Abstract
Coenzyme A transferases (CoATs) are important enzymes involved in carbon chain elongation, contributing to medium-chain fatty acid (MCFA) biosynthesis. For example, butyryl-CoA:acetate CoA transferase (BCoAT) is responsible for the final step of butyrate synthesis from butyryl-CoA. However, little is known about caproyl-CoA:acetate CoA-transferase (CCoAT), which is responsible for the final step of caproate synthesis from caproyl-CoA. In the present study, two CoAT genes from Ruminococcaceae bacterium CPB6 and Clostridium tyrobutyricum BEY8 were identified by gene cloning and expression analysis. Enzyme assays and kinetic studies were carried out using butyryl-CoA or caproyl-CoA as the substrate. CPB6-CoAT can catalyze the conversion of both butyryl-CoA into butyrate and caproyl-CoA into caproate, but its catalytic efficiency with caproyl-CoA as the substrate was 3.8-times higher than that with butyryl-CoA. In contrast, BEY8-CoAT had only BCoAT activity, not CCoAT activity. This demonstrated the existence of a specific CCoAT involved in chain elongation via the reverse β-oxidation pathway. Comparative bioinformatics analysis showed the presence of a highly conserved motif (GGQXDFXXGAXX) in CoATs, which is predicted to be the active center. Single point mutations in the conserved motif of CPB6-CoAT (Asp346 and Ala351) led to marked decreases in the activity for butyryl-CoA and caproyl-CoA, indicating that the conserved motif is the active center of CPB6-CoAT and that Asp346 and Ala351 have a significant impact on the enzymatic activity. This work provides insight into the function of CCoAT in caproic acid biosynthesis and improves understanding of the chain elongation pathway for MCFA production.
Keywords: Caproic acid, Chain elongation, CoA-transferase, Medium-chain fatty acids, Ruminococcaceae bacterium
Introduction
Medium-chain fatty acids (MCFAs, C6–C12) are widely utilized in agriculture and industry. For example, n-caproic acid (C6) is used as a precursor for the production of fragrances [20], antimicrobial agents [11], and drop-in biofuels [19]. Recent studies have shown that MCFAs produced from renewable feedstock by anaerobic fermentation hold promise for replacing fossil resources and botanical oils, such as palm kernel oil, to meet the requirements for sustainable development [26]. A few microorganisms, such as Megasphaera elsdenii [32], Ruminococcaceae bacterium CPB6 [50], Acinetobacter spp. [21], and Clostridium kluyveri [46], have been reported to be able to synthesize MCFAs from renewable feedstock via the carbon chain elongation pathway [1]. In the process of chain elongation, intermediates of acidogenesis, such as acetate (C2) and n-butyrate (C4), act as substrates and are elongated to caproic acid (C6) and octanoic acid (C8) by addition of acetyl-CoA in reverse β-oxidation cycles [40,34]. C2 or C4, transformed to acetyl-CoA or butyryl-CoA, respectively, represents the initial substrate for elongation via reverse β-oxidation. This pathway has been identified as a key metabolic process in MCFA biosynthesis [36].
The production of high concentrations of butyrate (>10 mM) in vitro has been reported in some anaerobes, such as Roseburia [13] and Faecalibacterium [27]. Butyrate is normally generated from two molecules of acetyl-CoA, yielding acetoacetyl-CoA, which is then converted into butyryl-CoA [47]. In the latter reaction, butyryl-CoA is exchanged with exogenously derived acetate to yield acetyl-CoA and butyrate [35]. The enzymes responsible for butyrate production in the reverse β-oxidation pathway are acetyl-CoA acetyltransferase (AtoB, EC 2.3.1.9), 3-hydroxybutyryl-CoA dehydrogenase (Hbd, EC 1.1.1.157), enoyl-CoA hydratase (Crt, EC 4.2.1.17), butyryl-CoA dehydrogenase (Bcd, EC 1.3.2.1), and butyryl-CoA:acetate CoA transferase (BCoAT, EC 2.8.3.8) [18]. Among them, BCoAT is a well-known coenzyme A transferase (CoAT) responsible for the final step of butyric acid synthesis, transforming the CoA moiety from butyryl-CoA to an exogenous acetate molecule, which results in the formation of butyrate and acetyl-CoA [17,8]. CoATs are abundant in anaerobic fermenting bacteria that cope with low ATP yields, but they are also found in aerobic bacteria and in the mitochondria of humans and other mammals [44]. The synthesis pathway and key genes associated with butyric acid in MCFA biosynthesis via reverse β-oxidation are well understood. However, little is known about key genes involved in the conversion of butyric acid (C4) into caproic acid (C6). Although most genes responsible for butyric acid production are suggested to function in further chain elongation of MCFAs [18], the fact that many butyrate-producing bacteria, such as Clostridium tyrobutyricum, produce only butyric acid instead of caproic acid via the reverse β-oxidation pathway suggests that there may be different functional genes involved in the production of caproic acid.
Recently, our study showed that Ruminococcaceae bacterium CPB6 is a caproic acid-producing bacterium with a highly prolific ability to perform chain elongation and can produce caproic acid (C6) from lactate (as an electron donor) with C2–C4 carboxylic acids and heptanoic acid (C7) with C3–C5 carboxylic acids as electron acceptors (EAs) [51,45]. Moreover, a set of genes correlated with chain elongation were identified by sequencing and annotating the entire genome of the CPB6 strain [48]. However, very little information is available on enzymes involved in the conversion of C4 into C6, especially the gene responsible for the conversion of caproyl-CoA into caproic acid.
In the present study, we cloned a predicted caproyl-CoA:acetate CoA-transferase (CCoAT) gene from the caproic acid-producing strain CPB6 [51] and a BCoAT gene from the butyric acid-producing C. tyrobutyricum BEY8 and expressed the two proteins in Escherichia coli BL21 (DE3) using pET28a. The aims of the present study were to: (i) compare differences in sequence, structure, enzymatic activity, and substrate specificity between CCoAT and BCoAT; (ii) identify the active center of CCoAT and its effects on the activities of enzymes with different structures; and (iii) verify the existence of CCoAT in the caproic acid biosynthesis pathway.
Materials and methods
Strain growth conditions
E. coli DH5α (TsingKe, Chengdu, China) and E. coli BL21 (DE3) (Transgene, Beijing, China) were cultured in Luria broth (LB) medium supplemented with 50 µg/ml kanamycin (Sangon Biotech, Shanghai, China) at 37°C. Ruminococcaceae bacterium CPB6 was grown anaerobically at 37°C in modified reinforced Clostridium medium (Binder, Qingdao, China). C. tyrobutyricum BEY8 was grown anaerobically at 37°C in TGY medium (30 g/l tryptone, 20 g/l glucose, 10 g/l yeast extract, and 1 g/l l-cysteine hydrochloric acid; pH 7).
Gene cloning and plasmid construction
The CoAT genes were amplified from the genomic DNA of Ruminococcaceae bacterium CPB6 [48] or C. tyrobutyricum BEY8 [22] through PCR using the primers listed in Table 1. During amplification, the following conditions were used: initial denaturation (5 min at 98°C); followed by 30 cycles of denaturation (10 s at 98°C), annealing (30 s at 52°C), and elongation (1 min at 72°C); and a final extension (5 min at 72°C). The PCR products were verified by agarose gel electrophoresis, recovered using a PCR purification kit (Fuji, Chengdu, China), and seamlessly inserted into pET28a double digested with Not I and Sal I (Thermo, Waltham, U.S.A.) to construct the recombinant plasmid using a seamless cloning kit (Biomed, Beijing, China). The recombinant plasmids were verified by Sanger sequencing and then used to transform E. coli BL21 (DE3) cells.
Table 1. Bacterial strains, plasmids, and primers used in the present study.
| Description | Reference or source | |
|---|---|---|
| Strains | ||
| CPB6 | Ruminococcaceae bacterium CPB6 | Zhu et al. |
| BEY8 | C. tyrobutyricum BEY8 | Hu et al. |
| E. coli DH5α | TreliefTtMm 5α chemically competent cells | TsingKe |
| E. coli BL21 (DE3) | Expression chemically competent cells | Transgene |
| Plasmids | ||
| pET28a | E. coli expression vector (Kan, T7 promoter) | The present study |
| pET28a-CoAT-CPB6 | pET28a carrying the CoAT gene from CPB6 fused with a His tag at the N-terminus | The present study |
| pET28a-CoAT-BEY8 | pET28a carrying the CoAT gene from BEY8 fused with a His tag at the N-terminus | The present study |
| D346H-mutant | pET28a-CoAT-CPB6 with the Asp346 aa mutation in CoAT | The present study |
| A351P-mutant | pET28a-CoAT-CPB6 with the Ala351 aa mutation in CoAT | The present study |
| Primers | Sequence (5′–3′) | |
| YT43-SalΙ-fw | TAATACGACTCACTATAGGG | The present study |
| YT44-NotΙ-rv | GCTAGTTATTGCTCAGCGG | The present study |
| YT50-CPB6-fw1 | GTGGTGCTCGAGTGCATGAGTTTTCAAGAAGAATATGCACAAAAACTGAC | The present study |
| YT51-CPB6-rv1 | CGAATTCGAGCTCCGTTAAATTTTATTGCTTCTGCGCCAGATGC | The present study |
| YT52-BEY8-fw1 | CGAATTCGAGCTCCGTCGACATGAGTTTTGAGGAATTGTATAAGAGTAAAGTTGTTAGT | The present study |
| YT53-BEY8-rv1 | GTGGTGCTCGAGTGCGGCCGCTTTTATAAGTTCTCTAGCTCTTTGTTTTAATGTCTTACCTCTAAG | The present study |
| YT60-A351P-fw2 | AGCTGGATTTTGTTCTGGGTCCCTATCTGAGCCACGGT | The present study |
| YT61-A351P-rv2 | GACCCAGAACAAAATCCAGCTGACCGGCAC | The present study |
| YT62-D346H-fw2 | AGCGGTGCCGGTGGTCAGCTGCATTTTGTTCT | The present study |
| YT63-D346H-rv2 | GCAGCTGACCACCGGCACCGCTAATCTGACGAAA | The present study |
Underscored letters match the sequence of vectors for seamless cloning.
The sequences corresponding to the mutated codons are written in bold.
Expression and purification of the CoA-transferases
E. coli BL21 (DE3) were transformed with the recombinant plasmids pET28-CoAT-CPB6 (pET28-CCoAT) and pET28-CoAT-BEY8 (pET28-BCoAT). The transformed cells were cultured in LB medium containing 50 µg/ml kanamycin at 37 °C until the OD600 reached 0.5 and then further cultured at 22 °C for 12 h with 0.4 mM IPTG. The cultured cells were harvested by centrifugation (8000×g, 10 min) at 4°C, and the cell pellet was resuspended in 50 mM potassium phosphate (pH 8.0). The cells were then disrupted with an ultrasonicator (Huxi, Shanghai, China) for 30 min (200 W, 4 s, interval 6 s) and centrifuged at 8000×g for 30 min to remove the insoluble material. Then, the enzyme was purified with Ni-NTA Sepharose (Genscript, Nanjing, China) and eluted with 50 mM sodium phosphate (pH 8) containing 300 mM NaCl and 250 mM imidazole. Finally, the purity and MW of the enzyme were assessed using SDS/PAGE analysis. Moreover, the enzyme was analyzed via Western blotting with anti-6× His rabbit polyclonal antibody (Sangon Biotech, Shanghai, China) to determine whether the target protein was obtained. The protein concentrations were determined using a BCA protein assay kit (Solarbio, Beijing, China).
Enzymatic characterization
The CoAT activity in crude enzyme extracts and of purified recombinant proteins was measured by determining the concentration of acetyl-CoA, a reaction byproduct, using a citrate synthase assay described in previous studies with minor modifications [37,25]. In brief, the reaction was initiated by the addition of enzyme (up to 20 ng/ml) and was performed in a total volume of 1 ml at 25°C: 100 mM potassium phosphate buffer (pH 7), 200 mM sodium acetate, 1 mM 5,5′-dithiobis(2-nitrobenzoate), 1 mM oxaloacetate, 8.4 nkat citrate synthase (Sigma, St. Louis, U.S.A.), and 0.5 mM CoA derivatives (Sigma, St. Louis, U.S.A.). The released CoA, corresponding to the formed amount of acetyl-CoA, was detected by measuring the absorbance at 412 nm. One unit of activity is defined as the amount of enzyme that converts 1 µmol of acetyl-CoA per min under these conditions.
The kinetic parameters of the recombinant protein were also calculated by using a coupled spectrophotometric enzyme assay through citrate synthesis [35]. The reaction mixture was the same as that mentioned above, and the concentrations of butyryl-CoA or caproyl-CoA were varied from 0.5 to 5 mM. The kinetic parameters were computed using the Lineweaver–Burk transformation of the Michaelis–Menten equation, in which velocity is a function of the substrate [5,6]. The catalytic constant (kcat) was defined as the number of CoAT molecules formed by one molecule of enzyme in a single second. All measurements were performed in triplicate for each biological replication.
Sequence alignment and phylogenetic reconstruction
Multiple alignment of CoAT amino acid sequences was performed using ESPript [9]. With the Akaike information criterion (AIC), the amino acid substitutions were predicted using ProtTest (version 3.4.2). We constructed a phylogenetic tree of the whole genomes of strains containing CoAT [20,12,3]. The construction followed the general approach of [15,49] and employed sequences downloaded from the online database NCBI (https://www.ncbi.nlm.nih.gov/). MEGA-X software was used to construct the whole genome phylogenetic tree [43]. The phylogenetic relationships of CoATs from different species were obtained by using OrthoFinder [15], and MUSCLE (version 3.8.31) was used to calibrate the 119 shared single-copy genes [14]. The phylogenomic tree was derived from a supermatrix comprising these shared single-copy genes with 41213 unambiguously aligned amino acids using the maximum likelihood (ML) method in RAxML (version 8.2.10) [41] under the PROTGAMMAAUTO model, with 100 bootstrap replicates.
Prediction of tertiary structures of CoAT proteins
The online modeling tool ScanProsite was used for protein homology comparisons, and SWISS-MODEL [2] was used to predict the active sites, tertiary structures and corresponding functions of CoAT proteins in multiple strains (Ruminococcaceae bacterium CPB6, Clostridium kluyveri, Megasphaera elsdenii, Clostridium tyrobutyricum BEY8, Lachnospiraceae bacterium, and Anaerostipes hadrus) to confirm possible variations in the tertiary structures. The targeted sequence was uploaded to search for the best matched template on the basis of data coverage and identity. We subsequently conducted model–template alignment for structural comparisons. Finally, the predicted CoAT protein tertiary structures were embellished and labeled using PyMOL (version 2.3.3) [31] and were adjusted in a similar pattern to identify variations.
Identification of putative positively selected sites
According to the above analysis, relatively conserved regions were identified, and the differences in properties and structures among different amino acids were compared with find two amino acids that might be key sites for site-directed mutagenesis. The point mutation vectors were constructed with the Fast Mutagenesis System (Transgene, Beijing, China). The QuikChange PCR method using pfu DNA polymerase was performed to generate the D346H mutant and A351P mutant. The recombinant plasmid (pET28a-CoAT-CPB6) was used as template DNA, and the complementary mutagenic oligonucleotides used as primers are shown in Table 1. After PCR amplification, the mixture was digested with restriction enzymes using DpnI to remove methylated template DNA and then sequenced (TsingKe, Chengdu, China) to verify site mutagenesis before being used to transform E. coli BL21 (DE3) (Transgene, Beijing, China). After purification, the enzymatic activities in the presence of butyryl-CoA and caproyl-CoA were measured following the method described above for the wildtype.
Results
Cloning, expression, and purification of CoA-transferase
According to the genome sequences of strains CPB6 and C. tyrobutyricum BEY8, specific primers targeting CoAT genes were designed and synthesized (Table 1). Agarose gel electrophoresis showed that the size of the PCR products and the double-digestion products was approximately 1300 bp, consistent with the expected sizes of the CPB6-CoAT (1344 bp) (Supplementary Figure S1) and the BEY8-CoAT (1233 bp) genes (Supplementary Figure S2). Sequence analysis of the recombinant CoAT plasmids showed that the cloned genes shared 100% similarity with the predicted CoAT genes of strains CPB6 (a CCoAT) and BEY8 (a BCoAT). This finding indicated that the recombinant E. coli/pET28a-CCoAT and E. coli/pET28a-BCoAT were successfully constructed.
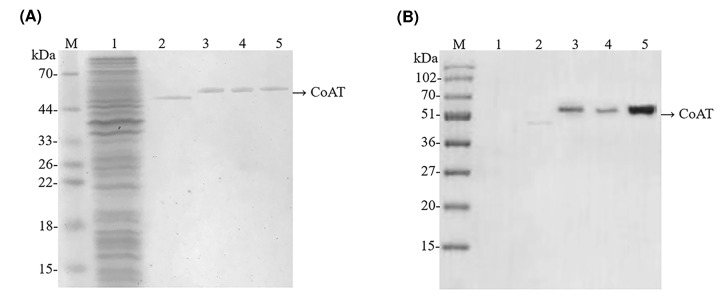
To characterize the functions of CoAT proteins, two recombinant plasmids (pET28a-CCoAT and pET28a-BCoAT) were expressed in E. coli BL21 (DE3). Single bands of the purified proteins were detected on SDS/polyacrylamide gels after affinity chromatography (Figure 1A). As shown in Figure 1A, there was no obvious protein band approximately the size of the target protein in E. coli/pET28a (control), while a single band was observed in E. coli/pET28a-BCoAT (lane 2) and E. coli/pET28a-CCoAT (lane 3), and the band sizes were consistent with the expected sizes of BEY8-CoAT (46 kDa) and CPB6-CoAT (49 kDa). Furthermore, Western blotting analysis with a His antibody (Figure 1B) also demonstrated that the observed bands were consistent with the expected molecular mass of BEY8-CoAT and CPB6-CoAT (approximately 46–49 kDa).
Figure 1. Purification and Western blot analysis of CPB6-CoAT (a CCoAT) and BEY8-CoAT (a BCoAT).
Analysis of purified CCoAT and BCoAT via SDS/PAGE (A). Analysis of purified CCoAT and BCoAT via Western blotting with an anti-His-tag antibody (B). M, molecular mass marker. Lanes: 1, pET28a; 2, BCoAT; 3, CCoAT; 4, CCoAT-D346H mutant; 5, CCoAT-A351P mutant. Samples (∼2 µg) were visualized by Coomassie Brilliant Blue staining after electrophoresis. Molecular mass positions are shown by markers (kDa).
Enzyme assay
The CoAT activity of crude enzyme extracts was determined by measuring the production of acetyl-CoA from butyryl-CoA or caproyl-CoA [37]. Previously, the key reactions for butyrate and caproate production have been reported to be (1) butyryl-CoA + acetate → butyrate + acetyl-CoA and (2) caproyl-CoA + acetate → caproate + acetyl-CoA [40,16,51]. As shown in Table 2, the crude and purified BEY8-CoAT activities with butyryl-CoA and sodium acetate as substrates were 6.91 ± 0.12 and 26.2 ± 0.09 U/mg of protein, respectively. However, this enzyme showed no activity for caproyl-CoA. This result suggests that BEY8-CoAT is a BCoAT, similar to the CoAT from Clostridium acetobutylicum ATCC 824 that is able to produce butyrate instead of caproate, and its purified enzyme activity was 29.1 U/mg of protein [10]. Moreover, the butyrate-producing bacterium Coprococcus sp. strain L2-50 from the human large intestine showed very high BCoAT activity (118.39 ± 5.02 U/mg of protein) but no CCoAT activity [13]. This result indicates that the BCoAT probably has substrate specificity for butyryl-CoA. In contrast, the activities of crude and purified CPB6-CoAT with butyryl-CoA and sodium acetate as substrates were 2.07 ± 0.06 and 10.8 ± 0.02 U/mg of protein, and the activities with caproyl-CoA and sodium acetate as substrates were 5.11 ± 0.08 and 27.6 ± 0.15 U/mg of protein, respectively (Table 2), indicating that CPB6-CoAT can catalyze the conversion of both butyryl-CoA into butyrate and caproyl-CoA into caproate. Notably, the crude and purified CPB6-CoAT activity for caproyl-CoA was 2.5–2.6-times higher (5.11 vs 2.07, 27.56 vs 10.28 U/mg of protein) than that for butyryl-CoA, suggesting that CPB6-CoAT specifically prefers caproyl-CoA as a substrate instead of butyryl-CoA.
Table 2. Purification and specific activities of the CoATs1.
| Enzyme | Total protein, mg | Total activity, U | Specific activity, U/mg of protein | Purification fold | |
|---|---|---|---|---|---|
| Butyryl-CoA | Caproyl-CoA | ||||
| CPB6-CoAT | |||||
| Crude extract | 87.04 | 180.2 | 2.07 ± 0.06 | 5.11 ± 0.08 | 1 |
| Purified protein2 | 22.61 | 244.2 | 10.8 ± 0.02 | 27.6 ± 0.15 | 5.4 |
| BEY8-CoAT | |||||
| Crude extract | 63.10 | 436.0 | 6.91 ± 0.12 | ND1 | 1 |
| Purified protein2 | 17.66 | 462.7 | 26.2 ± 0.09 | ND1 | 3.8 |
The purification data in the table were obtained from 300 ml of culture medium. Abbreviations: ND, not detectable; U, μmol/min.
Protein was purified via affinity chromatography.
Kinetics of CoA-transferases
The kinetic parameters of the recombinant proteins were investigated using a colorimetric assay according to a previous study [35]. Initial velocities were determined at fixed sodium acetate concentrations with different butyryl-CoA or caproyl-CoA concentrations. Km and Vm values were estimated from secondary plots (‘Materials and methods’ section). Additionally, kcat values were calculated from enzyme concentrations in the reaction mixtures. The double-reciprocal enzyme kinetics plot showed that the reactions of the two CoATs follow a ternary-complex mechanism (Supplementary Figure S4).
As kcat/Km can be used to compare the catalytic efficiency of different substrates catalyzed by the same enzyme [24], a lower Km value indicates that the enzyme has a higher affinity for the substrate, and vice versa [30]. In this study, the Km, kcat and kcat/Km values for CPB6-CoAT with caproyl-CoA were 359 ± 5.3 µM, 14.7 ± 0.9 min−1 and 41.1 ± 0.2 mM−1.min−1, respectively, and those with butyryl-CoA were 537 ± 10 µM, 5.81 ± 1.5 min−1 and 10.8 ± 0.2 mM−1.min−1, respectively (Table 3). The catalytic efficiency of CPB6-CoAT for caproyl-CoA was 3.8-times (41.1 ± 0.2 vs 10.8 ± 0.2 mM−1.min−1) higher than that for butyryl-CoA, consistent with our previous result showing that the CCoAT activity is predominantly higher than the BCoAT activity [51]. The Km of CPB6-CoAT for caproyl-CoA was significantly lower than that for butyryl-CoA (359 ± 5.3 vs 537 ± 10 µM), illustrating the higher affinity of this enzyme for caproyl-CoA relative to butyryl-CoA. These results also partly explain why caproate instead of butyrate is always the predominant product in the fermentation broth of strain CPB6 [48,45]. BEY8-CoAT had only BCoAT activity, with Km, kcat and kcat/Km values of 370 ± 4.1 µM, 13.9 ± 0.7 min−1 and 37.7 ± 0.2 mM−1.min−1, respectively, and there was no detectable CCoAT activity (Tables 2 and 3), supporting our previous results showing that strain BEY8 produces only butyric acid as the predominant product. Statistical gap analysis of the above enzymatic experimental data showed P-values that were less than 0.01, indicating a significant difference between them. Similar to the results of Lee et al. [22], the CoAT from C. tyrobutyricum only catalyzes the conversion of butyryl-CoA into butyrate and is not responsible for chain elongation of larger or higher carbon-numbered (>C5) fatty acids.
Table 3. Kinetic parameters for the CoATs.
| Enzyme | Butyryl-CoA | Caproyl-CoA | Reference | ||||
|---|---|---|---|---|---|---|---|
| Km (μM) | kcat (min−1) | kcat/Km (mM−1.min−1) | Km (μM) | kcat (min−1) | kcat/Km (mM−1.min−1) | ||
| BEY8-CoAT | 370 ± 4.1 | 13.9 ± 0.7 | 37.7 ± 0.2 | ND3 | ND3 | ND3 | The present study |
| CPB6-CoAT | 537 ± 10 | 5.81 ± 1.5 | 10.8 ± 0.2 | 359 ± 5.3 | 14.7 ± 0.9 | 41.1 ± 0.2 | The present study |
| CPB6-CoAT-D346H-mutant | 747 ± 2.8 | 1.30 ± 0.2 | 1.73 ± 0.1 | 748 ± 7.9 | 4.24 ± 0.2 | 5.66 ± 0.1 | The present study |
| CPB6-CoAT-A351P-mutant | 623 ± 4.4 | 2.99 ± 0.9 | 4.80 ± 0.2 | 532 ± 2.5 | 6.78 ± 1.2 | 12.8 ± 0.5 | The present study |
| PGN_07251 | 520 ± 10 | 9.33 ± 0.7 | 17.95 ± 0.1 | NR3 | NR3 | NR3 | [35] |
| CoA transferase2 | 21.0 ± 0.1 | NR3 | NR3 | NR3 | NR3 | NR3 | [10] |
Butyryl-CoA:acetate CoA transferase from Porphyromonas gingivalis.
Butyryl-CoA:acetate CoA transferase from Clostridium acetobutylicum ATCC 824.
ND is defined as not determined. Abbreviation: NR, not reported.
Phylogenetics of the whole genome and multiple amino acid sequence alignment
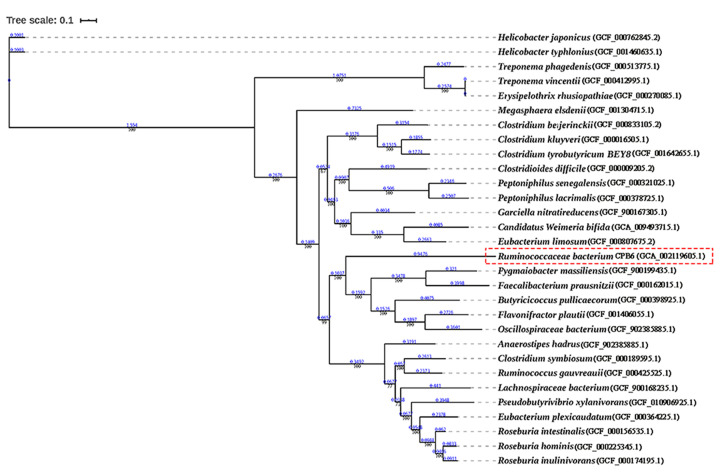
A phylogenetic tree of CoATs from different strains was constructed, as shown in Supplementary Figure S5. The whole-genome phylogenetic tree was constructed based on 119 single-copy genes (including CoATs) that were common among 29 strains (Figure 2). These strains have a wide range of butyrate metabolic pathways [28], for example, Roseburia sp., Faecalibacterium prausnitzii, and Coprococcus sp. from the human gut exhibit BCoAT activity values of 38.95, 18.64, and 118.39 U/mg of protein (crude extracts), respectively [13]. The two species closest to strain CPB6 were Pygmaiobacter massiliensis [4] and F. prausnitzii [39], which are also butyric acid-producing bacteria in human feces. Interestingly, the species closest to C. tyrobutyricum BEY8 was C. kluyveri, which is a well-known caproic acid-producing bacterium. This close relationship may be because they belong to the same genus, Clostridium.
Figure 2. Phylogenetic tree of the whole genomes of 29 strains containing the CoA-transferase.
Numbers at the nodes indicate the levels of bootstrap values. The scale bar for the tree represents a distance of 0.1 substitutions per site.
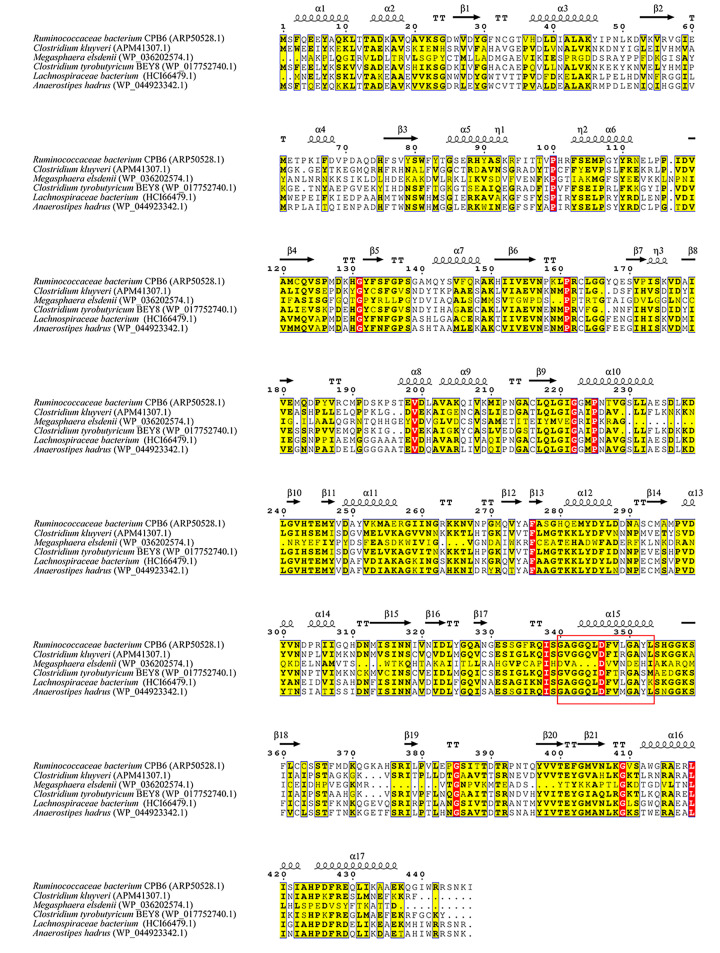
Based on the alignment results generated from CoAT protein sequences from six different species, 14 amino acids (GXGGQXDFXXGAXX, positions 340–353) of the CoATs in all the microbes were highly conserved (except in M. elsdenii), and their secondary structures consisted of 17 α-helices and 21 β-sheets (Figure 3). The sequence similarities between CPB6-CoAT and the analyzed CoATs were as follows: C. kluyveri (37.67%), M. elsdenii (10.27%), C. tyrobutyricum BEY8 (38.04%), Lachnospiraceae bacterium (60.59%), and Anaerostipes hadrus (58.52%). Among the six bacteria, strains CPB6, C. kluyveri, and M. elsdenii are caproic acid-producing bacteria, while C. tyrobutyricum BEY8, Lachnospiraceae bacterium, and A. hadrus are butyric acid-producing bacteria. The alignment results showed that CPB6-CoAT shared lower similarity (10.27–37.67%) with the CoATs of C. kluyveri and M. elsdenii and higher similarity (58.52–60.59%) with the CoATs of Lachnospiraceae bacterium and A. hadrus. This may be because strain CPB6 belongs to the family Ruminococcaceae, which is closer to Lachnospiraceae and Anaerostipes at the taxonomic phylogeny level than to Megasphaera and Clostridium.
Figure 3. Multiple amino acid sequence alignment for CoATs.
The structure contained 17 α-helices and 21 β-pleated sheets, which are represented with symbols. Nonconserved, 60% conserved, and 100% conserved residues are marked with white, yellow, and red font, respectively. Conserved motifs are boxed with a red frame.
Prediction and comparison of the three-dimensional structure and active site
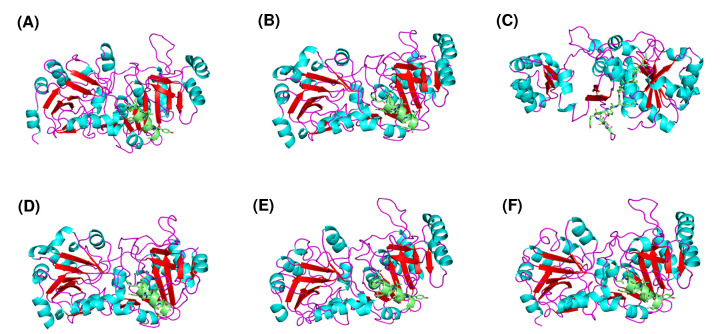
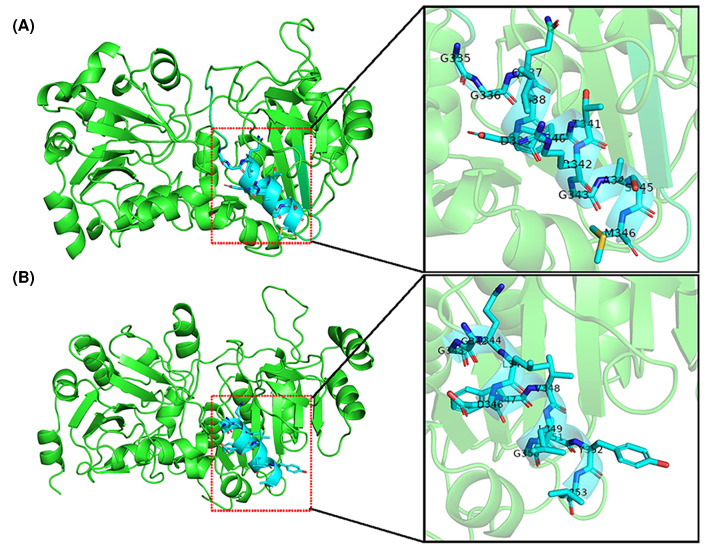
As shown in Figure 4A, the three-dimensional (3D) structure of CPB6-CoAT has one subunit that may consist of two main domains, resulting in a characteristic two-domain fold in a homotetrameric structure. A comparison of the 3D structures of the six CoAT proteins (Figure 4) showed that these CoATs shared similar conformations of their structural elements (α-helices and β-strands) with slight structural modifications in the loop regions and active centers, with the exception of the CoAT from M. elsdenii (Figure 4C). The 3D structure of the M. elsdenii protein was obviously different from that of other CoATs, and the divergences were located not only in the structural elements of α-helices and β-strands but also in the loops. This may be attributed to the distant genetic relationship between M. elsdenii and the other five bacteria. Although M. elsdenii produces caproic acid via acetyl-CoA and succinate [23], the functions of the CoATs may differ between strain CPB6 and M. elsdenii. The 3D structures of CoATs among C. tyrobutyricum BEY8, Lachnospiraceae bacterium, and A. hadrus shared almost the same conformation of α-helices and β-strands except for some slight variation in the loops (Figure 4D–F). The protein structure and active center structures were further compared between CPB6-CoAT and BEY8-CoAT, as shown in Figure 5, and both showed similar 3D structures except for the location and structure of the active center. The predicted active sites of the six CoATs are shown in Table 4. The predicted active center of the CPB6-CoAT protein was located between amino acids 342 and 353 (GGQLDFVLGAYL), while the active center of the BEY8-CoAT protein (GGQIDFTRGASM) was located at amino acids 335–346, and both active sites contained phenylalanine and tyrosine (Figure 5).
Figure 4. Predicted 3D structures of representative CoAT proteins.
3D structures of CoATs from Ruminococcaceae bacterium CPB6 (A), C. kluyveri (B), M. elsdenii (C), C. tyrobutyricum BEY8 (D), Lachnospiraceae bacterium (E), and A. hadrus (F). Helices of the catalytic domains, β-pleated sheets, loop regions, and active centers are colored sky blue, red, purple, and green, respectively.
Figure 5. Predicted 3D structures of the CoAT proteins (left) and the active center (right).
(A) BEY8-CoAT; (B) CPB6-CoAT.
Table 4. Prediction of the active sites of CoATs in different strains.
| Strain | Location of active site | Sequence of active site |
|---|---|---|
| Ruminococcaceae bacterium CPB6 (ARP50528.1) | 342–353 | GGQLDFVLGAYL |
| C. kluyveri (APM41307.1) | 335–346 | GGQVDFIRGANL |
| M. elsdenii (WP_036202574.1) | 356–367 | ADSYTYKKAPTL |
| C. tyrobutyricum BEY8 (WP_017752740.1) | 335–346 | GGQIDFTRGASM |
| Lachnospiraceae bacterium (HCI66479.1) | 340–351 | GGQLDFVLGAYK |
| A. hadrus (WP_044923342.1) | 342–353 | GGQLDFVMGAYL |
Site-directed mutagenesis
Site-directed mutagenesis was used to verify the active site of the proteins [38]. According to the predicted active center of CPB6-CoAT (GGQLDFVLGAYL, 342–353 aa) and compared with others (Table 4), site-directed mutagenesis targeting Asp346 and Ala351 was carried out to identify the effects of the two residues on the catalytic activity of CPB6-CoAT. Specifically, Asp346 was replaced by His and Ala351 was replaced by Pro via site-directed mutagenesis. The nucleotide substitutions were confirmed by Sanger sequencing of the DNA (Supplementary Figure S6). Enzyme assays showed that compared with wildtype CPB6-CoAT, the Asp346 substitution led to an approximately 76% loss of BCoAT activity and 72% loss of CCoAT activity, while the Ala351 substitution resulted in an almost 50% loss of BCoAT activity and 55% loss of CCoAT activity (Supplementary Figure S7). Statistical gap analysis of the above enzymatic experimental data (P-values <0.01) indicated a significant difference between them. The initial velocity of the reaction in different samples at different substrate concentrations can be seen in Supplementary Figure S8. Moreover, as shown in Table 3, the kcat/Km values for the D346H mutant with butyryl-CoA and caproyl-CoA (1.73 ± 0.1 and 5.66 ± 0.1 mM−1.min−1) and the A351P mutant (4.80 ± 0.2 and 12.8 ± 0.5 mM−1.min−1) were significantly lower than that for the wildtype CPB6-CoAT (10.8 ± 0.2 and 41.1 ± 0.2 mM−1.min−1), indicating that the Asp346 and Ala351 residues have significant effects on the enzyme activity, but the effect of Ala on the enzyme activity was lower than that of Asp.
Discussion
In the reverse β-oxidation pathway contributing to MCFA biosynthesis, BCoAT is required for butyrate biosynthesis in C. kluyveri [13] and C. tyrobutyricum [22]. This enzyme is responsible for the final step of butyrate production, catalyzing the conversion of butyryl-CoA and acetate into butyrate and releasing acetyl-CoA [35]. As reported in previous studies, this enzyme is considered to be a biomarker for identifying butyrate-producing bacteria [10,28,35] and may be involved in the conversion of caproyl-CoA into caproate in C. kluyveri, similar to the conversion of butyryl-CoA into butyrate [18]. In our present study, the BCoAT from C. tyrobutyricum only has activity for butyryl-CoA but has no activity for caproyl-CoA, suggesting that BCoAT in C. tyrobutyricum is not responsible for chain elongation of larger or higher carbon-numbered (>C5) fatty acids [22]. Moreover, the Km of BEY8-CoAT for butyryl-CoA (370 ± 4.1 µM) was obviously greater than that of CPB6-CoAT (537 ± 10 µM), indicating that BEY8-CoAT had a higher enzymatic affinity for butyryl-CoA than CPB6-CoAT. Similarly, the CoAT (PGN_0725) from Porphyromonas gingivalis [35,47] and CoAT from C. acetobutylicum ATCC 824 [10] both catalyze the conversion of butyryl-CoA into butyrate, with Km values of 520 ± 10 and 21.0 ± 0.1 µM, respectively. This indicates that BCoAT generally has higher affinity and catalytic activity for butyryl-CoA than CCoAT, while no BCoAT from butyric acid bacteria displayed affinity and catalytic activity for caproyl-CoA. These results suggest that BCoAT is only involved in chain elongation of C2–C4, not in that of C4 to C6 or C8.
Our previous study showed that the rate of caproate production with caproyl-CoA as the substrate in strain CPB6 was 3.5-times higher than that observed with butyryl-CoA as the substrate and suggested the existence of a CCoAT that specifically prefers caproyl-CoA instead of butyryl-CoA as the substrate [51]. In this study, CPB6-CoAT was confirmed for the first time to be a CCoAT responsible for the final step of caproate formation, although it has low BCoAT activity for butyryl-CoA. These data demonstrated the existence of a specific CCoAT involved in the chain elongation of MCFAs, which is significantly different from the function of BCoAT. The CPB6-CoAT protein catalyzed transferase reactions via a ternary-complex kinetic mechanism, whereas some other CoA transferases from Acidaminococcus fermentans [6], C. acetobutylicum ATCC 824 [10] and Clostridium propionicum [38], which belong to family I transferases, were reported to catalyze a transferase reaction via a ping-pong bi–bi mechanism. Thus, CPB6-CoAT was different from them in terms of substrate specificity and kinetic mechanism. The detailed mechanism underlying this functional difference needs to be further studied.
The structure of proteins plays an important role in their functional properties and catalytic efficiency [52]; for example, succinyl CoA:3-ketoate CoA transferase from pig heart [42] and 4-hydroxybutyrate CoA-transferase from Clostridium aminobutyricum [29] showed unexpected changes in protein modification and specific activity when their crystal structures changed. In the present study, a comparison of the 3D and active center structures showed similarities and differences between CPB6-CoAT and other CoATs (Figure 4 and Supplementary Figure S5), which may have affected enzyme catalytic function and activity. On the basis of these results, studying the functional differences caused by the structural changes in CoATs is of great significance. The exact structure and function of the active center of the CPB6-CoAT protein remains to be determined through subsequent comprehensive experiments and analysis. Additionally, site-directed mutagenesis showed that two residues (Asp346 and Ala351) in the conserved motif (GGQLDFVLGAYL, 342–353 aa) had significant effects on the enzymatic activity of CPB6-CoAT, but the effect of Ala351 on the enzyme activity was lower than that of Asp346. Generally, the exchange of Asp (an acidic amino acid) to His led to loss of a carboxy group and the introduction of two amidogens, while the replacement of Ala with Pro led to loss of an amidogen and the introduction of a carboxy group. Ala lacks a bulky side chain and therefore would likely not have any steric or electrostatic effects, and this change would not destroy the conformation of the main chain [7]. Differences in structures and properties among the sequences may be the reason for the differences in CoAT activity [33]. These results demonstrate that the conserved motif of CPB6-CoAT is directly linked to enzymatic activity. However, the effects of other residues on enzyme activity require further study to elucidate the function of the conserved motif in CPB6-CoAT. Clarification of the precise enzymatic mechanisms underlying enzyme binding of the butyryl-CoA or caproyl-CoA substrates might require crystallographic analyses.
In conclusion, these results confirmed the existence of a CCoAT involved in the production of caproic acid, and the enzyme is apparently different from the BCoAT responsible for the production of butyric acid. The present study improves our understanding of the metabolic reactions underlying chain elongation via the reverse β-oxidation pathway. However, determination of the detailed CCoAT structure and its function in MCFA biosynthesis require further study through protein crystallization and X-ray crystal structure analyses.
Supporting Information
The Supporting Information is available free of charge on the Publications website.
PCR of the CPB6-CoAT gene (encoding a CCoAT) and identification of the recombinant plasmid are shown in Supplementary Figure S1.
PCR of the BEY8-CoAT gene (encoding a BCoAT) and identification of the recombinant plasmid are shown in Supplementary Figure S2.
Western blot analysis of CPB6-CoAT (a CCoAT) and BEY8-CoAT (a BCoAT) is shown in Supplementary Figure S3.
The double-reciprocal enzyme kinetics (Lineweaver–Burk) plot is shown in Supplementary Figure S4.
The phylogenetic tree of CoATs from different strains is shown in Supplementary Figure S5.
The sequencing peak diagram of site-directed mutant and wildtype CPB6-CoAT is shown in Supplementary Figure S6.
The comparison of CoA-transferase activities (Mutant 1, D346H-mutant; Mutant 2, A351P-mutant) is shown in Supplementary Figure S7.
The initial velocity of the reaction in different samples at different substrate concentrations is shown in Supplementary Figure S8.
Supplementary Material
Acknowledgements
We would like to thank Dr. Su Dan for her advice and assistance with the present study.
Abbreviations
- BCoAT
butyryl-CoA:acetate CoA transferase
- CCoAT
caproyl-CoA:acetate CoA transferase
- CoAT
coenzyme A transferase
- k cat
catalytic constant
- LB
Luria broth
- MCFA
medium-chain fatty acid
- 3D
three-dimensional
Data Availability
All data generated or analyzed during the present study are included in this article and its supporting information.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the Natural Science Foundation of China [grant number 31770090]; the Sichuan Science and Technology Support Program [grant number 2021YJ0022] and the Open-Foundation Project of CAS Key Laboratory of Environmental and Applied Microbiology [grant number KLCAS-2017-01].
CRediT Author Contribution
Qingzhuoma Yang: Conceptualization, Resources, Data curation, Software, Formal analysis, Methodology, Writing—original draft, Writing—review and editing. Shengtao Guo: Conceptualization, Software, Formal analysis, Validation, Investigation. Qi Lu: Resources, Data curation, Validation, Visualization, Methodology. Yong Tao: Conceptualization, Data curation, Formal analysis, Funding acquisition, Project administration, Writing—review and editing. Decong Zheng: Conceptualization, Software, Visualization. Qinmao Zhou: Software, Investigation, Methodology. Jun Liu: Resources, Supervision, Methodology.
References
- 1.Angenent L.T., Richter H., Buckel W., Spirito C.M., Steinbusch K.J., Plugge C.M.et al. (2016) Chain elongation with reactor microbiomes: open-culture biotechnology to produce biochemicals. Environ. Sci. Technol. 50, 2796–2810 10.1021/acs.est.5b04847 [DOI] [PubMed] [Google Scholar]
- 2.Arnold K., Bordoli L., Kopp J. and Schwede T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 10.1093/bioinformatics/bti770 [DOI] [PubMed] [Google Scholar]
- 3.Bajaj J.S., Ridlon J.M., Hylemon P.B., Thacker L.R., Heuman D.M., Smith S.et al. (2012) Linkage of gut microbiome with cognition in hepatic encephalopathy. Am. J. Physiol. Gastrointest. Liver Physiol. 302, G168–G175 10.1152/ajpgi.00190.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bilen M., Mbogning M.D., Cadoret F., Dubourg G., Daoud Z., Fournier P.E.et al. (2017) ‘Pygmaiobacter massiliensis’ sp. nov., a new bacterium isolated from the human gut of a Pygmy woman. New Microbes New Infect. 16, 37–38 10.1016/j.nmni.2016.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bloess S., Beuel T., Kruger T., Sewald N., Dierks T. and Fischer von Mollard G. (2019) Expression, characterization, and site-specific covalent immobilization of an L-amino acid oxidase from the fungus Hebeloma cylindrosporum. Appl. Microbiol. Biotechnol. 103, 2229–2241 10.1007/s00253-018-09609-7 [DOI] [PubMed] [Google Scholar]
- 6.Buckel W., Dorn U. and Semmler R. (1981) Glutaconate CoA-Transferase from Acidaminococcus fermentans. Eur. J. Biochem. 118, 315–321 10.1111/j.1432-1033.1981.tb06404.x [DOI] [PubMed] [Google Scholar]
- 7.Cao J., Dang G., Li H., Li T., Yue Z., Li N.et al. (2015) Identification and characterization of lipase activity and immunogenicity of LipL from Mycobacterium tuberculosis. PLoS ONE 10, e0138151 10.1371/journal.pone.0138151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charrier C., Duncan G.J., Reid M.D., Rucklidge G.J., Henderson D., Young P.et al. (2006) A novel class of CoA-transferase involved in short-chain fatty acid metabolism in butyrate-producing human colonic bacteria. Microbiology 152, 179–185 10.1099/mic.0.28412-0 [DOI] [PubMed] [Google Scholar]
- 9.Cuff J.A. and Barton G.J. (2000) Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins 40, 502–511 [DOI] [PubMed] [Google Scholar]
- 10.Wiesenborn D.P., Rudolph F.B. and Papoutsakis E.T. (1989) Coenzyme A transferase from Clostridium acetobutylicum ATCC 824 and its role in the uptake of acids. Appl. Environ. Microbiol. 55, 323–329 10.1128/aem.55.2.323-329.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desbois A.P. (2012) Potential applications of antimicrobial fatty acids in medicine, agriculture and other industries. Recent Pat. Anti Infect. Drug Discov. 7, 111–122 10.2174/157489112801619728 [DOI] [PubMed] [Google Scholar]
- 12.Dowd S.E., Callaway T.R., Wolcott R.D., Sun Y., McKeehan T., Hagevoort R.G.et al. (2008) Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol. 8, 1–8 10.1186/1471-2180-8-125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duncan S.H., Barcenilla A., Stewart C.S., Pryde S.E. and Flint H.J. (2002) Acetate utilization and butyryl coenzyme A (CoA):acetate-CoA transferase in butyrate-producing bacteria from the human large intestine. Appl. Environ. Microbiol. 68, 5186–5190 10.1128/AEM.68.10.5186-5190.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edgar R.C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emms D.M. and Kelly S. (2015) OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16, 1–14 10.1186/s13059-015-0721-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.González-Cabaleiro R., Lema J.M., Rodríguez J. and Kleerebezem R. (2013) Linking thermodynamics and kinetics to assess pathway reversibility in anaerobic bioprocesses. Energy Environ. Sci. 6, 3780–3789 10.1039/c3ee42754d [DOI] [Google Scholar]
- 17.Heider J. (2001) A new family of CoA-transferases. FEBS 509, 345–349 10.1016/S0014-5793(01)03178-7 [DOI] [PubMed] [Google Scholar]
- 18.Seedorf H., Fricke W.F., Veith B., Bruggemann H., Liesegang H., Strittmatter A.et al. (2008) The genome of Clostridium kluyveri, a strict anaerobe with unique metabolic features. Proc. Natl. Acad. Sci. U.S.A. 105, 2128–2133 10.1073/pnas.0711093105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herman N.A., Li J., Bedi R., Turchi B., Liu X., Miller M.J.et al. (2017) Development of a high-efficiency transformation method and implementation of rational metabolic engineering for the industrial butanol hyperproducer Clostridium saccharoperbutylacetonicum Strain N1-4. Appl. Environ. Microbiol. 83, 02942–02916 10.1128/AEM.02942-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kenealy W.R., Cao Y. and Weimer P.J. (1995) Production of caproic acid by cocultures of ruminal cellulolytic bacteria and Clostridium kluyveri grown on cellulose and ethanol. Appl. Microbiol. Biotechnol. 44, 507–513 10.1007/BF00169952 [DOI] [PubMed] [Google Scholar]
- 21.Kucek L.A., Nguyen M. and Angenent L.T. (2016) Conversion of L-lactate into n-caproate by a continuously fed reactor microbiome. Water Res. 93, 163–171 10.1016/j.watres.2016.02.018 [DOI] [PubMed] [Google Scholar]
- 22.Lee J., Jang Y.S., Han M.J., Kim J.Y. and Lee S.Y. (2016) Deciphering Clostridium tyrobutyricum metabolism based on the whole-genome sequence and proteome analyses. mBio 7, 1–12 10.1128/mBio.00743-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee N.R., Lee C.H., Lee D.Y. and Park J.B. (2020) Genome-scale metabolic network reconstruction and in silico analysis of hexanoic acid producing Megasphaera elsdenii. Microorganisms 8, 1–12 10.3390/microorganisms8040539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li T.B., Zhao F.J., Liu Z., Jin Y., Liu Y., Pei X.Q.et al. (2019) Structure-guided engineering of ChKRED20 from Chryseobacterium sp. CA49 for asymmetric reduction of aryl ketoesters. Enzyme Microb. Technol. 125, 29–36 10.1016/j.enzmictec.2019.03.001 [DOI] [PubMed] [Google Scholar]
- 25.Lindenkamp N., Schurmann M. and Steinbuchel A. (2013) A propionate CoA-transferase of Ralstonia eutropha H16 with broad substrate specificity catalyzing the CoA thioester formation of various carboxylic acids. Appl. Microbiol. Biotechnol. 97, 7699–7709 10.1007/s00253-012-4624-9 [DOI] [PubMed] [Google Scholar]
- 26.Liu B., Kleinsteuber S., Centler F., Harms H. and Strauber H. (2020) Competition between butyrate fermenters and chain-elongating bacteria limits the efficiency of medium-chain carboxylate production. Front. Microbiol. 11, 1–13 10.3389/fmicb.2020.00336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Louis P. and Flint H.J. (2009) Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 294, 1–8 10.1111/j.1574-6968.2009.01514.x [DOI] [PubMed] [Google Scholar]
- 28.Louis P., Young P., Holtrop G. and Flint H.J. (2010) Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA:acetate CoA-transferase gene. Environ. Microbiol. 12, 304–314 10.1111/j.1462-2920.2009.02066.x [DOI] [PubMed] [Google Scholar]
- 29.Macieira S., Zhang J., Velarde M., Buckel W. and Messerschmidt A. (2009) Crystal structure of 4-hydroxybutyrate CoA-transferase from Clostridium aminobutyricum. Biol. Chem. 390, 1251–1263 10.1515/BC.2009.147 [DOI] [PubMed] [Google Scholar]
- 30.Makabe K., Hirota R., Shiono Y., Tanaka Y. and Koseki T. (2020) Biochemical and structural investigation of rutinosidase from Aspergillus oryzae. Appl. Environ. Microbiol. 87, 1–27 10.1128/AEM.02438-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marchler-Bauer A., Anderson J.B., Chitsaz F., Derbyshire M.K., DeWeese-Scott C., Fong J.H.et al. (2009) CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37, D205–D210 10.1093/nar/gkn845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marounek M., Fliegrova K. and Bartos S. (1989) Metabolism and some characteristics of ruminal strains of Megasphaera elsdenii. Appl. Environ. Microbiol. 55, 1570–1573 10.1128/aem.55.6.1570-1573.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park S.H., Kim S.J., Park S. and Kim H.K. (2019) Characterization of organic solvent-tolerant lipolytic enzyme from Marinobacter lipolyticus isolated from the Antarctic Ocean. Appl. Biochem. Biotechnol. 187, 1046–1060 10.1007/s12010-018-2865-5 [DOI] [PubMed] [Google Scholar]
- 34.San-Valero P., Abubackar H.N., Veiga M.C. and Kennes C. (2020) Effect of pH, yeast extract and inorganic carbon on chain elongation for hexanoic acid production. Bioresour. Technol. 300, 1–7 10.1016/j.biortech.2019.122659 [DOI] [PubMed] [Google Scholar]
- 35.Sato M., Yoshida Y., Nagano K., Hasegawa Y., Takebe J. and Yoshimura F. (2016) Three CoA transferases involved in the production of short chain fatty acids in Porphyromonas gingivalis. Front. Microbiol. 7, 1–13 10.3389/fmicb.2016.01146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scarborough M.J., Myers K.S., Donohue T.J. and Noguera D.R. (2020) Medium-chain fatty acid synthesis by “Candidatus Weimeria bifida” gen. nov., sp. nov., and “Candidatus Pseudoramibacter fermentans” sp. nov. Appl. Environ. Microbiol. 86, 1–19 10.1128/AEM.02242-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scherf U. and Buckel W. (1991) Purification and properties of4-hydroxybutyrate coenzyme a transferase from Clostridium aminobutyricum. Appl. Environ. Microbiol. 57, 2699–2702 10.1128/aem.57.9.2699-2702.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Selmer T., Willanzheimer A. and Hetzel M. (2002) Propionate CoA-transferase from Clostridium propionicum: cloning of the gene and identification of glutamate 324 at the active site. Eur. J. Biochem. 269, 372–380 10.1046/j.0014-2956.2001.02659.x [DOI] [PubMed] [Google Scholar]
- 39.Sitkin S. and Pokrotnieks J. (2019) Clinical potential of anti-inflammatory effects of Faecalibacterium prausnitzii and butyrate in inflammatory bowel disease. Inflamm. Bowel Dis. 25, e40–e41 10.1093/ibd/izy258 [DOI] [PubMed] [Google Scholar]
- 40.Spirito C.M., Richter H., Rabaey K., Stams A.J. and Angenent L.T. (2014) Chain elongation in anaerobic reactor microbiomes to recover resources from waste. Curr. Opin. Biotechnol. 27, 115–122 10.1016/j.copbio.2014.01.003 [DOI] [PubMed] [Google Scholar]
- 41.Stamatakis A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tammam S.D., Rochet J.-C. and Fraser M.E. (2007) Identification of the cysteine residue exposed by the conformational change in pig heart succinyl-CoA:3-ketoacid coenzyme A transferase on binding coenzyme A. Biochemistry 46, 10852–10863 10.1021/bi700828h [DOI] [PubMed] [Google Scholar]
- 43.Tamura K., Peterson D., Peterson N., Stecher G., Nei M. and Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jacob U., Mack M., Clausen T., Huber R., Buckel W. and Messerschmidt A. (1996) Glutaconate CoA-transferase from Acidaminococcus fermentans the crystal structure reveals homology with other CoA-transferases. Structure 5, 415–426 10.1016/S0969-2126(97)00198-6 [DOI] [PubMed] [Google Scholar]
- 45.Wang H., Li X., Wang Y., Tao Y., Lu S., Zhu X.et al. (2018) Improvement of n-caproic acid production with Ruminococcaceae bacterium CPB6: selection of electron acceptors and carbon sources and optimization of the culture medium. Microb. Cell Fact. 17, 99–108 10.1186/s12934-018-0946-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y., Li B., Dong H., Huang X., Chen R., Chen X.et al. (2018) Complete genome sequence of Clostridium kluyveri JZZ applied in Chinese strong-flavor liquor production. Curr. Microbiol. 75, 1429–1433 10.1007/s00284-018-1539-4 [DOI] [PubMed] [Google Scholar]
- 47.Yoshida Y., Sato M., Nagano K. and Hasegawa Y. (2015) Production of 4-hydroxybutyrate from succinate semialdehyde in butyrate biosynthesis in Porphyromonas gingivalis. Biochim. Biophys. Acta Gen. Subj. 1850, 2582–2591 10.1016/j.bbagen.2015.09.019 [DOI] [PubMed] [Google Scholar]
- 48.Tao Y., Zhu X., Wang H., Wang Y., Li X., Jin H.et al. (2017) Complete genome sequence of Ruminococcaceae bacterium CPB6: a newly isolated culture for efficient n-caproic acid production from lactate. J. Biotechnol. 259, 91–94 10.1016/j.jbiotec.2017.07.036 [DOI] [PubMed] [Google Scholar]
- 49.Zhang B., Bowman C. and Hackmann T.J. (2021) A new pathway for forming acetate and synthesizing ATP during fermentation in bacteria. Appl. Environ. Microbiol. 87, 02959–20 10.1101/2020.04.13.039867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu X., Tao Y., Liang C., Li X., Wei N., Zhang W.et al. (2015) The synthesis of n-caproate from lactate: a new efficient process for medium-chain carboxylates production. Sci. Rep. 5, 14360–14369 10.1038/srep14360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu X., Zhou Y., Wang Y., Wu T., Li X., Li D.et al. (2017) Production of high-concentration n-caproic acid from lactate through fermentation using a newly isolated Ruminococcaceae bacterium CPB6. Biotechnol. Biofuels 10, 102–114 10.1186/s13068-017-0788-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zoraghi R., See R.H., Gong H., Lian T., Swayze R., Finlay B.B.et al. (2010) Functional analysis, overexpression, and kinetic characterization of pyruvate kinase from methicillin-resistant Staphylococcus aureus. Biochemistry 49, 7733–7747 10.1021/bi100780t [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during the present study are included in this article and its supporting information.