Abstract
We previously reported Israa (immune-system-released activating agent), a novel gene nested in intron 6 of the mouse Zmiz1 gene. Zmiz1 is involved in several functions such as fertility and T cell development and its knockout leads to non-viable embryos. We also reported ISRAA's expression in lymphoid organs, particularly in the thymus CD3+ T cells during all developmental stages. In addition, we showed that ISRAA is a binding partner of Fyn and Elf-1 and regulates the expression of T cell activation-related genes in vitro. In this paper, we report the generation and characterization of an Israa−/− constitutive knockout mouse. The histological study shows that Israa−/− mice exhibit thymus and spleen hyperplasia. Israa−/− derived T cells showed increased proliferation compared to the wild-type mice T cells. Moreover, gene expression analysis revealed a set of differentially expressed genes in the knockout and wild-type animals during thymus development (mostly genes of T cell activation pathways). Immunological phenotyping of the thymocytes and splenocytes of Israa−/- showed no difference with those of the wild-type. Moreover, we observed that knocking out the Zmiz1 intron embedded Israa gene does not affect mice fertility, thus does not disturb this Zmiz1 function. The characterization of the Israa−/- mouse confirms the role ISRAA plays in the expression regulation of genes involved in T cell activation established in vitro. Taken together, our findings point toward a potential functional interrelation between the intron nested Israa gene and the Zmiz1 host gene in regulating T cell activation. This constitutively Israa−/− mice can be a good model to study T cell activation and to investigate the relationship between host and intron-nested genes.
Keywords: T cell activation, Nested gene, Knock-out, Fyn, Elf-1
Highlights
-
•
Israa gene is the first reported intron-nested gene involved in T cell activation in mouse.
-
•
Israa−/− derived T cells showed increased proliferation compared to the wild-type mice T cells.
-
•
Israa knockdown perturbates expression of genes involved in T cell activation.
1. Introduction
Israa (GenBank: EU552928) is an intron 6-nested gene of Zmiz1 on mouse chromosome 14. Following T. brucei parasite inoculation to mice, Israa was found to be upregulated in splenocytes [1]. The gene encodes a novel FYN-binding protein that might be involved in the regulation of T cell activation [2]. The protein also binds the transcription factor ELF-1 involved in the expression regulation of several genes related to T cell signalling. Israa is also expressed at all stages of T-lymphocyte development and is downregulated in activated T cells, which suggests a negative regulatory role during T cell activation. Moreover, induced ISRAA upregulation in mouse lymphocytes modulates the expression of several genes that are involved in this process [3]. Meanwhile, the Zmiz1 (also known as Zimp10) gene, that hosts the Israa open reading frame, encodes for a member of the PIAS (Protein Inhibitor of Activated STAT) family of proteins. PIAS proteins are known STAT transcription factors repressors and involved in transcriptional co-regulation to modulate the activity of a diverse set of transcription factors, such as SMADs, p53, and steroid hormone receptors [4]. Zmiz1-deficient embryos die at the embryonic day E10.5 due to severe defects in yolk-sac vessel organization, indicating the essential role that Zmiz1 plays in the extra-embryonic vascular development process [5]. Most importantly, Zmiz1 is widely expressed in the thymus [4], mainly in the earliest thymic precursors [6] and plays an important role in T cell development by controlling the expression of certain Notch1 target genes, such as Myc, Lef 1 and Hes1 [7], through interacting with ETS1, a member of the Ets transcription factors family. Indeed, Zmiz1 was shown to be a specific cofactor for Notch1 during Notch/Myc-dependent thymocyte proliferation [8].
Therefore, it seems interesting to study if the functions carried out by both the embedded and host genes are interrelated, which warrant the generation and characterization of an Israa Knock-out (KO) mice. Such animal model would be of interest in investigating any functional relevance between the host and the hosted gene. In this paper we describe the generation as well as the physiological and immunological phenotyping of a constitutive Israa knock-out mouse model. Using this mouse model of knocked-out intron-nested gene, we confirm that the Israa gene plays a role in the in vivo expression and regulation of genes involved in T cell activation. Furthermore, we show that knocking out of Israa does not disturb the function of the Zmiz1 host gene. In addition, our findings are in favour of interrelated actions of the host and intron embedded gene, particularly those related to T cell regulation. This mouse model of knocked out intron-nested gene is of interest for the studies of the relationship between host and intron embedded gene and T cell biology.
2. Materials and methods
Plasmids and Primers—Primers and probes used for genotyping, Southern blotting and Real-time PCR and are listed in Table 1 and Table 2.
Table 1.
List of primers and probes used for genotyping and Southern blot.
| Primer | Primer sequence 5′ to 3′ | |
|---|---|---|
| PCR Genotyping | 125374cre-BKN1 | CTGAGACCCACATCATAAGCCATAATCAGG |
| 125375cre-BKN1 | ACTCCTTGTTTGACAGAAGTCGTTAGCAGG | |
| 5′-SA-E-A probe | 125370PRO-BKN1 | GCCAGAGGCAGAGAATACAGTTTAGAGACG |
| 125371PRO-BKN1 | CCTTTCCCGGAGCTTATTAATGAGGTTCTA | |
| 3′-LA-I-C Probe | 125403PRO-BKN1 | GAGCCCACCTCATGTCAGCATAAGC |
| 125404PRO-BKN1 | CCATTTGCCAGGTATCCAGACCTCC |
Table 2.
List of primers used for Real Time PCR.
| Gene | Primer | Primer Sequence 5′ to 3′ |
|---|---|---|
| Il2ra | IL2RAF | CTCCCATGACAAATCGAGAAAGC |
| IL2RAR | ACTCTGTCCTTCCACGAAATGAT | |
| Foxj1 | FOXJ1F | CCCTGACGACGTGGACTATG |
| FOXJ1R | GCCGACAGAGTGATCTTGGT | |
| Prkcb | PRKCBF | GTGTCAAGTCTGCTGCTTTGT |
| PRKCBR | GTAGGACTGGAGTACGTGTGG | |
| Ndfip2 | NDFIP2F | GCAGCCGTCAACTTCTAGCTT |
| NDFIP2R | TAGCAACACTGTACGGAGGTG | |
| Gata3 | GATA3F | CTCGGCCATTCGTACATGGAA |
| GATA3R | GGATACCTCTGCACCGTAGC | |
| B2m | B2MF | TTCTGGTGCTTGTCTCACTGA |
| B2MR | CAGTATGTTCGGCTTCCCATTC | |
| Gapdh | GAPHF | AGGTCGGTGTGAACGGATTTG |
| GAPDHR | TGTAGACCATGTAGTTGAGGTCA | |
| Socs3 | SOCS3F | ATGGTCACCCACAGCAAGTTT |
| SOCS3R | TCCAGTAGAATCCGCTCTCCT | |
| Cd247 | CD247F | TTCAGAACTCACAAGGACCCT |
| CD247R | GCTACTCTGCTGGGTGCTTTC | |
| Fos | FOSF | CGGGTTTCAACGCCGACTA |
| FOSR | TTGGCACTAGAGACGGACAGA | |
| Israa | ISRAAF | TGACCATGCAGAAGGAGACA |
| ISRAAR | GGAAGCGTGGAGAAGAACAA | |
| Zmiz1 | ZMIZ1F | CCTGGGCTACAGACTCTTGG |
| ZMIZ1R | ATGGAGCTCATGGAGCTGAG |
Generation of Israa knockout mice by concurrent gene targeting— All procedures and protocols used in the experiments involving animals were reviewed and approved by the Arabian Gulf University Ethics Committee and are in accordance with the National Institutes of Health guide for the care and use of Laboratory animals (NIH Publications No. 8023, revised 1978) and complying with the ARRIVE guidelines [9]. The cloning and the initial generation of mice were conducted with in collaboration with Genoway, Inc. (Lyon, France). The targeting strategy was previously described [3]. In brief, an IRES-eGFP cassette was inserted downstream of the Israa coding sequence (CDS) along with insertion of LoxP sites flanking the entire gene (exon 1 and exon 2) (Fig. 1A). The correctly targeted ES cell clones were injected into C57BL/6J blastocysts to generate chimaeric mice. To generate Israa ± mice, chimaeras were then crossed with β-actin–Cre transgenic mice [strain: FVB/N-Tg (ACTB-cre)2 Mrt/J; stock no.: 003376; The Jackson Laboratory]. F1 heterozygous males were further validated by Southern blot as previously described [3] and backcrossed for 8 generations with C57BL/6J wild-type mice. Backcrossed heterozygous mice were inter-crossed to generate homozygous knockout Israa−/− mice and validated by Southern Blot (Fig. 1B), wild type (WT) C57BL/6J DNA was used as negative control.
Fig. 1.
Israa gene targeting strategy. Hatched rectangles represent Israa coding sequences, grey rectangles indicate non-coding exon portions, red and green rectangles represent respectively IRES and reporter (eGFP, ref: addgene 24,557) cassettes, solid lines represent chromosome sequences. LoxP sites are represented by blue triangles and FRT sites by double red triangles. The initiation (ATG) and Stop (Stop) codons are indicated. The size of the flanked Israa sequence to be deleted is specified. (A) Schematic representation of the wild-type, recombined, Cre-mediated excised and Flp-mediated excised Israa alleles with the relevant restriction sites for the Southern blot analysis. (B) Southern blot analysis of the homozygous constitutive Knock-out mice (4 kb) compared with C57BL/6 wild-type genomic DNA (WT, 15.7 kb). (C) PCR identification of the Cre-mediated excision event within the recombined Israa locus. The figure indicates the PCR screening strategy for the Cre-mediated excision event. Blue arrows illustrate the primers localisation. (D) Cropped gel showing the result of PCR testing the primers for detection of Cre-excision event. The optimised PCR screen was tested on wild-type mice (lane 1–3) and homozygous knock-out (lane 4–7). Expected sizes of 4 Kb and 1 Kb in Wild type C57BL/6 and Israa−/− mice, respectively, were detected. DTA, Diphtheria Toxin A negative selection marker, FRT, flippase recognition target sites; FLPe, flippase; IRES, internal ribosome entry site, neo, neomycin resistance gene. Diagram is not drawn to scale. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Genotyping— PCR for genotyping mice was performed on genomic DNA prepared from tail snips or ear clippings using the DNeasy Blood and Tissue kit® (QIAGEN, USA). Briefly, mouse ear biopsies were lysed, and genomic DNA was isolated and used for genotyping reaction. For the detection of the Cre-mediated recombination, the 125374cre-BKN1/125375cre-BKN1 primer set was used to amplify Cre-excised (984 bp) and wild-type (4444 bp) alleles (Fig. 1C and D).
Histological analysis of Israa−/−mice— For histopathological evaluation of genetically engineered mice, four female C57BL/6J mice of different ages (2, 4, 6 and 8 weeks) from each strain (Israa−/− and wild-type) were used. Upon necropsy, the brain, liver, kidneys, ovaries, heart, spleen, lymph nodes, thymus and lungs were collected and fixed in 10% formalin solution and embedded in paraffin before being cut into 5-μm-thick sections. Three sections from each sample were stained with haematoxylin and eosin (H&E) for standard histological investigations. In addition, four adult male C57BL/6J mice (3 Israa−/− and 1 wild-type) were used for the testicular histopathological evaluation. Testes and epididymis were collected after necropsy, fixed in Bouin's solution and embedded in paraffin before being cut into 5-μm-thick sections. For each sample, one section was stained with H&E for standard histology and one section was stained using Periodic Acid Schiff's reaction [10]. Two additional adult Israa−/− and wild-type males were used for the spermogram evaluation; each animal was evaluated separately. Sperm was collected from the epididymis cauda and analysed with a sperm analyser (IVOS Animal, Mouse Traxx software, Hamilton Thorne).
Fertility analysis. Crossbreeding was run for mice 8 weeks of age between Israa−/− and wild-type male and female, with 13 couples for each combination, for a period of three months. Wild-type couples were used as the control group. Pups were counted for each delivery and weaned after three weeks of delivery.
Isolation of thymocytes and splenocytes—Spleens and thymus were dissected from six-weeks old wild-type or Israa−/− female mice. Cells were prepared by forcing the tissues through a 70 μm Falcon™ Cell Strainer (Thermo Fisher, USA). The dissociated cells were washed once in PBS. Haemolysis of erythrocytes in the cell pellets was completed by adding 1 ml of RBC lysis buffer (BioLegend, USA) for 20 min at 4 °C. The cells were washed twice in RPMI medium supplemented with 2 mM l-glutamine, 50 units per ml penicillin, and 50 μg per ml streptomycin (Sigma-Aldrich, MO, USA) and then re-suspended in the same medium supplemented with 10% Foetal Calf Serum FCS (Life Technologies, CA, USA) to obtain a concentration of 2 × 106 cells/ml. Cells were then used for Flow cytometry, proliferation assay and total RNA extraction.
Flow cytometry analysis—Flow cytometry was used to analyse the lymphocyte cellularity and composition of thymocytes and splenocytes. Cells were isolated from two six-weeks old wild-type and two Israa−/− female mice, as described previously in this section. In brief, cells were harvested, and viability was checked by Trypan blue dye incorporation. Cells were then resuspended at a concentration of 2 × 106 cells/ml and blocked by adding Rat Anti-Mouse CD16/CD32 (#553141, BD Biosciences, USA) for 20 min at 4 °C at a final concentration of 5 μg/ml. Labelled antibodies were then added at respective concentrations and mixtures were incubated 20 min in the dark. Cells were then washed 3 times in 200 μl PBS1x and resuspended in 500 μl of PBS1x for FACS analysis on FACSCalibur Cell analyser (BD Biosciences, USA), after standardization with BD Calibrite beads (BD Biosciences, USA). For T cell populations, FITC Rat Anti-mouse CD8a (#553030, BD Biosciences, USA), PE Rat-Anti-mouse CD4 (#553730, BD Biosciences, USA) and PerCP Rat-Anti-mouse CD3e (#553067, BD Biosciences, USA) antibodies were used at a final concentration of 5 μg/ml. For each panel, 104 cellular events were recorded in two independent experiments. Data acquisition, analysis and statistical interpretation was performed using the CellQuest Pro Software (BD Biosciences, USA).
T cell isolation and proliferation assay— T cells were isolated from six-week-old wild-type and Israa−/− female mice splenocytes (five mice from each group) using Dynabeads™ FlowComp™ Mouse Pan T (CD90.2) Kit (Thermo Fischer Scientific, USA) according to the manufacturer recommendation. For the activation assay, Dynabeads® Mouse T-Activator CD3/CD28 kit (Thermo Fischer, USA) was used. Briefly, 8 × 104 purified T cells were incubated with 2 μl of pre-washed magnetic beads and incubated at 37 °C, 5% CO2 in a humidified incubator in presence and absence of Concanavalin A (ConA) (5 μg/ml, Merck-Sigma, USA)). Proliferation was assayed using CellTiter-Glo® Luminescent Cell Viability Assay (Promega, USA) after 72 h of incubation, according to the manufacturer instructions, luminescence was measured using GloMax® Discover Multimode Microplate Reader (Promega, USA). Experiments were run in triplicates and results were expressed as stimulation index using unstimulated cells as reference. Stimulation Index (SI) was calculated according to the following formula:
SI = (luminescence of stimulated cells / luminescence of untreated cells)Real time PCR for gene expression analysis— Four wild-type and Israa−/− female mice of different ages (4, 6 and 8 weeks) were used to extract thymus T cells as described previously. Total RNA was extracted from cell suspensions using the RNeasy RNA extraction kit (QIAGEN) and reverse transcribed using the ProtoScript® First Strand cDNA Synthesis Kit (NEB, UK) according to the manufacturer's instructions. Primers for real-time PCR (Table 2) were designed to specifically amplify target genes using the SybrGreen Gene Expression Master Mix (Thermo Fischer, USA). The experiments were run in triplicates and the results were expressed as the 2−ΔΔCq fold change [11] using GAPDH as a housekeeping gene.
2.1. Statistical data analysis
Statistical significance was calculated with t-test for comparing means with unequal variance for the fertility study and T cell proliferation assay. For all gene expression experiments, one-way ANOVA followed by Tukey's post hoc test was used to compare experimental groups to the control. All analyses were performed using SPSS statistics software version 27 (IBM Corp.). For all tests, p < 0.05 was considered to indicate a statistically significant difference.
3. Results
Generation of a constitutive Israa−/−mouse strain—To allow for constitutive deletion of Israa, LoxP recombination sites along with an FRT-flanked neomycin-selection cassette were inserted upstream of exon 1 and downstream of exon 2 (Fig. 1A). The gene targeting construct was introduced into ES cells to obtain heterozygous Israa-eGFP+/flox ES clones by homologous recombination. Israa-eGFP+/flox ES cells were then injected into mouse blastocysts and homozygous Israa-eGFPflox/flox mice were obtained using standard procedures as previously described [3]. Israa-eGFPflox/flox mice were crossed with β-actin-Cre mice to obtain Israa ± mice that were back-crossed with C57BL/6 wild type mice for 8 generations to stabilize the genetic background. Heterozygous mice were then intercrossed to obtain homozygous knockout Israa−/− mice. Genotypes of Israa−/− mice were validated by Southern Blot (Fig. 1B) and PCR genotyping (Fig. 1C and D).
Fertility and sperm analysis.Israa−/− mice were obtained at the expected Mendelian frequencies and displayed a normal lifespan when breeding homozygous mice together. Fertility analysis was carried on 13 couples of Israa−/− males and females crossed with wild-type mice; control was wild-type breeding mice. Couples were put in the same condition and deliveries monitored for six months. Average pups per delivery was calculated for each group (Fig. 2). Female Israa−/− mice were shown to be significantly more fertile (♂wt ♀Israa−/−, p < 0.001), with an average delivery of 8.17 pups per delivery, in comparison to control mice (♂wt ♀wt, average of 5.38 pups per delivery). Male Israa−/− mice showed no significant difference compared to control (♂Israa−/− ♀wt, average of 5.58 pups per delivery). Sperm analysis in two Israa−/− mice showed high cell count compared to standards with 306 × 106 and 163 × 106 sperm/ml, in addition to a high mobility rate (motile cells representing 85% of total counted cells).
Fig. 2.
Fertility analysis of Israa−/− mice. Average pups obtained per delivery are plotted for every group of 13 couples each. HW: ♂Israa−/− ♀wt (male Israa−/− x female wild type), WH: ♂wt ♀Israa−/− (female Israa−/− x male wild type) and WW: ♂wt ♀wt (male wild type x female wild type). Error bars represent the standard deviation. (*** for Student's test p < 0.001). Wt for wild type, Israa−/− for the Israa gene knocked-out mice.
Histological analysis of Israa−/−mice—Histopathological analysis revealed thymus hyperplasia with cell hyperactivation in the cortical zone in all considered Israa−/− mice compared to control mice. In addition, we noticed a splenic white pulp hyperplasia in the Israa−/− mice compared to control. Representative pictures are presented in Fig. 3A. No major histopathological differences were noticed in the other organs. However, in spite of high expression of Israa in testis as shown previously [3], its deletion did not result in any phenotypic changes and testis from Israa−/− mice displayed complete spermatogenesis (Fig. 3B).
Fig. 3.
Histopathological analysis of Israa−/− mice. (A) Representative sample pictures for H&E staining of thymus (2-week-old mice, w2) and spleen (6-week-old mice, w6) sections from Israa−/− and wild type mice. Arrows point to hyperplasia with cell hyperactivation in the thymus cortical zone and splenic white pulp hyperplasia in comparison to control. (B) Testes from 8-week-old (w8) Israa−/− and wild-type mice stained with H&E. Israa−/− testes showed no histological differences when compared to the wild-type testes.
T cell subpopulation counts in lymphatic organs. Flow Cytometry analysis covered T cell subpopulations (SP CD4+CD8−, SP CD4−CD8+, DP CD4+CD8+ and DN CD4−CD8−) in the thymus and the spleen. The analysis showed there are no significant differences in T cells sub-population distribution in the thymus and spleen in six-week aged Israa−/− mice (KO) compared to controls (WT) (Fig. 4).
Fig. 4.
Flow Cytometry analysis of T cell composition. (A) Thymus and spleen T cells sub-population of Israa−/− (KO) and control mice (WT) were analysed. CD3+ cells were gated then CD4+ vs CD8+ events were plotted to show different T cell populations in each sample: Single positive; SP CD4+CD8− (lower right, LR) and SP CD4−CD8+(upper left, UL); Double positive: DP CD4+CD8+ (upper right, UR) and DN CD4−CD8− (lower left, LL). (B) Cell percentage of each sub-population is represented for spleen and thymus from two independent experiments. Error bars represent the standard deviation for each group.
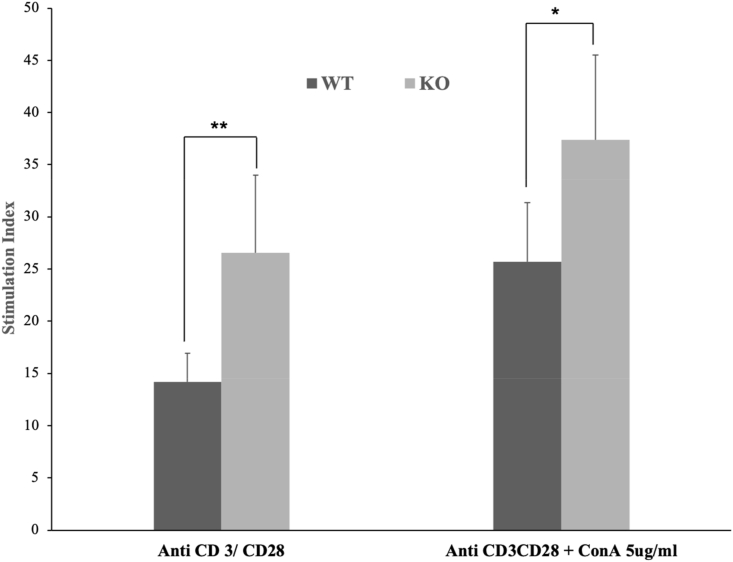
T cells proliferation assay — To further test for T cell activation ability, purified T cells were isolated from Israa−/− mice spleen and used for stimulation with anti-CD3/anti-CD28 antibody-coated beads in presence and absence of ConA (5 μg/ml). Israa−/− splenocytes showed significantly high proliferation index at 72 h compared to wild-type cells (Fig. 5). Stimulation index was 26.54 for Israa−/− vs 14.18 for the wild type, at 72 h post stimulation in absence of ConA, and 37.36 vs. 25.68 respectively at 72 h post stimulation in the presence of ConA (5 μg/ml). Detailed results of the proliferation assay are provided in the Supplementary Material 1.
Fig. 5.
T cell proliferation assay. Purified T Cells from six-week-old Israa−/− (KO) and control mice (WT) (Five from each group), were tested for proliferation with anti-CD3/anti-CD28 stimulation with or without ConA (5 μg/ml). Average stimulation index following 72 h is represented for each condition in comparison to untreated cells. Error bars represent the standard deviation for each group. (** for Student's t-test p < 0.01, * for Student's t-test p < 0.05).
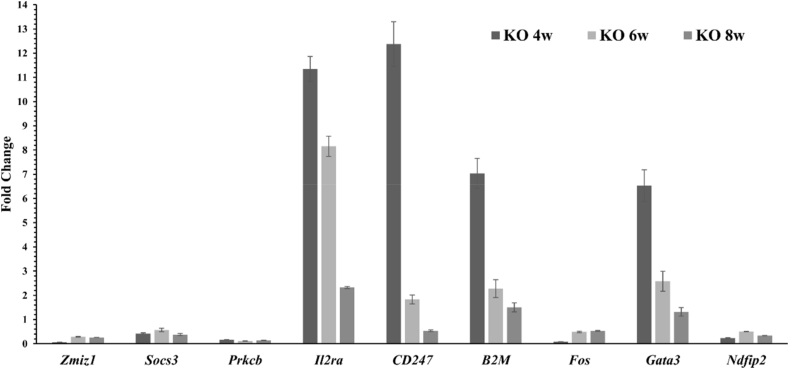
Gene expression analysis—Using real time PCR we validated several genes involved in T cell signalling, activation and differentiation to be differentially regulated in Israa−/− mice compared to control across different thymus maturation stages (4, 6 and 8 weeks of age). Among these genes, Il2ra, Cd247, B2m, Gata3 were upregulated mainly at weeks 4 and 6 in Israa−/− mice thymus, whereas Prkcb, Socs3, Fos, Ndfip2 were downregulated in all stages (Fig. 6). We also noticed a downregulation of Zmiz1 expression at the considered stages. Israa gene product was also shown, as expected, to be completely absent in Israa−/− mice thymocytes and present in the control mice.
Fig. 6.
Gene expression analysis. Real time PCR analysis of target genes were performed on thymocytes extracted mRNA extracts of Israa−/− (KO) and control mice (WT). Expression was monitored in four mice at 4, 6 and 8 weeks of age of each strain and average fold change compared to the control mice is represented for each target gene. Israa gene product was completely absent in Israa−/− mice thymocytes and present in the control mice (not shown). Error bars represent the standard error for each group.
4. Discussion
Intron-nested genes are genes whose entire coding sequences lie within a non-coding region of a host gene. In Drososphila melanogaster, nested genes account for approximately 10% of the organism's total number of genes, and 85% of these nested genes are predicted to encode proteins [12]. In the human genome, Yu et al. [13] identified 158 predicted protein-coding sequences embedded in the introns of various human genes. Several reports have progressively shown that intron-nested genes have major biological functions other than self-regulation [[14], [15], [16]]. We previously showed in vitro that Israa, a novel gene nested within the 50 Kb-long intron 6 of the Zmiz1 gene [2], is involved in T cell signalling through interacting with FYN and ELF-1 [2]. We showed Israa's expression in lymphoid organs (spleen and thymus) and observed that its in vitro overexpression modulates the expression of several genes involved in T cell activation [3].
To get insights into the role played by this gene in the mouse immune system regulation as well as its relationship with the Zmiz1 host gene, we generated an Israa−/− constitutive knockout mouse model. The generated mice were viable and fertile, with no major defect in organ anatomy, which contrasts with the Zimz1 knockout mouse. Nevertheless, knocking out the Israa coding sequence did not alter Zmiz1 gene function in embryonic development. Indeed, the Zmiz1 gene is essential for embryos viability and proper vascular development [5]. Interestingly, female Israa−/− mice showed increased fertility compared to wild-type animals. Noteworthy is the study by Liu et al. [17] who reported the Zmiz1 as a candidate gene for fertility modulation in females Holstein cattle.
Moreover, the observation of hyperplasia and cellular hyperactivation in the developing thymus and spleen of Israa−/− mice is in line with our previous findings in vitro on the role Israa plays in T cell activation [2]. Moreover, the identical T cell composition in both organs observed through FACS analysis in Israa−/− and the wild-type mice, rules out a lymphoma-related pathology in these organs. This also suggests that the maturation process is not affected. Meanwhile, the observation of the upregulation of Gata3 among the genes previously shown [3] to be downregulated by ISRAA overexpression, during thymus development is of particular interest. Indeed, Gata3 regulates the development and functions of naive lymphoid cell subsets at multiple stages [18] and is essential for T cell development [19]. Gata3 controls also the expression of TCRβ at a post-transcriptional level and inappropriate expression of GATA factors can divert the development of progenitors into alternative lineages [19]. In addition, B2m is another interesting gene found to be upregulated in Israa−/− mice. Indeed, B2m is involved in the regulation of CD49f, an essential factor in thymocyte development [20]. In addition, B2m deficient mice were reported to lack CD8+ T cells [21]. Even though cellularity analysis in the current work indicated that Israa depletion did not interfere with the composition of T cells, we cannot discard an eventual effect of these two important genes on the development of T cell lineages in the Israa−/− model. The role of Zmiz1 downregulation in the early stages of thymus maturation should also be highlighted, taking into account that Zmiz1 is co-expressed with activated Notch1 across a broad range of T-acute leukaemia oncogenic subgroups and is involved in T cell maturation [7]. It is important to note here that Notch1 represses Gata3 mRNA [8]. In addition, ChipSeq analysis showed Cd247, B2m, Gata3, Prkcb, Socs3, Fos and Ndfip2 are target genes for Zmiz1 [22]. In addition, it is reported in the GWAS database [23]that the Zmiz1 polymorphism is associated with at least eleven clinical phenotypes, including multiple autoimmune diseases associated to T cell activation such as sclerosis, psoriasis, Crohn's disease, inflammatory bowel disease, as well as the response to the drug methotrexate in rheumatoid arthritis [24]. Zmiz1 is also highly expressed in inflammatory monocytes and plasmacytoid cells [24]. Furthermore, the downregulation of Zimz1 combined with the upregulation of Gata3 and B2m in the Israa−/− mice thymus indicate that the deletion of Israa gene affected host gene expression and function; however, this did not alter T cell development but might have interfered with the process of T cell activation. In support of this hypothesis, is the fact that CD247 and Il2ra (CD25) genes are upregulated in the thymus of the Israa −/− mice. CD25 deficiency was associated with lymphoproliferation and increased T cell activation markers [25], and CD247 was reported to play an important role in coupling antigen recognition by T cell receptor to several intracellular signal-transduction pathways [26]. Israa−/− isolated T cells show increased proliferation upon CD3/CD28 stimulation as compared to control. In this work we show that ISRAA downregulates CD25 and CD247 expression which negatively regulates T cell activation. Indeed CD25 and CD247 are regulated by ISRAA's partner ELF-1 [27,28], which is an Ets-related transcription factor that is expressed at high levels in T cells [29] and is known to regulate the expression of several T cell genes involved in cellular signalling and activation, directly or through interaction with other transcription factors like NFκB [26,[30], [31], [32], [33]].
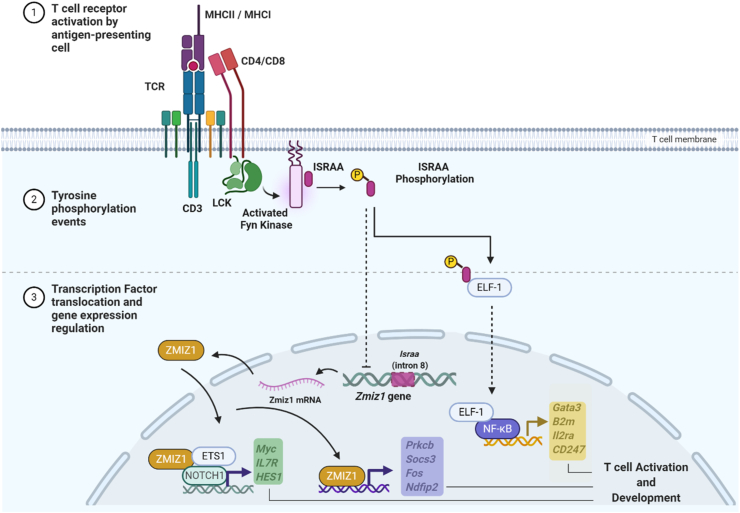
Interestingly, the Zmiz1, Israa's host gene, interacts with ETS1 [7], a member of the Ets family, and this interaction is important for regulating Notch1 target genes like Hes1, Myc and Il7r, all involved in T cell development and leukaemia [7]. These observations are in line with our previous findings where recombinant ISRAA protein caused a negative regulation of cell activation [2]. In addition, ISRAA is a substrate of FYN kinase, that is involved in terminating TCR signals, through phosphorylation of several negative regulators of T cell signalling, including PAG [34], SLAM-associated protein [35], and Cbl [36]. Therefore, it is likely that ISRAA's phosphorylation by FYN triggers its interaction with transcription factors like ELF-1 and NFκB to down-regulate T cell activation. Fig. 7 depicts integration of the data generated in this study with our previous in vitro observations and data from the literature to present a model that shows a likely interrelated function of Israa and its host gene Zmiz1 in regulating T cell activation and signalling following TCR engagement. The use of Israa knock-out to establish T cell mediated disease models would be of a great benefit to further investigate the role played by Fyn and its cascade of interactions and to get more insights on other interactions that regulate T cell functions.
Fig. 7.
A potential model for Israa and Zimz1 in the T cell activation pathway. Following TCR engagement, activated Fyn phosphorylates ISRAA which binds to ELF-1 and triggers the expression of genes regulated by NFκB and involved in T cell activation (Yellow Box). The observed down regulation of the Zmiz1 gene expression and that of Prkcb, Socs3, Fos, and Ndfip2 (Purple Box), involved in T cell activation and under the control of Zimz1, following Israa knockout, suggests that ISRAA inhibits the Zmiz1 expression. The latter is known to interact with Notch1 and Ets1 and regulates T cell activation through the control of the Myc, IL7R and HES1 genes (Green Box). The interactions shown by the dotted arrows are putative interactions that needs further experimental work. Created with BioRender.com. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work has been funded by Research Grant ISRAA of the Arabian Gulf University. We acknowledge the technical support of Ms. Nanitah Rachel in the Arabian Gulf University Animal Facility for the maintenance of the animal models in this study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2021.101100.
Contributor Information
Noureddine Ben Khalaf, Email: noureddinek@agu.edu.bh.
M. Dahmani Fathallah, Email: d.fathallah@agu.edu.bh.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Bakhiet M., Taha S. A novel nervous system-induced factor inducing immune responses in the spleen. Immunol. Cell Biol. 2008;86:688–699. doi: 10.1038/icb.2008.57. [DOI] [PubMed] [Google Scholar]
- 2.Ben Khalaf N., Taha S., Bakhiet M., Fathallah M.D. A central nervous system-dependent intron-embedded gene encodes a novel murine Fyn binding protein. PloS One. 2016;11 doi: 10.1371/journal.pone.0149612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ben Khalaf N., Al-Mashoor W., Saeed A., Al-Mehatab D., Taha S., Bakhiet M., Fathallah M.D. The mouse intron-nested gene, Israa, is expressed in the lymphoid organs and involved in T-cell activation and signaling. Mol. Immunol. 2019;111:209–219. doi: 10.1016/j.molimm.2019.04.006. [DOI] [PubMed] [Google Scholar]
- 4.Sharma M., Li X., Wang Y., Zarnegar M., Huang C.Y., Palvimo J.J., Lim B., Sun Z. hZimp10 is an androgen receptor co-activator and forms a complex with SUMO-1 at replication foci. EMBO J. 2003;22:6101–6114. doi: 10.1093/emboj/cdg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beliakoff J., Lee J., Ueno H., Aiyer A., Weissman I.L., Barsh G.S., Cardiff R.D., Sun Z. The PIAS-like protein Zimp10 is essential for embryonic viability and proper vascular development. Mol. Cell Biol. 2008;28:282–292. doi: 10.1128/MCB.00771-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rakowski L.A., Garagiola D.D., Li C.M., Decker M., Caruso S., Jones M., Kuick R., Cierpicki T., Maillard I., Chiang M.Y. Convergence of the ZMIZ1 and NOTCH1 pathways at C-MYC in acute T lymphoblastic leukemias. Canc. Res. 2013;73:930–941. doi: 10.1158/0008-5472.CAN-12-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pinnell N., Yan R., Cho H.J., Keeley T., Murai M.J., Liu Y., Alarcon A.S., Qin J., Wang Q., Kuick R., Elenitoba-Johnson K.S., Maillard I., Samuelson L.C., Cierpicki T., Chiang M.Y. The PIAS-like coactivator Zmiz1 is a direct and selective cofactor of Notch1 in T cell development and leukemia. Immunity. 2015;43:870–883. doi: 10.1016/j.immuni.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Q., Yan R., Pinnell N., McCarter A.C., Oh Y., Liu Y., Sha C., Garber N.F., Chen Y., Wu Q., Ku C.J., Tran I., Serna Alarcon A., Kuick R., Engel J.D., Maillard I., Cierpicki T., Chiang M.Y. Stage-specific roles for Zmiz1 in Notch-dependent steps of early T-cell development. Blood. 2018;132:1279–1292. doi: 10.1182/blood-2018-02-835850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Percie du Sert N., Hurst V., Ahluwalia A., Alam S., Avey M.T., Baker M., Browne W.J., Clark A., Cuthill I.C., Dirnagl U., Emerson M., Garner P., Holgate S.T., Howells D.W., Karp N.A., Lazic S.E., Lidster K., MacCallum C.J., Macleod M., Pearl E.J., Petersen O.H., Rawle F., Reynolds P., Rooney K., Sena E.S., Silberberg S.D., Steckler T., Wurbel H. The ARRIVE guidelines 2.0: updated guidelines for reporting animal research. BMJ Open Sci. 2020;4 doi: 10.1136/bmjos-2020-100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aterman K., Norkin S. The periodic acid-Schiff reaction. Nature. 1963;197:1306. doi: 10.1038/1971306a0. [DOI] [PubMed] [Google Scholar]
- 11.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 12.Hube F., Francastel C. Mammalian introns: when the junk generates molecular diversity. Int. J. Mol. Sci. 2015;16:4429–4452. doi: 10.3390/ijms16034429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu P., Ma D., Xu M. Nested genes in the human genome. Genomics. 2005;86:414–422. doi: 10.1016/j.ygeno.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 14.Baggio F., Bozzato A., Benna C., Leonardi E., Romoli O., Cognolato M., Tosatto S.C., Costa R., Sandrelli F. 2 mit, an intronic gene of Drosophila melanogaster timeless 2, is involved in behavioral plasticity. PloS One. 2013;8 doi: 10.1371/journal.pone.0076351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cawthon R.M., O'Connell P., Buchberg A.M., Viskochil D., Weiss R.B., Culver M., Stevens J., Jenkins N.A., Copeland N.G., White R. Identification and characterization of transcripts from the neurofibromatosis 1 region: the sequence and genomic structure of EVI2 and mapping of other transcripts. Genomics. 1990;7:555–565. doi: 10.1016/0888-7543(90)90199-5. [DOI] [PubMed] [Google Scholar]
- 16.Salmeron J.M., Oldroyd G.E., Rommens C.M., Scofield S.R., Kim H.S., Lavelle D.T., Dahlbeck D., Staskawicz B.J. Tomato Prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the Pto kinase gene cluster. Cell. 1996;86:123–133. doi: 10.1016/s0092-8674(00)80083-5. [DOI] [PubMed] [Google Scholar]
- 17.Liu A., Wang Y., Sahana G., Zhang Q., Liu L., Lund M.S., Su G. Genome-wide association studies for female fertility traits in Chinese and Nordic Holsteins. Sci. Rep. 2017;7:8487. doi: 10.1038/s41598-017-09170-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu J. GATA3 regulates the development and functions of innate lymphoid cell subsets at multiple stages. Front. Immunol. 2017;8:1571. doi: 10.3389/fimmu.2017.01571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho I.C., Tai T.S., Pai S.Y. GATA3 and the T-cell lineage: essential functions before and after T-helper-2-cell differentiation. Nat. Rev. Immunol. 2009;9:125–135. doi: 10.1038/nri2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Golbert D.C.F., Santana-Van-Vliet E., Ribeiro-Alves M., Fonseca M., Lepletier A., Mendes-da-Cruz D.A., Loss G., Cotta-de-Almeida V., Vasconcelos A.T.R., Savino W. Cell adhesion & migration; 2017. Small Interference ITGA6 Gene Targeting in the Human Thymic Epithelium Differentially Regulates the Expression of Immunological Synapse-Related Genes; pp. 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bevan M.J. The earliest knockouts. J. Immunol. 2010;184:4585–4586. doi: 10.4049/jimmunol.1090023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rouillard A.D., Gundersen G.W., Fernandez N.F., Wang Z., Monteiro C.D., McDermott M.G., Ma'ayan A. Database; Oxford: 2016. The Harmonizome: a Collection of Processed Datasets Gathered to Serve and Mine Knowledge about Genes and Proteins; p. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buniello A., MacArthur J.A.L., Cerezo M., Harris L.W., Hayhurst J., Malangone C., McMahon A., Morales J., Mountjoy E., Sollis E., Suveges D., Vrousgou O., Whetzel P.L., Amode R., Guillen J.A., Riat H.S., Trevanion S.J., Hall P., Junkins H., Flicek P., Burdett T., Hindorff L.A., Cunningham F., Parkinson H. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47:D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fewings N.L., Gatt P.N., McKay F.C., Parnell G.P., Schibeci S.D., Edwards J., Basuki M.A., Goldinger A., Fabis-Pedrini M.J., Kermode A.G., Manrique C.P., McCauley J.L., Nickles D., Baranzini S.E., Burke T., Vucic S., Stewart G.J., Booth D.R. The autoimmune risk gene ZMIZ1 is a vitamin D responsive marker of a molecular phenotype of multiple sclerosis. J. Autoimmun. 2017;78:57–69. doi: 10.1016/j.jaut.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 25.Goudy K., Aydin D., Barzaghi F., Gambineri E., Vignoli M., Ciullini Mannurita S., Doglioni C., Ponzoni M., Cicalese M.P., Assanelli A., Tommasini A., Brigida I., Dellepiane R.M., Martino S., Olek S., Aiuti A., Ciceri F., Roncarolo M.G., Bacchetta R. Human IL2RA null mutation mediates immunodeficiency with lymphoproliferation and autoimmunity. Clin. Immunol. 2013;146:248–261. doi: 10.1016/j.clim.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rellahan B.L., Jensen J.P., Howcroft T.K., Singer D.S., Bonvini E., Weissman A.M. Elf-1 regulates basal expression from the T cell antigen receptor zeta-chain gene promoter. J. Immunol. 1998;160:2794–2801. [PubMed] [Google Scholar]
- 27.Lecine P., Algarte M., Rameil P., Beadling C., Bucher P., Nabholz M., Imbert J. Elf-1 and Stat 5 bind to a critical element in a new enhancer of the human interleukin-2 receptor alpha gene. Mol. Cell Biol. 1997;17:2351. doi: 10.1128/mcb.17.4.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang J., Yang W., Hirankarn N., Ye D.Q., Zhang Y., Pan H.F., Mok C.C., Chan T.M., Wong R.W., Mok M.Y., Lee K.W., Wong S.N., Leung A.M., Li X.P., Avihingsanon Y., Rianthavorn P., Deekajorndej T., Suphapeetiporn K., Shotelersuk V., Baum L., Kwan P., Lee T.L., Ho M.H., Lee P.P., Wong W.H., Zeng S., Zhang J., Wong C.M., Ng I.O., Garcia-Barcelo M.M., Cherny S.S., Tam P.K., Sham P.C., Lau C.S., Lau Y.L. ELF1 is associated with systemic lupus erythematosus in Asian populations. Hum. Mol. Genet. 2011;20:601–607. doi: 10.1093/hmg/ddq474. [DOI] [PubMed] [Google Scholar]
- 29.Bassuk A.G., Barton K.P., Anandappa R.T., Lu M.M., Leiden J.M. Expression pattern of the Ets-related transcription factor Elf-1. Mol. Med. 1998;4:392–401. [PMC free article] [PubMed] [Google Scholar]
- 30.Wang C.Y., Bassuk A.G., Boise L.H., Thompson C.B., Bravo R., Leiden J.M. Activation of the granulocyte-macrophage colony-stimulating factor promoter in T cells requires cooperative binding of Elf-1 and AP-1 transcription factors. Mol. Cell Biol. 1994;14:1153–1159. doi: 10.1128/mcb.14.2.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Serdobova I., Pla M., Reichenbach P., Sperisen P., Ghysdael J., Wilson A., Freeman J., Nabholz M. Elf-1 contributes to the function of the complex interleukin (IL)-2-responsive enhancer in the mouse IL-2 receptor alpha gene. J. Exp. Med. 1997;185:1211–1221. doi: 10.1084/jem.185.7.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarafova S., Siu G. A potential role for Elf-1 in CD4 promoter function. J. Biol. Chem. 1999;274:16126–16134. doi: 10.1074/jbc.274.23.16126. [DOI] [PubMed] [Google Scholar]
- 33.Finco T.S., Justice-Healy G.E., Patel S.J., Hamilton V.E. Regulation of the human LAT gene by the Elf-1 transcription factor. BMC Mol. Biol. 2006;7:4. doi: 10.1186/1471-2199-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yasuda K., Nagafuku M., Shima T., Okada M., Yagi T., Yamada T., Minaki Y., Kato A., Tani-Ichi S., Hamaoka T., Kosugi A. Cutting edge: Fyn is essential for tyrosine phosphorylation of Csk-binding protein/phosphoprotein associated with glycolipid-enriched microdomains in lipid rafts in resting T cells. J. Immunol. 2002;169:2813–2817. doi: 10.4049/jimmunol.169.6.2813. [DOI] [PubMed] [Google Scholar]
- 35.Latour S., Gish G., Helgason C.D., Humphries R.K., Pawson T., Veillette A. Regulation of SLAM-mediated signal transduction by SAP, the X-linked lymphoproliferative gene product. Nat. Immunol. 2001;2:681–690. doi: 10.1038/90615. [DOI] [PubMed] [Google Scholar]
- 36.Tsygankov A.Y., Mahajan S., Fincke J.E., Bolen J.B. Specific association of tyrosine-phosphorylated c-Cbl with Fyn tyrosine kinase in T cells. J. Biol. Chem. 1996;271:27130–27137. doi: 10.1074/jbc.271.43.27130. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.