Abstract
Objective
CRISPR/Cas9 technology has revolutionized gene editing and fast tracked our capacity to manipulate genes of interest for the benefit of both research and therapeutic applications. Whilst many advances have, and continue to be made in this area, perhaps the most utilized technology to date has been the generation of knockout cells, tissues and animals. The advantages of this technology are many fold, however some questions still remain regarding the effects that long term expression of foreign proteins such as Cas9, have on mammalian cell function. Several studies have proposed that chronic overexpression of Cas9, with or without its accompanying guide RNAs, may have deleterious effects on cell function and health. This is of particular concern when applying this technology in vivo, where chronic expression of Cas9 in tissues of interest may promote disease-like phenotypes and thus confound the investigation of the effects of the gene of interest. Although these concerns remain valid, no study to our knowledge has yet to demonstrate this directly.
Methods
In this study we used the lox-stop-lox (LSL) spCas9 ROSA26 transgenic (Tg) mouse line to generate four tissue-specific Cas9-Tg models that express Cas9 in the heart, liver, skeletal muscle or adipose tissue. We performed comprehensive phenotyping of these mice up to 20-weeks of age and subsequently performed molecular analysis of their organs.
Results
We demonstrate that Cas9 expression in these tissues had no detrimental effect on whole body health of the animals, nor did it induce any tissue-specific effects on whole body energy metabolism, liver health, inflammation, fibrosis, heart function or muscle mass.
Conclusions
Our data suggests that these models are suitable for studying the tissue specific effects of gene deletion using the LSL-Cas9-Tg model, and that phenotypes observed utilizing these models can be confidently interpreted as being gene specific, and not confounded by the chronic overexpression of Cas9.
Keywords: CRISPR, Cas9, Transgenic mice, Metabolism, Tissue specific, Phenotype
Highlights
-
•
Detailed characterization of 4 tissue specific Cas9 TG mice in relevant metabolic tissues.
-
•
Demonstration that these models express robust Cas9 in a tissue specific manner.
-
•
Detailed phenotyping demonstrates that chronic Cas9 expression has no impact on tissue weight, body composition or body weight.
-
•
Metabolic phenotyping demonstrates that Cas9 expression does not impact whole body glucose tolerance, or heart function.
-
•
Tissue specific characterization confirms that there is no discernible effect on tissue health or function.
1. Introduction
Since the discovery and proven utility of CRISPR/Cas9-based gene editing technologies, there has been a deluge of publications that take advantage of this groundbreaking technology. Whilst the potential for this relatively simple but precise genetic manipulation tool is obvious, the speed at which the field is developing often means that subtle off-target and deleterious effects of such an approach can be overlooked. Studies over the past five years have demonstrated that each system requires important optimization to ensure accurate gene editing whilst minimizing off-target editing and potential toxicity induced by the introduction of foreign genetic machinery [1,2].
A major advantage of CRISPR-based editing in the pre-clinical biomedical arena is the rapid development of animal models that harbor global gene deletions or conditional targeting of alleles. These models historically took 2–3 years to generate, whereas now a global deletion model using CRISPR can take 3 months or less to generate [3]. Moreover, CRISPR overcomes the need to generate one mouse model per gene of interest, as is the case with floxed alleles. Indeed, by overexpressing Cas9 globally or in a tissue-specific manner in mice, one can generate a model where almost any gene can be manipulated simply by introducing a guide RNA that targets the gene of interest. This flexibility of manipulation has made it feasible to use one mouse model, or even an existing disease model, to study the effects of manipulating one or many genes in combination.
One such model is the loxP-STOP-loxP spCas9 transgenic (LSL-spCas9Tg) mouse [4]. This model harbors the spCas9 gene at the ROSA26 locus, which is silenced in the basal state by commonly applied repressor elements. By flanking the repressor or “STOP” element with loxP sites, the construct becomes inducible in the presence of Cre recombinase. This model has been used by a number of groups to demonstrate robust Cas9 expression induced by Cre recombinase as well as subsequent CRISPR-mediated gene editing upon the administration of a guide RNA [[5], [6], [7]].
One concern with such an approach is whether chronic expression of the foreign CRISPR machinery mediates phenotypic effects per se [1]. To this end, we have successfully developed four tissue-specific transgenic mouse models that express Cas9 in key tissues of interest in the field of metabolism––skeletal muscle, liver, adipose tissue, and heart. We phenotyped these animals for readouts of whole-body metabolism, adiposity, glucose tolerance, toxicity, and cardiac function and demonstrated that none of the models were impacted by the chronic (~12 weeks) presence of Cas9. These findings provide important and much-needed confidence for researchers who wish to use these mouse models in the future, who can now confidently ascribe any observed metabolic phenotype to their gene of interest and not to underlying, unwanted side effects of the model itself.
2. Results
2.1. Tissue-specific and inducible expression of Cas9 in four mouse models
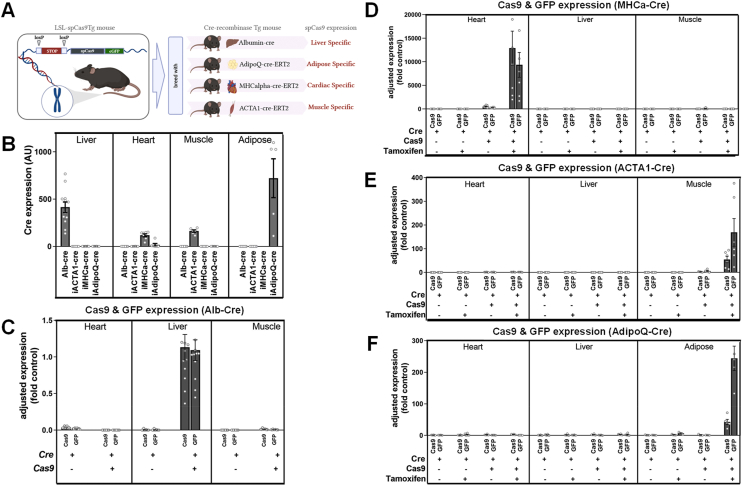
To generate tissue-specific Cas9 transgenic mice, we crossed the LSL-spCas9Tg mouse [4] with four different tissue-specific Cre recombinase transgenic mouse lines. Three of these models (ACTA1-Cre, AdipoQ-Cre, and MHCα-Cre) were tamoxifen-inducible via the use of Cre recombinase that was fused to the modified estrogen receptor (mER), often referred to as ERT2. The other model (albumin-Cre) was constitutively active and expressed from the albumin promoter (Alb-Cre). These breeding strategies resulted in four separate mouse models with an expected tissue-specific expression of Cas9 in liver (Albumin), heart (MHCa), skeletal muscle (ACTA1), and adipose tissue (AdipoQ) (Figure 1A).
Figure 1.
Tissue-Specific and Inducible Expression of Cas9 in four mouse models. A. Schematic outlining the breeding strategy and generation of the four tissue-specific Cas9 transgenic mouse lines. The LSL-spCas9Tg mouse was bred with four different Cre lines (Albumin-Cre = liver, AdipoQ-Cre-ERT2 = adipose, MHCa-Cre-ERT2 = cardiac, ACTA1-Cre-ERT2 = skeletal muscle) to generate lines that were independently maintained and studied. B. Relative Cre recombinase expression as determined by qPCR for each line (annotated across the bottom) in the various tissues (annotated across the top) for each mouse line. Data are normalized to a housekeeping gene (Rplp0) and presented as arbitrary unit (AU) for “Cas9+Cre Active” compared to “Cre active” groups; Adjusted expression of Cas9 and GFP as determined by qPCR and presented as fold change from control in the heart, liver, and muscle of C. spCas9Tg+Alb-Cre constitutively active Cre line, showing just the two groups per tissue = Cre-Cas9 and Cre+Cas9 as indicated by the (+) and (−) signs at the bottom of the graph; Cas9 and GFP in the D. spCas9Tg+MHCα-Cre-ERT2 mice, E. spCas9Tg+ACTA1-Cre-ERT2 mice, and F. spCas9Tg+AdipoQ-Cre-ERT2 mice. Because the MHCα-, ACTA1-and AdipoQ-Cre are inducible (tamoxifen, TAM) lines, there are four groups per tissue = Cre-Cas9 (no TAM), Cre-Cas9 (plus TAM), Cre+Cas9 (no TAM) and Cre+Cas9 (plus TAM) as indicated by the (+) and (−) signs at the bottom of the graphs. All data are presented as mean ± SEM, n = 4–12/group. LSL = lox-STOP-lox, spCas9 = Cas9 from S. Pyogenes, Tg = transgenic, eGFP = enhanced green fluorescent protein, ERT2 = 2 x tamoxifen sensitive mutant estrogen receptor.
Because three of the Cre models were inducible (with tamoxifen), we administered tamoxifen or a vehicle (oil) using specific regimens for each model (see methods) between 6 and 8 weeks of age, then allowed 2 weeks for maximal gene expression before any phenotyping was performed. For the inducible models, we studied four cohorts of mice per model, which included: Cre+OIL (Cre-inactive), Cas9+Cre+OIL (Cas9+Cre-inactive), Cre+Tamoxifen (Cre-active), and Cas9+Cre+Tamoxifen (Cas9+Cre-active) mice. For the constitutively active Albumin-Cre model, there was no oil/tamoxifen treatment; thus, there were only two cohorts: Cre-active and Cas9+Cre-active.
To characterize the different models, we performed phenotyping that included assessment of body weight, fat mass, and lean mass by EchoMRI and glucose tolerance tests (GTTs), which were performed at various times for different models over the subsequent 12 weeks. We also performed model-specific phenotyping, including echocardiography of the MHCα (designated as MHCa in the figures) model to assess heart function.
At the completion of each study (mice up to approximately 20–22 weeks of age), tissues were collected, processed, and analyzed to first confirm that each model displayed the expected expression profiles. Using qPCR analysis, we demonstrated that each model exhibited tissue-specific Cre recombinase expression, with robust expression in the expected tissue, but no expression detected in other tissues (Figure 1B). Importantly, we also demonstrated using qPCR that this expression was dependent on both Cre expression and the administration of tamoxifen in the inducible models. As a secondary confirmation, we also used qPCR to determine the abundance of GFP, which is co-expressed from the same transgene cassette as Cas9, but is independently processed by the ribosome (i.e., not tagged to Cas9). We demonstrated that both Cas9 and GFP exhibited the expected tissue-specific expression profiles in liver (Figure 1C), heart (Figure 1D), muscle (Figure 1E), and white adipose tissue (WAT, Figure 1F). The level of Cas9 induction varied slightly across the lines, which is likely a reflection of local transcriptional machinery, number of cells per unit of tissue, and the differential activity of Cre recombinase in each tissue.
2.2. Tissue-specific expression of Cre Recombinase, Cas9, and GFP does not impact animal body weight or tissue weights
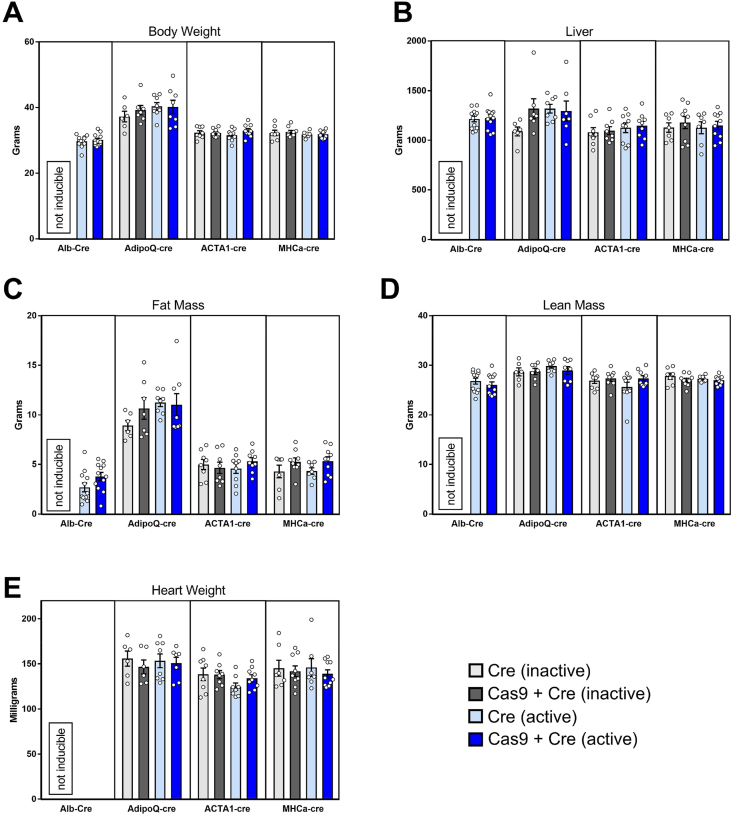
Upon demonstrating that each model expressed Cas9 in a tissue-specific manner, we next sought to test previously raised concerns that chronic overexpression of “foreign” enzymes such as Cre and Cas9 in metabolic tissues might lead to phenotypic differences in animal growth and development. A simple way of testing for toxicity or growth inhibition is to compare body weight and individual tissue weights from each model at study end. We demonstrated that body weights for each model were comparable between cohorts at the time of cull, with no significant differences in body weight, whether they were expressing Cre or Cas9 or had been treated with tamoxifen/oil (Figure 2A). Moreover, the assessment of liver, WAT, muscle (tibialis anterior; TA), and heart weights at study end demonstrated no difference in the weight of any of these tissues between the various groups within each model (Figure 2B–E).
Figure 2.
Tissue-Specific Expression of Cre Recombinase, Cas9, and GFP Does Not Impact Body Weight or Tissue Weights. All four tissue-specific Cas9 mouse lines were aged to approximately 18–20 weeks of age and analyzed for A. body weight, B. liver weight at cull, C. fat mass by EchoMRI (final two weeks), D. lean mass by EchoMRI (final 2 weeks) and E. heart weight at study end. The four groups per line are as follows: Cre (inactive––light grey), Cre+Cas9 (inactive––dark grey), Cre (active––light blue), and Cre+Cas9 (active––dark blue). All data are presented as mean ± SEM, n = 6–11/group. Albumin-Cre line is constitutively active, so the inactive groups were designated as “not inducible.”
2.3. Tissue-specific expression of Cre recombinase, Cas9, and GFP does not alter molecular or physiological readouts of tissue function
Whilst it was important to demonstrate that there was no effect of Cas9 expression on gross tissue weights or animal growth, we also sought to investigate whether tissue-specific pathways were being impacted by chronic overexpression of Cas9 in each tissue. Therefore, we performed a series of analyses on each tissue, utilizing qPCR, histology, and functional assessment.
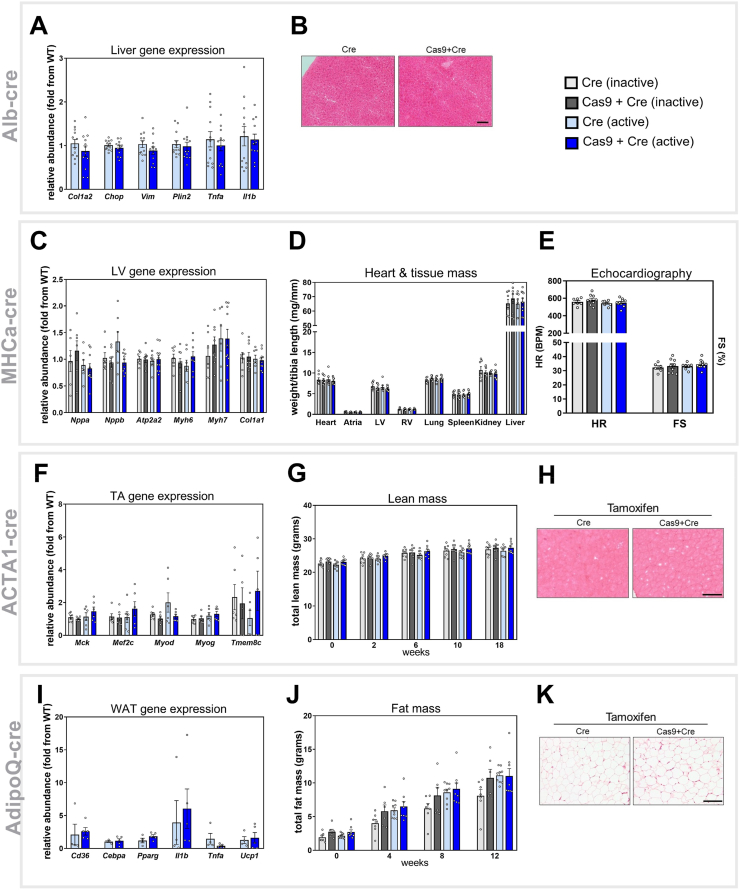
In the liver-specific Cas9 model (Alb-Cre), we used qPCR to analyze the expression of genes that were representative of pathways that provided insight into the health and activity of the liver. These included Col1a2 and Vim as markers of fibrosis, Chop for ER stress, Plin2 for lipid storage, and Tnfa and Il1b for inflammation (Figure 3A). We demonstrated that none of these genes were differentially expressed in mice with livers expressing Cas9 compared to control mice, indicating that the expression of Cas9 in the liver was not impacting liver inflammation, fibrosis, or lipid handling. Moreover, representative histological sections stained with hematoxylin and eosin (H&E) demonstrated no gross changes in liver morphology (Figure 3B).
Figure 3.
Tissue-Specific Expression of Cre Recombinase, Cas9, and GFP Does Not Alter Molecular or Physiological Readouts of Tissue Function. Molecular and functional readouts of tissue-specific function were performed on each of the strains. Alb-Cre line (liver-specific) was investigated for changes in A. hepatic mRNA expression for pathways indicative of fibrosis (Col1a2, Vim), ER stress (Chop), lipid metabolism (Plin2), and inflammation (Tnfa, Il1b) as analyzed by qPCR (n = 12/group), and B. representative images of liver tissue morphology as assessed by histology with H&E staining. The MHCα -Cre-ERT2 line (cardiac-specific) was analyzed for changes in C. cardiac mRNA expression for genes indicative of cardiac pathology (Nppa, Nppb, Atp2a2, Myh6 and Myh7) and fibrosis (Col1a1), D. mass of tissues pertinent to cardiac pathology (whole heart, atria, left ventricle (LV), right ventricle (RV), lung) and whole-body animal health (spleen, kidney, liver) and E. measurement of cardiac function including heart rate (HR) and fractional shortening (FS%) as analyzed by echocardiography, n = 6–10/group. The ACTA1-Cre-ERT2 line (muscle-specific) was investigated for changes in F.tibialisanterior (TA) mRNA expression for pathways indicative of muscle maturation (Mck), regeneration (Mef2c, Myod, and Myog), and fusion (Tmem8c) as analyzed by qPCR, G. lean (muscle) mass over the duration of the study (at timepoints indicated on graph) as analyzed by EchoMRI, and H. muscle (TA) morphology as assessed by histology with H&E staining n = 5–8/group. The AdipoQ-Cre-ERT2 line (adipose specific) was investigated for changes in I. white adipose tissue (WAT) gene expression for pathways indicative of lipid uptake (Cd36), adipocyte differentiation (Cebpa, Pparg) inflammation (Tnfa, Il1b) and WAT browning (Ucp1) as analyzed by qPCR, J. fat (adipose) mass over the duration of the study (at timepoints indicated on graph) as analyzed by EchoMRI, and K. WAT morphology as analyzed by histology with H&E staining, n = 4–9/group. All data are presented as mean ± SEM, scale bar represents 100 μm.
In the heart-specific Cas9 model (MHCa-Cre), we used qPCR to measure markers of pathways in the heart that are indicative of cardiac health and function. These included classic molecular markers of heart failure, including Atrial Natriuretic Peptide (ANP; Nppa) and B-type Natriuretic Peptide (BNP; Nppb) as well as αMHC (Myh6) and βMHC (Myh7). We also measured a marker of fibrosis (Col1a1) and a marker of cardiac contractile function via the sarcoplasmic reticulum/endoplasmic reticulum Ca2+ ATPase 2a (SERCA; Atp2a2) (Figure 3C). We demonstrated that there were no differences in the expression of any of these markers in the left ventricle (LV) across the four groups of mice, implying that these pathways were not altered by the expression of Cas9 (or Cre recombinase). This was supported by data demonstrating that the weight of the whole heart and the different regions of the heart, including atria, LV, and right ventricle (RV), were also not different between groups of these mice (Figure 3D). Consistent with this finding, the lung, spleen, and kidney weights (which can also reflect cardiac health) were all comparable across groups, demonstrating no peripheral effects of cardiac-specific overexpression of Cas9. Lastly, we assessed heart function in these mice using echocardiography. We demonstrated that at comparable heart rates (HR) under anesthesia, there were no differences in fractional shortening (FS%)––a measure of systolic function––between the four groups (Figure 3E). Collectively, these data demonstrate that overexpression of Cas9 in cardiomyocytes has no impact on heart health and function.
Phenotyping of the muscle-specific Cas9 model (ACTA1-Cre) was also performed using qPCR. We measured the expression of muscle-specific genes that are known readouts of muscle development and growth in the TA muscle. These included myogenic transcription factors Myod, Myog and Mef2c, as well as the pro-fusion gene, myomaker (Tmem8c), and mature muscle marker Mck (Figure 3F). As with our previous models, we demonstrated no difference in the expression of any of these genes between the four groups of mice, indicating that there were no major differences in the growth and function of adult skeletal muscle in the presence of Cas9 expression. In support of this data, by using EchoMRI, we demonstrated that there was no difference in lean muscle mass across the four groups at any time point throughout the study period (Figure 3G). Finally, histological analyses of TA muscle sections using H&E staining indicated that there were no major morphological differences in the muscle structure between Cas9-positive and Cas9-negative mice (Figure 3H). Collectively, these data indicate that Cas9 expression in skeletal muscle has no impact on muscle health and maturation.
The final model we characterized was the adipose-specific Cas9 mouse (AdipoQ-Cre). As with previous models, using qPCR, we demonstrated that there was no major difference in the expression of genes related to adipocyte differentiation and health in WAT, including the adipogenic transcription factors PPARγ (Pparg) and C/EBPα (Cebpa), the lipid transporter Cd36, inflammatory markers Tnfa and Il1b, and the browning marker Ucp1, in the absence or presence of Cas9 (Figure 3I). Moreover, using EchoMRI, we demonstrated that there was no difference in fat mass between the four groups in this AdipoQ-Cre model at any time throughout the study period (Figure 3J). Finally, histological assessment of WAT sections using H&E staining indicated that there were no major morphological differences in adipocyte size or structure between Cas9-positive and Cas9-negative mice (Figure 3K). Collectively, these data indicate that Cas9 expression in adipose tissue has no impact on WAT health and development.
2.4. Tissue specific expression of Cre recombinase, Cas9, and GFP does not alter whole-body glucose homeostasis
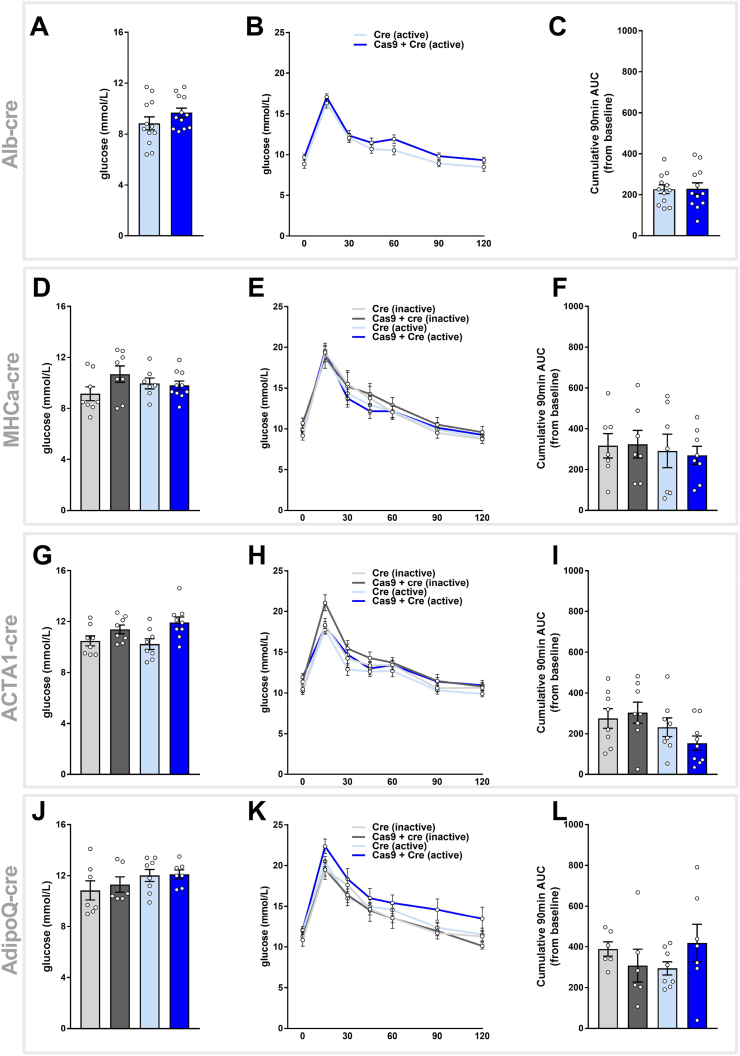
Given that many groups that study metabolism have an interest in glucose homeostasis and how it relates to tissue types such as liver, adipose, skeletal muscle, and heart, we sought to determine whether the expression of Cas9 in these tissues led to any changes in whole-body glucose handling. To investigate this, we performed fasting blood glucose measurements and oral glucose tolerance tests on all groups and models within the final two weeks of the study period. We demonstrated that there was no difference in fasting blood glucose levels between Cas9-positive and Cas9-negative mice in each of the four tissue-specific mouse models (Figure 4A, D, G, and J). To test the glucose tolerance of these models, we challenged each cohort with a standardized oral dose of glucose (2 mg/kg lean mass) and subsequently measured their blood glucose concentration over two hours by way of an oral glucose tolerance test (oGTT). We demonstrated that all groups and models had a peak glucose concentration of approximately 18–20 mmol/L at 15 minutes post-glucose delivery, which mostly returned to baseline by 60 minutes after delivery of the glucose bolus (Figure 4B, E, H, and K). These data suggest that there was no difference in the clearance of glucose across any of the groups and in each of the models, indicating that there was no difference in the glucose tolerance of these animals. This is further demonstrated quantitatively by assessing the 90-minute cumulative area under the curve (AUC) for the tolerance test (Figure 4C, F, I, and L), which confirms that there was no difference in glucose tolerance between the groups for each of the tissue-specific Cas9 models.
Figure 4.
Tissue-Specific Expression of Cre Recombinase, Cas9, or GFP Does Not Alter Whole-Body Glucose Homeostasis. All four Cas9 lines were phenotyped for parameters of whole-body glucose homeostasis at the end of the study. This included assessment of fasting blood glucose for A. Alb-Cre (n = 11–12/group), D. MHCα-Cre-ERT2 (n = 7–10/group), G. ACTA1-Cre-ERT2 (n = 8–9/group), and J. AdipoQ-Cre-ERT2 (n = 6–8/group) and 2-hour glucose tolerance as performed by oral glucose tolerance tests (oGTT) on B. Alb-Cre, E. MHCα-Cre-ERT2, H. ACTA1-Cre-ERT2, and K. AdipoQ-Cre-ERT2 mice as quantified by 90-minute cumulative area under the curve (AUC) analysis for C. Alb-Cre, F. MHCα-Cre-ERT2, I. ACTA1-Cre-ERT2, and L. AdipoQ-Cre-ERT2. All data are presented as mean ± SEM.
Collectively, the data presented above demonstrate that long-term expression of Cas9 in four different tissue-specific models does not lead to any effects on body weight, tissue weight, the expression of markers of pathological pathways, or readouts of whole-body glucose metabolism. These findings provide an important foundation for future studies that wish to use the LSL-Cas9 mouse model to study their gene of interest and afford researchers confidence that metabolic phenotypes they measure are unlikely to be impacted by the chronic expression of Cas9 or Cre recombinase in these models.
3. Discussion
The discovery and implementation of CRISPR/Cas9 as a gene editing tool has far-reaching implications for furthering knowledge gain in biomedical science. The flexibility and comparatively simple execution of this technology means it can be utilized by most research laboratories around the world, accelerating the opportunity for discovery several fold over existing technologies. Whilst CRISPR/Cas9 has indeed been adopted quickly and efficiently by the scientific community, this has often been accomplished without due consideration for the potential negative effects on metabolic readouts that might arise from such methodologies, particularly when appropriate optimization has not been performed. Many studies have shown that spurious gene editing can occur in the setting of chronic high-level expression of Cas9 [8] and their accompanying single guide RNAs (sgRNAs) [9,10], whilst others have expressed concern over the impacts of long-term exogenous expression of CRISPR machinery (Cas9/sgRNAs) causing unwanted effects on target cells [11,12]. This is analogous to when models for Cre recombinase-mediated gene editing were being optimized, when it was demonstrated that exceptionally high levels of Cre expression were shown to lead to spurious and unintended chromosomal rearrangements in male spermatids [13]. Such unwanted effects on target cells would be particularly concerning in the in vivo setting, where even minor disruptions to tissue function over many months have the potential to substantially impact animal health and disease risk. Unfortunately, the impact of potential unwanted effects in Cas9-Tg models is mostly unknown and is difficult to predict without performing validation of such models directly. This is likely time consuming and laborious, and thus, these important “control group comparisons” are often the first experiments to be overlooked when designing new CRISPR editing experiments.
With regard to in vivo CRISPR editing, the generation of the inducible spCas9 transgenic mouse (LSL-spCas9Tg) by Zhang and colleagues has been an important tool to enable tissue- and temporal-specific Cas9 expression in mice [4]. This model has been used in several labs around the world to successfully delete genes of interest, most commonly in myeloid or neuronal cell lineages [[5], [6], [7]]. Whilst there are obvious advantages to using this LSL-spCas9Tg mouse model, it is less obvious what the potential disadvantages are, if any. This is particularly true when generating tissue-specific Cas9 models for the first time, as there are no data available regarding whether chronic Cas9 overexpression will impact the tissue and whole-body phenotypes of interest.
Given that our group has a major interest in metabolism and the organs that regulate whole-body energy status, we are frequently performing studies in pertinent metabolic tissues such as the liver, muscle, adipose and heart. Unfortunately, to date, few studies have performed in vivo CRISPR editing in these tissues using the LSL-spCas9Tg mouse, and thus, it is unclear whether this model would be suitable for investigating CRISPR-mediated gene deletion in the aforementioned tissues without the risk of unwanted side effects due to chronic Cas9 overexpression. Thus, we generated liver-, muscle-, adipose-, and cardiac-specific Cas9-expressing mice using the LSL-Cas9Tg model and demonstrated that these four models appeared to be unaffected by the chronic expression of Cas9 and/or Cre recombinase. We performed comprehensive metabolic phenotyping of all four lines, including body composition, glucose tolerance, molecular, and biochemical measurements and functional readouts on various tissues. These analyses demonstrated clear tissue specificity of Cas9 expression, driven by temporal activation of Cre recombinase using tamoxifen in all three of the inducible lines (muscle, adipose, and heart), as well as the constitutive albumin (liver) line. Importantly, we were unable to detect any deleterious effects on metabolic pathways, morphology of tissues, body composition, or glucose tolerance in any lines overexpressing Cas9, supporting the notion that there is no negative impact of chronic Cas9 expression in these tissues. Moreover, echocardiography also demonstrated no impact on systolic heart function in cardiac-specific Cas9-expressing mice after 12 weeks of induction, providing evidence that heart function in these mice was not affected by chronic Cas9 expression.
An observation worth noting was that there appeared to be varying levels of Cas9 mRNA expression detected across the different models. This fluctuated from a ~200-fold increase in Cas9 expression in adipose tissue, up to 10,000-fold in the heart. There were also discrepancies in the level of GFP versus Cas9 expression within given tissues, which was unexpected given they are expressed from the same promoter construct. Various reasons could account for the differences that were observed, including differential activities of CAG promoters across cells and tissues [14,15], efficiency of the Cre recombinase to remove the STOP cassette, or tissue-specific inhibitors/transcriptional machinery that suppresses certain mRNAs. However, we believe this variation most likely stems from the method by which gene expression was determined. Because we used the ΔΔCT method to determine gene expression, the level of Cas9 expression is reported relative to the expression of the control or “housekeeping” gene. Whilst we see very tight expression of the housekeeping gene within a given tissue across all genotypes, the abundance of this housekeeping gene differs between various tissues. Moreover, because Cas9 and GFP are exogenous genes in mammalian systems, the assays are determining robust expression of these genes versus effectively no expression in non-transgene tissues. Hence, the coupling of ΔΔCT-determined gene expression with a binary expression outcome likely results in deceptively large variability between tissues. This is supported by the fact that we observe very consistent values for the absolute cycle threshold (CT) for Cas9 expression across the tissues (CT ~20), except for adipose tissue, in which we observed CT ~24 (data not shown). Although the current results successfully demonstrated that the transgenic mouse lines in this study were functioning as expected, for a more quantitative measure of gene expression one might wish to pursue other technologies such as digital PCR. Nevertheless, our data do demonstrate that the inducible adipose-specific Cas9-Tg model likely expresses less Cas9 than other tissue-specific models, which may be an important consideration for those wanting to use this model for gene editing experiments.
In summary, we provide critical evidence that whole-body metabolism of four different metabolic tissue-specific mouse lines is unaffected by the chronic expression of Cas9. These findings provide confidence moving forward for researchers who wish to use these Cas9 mouse models to manipulate the expression of genes in these particular tissues. The minimal impact of Cas9 in these studies will likely reduce the need for future studies to perform specific control groups, reducing animal numbers and sparing expensive resources. Our data will also provide confidence that observed phenotypes related to gene deletions in these models in future studies are likely to be specific to the gene of interest rather than being related to the chronic overexpression of Cas9. Thus, these findings provide an important resource for the research community.
4. Methods
4.1. Generation of tissue-specific Cas9 animal models
All animal experiments were approved by the Alfred Research Alliance (ARA) Animal Ethics Committee (E/1756/2017/B) and performed in accordance with the research guidelines of the National Health and Medical Research Council of Australia. Tissue-specific expression of Cas9 was achieved using the Cre-Lox system, where Cre recombinase was used to remove the “STOP” sequence from the lox-stop-lox (LSL) cassette separating the promoter and Cas9 genes in the mouse described by Zhang et al. [4]. The four different mouse lines were generated from base strains sourced from the Jackson Laboratory. This included crossing the LSL-spCas9Tg mouse (JAX Stock Number #026175) with either the Albumin-Cre (Stock Number #003574), ACTA1-Cre-ERT2 (Stock Number #025750), AdipoQ-Cre-ERT2 (Stock Number #024671) or MHCα-Cre-ERT2 (Stock Number #005657) mouse. All mice were on a C57BL/6J background. We generated two cohorts of n = 8–10 male Alb-Cre-Cas9Tg mice (Cre & Cre+Cas9) and two cohorts of n = 16–20 male mice of the inducible Cre-Cas9Tg mouse lines (Cre & Cre+Cas9), the latter of which were split further into two groups each and treated with either the vehicle (sunflower oil) or tamoxifen (Sigma T5648) in sunflower oil, generating the following four groups: Cre inactive (OIL), Cre+Cas9 inactive (OIL), Cre active (tamoxifen), and Cre+Cas9 active (tamoxifen). Only the final group (Cre+Cas9 active) was expected to express Cas9, with the others serving as either Cre or tamoxifen control groups.
4.2. Animal treatments and husbandry
All mice were bred through the ARA Precinct Animal Centre and offspring were genotyped (Transnetyx) and randomly allocated into their respective groups. For tamoxifen-inducible models, they were treated as follows. For ACTA1-Cre-ERT2 and AdipoQ-Cre-ERT2 models, mice were aged to 6–8 weeks old before being gavaged with either tamoxifen (80 mg/kg) in sunflower oil or sunflower oil alone for 3 consecutive days. For the MHCα-Cre-ERT2 model, mice were IP injected once with 40 mg/kg of tamoxifen in sunflower oil or sunflower oil alone. Following tamoxifen treatment, mice were left to recover for 2 weeks, after which they were maintained for the remainder of the study (12 weeks) in group housing on a normal chow diet (Normal rodent chow, Specialty feeds, Australia) at 22 °C on a 12 hr light/dark cycle with access to food and water ad libitum and cages changed weekly. Cohorts of mice were subjected to EchoMRI and body weight analysis throughout the study period. In the last two weeks of the study period, all animals underwent oral glucose tolerance tests (see detailed method below) whilst the MHCα mice were also subjected to cardiac function assessment via echocardiography (see detailed method below). At the end of the study, mice were fasted for 4–6 hours and then anesthetized with a lethal dose of ketamine/xylazine (Ceva Animal Health #1867-66-9 at 100 mg/kg, and Troy Laboratories #23076-35-9 at 20 mg/kg, respectively) before blood and tissues were collected, weighed, and snap frozen for subsequent analysis.
4.3. Glucose tolerance tests
Oral glucose tolerance tests (oGTT) were performed as previously described [16,17]. Briefly, mice were fasted for 4–6 hours in the morning, and the procedure was performed around midday. A basal blood glucose measurement was recorded using a glucometer (Accu Check Performa, Roche Diabetes Care), from a sample taken from the tail vein before mice were gavaged with glucose (25% in saline) at a dose of 2 g/kg of lean mass as determined by EchoMRI. Blood glucose was subsequently determined at the following time points after gavage: 15, 30, 45, 60, 90, and 120 minutes.
4.4. EchoMRI
Body composition was analyzed using the 4-in-1 NMR Body Composition Analyzer for Live Small Animals, according to the recommendations of the manufacturer (EchoMRI LLC, Houston, TX, USA). This analysis provides measurements of lean mass and fat mass in living animals as previously described [16,17].
4.5. Histology
Liver and muscle were embedded cut side–down in OCT before being frozen in a bath of isopentane submerged in liquid nitrogen. After freezing, blocks were brought to −20 °C and 5μm sections were cut using a Leica Cryostat. Sections were mounted and dried overnight at room temperature before being fixed in 100% methanol. WAT samples were fixed in formalin and mounted in paraffin before 7μm sections were cut on a Leica microtome. All sections were stained with hematoxylin and eosin and slide images were captured using an Olympus Slide scanner VS120 (Olympus, Japan) and viewed in the accompanying software (OlyVIA Build 13771, Olympus, Japan).
4.6. Quantitative PCR (qPCR)
RNA was isolated from tissues using RNAzol reagent and isopropanol precipitation as previously described [17,18]. cDNA was generated from RNA using MMLV reverse transcriptase (Invitrogen) according to the manufacturer's instructions. qPCR was performed on 10 ng of cDNA using the SYBR Green (BioRad) method on a Quant Studio 7 real-time PCR machine (Applied Biosystems), using primer sets outlined in Table 1. Primers were designed to span exon-exon junctions where possible and were tested for specificity using BLAST (Basic Local Alignment Search Tool; National Centre for Biotechnology Information). Amplification of a single amplicon was estimated from melt curve analysis, ensuring that only a single peak and an expected temperature dissociation profile were observed. Quantification of a given gene was determined by the relative mRNA level compared with the control using the ΔΔCT method, which was calculated after normalization to the housekeeping gene cyclophilin A (Ppia) or 36B4 (Rplp0).
Table 1.
Forward and reverse primer sets for the detection of the designated mouse genes using qPCR.
| Gene | Forward primer (5′–3′) | Reverse primer (5′–3′) |
|---|---|---|
| Cre | AGGGCGCGAGTTGATAGCT | GAGCGATGGATTTCCGTCTCT |
| spCas9 | CCAAGAGGAACAGCGATAAG | CACCACCAGCACAGAATAG |
| GFP | CAGGAGCGCACCATCTTCTT | CTTGTGCCCCAGGATGTTG |
| Col1a2 | GGGAATGGAGCAAGACAGTCTT | TGCGATATCTATGATGGGTAGTCTCA |
| Chop | AGGAGCCAGGGCCAACA | TCTGGAGAGCGAGGGCTTT |
| Vim | GAAATTGCAGGAGGAGATGC | GGATTCCACTTTCCGTTCAA |
| Plin2 | CCCGTATTTGAGATCCGTGT | TAGGTATTGGCAACCGCAAT |
| Tnfa | CCAGACCCTCACACTCAGATC | CACTTGGTGGTTTGCTACGAC |
| Il1b | GACGGCACACCCACCCT | AAACCGTTTTTCCATCTTCTTT |
| Nppa | GGGGGTAGGATTGACAGGAT | AGGGCTTAGGATCTTTTGCG |
| Nppb | ACAAGATAGACCGGATCGGA | AAGAGACCCAGGCAGAGTCA |
| Atp2a2 | AATATGAGCCTGAAATGGGC | TCAGCAGGAACTTTGTCACC |
| Myh6 | AAGATAGTGGAACGCAGGGA | CTCTTCAGCAGCGGTTTGAT |
| Myh7 | AGCATTCTCCTGCTGTTTCC | GAGCCTTGGATTCTCAAACG |
| Col1a1 | GGTTTCCACGTCTCACCATT | ACATGTTCAGCTTTGTGGACC |
| Mck | TGAGGTCTGGGTACTCCTCC | CCTCCACAGCACAGACAGAC |
| Tmem8c | GGAGGCCATGGTCTACCTCT | GGGCTGTTCCATAGATGCTG |
| Mef2c | GCCGGACAAACTCAGACATTG | GGGTTTCCCAGTGTGCTGAC |
| Myog | CAACCAGGAGGAGCGCGATCTCCG | AGGCGCTGTGGGAGTTGCATTCACT |
| Myod | AGGCCGTGGCAGCGA | GCTGTAATCCATCATGCCATCA |
| Cd36 | TTGTACCTATACTGTGGCTAAATGAGA | CTTGTGTTTTGAACATTTCTGCTT |
| Cebpa | TGGACAAGAACAGCAACGAG | GTCACTGGTCAACTCCAGCA |
| Pparg | GTTTTATGCTGTTATGGGTG | GTAATTTCTTGTGAAGTGCTCATAG |
| Ucp1 | ACTGCCACACCTCCAGTCATT | CTTTGCCTCACTCAGGATTGG |
| Rplpo | ACCCTGAAGTGCTCGACATC | ATTGATGATGGAGTGTGGCA |
| Ppia | AGCCAAATCCTTTCTCTCCAG | CACCGTGTTCTTCGACATCA |
4.7. Echocardiography
Echocardiography was performed on MHCa-cre-ERT2 mice anaesthetised with isoflurane (Abbvie, #26675-46-7, 1.5–2%) at the end of the 12-week period following tamoxifen induction, using a 15-MHz linear transducer L15-7io with a Philips iE33 Ultrasound Machine (North Ryde, NSW, Australia). Data were analyzed and verified by two independent researchers according to QC procedures and validation measures as outlined previously [19].
4.8. Data inclusion and exclusion criteria
For animal experiments, phenotyping data points were excluded using the following pre-determined criteria: if the animal was unwell at the time of analysis, there were technical issues identified (such as unclear signal from echocardiography), or data points were identified as outliers using Tukey's Outlier Detection Method (Q1 – 1.5 IQR or Q3 + 1.5 IQR). If repeated data points from the same mouse failed QC based on predetermined criteria or several data points were outliers as per Tukey's rule, the entire animal was excluded from that given analysis (i.e., during glucose tolerance tests, indicating inappropriate gavage). For in vivo and in vitro tissue and molecular analyses, data points were only excluded if there was a technical failure (i.e., poor RNA quality, failed amplification in qPCR) or the value was biologically improbable. This was performed in a blinded fashion (i.e., on grouped datasets before genotypes were known).
Acknowledgements
We acknowledge funding support from the Victorian State Government OIS program to Baker Heart & Diabetes Institute. These studies were supported by funding from the Baker Heine Trust through both the Obesity & Lipid and the Bioinformatics Programs, as well as the Bertalli mini-grant scheme at Baker. During the period of this study, some authors were supported by fellowship funding as follows: BGD and ACC, National Heart Foundation of Australia (101789 and 100067, respectively); JRM and PG, NHMRC of Australia (APP1078985 and APP1117835, respectively). We thank members of the MMA, LMCD, Cardiac Hypertrophy, Metabolomics, and Hematopoiesis & Leukocyte Biology laboratories at BHDI for their contributions. We also acknowledge the use of the facilities and technical assistance of the Monash Histology Platform, Department of Anatomy and Developmental Biology, Monash University. Figure 1A was created with Biorender (Biorender.com).
Contributor Information
Anna C. Calkin, Email: anna.calkin@baker.edu.au.
Brian G. Drew, Email: brian.drew@baker.edu.au.
Author contributions
BGD and ACC designed and conceived the study. BGD wrote the manuscript and all other authors read and/or edited the manuscript. BGD, STB, DCH, AZ, CY, EAMG, YL, HK, KIW, and GIL performed animal experiments and phenotyping. BGD, STB, AZ, TS, YL, YF, and YT analyzed data, processed tissue samples, and performed molecular and biochemical experiments. PG, JRM, PJM, and ACC provided reagents, experimental advice, and access to infrastructure and resources.
Conflict of interest
The authors declare that they have no conflicts of interest.
References
- 1.Broeders M., Herrero-Hernandez P., Ernst M.P.T., van der Ploeg A.T., Pijnappel W. Sharpening the molecular scissors: advances in gene-editing technology. iScience. 2020;23:100789. doi: 10.1016/j.isci.2019.100789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Molla K.A., Yang Y. CRISPR/Cas-Mediated base editing: technical considerations and practical applications. Trends in Biotechnology. 2019;37:1121–1142. doi: 10.1016/j.tibtech.2019.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Singh P., Schimenti J.C., Bolcun-Filas E. A mouse geneticist's practical guide to CRISPR applications. Genetics. 2015;199:1–15. doi: 10.1534/genetics.114.169771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Platt R.J., Chen S., Zhou Y., Yim M.J., Swiech L., Kempton H.R. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laidlaw B.J., Duan L., Xu Y., Vazquez S.E., Cyster J.G. The transcription factor Hhex cooperates with the corepressor Tle3 to promote memory B cell development. Nature Immunology. 2020;21:1082–1093. doi: 10.1038/s41590-020-0713-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shamsi F., Xue R., Huang T.L., Lundh M., Liu Y., Leiria L.O. FGF6 and FGF9 regulate UCP1 expression independent of brown adipogenesis. Nature Communications. 2020;11:1421. doi: 10.1038/s41467-020-15055-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu C., Jiang Z., Xu Y., Cai Z.L., Jiang Q., Xu Y. Profound and redundant functions of arcuate neurons in obesity development. Nature Metabolism. 2020;2:763–774. doi: 10.1038/s42255-020-0229-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hendel A., Fine E.J., Bao G., Porteus M.H. Quantifying on- and off-target genome editing. Trends in Biotechnology. 2015;33:132–140. doi: 10.1016/j.tibtech.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nature Biotechnology. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Link R.W., Nonnemacher M.R., Wigdahl B., Dampier W. Prediction of human immunodeficiency virus type 1 subtype-specific off-target effects arising from CRISPR-Cas9 gene editing therapy. CRISPR J. 2018;1:294–302. doi: 10.1089/crispr.2018.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charlesworth C.T., Deshpande P.S., Dever D.P., Camarena J., Lemgart V.T., Cromer M.K. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nature Medicine. 2019;25:249–254. doi: 10.1038/s41591-018-0326-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enache O.M., Rendo V., Abdusamad M., Lam D., Davison D., Pal S. Cas9 activates the p53 pathway and selects for p53-inactivating mutations. Nature Genetics. 2020;52:662–668. doi: 10.1038/s41588-020-0623-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidt E.E., Taylor D.S., Prigge J.R., Barnett S., Capecchi M.R. Illegitimate Cre-dependent chromosome rearrangements in transgenic mouse spermatids. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:13702–13707. doi: 10.1073/pnas.240471297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qin J.Y., Zhang L., Clift K.L., Hulur I., Xiang A.P., Ren B.Z. Systematic comparison of constitutive promoters and the doxycycline-inducible promoter. PLoS One. 2010;5:e10611. doi: 10.1371/journal.pone.0010611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Q., Triplett A.A., Harms D.W., Lin W.C., Creamer B.A., Rizzino A. Temporally and spatially controlled expression of transgenes in embryonic and adult tissues. Transgenic Research. 2010;19:499–509. doi: 10.1007/s11248-009-9329-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bond S.T., Kim J., Calkin A.C., Drew B.G. The antioxidant moiety of MitoQ imparts minimal metabolic effects in adipose tissue of high fat fed mice. Frontiers in Physiology. 2019;10:543. doi: 10.3389/fphys.2019.00543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bond S.T., King E.J., Henstridge D.C., Tran A., Moody S.C., Yang C. Deletion of Trim28 in committed adipocytes promotes obesity but preserves glucose tolerance. Nature Communications. 2021;12:74. doi: 10.1038/s41467-020-20434-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bond S.T., Moody S.C., Liu Y., Civelek M., Villanueva C.J., Gregorevic P. The E3 ligase MARCH5 is a PPARgamma target gene that regulates mitochondria and metabolism in adipocytes. American Journal of Physiology Endocrinology and Metabolism. 2019;316:E293–E304. doi: 10.1152/ajpendo.00394.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donner D.G., Kiriazis H., Du X.J., Marwick T.H., McMullen J.R. Improving the quality of preclinical research echocardiography: observations, training, and guidelines for measurement. American Journal of Physiology - Heart and Circulatory Physiology. 2018;315:H58–H70. doi: 10.1152/ajpheart.00157.2018. [DOI] [PubMed] [Google Scholar]