Abstract
Ovarian cancer (OC) is one of the most lethal type of cancer in women due to a lack of effective targeted therapies and high rates of treatment resistance and disease recurrence. Recently Poly (ADP-ribose) polymerase inhibitors (PARPi) have shown promise as chemotherapeutic agents; however, their efficacy is limited to a small fraction of patients with BRCA mutations. Here we show a novel function for the Hedgehog (Hh) transcription factor Glioma associated protein 1 (GLI1) in regulation of key Fanconi anemia (FA) gene, FANCD2 in OC cells. GLI1 inhibition in HR-proficient OC cells induces HR deficiency (BRCAness), replication stress and synergistic lethality when combined with PARP inhibition. Treatment of OC cells with combination of GLI1 and PARP inhibitors shows enhanced DNA damage, synergy in cytotoxicity, and strong in vivo anticancer responses.
Keywords: GLI1, Ovarian Cancer, FANCD2, GANT61, Olaparib, and Homologous recombination deficiency
INTRODUCTION
Physiological and balanced activation of hedgehog (Hh) signaling through the effector GLI transcription factors (GLI1, GLI2 and GLI3) regulates tissue patterning, stem cell maintenance and differentiation [1]. However, in tumors, GLI1 can be activated through canonical Hh ligand-dependent mechanisms or non-canonical oncogenic signaling cascades [2]. This aberrant (non-homeostatic) Hh/GLI1 signaling has been documented in many cancers including ovarian, breast, lung, colon, skin, brain tumors and correlates with aggressive disease [[3], [4], [5], [6], [7], [8], [9]]. We previously demonstrated that GLI1 regulates the S-phase checkpoint in tumor cells by regulating transcription of Bid, and inhibition of GLI1 by small molecule inhibitor GANT61 or siRNA blocks CHK1 phosphorylation and sensitizes cells to topoisomerase inhibitor [10]. Although GLI1 inhibition or downregulation in tumor cells alone elicits replication associated DNA damage response (DDR) [10], the role for aberrant GLI1 in promoting oncogenic replication has not been studied.
The Fanconi anemia (FA)-BRCA tumor suppressor pathway is a multi-gene component consists of multiple (at least 22) components (FA-BRCA proteins), including FANCD2, breast and ovarian cancer early onset genes BRCA1 and BRCA2, and eukaryotic recombinase RAD51, and disruption of this pathway greatly increases incidence of genomic instability and cancer [11]. FANCD2 is the effector protein, which upon monoubiquitination by FA-core complex localizes to the nucleus and forms discrete foci at stalled or damaged replication forks [[12], [13], [14], [15]], where it associates with several replication and repair factors and inhibits carcinogenesis [16]. However, upregulation of DDR and repair proteins including FANCD2 has been shown to promote tumor cell survival, progression and chemoresistance in many tumors [17,18].
OC is the most lethal forms of malignant diseases affecting women. Currently, cytotoxic chemotherapy is the major option for treating these patients. Recently, several PARP inhibitors (PARPi) were approved to treat ovarian, breast, prostate and pancreatic cancer patients with germline BRCA mutation or HR deficiency [19]. PARP is required to repair DNA single strand breaks and PARPi allow their conversion into lethal double strand breaks (DSB) during DNA replication [20]. BRCA-negative cells that are deficient in other HR genes cannot repair DSB making these cells highly susceptible to PARPi, a condition known as synthetic lethality. However, the major limitation of PARPi treatment is that only a small fraction of (10 to 15%) OC patients have germline BRCA mutations, and some BRCA mutations do not affect HR or cause synthetic lethality with PARPi [21,22]. Thus, the efficacy of PARPi would be greatly expanded if they could be combined with another agent that specifically induces HR deficiency in tumor cells, resulting in synthetic lethality.
Here, we demonstrate that the aberrantly activated Hh/GLI1 in OC promotes oncogenic replication and cell survival by transcriptionally regulating FANCD2. GLI1 downregulation or inhibition in these cells induces replication stress, spontaneous DNA lesions and cell death. Inhibition of Hh/GLI1 in BRCA-proficient ovarian tumor cells induces HR deficiency and synergistic lethality with PARPi, while not significantly impacting normal ovarian cells.
Materials and Methods
Cell culture
Normal immortalized human FTEC cells [23] and human OC cell line A2780 cells were grown in RPMI media. Human serous adenocarcinoma cell lines OV90, OVCAR-4 and OVCAR-5 were grown in DMEM media. Human ovarian adenocarcinoma cell line SKOV3 was cultured in DMEM:F12 media. All the cells were supplemented with 10% FBS, 100 μg/ml streptomycin sulfate and 100 U/ml penicillin. All the cells were authenticated and regularly monitored for mycoplasma contamination.
Transfection of siRNA and plasmids
The siRNAs used in this study were purchased from Dharmacon and transfected twice with 24 hours difference using Lipofectamine 2000 (Cat No: 11668019, Invitrogen) as described previously [10]. The siRNAs used in this study are as follows: siControl (non-targeting control siRNA; D-001210-01), siGLI1-1 (GGAAAUGACUGGCAAUGCA), siGLI1-2 (GUCCUCGACUUGAACAUUA), siGLI1-5 (GCACUGGUCUGUCCACUCU) [10]. For GLI1 down-regulation, the data using siGLI1-5 sequences were presented in these studies unless otherwise mentioned. GLI1 (Myc- tagged) plasmid (Cat No: RC201110) was procured from Origene and transfected using Lipofectamine 2000 according to the manufacturer's protocols.
Colony assay
For experiments using siRNA knockdown, 48 hours after siGLI1 or siControl transfection, cells were treated with indicated concentrations of olaparib for 24 hours. The small molecule inhibitors GANT61 and olaparib were administered alone or in combination for 72 hours. After drug treatment, cells were washed thrice with PBS and were allowed to form colonies for 10-14 days. Colonies were fixed in ice-cold methanol and stained with crystal violet and colonies with more than 25 cells were counted using an imaging system (Gene Tools, Syngene). The experiments were done in triplicate and repeated three times.
Sphere formation assay
A2780 cells were trypsinized, dispersed into single cells and approximately 10,000 cells were seeded per well in ultra-low attachment 6 well dishes (Cat No: 3471, Corning) and cultured in stem cell-specific serum free media (2 mL) [Serum free DMEM/F-12 media (1:1) supplemented with growth factors [recombinant human epidermal growth factor (EGF) (Cat No: PHG0313, Invitrogen) and fibroblast growth factor (FGF) (Cat No: RFGFB50, Invitrogen)] to form spheres. Cells were continuously monitored for the sphere formation and fresh media with supplements and DMSO or 20µM GANT61 were added every 72 hours. Images were captured using the Nikon TE2000 microscope.
Primary OC isolation from ascetic fluid
Ascetic fluids were collected from two different OC patients undergoing treatment at University Medical Centre at TTUHSC. Pathologists confirmed the presence of OC cells and genetic screening further confirmed these cells do not have any germline mutations that affects HR. De-identified, freshly isolated ascetic fluid (15ml) was transferred to a T-75 cm2 tissue culture flasks and supplemented with 15ml DMEM media with 20% FBS. Culture flasks were maintained in a humidified chamber with 5% CO2 at 37°C in an incubator undisturbed for 3–4 days prior to first change of complete medium with OCMI-Le media from USBiological Life Sciences (Cat No: 506391). Further culture of ascetic cells was grown in OCMI-Le media. Two cell lines established have been coded as TT-OC-A1 and TT-OC-A2.
Primary organoid OC Organoid assay
Ascetic cells (TT-OC-A1 and TT-OC-A2) isolated and established in our lab were trypsinized, dispersed into single cells and approximately 25,000 cells were seeded per well in ultra-low attachment 6 well dishes (Cat No: 3471, Corning) and cultured in OCMI-Le media to form spheres. Cells were continuously monitored for the sphere formation and fresh media containing DMSO or 20µM GANT61 were added every 72 hours. Images were captured using the Nikon TE2000 microscope.
Cell cycle analysis
Cells were treated with siGLI1 or siControl for 48 hours or treated with DMSO or 20µM GANT61 for 24 hours, trypsinized and fixed with ice-cold ethanol. The cells were then stained with propidium iodide (Cat No: P3566, Invitrogen) and analyzed by flow cytometry for the cell cycle profile using a BD FACSCanto™ II (BD Biosciences).
NanoString gene expression assay for DDR
To explore GLI1 induced regulation of DDR in OVCAR8 cells, we evaluated a panel consisting of 180 genes involved in the DDR using NanoString nCounter Technology. For this study, GLI1 was downregulated either using siRNA or GANT61 and RNA was isolated and 100ng RNA was used from each sample for gene expression profiling, performed with the digital multiplexed NanoString nCounter analysis system (NanoString Technologies, Seattle, WA, USA). Raw data was normalized against the six housekeeping genes. Normalized data were then analyzed and visualized using nSolver software (NanoString Technologies). The log2 fold change or log2 RNA count number was calculated using normalized RNA count numbers. Statistical significance between cases and the control was determined using an unpaired student's t-test with significance set at p ≤ 0.05. This experiment was done in two biological replicates.
Western blot
For protein analysis by western blotting, cells were washed with ice-cold PBS and lysed on ice using ice-cold cytoskeletal (CSK) buffer [10 mM PIPES (pH 6.8), 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, 1 mM dithiothreitol, 0.1 mM ATP, 1 mM Na3VO4, 10 mM NaF and 0.1% Triton X-100], freshly supplemented with protease (Cat No: 05892970001, Roche) and phosphatase inhibitors (Cat No: 04906837001, Roche). Lysed samples were normalized and diluted with 2X laemmli buffer (Cat No: 161-0737, Bio-Rad) and heated to 100°C for 10 minutes. Then the denatured samples were resolved by SDS-PAGE, transferred to nitrocellulose membranes and probed with the specific antibodies such as γH2AX (Cat No: 05636, Millipore), FANCD2 (Cat No: 20022, Santa Cruz), GLI1 (Cat No: 2553, Cell Signaling), PTCH1 (Cat No: 6147, Santa Cruz) and GAPDH (Cat No: 32233, Santa Cruz).
Luciferase promoter assay
Luciferase control (Cat No: S790005, Active Motif) and FANCD2 promoter (Cat No: S718877, Active Motif) constructs were purchased from Switch Gear Genomics. Luciferase assays were performed in A2780 cells using LightSwitch™ Luciferase Assay System kit (Cat No: LS100, Switch Gear Genomics) according to the manufacturer's protocol and as described previously [10]. In brief, for siRNA mediated experiment, A2780 cells were transfected with siControl or siGLI1 for 48 hours and the last 24 hours was co-transfected with the vectors expressing control Luciferase vector, or the Luciferase under the control of FANCD2 promoter. For GANT61 mediated experiment, A2780 cells were transfected with the above Luciferase constructs in the presence or absence of 20µM GANT61 for 24 hours after 24 hours of transfection. After transfection/treatment, luciferase activity in the cells were measured using LightSwitch Assay reagents from Switchgear Biosciences.
RNA isolation and RT-PCR
After the siRNA or drug treatment, total RNA was extracted from cells and 1 μg of RNA was used for reverse transcription reaction by High-Capacity cDNA Reverse Transcription Kit (Cat No: 4368814, Applied Biosystems) per the manufacturer's protocol. mRNAs were amplified and quantitated using SYBR green dye (Cat No: 172-5271, Bio-Rad), and fluorescence was monitored on a CFX96 Bio-Rad sequence detection system. Melting curve analysis was done for each amplicon. The 2−ΔΔCt method was used for quantitation with glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as an endogenous control. The analysis for each gene was done in triplicate and three independent biological replicates were performed. The gene specific primers used for the analysis were purchased from Bio-Rad.
Immunofluorescence
Cells transfected with siControl or siGLI1 siRNAs were seeded into glass-bottom 35 mm dishes. For drug treatment, cells were treated with DMSO or 20µM GANT61 or 25µM olaparib for 24 hours. Cells were fixed in 3% formaldehyde for 10 minutes and then in 100% methanol (-20°C) for 10 minutes at room temperature. Fixed cells were blocked in 10% goat serum for 30 minutes. After three washes with PBS, cells were incubated overnight at 4°C with primary antibodies [γH2AX (Cat No: 05636, Millipore), FANCD2 (Cat No: 20022, Santa Cruz), GLI1 (Cat No: 2553, Cell Signaling), BRCA1 (Cat No: 6954, Santa Cruz) and RAD51 (Cat No: 8439, Santa Cruz)] in PBS containing 5% bovine serum albumin (BSA) and 0.1% Triton X-100 (PBS-T). The slides were washed three times with PBS-T containing 1% BSA then incubated with fluorescence tagged secondary antibodies (Molecular Probes) for 2 hours at room temperature and mounted with Vectashield containing DAPI (Cat No: H-1500, Vector).
DNA Fiber assay
DNA fiber labeling analysis was used to assess DNA replication fork progression. 48 hours after siRNA transfection A2780 cells were labeled for 20 minutes with 25 µM IdU (Cat No: I7125, Sigma) followed by 20 minutes labeling with 250 µM CldU (Cat No: C6891, Sigma). Cells were harvested and re-suspended in ice-cold PBS. Then, 2µl of the cell suspension was deposited on a slide and 10 µl of lysis buffer (0.5% SDS, 200 mM Tris-HCl pH 7.4, 50 mM EDTA) was added. The slides were tilted to 15° to stretch the DNA fibers. Slides were then air-dried, fixed in 3:1 methanol:acetic acid, denatured in 2.5M HCl and blocked with 5% BSA in PBS. Then slides were incubated with mouse anti-BrdU (Cat No: 347580, BD) and rat anti-BrdU (Cat No: 6326, Abcam) for 1 hour followed by goat anti-mouse alexafluor 568 (Cat No: A11031, Molecular probes) and chicken anti-rat alexafluor 488 (Cat No: A21470, Molecular probes). Fork velocity and stalled replication fork (only red) were measured and statistical analysis was performed using Prism 5 (GraphPad Software) as described previously [24,25].
Immunohistochemistry
Normal and mice tumor tissues were stained for the expression of FANCD2, PTCH1 and GLI1 proteins by immunohistochemistry. Tissue sections were incubated with specific antibodies FANCD2 (Cat No: 20022, Santa Cruz), PTCH1 (Cat No: 6147, Santa Cruz) and GLI1 (Cat No: 20687, Santa Cruz) followed by a specific biotinylated secondary antibody (1:250 dilution), and then conjugated HRP streptavidin and DAB chromogen, and tissues were counterstained with hematoxylin. Stained sections were analyzed by Zeiss Axioscope microscope and as described previously [26] images were captured by AxioCam camera.
High-throughput neutral comet-ChIP assay
A2780 cells were used for high-throughput neutral comet-ChIP assay. Cells were exposed to DMSO or 20μM GANT61 or 25μM olaparib or 20μM GANT61 + 25μM olaparib for 8 hours. Comet assays were performed under alkaline conditions using the CometChip Assay Kit (Trevigen, Gaithersburg, MD) per manufacturer's instructions and as described previously (PMID F10). Drug treated cells were trypsinized and gravity loaded into 30-micron sized CometChip (Trevigen, Gaithersburg, MD) slide. The CometChip slide was then washed with PBS and sealed with low melting point agarose. The CometChip was then submerged in lysis solution for 40 minutes at 4 °C, followed by alkaline buffer (pH > 13, 200 mM NaOH, 1 mM EDTA, 0.1% Triton X-100) treatment for 30 minutes. Electrophoresis was conducted at 22 V for 50 minutes at 4 °C in an alkaline buffer. After electrophoresis, the CometChip was re-equilibrated to neutral pH using Tris buffer (0.4 M Tris•Cl, pH 7.4). Subsequently, DNA was stained with 1 × SYBR Gold diluted in Tris buffer (20 mM Tris•Cl, pH 7.4) for 30 minutes and de-stained for 1 hour in Tris buffer (20 mM Tris•Cl, pH 7.4). Image acquisition was performed using Zeiss microscope. Fluorescence images of more than 100 comets from 3 independent experiments in A2780 cells were analyzed using the ImageJ open comet program and comet tail area was measured. Prism 7 (GraphPad Prism) was used to perform t-test and to calculate statistical significance.
cDNA panel of OC
Ovarian Cancer cDNA Array IV Kit (Cat No: HORT104, Origene) was used to study the expression levels of FANCD2 and GLI1 in the different stages of OC according to the manufacturer's protocol.
Homologous recombination assay
Human lung cancer cells (H1299) were stably transfected with pDR-GFP and selected for puromycin resistance (10 μg/ml) as described previously [27]. Upon 60% confluence, the cells were transfected with siControl or siGLI1 for 24 hours, then transfected with plasmid expressing the restriction enzyme I-SceI (pCBASce1). This restriction enzyme cuts the reporter plasmid and when repaired by HR, GFP is expressed. Twenty-four hours after plasmid transfection, GFP was measured by flow cytometry using a BD FACSCanto™ II (BD Biosciences).
PTCH1 Heterozygous mouse model generation
We developed Ptch1+/−/ODCt/C57BL/6 murine model in which Ptch1 mutations drive aberrant activation of Sonic hedgehog (SHH) signaling [28]. These mice were generated by deletion of exons 1 and 2 of the Ptch1 gene. ODC transgene overexpression in the skin of these mice was driven by a K6 promoter near the hair follicles where basal skin cells presumably reside [29]. The male ODCt breeders (6–7 weeks old) were purchased from Taconic (Germantown, NY, USA). The detailed breeding protocols and genotyping of Ptch1+/−/ODCt/C57BL/6 animals are described earlier. Of note, none of the animals, whether Ptch1 wild type or Ptch1 mutated, studied herein were exposed to UVB or other pro-carcinogenic environmental stimuli. Phenotypically normal skin is defined here as untreated skin of animals with no visible neoplastic lesions. Hence, no difference in skin phenotype was observed between Ptch1+/−/ODCt/C57BL/6 and Ptch1+/+/ODCt/C57BL/6 mice. All animal care and experimental protocols were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Alabama at Birmingham. All experiments were carried out in accordance with relevant guidelines and regulations.
SKOV3 luciferase in-vivo tumor formation and treatment
Female athymic nu/nu mice were injected with 2.5 × 106 SKOV3 cells per flank in PBS and allowed the tumors to grow till it reached 100mm3. Mice-bearing xenograft tumors were treated with i.p injections of vehicle (control) or GANT61 (25mg/kg, body weight in 200μl PBS; daily) or olaparib (25mg/kg, body weight in 200μl PBS; daily) or combination for 31 days. D-luciferin was used to visualize the tumor size and measure the tumor volumes. All animal care and experimental protocols were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Alabama at Birmingham. All experiments were carried out in accordance with relevant guidelines and regulations.
Statistical analysis
All data are the representation of three independent experiments performed in triplicates unless mentioned. Statistical significance was analyzed by students’ t-test using GraphPad Prism 6 or Excel 2010.
Results
GLI1 inhibition downregulates FANCD2 expression in ovarian cancer cells and its inhibition causes replication stress-mediated DDR
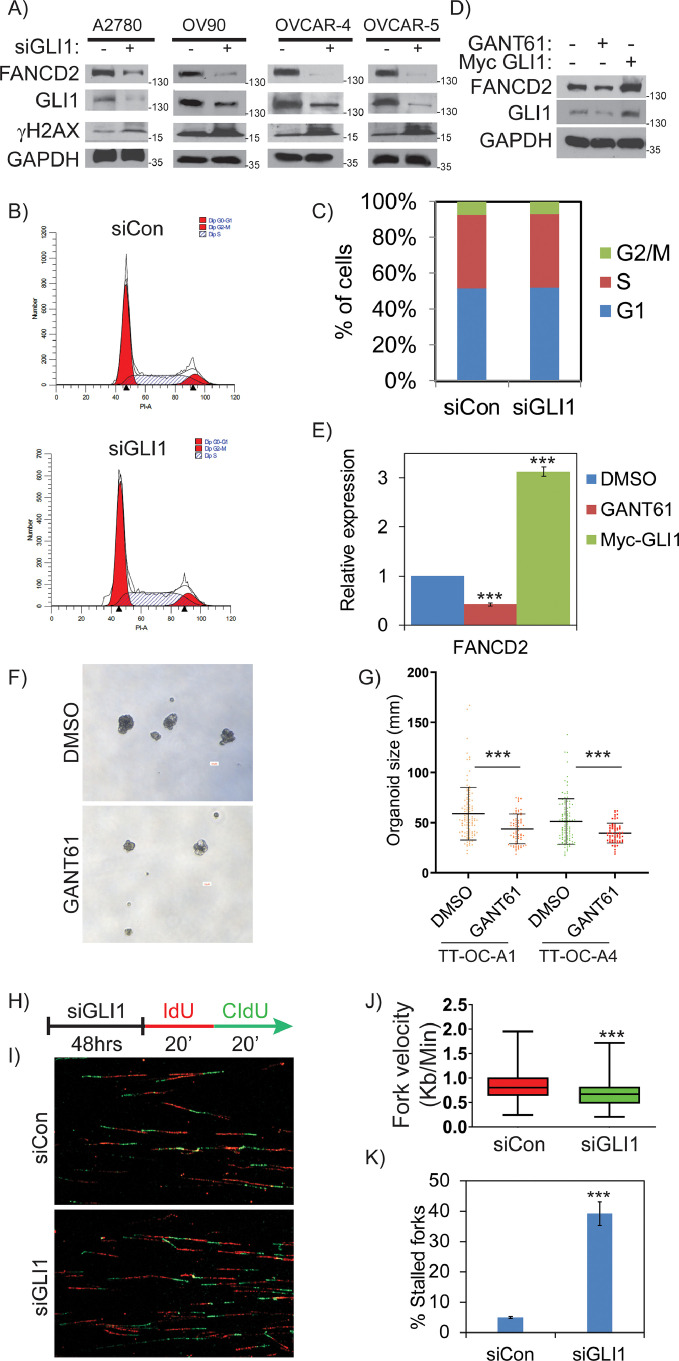
Aberrant expression of Hh/GLI1 signaling proteins are common in many advanced stage cancers and correlates with tumor progression, stem cell-like phenotype and therapeutic resistance [7,[30], [31], [32], [33]]. Previous studies indicated that GLI1 downregulation or inhibition induces replication-associated DDR in tumor cells [3,10]. Downregulation of GLI1 in ovarian cancer cell line A2780 reduces colony formation and increased comet tail, a marker for DNA damage (Figure 1A and 1B). Time course treatment of OVCAR8 cells with GANT61 increases sub-G0 cell population and apoptosis markers cleaved PARP and cleaved caspase-8 after 48 and 72 hours of treatment, suggests GLI1 inhibition causes apoptosis in ovarian cancer cells (Figure S1A, S1B and S1C). Furthermore, sphere forming ability of A2780 cells were inhibited upon GANT61 treatment (Figure S2A). However, the mechanisms behind GLI1 downregulation induced DNA damage in tumor cells were not evaluated. To explore the role of aberrant GLI1 in regulating replication-associated DNA damage response, we analyzed the expression of 180 genes that are involved in various pathways of DDR using nCounter® Vantage 3D™ DNA Damage and Repair Panel from NanoString Technologies (Human) in ovarian cancer cells (Figure 1C). We examined the expression of the genes involved in DDR and repair by both knocking down GLI1 using siRNAs and by inhibiting with GANT61, a pharmacological inhibitor that targets GLI transcription factors in OVCAR8 cells. Overall, we found that 35 genes were differentially expressed in GLI1 knockdown cells and 64 genes were differentially expressed in GANT61 treated cells compared to control siRNAs transfected cells and DMSO treated OVCAR8 cells respectively (Figure S2B and S2C). Among these, 17 genes were found to be commonly altered in both siGLI1 and GANT61 treated cells (Figure 1D and S2D), and most of them belong to Fanconi anemia tumor suppressor and cell cycle checkpoint pathways. Interestingly, FANCD2 was found to be the most significantly downregulated genes in both GLI1 knockdown and GANT61 treated OVCAR8 cells (Figure 1E and 1F). Our data suggest that aberrant GLI1 may regulate FA genes in order to suppress oncogenic replication stress and promotes tumor cell survival and progression of ovarian cancer. Interestingly, analysis of TCGA database for OC by UALCAN portal shows that upregulated FANCD2 (n=76) have low survival probability compared to OC patients with low/medium FANCD2 expression (n=227) (Figure 1G). These data indicate a role for upregulated FANCD2 in OC disease progression and clinical outcome.
Figure 1.
Inhibition of Hh/GLI1 by siRNA or inhibitor alters the expression of genes involved in various DNA repair pathways in OC cells. A) Colony assay in A2780 cells treated with DMSO or 20µM GANT61 B) High throughput comet assay in DMSO or GANT61 treated A2780 cells. C) Enriched KEGG pathways in OVCAR8 cells treated with siGLI1 or GANT61 compared to control cells. D) Set of differentially expressed genes that are common in both siGLI1 and GANT61 treated samples. E and F) Volcano plot shows FANCD2 is significantly downregulated in both siGLI1 and GANT61 treated OVCAR8 cells compared to their respective control (…. =p< 0.05, —– =p<0.01, -.-.-. =p<0.001). G) Statistically significant decrease in survival probability was observed in high FANCD2 expressing OC patients compared to low/medium FANCD2 expressing OC patients. *** p<0.001.
To examine GLI1 dependent expression of FANCD2 in OC cells, we transiently knocked down GLI1 using siRNAs and evaluated FANCD2 protein levels. Consistent with the expression array data, FANCD2 protein levels were significantly downregulated in GLI1 deficient cells compared to their respective controls in multiple ovarian cancer cell lines (Figure 2A). To rule out any off target effects of siRNAs, we used three individual siRNAs that target GLI1 at different coding regions. As shown in the figure S3A, downregulation of GLI1 by three different siRNAs downregulated FANCD2 in OC cells. However, FANCD2 expression normally increases during S and G2 phases; and it is possible that the GLI1 inhibition mediated FANCD2 downregulation could be due to altered cell cycle in these cells. Importantly, cell cycle analysis of control and GLI1 knockdown cells showed no significant differences in their overall cell cycle profiles, compared to controls cells, dismissing any cell cycle specific effects (Figure 2B and 2C). To further establish GLI1-dependent expression of FANCD2, we evaluated simultaneously, GLI1 inhibition and ectopically expressing myc-GLI1 in A2780 cells. As shown in Figure 2D and 2E, FANCD2 levels were downregulated in GANT61 treated cells and ectopic expression of myc-GLI1 upregulated the levels of FANCD2 compared to control cells, suggesting GLI1 dependent expression of FANCD2. Further to determine the importance of GLI1 in ovarian tumor growth, we treated the organoids developed from acetic fluids isolated from two individuals with high-grade serous ovarian cancer (TT-OC-A1 and TT-OC-A2) with GANT61 (Figure 2F). Addition of GANT61 attenuated organoids significantly (Figure 2G), confirming that OC cells depend on GLI1 mediated signaling for survival and progression.
Figure 2.
GLI1 regulates expression of FANCD2 and its inhibition induces replication stress in OC cells. A) Western blot analysis shows the expression profile of FANCD2 in different OC cells treated with siControl vs siGLI1. B and C) Cell cycle profile in A2780 cells transfected with sicontrol vs siGLI1 for 48 hours. D and E) Differential expression of FANCD2 in A2780 cells transfected with myc-GLI1 or treated with 20µM GANT61 for 24 hours. F and G) Inhibition of ovarian organoid formation in DMSO or 15µM GANT61 treatment in cells isolated from ascetic fluid of two different OC patients (TT-OC-A1 and TT-OC-A2). Scale bar represents 50μM. H) DNA combing studies was done in A2780 cells treated with siControl or siGLI1. I, J and K) Fiber assay reveal decreased fork velocity and increased stalled forks in GLI1 deficient A2780 cells. More than 100 replication structures from three independent experiments were scored and statistical significance were assessed using Mann-Whitney test and students “t” test was performed using Prism 5. All the experiments were repeated three times and their S.E. is denoted in the bar graph. ** p < 0.01 and *** p < 0.001.
Tumor cells are characterized by dysregulated DNA replication and cell division, driven by oncogenes and growth factors and resulting in metabolic and replication stress, which may cause DNA damage and cell death [34]. FANCD2, the key component of FA-BRCA pathway, associates with replication forks to promote their stability and timely progression during metabolic and exogenous damage-induced replication stress [35,36]. To determine that aberrant GLI1 promotes oncogenic replication or suppresses oncogenic replication stress by regulating FANCD2, we employed single cell DNA fiber assays and measured replication dynamics in A2780 cells after transfecting with control or GLI1 siRNAs. As represented in figures (2H and 2I), cells were sequentially treated with the nucleoside analogues IdU and CldU for 20 minutes each to label the newly synthesized DNA. Consistent with FANCD2 downregulation, GLI1 inhibited cells showed significantly reduced fork velocities comparing to control cells. The average fork velocities in control and GLI1-deficient cells were 0.85 and 0.67 kb/minutes (p>0.001) for A2780 cells, respectively (Figure 2J). Consistent with the increased pH2AX levels and DNA lesions (Figure 2A), there was a greater than 5-fold increase in stalled forks (red only) in GLI1-downregulated cells (Figure 2K). These data suggest that GLI1 inhibition causes FANCD2 deficiency in OC cells, leading to stalled or collapsed forks and DNA lesions. Since GLI1 is the downstream effector transcription factor [37], these results indicate that Hh/GLI1 may transcriptionally regulate FANCD2 to support the increased growth and differentiation potential of tumor cells.
GLI1 regulates expression of FANCD2 in transgenic mouse models of hedgehog signaling and basal cell carcinomas (BCC)
Hh/GLI1 signaling is highly regulated during development and tissue patterning. However, its expression is undetectable in most adult tissues and is only limited to certain cell populations such as stem cells that maintain tissue homeostasis and regeneration. Previously, Ptch1+/– heterozygous knockout C57BL/6 mice were generated by replacing exons 1 and 2 of the Ptch1 with the LacZ gene to study the Hh-dependent carcinogenesis mechanisms, particularly BCCs, as more than 90% of these tumors are due to aberrant Hh signaling [38]. These mice have reduced Ptch1 expression due to haplo-insufficiency that causes increased expression of GLI1 (Figure 3A). Consistent with previous studies, GLI1 protein expression was higher in skin tissue from Ptch1+/- and Ptch1+/− BCC compared to Ptch1+/+ mouse skin tissues, as shown by both western blots and IHC staining (Figure 3B and C). Reproducibly, the increased levels of GLI1 protein also correlated with FANCD2 protein expression and exhibited co-localization in Ptch1+/− BCC in co-immuno-staining (Figure 3D). These results further emphasize in-vitro results that GLI1-dependent expression of FANCD2 seen in OC cell lines is also true in genetic models of Hh signaling, and provides in-vivo evidence for the GLI1-dependent expression of FANCD2.
Figure 3.
GLI1 regulates expression of FANCD2 in normal skin and BCC mouse tissues. A) Expression of GLI1 in Ptch1+/+ vs Ptch1+/- mouse skin tissues. B) Expression of GLI1 and FANCD2 in Ptch1+/+ vs Ptch1+/- vs BCC Ptch1+/- mouse skin tissues analyzed by western blot. C) Expression of GLI1 and FANCD2 in Ptch1+/+ vs Ptch1+/- vs BCC Ptch1+/- mouse skin tissues analyzed by IHC. Scale bar represents 100μM. D) FANCD2 (green) and GLI1 (red), and overlay showing co-localization (yellow). Scale bar represents 50μm. *** indicates p<0.001.
GLI1 transcriptionally regulates FANCD2 and its inhibition induces HR deficiency in tumor cells
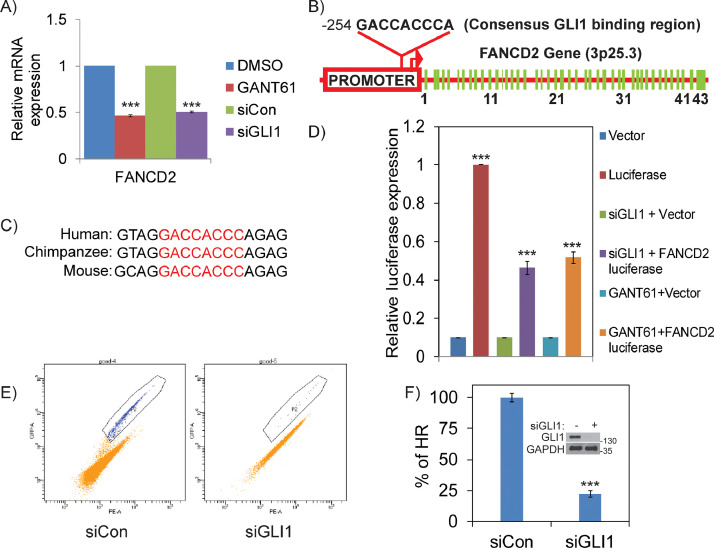
To examine the mechanisms by which GLI1 regulates FANCD2 in OC cells, we first analyzed the transcript levels of FANCD2 by quantitative RT-PCR. A greater than 50% reduction in the transcript levels of FANCD2 was observed in both siGLI1 and GANT61 treated A2780 (Figure 4A) cells. In Hh signaling cascade, GLI transcription factors regulate many genes directly through binding to consensus “GACCACCC” DNA sequences [39]. Interestingly, in silico analysis of the promoter region of FANCD2 revealed a consensus GLI1 binding sequence “GACCACCC”, which is highly conserved in other vertebrates (Figure 4B and 4C). To confirm that GLI1 directly regulates FANCD2 through its promoter, we performed luciferase reporter assays under the control of FANCD2 promoter (1 Kb upstream of transcription start site). Inhibition of GLI1 by siRNAs or by GANT61 in A2780 cells showed ∼ 50% decrease in luciferase activity compared to GLI1-proficient cells (Figure 4D). Together these results strongly indicate that GLI1 transcriptionally regulates HR pathway by direct regulation of FANCD2 through its promoter.
Figure 4.
GLI1 transcriptionally regulates FANCD2 and its inhibition induces HR deficiency in tumor cells. A) RT-PCR showing GLI1 transcriptionally regulates FANCD2 in A2780 cells. B) FANCD2 promoter in chromosome region 3p25.3 confirms GLI1 binding sequence “GACCACCC” and C) conserved among various species. D) Luciferase activity measured after transfection of Luciferase expression constructs either control or under the FANCD2 promoter in A2780 cells, that are co-transfected either control or GLI1 targeting siRNAs; or with DMSO or 20µM GANT61. E and F) Dr-GFP assay mediated analysis of HR efficiency in siGLI1 treated H1299 cells compared to siControl cells. ** p < 0.01 and *** p < 0.001.
Deficiency in FANCD2 protein causes defective HR-mediated DSB repair, a status called HR deficiency [40]. We previously generated a stable pool of clones that express DR-GFP reporter constructs in H1299 lung cancer cells and used them to evaluate the role of FA-BRCA pathway in HR [41]. Result shows that knocking down GLI1 by siRNAs downregulated FANCD2 levels as observed in ovarian cancer cells (Figure S3B). These assays revealed an approximately 70% decrease in HR efficiency in GLI1 knockdown cells compared to control cells (Figure 4E and 4F). Collectively, these data further confirm that aberrant Hh/GLI1 regulates FA-BRCA genes in tumors and its inhibition induces HR deficiency. This is particularly interesting, because tumors deficient in HR exhibit synthetic lethality with PARP inhibitors (PARPi), an important class of drugs showing promising efficacy in treating ovarian, breast and several other cancers [42].
Hh/GLI1 inhibition in combination with PARPi induces synergistic lethality in OC cells
PARP enzymes (PARP1 and PARP2) play important roles during replication by facilitating repair of DNA single strand breaks, that if not repaired in a timely manner may lead to replication fork collapse and lethal DSB [20]. PARP inhibitors show potent cytotoxic effects in FA-BRCA mutant cells, which are deficient in HR [43,44]. We then posited that inhibition of GLI1 in combination with PARP inhibitors in HR-proficient tumor cells should induce enhanced DNA lesions due to HR deficiency. As expected, GLI1 knockdown by siRNAs or inhibition by GANT61 attenuated olaparib induced FANCD2 mono-ubiquitination and increased phosphorylation of H2AX, a marker for increased DNA damage in A2780 and SKOV3 cells (Figure 5A). Similar to these western blots data, GLI1 inhibition attenuated FANCD2 repair foci formation in olaparib treated cells (Figure 5B and 5C). Moreover, repair deficiency in GLI1 inhibited cells caused increased γH2AX foci formation, as well as significantly increased DNA lesions as indicated by COMET assays (Figure 5D and 5E).
Figure 5.
Hh/GLI1 inhibition in combination with PARPi induces synergistic therapeutic effects in OC cells. A) Inhibiting Hh/GLI1 signaling by siGLI1 for 48 hours or 20μM GANT61 for 24 hours impairs FANCD2 protein expression induced by 25μM olaparib for 24 hours in A2780 and SKOV3 cells. B and C) Histogram and confocal images of γH2AX and FANCD2 in A2780 cells treated with DMSO or 20µM GANT61 or 25µM olaparib or 20µM GANT61 + 25µM olaparib for 24 hours. Scale bar represents 10μM. D and E) Analysis of DNA damage by high-throughput neutral comet-ChIP assay in GLI1 and PARP inhibited A2780 cells. Inhibition of GLI1 and PARP in combination shows increased DNA damage (tail length) compared to individual treatments in a dose dependent manner. F) Western blot analysis of sicontrol vs siGLI1 in normal human fallopian tube epithelial cells (FTECs). G) RT-PCR analysis of FANCD2 and GLI1 expression in different stages of OC using Ovarian Cancer cDNA Array IV Kit. ** p<0.01 and *** p<0.001.
It has been reported that in most of the normal cells and adult tissues that are differentiated, expression of GLI1 is minimal or undetectable [45]. Consistently, GLI1 expression in immortalized normal human fallopian tube epithelial cells (FTECs) expresses no or undetectable levels of GLI1, and GLI1-targeted siRNAs did not generate significant differences in expression of FANCD2 in these cells (Figure 5F). To find out the expression of GLI1 in ovarian tumors, we analyzed the expression levels of GLI1 and FANCD2 in an ovarian cDNA panel with different stages of epithelial ovarian cancers (n=60) and normal ovarian tissues (n=16). As reported in previous studies [46,47], GLI1 is significantly upregulated in ovarian tumors compared to normal ovarian tissues. Consistently, FANCD2 expression is also upregulated in the ovarian tumors compared to the normal ovarian tissues, and showed positive correlations with the GLI1 (Figure 5G).
Together these results confirm that aberrant GLI1 in OC cells induces expression of FANCD2 and efficient repair of DSB by HR, and suggests inhibition of GLI1 can cause HR deficiency and synergistic lethality with PARP inhibitors.
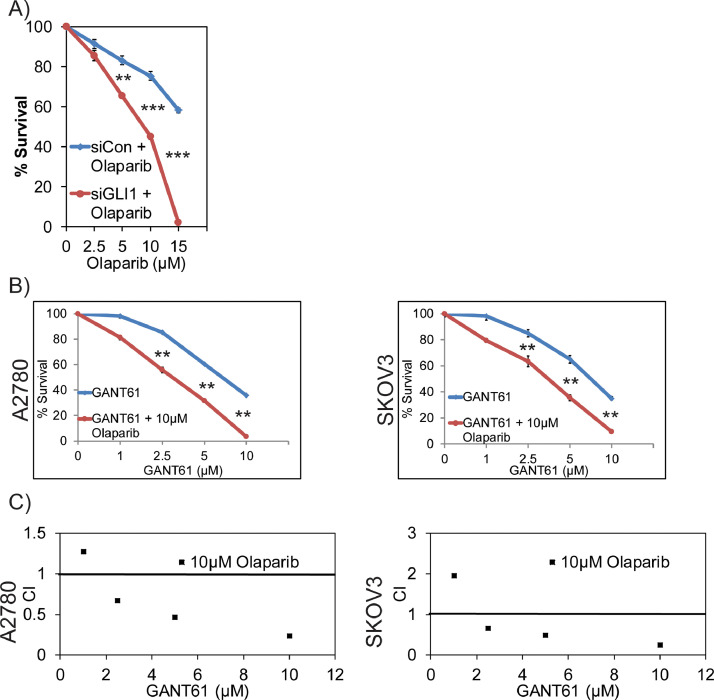
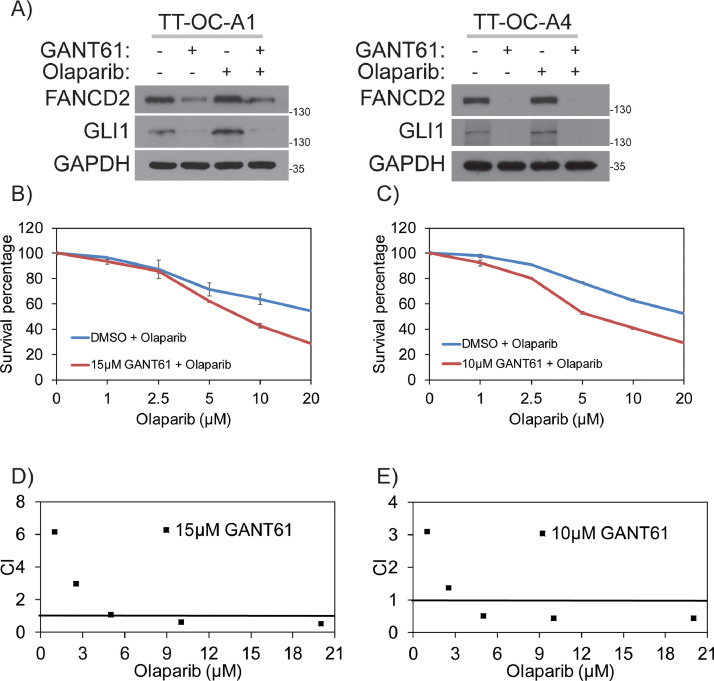
To evaluate GLI1 and PARP1 synergistic lethality interactions, first we exposed control and GLI1 knockdown A2780 cells to different concentrations of olaparib and assessed their survival by clonogenic survival assays. Consistent with the findings that GLI1 downregulation results in HR deficiency, these cells also showed hypersensitivity to PARPi olaparib compared to GLI1-proficient cells, suggesting synergistic lethality phenotype based on the defect in two compensatory DNA repair mechanisms (Figure 6A). Similarly, inhibition of GLI1 using GANT61 also showed synergistic lethality compared to individual drugs, as indicated by combination index (CI) values in A2780 and SKOV2 cell lines [Figure 6B and C). Particularly, concentrations of GANT61 (2.5 µM) and olaparib (2.5 µM); GANT61 (5 µM) and olaparib (5 µM); GANT61 (10 µM) and olaparib (10 µM) were most significant conditions for the indicated cell lines. To further confirm the results observed in established cell lines are reproducible in primary OC cells, we established two primary OC cell lines from ascetic fluids collected from two patients with high grade serous ovarian cancer. These two BRCA proficient patient derived cell lines express GLI1 and FANCD2. Consistent with the established cell lines data, inhibition of GLI1 in these cells downregulated both basal level (DMSO) and olaparib induced expression of FANCD2 [Figure 7A). Similarly, combinations of GANT61 and different concentration of olaparib show reduced survival compared to individual olaparib treatment (Figure 7B and 7C). Most of the combinations also showed synergistic lethality as indicated by CI values (Figure 7D and 7E).
Figure 6.
Combination index and clonogenic survival for Hh/GLI1 inhibition in combination with PARPi. A) Survival graph for various concentration of olaparib treatment in siControl and siGLI1 A2780 cells. B) Survival graph for various GANT61 and olaparib combinations in different ovarian cancer cell lines. C) Combination index for various GANT61 and olaparib combinations as calculated by CalcuSyn software. ** p < 0.01 and *** p < 0.001.
Figure 7.
Combination index and clonogenic survival for Hh/GLI1 inhibition in combination with PARPi. A) Inhibiting Hh/GLI1 signaling by 20μM GANT61 for 24 hours impairs FANCD2 protein expression induced by 25μM olaparib for 24 hours in TT-OC-A1 and TT-OC-A4 ascetic cells isolated from OC patients. B and D) Survival graph and combination index values for various GANT61 and olaparib combinations in TT-OC-A1 ascetic cells. C and E) Survival graph and combination index values for various GANT61 and olaparib combinations in TT-OC-A4 ascetic cells.
Overall, these in-vitro data from established and primary OC cell lines demonstrates a novel synergistic lethality interaction between GLI1i and PARP inhibitor combination in BRCA/HR-proficient OC cells. Furthermore, the cytotoxic effects of GANT61/olaparib combination therapy were limited to tumor cells with no significant impact on FTECs, suggesting this combination may have fewer side effects to normal tissues (Figure S3C).
Combination of GLI1 and PARP inhibitors treatment shows synergistic lethality in mouse ovarian cancer model
While many small molecule inhibitors of GLI1 developed, GANT61 is the only agent available that showed in-vitro and in-vivo efficacy in inhibiting GLI1 [48]. To evaluate the effectiveness of GLI1i and PARPi combination treatment in in-vivo ovarian cancer models, we subcutaneously implanted SKOV3-Luc cells in both the planks of athymic nu/nu mice. Once tumor size reached (100mm3), mice-bearing xenograft tumors were randomized into the following four groups (6 mice per group): Control group (vehicle/PBS); GANT61 group (GANT61 administered through i.p. injections of 25mg/kg, body weight in 200μl PBS; daily); olaparib group (received 25 mg/kg body weight of olaparib through parenteral administration daily); and combination treatment group (received both the drugs as administered in individual drug doses and times). Tumor growth was measured weekly by non-invasively measuring tumor size using IVIS imaging after administering luciferin for about four weeks (31 days), and bioluminescence intensity of tumors were quantitated and presented as the mean of photon counts per second per tumor. Consistent with the in-vitro combination cytotoxic data, combination of GANT61 and olaparib showed statistically significant reduction in tumor volume compared to the individual treatments (Figure 8A and 8B). GLI1-dependent expression of FANCD2 was measured by immunohistochemistry in cells from xenograft tumors treated with vehicle control, and GANT61, olaparib, and combination of GANT61 and olaparib (Figure 8C). Interestingly, olaparib-treated tumor tissues exhibited increased levels of GLI1 and FANCD2 expression compared to vehicle control and GANT61-treated tumor xenograft tissues. Tumor cells treated with both GANT61 and olaparib showed significantly reduced GLI1 and FANCD2 staining. Consistent with the in-vitro data, GANT61 treated tumors show reduced proliferation (Ki-67 expression) (Figure S3D). Overall, in-vitro and in-vivo data supports a novel synergistic lethality interaction between GLI1 and PARP1, and suggests a rational combination of GLI1 and PARP inhibitors as an effective therapy for advanced BRCA proficient OC.
Figure 8.
GLI1i in combination with PARPi induces synergistic lethality in mouse orthotopic ovarian model. A & B) Combination of GANT61/olaparib treatment slows growth of ovarian (SKOV3) xenograft tumors in athymic nu/nu mice. Representative mice (n=10, each) bearing SKOV3 xenograft tumors and average tumor volume (mm3) 31days post treatment. C) IHC analysis of (SKOV3) xenograft tumors in athymic nu/nu mice for FANCD2 and GLI1 expression. Scale bar represents 100μM. D) Model illustrates novel regulation of FA/BRCA and HR DNA repair proteins by Hh/GLI1 in response to DNA damage to promote acquired chemoresistance. * p<0.05 and *** p<0.001.
Discussion
In this study we show a novel function for Hh/GLI1 in regulation of FANCD2 tumor suppressor gene to support oncogenic replication and survival of OC cells. Mutations or deficiency in FA-BRCA genes leads to developmental abnormalities and increased cancer risk [49]. Similarly, mutations or aberrant regulation of Hh signaling molecules result in developmental defects and cancer, even though clinical features vary from FA-BRCA phenotype [2]. FANCD2 is monoubiquitinated in response to fork blocking lesions, and regulates replication fork stability, and promotes repair and restart of collapsed forks (DSB) by error free HR in association with BRCA1, BRCA2 and RAD51 [16]. Thus, cells deficient in FA-BRCA genes exhibit replication stress and genomic instability. Based on these observations, reduced fork velocity, increased stalled or collapsed forks observed in GLI1-inhibited cells (Figure 2G-2F) could be due to FANCD2 deficiency. Previously, a functional link between Hh/GLI1 signaling and DDR was documented [3,10]; however, a role for GLI1 in promoting oncogenic replication has not been studied. Data presented here supports an important function for aberrant GLI1 in replication progression and survival of ovarian cancer cells.
GLI1 transcription factor bind to the promoters through an 8 base pair consensus binding site “GACCACCC” and regulate gene transcription [39]. The presence of GLI1 consensus binding site in FANCD2 promoter, and GLI1-dependent promoter activation in luciferase reporter assays confirm that FANCD2 is a direct GLI1 transcriptional target (Figure 4B, C and D). Additionally, GLI1-dependent expression of FANCD2 in both normal and BCC skin tissues of Ptch1+/- mice further supports a novel role for Hh/GLI1 signaling in regulation of FA-BRCA genes during development and in cancer (Figure 3]. This GLI1-mediated FA-BRCA gene regulation may be at least in part an adaptive response to maintain genomic integrity during oncogenic replication in tumors and to promote their survival from DNA lesions induced by replication and metabolic stress. These noteworthy observations suggest further evaluation of Hh/GLI1 and FANCD2 relationship during development.
GLI1-dependent expression of FANCD2 genes in OC cells attributes novel function for Hh/GLI1 and FA-BRCA pathway in tumorigenesis and chemoresistance. However, FANCD2 upregulation is observed in many BRCA (BRCA1/BRCA2) deficient tumors/cells and is implicated in regulation of alternative DNA repair mechanisms and correlates with poor prognosis [50].
BRCA-deficient tumors show better response to chemotherapy and exhibit synthetic lethality with PARP inhibitors (PARPi), a promising class of therapeutic agents [22]. Recently, at least four PARPi (olaparib, rucaparib, niraparib and talazoparib) were FDA approved for treatment of primarily ovarian, breast and later other cancers in patients with BRCA mutations or HR deficiency [51]. Although PARPi are promising anticancer agents, majority of OC patients are BRCA-proficient and cannot benefit from this class of drugs [52,53]. Since Hh/GLI1 signaling has been an effective therapeutic target for cancer treatment, GLI1 depletion or inhibition causes a state of HR deficiency in OC cells, representing a rational therapeutic approach for expanding the effectiveness of PARP inhibitors to BRCA-proficient patients (Figure 8D). Although many small molecule GLI1 inhibitors have been developed, presently most of them are still in preclinical testing [54]. Agents that target members of the Hh pathway upstream of GLI1, like SMO have been developed and evaluated them in many indications [55]. While many SMO inhibitors show initial promise, tumors often develop resistance to these agents and the patients relapse [56]. It is important to note that GLI1 can be activated non-canonically by signals like TGFβ, K-RAS, WNT, β-catenin and C-MYC [3]. SMO is an upstream regulator of GLI1 in canonical Hh signaling; thus, non-canonically activated GLI1 cannot be inhibited by SMO-targeting agents. Recent studies identified novel non-canonical regulation of GLI in various cancers. Especially, nuclear myocardin-related transcription factor in basal cell carcinoma [57], deregulated GSK3β in colon cancer cells [58], Ras–Raf–MEK ERK pathway in hepatocellular carcinoma [59], fibroblast-derived insulin-like growth factor-1 in oral squamous cell carcinoma [60], MAPK/ERK signaling in lung adenocarcinoma [61], transcriptionally-active androgen receptors in pancreatic cancer [62] and SOX2-BRD4 transcriptional complex in melanoma [63]. Additionally, studies in breast cancer models, EMT cells induce increased metastasis of weakly metastatic, non-EMT tumor cells in a paracrine manner, in part by non-cell autonomous activation of the GLI transcription factor. Although all Hh inhibitors may act against tumors with canonical Hh/GLI signaling, only GLI1 inhibitors would act against non-canonical EMT-induced GLI1 activation [64]. The non-canonical activation of GLI1 and subsequent resistance to SMO inhibitors were reported in multiple cancers. However, the possibility of similar mechanisms active in OC is very high and needs further studies to validate.
Overall, our results show a novel function for aberrantly activated Hh/GLI1 in regulation of HR meditated repair of DSB to support oncogenic replication and survival of OC cells. Mechanistically, we found that GLI1 transcriptionally regulates FANCD2 expression, an important member of FA-BRCA and HR pathway. Inhibition of GLI1 in OC attenuates FANCD2 expression and induces HR deficiency. HR deficient OC cells were then can be sensitized by using FDA approved PARP inhibitor olaparib. Therefore, our studies emphasize the urgent need to develop novel GLI1 specific inhibitors that can be used to treat lethal malignancies such as OC in combination with PARP targeted therapies.
Acknowledgments
Abbreviations
Ovarian cancer (OC)
Hedgehog (Hh)
Glioma associated protein 1 (GLI1)
Fanconi anemia (FA)
Poly (ADP-ribose) polymerase inhibitors (PARPi)
Homologous recombination (HR)
DNA damage response (DDR)
Human epidermal growth factor (EGF)
Fibroblast growth factor (FGF)
Funding
This work was supported by National Cancer Institute (R01CA219187).
Author contributions
C.M., K.T., and K.P. Conceptualization, Data curation; C.M., K.T., S.C., R.S., and K.P. Investigation, Methodology. C.M., K.T., S.C., R.S., R.P., M.R., M.A., and K.P. analyzed the data. C.M., K.T., S.C., R.S., R.P., M.R., M.A., and K.P. Visualization, Writing. C.M., K.T., S.C., R.S., R.P., M.R., M.A., and K.P. Investigation, Validation. C.C. Software. M.A and K.P. Resources, Supervision.
Ethics approval and consent to participate
All animal experiments have been approved by the Institutional review board at University of Alabama at Birmingham, USA.
Consent for publication
All authors have agreed to publish this manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Palle lab members for their help towards submission of the manuscript.
Data Availability
All data that support the findings of this study are available from the corresponding authors upon reasonable request.
Footnotes
One Sentence Summary: GLI1 inhibition in BRCA proficient OC cells induces HR deficiency and synergistic lethality with PARP inhibition.
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2021.06.010.
Appendix. Supplementary materials
References
- 1.Briscoe J, Thérond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol. 2013;14(7):416–429. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- 2.Jiang J, Hui C-C. Hedgehog signaling in development and cancer. Dev Cell. 2008;15(6):801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palle K, Mani C, Tripathi K, Athar M. Aberrant GLI1 Activation in DNA Damage Response, Carcinogenesis and Chemoresistance. Cancers. 2015;7(4):2330–2351. doi: 10.3390/cancers7040894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abula Y, Yi C, Wang X-Y, Wang M, Qin R-Y, Guo Y-Q. Gli1 expression in pancreatic ductal adenocarcinoma and its clinical significance. Genet Mol Res GMR. 2015;14(4):12323–12329. doi: 10.4238/2015.October.9.21. [DOI] [PubMed] [Google Scholar]
- 5.Cui W, Wang L-H, Wen Y-Y, Song M, Li B-L, Chen X-L. Expression and regulation mechanisms of Sonic Hedgehog in breast cancer. Cancer Sci. 2010;101(4):927–933. doi: 10.1111/j.1349-7006.2010.01495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hong Z, Bi A, Chen D, Gao L, Yin Z, Luo L. Activation of hedgehog signaling pathway in human non-small cell lung cancers. Pathol Oncol Res POR. 2014;20(4):917–922. doi: 10.1007/s12253-014-9774-x. [DOI] [PubMed] [Google Scholar]
- 7.McCann CK, Growdon WB, Kulkarni-Datar K, Curley MD, Friel AM, Proctor JL. Inhibition of Hedgehog signaling antagonizes serous ovarian cancer growth in a primary xenograft model. PloS One. 2011;6(11):e28077. doi: 10.1371/journal.pone.0028077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Athar M, Tang X, Lee JL, Kopelovich L, Kim AL. Hedgehog signalling in skin development and cancer. Exp Dermatol. 2006;15(9):667–677. doi: 10.1111/j.1600-0625.2006.00473.x. [DOI] [PubMed] [Google Scholar]
- 9.Cui D, Xu Q, Wang K, Che X. Gli1 is a potential target for alleviating multidrug resistance of gliomas. J Neurol Sci. 2010;288(1–2):156–166. doi: 10.1016/j.jns.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 10.Tripathi K, Mani C, Barnett R, Nalluri S, Bachaboina L, Rocconi RP. Gli1 protein regulates the S-phase checkpoint in tumor cells via Bid protein, and its inhibition sensitizes to DNA topoisomerase 1 inhibitors. J Biol Chem. 2014;289(45):31513–31525. doi: 10.1074/jbc.M114.606483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.FA complementation groups | HUGO Gene Nomenclature Committee [Internet]. [cited 2020 Dec 17]. Available from: https://www.genenames.org/data/genegroup/#!/group/548
- 12.Thompson EL, Yeo JE, Lee E-A, Kan Y, Raghunandan M, Wiek C. FANCI and FANCD2 have common as well as independent functions during the cellular replication stress response. Nucleic Acids Res. 2017;45(20):11837–11857. doi: 10.1093/nar/gkx847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okamoto Y, Iwasaki WM, Kugou K, Takahashi KK, Oda A, Sato K. Replication stress induces accumulation of FANCD2 at central region of large fragile genes. Nucleic Acids Res. 2018;46(6):2932–2944. doi: 10.1093/nar/gky058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwab RA, Nieminuszczy J, Shah F, Langton J, Lopez Martinez D, Liang C-C. The Fanconi Anemia Pathway Maintains Genome Stability by Coordinating Replication and Transcription. Mol Cell. 2015;60(3):351–361. doi: 10.1016/j.molcel.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D'Andrea AD, Grompe M. The Fanconi anaemia/BRCA pathway. Nat Rev Cancer. 2003;3(1):23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- 16.Grompe M, D'Andrea A. Fanconi anemia and DNA repair. Hum Mol Genet. 2001;10(20):2253–2259. doi: 10.1093/hmg/10.20.2253. [DOI] [PubMed] [Google Scholar]
- 17.Chen CC, Taniguchi T, D'Andrea A. The Fanconi anemia (FA) pathway confers glioma resistance to DNA alkylating agents. J Mol Med Berl Ger. 2007;85(5):497–509. doi: 10.1007/s00109-006-0153-2. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Wiltshire T, Senft J, Wenger SL, Reed E, Wang W. Fanconi anemia D2 protein confers chemoresistance in response to the anticancer agent, irofulven. Mol Cancer Ther. 2006;5(12):3153–3161. doi: 10.1158/1535-7163.MCT-06-0427. [DOI] [PubMed] [Google Scholar]
- 19.Bose CK, Basu N. PARP inhibitors and more. J Turk Ger Gynecol Assoc. 2015;16(2):107–110. doi: 10.5152/jtgga.2015.15029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morales J, Li L, Fattah FJ, Dong Y, Bey EA, Patel M. Review of poly (ADP-ribose) polymerase (PARP) mechanisms of action and rationale for targeting in cancer and other diseases. Crit Rev Eukaryot Gene Expr. 2014;24(1):15–28. doi: 10.1615/critreveukaryotgeneexpr.2013006875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petrucelli N, Daly MB, Pal T. BRCA1- and BRCA2-Associated Hereditary Breast and Ovarian Cancer. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJ, editors. GeneReviews(®) [Internet]. Seattle (WA) University of Washington, Seattle; 1993. http://www.ncbi.nlm.nih.gov/books/NBK1247/ [cited 2017 Jan 14]. Available from: [PubMed] [Google Scholar]
- 22.Weil MK, Chen A. PARP Inhibitor Treatment in Ovarian and Breast Cancer. Curr Probl Cancer. 2011;35(1):7–50. doi: 10.1016/j.currproblcancer.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Somasagara RR, Tripathi K, Spencer SM, Clark DW, Barnett R, Bachaboina L. Rad6 upregulation promotes stem cell-like characteristics and platinum resistance in ovarian cancer. Biochem Biophys Res Commun. 2016 Jan 15;469(3):449–455. doi: 10.1016/j.bbrc.2015.11.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tripathi K, Hussein UK, Anupalli R, Barnett R, Bachaboina L, Scalici J. Allyl isothiocyanate induces replication-associated DNA damage response in NSCLC cells and sensitizes to ionizing radiation. Oncotarget. 2015 Mar 10;6(7):5237–5252. doi: 10.18632/oncotarget.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tripathi K, Mani C, Somasagara RR, Clark DW, Ananthapur V, Vinaya K. Detection and evaluation of estrogen DNA-adducts and their carcinogenic effects in cultured human cells using biotinylated estradiol. Mol Carcinog. 2016;56(3):1010–1020. doi: 10.1002/mc.22566. [DOI] [PubMed] [Google Scholar]
- 26.Somasagara RR, Spencer SM, Tripathi K, Clark DW, Mani C, Madeira da Silva L. RAD6 promotes DNA repair and stem cell signaling in ovarian cancer and is a promising therapeutic target to prevent and treat acquired chemoresistance. Oncogene. 2017;36(48):6680–6690. doi: 10.1038/onc.2017.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clark DW, Tripathi K, Dorsman JC, Palle K. FANCJ protein is important for the stability of FANCD2/FANCI proteins and protects them from proteasome and caspase-3 dependent degradation. Oncotarget. 2015 Oct 6;6(30):28816–28832. doi: 10.18632/oncotarget.5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang X, Kim AL, Feith DJ, Pegg AE, Russo J, Zhang H. Ornithine decarboxylase is a target for chemoprevention of basal and squamous cell carcinomas in Ptch1+/- mice. J Clin Invest. 2004;113(6):867–875. doi: 10.1172/JCI20732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arumugam A, Weng Z, Chaudhary SC, Afaq F, Elmets CA, Athar M. Keratin-6 driven ODC expression to hair follicle keratinocytes enhances stemness and tumorigenesis by negatively regulating Notch. Biochem Biophys Res Commun. 2014;451(3):394–401. doi: 10.1016/j.bbrc.2014.07.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cochrane CR, Szczepny A, Watkins DN, Cain JE. Hedgehog Signaling in the Maintenance of Cancer Stem Cells. Cancers. 2015;7(3):1554–1585. doi: 10.3390/cancers7030851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merchant A, Matsui W. Targeting Hedgehog - a Cancer Stem Cell Pathway. Clin Cancer Res Off J Am Assoc Cancer Res. 2010;16(12):3130–3140. doi: 10.1158/1078-0432.CCR-09-2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noman AS, Uddin M, Rahman MZ, Nayeem MJ, Alam SS, Khatun Z. Overexpression of sonic hedgehog in the triple negative breast cancer: clinicopathological characteristics of high burden breast cancer patients from. Bangladesh. Sci Rep. 2016;6:18830. doi: 10.1038/srep18830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steg AD, Katre AA, Bevis KS, Ziebarth A, Dobbin ZC, Shah MM. Smoothened antagonists reverse taxane resistance in ovarian cancer. Mol Cancer Ther. 2012;11(7):1587–1597. doi: 10.1158/1535-7163.MCT-11-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaillard H, García-Muse T, Aguilera A. Replication stress and cancer. Nat Rev Cancer. 2015;15(5):276–289. doi: 10.1038/nrc3916. [DOI] [PubMed] [Google Scholar]
- 35.Raghunandan M, Chaudhury I, Kelich SL, Hanenberg H, Sobeck A. FANCD2, FANCJ and BRCA2 cooperate to promote replication fork recovery independently of the Fanconi Anemia core complex. Cell Cycle Georget Tex. 2015;14(3):342–353. doi: 10.4161/15384101.2014.987614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu J, Su F, Mukherjee S, Mori E, Hu B, Asaithamby A. FANCD2 influences replication fork processes and genome stability in response to clustered DSBs. Cell Cycle Georget Tex. 2015;14(12):1809–1822. doi: 10.1080/15384101.2015.1036210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carpenter RL, Lo H-W. Hedgehog pathway and GLI1 isoforms in human cancer. Discov Med. 2012;13(69):105–113. [PMC free article] [PubMed] [Google Scholar]
- 38.Aszterbaum M, Epstein J, Oro A, Douglas V, LeBoit PE, Scott MP. Ultraviolet and ionizing radiation enhance the growth of BCCs and trichoblastomas in patched heterozygous knockout mice. Nat Med. 1999;5(11):1285–1291. doi: 10.1038/15242. [DOI] [PubMed] [Google Scholar]
- 39.Yoon JW, Kita Y, Frank DJ, Majewski RR, Konicek BA, Nobrega MA. Gene expression profiling leads to identification of GLI1-binding elements in target genes and a role for multiple downstream pathways in GLI1-induced cell transformation. J Biol Chem. 2002;277(7):5548–5555. doi: 10.1074/jbc.M105708200. [DOI] [PubMed] [Google Scholar]
- 40.Lord CJ, Ashworth A. BRCAness revisited. Nat Rev Cancer. 2016;16(2):110–120. doi: 10.1038/nrc.2015.21. [DOI] [PubMed] [Google Scholar]
- 41.Tripathi K, Mani C, Clark DW, Palle K. Rad18 is required for functional interactions between FANCD2, BRCA2, and Rad51 to repair DNA topoisomerase 1-poisons induced lesions and promote fork recovery. Oncotarget. 2016;7(11):12537–12553. doi: 10.18632/oncotarget.7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown JS, Kaye SB, Yap TA. PARP inhibitors: the race is on. Br J Cancer. 2016;114(7):713–715. doi: 10.1038/bjc.2016.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen A. PARP inhibitors: its role in treatment of cancer. Chin J Cancer. 2011;30(7):463–471. doi: 10.5732/cjc.011.10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krajewska M, Fehrmann RSN, de Vries EGE, van Vugt MATM. Regulators of homologous recombination repair as novel targets for cancer treatment. Front Genet. 2015;6:96. doi: 10.3389/fgene.2015.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruiz i Altaba A, Sánchez P, Dahmane N. Gli and hedgehog in cancer: tumours, embryos and stem cells. Nat Rev Cancer. 2002;2(5):361–372. doi: 10.1038/nrc796. [DOI] [PubMed] [Google Scholar]
- 46.Chen Q, Gao G, Luo S. Hedgehog signaling pathway and ovarian cancer. Chin J Cancer Res Chung-Kuo Yen Cheng Yen Chiu. 2013;25(3):346–353. doi: 10.3978/j.issn.1000-9604.2013.06.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Szkandera J, Kiesslich T, Haybaeck J, Gerger A, Pichler M. Hedgehog signaling pathway in ovarian cancer. Int J Mol Sci. 2013;14(1):1179–1196. doi: 10.3390/ijms14011179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Agyeman A, Jha BK, Mazumdar T, Houghton JA. Mode and specificity of binding of the small molecule GANT61 to GLI determines inhibition of GLI-DNA binding. Oncotarget. 2014;5(12):4492–4503. doi: 10.18632/oncotarget.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mathew CG. Fanconi anaemia genes and susceptibility to cancer. Oncogene. 2006;25(43):5875–5884. doi: 10.1038/sj.onc.1209878. [DOI] [PubMed] [Google Scholar]
- 50.Kais Z, Rondinelli B, Holmes A, O'Leary C, Kozono D, D'Andrea AD. FANCD2 maintains fork stability in BRCA1/2-deficient tumors and promotes alternative end-joining DNA repair. Cell Rep. 2016;15(11):2488–2499. doi: 10.1016/j.celrep.2016.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meehan RS, Chen AP. New treatment option for ovarian cancer: PARP inhibitors. Gynecol Oncol Res Pract. 2016;3:3. doi: 10.1186/s40661-016-0024-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. N Engl J Med. 2017 doi: 10.1056/NEJMoa1706450. [DOI] [PubMed] [Google Scholar]
- 53.Munroe M, Kolesar J. Olaparib for the treatment of BRCA-mutated advanced ovarian cancer. Am J Health-Syst Pharm AJHP Off J Am Soc Health-Syst Pharm. 2016;73(14):1037–1041. doi: 10.2146/ajhp150550. [DOI] [PubMed] [Google Scholar]
- 54.Rimkus TK, Carpenter RL, Qasem S, Chan M, Lo H-W. Targeting the Sonic Hedgehog Signaling Pathway: Review of Smoothened and GLI Inhibitors. Cancers. 2016;8(2) doi: 10.3390/cancers8020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taipale J, Chen JK, Cooper MK, Wang B, Mann RK, Milenkovic L. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature. 2000;406(6799):1005–1009. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- 56.Das S, Samant RS, Shevde LA. Nonclassical activation of Hedgehog signaling enhances multidrug resistance and makes cancer cells refractory to Smoothened-targeting Hedgehog inhibition. J Biol Chem. 2013;288(17):11824–11833. doi: 10.1074/jbc.M112.432302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yao CD, Haensel D, Gaddam S, Patel T, Atwood SX, Sarin KY. AP-1 and TGFß cooperativity drives non-canonical Hedgehog signaling in resistant basal cell carcinoma. Nat Commun. 2020;11(1):5079. doi: 10.1038/s41467-020-18762-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chien T, Weng Y-T, Chang S-Y, Lai H-L, Chiu F-L, Kuo H-C. GSK3β negatively regulates TRAX, a scaffold protein implicated in mental disorders, for NHEJ-mediated DNA repair in neurons. Mol Psychiatry. 2018 doi: 10.1038/s41380-017-0007-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harada K, Ohashi R, Naito K, Kanki K. Hedgehog Signal Inhibitor GANT61 Inhibits the Malignant Behavior of Undifferentiated Hepatocellular Carcinoma Cells by Targeting Non-Canonical GLI Signaling. Int J Mol Sci. 2020;21(9) doi: 10.3390/ijms21093126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ferreira Mendes JM, de Faro Valverde L, Torres Andion Vidal M, Paredes BD, Coelho P, Allahdadi KJ. Effects of IGF-1 on Proliferation, Angiogenesis, Tumor Stem Cell Populations and Activation of AKT and Hedgehog Pathways in Oral Squamous Cell Carcinoma. Int J Mol Sci. 2020;21(18) doi: 10.3390/ijms21186487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Po A, Silvano M, Miele E, Capalbo C, Eramo A, Salvati V. Noncanonical GLI1 signaling promotes stemness features and in vivo growth in lung adenocarcinoma. Oncogene. 2017;36(32):4641–4652. doi: 10.1038/onc.2017.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li N, Truong S, Nouri M, Moore J, Al Nakouzi N, Lubik AA. Non-canonical activation of hedgehog in prostate cancer cells mediated by the interaction of transcriptionally active androgen receptor proteins with Gli3. Oncogene. 2018;37(17):2313–2325. doi: 10.1038/s41388-017-0098-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pietrobono S, Gaudio E, Gagliardi S, Zitani M, Carrassa L, Migliorini F. Targeting non-canonical activation of GLI1 by the SOX2-BRD4 transcriptional complex improves the efficacy of HEDGEHOG pathway inhibition in melanoma. Oncogene. 2021;40(22):3799–3814. doi: 10.1038/s41388-021-01783-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Neelakantan D, Zhou H, Oliphant MUJ, Zhang X, Simon LM, Henke DM. EMT cells increase breast cancer metastasis via paracrine GLI activation in neighbouring tumour cells. Nat Commun. 2017;8:15773. doi: 10.1038/ncomms15773. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data that support the findings of this study are available from the corresponding authors upon reasonable request.