Correction to: Scientific Reports 10.1038/s41598-018-22183-2 published online 01 March 2018
The original version of this Article contained errors. Panels upf1 and dom34 in Figure 1 looked to have originated from the same sample. The Authors now reviewed the original data and for clarity all representative images in Figure 1 have been replaced. Additionally, the Authors recalculated the results shown in Figure 1B using the original data and the graph has also been updated. The original Figure 1 is shown below, for reference:
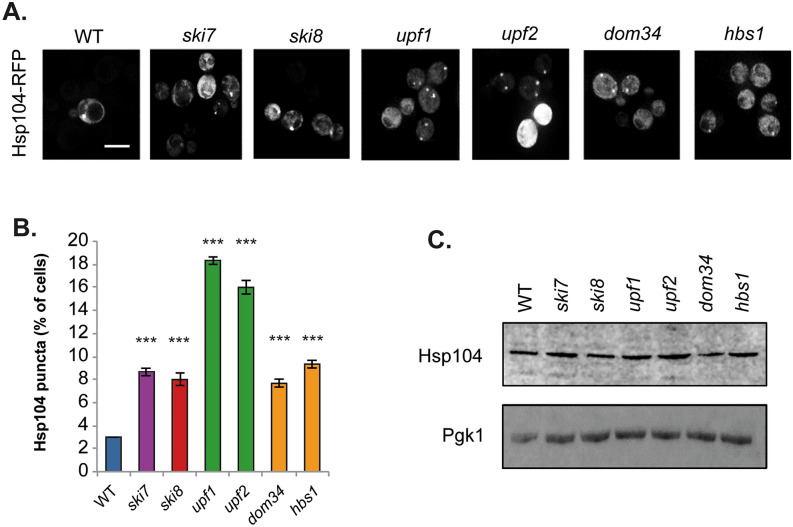
Figure 1.
Strains lacking components of mRNA surveillance pathways have higher levels of protein aggregation. (A) Hsp104-RFP was visualized in wild-type and mutant strains disrupted for NGD (dom34, hbs1), NMD (upf1, upf2), NSD (ski7) and the Ski complex (ski8). Examples of cells containing visible puncta are shown. (B) The percentage of cells containing visible Hsp104-RFP puncta is quantified for each strain. Data shown are the means of three independent biological repeat experiments ± SD. Significance is shown compared with the wild-type strain; ***p < 0.001. (C) Western blot analysis of Hsp104 protein levels. Blots were probed with a Pgk1 antibody as a loading control. The full blots are shown in Supplementary Fig. 1.
The original version of the Article has been corrected.