Abstract
Staphylococcus aureus is a well-known foodborne pathogen. The aim of this study was to investigate the presence of S. aureus isolated from serving utensils in food processing environments in Mymensingh city, Bangladesh and to determine their antibiogram and resistance determinants. A total of 120 environmental samples were collected from different food settings. Isolation and identification were conducted using conventional biochemical tests. Molecular identification of isolates and detection of methicillin and vancomycin resistance were done using primer-specific polymerase chain reaction (PCR) targeting Tuf, nuc, mecA, and mecC genes. Antibiotic sensitivity tests were performed, and resistance genes were also detected by amplifying blaTEM, vanA, vanB, and vanC genes. Among the 120 samples, 81 (67.5%) were positive for Staphylococcus spp. and 41 (50.62%) were positive for the nuc-gene. Among the 41 isolates, 5 (12.20%) were positive for mecA, but none were positive for the mecC gene. A total of 12.2% of the isolates were vanC-positive, of which 4 isolates (9.76%) were also positive for the mecA gene. Antibiotic sensitivity testing revealed that all S. aureus isolates (100%) from hotel samples were sensitive to ciprofloxacin and chloramphenicol, 90.32% were sensitive to doxycycline, and 80.65% were sensitive to streptomycin. Conversely, all isolates (100%) were resistant to ampicillin, and 29.03% were resistant to vancomycin. All S. aureus isolates obtained from non-hotel samples were susceptible to chloramphenicol, ceftriaxone, ciprofloxacin, doxycycline, meropenem, and vancomycin; however, 40% of isolates were resistant to novobiocin. Among the hotel isolates, 29 (93.55%) of the ampicillin-resistant isolates harbored the blaTEM gene while 5 (55.55%) of the vancomycin-resistant isolates harbored the vanC gene. Four of the five vanC positive isolates were also positive for the mecA gene. The presence of methicillin-resistant S. aureus (MRSA) which is also vancomycin-resistant in food processing environments is a threat to public health. This is the first report on the molecular detection of methicillin and vancomycin-resistant S. aureus isolated from food processing environments in Bangladesh.
Keywords: Staphylococcus aureus, Vancomycin, Methicillin, One health, antimicrobial resistance
1. Introduction
Foodborne diseases represent a significant public health problem worldwide [1,2]. Good personal hygiene and proper sanitation are essential measures for protecting against foodborne diseases. Plates, dishes, glasses, cups, spoons, cutlery, and other serving utensils used in food service establishments can potentially spread foodborne diseases [3]. It is crucial to make sure that serving utensils are kept clean [4]. The apparent cleanliness of surfaces and utensils in a food processing environment does not necessarily indicate the absence of bacterial contamination [5]. Previous studies also reported that cleaning utensils and food contact surfaces contributed to cross-contamination in food service establishments [6].
Staphylococcal foodborne diseases are known to be significant threats to public health worldwide [7]. Staphylococci remain viable on hands and surfaces for a long time following the initial contact [8]. A minimal dose of staphylococcal enterotoxins can cause food poisoning in humans [2]. Staphylococcal enterotoxins caused more than 300 foodborne outbreaks in the European Union in 2012 [9]. Staphylococcal food poisoning is also a major cause of hospitalization in the U.S. every year [9,10]. Similarly, Staphylococcus aureus is responsible for a significant proportion of foodborne outbreaks in China [11].
Because of the acquisition of mecA gene, S. aureus can sometimes display methicillin-resistance, which may complicate therapy of infections in animals and humans [12]. Methicillin-resistant S. aureus (MRSA) strains have been detected in raw and cooked meat, milk, cheese, and other food products [[13], [14], [15], [16], [17]]. Food handlers in food service establishments are in close contact with food products, which can lead to contamination of the serving utensils.
Vancomycin is the recommended antibiotic for infections caused by MRSA. Vancomycin-resistant MRSA has been isolated for the first time more than two decades ago [18]. After this, vancomycin-resistant S. aureus (VRSA) has been isolated from more countries in North America, Europe, Asia, Africa, and South America [[19], [20], [21], [22], [23]].
There is very limited data on the microbiological safety of serving utensils in Bangladesh. Mymensingh is a densely populated city that has many educational institutions and historic places that attract local and international visitors. Due to its frequent food processing environments, it was selected as a suitable location for this study on the microbiological safety of serving utensils.
Staphylococcus aureus is not only a threat to human health but also to animals. Given that samples were taken from the environment, different aspects of the One Health concept were investigated in the present study. In addition to the isolation and molecular identification of S. aureus, we aimed to determine the antimicrobial resistance pattern of the isolated bacteria from serving utensils in food service establishments in Mymensingh city. We also detected the resistance genes using PCR. Results of this study could be used to develop a food sanitation strategy in the city, which involves both prevention and treatment. Results could also be used to plan a strategy for the use of drugs that can minimize resistant bacteria in the future.
2. Materials and methods
2.1. Sample collection and transportation
For sample collection, ten food service establishments were selected in Mymensingh city. A total of 120 samples were collected, including 40 plate samples, 40 glass samples, and 40 spoon samples. The term “hotel isolates” refers to S. aureus isolates obtained from food establishments that are located within hotels. The term “non-hotel isolates” refers to S. aureus isolates obtained from food establishments elsewhere. All utensils were ready to be served to the customers and were clean to the best of the knowledge of the owners of the food service establishment. We took precautions to minimize self-contamination by using sterile cotton buds to wipe the upper surfaces of plates, spoons, and inner surfaces of glass followed by dipping the cotton buds into sterile nutrient broth. In order to maintain a temperature of 4°C, the samples were immediately transferred to the laboratory and were incubated overnight at 37°C.
2.2. Isolation and identification
Following overnight incubation, the preliminary cultures were streaked onto Mannitol Salt agar (HiMedia, India) plates, which is specific for Staphylococcus spp. followed by incubation at 37°C for 24 h. All isolates were sub-cultured on Mannitol Salt agar to obtain pure colonies. For morphological confirmation, Gram staining was performed. For biochemical confirmation, catalase and coagulase tests were performed.
2.3. Molecular detection using genus-specific and species-specific primers
Bacterial DNA was extracted by the conventional boiling method [24], and the extracted DNA was used as a template DNA for the molecular detection of Staphylococcus spp. and S. aureus using specific primers (Table 1). To detect MRSA, the extracted DNA was amplified with specific primers as shown in Table 1. In all cases, the amplified products were electrophoresed at 60 V for one hour in 1.5% agarose gel [25]. After staining with ethidium bromide, the gel was examined under a UV transilluminator using the 100-bp/1-kb DNA ladder (Promega, USA).
Table 1.
Oligonucleotide primers used for the amplification of Staphylococcus aureus genes.
| Primer | Target gene |
Primer sequence (5′-3′) | Amplicon size (bp) | Reference |
|---|---|---|---|---|
| Tseq271 | Tuf | 5′-AAYATGATIACIGGIGCIGCICARATGGA-3′ | 884 | [28] |
| Tseq1138 | 5′-CCIACIGTICKICCRCCYTCRCG-3′ | |||
| nuc-F | nuc | 5′-GCGATTGATGGTGATACGGT-3′ | 279 | [29] |
| nuc-R | 5′-AGCCAAGCCTTGACGAACTAAAGC-3′ | |||
| mecA-F | mecA | 5′-AAAATCGATGGTAAAGGTTGG-3′ | 533 | [30] |
| mecA-R | 5′-AGTTCTGGCACTACCGGATTTTGC-3′ | |||
| mecC-P1 | mecC | 5′-GAAAAAAAGGCTTAGAACGCCTC-3′ | 138 | [31] |
| mecC-P2 | 5′-GAAGATCTTTTCCGTTTTCAGC-3′ | |||
| blaTEM-1-F | blaTEM | 5′-CATTTCCGTGTCGCCCTTAT-3′ | 793 | [32] |
| blaTEM-1-R | 5′-TCCATAGTTGCCTGACTCCC-3′ | |||
| vanA-F2 | vanA | 5′-AATGTGCGAAAAACCTTGCG-3′ | 677 | [33] |
| vanA-R2 | 5′-CCGTTTCCTGTATCCGTCC-3′ | |||
| vanB-F2 | vanB | 5′-GCTCCGCAGCCTGCATGGA-3′ | 463 | [34] |
| vanB-R2 | 5′-ACGATGCCGCCATCCTCCT-3′ | |||
| vanC1-F | vanC | 5′-GAAAGACAACAGGAAGACCGC-3′ | 796 | [34] |
| vanC1-R | 5′-TCGCATCACAAGCACCAATC-3′ |
2.4. Antibiogram test
The following ten commonly-prescribed antibiotics (HiMedia, India) were used for antimicrobial susceptibility testing: ampicillin (AMP, 10 mg), chloramphenicol (C, 30 mg), ceftriaxone (CTR, 30 mg), ciprofloxacin (CIP, 30 mg), doxycycline (DO, 30 mg), meropenem (MEM, 10 mg), novobiocin (NV, 30 mg), streptomycin (S, 10 mg), tetracycline (TE, 30 mg), and vancomycin (VA, 30 mg). For each culture suspension of the bacterial isolates, a McFarland 0.5 standard was maintained [26]. Antibiogram phenotyping was performed using the disk diffusion method in Mueller Hinton agar media (HiMedia, India). Results were recorded as sensitive, intermediate, or resistant in accordance with the CLSI recommendations [27].
2.5. Molecular detection of resistance genes
Phenotypically-resistant S. aureus isolates were screened for antibiotic resistance genes using PCR using specific primers (Table 1). The amplified products were processed as in section 2.3.
3. Results
Out of the 120 samples, 81 (67.5%) were positive for Staphylococcus spp. by observing the color change on Mannitol Salt agar plates, microscopic examination, and PCR using genus-specific Tuf gene. Using PCR amplification of the nuc gene, 41 (50.62%) out of the 81 isolates were positive for the nuc gene (Table 2). The highest rate was obtained from the hotel glass samples (18.52%), and the lowest rate was obtained from the non-hotel spoons (1.23%)
Table 2.
The percentage of nuc-gene positive samples in Staphylococcus aureus isolates in the current study.
| Study area | Staphylococcus-positive | Number (%) |
S. aureus-positive (nuc-positive) |
Number (%) |
|---|---|---|---|---|
| Hotel Food Establishments (N = 60) |
15 (Spoons) | 15/120 (12.5) | 10 | 10/81 (12.35) |
| 20 (Glass) | 20/120 (16.67) | 15 | 15/81 (18.52) | |
| 18 (Plates) | 18/120 (15.0) | 6 | 6/81 (7.41) | |
| Non-hotel Food Establishments (N = 60) |
6 (Spoons) | 6/120 (5.0) | 1 | 1/81 (1.23) |
| 12 (Glass) | 12/120 (10.0) | 4 | 4/81 (4.94) | |
| 10 (Plates) | 10/120 (12.0) | 5 | 5/81 (6.17) | |
| Total samples = 120 | 81 | 81/120 (67.5) | 41 | 41/81 (50.62) |
The 41 isolates were further analyzed by PCR using the specific primers mecA and mecC for MRSA and five were found to be positive for the mecA gene (Table 3). None were positive for the mecC gene. Among the five isolates positive for mecA, four (9.76%) were obtained from hotel glass samples and one (2.44%) was obtained from spoon samples (Table 3). Among the non-hotel nuc-positive isolates, none was positive for mecA or vanC. Among the hotel nuc-positive isolates, four isolates were positive for both mecA and vanC, while one isolate was only positive for mecA, and another isolate was only positive for vanC (Table 3).
Table 3.
The percentage of mecA-positive Staphylococcus aureus isolates in the current study.
| Study area | nuc-positive | mecA-positive (%) | vanC-positive (%) |
|---|---|---|---|
| Hotel Food Establishments |
10 (Spoons) | 1 (2.44) | 0 (0) |
| 15 (Glass) | 4 (9.76) | 4 (9.76) | |
| 6 (Plates) | 0 (0) | 1 (2.44) | |
| Non-hotel Food Establishments |
1 (Spoons) | 0 (0) | 0 (0) |
| 4 (Glass) | 0 (0) | 0 (0) | |
| 5 (Plates) | 0 (0) | 0 (0) | |
| Total | 41 | 5 (12.20) | 5 (12.20) |
The nuc-gene positive isolates were tested for antibiotic susceptibility using ten commonly prescribed antibiotics. Among 31 S. aureus isolates from hotel samples, 100% were resistant to ampicillin, 45.16% were resistant to novobiocin, and 29.03% were resistant to vancomycin. All isolates were sensitive to chloramphenicol and ciprofloxacin while 90.32% and 80.65% of the isolates were sensitive to doxycycline and streptomycin, respectively. Among the 10 nuc-gene positive non-hotel isolates, four (40%) were resistant to novobiocin, while six (60%) were sensitive to novobiocin. All other isolates exhibited sensitive or intermediate resistance to chloramphenicol, ceftriaxone, ciprofloxacin, doxycycline, meropenem, and vancomycin (Table 4, Fig. 1, Fig. 2). A total of 29 out of the 31 (93.55%) ampicillin-resistant isolates (as detected phenotypically) were positive for the blaTEM gene. Vancomycin resistance genes were detected by PCR using vanA, vanB, and vanC primers. Results showed that none of the isolates harbored vanA or vanB genes. Five (55.55%) isolates harbored the vanC-resistance gene. Four isolates which were methicillin resistant (as detected phenotypically) were found to harbor the vanC gene. To the best of our knowledge, this is the first report to detect the vanC resistance gene in S. aureus isolated from food service settings in Bangladesh.
Table 4.
Antibiotic resistance profile of Staphylococcus aureus isolates in the current study
| Antimicrobial agent | Number of isolates and their antibiotic susceptibility patterns (%) |
|||||
|---|---|---|---|---|---|---|
| Resistant |
Intermediate |
Sensitive |
||||
| Hotel (n = 31) |
Non-hotel (n = 10) |
Hotel (n = 31) |
Non-hotel (n = 10) |
Hotel (n = 31) |
Non-hotel (n = 10) |
|
| Ampicillin | 31 (100%) | – | – | 6 (60%) | – | 4 (40%) |
| Chloramphenicol | – | – | – | – | 31 (100%) | 10 (100%) |
| Ceftriaxone | 12 (38.71%) | – | – | – | 19 (61.29%) | 10 (100%) |
| Ciprofloxacin | – | – | – | – | 31 (100%) | 10 (100%) |
| Doxycycline | 1 (3.23%) | – | 2 (6.45%) | – | 28 (90.32%) | 10 (100%) |
| Meropenem | – | – | 10 (32.26%) | – | 21 (67.74%) | 10 (100%) |
| Novobiocin | 14 (45.16%) | 4 (40%) | 3 (9.67%) | – | 14 (45.16%) | 6 (60%) |
| Streptomycin | – | – | 6 (19.35%) | 2 (20%) | 25 (80.65%) | 8 (80%) |
| Tetracycline | – | – | 14 (45.16%) | 5 (50%) | 17 (54.84%) | 5 (50%) |
| Vancomycin | 9 (29.03%) | – | – | – | 22 (70.97%) | 10 (100%) |
Fig. 1.

Antibiotic Susceptibility patterns of hotel Staphylococcus aureus isolates.
Fig. 2.
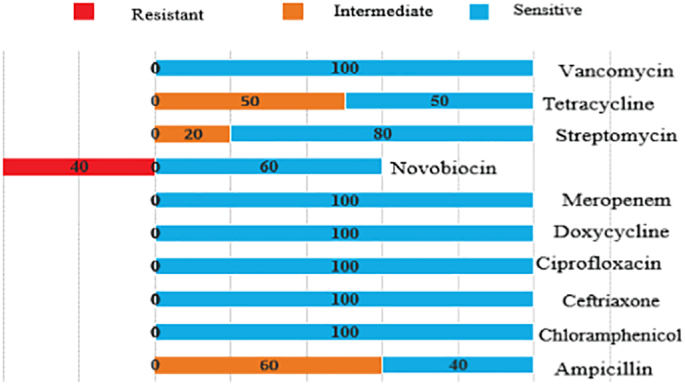
Antibiotic susceptibility patterns of non-hotel Staphylococcus aureus isolates.
Multidrug-resistance (MDR) to two, three, four, or five antibiotic classes was detected in 19 out of 31 (61.3%) nuc-gene positive isolates. Only one isolate (3.2%) was resistant to five classes of antibiotics, three (9.67%) isolates were resistant to four classes of antibiotics, seven (22.58%) isolates were resistant to three classes of antibiotics, and eight (25.81%) isolates were resistant to two classes of antibiotics.
4. Discussion
Methicillin-resistant S. aureus (MRSA) causes zoonotic infections that have both public health and veterinary importance, especially with the MDR patterns that can be associated with MRSA. MRSA causes food poisoning, suppurative pneumonia, pyogenic endocarditis, and otitis media infections in humans [35]. It causes a variety of infections in animals including botryomycosis and localized purulent infection in horses, localized pyogenic infection and severe acute mastitis in cattle, pustular dermatitis and food poisoning in dogs and cats [35]. In addition, it causes greasy pig disease in swine and bumble-foot disease in birds [35]. Through skin infections and other routes, there can be human-to-animal and animal-to-human transmission of MRSA. The excessive use of antibiotics in the veterinary sector can result in MRSA-infected animal reservoirs that can cause zoonotic MRSA infections in humans [35]. To fully control MRSA infections, a One Health approach that considers animals and humans in addition to the environment needs to be implemented.
Multidrug-resistant MRSA has been detected in animals and animal-derived food products (such as frozen chicken meat and processed raw meat) in Bangladesh. [36, 37]. In addition, S. aureus has been isolated from raw milk samples in Bangladesh, in which mastitis is known to be a major problem [38]. Contamination of milk can happen while milking and is thus dependent on the cleanliness of the environment and the utensils that are used for the milking process. This highlights the impact of the environment, which is the third component of the One Health triangle.
MRSA can also be transmitted between humans and their companion animals or even by stray dogs and cats. MRSA was isolated from dogs and cats [39,40] and from ornamental birds [41] in Bangladesh. Some of the isolates from ornamental birds were also found to be resistant to vancomycin [41].
Cross-contamination of food due to improper handling and non-hygienic utensils can cause foodborne outbreaks and infections [42]. Sources of contamination include human hands, breath, hair, raw food products, and food processing environments. People suffering from respiratory tract infections, or gastrointestinal disorders should not handle food. As S. aureus can survive for a long period in the respiratory tract of infected patients, on cloths, and in other environments, there is a possibility of contaminating food processing environments as well as the serving utensils. The pathogenic S. aureus can also adhere to gloves of employees in food processing establishments and can be transmitted to other humans or to the environment if gloves are not changed frequently [43]. It is important to ensure that the nasopharyngeal and oropharyngeal tracts of food handlers are free of S. aureus in order to prevent any foodborne outbreaks [[44], [45], [46]]. Utensils can also become contaminated with S. aureus from food products including raw meat and vegetables. Dairy cows, chickens, and pigs are known to have the ability to transmit MRSA to humans [47]. Improper food handling by infected personnel is responsible for 20% of bacterial foodborne illnesses [48]. An earlier study reported a carriage rate of 79% in S. aureus among food handlers [49].
In order to prevent infections, there needs to be daily cleaning and disinfection of tables and environments in which food is served A major concern with S. aureus is its biofilm production ability, which increases its tolerance to dryness and dehydration, which helps it to persist and multiply [50,51]. A study reported that 34.3% of kitchen sponges from food establishments were positive for S. aureus. Moreover, S. aureus was isolated from 30% of the kitchen sponges of restaurants, 36.4% of the kitchen sponges of hotels, 33.3% of the kitchen sponges of pastry shops, and 34% of the kitchen sponges of cafeterias [52].
Other potential sources of cross-contamination include the use of paper bills and coins. The older the paper bill or coin is, the more the microbial contamination [53]. A study conducted in Bangladesh reported that the majority of paper bills were contaminated with Staphylococcus spp. [54] Another report indicated that bacterial contamination of paper bills in Bangladesh was as high as 93.7% [55]. In Nigeria, the rate of contamination of paper bills with S. aureus was reproted to be 22.5% [56]. Contaminated paper bills can lead to cross-contamination of food items or serving utensils. Flies can act as mechanical vectors for food contamination, and microorganisms such as E. coli, Vibrio cholera, and S. aureus have been isolated from house flies. It is important to note that dishwashing detergents were found to have different capacities to eliminate bacteria in kitchen sponges [57]. Bacteria can remain on hands and utensils for up to several days after the initial contact.
The emergence of antibiotic-resistant S. aureus such as MRSA and VRSA is a worldwide public health threat [58,59]. Our data showed that S. aureus is prevalent in serving utensils in food processing environments. Many isolates in our study were found to be antibiotic-resistant and can be a potential source of transmission of resistance to humans. Serving utensils played a significant role in transmission of multidrug-resistant staphylococci detected in the present study. Continuous monitoring and surveillance for antibiotic resistance are key for the control and treatment of MRSA infections. A multifaceted One-Health approach involving public health and veterinary consultants in addition to environmental microbiologists needs to be implemented for the effective control of MRSA infections.
Funding
This research received no external funding.
Declaration of Competing Interest
The authors declare no conflict of interest.
Contributor Information
Ahosanul H. Shahid, Email: shahid41192@bau.edu.bd.
K.H.M. Nazmul Hussain Nazir, Email: nazir@bau.edu.bd.
Mohamed E. El Zowalaty, Email: elzow005@gmail.com.
Ajran Kabir, Email: dvm47012@bau.edu.bd.
Shahjahan A. Sarker, Email: msarker24297@bau.edu.bd.
Hossam M. Ashour, Email: hossamking@mailcity.com.
References
- 1.Balaban N., Rasooly A. Staphylococcal enterotoxins. Int. J. Food Microbiol. 2000;61(1):1–10. doi: 10.1016/s0168-1605(00)00377-9. [DOI] [PubMed] [Google Scholar]
- 2.Le Loir Y., Baron F., Gautier M. Staphylococcus aureus and food poisoning. Genet. Mol. Res. 2003;2(1):63–76. [PubMed] [Google Scholar]
- 3.Fawole M., Oso B. Spectrum Book Ltd; Ibadan: 1988. Laboratory Manual of Microbiology; pp. 22–45. [Google Scholar]
- 4.Nizame F., Alam M.U., Masud A.A., Shoab A.K., Opel A., Islam M.K.…Unicomb L. Hygiene in restaurants and among street food vendors in Bangladesh. The American journal of tropical medicine and hygiene. 2019;101(3):566–575. doi: 10.4269/ajtmh.18-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garden-Robinson J. North Dakota State University; ND, USA: 2007. Food Safety Basics: A Reference Guide for Foodservice Operators.https://library.ndsu.edu/ir/bitstream/handle/10365/5187/fn572.pdf?sequence=1 Availabe at: [Google Scholar]
- 6.Scott E., Bloomfield S.F., Barlow C. An investigation of microbial contamination in the home. Epidemiol. Infect. 1982;89(2):279–293. doi: 10.1017/s0022172400070819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hennekinne J.-A., De Buyser M.-L., Dragacci S. Staphylococcus aureus and its food poisoning toxins: characterization and outbreak investigation. FEMS Microbiol. Rev. 2012;36(4):815–836. doi: 10.1111/j.1574-6976.2011.00311.x. [DOI] [PubMed] [Google Scholar]
- 8.Kusumaningrum H., Van Putten M., Rombouts F., Beumer R. Effects of antibacterial dishwashing liquid on foodborne pathogens and competitive microorganisms in kitchen sponges. J. Food Prot. 2002;65(1):61–65. doi: 10.4315/0362-028x-65.1.61. [DOI] [PubMed] [Google Scholar]
- 9.Macori G., Bellio A., Bianchi D.M., Gallina S., Adriano D., Zuccon F., Chiesa F., Acutis P.L., Casalinuovo F., Decastelli L. Molecular typing of Staphylococcus aureus isolate responsible for staphylococcal poisoning incident in homemade food. Ital. J. Food Saf. 2016:5(2). doi: 10.4081/ijfs.2016.5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Byrd-Bredbenner C., Berning J., Martin-Biggers J., Quick V. Food safety in home kitchens: a synthesis of the literature. Int. J. Environ. Res. Public Health. 2013;10(9):4060–4085. doi: 10.3390/ijerph10094060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X., Li G., Xia X., Yang B., Xi M., Meng J. Antimicrobial susceptibility and molecular typing of methicillin-resistant Staphylococcus aureus in retail foods in Shaanxi, China. Foodborne Pathog. Dis. 2014;11(4):281–286. doi: 10.1089/fpd.2013.1643. [DOI] [PubMed] [Google Scholar]
- 12.Fooladi A.A.I., Ashrafi E., Tazandareh S.G., Koosha R.Z., Rad H.S., Amin M., Soori M., Larki R.A., Choopani A., Hosseini H.M. The distribution of pathogenic and toxigenic genes among MRSA and MSSA clinical isolates. Microb. Pathog. 2015;81:60–66. doi: 10.1016/j.micpath.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 13.Otalu O., Junaidu K., Chukwudi O.E., Jarlath U.V. Multi-drug resistant coagulase positive Staphylococcus aureus from live and slaughtered chickens in Zaria, Nigeria. Int. J. Poult. Sci. 2011;10(11):871–875. [Google Scholar]
- 14.Attien P., Sina H., Moussaoui W., Dadié T., Djéni T., Bankole H., Kotchoni S., Edoh V., Prévost G., Baba-Moussa L. Prevalence and antibiotic resistance of Staphylococcus strains isolated from meat products sold in Abidjan streets (Ivory Coast) Afr. J. Microbiol. Res. 2013;7(26):3285–3293. [Google Scholar]
- 15.Chairat S., Gharsa H., Lozano C., Gómez-Sanz E., Gómez P., Zarazaga M., Boudabous A., Torres C., Ben Slama K. Characterization of Staphylococcus aureus from raw meat samples in Tunisia: detection of clonal lineage ST398 from the African continent. Foodborne Pathog. Dis. 2015;12(8):686–692. doi: 10.1089/fpd.2015.1958. [DOI] [PubMed] [Google Scholar]
- 16.Al-Ashmawy M.A., Sallam K.I., Abd-Elghany S.M., Elhadidy M., Tamura T. Prevalence, molecular characterization, and antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus isolated from milk and dairy products. Foodborne Pathog. Dis. 2016;13(3):156–162. doi: 10.1089/fpd.2015.2038. [DOI] [PubMed] [Google Scholar]
- 17.Ben Said M., Abbassi M., Bianchini V., Sghaier S., Cremonesi P., Romanò A., Gualdi V., Hassen A., Luini M. Genetic characterization and antimicrobial resistance of Staphylococcus aureus isolated from bovine milk in Tunisia. Lett. Appl. Microbiol. 2016;63(6):473–481. doi: 10.1111/lam.12672. [DOI] [PubMed] [Google Scholar]
- 18.Chambers H.F. The changing epidemiology of Staphylococcus aureus? Emerg. Infect. Dis. 2001;7(2):178. doi: 10.3201/eid0702.010204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chesneau O., Morvan A., Solh N.E. Retrospective screening for heterogeneous vancomycin resistance in diverse Staphylococcus aureus clones disseminated in French hospitals. J. Antimicrob. Chemother. 2000;45(6):887–890. doi: 10.1093/jac/45.6.887. [DOI] [PubMed] [Google Scholar]
- 20.Ferraz V., Duse A., Kassel M., Black A., Ito T., Hiramatsu K. Vancomycin-resistant Staphylococcus aureus occurs in South Africa. South African medical journal= Suid-Afrikaanse tydskrif vir geneeskunde. 2000;90(11):1113. [PubMed] [Google Scholar]
- 21.Hood J., Edwards G.F., Cosgrove B., Curran E., Morrison D., Gemmell C.G. Vancomycin-intermediate Staphylococcus aureus at a Scottish hospital. J. Infect. 2000;40(2):A11. [Google Scholar]
- 22.Kim M.-N., Pai C.H., Woo J.H., Ryu J.S., Hiramatsu K. Vancomycin-intermediate Staphylococcus aureus in Korea. J. Clin. Microbiol. 2000;38(10):3879–3881. doi: 10.1128/jcm.38.10.3879-3881.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ploy M.C., Grélaud C., Martin C., de Lumley L., Denis F. First clinical isolate of vancomycin-intermediate Staphylococcus aureus in a French hospital. Lancet. 1998;351(9110):1212. doi: 10.1016/s0140-6736(05)79166-2. [DOI] [PubMed] [Google Scholar]
- 24.Queipo-Ortuño M.I., Colmenero J.D.D., Macias M., Bravo M.J., Morata P. Preparation of bacterial DNA template by boiling and effect of immunoglobulin G as an inhibitor in real-time PCR for serum samples from patients with brucellosis. Clin. Vaccine Immunol. 2008;15(2):293–296. doi: 10.1128/CVI.00270-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J., Russell D., Russell D. Molecular cloning, a laboratory manual (3-volume set) Cold spring harbor laboratory press Cold Spring Harbor, New York (2001) In: Sterer N., Hendler A., Davidi M.P., Rosenberg M.A., editors. novel microscopic assay for oral malodour-related microorganisms. J. Breath. Res. 2008. 2(026003. [DOI] [PubMed] [Google Scholar]
- 26.Hudzicki J. 2009. Kirby-Bauer Disk Diffusion Susceptibility Test Protocol. [Google Scholar]
- 27.CLSI. Performance Standards for Antimicrobial Susceptibility Testing: Wayne, PA, USA, 2016., 17th Informational Supplement; ed.; Clinical and Laboratory Standards Institute.
- 28.Martineau F., Picard F.J., Ke D., Paradis S., Roy P.H., Ouellette M., Bergeron M.G. Development of a PCR assay for identification of staphylococci at genus and species levels. J. Clin. Microbiol. 2001;39(7):2541–2547. doi: 10.1128/JCM.39.7.2541-2547.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kalorey D.R., Shanmugam Y., Kurkure N.V., Chousalkar K.K., Barbuddhe S.B. PCR-based detection of genes encoding virulence determinants in Staphylococcus aureus from bovine subclinical mastitis cases. J. Vet. Sci. 2007;8(2):151–154. doi: 10.4142/jvs.2007.8.2.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee J.H. Methicillin (oxacillin)-resistant Staphylococcus aureus strains isolated from major food animals and their potential transmission to humans. Appl. Environ. Microbiol. 2003;69(11):6489–6494. doi: 10.1128/AEM.69.11.6489-6494.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stegger M., Andersen P.S., Kearns A., Pichon B., Holmes M.A. Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecALGA251. Clin. Microbiol. Infect. 2003;18:395–400. doi: 10.1111/j.1469-0691.2011.03715.x. [DOI] [PubMed] [Google Scholar]
- 32.Walker R.A., Lindsay E., Woodward M.J., Ward L.R., Threlfall E.J. Variation in clonality and antibiotic-resistance genes among multiresistant Salmonella enterica serotype typhimurium phage-type U302 (MR U302) from humans, animals, and foods. Microb. Drug Resist. 2001;7(1):13–21. doi: 10.1089/107662901750152701. [DOI] [PubMed] [Google Scholar]
- 33.Lu J.J., Perng C.L., Chiueh T.S., Lee S.Y., Chen C.H., Chang F.Y., Wang C.C., Chi W.M. Detection and typing of vancomycin-resistance genes of enterococci from clinical and nosocomial surveillance specimens by multiplex PCR. Epidemiol. & Infect. 2001;126(3):357–363. doi: 10.1017/s0950268801005453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lemcke R., Bülte M. Occurrence of the vancomycin-resistant genes vanA, vanB, vanC1, vanC2 and vanC3 in Enterococcus strains isolated from poultry and pork. Int. J. Food Microbiol. 2000;60(2–3):185–194. doi: 10.1016/s0168-1605(00)00310-x. [DOI] [PubMed] [Google Scholar]
- 35.Algammal A.M., Hetta H.F., Elkelish A., Alkhalifah D.H., Hozzein W.N., Batiha G.E., El Nahhas N., Mabrok M.A. Methicillin-resistant Staphylococcus aureus (MRSA): one health perspective approach to the bacterium epidemiology, virulence factors, antibiotic-resistance, and zoonotic impact. Infect. and Drug Resist. 2020;13:3255. doi: 10.2147/IDR.S272733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parvin M., Ali M., Talukder S., Nahar A., Chowdhury E.H., Rahman M., Islam M. Prevalence and multidrug resistance pattern of methicillin resistant S. aureus isolated from frozen chicken meat in Bangladesh. Microorganisms. 2021;9(3):636. doi: 10.3390/microorganisms9030636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Islam M.A., Parveen S., Rahman M., Huq M., Nabi A., Khan Z.U., Ahmed N., Wagenaar J.A. Occurrence and characterization of methicillin resistant Staphylococcus aureus in processed raw foods and ready-to-eat foods in an urban setting of a developing country. Front. Microbiol. 2019;10:503. doi: 10.3389/fmicb.2019.00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Islam M.A., Kabir S.M., Rahman M.T. Molecular detection and characterization of Staphylococcus aureus isolated from raw milk sold in different markets of Bangladesh. Bangladesh J. Vet. Med. 2016;14(2):277–282. [Google Scholar]
- 39.Rahman M.M., Amin K.B., Rahman S.M., Khair A., Rahman M., Hossain A., Rahman A.K., Parvez M.S., Miura N., Alam M.M. Investigation of methicillin-resistant Staphylococcus aureus among clinical isolates from humans and animals by culture methods and multiplex PCR. BMC Vet. Res. 2018;14(1):1–6. doi: 10.1186/s12917-018-1611-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Habibullah A., Rahman A.M., Haydar M.R., Nazir K.H., Rahman M.T. Prevalence and molecular detection of methicillin-resistant Staphylococcus aureus from dogs and cats in Dhaka City. Bangladesh J. Vet. Med. 2017;15(1):51–57. [Google Scholar]
- 41.Zaman S.B., Sobur M.A., Hossain M.J., Pondit A., Khatun M.M., Choudhury M.A., Tawyabur M., Rahman M.T. Molecular detection of methicillin-resistant Staphylococcus aureus (MRSA) in ornamental birds having public health significance. J. Bangladesh Agri. Univ. 2020;18:415–420. [Google Scholar]
- 42.Tirado M.C., Schmidt K. Vol. 43. World Health Organization. The Journal of Infection; 2001. WHO surveillance programme for control of foodborne infections and intoxications: preliminary results and trends across greater Europe; pp. 80–84. [DOI] [PubMed] [Google Scholar]
- 43.Lues J., Van Tonder I. The occurrence of indicator bacteria on hands and aprons of food handlers in the delicatessen sections of a retail group. Food Control. 2007;18(4):326–332. [Google Scholar]
- 44.Todd E.C., Greig J.D., Bartleson C.A., Michaels B.S. Outbreaks where food workers have been implicated in the spread of foodborne disease. Part 5. Sources of contamination and pathogen excretion from infected persons. J. Food Prot. 2008;71(12):2582–2595. doi: 10.4315/0362-028x-71.12.2582. [DOI] [PubMed] [Google Scholar]
- 45.Todd E.C., Greig J.D., Michaels B.S., Bartleson C.A., Smith D., Holah J. Outbreaks where food workers have been implicated in the spread of foodborne disease. Part 11. Use of antiseptics and sanitizers in community settings and issues of hand hygiene compliance in health care and food industries. J. Food Prot. 2010;73(12):2306–2320. doi: 10.4315/0362-028x-73.12.2306. [DOI] [PubMed] [Google Scholar]
- 46.Gallina S., Bianchi D., Bellio A., Nogarol C., Macori G., Zaccaria T., Biorci F., Carraro E., Decastelli L. Staphylococcal poisoning foodborne outbreak: epidemiological investigation and strain genotyping. J. Food Prot. 2013;2093-2098:76(12). doi: 10.4315/0362-028X.JFP-13-190. [DOI] [PubMed] [Google Scholar]
- 47.Khanna T., Friendship R., Dewey C., Weese J. Methicillin resistant Staphylococcus aureus colonization in pigs and pig farmers. Vet. Microbiol. 2008;128(3–4):298–303. doi: 10.1016/j.vetmic.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 48.Food U. Silva Spring, MD; Author: 2000. Report of the FDA Retail Food Program Database of Foodborne Illness Risk Factors. [Google Scholar]
- 49.Sezer Ç., Özgür Ç., Aksem A., Leyla V. Food handlers: a bridge in the journey of enterotoxigenic MRSA in food. J. Verbr. Lebensm. 2015;10(2):123–129. [Google Scholar]
- 50.Møretrø T., Hermansen L., Holck A.L., Sidhu M.S., Rudi K., Langsrud S. Biofilm formation and the presence of the intercellular adhesion locus Ica among staphylococci from food and food processing environments. Appl. Environ. Microbiol. 2003;69(9):5648–5655. doi: 10.1128/AEM.69.9.5648-5655.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elkhatib W.F., Khairalla A.S., Ashour H.M. Evaluation of different microtiter plate-based methods for the quantitative assessment of Staphylococcus aureus biofilms. Future Microbiol. 2014;9(6):725–735. doi: 10.2217/fmb.14.33. [DOI] [PubMed] [Google Scholar]
- 52.Wolde T., Bacha K., Abate M., Sileshi H. Prevalence and antibiotics resistance patterns of Staphylococcus aureus Isolated from Kitchen Sponge’s at Jimma Town Food Estab-lishments, south West Ethiopia. Int J Res. Stud. Biosci. 2015;3(7):63–71. [Google Scholar]
- 53.El-Din El-Dars F.M., Hassan W.M. A preliminary bacterial study of Egyptian paper money. Int. J. Environ. Health Res. 2005;15(3):235–240. doi: 10.1080/09603120500105976. [DOI] [PubMed] [Google Scholar]
- 54.Barua N., Sabuj A.A.M., Haque Z.F., Das M., Hossain M.T., Saha S. Survey of bacterial contamination and antibiotic resistance pattern of Bangladeshi paper currency notes in Mymensingh city. Afr. J. Microbiol. Res. 2019;13(10):206–213. [Google Scholar]
- 55.Akond M.A., Alam S., Zohora F.T., Mutahara M., Rashed Noor M. Assessment of bacterial contamination of paper currency notes in Bangladesh. ESAIJ. 2015;10(3):114–120. [Google Scholar]
- 56.Yazah A., Yusuf J., Agbo A. Bacterial contaminants of Nigerian currency notes and associated risk factors. Res. J. Med. Sci. 2012;6(1):1–6. [Google Scholar]
- 57.Nielsen P., Brumbaugh E., Kananen L. Evaluation of the use of liquid dishwashing compounds to control bacteria in kitchen sponges. J. AOAC Int. 2002;85(1):107–112. [PubMed] [Google Scholar]
- 58.Khairalla A.S., Wasfi R., Ashour H.M. Carriage frequency, phenotypic, and genotypic characteristics of methicillin-resistant Staphylococcus aureus isolated from dental health-care personnel, patients, and environment. Sci. Rep. 2017;7(1):1–16. doi: 10.1038/s41598-017-07713-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ashour H.M., El-Sharify A. 25(36) 2007. Microbial spectrum and antibiotic susceptibility profile of Gram-positive aerobic bacteria isolated from cancer patients; pp. 5763–5769. [DOI] [PubMed] [Google Scholar]


