Summary
In self‐incompatible Petunia species, the pistil S‐RNase acts as cytotoxin to inhibit self‐pollination but is polyubiquitinated by the pollen‐specific nonself S‐locus F‐box (SLF) proteins and subsequently degraded by the ubiquitin‐proteasome system (UPS), allowing cross‐pollination. However, it remains unclear how S‐RNase is restricted by the UPS.
Using biochemical analyses, we first show that Petunia hybrida S3‐RNase is largely ubiquitinated by K48‐linked polyubiquitin chains at three regions, R I, R II and R III. R I is ubiquitinated in unpollinated, self‐pollinated and cross‐pollinated pistils, indicating its occurrence before PhS3‐RNase uptake into pollen tubes, whereas R II and R III are exclusively ubiquitinated in cross‐pollinated pistils.
Transgenic analyses showed that removal of R II ubiquitination resulted in significantly reduced seed sets from cross‐pollination and that of R I and R III to a lesser extent, indicating their increased cytotoxicity. Consistent with this, the mutated R II of PhS3‐RNase resulted in a marked reduction of its degradation, whereas that of R I and R III resulted in less reduction.
Taken together, we demonstrate that PhS3‐RNase R II functions as a major ubiquitination region for its destruction and R I and R III as minor ones, revealing that its cytotoxicity is primarily restricted by a stepwise UPS mechanism for cross‐pollination in P. hybrida.
Keywords: Petunia hybrida , S‐RNase, self‐incompatibility, SLF, ubiquitination
Introduction
Self‐incompatibility (SI), an inability of a fertile seed plant to produce a zygote after self‐pollination, represents a reproductive barrier adopted by nearly 40% of flowering plant species to prevent self‐fertilisation and to promote outcrossing (De Nettancourt, 2001). In many species, SI is usually controlled by a single multiallelic S‐locus encoding both male and female S‐determinants (De Nettancourt, 2001; Takayama & Isogai, 2005; Franklin‐Tong, 2008; Zhang et al., 2009). Their molecular interaction confers the pistil with the ability to distinguish between genetically related self‐pollen and nonself‐pollen. In general, SI can be classified into self‐recognition and nonself‐recognition systems based on their distinct molecular mechanisms (Fujii et al., 2016). In the self‐recognition system of Papaveraceae and Brassicaceae, self‐pollen rejection occurs as a specific interaction between the S‐determinants from the same S haplotype. In Papaver rhoeas, the female S‐determinant Prs S (P. rhoeas stigmatic S) interacts with its cognate Prp S (P. rhoeas pollen S) to stimulate a signalling cascade leading to programmed cell death (PCD) of self‐pollen (Foote et al., 1994; Thomas & Franklin‐Tong, 2004; Wheeler et al., 2009; Wilkins et al., 2014). In Brassicaceae, the SI response is initiated by the specific interaction of the stigma S‐locus receptor kinase (SRK) and its cognate pollen‐coat‐localised ligand S‐locus cysteine‐rich protein (SCR/SP11), triggering a phosphorylation‐mediated signalling pathway and resulting in the destruction of factors indispensable for pollen compatibility by the UPS (Schopfer et al., 1999; Suzuki et al., 1999; Takasaki et al., 2000; Takayama et al., 2000; Kakita et al., 2007; Samuel et al., 2008, 2009; Ma et al., 2016). S‐RNase‐based SI, also termed as Solanaceae‐type SI, is a well studied nonself‐recognition system widely present in Solanaceae, Plantaginaceae, Rosaceae and Rutaceae (Anderson et al., 1986; McClure et al., 1989; Sassa et al., 1996; Xue et al., 1996; Lai et al., 2002; Ushijima et al., 2003; Sijacic et al., 2004; Liang et al., 2020). In self‐incompatible Antirrhinum and Petunia species, the pistil S‐determinant S‐RNase serving as a cytotoxin can be recognised and ubiquitinated by multiple pollen S‐determinant SLFs to form functional SCF ubiquitin ligases in a collaborative nonself‐recognition manner, therefore restricting cytotoxicity of nonself S‐RNases and resulting in cross‐pollination (Qiao et al., 2004b; Hua & Kao, 2006, 2008; Hua et al., 2008; Zhang et al., 2009; Kubo et al., 2010; Liu et al., 2014).
However, it remains largely unclear how S‐RNases are specifically regulated in the nonself‐recognition system. Currently, two models, the S‐RNase degradation model and the S‐RNase compartmentalisation model, have been proposed to explain how S‐RNase cytotoxicity is restricted for cross‐pollination (Qiao et al., 2004b; Goldraij et al., 2006; McClure, 2009, 2011; Liu et al., 2014). In the degradation model proposed in Petunia hybrida, both self and nonself S‐RNases taken up by pollen tubes are mainly localised in the cytosol, where they are further recognised by SLFs. Entani et al. (2014) showed that SCFSLF complexes can specifically polyubiquitinate nonself S‐RNases rather than self S‐RNases in vitro in P. hybrida, providing evidence for S‐RNase ubiquitination by cross‐pollen. Alternatively, in self‐pollen tubes, the binding of self S‐RNase and SLF leads to the formation of the nonfunctional SCFSLF complex, therefore resulting in the survival of self S‐RNase to inhibit pollen‐tube growth. Together with the discoveries of SCFSLF complex components such as SLF‐interacting SKP1‐like 1 (SSK1) and Cullin1 in species from Solanaceae, Plantaginaceae and Rosaceae (Huang et al., 2006; Zhao et al., 2010; Xu et al., 2013; Entani et al., 2014; Li & Chetelat, 2014), the degradation model appears to function in several species of flowering plants that possess S‐RNase‐based SI. In Nicotiana species, Goldraij et al. (2006) proposed that the majority of self S‐RNases and nonself S‐RNases would be sequestered in vacuole‐like structures once imported into pollen tubes. Subsequently, self‐recognition between SLFs and a small fraction of S‐RNases localised in the cytosol would break the structures, releasing S‐RNases in a late stage of self‐pollination, and triggering the SI response. By contrast, nonself‐recognition could stabilise S‐RNases and maintain their sequestration. Most previous studies have shown that S‐RNase degradation rather than compartmentalisation acts as the major strategy to restrict S‐RNase cytotoxicity in P. hybrida (Liu et al., 2014). Nevertheless, little information is known about the linkage type of the polyubiquitin chains and the specific residue of S‐RNase ubiquitinated by nonself SCFSLF complexes in cross‐pollen tubes.
To address these questions, in this study, we first established an in vivo assay to examine the polyubiquitination of PhS3‐RNase in cross‐pollinated pistils and, together with in vitro ubiquitination analyses, we found that PhS3‐RNase was mainly ubiquitinated by K48‐linked polyubiquitin chains in three regions named R I, R II and R III. Among them, R I ubiquitination occurred before PhS3‐RNase entry into pollen tubes and is likely to be mediated by an unknown E3 ligase, whereas R II and III were specifically ubiquitinated by SCFSLF. Second, the ubiquitination removal of those three regions had little effect on the physicochemical properties of PhS3‐RNase, but negatively impacted their functions in cross‐pollen tubes. The transgene with a mutated R II led to a significant reduction of seed sets from cross‐pollination, whereas in mutated R I and R III seeds sets were reduced to a much lesser extent in P. hybrida, showing that R II ubiquitination of PhS3‐RNase played a major role in its destruction and cytotoxicity restriction, whereas R I and III had minor roles. Furthermore, the ubiquitination removal of all three regions did not completely inhibit PhS3‐RNase degradation and cross seed sets, suggesting that UPS was not the exclusive mechanism to restrict S‐RNase cytotoxicity. Taken together, our results demonstrated a stepwise UPS mechanism for primary restriction of S‐RNase cytotoxicity during cross‐pollination in P. hybrida, providing novel mechanistic insight into a dynamic regulation of S‐RNases.
Materials and Methods
Plant materials
Self‐incompatible wild‐type lines of PhS3S3 , PhSVSV and PhS3S3L have been previously described (Robbins et al., 2000; Sims & Ordanic, 2001). The transgenic plants PhS3S3L /PhS3LSLF1‐FLAG were constructed by transforming PhS3S3L with pBI101‐PhS3A‐SLF::PhS3LSLF1‐FLAG. PhS3A‐SLF is a native promotor used for PhSLFs expression as previously described (Qiao et al., 2004a; Liu et al., 2014).
Ti plasmid construction and transgenic plant generation
PhS3‐RNase cDNA and its FLAG‐tagged form were amplified by primers listed in Supporting Information Table S1 to introduce XhoI and SacI restriction sites at their 5′ and 3′ ends, respectively. PhS3‐RNase point mutations were generated by polymerase chain reaction (PCR) using site‐directed mutagenesis primers listed in Table S1. Ti plasmid constructs were separately electroporated into Agrobacterium tumefaciens strain LBA4404 and introduced into PhS3S3L using the leaf disc transformation method as described previously (Lee et al., 1994; Qiao et al., 2004a).
Protein structure prediction and electrostatic potential analysis
PhS3‐RNase protein structure was modelled using the I‐TASSER server (http://zhanglab.ccmb.med.umich.edu/I‐TASSER/) according to its instructions (Yang et al., 2015; Li et al., 2017). The first model generated by iterative simulations was selected for further analysis based on model quality evaluation using the Vadar v.1.8 program (http://vadar.wishartlab.com; Willard et al., 2003) and the prosa‐web program (https://prosa.services.came.sbg.ac.at/prosa.php; Wiederstein & Sippl, 2007). Structures of point‐mutated S‐RNases were generated by mutagenesis in PyMol, and electrostatic potential analyses of PhS3‐RNase and its point‐mutated structures were performed using plug‐in APBS tools as previously described (Baker et al., 2001; Li et al., 2017).
Quantitative (q) RT‐PCR analysis
Total RNAs were separately isolated from pistils derived from PhS3S3L /PhS3 ‐RNase‐ or PhS3 ‐RNase (Mutant) (M)‐(FLAG) using TRIzol reagent (Ambion) according to the manufacturer’s instructions. cDNA was subsequently synthesised using cDNA synthesis supermix (AU311‐02; Transgen, Beijing, China). qRT‐PCR reaction mixes were prepared according to manufacturer’s guidelines of ChamQ™ Universal SYBR qPCR Master Mix (Q711‐02/03, Vazyme, Nanjing, China). Relevant primer sequences are listed in Table S1. qRT‐PCR assays were performed using the CFX96™ Real‐Time System (Bio‐Rad). Petunia hybrida 18S rRNA gene transcripts were used as an internal control. The data were analysed following the method of Livak () (Livak & Schmittgen, 2001).
Mass spectrometry analysis for ubiquitination sites
PhS3S3L plants were self‐pollinated or cross‐pollinated with PhS3S3L /PhS3LSLF1. Then the pollinated pistils collected after 2, 6, 12 and 24 h, respectively, were mixed up, minced and lysed in buffer containing 7 M urea, 2 M thiourea and 0.1% CHAPS, and followed by 5‐min ultrasonication on ice. Samples of unpollinated pistils were prepared as controls. The lysate was centrifuged at 14 000 g for 10 min at 4°C and the supernatant was reduced with 10 mM DTT for 1 h at 56°C, and subsequently alkylated with sufficient iodoacetamide for 1 h at room temperature in the dark (Udeshi et al., 2013b). Then the supernatant containing 10 mg protein was digested with Trypsin Gold (Promega) at enzyme : substrate ratio 1 : 50 at 37°C for 16 h. Peptides were desalted with C18 cartridge and dried by vacuum centrifugation (Udeshi et al., 2013b), and then resuspended in MOPS IAP buffer (50 mM MOPS, 10 mM KH2PO4, 50 mM NaCl, pH 7.0) and centrifuged for 5 min at 12 000 g. The supernatants were incubated with anti‐Ubiquitin Remnant Motif (K‐ε‐GG) beads (CST #5562; Cell Signaling Technology, Beverly, MA, USA) for 2.5 h at 4°C and centrifuged for 30 s at 3000 g at 4°C. Beads were washed in MOPS IAP buffer, then in water, before elution of the peptides with 0.15% TFA (Udeshi et al., 2013a). Then desalted by peptide desalting spin columns (89852; Thermo Fisher, Waltham, MA, USA) before LC‐MS/MS analysis using an Orbitrap Fusion mass spectrometer (Thermo Fisher). The resulting spectra from each fraction were searched separately against PhS3‐RNase amino acid sequences using the Maxquant search engines. Precursor quantification based on intensity was used for label‐free quantification.
S‐RNase activity assay
The coding sequences of PhS3‐RNase (without signal peptide) and PhS3 ‐RNase (M) were separately cloned into the pCold‐TF vector (TaKaRa, Kusatsu, Japan). Relevant primer sequences used are listed in Table S1. Trigger Factor (TF) is a 48 kDa soluble tag located at the N‐terminus of His. The His‐fused proteins were expressed in Escherichia coli Trans BL21 (DE3) plysS (Transgen) at 16°C for 24 h at 180 rpm and purified using Ni Sepharose 6 Fast Flow beads (10249123; GE Healthcare, Waukesha, WI, USA) according to the manufacturer’s instructions. Protein concentration was determined by Bradford protein assays. The relative fluorescence units (RFU) of the recombinant proteins were monitored according to the manufacturer’s instructions of RNase Alert Lab Test Kit (Ambion) using Synergy 2 (Biotek, Winooski, VT, USA).
Ubiquitination assay and immunoblotting
The SCFSLF‐FLAG complex attached to anti‐FLAG M2 affinity gel (Sigma‐Aldrich) was purified from PhS3S3L /PhS3LSLF1‐FLAG pollen tubes as described (Li et al., 2017), with that from PhS3S3L as a negative control. PhS3‐RNase was purified from PhS3S3 pistils through Fast protein liquid chromatography (FPLC) as described previously (Entani et al., 2014; Li et al., 2017), recombinant His‐PhS3‐RNase and ‐PhS3‐RNase (M) were used separately as a substrate for the ubiquitination reactions (Li et al., 2017). For immunoblotting, PVDF membranes (Millipore, Burlington, MA, USA) were probed with primary antibodies, including mouse monoclonal anti‐PhS3‐RNase, anti‐ubiquitin (Abgent, San Diego, CA, USA), and anti‐His (Sigma) antibodies at a 1 : 2000 dilution for western blot detection. ImageJ software was used to quantify the immunosignal of the ubiquitinated proteins. K48‐ or K63‐linkage specific polyubiquitin rabbit mAb (Cell Signaling) at a 1 : 1000 dilution was used to analyse the linkage type of PhS3‐RNase ubiquitination.
Subcellular fractionation and pulse–chase assays
For the subcellular fractionation assay, mature pollen grains of PhSVSV were cultured, germinated and collected as described previously (Liu et al., 2014). Style extracts of transgenic pistils containing PhS3‐RNase‐FLAG or PhS3‐RNase (M)‐FLAG were separately used to incubate the germinated pollen tubes. Then the pollen tubes were harvested for fractionation and equal amounts of protein samples derived from each centrifugation step were applied to immunoblots, as described previously (Liu et al., 2014). For the pulse–chase assay, the germinated pollen tubes of PhSVSV or PhS3S3 were incubated separately for 1 h with style extracts from the transgenic pistils containing PhS3‐RNase‐FLAG or PhS3‐RNase (M)‐FLAG that were collected and rinsed and allowed to grow further in fresh medium with or without MG132 for c. 5 h. Equal amounts of pollen‐tube samples were loaded and detected by immunoblotting using anti‐FLAG antibodies (Sigma).
Cell‐free degradation and ubiquitination assays
Total proteins of germinated PhSVSV pollen tubes were extracted on ice using cell‐free degradation buffer containing 25 mM Tris‐HCl (pH 7.5), 10 mM NaCl, 10 mM MgCl2, 1 mM DTT, 10 mM ATP and 1 mM PMSF. Then, equal amounts of extracts were applied to react with recombinant SUMO‐His‐PhS3‐RNase or its ‐PhS3‐RNase (M) with or without MG132 at 30°C. Equal amounts of samples were taken out at indicated time points for immunoblots. ImageJ software was used to quantify the immunosignals. The SUMO‐His‐tagged fusion proteins were generated as follows. The coding sequences of PhS3‐RNase (without signal peptide) and PhS3‐RNase (M) were separately cloned into engineered pET‐30a (Novagen, Madison, WI, USA) containing N‐terminal SUMO tag to produce SUMO‐His‐tagged proteins. Relevant primer sequences are listed in Table S1. The fusion proteins were, respectively, expressed in E. coli Trans BL21 (DE3) plysS (Transgen) at 16°C for 24 h, and then purified using Ni Sepharose 6 Fast Flow beads. For the cell‐free ubiquitination assays, equal amounts of recombinant His‐PhS3‐RNase and PhS3‐RNase (M) were separately incubated with PhS3S3 pollen‐tube extracts using ubiquitin reaction buffer as described previously (Hua & Kao, 2006). Then the His‐fused substrates and their ubiquitinated forms were purified from the reaction products through Ni Sepharose 6 Fast Flow beads for immunoblots as described above.
Pull‐down assay
The coding sequences of PhS3 ‐RNase (without signal peptide) and PhS3‐RNase (M) were cloned separately into pMAl‐c2x (Novagen) to generate MBP‐tagged fusion proteins. The full length of PhS3LSLF1 was cloned into engineered pET‐30a (Novagen) described above to produce SUMO‐His‐PhS3LSLF1. Relevant primer sequences used are listed in Table S1. All the recombinant proteins were induced overnight at 16°C at 180 rpm, with MBP‐tagged proteins expressed in E. coli Trans BL21 (DE3) plysS cells described above and SUMO‐His‐PhS3LSLF1 in E. coli Trans BL21 (DE3) (Transgen). Cells were collected and resuspended using binding buffer (20 mM Tris‐HCl (pH 8.0), 200 mM NaCl, 1 mM DTT and 1 mM EDTA (pH 8.0)) for ultrasonication on ice. Then equal lysates containing SUMO‐His‐PhS3LSLF1 were incubated with the same amount of lysates containing MBP or MBP‐tagged fusion protein (PhS3‐RNase or its six mutant forms), respectively, for 2 h at 4°C. The mixed lysates were subsequently immobilised on Dextrin Sepharose High Performance medium (GE HealthCare, 10284602) following the manufacturer’s instructions and eluted by binding buffer supplemented with 10 mM maltose for immunoblots using anti‐MBP (NEB) and anti‐His (Sigma) antibodies.
Bimolecular fluorescence complementation (BiFC) and split firefly luciferase complementation (SFLC) assays
For the BiFC assay, the coding sequences of PhS3 ‐RNase (without signal peptide) and PhS3‐RNase (M) were amplified separately and inserted into pSY‐735‐35S‐cYFP‐HA and the full‐length cDNA of PhS3LSLF1 was cloned into pSY‐736‐35S‐nYFP‐EE as described previously (Li et al., 2018). Relevant primer sequences used are listed in Table S1. Different construct combinations (e.g. nYFP‐PhS3LSLF1 and cYFP‐PhS3 ‐RNase), together with the p19 silencing plasmid, were cotransfected into tobacco leaf epidermal cells using Agrobacterium (GV3101)‐mediated infiltration to generate fusion proteins (e.g. nYFP‐PhS3LSLF1 and cYFP‐PhS3‐RNase) for their interaction test. After culture for another 48 h in the dark, a portion of the injected leaf was cut off and examined by confocal microscope (Zeiss LSM710) to capture the YFP signal. For the SFLC assay, PhS3LSLF1 (or PhS3SLF1) and PhS3 ‐RNase (or its mutant forms) were cloned into pCAMBIA1300‐35S‐HA‐nLUC‐RBS and pCAMBIA1300‐35S‐cLUC‐RBS vectors, respectively, as described previously (Liu et al., 2018). At 48 h post‐injection, 1 mM luciferin was sprayed on the injected leaves and the LUC signals were captured using a cooled CCD imaging system (LB985; Berthold, Bad Wildbad, Germany).
Aniline blue staining of pollen tubes within pistils
Pollinated pistils were chemically fixed and treated for observation as described (Liu et al., 2014).
Results
S‐RNase polyubiquitination mainly occurs through K48 linkages at three conserved spatial regions among S‐RNases
Previous studies have revealed that S‐RNase is ubiquitinated in cross‐pollen tubes, but the linkage type or site of this ubiquitination remains unclear. To investigate these questions, we performed an in vitro ubiquitination assay and showed that both oligoubiquitinated and polyubiquitinated PhS3‐RNase were detected by anti‐ubiquitin, anti‐PhS3‐RNase and anti‐ubiquitin‐K48 antibodies compared with the PhS3S3L wild‐type control, indicating that nonself PhS3LSLF1 is capable of forming an SCFSLF complex to ubiquitinate PhS3‐RNase mainly through K48‐linked polyubiquitin chains (Fig. 1a). To further detect the ubiquitination site of S‐RNase, we used LC‐MS/MS and identified six ubiquitinated residues at T102, K103, C118, T153, K154 and K217 positions of PhS3‐RNase from wild‐type pistils cross‐pollinated with the transgenic pollen containing the pollen‐specific PhS3LSLF1 in P. hybrida (Figs 1b, S1). Furthermore, we found that the ubiquitinated C118, T153, K154 and K217 residues were exclusively detected in cross‐pollinated pistils, suggesting that they are specific for cross‐pollination, whereas the ubiquitinated T102 and K103 residues were detected in both unpollinated and self‐pollinated pistils (Fig. S2), suggesting that they are likely to occur before S‐RNase uptake into pollen tubes.
Fig. 1.
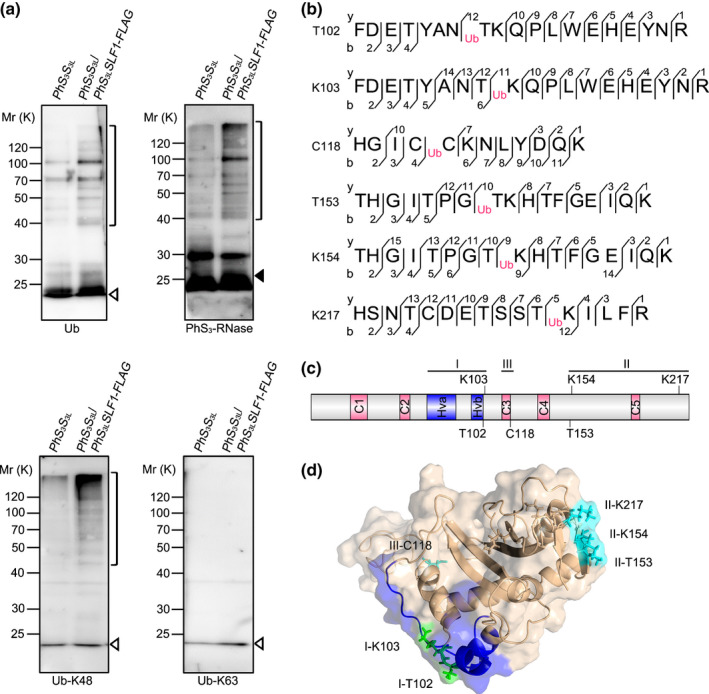
Six amino acid residues of Petunia hybrida S3‐RNase are ubiquitinated by K48‐linked polyubiquitin chains through SCFPhS3LSLF1. (a) Immunoblot detection of in vitro ubiquitinated products of PhS3‐RNase by PhS3LSLF1. The pollen genotypes and the transgene are indicated on top and PhS3S3L used as a negative control. Brackets indicate polyubiquitinated PhS3‐RNases. Open and closed arrowheads indicate ubiquitin and unubiquitinated PhS3‐RNase monomers, respectively. Antibodies used are indicated in the bottom as ubiquitin (Ub), PhS3‐RNase, Ub‐K48 and Ub‐K63, respectively. K, lysine; FLAG, a protein tag. (b) Ubiquitination sites of PhS3‐RNase identified by LC‐MS/MS. Ub: the amino acid residue on its right within the peptide sequence of PhS3‐RNase is ubiquitinated. The b‐type and y‐type product ions are indicated. (c) The secondary structural features of PhS3‐RNase with the locations of the six ubiquitination sites. C1–C5, five conserved regions; Hva and Hvb, hypervariable region a and b. K, C and T: lysine, cysteine and threonine, respectively. (d) Spatial locations of the six ubiquitination sites on the three‐dimensional (3D) structure of PhS3‐RNase. The dark blue region indicates the Hv regions of PhS3‐RNase, the green region indicates the residues identified by LC‐MS/MS in unpollinated, self‐pollinated and cross‐pollinated pistils, and the cyan region indicates the residues identified specifically in cross‐pollinated pistils. I, II and III: three regions containing the ubiquitination sites shown in the predicted 3D structure of PhS3‐RNase.
To determine the locations of these ubiquitinated amino acid residues in S‐RNases, we compared a total of 36 S‐RNases from Solanaceae and found that C118 was within the conserved (C) 3 region, T102 and K103 were adjacent to hypervariable (Hv) b, T153 and K154 were between C4 and C5 and K217 was found at the C‐terminal region, implying they are located in three largely conserved S‐RNase regions (Figs 1c, S3; Table S2). Next, we reasoned that the ubiquitination sites should be spatially close to E2. To examine this possibility, we determined the spatial localisation of six ubiquitinated residues on the predicted spatial structure of PhS3‐RNase and found that T102 and K103 residues were located near the Hvb region on an interface between S‐RNase and SLF and were termed region (R) I, T153, K154 and K217 were found in a region close to E2 and termed R II, whereas C118 was found inside the predicted spatial structure and was named R III (Fig. 1d). Taken together, our results demonstrated that S‐RNases are ubiquitinated mainly through K48 linkage at three largely conserved spatial regions among Solanaceae S‐RNases.
Two ubiquitinated amino acids from R I are partially involved in PhS3‐RNase degradation during cross‐pollination
To examine how six ubiquitinated amino acids from three spatial regions mediate the S‐RNase ubiquitination, we first designed a mutant construct named MI containing T102A and K103R substitutions that was incapable of ubiquitination at R I of PhS3‐RNase and showed that its RNase activity increased with time, similar to the wild‐type (Fig. S4a), suggesting that MI possessed normal ribonuclease activity. To examine whether the substitutions affected the subcellular location of PhS3‐RNase, we performed fractionation experiments and found that MI was predominantly enriched in the S160 fraction derived from the pollen‐tube cytosol, similar to wild‐type PhS3‐RNase (S3R) (Fig. S4b). Furthermore, we performed pull‐down, SFLC and BiFC assays and found that MI was capable of interacting with nonself PhS3LSLF1 (Fig. S4c–e). Nevertheless, we also found that it displayed a weak interaction with self PhS3SLF1 (Fig. S5), similar to previous studies (Hua & Kao, 2006; Kubo et al., 2010). Consistent with these findings, we found that the predicted structure and electrostatic potentials of MI remain essentially unaltered (Fig. S6). Taken together, these results indicated that MI has an enzymatic activity and structure similar to wild‐type S3R.
To examine the in vivo function of MI, we transformed S3R and MI driven by the pistil‐specific Chip promotor into SI PhS3S3L plants, respectively and also transformed their FLAG‐tagged forms into PhS3S3L . For each construct, we identified at least 24 T0 transgenic lines by PCR analysis (Figs S7, S8) and found that MI was expressed normally in the transgenic lines (Figs S9, S10a,b). Furthermore, self‐pollination assays showed that each construct did not alter the SI phenotypes of the transgenic plants (Tables S3, S4). To examine their roles in cross‐pollination, we further identified several lines with similar transgene expression levels. Compared with c. 398 seed sets per capsule from PhS3S3L carrying the transgenic S3R (S3S3L /S3R‐60) pollinated with cross‐pollen of PhSVSV , S3S3L /MI had a reduced seed set of 298 with a reduction of 25% (Fig. S10c,d; Table S3). Consistent with this, we also found a substantial reduction of seed sets derived from cross‐pollination of the FLAG‐tagged transgenic line S3S3L /MI‐FLAG‐24 (292 per capsule) with a 30% reduction compared with 421 seeds per capsule from S3S3L /S3R‐FLAG‐34 (Fig. S10c,d; Table S4). Taken together, these results suggested that ubiquitinated R I was involved in cross‐pollination.
To verify this role, we assessed the degradation rates of recombinant SUMO‐His‐tagged S3R and MI proteins by cell‐free degradation assays using self‐ (PhS3S3 ) or cross‐ (PhSVSV ) pollen‐tube extracts (PTE). The results showed that both SUMO‐His‐tagged S3R and MI remained essentially stable in PhS3S3 PTE but SUMO‐His‐S3R degraded rapidly in the PhSVSV PTE without MG132 and only 7% remained after a 10‐min treatment (Fig. S10e,f). However, the degradation rates decreased slightly and c. 25% remained after a 10‐min treatment for SUMO‐His‐MI protein (Fig. S10f), showing that MI degradation was partially inhibited and, therefore, resulted in its accumulation in cross‐pollen tubes. Consistently, both S3R‐FLAG and MI‐FLAG remained largely stable in self‐ (PhS3S3 ) pollen tubes, but their amounts significantly decreased in cross‐ (PhSVSV ) pollen tubes during semi in vivo pulse–chase experiments using style extracts of S3S3L /MI‐FLAG‐24 or S3S3L /S3R‐FLAG‐34 to mimic self‐pollination and cross‐pollination (Fig. S10g). Nevertheless, an additional c. 12% remained for MI‐FLAG in cross‐pollen tubes compared with S3R‐FLAG, suggesting that the cross‐pollen rejections by the transgenic pistils containing MI‐FLAG mainly resulted from its decreased degradation. In addition, degradation of these proteins by cross‐pollen was significantly delayed by MG132 treatment (Fig. S10e–g), indicating the degradation of MI by the UPS pathway similar to that found in wild‐type.
To confirm the role of the ubiquitinated R I in S‐RNase degradation, we further performed ubiquitination assays and found that by contrast with self‐ (PhS3S3 ) PTE, both polyubiquitinated His‐tagged S3R and MI proteins were detected by anti‐ubiquitin and anti‐His antibodies (Figs S11, S12) after incubation with SCFPhS3LSLF1 serving as E3, and indicating that they both could be specifically ubiquitinated by nonself SCFPhS3LSLF1. Nevertheless, the ubiquitinated products of MI reduced to c. 60% that of S3R (Fig. S11), suggesting that the ubiquitinated residues located in R I were partially responsible for the ubiquitination of PhS3‐RNase by cross‐pollen. Taken together, these results revealed that two ubiquitinated amino acids from R I were partially involved in PhS3‐RNase degradation during cross‐pollination.
The R II from PhS3‐RNase serves as a major region for its ubiquitination and degradation in cross‐pollen tubes
To examine the function of three ubiquitinated amino acids from R II, we followed a similar strategy to that used for R I by creating MII with T153A, K154R and K217R substitutions of PhS3‐RNase. We found that its RNase activity increased with time, similarly to that found for wild‐type (Fig. 2a), suggesting that it possessed normal ribonuclease activity. Second, it showed that MII was also predominantly located in the pollen‐tube cytosol (Fig. 2b), capable of interacting with nonself PhS3LSLF1 (Fig. 2c–e), also with a weak interaction with self PhS3SLF1 (Fig. S5) and had unaltered predicted structure and electrostatic potentials (Fig. S6). Furthermore, MII and its FLAG‐tagged transgenes were expressed normally in SI PhS3S3L plants, and the transgenic lines also maintained an SI phenotype (Figs 2f,g, S7–S9; Tables S3, S4). Compared with MI transgenic lines, a significant difference observed for S3S3L /MII was the seed sets derived from pollination with cross‐pollen of PhSVSV (c. 75 seed sets per capsule), with a significant reduction of 81% compared with S3S3L /S3R‐60 (398 seed sets per capsule) (Figs 2h, S13a; Table S3). Consistent with this result, compared with 421 seeds per capsule from S3S3L /S3R‐FLAG‐34, c. 113 seeds were set for the FLAG‐tagged transgenic lines with a significant 73% reduction (Figs 2h, S13b; Table S4). We further found that the MII‐FLAG transgene led to the production of much fewer seed sets per capsule than MI‐FLAG whereas their protein levels remained similar (Figs 2g,h, S13b), indicating that the ubiquitinated R II plays a major role in cross‐pollination. Furthermore, cell‐free degradation assays and pulse–chase experiments mimicking cross‐pollination showed that the degradation of MII protein was mainly through the UPS pathway and was severely inhibited, with a significantly decreased difference between its remaining amounts in self‐pollen and cross‐pollen tubes compared with MI (Figs 2i,j, S10f). Consistently, in vitro ubiquitination assays showed that the ubiquitination amount of MII with SCFPhS3LSLF1 serving as E3 significantly reduced to 40% of S3R, with a reduction of 20% compared with MI (Fig. S11). In addition, both the attenuated degradation and ubiquitination of MII specifically occurred in cross‐pollen tubes (Figs 2i,j, S11, S12). Taken together, these results suggested that R II of PhS3‐RNase acts as a major ubiquitination region for its degradation resulting in cross‐pollination.
Fig. 2.
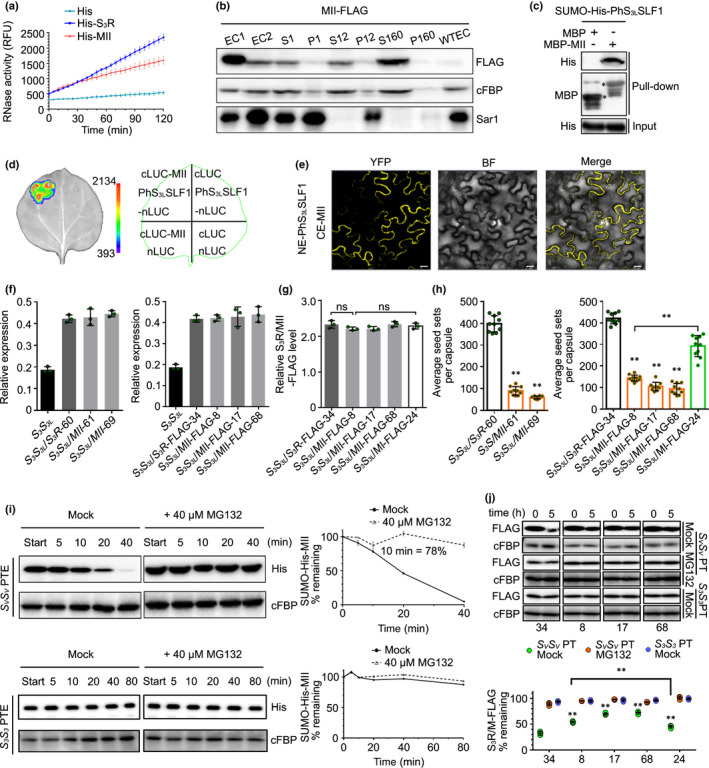
Petunia hybrida S3‐RNase with mutated region (R) II significantly inhibits cross seed sets. (a) RNase activity detection of His‐S3R and mutant (M) II expressed by pCold‐TF vectors. The relative fluorescence unit (RFU) indicating RNase activity during a time‐course experiment is shown as mean ± SD (n = 3). II: the ubiquitinated region II of PhS3‐RNase. (b) Immunoblot detection of FLAG‐tagged MII in subcellular fractions of in vitro germinated pollen tubes. EC1, EC2 and WTEC indicate entire cell homogenates of the pistils from the transgenic plants containing MII‐FLAG, the pollen tubes of PhSVSV treated with EC1 and the pistils from wild‐type PhS3S3L . WTEC was a negative control. S1 and P1, S12 and P12, S160 and P160 indicate supernatant and pellet fractions obtained by centrifugation of EC2 at 1000 g, 12 000 g and 160 000 g, respectively. cFBP and Sar1 are respective marker antibodies of cytosol and endoplasmic reticulum (ER). (c) Physical interactions between PhS3LSLF1 and MII detected by pull‐down assay. Input and pull‐down: bait protein SUMO‐His‐PhS3LSLF1 and prey proteins detected by immunoblots, respectively. MBP and SUMO‐His are protein tags. Asterisks indicate bands of target proteins. (d) Split firefly luciferase complementation (SFLC) assay. The numbers on the left side of the colour signal bars represent the values of the fluorescent signal. The injection positions of each component on tobacco leaves are indicated in the contour diagram of leaf margin. nLUC and cLUC indicate transiently expressed N‐terminal and C‐terminal regions of luciferase. (e) Bimolecular fluorescence complementation (BiFC) assay. NE and CE: transiently expressed N‐terminal and C‐terminal regions of YFP by pSPYNE and pSPYCE vectors. YFP, BF and Merge represent the YFP fluorescence, bright field and their merged field, respectively. Bars, 20 μm. (f) Transcripts of the transgene and native PhS3 ‐RNase detected by qRT‐PCR. The T0 transgenic lines are indicated below the horizontal axes. S3S3L is a wild‐type. Data are shown as mean ± SD (n = 3). (g) Quantitative analyses of S3R‐ and MII‐FLAG proteins. The T0 transgenic lines are indicated below the horizontal axes. Data are shown as mean ± SD (n = 3). Student’s t‐test was used to generate the P‐values. ns (not significant), P > 0.05. (h) Statistical analyses of seed sets per capsule from T0 transgenic plants pollinated with cross‐pollen of PhSVSV . Data are shown as mean ± SD (n ≥ 9). Student’s t‐test was used to generate the P‐values. **, P < 0.01. (i) Cell‐free degradation of recombinant SUMO‐His‐MII by pollen‐tube extracts (PTE) of PhSVSV or PhS3S3 . Left, immunoblots of the reaction products incubated with or without MG132 (Mock). Start, time point zero in each degradation assay. cFBP antibody was used to detect nondegraded loading control. Right, quantitative analyses of the degradation rates. Data are shown as mean ± SD (n = 3). The remaining amount at 10 min is indicated. (j) Time‐course analyses of PhS3R‐FLAG and MII‐FLAG levels in the cross‐pollen tubes (PTs) (PhSVSV ) or self‐PTs (PhS3S3 ) incubated with or without MG132 (Mock). PhSVSV and PhS3S3 PTs were challenged with style extracts of PhS3S3L /PhS3R‐FLAG or PhS3S3L /MII‐FLAG for 5 h to mimic cross‐pollination and self‐pollination, respectively. Top, immunoblots of PhS3R‐FLAG or MII‐FLAG in the PT using FLAG antibody. cFBP was detected as a loading control. The numbers at the bottom indicate the transgenic line numbers corresponding to those in (h). Bottom, quantitative analyses of the immunoblots. Data are shown as mean ± SD (n = 3). Student’s t‐test was used to generate the P‐values. **, P < 0.01.
K154 and K217 from R II act as two major ubiquitination residues for PhS3‐RNase degradation in cross‐pollen tubes
To explore the function of three lysines (K103, K154 and K217) and two threonines (T102 and T153) residues of PhS3‐RNase in its degradation, we designed two mutant constructs termed MK (K103R, K154R and K217R) and MT (T102A and T153A), which showed similar enzymatic activities, subcellular localisations, SLF interactions, predicted structures and electrostatic potentials to wild‐type S3R as well as normal pistil expressions and SI phenotypes similar to SI PhS3S3L plants (Figs 3a–g, S5–S9; Tables S3, S4). However, MK and MT transgenic lines showed differential seed sets of 207 and 356 per capsule after pollination with cross‐pollen of PhSVSV , with a significant reduction of 48% and 15%, respectively, compared with S3S3L /S3R‐60 (398 seeds per capsule) (Figs 3h, S14a; Table S3). This was consistent with the S3S3L /MK‐FLAG‐16 set with 113 seeds per capsule with a significant reduction of 73% and 66% compared with S3S3L /S3R‐FLAG‐34 (421 seeds per capsule) and S3S3L /MT‐FLAG‐44 (334 seeds per capsule), respectively (Figs 3h, S14b; Table S4). In addition, we showed that, compared with the MT transgene, MK resulted in a greater seed set reduction similar to that found for MII when the transgene expression levels were similar (Fig. 3g,h), suggesting that the identified lysine amino acids, especially K154 and K217, played a major role in the ubiquitination and degradation of PhS3‐RNase. Furthermore, cell‐free degradation and pulse–chase assays showed that MK degradation by the 26S proteasome in cross‐ (PhSVSV ) pollen tubes had been significantly delayed compared with MT in the absence of MG132 (Figs 3i,S14c,d). Ubiquitination assays also indicated that lysine residues rather than threonine residues act as the major sites for PhS3‐RNase ubiquitination by nonself SCFPhS3LSLF1 (Figs 3j, S12). Taken together, our results suggested that K154 and K217 from R II functioned as two major ubiquitination residues of PhS3‐RNase for cross‐pollination.
Fig. 3.
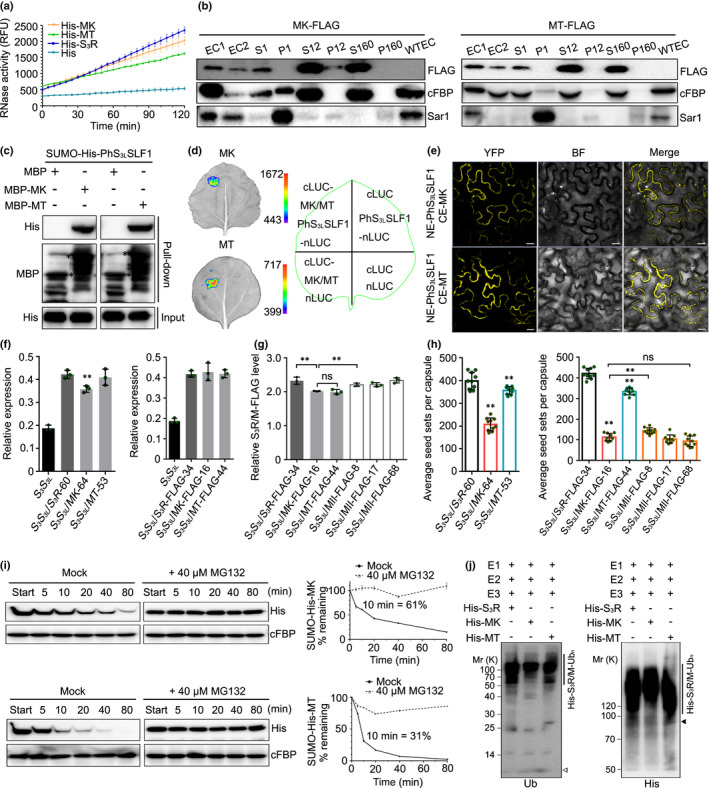
K154 and K217 of the region (R) II of Petunia hybrida S3‐RNase serve as two major ubiquitination sites for its degradation in cross‐pollen tubes. (a) RNase activity detection of His‐mutant (M) K and MT expressed by pCold‐TF vectors. RFU: relative fluorescence unit. K and T: lysine and threonine within six ubiquitinated residues of PhS3‐RNase. Data represent mean ± SD (n = 3). (b) Immunoblot detection of FLAG‐tagged MK and MT in subcellular fractions of in vitro germinated pollen tubes. EC1, EC2 and WTEC: entire cell homogenates of the pistils from the transgenic plants containing MK‐FLAG or MT‐FLAG, the pollen tubes of PhSVSV treated with EC1 and the pistils from wild‐type PhS3S3L . S1, P1, S12, P12, S160 and P160: supernatant and pellet fractions obtained by centrifugation of EC2 at 1000 g, 12 000 g and 160 000 g, respectively. cFBP and Sar1: respective marker antibodies of cytosol and endoplasmic reticulum (ER). (c) Physical interactions between PhS3LSLF1 and MK and MT detected by pull‐down assays. MBP and SUMO‐His: protein tags. (d) SFLC assays. nLUC and cLUC: transiently expressed N‐terminal and C‐terminal regions of luciferase. (e) BiFC assays. NE and CE: transiently expressed N‐terminal and C‐terminal regions of YFP. YFP, BF and Merge: the YFP fluorescence, bright field and their merged field, respectively. Bars, 20 μm. (f) Transcripts of transgene and native PhS3 ‐RNase detected by qRT‐PCR. Data are shown as mean ± SD (n = 3). Student’s t‐test: **, P < 0.01. (g) Quantitative analyses of S3R‐FLAG, MK‐FLAG and MT‐FLAG proteins. Data are shown as mean ± SD (n = 3). Student’s t‐test: ns, not significant; P > 0.05; **, P < 0.01. (h) Statistical analyses of seed sets per capsule from T0 transgenic plants pollinated with cross‐pollen of PhSVSV . Data are shown as mean ± SD (n = 10). Student’s t‐test: ns (not significant), P > 0.05; **, P < 0.01. (i) Immunoblots of recombinant SUMO‐His‐MK and ‐MT in the cell‐free degradation products incubated by PhSVSV pollen‐tube extracts (PTE) with or without MG132 (Mock). Data are shown as mean ± SD (n = 3). (j) Immunoblot detection of in vitro ubiquitination products of His‐S3R, ‐MK and ‐MT by SCFPhS3LSLF1‐FLAG (E3) using anti‐ubiquitin (Ub) and anti‐His antibodies. The vertical lines illustrate the ubiquitinated substrates. Open and closed arrowheads indicate ubiquitin and unubiquitinated substrate monomers, respectively. Annotations of this figure are identical to those of Fig. 2.
R III functions as the second major ubiquitination region for PhS3‐RNase degradation allowing cross‐pollination
To investigate the function of the ubiquitination site C118 from the internal R III, we designed MIII (C118A) and found that it also maintained ribonuclease activity, subcellular localisation and predicted structure similar to the wild‐type S3R (Figs S5, S6, S15). We further transformed MIII and its FLAG‐tagged form into SI PhS3S3L plants and detected significantly reduced numbers of seed sets of c. 160 and 261 per capsule from S3S3L /MIII‐84 and S3S3L /MIII‐FLAG‐18 after pollination with cross‐pollen of PhSVSV , with a respective reduction of 59% and 38% compared with S3S3L /S3R‐60 and S3S3L /S3R‐FLAG‐34 (Figs S7–S9, S16a–d; Tables S3, S4). Furthermore, the average seed set number per capsule was much fewer than that for S3S3L /MI‐FLAG when they showed similar transgene expression levels (Fig. S16b,d), supporting a role for R III in the degradation of PhS3‐RNase. In addition, we detected a marked accumulation of MIII in cross‐pollen tubes compared with S3R and MI in the absence of MG132 (Figs S10e–g, S16e,f) and a significantly decreased level of ubiquitination by nonself SCFPhS3LSLF1 compared with MI (Figs S11, S12). Taken together, these results suggested that R III acts as a second major ubiquitination region for the degradation of PhS3‐RNase, therefore leading to cross‐pollination.
R I, R II and R III of PhS3‐RNase function additively in its degradation during cross‐pollination
To examine the function of the three ubiquitination regions together, we made MI/II/III (T102A, K103R, T153A, K154R, K217R and C118A). Similar to wild‐type PhS3‐RNase, the mutant form exhibited normal physicochemical properties (Figs S5, S6, S17) but resulted in 197 and 93 cross seeds per capsule derived from S3S3L /MI/II/III‐45 and S3S3L /MI/II/III‐FLAG‐49 with PhSVSV pollen, respectively, a significant reduction of 50% and 77%, and similar to the lines containing mutated R II (Figs 4a–c, S7–S9, S18; Tables S3, S4). Furthermore, the degradation of MI/II/III in cross‐pollen tubes was strongly inhibited in the absence of MG132 (Fig. 4d,e), indicating a significantly reduced ubiquitination by nonself SCFPhS3LSLF1 in cross‐pollen (Figs 4f, S12). Taken together, these results suggested that the degradation of PhS3‐RNase was largely dependent on an additive role of its three ubiquitination regions during cross‐pollination.
Fig. 4.
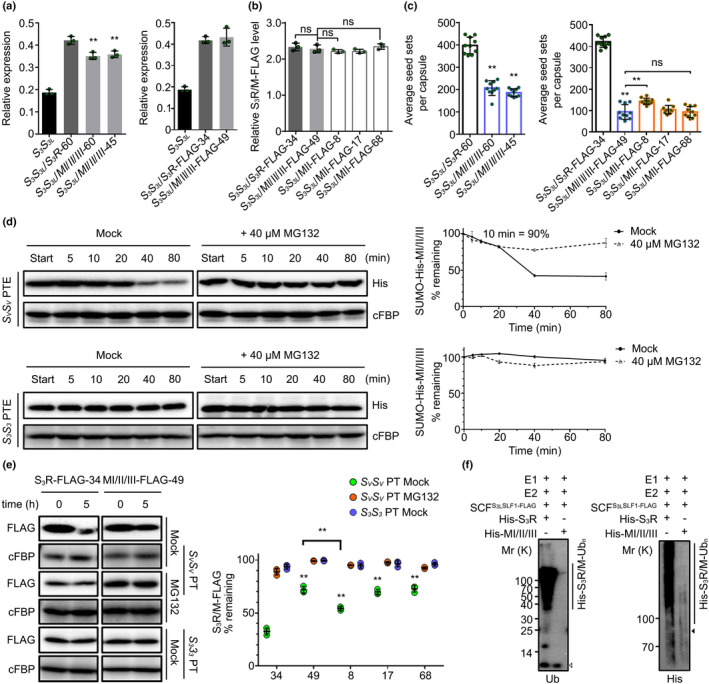
Petunia hybrida S3‐RNase with mutated region (R) I, II and III regions significantly reduces cross seed sets. (a) Transcripts of the transgene (PhS3 ‐RNase or mutant (M) I/II/III) and native PhS3 ‐RNase detected by qRT‐PCR. Data are shown as mean ± SD (n = 3). Student’s t‐test: **, P < 0.01. I, II and III, three ubiquitination regions of PhS3‐RNase. FLAG, a protein tag. (b) Quantitative analyses of S3R‐FLAG and MI/II/III‐FLAG proteins. Data are shown as mean ± SD (n = 3). Student’s t‐test: ns (not significant), P > 0.05. (c) Statistical analyses of seed sets per capsule from T0 transgenic lines containing MI/II/III pollinated with cross‐pollen of PhSVSV . Data are shown as mean ± SD (n = 10). Student’s t‐test: ns (not significant), P > 0.05; **, P < 0.01. (d) Immunoblots of recombinant SUMO‐His‐MI/II/III in the cell‐free degradation products incubated in the pollen‐tube extracts (PTE) of PhSVSV or PhS3S3 with or without MG132 (Mock). Data represent mean ± SD (n = 3). (e) Time‐course analyses of MI/II/III‐FLAG amounts in the cross‐ (PhSVSV ) or self‐ (PhS3S3 ) pollen tubes (PT) incubated with or without MG132. Data are shown as mean ± SD (n = 3). Student’s t‐test: **, P < 0.01. (f) Immunoblot detections of in vitro ubiquitinated recombinant His‐tagged S3R and MI/II/III. Annotations of this figure are identical to those of Figs 2 and 3.
Discussion
Previous studies have shown that nonself S‐RNases are collaboratively recognised by multiple nonself SLFs leading to the formation of canonical SCFSLF complexes for their ubiquitination and subsequent degradation by the 26S proteasome during cross‐pollination, but the ubiquitination linkage type and site remain unclear. In this study, we found that nonself S‐RNase is mainly polyubiquitinated through K48 linkages by SCFSLF at three spatial regions (R I, R II and R III) in P. hybrida. Among them, R I ubiquitination appears to occur before S‐RNase uptake into pollen tubes with a minor role, if any, in cross‐pollen tubes, whereas R II and R III act as two major ubiquitination regions for S‐RNase degradation. Consistently, Hua & Kao (2008) identified six lysine residues mainly responsible for the ubiquitination and degradation of S3‐RNase in P. inflata, which are located in the same region of R II identified in our study, reinforcing the major role of R II in Petunia S3‐RNase degradation for cross‐pollination. Based on our results, we propose a stepwise UPS model for S‐RNases cytotoxicity restriction allowing cross‐pollination in P. hybrida (Fig. 5). In this model, both self and nonself S‐RNases with a small fraction of R I ubiquitinated forms are likely to be mediated by an unknown E3 ligase and are taken up into the cytosols of either self‐pollen or cross‐pollen tubes. First, the R I ubiquitinated forms would make them unable to be recognised by SLFs but degraded by the 26S proteasome. Second, other S‐RNases could be recognised by SLFs on the basis of ‘like charges repel and unlike charges attract’, and the like electrostatic potentials together with other unknown forces between self S‐RNase and its cognate SLF would result in the formation of nonfunctional SCFSLF complexes as demonstrated previously (Li et al., 2017). By contrast, nonself S‐RNase would be attracted by unlike electrostatic potentials and other unknown factors and polyubiquitinated by functional SCFSLF complexes (Li et al., 2017; Sun & Kao, 2018) at R II, leading to its degradation by the 26S proteasome. Thirdly, the internal R III of nonself S‐RNase could be exposed by a conformational change for its further ubiquitination by SLFs and degradation resulting in cross‐pollination. Our studies have revealed that the ubiquitination and degradation of nonself S‐RNases depend on at least three regions with distinct ubiquitination sites, including lysine, threonine and cysteine, reinforcing the notion that the restriction of S‐RNase cytotoxicity occurs mainly by the ubiquitination‐mediated degradation mechanism in P. hybrida (Liu et al., 2014).
Fig. 5.
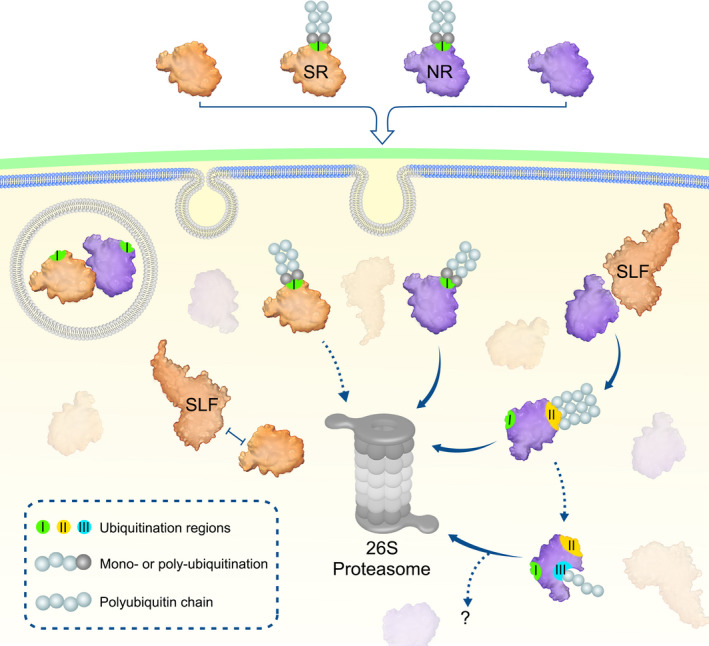
Proposed model for a stepwise ubiquitination and degradation mechanism of Petunia hybrida S‐RNases. Self and nonself S‐RNases (SR and NR) and their region (R) I ubiquitinated forms can enter the cytoplasm through the pollen‐tube membrane. The R I ubiquitinated S‐RNases could be degraded by the 26S proteasome, whereas their unubiquitinated forms are identified by SLFs. SR and its cognate SLF repel each other and results in an inability of SCFSLF to ubiquitinate SR. By contrast, NR is attracted by nonself SLF for R II ubiquitination by SCFSLF and its subsequent degradation by the 26S proteasome. Subsequently, internal R III could be further exposed for ubiquitination, leading to degradation of NR by the 26S proteasome or other unknown pathways. In addition, S‐RNase compartmentalisation could occur in the vacuole and contribute to its sequestration. I, II, and III: three ubiquitination regions in S‐RNases.
Nevertheless, the underlying mechanisms of R I and R III ubiquitination remain to be further elucidated. Notably, newly synthesised secretory proteins are constantly scrutinised and destructed by protein quality control systems such as ER‐associated degradation (ERAD) or autophagy to maintain proteostasis once they are misfolded or aggregated (Anelli & Sitia, 2008). The ubiquitination of R I in the unpollinated pistils suggested that it might result from the polyubiquitination of misfolded PhS3‐RNases. Ubiquitination of closely spaced residues is predicted to be important for polyubiquitin chain assembly (Wang et al., 2012). Here, we found that the identified threonine ubiquitination residues paired with lysine also contributed to the ubiquitination and degradation of nonself S‐RNase for cross‐pollination, suggesting that they may play a role in building polyubiquitin chains long enough for proteasome recognition (Thrower et al., 2000). In addition, ubiquitin often serves as a critical signal governing the membrane traffic system. Monoubiquitination is sufficient to initiate the internalisation of plasma membrane proteins, and K63‐linked polyubiquitination is frequently involved in their subsequent sorting and trafficking (Hicke & Dunn, 2003; Clague & Urbé, 2010; Paez Valencia et al., 2016). It is therefore possible that R I could also be monoubiquitinated leading to S‐RNase entry into pollen tubes by endocytosis, consistent with the results showing that a small fraction of S‐RNases is sequestered in microsome fractions (Liu et al., 2014). As for the internal R III, its ubiquitination is likely to occur only after being exposed. Therefore, the ubiquitination of R II might lead to the conformational change of nonself S‐RNase and exposure of R III, and the subsequent ubiquitination of R III would further block the enzymatic activity of the S‐RNase (Sagar et al., 2007). Previous simulations have demonstrated that the conserved complementary electrostatic patterns and hydrophobic patches of Rpn10, a recognition subunit of proteasome, and K48‐linked tetraubiquitin of the substrates, are critical for their interaction (Zhang et al., 2016). Similarly, the ubiquitination of R III might further enhance the electrostatic potentials and hydrophobicity to strengthen the recognition of ubiquitinated S‐RNase by the proteasome as well as its degradation.
It remains unclear why the additive action of the three regions did not completely restrict nonself S‐RNase cytotoxicity in cross‐pollen tubes. We suggest that there might be additional mechanism (s) occurring either after the R III‐mediated ubiquitination of nonself S‐RNase or during S‐RNase uptake into pollen tubes by endocytosis resulting in its compartmentalisation (Goldraij et al., 2006). In Nicotiana, S‐RNases appeared to be sequestered early in the vacuole but released out late to the cytosol in self‐pollen tubes (Goldraij et al., 2006). In P. hybrida, a small amount of S‐RNases was detected in the microvesicles of pollen tubes (Liu et al., 2014). Furthermore, it has been shown that S‐RNases were compartmentalised in the cross‐pollen tubes even 36 h after cross‐pollination in Nicotiana (Goldraij et al., 2006), longer than 24 h, the longest post‐pollination time used for identification of R I/R II/R III ubiquitination in P. hybrida. Together, these findings suggested that S‐RNase compartmentalisation and its stepwise ubiquitination and degradation might function synergistically from the beginning to the late phases of S‐RNase action in the cross‐pollen tubes. In this scenario, it is possible that the mutant S‐RNase evades ubiquitination and degradation and that would partially reverse the downstream response derived from nonself‐recognition between SLFs and S‐RNases to that of their self‐recognition. This would further lead to S‐RNase release from the vacuole to varying degrees, similar to the late stage of self‐pollen rejection proposed in Nicotiana (Goldraij et al., 2006). In this view, the increased S‐RNase cytotoxicity could result from the total effects of S‐RNases, both originally present in and later released from the vacuole into the cross pollen‐tube cytosol. Nevertheless, removal of the additive ubiquitination of R I/R II/R III could not lead to sufficient cytosolic S‐RNases, therefore allowing the cross‐pollen to escape and to seed sets. Further investigation into the S‐RNase uptake mechanism and the relationship between the stepwise ubiquitination and degradation of S‐RNases and their compartmentalisation would provide the answer to these possibilities.
T2 RNases are widespread in every organism except Archaea and are involved in a variety of biological processes, including phosphate starvation, viral infection, self‐fertilisation, tumour growth control and cell death (Löffler et al., 1992; Bariola et al., 1994; Meyers et al., 1999; Thompson & Parker, 2009; Ramanauskas & Igić, 2017). However, our understanding of their function remains largely incomplete, especially when their roles appear to be independent of their enzymatic activity. In Saccharomyces cerevisiae, T2 RNase Rny1 can be released from the vacuole to cleave tRNA and rRNA under superoxygen stress (Thompson & Parker, 2009). Rny1 is indispensable for cell viability, but overexpressed Rny1 can act as a cytotoxin during oxidative stress (MacIntosh et al., 2001; Thompson & Parker, 2009). Moreover, its inactivation strikingly has no effects on cell viability (MacIntosh et al., 2001), but the underlying mechanism remains elusive. In human, RNASET2 is not only implicated to regulate neurodevelopment downstream of the immune response, but also serve as a tumour suppresser (Henneke et al., 2009), whereas how it contributes to this process in a cleavage‐independent manner is poorly defined. In addition, the catalytic‐independent function of T2 RNase has also been confirmed for ACTIBIND from Aspergillus niger that can bind to and destroy the normal actin networks, and is supposed to be conserved in other T2 RNase family members including S‐RNases (Roiz et al., 2006). Therefore, T2 RNase may act as a molecular signal mediating multiple biological settings, revealing that diverse T2 RNase roles could be derived through neofunctionalisation in these lineages.
In S‐RNase‐based SI, Yang et al. (2018) reported an S‐RNase‐mediated actin disruption in apple (Malus × domestica). The disrupted cytoskeleton dynamic served as a major cause of PCD (Thomas et al., 2006), which is also proposed to occur in self‐pollen tubes of Pyrus bretschneideri (Chen et al., 2018). Moreover, self S‐RNase can disrupt Ca2+ gradients at the pollen‐tube apex by inhibiting phospholipase C (PLC) (Qu et al., 2017). In addition, heat‐inactivated S‐RNase surprisingly exerts a more severe inhibition of pollen tubes (Gray et al., 1991). These studies suggested that S‐RNase could function in a signalling pathway independent of its enzymatic activity. Our results indicated that self S‐RNase could be partially ubiquitinated extracellularly and destroyed during its uptake, but we cannot rule out the possibility that its ubiquitination could act as an initial signal for the SI response.
In addition to ubiquitination, a recent study in Solanum chacoense showed that the number of carbohydrate chains of S‐RNase may influence its threshold for pollen rejection (Liu et al., 2008). Torres‐Rodriguez et al. (2020) found that the ribonuclease activity of SC10‐RNase could be significantly enhanced if its conserved Cys155–Cys185 disulphide bond was reduced by Nicotiana alata thioredoxin type h (Natrxh) in the pollen‐tube cytosols. Furthermore, the disulphide bond of SC10‐RNase is highly conserved among S‐RNases (Anderson et al., 1989), and it is likely that S‐RNase reduction occurs after incompatible recognition between SLFs and S‐RNases, leading to RNA degradation necessary for self‐pollen rejection. In this case, the mutant S‐RNases incapable of ubiquitination and degradation in the cytosols of cross‐pollen tubes could induce downstream responses, including S‐RNase reduction, resulting in its increased ribonuclease activity as well as cytotoxicity for cross‐pollen inhibition. Moreover, as phosphorylation serves as a critical modification modulating multiple cellular events, it may also be involved in S‐RNase activity regulation and the downstream signalling transduction in Solanaceae‐type SI. In addition, previous studies have shown that other factors, except electrostatic potentials, should exist that contribute to the recognition between SLF and S‐RNase (Li et al., 2017). Therefore, future studies on the structure of SLF bound to S‐RNase, and other post‐translational modifications such as glycosylation, reduction and phosphorylation of S‐RNase and their relationships with its ubiquitination should shed light on how S‐RNase functions and stimulates downstream signalling networks in the pollen tubes.
In conclusion, our results have revealed a novel stepwise UPS mechanism for S‐RNase cytotoxicity restriction resulting in cross‐pollination in P. hybrida. Our findings also indicated a possible mechanism for dynamic regulation of secreted cytotoxin activities including other T2 ribonuclease members. Further validation of this mechanism using biochemical and cytological approaches is expected to provide additional insights into UPS and Solanaceae‐type SI.
Author contributions
YX conceived and designed the project. HZ and YS performed the experiments. JL and YZ conducted functional analyses of PhS3LSLF1. HH assisted transgenic plant construction. QL and YZ provided technical support. HZ and YX analysed data and wrote the manuscript. All authors commented on the article. HZ and YS contributed equally to this work.
Supporting information
Fig. S1 Identification of six ubiquitinated residues of Petunia hybrida S3‐RNase by LC‐MS/MS analysis of cross‐pollinated pistils.
Fig. S2 Two ubiquitinated residues of Petunia hybrida S3‐RNase identified by LC‐MS/MS analysis of self‐pollinated and unpollinated pistils.
Fig. S3 Locations of six ubiquitinated residues of Petunia hybrida S3‐RNase in Solanaceous S‐RNases.
Fig. S4Petunia hybrida S3‐RNase with the mutated region (R) I displays largely unaltered biochemical and physical properties.
Fig. S5 Physical interactions between Petunia hybrida S3R/Mutant (M) and PhS3SLF1.
Fig. S6 Predicted three‐dimensional (3D) structures and surface electrostatic potentials of Petunia hybrida S3‐RNases with mutated ubiquitinated residues.
Fig. S7Petunia hybridaS3R and S3R (Mutant) (M) transgenes identification by PCR analysis.
Fig. S8 Identification of FLAG‐tagged Petunia hybrida S3R and S3R (Mutant) (M) transgenes by PCR analysis.
Fig. S9 Detection of FLAG‐tagged Petunia hybrida S3R and S3R (Mutant) (M) transgenes expression by immunoblots.
Fig. S10Petuniahybrida S3‐RNase with the mutated region (R) I slightly reduces cross seed sets.
Fig. S11 Decreased ubiquitination amount of mutant (M) I, II and III mediated by SCFS3LSLF1 of Petunia hybrida.
Fig. S12 Ubiquitination of Petunia hybrida S3R and its mutant forms by self‐ (PhS3S3 ) pollen‐tube extracts (PTE).
Fig. S13 Reduced seed set per capsule from T0 transgenic lines with mutated region (R) II of Petunia hybrida S3‐RNase.
Fig. S14 Reduced seed set per capsule from T0 transgenic lines with mutated lysine or threonine within the six ubiquitinated residues of Petunia hybrida S3‐RNase.
Fig. S15 Mutant (M) III largely maintains the biochemical and physical properties of Petunia hybrida S3‐RNase.
Fig. S16Petuniahybrida S3‐RNase with mutated region (R) III markedly reduces cross seed sets.
Fig. S17 Largely unaltered physicochemical properties of Petunia hybrida S3‐RNase with mutated region (R) I, II and III.
Fig. S18 Reduced seed set per capsule from T0 transgenic lines with mutated region (R) I/II/III of Petunia hybrida S3‐RNase.
Table S1 List of primer sequences.
Table S2 Names and accession numbers of S‐RNases used in this study.
Table S3 Seed sets of S3S3L /S3R and S3S3L /S3R (Mutant) (M) T0 transgenic plants of Petunia hybrida.
Table S4 Seed sets of S3S3L /S3R‐FLAG and S3S3L /S3R (Mutant) (M)‐FLAG T0 transgenic plants of Petunia hybrida.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
This work was supported by the Strategic Priority Research Programme of the Chinese Academy of Sciences (XDB27010302) and the National Natural Science Foundation of China (32030007).
Data availability
The data supporting the findings of this work are available from the corresponding author upon request.
References
- Anderson MA, Cornish EC, Mau SL, Williams EG, Hoggart R, Atkinson A, Bonig I, Grego B, Simpson R, Roche PJ. 1986. Cloning of cDNA for a stylar glycoprotein associated with expression of self‐incompatibility in Nicotiana alata . Nature 321: 38–44. [Google Scholar]
- Anderson MA, McFadden GI, Bernatzky R, Atkinson A, Orpin T, Dedman H, Tregear G, Fernley R, Clarke AE. 1989. Sequence variability of three alleles of the self‐incompatibility gene of Nicotiana alata . Plant Cell 1: 483–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anelli T, Sitia R. 2008. Protein quality control in the early secretory pathway. EMBO Journal 27: 315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proceedings of the National Academy of Sciences, USA 98: 10037–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bariola PA, Howard CJ, Taylor CB, Verburg MT, Jaglan VD, Green PJ. 1994. The Arabidopsis ribonuclease gene RNS1 is tightly controlled in response to phosphate limitation. The Plant Journal 6: 673–685. [DOI] [PubMed] [Google Scholar]
- Chen J, Wang P, de Graaf BHJ, Zhang H, Jiao H, Tang C, Zhang S, Wu J. 2018. Phosphatidic acid counteracts S‐RNase signaling in pollen by stabilizing the actin cytoskeleton. Plant Cell 30: 1023–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clague MJ, Urbé S. 2010. Ubiquitin: same molecule, different degradation pathways. Cell 143: 682–685. [DOI] [PubMed] [Google Scholar]
- Entani T, Kubo K‐I, Isogai S, Fukao Y, Shirakawa M, Isogai A, Takayama S. 2014. Ubiquitin‐proteasome‐mediated degradation of S‐RNase in a Solanaceous cross‐compatibility reaction. The Plant Journal 78: 1014–1021. [DOI] [PubMed] [Google Scholar]
- Foote HC, Ride JP, Franklin‐Tong VE, Walker EA, Lawrence MJ, Franklin FC. 1994. Cloning and expression of a distinctive class of self‐incompatibility (S) gene from Papaver rhoeas L. Proceedings of the National Academy of Sciences, USA 91: 2265–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin‐Tong VE. 2008. Self‐incompatibility in flowering plants. Berlin/Heidelberg, Germany: Springer. [Google Scholar]
- Fujii S, Kubo K‐I, Takayama S. 2016. Non‐self‐ and self‐recognition models in plant self‐incompatibility. Nature Plants 2: 16130. [DOI] [PubMed] [Google Scholar]
- Goldraij A, Kondo K, Lee CB, Hancock CN, Sivaguru M, Vazquez‐Santana S, Kim S, Phillips TE, Cruz‐Garcia F, McClure B. 2006. Compartmentalization of S‐RNase and HT‐B degradation in self‐incompatible Nicotiana . Nature 439: 805–810. [DOI] [PubMed] [Google Scholar]
- Gray JE, McClure BA, Bonig I, Anderson MA, Clarke AE. 1991. Action of the style product of the self‐incompatibility gene of Nicotiana alata (S‐RNase) on in vitro‐grown pollen tubes. Plant Cell 3: 271–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henneke M, Diekmann S, Ohlenbusch A, Kaiser J, Engelbrecht V, Kohlschütter A, Krätzner R, Madruga‐Garrido M, Mayer M, Opitz Let al. 2009. RNASET2‐deficient cystic leukoencephalopathy resembles congenital cytomegalovirus brain infection. Nature Genetics 41: 773–775. [DOI] [PubMed] [Google Scholar]
- Hicke L, Dunn R. 2003. Regulation of membrane protein transport by ubiquitin and ubiquitin‐binding proteins. Annual Review of Cell and Developmental Biology 19: 141–172. [DOI] [PubMed] [Google Scholar]
- Hua Z, Fields A, Kao T‐H. 2008. Biochemical models for S‐RNase‐based self‐incompatibility. Molecular Plant 1: 575–585. [DOI] [PubMed] [Google Scholar]
- Hua Z, Kao T‐H. 2006. Identification and characterization of components of a putative Petunia S‐locus F‐box‐containing E3 ligase complex involved in S‐RNase‐based self‐incompatibility. Plant Cell 18: 2531–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z, Kao T‐H. 2008. Identification of major lysine residues of S3‐RNase of Petunia inflata involved in ubiquitin‐26S proteasome‐mediated degradation in vitro . The Plant Journal 54: 1094–1104. [DOI] [PubMed] [Google Scholar]
- Huang J, Zhao L, Yang Q, Xue Y. 2006. AhSSK1, a novel SKP1‐like protein that interacts with the S‐locus F‐box protein SLF. The Plant Journal 46: 780–793. [DOI] [PubMed] [Google Scholar]
- Kakita M, Murase K, Iwano M, Matsumoto T, Watanabe M, Shiba H, Isogai A, Takayama S. 2007. Two distinct forms of M‐locus protein kinase localize to the plasma membrane and interact directly with S‐locus receptor kinase to transduce self‐incompatibility signaling in Brassica rapa . Plant Cell 19: 3961–3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo K‐I, Entani T, Takara A, Wang N, Fields AM, Hua Z, Toyoda M, Kawashima S‐I, Ando T, Isogai Aet al. 2010. Collaborative non‐self recognition system in S‐RNase‐based self‐incompatibility. Science 330: 796–799. [DOI] [PubMed] [Google Scholar]
- Lai Z, Ma W, Han B, Liang L, Zhang Y, Hong G, Xue Y. 2002. An F‐box gene linked to the self‐incompatibility (S) locus of Antirrhinum is expressed specifically in pollen and tapetum. Plant Molecular Biology 50: 29–42. [DOI] [PubMed] [Google Scholar]
- Lee HS, Huang S, Kao T‐H. 1994. S proteins control rejection of incompatible pollen in Petunia inflata . Nature 367: 560–563. [DOI] [PubMed] [Google Scholar]
- Li J, Zhang Y, Song Y, Zhang H, Fan J, Li Q, Zhang D, Xue Y. 2017. Electrostatic potentials of the S‐locus F‐box proteins contribute to the pollen S specificity in self‐incompatibility in Petunia hybrida . The Plant Journal 89: 45–57. [DOI] [PubMed] [Google Scholar]
- Li S, Tian Y, Wu K, Ye Y, Yu J, Zhang J, Liu Q, Hu M, Li H, Tong Yet al. 2018. Modulating plant growth‐metabolism coordination for sustainable agriculture. Nature 560: 595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Chetelat RT. 2014. The role of a pollen‐expressed cullin1 protein in gametophytic self‐incompatibility in Solanum . Genetics 196: 439–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M, Cao Z, Zhu A, Liu Y, Tao M, Yang H, Xu Q Jr, Wang S, Liu J, Li Yet al. 2020. Evolution of self‐compatibility by a mutant Sm‐RNase in citrus. Nature Plants 6: 131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Morse D, Cappadocia M. 2008. Glycosylation of S‐RNases may influence pollen rejection thresholds in Solanum chacoense . Journal of Experimental Botany 59: 545–552. [DOI] [PubMed] [Google Scholar]
- Liu Q, Han R, Wu K, Zhang J, Ye Y, Wang S, Chen J, Pan Y, Li Q, Xu Xet al. 2018. G‐protein βγ subunits determine grain size through interaction with MADS‐domain transcription factors in rice. Nature Communications 9: 852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Fan J, Li J, Song Y, Li Q, Zhang Yu'e, Xue Y. 2014. SCFSLF‐mediated cytosolic degradation of S‐RNase is required for cross‐pollen compatibility in S‐RNase‐based self‐incompatibility in Petunia hybrida . Frontiers in Genetics 5: 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real‐time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Löffler A, Abel S, Jost W, Beintema JJ, Glund K. 1992. Phosphate‐regulated induction of intracellular ribonucleases in cultured tomato (Lycopersicon esculentum) cells. Plant Physiology 98: 1472–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma R, Han Z, Hu Z, Lin G, Gong X, Zhang H, Nasrallah JB, Chai J. 2016. Structural basis for specific self‐incompatibility response in Brassica . Cell Research 26: 1320–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIntosh GC, Bariola PA, Newbigin E, Green PJ. 2001. Characterization of Rny1, the Saccharomyces cerevisiae member of the T2 RNase family of RNases: unexpected functions for ancient enzymes? Proceedings of the National Academy of Sciences, USA 98: 1018–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure B. 2009. Darwin's foundation for investigating self‐incompatibility and the progress toward a physiological model for S‐RNase‐based SI. Journal of Experimental Botany 60: 1069–1081. [DOI] [PubMed] [Google Scholar]
- Mcclure B, Cruz‐García F, Romero C. 2011. Compatibility and incompatibility in S‐RNase‐based systems. Annals of Botany 108: 647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure BA, Haring V, Ebert PR, Anderson MA, Simpson RJ, Sakiyama F, Clarke AE. 1989. Style self‐incompatibility gene products of Nicotiana alata are ribonucleases. Nature 342: 955–957. [DOI] [PubMed] [Google Scholar]
- Meyers G, Saalmüller A, Büttner M. 1999. Mutations abrogating the RNase activity in glycoprotein Erns of the pestivirus classical swine fever virus lead to virus attenuation. Journal of Virology 73: 10224–10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nettancourt D. 2001. Incompatibility and incongruity in wild and cultivated plants. Berlin, Germany: Springer Verlag. [Google Scholar]
- Paez Valencia J, Goodman K, Otegui MS. 2016. Endocytosis and endosomal trafficking in plants. Annual Review of Plant Biology 67: 309–335. [DOI] [PubMed] [Google Scholar]
- Qiao H, Wang F, Zhao L, Zhou J, Lai Z, Zhang Y, Robbins TP, Xue Y. 2004a. The F‐box protein AhSLF‐S2 controls the pollen function of S‐RNase‐based self‐incompatibility. Plant Cell 16: 2307–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H, Wang H, Zhao L, Zhou J, Huang J, Zhang Y, Xue Y. 2004b. The F‐box protein AhSLF‐S2 physically interacts with S‐RNases that may be inhibited by the ubiquitin/26S proteasome pathway of protein degradation during compatible pollination in Antirrhinum . Plant Cell 16: 582–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu H, Guan Y, Wang Y, Zhang S. 2017. PLC‐mediated signaling pathway in pollen tubes regulates the gametophytic self‐incompatibility of Pyrus species. Frontiers in Plant Science 8: 1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanauskas K, Igić B. 2017. The evolutionary history of plant T2/S‐type ribonucleases. PeerJ 5: e3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins TP, Harbord RM, Sonneveld T, Clarke K. 2000. The molecular genetics of self‐incompatibility in Petunia hybrida . Annals of Botany 85: 105–112. [Google Scholar]
- Roiz L, Smirnoff P, Bar‐Eli M, Schwartz B, Shoseyov O. 2006. ACTIBIND, an actin‐binding fungal T2‐RNase with antiangiogenic and anticarcinogenic characteristics. Cancer 106: 2295–2308. [DOI] [PubMed] [Google Scholar]
- Sagar GDV, Gereben B, Callebaut I, Mornon J‐P, Zeöld A, da Silva WS, Luongo C, Dentice M, Tente SM, Freitas BCGet al. 2007. Ubiquitination‐induced conformational change within the deiodinase dimer is a switch regulating enzyme activity. Molecular and Cellular Biology 27: 4774–4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel MA, Chong YT, Haasen KE, Aldea‐Brydges MG, Stone SL, Goring DR. 2009. Cellular pathways regulating responses to compatible and self‐incompatible pollen in Brassica and Arabidopsis stigmas intersect at Exo70A1, a putative component of the exocyst complex. Plant Cell 21: 2655–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel MA, Mudgil Y, Salt JN, Delmas F, Ramachandran S, Chilelli A, Goring DR. 2008. Interactions between the S‐domain receptor kinases and AtPUB‐ARM E3 ubiquitin ligases suggest a conserved signaling pathway in Arabidopsis. Plant Physiology 147: 2084–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassa H, Nishio T, Kowyama Y, Hirano H, Koba T, Ikehashi H. 1996. Self‐incompatibility (S) alleles of the Rosaceae encode members of a distinct class of the T2/S ribonuclease superfamily. Molecular & General Genetics 250: 547–557. [DOI] [PubMed] [Google Scholar]
- Schopfer CR, Nasrallah ME, Nasrallah JB. 1999. The male determinant of self‐incompatibility in Brassica . Science 286: 1697–1700. [DOI] [PubMed] [Google Scholar]
- Sijacic P, Wang X, Skirpan AL, Wang Y, Dowd PE, McCubbin AG, Huang S, Kao T‐H. 2004. Identification of the pollen determinant of S‐RNase‐mediated self‐incompatibility. Nature 429: 302–305. [DOI] [PubMed] [Google Scholar]
- Sims TL, Ordanic M. 2001. Identification of a S‐ribonuclease‐binding protein in Petunia hybrida . Plant Molecular Biology 47: 771–783. [DOI] [PubMed] [Google Scholar]
- Sun L, Kao T‐H. 2018. CRISPR/Cas9‐mediated knockout of PiSSK1 reveals essential role of S‐locus F‐box protein‐containing SCF complexes in recognition of non‐self S‐RNases during cross‐compatible pollination in self‐incompatible Petunia inflata . Plant Reproduction 31: 129–143. [DOI] [PubMed] [Google Scholar]
- Suzuki G, Kai N, Hirose T, Fukui K, Nishio T, Takayama S, Isogai A, Watanabe M, Hinata K. 1999. Genomic organization of the S locus: identification and characterization of genes in SLG/SRK region of S9 haplotype of Brassica campestris (syn. rapa). Genetics 153: 391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasaki T, Hatakeyama K, Suzuki G, Watanabe M, Isogai A, Hinata K. 2000. The S receptor kinase determines self‐incompatibility in Brassica stigma. Nature 403: 913–916. [DOI] [PubMed] [Google Scholar]
- Takayama S, Isogai A. 2005. Self‐incompatibility in plants. Annual Review of Plant Biology 56: 467–489. [DOI] [PubMed] [Google Scholar]
- Takayama S, Shiba H, Iwano M, Shimosato H, Che F‐S, Kai N, Watanabe M, Suzuki G, Hinata K, Isogai A. 2000. The pollen determinant of self‐incompatibility in Brassica campestris . Proceedings of the National Academy of Sciences, USA 97: 1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SG, Franklin‐Tong VE. 2004. Self‐incompatibility triggers programmed cell death in Papaver pollen. Nature 429: 305–309. [DOI] [PubMed] [Google Scholar]
- Thomas SG, Huang S, Li S, Staiger CJ, Franklin‐Tong VE. 2006. Actin depolymerization is sufficient to induce programmed cell death in self‐incompatible pollen. The Journal of Cell Biology 174: 221–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DM, Parker R. 2009. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae . The Journal of Cell Biology 185: 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. 2000. Recognition of the polyubiquitin proteolytic signal. EMBO Journal 19: 94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres‐Rodriguez MD, Cruz‐Zamora Y, Juarez‐Diaz JA, Mooney B, McClure BA, Cruz‐Garcia F. 2020. NaTrxh is an essential protein for pollen rejection in Nicotiana by increasing S‐RNase activity. The Plant Journal 103: 1304–1317. [DOI] [PubMed] [Google Scholar]
- Udeshi ND, Mertins P, Svinkina T, Carr SA. 2013a. Large‐scale identification of ubiquitination sites by mass spectrometry. Nature Protocols 8: 1950–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udeshi ND, Svinkina T, Mertins P, Kuhn E, Mani DR, Qiao JW, Carr SA. 2013b. Refined preparation and use of anti‐diglycine remnant (K‐ε‐GG) antibody enables routine quantification of 10,000s of ubiquitination sites in single proteomics experiments. Molecular & Cellular Proteomics 12: 825–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushijima K, Sassa H, Dandekar AM, Gradziel TM, Tao R, Hirano H. 2003. Structural and transcriptional analysis of the self‐incompatibility locus of almond: identification of a pollen‐expressed F‐box gene with haplotype‐specific polymorphism. Plant Cell 15: 771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Herr RA, Hansen TH. 2012. Ubiquitination of substrates by esterification. Traffic 13: 19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler MJ, de Graaf BHJ, Hadjiosif N, Perry RM, Poulter NS, Osman K, Vatovec S, Harper A, Franklin FCH, Franklin‐Tong VE. 2009. Identification of the pollen self‐incompatibility determinant in Papaver rhoeas . Nature 459: 992–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederstein M, Sippl MJ. 2007. ProSA‐web: interactive web service for the recognition of errors in three‐dimensional structures of proteins. Nucleic Acids Research 35: W407–W410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins KA, Poulter NS, Franklin‐Tong VE. 2014. Taking one for the team: self‐recognition and cell suicide in pollen. Journal of Experimental Botany 65: 1331–1342. [DOI] [PubMed] [Google Scholar]
- Willard L, Ranjan A, Zhang H, Monzavi H, Boyko RF, Sykes BD, Wishart DS. 2003. VADAR: a web server for quantitative evaluation of protein structure quality. Nucleic Acids Research 31: 3316–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Li M, Wu J, Guo H, Li Q, Zhang YE, Chai J, Li T, Xue Y2013. Identification of a canonical SCFSLF complex involved in S‐RNase‐based self‐incompatibility of Pyrus (Rosaceae). Plant Molecular Biology 81: 245–257. [DOI] [PubMed] [Google Scholar]
- Xue Y, Carpenter R, Dickinson HG, Coen ES. 1996. Origin of allelic diversity in Antirrhinum S locus RNases. Plant Cell 8: 805–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. 2015. The I‐TASSER Suite: protein structure and function prediction. Nature Methods 12: 7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Meng D, Gu Z, Li W, Chen Q, Li Y, Yuan H, Yu J, Liu C, Li T. 2018. Apple S‐RNase interacts with an actin‐binding protein, MdMVG, to reduce pollen tube growth by inhibiting its actin‐severing activity at the early stage of self‐pollination induction. The Plant Journal 95: 41–56. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Vuković L, Rudack T, Han W, Schulten K. 2016. Recognition of poly‐ubiquitins by the proteasome through protein refolding guided by electrostatic and hydrophobic interactions. Journal of Physical Chemistry 120: 8137–8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhao Z, Xue Y. 2009. Roles of proteolysis in plant self‐incompatibility. Annual Review of Plant Biology 60: 21–42. [DOI] [PubMed] [Google Scholar]
- Zhao L, Huang J, Zhao Z, Li Q, Sims TL, Xue Y. 2010. The Skp1‐like protein SSK1 is required for cross‐pollen compatibility in S‐RNase‐based self‐incompatibility. The Plant Journal 62: 52–63. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Identification of six ubiquitinated residues of Petunia hybrida S3‐RNase by LC‐MS/MS analysis of cross‐pollinated pistils.
Fig. S2 Two ubiquitinated residues of Petunia hybrida S3‐RNase identified by LC‐MS/MS analysis of self‐pollinated and unpollinated pistils.
Fig. S3 Locations of six ubiquitinated residues of Petunia hybrida S3‐RNase in Solanaceous S‐RNases.
Fig. S4Petunia hybrida S3‐RNase with the mutated region (R) I displays largely unaltered biochemical and physical properties.
Fig. S5 Physical interactions between Petunia hybrida S3R/Mutant (M) and PhS3SLF1.
Fig. S6 Predicted three‐dimensional (3D) structures and surface electrostatic potentials of Petunia hybrida S3‐RNases with mutated ubiquitinated residues.
Fig. S7Petunia hybridaS3R and S3R (Mutant) (M) transgenes identification by PCR analysis.
Fig. S8 Identification of FLAG‐tagged Petunia hybrida S3R and S3R (Mutant) (M) transgenes by PCR analysis.
Fig. S9 Detection of FLAG‐tagged Petunia hybrida S3R and S3R (Mutant) (M) transgenes expression by immunoblots.
Fig. S10Petuniahybrida S3‐RNase with the mutated region (R) I slightly reduces cross seed sets.
Fig. S11 Decreased ubiquitination amount of mutant (M) I, II and III mediated by SCFS3LSLF1 of Petunia hybrida.
Fig. S12 Ubiquitination of Petunia hybrida S3R and its mutant forms by self‐ (PhS3S3 ) pollen‐tube extracts (PTE).
Fig. S13 Reduced seed set per capsule from T0 transgenic lines with mutated region (R) II of Petunia hybrida S3‐RNase.
Fig. S14 Reduced seed set per capsule from T0 transgenic lines with mutated lysine or threonine within the six ubiquitinated residues of Petunia hybrida S3‐RNase.
Fig. S15 Mutant (M) III largely maintains the biochemical and physical properties of Petunia hybrida S3‐RNase.
Fig. S16Petuniahybrida S3‐RNase with mutated region (R) III markedly reduces cross seed sets.
Fig. S17 Largely unaltered physicochemical properties of Petunia hybrida S3‐RNase with mutated region (R) I, II and III.
Fig. S18 Reduced seed set per capsule from T0 transgenic lines with mutated region (R) I/II/III of Petunia hybrida S3‐RNase.
Table S1 List of primer sequences.
Table S2 Names and accession numbers of S‐RNases used in this study.
Table S3 Seed sets of S3S3L /S3R and S3S3L /S3R (Mutant) (M) T0 transgenic plants of Petunia hybrida.
Table S4 Seed sets of S3S3L /S3R‐FLAG and S3S3L /S3R (Mutant) (M)‐FLAG T0 transgenic plants of Petunia hybrida.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Data Availability Statement
The data supporting the findings of this work are available from the corresponding author upon request.


