Summary
ESC/iPSC-retinal sheet transplantation, which supplies photoreceptors as well as other retinal cells, has been shown to be able to restore visual function in mice with end-stage retinal degeneration. Here, by introducing a novel type of genetically engineered mouse ESC/iPSC-retinal sheet with reduced numbers of secondary retinal neurons but intact photoreceptor cell layer structure, we reinforced the evidence that ESC/iPSC-retinal sheet transplantation can establish synaptic connections with the host, restore light responsiveness, and reduce aberrant retinal ganglion cell spiking in mice. Furthermore, we show that genetically engineered grafts can substantially improve the outcome of the treatment by improving neural integration. We speculate that this leads to reduced spontaneous activity in the host which in turn contributes to a better visual recovery.
Subject areas: Neuroscience, Cellular neuroscience, Bioengineering, Tissue engineering, Stem cells research
Graphical Abstract
Highlights
-
•
Photoreceptors in organoids with bipolar cell-KO can functionally mature in vivo
-
•
Photoreceptors in KO grafts form more synapses with host bipolar cells
-
•
KO grafts better suppress heightened spontaneous spiking in degenerated retina
-
•
Mice transplanted with KO grafts perform better in light-guided behavior test
Neuroscience; Cellular neuroscience; Bioengineering; Tissue engineering; Stem cells research
Introduction
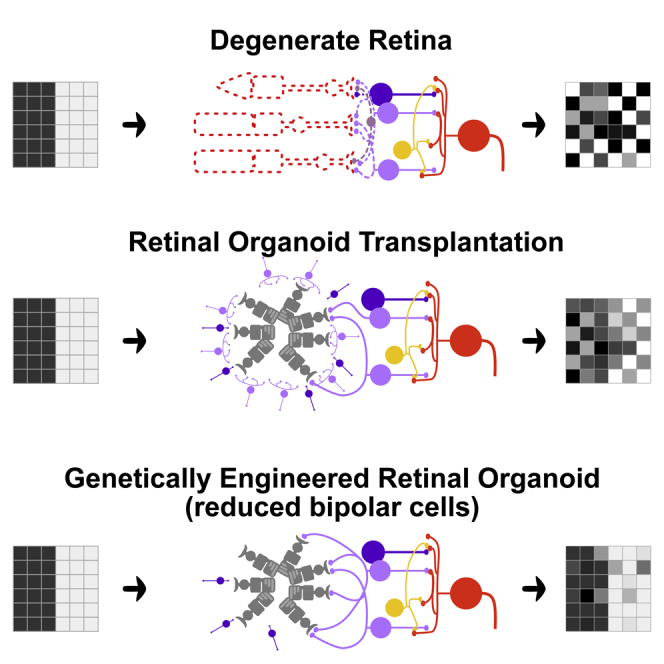
Several cell-based therapies are currently under development to treat photoreceptor cell loss (Aghaizu et al., 2017; Foik et al., 2018; Santos-Ferreira et al., 2016b). While transplantations may be conducted on hosts with varying degrees of retinal degeneration using donor or pluripotent-stem-cell-derived materials, the overall strategy can be categorized into either single-cell photoreceptor suspensions or retinal sheet transplantation. Cell suspension strategies consist of transplanting purified photoreceptor precursor cells (Bartsch et al., 2008; MacLaren et al., 2006; Pearson et al., 2012), whereas retinal sheet transplantations engraft retinal organoids containing both photoreceptor cells and inner retinal neurons (Assawachananont et al., 2014; Radtke et al., 2008; Shirai et al., 2016). Both methods have shown cellular integration, maturation of the photoreceptor cells, and some recovery of light responsiveness (Barnea-Cramer et al., 2016; Iraha et al., 2018; Mandai et al., 2017; McLelland et al., 2018; Singh et al., 2013; Tu et al., 2018).
The inner neurons in the retinal sheet transplantation can serve as a scaffold and nurturing microenvironment for photoreceptor cells to differentiate and mature, producing an organized layer structure resembling the retina and well-developed photoreceptor morphology (Assawachananont et al., 2014; Mandai et al., 2017; Shirai et al., 2016), whereas single-cell photoreceptor transplantations seldom form organized layered structures or complete outer segments. On the other hand, single-cell photoreceptor suspension preparations may have a better chance to contact host cells with directly exposed photoreceptor cells to host bipolar cells, whereas inner neurons in retinal sheet transplantation may become a hindrance for graft-host neural integration, as graft photoreceptors form synapses with concomitantly developing bipolar cells, resulting in numerous intragraft synapses.
In this study, we attempted to bring the best of the two approaches together by using genetically engineered cell lines to prepare retinal sheets with fewer bipolar cells that directly receive the output from the photoreceptor cells. To this end, we prepared two cell lines, Bhlhb4−/− and Islet1−/−. The basic-helix-loop-helix family member e23 (Bhlhe23, also known as Bhlhb4) is a transcription factor (Bramblett et al., 2002) expressed in rod bipolar cells, and it is required for maintenance and neural maturation of rod bipolar cells (Bramblett et al., 2004). Bhlhb4 functions downstream of factors that determine bipolar cell specification, and thus, Bhlhb4−/− retinas can engage rod bipolar cell fate; however, nascent cells that fail to mature eventually succumb to apoptosis (Bramblett et al., 2004). Islet1 (or Isl1), a LIM-homeodomain transcription factor mediates neuronal differentiation of rod and cone bipolar, cholinergic amacrine, and ganglion cells in the retina. Deletion of Islet1 results in the loss of the majority of bipolar, cholinergic amacrine, and ganglion cells (Elshatory et al., 2007). Owing to the relatively late expression of Bhhb4 and Islet1, we speculated that deletion of these genes would not largely affect retinal development before bipolar differentiation, but grafts would have fewer bipolar cells around the time photoreceptors start to form synapses. We prepared retinal sheets from wild-type (wt), Bhlhb4−/−, and Islet1−/− pluripotent cell lines (both iPS and ES cells) and transplanted them to end-stage rd1 mouse retinas, in which rod photoreceptors are virtually abolished by postnatal day 30 and surviving cone photoreceptors degenerate secondarily. These genetically engineered cell lines can be differentiated to retinal organoids with similar potency as wt cell lines and integrate with host retinas after transplantation. Our analyses of posttransplantation retinas show that Bhlhb4−/− and Islet1−/− result in a drastically reduced bipolar cell population upon graft maturation and a concomitant improved neural integration. We further conducted detailed electrophysiological analyses that indicate the improved neural integration may be manifested as a decrease in spontaneous activity, which in turn is observed as an enhanced performance in behavior tests. Overall, our results show that Bhlhb4−/− and Islet1−/− can be used to improve neural integration of retinal sheet transplantation.
Results
Bhlhb4−/− and Islet1−/− cells differentiate to retinal organoids similarly to wt cells
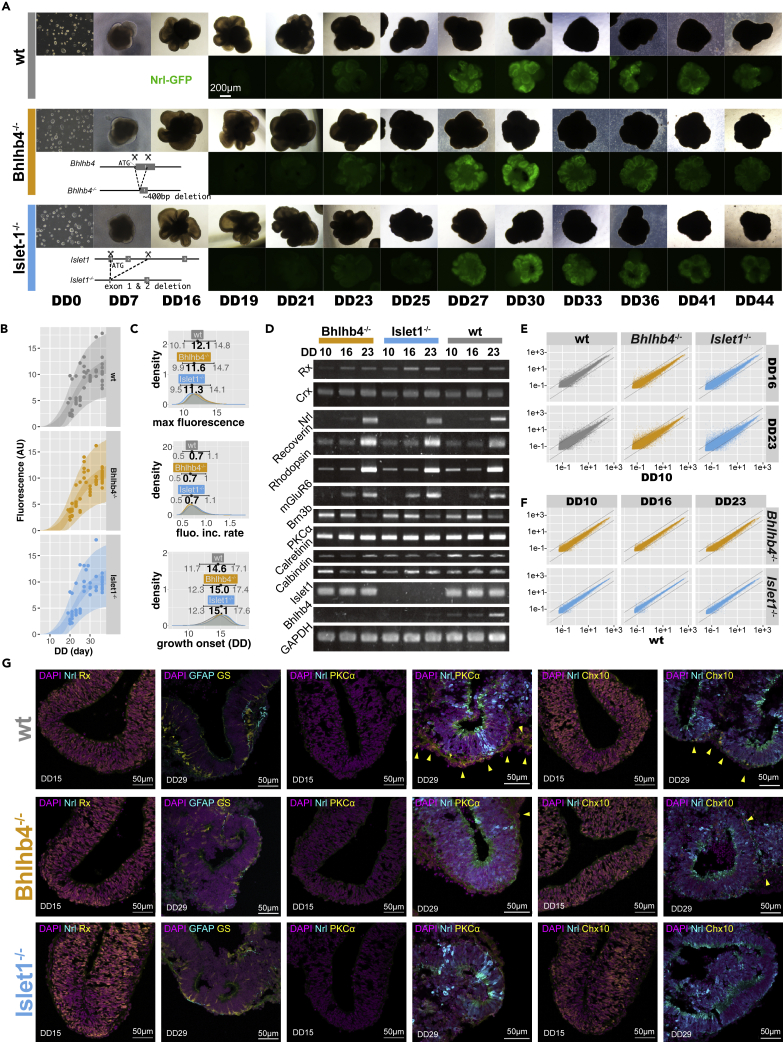
Both Bhlhb4−/− and Islet1−/− cell lines were able to differentiate into retinal structures similarly to wt cell lines. All the Bhlhb4−/− (6 clones) and Islet1−/− (4 clones) cells (iPS and ES cells) self-reorganized to homogeneous aggregates and formed hollowed structures resembling the optic vesicle by differentiation day (DD) 7. These retinal organoids grew bigger by sprouting more vesicular structures through DD23 (Figure 1A). Transgenic cell lines expressing green fluorescent protein (GFP) under the Nrl promoter, which is specifically activated in rod photoreceptors shortly after terminal cell division (Akimoto et al., 2006), started to show signs of detectable fluorescence around DD19. The GFP signal consistently grew stronger until DD33, and then the fluorescence became weaker. The Nrl-GFP fluorescence signal of representative clones between DD19 and DD33 was modeled with an exponential growth curve revealing that Bhlhb4−/− and Islet1−/− cell lines have broadly similar time courses and maximum intensities to wt lines (Figures 1B, 1C, and S1), indicating that these gene knockouts do not affect early retinal differentiation.
Figure 1.
Differentiation of retinal organoids
(A) Phase-contrast and fluorescence images of differentiating retinal organoids at different differentiation days (DD).
(B) Modeling of Nrl-GFP signal with a growth curve. Points represent the intensity value for individual retinal organoids, the light shaded area shows the range of expected values, and the dark shaded area shows the expected mean signal intensity with 95% compatibility intervals. Full parameter estimates for the model are provided in Figure S1.
(C) Estimated parameter values for maximum fluorescence value (top), maximum rate of fluorescence increase (middle), and onset of fluorescence increase (bottom). Estimates (shown with mode and 95% compatibility interval on top) largely overlap, showing no differences among organoids. A total of 170 organoids (54 wt, 61 Bhlhb4−/−, and 55 Islet1−/−) were used for this analysis.
(D) RT-PCR of key genes from organoids differentiated from a clone of Tg(Nrl-GFP) iPS cell lines. Similar results were obtained for ES-derived organoids (Figure S1).
(E and F) Scatterplots of microarray expression pattern of retinal organoids show no large differences among retinal organoids. (E) shows the gene expression of DD16 and DD23 compared with DD10, and (F) shows the comparison between Bhlhb4−/− and Iselt1−/− with wt. Diagonal lines correspond to fourfold expression differences. GO analysis of differentially expressed genes is provided in Figure S2.
(G) Immunohistochemistry characterization of DD15 and DD29 organoids. Arrow heads show PKCα and Chx10 positive bipolar cells.
Bhlhb4 expression begins around postnatal day 5 (P5) in mouse retinas, which is roughly equivalent to DD25 of the retinal organoids and is restricted to developing rod bipolar cells (Bramblett et al., 2004). Consistently, we observed Bhlhb4 mRNA expression gradually increase on wt and Islet1−/− retinal organoids from DD10 to DD23, whereas the band was absent on Bhlhb4−/− organoids (Figures 1D and S1). Similarly, Islet1 expression in bipolar cells begins around P5, although it is observed at earlier stages in retinal ganglion cells (RGCs) (Elshatory et al., 2007). Consequently, Islet1 is detected on DD10, DD16, and DD23 in wt and Bhlhb4−/− retinal organoids but is absent in Islet1−/− (Figures 1D and S1). We saw no differences between ES- and iPS cell-derived organoids in the temporal expression pattern of key genes, and the effect of Bhlhb4 or Islet1 deletion was also identical (Figures 1D and S1). In addition to real-time polymerase chain reaction (RT-PCR), we conducted the microarray analyses of DD10, DD16, and DD23 organoids derived from Nrl-GFP; Ribeye-reporter iPSC lines (Mandai et al., 2017). Figure S1 shows some key genes specific to retinal progenitors, bipolar, and photoreceptor cells. The expression pattern of these genes is largely unaltered. Although Bhlhb4−/− retinal organoids seem to have lower expression of some photoreceptor markers (Cnga1, Cnga2, Cnga3, Gucy2d, Rho), and Islet1−/− seem to have higher expression (Cabp4, Cnga1, Cngb1, Gnat2, Gngt1, Rho, Rcvrn, Sag), photoreceptor markers are generally low at this early developmental stage. Expression patterns for retinal pigment epithelium, glial cells, retinal ganglion cells, amacrine cells, horizontal cells, as well as other controls (pluripotency, neuron, apoptosis, and expression reference marker genes) were also largely identical among the three lines. Overall gene expression across cell lines was very similar, with few genes upregulated or downregulated compared with the wt line (Figures 1E and 1F). Gene ontology (GO) enrichment analysis shows similar genes, mostly pertaining to localization, metabolic, and developmental processes, are upregulated or downregulated across development (Figure S2).
Figure 1G shows immunostaining for PKCα, Chx10, and Rx, as well as the Nrl-GFP reporter fluorescence of DD15 and DD29 retinal organoids in vitro. At DD15, a widespread expression of retinal progenitor markers Rx and Chx10, but not much of Nrl-GFP or PKCα, were observed in all organoids (wt, Bhlhb4−/−, and Islet1−/−), reflecting the early stage of differentiation. At DD29, an Nrl-GFP+ outer-nuclear-layer (ONL)-like structure with regularly lined up GS+ Müller cells was the major composite, while PKCα+ or Chx10+ cells, which are typically suggestive of retinal bipolar cells, were occasionally observed localized on the outer border of the ONL-like structure, or in a very thin layer of presumptive inner nuclear layer (INL). While the overall trend was similar across all lines, inner cells in Bhlhb4−/− or Islet1−/− organoids seemed fewer and showed weaker bipolar cell marker signals than those in wt organoids. Overall, our data show that Bhlhb4−/− and Islet1−/− cells can differentiate to retinal organoids without any apparent differences to wt cells at least up to DD15, around the time of transplantation.
Bipolar cells are markedly reduced in Bhlhb4−/− and Islet1−/− grafts after transplantation
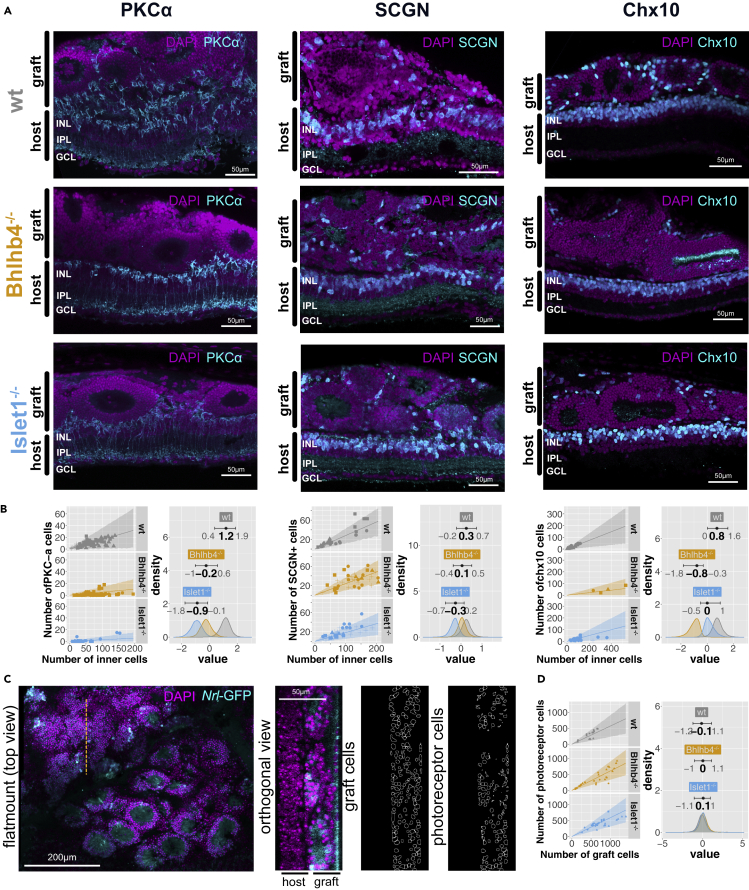
We transplanted early developmental stage (DD10-DD15) retinal organoids to the subretinal space of rd1 mouse and observed maturation and integration with the host. Grafts were examined at 4-5 weeks after transplantation, that is, 6- to 7-week-old grafts. Transplanted grafts of all genotypes consistently developed mature photoreceptor layers as a major composite, characterized by small and dense nuclei, surrounded by inner retinal cells. We characterized graft inner cell composition by their PKCα (rod bipolar cells), secretagogin (SCGN) (subsets of cone bipolar cells), Chx10 (pan bipolar cells), Calretinin (subsets of RGCs and amacrine cells), and Calbindin (horizontal cells and subsets of amacrine cells) expression (Figures 2 and S3A). We first assessed if PKCα+ rod bipolar cells were actually reduced as primarily aimed in this study. In grafts derived from all the Bhlhb4−/− and Islet1−/− clones, the number of PKCα+ cells was consistently and markedly reduced (Figures 2A and 2B). Our data show that PKCα+ cells account for about 0.14 [95% confidence interval (CI) = 0.04–0.38] of the inner cells in wt grafts and about 0.04 [95% CI = 0.01–0.16] and 0.02 [95% CI = 0.00–0.09] in Bhlhb4−/− and Islet1−/− grafts, respectively (confidence in the difference in the effect of graft genotype, i.e., the fraction of area over zero of the difference in the distributions, was 97% for Bhlhb4−/−-wt, 99% for Islet1−/−-wt, and 84% for Bhlhb4−/−- Islet1−/−). On the other hand, cone bipolar cells (SCGN+) seemed to decrease slightly in Islet1−/− (0.19 [95% CI = 0.09–0.30]) compared with the wt (0.28 [95% CI = 0.16–0.42]), whereas the decrease in Bhlhb4−/−(0.25 [95% CI = 0.14–0.36]) is not as evident (confidence in the difference in the effect of graft genotype for SCGN+ cells were 72% for Bhlhb4−/−-wt, 93% for Islet1−/−-wt, and 86% for Islet1−/−-Bhlhb4−/−). These results are consistent with the notion that Bhlhb4 affects rod bipolar cells specifically, whereas Islet1 affects both rod and cone bipolar cells. Chx10 immunoreactivities, which are typically suggestive of bipolar cells in the mature retina but also present in some Müller cells, were also reduced in Bhlhb4−/− and slightly less in Islet1−/− (confidence in the difference in the effect of graft genotype for Chx10+ cells were 99% for Bhlhb4−/−-wt, 84% for Islet1−/−-wt, and 99% for Bhlhb4−/−- Islet1−/−). It is noteworthy to mention that Chx10+ cells in Islet1−/− were characterized by a weaker signal, indicating these cells were different from typical bipolar cells. The number of Calbindin+ and Calretinin+ cells were similar among wt, Bhlhb4−/−, and Islet1−/− grafts (Figure S3). Additionally, we investigated if photoreceptor population was changed in Bhlhb4−/− and Islet1−/− grafts owing to the reduction in the number of bipolar cells. Photoreceptor rosettes varied in size but consistently presented a strong rhodopsin expression at segment-like structures inside rosettes. The number of rod photoreceptor cells seems to be proportional to the number of graft cells (Figures 2C and 2D). We found no significant differences in the photoreceptor content, with all cell lines producing grafts of about 0.6 rod photoreceptor content (Figures 2D and S3). Finally, while most of the grafted cells formed spherical rosette structures, substantial organization of the highly polarized ONL structure was observed in some cases (Figure 3A, Bhlhb4−/−). This was sporadically observed and seemed to be independent of the graft genotype (wt, Bhlhb4−/−, Iselet1−/−). Although we do not yet know the specific conditions leading to these cases, they showcase the regenerative potential after transplantation in the end-stage degenerate retina.
Figure 2.
Immunohistochemistry characterization of wt and Bhlhb4−/− and Islet1−/− grafts after subretinal transplantation
(A) PKCα+ (rod bipolar), SCGN+ (cone bipolar), and Chx10+ (pan-bipolar) cells in wt, Bhlb4−/−, and Islet1−/− grafts.
(B) Quantification of cell markers. Left plots show the number of PKCα+, SCGN+, and Chx10+ cells (vertical axis) against the number of inner cells in the graft per section (horizontal axis). Dots indicate the count value for a given section, with different shapes representing samples from different animals. The line represents the most likely value, and the shaded area represents the 95% compatibility interval assuming that the number of positive cells for a positive marker follows the binomial distribution given the number of total (inner) cells. Right plots show the estimated effect of graft genotype on the probability of the marker being positive, with mode and 95% compatibility intervals indicated on top. Note that the horizontal scale (value) is logit, i.e., log odds, with higher values indicating higher probability of observing a cell positive for a particular marker. Distribution for the rest of the parameters, as well as results for a similar analysis on Calbindin+ and Calretinin+ cells are provided in Figure S2.
(C) Flat-mounted retinas were imaged, and orthogonal sections intersecting rosettes were constructed to analyze the number of photoreceptor cells. The images to the right correspond to orthogonal views at the dotted orange line.
(D) The number of photoreceptor cells (vertical axis) was modeled as a function of the number of graft cells (horizontal axis), similar to (B). The graph on the right shows there is no difference in photoreceptor ratio by graft genotype as the posterior distributions largely overlap. Distribution for the rest of the parameters is provided in Figure S2 as well as predicted values from the model. Eleven mice were used for PKCα+ analysis (3 wt, 3 Bhlhb4−/−, and 4 Islet1−/−), 11 for SCGN+ analysis (3 wt, 4 Bhlhb4−/−, and 4 Islet1−/−), 9 for Chx10+ cells (2 wt, 3 Bhlhb4−/−, and 4 Islet1−/−), and 8 for the photoreceptor cell number analysis (2 wt, 3 Bhlhb4−/−, and 3 Islet1−/−). GCL: ganglion cell layer; INL: inner nuclear layer; ONL: outer nuclear layer.
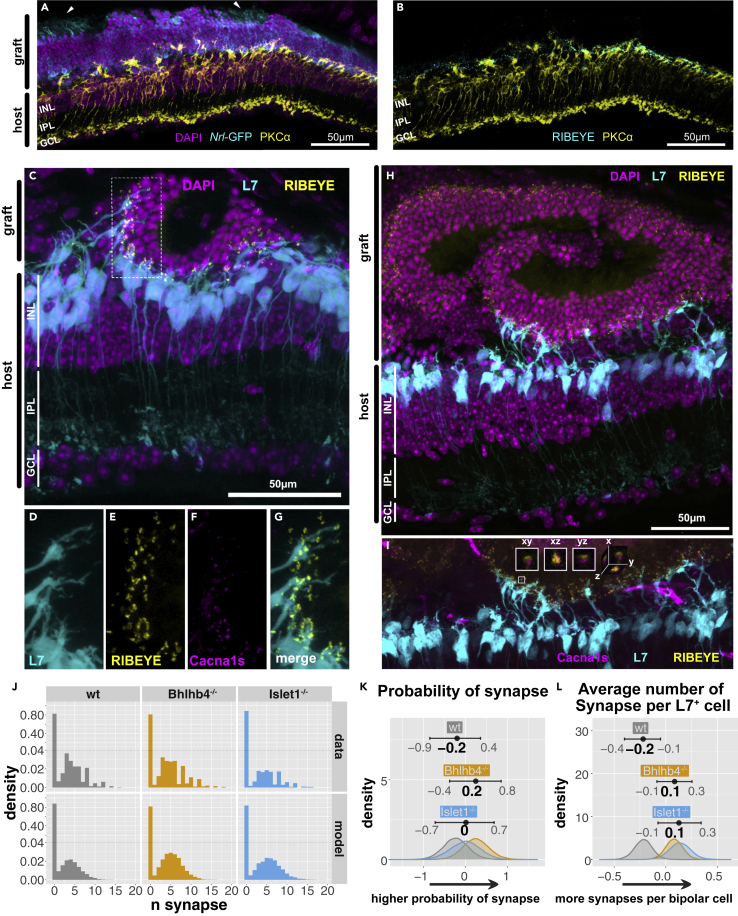
Figure 3.
Host-graft synapse formation
(A and B) Example of graft (Bhlhb4−/−) contacting host forming an outer nuclear layer (ONL) reminiscent of wildtype retina. PKCα-positive host bipolar cells extend dendrites with the presynaptic marker (RIBEYE) at their tips and graft ONL shows oriented inner/outer segment-like structures (arrow heads). (C–I) Representative images of synapse formation from Bhlhb4−/− grafts.
(C) L7-GFP host rod bipolar cells (cyan) are extending dendrites toward graft photoreceptor cells.
(D–G, and I) Example of synapse quantification. (D) Shows the L7-GFP host bipolar cell dendrites. RIBEYE (from Ribeye-reporter graft cell line) (E) and Cacna1s (immunolabeled) (F) pairs at the tip ofL7-GFP cells were treated as host-graft synapses (G).
(H and I) Heterogeneity in synaptic integration. The right part of the image shows extensive synaptic contact, with L7-GFP bipolar aggressively extending toward graft photoreceptor cells while host bipolar cells on the left are completely retracted and show no synapse formation.
(J–L) (J) Histogram of number synapse per L7-GFP cell (host bipolar). Note that the scaling of the vertical axis has been altered to facilitate visualization (dotted line). Top panels represent the raw data while bottom rows represent the modeled data. Most of the host bipolar cells do not form synapses with the graft, resulting in a large zero-count. The effect of the graft on the probability of forming synapse (K) and the average number of synapses per L7-GFP cell (L), respectively, was estimated using a zero-inflated Poisson model and shown with 95% compatibility intervals and modes indicated. Note that the horizontal scale for (K) is in log odds and (L) in log. Larger values in (K) indicate larger probability of synapse and higher values in (L) indicate more synapses per bipolar cell. GCL: ganglion cell layer, INL: inner nuclear layer, ONL: outer nuclear layer. 10,354 observation (L7-GFP bipolar cells) from 27 mice (9 wt, 9 Bhlhb4−/−, 9 Islet1−/−) were used for this analysis. Per mouse posterior predictions are provided in Figure S4.
Bhlhb4−/− and Islet1−/− grafts have more synapses per host bipolar cell
We have often seen images indicative of robust neural integration, such as in Figures 3A and 3B, with host bipolar dendrites extending toward graft cells and surrounded by photoreceptor presynaptic marker RIBEYE (Akiba et al., 2019). It is, however, difficult to confirm bona fide synapses formed between host bipolar cells and graft photoreceptor cells (hereinafter referred to as “de novo” synapse), as graft bipolar cells may also form synapses. In order to identify de novo synapses, we used rd1;L7-GFP mice as host and grafts derived from the Ribeye-reporter cell line (Mandai et al., 2017). Additionally, we immunostained the postsynaptic marker Cacna1s and counted synapses on the dendritic tips of host L7-GFP+ rod bipolar cells, ensuring that we are counting genuine synapses and that these are between host and graft cells. Figures 3C–3G show an example of host-graft synapse formation in Bhlhb4−/− line. L7-GFP+ bipolar cells forming de novo synapses exhibited the typical candelabra-like dendritic arbor of normal rod bipolar cells (Figure 3C, Video S1) in contrast to the dendrite-less morphology of the degenerate retina. Notably, host rod bipolar cells sometimes extended their dendrites far into the graft area (Figures 3H and 3I, right area). The majority of host bipolar cells do not make contact at all, but when they do, multiple synapses are formed between a host bipolar cell and graft photoreceptor cells, resulting in synapse distribution with a sharp zero count. Figure 3J shows the distribution of the number of synapses per host bipolar cell (L7-GFP+). These data are best described as a mixture of two distributions with two parameters, known as “zero inflated Poisson.” One distribution, the large zero counts, describes the probability of forming a synapse at all , and the other one, the broad peak, describes the average number of synapses once synaptic contact is established (Figure S4A). The probability of host bipolar cells to form synapses varied greatly. For example, the modes of predicted value (Figure S4B: predicted value) range from −2.4 to −1.2 (log odds) corresponding to probabilities ranging from 0.08 to 0.23. Except for a modest improvement in Bhlhb4−/−, the effect of different graft genotype was unclear with estimated effect ranges largely overlapping (Figures 3K and S4B; confidence in the difference in the effect of graft genotype was 84% for wt-Bhlhb4−/−, 66% for wt-Islet1−/−, and 66% for Islet1−/−-Bhlhb4−/−). On the other hand, there was a notable increase in the average number of synapses per host bipolar cell in the KO lines (Figure 3L; confidence in the difference in effect of graft genotype was 98% for Bhlhb4−/−-wt, 98% for Islet1−/−-wt, and 64% for Bhlhb4−/−-Islet1−/−). Interestingly, we also found that sex of the host has a significant effect on the outcome resulting in more synapses per bipolar in females (Figure S4C; confidence in the difference in the effect of sex was 91% for male-female). In addition to these main effects (overall effects), we estimated the interactions, conditional effects that are present under different combinations, of graft genotype and animal sex (Figure S4, genotype x sex panels). Together the effects of line and sex and their interaction indicate that wt grafts produce 3-6 synapses (male: 3.2 [95% CI = 2.4–4.3], female: 5.1 [95% CI = 4.0–6.5]), Bhlhb4−/− grafts produce 4-7 synapses (male: 5.3 [95% CI = 3.9–7.2], female: 5.5 [95% CI = 4.5–7.0]), and Islet1−/− grafts produce 4-9 synapses (male: 6.0 [95% CI = 3.4–9.6), female: 5.7 [95% CI = 4.7–7.0]) per L7+ bipolar cell. The large variability in the probability of synapse, compared with the more stable and narrower estimates for the average number of synapses, perhaps reflects the stochastic nature of transplantation procedures. Finally, it is noteworthy that some rod bipolar cells were able to make up to 20 synapses. This value is similar to the number of synapses for a single bipolar cell in the wt retina, with reported values varying from 25 (Tsukamoto and Omi, 2013) to 35 (Behrens et al., 2016). Although such robust de novo synaptic formation was rare, it highlights the potential for extensive synaptic formation of transplanted grafts.
A host rod bipolar cell, extending its axon toward the IPL is show (cyan) with several ribbon synapses (yellow), exhibiting a typical candelabra-like morphology of rod bipolar cells. Photoreceptor cells can be distinguished by their more concentrated and brighter signal (magenta). Note that there is a small gap between graft and host cells, however the host bipolar dendrites extend through the graft inner cells and make direct contact with graft photoreceptor cells.
De novo host-graft connection remains steady while intragraft connection decays
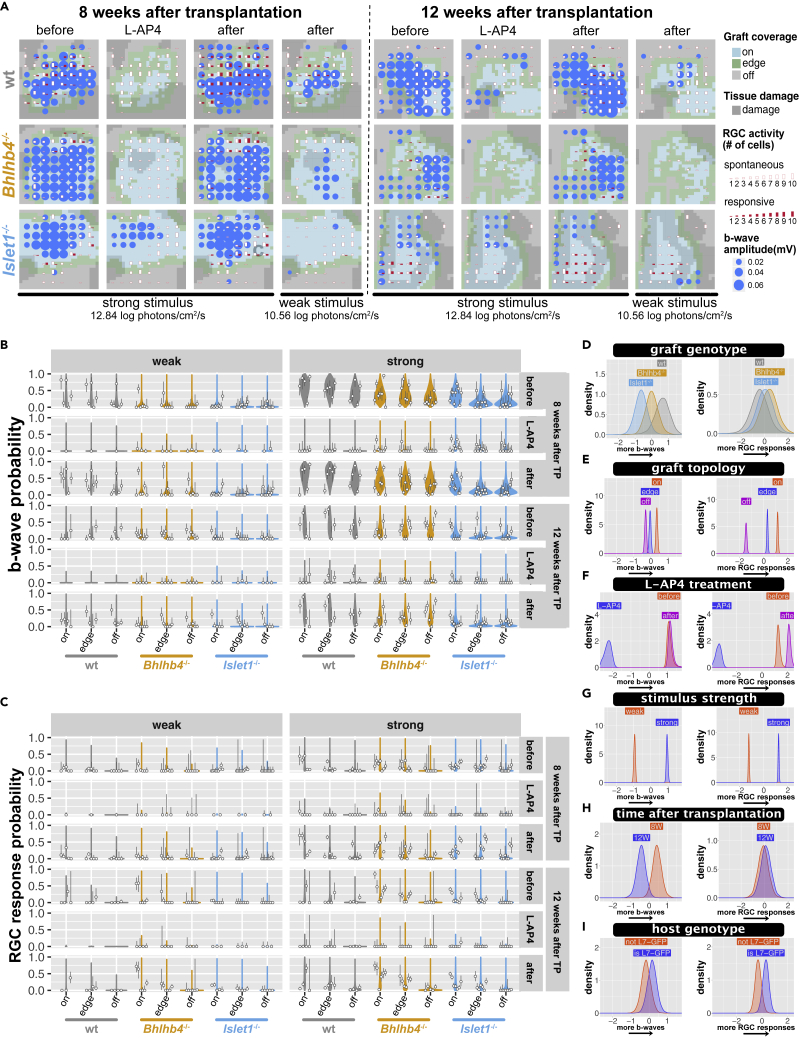
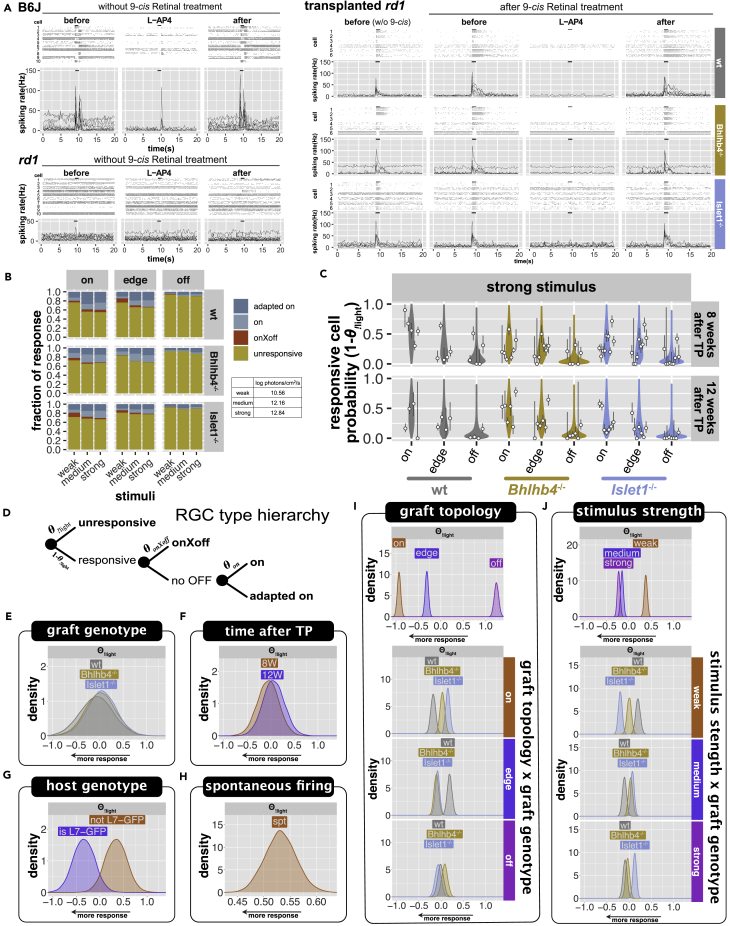
Transplanted retinas were examined by multielectrode array (MEA) recording at 8 and 12 weeks after transplantation in male rd1 or rd1;L7-GFP mice that were transplanted at 8-12 weeks old. Although no obvious mERG a-waves were detected with the 10-ms pulse stimulation, we consistently observed mERG b-waves as upward peaks of field potential near and over the grafted area, sometimes accompanied by RGC responses (Figure 4A; see Materials and Methods for details). The summary of data (circles) and model predictions (violin plots) for the mERG b-wave probability (Figure 4B) and the response probability of all detected RGCs (Figure 4C) at different conditions indicate the model is a reasonable description of collected data. The recorded data are multidimensional, with observations being influenced by several predictors (parameters), including graft genotype, graft topology (whether the tissue on top of the recording electrode was covered by graft cells), L-AP4 treatment, stimulus strength, time after transplantation, host genotype, tissue damage, and individual mouse biases. The effect of these predictors was estimated using hierarchical Bayesian inference, considering the hierarchy between predictors. We also included b-wave amplitude and spontaneous spiking frequency as covariates to estimate the probability of RGC response. Our model shows the RGC response probability is anticorrelated to both the b-wave amplitude and the spontaneous spiking frequency (Figure S7 spontaneous firing frequency). Interestingly, KO grafts exhibited similar RGC responsiveness to wt grafts despite their reduced b-wave activity (Figure 4D; the confidence in the difference in the effect of graft genotype on b-wave amplitude was 80% for wt-Bhlhb4−/−, 95% for wt-Islet1−/−, and 87% for Islet1−/−-Bhlhb4−/−), possibly reflecting the presence of a greater number of PKCα+ rod bipolar cells in wt grafts than in KO grafts (Figure 2B). Channels within or at the edge of the graft-covered areas have both higher b-wave probability and RGC response probability, indicating these responses are graft-driven (Figures 4E, S6, and S7 graft coverage; confidence in the effect of graft topology were 100% for on-edge, on-off, and edge-off for both the b-wave and RGC response). L-AP4, the agonistic blocker of glutamate receptor mGluR6 known to be specifically expressed by ON-bipolar cells, largely abolished both b-wave and RGC responses (Figure 4F; confidence in the effect of L-AP4 was 100% for before-L-AP4 and L-AP4-after for both b-wave and RGC response), suggesting the requirement of photoreceptor-bipolar synaptic connection for these light responses. The intensity of light stimulus has a clear and significant effect, with stronger stimuli increasing both b-wave and RGC response probability (Figure 4G; confidence in the effect of stimuli was 100% for weak-strong for both the RGC response and b-wave) although the effect is less pronounced for wt graft b-wave and more apparent in wt RGC response (Figures S6 and S7 graft genotype and stimulus interaction). Intriguingly, the b-wave probability but not RGC responsiveness clearly decreases from 8 weeks after transplantation to 12 weeks after transplantation (Figure 4H; confidence for the effect of time after transplantation was 96% for 8W–12W for the b-wave), possibly indicating that the established host-graft connectivity by 8 weeks after transplantation remains steady while the intragraft connections may be lost with time. Unexpectedly, we found a small but noticeable effect on RGC response probability when using the mice with L7-GFP labeling as host (Figure 4I; confidence in the effect of L7-GFP was 89% for the RGC response), especially when transplanted with wt and Islet1−/− grafts (Figure S7; confidence in the interaction of graft genotype and host genotype was 89% for wt-Bhlhb4−/− and 94% for Bhlhb4−/−-Islet1−/−, respectively). We also confirmed that these RGC responses of transplanted retinas derived from host RGCs by two-photon calcium imaging (Figure S8).
Figure 4.
mERG and RGC response to 10-ms pulse stimulus
(A) Examples of MEA recording with graft location, mERG b-wave-like response, and RGC response overlaid for each graft genotype. Color tiles represent the graft location (on, edge, off). mERG b-wave-like activity by amplitude (>3-sd) and RGC activity by cell number are shown as circles and bars, respectively, over the 8 x 8 electrode locations. Red bars indicate the number of light-responsive cells, and white bars show the number of cells with spontaneous spiking >3Hz. See Figure S5 for more examples of recordings.
(B and C) Summary of collected data and posterior predictive check. Circles indicate the observed probability with bars showing the 95% compatibility interval of binomial trials as calculated by the exact method (Clopper and Pearson). The predictions from our model are shown as violin plots, with wider regions indicating more likely values. (B) shows the probability of observing a b-wave peak on a channel, and (C) shows the probability of RGCs to respond to light at 8 and 12 weeks after transplantation (TP).
(D–I) Posterior distribution (main effect) of mERG b-wave and RGC response models. Plots show: graft genotype (D), graft topology (E), L-AP4 treatment (F), stimulus strength (G), time after transplantation (H), and host genotype (I). Full description of the model parameters, including covariates and interaction terms is provided in Figure S6 (b-wave model) and Figure S7 (RGC response model). Horizontal axes are in log odds, with higher values indicating higher probability of response. n = 30,167 observations, from 6827 cells, collected from 38 animals (9 wt, 13, Bhlhb4−/−, and 16 Islet1−/−) mice (male only), the same mice cohort from the RGC response analysis (Figures 5 and 6).
ON-pathway activity dominates the host RGC responses in transplanted retinas
Figure 5A shows representative raster plots and peristimulus time histograms of recordings from a normal mouse, an rd1 mouse transplanted with wt and KO grafts, and an age-matched rd1 mouse without transplantation. It is evident that grafts can restore some of the light responsiveness lost in the end-stage rd1 mice. While recordings were carried out with 9-cis-retinal for a more stable response, the light response was clearly present even without supplemental 9-cis-retinal, indicating that graft photoreceptors can potentially function in vivo without direct retinal pigment epithelium contact in spite of the rosette formation. We further characterized RGC activity (both responsive and spontaneous) taking into account graft genotype (wt, Bhlhb4−/−, and Islet1−/−), L-AP4 treatment, stimulus intensity, and graft coverage. We examined the response pattern (ON, OFF, etc.) of individual cells to the 1-s light stimulus through the full before-during-after L-AP4 treatment procedure and separated RGCs to 4 functional types: not responding to light (unresponsive) or being responsive with robust ON responses from the beginning (on) or after L-AP4 (adapted on), or with response containing the OFF-pathway component (onXoff) (see Materials and Methods for detailed description and categorization). Surprisingly, we found the majority of responding RGCs were on or adapted on types (Figure 5B), with the latter showing ON responses enhanced or emerging after L-AP4 treatment, suggesting the potential of newly formed host-graft synapses to be sensitized. These adapted on responses seemed to be more common in Islet1−/− than in the other grafts, although we cannot be very confident of these differences owing to the wide CIs (Figure S10 graft genotype; confidence in the effect of graft genotype for adapted on response probability was 76% for wt-Islet1−/− and 84% for Bhlhb4−/−-Islet1−/−). The onXoff RGCs, covering only a small portion of responsive RGCs, are suggested to have inputs from the OFF pathways as they responded to the light offset. However, these OFF responses were largely diminished by L-AP4 blockade, implying their dependence of ON-pathway activities, potentially via cross-inhibition. Such ON-OFF crossover activities are more often seen in the Islet1−/− and less frequently in Bhlhb4−/− lines (Figure S10 graft genotype; confidence in the effect of graft genotype for onXoff response probability was 92% for wt-Bhlhb4−/−, 89 for wt-Islet1−/−, and 100% for Bhlhb4−/−-Islet1−/−) and are more strongly associated with graft coverage in KO lines (Bhlhb4−/− > Islet1−/− > wt, Figure S10 graft genotype and topology interaction).
Figure 5.
RGC response to 1-s continuous light stimulus
(A) Representative raster plots and peristimulus time histograms of normal mouse, rd1 mouse, and rd1 mouse transplanted with retinal organoids. Raster plots show three repeat recordings from some representative cells. Histogram lines show the spiking frequency of each cell, with bars on top indicating the 1-s light stimulation (strong stimulus for normal and rd1, and medium stimulus for transplanted mice).
(B) Summary of response type ratios of different graft types under different light stimulation and graft coverage from all the recorded samples.
(C) Response probability () of collected data for strong light stimulation at 8 and 12 weeks after transplantation (TP) is shown along with model predictions. Circles indicate the observed probability with bars showing the 95% compatibility interval of binomial trials as calculated by the exact method (Clopper and Pearson). The predictions from our model are shown as violin plots, with wider regions indicating more likely values.
(D) Hierarchy of RGC response categories for light response pattern.
(E–I) Posterior distributions of the parameters for the “no light response” probability () for RGC types. Panels show: graft genotype (E), time after TP (F), host genotype (G), spontaneous firing (H), graft topology (I), and stimulus strength (J). For the posterior distributions of the full model parameters including all the response categories, refer to Figure S10. Horizontal axes, compatibility intervals, and modes are in log odds. n = 16,353 observations, from 5738 cells, collected from 38 mice (9 wt, 13, Bhlhb4−/−, and 16 Islet1−/−).
Host RGC responses are strongly associated with graft
Figure 5C shows the summary of collected data and model predictions for the probability of individual RGCs to respond to light. We estimated the effect of predictors using hierarchical Bayesian linear regression, taking into account the dependence between predictors. We used a conditional logit model to estimate the probability of the unresponsive and three responsive types (on, adapted on, and onXoff). In this model, a simple hierarchical structure among these RGC functional types is assumed as shown in Figure 5D. First, the probability of unresponsive RGC () is estimated (the exclamation symbol in “!light” denotes the logical NOT operator). Note that is the probability of cells to respond to light, and therefore, lower values of indicate higher responsiveness. Of the cells that respond to light (excluding the unresponsive RGCs), cells responding to light offset have a probability to be onXoff type, and the rest of responsive cells (responsive cells excluding onXoff type) have either a probability to be “on” or to be adapted on type. Figures 5E–5J show the posterior distribution of parameters affecting the probability of unresponsive RGC (). Consistent with RGC response to 10-ms stimuli, we did not find significant differences between graft genotypes for RGC responsiveness (Figure 5E). There is a clear correlation with graft coverage for response probability (Figure 5I; confidence in the effect of graft topology on the probability of being unresponsive was 100% for on-edge, on-off, and edge-off). We estimate that upon weak light stimulation, cells have approximately 0.03 [95% CI = 0-0.23] response probability on areas not covered by the graft (off), whereas cells below the graft (on) have a 0.24 [95% CI = 0.04–0.72] response probability. In addition to the overall effects of the predictors (the main effects), we further characterize the effect of different graft types by estimating interaction terms with graft genotype. The interaction terms are effects additional to the main effect, under particular combination of predictors. Consequently, although there is a strong overall preference for responses on areas under and near the graft (on > edge > off, Figure 5I top), wt grafts were characterized by more responses on the “on” graft areas than the other grafts, whereas KO grafts show more responses on “edge” graft areas than wt grafts (Figure 5I genotype x topology; confidence in the effect of interaction of genotype and topology was higher than 99% for wt-Bhlhb4−/− and wt-Islet1−/− for both on and edge locations). Thus, although the overall responsiveness is similar among graft types, KO line responses are more dispersed and far-reaching. These light responses were intensity-dependent, with stronger stimuli eliciting more responses (Figure 5J; confidence in the effect of stimulus strength was 100% for weak-medium and weak-strong and 93% for medium-strong). Interestingly, KO graft-transplanted retinas seem to be more sensitive to low-intensity light (Figure S10; confidence in the effect of interaction between graft genotype and stimulus strength was 99% for wt-Bhlhb4−/− and 100% for wt-Islet1−/− and Bhlhb4−/−-Islet1−/− for the weak stimulus), whereas the wt had more robust responses under stronger light stimulation. RGC responses remained largely identical at 8 weeks and 12 weeks after transplantation (Figure 5F). Notably, cells with higher spontaneous activity tend to respond less to light (Figure 5H). Lastly, there was an unexpected effect of host genotype, wherein L7-GFP hosts seem to respond better than non-L7-GFP hosts (Figure 5G; confidence in the effect of host genotype on responsiveness was 93%).
Inputs from Bhlhb4−/− grafts drive host RGC ON responses with better signal-to-noise ratio
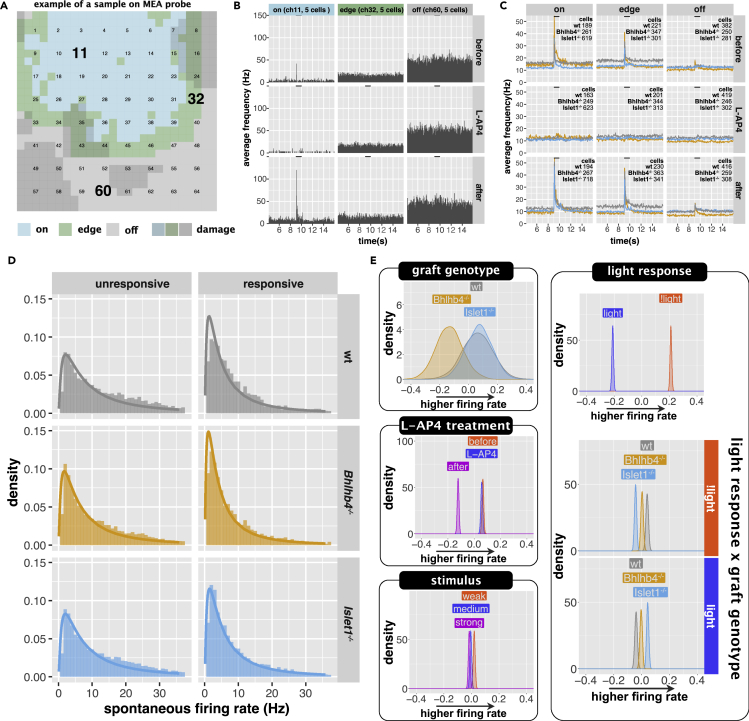
Since the increased spontaneous activity in degenerating retina may interfere with light responses, we also analyzed the spontaneous firing of RGCs using the spiking rate of cells from the 9-s recording before light stimulation. Figures 6A and 6B show an example from a sample, illustrating the effect of lowered spontaneous activity by grafts. Figure 6C shows population averages for the three graft types. As the distribution of spontaneous firing follows a lognormal distribution, we estimated the effect of different parameters on the logmean. Figure 6D shows the distribution of RGC firing frequency for responsive and unresponsive cells for different graft genotype, overlaid with model predictions (see Figure S11 for the same plot in log frequency). In this analysis, we used light responsiveness instead of graft topology, as light responsiveness is a more accurate indicator that RGCs have connections that lead to graft photoreceptors. Our analysis indicates that higher spontaneous activity is reduced in host retina transplanted with Bhlhb4−/− (Figure 6E, confidence in the effect of graft genotype was 87% for wt-Bhlhb4−/− and 92% for Bhlhb4−/−-Iselet1−/−). Spontaneous activity clearly decreased after L-AP4 washout (Figure 6G, confidence in the effect of L-AP4 was 100% for before-after and L-AP4-after), and we speculate that this is related to the sensitization of host-graft synapses by L-AP4, followed by a dis-inhibitive potentiation of bipolar cells after washout. Similar to RGC responses, spontaneous firing remained largely identical at 8 weeks and 12 weeks after transplantation (Figure S11 time after TP). There is a clear and substantial difference between cells that respond to light (on, adapted on, onXoff) and cells that do not (unresponsive), with responsive cells having lower spontaneous activity (Figures 6E and S11 responsiveness; confidence in the effect of light responsiveness was 100%). Interestingly, there was a small but distinctive effect of graft genotype on the effect of light responsiveness, with KO grafts having lower spontaneous firing rates in unresponsive cells (Figures 6E and S11 graft genotype responsive interaction; confidence in the effect of graft genotype was higher than 99% for wt-Bhlhb4−/−, wt-Islet1−/−, and Bhlhb4−/−-Islet1−/− for both responsive and unresponsive cells).
Figure 6.
RGC spontaneous firing before light stimulation
(A and B) Examples of MEA recordings. (A) shows the position of graft (on, edge, off) on the 64 electrodes of the MEA probe. The dark gray areas indicate areas where tissue damage was apparent (see Materials and Methods for details). (B) shows the peristimulus histogram of three channels (11, 32, and 60, each with 5 cells detected) which correspond to on, edge, and off graft areas, respectively.
(C) shows population averages of spontaneous firing rate (the height of the baseline) and light responses (peaks triggered by onset and offset of light stimulus) on different graft areas (on, edge, and off) and before, during, and after L-AP4 treatment. The bar on top indicates the 1-s light stimulation. This is the average of responses to samples collected 12 weeks after transplantation and stimulated with the strong light condition. See Figure S9 for population averages of other conditions. The spontaneous activity (defined as the average firing frequency prior to light stimulation) was modeled with a lognormal distribution.
(D) shows the histogram of the spontaneous firing rate overlayed with the modeled lognormal distribution for responsive and unresponsive of host transplanted with different graft genotype. The same plot but in with the horizontal axis in log is shown in Figure S10.
(E) Posterior distributions of the parameters affecting the logmean. For the posterior distributions of the full model parameters, refer to Figure S11. n = 44,213 observations, from 5738 cells, collected from 38 mice (9 wt, 13, Bhlhb4−/−, and 16 Islet1−/−).
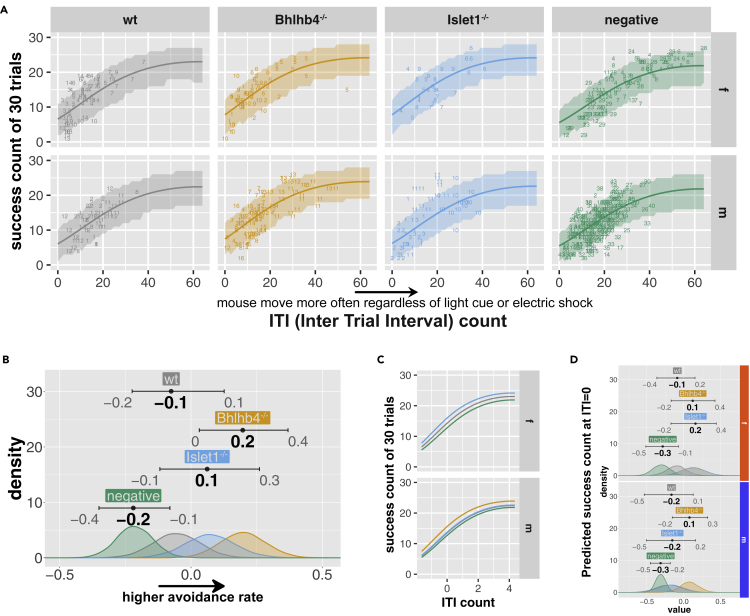
Mice transplanted with Bhlhb4−/− and Islet1−/− grafts perform better at light avoidance response
While KO grafts did not show a noticeable improvement on the RGC light response over wt grafts, they seemed to better suppress high-frequency spontaneous spiking. We speculated that KO graft transplantation would result in better functional recovery owing to its reduced noise. To test this, we conducted a behavior examination of light avoidance (Figure 7, see Materials and Methods for details). Mice were placed in a cage with two chambers which are randomly illuminated by mesopic light proceeded by a mild electric shock. Mice that move often, indicated by higher inter trial interval (ITI) counts, can avoid the electric shock purely by chance as can be seen in Figure 7A for the negative control. However, mice that can perceive the light learn to avoid the electric shock using the light as a cue, shifting this curve upward. We modeled the effect of each group (wt, Bhlhb4−/−, Islet1−/−, and negative control) on the success probability while accounting for the ITI and other factors (Figure S12). Consistent with our hypothesis, we found that mice transplanted with KO grafts performed better than wt grafts (Bhlhb4−/− > Islet1−/− > wt, Figures 7B and 7C; confidence in the effect of graft/group was 90% for wt-negative, 98% for Islet1−/−-negative, 100% for Bhlhb4−/−-negative, 80% for wt-Islet1−/−, 96% for wt-Bhlhb4−/−, and 80% for Bhlhb4−/−-Islet1−/−). We found that females performed somewhat better than males (Figure S12; confidence in the effect of sex was 89%). In females, both Bhlhb4−/− and Islet1−/− grafts show a modest improvement over wt grafts, whereas the improvement is only apparent in Bhlhb4−/− in males (Figure 7D).
Figure 7.
Light avoidance test
Mice are placed in a dark box and presented with a light cue before a mild electric shock. Mice that can sense the light learn to avoid the electric shock using the light cue.
(A) The number of successful avoidances out of 30 trials is plotted against intertrial interval (ITI), which represent the sporadic movements of the mouse. Numbers indicate the id of a mouse for each group. Success count increases with ITI as mice that move often may avoid the shock purely by chance. Mice that perceive light and learn to use it as a warning can increase their success count with lower ITI. The number of success counts was modeled using the binomial distribution using group, sex, mouse, ITI, and trail number as predictors, and the shaded area shows the 95% predicted success count.
(B) Estimated effect of each group () with 95% compatibility intervals. Distribution for the rest of the parameters is provided in Figure S12. Note the horizontal axis is the log odds.
(C) Overlay of predicted mean avoidance.
(D) Predicted success rate, at ITI = 0, for male and female mice. Note that the bottom axis is logit (log odds). The number of mice used for this analysis was 86 mice (14 wt, 15 Bhlhb4−/−, 11 Islet1−/−, and 45 age-matched negative control (rd1)).
Discussion
In this study, we prepared Bhlhb4−/− and Islet1−/− grafts aiming to reduce the population of graft bipolar cells to improve the first synaptic transmission in the reconstructed retina from graft photoreceptors to host bipolar cells. An alternative approach would be to use organoids that are further along the retinal development with a clearly defined ONL and mechanically eliminating the inner part of the retina. However, we have shown that DD17 grafts (approximately equivalent to embryonic day 17) or earlier are more suited for transplantation as they are able to develop structured ONL with functional reconstruction after transplantation, whereas transplantation of older grafts tend to result in disorganized structures (Assawachananont et al., 2014; Mandai et al., 2017). Our approach of using genetically engineered grafts allows us to properly time photoreceptor development and transplantation, while drastically reducing the population of graft bipolar cells.
The PKCα+ rod bipolar cell population was markedly reduced as expected; however, this did not translate to concomitant photoreceptor enrichment. It is possible that nascent progenitor cells are diverted to alternative cell fates or remain partially differentiated, unable to commit to a final retinal cell fate in Bhlhb4−/−and Islet1−/− grafts (Elshatory et al., 2007; Pan et al., 2008). Overall, however, Bhlhb4−/− and Islet1−/− grafts displayed typical photoreceptor differentiation and maturation, with a prominent photoreceptor rosette and/or layered structure surrounded by some inner retinal cells, essentially identical to wt grafts except for the reduced bipolar cell population.
KO grafts did not substantially increase the number of host bipolar cells forming synapses, or improved RGC light responsiveness. Instead we found that KO grafts allow a higher number of synapses per host bipolar cells, possibly owing to the higher number of graft photoreceptors without intragraft partners. The higher number of synapses per rod bipolar is also consistent with the higher light responsiveness to weak light stimuli observed in KO-graft-transplanted retinas. On the other hand, KO graft effects were characterized by reduced spontaneous activity in the host retinas. Degenerated retinas are characterized by increased spontaneous activity owing to the lack of photoreceptor input (Biswas et al., 2014; Trenholm and Awatramani, 2015; Tu et al., 2016). It is tempting to speculate that host bipolar cells were able to establish more connections in grafts deprived of bipolar cells, resulting in sufficient photoreceptor input to restore inner retinal signaling and to reduce aberrant RGC activity. This in turn may be reflected in the better performance of KO-graft-transplanted animals in behavior tests. The reduction in spontaneous activity (Figure 6) and better performance in behavior test (Figure 7) seem to indicate Bhlhb4−/− is more advantageous than Islet1−/−, at least in male mice, although synaptic connectivity was similar. It may be that we were unable to observe differences in connectivity owing to the large sample variation, or alternatively it may be that although the number of synapses is similar, the quality, i.e. the strength of synaptic connectivity (Akiba et al., 2019) or the distribution of synapse across rosettes differs. Although we are uncertain of the underlying mechanism, reduction in aberrant RGC activity seems to substantially improve the performance after transplantation. This highlights the importance of future approaches to focus on the reconstruction of the inner retina circuitry in addition to neural integration of transplanted cells, as degenerated retinas undergo neural remodeling (Jones and Marc, 2005; Marc et al., 2007). Our results indicate the possibility that remodeled inner retinal circuitry can still be reengaged with sufficient photoreceptor input.
KO-graft-transplanted retinas display reduced b-wave activity despite the similar RGC light responsiveness, consistent with the reduction in bipolar cell population in the graft (wt > Bhlhb4−/− > Islet1−/− for both b-wave and bipolar cells; see Figures 2 and 4). We think the difference between b-wave and RGC responsiveness is reflective of intragraft photoreceptor-bipolar connections. These intragraft connections not only deprive the graft photoreceptor cells from forming contacts with host retina but also may have some adverse effects on the host retinal activities. Our analysis of the spontaneous activity shows that RGCs that are light responsive with synaptic inputs originating from graft photoreceptors have a clear reduction in spontaneous activity. However, in unresponsive host cells without effective inputs from grafts, the spontaneous activity is higher wherein more presumptive intragraft photoreceptor-bipolar connections exist. Although we are not certain of the mechanism that leads to more spontaneous activity in grafted areas with more intragraft connections, these results further highlight the benefits of KO grafts.
During the course of our experiments, we found unexpected effects of host genetic background. First, host sex seems to affect synapse formation and possibly the behavior response, with females performing better than males. Second, rd1;L7-GFP mice seem to perform better in MEA recordings than rd1 mice although these mice have the same C57BL/6J background. It is possible that the L7-GFP may retain some FVB/N background. We do not currently understand how these differences originate, but it underscores the need to control for these variables.
The requirement of establishing synaptic integration is a significant challenge for photoreceptor cell therapies. We have re-enforced our previous report showing conclusively that stem-cell-derived retinas form synaptic connections with the host to restore light responsiveness (Mandai et al., 2017). Furthermore, we have expanded our analysis and shown that aberrant firing is reduced by grafts. This effect is further enhanced in genetically engineered cell lines, perhaps owing to improved input to bipolar cells and therefore improved outcome of transplantation. Our genetically engineered cell lines are unlikely to result in deleterious effects, as they are not introducing an exogenous gene, but rather deleting part of a gene that is expressed in a subset of cells in the graft. Furthermore, in the reported Bhlhb4−/− or Islet1−/− mice, there are no known unfavorable ocular abnormalities other than the reduction or absence of the b-wave, and both morphology and function of photoreceptors seem normal. Local transplantation of these tissues is therefore well justified considering the potential benefits in patients at the risk of becoming blind.
Limitations of the study
In this study, we transplanted to end-stage degenerated retinas (8- to 12-week-old rd1 mice), minimizing the likelihood of observing the effects of remaining host photoreceptor cells or cytoplasmic material transfer rather than neural integration of grafted cells (Pearson et al., 2016; Santos-Ferreira et al., 2016a; Singh et al., 2016; Waldron et al., 2018). Furthermore, the use of end-stage rd1;L7-GFP model conclusively and unequivocally proves the neural integration of grafted cells. However, while we are convinced that the observed light responses, which are highly correlated to graft coverage, do not originate from so-called “remaining cone cells,” we cannot rule out the possibility of remaining cones being rescued by cytoplasmic material transfer or some other means. We, however, think this is unlikely as responses are observed even under scotopic-mesopic light, more consistent with a rod-driven response. Furthermore, the number of synapse-forming graft photoreceptor cells seems to largely excel the number of any potentially rescued remaining host cone photoreceptor.
In our analyses, we found that host sex may play a role in the outcome of retinal cell transplantation. Given the effect of sex on synapse numbers and performance in behavior tests, there may also be a significant difference on the light response and/or the activity of cells observed by MEA recordings. However, owing to resource limitations, we were not able to analyze the effect of sex on MEA recordings.
The effect of mice is substantially bigger than other predictors, such as graft genotype, in all analyses. This large mouse-to-mouse variance is reflective of the innate stochastic nature in the transplantation procedure. For example, the extent of damage to the host retina, which is already fragile owing to degeneration, during transplantation, and/or dosage of successfully transplanted graft cells are factors that are challenging to control and/or account for in mice. As we can visually locate and map the location of the graft on the host retina, this stochastic nature is less problematic for analyses of the local effects of the transplantation, such as cell types, synapse formation, and cell activities (light response and spontaneous firing). On the other hand, the large mouse-to-mouse variability may have obscured the effect of graft genotype on the behavior test (Shuttle Avoidance System), as performance may well depend on graft size, location, and integrating pattern. While these are challenging to control in mice, owing to its small size and limitations on ophthalmological surgical equipment, these are issues that are less likely to be problematic on primates.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-PKCα | NOVUS | cαt#NB600-201; RRID:AB_10003372 |
| Mouse monoclonal anti-Chx10 | Santa cruz Biotechnology | cat# sc-365519;RRID:AB_10842442 |
| Guinea pig polyclonal anti-human/mouse Rx | Takara | Cat# M229; RRID:AB_2783559 |
| Mouse anti-glutamine synthetase | Millipore | Cat# MAB302; RRID:AB_2110656 |
| Rabbit anti-glial fibrillary acidic protein | Dako | Cat# Z0334; RRID:AB_10013382 |
| Sheep polyclonal anti-human secretagogin | BioVender | Cat# RD184120100; RRID:AB_2034062 |
| Mouse anti-CtBP2 | BD Bioscience | Cat# 612044; RRID:AB_399431 |
| Mouse anti-Cacna1s | Millipore | Cat# MAB427; RRID:AB_2069582 |
| Rabbit anti-calbindin D-28K | Millipore | Cat# AB1778; RRID:AB_2068336 |
| Rabbit anti-calretinin | Millipore | Cat# AB5054; RRID:AB_2068506 |
| Mouse monoclonal anti-rhodopsin (RetP1) | Sigma-Aldrich | Cat# O4886; RRID:AB_260838 |
| Chemicals, peptides, and recombinant proteins | ||
| L-AP4 | Wako | 016-22083 |
| 9-cis-Retinal | Sigma-Aldrich | R5754 |
| Opsinamide | Sigma-Aldrich | AA92593 |
| Critical commercial assays | ||
| RNeasy kit | QUIAGEN | 74004 |
| Mouse Embryonic Stem Cell Nucleofector™ Kit | Lonza | VPH-1001 |
| Deposited data | ||
| Retinal organoid microarray sequencing data | NCBI's Gene Expression Omnibus | GSE:178653 |
| Experimental models: Cell lines | ||
| Mouse Nrl-GFP iPS-cell line | Assawachananont et al., 2014 | N/A |
| Tg(Nrl-GFP);Ribeye-reporter iPS cells | Mandai et al., 2017 | N/A |
| Thy1-GCaMP6f;Ribeye-reporter ES cells | This paper | N/A |
| Mouse Thy1-GCaMP6f ES cells | This paper | N/A |
| Experimental models: Organisms/strains | ||
| C57BL/6J-Pde6brd1-2J/J | The Jackson Laboratory | JAX004766 |
| B6;FVB-Tg(Pcp2-EGFP)2Yuza/J | The Jackson Laboratory | JAX004690 |
| B6;CBA-Tg(Thy1-GCaMP3)6Gfng/J | The Jackson Laboratory | JAX 0017893 |
| Recombinant DNA | ||
| pSpCas9(BB)-2A-Puro (PX459) | Addgene | 48139 |
| Software and algorithms | ||
| Fiji/ImageJ | Schindelin et al., 2012 | https://fiji.sc |
| IMARIS 8.4 | Oxford Instruments | https://imaris.oxinst.com |
| R | R Core Team, 2020 | https://cran.r-project.org |
| Rstan | Carpenter et al., 2017 | http://mc-stan.org |
| Other | ||
| MED64 system | Alphamed Scientific Inc. | MED64-Basic |
| MED probe | Alphamed Scientific Inc. | MED-P515A |
| shuttle-avoidance system | Muromachi Kikai | CompACT SAS/W |
| Nucleofector Device | Lonza | Amaxa Nucleofector II |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Mandai Michiko (michiko.mandai@riken.jp).
Materials availability
Materials generated on this study are available from the corresponding author on request.
Experimental model and subject details
ES/iPS cell lines
Tg(Nrl-GFP) is a transgenic iPS cell line expressing GFP under the transcriptional control of Nrl promoter (Akimoto et al., 2006; Homma et al., 2013). ROSA26+/Nrl-CtBP2:tdTomato is a Rosa26 knock-in expressing a CtBP2 and tdTomato fusion protein under the Nrl promoter (Mandai et al., 2017). These ROSA26+/Nrl-CtBP2:tdTomato constructs will be referred to as Ribeye-reporter. We also constructed a Ribeye-reporter line on the C57BL/6J-Tg(Thy1-GCaMP6f)GP5.5Dkim/J ES cell line (Thy1-GCaMP6f;Ribeye-reporter) (Dana et al., 2014). Synapse count analysis and behavior tests were conducted with retinal organoids derived from the Ribeye-reporter ES cell lines. Immunohistochemistry characterization (except for synapse analysis), microarray analyses, and electrophysiology assays were conducted with grafts derived from the Tg(Nrl-GFP) lines.
Animals
All the experimental protocols were approved by the animal care committee of the RIKEN Center for Biosystems Dynamics Research (BDR) and were conducted in accordance with local guidelines and the ARVO statement on the use of animals in ophthalmic and vision research.
C57BL/6J-Pde6brd1-2/rd1-2J/J (JAX stock #004766, hereinafter referred as rd1) mice were used as the end-stage retinal degeneration model. B6;CBA-Tg(Thy1-GCaMP3)6Gfng/J (JAX stock #017893, hereinafter referred as Thy1-GCaMP3) mice (Chen et al., 2012) were crossed with rd1 mice to generate rd1;Thy1-GCaMP3 animals for live imaging using two photon microscopy. B6;FVB-Tg(Pcp2-EGFP)2Yuza/J (JAX stock #004690) (Tomomura et al., 2001) crossed with rd1-2J (hereinafter referred as rd1;L7-GFP) were used for host graft synapse analysis and electrophysiology. Eight- to 12-week-old mice of both sexes were used for transplantation.
Method details
Generation of Bhlhb4-/- and Islet1-/- cell lines
Bhlhb4-/- lines were constructed by targeting sites upstream and downstream of the start codon as shown on the schematics in Figure1A. Islet1-/- lines were constructed by targeting an upstream site of the start codon in exons 1 and the flanking region at the 3’-end of exon 2. Guide RNAs were designed using the CRISPR design tool (http://crispr.mit.edu). The oligo pairs were cloned into the BbsI-BbsI site in the plasmid containing Cas9 and the single-guide RNA (sgRNA) scaffold (pSpCas9(BB)-2A-Puro, Addgene plasmid ID: 481398), to generate puromycin-Cas9 fusion plasmids coexpressing sgRNAs (Ran et al., 2013).
Tg(Nrl-GFP);Ribeye-reporter iPS cells and Thy1-GCaMP6f;Ribeye-reporter ES cells were transfected with Cas9-sgRNA complexes targeting Bhlhb4 or Islet1, using the mouse ES Cell nucleofector kit (Lonza) and Amaxa Nucleofector II device (Lonza) following the manufacturer’s instructions. Transfected cells were treated with antibiotic (0.5-1 ug/mL puromycin) 24 hours after transfection for 48 hours. Four to 10 days after antibiotic selection, antibiotic-resistant colonies were collected for passage and genotyping. Genotyping was performed by PCR using specific primers to amplify fragments from the wt allele and of a shorter fragment from the deleted alleles (Figure 1). The PCR products from genotyping were purified (Macherey-Nagel) and sequenced (Thermo Fisher Scientific) to confirm the deletion. At least 2 or 3 clones were obtained for each KO line.
Transplantation of 3D retinas
Maintenance, differentiation, and optic vesicle structure preparation for transplantation of ES/iPS-cell-derived retinas were as previously described (Assawachananont et al., 2014). Briefly, neural retina (DD10-15), a characteristic continuous homogeneous epithelium region on the optic vesicle structure of organoids, was cut out in a small piece (around 400 μm × 500-800 μm), on the day of transplantation, and inserted subretinally into the eye, at approximately 0.5-1 mm from the disk at the ventrotemporal side, of the 8- to 12-week-old rd1 and rd1;L7-GFP mice with 1 mM VPA using a glass micropipette with a tip diameter of approximately 500 μm. Indomethacin (10 mg/L) was added to the drinking water of all transplanted mice starting on the day of transplantation. OCT and fundus images of transplanted eyes were acquired before sampling, and samples with unsuccessful transplantation (for example, leakage) were excluded from the analyses.
RT-PCR of retinal organoids
Total RNA was extracted from the differentiated retinal organoids using TRIzol reagent (Invitrogen), and cDNA was synthesized using Oligo(dT) primers and SuperScript III reverse transcriptase (Invitrogen). Target genes were amplified using SapphireAmp Fast PCR Master Mix (TaKaRa) with the specific primers detailed in the supplemental materials.
Microarray analysis
Total RNA was extracted from the differentiated retinal organoids (5 retinal organoids for each condition) using TRIzol reagent (Invitrogen) and was purified using RNeasy kit (QUIAGEN) following manufactures’ instructions. RNA quality check, concentration, and data collection using the SurePrint G3 Mouse GE microarray 8x60K Ver. 2.0 (Agilent technologies) were performed by Hokkaido System Corporation (Hokkaido, Japan), according to Agilent protocols. The array contained 56,605 probe sets covering 37,074 genes. The data have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE178653. Data processing, analyses, and visualization were conducted in R (R Core Team, 2020) using custom scripts. Probes below background level were discarded. For GO analysis, GO annotations for the microarray (downloaded from Agilent’s site) were correlated with GO annotations in the Mouse (GO.db: a set of annotation maps describing the entire Gene Ontology. R package, version 3.6.0.).
Immunohistochemistry
After mice were sacrificed by cervical dislocation, the eyes were enucleated and immediately fixed with 4% paraformaldehyde (PFA) for an hour at 4°C. The eyes were then washed with phosphate buffered saline (PBS), followed by the removal of the cornea, iris, and lens under a microscope. For cryosection preparation, the eyes were infiltrated with 30% sucrose overnight at 4°C and embedded in optimum cutting temperature (OCT) compound (4583, Sakura Finetek Japan, Tokyo) and stored at -30 °C. Coronal plane cryosections of 10-14 μm thickness were made with a cryostat (Leica CM3050S). After two rounds of PBS wash, sections were exposed to 0.1% Triton X-100 in PBS (vol/vol) for 30 min to permeabilize cellular membranes. Cells were then blocked with blocking solution for 1 h at room temperature. Primary antibodies were diluted in blocking solution and incubated with samples overnight. Samples were then rinsed three times with 0.05% Tween-20 in PBS (vol/vol) and incubated with the appropriate secondary antibodies for 1 h at room temperature and counterstained with DAPI (Life Technologies).
For free-floating immunostaining, 50-μm-thick retinal flat mount sections were collected into 12-well tissue culture plates containing cold PBS (5-8 sections/well) and blocked with staining solution (3% Triton X-100 and 1% BSA-PBS) at 4°C overnight. The samples were then incubated with primary and secondary antibodies in staining solution at 4°C for three to four days and two days, respectively, and mounted with 2,20-thiodiethanol (Sigma) or VECTASHIELD (Vector). All staining procedures were performed with gentle mixing on a rocker.
Images were acquired with a Leica TCS SP8 confocal microscope and reconstructed in 3D Imaris Microscopy Image Analysis Software (http://www.bitplane.com/) and Fiji (ImageJ).
Electrophysiology: MEA recording
Micro electroretinogram (mERG) and RGC spiking were recorded using the MED64 system (Alphamed Scientific Inc., Osaka, Japan) with a standard 8 x 8 probe (MED-P515A) as described in detail in the following (Fujii et al., 2016; Iraha et al., 2018; Tu et al., 2018; Tu and Matsuyama, 2020). After 1-3 days of dark adaptation, mice were anesthetized with sevoflurane (Abbott Japan, Osaka, Japan) and sacrificed by cervical dislocation. After enucleation, the transplanted retina was carefully isolated and vitreous was gently but thoroughly cleaned. The position of the graft was visually targeted in between the optic nerve disc and the transplantation entry site to be mounted over the probe with the ganglion cell side contacting the electrodes. A dim LED light source with the peak wavelength at 690 nm was used during sample preparation. The flat-mounted retinas were constantly supplied with warmed (34 ± 0.5°C), carbonated (95% O2 and 5% CO2) Ames’ medium (A1420, Sigma-Aldrich) perfused at 3-3.5 mL/min. Opsinamide (10 μM; AA92593, Sigma-Aldrich) was added in the perfusion medium to suppress the melanopsin-driven RGC light responses during recording. Retinas were kept in the dark, allowed to recover in the chamber for at least 20 min before the first set of stimulation while spontaneous spikes were monitored from the beginning. Full-field stimuli with intensities of 10.56 (weak), 12.16 (medium), and 12.84 (strong) log photons/cm2/s (equivalent to approximately 0.03 to 6.5 lux or 1.7 x 102 to 3.4 x 104 Rh∗/rod/s) at the focal plane of probe were generated with a white LED (NSPW500C, Nichia Corp., Tokushima, Japan) without background illumination. An electronic pulse generator (DSP-420, Dia-Medical System, Tokyo, Japan) was used to control the LED intensity under the external command from MED64 system using scheduled TTL pulses to turn on and off the LED. Pulse stimuli of 10 ms were used to elicit the mERGs and 1 s stimuli were used to separate the RGC response types. In our standard protocol, three-time serial recordings were repeated with an interval of 1 min, and a single recording time was 10 s for the 10-ms stimuli and 20 s for the 1-s stimuli. Data were collected at 20 kHz sampling rate and filtered at 1 Hz (10 ms stimuli) or 100 Hz (1 s stimuli) online. Recordings were repeated before and after 9-cis-retinal (100 μM for 10 min incubation in the recording probe; R5754, Sigma-Aldrich) replenishment, as well as in the presence and absence of L-AP4 (20 μM; 016-22083, Wako) blockade to confirm the reproducibility.
Only male animals were analyzed in these experiments, for logistic reasons. Through the course of the study, it became apparent that sex maybe an important factor; however, we had very few female samples and opted to limit this experiment to males only to avoid obfuscating results by mixing females since we could not gather enough samples to analyze the effect of sex.
Electrophysiology: two-photon imaging
rd1;Thy1-GCaMP3 mice transplanted with retinal sheets (wt, Bhlhb4-/-, Islet1-/-) were sacrificed at various time points after transplantation (7-26 weeks) by cervical dislocation, after at least 12 hours of dark adaptation before recordings. Areas away from the graft (designated as “off”) served as negative controls, as the grafted cells only covered a small fraction of the retina. Age-matched Thy1-GCaMP3 animals were used as positive controls (“normal”). Eyes were enucleated, and the retinas were flat mounted using a mesh-covered anchor with the ganglion cell layer on top. Retinas were constantly supplied with warmed (28 ± 0.5°C), carbonated (95% O2 and 5% CO2) Ames’ medium (A1420, Sigma-Aldrich) perfused at about 10 mL/min. Sulforhodamine 101 (500 nM; S7635 Sigma) was added in the perfusion medium to confirm ganglion cell viability and to help locate graft. The ganglion cell layer plane was identified using sulforhodamine counterstaining and the basal fluorescence signal from GCaMP3. Images were acquired with an upright multiphoton laser scanning microscope (Olympus, FV1000MPE) using a water-immersion 25x objective lens (Olympus, XLPLN25XWMP). The light source was a Mai Tai femtosecond-pulse laser (Spectra-Physics) running at 920 nm. The graft position was identified by the fluorescence of the graft (Nrl-GFP). Images were acquired at 0.066 s/frames on an area of 254.46 μm x 254.46 μm at 128 px x 128 px resolution for approximately 152 s (2300 frames). Retinas were recorded for at least a minute before a brief pulse light stimulation, at around 90 s, to allow transient fluorescent changes to equilibrate. The light source for the stimulus was a homogenous green light (λ=505 nm LED passed through a collimator lens) of 5.78 x 10-13 W/μm2 which would be equivalent to 1.28 x 106 Rh∗/Rod/s in a normal mouse retina.
Behavior: light avoidance
The shuttle-avoidance system (CompACT SAS/W, Muromachi Kikai) was used to evaluate the light responsiveness of transplanted mice as previously described (Mandai et al., 2017). Briefly, a dark-adapted mouse is placed in a sound- and light-insulated box with two chambers and able to freely move between chambers. The mouse is then presented with stimulus at random intervals at one of the chambers. If the mouse is stationary, the stimulus is proceeded by a mild electric shock (0.08 mA) delivered on the floor bars. The mouse can avoid the shock if it moves away from the chamber within 5 s of stimulus presentation. Each experiment consisted of 30 such trials, and the number of successful avoidances is recorded. Mice able to sense light (3 cd) can learn to use light as a warning and avoid the shock. Mice may also increase their success rate by frequently moving between chambers, so the number of times the mouse moves before light stimuli (ITI) is used as a proxy to account for this behavior. Initially mice were trained with a dual sound and light stimuli, and only mice that successfully learn to respond to the dual stimuli were subsequently tested with a light stimulus only. Mice were tested daily, except for weekends, until their response appeared to plateau.
Quantification and statistical analysis
Quantification of graft rods
The number of rods in grafts was quantified from Z-stack images of flat mounted retinas from mice sampled roughly 60 days (8-9 weeks) after transplantation. Orthogonal sections at coordinates that intersected grafts were resliced in Fiji, and the graft area was traced manually. Using grafts with the Nrl-GFP reporter, rod photoreceptors can easily be identified by the GFP signal, as well as by their size, localization, and DAPI signal pattern. We delineated the area of the graft manually and counted the number of cells in the graft (DAPI+ cells) and the number of rod photoreceptor cells (GFP+ cells) using a Fiji macro. Figures 2A–2D show a typical image of this quantification process. Fiji macro scripts are provided in GitHub repository (https://github.com/matsutakehoyo/KO-graft).
Quantification of graft inner cells
The numbers of inner cells, Calbindin+, Calretinin+, PKCα+, and Chx10+ cells in the grafts were counted manually from retinal sections of mice sampled roughly 30 days (4 weeks) after transplantation. The total population of graft inner cells was determined by elimination. First, host area, characterized by the laminar structure of the host INL, was identified. Second, graft photoreceptors were identified by Nrl-GFP, nucleus morphology, and local distribution within rosettes. The remaining cells were considered inner cells, and the total number of inner cells and the number of cells positive for a particular marker were counted.
Host-graft synapse count
For the host-graft synapse count, Thy1-GCaMP6f;Ribeye-reporter ESC-retina was transplanted to rd1;L7-GFP mice and the eyes were collected at DD50 (approximately 5 weeks after transplantation). Regions containing part of the graft rosettes were cropped, and the number of L7-GFP-positive bipolar cells was manually counted by one observer in a blind manner. Images were acquired on 50-μm sections, which allowed us to trace the entire L7-GFP-positive bipolar cell, from its axon extending toward host IPL to synapses at the dendritic tips, ensuring the genuine counting of synapses with host bipolar cells with potential synaptic inputs to downstream cells. An example of a host bipolar (L7-GFP+) cell extending an axon to the host IPL and forming multiple synapses with graft photoreceptors at its dendrites is provided in Video S1. A synapse was defined as a pairing of the tdTomato signal from the RIBEYE reporter at the graft rod presynaptic terminal (Thy1-GCaMP6f;Ribeye-reporter) and the Cacna1s signal (stained) at the postsynaptic terminal. Importantly, only Cacna1s signals at the dendritic tips of L7-GFP cells were counted, thus ensuring that counted synapses were strictly between graft photoreceptors and host bipolar cells. Furthermore, the pairing of synaptic markers was judged from three orthogonal sectional views (X-Y, X-Z, and X-Z) independently to avoid optical artifacts.
Quantification of MEA recordings
Graft locations were manually mapped on a 26 x 26 grid overlaid on the MEA probe. Our MEA probe consisted of 50 x 50 μm2 electrodes separated by 100 μm in an 8 x 8 array. The size of a channel corresponds to a square, i.e. a pixel, in our 26 x 26 map, and the separation between channels correspond to two squares. On each square, we mapped the presence or absence of graft (on, off) and applied a morphology transformation to dilate the graft area to define graft edges. In addition to graft location, areas where tissue damage was apparent or covered by the optic disc were marked as “damage,” assuming these areas would have some kind of impairment to respond.
mERG traces were processed and analyzed in R (R Core Team, 2020). A band-pass Butterworth filter (1 to 50 Hz) was applied to traces to remove low-frequency fluctuations and high-frequency jitter. Local minima within 55 ms from light stimulation were flagged as a-wave, and local maxima within 150 ms from light stimulation were flagged as b-wave. The a-wave amplitude was calculated from the baseline, and the b-wave amplitude was calculated from the a-wave, or from the baseline when the a-wave was not detected. Replicates from three repeated stimulations were averaged.
RGC spike sorting was conducted using the CED Spike 2 software (version 7.2). Recordings taken at the same stimulus intensity were processed together by merging recordings taken before 9-cis-retinal treatment and before, during, and after L-AP4 blockade, to follow spike trains from the same set of cells across the different conditions. Recording traces were first cleaned up by applying a Butterworth band-pass filter (200 to 2800 Hz) and DC offset removal. The spike templates were generated for each channel independently, based on the automatic offline template formation and spike matching algorithm of Spike 2 with a few minor modifications. Spikes with amplitudes smaller than 64 μV (negative) were excluded; this threshold level was about 10-sd of recording without retina and 4-sd of recording with robust spontaneous spiking of degenerated retina. As the whole recording procedure lasted for more than 3-4 hours, sometimes the spike amplitudes were found amplified over time. A 5% tolerance for maximal spike amplitude change was therefore included in the template matching process so that each spike would be slightly resized (±5%) to see if its shape matches any established template. This process allowed us to isolate the individual spike sources (cells) with characteristic spike size and shape from each sample for the given stimulus intensity. The average number of cells detected per channel was 2.4 ± 2.9 (mean±sd) for 1-s stimulus recordings (4 ± 2.5 when excluding channels with no cells) and 2.4 ± 2.8 for 10-ms stimulus recordings (4 ± 0.26 when excluding channels with no cells).
The assignment of light responsiveness (10-ms stimuli) or light response types (1-s stimuli) was determined arbitrarily by comparing the spiking patterns following light stimulus to the basal frequency. Cells with basal spiking frequency lower than 1 Hz were considered inactive and excluded from the following analysis. In contrast, active cells with basal spiking frequency higher than 3 Hz were determined as “spontaneous firing.” In the 10-s recordings, cells were simply defined as light responsive if showing two fold or more spiking than resting state within 500 ms after the 10-ms stimuli. The response types to 1-s stimulus for each cell in each condition were first described as non-responsive, transient ON, sustained ON, delayed ON, ON suppression, OFF, ON-OFF, and hypersensitive separately (see also (Iraha et al., 2018)), by comparing the onset and termination timing of their spiking frequency changes to the stimulation. Cells were then grouped as “!light” (unresponsive), “on,” “adapted on,” or “onXoff” types based on their response patterns across all three conditions (before, during, and after L-AP4). In principle, “on” cells showed L-AP4-sensitive ON (including sustained ON) responses throughout the whole recording, while “adapted on” cells had their ON responses (mostly sustained ON) seen only after recovery from L-AP4. Although cells with OFF responses were also included, this rarely found “onXoff” type mostly consists of cells showing ON and OFF responses that were both sensitive to L-AP4 blockade, suggesting an ON-dependent OFF pathway involvement in the synaptic inputs to these cells. Note that delayed ON and hypersensitive responses were seldom observed and proved independent to the synaptic inputs and therefore assigned to the “!light” unresponsive type. For both recordings with 10-ms and 1-s stimuli, responses that were not consistent across the three replicate recordings were disregarded. For the spontaneous firing analyses, we averaged the spiking activity before light stimulation (9-s duration from the 20-s recording) of the three replicate recordings.
Quantification of two-photon imaging
Calcium imaging images were processed in Fiji (ImageJ) using custom scripts by Z-projecting time series images and extracting regions of interest (ROIs) using the analyze particles option. The fluorescence intensity of each ROI along the time axis was then exported to be processed in R. We defined a light response index (LRI) as follows, to compare fluorescence intensity changes before and after light stimulation.
and represent the fluorescence changes before and after light stimulation, respectively, which are calculated relative to the baseline fluorescence.
Finally, the light response of each ROI was weighted by the total area of detected ROIs and aggregated to obtain the light response for each recording.
Statistical analysis
We used full Bayesian statistical inference with Markov Chain Monte Carlo sampling for statistical modeling using Rstan (Stan Development Team. 2017. RStan: the R interface to Stan. R package version 2.16.2. http://mc-stan.org). We estimated population effects, such as the effect of different graft genotype (wt, Bhlhb4-/-, Islet1-/-), group effects such as the effect of different treatments or conditions, intermouse variation, and others, using linear regression models. In this framework, an observation from a particular set of treatments or conditions, coming from a certain mouse in a certain group, is simultaneously used to estimate the effect of that group, mouse, and population. Our estimates, therefore, do not represent a simple pooling of data for a particular set of predictor combinations. Instead, linear regression analyses can isolate the effect of predictors while considering the data as a whole.
During the course of the study, it became apparent that some of the outcomes could potentially be influenced by the sex of the host animal, and so whenever possible, we included the host sex as a predictor; this however was not feasible in all of the analyses because of logistics, and in such cases, the analysis was conducted on males only discarding the data for the females.
Along with the estimation of main effects, the overall effect of predictors, we estimated interactions, the conditional effects that manifest under a particular combination of predictors in addition to main effects. We mainly estimated interactions of genotype with other predictors (e.g., interaction of genotype and sex, denoted genotype x sex), in order to characterize the genotype effect in more detail. Whenever an interaction term was considered, we present the main effects and interaction effects together, as interaction terms must be considered alongside their main effects.
Posterior distribution of parameters of interest, which show the probability for the value of the parameter given the data, is shown with 95% compatibility intervals (CIs) and mean indicated on top. These marginal distributions of parameters do not necessarily indicate the relationship between parameter values as parameters maybe correlated. In order to judge if there is a difference between parameters, we must look at the joint distribution of parameters, i.e., the distribution of the difference between parameter values. Whenever a difference is indicated, we therefore indicate the fraction of the area over zero, which indicates the confidence that we have that there is a difference.
Many of the models use a link function (logistic, exp, etc.), and so the interpretation of the results is not intuitive. In order to facilitate interpretation, distribution of the posterior predictions is shown in the supplemental data with compatibility intervals translated to the original data scale. Stan scripts for the models are provided in a GitHub repository (https://github.com/matsutakehoyo/KO-graft).
Analysis of Nrl-GFP fluorescence of retinal organoids
Image analysis was conducted using EBImage (Pau et al., 2010), and statistical modeling was implemented using Stan (Carpenter et al., 2017) in R (Team, n.d.). Acquired fluorescence images were recorded as RGB images. Pixels containing the GFP signal were identified from background by thresholding using Otsu’s method, and the mean signal and background signal of the green channel were calculated for each image. Signal and background were also calculated for the blue channel to account for differing recording conditions by normalizing the fluorescence intensity on the blue channel. The relative fluorescence intensity is thus expressed as
The relative fluorescence intensity was fitted with a modified Gompertz growth curve (Zwietering et al., 1990) with the Gamma distribution as likelihood, as relative fluorescence is a continuous value that is always positive. The modified Gompertz growth curve is defined by three parameters representing maximum fluorescence intensity , maximum rate of fluorescence increase , and onset of fluorescence increase (see Figure S1). Thus, the fluorescence intensity for a (wt, Bhlhb4-/- or Islet1-/-) on time is
where and represent the shape and rate parameters of the Gamma distribution, respectively, and represents the average fluorescence intensity. The data are very noisy, and estimates resulted in either an extremely short or long delay onset (with extremely fast or slow increase rate), with the data mostly being accounted for by the noise component (the spread of the Gamma distribution) without some kind of informative prior on the parameters.
We therefore used the following informative priors:
And a uniform prior for ().
These priors reflect our beliefs, based on our previous experience, that the fluorescence intensity is well contained between 7 and 16 once organoids have differentiated. Moreover, cell aggregation starts at DD7 and fluorescence is observed a few days after that at the earliest. Similarly, the fluorescence increase rate is based on our experience that fluorescence increases gradually over the course of about 20 days.
Graft cell number analysis
The number of photoreceptor cells in the graft and the number of cells positive for PKCα, Chx10, Calbindin, and Calretinin, within the inner cells of the graft, were modeled with the Binomial distribution, as these represent discrete count data with a binary outcome. Thus, the number of positive cells for a particular marker in a given sample is given by
where represents the total number of cells present in the sample and represents the probability of cells being positive for the marker. We implemented robustness in our model using a parameter (Kruschke, 2014). In this model, is given by two parts, a purely random part () and the logistic regression of the predictors . A large value indicates the data contain many points not conforming to the regression. For the logistic regression, we considered three predictors: the genotype of the cell (either wt, Bhlhb4-/-, or Islet1-/-) , the sex of the host , and the mouse from which the sample was derived. was omitted from the analysis of photoreceptor cell number and SCGN-positive cells (cone bipolar) as samples consisted of single sex. represents the overall mean, and the effect of a particular predictor is modeled as a deviation from the overall mean with a sum-to-zero constraints for each predictor (). For the effect of mouse (), we estimated hyperparameters from the data; otherwise, we used generic weakly informative priors ().
For the guess parameter, we followed the recommendation from Kruschke (Kruschke, 2014) for robust logistic regression (Chapter 21.3). Using which emphasizes small values, reflecting our expectation that the proportion of outliers is small. In particular, gives values greater than 0.5, very small but nonzero probability.
Number of synapses
Bipolar cells typically make multiple synapses to photoreceptor cells. We therefore counted the number of synapses per bipolar cell. The collected data contained a large zero count peak, indicating that there was a large population of bipolar cells that do not make connections at all and a second population that makes multiple synapses as shown in Figure 3J. The data were therefore analyzed with a zero-inflated Poisson model, assuming that the probability of forming a synapse (with probability ) is separate from the distribution of synapses (with mean ). Therefore, whether a synapse is formed at all is modeled as a logit model (either success or failure), and the number of synapses per L7-GFP bipolar cell is modeled as a Poisson count model with the zeros being generated by both processes. A host bipolar cell (L7-GFP) therefore has a probability to form a chemical synapse with graft photoreceptor cells, and the number of connections it makes is described by . The probability function of synapse numbers is given by
where represents the probability of nonzero (probability of synapse) of the -th data and represents the average number of synapses, with the parameter similarly implemented as in the cell type analysis. Predictors include (wt, Bhlhb4-/-, or Islet1-/- ), , and (mouse), as well as the interaction between and (). The priors for the overall mean were estimated from the data (in log odd for and log for )
where is the fraction of data that is higher than zero.
where is the mean number of synapses. For the effect of mouse, we estimated hyperparameters from the data; otherwise, we used generic weakly informative priors () for predictors.
So for ,
and similarly, for ,
We examine the impact of these priors on posteriors by replacing the above priors with more diffuse priors. The model was able to converge, and the posteriors were practically identical. For the guess parameter, we followed the recommendation from Kruschke (Kruschke, 2014) for robust logistic regression (Chapter 21.3). Using which emphasizes small values, reflecting our expectation that the proportion of outliers is small. In particular, gives values greater than 0.5, very small but nonzero probability.
Two-photon imaging
The LRI was analyzed with a Gamma hurdle model, a mixture of response probability and response amplitude. Whether a response is detected or not (>0) is modeled as a Bernoulli process, and the amplitude of the response is modeled with the Gamma distribution. The probability function of response amplitude is given by
where represents the probability of nonzero (probability of response) and represents the average response amplitude. Predictors include (wt, Bhlhb4-/-, Islet1-/-, off, normal) and . Furthermore, and account for the varied sampled times, with normalized and standardized so that its mean=0 and sd=1. The priors for the overall mean were estimated from the data as follows.
For the rest of the regression parameters, we used weakly informative priors ( for the Gamma process and for the Bernoulli process (logistic regression)),
and a uniform prior for the shape parameter of the gamma distribution (). We examine the impact of these priors on posteriors by replacing the above and priors with more diffuse and priors. The model with more diffuse priors was able to converge, and the posteriors were practically identical.
b-wave analysis (10-ms stimulus)
Field potential recordings from individual channels in the MEA probe were processed individually. First, positive peaks within 120 ms of the light pulse were selected as candidate b-wave peaks using a custom program in R. We then calculate the standard deviation of the baseline (2 s before the light stimulation) for each trace, and channels with a peak amplitude higher than three times the baseline standard deviation were considered to have detected a b-wave. We obtained 12736 recordings from 38 animals transplanted with retinal organoids (excluding the channels assigned to be covered by “damage” retinal areas). We then modeled the probability of observing a b-wave in each channel under different conditions using robust logistic regression. The probability of observing a b-wave is then given by
where is either 0 (no b-wave) or 1 (b-wave) and is the probability of observing a b-wave in that channel. represents the overall mean, and the effect of different predictors was calculated as a deviation from this mean with the sum-to-zero constraint on each of the predictors (). Table1 shows the meaning and value of each of the predictors.
Table 1.
Predictors for analysis of b-wave
| Predictor | Meaning | Value |
|---|---|---|
| Line genotype | wt, Bhlhb4−/−, Islet1−/− | |
| Stimulus strength | Weak, strong | |
| Time after transplantation | 8 weeks, 12 weeks | |
| Host genotype | rd1, rd1;L7-GFP | |
| Condition | Before L-AP4, L-AP4, after L-AP4 | |
| Graft topology | On graft, edge of graft, off graft | |
| Tissue damage | No damage, damage | |
| Mouse | Mouse id 1 … 38 | |
| Interaction of line and time | (wt, Bhlhb4−/−, Islet1−/−) x (8 weeks, 12 weeks) | |
| Interaction of line and topology | (wt, Bhlhb4−/−, Islet1−/−) x (on, edge, off) | |
| Interaction of line and stimulus | (wt, Bhlhb4−/−, Islet1−/−) x (weak, strong) | |
| Interaction of line and host | (wt, Bhlhb4−/−, Islet1−/−) x (not L7, L7) | |
| Interaction of line and condition | (wt, Bhlhb4−/−, Islet1−/−) x (before, L-AP4, after) |
The prior for the overall mean () was estimated from the data (in log odds, probability of response: ) with sd of 1.
For the effect of mouse, we estimated the hyperparameter () from the data, using a Gamma distribution with mode 2 and sd 4 () following the recommendation of Kruschke (Kruschke, 2014).
For the rest of the parameters (, , , , , , , , , , , ), we used generic weakly informative priors for logistic regression . We checked the impact of these priors on model convergence and posterior distributions by running the same model with more diffuse priors ( instead of and instead of ). The model with more diffuse priors resulted in very wide posterior tails for some of the parameters (, ,, and ), while the other parameters converged to similar values.
For the guess parameter, we followed the recommendation from Kruschke (Kruschke, 2014) for robust logistic regression (Chapter 21.3). Using which emphasizes small values, reflecting our expectation that the proportion of outliers is small. In particular, gives values greater than 0.5 very small but nonzero probability.
RGC response (10-ms stimuli)
The spikes from RGC upon the 10-ms stimuli allow us to examine the correlation of b-wave and RGC response. Following spike sorting and response assignment, we modeled the probability of each spike train to be light responsive, i.e., the probability of each RGC to respond to light.
We obtained 30167 observations, from 6827 cells, collected from 38 animals transplanted with retinal organoids. The number of observations roughly corresponds to the number of cells multiplied by six, for the two light stimulus conditions (weak and strong) and the three L-AP4 treatment conditions (before, L-AP4, and after). As the outcome is dichotomous (either light responsive or not), we used a similar Bernoulli model as in the b-wave analysis with some modifications (robust logistic regression). In addition to the predictors in the b-wave model, we used the spontaneous frequency rate of each cell and the b-wave amplitude recorded on that channel as covariates. These covariates were normalized and standardized so that their mean=0 and sd=1. The probability of an RGC to respond to light is given by
where is either 0 (unresponsive) or 1 (light responsive) for a particular cell and is the probability of observing a light response in that cell. is a coefficient for the effect of the frequency of the spontaneous activity, and is the spontaneous spiking frequency (normalized and standardized) of the cell . Similarly, is a coefficient for the effect of the b-wave amplitude on that channel and is the amplitude of the recorded b-wave (normalized and standardized).
The prior for the overall mean () was estimated from the data (in log odds, probability of response: ) with sd of 1.
For the effect of mouse, we estimated the hyperparameter () from the data, using a Gamma distribution with mode 2 and sd 4 () following the recommendation of Kruschke (Kruschke, 2014).
For the rest of the regression coefficients (, , , , , , , , , , , , , ), we used generic weakly informative priors for logistic regression . We checked the impact of these priors on model convergence and posterior distributions by running the same model with more diffuse priors ( instead of and instead of ). The model with more diffuse priors resulted in very wide posterior tails for some of the parameters (, , ,, and ), while the other parameters converged to similar values.
RGC response (1-s stimuli)
The 1-s stimuli allow us to analyze the light response in more detail taking into account different response subtypes and the response patterns under different conditions. For these analyses, we used responses from 16353 observations, from 5738 cells, collected across 38 animals transplanted with retinal organoids. The number of observations is roughly three times of the cell numbers for there are three different stimulation conditions.
We categorized response patterns and modeled recorded data with a conditional logit model, assuming that response categories are hierarchical. For the light response, we assumed that cells are either responsive or not responsive to light (!light). Of the responsive cells, i.e., excluding unresponsive cells, we assumed that the response either intersects the off pathway (“onXoff” type) or not. Finally, responses that are independent of the OFF pathway are divided into “on” type or “adapted on” depending on whether they show some sort of potentiation (light response appearing only after several flashes or L-AP4 treatment) or not. A diagram of the category hierarchy is provided in Figure S10. The light response can therefore be described as follows:
where represents the probability of each response category, adding up to 1 (). Note that corresponds to the response probability excluding previous categories and therefore does not add up to 1. So represents the probability of cells not to respond to light, represents the probability of light responding cells to be , and corresponds to the probability of non- light responsive cells to be . represents the overall mean probability of response category, and the effect of different predictors was calculated as a deviation from this mean with the sum-to-zero constraint on each of the predictors (). Table2 shows the meaning and value of each of the predictors.
Table 2.
Predictors for conditional categorical model of RGC light response to 1s stimuli
| Predictor | Meaning | Value |
|---|---|---|
| Line genotype | wt, Bhlhb4−/−, Islet1−/− | |
| Stimulus strength | Weak, middle, strong | |
| Time after transplantation | 8 weeks, 12 weeks | |
| Host genotype | rd1, rd1;L7-GFP | |
| Graft topology | On graft, edge of graft, off graft | |
| Tissue damage | No damage, damage | |
| Mouse | Mouse id 1 … 38 | |
| Interaction of line and time | (wt, Bhlhb4−/−, Islet1−/−) x (8 weeks, 12 weeks) | |
| Interaction of line and topology | (wt, Bhlhb4−/−, Islet1−/−) x (on, edge, off) | |
| Interaction of line and stimulus | (wt, Bhlhb4−/−, Islet1−/−) x (weak, middle, strong) | |
| Interaction of line and host | (wt, Bhlhb4−/−, Islet1−/−) x (not L7, L7) |
The prior for the overall mean () was estimated from the data (in log odds, probability of response for category : ) with sd of 1.
For the effect of mouse, we estimated the hyperparameter () from the data, using a Gamma distribution with mode 2 and sd 4 () following the recommendation of Kruschke (Kruschke, 2014).
For the rest of the regression coefficients (, , , , , , , , , ), we used generic weakly informative priors for logistic regression . We checked the impact of these priors on model convergence and posterior distributions by running the same model with more diffuse priors ( instead of ). The model with more diffuse priors resulted in very wide posterior tails specially for parameters with interactions.
RGC spontaneous firing (9-s recording before the 1-s stimuli)
We noticed that there was an inverse correlation between light responsiveness and the spontaneous firing frequency. We therefore analyzed the distribution of spontaneous firing, by calculating the spontaneous firing rate before light stimulation (9 s). The distribution of spontaneous firing rate closely follows a lognormal distribution, as shown in Figures 6 and S11. We therefore modeled the influence of parameters on the mean log spontaneous firing. For the spontaneous firing analysis, we used 44213 observations from 5738 cells collected across 38 animals transplanted with retinal organoids. The number of observations in this analysis is higher than the number in the light response analysis, as we consider the spontaneous activity before, during, and after L-AP4 treatment, in addition to the different stimuli.
where is the log spontaneous firing rate for a particular observation . and are the overall logmean and logsd of the lognormal distribution, respectively. We assume a common to simplify the model. The effect of different predictors was calculated as a deviation from this mean with the sum-to-zero constraint on each of the predictors (). Table 3 shows the meaning and value of each of the predictors.
Table 3.
Predictors for analysis of spontaneous activity
| Predictor | Meaning | Value |
|---|---|---|
| Line genotype | wt, Bhlhb4−/−, Islet1−/− | |
| Light responsiveness | Unresponsive, responsive | |
| Stimulus strength | Weak, strong | |
| Time after transplantation | 8 weeks, 12 weeks | |
| Host genotype | rd1, rd1;L7-GFP | |
| Condition | Before L-AP4, L-AP4, after L-AP4 | |
| Tissue damage | No damage, damage | |
| Mouse | Mouse id 1 … 38 | |
| Interaction of line and time | (wt, Bhlhb4−/−, Islet1−/−) x (8 weeks, 12 weeks) | |
| Interaction of line and host | (wt, Bhlhb4−/−, Islet1−/−) x (not L7, L7) | |
| Interaction of line and light responsiveness | (wt, Bhlhb4−/−, Islet1−/−) x (unresponsive, responsive) |
The prior for the overall mean () was estimated from the data with sd of 0.5.
For the effect of mouse, we estimated the hyperparameter () from the data, using a Gamma distribution with mode 2 and sd 4 () following the recommendation of Kruschke (Kruschke, 2014).
For the rest of the regression coefficients (,, , , , , , , , ), we used generic weakly informative priors .
We checked the impact of these priors on model convergence and posterior distributions by running the same model with more diffuse priors ( instead of ). The model with more diffuse priors was able to converge and resulted in very similar posteriors. While most of the parameters were practically identical, there was a small difference in the interaction term of graft genotype and time after transplantation () which resulted in slightly different posteriors. On both models, the posteriors for this parameter can be considered to mostly overlap but the order of the peaks was slightly different (wt>Bhlhb-/->Istlet1-/- for 8W and wt<Bhlhb-/-<Istlet1-/-for 12W with diffuse priors).
Behavior: Light avoidance
The number of successful avoidances out of 30 trials was modeled with the Binomial likelihood.
where represents the number of successful avoidances out of 30 trials and is the probability of success. is the effect of different groups (wt, Bhlhb4-/-, Islet1-/-, and age matched rd control without treatment), is the sex of the animal, and is the interaction term of sex and group. represents the overall mean (at mean and ), and the effect of a particular predictor is modeled as a deviation from this overall mean with a sum-to-zero constraints for each predictor (). (trial) is a covariate to account for the accumulated experience, as more experienced mice would be expected to perform better. is a proxy for the locomotion during the experiment, which greatly influences the success rate as mice randomly moving may avoid the electric shock merely by chance. Both and were normalized and standardized so that their mean=0 and sd=1. We assume a quadratic relationship for rather than linear; however, the linear model produced similar results. We believe the quadratic model better describes the data as the random moves of the mouse payoff better at lower but the benefit of moving faster decreases at high as the animal tires. The prior for the overall mean () was estimated from the data with sd=1.
For the effect of mouse, we estimated the hyperparameter () from the data, using a Gamma distribution with mode 2 and sd 4 () following the recommendation of Kruschke (Kruschke, 2014).
For the rest of the regression coefficients (, , , , ), we used generic weakly informative priors (. We checked the impact of these priors on model convergence and posterior distributions by running the same model with more diffuse priors ( instead of ). The model with more diffuse priors was able to converge and resulted in practically identical posteriors.
Acknowledgments
We thank Dr. Genshiro A Sunagawa and Prof. Chuan-Chin Chiao for critical reading and comments on the manuscript. This work was supported by JSPS KAKENHI Grant Number 15K10913, JK19K09942, and 17K16994.
Author contributions
MM and MT conceived the study. HYT and TM performed electrophysiological experiments. TH and JSun prepared 3D retinas. MM and JSun designed, constructed cell lines, and transplanted 3D retinas. AO procured the Thy1-GCaMP3 mouse line. JSho maintained animals and assisted on transplantation. TH and MF performed RT-PCR. TM conducted statistical analysis. TH, RA, MM, JSun, and HYT performed immunostaining, imaging, and quantification. TM, MM, and HYT wrote the manuscript
Declaration of interests
There is potential competing interest. The corresponding author is currently filing for a patent regarding the genetically modified retinal organoids.
Information of Patent applicant: RIKEN, SUMITOMO DAINIPPON PHARMA CO., LTD name of inventors: Michiko MANDAI, Masayo TAKAHASHI, Suguru YAMASAKI application number: PCT/JP2017/042238 status of application: national phase specific aspect of manuscript covered in patent application: The phenotype of retinal grafts with reduced inner cell population and the probability of improved results following retinal transplantation are covered by the above patent.
Published: August 20, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102866.
Supplemental information
Data and code availability
-
•
The microarray data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE178653 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE178653).
-
•
The code used for statistical analyses is available in a GitHub repository (https://github.com/matsutakehoyo/KO-graft) . The datasets supporting the current study have not been deposited in a public repository because of the large size of the data but are available from the corresponding author on request.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Aghaizu N.D., Kruczek K., Gonzalez-Cordero A., Ali R.R., Pearson R.A. Pluripotent stem cells and their utility in treating photoreceptor degenerations. Prog. Brain Res. 2017;231:191–223. doi: 10.1016/bs.pbr.2017.01.001. [DOI] [PubMed] [Google Scholar]
- Akiba R., Matsuyama T., Tu H.-Y., Hashiguchi T., Sho J., Yamamoto S., Takahashi M., Mandai M. Quantitative and qualitative evaluation of photoreceptor synapses in developing, degenerating and regenerating retinas. Front Cell Neurosci. 2019;13:16. doi: 10.3389/fncel.2019.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akimoto M., Cheng H., Zhu D., Brzezinski J.A., Khanna R., Filippova E., Oh E.C.T., Jing Y., Linares J.-L., Brooks M. Targeting of GFP to newborn rods by Nrl promoter and temporal expression profiling of flow-sorted photoreceptors. PNAS. 2006;103:3890–3895. doi: 10.1073/pnas.0508214103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assawachananont J., Mandai M., Okamoto S., Yamada C., Eiraku M., Yonemura S., Sasai Y., Takahashi M. Transplantation of embryonic and induced pluripotent stem cell-derived 3D retinal sheets into retinal degenerative mice. Stem Cell Rep. 2014;2:662–674. doi: 10.1016/j.stemcr.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnea-Cramer A.O., Wang W., Lu S.-J., Singh M.S., Luo C., Huo H., McClements M.E., Barnard A.R., MacLaren R.E., Lanza R. Function of human pluripotent stem cell-derived photoreceptor progenitors in blind mice. Sci. Rep. 2016;6:29784. doi: 10.1038/srep29784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch U., Oriyakhel W., Kenna P.F., Linke S., Richard G., Petrowitz B., Humphries P., Farrar G.J., Ader M. Retinal cells integrate into the outer nuclear layer and differentiate into mature photoreceptors after subretinal transplantation into adult mice. Exp. Eye Res. 2008;86:691–700. doi: 10.1016/j.exer.2008.01.018. [DOI] [PubMed] [Google Scholar]
- Behrens C., Schubert T., Haverkamp S., Euler T., Berens P. Connectivity map of bipolar cells and photoreceptors in the mouse retina. Elife. 2016;5:e20041. doi: 10.7554/eLife.20041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S., Haselier C., Mataruga A., Thumann G., Walter P., Müller F. Pharmacological analysis of intrinsic neuronal oscillations in rd10 retina. PLoS ONE. 2014;9:e99075. doi: 10.1371/journal.pone.0099075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramblett D.E., Copeland N.G., Jenkins N.A., Tsai M.-J. BHLHB4 is a bHLH transcriptional regulator in pancreas and brain that marks the dimesencephalic boundary. Genomics. 2002;79:402–412. doi: 10.1006/geno.2002.6708. [DOI] [PubMed] [Google Scholar]
- Bramblett D.E., Pennesi M.E., Wu S.M., Tsai M.-J. The transcription factor Bhlhb4 is required for rod bipolar cell maturation. Neuron. 2004;43:779–793. doi: 10.1016/j.neuron.2004.08.032. [DOI] [PubMed] [Google Scholar]
- Carpenter B., Gelman A., Hoffman M.D., Lee D., Goodrich B., Betancourt M., Brubaker M., Guo J., Li P., Riddell A. Stan : a probabilistic programming language. J. Stat. Softw. 2017;76 doi: 10.18637/jss.v076.i01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Cichon J., Wang W., Qiu L., Lee S.-J.R., Campbell N.R., Destefino N., Goard M.J., Fu Z., Yasuda R. Imaging neural activity using Thy1-GCaMP transgenic mice. Neuron. 2012;76:297–308. doi: 10.1016/j.neuron.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana H., Chen T.-W., Hu A., Shields B.C., Guo C., Looger L.L., Kim D.S., Svoboda K. Thy1-GCaMP6 transgenic mice for neuronal population imaging in vivo. PLoS ONE. 2014;9:e108697. doi: 10.1371/journal.pone.0108697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshatory Y., Elshatory Y., Everhart D., Everhart D., Deng M., Deng M., Xie X., Xie X., Barlow R.B., Barlow R.B. Islet-1 controls the differentiation of retinal bipolar and cholinergic amacrine cells. J. Neurosci. 2007;27:12707–12720. doi: 10.1523/JNEUROSCI.3951-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foik A.T., Lean G.A., Scholl L.R., McLelland B.T., Mathur A., Aramant R.B., Seiler M.J., Lyon D.C. Detailed visual cortical responses generated by retinal sheet transplants in rats with severe retinal degeneration. J. Neurosci. 2018;38:10709–10724. doi: 10.1523/JNEUROSCI.1279-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii M., Sunagawa G.A., Kondo M., Takahashi M., Mandai M. Evaluation of micro electroretinograms recorded with multiple electrode array to assess focal retinal function. Sci. Rep. 2016;6:30719. doi: 10.1038/srep30719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma K., Okamoto S., Mandai M., Gotoh N., Rajasimha H.K., Chang Y.-S., Chen S., Li W., Cogliati T., Swaroop A., Takahashi M. Developing rods transplanted into the degenerating retina of Crx-knockout mice exhibit neural activity similar to native photoreceptors. Stem Cells. 2013;31:1149–1159. doi: 10.1002/stem.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iraha S., Tu H.-Y., Yamasaki S., Kagawa T., Goto M., Takahashi R., Watanabe T., Sugita S., Yonemura S., Sunagawa G.A. Establishment of Immunodeficient retinal degeneration model mice and functional maturation of human ESC-derived retinal sheets after transplantation. Stem Cell Rep. 2018;10:1059–1074. doi: 10.1016/j.stemcr.2018.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B.W., Marc R.E. Retinal remodeling during retinal degeneration. Exp. Eye Res. 2005;81:123–137. doi: 10.1016/j.exer.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Kruschke J. Doing Bayesian data analysis: a tutorial with R, JAGS, and Stan. Phys. Rev. E. 2014 doi: 10.1103/PhysRevE.70.046127. [DOI] [Google Scholar]
- MacLaren R.E., Pearson R.A., MacNeil A., Douglas R.H., Salt T.E., Akimoto M., Swaroop A., Sowden J.C., Ali R.R. Retinal repair by transplantation of photoreceptor precursors. Nature. 2006;444:203–207. doi: 10.1038/nature05161. [DOI] [PubMed] [Google Scholar]
- Mandai M., Fujii M., Hashiguchi T., Sunagawa G.A., Ito S.-I., Sun J., Kaneko J., Sho J., Yamada C., Takahashi M. iPSC-derived retina transplants improve vision in rd1 end-stage retinal-degeneration mice. Stem Cell Rep. 2017;8:69–83. doi: 10.1016/j.stemcr.2016.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marc R.E., Jones B.W., Anderson J.R., Kinard K., Marshak D.W., Wilson J.H., Wensel T., Lucas R.J. Neural reprogramming in retinal degeneration. Invest Ophthalmol. Vis. Sci. 2007;48:3364–3371. doi: 10.1167/iovs.07-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLelland B.T., Lin B., Mathur A., Aramant R.B., Thomas B.B., Nistor G., Keirstead H.S., Seiler M.J. Transplanted hESC-derived retina organoid sheets differentiate, integrate, and improve visual function in retinal degenerate rats. Invest Ophthalmol. Vis. Sci. 2018;59:2586–2603. doi: 10.1167/iovs.17-23646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L., Deng M., Xie X., Gan L. ISL1 and BRN3B co-regulate the differentiation of murine retinal ganglion cells. Development. 2008;135:1981–1990. doi: 10.1242/dev.010751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pau G., Fuchs F., Sklyar O., Boutros M., Huber W. EBImage--an R package for image processing with applications to cellular phenotypes. Bioinformatics. 2010;26:979–981. doi: 10.1093/bioinformatics/btq046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson R.A., Barber A.C., Rizzi M., Hippert C., Xue T., West E.L., Duran Y., Smith A.J., Chuang J.Z., Azam S.A. Restoration of vision after transplantation of photoreceptors. Nature. 2012;485:99–103. doi: 10.1038/nature10997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson R.A., Gonzalez-Cordero A., West E.L., Ribeiro J.R., Aghaizu N., Goh D., Sampson R.D., Georgiadis A., Waldron P.V., Duran Y. Donor and host photoreceptors engage in material transfer following transplantation of post-mitotic photoreceptor precursors. Nat. Commun. 2016;7:13029. doi: 10.1038/ncomms13029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2020. R: A language and environment for statistical computing.https://www.R-project.org/ [Google Scholar]
- Radtke N.D., Aramant R.B., Petry H.M., Green P.T., Pidwell D.J., Seiler M.J. Vision improvement in retinal degeneration patients by implantation of retina together with retinal pigment epithelium. Am. J. Ophthalmol. 2008;146:172–182. doi: 10.1016/j.ajo.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Ferreira T., Llonch S., Borsch O., Postel K., Haas J., Ader M. Retinal transplantation of photoreceptors results in donor-host cytoplasmic exchange. Nat. Commun. 2016;7:13028. doi: 10.1038/ncomms13028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Ferreira T.F., Borsch O., Ader M. Rebuilding the missing part-A review on photoreceptor transplantation. Front Syst. Neurosci. 2016;10:105. doi: 10.3389/fnsys.2016.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Cardona A. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirai H., Mandai M., Matsushita K., Kuwahara A., Yonemura S., Nakano T., Assawachananont J., Kimura T., Saito K., Terasaki H. Transplantation of human embryonic stem cell-derived retinal tissue in two primate models of retinal degeneration. Proc. Natl. Acad. Sci. USA. 2016;113:E81–E90. doi: 10.1073/pnas.1512590113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M.S., Balmer J., Barnard A.R., Aslam S.A., Moralli D., Green C.M., Barnea-Cramer A., Duncan I., MacLaren R.E. Transplanted photoreceptor precursors transfer proteins to host photoreceptors by a mechanism of cytoplasmic fusion. Nat. Commun. 2016;7:13537. doi: 10.1038/ncomms13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M.S., Issa P.C., Butler R., Martin C., Lipinski D.M., Sekaran S., Barnard A.R., MacLaren R.E. Reversal of end-stage retinal degeneration and restoration of visual function by photoreceptor transplantation. PNAS. 2013;110:1101–1106. doi: 10.1073/pnas.1119416110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomomura M., Rice D.S., Morgan J.I., Yuzaki M. Purification of Purkinje cells by fluorescence-activated cell sorting from transgenic mice that express green fluorescent protein. Eur. J. Neurosci. 2001;14:57–63. doi: 10.1046/j.0953-816x.2001.01624.x. [DOI] [PubMed] [Google Scholar]
- Trenholm S., Awatramani G.B. Origins of spontaneous activity in the degenerating retina. Front Cell Neurosci. 2015;9:e99075. doi: 10.3389/fncel.2015.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto Y., Omi N. Functional allocation of synaptic contacts in microcircuits from rods via rod bipolar to AII amacrine cells in the mouse retina. J. Comp. Neurol. 2013;521:3541–3555. doi: 10.1002/cne.23370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu H.-Y., Chen Y.-J., McQuiston A.R., Chiao C.-C., Chen C.-K. A novel retinal oscillation mechanism in an autosomal dominant photoreceptor degeneration mouse model. Front Cell Neurosci. 2016;9:2615. doi: 10.3389/fncel.2015.00513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu H.-Y., Matsuyama T. Retinal Development, Methods and Protocols. Humana; 2020. Multielectrode array recording of mouse retinas transplanted with stem cell-derived retinal sheets; pp. 207–220. [DOI] [PubMed] [Google Scholar]
- Tu H.-Y., Watanabe T., Shirai H., Yamasaki S., Kinoshita M., Matsushita K., Hashiguchi T., Onoe H., Matsuyama T., Kuwahara A. Medium- to long-term survival and functional examination of human iPSC-derived retinas in rat and primate models of retinal degeneration. EBioMedicine. 2018 doi: 10.1016/j.ebiom.2018.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron P.V., Di Marco F., Kruczek K., Ribeiro J., Graca A.B., Hippert C., Aghaizu N.D., Kalargyrou A.A., Barber A.C., Grimaldi G. Transplanted donor- or stem cell-derived cone photoreceptors can both integrate and undergo material transfer in an environment-dependent manner. Stem Cell Rep. 2018;10:406–421. doi: 10.1016/j.stemcr.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwietering M.H., Jongenburger I., Rombouts F.M., van 't Riet K. Modeling of the bacterial growth curve. Appl. Environ. Microbiol. 1990;56:1875–1881. doi: 10.1128/aem.56.6.1875-1881.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A host rod bipolar cell, extending its axon toward the IPL is show (cyan) with several ribbon synapses (yellow), exhibiting a typical candelabra-like morphology of rod bipolar cells. Photoreceptor cells can be distinguished by their more concentrated and brighter signal (magenta). Note that there is a small gap between graft and host cells, however the host bipolar dendrites extend through the graft inner cells and make direct contact with graft photoreceptor cells.
Data Availability Statement
-
•
The microarray data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE178653 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE178653).
-
•
The code used for statistical analyses is available in a GitHub repository (https://github.com/matsutakehoyo/KO-graft) . The datasets supporting the current study have not been deposited in a public repository because of the large size of the data but are available from the corresponding author on request.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.