cut in a Petri dish, disrupted in the presence of Collagenase D (1X) (Roche) using a MACs dissociator (Miltenyi Biotec) and incubated for 45min at 37°C under agitation. After incubation, lung cells were disrupted a second time on a MACs dissociator (Miltenyi Biotec) and processed as above. After mechanical disruption onto a 70μM strainer, lymphocytes from livers were enriched using Percoll (GE Healthcare) density gradient separation (80%/40%), washed two times with cRPMI and processed as above. Blood samples were collected in 1ml of PBS plus 2mM EDTA. RPMI was added and samples were underlaid with Histopaque 1083 (Sigma Aldrich) for lymphocyte enrichment using density gradient concentration. Remaining red blood cells were lysed using ACK lysing buffer (GIBCO) for 3min at RT. For enrichment of mouse TILs, tumors were cut in pieces using a scalpel and incubated in serum free RPMI containing Collagenase D Type 4 (1mg/mL, Worthington) and 0.02mg/mL of DNase I (Sigma) 45 min at 37°C under agitation. Cell suspensions were filtered again on a 70μM strainer and processed as above (Ngiow et al., 2016). Equal number of cells were stained with extracellular antibodies for 30min on ice in FACs buffer (PBS1X, 1%FBS, 2mMEDTA) in the presence of Live/Dead Fixable Aqua Cell Stain (ThermoFisher Scientific). For tumor samples staining, anti-mouse CD16/32 (Biolegend) was added (1/100) at the time of extra-cellular staining as previously described. (Muroyama et al., 2017) Cells were then fixed for 20 min on ice with Cytofix/Cytoperm (BD bioscience) for intracellular proteins detection (active-caspase3, granzyme B, IFNγ, TNFα), incubated 30 min on ice in Perm/Wash buffer (BD bioscience) and stained for an extended 1 h on ice in Perm/Wash buffer (BD bioscience) containing antibodies for intracellular proteins. For TFs detection, cells were fixed and permeabilized using the Foxp3 Transcription Factor buffer set (ThermoFisher Scientific) and incubated for an additional 1 h with TF antibodies. Samples were acquired on an LSR II or BD FACSymphony and analyzed with FlowJo v.10 software (Tree Star Inc).
For cell sorting, CD8+ T cells were enriched from total splenocytes using the EasySeptm CD8+ T cell isolation Kit (StemCell) (routinely > 90% purity). CD8+ T cells were stained with Live/Dead Fixable Aqua Cell Stain (ThermoFisher Scientific) and Tex subsets were sorted based on CD8, CD45.1, CD45.2, CD69 and Ly108 for in vitro CFSE proliferation assays, killing assays, RNaseq and ATACseq experiments. For in vivo CFSE tracing, Tex subsets were sorted from activated CD8+ T cells using CD4 and CD19 as exclusion markers and PD-1 to gate on activated cells and CD69 and Ly108 to identify each Tex subset. Cells were sorted on an BD FACSARIA (BD Bioscience) using a 70 micron nozzle in RPMI 50%FBS maintained at 4°C using a circulating cool-down system. Purity was routinely > 94%.
Intracellular cytokine staining
Splenocytes (1-2x106) were re-stimulated in vitro for 5 h at 37°C 5% CO2 in cRPMI supplemented with GolgiStop (1/250; BD bioscience), GolgiPlug (1/500; BD bioscience), gp33-41 peptide (NIH, 0.4μg/mL) and CD107a antibodies (1/500). Cells were then washed and stained using the BD Fixation/permeabilization kit (BD Bioscience).
CFSE tracing experiments
C57BL/6 mice (CD45.2+) were infected with LCMV clone 13 and spleens were collected at day 21 pi. Mice with low viral titers in the serum at day 15pi were excluded. CD8+ T cells were enriched using EasySeptm CD8+ T cell isolation Kit (StemCell) (routinely > 90% purity) and Tex subsets were sorted from endogenous activated CD8+ T cells. Briefly, Live/Dead Fixable Aqua Cell Stain (ThermoFisher Scientific), CD4 and CD19 were used as exclusion markers and PD-1, Ly108 and CD69 were used to discriminate and sort Tex subsets among endogenous activated (PD-1+) CD8+ T cells (purity was routinely > 94% for each subset). Sorted cells were stained with Carboxyfluorescein succinimidyl ester (CFSE; ThermoFisher Scientific) for 8min at RT in PBS containing the CFSE dye (5μM). Reaction was stopped by adding an equal volume of cold FBS and cells were subsequently washed two times in cRPMI. Cells were counted and 1x105 of each subset was adoptively transferred into infection-matched CD45.1+ recipient mice. Proliferation and phenotypic changes were assessed in the spleen seven days post-transfer.
Intravascular staining
Intravascular staining was performed as previously described. (Anderson et al., 2014) At day 30 pi, recipient mice were injected i.v. with 200μl of PBS containing 3μg of eF-450-conjugated anti-CD8 antibody (ThermoFisher Scientific). Three minutes later, mice were sacrificed and spleens were collected and processed as above.
Antibody treatment
Where indicated, mice were depleted of CD4+ T cells using two i.p. injections of 200μL of PBS containing 200μg of monoclonal anti-CD4 antibody (clone GK1.5, BioXcell) one day prior and post infection with LCMV clone 13.
PD-L1 blockade was performed in CD4-depleted mice as previously described. (Pauken et al., 2016) Sequential i.p. injections of 200μl of PBS containing or not rat anti-mouse PD-L1 monoclonal antibody (200μg/injection, clone 10F.9G2, BioXcell) were performed every three days between days 22 and 34 for a total of five injections.
CFSE Proliferation assay in vitro
In vitro CFSE proliferation assays were performed as previously described (Beltra et al., 2016). Sorted Tex subsets were re-suspended in 1ml of PBS and stained for 6min with CFSE by addition of an equal volume of PBS containing 4μM of CFSE (2μM final). The reaction was stopped by addition of an equal volume of cold FBS and cells were subsequently washed two times in cRPMI. Dendritic cells (DCs) were enriched from the spleen of naive C57B/L6 mice using CD11c+ MicroBeads Ultra pure mouse kit (Miltenyi) and loaded with gp33-41 peptide (0.1 μM) for 30min at 37°C 5% CO2 in cRPMI. Each Tex subset was co-cultured with peptide loaded DCs (104 each) for three days in 96-round bottom well plates.
Killing assay
Splenocytes from naive CD45.2+ C57BL/6 mice were coated with 0.2μM of gp33-41 peptide (targets) or the irrelevant OVA257-264 SIINFEKL peptide (non-target). Target and non-target cells were stained with CFSE at 0.1 and 10μM respectively. Target and non-target cells were incubated or not with sorted Tex subsets (E:T 3/1) for 16 h in cRPMI. Percentage of specific lysis was calculated as follows: 100-((100X % gp33 targets)/% gp33 targets without effectors).
Retroviral (RV) transduction
RV transduction of P14 CD8+ T cells was performed as described (Kurachi et al., 2017). P14 CD8+ T cells were enriched from total splenocytes using EasySeptm CD8+ T cell isolation Kit (StemCell) and activated in vitro in cRPMI supplemented with αCD3 (1 μg/mL), αCD28 (0.5μg/mL) antibodies and IL-2 (100U/mL) (PeproTech). One day post activation (between 24-27 h), activated CD8+ T cells were enriched using Percoll (GE Healthcare) density gradient (30%/60%) and spin-transduced during 60-75min at 2000g 30°C with RV supernatant containing polybrene (4μg/mL). Transduced cells were then incubated for 6 h, washed twice in cRPMI, counted and injected (1x105 per mouse) into LCMV clone 13 infected mice at day 1.5pi.
Active caspase-3 detection
Splenocytes from infected mice or TILs isolated from Human melanoma tumors were incubated for 5 h at 37°C 5%CO2 in cRPMI prior intra-cytoplasmic detection of active-caspase 3 (BD Bioscience) using BD Fixation/Permeabilization kit (BD Bioscience).
Sample preparation for RNaseq
Sorted cells were centrifuged, resuspended in 350μl of RLT buffer (RNeasy Plus Micro kit, QIAGEN) in DNA LoBind tubes (Eppendorf), vortexed for 30 s, flash-frozen on dry ice and stored at −80°C. mRNA was extracted using RNeasy Plus Micro kit (QIAGEN) following manufacturer’s protocol and eluted in 14μl of RNase free water. mRNA quality was assessed on a 2200 TapeStation (Agilent Technologies) using High Sensitivity RNA ScreenTape (Agilent Technologies). mRNA from each biological replicate (5ng/sample) was converted into cDNA and amplified (9 cycles) using the SMART-Seq V4 Ultra Low Input RNA Kit (Takara Bio). 10ng of cDNA was used to prepare DNA libraries using Nextera XT DNA Library Prep Kit (Illumina). Barcoded samples were pooled, diluted at 1.8pM, loaded onto a TG NextSeq 500/550 High Output Kit v2 (150 cycles, 400M reads, Illumina) and paired-end sequencing was performed on a NextSeq 550 (Illumina).
Sample preparation for ATACseq
ATACseq sample preparation was performed as described with minor modifications. (Buenrostro et al., 2013) Sorted cells (2.5-to-5x104) were washed twice in cold PBS and resuspended in 50μl of cold lysis buffer (10nM Tris-HCl, pH 7.4, 10mM NaCl, 3mM MgCl2, 0.1% Tween). Lysates were centrifuge (750xg, 10min, 4°C) and nuclei were resuspended in 50μl of transposition reaction mix (TD buffer [25μl], Tn5 Transposase [2.5μl], nuclease-free water [22.5μl]; (Illumina)) and incubated for 30min at 37°C. Transposed DNA fragments were purified using a QIAGEN Reaction MiniElute Kit, barcoded with NEXTERA dual indexes (Illumina) and amplified by PCR for 11 cycles using NEBNext High Fidelity 2x PCR Master Mix (New England Biolabs). PCR products were purified using a PCR Purification Kit (QIAGEN) and amplified fragments size was verified on a 2200 TapeStation (Agilent Technologies) using High Sensitivity D1000 ScreenTapes (Agilent Technologies). Libraries were quantified by qPCR using a KAPA Library Quant Kit (KAPA Biosystems). Normalized libraries were pooled, diluted to 1.8pg/mL loaded onto a TG NextSeq 500/550 High Output Kit v2 (150 cycles, 400M reads, Illumina) and paired-end sequencing was performed on a NextSeq 550 (Illumina).
QUANTIFICATION AND STATISTICAL ANALYSIS
Spice charts analysis
Positive gates for each parameter were created in FlowJo (V10.4.2, Tree Star) and the Boolean tool was used to evaluate the frequency of cells in each possible combination of markers for each sample of interest. Excel sheet containing all the samples and relative frequencies of cells in each combination was created from FlowJo, converted to a spice-compatible file using Pestle and pie charts were realized with Spice (v5.35).
RNA seq
Raw FASTQ files from RNaseq paired-end sequencing were aligned to the GRCm38/mm10 reference genome using Kallisto (https://pachterlab.github.io/kallisto/), filtered to remove transcripts with low counts and trimmed using the TMM normalization method. Genes with differential expression across samples (DEGs) were assessed using the Limma package of R. A FDR of 0.01 and Log2 fold change cut-off of 1 were imposed. PCA plots were built using normalized and filtered log2 cpm. Unsupervised clustering was performed using K-means to cluster heatmap rows. ComBat in the R package svaseq was used to correct for non-biological variations accounting for unwanted differences when merging the RNaseq dataset from this paper and the Scott-Browne et al. (2016) (Scott-Browne et al., 2016) RNaseq dataset for comparisons. (Leek, 2014)
Gene ontology
Gene ontology of gene sets of interest were obtained using the Metascape online tool (http://metascape.org/gp/index.html#/main/step1). Pathway enrichment analysis (GO Biological processes) was set for a minimum overlap of 3, a p-value cut-off of 0.01 and a minimum enrichment score of 1.5. The five pathways with the most significant enrichment score by log (q-value) were presented and plotted onto a bubble graph using Datagraph (https://www.visualdatatools.com/DataGraph/).
Gene set enrichment analysis
GSEA was performed using the Broad Institute software (https://www.broadinstitute.org/gsea/index.jsp). Enrichment scores were calculated by comparing each Tex subset (Texprog1, Texprog2, Texint and Texterm) to the three others. Multiple comparative analysis were performed using datasets publicly available through the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/gds) under the following accession numbers: CXCR5+Tim3− and CXCR5−Tim3+ (GSE84105)(Im et al., 2016), MPEC and SLEC (GSE8678)(Joshi et al., 2007), state 1 (plastic) and state 2 (fixed)(GSE89307)(Philip et al., 2017) or using gene signatures listed as supplemental material: TRM and TCIRC (Milner et al., 2017), Texhausted (Bengsch et al., 2018), CD8-G, CD8-B and CD8-1_to_6.(Sade-Feldman et al., 2019)
ATACseq
Raw ATACseq FASTQ files from paired-end sequencing were processed using the script available at the following repository (https://github.com/wherrylab/jogiles_ATAC). DWe used samtools to remove unmapped, unpaired, mitochondrial reads and ENCODE blacklist regions were also removed (https://sites.google.com/site/anshulkundaje/projects/blacklists). PCR duplicates were removed using Picard. Peak calling was performed using MACS v2 (FDR q-value 0.01). For each experiment, we combined peaks of all samples to create a union peak list and merged overlapping peaks with BedTools merge. The number of reads in each peak was determined using BedTools coverage. Differentially accessible regions were identified following DESeq2 normalization using a FDR cut-off < 0.05 or 0.01 unless otherwise indicated. ComBat in the R package svaseq was used to correct for non-biological variations accounting for unwanted differences when merging the ATACseq dataset from this paper and the Pauken et al. (2016)(Pauken et al., 2016) ATACseq dataset for comparisons.(Leek, 2014)
Correlation network analysis
For all pairwise comparisons, a correlation coefficient was calculated using the Spearman method. A correlation network was created based on correlation scores and projected using Cytoscape.
Motif enrichment analysis
Motif enrichment was calculated using HOMER (default parameters) on peaks differentially accessible during indicated Tex subset transitions (Figure 5e; Table. S6). Transcription binding site prediction analysis was performed using the de novo motif discovery strategy. For each predicted motif, top 10 TF matches were selected and shown in Table. S7 and the top match was highlighted in Figure 5g.
Sample distance analysis
A distance matrix was calculated using the “euclidean” measure using the top 25% peak by variance and plotted as a heatmap in Figure S6E.
Biclustering analysis
Differential expression of genes was calculated for all pairwise comparisons of cell states using DESeq2_1.22.2. Normalized expression values of genes which are differential in any of the pairwise comparison were used as input to bicluster the data using the SpectralBiclustering method from the scikit-learn_0.21.3 package. Seven gene modules were calculated for the four sample groups.
Gene signature analysis on scRNaseq data
The single cell expression data used was from GSE122712 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE122712)(Miller et al., 2019), the supplementary files from the repository were used as input and processed using the Seurat_3.1.1 package. The above-calculated seven gene modules were used as features to calculate module scores of single cells using the AddModuleScore function from the Seurat_3.1.1 package. Module scores for each of the gene signatures was used to color the cells on the UMAP projection created from the Seurat processing.
Statistical analysis
Statistics on flow cytometry data were performed using unpaired two-tailed Student’s t test (GraphPad Prism v6; *p < 0.05, **p < 0.005, ***p < 0.0005).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies (Mouse, Human, Intracellular, In Vivo) | ||
| FITC mouse anti-mouse CD244 (clone 2B4) | BD Biosciences | Cat# 553305; RRID: AB_394769 |
| FITC Hamster anti-mouse KLRG1 (clone 2F1) | SouthernBiotech | Cat# 1807-02; RRID: AB_2795367 |
| Alexa Fluor 488 anti-mouse CD45 (clone 30-F11) | Biolegend | Cat# 103122; RRID: AB_493531 |
| Alexa Fluor 488 anti-mouse Ly-6C (clone HK1.4) | Biolegend | Cat# 128022; RRID: AB_10639728 |
| PE/Dazzle 594 anti-mouse TIGIT (clone 1G9) | Biolegend | Cat# 142110; RRID: AB_2566573 |
| PE-CF594 rat anti-mouse CD127 (clone SB/199) | BD Biosciences | Cat# 562419; RRID: AB_11153131 |
| PE-Cyanine 5 anti-mouse/rat ICOS (clone C398.4A) | Ebioscience | Cat# 15-9949-82; RRID: AB_468828 |
| PE/Cy5 anti-mouse CD69 (clone H1.2F3) | Biolegend | Cat# 104510; RRID: AB_313113 |
| PE-Cyanine7 anti-mouse CD28 (clone 37.51) | Ebioscience | Cat# 25-0281-82; RRID: AB_469612 |
| PE/Cy7 anti-mouse CD279 (PD-1, clone RMP1-30) | Biolegend | Cat# 109110; RRID: AB_572017 |
| PE anti-mouse Ly108 (clone 330-AJ) | Biolegend | Cat# 134606; RRID: AB_2188095 |
| PE anti-mouse CD160 (clone 7H1) | Biolegend | Cat# 143004; RRID: AB_10960743 |
| APC anti-mouse CD223 (Lag-3, clone eBioC9B7W) | Ebioscience | Cat# 17-2231-82; RRID: AB_2573184 |
| Alexa Fluor 647 anti-mouse/human CD44 (clone IM7) | Biolegend | Cat# 103018; RRID: AB_493681 |
| APC-eFluor 780 anti-mouse CD62L (clone MEL-14) | Ebioscience | Cat# 47-0621-82; RRID: AB_1603256 |
| APC-eFluor 780 anti-mouse KLRG1 (clone 2F1) | Ebioscience | Cat# 47-5893-82; RRID: AB_2573988 |
| APC-eFluor 780 anti-human/mouse CD44 (clone IM7) | Ebioscience | Cat# 47-0441-82; RRID: AB_1272244 |
| APC-Cy7 rat anti-mouse CD25 (clone PC61) | BD Biosciences | Cat# 557658; RRID: AB_396773 |
| APC-eFluor 780 anti-mouse CD4 (clone RM4-5) | Ebioscience | Cat# 47-0042-82; RRID: AB_1272183 |
| Alexa Fluor 700 anti-mouse CD45.1 (clone A20) | Biolegend | Cat# 110724; RRID: AB_493733 |
| eFluor 450 anti-mouse CD223 (Lag-3, clone eBioC9B7W) | Ebioscience | Cat# 48-2231-82; RRID: AB_11149866 |
| eFluor 450 anti-mouse CD8a (clone 53-6.7) | Ebioscience | Cat# 48-0081-82; RRID: AB_1272198 |
| Pacific Blue anti-mouse Ly108 (clone 330-AJ) | Biolegend | Cat# 134608; RRID: AB_2188093 |
| Brillant Violet 605 anti-mouse CD4 (clone RM4-5) | Biolegend | Cat# 100548; RRID: AB_2563054 |
| Brillant Violet 605 anti-mouse CX3CR1 (clone SA011F1) | Biolegend | Cat# 149027; RRID: AB_2565937 |
| Brillant Violet 605 anti-mouse CD366 (Tim-3, clone RMT3-23) | Biolegend | Cat# 119721; RRID: AB_2616907 |
| Brillant Violet 650 anti-mouse CD8a (clone 53-6.7) | Biolegend | Cat# 100742; RRID: AB_2563056 |
| Brillant Violet 785 anti-mouse CD45.2 (clone 104) | Biolegend | Cat# 109839; RRID: AB_2562604 |
| Purified anti-mouse CD16/32 Antibody | Biolegend | Cat#101302; RRID: AB_312801 |
| Ultra-LEAF purified anti-mouse CD28 (clone 37.51) | Biolegend | Cat#102116; RRID: AB_11147170 |
| LEAF purified anti-mouse CD3ε (clone 145-2C11) | Biolegend | Cat#100331; RRID: AB_1877073 |
| BUV395 Mouse Anti-Human CD45RA (clone HI100) | BD Biosciences | Cat#740298; RRID: AB_2740037 |
| BUV496 Mouse Anti-Human CD8 (clone RPA-T8) | BD Biosciences | Cat#564805; RRID: AB_2744460 |
| BUV737 Mouse Anti-Human CD27 (clone L128) | BD Biosciences | Cat#564302; RRID: AB_2744350 |
| BUV805 Mouse Anti-Human CD3 (clone UCHT1) | BD Biosciences | Cat#565515; RRID: AB_2739277 |
| PE/Cy5 anti-human CD69 Antibody (clone FN50) | Biolegend | Cat#310908; RRID: AB_314843 |
| PE Anti-TOX antibodies, human and mouse (clone REA473) | Miltenyi | Cat#130-107-785; RRID: AB_2654225 |
| Alexa Fluor® 647 anti-TCF1 (TCF7) Antibody (clone C63D9) | Cell Signaling Technology | Cat#6709S; RRID: AB_2797631 |
| Brilliant Violet 605™ anti-T-bet Antibody (clone 4B10) | Biolegend | Cat#644817; RRID: AB_11219388 |
| Alexa Fluor 700 Mouse anti-Ki-67 (clone B56) | BD Biosciences | Cat# 561277; RRID: AB_10611571 |
| FITC Rabbit Anti-Active Caspase-3 (clone C92-605) | BD Biosciences | Cat# 560901; RRID: AB_10563896 |
| PE/Cy7 anti-human CD279 (PD-1) Antibody (clone EH12.2H7) | Biolegend | Cat#329918; RRID: AB_2159324 |
| Eomes Monoclonal Antibody, PEeFluor 610, eBioscience™ (clone WD1928) | Ebioscience | Cat# 61-4877-42; RRID: AB_2574616 |
| Alexa Fluor 488 anti-mouse CD107a (LAMP-1, clone 1D4B) | Biolegend | Cat# 121608; RRID: AB_571983 |
| FITC mouse anti-Ki67 (clone B56) | BD Biosciences | Cat# 51-36524X; RRID: AB_396302 |
| FITC anti-T-bet (clone 4B10) | Biolegend | Cat# 644812; RRID: AB_2200540 |
| R-PE conjugate anti-human Granzyme B (clone GB12) | Invitrogen | Cat# MHGB04; RRID: AB_1500191 |
| PE Rabbit anti-active Caspase-3 (clone C92-605) | BD Biosciences | Cat# 51-68655X; RRID: AB_393957 |
| PE mouse anti-TCF-7/TCF-1 (clone S33-966) | BD Biosciences | Cat# 564217; RRID: AB_2687845 |
| PE-Cy7 anti-mouse TNFa (clone MP6-XT22) | Ebioscience | Cat# 25-7321-82; RRID: AB_11042728 |
| APC anti-mouse IFNg (clone XMG1.2) | Ebioscience | Cat# 17-7311-82; RRID: AB_469504 |
| APC anti-TOX human and mouse (clone REA473) | Miltenyi Biotech | Cat# 130-118-335; RRID: AB_2751485 |
| In vivo Mab anti-mouse CD4 (clone GK1.5) | BioXcell | Cat# BE0003-1; RRID: AB_1107636 |
| In vivo Mab anti-mouse PD-L1 (clone 10F.9G2) | BioXcell | Cat# BE0101; RRID: AB_10949073 |
| Bacterial and Virus Strains | ||
| LCMV clone 13 | Rafi Ahmed | Grew up in house |
| LCMV Armstrong (Arm) | Rafi Ahmed | Grew up in house |
| Biological Samples | ||
| Patients samples used in this study are detailed elsewhere (Huang et al., 2019) | N/A | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Live/Dead Fixable Aqua Dead cell stain Kit | ThermoFisher Scientific | Cat# L34957 |
| Zombie NIR Fixable viability kit | Biolegend | Cat# 423105 |
| CellTrace CFSE Cell Proliferation Kit | ThermoFisher Scientific | Cat# C34554 |
| Recombinant Murine IL-2 | Peprotech | Cat#212-12 |
| ACK Lysing Buffer | ThermoFisher Scientific | Cat#A1049201 |
| Collagenase, Type 4 | Worthington | Cat#LS004189 |
| Deoxyribonuclease I from bovine pancreas | Sigma-Aldrich | Cat#D5025 |
| Collagenase D | Roche | Cat#11088866001 |
| Golgi Stop | BD Biosciences | Cat#554724 |
| Golgi Plug | BD Biosciences | Cat#555029 |
| gp (33-41) peptide KAVYNFATM | GenScript | Custom |
| OVA (257-264) peptide SIINFEKL | GenScript | Custom |
| LCMV tetramers (gp33, gp276) | NIH | N/A |
| Percoll® | Sigma-Aldrich | Cat#GE17-0891-01 |
| Histopaque®-1083 | Sigma-Aldrich | Cat#10831 |
| Tat-cre Recombinase | Protein and Proteomics Core Facility, The Children’s Hospital of Philadelphia | N/A |
| NEBNext High-Fidelity 2X PCR Master Mix | New England Biolabs | Cat#M0541 |
| Cell Culture Reagents | ||
| RPMI-1640 medium | Corning/Mediatech | Cat#10-040-CV |
| HI Fetal Bovine Serum | ThermoFisher | Cat#26170-043 |
| HEPES | ThermoFisher | Cat#15630080 |
| Non-Essential Amino Acids | ThermoFisher | Cat#11140050 |
| Penicillin-Streptomycin | ThermoFisher | Cat#15140122 |
| β-mercaptoethanol | Sigma-Aldrich | Cat#M6250-500ML |
| Opti-MEM | ThermoFisher | Cat#31985088 |
| Polybrene | Sigma-Aldrich | Cat#TR-1003-G |
| Lipofectamine 3000 transfection reagent | ThermoFisher | Cat#L3000001 |
| Critical Commercial Assays | ||
| Fixation/Permeabilization Solution Kit | BD Biosciences | Cat#554714 |
| Foxp3 / Transcription Factor Staining Buffer Set | BD Biosciences | Cat#00-5523-00 |
| RNeasy Micro Kit | QIAGEN | Cat#74004 |
| High Sensitivity RNA ScreenTape | Agilent | Cat#5067-5579 |
| Agencourt AMPure XP | Beckman | Cat#A63880 |
| High sensitivity D5000 screentape | Agilent | Cat#5067-5592 |
| Library Quantification Kit | Kapabiosystems | Cat#KK4824 |
| EasySep Mouse CD8+ T Cell Isolation Kit | StemCell Technologies | Cat#19853 |
| MinElute Reaction Cleanup Kit | QIAGEN | Cat#28204 |
| QIAquick PCR Purification Kit | QIAGEN | Cat#28104 |
| Nextera DNA Library Preparation Kit | Illumina | Cat#FC-121-1030 |
| SMART-Seq® v4 Ultra® Low Input RNA Kit for Sequencing | Takara/Clontech | Cat#634889 |
| Nextera XT DNA Library Preparation Kit | Illumina | Cat#FC-131-1024 |
| NSQ 500 Hi Output KT v2 (150 CYS) | Illumina | Cat#FC-404-2002 |
| D1000 ScreenTape | Agilent | Cat#5067-5582 |
| Deposited Data | ||
| RNAseq and ATACseq for the Ly108- and CD69-defined Tex subsets (d30pi) | In this paper | GEO: GSE149879 |
| RNaseq for Cxcr5+Tim3− and Cxcr5−Tim3+ CD8s | Im et al., 2016 | GEO: GSE84105 |
| RNaseq for MPEC and SLEC | Joshi et al., 2007 | GEO: GSE8678 |
| Teff and Tmem transcriptional signature | Scott-Browne et al., 2016 | GEO: GSE88987 |
| Mouse TILs transcriptional signature | Philip et al., 2017 | GEO: GSE89307 |
| ATACseq profile of Teff and Tmem | Pauken et al., 2016 | GEO: GSE86797 |
| 10X single-cell RNASeq profiling of GP33-tetramer+ CD8+ T cells from mice chronically infected with LCMV Clone 13 | Miller et al., 2019 | GEO: GSE122712 |
| Experimental Models: Cell Lines | ||
| B16 melanoma cell line | Grown in house | N/A |
| MC-38 colon adenocarcinoma cell line | Grown in house | N/A |
| Experimental Models: Organisms/Strains | ||
| C57BL/6 mice | NCI/Charles River | Cat#027 |
| B6-Ly5.1/Cr | NCI/Charles River | Cat#564 |
| B6.SJL-Ptprca Pepcb/BoyJ | The Jackson Laboratory | Cat#002014 |
| P14 Tg mice | Bred in house (NCI background) | N/A |
| CD4-cre | The Jackson Laboratory | Cat#022071 |
| P14 Tcf7flox/flox CD4-Cre | In this paper | N/A |
| P14 Tbx21flox/flox ERT2-Cre+/− Rosa-YFP+/− mice | In this paper | N/A |
| P14 Tox+/− | In this paper | N/A |
| Recombinant DNA | ||
| TCF-1 p33 cDNA | OriGene | Cat#MR226713 |
| TCF-1 p33 overexpression vector | In this paper | N/A |
| T-bet overexpression vector | In this paper | N/A |
| Empty-VEX retroviral vector | Kurachi et al., 2017 | N/A |
| Software and Algorithms | ||
| FlowJo V10.4.2 | TreeStar | https://www.flowjo.com/solutions/flowjo/downloads |
| GraphPad Prism v6 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| Biorender | Biorender Software | https://biorender.com/ |
| Pestle v1.8 | NIH/NIAID (Roederer et al., 2011) | On demand |
| Spice v5.35 | NIH/NIAID (Roederer et al., 2011) | https://niaid.github.io/spice/help/dataformat |
| IGV v2.4.16 | The Broad Institute | https://software.broadinstitute.org/software/igv/download |
| Datagraph | Visual Data Tools | https://www.visualdatatools.com/DataGraph/Download/ |
| Cytoscape (v3.4) | Cytoscape | https://cytoscape.org/download.html |
| GSEA (v3.0) | The Broad Institute | http://software.broadinstitute.org/gsea/index.jsp |
| Monocle (v2) | Qiu et al., 2017 | http://cole-trapnell-lab.github.io/monocle-release/docs/ |
| Homer (v4.6) | Heinz et al., 2010 | http://homer.ucsd.edu/homer/download.html |
| Bowtie (v2) | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/index.html |
| Samtools (v1.1) | Li et al., 2009 | http://samtools.sourceforge.net/ |
| Kallisto (v0.44) | Bray et al., 2016 | https://pachterlab.github.io/kallisto/download |
| Deseq2 1.22.2 | Love et al., 2014 | http://www.bioconductor.org/packages/release/bioc/html/DESeq2.html |
| svaseq | Leek, 2014 | https://bioconductor.org/packages/release/bioc/html/sva.html |
| Pheatmap | Bioconductor | https://cran.r-project.org/web/packages/pheatmap/index.html |
| Great (v3.0) | McLean et al., 2010 | http://great.stanford.edu/public/html/ |
| ggplot2 (v3.2.0) | R package | https://cran.r-project.org/web/packages/ggplot2/index.html |
| Seurat_3.1.1 | Butler et al., 2018 | https://cran.r-project.org/web/packages/Seurat/index.html |
Highlights.
Ly108 and CD69 define four Tex subsets linked in a hierarchical developmental pathway
Two TCF1+ subsets, effector-like and terminally exhausted subsets, are identified
Key transcriptional, epigenetic, and biological changes define subset transitions
TCF1, T-bet, and Tox coordinate Tex subset development and dynamics
ACKNOWLEDGMENTS
We thank the Wherry lab for input. This work was funded by NIH grants N2T32CA009615; CA230157 (A.C.H.); CA234842 (Z.C.); and AI105343, AI108545, AI117950, AI082630, and CA210944 (E.J.W.). This work was also supported by the Tara Miller Foundation (A.C.H.), the Melanoma Research Alliance (E.J.W.), the David and Hallee Adelman Immunotherapy Research Fund (E.J.W.), and the Parker Institute for Cancer Immunotherapy Bridge Scholar Award (A.C.H.). S.F.N. is supported by the NHMRC CJ Martin Fellowship GNT1111469 and the Mark Foundation for Cancer Research Momentum Fellowship. M.S.A.H. is a Cancer Research Institute (CRI) Irvington Postdoctoral fellows. J.R.G. is a Cancer Research Institute-Mark Foundation Fellow and was previously supported by T32CA009140. We thank the Human Immunology Core and the Tumor Tissue and Biospecimen Bank of the University of Pennsylvania (supported by P30-CA016520 and P50-CA174523). Work in the Wherry lab is supported by the Parker Institute for Cancer Immunotherapy.
Footnotes
DECLARATION OF INTERESTS
E.J.W. is a member of the Parker Institute for Cancer Immunotherapy which supported these studies. E.J.W. has consulting agreements with and/or is on the scientific advisory board for Merck, Roche, Pieris, Elstar, and Surface Oncology. E.J.W. has a patent licensing agreement on the PD-1 pathway with Roche/Genentech. E.J.W. is a founder of Arsenal Biosciences.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.immuni.2020.04.014.
REFERENCES
- Alfei F, Kanev K, Hofmann M, Wu M, Ghoneim HE, Roelli P, Utzschneider DT, von Hoesslin M, Cullen JG, Fan Y, et al. (2019). TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature 571, 265–269. [DOI] [PubMed] [Google Scholar]
- Anderson KG, Mayer-Barber K, Sung H, Beura L, James BR, Taylor JJ, Qunaj L, Griffith TS, Vezys V, Barber DL, and Masopust D (2014). Intravascular staining for discrimination of vascular and tissue leukocytes. Nat. Protoc 9, 209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltra JC, Bourbonnais S, Bédard N, Charpentier T, Boulangé M, Michaud E, Boufaied I, Bruneau J, Shoukry NH, Lamarre A, and Decaluwe H (2016). IL2Rβ-dependent signals drive terminal exhaustion and suppress memory development during chronic viral infection. Proc. Natl. Acad. Sci. USA 113, E5444–E5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengsch B, Ohtani T, Khan O, Setty M, Manne S, O’Brien S, Gherardini PF, Herati RS, Huang AC, Chang KM, et al. (2018). Epigenomic-Guided Mass Cytometry Profiling Reveals Disease-Specific Features of Exhausted CD8 T Cells. Immunity 48, 1029–1045.e5, e1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrien-Elliott MM, Yuan J, Swier LE, Jackson SR, Chen CL, Donlin MJ, and Teague RM (2015). Checkpoint blockade immunotherapy relies on T-bet but not Eomes to induce effector function in tumor-infiltrating CD8+ T cells. Cancer Immunol. Res 3, 116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beura LK, Anderson KG, Schenkel JM, Locquiao JJ, Fraser KA, Vezys V, Pepper M, and Masopust D (2015). Lymphocytic choriomeningitis virus persistence promotes effector-like memory differentiation and enhances mucosal T cell distribution. J. Leukoc. Biol 97, 217–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn SD, Shin H, Freeman GJ, and Wherry EJ (2008). Selective expansion of a subset of exhausted CD8 T cells by alphaPD-L1 blockade. Proc. Natl. Acad. Sci. USA 105, 15016–15021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, and Pachter L (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol 34, 525–527. [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, and Greenleaf WJ (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Ji Z, Ngiow SF, Manne S, Cai Z, Huang AC, Johnson J, Staupe RP, Bengsch B, Xu C, et al. (2019). TCF-1-Centered Transcriptional Network Drives an Effector versus Exhausted CD8 T Cell-Fate Decision. Immunity 51, 840–855.e5, e845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daud AI, Loo K, Pauli ML, Sanchez-Rodriguez R, Sandoval PM, Taravati K, Tsai K, Nosrati A, Nardo L, Alvarado MD, et al. (2016). Tumor immune profiling predicts response to anti-PD-1 therapy in human melanoma. J. Clin. Invest 126, 3447–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehart H, and Clevers H (2019). Tales from the crypt: new insights into intestinal stem cells. Nat. Rev. Gastroenterol. Hepatol 16, 19–34. [DOI] [PubMed] [Google Scholar]
- Gordon SM, Chaix J, Rupp LJ, Wu J, Madera S, Sun JC, Lindsten T, and Reiner SL (2012). The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity 36, 55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grompe M (2012). Tissue stem cells: new tools and functional diversity. Cell Stem Cell 70, 685–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R, Hou S, Liu C, Zhang A, Bai Q, Han M, Yang Y, Wei G, Shen T, Yang X, et al. (2016). Follicular CXCR5− expressing CD8(+) T cells curtail chronic viral infection. Nature 537, 412–428. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang AC, Postow MA, Orlowski RJ, Mick R, Bengsch B, Manne S, Xu W, Harmon S, Giles JR, Wenz B, et al. (2017). T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature 545, 60–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang AC, Orlowski RJ, Xu X, Mick R, George SM, Yan PK, Manne S, Kraya AA, Wubbenhorst B, Dorfman L, et al. (2019). A single dose of neoadjuvant PD-1 blockade predicts clinical outcomes in resectable melanoma. Nat. Med 25, 454–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson WH, Gensheimer J, Hashimoto M, Wieland A, Valanparambil RM, Li P, Lin JX, Konieczny BT, Im SJ, Freeman GJ, et al. (2019). Proliferating Transitory T Cells with an Effector-like Transcriptional Signature Emerge from PD-1+ Stem-like CD8+ T Cells during Chronic Infection. Immunity 51, 1043–1058.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im SJ, Hashimoto M, Gerner MY, Lee J, Kissick HT, Burger MC, Shan Q, Hale JS, Lee J, Nasti TH, et al. (2016). Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537, 417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, Mullen AC, Gasink CR, Kaech SM, Miller JD, et al. (2005). Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat. Immunol 6, 1236–1244. [DOI] [PubMed] [Google Scholar]
- Intlekofer AM, Banerjee A, Takemoto N, Gordon SM, Dejong CS, Shin H, Hunter CA, Wherry EJ, Lindsten T, and Reiner SL (2008). Anomalous type 17 response to viral infection by CD8+ T cells lacking T-bet and eomesodermin. Science 327, 408–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen CS, Prokhnevska N, Master VA, Sanda MG, Carlisle JW, Bilen MA, Cardenas M, Wilkinson S, Lake R, Sowalsky AG, et al. (2019). An intra-tumoral niche maintains and differentiates stem-like CD8 T cells. Nature 576, 465–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, Hagman J, Gapin L, and Kaech SM (2007). Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity 27, 281–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphorst AO, Wieland A, Nasti T, Yang S, Zhang R, Barber DL, Konieczny BT, Daugherty CZ, Koenig L, Yu K, et al. (2017). Rescue of exhausted CD8 T cells by PD-1-targeted therapies is CD28-dependent. Science 355, 1423–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao C, Oestreich KJ, Paley MA, Crawford A, Angelosanto JM, Ali MA, Intlekofer AM, Boss JM, Reiner SL, Weinmann AS, and Wherry EJ (2011). Transcription factor T-bet represses expression of the inhibitory receptor PD-1 and sustains virus-specific CD8+ T cell responses during chronic infection. Nat. Immunol 12, 663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan O, Giles JR, McDonald S, Manne S, Ngiow SF, Patel KP, Werner MT, Huang AC, Alexander KA, Wu JE, et al. (2019). TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Nature 571, 211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurachi M, Kurachi J, Chen Z, Johnson J, Khan O, Bengsch B, Stelekati E, Attanasio J, McLane LM, Tomura M, et al. (2017). Optimized retroviral transduction of mouse T cells for in vivo assessment of gene function. Nat. Protoc 12, 1980–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtulus S, Madi A, Escobar G, Klapholz M, Nyman J, Christian E, Pawlak M, Dionne D, Xia J, Rozenblatt-Rosen O, et al. (2019). Checkpoint Blockade Immunotherapy Induces Dynamic Changes in PD-1−CD8+ Tumor-Infiltrating T Cells. Immunity 50, 181–194.e6, e186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT (2014). svaseq: removing batch effects and other unwanted noise from sequencing data. Nucleic Acids Res. 42, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R; 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, van der Leun AM, Yofe I, Lubling Y, Gelbard-Solodkin D, van Akkooi ACJ, van den Braber M, Rozeman EA, Haanen JBAG, Blank CU, et al. (2019). Dysfunctional CD8 T Cells Form a Proliferative, Dynamically Regulated Compartment within Human Melanoma. Cell 176, 775–789.e18, e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken MN, George BM, Kao KS, Marjon KD, Raveh T, and Weissman IL (2016). Normal and Neoplastic Stem Cells. Cold Spring Harb. Symp. Quant. Biol 81, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney EF, Lee JC, Jayne DR, Lyons PA, and Smith KG (2015). T-cell exhaustion, co-stimulation and clinical outcome in autoimmunity and infection. Nature 523, 612–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLane LM, Abdel-Hakeem MS, and Wherry EJ (2019). CD8 T Cell Exhaustion During Chronic Viral Infection and Cancer. Annu. Rev. Immunol 37, 457–495. [DOI] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, and Bejerano G (2010). GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol 28, 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BC, Sen DR, Al Abosy R, Bi K, Virkud YV, LaFleur MW, Yates KB, Lako A, Felt K, Naik GS, et al. (2019). Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat. Immunol 20, 326–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner JJ, Toma C, Yu B, Zhang K, Omilusik K, Phan AT, Wang D, Getzler AJ, Nguyen T, Crotty S, et al. (2017). Runx3 programs CD8+ T cell residency in non-lymphoid tissues and tumours. Nature 552, 253–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muroyama Y, Nirschl TR, Kochel CM, Lopez-Bujanda Z, Theodros D, Mao W, Carrera-Haro MA, Ghasemzadeh A, Marciscano AE, Velarde E, et al. (2017). Stereotactic Radiotherapy Increases Functionally Suppressive Regulatory T Cells in the Tumor Microenvironment. Cancer Immunol. Res 5, 992–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngiow SF, Young A, Blake SJ, Hill GR, Yagita H, Teng MW, Korman AJ, and Smyth MJ (2016). Agonistic CD40 mAb-Driven IL12 Reverses Resistance to Anti-PD1 in a T-cell-Rich Tumor. Cancer Res. 76, 6266–6277. [DOI] [PubMed] [Google Scholar]
- Odorizzi PM, Pauken KE, Paley MA, Sharpe A, and Wherry EJ (2015). Genetic absence of PD-1 promotes accumulation of terminally differentiated exhausted CD8+ T cells. J. Exp. Med 212, 1125–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paley MA, Kroy DC, Odorizzi PM, Johnnidis JB, Dolfi DV, Barnett BE, Bikoff EK, Robertson EJ, Lauer GM, Reiner SL, and Wherry EJ (2012). Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science 338, 1220–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauken KE, Sammons MA, Odorizzi PM, Manne S, Godec J, Khan O, Drake AM, Chen Z, Sen DR, Kurachi M, et al. (2016). Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science 354, 1160–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, Zediak VP, Banica M, DiCioccio CB, Gross DA, Mao CA, et al. (2003). Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science 302, 1041–1043. [DOI] [PubMed] [Google Scholar]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, et al. (2011). Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res 12, 2825–2830. [Google Scholar]
- Philip M, Fairchild L, Sun L, Horste EL, Camara S, Shakiba M, Scott AC, Viale A, Lauer P, Merghoub T, et al. (2017). Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature 545, 452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, and Trapnell C (2017). Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roederer M, Nozzi JL, and Nason MC (2011). SPICE: exploration and analysis of post-cytometric complex multivariate datasets. Cytometry A 79, 167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sade-Feldman M, Yizhak K, Bjorgaard SL, Ray JP, de Boer CG, Jenkins RW, Lieb DJ, Chen JH, Frederick DT, Barzily-Rokni M, et al. (2019). Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma. Cell 176, 404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott AC, Dündar F, Zumbo P, Chandran SS, Klebanoff CA, Shakiba M, Trivedi P, Menocal L, Appleby H, Camara S, et al. (2019). TOX is a critical regulator of tumour-specific T cell differentiation. Nature 571, 270–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott-Browne JP, López-Moyado IF, Trifari S, Wong V, Chavez L, Rao A, and Pereira RM (2016). Dynamic Changes in Chromatin Accessibility Occur in CD8+ T Cells Responding to Viral Infection. Immunity 45, 1327–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen DR, Kaminski J, Barnitz RA, Kurachi M, Gerdemann U, Yates KB, Tsao HW, Godec J, LaFleur MW, Brown FD, et al. (2016). The epigenetic landscape of T cell exhaustion. Science 354, 1165–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo H, Chen J, González-Avalos E, Samaniego-Castruita D, Das A, Wang YH, López-Moyado IF, Georges RO, Zhang W, Onodera A, et al. (2019). TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8+ T cell exhaustion. Proc. Natl. Acad. Sci. USA 116, 12410–12415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui I, Schaeuble K, Chennupati V, Fuertes Marraco SA, Calderon-Copete S, Pais Ferreira D, Carmona SJ, Scarpellino L, Gfeller D, Pradervand S, et al. (2019). Intratumoral Tcf1+PD-1+CD8+ T Cells with Stem-like Properties Promote Tumor Control in Response to Vaccination and Checkpoint Blockade Immunotherapy. Immunity 50, 195–211.e10, e110. [DOI] [PubMed] [Google Scholar]
- Taqueti VR, Grabie N, Colvin R, Pang H, Jarolim P, Luster AD, Glimcher LH, and Lichtman AH (2006). T-bet controls pathogenicity of CTLs in the heart by separable effects on migration and effector activity. J. Immunol 177, 5890–5901. [DOI] [PubMed] [Google Scholar]
- Thommen DS, Koelzer VH, Herzig P, Roller A, Trefny M, Dimeloe S, Kiialainen A, Hanhart J, Schill C, Hess C, et al. (2018). A transcriptionally and functionally distinct PD-1+ CD8+ T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat. Med 24, 994–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utzschneider DT, Charmoy M, Chennupati V, Pousse L, Ferreira DP, Calderon-Copete S, Danilo M, Alfei F, Hofmann M, Wieland D, et al. (2016). T Cell Factor 1-Expressing Memory-like CD8(+) T Cells Sustain the Immune Response to Chronic Viral Infections. Immunity 45, 415–427. [DOI] [PubMed] [Google Scholar]
- Wu T, Ji Y, Moseman EA, Xu HC, Manglani M, Kirby M, Anderson SM, Handon R, Kenyon E, Elkahloun A, et al. (2016). The TCF1-Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Sci. Immunol 1, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Madireddi S, de Almeida PE, Banchereau R, Chen YJ, Chitre AS, Chiang EY, Iftikhar H, O’Gorman WE, Au-Yeung A, et al. (2020). Peripheral T cell expansion predicts tumour infiltration and clinical response. Nature 579, 274–278. [DOI] [PubMed] [Google Scholar]
- Wykes MN, and Lewin SR (2018). Immune checkpoint blockade in infectious diseases. Nat. Rev. Immunol 18, 91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C, Sun HW, Lacey NE, Ji Y, Moseman EA, Shih HY, Heuston EF, Kirby M, Anderson S, Cheng J, et al. (2019). Single-cell RNA-seq reveals TOX as a key regulator of CD8+ T cell persistence in chronic infection. Nat. Immunol 20, 890–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yost KE, Satpathy AT, Wells DK, Qi Y, Wang C, Kageyama R, McNamara KL, Granja JM, Sarin KY, Brown RA, et al. (2019). Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat. Med 25, 1251–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander R, Schauder D, Xin G, Nguyen C, Wu X, Zajac A, and Cui W (2019). CD4+ T Cell Help Is Required for the Formation of a Cytolytic CD8+ T Cell Subset that Protects against Chronic Infection and Cancer. Immunity 51, 1028–1042.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yu X, Zheng L, Zhang Y, Li Y, Fang Q, Gao R, Kang B, Zhang Q, Huang JY, et al. (2018). Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 564, 268–272. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the RNAseq and ATACseq reported in this paper is GEO:GSE149879.