Abstract
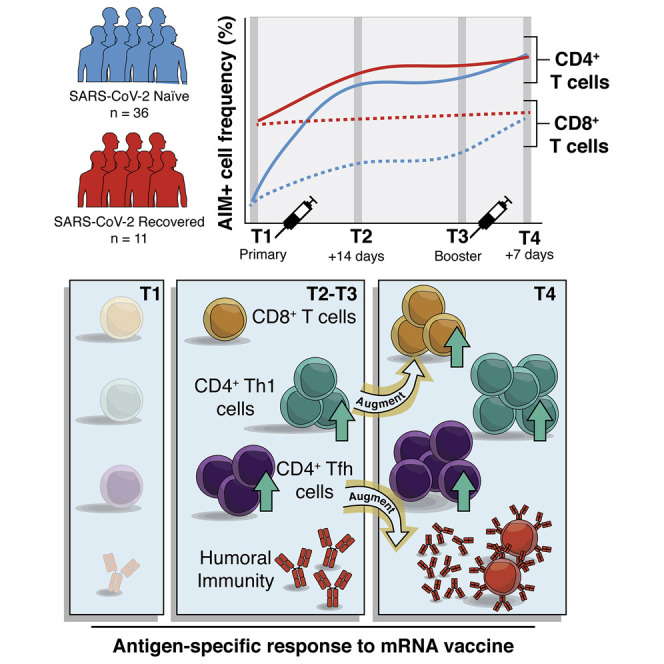
SARS-CoV-2 mRNA vaccines have shown remarkable clinical efficacy, but questions remain about the nature and kinetics of T cell priming. We performed longitudinal antigen-specific T cell analyses on healthy SARS-CoV-2-naive and recovered individuals prior to and following mRNA prime and boost vaccination. Vaccination induced rapid antigen-specific CD4+ T cell responses in naive subjects after the first dose, whereas CD8+ T cell responses developed gradually and were variable in magnitude. Vaccine-induced Th1 and Tfh cell responses following the first dose correlated with post-boost CD8+ T cells and neutralizing antibodies, respectively. Integrated analysis revealed coordinated immune responses with distinct trajectories in SARS-CoV-2-naive and recovered individuals. Last, whereas booster vaccination improved T cell responses in SARS-CoV-2-naive subjects, the second dose had little effect in SARS-CoV-2-recovered individuals. These findings highlight the role of rapidly primed CD4+ T cells in coordinating responses to the second vaccine dose in SARS-CoV-2-naive individuals.
Keywords: SARS-CoV-2, mRNA vaccine, T cells, activation induced markers, AIM, T follicular helper, Tfh, Th1, immunological memory
Graphical abstract
SARS-CoV-2 mRNA vaccines have demonstrated remarkable efficacy, but T cell responses to vaccination have not been well studied. In a longitudinal cohort, Painter et al. show that mRNA vaccines activate SARS-CoV-2-specific T cells that could contribute to durable immunity. The findings highlight the central role of T cells in the two-dose vaccine regimen for individuals not previously infected with SARS-CoV-2.
Introduction
The coronavirus disease 2019 (COVID-19) pandemic has had a profound global toll on human life and socioeconomic well-being, prompting emergency use authorization of prophylactic mRNA vaccines (Cutler and Summers, 2020). Recent studies have documented strong memory B cell and antibody responses post-vaccination that neutralize SARS-CoV-2, including variants of concern (VOCs) such as B.1.351 (Beta) (Goel et al., 2021; Krammer et al., 2021; Sahin et al., 2020; Widge et al., 2021). B cells and antibodies are important components of immunological memory, and antibody responses are the surrogate of protection for most licensed vaccines. However, patients who failed to develop neutralizing antibodies, in some cases because of inherited or treatment-induced B cell deficiencies, have recovered from COVID-19 (Soresina et al., 2020; Wu et al., 2020; Wurm et al., 2020). Moreover, in patients with hematological malignancy, CD8+ T cells appear to compensate for lack of humoral immunity and were associated with improved outcomes, indicating a role for T cells in protection against SARS-CoV-2 infection (Bange et al., 2021). The T cell response to mRNA vaccination is less well characterized than the humoral response, though initial reports indicate that T cells, particularly CD4+ T cells, are primed by the vaccine (Anderson et al., 2020; Angyal et al., 2021; Camara et al., 2021; Jackson et al., 2020; Kalimuddin et al., 2021; Lederer et al., 2020; Mazzoni et al., 2021; Prendecki et al., 2021; Sahin et al., 2020; Stamatatos et al., 2021; Tarke et al., 2021b; Woldemeskel et al., 2021). However, the details of antigen-specific T cell induction following vaccination remain incompletely understood, and questions remain about the trajectory of the adaptive immune response following vaccination.
T cell immunity is functionally heterogeneous, with subsets of both CD4+ and CD8+ T cells contributing to protective immunity and long-term immunological memory. Specifically, CD4+ T follicular helper (Tfh) cells have key roles in the development of memory B cells, plasma cells, and antibodies, whereas Th1 cells support and enhance the quality of memory CD8+ T cell responses (Crotty, 2011; Krawczyk et al., 2007; Luckheeram et al., 2012; Williams et al., 2006). In addition, the central memory (CM) or effector memory (EM) differentiation states of CD4+ and CD8+ T cells have implications for durability, recirculation, tissue access, and responses upon antigen re-exposure (Kaech et al., 2002; Martin and Badovinac, 2018; Wherry et al., 2003). In the context of mRNA vaccination, relatively little is known about the nature and differentiation state of antigen-specific CD4+ and CD8+ T cells. For example, it is unclear whether Tfh cells are efficiently primed and whether these cells relate to vaccine-induced antibodies or memory B cells. It is also unclear whether the kinetics of T cell priming differs for CD4+ and CD8+ T cells and how such T cell priming events might compare for SARS-CoV-2-naive versus recovered subjects. Overall, the orchestration of different vaccine-induced immune responses remains to be fully understood.
In this study we sought to address these questions and define the kinetics and differentiation state of vaccine-induced CD4+ and CD8+ T cells following mRNA vaccination. All SARS-CoV-2-naive subjects mounted robust CD4+ T cell responses following the first vaccine dose, and the second dose further boosted both CD4+ and CD8+ T cell responses. In contrast, SARS-CoV-2-recovered individuals had maximal CD4+ and CD8+ T cell responses following the first dose of mRNA vaccine, and there was little additional T cell boosting after the second dose. Both CD4+ and CD8+ T cell responses were dominated by CM-like cells, similar to memory T cells generated following natural infection. For CD4+ T cells, both Tfh and Th1 responses were efficiently generated following primary vaccination and strongly correlated with post-boost neutralizing antibody and CD8+ T cell responses, respectively. Finally, integrated analysis of 26 individual measures of antigen-specific T and B cells revealed coordinated immune response patterns and provided a detailed assessment of how antigen-specific adaptive immunity is shaped by mRNA vaccination.
Results
Activation-induced marker (AIM) assay enables detection of SARS-CoV-2-specific T cells
We acquired longitudinal peripheral blood samples from a cohort of 36 SARS-CoV-2-naive and 11 SARS-CoV-2-recovered individuals who received mRNA vaccines through the University of Pennsylvania Health System (Table S1). We obtained peripheral blood mononuclear cells (PBMCs) at four key time points (Figure 1 A): pre-vaccine baseline (time point 1), 2 weeks post-primary vaccination (time point 2), the day of the booster vaccination (time point 3), and 1 week post-boost (time point 4). PBMCs from each of these time points were stimulated with peptide megapools containing SARS-CoV-2 Spike epitopes optimized for presentation by MHC-I (CD8-E) or MHC-II (CD4-S) (Grifoni et al., 2020; Tarke et al., 2021a). We then assessed peptide-dependent AIM expression by flow cytometry compared with unstimulated control samples (Figure 1A; Figure S1A; Betts et al., 2003; Reiss et al., 2017). AIM+ CD4+ T cells were defined by dual expression of CD200 and CD40L. Although dual expression of IFN-γ and 41BB was useful to visualize AIM+ CD8+ T cell populations (Figure 1A), a two-marker strategy alone was sub-optimal for detecting vaccine-elicited responses because of high baseline signals (Figure S1B). These responses at baseline likely represent cross-reactive T cells, possibly primed during a prior seasonal coronavirus infection, that can mask low frequencies of vaccine-induced T cells (Grifoni et al., 2020). As we sought to study vaccine responses, AIM+ CD8+ T cells were defined by expression of at least four of five markers: CD200, CD40L, 41BB, CD107a, and intracellular IFN-γ (Figure S1C). These distinct approaches for defining AIM+ CD4+ and CD8+ T cells provided optimal detection of vaccine-elicited responses relative to background signals in unstimulated controls (Figures S2A–S2C). Alternative combinations of AIMs revealed similar kinetics of antigen-specific T cell responses, though in some cases with higher background (Figures S2D and S2E). We further confirmed that the frequency of AIM+ CD4+ and CD8+ T cells correlated strongly with the frequency of activated Ki67+CD38+ T cells (Figure S1D), another method for quantifying antigen-specific responses, after each vaccine dose (Miller et al., 2008; Ndhlovu et al., 2015).
Figure 1.
mRNA vaccination elicits antigen-specific CD4+ and CD8+ T cell responses
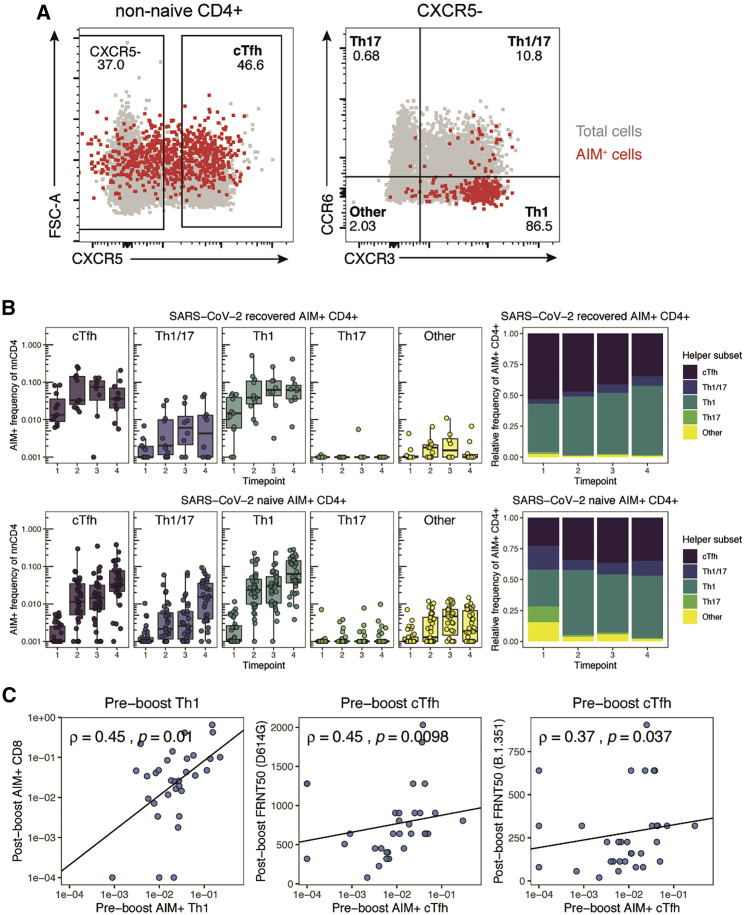
(A) Longitudinal study design and representative flow cytometry plots for identifying AIM+ CD4+ T cells (left) and visualizing AIM+ CD8+ T cells (right). Numbers represent the frequency of total non-naive CD4+ or CD8+ T cells. The CD4-S peptide megapool was used for analysis of CD4+ T cells, while the CD8-E peptide megapool was used for analysis of CD8+ T cells.
(B) Summary plots of AIM+ CD4+ (left) and CD8+ (right) T cells defined as indicated above each plot. AIM+ CD8+ T cells were quantified throughout the study on the basis of expression of at least four of five activation induced markers (CD200, CD40L, 41BB, CD107a, and intracellular IFN-γ), as in Figure S1C. Values represent the frequency of AIM+ non-naive cells after subtracting the frequency from paired unstimulated samples. Solid lines connect individual donors sampled longitudinally. Statistics were calculated using unpaired Wilcoxon test. n.s., not significant. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Blue indicates SARS-CoV-2-naive, red indicates SARS-CoV-2-recovered individuals.
Longitudinal samples from 36 SARS-CoV-2-naive and 11 SARS-CoV-2-recovered individuals were used for each experiment, analyzed in nine independent batches. All paired longitudinal samples were analyzed within a single batch. See also Figures S1 and S2.
Antigen-specific CD4+ T cell responses to the first vaccine dose are boosted by the second dose in SARS-CoV-2-naive individuals
As expected, most SARS-CoV-2-recovered donors had clearly detectable antigen-specific CD4+ and CD8+ T cell populations at baseline (Figure 1B). In contrast, pre-vaccination responses to peptide stimulation were undetectable in many SARS-CoV-2-naive individuals, though some subjects did have low frequencies of pre-vaccination AIM+ T cells that may be attributed to cross-reactive cells from a prior seasonal coronavirus infection (Grifoni et al., 2020; Figure 1B). SARS-CoV-2 Spike-specific CD4+ T cells were robustly primed in SARS-CoV-2-naive and recovered individuals following the first dose of mRNA vaccine, with all participants generating detectable responses to the first dose (Figure 1B). SARS-CoV-2-naive individuals, but not recovered individuals, received an additional boost to antigen-specific CD4+ T cells following the second vaccine dose (Figure 1B). Overall, mRNA vaccination induced a universal CD4+ T cell response, as all individuals, regardless of prior infection with SARS-CoV-2, had greater frequencies of AIM+ CD4+ T cells post-boost than at pre-vaccine baseline (Figure S1E).
Prime-boost vaccination induces antigen-specific CD8+ T cells in most SARS-CoV-2-naive individuals
In contrast to the rapid and universal induction of Spike-specific CD4+ T cells, SARS-CoV-2-specific CD8+ T cell responses developed more gradually and with greater variability in naive individuals. Only 24 of 34 SARS-CoV-2-naive subjects (71%) generated detectable antigen-specific CD8+ T cell responses following the first dose. These CD8+ T cell responses were boosted by the second dose, and though the magnitude of response was variable, 29 of 33 SARS-CoV-2-naive individuals (88%) had post-boost CD8+ T cell responses that were detectable above their individual pre-vaccine baseline (Figure 1B; Figure S1E). Individuals who had previously recovered from SARS-CoV-2 infection experienced no significant increase in the frequency of AIM+ CD8+ T cells from either dose of vaccine (Figure 1B; Figure S1E). A subset of recovered individuals (70%) did appear to have increased AIM+ CD8+ T cell frequencies compared with baseline, but as a group this increase did not reach statistical significance (Figure S1E). In contrast to the modestly weaker induction of antibodies and memory B cells with increasing age observed in this cohort and others (Abu Jabal et al., 2021; Goel et al., 2021; Levi et al., 2021; Prendecki et al., 2021), T cell responses upon mRNA vaccination were not correlated with age (Figure S1F). Taken together, these data demonstrate robust induction of antigen-specific T cell responses following mRNA vaccination, with more consistent induction of CD4+ T cell responses compared with CD8+ T cell responses.
Vaccine-induced T cells have CM characteristics and resemble memory T cells generated by SARS-CoV-2 infection
We next sought to define the differentiation state of vaccine-induced AIM+ T cells. We first examined subsets of central and effector memory populations using CD45RA, CD27, and CCR7 (Hamann et al., 1997; Sallusto et al., 1999). With these markers, we defined CM, effector memory types 1, 2, and 3 (EM1, EM2, and EM3), and terminally differentiated effector memory (EMRA) cells (Figures 2 A, 2C, and S1A; Mathew et al., 2020). Total non-naive CD4+ T cells were predominantly CM (CD45RA− CD27+ CCR7+) in this cohort, and the overall frequencies of these subsets were unchanged by vaccination (Figure S3A). The baseline AIM+ CD4+ T cell response in SARS-CoV-2-recovered individuals, presumably generated during prior SARS-CoV-2 infection, was composed mainly of EM1 (CD45RA− CD27+ CCR7−) and CM cells (Figures 2A and 2B). The memory T cell subset distribution of these SARS-CoV-2-specific CD4+ T cells did not change substantially following vaccination (Figure 2B). In SARS-CoV-2-naive individuals, the first dose of vaccine primarily induced AIM+ CD4+ T cells in the EM1 and CM subsets, similar to the response in recovered donors (Figure 2B). Antigen-specific CD4+ EM2 (CD45RA− CD27− CCR7+) and EM3 (CD45RA− CD27− CCR7−) T cells, which share more effector-like properties (Romero et al., 2007), were also boosted by the vaccine but remained minority populations compared with CM and EM1 (Figure 2B).
Figure 2.
mRNA vaccination induces antigen-specific memory T cells that mirror memory T cell responses from natural infection
(A and C) Representative flow cytometric plots depicting the gating of AIM+ CD4+ (CD200+CD40L+; A) and CD8+ (four of five markers; C) T cells to identify the indicated memory T cell subsets in a SARS-CoV-2-naive donor at time point 4. Red events depict AIM+ cells, gray events depict total CD4+ (A) or CD8+ (C) T cells from the same donor. Numbers indicate the frequency of AIM+ cells falling within each gate.
(B and D) Frequency of memory T cell subsets in AIM+ CD4+ (B) and AIM+ CD8+ (D) T cells. Top panels depict SARS-CoV-2-recovered donors. Bottom panels depict SARS-CoV-2-naive donors. Left panels depict the background-subtracted percentage of non-naive T cells that are AIM+ cells of each subset. Right panels depict the relative frequency of each memory T cell subset in the background-subtracted AIM+ population. CM, CD45RA− CD27+ CCR7+; EM1, CD45RA− CD27+ CCR7−; EM2, CD45RA− CD27− CCR7+; EM3, CD45RA− CD27− CCR7−; EMRA, CD45RA+ CD27− CCR7−. Time points are as defined in Figure 1A. Boxplots represent median with interquartile range.
Longitudinal samples from 36 SARS-CoV-2-naive and 11 SARS-CoV-2-recovered individuals were used for each experiment, analyzed in nine independent batches. All paired longitudinal samples were analyzed within a single batch. See also Figure S3.
Total non-naive CD8+ T cells were distributed throughout memory T cell subsets, and the frequencies of these subsets were unchanged by vaccination (Figure S3B). AIM+ CD8+ T cells had a similar subset distribution to AIM+ CD4+ T cells. The baseline antigen-specific CD8+ T cell response in recovered subjects was composed of similar proportions of EM1, CM, and terminally differentiated CD8+ EMRA (CD45RA+ CD27− CCR7−) T cells (Figures 2C and 2D). A smaller proportion of AIM+ EM2 and EM3 CD8+ T cells was observed at baseline in recovered subjects. These proportions stayed relatively consistent throughout the course of vaccination in recovered subjects, and there were no statistically significant changes from baseline (Figure 2D). In contrast, in SARS-CoV-2-naive individuals, few AIM+ EMRA CD8+ T cells were observed at any time point (Figure 2D). Rather, vaccine-primed AIM+ CD8+ T cells in these subjects were largely EM1, with minority populations of CM and EM3 cells (Figure 2D). With the exception of the EMRA population, the antigen-specific AIM+ CD8+ T cell response in SARS-CoV-2-naive donors following vaccination resembled that observed in recovered donors (Figure 2D). These data indicate that the vaccine-elicited T cell response has a similar memory T cell subset distribution to the response generated following SARS-CoV-2 infection and is composed of primarily CD45RA− CD27+ memory T cells.
Vaccine-induced CD4+ T cells are predominantly Th1 and cTfh cells, resembling cells generated by SARS-CoV-2 infection
Given the role of helper T cell subsets such as CD4+ Tfh cells to help B cell responses and the importance of Th1 cells in viral infections, we next explored the differentiation state of AIM+ CD4+ T cells. To this end, we examined antigen-specific CXCR5+ Tfh in circulation (cTfh) as well as CXCR5− Th1 (CXCR3+CCR6−), Th17 (CXCR3−CCR6+), Th1/17 (CXCR3+CCR6+), and CXCR3−CCR6− cells (likely to include Th2) (Figures 3 A and S1A; Acosta-Rodriguez et al., 2007; Schmitt et al., 2014; Trifari et al., 2009). Total non-naive CD4+ T cell populations predominantly had Th1 and CXCR3−CCR6− phenotypes (Figure S3C). The baseline AIM+ CD4+ T cell response in recovered individuals, however, was dominated by cTfh and Th1 cells (Figures 3A and 3B). The first dose of vaccine led to further expansion of AIM+ cTfh and Th1 cells in these recovered subjects, and this pattern was largely maintained through the course of vaccination (Figure 3B). In SARS-CoV-2-naive subjects, the first vaccine dose also elicited predominantly antigen-specific Th1 and cTfh cells (Figure 3B). This distribution was sustained through booster vaccination in SARS-CoV-2-naive individuals, with these AIM+ subsets being further boosted by the second vaccine dose (Figure 3B). The magnitude of AIM+ cTfh responses was correlated with activated (Ki67+CD38+) cTfh (CXCR5+PD-1+) analyzed in parallel after each vaccine dose (Figure S3D), indicating that the AIM assay accurately captures cTfh known to contain antigen-specific T cells in other settings (Herati et al., 2017). Thus, the vaccine-elicited AIM+ CD4+ T cell response to mRNA vaccination qualitatively resembled the response to natural infection and was characterized by robust induction of antigen-specific cTfh and Th1 cells.
Figure 3.
Early antigen-specific CD4+ helper T cell responses shape humoral and cellular adaptive immune responses to mRNA vaccination
(A) Representative flow cytometric plots depicting the gating of AIM+ (CD200+CD40L+) CD4+ T cells to identify the indicated helper subsets in a SARS-CoV-2-naive donor at time point 4. Red events depict AIM+ T cells, gray events depict total CD4+ T cells from the same donor.
(B) Frequency of T helper subsets in AIM+ CD4+ T cells. Top panel depicts SARS-CoV-2-recovered donors. Bottom panel depicts SARS-CoV-2-naive donors. Left panel depicts the background-subtracted percentage of non-naive CD4+ T cells that are AIM+ helper T cells in each subset. Right panel depicts the relative frequency of each helper T cell subset in the background-subtracted AIM+ population. cTfh, CXCR5+ of non-naive CD4+ T cells; Th1, CXCR5− CXCR3+ CCR6−; Th17, CXCR5− CXCR3− CCR6+; Th1/17, CXCR5− CXCR3+ CCR6+; Other, CXCR5− CXCR3− CCR6−. Boxplots represent median with interquartile range.
(C) Correlations between the frequency of pre-boost (time point 2) AIM+ Th1 or AIM+ cTfh cells with post-boost (time point 4) AIM+ CD8+ T cells or neutralizing titers against dominant (D614G) or variant (B.1.351) strains of SARS-CoV-2 as published in a previous study of the same cohort (Goel et al., 2021). FRNT50, focus reduction neutralization titer 50%. Only SARS-CoV-2-naive donors were considered for these correlations. Associations were calculated using Spearman rank correlation and are shown with Pearson trend lines for visualization. Time points are as defined in Figure 1A.
Longitudinal samples from 36 SARS-CoV-2-naive and 11 SARS-CoV-2-recovered individuals were used for each experiment, analyzed in nine independent batches. All paired longitudinal samples were analyzed within a single batch. See also Figure S3.
Antigen-specific Th1 and cTfh cells induced by the first dose correlate with CD8+ T cell and humoral responses to the second dose
The rapid induction of antigen-specific CD4+ T cells following the first mRNA vaccine dose, particularly Th1 and cTfh cells, may provide a population of helper T cells available to enhance immune responses to the second vaccine dose. Th1 cells predominantly facilitate the CD8+ T cell response, whereas Tfh cells help foster optimal B cell, germinal center, and antibody responses (Crotty, 2011; Krawczyk et al., 2007; Luckheeram et al., 2012; Williams et al., 2006). Indeed, we observed a strong correlation between the frequency of pre-boost AIM+ Th1 cells and the frequency of post-boost AIM+ CD8+ T cells in SARS-CoV-2-naive individuals (Figure 3C), consistent with a role for Th1 cells generated by primary vaccination in enhancing the CD8+ T cell responses following booster vaccination. Similarly, the frequency of pre-boost antigen-specific cTfh cells correlated with post-boost neutralizing antibody titers against both the dominant strain of SARS-CoV-2 (D614G, dominant at the time of study) and the Beta variant (B.1.351) (Figure 3C). Despite a strong correlation between AIM+ Th1 and cTfh (Figure S3E), pre-boost Th1 were less well correlated with post-boost neutralizing titers than were cTfh, and pre-boost cTfh did not significantly correlate with post-boost CD8+ T cell responses, supporting the distinct associations of these pre-boost immune cell types to post-boost vaccine-elicited immune responses (Figure S3F). Notably, the pre-boost Th1/cTfh ratio within the AIM+ cells did not correlate with post-boost humoral or CD8+ T cell responses, suggesting that the independent magnitudes of pre-boost AIM+ cTfh and Th1, rather than the relative skewing between these responses, contribute to humoral and CD8+ T cell responses to the second dose (Figure S3G). Furthermore, baseline AIM+ Th1 and cTfh cells in SARS-CoV-2-naive subjects did not correlate with post-boost CD8+ T cell or neutralizing responses, respectively, suggesting minimal contribution of pre-existing cross-reactive CD4+ T cells to the immune response to SARS-CoV-2 mRNA vaccines (Figure S3H). These observations highlight a key functional role for vaccine-elicited CD4+ T cells and suggest possible downstream effects of skewed antigen-specific CD4+ T cell responses. Moreover, these data highlight one of the potential benefits of a two-dose vaccination regimen, whereby CD4+ T cells primed by the first vaccine dose may augment and coordinate responses following the booster vaccination.
Integrated analysis reveals coordinated humoral and cellular responses to mRNA vaccination with distinct trajectories in SARS-CoV-2-naive and recovered individuals
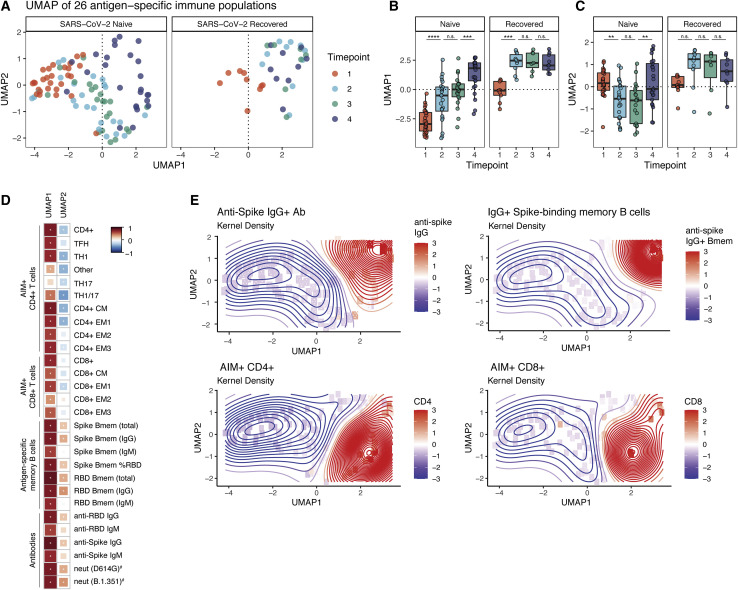
These CD4+ T cell data suggested interrelationships between distinct immune responses generated by mRNA vaccination. To further examine this notion of coordinated immune responses following vaccination, we compiled the antigen-specific T cell data described above with a previously reported dataset of antibody and memory B cell responses from this cohort (Goel et al., 2021). Using these data, we integrated 26 antigen-specific features of the immune response to mRNA vaccination into high-dimensional uniform manifold approximation and projection (UMAP) space (Figure 4 A). Correlating individual antigen-specific features with the UMAP coordinates revealed that UMAP1 is a measure of the anti-SARS-CoV-2 immune response to vaccination (Figure 4D). UMAP1 also revealed a signal of previous SARS-CoV-2 infection, as recovered subjects occupied a location with increased UMAP1 signal at baseline (Figures 4A and 4B). Specifically, UMAP1 captured a coordinated immune response in which antigen-specific CD8+ T cells and CD4+ Th1 and cTfh cells were increased coordinately with antibodies, IgG+ memory B cells, RBD-focused humoral responses, and increased neutralizing antibody titers (Figures 4D and 4E). UMAP2 captures the relative balance between humoral and cellular immune responses, especially CD4+ T cell responses (Figure 4D). Total non-naive lymphocyte populations were not altered and did not correlate with the antigen-specific responses, consistent with induction of a targeted vaccine-elicited response (Figure S4A). This UMAP projection revealed trajectory shifts that were notable between naive and recovered subjects. For example, in SARS-CoV-2-recovered individuals, there was an increase in both UMAP1 and UMAP2 following primary vaccination but essentially no change following the second vaccine dose, indicating that both the magnitude and relative balance of the antigen-specific response is stabilized after a single dose (Figures 4A–4C). In SARS-CoV-2-naive subjects there was a more dynamic trajectory over time with an initial increase in UMAP1 and decrease in UMAP2 signal at time points 2 and 3, followed by a coalescence toward increased UMAP1 and UMAP2 after the second vaccine dose (Figures 4A–4C). UMAP2 captures the different kinetics of T cell and humoral responses in SARS-CoV-2-naive and recovered individuals. A rapid antigen-specific CD4+ T cell and, to a lesser extent, CD8+ T cell response drives the UMAP2 coordinate in a negative direction post-primary in naive individuals, whereas robust humoral immunity drives UMAP2 in a positive direction post-boost in naive individuals and post-primary in recovered individuals (Figures 4C–4E). This integrated analysis highlights the differential effects of the first vaccine dose in SARS-CoV-2-recovered and naive individuals. Finally, correlations of key antigen-specific parameters of the vaccine response over time revealed relationships between post-primary and post-boost immunity within and between arms of the adaptive immune system, highlighting correlations within the T cell, memory B cell, and antibody responses (Figure S4B). This analysis also highlights pre-boost immune response features such as cTfh and Th1 that correlate with post-boost humoral and cellular responses (Figure S4B). In summary, this unbiased integrated analysis of 26 antigen-specific immune responses illustrates the coordinated immunological underpinnings of the immunity induced by mRNA vaccines.
Figure 4.
mRNA vaccination provokes a coordinated immune response in SARS-CoV-2-naive and recovered individuals
(A) UMAP projections of aggregated antigen-specific data for T cell, memory B cell, and antibody responses over time. Memory B cell and antibody data were taken from a previously published dataset using 29 SARS-CoV-2-naive and 10 SARS-CoV-2-recovered subjects from the same cohort (Goel et al., 2021). Colors represent time points at which PBMCs were collected throughout the study. Parameters were considered as frequency of non-naive T cells or memory B cells, capturing both the magnitude and skewing of responses.
(B and C) Summary plots of UMAP1 (B) and UMAP2 (C) coordinates over time. Individual points represent individual participants. Statistics were calculated using unpaired Wilcoxon test. n.s., not significant. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Boxplots represent median with interquartile range.
(D) Correlations of the individual antigen-specific features used to train the UMAP against the UMAP1 and UMAP2 axes. Red indicates positive correlations and blue indicates negative correlations. ∗ = FDR < 0.05. #Features that were not used to train the original UMAP.
(E) Kernel density plots displaying the variation of selected antigen-specific features across UMAP space. Time points are as defined in Figure 1A.
Longitudinal samples from 29 SARS-CoV-2-naive and 10 SARS-CoV-2-recovered individuals were used for each experiment, analyzed in eight independent batches. All paired longitudinal samples were analyzed within a single batch. See also Figure S4.
Discussion
In this study, we interrogated the antigen-specific CD4+ and CD8+ T cell responses induced by SARS-CoV-2 mRNA vaccination in a longitudinal cohort of SARS-CoV-2-naive and recovered individuals. Our data demonstrate robust induction of antigen-specific T cells by mRNA vaccination that may contribute, in addition to previously defined humoral responses, to durable protective immunity. In particular, antigen-specific memory CD4+ and CD8+ T cells are likely to be less affected by antibody escape mutations in variant viral strains, as T cells can recognize peptide epitopes distributed throughout the SARS-CoV-2 Spike protein (Angyal et al., 2021; Tarke et al., 2021b; Woldemeskel et al., 2021). Moreover, unlike vaccine-induced B cell and antibody responses, which have been noted to decrease with age (Abu Jabal et al., 2021; Goel et al., 2021; Levi et al., 2021; Prendecki et al., 2021), substantial age-associated changes in the induction of antigen-specific T cell responses were not observed. Finally, the generation of robust T cell responses by mRNA vaccines may have implications for long-term protective immunity, as memory CD4+ and CD8+ T cells can be exceptionally durable in other vaccine settings (Akondy et al., 2017; Hammarlund et al., 2003).
Vaccine-induced CD4+ and CD8+ T cells specific for SARS-CoV-2 were qualitatively similar to baseline memory T cell responses generated following natural SARS-CoV-2 infection and mainly mapped to CM and EM1 memory T cell subsets. These two subsets share many functional and memory-like attributes but differ in CCR7 expression. As CCR7 promotes homing to secondary lymphoid tissues, EM1 may represent memory T cells that can survey blood and peripheral tissues, whereas CM can home efficiently to lymphoid tissues (Romero et al., 2007). These memory T cell subsets are longer lived compared with effector T cells, and access to secondary lymphoid tissues may allow CM cells to contribute to recall responses upon booster vaccination or future infection. Although we await follow-up studies to directly interrogate longevity, the observed induction of memory T cell subsets with capacity for durability by mRNA vaccination supports the hypothesis that vaccine-induced CD4+ and CD8+ T cell responses will be long lived and capable of contributing to future recall responses.
One key observation was the rapid and universal induction of SARS-CoV-2-specific CD4+ T cells following the first vaccine dose in SARS-CoV-2-naive individuals. This observation may be noteworthy given the gradual development of antigen-specific CD8+ T cells observed here and the previous observations for humoral responses (Goel et al., 2021; Jackson et al., 2020), which only consistently reach maximal levels after the second vaccine dose. These data point to the early induction of antigen-specific CD4+ T cells as a possible contributor to the protection observed in clinical trials as early as two weeks after the first vaccine dose (Baden et al., 2021; Polack et al., 2020), when neutralizing antibody titers are still low in many individuals (Goel et al., 2021). Indeed, CD4+ T cells can prevent symptomatic SARS-CoV infection in animal models (Zhao et al., 2016), and the rapid induction of antigen-specific CD4+ T cells after only a single vaccine dose may explain the disconnect between low neutralizing responses and vaccine-induced protective immunity following the first dose.
The notion that early CD4+ T cell responses have a functional role in immunity is also supported by the correlation between pre-boost Th1 and cTfh cells with post-boost CD8+ T cell and neutralizing antibody responses, respectively. In addition to strongly correlating with post-boost neutralizing antibody titers, pre-boost antigen-specific cTfh cells also correlated with post-boost memory B cell responses. When examining multiple individual antigen-specific responses over time, pre-boost cTfh were better predictors of post-boost humoral responses than many pre-boost readouts of humoral immunity, pointing to the critical role of Tfh in coordinating humoral immunity. Likewise, pre-boost antigen-specific Th1 cells were as strongly correlated with post-boost CD8+ T cell responses as pre-boost CD8+ T cells. These findings suggest that the CD4+ T cell response generated by the first vaccine dose guides multiple arms of the adaptive immune response to booster vaccination and highlight the benefits of a prime-boost strategy to amplify a coordinated vaccine-induced immune response.
Previous studies have demonstrated that individuals who have recovered from SARS-CoV-2 infection achieve maximum antigen-specific humoral immune responses after only a single vaccine dose, raising the question of whether a second vaccine dose is necessary in these individuals (Angyal et al., 2021; Bradley et al., 2021; Camara et al., 2021; Goel et al., 2021; Mazzoni et al., 2021; Saadat et al., 2021; Samanovic et al., 2021; Stamatatos et al., 2021). Our present studies now provide information on both CD4+ and CD8+ T cell responses in naive and recovered subjects and support the idea that the second dose of vaccine has minimal impact on the magnitude, memory phenotype, or helper subset distribution of antigen-specific CD4+ or CD8+ T cell responses in SARS-CoV-2-recovered subjects. Moreover, an integrated analysis of 26 antigen-specific features of the immune response to vaccination highlighted the immunological benefit of the first dose in recovered subjects while also illustrating the relative stability of the immune landscape in response to the second vaccine dose. In contrast, in SARS-CoV-2-naive subjects, there was robust and dynamic change in the coordination and evolution of the antigen-specific immune response following the first as well as the second vaccine dose. These data point to the immunological benefit of two vaccine doses in SARS-CoV-2-naive subjects and highlight the coordination between different arms of the adaptive immune response following mRNA vaccination. In concert with robust humoral immunity, the preferential induction of Th1, Tfh, and CM-like T cells indicates that the vaccine-elicited immune response is specifically focused on the key hallmarks of long-term antiviral immunity that are likely to confer lasting protection against SARS-CoV-2 infection.
Limitations of the study
The scope of these findings is limited by the nature of the cohort. Larger studies will be valuable to confirm and extend the findings, especially for SARS-CoV-2-recovered individuals. In particular, all of the recovered participants in this study experienced mild disease, and it is unclear how mRNA vaccination would affect immune responses in those who have recovered from severe COVID-19. Although we observed no decrease in T cell responses with age, the cohort examined here had relatively few participants of advanced age, necessitating larger follow-up studies. The AIM assay used to identify antigen-specific T cells relies on stimulation with peptide megapools. Despite the utility of this approach for capturing the diversity of the T cell response to epitopes throughout the Spike protein and in individuals with diverse HLA allotypes, the sensitivity of the assay may differ on the basis of the individual immunodominant responses and HLA types. A further limitation is that stimulation of T cells may alter surface marker expression. Although on the basis of our assay optimization studies, the differentiation markers used to separate memory T cell and helper T cell subsets appear to remain stable during the time frame of the AIM assay, it is possible that subtle changes following stimulation might influence precise differentiation state assignment. In the future it will be informative to perform epitope mapping and use HLA class I and class II tetramers to examine antigen-specific cells without stimulation. Finally, assessing long-term durable immune memory remains a key goal. Reliable assessments of durable T cell memory will require analysis at extended time points post-vaccination that were not available at the time of the study.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| BUV395 CD4 | BD Biosciences | Cat#563550; RRID: AB_2738273 |
| BUV496 CD8 | BD Biosciences | Cat#612943; RRID: AB_2870223 |
| BUV615 CD45RA | BD Biosciences | Cat#751555; RRID: AB_2875550 |
| BUV737 CD27 | BD Biosciences | Cat#612829; RRID: AB_2870151 |
| BUV805 CD3 | BD Biosciences | Cat#612896; RRID: AB_2870184 |
| BV421 CXCR3 | Biolegend | Cat#353716; RRID: AB_2561448 |
| BV650 CCR7 | Biolegend | Cat#353234; RRID: AB_2563867 |
| BV605 CD69 | Biolegend | Cat#310938; RRID: AB_2562307 |
| BV711 CD40L | Biolegend | Cat#310838; RRID: AB_2563845 |
| BV785 CD107a | Biolegend | Cat#328644; RRID: AB_2565968 |
| FITC IFNy | Biolegend | Cat#502515; RRID: AB_493029 |
| PE CD200 | Biolegend | Cat#399804; RRID: AB_2861016 |
| PE-Cy7 OX40 | Biolegend | Cat#350012; RRID: AB_10901161 |
| AF647 41BB | Biolegend | Cat#309810; RRID: AB_830672 |
| APC-R700 CXCR5 | BD Biosciences | Cat#565191; RRID: AB_2739103 |
| APC-Cy7 CCR6 | Biolegend | Cat#353432; RRID: AB_2566274 |
| Chemicals, peptides, and recombinant proteins | ||
| CD4-S peptide megapool | Synthetic Biomolecules (aka A&A) | http://www.syntheticbiomolecules.com/ |
| CD8-E peptide megapool | Synthetic Biomolecules (aka A&A) | http://www.syntheticbiomolecules.com/ |
| Biological samples | ||
| Human peripheral blood samples from SARS-CoV-2 mRNA vaccine recipients | Collected at the University of Pennsylvania | N/A |
| Other | ||
| Ghost Dye Violet 510 | Tonbo | Cat#13-0870-T500 |
| GolgiStop (Containing Monensin) | BD Biosciences | Cat#51-2092K7 |
| CD40 Antibody, anti-human, pure-functional grade | Miltenyi Biotech | Cat#130-094-133; RRID: AB_10839704 |
| Anti-Human CD28/CD49d Purified | BD Biosciences | Cat#347690; RRID: AB_647457 |
| Human TruStain FcX (Fc Receptor Blocking Solution) | Biolegend | Cat#422302; RRID: AB_2818986 |
| Foxp3 / Transcription Factor Fixation/Permeabilization Concentrate and Diluent | eBioscience | Cat#00-5521-00 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, E. John Wherry (wherry@pennmedicine.upenn.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Human subjects
47 individuals (36 SARS-CoV-2 naive, 11 SARS-CoV-2 recovered) provided informed consent and were enrolled in the study with approval from the University of Pennsylvania Institutional Review Board (IRB# 844642). All participants were otherwise healthy and did not report any history of chronic health conditions. Subjects were identified as SARS-CoV-2 naive or recovered via combined self-reporting and laboratory evidence of a prior SARS-CoV-2 infection. All subjects received either Pfizer (BNT162b2) or Moderna (mRNA-1273) mRNA vaccines and were enrolled irrespective of which mRNA vaccine they received. Samples were collected at 4 time points: pre-vaccine baseline (time point 1), two weeks post-primary vaccination (time point 2), the day of the booster vaccination (time point 3), and one week post-boost (time point 4). Each study visit included collection of clinical questionnaire data and 80-100mL of peripheral blood. Full cohort and demographic information is provided in Table S1.
Method details
Sample processing
Venous blood was collected into sodium heparin and EDTA tubes by standard phlebotomy. Blood tubes were centrifuged at 3000rpm for 15 minutes to separate plasma. Heparin and EDTA plasma were stored at −80°C for paired serological analyses. Remaining whole blood was diluted 1:1 with RPMI 1640 (Corning) supplemented with 1% fetal bovine serum (FBS), 2mM L-Glutamine, 100 U/mL Penicillin, and 100 μg/mL Streptomycin (R1 medium) and layered onto SEPMATE tubes (STEMCELL Technologies) containing lymphoprep gradient (STEMCELL Technologies). SEPMATE tubes were centrifuged at 1200 g for 10 minutes and the PBMC fraction was collected into new tubes and washed with R1. PBMCs were then treated with ACK lysis buffer (Thermo Fisher) for 5 minutes to lyse red blood cells. Samples were washed again with R1, passed through a 70μm cell strainer, and cell counts were acquired with a Countess automated cell counter (Thermo Fisher). PBMCs were cryopreserved in 10% DMSO in FBS.
Activation induced marker (AIM) expression assay
PBMCs were thawed by warming frozen cryovials in a 37°C water bath and resuspending cells in 10mL of RPMI supplemented with 10% FBS, 2mM L-Glutamine, 100 U/mL Penicillin, and 100 μg/mL Streptomycin (R10). Cells were washed once in R10, counted using a Countess automated cell counter (Thermo Fisher), and resuspended in fresh R10 to a density of 5x106 cells/mL. For each condition, duplicate wells containing 1x106 cells in 200 μL were plated in 96-well round-bottom plates and rested overnight in a humidifed incubator at 37°C, 5% CO2. After 16 hours, CD40 blocking antibody (0.5 μg/mL final concentration) was added to cultures for 15 minutes prior to stimulation. Cells were then stimulated for 24 hours with costimulation (anti-human CD28/CD49d, BD Biosciences) and peptide megapools (CD4-S for all CD4+ T cell analyses, CD8-E for all CD8+ T cell analyses) at a final concentration of 1 μg/mL. Peptide megapools were prepared as previously described (Grifoni et al., 2020; Tarke et al., 2021a). Matched unstimulated samples for each donor at each time point were treated with costimulation alone. 20 hours post-stimulation, antibodies targeting CXCR3, CCR7, CD40L, CD107a, CXCR5, and CCR6 were added to the culture along with monensin (GolgiStop, BD Biosciences) for a four-hour stain at 37°C. After four hours, duplicate wells were pooled and cells were washed in PBS supplemented with 2% FBS (FACS buffer). Cells were stained for 10 minutes at room temperature with Ghost Dye Violet 510 and Fc receptor blocking solution (Human TruStain FcX, BioLegend) and washed once in FACS buffer. Surface staining for 30 minutes at room temperature was then performed with antibodies directed against CD4, CD8, CD45RA, CD27, CD3, CD69, CD40L, CD200, OX40, and 41BB in FACS buffer. Cells were washed once in FACS buffer, fixed and permeabilizied for 30 minutes at room temperature (eBioscience Foxp3 / Transcription Factor Fixation/Permeabilization Concentrate and Diluent), and washed once in 1X Permeabilization Buffer prior to staining for intracellular IFN-γ overnight at 4°C. Cells were then washed once and resuspended in 1% paraformaldehyde in PBS prior to data acquisition.
All data from AIM expression assays were background-subtracted using paired unstimulated control samples. For memory T cell and helper T cell subsets, the AIM+ background frequency of non-naive T cells was subtracted independently for each subset. AIM+ cells were identified from non-naive T cell populations. AIM+ CD4+ T cells were defined by dual-expression of CD200 and CD40L. AIM+ CD8+ T cells were defined by a boolean analysis identifying cells expressing at least four of five markers: CD200, CD40L, 41BB, CD107a, and intracellular IFN-γ.
Flow cytometric quantification of activated (Ki67+CD38+) T cells
PBMCs were thawed as for the AIM assay and stained with antibodies immediately post-thaw, including CD3, CD4, CD8, CD27, CD45RA, CXCR5, CD38, PD-1, and Ki67. Surface staining was performed as for the AIM assay.
Flow cytometry
Data were acquired on a BD Symphony A5 instrument. Standardized SPHERO rainbow beads (Spherotech) were used to track and adjust photomultiplier tube voltages over time. Compensation was performed using UltraComp eBeads (Thermo Fisher). Up to 2x106 events were acquired per sample. Data were analyzed using FlowJo v10 (BD Bioscience). A full gating strategy for segregation of T cell subsets is shown in Figure S1A.
Antibody and memory B cell responses
The dataset of antibody and memory B cell responses from the same cohort of individuals was published previously (Goel et al., 2021).
Quantification and statistical analysis
Data visualization and statistics
All data were analyzed using custom scripts in R and visualized using RStudio. Boxplots represent median with interquartile range. The 26 parameters used to train the UMAP were scaled by column (z-score normalization) prior to generating UMAP coordinates. Statistical tests are indicated in the corresponding figure legends. All tests were performed two-sided with a nominal significance threshold of p < 0.05. In all cases of multiple comparisons, adjustment was performed using Holm correction. Unpaired tests were used for comparisons between time points, as some participants lacked samples from individual time points. ∗ indicates p < 0.05, ∗∗ indicates p < 0.01, ∗∗∗ indicates p < 0.001, ∗∗∗∗ indicates p < 0.0001. Source code and data files are available upon request from the authors.
Acknowledgments
We would like to thank the study participants for their generosity in making the study possible. We also thank Wenzhao Meng, Aaron M. Rosenfeld, Eline T. Luning Prak, the University of Pennsylvania Flow Cytometry Core, and the members of the Wherry lab for helpful discussions and feedback. This work was supported by grants from the NIH (AI105343, AI082630, AI108545, AI155577, and AI149680 to E.J.W.; AI152236 to P.B.; AI082630 to R.S.H.; HL143613 to J.R.G.; T32 AR076951-01 to S.A.A.; T32 CA009140 to J.R.G. and D.M.; T32 AI055400 to P.H.; and U19AI082630 to S.E.H. and E.J.W.), funding from the Allen Institute for Immunology (to S.A.A. and E.J.W.), a Cancer Research Institute-Mark Foundation Fellowship (to J.R.G.), the Chen Family Research Fund (to S.A.A.), the Parker Institute for Cancer Immunotherapy (to J.R.G. and E.J.W.), the Penn Center for Research on Coronavirus and Other Emerging Pathogens (to P.B.), the University of Pennsylvania Perelman School of Medicine COVID Fund (to R.R.G. and E.J.W.), the University of Pennsylvania Perelman School of Medicine 21st Century Scholar Fund (to R.R.G.), and a philanthropic gift from Jeffrey Lurie, Joel Embiid, Josh Harris, and David Blitzer (to S.E.H.). Work in the Wherry lab is supported by the Parker Institute for Cancer Immunotherapy. This work was also supported by NIH contract 75N9301900065 (to D.W. and A.S.).
Author contributions
M.M.P., D.M., and E.J.W. conceived the study. M.M.P., D.M., R.R.G., and S.A.A. carried out experiments. S.A.A. and O.K. were involved in clinical recruitment. D.A.O., and J.R.G. provided input on statistical analyses. D.M., A.E.B., and R.S.H. contributed to the methodology. M.M.P., D.M., R.R.G., S.A., A.H., S.K., K.D., and J.T.H. processed peripheral blood samples and managed the sample database. R.R.G., O.K., J.D., and S.L. performed phlebotomy. A.P., S.G., P.H., S.D., K.A.L., L.K.-C., A.E.B., M.E.W., C.M.M., A.G., D.W., and A.S. provided data and materials. A.R.G. and E.J.W. supervised the study. All authors participated in data analysis and interpretation. M.M.P., D.M., and E.J.W. wrote the manuscript.
Declaration of interests
S.E.H. has received consultancy fees from Sanofi Pasteur, Lumen, Novavax, and Merk for work unrelated to this study. E.J.W. is a consultant or an adviser for Merck, Elstar, Janssen, Related Sciences, Synthekine, and Surface Oncology. E.J.W. is a founder of Surface Oncology and Arsenal Biosciences. E.J.W. is an inventor on a patent (U.S. Patent No. 10,370,446) submitted by Emory University that covers the use of PD-1 blockade to treat infections and cancer. A.S. is a consultant for Gritstone, Flow Pharma, CellCarta, Arcturus, Oxfordimmunotech, and Avalia. La Jolla Institute for Immunology has filed for patent protection for various aspects of T cell epitope and vaccine design work.
Published: August 13, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.immuni.2021.08.001.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abu Jabal K., Ben-Amram H., Beiruti K., Batheesh Y., Sussan C., Zarka S., Edelstein M. Impact of age, ethnicity, sex and prior infection status on immunogenicity following a single dose of the BNT162b2 mRNA COVID-19 vaccine: real-world evidence from healthcare workers, Israel, December 2020 to January 2021. Euro Surveil. 2021;26:2100096. doi: 10.2807/1560-7917.ES.2021.26.6.2100096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acosta-Rodriguez E.V., Rivino L., Geginat J., Jarrossay D., Gattorno M., Lanzavecchia A., Sallusto F., Napolitani G. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat. Immunol. 2007;8:639–646. doi: 10.1038/ni1467. [DOI] [PubMed] [Google Scholar]
- Akondy R.S., Fitch M., Edupuganti S., Yang S., Kissick H.T., Li K.W., Youngblood B.A., Abdelsamed H.A., McGuire D.J., Cohen K.W., et al. Origin and differentiation of human memory CD8 T cells after vaccination. Nature. 2017;552:362–367. doi: 10.1038/nature24633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E.J., Rouphael N.G., Widge A.T., Jackson L.A., Roberts P.C., Makhene M., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., et al. mRNA-1273 Study Group Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N. Engl. J. Med. 2020;383:2427–2438. doi: 10.1056/NEJMoa2028436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angyal A., Longet S., Moore S., Payne R.P., Harding A., Tipton T., Rongkard P., Ali M., Hering L.M., Meardon N., et al. T-cell and antibody responses to first BNT162b2 vaccine dose in previously SARS-CoV-2-infected and infection-naive UK healthcare workers: a multicentre, prospective, observational cohort study. 2021. https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3820576 [DOI] [PMC free article] [PubMed]
- Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. COVE Study Group Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bange E.M., Han N.A., Wileyto P., Kim J.Y., Gouma S., Robinson J., Greenplate A.R., Hwee M.A., Porterfield F., Owoyemi O., et al. CD8+ T cells contribute to survival in patients with COVID-19 and hematologic cancer. Nat. Med. 2021;27:1280–1289. doi: 10.1038/s41591-021-01386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts M.R., Brenchley J.M., Price D.A., De Rosa S.C., Douek D.C., Roederer M., Koup R.A. Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. J. Immunol. Methods. 2003;281:65–78. doi: 10.1016/s0022-1759(03)00265-5. [DOI] [PubMed] [Google Scholar]
- Bradley T., Grundberg E., Selvarangan R. Antibody responses boosted in seropositive healthcare workers after single dose of SARS-CoV-2 mRNA vaccine. medRxiv. 2021 doi: 10.1056/NEJMc2102051. 2021.02.03.21251078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camara C., Lozano-Ojalvo D., Lopez-Granados E., Paz-Artal E., Pion M., Correa-Rocha R., Ortiz A., Lopez-Hoyos M., Iribarren M.E., Portoles J., et al. Differential effects of the second SARS-CoV-2 mRNA vaccine dose on T cell immunity in naïve and COVID-19 recovered individuals. bioRxiv. 2021 doi: 10.1016/j.celrep.2021.109570. 2021.03.22.436441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. Follicular helper CD4 T cells (TFH) Annu. Rev. Immunol. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- Cutler D.M., Summers L.H. The COVID-19 pandemic and the $16 trillion virus. JAMA. 2020;324:1495–1496. doi: 10.1001/jama.2020.19759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel R.R., Apostolidis S.A., Painter M.M., Mathew D., Pattekar A., Kuthuru O., Gouma S., Hicks P., Meng W., Rosenfeld A.M., et al. Distinct antibody and memory B cell responses in SARS-CoV-2 naïve and recovered individuals following mRNA vaccination. Sci. Immunol. 2021;6:eabi6950. doi: 10.1126/sciimmunol.abi6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann D., Baars P.A., Rep M.H., Hooibrink B., Kerkhof-Garde S.R., Klein M.R., van Lier R.A. Phenotypic and functional separation of memory and effector human CD8+ T cells. J. Exp. Med. 1997;186:1407–1418. doi: 10.1084/jem.186.9.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarlund E., Lewis M.W., Hansen S.G., Strelow L.I., Nelson J.A., Sexton G.J., Hanifin J.M., Slifka M.K. Duration of antiviral immunity after smallpox vaccination. Nat. Med. 2003;9:1131–1137. doi: 10.1038/nm917. [DOI] [PubMed] [Google Scholar]
- Herati R.S., Muselman A., Vella L., Bengsch B., Parkhouse K., Del Alcazar D., Kotzin J., Doyle S.A., Tebas P., Hensley S.E., et al. Successive annual influenza vaccination induces a recurrent oligoclonotypic memory response in circulating T follicular helper cells. Sci. Immunol. 2017;2:eaag2152. doi: 10.1126/sciimmunol.aag2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., et al. mRNA-1273 Study Group An mRNA vaccine against SARS-CoV-2—preliminary report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S.M., Wherry E.J., Ahmed R. Effector and memory T-cell differentiation: implications for vaccine development. Nat. Rev. Immunol. 2002;2:251–262. doi: 10.1038/nri778. [DOI] [PubMed] [Google Scholar]
- Kalimuddin S., Tham C.Y.L., Qui M., de Alwis R., Sim J.X.Y., Lim J.M.E., Tan H.-C., Syenina A., Zhang S.L., Le Bert N., et al. Early T cell and binding antibody responses are associated with COVID-19 RNA vaccine efficacy onset. Med. (N Y) 2021;2:682–688.e4. doi: 10.1016/j.medj.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F., Srivastava K., Alshammary H., Amoako A.A., Awawda M.H., Beach K.F., Bermúdez-González M.C., Bielak D.A., Carreño J.M., Chernet R.L., et al. Antibody responses in seropositive persons after a single dose of SARS-CoV-2 mRNA vaccine. N. Engl. J. Med. 2021;384:1372–1374. doi: 10.1056/NEJMc2101667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczyk C.M., Shen H., Pearce E.J. Memory CD4 T cells enhance primary CD8 T-cell responses. Infect. Immun. 2007;75:3556–3560. doi: 10.1128/IAI.00086-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederer K., Castaño D., Gómez Atria D., Oguin T.H., 3rd, Wang S., Manzoni T.B., Muramatsu H., Hogan M.J., Amanat F., Cherubin P., et al. SARS-CoV-2 mRNA vaccines foster potent antigen-specific germinal center responses associated with neutralizing antibody generation. Immunity. 2020;53:1281–1295.e5. doi: 10.1016/j.immuni.2020.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi R., Azzolini E., Pozzi C., Ubaldi L., Lagioia M., Mantovani A., Rescigno M. A cautionary note on recall vaccination in ex-COVID-19 subjects. medRxiv. 2021 [Google Scholar]
- Luckheeram R.V., Zhou R., Verma A.D., Xia B. CD4+T cells: differentiation and functions. Clin. Dev. Immunol. 2012;2012 doi: 10.1155/2012/925135. 925135–925135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M.D., Badovinac V.P. Defining memory CD8 T cell. Front. Immunol. 2018;9:2692. doi: 10.3389/fimmu.2018.02692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew D., Giles J.R., Baxter A.E., Oldridge D.A., Greenplate A.R., Wu J.E., Alanio C., Kuri-Cervantes L., Pampena M.B., D’Andrea K., et al. UPenn COVID Processing Unit Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369:eabc8511. doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoni A., Di Lauria N., Maggi L., Salvati L., Vanni A., Capone M., Lamacchia G., Mantengoli E., Spinicci M., Zammarchi L., et al. First dose mRNA vaccination is sufficient to reactivate immunological memory to SARS-CoV-2 in ex COVID-19 subjects. medRxiv. 2021 doi: 10.1172/JCI149150. 2021.03.05.21252590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.D., van der Most R.G., Akondy R.S., Glidewell J.T., Albott S., Masopust D., Murali-Krishna K., Mahar P.L., Edupuganti S., Lalor S., et al. Human effector and memory CD8+ T cell responses to smallpox and yellow fever vaccines. Immunity. 2008;28:710–722. doi: 10.1016/j.immuni.2008.02.020. [DOI] [PubMed] [Google Scholar]
- Ndhlovu Z.M., Kamya P., Mewalal N., Kløverpris H.N., Nkosi T., Pretorius K., Laher F., Ogunshola F., Chopera D., Shekhar K., et al. Magnitude and kinetics of CD8+ T cell activation during hyperacute HIV infection impact viral set point. Immunity. 2015;43:591–604. doi: 10.1016/j.immuni.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. C4591001 Clinical Trial Group Safety and efficacy of the BNT162b2 mRNA COVID-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendecki M., Clarke C., Brown J., Cox A., Gleeson S., Guckian M., Randell P., Pria A.D., Lightstone L., Xu X.-N., et al. Effect of previous SARS-CoV-2 infection on humoral and T-cell responses to single-dose BNT162b2 vaccine. Lancet. 2021;397:1178–1181. doi: 10.1016/S0140-6736(21)00502-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss S., Baxter A.E., Cirelli K.M., Dan J.M., Morou A., Daigneault A., Brassard N., Silvestri G., Routy J.-P., Havenar-Daughton C., et al. Comparative analysis of activation induced marker (AIM) assays for sensitive identification of antigen-specific CD4 T cells. PLoS ONE. 2017;12:e0186998. doi: 10.1371/journal.pone.0186998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero P., Zippelius A., Kurth I., Pittet M.J., Touvrey C., Iancu E.M., Corthesy P., Devevre E., Speiser D.E., Rufer N. Four functionally distinct populations of human effector-memory CD8+ T lymphocytes. J. Immunol. 2007;178:4112–4119. doi: 10.4049/jimmunol.178.7.4112. [DOI] [PubMed] [Google Scholar]
- Saadat S., Tehrani Z.R., Logue J., Newman M., Frieman M.B., Harris A.D., Sajadi M.M. Single dose vaccination in healthcare workers previously infected with SARS-CoV-2. medRxiv. 2021 doi: 10.1001/jama.2021.3341. 2021.01.30.21250843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin U., Muik A., Derhovanessian E., Vogler I., Kranz L.M., Vormehr M., Baum A., Pascal K., Quandt J., Maurus D., et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature. 2020;586:594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- Sallusto F., Lenig D., Förster R., Lipp M., Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- Samanovic M.I., Cornelius A.R., Wilson J.P., Karmacharya T., Gray-Gaillard S.L., Allen J.R., Hyman S.W., Moritz G., Ali M., Koralov S.B., et al. Poor antigen-specific responses to the second BNT162b2 mRNA vaccine dose in SARS-CoV-2-experienced individuals. medRxiv. 2021 doi: 10.1126/scitranslmed.abi8961. 2021.2002.2007.21251311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt N., Bentebibel S.-E., Ueno H. Phenotype and functions of memory Tfh cells in human blood. Trends Immunol. 2014;35:436–442. doi: 10.1016/j.it.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soresina A., Moratto D., Chiarini M., Paolillo C., Baresi G., Focà E., Bezzi M., Baronio B., Giacomelli M., Badolato R. Two X-linked agammaglobulinemia patients develop pneumonia as COVID-19 manifestation but recover. Pediatr. Allergy Immunol. 2020;31:565–569. doi: 10.1111/pai.13263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatatos L., Czartoski J., Wan Y.-H., Homad L.J., Rubin V., Glantz H., Neradilek M., Seydoux E., Jennewein M.F., MacCamy A.J., et al. mRNA vaccination boosts cross-variant neutralizing antibodies elicited by SARS-CoV-2 infection. Science. 2021 doi: 10.1126/science.abg9175. Published online March 25, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarke A., Sidney J., Kidd C.K., Dan J.M., Ramirez S.I., Yu E.D., Mateus J., da Silva Antunes R., Moore E., Rubiro P., et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep Med. 2021;2:100204. doi: 10.1016/j.xcrm.2021.100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarke A., Sidney J., Methot N., Zhang Y., Dan J.M., Goodwin B., Rubiro P., Sutherland A., da Silva Antunes R., Frazier A., et al. Negligible impact of SARS-CoV-2 variants on CD4+ and CD8+ T cell reactivity in COVID-19 exposed donors and vaccinees. bioRxiv. 2021 doi: 10.1016/j.xcrm.2021.100355. 2021.02.27.433180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifari S., Kaplan C.D., Tran E.H., Crellin N.K., Spits H. Identification of a human helper T cell population that has abundant production of interleukin 22 and is distinct from T(H)-17, T(H)1 and T(H)2 cells. Nat. Immunol. 2009;10:864–871. doi: 10.1038/ni.1770. [DOI] [PubMed] [Google Scholar]
- Wherry E.J., Teichgräber V., Becker T.C., Masopust D., Kaech S.M., Antia R., von Andrian U.H., Ahmed R. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat. Immunol. 2003;4:225–234. doi: 10.1038/ni889. [DOI] [PubMed] [Google Scholar]
- Widge A.T., Rouphael N.G., Jackson L.A., Anderson E.J., Roberts P.C., Makhene M., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., et al. mRNA-1273 Study Group Durability of responses after SARS-CoV-2 mRNA-1273 vaccination. N. Engl. J. Med. 2021;384:80–82. doi: 10.1056/NEJMc2032195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams M.A., Tyznik A.J., Bevan M.J. Interleukin-2 signals during priming are required for secondary expansion of CD8+ memory T cells. Nature. 2006;441:890–893. doi: 10.1038/nature04790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldemeskel B.A., Garliss C.C., Blankson J.N. SARS-CoV-2 mRNA vaccines induce broad CD4+ T cell responses that recognize SARS-CoV-2 variants and HCoV-NL63. J. Clin. Invest. 2021;131:e149335. doi: 10.1172/JCI149335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Wang A., Liu M., Wang Q., Chen J., Xia S., Ling Y., Zhang Y., Xun J., Lu L., et al. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. medRxiv. 2020 2020.03.30.20047365. [Google Scholar]
- Wurm H., Attfield K., Iversen A.K., Gold R., Fugger L., Haghikia A. Recovery from COVID-19 in a B-cell-depleted multiple sclerosis patient. Mult. Scler. 2020;26:1261–1264. doi: 10.1177/1352458520943791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhao J., Mangalam A.K., Channappanavar R., Fett C., Meyerholz D.K., Agnihothram S., Baric R.S., David C.S., Perlman S. Airway memory CD4(+) T cells mediate protective immunity against emerging respiratory coronaviruses. Immunity. 2016;44:1379–1391. doi: 10.1016/j.immuni.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.