Abstract
Innate Lymphoid Cells (ILCs) are a class of innate immune cells that form the first line of defense against internal or external abiotic and biotic challenges in the mammalian hosts. As they reside in both the lymphoid and non-lymphoid tissues, they are involved in clearing the pathogens through direct killing or by secretion of cytokines that modulate the adaptive immune responses. There is burgeoning evidence that these cells are important in clearing viral infections; therefore, it is critical to understand their role in the resolution or exacerbation of the disease caused by severe acute respiratory syndrome coronavirus (SARS-CoV-2). In this review, we summarize the recent findings related to ILCs in response to SARS-CoV-2 infections.
Keywords: Innate lymphoid cells, SARS CoV-2, NK cells
Abbreviations
- HSC
hematopoietic stem cell
- CMP
common myeloid progenitor
- CLP
common lymphoid progenitor
- CHILP
common helper innate lymphoid precursors
- ILCp
Innate lymphoid cells progenitor
- NKmem
Natural killer memory cells
- NCR
natural cytotoxicity receptor
- CRTH2
Chemoattractant receptor expressed on Th2 cells
- Eomes
Eomesodermin
- GATA3
GATA binding protein 3
- IFN
Interferon
- ILCs
Innate lymphoid cells
- LTi
Lymphoid tissue-inducer cells
- NKs
Natural killer cells
- PLZF
Promyelocytic leukemia zinc finger
- ROR-α,
Retinoic acid receptor-related orphan receptor-α;
- ROR-γt
Retinoic acid receptor-related orphan receptor-γt
- TCF1
T cell factor 1
- Th2
T helper 2 cells
- TNF
Tumor necrosis factor
- TSLP
Thymic stromal lymphopoietin
1. Introduction
Innate lymphoid cells (ILCs) arise from the Hemopoietic stem cells (HSCs) in the bone marrow and perform similar functions as the cells of the adaptive immune system but do not express the specific antigen receptors (Constantinides et al., 2014; Klose et al., 2014; Eberl et al., 2015; Kiessling et al., 1976). Like the CD8+ cytotoxic T cells of the adaptive immune system, one subset of the ILCs, the natural killer (NK) subset, perform lytic functions, and like the type 1 helper (Th1), Th2, and Th17 subsets of CD4+ T cells, ILC1, ILC2, and ILC3 cell subsets perform helper functions through secretion of cytokines (Klose and Artis, 2020; Mortha and Burrows, 2018). Similar to regulatory T cells (Tregs), regulatory subsets of ILC (ILCregs) that can modulate ongoing adaptive immune responses are now being described in the literature (Crome et al., 2017; O'Connor et al., 2021). While some mature NK subsets and ILC precursors circulate through the blood, ILCs, for the most part, reside in the mucosal epithelia (Klose and Diefenbach, 2014). They are the first responders who encounter the incoming pathogen to initiate and modulate innate and adaptive immune responses soon after pathogen recognition. Thus, ILCs play a pivotal role in immune surveillance and form the front-line of immune defense (Nabekura and Shibuya, 2021a, 2021b).
The ongoing coronavirus disease 2019 (COVID-19) pandemic, caused by a highly transmissible and pathogenic severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has been posing a significant challenge to humanity worldwide. The disease is prevalent globally, with 123 million confirmed cases and 2.70 million deaths (Dong et al., 2020). There have been 36.1 million confirmed cases in the United States alone, with 616,459 deaths as August 10, 2021 (CDC, 2021). SARS-CoV-2, a respiratory pathogen, is a positive-sense single-stranded RNA virus that attacks the host respiratory tract by primarily engaging the angiotensin-converting enzyme 2 (ACE2) receptor expressed on the airway epithelial cells (Ehre, 2020). The ACE2 receptor is identified as the primary receptor for viral entry, although the tyrosine-protein kinase receptor UFO (AXL) can also function as a receptor in the absence of ACE2 (Wang et al., 2021). Within hours after viral entry, numerous virions are produced, which bud into the airways leading to the virus's spread in the lung tissue (Cevik et al., 2020). As the first line of defense, epithelial cells, neutrophils, dendritic cells, and macrophages actively participate in mounting innate and facilitate adaptive responses to SARS-CoV-2 (de Candia et al., 2021; O'Connell and Aldhamen, 2020; Sette and Crotty, 2021). The ILCs resident in the respiratory epithelial tissue play a seminal role in immune surveillance and contribute substantially to lung defense, pathology, and disease in response to an incoming pathogen or microenvironmental changes (Stehle et al., 2018). Since ILCs lack antigen-specific receptors, it is speculated that they can be activated by inflammatory mediators or by r some yet to be identified specific receptor/s. It is possible that ILC are activated by direct or indirect interactions of pathogen-associated molecular patterns (PAMPs) with pathogen recognition receptors (PRRs) that are either membrane-bound, vesicular or cytosolic (Ranjan et al., 2009).
1.1. Overview of the ILC subsets
In humans, based on the expression of the transcription factors, cell surface phenotypic markers, and signature cytokine production profiles the mature subsets of ILCs have classically been categorized into the cytokine-secreting or cytotoxic NK subset, helper ILC1, ILC2, ILC3 subsets, and the lymphoid tissue inducer (LTi) subset (Krabbendam et al., 2021). In contrast to these ILCs, the regulatory ILCs have not yet been classified into a separate subset (Crome et al., 2017; O'Connor et al., 2021). We have denoted the ILC (regs) as a class of its own for the purposes of this review. The origin of all classes of ILCs can be traced to the HSCs in the bone marrow, which in response to microenvironmental cues, differentiate into several subsets of ILCs through multiple stages as schematized in Fig. 1 . ILC differentiation begins with the mobilization of the bone marrow-derived CD34+CD45RA+ HSCs circulating in the blood to the lymphoid organs, where they differentiate into the common lymphoid progenitor (CLP) and the common myeloid progenitor (CMP) cells. The CLPs give rise to T and B lymphocytes, NK cells, and ILCs subsets, while the CMPs differentiate into granulocytes, monocytes, dendritic cells, erythrocytes, and platelets (Zipori, 1992). Further differentiation of these progenitor cells into the different subsets of ILCs is regulated by the expression of several transcription factors highlighted in Fig. 1. Increasing evidence provides proof that the classification of the ILC subsets into the five classes described below are plastic in nature because, in response to changes in the inflammatory milieu or viral and bacterial challenges, ILC subsets can transdifferentiate (Bal et al., 2020).
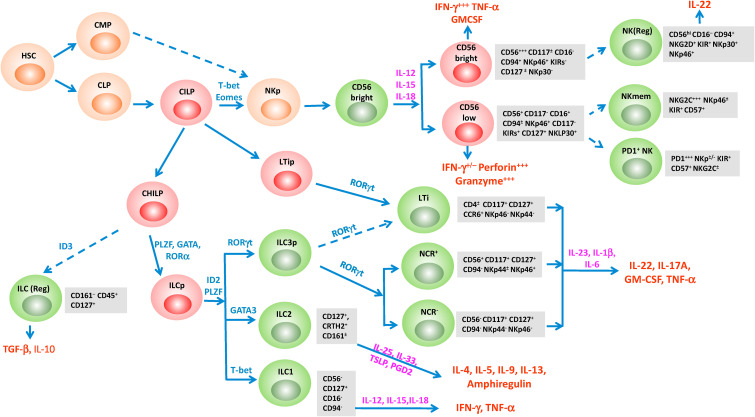
Fig. 1.
Human innate lymphoid cells (ILCs) development. Differentiation from the hemopoietic stem cells (HSCs) giving rise to natural killer (NK) cells and various subsets of ILC occurs through multiple steps as detailed in the text. The subsets are color-coded to indicate their localization in the bone marrow (orange), in the peripheral blood (red), or the tissue (green). The indicated transcription factors and cytokines facilitate the differentiation into specific subsets with distinct functions. Broken arrows indicate a potential pathway of differentiation.
The NK cells, the first identified ILC subsets, develop from the common innate lymphoid cell precursor (CILP) that arises from the CLPs in the bone marrow. The CILPs, under the influence of transcription factors, T-bet and eomesodermin (Eomes), differentiate into the NK cell precursors (NKp), which gives rise to CD56bright and further to CD56brightCD16lowand CD56lowCD16high-subsets. While CILPs are considered to be the primary precursors to NKp, there is evidence that CMPs give rise to NKp cells as well (Grzywacz et al., 2011). NK cells originally detected in the spleen, peripheral blood, and bone marrow are found in the liver, uterus, and lymph nodes. NK cells primarily originate in the bone marrow. They can rapidly migrate to the site of infection or inflammation, where they are involved in defensive roles against pathogens or immunosurveillance against tumors (Del Zotto et al., 2017). NK cells also circulate in the blood of which 90% are CD56lowCD16high and 10% are CD56brightCD16low (Simoni and Newell, 2018; Swann et al., 2007). These subsets can be activated by PRR ligands, and their levels and functions under homeostasis are regulated by type I interferon (IFN) (Pende et al., 2019a; Walzer et al., 2005). The functionality of the NK cells is regulated by the expression of several inhibitory and activation receptors that integrate signals to govern NK cells' responses. The inhibitory receptors expressed on the surface of NK cells, primarily members of killer immunoglobulin-like receptors (KIRs), namely 2DL1/L2/L3, 3DL1/L2/L3, NKG2A/CD94, and CD85j (ILT2, LIR1), can recognize the expression of class I MHC (HLA A, B, C, E, F, and G) on normal cells to attenuate NK cell response thereby preventing NK cells from killing normal cells (Del Zotto et al., 2017; Pende et al., 2019b). On the other hand, virus-infected and tumor cells express reduced levels of class I MHC, leading to failure to engage inhibitory receptors resulting in NK cell-mediated killing through the release of perforin and granzymes (Ikram et al., 2021). In addition to expressing inhibiting receptors, NK cells also express killer activation receptors (KAR) which include some members of the killer cell Ig-like receptor (KIR) family, namely 2DS1/S2/S4, 3DS1, and NKG2C (Rajalingam, 2011). KARs recognize MHC class I polypeptide–related sequences A (MICA) and B (MICB) expressed only on infected or tumor cells but not on normal cells (Liu et al., 2019). CD56brightCD16low KIR + NK subsets predominantly secrete IFN-γ and do not exhibit cytotoxicity (Poli et al., 2009) after receptor activation. On the other hand, CD56lowCD16highKIR− NK subsets express FcRγΙΙΙ (CD16), engage in antibody-dependent cellular toxicity (ADCC)-mediated clearance of virus-infected cells by expressing high levels of granzyme and perforin with little or no expression of IFN-γ (Wang et al., 2015) when stimulated with cytokines, interleukin (IL)-12, IL-15, and IL-18. In human lungs, the vast majority of the NK cells display the CD56dimCD16- phenotype like the circulating subset, and express CD69, CD49a, and CD103 once they become tissue-resident (Marquardt et al., 2017; Dogra et al., 2020; Cooper et al., 2018). A subset of programmed cell death protein-1 (PD-1+) NK cells were detected in the peripheral blood of approximately one fourth of healthy subjects which is characterized by the CD56dimNKG2A−KIR+CD57+ phenotype (Pesce et al., 2017). This NK subset displays low proliferative responses and impaired antitumor activity (Pesce et al., 2017; Della Chiesa et al., 2016). PD-1 expression can also be induced on NK cells upon many viral infections including murine hepatitis virus strain-3 (MHV-), chronic HIV-1 and murine CMV infection (Quatrini et al., 2018; Norris et al., 2012; Chen et al., 2011). An additional NK subset with regulatory functions has been identified from high-grade serous tumors as CD56hiCD16−CD94+NKG2D+KIR+NKP30+NKp46+ that can produce IL-22 and limit T cell cytokine production and expansion (Crome et al., 2017). Viral infections with human cytomegalovirus (HCMV), human immunodeficiency virus (HIV), vesicular stomatitis virus (VSV), influenza virus generate tissue-resident memory/adaptive NK cells under the influence of cytokines IL-12, IL-15, and IL-18 (Cooper et al., 2009, 2018). (Crome et al., 2017; Della Chiesa et al., 2016; Quatrini et al., 2020)- NK memory cells, like memory T cells, undergo clonal-like expansion upon subsequent encounter with the same antigen to generate memory cells that are activated to lyse the target cells (Gabrielli et al., 2016).
The CILPs also give rise to the common helper innate lymphoid cell precursors (CHILP) that further differentiate into three distinct lineages, ILC1, ILC2, and ILC3 subsets via a common precursor, ILCp. ILC1, ILC2, and ILC3 subsets require distinct cytokines for maturation to secrete Th1, Th2, and Th17 cytokines, respectively. These subsets primarily populate mucosal surfaces and are replenished from the bone marrow-derived precursors (Klose and Artis, 2016). ILC1 constitutively expresses the T-box transcription factor (T-bet) and responds to IL-12, IL-15, and IL-18 to produce IFN-γ and tumor necrosis factor (TNF-α) (Fuchs et al., 2013). ILC1 cells are present in the liver, gut, salivary glands, and adipose tissue and mediate immune responses to intracellular pathogens like CD8+ and CD4+ Th1 cells. In addition to the origination of ILCs in the lymphoid organ, studies in mouse models present evidence that there is a feedback loop between the ILC1 development and the HSC residing in the liver controlled by ICL-1 driven interferon−γ production (IFN-γ) (Bai et al., 2021). The ILC2 development from the ILCp cells requires the IL-7 mediated expression of the transcription factor GATA binding protein 3 (GATA3) (Hoyler et al., 2012; Sheikh and Abraham, 2019). ILC2 are found at the mucosal tissue sites, including lung, small intestine, colon, and mesenteric lymph node (MLN), as well as in the bone marrow, spleen, liver, kidney, and adipocyte tissue and are activated by cytokines including IL-33, IL-25, thymic stromal lymphopoietin (TSLP), prostaglandin D2 (PGD2) to produce high levels of IL-4, IL-5, IL-9, IL-15. IL-13 and the epidermal growth factor, amphiregulin (Herbert et al., 2019; Mjosberg et al., 2012). Like CD4 T helper 2 (Th2) cells, ILC2 promotes protective immunity to helminth infections, allergic reaction, tissue remodeling, and metabolic homeostasis (Moro et al., 2010; Price et al., 2010). ILC3 express the transcription factor retinoic acid receptor-related orphan receptor γt (ROR-γt) and produce IL-17A, IL-22, granulocyte-macrophage colony-stimulating factor (GM-CSF), and/or tumor necrosis factor α (TNF-α) in response to IL-1β, IL-6, IL-23, and aryl hydrocarbon receptor (AHR) ligands (Ardain et al., 2019). ILC3 ranks the second most frequent subsets of the human pulmonary ILCs (Simoni et al., 2017). There are two subsets of ILC3 populations which express different levels of natural cytotoxicity receptor (NCR). NCR− and NCR+ subsets are the innate equivalent of Th17 and Th22 cells, respectively (Vacca et al., 2015). Like Th17 cells, these cells respond to extracellular microbial (fungi, bacteria) infections (Ardain et al., 2019).
The CLIPs are also precursors to the LTi precursor (LtiP) cells which ultimately give rise to the LTi subset (Cherrier and Eberl, 2012). Like the ILC3 subset, the LTi subset strictly depends on transcription factor RORγt for development but functions mainly in the fetal stages (Eberl et al., 2004), although LTi-like cells can also be generated during adulthood (Cherrier and Eberl, 2012; van de Pavert). LTi cells can produce cytokine IL-22 and provide protective immune responses against extracellular bacteria (Zhong et al., 2018). LTi cells are required for lymphoid organogenesis (Scandella et al., 2008) and directly influence the development and function of adaptive immune cells (Sonnenberg and Hepworth, 2019).
In addition to the above classified sub types of ILCs that mirror the Th1/Th2 type T cells, another category of innate cells that are becoming increasingly important are the ILC (regs). ILC (regs) initially identified in the lamina propria of small intestine are Lin−CD45+CD127+IL-10+ cells, do not express markers associated with ILC1, ILC2, and IL3 subsets, and produce IL-10. The lineage of these cells can be tracked to CHILP (Wang et al., 2017). A recent study reports describes a similar type of regulatory cells in the tonsils (O'Connor et al., 2021). Follicular regulatory ILC (ILCFR) is a unique population identified by flow cytometry of tonsillar mononuclear cells from individuals with repeated chronic tonsillar infections, but who were uninfected at the time of tissue collection (O'Connor et al., 2021). These cells are Lin−CD161−CD45+CD127loCD74+ ID3+CXCR5+ and lacking IL7RA), exhibiting very low expression of ILC1 (NKp44, CD56, Tbet), ILC2 (KLRG1, GATA3), and ILC3 (NKp44) markers, therefore, these cells are now classified in their own class. ILCFR while identified in tonsils are also found in lymph nodes with primary residence in the germinal center (GC) follicles where they modulate T cell-B cell interactions to attenuate IgG production from GC-B cells. Comparison of transcriptional profiles ILCFR with other ILC subset showed that ILCFR lack expression of transcriptional factors, RORγt, Tbet, GATA3, associated with development of the ILC1, ILC2, and ILC3 subsets but express transcriptions factors inhibitor of differentiation (ID)3, ID2 and NFIL3. These cells produce robust amounts of TGF-β upon activation and are expanded during HIV infection in the lymph nodes. An expansion of ILCFR has been observed during chronic HIV infection suggesting that they could play a key role in the dysfunction observed in virus-modulated microenvironments. Since the origin of the cells is not delineated, we have denoted this category of cells as originating the CHILP by broken lines in Fig. 1, however, additional research needs to be conducted with these cells for further elaboration on their origin, functions and markers.
The majority of ILCs found in the lung are bona fide tissue-resident cells locally maintained through self-renewal (Gasteiger et al., 2015). In human lungs, under steady-state conditions, ILCs exist in much lower numbers and are characterized as CD45+, Lin−(i.e., CD3, CD19, CD11c, CD11b), and CD127+ cells (De Grove et al., 2016; Yu et al., 2016). The majority of the ILC subsets in the human lungs comprise the NCR−ILC3 followed by the ILC2 and ILC1 subsets (De Grove et al., 2016). Although ILC2 is detectable in human fetal lungs as early as gestational week 14, they are rare in newborns' lungs. Still, in response to IL-33 secreted by lung epithelial and stromal cells, they can quickly expand to reach the levels detected in adults by postnatal day 8 (Saluzzo et al., 2017; Dahlgren et al., 2019). In addition, these cytokines enable the neonatal ILC2s to reside longer in the adult lung and respond efficiently to challenges in adult life ((Nussbaum et al., 2013), (Molofsky et al., 2015)). Resting lung ILC2s can serve as a rapid source of IL-5, IL-13, IL-17A, IL-22, or IFN-γ which is important to manage allergic lung diseases (Nussbaum et al., 2013; Barlow and McKenzie, 2019).
1.2. Clinical symptoms of SARS-CoV-2 infections
The ongoing pandemic by the SARS-CoV-2 virus causes a range of clinical symptoms, from asymptomatic, to moderate cases who recover within weeks, to severe pneumonia, acute respiratory distress syndrome (ARDS), multi-organ failure, and an in-hospital mortality reported to range from 12% to 28% (Wiersinga et al., 2020; Finelli et al., 2021). The observed disparity in clinical symptoms can be in part explained by age of the patients, a hyperactive immune response, viral loads or underlying medical conditions. Various inflammatory cytokines and chemokines (IL-1, IP-10, and MCP-1) are upregulated in the serum from patients with COVID-19 in proportions with viral load and lung injury (Jamal et al., 2021; Ombrello and Schulert, 2021; Xu et al., 2020). Patients with the severe progression of COVID-19 also demonstrate lymphopenia, increased serum ferritin, D-dimer, C-reactive proteins (CRP), and lactic dehydrogenase (LDH) with severe elevation in TNF-α, IL-1, IL-6, IL-18, IL-8, IL-10, and MCP-1 (Ponti et al., 2020). In addition to the dysregulation in cytokines, using a cohort of 32 patients, Galani et al. demonstrate that IFN-γ and type I IFN production are both diminished and delayed and induced in only a fraction of the critically ill patients (Galani et al., 2021). The altered cytokine pattern correlates with longer hospitalization and higher incidences of severe disease and mortality (Galani et al., 2021). As patients recover, the observed imbalance in immune cells and cytokine profiles are reset to levels as observed in healthy individuals (Kuri-Cervantes et al., 2020; Jesenak et al., 2020). The altered expression of cytokines reveals important insights into immune dysregulation and suggests different ILC subsets in the disease pathology and severity outcomes. Alterations in the frequency of circulating innate and adaptive immune cells are the hallmark of severe disease caused by SARS-CoV-2 (de Candia et al., 2021; Chen and John Wherry, 2020; Diao et al., 2020; Mathew et al., 2020). COVID-19 severity is associated with an about 1.8-fold reduction in ILCs and 2.31-fold reduction in CD16+ NK cells. Further, ILCs abundance negatively correlates with the odds of being hospitalized and other observed correlates of clinical symptoms such as decreased blood C-reactive protein, D-dimer, and erythrocyte sedimental rate (Silverstein et al., 2021). Fig. 2 and the section below highlights the observed changes in the ILC subsets in response to SARS-CoV-2 infection.
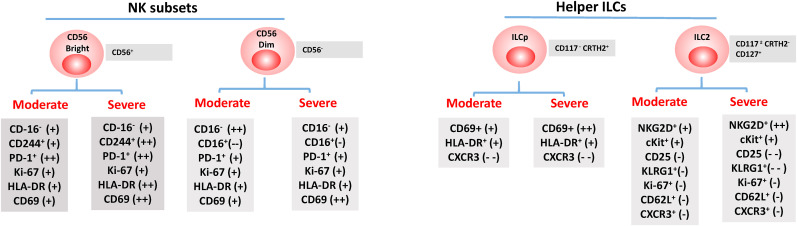
Fig. 2.
Human ILCs in COVID-19. Subsets of ILCs identified in moderately and severely ill COVID-19 patients that are differentially regulated when compared to healthy individuals. Inhibitory (PD-1), cytotoxic (CD16), chemokine receptors (CXCR3), homing (CD62L), activation (CD69, HLA-DR, Ki-67), and markers identified within each subset are shown in a box below each subset. Differentiation markers used to mark the populations in COVID-19 patients are displayed to the right of each subset. The plus (+) and minus (−) indicate the level of change observed in severe and moderate COVID-19 patient samples compared to the levels in healthy individuals.
1.3. NK cells in COVID-19
The importance of NK cells in clearing viral infection is supported by observations that patients with NK cell deficiencies are predisposed to severe recurrent viral infections (Biron et al., 1989; Orange, 2002). The role of NK cells indirectly killing infected cells or influencing T cell responses is also established with several other viruses like influenza A, dengue virus, or after vaccination with live-attenuated yellow fever virus (van Erp et al., 2019; Hammer et al., 2018). While some activated NK cells can directly recognize viral-infected cells to kill and promote viral clearance, NK cells have the potential to secrete cytokines such as IFN-γ, TNF-α, IL-10, IL-5, and IL-13, and chemokines, IL-8, MIP-1α, and MIP-1β that can drive inflammation (Horowitz et al., 2011; Zitti and Bryceson, 2018; Fauriat et al., 2010). Patients with severe SARS-CoV-2 infection show reduced levels of circulating NK cells, particularly CD56bright cells, which correlates with disease severity 4–11 days post-onset compared to healthy individuals (Maucourant et al., 2020). Since NK cells do not express the ACE2 receptor, it is debatable how the virus causes the depletion or sequestration or altered circulation of the NK subset. While CD56bright cells have an increased capacity to produce cytokines and proliferate, CD56dim cells are primarily cytolytic and have a role in directly killing the virus-infected cells. A sharp decline in the CD56dimCD16- subset is observed in pediatric patients infected with SARS-CoV-2 compared to adults (Vella et al., 2021). In patients with late-stage melanoma or HIV infection it is observed that hyperstimulation of the CD56dim population causes terminal differentiation of the cells into a phenotype that mimics the phenotype of exhausted cells and memory T cells (Amand et al., 2017; Hong et al., 2010). These terminally differentiated NK cells (PD1+NK) express markers such as PD-1 and CD57 and have lower effector functions (Fig. 1). Consistent with this finding, an increase in CD56dimCD57+ NK cells are observed in fatal cases with COVID-19 (Varchetta et al., 2021). NK cells expressing CD16 mediate ADCC; however, a marked decrease in the CD56+CD16+ populations correlate with the severity of the disease (Maucourant et al., 2020; Varchetta et al., 2021; Hu et al., 1995; Li et al., 2020). Increased frequency of KIR+ (especially NKG2A) NK cells are observed in COVID-19 patients, which returns to normal frequency after clinical recovery (Zheng et al., 2020a). In the future, it would be interesting to investigate if the reduced frequency of CD16+ NK cells that participate in ADCC and the increased expression of KIR+ NK cells which inhibit the direct killing of virus-infected cells, are one of the elusive viral mechanisms or unintended consequence of the host cytokine storm observed in COVID-19 patients.
Alterations in levels of activation and homing markers in the circulating NK populations correlate with the severity of the disease. This suggests that NK cells migrate from the peripheral blood to the lungs during infection (Zheng et al., 2020a). In addition to changes in activation makers, NK cells in patients with advanced stages of the disease are functionally defective and exhibit exhaustion phenotype, expressing PD1 (Li et al., 2020). Varchetta et al. (2021) demonstrate that in patients, IFN-γ production by NK cells was significantly downregulated, along with a reduction in degranulation of cytotoxic granules in CD56bright cells. Further, there was a negative correlation between degranulating (CD107a+), and IFN-γ-producing NK cells and serum CRP values (Varchetta et al., 2021). Similarly, negative correlation between the cytotoxic activity of NK cells and serum IL6 levels was reported in ICU patients and off-label treatment of tocilizumab, a humanized monoclonal antibody against the IL-6 receptor restored the cytotoxic potential of NK cells (Mazzoni et al., 2020). These findings suggest that SARS-CoV-2 infection leads to reduced circulating NK, altered subset composition and function, which impact the severity of the disease resulting in fatalities. This notion is further supported by the evidence that SARS-CoV-2 survivors had higher levels of NK cells than the non-survivors (Yan et al., 2020).
1.4. Helper ILCs in COVID-19
Unlike several studies that have characterized the populations of NK cells in COVID-19 patients, studies describing the impact of SARS-CoV-2 on the frequency and functionality of other ILCs subsets are limited. Although helper ILC subsets are primarily tissue-resident, they can also circulate in the blood. Therefore, our understanding of these cells during COVID-19 is mainly limited to population changes in the peripheral blood. In the lungs, the ILC2 subsets limit the viral- and allergen-induced type 2 responses, recruitment of eosinophils, termination of inflammatory responses, and tissue repair (van der Ploeg et al., 2020). Similar to the NK subset, there is a decrease in total levels of the three ILCs subsets and the ILCp subsets in peripheral blood of moderate and severe COVID‐19 patients; however, when calculated as percent of total ILCs, only the ILC2 subset increases in peripheral blood of patients with the moderate disease when compared to healthy controls (Garcia et al., 2020; Gomez-Cadena et al., 2021) The levels of the helper ILC subsets in samples from patients who have recovered are similar to those observed in healthy controls (Garcia et al., 2020; Gomez-Cadena et al., 2021). The SARS-CoV-2 genome relies on the activity of papain-like proteases (PLpro) to generate a functional replicase complex that regulates viral spread and innate immunity (Shin et al., 2020). Papain is shown to induce pulmonary fitness of ILC2 cells in the lungs during allergic inflammation and asthma (Halim et al., 2014). Administration of SARS-CoV-2 PLpro in mice lungs increases the frequency of IL5-secreting ILC2 in the lungs (Garcia et al., 2020). Like these observations in mice, patients exhibiting moderate disease have an increased level of IL-33, IL-5, IL-13 and an increase in the levels of ILC2 (Gomez-Cadena et al., 2021).
The ILC2 population is further divided based on the expression of CD117 (Garcia et al., 2020) into CD117high and CD117lo subsets. The CD117lo subset secrete more type 2 cytokines and is expanded in COVID-19 patients (Gomez-Cadena et al., 2021; Hochdorfer et al., 2019). There is not only a general decrease in the total ILC population but also alternations in the expression of activation, migration, and differentiation markers that correlate with the severity of the disease in the COVID-19 patients (Silverstein et al., 2021). ILC2 and ILCp express enhanced levels of CD69 expression and reduced levels of CXCR3 and CCR4 (Garcia et al., 2020; Gomez-Cadena et al., 2021). In severe COVID-19 patients, there is an increase in the activating receptors NKG2D+ in the ILC2 subset and a significant decrease in inhibitory receptors CD25 and KLRG1 (Garcia et al., 2020; Gomez-Cadena et al., 2021). These data suggest that COVID-19 modulates the levels of the total ILC population in the peripheral blood, with most changes occurring in the ILC2 subset. These changes in the ILC subsets in the peripheral blood correlate with increased expression of cytokines, IL-5, IL-13 that are secreted by the ILC2 (Gomez-Cadena et al., 2021) (See Fig. 1). Further, the severity of disease, requirement for hospitalization, and increased duration of hospital stay correlate with reduced numbers of ILCs, indicating the vital role played by ILCs in SARS-CoV-2 infections (Silverstein et al., 2021). It is not known yet, if SARS-CoV-2 infection impacts the phenotype, differentiation, migration and functionality of the LTi subset.
1.5. Changes in molecular signatures of ILCs populations by scRNA-seq
In addition to the changes in ILC subsets explored using the traditional flow cytometry methods described above, single-cell RNA sequencing (scRNA-seq) is also considerably advancing our knowledge of ILC subsets and providing important insights into the biology and role of ILCs in infectious diseases (Ziegenhain et al., 2017; Stuart et al., 2019; Papalexi and Satija, 2018). The scRNA-seq analysis enables an unbiased alternative workflow grouping of cells based on their transcriptional signatures, which allows the sequencing of cells without prior knowledge of genes and/or proteins. The analysis depends on a robust understanding of the transcriptional signatures of distinct developmental stages and categories of ILCs across different tissues. However, the transcriptional markers that can be used to identify the ILC subsets are not identified and defined.
The vast majority of studies published thus far with the scRNA-seq analysis used peripheral blood mononuclear cells (PBMC) or bronchoalveolar lavage fluid (BALF) from COVID-19 patients and found a reduction in the transcriptional expression of tyrosine kinase binding protein (TYRPBP) and Fc Fragment Of IgG Receptor IIIa (FCGr3A) (Yao et al., 2021), granulysin (GNLY)- CD56 (Bernardes et al., 2020), killer cell lectin-like receptorF1 (KLRF1)+ (Zhu et al., 2020), killer cell lectin-like receptorsC1, C2 and KLRC1, and Spondin-2 (SPON2) (Zheng et al., 2020b) in NK cells; however, the populations were identified without corresponding flow cytometric analyses or with barcoded antibodies against surface antigens, and hence transcriptional markers for NK grouping are not conclusive. Furthermore, there is considerable overlap between transcriptional profiles within ILC subsets and T-cells (Zheng et al., 2020b; Mazzurana et al., 2021; Li et al., 2019). Hence, we limit our discussions to the studies in which barcoded antibodies are used on sorted ILC populations in conjunction with the scRNA-seq analysis.
A study in which scRNA-seq analysis was conducted on CD3−/+ sorted cells from the BALF and PBMC from 9 patients with COVID-19 and 5 patients with non-viral infection-associated pneumonia, innate-like cells were identified in the blood and BALF of COVID-19 patients. Still, differences in the levels of these populations were not established in the study (Zhao et al., 2021). The presence of NK cells in BALF suggests a role for these cell types in COVID-19. Furthermore, NK cells from infected patients were more responsive to type 1 interferon and showed a transcriptome correlated with an increased viral response (Yao et al., 2021). These data, although preliminary, suggest that NK and ILC subsets are present in the lungs of the COVID-19 patients, where they may contribute to the observed inflammation and recovery/injury.
1.6. Activation of ILC as a prevention strategy for COVID-19
The severity of the COVID-19 illness, requirement for hospitalization, duration of symptoms, and increased duration of hospital stay correlate with reduced numbers of ILCs (Silverstein et al., 2021). Conversely, convalescent COVID-19 patients tend to have higher numbers of ILCs when compared to those who succumbed to infection (Garcia et al., 2020; Gomez-Cadena et al., 2021). These findings suggest that induction and activation of ILCs could potentially prevent and reduce disease severity with favorable outcomes. Studies conducted in our laboratory using either the in vivo animal model or with human PBMCs in vitro show that ligands of pathogen sensors as well as replication-defective adenoviruses induce/activate different ILC subsets as compared to unstimulated controls (unpublished results). TLR7 agonist, imiquimod, is a FDA approved drug for external application against actinic keratosis, external genital warts and certain forms of skin cancer (Baird et al., 2017; Eisen et al., 2021). Additionally, in influenza mouse model of viral infection, intranasally administered imiquimod reduces viral replication, airway and pulmonary inflammation, weight loss, and lung neutrophils levels (To et al., 2019). Furthermore, imiquimod also reduced cytokines and chemokines and prevented influenza virus-induced lung pathology. In our assays, TLR7 agonist stimulated activation of the ILC subsets, ILC1, ILC2, ILC3, and NK cells, in human PBMCs (unpublished results). Additionally, using a mouse model of infection we observed that ILCs confers protection against influenza infection in the absence of influenza-specific adaptive immunity (unpublished results). Since there are no specific drugs against SARS-CoV-2 either for prophylactic or therapeutic use and the Phase III clinical trials with remdesivir in patients with SARS-Cov-2 produced mixed results (Goldman et al., 2020; Wang et al., 2020), repurposing imiquimod for prophylactic and therapeutic intervention against SARS-CoV-2 infection is an attractive option.
2. Conclusions
We are making strides in understanding the role of ILC cell populations in infectious diseases, cancer, autoimmune diseases, allergy, and homeostasis. As the first line of defense, ILCs play an important role in eliminating the insult and initiation of adaptive immune responses. While ILCs are required to clear the virus, the viruses, in turn, must have developed strategies to counteract the functionality of the ILCs through several mechanisms, highlighting the complex interplay between viruses and ILCs. The mechanism producing lymphopenia in COVID-19 and altered ILC activation and/or function leading to disease severity are yet to be determined. Detailed investigations are required to address the role of host factors, such as age, genetic conditions, nutrition, diet, underlying medical conditions, on ILCs function and COVID-19 severity outcomes.
Acknowledgment
This work was partly supported by Public Health Service grant AI158177 from the National Institute of Allergy and Infectious Diseases.
Footnotes
The conclusions, findings, and opinions expressed by authors contributing to this review do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions.
References
- Amand M., Iserentant G., Poli A., et al. Human CD56(dim)CD16(dim) cells as an individualized natural killer cell subset. Front. Immunol. 2017;8:699. doi: 10.3389/fimmu.2017.00699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardain A., Porterfield J.Z., Kloverpris H.N., et al. Type 3 ILCs in lung disease. Front. Immunol. 2019;10:92. doi: 10.3389/fimmu.2019.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L., Vienne M., Tang L., et al. Liver type 1 innate lymphoid cells develop locally via an interferon-gamma-dependent loop. Science. 2021;(6536):371. doi: 10.1126/science.aba4177. [DOI] [PubMed] [Google Scholar]
- Baird J.R., Monjazeb A.M., Shah O., et al. Stimulating innate immunity to enhance radiation therapy-induced tumor control. Int. J. Radiat. Oncol. Biol. Phys. 2017;99(2):362–373. doi: 10.1016/j.ijrobp.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bal S.M., Golebski K., Spits H. Plasticity of innate lymphoid cell subsets. Nat. Rev. Immunol. 2020;20(9):552–565. doi: 10.1038/s41577-020-0282-9. [DOI] [PubMed] [Google Scholar]
- Barlow J.L., McKenzie A.N.J. Innate lymphoid cells of the lung. Annu. Rev. Physiol. 2019;81:429–452. doi: 10.1146/annurev-physiol-020518-114630. [DOI] [PubMed] [Google Scholar]
- Bernardes J.P., Mishra N., Tran F., et al. Longitudinal multi-omics analyses identify responses of megakaryocytes, erythroid cells, and plasmablasts as hallmarks of severe COVID-19. Immunity. 2020;53(6):1296–12314 e9. doi: 10.1016/j.immuni.2020.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biron C.A., Byron K.S., Sullivan J.L. Severe herpesvirus infections in an adolescent without natural killer cells. N. Engl. J. Med. 1989;320(26):1731–1735. doi: 10.1056/NEJM198906293202605. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control . CDC; 2021. COVID Data Tracker.https://covid.cdc.gov/covid-data-tracker/#datatracker-home [Google Scholar]
- Cevik M., Kuppalli K., Kindrachuk J., et al. Virology, transmission, and pathogenesis of SARS-CoV-2. BMJ. 2020;371:m3862. doi: 10.1136/bmj.m3862. [DOI] [PubMed] [Google Scholar]
- Chen Z., John Wherry E. T cell responses in patients with COVID-19. Nat. Rev. Immunol. 2020;20(9):529–536. doi: 10.1038/s41577-020-0402-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Wu S., Guo G., et al. Programmed death (PD)-1-deficient mice are extremely sensitive to murine hepatitis virus strain-3 (MHV-3) infection. PLoS Pathog. 2011;7(7) doi: 10.1371/journal.ppat.1001347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherrier M., Eberl G. The development of LTi cells. Curr. Opin. Immunol. 2012;24(2):178–183. doi: 10.1016/j.coi.2012.02.003. [DOI] [PubMed] [Google Scholar]
- Constantinides M.G., McDonald B.D., Verhoef P.A., et al. A committed precursor to innate lymphoid cells. Nature. 2014;508(7496):397–401. doi: 10.1038/nature13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper M.A., Elliott J.M., Keyel P.A., et al. Cytokine-induced memory-like natural killer cells. Proc. Natl. Acad. Sci. U. S. A. 2009;106(6):1915–1919. doi: 10.1073/pnas.0813192106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper G.E., Ostridge K., Khakoo S.I., et al. Human CD49a(+) lung natural killer cell cytotoxicity in response to influenza A virus. Front. Immunol. 2018;9:1671. doi: 10.3389/fimmu.2018.01671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crome S.Q., Nguyen L.T., Lopez-Verges S., et al. A distinct innate lymphoid cell population regulates tumor-associated T cells. Nat. Med. 2017;23(3):368–375. doi: 10.1038/nm.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlgren M.W., Jones S.W., Cautivo K.M., et al. Adventitial stromal cells define group 2 innate lymphoid cell tissue niches. Immunity. 2019;50(3):707–722 e6. doi: 10.1016/j.immuni.2019.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Candia P., Prattichizzo F., Garavelli S., et al. T cells: warriors of SARS-CoV-2 infection. Trends Immunol. 2021;42(1):18–30. doi: 10.1016/j.it.2020.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Grove K.C., Provoost S., Verhamme F.M., et al. Characterization and quantification of innate lymphoid cell subsets in human lung. PloS One. 2016;11(1) doi: 10.1371/journal.pone.0145961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Zotto G., Marcenaro E., Vacca P., et al. Markers and function of human NK cells in normal and pathological conditions. Cytometry B Clin Cytom. 2017;92(2):100–114. doi: 10.1002/cyto.b.21508. [DOI] [PubMed] [Google Scholar]
- Della Chiesa M., Pesce S., Muccio L., et al. Features of memory-like and PD-1(+) human NK cell subsets. Front. Immunol. 2016;7:351. doi: 10.3389/fimmu.2016.00351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao B., Wang C., Tan Y., et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front. Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogra P., Rancan C., Ma W., et al. Tissue determinants of human NK cell development, function, and residence. Cell. 2020;180(4):749–763 e13. doi: 10.1016/j.cell.2020.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20(5):533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl G., Marmon S., Sunshine M.J., et al. An essential function for the nuclear receptor RORgamma(t) in the generation of fetal lymphoid tissue inducer cells. Nat. Immunol. 2004;5(1):64–73. doi: 10.1038/ni1022. [DOI] [PubMed] [Google Scholar]
- Eberl G., Colonna M., Di Santo J.P., et al. Innate lymphoid cells. Innate lymphoid cells: a new paradigm in immunology. Science. 2015;348(6237):aaa6566. doi: 10.1126/science.aaa6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehre C. SARS-CoV-2 infection of airway cells. N. Engl. J. Med. 2020;383(10):969. doi: 10.1056/NEJMicm2023328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen D.B., Asgari M.M., Bennett D.D., et al. Guidelines of care for the management of actinic keratosis. J. Am. Acad. Dermatol. 2021;S0190-9622(21) doi: 10.1016/j.jaad.2021.02.082. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauriat C., Long E.O., Ljunggren H.G., et al. Regulation of human NK-cell cytokine and chemokine production by target cell recognition. Blood. 2010;115(11):2167–2176. doi: 10.1182/blood-2009-08-238469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finelli L., Gupta V., Petigara T., et al. Mortality among US patients hospitalized with SARS-CoV-2 infection in 2020. JAMA Netw Open. 2021;4(4) doi: 10.1001/jamanetworkopen.2021.6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs A., Vermi W., Lee J.S., et al. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-gamma-producing cells. Immunity. 2013;38(4):769–781. doi: 10.1016/j.immuni.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrielli S., Ortolani C., Del Zotto G., et al. The memories of NK cells: innate-adaptive immune intrinsic crosstalk. J Immunol Res. 2016;2016 doi: 10.1155/2016/1376595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galani I.E., Rovina N., Lampropoulou V., et al. Untuned antiviral immunity in COVID-19 revealed by temporal type I/III interferon patterns and flu comparison. Nat. Immunol. 2021;22(1):32–40. doi: 10.1038/s41590-020-00840-x. [DOI] [PubMed] [Google Scholar]
- Garcia M., Kokkinou E., Carrasco Garcia A., et al. Innate lymphoid cell composition associates with COVID-19 disease severity. Clin Transl Immunology. 2020;9(12):e1224. doi: 10.1002/cti2.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger G., Fan X., Dikiy S., et al. Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science. 2015;350(6263):981–985. doi: 10.1126/science.aac9593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman J.D., Lye D.C.B., Hui D.S., et al. Remdesivir for 5 or 10 Days in patients with severe covid-19. N. Engl. J. Med. 2020;383(19):1827–1837. doi: 10.1056/NEJMoa2015301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Cadena A., Spehner L., Kroemer M., et al. Severe COVID-19 patients exhibit an ILC2 NKG2D(+) population in their impaired ILC compartment. Cell. Mol. Immunol. 2021;18(2):484–486. doi: 10.1038/s41423-020-00596-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzywacz B., Kataria N., Kataria N., et al. Natural killer-cell differentiation by myeloid progenitors. Blood. 2011;117(13):3548–3558. doi: 10.1182/blood-2010-04-281394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halim T.Y., Steer C.A., Matha L., et al. Group 2 innate lymphoid cells are critical for the initiation of adaptive T helper 2 cell-mediated allergic lung inflammation. Immunity. 2014;40(3):425–435. doi: 10.1016/j.immuni.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer Q., Ruckert T., Romagnani C. Natural killer cell specificity for viral infections. Nat. Immunol. 2018;19(8):800–808. doi: 10.1038/s41590-018-0163-6. [DOI] [PubMed] [Google Scholar]
- Herbert D.R., Douglas B., Zullo K. Group 2 innate lymphoid cells (ILC2): type 2 immunity and helminth immunity. Int. J. Mol. Sci. 2019;20(9) doi: 10.3390/ijms20092276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochdorfer T., Winkler C., Pardali K., et al. Expression of c-Kit discriminates between two functionally distinct subsets of human type 2 innate lymphoid cells. Eur. J. Immunol. 2019;49(6):884–893. doi: 10.1002/eji.201848006. [DOI] [PubMed] [Google Scholar]
- Hong H.S., Eberhard J.M., Keudel P., et al. HIV infection is associated with a preferential decline in less-differentiated CD56dim CD16+ NK cells. J. Virol. 2010;84(2):1183–1188. doi: 10.1128/JVI.01675-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz A., Stegmann K.A., Riley E.M. Activation of natural killer cells during microbial infections. Front. Immunol. 2011;2:88. doi: 10.3389/fimmu.2011.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyler T., Klose C.S., Souabni A., et al. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity. 2012;37(4):634–648. doi: 10.1016/j.immuni.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P.F., Hultin L.E., Hultin P., et al. Natural killer cell immunodeficiency in HIV disease is manifest by profoundly decreased numbers of CD16+CD56+ cells and expansion of a population of CD16dimCD56- cells with low lytic activity. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 1995;10(3):331–340. [PubMed] [Google Scholar]
- Ikram S., Ahmad F., Ahmad J., et al. Screening of small molecule libraries using combined text mining, ligand- and target-driven based approaches for identification of novel granzyme H inhibitors. J. Mol. Graph. Model. 2021;105 doi: 10.1016/j.jmgm.2021.107876. [DOI] [PubMed] [Google Scholar]
- Jamal M., Bangash H.I., Habiba M., et al. Immune dysregulation and system pathology in COVID-19. Virulence. 2021;12(1):918–936. doi: 10.1080/21505594.2021.1898790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jesenak M., Brndiarova M., Urbancikova I., et al. Immune parameters and COVID-19 infection - associations with clinical severity and disease prognosis. Front Cell Infect Microbiol. 2020;10:364. doi: 10.3389/fcimb.2020.00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiessling R., Petranyi G., Karre K., et al. Killer cells: a functional comparison between natural, immune T-cell and antibody-dependent in vitro systems. J. Exp. Med. 1976;143(4):772–780. doi: 10.1084/jem.143.4.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose C.S., Artis D. Innate lymphoid cells as regulators of immunity, inflammation and tissue homeostasis. Nat. Immunol. 2016;17(7):765–774. doi: 10.1038/ni.3489. [DOI] [PubMed] [Google Scholar]
- Klose C.S.N., Artis D. Innate lymphoid cells control signaling circuits to regulate tissue-specific immunity. Cell Res. 2020;30(6):475–491. doi: 10.1038/s41422-020-0323-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose C.S., Diefenbach A. Transcription factors controlling innate lymphoid cell fate decisions. Curr. Top. Microbiol. Immunol. 2014;381:215–255. doi: 10.1007/82_2014_381. [DOI] [PubMed] [Google Scholar]
- Klose C.S.N., Flach M., Mohle L., et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell. 2014;157(2):340–356. doi: 10.1016/j.cell.2014.03.030. [DOI] [PubMed] [Google Scholar]
- Krabbendam L., Bernink J.H., Spits H. Innate lymphoid cells: from helper to killer. Curr. Opin. Immunol. 2021;68:28–33. doi: 10.1016/j.coi.2020.08.007. [DOI] [PubMed] [Google Scholar]
- Kuri-Cervantes L., Pampena M.B., Meng W., et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci Immunol. 2020;5(49) doi: 10.1126/sciimmunol.abd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Morita H., Sokolowska M., et al. Gene expression signatures of circulating human type 1, 2, and 3 innate lymphoid cells. J. Allergy Clin. Immunol. 2019;143(6):2321–2325. doi: 10.1016/j.jaci.2019.01.047. [DOI] [PubMed] [Google Scholar]
- Li M., Guo W., Dong Y., et al. Elevated exhaustion levels of NK and CD8(+) T cells as indicators for progression and prognosis of COVID-19 disease. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.580237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Wang S., Xin J., et al. Role of NKG2D and its ligands in cancer immunotherapy. Am J Cancer Res. 2019;9(10):2064–2078. [PMC free article] [PubMed] [Google Scholar]
- Marquardt N., Kekalainen E., Chen P., et al. Human lung natural killer cells are predominantly comprised of highly differentiated hypofunctional CD69(-)CD56(dim) cells. J. Allergy Clin. Immunol. 2017;139(4):1321–13230 e4. doi: 10.1016/j.jaci.2016.07.043. [DOI] [PubMed] [Google Scholar]
- Mathew D., Giles J.R., Baxter A.E., et al. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369(6508) doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maucourant C., Filipovic I., Ponzetta A., et al. Natural killer cell immunotypes related to COVID-19 disease severity. Sci Immunol. 2020;5(50) doi: 10.1126/sciimmunol.abd6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoni A., Salvati L., Maggi L., et al. Impaired immune cell cytotoxicity in severe COVID-19 is IL-6 dependent. J. Clin. Invest. 2020;130(9):4694–4703. doi: 10.1172/JCI138554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzurana L., Czarnewski P., Jonsson V., et al. Tissue-specific transcriptional imprinting and heterogeneity in human innate lymphoid cells revealed by full-length single-cell RNA-sequencing. Cell Res. 2021;31(5):554–568. doi: 10.1038/s41422-020-00445-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mjosberg J., Bernink J., Golebski K., et al. The transcription factor GATA3 is essential for the function of human type 2 innate lymphoid cells. Immunity. 2012;37(4):649–659. doi: 10.1016/j.immuni.2012.08.015. [DOI] [PubMed] [Google Scholar]
- Molofsky A.B., Van Gool F., Liang H.E., et al. Interleukin-33 and interferon-gamma counter-regulate group 2 innate lymphoid cell activation during immune perturbation. Immunity. 2015;43(1):161–174. doi: 10.1016/j.immuni.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro K., Yamada T., Tanabe M., et al. Innate production of T(H)2 cytokines by adipose tissue-associated c-Kit(+)Sca-1(+) lymphoid cells. Nature. 2010;463(7280):540–544. doi: 10.1038/nature08636. [DOI] [PubMed] [Google Scholar]
- Mortha A., Burrows K. Cytokine networks between innate lymphoid cells and myeloid cells. Front. Immunol. 2018;9:191. doi: 10.3389/fimmu.2018.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabekura T., Shibuya A. ILC1: guardians of the oral mucosa against enemy viruses. Immunity. 2021;54(2):196–198. doi: 10.1016/j.immuni.2021.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabekura T., Shibuya A. Type 1 innate lymphoid cells: soldiers at the front line of immunity. Biomed. J. 2021;44(2):115–122. doi: 10.1016/j.bj.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris S., Coleman A., Kuri-Cervantes L., et al. PD-1 expression on natural killer cells and CD8(+) T cells during chronic HIV-1 infection. Viral Immunol. 2012;25(4):329–332. doi: 10.1089/vim.2011.0096. [DOI] [PubMed] [Google Scholar]
- Nussbaum J.C., Van Dyken S.J., von Moltke J., et al. Type 2 innate lymphoid cells control eosinophil homeostasis. Nature. 2013;502(7470):245–248. doi: 10.1038/nature12526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell P., Aldhamen Y.A. Systemic innate and adaptive immune responses to SARS-CoV-2 as it relates to other coronaviruses. Hum. Vaccines Immunother. 2020;16(12):2980–2991. doi: 10.1080/21645515.2020.1802974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor M.H., Muir R., Chakhtoura M., et al. A follicular regulatory Innate Lymphoid Cell population impairs interactions between germinal center Tfh and B cells. Commun Biol. 2021;4(1):563. doi: 10.1038/s42003-021-02079-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ombrello M.J., Schulert G.S. COVID-19 and cytokine storm syndrome: are there lessons from macrophage activation syndrome? Transl. Res. 2021;232:1–12. doi: 10.1016/j.trsl.2021.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orange J.S. Human natural killer cell deficiencies and susceptibility to infection. Microb. Infect. 2002;4(15):1545–1558. doi: 10.1016/s1286-4579(02)00038-2. [DOI] [PubMed] [Google Scholar]
- Papalexi E., Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat. Rev. Immunol. 2018;18(1):35–45. doi: 10.1038/nri.2017.76. [DOI] [PubMed] [Google Scholar]
- Pende D., Falco M., Vitale M., et al. Killer ig-like receptors (KIRs): their role in NK cell modulation and developments leading to their clinical exploitation. Front. Immunol. 2019;10:1179. doi: 10.3389/fimmu.2019.01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pende D., Falco M., Vitale M., et al. Killer ig-like receptors (KIRs): their role in NK cell modulation and developments leading to their clinical exploitation. Front. Immunol. 2019;10:1179. doi: 10.3389/fimmu.2019.01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesce S., Greppi M., Tabellini G., et al. Identification of a subset of human natural killer cells expressing high levels of programmed death 1: a phenotypic and functional characterization. J. Allergy Clin. Immunol. 2017;139(1):335–346 e3. doi: 10.1016/j.jaci.2016.04.025. [DOI] [PubMed] [Google Scholar]
- Poli A., Michel T., Theresine M., et al. CD56bright natural killer (NK) cells: an important NK cell subset. Immunology. 2009;126(4):458–465. doi: 10.1111/j.1365-2567.2008.03027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponti G., Maccaferri M., Ruini C., et al. Biomarkers associated with COVID-19 disease progression. Crit. Rev. Clin. Lab Sci. 2020;57(6):389–399. doi: 10.1080/10408363.2020.1770685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A.E., Liang H.E., Sullivan B.M., et al. Systemically dispersed innate IL-13-expressing cells in type 2 immunity. Proc. Natl. Acad. Sci. U. S. A. 2010;107(25):11489–11494. doi: 10.1073/pnas.1003988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quatrini L., Wieduwild E., Escaliere B., et al. Endogenous glucocorticoids control host resistance to viral infection through the tissue-specific regulation of PD-1 expression on NK cells. Nat. Immunol. 2018;19(9):954–962. doi: 10.1038/s41590-018-0185-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quatrini L., Mariotti F.R., Munari E., et al. The immune checkpoint PD-1 in natural killer cells: expression, function and targeting in tumour immunotherapy. Cancers. 2020;12(11) doi: 10.3390/cancers12113285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajalingam R. Human diversity of killer cell immunoglobulin-like receptors and disease. Korean J Hematol. 2011;46(4):216–228. doi: 10.5045/kjh.2011.46.4.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan P., Bowzard J.B., Schwerzmann J.W., et al. Cytoplasmic nucleic acid sensors in antiviral immunity. Trends Mol. Med. 2009;15(8):359–368. doi: 10.1016/j.molmed.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Saluzzo S., Gorki A.D., Rana B.M.J., et al. First-breath-induced type 2 pathways shape the lung immune environment. Cell Rep. 2017;18(8):1893–1905. doi: 10.1016/j.celrep.2017.01.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scandella E., Bolinger B., Lattmann E., et al. Restoration of lymphoid organ integrity through the interaction of lymphoid tissue-inducer cells with stroma of the T cell zone. Nat. Immunol. 2008;9(6):667–675. doi: 10.1038/ni.1605. [DOI] [PubMed] [Google Scholar]
- Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184(4):861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh A., Abraham N. Interleukin-7 receptor alpha in innate lymphoid cells: more than a marker. Front. Immunol. 2019;10:2897. doi: 10.3389/fimmu.2019.02897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D., Mukherjee R., Grewe D., et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020;587(7835):657–662. doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstein N.J., Wang Y., Manickas-Hill Z., et al. Innate lymphoid cells and disease tolerance in SARS-CoV-2 infection. medRxiv. 2021 doi: 10.1101/2021.01.14.21249839. [Preprint] [DOI] [Google Scholar]
- Simoni Y., Newell E.W. Dissecting human ILC heterogeneity: more than just three subsets. Immunology. 2018;153(3):297–303. doi: 10.1111/imm.12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoni Y., Fehlings M., Kloverpris H.N., et al. Human innate lymphoid cell subsets possess tissue-type based heterogeneity in phenotype and frequency. Immunity. 2017;46(1):148–161. doi: 10.1016/j.immuni.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg G.F., Hepworth M.R. Functional interactions between innate lymphoid cells and adaptive immunity. Nat. Rev. Immunol. 2019;19(10):599–613. doi: 10.1038/s41577-019-0194-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehle C., Hernandez D.C., Romagnani C. Innate lymphoid cells in lung infection and immunity. Immunol. Rev. 2018;286(1):102–119. doi: 10.1111/imr.12712. [DOI] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–1902 e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swann J.B., Hayakawa Y., Zerafa N., et al. Type I IFN contributes to NK cell homeostasis, activation, and antitumor function. J. Immunol. 2007;178(12):7540–7549. doi: 10.4049/jimmunol.178.12.7540. [DOI] [PubMed] [Google Scholar]
- To E.E., Erlich J., Liong F., et al. Intranasal and epicutaneous administration of Toll-like receptor 7 (TLR7) agonists provides protection against influenza A virus-induced morbidity in mice. Sci. Rep. 2019;9(1):2366. doi: 10.1038/s41598-019-38864-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacca P., Montaldo E., Croxatto D., et al. Identification of diverse innate lymphoid cells in human decidua. Mucosal Immunol. 2015;8(2):254–264. doi: 10.1038/mi.2014.63. [DOI] [PubMed] [Google Scholar]
- van de Pavert SA. Lymphoid tissue inducer (LTi) cell ontogeny and functioning in embryo and adult,. [DOI] [PMC free article] [PubMed]
- van der Ploeg E.K., Carreras Mascaro A., Huylebroeck D., et al. Group 2 innate lymphoid cells in human respiratory disorders. J Innate Immun. 2020;12(1):47–62. doi: 10.1159/000496212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Erp E.A., van Kampen M.R., van Kasteren P.B., et al. Viral infection of human natural killer cells. Viruses. 2019;11(3) doi: 10.3390/v11030243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varchetta S., Mele D., Oliviero B., et al. Unique immunological profile in patients with COVID-19. Cell. Mol. Immunol. 2021;18(3):604–612. doi: 10.1038/s41423-020-00557-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella L.A., Giles J.R., Baxter A.E., et al. Deep immune profiling of MIS-C demonstrates marked but transient immune activation compared to adult and pediatric COVID-19. Sci Immunol. 2021;6(57) doi: 10.1126/sciimmunol.abf7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walzer T., Dalod M., Robbins S.H., et al. Natural-killer cells and dendritic cells: "l'union fait la force. Blood. 2005;106(7):2252–2258. doi: 10.1182/blood-2005-03-1154. [DOI] [PubMed] [Google Scholar]
- Wang W., Erbe A.K., Hank J.A., et al. NK cell-mediated antibody-dependent cellular cytotoxicity in cancer immunotherapy. Front. Immunol. 2015;6:368. doi: 10.3389/fimmu.2015.00368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Xia P., Chen Y., et al. Regulatory innate lymphoid cells control innate intestinal inflammation. Cell. 2017;171(1):201–216 e18. doi: 10.1016/j.cell.2017.07.027. [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhou F., Zhang D., et al. Evaluation of the efficacy and safety of intravenous remdesivir in adult patients with severe COVID-19: study protocol for a phase 3 randomized, double-blind, placebo-controlled, multicentre trial. Trials. 2020;21(1):422. doi: 10.1186/s13063-020-04352-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Qiu Z., Hou Y., et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 2021;31(2):126–140. doi: 10.1038/s41422-020-00460-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiersinga W.J., Rhodes A., Cheng A.C., et al. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. J. Am. Med. Assoc. 2020;324(8):782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- Xu Z., Shi L., Wang Y., et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Cai B., Li Y., et al. Dynamics of NK, CD8 and Tfh cell mediated the production of cytokines and antiviral antibodies in Chinese patients with moderate COVID-19. J. Cell Mol. Med. 2020;24(24):14270–14279. doi: 10.1111/jcmm.16044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C., Bora S.A., Parimon T., et al. Cell-type-specific immune dysregulation in severely ill COVID-19 patients. Cell Rep. 2021;34(1):108590. doi: 10.1016/j.celrep.2020.108590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y.R., O'Koren E.G., Hotten D.F., et al. A protocol for the comprehensive flow cytometric analysis of immune cells in normal and inflamed murine non-lymphoid tissues. PloS One. 2016;11(3) doi: 10.1371/journal.pone.0150606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Kilian C., Turner J.E., et al. Clonal expansion and activation of tissue-resident memory-like Th17 cells expressing GM-CSF in the lungs of severe COVID-19 patients. Sci Immunol. 2021;6(56) doi: 10.1126/sciimmunol.abf6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Gao Y., Wang G., et al. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol. 2020;17(5):533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Liu X., Le W., et al. A human circulating immune cell landscape in aging and COVID-19. Protein Cell. 2020;11(10):740–770. doi: 10.1007/s13238-020-00762-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong C., Zheng M., Zhu J. Lymphoid tissue inducer-A divergent member of the ILC family. Cytokine Growth Factor Rev. 2018;42:5–12. doi: 10.1016/j.cytogfr.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Yang P., Zhao Y., et al. Single-cell sequencing of peripheral mononuclear cells reveals distinct immune response landscapes of COVID-19 and influenza patients. Immunity. 2020;53(3):685–696 e3. doi: 10.1016/j.immuni.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegenhain C., Vieth B., Parekh S., et al. Comparative analysis of single-cell RNA sequencing methods. Mol. Cell. 2017;65(4):631–643 e4. doi: 10.1016/j.molcel.2017.01.023. [DOI] [PubMed] [Google Scholar]
- Zipori D. The renewal and differentiation of hemopoietic stem cells. Faseb. J. 1992;6(9):2691–2697. doi: 10.1096/fasebj.6.9.1612293. [DOI] [PubMed] [Google Scholar]
- Zitti B., Bryceson Y.T. Natural killer cells in inflammation and autoimmunity. Cytokine Growth Factor Rev. 2018;42:37–46. doi: 10.1016/j.cytogfr.2018.08.001. [DOI] [PubMed] [Google Scholar]