Abstract
Transition metal homeostasis ensures that cells and organisms obtain sufficient metal to meet cellular demand while dispensing with any excess so as to avoid toxicity. In bacteria, zinc restriction induces the expression of one or more Zur (zinc-uptake repressor)-regulated Cluster of Orthologous Groups (COG) COG0523 proteins. COG0523 proteins encompass a poorly understood sub-family of G3E P-loop small GTPases, others of which are known to function as metallochaperones in the maturation of cobalamin (CoII) and NiII cofactor-containing metalloenzymes. Here, we use genomic enzymology tools to functionally analyse over 80 000 sequences that are evolutionarily related to Acinetobacter baumannii ZigA (Zur-inducible GTPase), a COG0523 protein and candidate zinc metallochaperone. These sequences segregate into distinct sequence similarity network (SSN) clusters, exemplified by the ZnII-Zur-regulated and FeIII-nitrile hydratase activator CxCC (C, Cys; X, any amino acid)-containing COG0523 proteins (SSN cluster 1), NiII-UreG (clusters 2, 8), CoII-CobW (cluster 4), and NiII-HypB (cluster 5). A total of five large clusters that comprise ≈ 25% of all sequences, including cluster 3 which harbors the only structurally characterized COG0523 protein, Escherichia coli YjiA, and many uncharacterized eukaryotic COG0523 proteins. We also establish that mycobacterial-specific protein Y (Mpy) recruitment factor (Mrf), which promotes ribosome hibernation in actinomycetes under conditions of ZnII starvation, segregates into a fifth SSN cluster (cluster 17). Mrf is a COG0523 paralog that lacks all GTP-binding determinants as well as the ZnII-coordinating Cys found in CxCC-containing COG0523 proteins. On the basis of this analysis, we discuss new perspectives on the COG0523 proteins as cellular reporters of widespread nutrient stress induced by ZnII limitation.
Keywords: metallochaperone, metalloproteome, nutritional immunity, metallostasis, zinc, cobW, ribosome remodeling
Graphical Abstract
Graphical Abstract.
A sequence similarity network (SSN) analysis is used to provide new perspectives on COG0523 metallochaperone structure and function.
Introduction
Late 3d-block, first-row metals from manganese (Mn) to zinc (Zn) play myriad and often highly specialized catalytic, structural, and regulatory roles in cellular physiology. Whether living in the soil or colonizing an infected host, all bacterial cells must acquire sufficient transition metal from the surrounding milieu to meet significant cellular demands, while avoiding toxicity associated with too much metal.1 More than 30% of all proteins encoded by a typical bacterial genome require transition metals to function.2 Bacterial transcriptional regulators ensure the bioavailable concentrations of transition metals to meet cellular metal needs, defined as sufficient metal to fill all metal binding sites of the metalloproteome.3 This allows, for example, a ZnII enzyme to access only the bioavailable pool of ZnII and not other noncognate metals. Some metalloenzymes, however, cannot assemble an active site spontaneously within the cellular milieu and require one or more accessory proteins to build the metallocenter. While the necessity for accessory proteins in the biogenesis of complex metallocenters like, for example the organometallic NiII-pincer nucleotide cofactor in Lactobacillus plantarum lactate racemase4 is easy to understand, it is not as clear for other enzymes, including urease which simply incorporates a binuclear NiII active center bridged by a carbamylated Lys residue.5 Nonetheless, both enzymes require accessory proteins for metallocofactor assembly and often require the activity of one or more dedicated metallochaperones.6
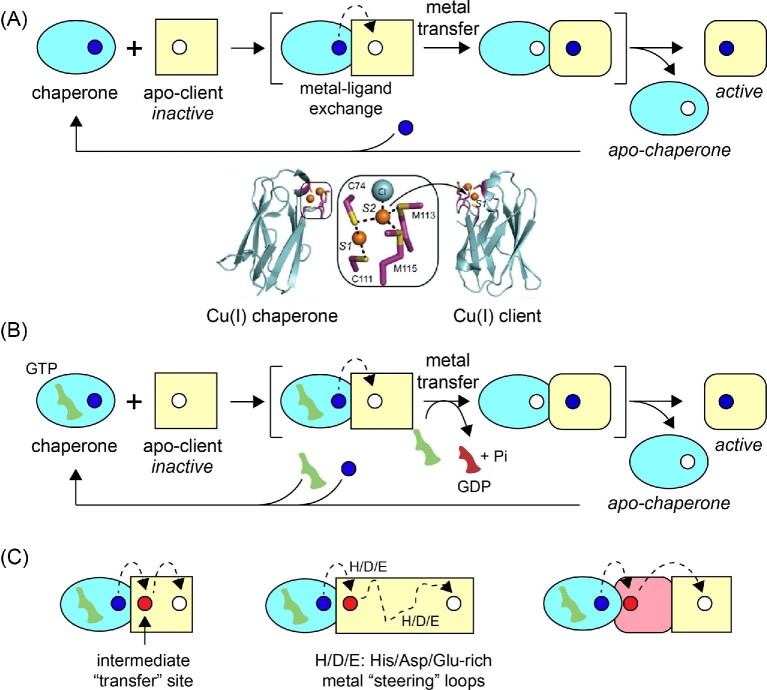
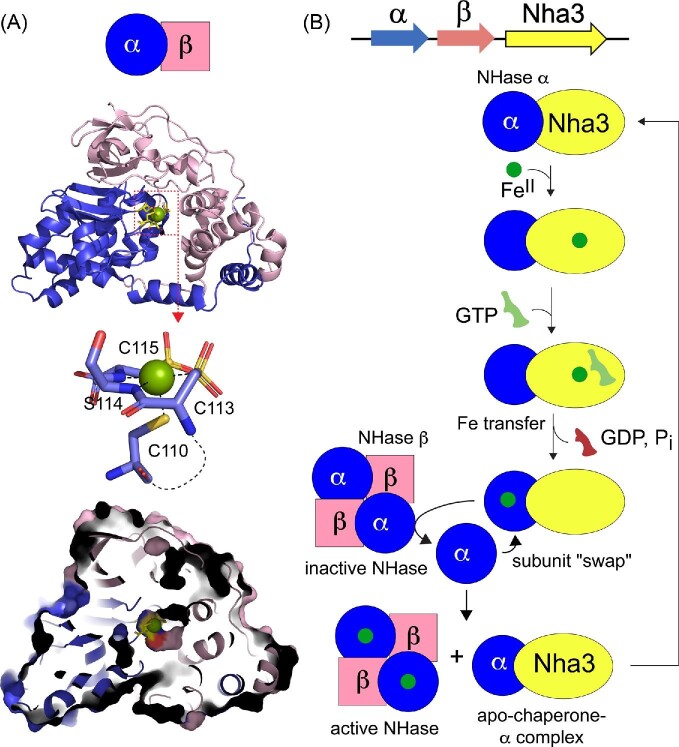
A metallochaperone is defined as a metal-binding protein that acquires a specific metal within the context of the cellular milieu, physically interacts with an unmetallated client or intermediary protein, and transfers the metal to the acceptor protein. The concept of a metallochaperone originated in the copper (Cu) homeostasis and resistance fields.7–9 Cu chaperones are generally small single domain proteins that bind CuI with high affinity, transiently dock with apo or metal-free client protein, and, via metal–ligand exchange and without dissociation into bulk solvent, transfer the metal to an acceptor protein down a shallow thermodynamic gradient, regenerating the apo-chaperone and a metalated client (Fig. 1A).10,11 In the case of bacterial Cu resistance, the chaperone and the client adopt isostructural folds of opposite electrostatic potential and form a specific, yet transient protein-protein complex; the ‘client’ is an appended domain of a CuI efflux pump, which removes the CuI from the cytoplasm.10,12 A physiologically distinct, yet mechanistically analogous model of chaperone-dependent enzyme metalation underpins the maturation of superoxide dismutase in eukaryotic cells, which is required only under conditions of intracellular Cu limitation.13 Here, a metallochaperone can be expected to traffic a specific metal to an acceptor as a result of highly specific coordination chemistry and protein–protein interactions but becomes dispensable under conditions where the cognate metal is replete in the cell.13
Fig. 1.
Mechanistic metallochaperone models. (A) Ligand exchange-driven model of metallochaperone function as shown for CupA CuI delivery to CopA, where ligand exchange between metallochaperone and client protein is proposed to drive metal transfer.10,12 (B) GTPase- or nucleotide exchange-driven model of metallochaperone function as proposed for COG0523 metallochaperones, where GTP hydrolysis induces a conformational change that allows for intermolecular transfer of metal from metallochaperone to client protein.28 (C) Mechanisms for COG0523-mediated metal transfer to metal binding sites that are not readily accessible to the bulk solvent. Metal is first delivered to an intermediate site prior to moving down a thermodynamic gradient to the active site (left). Metal is transiently passed through loops containing stretches of His/Asp/Glu residues before reaching an active site (center). Metal is delivered first to an accessory protein, before moving to the active site of the client (right).
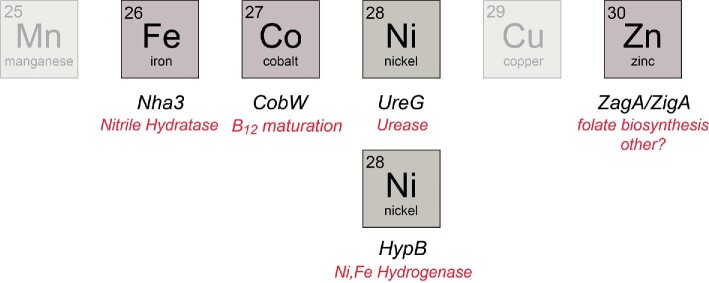
One important group of metallochaperones is the P-loop G3E GTPase family, which is named for a D-to-E substitution relative to a canonical P-loop GTPase, e.g. human Ras and is thus denoted G3E.14 Rather than relying solely on metal–ligand exchange like CuI chaperones, the G3E family is thought to utilize GTP hydrolysis to thermodynamically drive and/or regulate metallocofactor assembly (Fig. 1B). These are known to include subfamilies for the NiII chaperone HypB, which functions in the pathway of NiFe-hydrogenase cofactor assembly,15–17 UreG, the NiII chaperone required for the assembly of the binuclear NiII site in urease,18,19 and MeaB (CblA20), the cobalamin (vitamin B12) chaperone for methylmalonyl-coenzyme A mutase (Fig. 2).21,22 Although all are metallochaperones, the MeaB subfamily is not considered further here because it functions to gate CoIII-corrinoid cofactor, 5′-deoxyadenosylcobalamin, to an apoprotein client, rather than inserting the free metal itself.21 G3E GTPases have five signature G (GTP-binding) loops required for GTP binding and hydrolysis. G1 consists of the Walker A GxGKTT motif or P-loop that binds the β-phosphate group of GTP.15,23 G2 comprises Switch 1, which interacts with the γ-phosphate and is responsible for molecular switching upon GTP hydrolysis. The G3 or Walker B (Switch 2) loop harbors the D-to-E substitution that coordinates the MgII in the MgII-GTP substrate, while G4 and G5 are guanine binding motifs that dictate GTP specificity (Fig. 3A). Mechanistically, UreG and HypB operate through nucleotide- or metal-inducible oligomers, interactions with various accessory proteins, and a subunit-bridging metal binding site, to fulfill GTP hydrolysis-dependent metallochaperone function.24–27 The Cluster of Orthologous Groups (COG) COG0523 subfamily, defined by the conserved CxCC (C, Cys; x, any amino acid) primary structure motif responsible for high affinity metal binding,28,29 comprises the last known subfamily of the G3E P-loop superfamily, and is far less understood.
Fig. 2.
Known G3E P-loop metallochaperones and client proteins assigned by cognate, late d-block transition metals. See text for details.
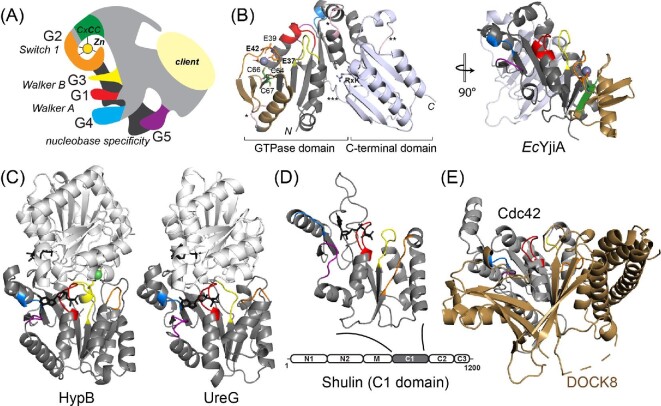
Fig. 3.
COG0523 structural models. (A) ‘Elephant’ representation of COG0523 metallochaperones with G-loops and nucleotide binding and hydrolysis motifs highlighted.1 The variable C-terminal domain is thought to harbor sequence-specific determinants for specific interactions with a client protein(s). (B) The only metal-bound COG0523 structure, of E. coli YjiA (PDB: 4ixm). The GTPase and C-terminal domains are indicated and the G-loops are shaded as in panel A. The coordination structure of the high affinity, regulatory ZnII site is shown, proposed to involve E37, E42, and C66; the side chain of E39, also highlighted, appears to point away from the metal, but could not be modeled past Cβ.29 This side chain also points away from the metal binding cleft in the apo-YjiA structure.63 This metal site localizes to the G2 loop and the β-meander63 (shaded gold) defines an insertion in a canonical G3E P-loop GTPase fold (right). (C) Ribbon structures of Helicobacter pylori HypB (4lps)24 and H. pylori UreG (4hi0)112 dimers (one subunit shaded gray, the other white) with the G-loops in the gray protomer shaded as in panels A and B in the same orientation as EcYjiA, (panel B, right). GTP analog is shown in black stick. (D) Structure of Tetrahymena Shulin C1 domain illustrating structural similarity to G3E P-loop GTPases;81 however, sequence similarity to EcYjiA is undetectable. (E) Ribbon representation of the vertebrate Cdc42-DOCK8 complex (3vh1), with Cdc42 shaded gray and G-loops colored as above, and DOCK8, a Cdc42-specific guanine nucleotide exchange factor, (GEF) shaded gold.65
COG0523 proteins as candidate metallochaperones
An initial phylogenetic analysis of COG0523 proteins showed that many bacterial representatives were expressed under the transcriptional control of Zur (ZnII uptake repressor) and derepressed under conditions of low ZnII, although many were not.30 That work made the prediction that these proteins may function as zinc metallochaperones since they are evolutionarily related to other small P-loop G3E GTPases, e.g. UreG or HypB, that while not COG0523 proteins, are clearly evolutionarily related (vide infra), and are known to function in the metallocofactor assembly and maturation in NiII-containing enzymes (see Fig. 2). However, the first COG0523 protein to be discovered was associated with cobalamin (vitamin B12) maturation, from Pseudomonas denitrificans, denoted CobW.31 The result of this discovery was that the designation CobW came to represent the entire COG0523 superfamily and many have been misannotated as CobW or CobW-domains although they are not involved in cobalamin biosynthesis.32 Rhodobacter capsulatus CobW has only recently been biochemically characterized as a putative CoII chaperone for a single known client enzyme, the multi-subunit cobalt chelatase and ATPase (CobNST) which inserts CoII into the corrinoid ring in the aerobic cobalamin biosynthesis pathway (Fig. 2).33,34 Another member of the COG0523 family of candidate metallochaperones is the FeIII nitrile hydratase (NHase) activator protein, exemplified by Nha3 from Rhodococcus equi or NhpC/P47K from Pseudomonas chlororaphis.35,36 It has recently been established that GTPase activity of NhpC in P. chlororaphis is required to incorporate the FeIII into the α2β2 nitrile hydratase heterotetramer in cells.36
In contrast to CobW and Nha3, specific client or target proteins for virtually all other COG0523 proteins are unknown. While many bacterial COG0523 proteins have been linked to ZnII enzyme maturation due to their connection to Zur regulation, only Acinetobacter baumannii ZigA and Bacillus subtilis ZagA have been studied in any detail.37–39 Nonetheless, physiological ZnII limitation would be expected to create conditions that require ZnII metallochaperone-mediated ZnII delivery to specific undermetallated enzyme(s), analogous to the CCS-dependence of superoxide dismutase metalation only under conditions of low Cu.13 This happens during infections, where bacterial pathogens are exposed to host defenses designed to limit the availability of transition metals in an effort to broadly cripple cellular metabolism, in a process known as nutritional immunity.40 The calcium-activated transition-metal chelating protein, calprotectin (CP), is a major component of metal-withholding during infections.41,42
Metalloproteome status under conditions of transition metal restriction
When transition metal bioavailability becomes restricted, it is generally believed that cells are no longer capable of fully metalating their metalloproteome.3 Experiments in B. subtilis and Streptomyces coelicolor reveal that bacterial cells effect a ‘graded’ response to chronic ZnII limitation induced by the cell permeable metal chelator TPEN.43,44 This response appears orchestrated by distinct metallostates of the zinc uptake repressor (Zur) that become differentially populated as the bioavailable ZnII falls. Although the details differ in the two organisms, a common feature is the upregulation of non-ZnII-requiring ribosomal protein paralogs, known as ‘C—’ ribosomal proteins since they lack the cysteine residues that coordinate the structural ZnII in the bona fide C + ribosomal subunits.45,46 This is a specific aspect of a more general process, termed ZnII sparing. Early-to-intermediate phases of ZnII limitation involve the expression of C—paralogs of ribosomal proteins that are added late in the assembly process, e.g. L31 or L33 which are replaced by L31B and L33B, respectively, such that ribosomes are not required to be completely disassembled to incorporate these C—subunits.43,47 This substitution results in intracellular ZnII mobilization. Later stages of the ZnII starvation response involving mobilization of ribosomal ZnII stores also occurs with other C—paralogs, such as S14B, that are incorporated into ribosomes early in the assembly process and thus are needed for de novo ribosome synthesis under conditions of chronic ZnII deficiency.48,49 Some bacteria, such as A. baumannii, contain only a single C—ribosomal protein paralog, which is typically L31B, while many Actinomycete spp., including the causative agent of tuberculosis, Mycobacterium tuberculosis, encode five C—ribosomal protein paralogs which are found throughout the 70S ribosome structure.46 This allows re-allocation of a scarce resource, ZnII, to obligatory zinc metalloenzymes without compromising translation or engaging direct competition with calprotectin on the outside of the cell.49
During or following mobilization of intracellular ZnII from the ribosome, bacterial cells upregulate the expression of high affinity transport systems to acquire extracellular ZnII.50,51 Moreover, metallophores, analogous to siderophores, are synthesized and sent out into the extracellular milieu where they bind ZnII and are taken up as ZnII-metallophore complexes by dedicated uptake systems.52–55 Metabolic adaptation is typically a later-stage response and occurs through additional mechanisms of ZnII sparing beyond ribosome remodeling, and through ZnII prioritization. ZnII sparing is a process where ‘metal-promiscuous’ or metal-independent paralogs of obligatory ZnII enzymes and proteins are expressed, often as part of the Zur regulon; these enzymes are thought to be active with another metal(s) or no metal at all, respectively, and thus can be used to sustain a specific metabolic pathway upon failure of the obligatory ZnII enzyme under these conditions. In ZnII prioritization, a Zur-regulated COG0523 metallochaperone directs or ‘prioritizes’ that ZnII to a specific ZnII metalloenzyme client to sustain an otherwise failing metabolic pathway(s) (see below). In our view, ZnII sparing and ZnII prioritization are not necessarily mutually exclusive and can be used by the cell to target the same metabolic step, e.g. in the rate-determining step of de novo folate biosynthesis catalyzed by two GTP cyclohydrolases IA (FolE, strictly ZnII-dependent) and IB (FolEB; metal-promiscuous56,57). Many ZnII-independent or metal-promiscuous paralogs have been identified in Zur regulons in various bacteria and include DksA2, a global transcriptional regulator that functions under conditions of nutrient limitation (see below), and enzymes that catalyze key, often committed, steps in queuosine biosynthesis (QueD2), pyrimidine biosynthesis (PyrC2), and tetrapyrrole biosynthesis (porphobilinogen synthase, HemB2),30,58,59 as well as the third biosynthetic step in histidine biosynthesis, catalyzed by phosphoribosyl-AMP cyclohydrolase (HisI2).60 It is important to emphasize that aside from FolEB and HemB2, the metal specificity and other potential factors that endow these so-designated ‘metal-promiscuous’ or ‘ZnII-independent’ enzyme paralogs with significant catalytic activity under conditions of extreme metal restriction have not been systematically investigated.
COG0523 GTPases are proposed to function in metallocofactor assembly and maturation
Bacillus subtilis ZagA (ZTP-activated GTPase A)39,61 and A. baumannii ZigA (Zur-induced GTPase A)37,38,62 are recently characterized Zur-regulated COG0523 proteins, and we and others have postulated that they function in metalating a metalloprotein target(s) under conditions of extreme ZnII and/or FeII restriction by functioning as metallochaperones. Acinetobacter baumannii ZigA has been described as playing a role in cytoplasmic zinc mobilization in the activation of the histidine catabolic pathway, while also playing a role in sustaining flavin biosynthesis and perhaps other metabolic pathways under conditions of CP-mediated metal restriction.37,38 In striking contrast, ZagA appears to function to sustain folate biosynthesis under conditions of EDTA-mediated metal restriction in B. subtilis.39 ZagA senses and hydrolyzes the cellular alarmone ZTP, which accumulates under conditions of purine deficiency.39 In so doing, ZagA sustains the activity of GTP cyclohydrolase IA (FolE), which catalyzes the rate-determining step in de novo folate biosynthesis. This activation is postulated to occur via a physical interaction between the two proteins although molecular-level insights remain lacking.39 In contrast, Cupriavidus metallidurans CH34 CobW1, closely related to ZigA/ZagA (see below) has been reported to interact with CmFolEB, rather than FolE, although the physiological importance of this interaction was not demonstrated.32 Interestingly, A. baumannii ZigA is unable to function in place of B. subtilis ZagA in sustaining folate biosynthesis in B. subtilis, consistent with a non-overlapping specificity in client protein recognition by ZigA.39 Acinetobacter baumannii and Staphylococcus aureus ZigAs each bind one ZnII with high affinity (KZn ≥ 1011 M–1) to a Cys-containing site, and ZnII coordination stimulates GTP hydrolysis by ≈5-fold.28 A mechanistic framework for ZnII chaperone activity has been developed, the essential tenets of which are a kinetically ‘locked’ GTP and ZnII bound conformation i.e. poised to interact with an apo-client to deliver the metal cargo.28
Structural insights into COG0523 proteins from Escherichia coli YjiA
There is as yet no physical evidence that any COG0523 protein inserts a metal into an active site of a client protein, directly or indirectly. Furthermore, only one structure is available, of E. coli YjiA (Fig. 3),29,63 a protein of unknown function that is unlikely to capture what is anticipated to be a wide range of biological activities ascribed to individual COG0523 proteins in metallocenter maturation. A cartoon representation of a COG0523 protein depicts the five family-defining G loops, labeled G1–G5 from the N-terminus, that collectively define nucleotide specificity and hydrolysis determinants required for the hydrolysis of a nucleotide triphosphate, thought to be GTP (Fig. 3A). The overall structure of the ZnII-bound, nucleotide free YjiA protomer (YjiA is dimeric, with at least one subunit-bridging ZnII site) is shown in Fig. 3B and reveals a canonical N-terminal small GTPase domain of ≈ 230 residues that is connected to a highly variable α/β C-terminal domain by a long linker between the α9 (GTPase) and β10 (C-terminal domain) secondary structural elements.29 His/Asp/Glu-rich insertions, if present, tend to be found here or between the α5 and α6 helical elements of the GTPase domain itself (Fig. 3B). The variable sequence of the C-terminal domain suggests a possible role in client protein recognition. The only conserved distinguishing feature of the C-terminal domain that is common to all GOG0523 proteins is an RxK motif between β11 and α10 in E. coli YjiA that appears to make salt bridges to two conserved Asp/Glu residues, one positioned at the base of the α6 helix, and one in the middle of the long connector sequence;29 these may serve as ‘transducers’ of client protein docking and activation of GTP hydrolysis.
The primary ZnII site in E. coli YjiA specifically engages the second Cys (C66) in the signature CxCC motif located between loops G2 and G3 in the short β5 strand. The immediately adjacent C64 and C67 are shielded from solvent in this structure. The remaining two ligands in the high affinity ZnII site, E37 and E42, are coincident with the G2 (Switch 1) motif found between the β2 and β3 strands, which provides an explanation for allosteric activation of GTPase activity by ZnII observed by a number of groups.28,29,64 We find that E37 is poorly conserved, and E42 only somewhat better conserved, suggesting that this coordination model may not characterize all COG0523 proteins (vide infra). Another acidic residue, E39, is near the bound ZnII but its side chain points away from the metal and could not be modeled beyond the Cβ carbon in this structure (Fig. 3B); further, this residue has been suggested to function in MgII coordination of the MgII•GTP substrate, thus physically linking the two metal sites.29 Consistent with an important catalytic role, E39 is conserved as a Glu/Asp residue (vide infra). Importantly, this ZnII site is essentially solvent-exposed, at the protein surface, coordinatively unsaturated (with just three protein-derived ligands), and not well ordered in this particular structure.29 These are features that one might expect to see in a metallochaperone site mediated by ligand exchange or by conformational changes in the surrounding G loops, that may well alter the ZnII coordination sphere in different nucleotide or client-bound states. The three-stranded β3-β4-β5 β-meander harboring the metal liganding Cys defines a unique insertion into the canonical G3E P-loop fold and occupies space where a guanine nucleotide exchange factor (GEF) docks to mammalian Ras and mediates exchange of product GDP for substrate GTP (Fig. 3C, right).65 In bacterial ZigAs, ZnII allosterically lowers the affinity for product GDP and thus may well function in part as a GEF to facilitate nucleotide exchange during multiple catalytic turnovers.28 This β-meander insert is precisely what distinguishes COG0523 proteins from other PF02492 proteins discussed here, UreG and HypB (Fig. 3C). Remarkably, the YjiA structure provides no insights into the strict conservation of the other two Cys in the CxCC motif of COG0523 proteins; we favor as yet untested model that these Cys function in the metal-ligand exchange process following GTP hydrolysis to facilitate transfer of the metal bound here to an acceptor site via ligand exchange (Fig. 1B).
This ligand exchange reaction can be direct, from the metallochaperone site to the client protein active site, or perhaps more likely, to an intermediate site at the protein surface or on a ligand-exchange ‘highway’ in which the metal ultimately moves down a thermodynamic gradient to the client protein active site (Fig. 1C). This line of thinking is consistent with what is known about HypB and UreG, where metals are delivered to an intermediary protein(s), which ultimately donates metal to assemble the enzyme cofactor on the apo-enzyme (Fig. 1C).18,25,27,66 We further suggest that these His/Asp/Glu (H/D/E)-rich insertions that are found in many bacterial G3E P-loop GTPases (see below) function not as metal sinks,32 but to retain the cognate metal in structurally heterogeneous, highly dynamic and coordinately unsaturated exchange-labile complexes, analogous to electrostatic ‘steering’ used to retain substrates in multi-enzyme metabolons, without dissociation of the metal into bulk solvent.67,68 The two-domain solute binding protein, AdcA, of the ATP binding cassette (ABC) transporter in Gram-positive bacteria may likewise employ a His-rich sequence that caps the ZnuA metal site between component ZinT and ZnuA domains;69 intermolecular ZnII transfer in the periplasm of some Gram-negative bacteria may also involve exchange via transfer intermediates.70 In these cases, nature seems to leverage a key characteristic of first row, late d-block divalent metals to form thermodynamically stable, yet kinetically exchange-labile, complexes with biological ligands.71 Solution structural snapshots of various ligand-bound states35 and mechanistic intermediates are clearly needed to shed more light on COG0523 function.
Sequence similarity network (SSN) analysis of G3E P-loop GTPases
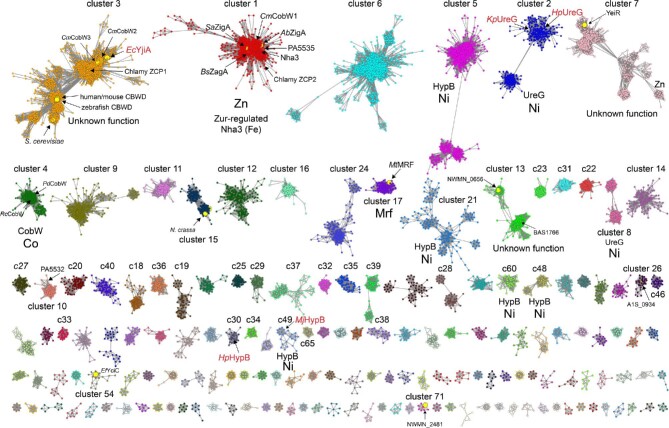
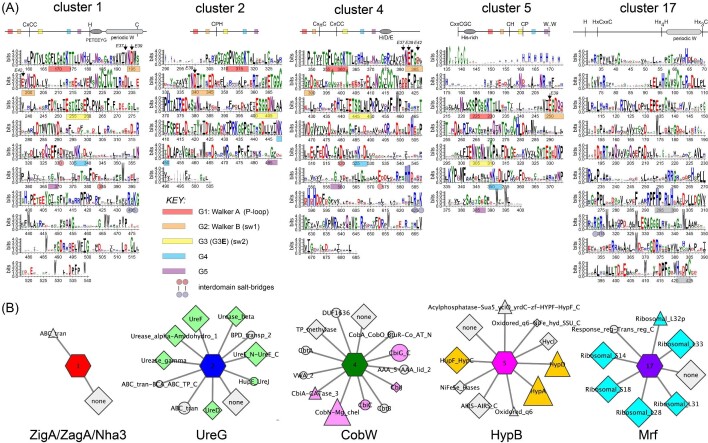
We used genomic enzymology (GE) tools72,73 to segregate over 80 000 protein sequences in an effort to gain better insights into COG0523 structure and physiological function (Fig. 4). These sequences originated from a sequence BLAST of A. baumannii ZigA and include protein sequences that range from 100 to 650 amino acids (see Supplementary methods). We analysed the top 40 SSN clusters (labeled 1–40 and ranked according to number of sequences), which correspond to 76 928 sequences or ≈95% of total sequences, or 26 688 sequences that are 90% identical over 80% of the sequence (Figs. S1 and S2; Tables S1 and S2). We also discuss selected smaller clusters beyond these top 40 that include COG0523 proteins previously discussed in the literature. Clusters 1, 2, and 3 comprise ≈ 46% of all sequences, and we further suggest that, along with MeaB, clusters 1, 2, 4, ,5 and 17 comprise the six major functionally characterized groupings of all P-loop G3E enzymes (see Fig. 5).
Fig. 4.
SSN of G3E P-loop GTPases assembled from Pfam families PF02492 (cobw) and PF07683 (cobw_c) using A. baumannii ZigA37 as query (see Supplementary Methods). Each node corresponds to proteins with >60% sequence identity over >80% sequence coverage and individual clusters (arranged according to the number of nodes, from left to right, from top to bottom) are separated and colored accordingly. Cluster numbers are ranked according to the sequence cluster count (see Fig. S1 and Table S1) with SSN cluster 1 containing the largest number of sequences. Characterized proteins or genes that encode the indicated protein are highlighted within each cluster, with known functions assigned to each cluster, along with the corresponding cognate metal. Those nodes that harbor known protein structures are highlighted in red text (see Fig. S3 for additional information). The small node grouping in cluster 7 (YeiR) marked ‘Zn’ are derived from candidate Zur-regulated operons in β-proteobacteria. Only SSN clusters with larger than seven (7) nodes are shown here for clarity (see Tables S1 and S2 for a complete listing of all clusters and singletons, and associated uniprot identifiers associated with each).
Fig. 5.
Sequence and genomic neighborhood determinants of SSN clusters 1 (ZigA/ZagA/Nha3), 2 (UreG), 4 (CobW), 5 (HypB), and 17 (Mrf). (A) WebLogo schematic cartoon representations (top) and corresponding plots (bottom) of all sequences contained within each cluster (see Table S3 for aligned sequences). G-loops and the interdomain salt bridges are highlighted as indicated; G5 assignments are typically tentative. The residue positions corresponding to EcYjiA acidic residues E37, E39, and E42 in loop G2 are highlighted (arrows) revealing a general lack of strict conservation of YjiA ZnII ligands E37 and E42 across the dataset. (B) GNN for each cluster shown in a ‘wheel’ and ‘spoke’ representation.73 The wheel is identified by sequence cluster number and colored as in Fig. 4, while spokes represent neighboring genes (±10 genes around the target gene) found with a co-occurrence frequency of ≥ 20% in all genomes within which that particular wheel appears. Neighboring genes are sized by co-occurrence frequency and selected genes colored according to function: Zinc homeostasis (light purple), urease maturation (forest green), NiFe-hydrogenase maturation (yellow-orange), ribosomal subunits (cyan), and cobalamin maturation (magenta).
Cluster 1 appears dominated by bacterial Zur-regulated COG0523 proteins closely related to A. baumannii ZigA,37 S. aureus ZigA,28 B. subtilis ZagA,39 Acinetobacter baylii Zur-regulated ZagA homolog (ACIAD1714),30,39 and Cupriavidus metallidurans CobW1, but also contains subclusters of nodes from lower eukaryotes (Fig. S3).32 Interestingly, cluster 1 also harbors the FeIII nitrile hydratase activator protein Nha3/NhpC, consistent with high pairwise similarity to ZigA.28 Both the Zur-regulated cluster 1 proteins and Nha3 bind ZnII or FeII at the CxCC motif (A. baumannii ZigA notably binds FeII very weakly, KFe ≤ 104 M–1), but it is not yet possible to distinguish one from the other on the basis of sequence analysis alone.28,35 Cluster 2 is the largest collection of NiII UreG sequences, which are also found in clusters 8, 19, 28, and 44 and conserve the NiII binding CPH sequence positioned between the G2 and G3 (Switch 2) loops (Fig. S4). Interestingly, the sequences in the sub-cluster shown at the lower left of cluster 2 are from eukaryotes such as fungi, algae, and higher plants, which are all known to encode urease (Fig. S3).5,74 Cluster 4 comprises all known authentic CobW proteins, which harbor an invariant C-x2-4-C pair just N-terminal to the CxCC motif in the anticipated β-meander, and likely donates thiolate ligands to the CoII ion in R. capsulatus CobW.34 Other G3E P-loop proteins that are genomically linked to genes involved in cobalamin maturation or delivery are only distantly related (cluster 35) or entirely unrelated (cluster 46) to authentic CobW on the basis of sequence (Fig. S5); this tight clustering of authentic CobW sequences is consistent with a single metalloenzyme client in most organisms. Cluster 5 represents that largest collection of NiII HypB sequences, which conserves the N-terminal CGC cluster and the CH motif between the G2 and G3 loops (as in UreG) and a more distal Cys residue in G3, one or both of which play a role in NiFe hydrogenase maturation depending on the bacterial species.75 Other HypB-containing clusters include clusters 14, 21, 25, 29, 30, 34, and 48 based both on sequence conservation and genomic neighborhood network (GNN) analysis (Fig. S4).
We identified a previously undocumented functional grouping of COG0523-related sequences in cluster 17, represented by mycobacterial-specific protein Y (Mpy) recruitment factor (MtMRF, Rv0106; Fig. 4). Mrf is reported to load Mpy onto the 70S ribosome at the decoding center to create a ‘hibernating’ or inhibited ribosome under conditions of ZnII restriction,46,76 although this finding is somewhat controversial.77 While the mechanism of Mpy loading is unclear, Mrf interacts with the ribosome, primarily with the 30S subunit. Mrf is Zur-regulated in M. tuberculosis (Rv0106),78 and a GNN analysis reveals that Mrf genomically groups with genes encoding C—ribosomal protein paralogs (Figs. 5 and S6). Remarkably, the signature G-loops of G3E and COG0523 proteins are entirely lost in Mrf, with no recognizable Walker A or Walker B sequence (Fig. 5). In addition, the metal coordinating Cys of the CxCC motif29 is not conserved (Fig. 5). Despite the loss of the liganding Cys, recent work reveals that Zur-regulated M. smegmatis Mrf binds ZnII; this in turn leads to rapid proteolysis by the Clp system thus preventing Mrf from loading constitutively expressed Mpy onto the ribosome under ZnII-replete conditions.76 ZnII ligands are proposed to localize to the GTPase domain since a mutant Mrf harboring six Ala substitutions of four His and two Cys in the remaining CxxC motif, denoted Mrf*, appears to bind ZnII more weakly; this in turn leads to a weaker interaction with ClpS and decreased rates of cellular clearance by the Clp system.46 We suggest the alternative possibility that several conserved residues in the Mrf C-terminal domain may be involved in regulatory ZnII binding, including a Hx4H sequence and a Hx3C motif near the C-terminus (Fig. 5). Clusters 20 and 55 also comprise Mrf proteins, from distantly related actinomycetes, including Streptomyces spp. (cluster 20) and Actinoplanes and Glycomyces spp. (cluster 55; Fig. S6). Both cluster 20 and cluster 55 proteins, like authentic Mrfs, have lost the G-loops, but a subset of cluster 20 proteins retain the COG0523-liganding Cys residue. These findings with Mrf raise the intriguing possibility that bona fide COG0523 proteins beyond Mrf interact with the ribosome.
Other SSN clusters of COG0523 proteins
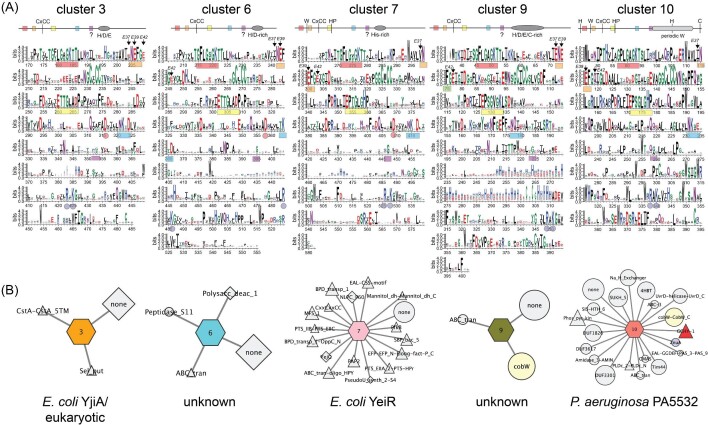
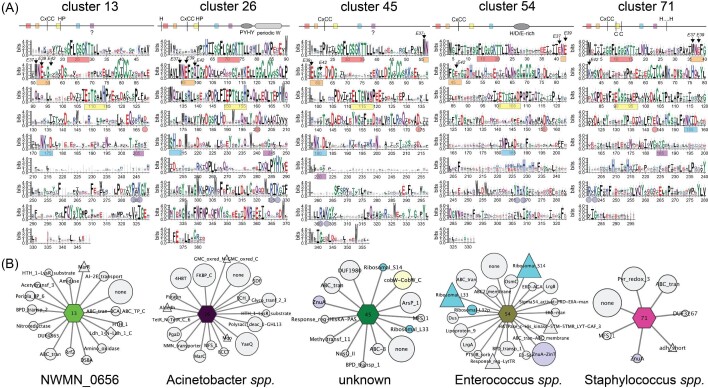
Ten additional SSN clusters comprise functionally uncharacterized CxCC COG0523 proteins that may well play distinct roles in cellular metabolism (Figs 6 and 7). These include 5 of the 10 largest clusters, 3, 6, 7, 9, and 10 (Fig. 6), as well as clusters 13, 26, 45, 54, and 71 (Fig. 7), discussed here because representatives of these clusters have been previously described in the literature. A comparison of these sequences, alongside cluster 1 (Fig. 5) reveals a remarkable degree of sequence diversity in the C-terminal domains superimposed on only short stretches of high sequence conservation within a cluster, e.g., the PETEEYG motif found in cluster 1 proteins (Fig. 5). Furthermore, inspection of the GNN diagrams associated with COG0523 proteins in clusters 1, 3, 6, and 9 provide virtually no clues as to physiological function, in striking contrast to UreG, CobW, and HypB metallochaperones (Figs. 5 and 6).
Fig. 6.
Sequence and genomic neighborhood determinants of SSN clusters 3 (E. coli YjiA and many eukaryotic COG0523 proteins; Fig. S3), 6 (unknown function), 7 (E. coli YeiR64), 9 (unknown function), and 10 (P. aeruginosa PA553258). (A) WebLogo schematic cartoon representations (top) and corresponding plots (bottom) of all sequences contained within each cluster (see Table S3 for aligned sequences). P-loops, interdomain salt bridges and positions of EcYjiA ZnII ligands E37 and E42 are highlighted as in Fig. 5A. (B) GNN for each cluster shown in a ‘wheel’ and ‘spoke’ representation.73 The wheel is identified by sequence cluster number and colored as in Fig. 4, while spokes represent neighboring genes found, sized, and colored according to function as described in Fig. 5B.
Fig. 7.
Sequence and genomic neighborhood determinants of SSN clusters 13 (S. aureus NWMN_0656), 26 (unique to Acinetobacter spp.), 45 (unknown function), 54 (unique to Enterococcus spp.), and 71 (unique to Staphylococcus spp.). (A) WebLogo schematic cartoon representations (top) and corresponding plots (bottom) of all sequences contained within each cluster (see Table S3 for aligned sequences). P-loops, interdomain salt bridges and positions of EcYjiA ZnII ligands E37 and E42 are highlighted as in Fig. 5A. (B) GNN for each cluster shown in a ‘wheel’ and ‘spoke’ representation.73 The wheel is identified by sequence cluster number and colored as in Fig. 4, while spokes represent neighboring genes found, sized, and colored according to function as described in Fig. 5B.
Cluster 3 proteins are unique in that they are derived from the most diverse group of organisms, from bacteria to man, and exhibit the largest overall pairwise variability in both sequence and length (Figs. 4 and S2). These proteins split into two large sub-clusters that do not cleanly segregate under these conditions. One sub-cluster contains largely eukaryotic COG0523 proteins, from Saccharomyces cerevisiae, zebrafish, Chlamydomonas reinhardtii (10 orthologs;79 two of which become cell-abundant under ZnII restriction and are highlighted in Fig. 480) and the human and mouse CBWD proteins, which remain functionally uncharacterized (Figs. 4 and S3). Tetrahymena Shulin, which inhibits dynein motor activity as part of a large 1.8 MDa trafficking complex involved in ciliogenesis, is reported to contain a central EcYjiA-like domain (Fig. 3D).81 However, this is based on structural homology alone since Shulin lacks the COG0523 signature β-meander insertion harboring the CxCC motif of EcYjiA, as well as recognizable G3, G4 and G5 loops; in addition, Shulin is not identified in the SSN analysis run specifically to detect it (data not shown). Thus, the structure and function of eukaryotic COG0523 proteins remain poorly understood.
A second sub-cluster within cluster 3 features a group of bacterial proteins and includes E. coli YjiA (≈320 residues; Fig. S2), and CobW2 and CobW3 from C. metallidurans, which have been partially characterized.32 Among these proteins there is a weak genomic association with the bacterial carbon starvation response, and consistent with this, YjiA is one of top ‘hits’ in a pulldown experiment in E. coli using derivatized guanosine pentaphosphate tetraphosphate [(p)ppGpp] as bait.82 (p)ppGpp is a cellular alarmone that accumulates in cells as part of the stringent response to nutrient-poor conditions and other stressors. While (p)ppGpp has myriad regulatory functions including inhibition of ribosome biogenesis,83 de novo purine biosynthesis,82 and down-regulation of respiratory chain activity,84 (p)ppGpp is also an allosteric effector of DksA, a ZnII protein and RNA polymerase regulatory factor; DksA and (p)ppGpp function synergistically to inhibit transcription initiation in proteobacteria.85 Some bacteria, including the major pathogen Pseudomonas aeruginosa, encode a Zur-regulated ZnII-free paralog of DksA, DksA258, that allows cells to ‘dial down’ transcription under conditions of extreme ZnII starvation where DskA cannot be metalated.86 In other bacteria, COG0523 proteins from clusters 32 (including some strains of E. coli, Salmonella and Klebsiella pneumoniae) and 65 (Moraxella and Psychrobacter genus) co-localize with a DskA paralog in the genome (Figs. 4 and S7); interestingly, cluster 32 COG0523 proteins replace the CxCC motif with an SxSS motif, known to greatly weaken metal binding in canonical SSN cluster 1 and cluster 3 COG0523 proteins.28,29
A strong connection between nutrient stress reported by elevated (p)ppGpp and transition metal (Mn, Fe) homeostasis has also been noted in Enterococcus faecalis and S. aureus in ways that impact pathogenicity, oxidative stress resistance and antibiotic susceptibility.84,87 The COG0523 protein in Enterococccus spp. and related firmicutes exemplifies cluster 54 (Fig. 7), and is genetically linked to a high-affinity ZnII uptake system (ZnuA-ZinT) and ribosomal protein C—paralogs L33B and S14B (Fig. 7).45 In the broadest possible sense, these studies raise the possibility that COG0523 proteins define a central feature of the nutrient stress response in bacteria, and may well bind and/or hydrolyze a wide range of nucleotide di- and triphosphate species. This finding is consistent with the ability of B. subtilis ZagA to hydrolyze the alarmone ZTP,39 GTP-like rates of hydrolysis of ATP and inorganic pyrophosphate (PPi) by A. baumannii ZigA,28 and the relatively poor conservation of the G5 loop in particular (Figs. 5 and 6), which dictates the nucleotide specificity of G-proteins along with loop G4.88 Alternatively, (p)ppGpp may inhibit GTP hydrolysis by COG0523 enzymes, thus switching its activity off, as found for the ribosome biogenesis GTPase, RbgA in S. aureus.83
Other CxCC COG0523 SSN clusters are 6, 7, and 9, many of which are characterized by a poorly conserved His/Asp/Glu-rich stretch of residues preceding the C-terminal domain that may aid in metal capture or channeling (Fig. 1C). We obtain few insights into cluster 6 and cluster 9 function from the GNN analysis (Fig. 6) nor have members of these clusters been biochemically characterized. Cluster 7 harbors YeiR from E. coli and Salmonella, both known to bind at least one ZnII or CoII with high affinity (KZn ≈ 1011 M–1; KCo ≈ 109 M–1) and metal appears to weakly activate GTP hydrolysis.28,64 Although both YeiRs have some connection to zinc homeostasis, and in the case of Salmonella YeiR appears to be Zur-regulated, a GNN analysis of cluster 7 entries reveals considerable diversity of function, with no single ubiquitous genomic neighbor; however, many genes are known to be involved in solute or simple carbohydrate uptake and metabolism (Fig. 6). Cluster 10 COG0523 proteins are derived nearly exclusively from Pseudomonas spp. and are also of unknown function, but are predicted to be Zur-regulated if findings from P. aeruginosa can be extended to other species.58 In P. aeruginosa, this cluster 10 COG0523 PA5532 is encoded alongside two ‘metal-promiscuous’ enzymes, GTP cyclohydrolase IB (FolEB)56 and dihydrooratase (PyrC2), and a cluster 1 P-loop G3E GTPase (PA5535; see Fig. 4) at high co-occurrence frequencies. Inspection of conserved sequence features reveals significant, yet distinct, regions of sequence conservation in the C-terminal domain, punctuated by a periodic disposition of nearly invariant Trp residues, features that characterize cluster 1 and cluster 26 COG0523 proteins, as well as Mrf (Figs. 5–7). Further investigations into cluster 10 COG0523 proteins are clearly warranted.
The major human pathogen S. aureus encodes three COG0523 proteins, including S. aureus ZigA (cluster 1), NWMN_0656 (cluster 13), and NWMN_2481 (cluster 71; Figs 5–7). None have well-defined functions and only ZigA and NWMN_2481 are bona fide Zur targets.28,89 Cluster 13 proteins are found in pathogenic Bacillus anthracis strains, and GNN analysis provides no clear insights beyond solute uptake; however, a recent report suggests this gene (BAS1786 in B. anthracis Sterne 34F2 strain) is regulated by Zur (Fig. 7).90 Cluster 71 proteins are only found in Staphylococcus spp. and in the S. aureus Newman strain it is part of an operon that is highly induced by calprotectin treatment and encodes a genomically linked pyridine nucleotide oxidoreductase (Fig. 7) and a transmembrane protein that is annotated as a FeoB homolog lacking the N-terminal GTPase domain, responsible for FeII uptake in other bacteria.91–93 The hypothesis, not yet tested, is that NWMN_2481 (cluster 71) and NWMN_2482 form a bipartite ferrous ion or FeII-complex transporter. Finally, cluster 26 proteins are only found in Acinetobacter ssp. (AC1AD1025 in A. baylii; A1S_0934 in A. baumannii ATCC 17978),30 and in A. baumannii, is not regulated by Zur nor induced by calprotectin treatment.37 Here, the GNN analysis reveals strong co-localization with YaeQ (a phosphodiesterase), FKBP_C (a peptidyl prolyl isomerase) and 4-HBT (a thioesterase), but the expression of these genes is not likely co-regulated with that of the COG0523 (Fig. 7).
Mechanistic perspectives
Multiple structures of UreG and HypB individually and as part of multiprotein subassemblies show that GTP binding and hydrolysis impact the coordination chemistry of these metallochaperones which in turn modulates the nature of protein-protein interactions as a means to orchestrate ordered metallocenter maturation.27,75 The bioinformatics analysis presented here outlines an important first step in developing hypotheses on how individual COG0523 proteins function while also identifying cluster-conserved features that will facilitate structure determination and biochemical characterization. These analyses suggest that while it might be tempting to consider UreG and HypB as templates for understanding COG0523 function, there are limitations to this perspective, since UreG and HypB function through a dimer-bridging metal binding site and COG0523 proteins likely function through the single CxCC site contained within a monomer28 or protomer.64 On the other hand, both embed key transition metal binding residues into or between the G2 (Switch 1) or G3 (Switch 2) regulatory loops (Fig. 5). One major conclusion from the analysis presented here is that the C-terminal domain is largely the sequence feature that distinguishes one COG0523 SSN cluster from another. Many harbor nearly invariant transition metal binding ligands in the GTPase domain, like that of authentic CobW,34 but most do not. Functional diversity is also found within individual SSN clusters, notably in clusters 1 and 3, but which may characterize other SSN clusters as well. A striking example of diversity of function in COG0523 proteins within a single organism is in the common phytoplankton, Emiliania huxleyi. Emiliania huxleyi has evolved in a chronically zinc-starved environment and encodes no fewer than 26 COG0523 proteins, 17 of which localize to SSN clusters 1 and 3, with just three increasing in cell-abundance under conditions of ZnII limitation.79
The finding that mycobacterial Mrf proteins are evolutionarily related to members of P-loop G3E G-protein superfamily (clusters 17, 20, and 55) that may have evolved a unique ZnII binding site76 is striking and suggests the possibility that some COG0523 proteins may interact with the ribosome, perhaps near the exit tunnel where they might facilitate co-translational metal association with the nascent chain as it folds.46,49 This would allow for metallochaperone mediated ZnII delivery to a specific ZnII binding site in a partially folded protein that would otherwise be entirely surface inaccessible and buried within a protein core once folded. Co-translational metal incorporation is poorly understood, but recent work shows that it is possible to ‘fold’ a single zinc-finger domain inside the exit tunnel, and that this folding is enhanced in the presence of ZnII.94 A number of other models of co-translational metal incorporation are also possible that do not require a specific interaction with a client(s). For example, transient association of a metal-loaded COG0523 protein may simply increase the local concentration of ZnII near the exit tunnel for metal-binding domains to access as they fold. Alternatively, a specific COG0523 protein simply activates a client protein or a metabolic process that leads to an increase in bioavailable ZnII that the ribosome simply accesses.37 In any case, extending this line of thinking to candidate metalloenzyme clients as they are discovered, particularly those with metal ligands closely spaced in the primary structure, as found in a zinc-finger, may well be insightful.
Two additional mechanistic models of COG0523 metallochaperone activity are worth considering in an effort to design experiments to investigate post-translational metallocofactor biogenesis. One model draws heavily on studies of CoIII- and FeIII-nitrile hydratases.95,96 These enzymes harbor isostructural coordination chelates involving three closely spaced cysteines in a Cx2CSC sequence in the α subunit to form an octahedral coordination site featuring an axial Cys thiolate ligand (C110) and four ligands in a plane: two backbone amidinate N atoms and two post-translationally modified (oxidized) Cys residues (C113 and C115; Fig. 8A). This metal site is buried at the α−β subunit interface in an α2β2 heterotetramer (Fig. 8A) and thus metal–ligand exchange between a chaperone and the intact α2β2 oligomer is not likely. Recent studies of the maturation of FeIII-nitrile hydratase suggest that the cluster 1 metallochaperone Nha3/NhpC forms a complex with the free α-subunit, which leads to metalation via an unknown mechanism; once metalated, Nha3/NhpC enables a subunit ‘swap’ or exchange between a metal-free α-subunit of the apo- or Fe1-tetramer (Fig. 8B).97 Bacterial strains lacking the chaperone give rise to high levels of apo α and β subunits in cells with no detectable NHase activity; in addition, a Walker A mutation in NhpC abolishes NHase maturation in vivo,36 as previously established for UreG and urease and HypB and NiFe-hydrogenase (vide supra). An analogous subunit swap model has also been reported for Rhodococcus rhodochrous J1 CoIII-NHase;98–100 here, however, the cluster 1 chaperone is replaced by an NhlE dimer that binds Co(II) but is otherwise unrelated to Nha3/NhpC and has no GTP binding or hydrolysis activity. A subunit exchange model requires that the α-β subunit interface be exchange labile, perhaps more so in the apo-state, despite the extensive inter-subunit contacts (Fig. 8A). The kinetics of subunit exchange under native-like solution conditions have been carefully investigated in only a few oligomeric proteins,101,102 and are clearly worth investigating as other COG0523 client proteins are discovered.
Fig. 8.
Ribbon representation of the structure of the αβ heterodimer of mature FeIII nitrile hydratase (NHase) from Rhodococcus erythropolis AJ270 (2qdy)95 (A) and a proposed mechanistic model for NHase activator protein, Nha3 (B).35,36,97,113
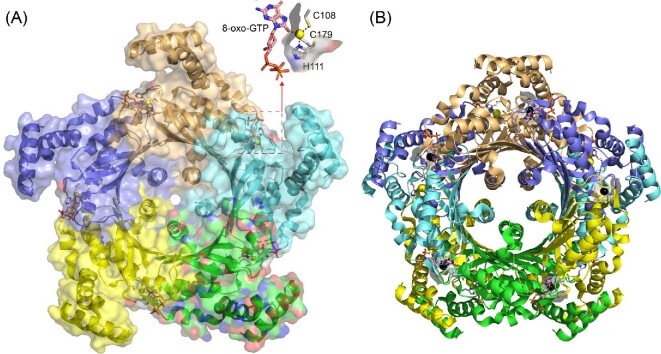
A third metallochaperone model posits direct metal exchange from the metallochaperone to the active site itself or an intermediate ‘transfer’ site on the client protein. To discuss this model in the context of the other models, we consider B. subtilis cyclohydrolase 1A (FolE) and its reported cluster 1 metallochaperone ZagA (Fig. 9A).39 FolE is a dimer of pentamers (Fig. 9B) and adopts a tunnel (T)-fold architecture, in which each protomer in the pentamer contributes four antiparallel β-strands to create a central 20-stranded β-barrel.103–106 An N-terminal α-helical bundle mediates contacts between pentamers in the decamer, while a C-terminal α-helix effectively plugs the hole in the β-barrel. In both the pentamer and decamer structures, the active site ZnII is found at the interface between protomers, with one of the ligands (H111 in the Thermus thermophilus structure shown; Fig. 9A) and the adjacent catalytically important histidine (H110) positioned at the protein surface.106 The quaternary structures of FolE and other T-fold proteins107 make it unlikely that subunit exchange would occur at any appreciable rate, particularly in the context of the FolE decamer, which is further stabilized by the N-terminal helical domain interactions across the pentamer interface. This has not been systematically investigated, however. On the other hand, direct ligand exchange with a metallochaperone could likely only occur upon displacement of the independently folded N-terminal domain. The bacterial two-hybrid experiment that provides the only evidence of a physical interaction between FolE and ZagA, however, can be explained by a minimal 1:1 FolE-ZagA complex, without invoking an oligomeric species.39 We also point out that two of the ligands to the ZnII (C108 and H111) are near one another in the primary structure, and thus the metal could potentially be incorporated co-translationally.
Fig. 9.
Ribbon representation of the structure of T. thermophilus GTP cyclohydrolase IA (FolE) bound to the inhibitor 8-oxo-GTP (1wuq).106 (A) TtFolE adopts a decameric stacked pentamer configuration i.e. representative of the Tunnel-fold family of proteins (GOG0720), with one of the pentamers shown, and each subunit shaded differently.103 The active site is positioned near the subunit interface with the ZnII (yellow sphere) tetrahedrally coordinated by the side chains of C108, H111, C179, and the 8-oxygen atom of 8-oxo-GTP. The H111 side chain is at surface, adjacent to the open coordination site that engages the inhibitor. (B) Decamer structure representation, with the pentamer in panel A in the ‘back’. ZnII ions from the ‘front’ pentamer are shaded black.
Conclusions
In this perspective, we provide a comprehensive comparative phylogenetic, biochemical, structural, and functional analysis of the two-domain COG0523 proteins placed in the context of our increasingly sophisticated understanding of the G3E P-loop family metallochaperones and more broadly, cellular nutrient restriction. We show that much of the phylogenetic and structural diversity of COG0523 proteins localizes to the C-terminal domain, which makes the prediction that this domain is responsible for mediating a specific protein-protein interaction that is necessary for intermolecular, directional metal transfer to one or a small number of closely related client proteins.108 More structures of closely related as well as more distantly related COG0523 proteins are needed to obtain a sense of this structural diversity, while the identification of client proteins with which individual COG0523 proteins interact is an important goal. Comparative structural, dynamical and mechanistic characterization of these complexes will elucidate those features that are common to COG0523 enzymes.
A striking finding from this work is an evolutionary link between COG0523 proteins and one class of actinomycete ribosome hibernation promotion factors (Mrf; SSN cluster 17),76 which suggests the intriguing possibility that some COG053 enzymes function co-translationally under conditions of metal nutrient restriction. Productive lines of investigation for future experiments are to systematically investigate whether cluster 1 COG0523 proteins reversibly associate with the ribosome under conditions of extreme ZnII limitation, and/or the extent to which ZagA might catalyze subunit exchange in a FolE oligomer in a ZTP/GTP-hydrolysis dependent manner. Quantification of the number of the cluster 1 COG0523 molecules per cell relative to the number of ribosomes will provide new insights into COG0523 function, while chemical cross-linking approaches promise to identify protein–protein interaction partners for individual members of this enigmatic family of P-loop G3E GTPases. Given the dramatic structural diversity of COG0523 proteins described here, it may well be that these three mechanistic models are not mutually exclusive, and that individual members may function differently, as implied by the large numbers of COG0523 proteins encoded in single genomes, particularly by marine phytoplankton.79 Likewise, the infected host represents a strongly zinc-restricted microenvironment where the high affinity ZnII uptake system of many bacterial pathogens is a documented virulence factor,109 novel Zur-regulated targets in general,110,111 and COG0523 proteins and their metalloenzyme clients specifically, represent highly expressed targets for the development of new antimicrobial strategies to combat multi-drug resistance in bacterial pathogens.38
Supplementary Material
Acknowledgements
We thank members of the Giedroc laboratory for their help with the preparation and critical review of this manuscript.
Contributor Information
Katherine A Edmonds, Department of Chemistry, Indiana University, Bloomington, IN 47405-7102, USA.
Matthew R Jordan, Department of Chemistry, Indiana University, Bloomington, IN 47405-7102, USA; Department of Molecular and Cellular Biochemistry, Indiana University, Bloomington, IN 47405, USA.
David P Giedroc, Department of Chemistry, Indiana University, Bloomington, IN 47405-7102, USA; Department of Molecular and Cellular Biochemistry, Indiana University, Bloomington, IN 47405, USA.
Dedication: This article is dedicated to the memory of Prof. Deborah Zamble, a friend, colleague and former Editorial Board member of Metallomics, in recognition of her ground-breaking studies of bacterial NiFe-hydrogenase maturation and ZnII-binding COG0523 proteins.
Funding
We gratefully acknowledge support by the US National Institutes of Health (R35 GM118157 and R01 AI110171) to DPG and fellowship support by the Indiana University Graduate Training Program in Quantitative and Chemical Biology (QCB; T32 GM109825 and GM131994) to MRJ.
Conflicts of interest
The authors declare no conflicts of interest.
Data availability
The data presented in this article are available in the article and in the online supplementary material.
References
- 1. Capdevila D. A., Edmonds K. A., Giedroc D. P., Metallochaperones and metalloregulation in bacteria, Essays Biochem., 2017, 61 (2), 177–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Waldron K. J., Robinson N. J., How do bacterial cells ensure that metalloproteins get the correct metal?, Nat. Rev. Microbiol., 2009, 7 (1), 25–35. [DOI] [PubMed] [Google Scholar]
- 3. Wang Y., Weisenhorn E., MacDiarmid C. W., Andreini C., Bucci M., Taggart J., Banci L., Russell J., Coon J. J., Eide D. J., The cellular economy of the Saccharomyces cerevisiae zinc proteome, Metallomics, 2018, 10 (12), 1755–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Desguin B., Zhang T., Soumillion P., Hols P., Hu J., Hausinger R. P., A tethered niacin-derived pincer complex with a nickel-carbon bond in lactate racemase, Science, 2015, 349 (6243), 66–69. [DOI] [PubMed] [Google Scholar]
- 5. Righetto R. D., Anton L., Adaixo R., Jakob R. P., Zivanov J., Mahi M. A., Ringler P., Schwede T., Maier T., Stahlberg H., High-resolution cryo-EM structure of urease from the pathogen Yersinia enterocolitica, Nat. Commun., 2020, 11 (1), 5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Olson J. W., Mehta N. S., Maier R. J., Requirement of nickel metabolism proteins HypA and HypB for full activity of both hydrogenase and urease in Helicobacter pylori, Mol. Microbiol., 2001, 39 (1), 176–182. [DOI] [PubMed] [Google Scholar]
- 7. Portnoy M. E., Rosenzweig A. C., Rae T., Huffman D. L., O'Halloran T. V., Culotta V. C., Structure-function analyses of the ATX1 metallochaperone, J. Biol. Chem., 1999, 274 (21), 15041–15045. [DOI] [PubMed] [Google Scholar]
- 8. Rosenzweig A. C., O'Halloran T. V., Structure and chemistry of the copper chaperone proteins, Curr. Opin. Chem. Biol., 2000, 4 (2), 140–147. [DOI] [PubMed] [Google Scholar]
- 9. Tottey S., Patterson C. J., Banci L., Bertini I., Felli I. C., Pavelkova A., Dainty S. J., Pernil R., Waldron K. J., Foster A. W., Robinson N. J., Cyanobacterial metallochaperone inhibits deleterious side reactions of copper, Proc. Natl. Acad. Sci., 2012, 109 (1), 95–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Banci L., Bertini I., Ciofi-Baffoni S., Kandias N. G., Robinson N. J., Spyroulias G. A., Su X. C., Tottey S., Vanarotti M., The delivery of copper for thylakoid import observed by NMR, Proc. Natl. Acad. Sci., 2006, 103 (22), 8320–8325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Banci L., Bertini I., Ciofi-Baffoni S., Kozyreva T., Zovo K., Palumaa P., Affinity gradients drive copper to cellular destinations, Nature, 2010, 465 (7298), 645–648. [DOI] [PubMed] [Google Scholar]
- 12. Fu Y., Tsui H. C., Bruce K. E., Sham L. T., Higgins K. A., Lisher J. P., Kazmierczak K. M., Maroney M. J., Dann C. E. 3rd, Winkler M. E., Giedroc D. P., A new structural paradigm in copper resistance in Streptococcus pneumoniae., Nat. Chem. Biol., 2013, 9 (3), 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rae T. D., Schmidt P. J., Pufahl R. A., Culotta V. C., O'Halloran T. V., Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase, Science, 1999, 284 (5415), 805–808. [DOI] [PubMed] [Google Scholar]
- 14. Hubbard P. A., Padovani D., Labunska T., Mahlstedt S. A., Banerjee R., Drennan C. L., Crystal structure and mutagenesis of the metallochaperone MeaB: insight into the causes of methylmalonic aciduria, J. Biol. Chem., 2007, 282 (43), 31308–31316. [DOI] [PubMed] [Google Scholar]
- 15. Mehta N., Benoit S., Maier R. J., Roles of conserved nucleotide-binding domains in accessory proteins, HypB and UreG, in the maturation of nickel-enzymes required for efficient Helicobacter pylori colonization, Microb. Pathog., 2003, 35 (5), 229–234. [DOI] [PubMed] [Google Scholar]
- 16. Douglas C. D., Ngu T. T., Kaluarachchi H., Zamble D. B., Metal transfer within the Escherichia coli HypB-HypA complex of hydrogenase accessory proteins., Biochemistry, 2013, 52 (35), 6030–6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Watanabe S., Sasaki D., Tominaga T., Miki K., Structural basis of [NiFe] hydrogenase maturation by Hyp proteins, Biol. Chem., 2012, 393 (10), 1089–1100. [DOI] [PubMed] [Google Scholar]
- 18. Farrugia M. A., Wang B., Feig M., Hausinger R. P., Mutational and computational evidence that a nickel-transfer tunnel in UreD is used for activation of Klebsiella aerogenes urease, Biochemistry, 2015, 54 (41), 6392–6401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fong Y. H., Wong H. C., Yuen M. H., Lau P. H., Chen Y. W., Wong K. B., Structure of UreG/UreF/UreH complex reveals how urease accessory proteins facilitate maturation of Helicobacter pylori urease, PLoS Biol., 2013, 11 (10), 6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Banerjee R., Gouda H., Pillay S., Redox-linked coordination chemistry directs vitamin B12 trafficking, Acc. Chem. Res., 2021, 54 (8), 2003–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Campanello G. C., Lofgren M., Yokom A. L., Southworth D. R., Banerjee R., Switch I-dependent allosteric signaling in a G-protein chaperone-B12 enzyme complex, J. Biol. Chem., 2017, 292 (43), 17617–17625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Padovani D., Banerjee R., A G-protein editor gates coenzyme B12 loading and is corrupted in methylmalonic aciduria, Proc. Natl. Acad. Sci., 2009, 106 (51), 21567–21572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moncrief M. B. C., Hausinger R. P., Characterization of UreG, identification of a UreD-UreF-UreG complex, and evidence suggesting that a nucleotide-binding site in UreG is required for in vivo metallocenter assembly of Klebsiella aerogenes urease, J. Bacteriol., 1997, 179 (13), 4081–4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sydor A. M., Lebrette H., Ariyakumaran R., Cavazza C., Zamble D. B., Relationship between Ni(II) and Zn(II) coordination and nucleotide binding by the Helicobacter pylori [NiFe]-hydrogenase and urease maturation factor HypB, J. Biol. Chem., 2014, 289 (7), 3828–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lacasse M. J., Douglas C. D., Zamble D. B., The mechanism of selective nickel transfer from HypB to HypA, E. coli [NiFe]-hydrogenase accessory proteins, Biochemistry, 2016, 55 (49), 6821–6831. [DOI] [PubMed] [Google Scholar]
- 26. Yang X., Li H., Lai T. P., Sun H., UreE-UreG complex facilitates nickel transfer and preactivates GTPase of UreG in Helicobacter pylori, J. Biol. Chem., 2015, 290 (20), 12474–12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yuen M. H., Fong Y. H., Nim Y. S., Lau P. H., Wong K. B., Structural insights into how GTP-dependent conformational changes in a metallochaperone UreG facilitate urease maturation, Proc. Natl. Acad. Sci., 2017, 114 (51), E10890–E10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jordan M. R., Wang J., Weiss A., Skaar E. P., Capdevila D. A., Giedroc D. P., Mechanistic insights into the metal-dependent activation of Zn(II)-dependent metallochaperones, Inorg. Chem., 2019, 58 (20), 13661–13672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sydor A. M., Jost M., Ryan K. S., Turo K. E., Douglas C. D., Drennan C. L., Zamble D. B., Metal binding properties of Escherichia coli YjiA, a member of the metal homeostasis-associated COG0523 family of GTPases, Biochemistry, 2013, 52 (10), 1788–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Haas C. E., Rodionov D. A., Kropat J., Malasarn D., Merchant S. S., de Crécy-Lagard V., A subset of the diverse COG0523 family of putative metal chaperones is linked to zinc homeostasis in all kingdoms of life, BMC Genomics, 2009, 10 (1), 470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Crouzet J., Levy-Schil S., Cameron B., Cauchois L., Rigault S., Rouyez M. C., Blanche F., Debussche L., Thibaut D., Nucleotide sequence and genetic analysis of a 13.1-kilobase-pair Pseudomonas denitrificans DNA fragment containing five cob genes and identification of structural genes encoding Cob(I)alamin adenosyltransferase, cobyric acid synthase, and bifunctional cobinamide kinase-cobinamide phosphate guanylyltransferase, J. Bacteriol., 1991, 173 (19), 6074–6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Butof L., Grosse C., Lilie H., Herzberg M., Nies D. H., Interplay between the Zur Regulon components and metal resistance in Cupriavidus metallidurans, J. Bacteriol., 2019, 201 (15), e00192–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Osman D., Cooke A., Young T. R., Deery E., Robinson N. J., Warren M. J., The requirement for cobalt in vitamin B12: a paradigm for protein metalation, Biochim. Biophys. Acta. Mol. Cell Res., 2021, 1868 (1), 118896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Young T. R., Martini M. A., Foster A. W., Glasfeld A., Osman D., Morton R. J., Deery E., Warren M. J., Robinson N. J., Calculating metalation in cells reveals CobW acquires Co(II) for vitamin B12 biosynthesis while related proteins prefer Zn(II), Nat. Commun., 2021, 12 (1), 1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gumataotao N., Lankathilaka K. P., Bennett B., Holz R. C., The iron-type nitrile hydratase activator protein is a GTPase, Biochem. J., 2017, 474 (2), 247–258. [DOI] [PubMed] [Google Scholar]
- 36. Hashimoto Y., Ube Y., Doi S., Kumano T., Kobayashi M., Metal chaperone, NhpC, involved in the metallocenter biosynthesis of nitrile hydratase, J. Gen. Appl. Microbiol., 2020, 67 (1), 24–32. [DOI] [PubMed] [Google Scholar]
- 37. Nairn B. L., Lonergan Z. R., Wang J., Braymer J. J., Zhang Y., Calcutt M. W., Lisher J. P., Gilston B. A., Chazin W. J., de Crecy-Lagard V., Giedroc D. P., Skaar E. P., The response of Acinetobacter baumannii to zinc starvation, Cell Host & Microbe, 2016, 19 (6), 826–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang J., Lonergan Z. R., Gonzalez-Gutierrez G., Nairn B. L., Maxwell C. N., Zhang Y., Andreini C., Karty J. A., Chazin W. J., Trinidad J. C., Skaar E. P., Giedroc D. P., Multi-metal restriction by calprotectin impacts de novo flavin biosynthesis in Acinetobacter baumannii, Cell Chem. Biol., 2019, 26 (5), 745–755.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chandrangsu P., Huang X., Gaballa A., Helmann J. D., Bacillus subtilis FolE is sustained by the ZagA zinc metallochaperone and the alarmone ZTP under conditions of zinc deficiency, Mol. Microbiol., 2019, 112 (3), 751–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kehl-Fie T. E., Skaar E. P., Nutritional immunity beyond iron: a role for manganese and zinc, Curr. Opin. Chem. Biol., 2010, 14 (2), 218–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zygiel E. M., Nolan E. M., Transition Metal Sequestration by the host-defense protein calprotectin, Annu. Rev. Biochem., 2018, 87 (1), 621–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Antelo G. T., Vila A. J., Giedroc D. P., Capdevila D. A., Molecular evolution of transition metal bioavailability at the host-pathogen interface, Trends Microbiol., 2021, 29 (5), 441–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shin J. H., Helmann J. D., Molecular logic of the Zur-regulated zinc deprivation response in Bacillus subtilis, Nat. Commun., 2016, 7 (1), 12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shin J.-H., Jung H. J., An Y. J., Cho Y.-B., Cha S.-S., Roe J.-H., Graded expression of zinc-responsive genes through two regulatory zinc-binding sites in Zur, Proc. Natl. Acad. Sci., 2011, 108 (12), 5045–5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Panina E. M., Mironov A. A., Gelfand M. S., Comparative genomics of bacterial zinc regulons: Enhanced ion transport, pathogenesis, and rearrangement of ribosomal proteins, Proc. Natl. Acad. Sci., 2003, 100 (17), 9912–9917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li Y., Sharma M. R., Koripella R. K., Yang Y., Kaushal P. S., Lin Q., Wade J. T., Gray T. A., Derbyshire K. M., Agrawal R. K., Ojha A. K., Zinc depletion induces ribosome hibernation in mycobacteria, Proc. Natl. Acad. Sci., 2018, 115 (32), 8191–8196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ueta M., Wada C., Wada A., YkgM and YkgO maintain translation by replacing their paralogs, zinc-binding ribosomal proteins L31 and L36, with identical activities, Genes Cells, 2020, 25 (8), 562–581. [DOI] [PubMed] [Google Scholar]
- 48. Natori Y., Nanamiya H., Akanuma G., Kosono S., Kudo T., Ochi K., Kawamura F., A fail-safe system for the ribosome under zinc-limiting conditions in Bacillus subtilis, Mol. Microbiol., 2007, 63 (1), 294–307. [DOI] [PubMed] [Google Scholar]
- 49. Chen Y. X., Xu Z. Y., Ge X., Sanyal S., Lu Z. J., Javid B., Selective translation by alternative bacterial ribosomes, Proc. Natl. Acad. Sci., 2020, 117 (32), 19487–19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Makthal N., Nguyen K., Do H., Gavagan M., Chandrangsu P., Helmann J. D., Olsen R. J., Kumaraswami M., A critical role of zinc importer AdcABC in Group A streptococcus-host interactions during infection and its implications for vaccine development, EBioMedicine, 2017, 21, 131–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hesse L. E., Lonergan Z. R., Beavers W. N., Skaar E. P., The Acinetobacter baumannii Znu system overcomes host-imposed nutrient zinc limitation, Infect. Immun., 2019, 87 (12), e00746–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lhospice S., Gomez N. O., Ouerdane L., Brutesco C., Ghssein G., Hajjar C., Liratni A., Wang S., Richaud P., Bleves S., Ball G., Borezee-Durant E., Lobinski R., Pignol D., Arnoux P., Voulhoux R., Pseudomonas aeruginosa zinc uptake in chelating environment is primarily mediated by the metallophore pseudopaline, Sci. Rep., 2017, 7 (1), 17132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ghssein G., Brutesco C., Ouerdane L., Izaute A., Foljcik C., Wang S., Hajjar C., Lobinski R., Lemaire D., Richaud P., Voulhoux R., Espaillet A., Cava F., Pignol D., Borezee-Durant E., Arnoux P., Biosynthesis of a broad-spectrum nicotianamine-like metallophore in Staphylococus aureus, Science, 2016, 352 (6289), 1105–1109. [DOI] [PubMed] [Google Scholar]
- 54. Grim K. P., San Francisco B., Radin J. N., Brazel E. B., Kelliher J. L., Parraga Solorzano P. K., Kim P. C., McDevitt C. A., Kehl-Fie T. E., The metallophore staphylopine enables Staphylococcus aureus to compete with the host for zinc and overcome nutritional immunity, mBio, 2017, 8 (5), e01281–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kallifidas D., Pascoe B., Owen G. A., Strain-Damerell C. M., Hong H. J., Paget M. S., The zinc-responsive regulator Zur controls expression of the coelibactin gene cluster in Streptomyces coelicolor, J. Bacteriol., 2010, 192 (2), 608–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sankaran B., Bonnett S. A., Shah K., Gabriel S., Reddy R., Schimmel P., Rodionov D. A., de Crécy-Lagard V., Helmann J. D., Iwata-Reuyl D., Swairjo M. A., Zinc-independent folate biosynthesis: genetic, biochemical, and structural investigations reveal new metal dependence for GTP cyclohydrolase IB, J. Bacteriol., 2009, 191 (22), 6936–6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Paranagama N., Bonnett S. A., Alvarez J., Luthra A., Stec B., Gustafson A., Iwata-Reuyl D., Swairjo M. A., Mechanism and catalytic strategy of the prokaryotic-specific GTP cyclohydrolase-IB, Biochem. J., 2017, 474 (6), 1017–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pederick V. G., Eijkelkamp B. A., Begg S. L., Ween M. P., McAllister L. J., Paton J. C., McDevitt C. A., ZnuA and zinc homeostasis in Pseudomonas aeruginosa., Sci. Rep., 2015, 5 (1), 13139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jaffe E. K., An unusual phylogenetic variation in the metal ion binding sites of porphobilinogen synthase, Chem. Biol., 2003, 10 (1), 25–34. [DOI] [PubMed] [Google Scholar]
- 60. Winkler M. E., Ramos-Montanez S., Biosynthesis of histidine, EcoSal Plus, 2009, 3 (2), 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gabriel S. E., Miyagi F., Gaballa A., Helmann J. D., Regulation of the Bacillus subtilis yciC gene and insights into the DNA-binding specificity of the zinc-sensing metalloregulator Zur, J. Bacteriol., 2008, 190 (10), 3482–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mortensen B. L., Rathi S., Chazin W. J., Skaar E. P., Acinetobacter baumannii response to host-mediated zinc limitation requires the transcriptional regulator zur, J. Bacteriol., 2014, 196 (14), 2616–2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Khil P. P., Obmolova G., Teplyakov A., Howard A. J., Gilliland G. L., Camerini-Otero R. D., Crystal structure of the Escherichia coli YjiA protein suggests a GTP-dependent regulatory function, Proteins Struct. Funct. Bioinf., 2004, 54 (2), 371–374. [DOI] [PubMed] [Google Scholar]
- 64. Blaby-Haas C. E., Flood J. A., Crecy-Lagard V., Zamble D. B., YeiR: a metal-binding GTPase from Escherichia coli involved in metal homeostasis, Metallomics, 2012, 4 (5), 488–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Harada Y., Tanaka Y., Terasawa M., Pieczyk M., Habiro K., Katakai T., Hanawa-Suetsugu K., Kukimoto-Niino M., Nishizaki T., Shirouzu M., Duan X., Uruno T., Nishikimi A., Sanematsu F., Yokoyama S., Stein J. V., Kinashi T., Fukui Y., DOCK8 is a Cdc42 activator critical for interstitial dendritic cell migration during immune responses, Blood, 2012, 119 (19), 4451–4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Farrugia M. A., Macomber L., Hausinger R. P., Biosynthesis of the urease metallocenter, J. Biol. Chem., 2013, 288 (19), 13178–13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bulutoglu B., Garcia K. E., Wu F., Minteer S. D., Banta S., Direct evidence for metabolon formation and substrate channeling in recombinant TCA cycle enzymes, ACS Chem. Biol., 2016, 11 (10), 2847–2853. [DOI] [PubMed] [Google Scholar]
- 68. Wu F., Minteer S., Krebs cycle metabolon: structural evidence of substrate channeling revealed by cross-linking and mass spectrometry, Angew. Chem. Int. Ed., 2015, 54 (6), 1851–1854. [DOI] [PubMed] [Google Scholar]
- 69. Luo Z., Morey J. R., Deplazes E., Motygullina A., Tan A., Ganio K., Neville S. L., Eleftheriadis N., Isselstein M., Pederick V. G., Paton J. C., Cordes T., Harmer J. R., Kobe B., McDevitt C. A., A Trap-Door mechanism for zinc acquisition by Streptococcus pneumoniae AdcA, mBio, 2021, 12 (1), e01958–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Neupane D. P., Fullam S. H., Chacon K. N., Yukl E. T., Crystal structures of AztD provide mechanistic insights into direct zinc transfer between proteins, Commun. Biol., 2019, 2 (1), 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zangger K., Oz G., Otvos J. D., Armitage I. M., Three-dimensional solution structure of mouse [Cd7]-metallothionein-1 by homonuclear and heteronuclear NMR spectroscopy, Protein Sci., 1999, 8 (12), 2630–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gerlt J. A., Genomic enzymology: web tools for leveraging protein family sequence-function space and genome context to discover novel functions, Biochemistry, 2017, 56 (33), 4293–4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zallot R., Oberg N., Gerlt J. A., The EFI web resource for genomic enzymology tools: leveraging protein, genome, and metagenome databases to discover novel enzymes and metabolic pathways, Biochemistry, 2019, 58 (41), 4169–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Berns D. S., Holohan P., Scott E., Urease activity in blue-green algae, Science, 1966, 152 (3725), 1077–1078. [DOI] [PubMed] [Google Scholar]
- 75. Higgins K., Nickel metalloregulators and chaperones, Inorganics (Basel), 2019, 7 (8), 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Li Y., Corro J. H., Palmer C. D., Ojha A. K., Progression from remodeling to hibernation of ribosomes in zinc-starved mycobacteria, Proc. Natl. Acad. Sci., 2020, 117 (32), 19528–19537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Tobiasson V., Dow A., Prisic S., Amunts A., Zinc depletion does not necessarily induce ribosome hibernation in mycobacteria, Proc. Natl. Acad. Sci., 2019, 116 (7), 2395–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Maciag A., Dainese E., Rodriguez G. M., Milano A., Provvedi R., Pasca M. R., Smith I., Palu G., Riccardi G., Manganelli R., Global analysis of the Mycobacterium tuberculosis Zur (FurB) regulon, J. Bacteriol., 2007, 189 (3), 730–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Shire D. M., Kustka A. B., Proteomic responses of the coccolithophore Emiliania huxleyi to zinc limitation and trace metal substitution, Environ. Microbiol., 2021, in press (doi: 10.1111/1462-2920.15644). [DOI] [PubMed] [Google Scholar]
- 80. Hsieh S. I., Castruita M., Malasarn D., Urzica E., Erde J., Page M. D., Yamasaki H., Casero D., Pellegrini M., Merchant S. S., Loo J. A., The proteome of copper, iron, zinc, and manganese micronutrient deficiency in Chlamydomonas reinhardtii, Mol. Cell. Proteomics, 2013, 12 (1), 65–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Mali G. R., Ali F. A., Lau C. K., Begum F., Boulanger J., Howe J. D., Chen Z. A., Rappsilber J., Skehel M., Carter A. P., Shulin packages axonemal outer dynein arms for ciliary targeting, Science, 2021, 371 (6532), 910–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wang B., Dai P., Ding D., Del Rosario A., Grant R. A., Pentelute B. L., Laub M. T., Affinity-based capture and identification of protein effectors of the growth regulator ppGpp, Nat. Chem. Biol., 2019, 15 (2), 141–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Pausch P., Steinchen W., Wieland M., Klaus T., Freibert S. A., Altegoer F., Wilson D. N., Bange G., Structural basis for ppGpp-mediated inhibition of the GTPase RbgA, J. Biol. Chem., 2018, 293 (51), 19699–19709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Fritsch V. N., Loi V. V., Busche T., Tung Q. N., Lill R., Horvatek P., Wolz C., Kalinowski J., Antelmann H., The alarmone ppGpp confers tolerance to oxidative stress during the stationary phase by maintenance of redox and iron homeostasis in Staphylococcus aureus, Free Radic. Biol. Med., 2020, 161, 351–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Molodtsov V., Sineva E., Zhang L., Huang X., Cashel M., Ades S. E., Murakami K. S., Allosteric effector ppGpp potentiates the inhibition of transcript initiation by DksA, Mol. Cell, 2018, 69 (5), 828–839.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Furman R., Biswas T., Danhart E. M., Foster M. P., Tsodikov O. V., Artsimovitch I., DksA2, a zinc-independent structural analog of the transcription factor DksA, FEBS Lett., 2013, 587 (6), 614–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Colomer-Winter C., Gaca A. O., Lemos J. A., Association of metal homeostasis and ppGpp Regulation in the pathophysiology of Enterococcus faecalis, Infect. Immun., 2017, 85 (7), e00260–00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Gasper R., Scrima A., Wittinghofer A., Structural insights into HypB, a GTP-binding protein that regulates metal binding, J. Biol. Chem., 2006, 281 (37), 27492–27502. [DOI] [PubMed] [Google Scholar]
- 89. Peng H., Shen J., Edmonds K. A., Luebke J. L., Hickey A. K., Palmer L. D., Chang F. J., Bruce K. A., Kehl-Fie T. E., Skaar E. P., Giedroc D. P., Sulfide homeostasis and nitroxyl intersect via formation of reactive sulfur species in Staphylococcus aureus, mSphere, 2017, 2 (3), e00082–00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kandari D., Gopalani M., Gupta M., Joshi H., Bhatnagar S., Bhatnagar R., Identification, functional characterization, and regulon prediction of the zinc uptake regulator (zur) of Bacillus anthracis - an insight into the zinc homeostasis of the pathogen, Front. Microbiol., 2018, 11 (9), 3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zhang Y., Sen S., Giedroc D., Iron acquisition by bacterial pathogens: beyond tris-Catecholate complexes, ChemBioChem, 2020, 21 (14), 1955–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Smith A. T., Sestok A. E., Expression and purification of functionally active ferrous iron transporter FeoB from Klebsiella pneumoniae, Protein Expr. Purif. 2018, 142, 1–7. [DOI] [PubMed] [Google Scholar]
- 93. Subashchandrabose S., Smith S., DeOrnellas V., Crepin S., Kole M., Zahdeh C., Mobley H. L., Acinetobacter baumannii genes required for bacterial survival during bloodstream infection, mSphere, 2016, 1 (1), e00013–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Nilsson O. B., Hedman R., Marino J., Wickles S., Bischoff L., Johansson M., Muller-Lucks A., Trovato F., Puglisi J. D., O'Brien E. P., Beckmann R., von Heijne G., Cotranslational protein folding inside the ribosome exit tunnel, Cell Rep., 2015, 12 (10), 1533–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Song L., Wang M., Shi J., Xue Z., Wang M. X., Qian S., High resolution X-ray molecular structure of the nitrile hydratase from Rhodococcus erythropolis AJ270 reveals posttranslational oxidation of two cysteines into sulfinic acids and a novel biocatalytic nitrile hydration mechanism, Biochem. Biophys. Res. Commun., 2007, 362 (2), 319–324. [DOI] [PubMed] [Google Scholar]
- 96. Dutta A., Flores M., Roy S., Schmitt J. C., Hamilton G. A., Hartnett H. E., Shearer J. M., Jones A. K., Sequential oxidations of thiolates and the cobalt metallocenter in a synthetic metallopeptide: Implications for the biosynthesis of nitrile hydratase, Inorg. Chem., 2013, 52 (9), 5236–5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Lankathilaka K. P. W., Bennett B., Holz R. C., The Fe-type nitrile hydratase from Rhodococcus equi TG328-2 forms an alpha-activator protein complex, J. Biol. Inorg. Chem., 2020, 25 (6), 903–911. [DOI] [PubMed] [Google Scholar]
- 98. Zhou Z., Hashimoto Y., Kobayashi M., Self-subunit swapping chaperone needed for the maturation of multimeric metalloenzyme nitrile hydratase by a subunit exchange mechanism also carries out the oxidation of the metal ligand cysteine residues and insertion of cobalt, J. Biol. Chem., 2009, 284 (22), 14930–14938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zhou Z., Hashimoto Y., Shiraki K., Kobayashi M., Discovery of posttranslational maturation by self-subunit swapping, Proc. Natl. Acad. Sci., 2008, 105 (39), 14849–14854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Zhou Z., Hashimoto Y., Cui T., Washizawa Y., Mino H., Kobayashi M., Unique biogenesis of high-molecular mass multimeric metalloenzyme nitrile hydratase: Intermediates and a proposed mechanism for self-subunit swapping maturation, Biochemistry, 2010, 49 (44), 9638–9648. [DOI] [PubMed] [Google Scholar]
- 101. Bhattacharyya M., Stratton M. M., Going C. C., McSpadden E. D., Huang Y., Susa A. C., Elleman A., Cao Y. M., Pappireddi N., Burkhardt P., Gee C. L., Barros T., Schulman H., Williams E. R., Kuriyan J., Molecular mechanism of activation-triggered subunit exchange in Ca(2+)/calmodulin-dependent protein kinase II, Elife, 2016, 5, e13405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Sugiyama M., Kurimoto E., Yagi H., Mori K., Fukunaga T., Hirai M., Zaccai G., Kato K., Kinetic asymmetry of subunit exchange of homooligomeric protein as revealed by deuteration-assisted small-angle neutron scattering, Biophys. J., 2011, 101 (8), 2037–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Phillips G., Grochowski L. L., Bonnett S., Xu H., Bailly M., Blaby-Haas C., El Yacoubi B., Iwata-Reuyl D., White R. H., de Crecy-Lagard V., Functional promiscuity of the COG0720 family, ACS Chem. Biol., 2012, 7 (1), 197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Schussler S., Haase I., Perbandt M., Illarionov B., Siemens A., Richter K., Bacher A., Fischer M., Grawert T., Structure of GTP cyclohydrolase I from Listeria monocytogenes, a potential anti-infective drug target, Acta. Crystallogr. F Struct. Biol. Commun., 2019, 75 (9), 586–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. El Yacoubi B., Bonnett S., Anderson J. N., Swairjo M. A., Iwata-Reuyl D., de Crecy-Lagard V., Discovery of a new prokaryotic type I GTP cyclohydrolase family, J. Biol. Chem., 2006, 281 (49), 37586–37593. [DOI] [PubMed] [Google Scholar]
- 106. Tanaka Y., Nakagawa N., Kuramitsu S., Yokoyama S., Masui R., Novel reaction mechanism of GTP cyclohydrolase I. High-resolution X-ray crystallography of Thermus thermophilus HB8 enzyme complexed with a transition state analogue, the 8-oxoguanine derivative, J. Biochem., 2005, 138 (3), 263–275. [DOI] [PubMed] [Google Scholar]
- 107. Miles Z. D., Roberts S. A., McCarty R. M., Bandarian V., Biochemical and structural studies of 6-carboxy-5,6,7,8-tetrahydropterin synthase reveal the molecular basis of catalytic promiscuity within the tunnel-fold superfamily, J. Biol. Chem., 2014, 289 (34), 23641–23652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Eide D. J., Zinc transporters and the cellular trafficking of zinc, Biochim. Biophys. Acta Mol. Cell Res., 2006, 1763 (7), 711–722. [DOI] [PubMed] [Google Scholar]
- 109. Lonergan Z. R., Skaar E. P., Nutrient zinc at the host-pathogen interface, Trends Biochem. Sci., 2019, 44 (12), 1041–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Lee E. K., Choi C. H., Oh M. H., Zur-regulated lipoprotein A contributes to the fitness of Acinetobacter baumannii, J. Microbiol., 2020, 58 (1), 67–77. [DOI] [PubMed] [Google Scholar]
- 111. Lonergan Z. R., Nairn B. L., Wang J., Hsu Y. P., Hesse L. E., Beavers W. N., Chazin W. J., Trinidad J. C., VanNieuwenhze M. S., Giedroc D. P., Skaar E. P., An Acinetobacter baumannii, zinc-regulated peptidase maintains cell wall integrity during immune-mediated nutrient sequestration, Cell Rep., 2019, 26 (8), 2009–2018.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Fong Y. H., Wong H. C., Yuen M. H., Lau P. H., Chen Y. W., Wong K. B., Structure of UreG/UreF/UreH complex reveals how urease accessory proteins facilitate maturation of Helicobacter pylori urease, PLoS Biol., 2013, 11 (10), e1001678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Xia Y., Peplowski L., Cheng Z., Wang T., Liu Z., Cui W., Kobayashi M., Zhou Z., Metallochaperone function of the self-subunit swapping chaperone involved in the maturation of subunit-fused cobalt-type nitrile hydratase, Biotechnol. Bioeng., 2019, 116 (3), 481–489. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this article are available in the article and in the online supplementary material.