Abstract
Objectives
To analyse nosocomial transmission in the early stages of the coronavirus 2019 (COVID-19) pandemic at a large multisite healthcare institution. Nosocomial incidence is linked with infection control interventions.
Methods
Viral genome sequence and epidemiological data were analysed for 574 consecutive patients, including 86 nosocomial cases, with a positive PCR test for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) during the first 19 days of the pandemic.
Results
Forty-four putative transmission clusters were found through epidemiological analysis; these included 234 cases and all 86 nosocomial cases. SARS-CoV-2 genome sequences were obtained from 168/234 (72%) of these cases in epidemiological clusters, including 77/86 nosocomial cases (90%). Only 75/168 (45%) of epidemiologically linked, sequenced cases were not refuted by applying genomic data, creating 14 final clusters accounting for 59/77 sequenced nosocomial cases (77%). Viral haplotypes from these clusters were enriched 1–14x (median 4x) compared to the community. Three factors implicated unidentified cases in transmission: (a) community-onset or indeterminate cases were absent in 7/14 clusters (50%), (b) four clusters (29%) had additional evidence of cryptic transmission, and (c) in three clusters (21%) diagnosis of the earliest case was delayed, which may have facilitated transmission. Nosocomial cases decreased to low levels (0–2 per day) despite continuing high numbers of admissions of community-onset SARS-CoV-2 cases (40–50 per day) and before the impact of introducing universal face masks and banning hospital visitors.
Conclusion
Genomics was necessary to accurately resolve transmission clusters. Our data support unidentified cases—such as healthcare workers or asymptomatic patients—as important vectors of transmission. Evidence is needed to ascertain whether routine screening increases case ascertainment and limits nosocomial transmission.
Keywords: Healthcare-associated infection, Molecular epidemiology, Nosocomial transmission, SARS-CoV-2, Whole-genome sequencing
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was first reported in Wuhan, China in December 2019 [1], and over 4 million deaths have since been reported worldwide [2]. Cases in the UK increased rapidly during March 2020, leading to social distancing policies [3,4]. On 23rd March, legislation compelled the UK population to stay at home, with only limited exceptions. Hospital admissions for coronavirus disease 2019 (COVID-19) peaked 1 week later (around 1st April) [5].
Nosocomial infection may account for 10–20% of all confirmed cases [[6], [7], [8]], with associated mortality of up to 30% [7]. Most SARS-CoV-2 transmission studies during the first wave of the pandemic utilized epidemiological analysis alone to identify outbreaks [[10], [11], [12], [13], [9]]. The main limitation to using epidemiology alone is that when point prevalence is high, for instance at 2.2% in London during April 2020 [4], this increases the chance that two people in epidemiological contact are independent cases. Furthermore, a wide incubation period of 2–14 days [14,15] means that infections arising several days after hospital admission may still have been acquired in the community.
Epidemiological data can be supplemented with SARS-CoV-2 genome sequencing to aid analysis of transmission [16,17]. Genomic analysis is complicated during the early stages of the pandemic due to low genetic diversity, with less than 200 mutations recognized by April 2020 [18]. Thus, two people infected with an identical strain may not be linked epidemiologically.
This study combines epidemiological and genomic data to analyse clusters of nosocomial SARS-CoV-2 transmission during the first weeks of the pandemic before infection control policies had been formalized, and when community incidence was high [3,4]. Understanding nosocomial transmission would help set priorities for future infection control planning.
Methods
Setting
Our institution comprises an acute hospital site (STH) admitting COVID-19 patients, an elective site including surgery and oncology (GUY), two long-stay community-care units, and multiple dialysis units. Diagnostics and infection control policies were uniform across sites. Major infection control policies introduced during this time are shown in Fig. 1 . Wards are all multi-bedded with a small allocation of side rooms.
Fig. 1.
Epidemiological description of cases diagnosed during the first wave of the pandemic. On the left-hand y-axis, the grey bar chart displays new cases over time between 10th March and 31st April 2020. Over the same period the right-hand y-axis shows incidence of nosocomial cases (maroon line). Overlaid are five key dates in public policy and infection control: (A) 13th March: testing recommended for all inpatients with cough and fever; use of aprons, gloves and surgical face masks for interactions with confirmed/suspected cases; (B) 16th March: strong government advice for social distancing; (C) 23rd March: implementation of national lockdown; (D) 25th March: exclusion of hospital visitors, and (E) 28th March: mandatory use of surgical masks for all patient interactions under 2 metres.
From 13th March only patients requiring admission and with cough, fever or shortness of breath were tested for SARS-CoV-2 infection, as per Public Health England (PHE) recommendations. Inpatients developing these symptoms were tested before isolation in side rooms whilst results were awaited. Confirmed cases were cohorted in wards with other confirmed cases only. Very exceptionally, confirmed cases stayed on a non-COVID ward in a side room due to capacity issues. Neither asymptomatic individuals nor patients/staff exposed to known cases were routinely screened for infection.
SARS-CoV-2 whole-genome sequencing
RNA extracts were processed using the ARTIC protocol v1.0 [19] and V3 primers set [20] using Oxford Nanopore Technology and the ARTIC bioinformatics pipeline v1.0 [21]. Lineages were assigned using Pangolin [22] v1.1.14 with lineages v2020-05-19.
Deduction of transmission clusters
Transmission clusters were deduced using combined epidemiological and genetic information. First, each case was classified based on the time between admission and symptom onset, according to European Centre for Disease Prevention and Control (ECDC) definitions [23]: community-onset (<3 d), indeterminate (3–7d), probable nosocomial (8–14d) and definite nosocomial (>14 d). Epidemiological clusters required at least two cases, including a probable or definite nosocomial case, and all required an overlapping ward stay with another case during the incubation period. Incubation period was calculated as symptom onset minus 14 days [15] (or sample collection date, if symptom onset unknown). Viral haplotypes were then used to exclude cases differing by two or more single-nucleotide polymorphisms (SNPs), or by three or more SNPs in the secondary analysis. Finally, there were two clusters (GUY4 and GUY5) where nosocomial cases were linked manually on adjacent wards due to the presence of a specific SNP that was highly enriched compared to community haplotypes (see Supplementary Material Methods).
Choice of SNP threshold for excluding transmission
Previous literature has discussed the probability of acquiring SNPs between cases based on the mutation rate of SARS-CoV-2, estimating a 24% chance of one new SNP and 4% of two new SNPs per generation [24]. Assuming that all relevant cases were captured, for our 77 sequenced nosocomial cases one would expect 0.04 × 77 = 3.1 cases to differ by two or more SNPs from their infection source (see Supplementary Material Methods). Other published literature also supports the lower SNP exclusion threshold of ≥2 [16]. Notably, however, one study found evidence that two SNPs could occur in institutional outbreaks in 17 days [25].
Construction of phylogenetic tree
Maximum likelihood phylogenetic trees were derived using phangorn (v2.5.5) and plotted with ggtree (v.2.41) in R (v4.0.2). Trees were fitted separately according to Pangolin lineage assignment using a generalized time reversible (GTR) + Γ(4) + I model.
Healthcare worker (HCW) symptomology and seroconversion
Two hundred and twenty-eight HCWs were followed up from 13th March until 10th June 2020 for self-reported COVID-19-compatible symptoms and SARS-CoV-2 seroconversion. Sequential serum samples were collected every 1–2 weeks and tested using ELISA [27]. The median time between symptom onset and seroconversion in symptomatic HCWs was used to infer the infectious period for asymptomatic cases. HCW absenteeism was retrieved from human resource records.
Results
Clinical characteristics and epidemiology
By 31st March there were 574 laboratory-confirmed cases (Supplementary Material Table S1). Most were admitted (483/574, 84%, Supplementary Material Table S2) with a median length of stay of 12 days (IQR 5–27, Table 1 ).
Table 1.
Demographics of the 574 cases diagnosed by the diagnostic lab until 31st March, separated by community-onset, indeterminate, probable nosocomial, and definite nosocomial infections
| Overall | Community | Indeterminate | Probable nosocomial | Definite nosocomial | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cases (n) | 574 | 471 | 82% | 17 | 3% | 27 | 5% | 59 | 10% | |
| In-hospital mortality | 114 | 20% | 81 | 17% | 4 | 24% | 6 | 22% | 23 | 39% |
| Inpatients | 483 | 84% | 380 | 81% | — | — | — | |||
| Length of stay (IQR) | 12 (5–27) | 9 (4–16) | 19 (11–24) | 23 (21–30) | 53 (36–94) | |||||
| Sex | ||||||||||
| Female | 251 | 44% | 208 | 44% | 4 | 24% | 9 | 33% | 30 | 51% |
| Male | 323 | 56% | 263 | 56% | 13 | 76% | 18 | 67% | 29 | 49% |
| Median age (IQR) | 61 (48–76) | 58 (45–73) | 73 (61–80) | 75 (69–81) | 73 (61–82) | |||||
| Ethnicity | ||||||||||
| Known | 455 | 79% | 377 | 80% | 10 | 59% | 18 | 67% | 50 | 85% |
| White | 230 | 51% | 174 | 46% | 5 | 50% | 14 | 78% | 37 | 74% |
| BAME | 225 | 49% | 203 | 54% | 5 | 50% | 4 | 22% | 13 | 26% |
| Pregnant | 13 | 2% | 13 | 3% | 0 | 0% | 0 | 0% | 0 | 0% |
| Charlson score (IQR) | 2 (1–5) | 1 (0–3) | 5 (4–6) | 5 (4–6) | 5 (4–6) | |||||
| Hypertension | 257 | 45% | 203 | 43% | 7 | 41% | 17 | 63% | 30 | 51% |
| Congestive cardiac failure | 28 | 5% | 13 | 3% | 1 | 6% | 4 | 15% | 10 | 17% |
| Myocardial infarction | 19 | 3% | 12 | 3% | 1 | 6% | 2 | 7% | 4 | 7% |
| Diabetes mellitus | 168 | 29% | 138 | 29% | 3 | 18% | 9 | 33% | 18 | 31% |
| End organ damage | 38 | 7% | 28 | 6% | 3 | 18% | 3 | 11% | 4 | 7% |
| Renal impairment | 111 | 19% | 87 | 18% | 4 | 24% | 6 | 22% | 14 | 24% |
| Mild | 49 | 9% | 34 | 7% | 3 | 18% | 3 | 11% | 9 | 15% |
| Moderate | 7 | 1% | 5 | 1% | 0 | 0% | 1 | 4% | 1 | 2% |
| Severe | 54 | 9% | 47 | 10% | 1 | 6% | 2 | 7% | 4 | 7% |
| Dementia | 50 | 9% | 31 | 7% | 2 | 12% | 4 | 15% | 13 | 22% |
| COPD | 46 | 8% | 30 | 6% | 3 | 18% | 3 | 11% | 10 | 17% |
| Immunosuppression | 35 | 6% | 24 | 5% | 0 | 0% | 3 | 11% | 8 | 14% |
| HIV/AIDS | 2 | 0% | 2 | 0% | 0 | 0% | 0 | 0% | 0 | 0% |
| Solid tumour | 71 | 13% | 45 | 10% | 6 | 35$ | 6 | 22% | 14 | 24% |
| Localized | 53 | 9% | 36 | 8% | 3 | 18% | 4 | 15% | 10 | 17% |
| Metastatic | 18 | 3% | 9 | 2% | 3 | 18% | 2 | 7% | 4 | 7% |
| Haematological malignancy | 14 | 2% | 4 | 1% | 2 | 12% | 2 | 7% | 6 | 10% |
| Lymphoma | 7 | 1% | 1 | 0% | 2 | 12% | 1 | 4% | 3 | 5% |
| Leukaemia | 7 | 1% | 3 | 1% | 0 | 0% | 1 | 4% | 3 | 5% |
IQR, interquartile range; COPD, chronic obstructive pulmonary disease.
New cases peaked between 31st March and 8th April before falling steadily through April (Fig. 1). The daily number of probable and definite nosocomial cases peaked earlier on 23rd March with 12 new cases. Nosocomial cases then rapidly declined to 0–2 cases per day during April (Fig. 1) and none in the following 4 months (data not shown).
Of the 574 SARS-CoV-2-positive patients, 471 (82%) were categorized based on ECDC definitions [23] as community-onset, with 59 (10%) definite nosocomial, 27 (5%) probable nosocomial, and 17 (4%) indeterminate cases. Demographics are shown in Table 1. The crude in-hospital mortality was 20%, highest in the definite nosocomial group (39%, 23/59).
Five hundred and forty-one of 574 cases were within the period for genomic analysis between 13th and 31st March. The SARS-CoV-2 genome sequence was obtained from 380/541 cases (70%), including 90% of all probable and definite nosocomial cases (77/86) and 72% of cases placed into epidemiological clusters (168/234) (Supplementary Material Table S4).
Linking epidemiology and genomics to define transmission clusters
Forty-four epidemiological clusters were formed involving 234 cases (including all 86 nosocomial acquisitions), with a median of six patients per cluster (IQR 2–10) (Fig. 2 a and b, Supplementary Material Table S4). These 44 clusters were resolved into 14 final clusters where genomic data was available and did not refute epidemiological linkage (Fig. 2a and c, Supplementary Material Table S5). These final clusters included 75 cases and 59/77 (77%) sequenced nosocomial cases.
Fig. 2.
(a) Haplotype representations of the 14 clusters that emerge after applying the clustering process using epidemiological and viral genetic data (see Methods). Clusters are named after the hospital site in which they occurred (leftmost column). Cluster haplotype lineages are shown in black (second column from left). Cluster haplotypes are depicted with a ‘1’ in a given position indicating the presence of the SNP relative to the reference genome shown above in vertical text, and a ‘.’ indicating its absence (wild-type sequence). Cluster rows are coloured based loosely on the similarity of the cluster haplotypes to one another. This same colour scheme is used to represent specific clusters in subsequent figures. (b) Epidemiological clusters 4–33, including cases where n > 2 (Supplementary Material Table S6), are coloured according to how many of their patients belong to a combined epidemiological plus genomics cluster, with the colour indicative of the viral haplotype (Fig. 2a). Patients with viral haplotypes not found in any combined cluster are coloured grey, and those patients for which sequence was unavailable are shown in black. Epidemiological cluster number is shown on the x-axis. Epidemiological clusters 1–3 are not displayed due to their large size. (c) Combined epidemiological plus genomic clusters from the acute and elective hospital sites. Clusters are coloured according to viral genomic haplotype (Fig. 2a). Clusters are shown broken down into ECDC patient nosocomial categories, with different shapes indicating the different categories. Enrichment of the cluster viral haplotype frequency in our study dataset versus the frequency in the community (Supplementary Material Table S7 and Methods) is shown on top of each cluster column.
These 14 final clusters are mapped onto the 44 epidemiology-only clusters to demonstrate the impact of introducing genomics (Fig. 2b, Supplementary Material Table S4). Of the 168 sequenced cases in epidemiological clusters, 93/168 (55)% were refuted as being part of a plausible transmission network with other sequenced cases in their epidemiological cluster, leaving 75 (45%) cases in the 14 final clusters (Fig. 2b, Supplementary Material Table S4). Thirteen epidemiological clusters (30%) had at least two cases from different final clusters, indicating multiple contemporaneous transmission clusters within an epidemiologically defined cluster (Fig. 2b, Supplementary Material Table S4).
Genomic clusters from different SARS-CoV-2 lineages were further assessed using maximum likelihood phylogenetic trees (Fig. 3 , Supplementary Material Fig. S1). This showed limited genetic diversity, with multiple community-onset cases showing genomic relatedness to nosocomial cases despite having no plausible epidemiological link. This illustrates the need for epidemiological linkage to postulate plausible transmission networks. No additional nosocomial cases could be linked to or excluded from existing clusters through review of the phylogenetic trees.
Fig. 3.
Phylogenetic tree (left panel) for sequences with Pangolin lineage assignment B.1. Tree tips are labelled with patient ID, colour-coded according to transmission cluster assigned in our combined epidemiological and genomic investigation. Symbols at the tree tips are displayed according to community-acquired or nosocomial infection classifications. Sequence sample dates are plotted in line with the tree tips using the same symbols in the right-hand panel; admission periods prior to the sample date for each patient are also displayed in this plot as horizontal lines.
Differences in final clusters with less stringent exclusion criteria of three or more SNP
Next, we reapplied our clustering method with a less stringent SNP threshold for excluding cases of three or more SNPs. This identified three further cases possibly linked to existing clusters (Supplementary Material Table S6): case 84 (probable nosocomial) and case 135 (community-onset) to STH3, and case 359 (definite nosocomial) to STH1. Including them in the final clusters would increase the proportion of sequenced nosocomial cases accounted for to 61/77 (79%).
Nosocomial cases not present in final clusters
Eighteen remaining sequenced nosocomial cases (18/77, 23%) are not present in the final clusters. We reviewed the epidemiological clusters in which these 18 remaining nosocomial cases were placed (Supplementary Material Table S7). In total, 265/344 cases (77%) in these epidemiological clusters were sequenced, and none shared a viral haplotype within less than two SNPs of a remaining nosocomial patient, excluding them from being part of a transmission network. Instead, it is plausible that non-sequenced cases in these epidemiological clusters (79/344, 23%) or other unidentified cases (e.g. point-source infectors like HCWs) could form a transmission cluster with our remaining nosocomial cases.
Originators of final transmission clusters, spatial distribution and enrichment of haplotypes
Seven of 14 cases (50%) did not include a community-onset or intermediate case that plausibly served as the potential originator of the cluster (Fig. 2, Fig. 4 , Supplementary Material Fig. S2), suggesting many clusters were originated by unidentified cases.
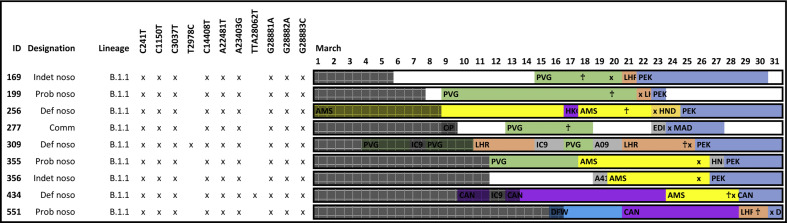
Fig. 4.
Pictorial representation of ward stays and movements for patients within cluster GUY1. Each row represents a different case. Patient ID, designation, lineage and single-nucleotide polymorphism (SNP) variants are marked. Ward movements between 1st and 31st March are displayed. Different wards are distinguished by given colours. Where there is more than one ward stay on 1 day, the longest ward stay is represented. The sample collection date is marked with an ‘x’. Symptom onset, where known, is marked with a cross ‘†’. Time periods outside of the acquisition period are shaded.
Of hospital clusters, 7/12 (58%) were contained within single wards and 5/12 (42%) spread across two or more wards (Supplementary Material Fig. S3). For in-depth ward movement data see Fig. 4 and Supplementary Material Fig. S2.
The validity of these 14 final clusters was supported by calculating haplotype enrichment compared to community sequences reported in the COG-UK CLIMB database [26] of between one- and 14-fold (median four-fold) (Fig. 2c, Supplementary Material Table S5).
Non-sequenced community-onset cases are unlikely to be originators of clusters
We assessed whether non-sequenced cases could have originated clusters which contained no community-onset or indeterminate cases by reviewing non-sequenced cases present in the same epidemiological cluster (Supplementary Material Table S8). We excluded cases as potential originators if (a) they were symptomatic after the first nosocomial case (or sampling date was later, if symptom onset not known), or (b) viral haplotype differed by two or more SNPs, or (c) if cases were not community-onset or indeterminate. Only 2/8 clusters without originators (25%) could have potentially been originated by a non-sequenced community-onset case with epidemiological linkage (case 50 or case 187 cluster GUY4, case 62 for cluster GUY5).
Delayed identification of cases may have contributed to transmission in five clusters
Conversely, where community-onset or indeterminate cases were found as possible originators, earlier testing after symptom onset could have identified possible originators in three clusters (Supplementary Material Fig. S2). For example, case 34 in STH2 was symptomatic for 3 days before sample collection, case 90 in STH3 for 5 days, and case 277 in GUY1 for 6 days. Additionally, other cases were tested several days after symptom onset, possibly facilitating onward transmission: for instance, case 173 in STH2, case 160 in GUY1, case 295 in GUY3 and case 471 in GUY5.
Evidence of cryptic transmission in four clusters
Four clusters had other evidence of cryptic transmission: clusters GUY1 and GUY2 involved different wards in the same building with an identical viral haplotype that was highly enriched compared with community haplotypes, suggesting that these clusters are linked by cryptic transmission. GUY4 and GUY5 both similarly involved neighbouring wards with high enrichment of viral haplotype. Of note, these neighbouring wards share multiple HCWs, including allied health professionals, cleaners, and visiting clinicians. These HCWs plausibly may have served as vectors for cryptic transmission between wards.
Representation of HCWs in transmission networks
HCWs were not offered SARS-CoV-2 testing when developing COVID-19-compatible symptoms; instead they self-isolated. As such, only 20 SARS-CoV-2 sequences from HCWs were available. Three (3/20) HCWs were sufficiently similar to plausibly link to our final clusters. The infectious diseases team undertook contact tracing, identifying on which wards they worked and for which patients they cared during the period of acquisition. One HCW (case 280) had cared for a nosocomial case (case 61) within cluster STH1, becoming symptomatic 5 days later. Case 280 can therefore be added to cluster STH1, although they are unlikely to be the originator of this cluster.
Given that HCWs were not routinely tested, we used HCW absenteeism due to COVID-19-related sickness or isolation as a marker for COVID-19 infection. Across the period, 337 working days were lost due to HCW COVID-19-related absenteeism across the nine main wards implicated in our hospital clusters (Supplementary Material Table S9), averaging 1.9 lost working days per ward each day.
From a hospital-wide cohort of 228 HCWs we collected information on COVID-19-compatible symptoms and judged seroconversion to SARS-CoV-2 IgG every 1–2 weeks (Supplementary Material Fig. S4 and Table S10); 43/228 (19%) seroconverted to SARS-CoV-2 IgG, with 13/43 (30%) being asymptomatic. Supplementary Material Fig. S4 presents the predicted period of peak HCW infectiousness based on a combination of ±2 days from date of symptom onset, or inferred from seroconversion where asymptomatic. The rapid rise in HCW infectiousness is predicted to occur between 16th and 25th March, overlapping with incidence of nosocomial cases.
Discussion
Applying epidemiological and genomic data described 14 transmission clusters, accounting for the vast majority of sequenced nosocomial cases. The majority of sequenced cases were refuted from being part of a plausible transmission network with other sequenced cases in their epidemiological cluster, and multiple contemporaneous clusters were found within clusters formed by epidemiology alone, emphasizing the importance of applying genomic data for transmission analysis. Haplotypes in these 14 final clusters were enriched 1–14-fold compared with the surrounding community, which increases confidence that they are true nosocomial clusters, an assessment which has not been used in other genomic studies of nosocomial transmission. Our final clusters contained a similar proportion of the probable (18/25, 72%) and definite (41/52, 79%, χ2 p=0.8) sequenced nosocomial cases, increasing confidence that probable nosocomial cases are genuine nosocomial acquisitions.
Furthermore, our analysis adds to the literature on SNP thresholds for analysis of nosocomial transmission. Importantly, relaxing the SNP threshold did not cause clustering of community-onset cases where transmission is unlikely to have occurred. We permitted epidemiological contact during previous admissions given the 14-day incubation time of SARS-CoV-2, which is not considered in published definitions of nosocomial cases [23]. Five patients in three clusters were epidemiologically linked during a previous admission (Supplementary Material Fig. S2).
Overlapping ward stays allow epidemiological linkage by inferring risk of exposure between cases. More granular information not available through our hospital computer systems may improve epidemiological linkage, for instance, a live bed state to determine the exact bed allocation and movements of patients between departments. This would allow environmental risk factors for acquisition to be identified, such as multi-bedded rooms, shared bathrooms, and air changes.
The presence of cryptic transmission and the absence of plausible originators in half of the clusters suggests that unidentified cases are involved in transmission: most likely HCWs or minimally symptomatic/asymptomatic patients. Even in clusters where a plausible originator is present, it is still possible that an unidentified case (e.g. an HCW) is responsible for introducing infection; however, this is less parsimonious. Cryptic transmission may also represent transmission in non-ward areas not covered by our epidemiological data. Lack of routine screening of patients and HCWs, along with poor recognition of milder yet relevant symptoms early in the pandemic (such as anosmia and upper respiratory coryzal illness) may have facilitated transmission as cases were missed. Our data suggest that routine screening of staff and patients may be beneficial to improve case ascertainment.
Importantly, nosocomial cases declined before any possible impact from universal surgical mask use by HCWs or banning of hospital visitors. This may be due to falling infection rates in the community after implementation of non-pharmacological measures, effectively social distancing, decreasing transmission to admitted patients in hospitals. Interestingly, community infections were predicted to peak around the same time as social distancing was introduced [4], with nosocomial cases beginning to fall around 7 days after this point, consistent with a delay of 5–7 days for incubation [28].
Moreover, nosocomial cases declined even whilst admission of community-onset cases continued to rise. This suggests that infection control measures can be effective at preventing transmission from admitted cases to other patients by rapid diagnosis, isolation and use of personal protective equipment. Community-onset cases may have passed peak viral shedding, often the first 4 days of illness [29], upon admission to hospital. Indeed in our cohort, admission was a median of 7 days after symptom onset. Instead, we hypothesize that infection is often introduced into the hospital by HCWs or patients who are minimally symptomatic/asymptomatic, who remain unidentified.
In summary, this study supports the role of genome sequencing in SARS-CoV-2 outbreak investigation. In addition, the presence of cryptic transmission and the implication of unidentified cases suggests that routine screening of both HCWs and patients may be valuable. It will be important to assess whether interventions such as universal mask use and intermittent screening limit nosocomial transmission.
Ethics
Favourable opinion to conduct this work was granted by the North West Preston Research Ethics Committee (Reference 18/NW/0584). The COVID-19 Genomics UK (COG-UK) consortium study protocol was approved by the Public Health England Research Ethics and Governance Group (reference: R&D NR0195).
Author contributions
LBS, CLF, JDE and ARA designed the study, participated in dry and wet laboratory, performed analysis and drafted the manuscript. UT curated metadata and performed epidemiological analysis of outbreaks. CLF, AA-M, TC, AWS, HDW and GB performed nanopore sequencing. BM provided epidemiological data. SP, RPG, SJDN, MHM and KJD provided supervisory support. EC, MTKI, PRC, RB, STD and GN managed patient samples, including PCR testing. COG-UK curated and maintained the CLIMB database, and provided input into manuscript. All authors approved the final manuscript.
Transparency declaration
The authors declare that they have no competing interests. This work was supported by the King's Together Multi and Interdisciplinary Research Scheme (Wellcome Trust Revenue Retention Award) and the National Institute for Health Research (NIHR) Biomedical Research Centre programme of Infection and Immunity (RJ112/N027) based at Guy's and St Thomas' National Health Service (NHS) Foundation Trust and King's College London. COG-UK is supported by funding from the Medical Research Council (MRC) part of UK Research & Innovation (UKRI), the National Institute of Health Research (NIHR) and Genome Research Limited, operating as the Wellcome Sanger Institute. This work was also supported by the Guy's and St Thomas' Charity (https://www.gsttcharity.org.uk/).
Editor: L. Kaiser
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2021.07.040.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.John Hopkins University . 2020. COVID-19 map-johns hopkins coronavirus resource center.https://coronavirus.jhu.edu/map.html [Google Scholar]
- 3.Birrell P., Blake J., van Leeuwen E. 2020. COVID-19: nowcast and forecast.https://joshuablake.github.io/public-RTM-reports/iframe.html [Google Scholar]
- 4.Edelstein M., Obi C., Chand M., Hopkins S., Brown K., Ramsay M. SARS-CoV-2 infection in London, England: changes to community point prevalence around lockdown time, March–May 2020. J Epidemiol Community Health. 2021;75:185–188. doi: 10.1136/jech-2020-214730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coronavirus (COVID-19) in the UK. GOV.uk; 2020. https://coronavirus.data.gov.uk/healthcare n.d. [Google Scholar]
- 6.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter B., Collins J.T., Barlow-Pay F., Rickard F., Bruce E., Verduri A., et al. Nosocomial COVID-19 infection: examining the risk of mortality. The COPE-Nosocomial Study (COVID in Older PEople) J Hosp Infect. 2020;106:376–384. doi: 10.1016/j.jhin.2020.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scientific Advisory Group for Emergencies . 2020. Dynamic CO-CIN report to SAGE and NERVTAG.https://www.gov.uk/government/publications/dynamic-co-cin-report-to-sage-and-nervtag-1-april-2020 [Google Scholar]
- 9.Rickman H.M., Rampling T., Shaw K., Martinez-Garcia G., Hail L., Coen P., et al. Nosocomial transmission of COVID-19: a retrospective study of 66 hospital-acquired cases in a London teaching hospital. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor J., Rangaiah J., Narasimhan S., Clark J., Alexander Z., Manuel R., et al. Nosocomial COVID-19: experience from a large acute NHS Trust in South-West London. J Hosp Infect. 2020;106:621–625. doi: 10.1016/j.jhin.2020.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao G., Tang S., Yang D., Shi W., Wang X., Wang H., et al. The potential transmission of SARS-CoV-2 from patients with negative RT-PCR swab tests to others: two related clusters of COVID-19 outbreak. Jpn J Infect Dis. 2020;73:399–403. doi: 10.7883/yoken.JJID.2020.165. [DOI] [PubMed] [Google Scholar]
- 12.Vanhems P. Fast nosocomial spread of SARS-CoV2 in a French geriatric unit Lyon Study Group on Covid-19 infection. Infect Contr Hosp Epidemiol. 2020:1–4. doi: 10.1017/ice.2020.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ji H., Liu L., Huang T., Zhu Y. Nosocomial infections in psychiatric hospitals during the COVID-19 outbreak. Eur J Psychiatr. 2020;34:177–179. doi: 10.1016/j.ejpsy.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lauer S.A., Grantz K.H., Bi Q., Jones F.K., Zheng Q., Meredith H.R., et al. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med. 2020;172:577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wiersinga W.J., Rhodes A., Cheng A.C., Peacock S.J., Prescott H.C. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;324:782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- 16.Meredith L.W., Hamilton W.L., Warne B., Houldcroft C.J., Hosmillo M., Jahun A.S., et al. Rapid implementation of SARS-CoV-2 sequencing to investigate cases of health-care associated COVID-19: a prospective genomic surveillance study. Lancet Infect Dis. 2020;20:1263–1271. doi: 10.1016/S1473-3099(20)30562-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lucey M., Macori G., Mullane N., Sutton-Fitzpatrick U., Gonzalez G., Coughlan S., et al. Whole-genome sequencing to track SARS-CoV-2 transmission in nosocomial outbreaks. Clin Infect Dis. 2021;72:e727–e735. doi: 10.1093/cid/ciaa1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Dorp L., Acman M., Richard D., Shaw L.P., Ford C.E., Ormond L., et al. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect Genet Evol. 2020;83:104351. doi: 10.1016/j.meegid.2020.104351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quick J. nCoV-2019 sequencing protocol v1 (protocols.io.bbmuik6w) n.d. 10.17504/protocols.io.bbmuik6w. [DOI]
- 20.artic-network https://github.com/artic-network/artic-ncov2019 artic-network/artic-ncov2019 n.d.
- 21.Artic Network https://artic.network/ncov-2019/ncov2019-bioinformatics-sop.html n.d.
- 22.Pangolin Github; n.d.
- 23.Surveillance definitions for COVID-19. https://www.ecdc.europa.eu/en/covid-19/surveillance/surveillance-definitions n.d.
- 24.Stirrup O., Hughes J., Parker M., Partridge D.G., Shepherd J.G., Blackstone J., et al. Rapid feedback on hospital onset SARS-CoV-2 infections combining epidemiological and sequencing data. Elife. 2021;10 doi: 10.7554/eLife.65828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rockett R.J., Arnott A., Lam C., Sadsad R., Timms V., Gray K.-A., et al. Revealing COVID-19 transmission in Australia by SARS-CoV-2 genome sequencing and agent-based modeling. Nat Med. 2020;26:1398–1404. doi: 10.1038/s41591-020-1000-7. [DOI] [PubMed] [Google Scholar]
- 26.Nicholls S.M., Poplawski R., Bull M.J., Underwood A., Chapman M., Abu-Dahab K., et al. MAJORA: continuous integration supporting decentralised sequencing for SARS-CoV-2 genomic surveillance. Cold Spring Harbor Lab. 2020:2020. doi: 10.1186/s13059-021-02395-y. 10.06.328328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seow J., Graham C., Merrick B., Acors S., Pickering S., Steel K.J.A., et al. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat Microbiol. 2020;5:1598–1607. doi: 10.1038/s41564-020-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McAloon C., Collins Á., Hunt K., Barber A., Byrne A.W., Butler F., et al. Incubation period of COVID-19: a rapid systematic review and meta-analysis of observational research. BMJ Open. 2020;10 doi: 10.1136/bmjopen-2020-039652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singanayagam A., Patel M., Charlett A., Lopez Bernal J., Saliba V., Ellis J., et al. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.32. 2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.