Abstract
Background
Prader–Willi syndrome is a rare genetic neurodevelopmental disorder caused by a paternal deficiency of maternally imprinted gene expression located in the chromosome 15q11–q13 region. Previous studies have demonstrated that several classes of neurodevelopmental disorders can be attributed to either over- or under-expression of specific genes that may lead to impairments in neuronal generation, differentiation, maturation and growth. Epigenetic changes that modify gene expression have been highlighted in these disorders. One recent study focused on epigenetic analysis and compared patients with PWS with patients with other imprinting disorders. No study, however, has yet focused on epigenetics in patients with PWS specifically by comparing the mutations associated with this syndrome.
Objective
This study investigated the epigenetic modifications in patients with PWS and patients with PWS-related disorders caused by inactivation of two genes of the PWS chromosomal region, SNORD116 and MAGEL2. Our approach also aimed to compare the epigenetic modifications in PWS and PWS-related disorders.
Methods
We compared genome-wide methylation analysis (GWAS) in seven blood samples from patients with PWS phenotype (five with deletions of the PWS locus, one with a microdeletion of SNORD116 and one with a frameshift mutation of MAGEL2 presenting with Schaaf–Yang syndrome), as well as two control patients. Controls were infants that had been studied for suspicion of genetic diseases that was not confirmed by the genetic analysis and the clinical follow-up.
Results
The analysis identified 29,234 differentially methylated cytosines, corresponding to 5,308 differentially methylated regions (DMRs), which matched with 2,280 genes. The DMRs in patients with PWS were associated with neurodevelopmental pathways, endocrine dysfunction and social and addictive processes consistent with the key features of the PWS phenotype. In addition, the separate analysis for the SNORD116 and MAGEL2 deletions revealed that the DMRs associated with the SNORD116 microdeletion were found in genes implicated in metabolic pathways and nervous system development, whereas MAGEL2 mutations mostly concerned genes involved in macromolecule biosynthesis.
Conclusion
The PWS is associated with epigenetic modifications with differences in SNORD116 and MAGEL2 mutations, which seem to be relevant to the different associated phenotypes.
Keywords: Neurodevelopmental disorder, Genome-wide methylation analysis, Prader–Willi, SNORD116, MAGEL2
Background
Prader–Willi syndrome (PWS) is a rare genetic neurodevelopmental disorder (NDD) caused by a paternal deficiency of maternally imprinted gene expression in the chromosome 15q11–q13 region [1]. This lack of gene expression occurs because of deletions of paternally inherited 15q11–q13 chromosomal region or because of the occurrence of maternal uniparental disomy (UPD). The 15q11.2–q13 region can be divided into distinct regions that are delineated by three common deletion breakpoints (BP), a proximal non-imprinted region between the two common proximal breakpoints (BP1 and BP2) containing four biparentally expressed genes, NIPA1, NIPA2, CYF1P1 and GCP5.94 and the “PWS paternal-only expressed region” between BP2 and BP3 containing five polypeptide coding genes (MKRN3, MAGEL2, NECDIN and the bicistronic SNURF-SNRPN); C15orf2; a cluster of C/D box small nucleolar RNA genes (snoRNAs); and several antisense transcripts (including the antisense transcript to UBE3A). These three breakpoints cause two classes of deletions commonly called type 1 or long deletion when they extend from BP1 to BP3 and type 2 or short deletion when they extend between BP2 and BP3. In childhood and adulthood, patients with PWS present severe obesity related to eating disorders and obsession for food, endocrine dysfunction (impaired sexual development and growth, central hypothyroidism, rare adrenal insufficiency) and intellectual disabilities. It is now acknowledged that an impaired development and function of the hypothalamus may explain the specific features of the PWS phenotype (art lancet endocrinol metab M Tauber 2021).
The PWS phenotype occurs with a specific nutritional trajectory from anorexia at birth to hyperphagia in children and adults. The literature on PWS is broad concerning the cognitive impairments, social deficits [2] and brain metabolism modifications [3] that occur during neurodevelopment, especially those involving hypothalamic dysfunction with structural abnormalities [4] and impaired hypothalamic brain connectivity [5]. In addition, this syndrome is associated with several psychiatric dimensions [6] that can be connected to NDDs such as the autism spectrum disorders (ASD) [7] and attention deficit hyperactivity disorder (ADHD) [8]. The role of epigenetics in NDDs has been highlighted, and some authors have suggested that these disorders be classified as “epigenetic” as they are at the interface between genetics and environmental risk factors [9].
Several studies have indicated that imprinted genes play important roles in the postnatal processes that may be particularly responsive to environmental influences [10–12]. Genomic imprinting is a form of epigenetic inheritance whereby the regulation of the imprinting-associated differentially methylated regions (iDMRs) is dependent on the sex of the transmitting parent [13]. However, DMRs can be identified in loci other than iDMRs. Indeed, a genome-wide methylation analysis (GWMA) study in Silver–Russell syndrome (SRS) patients showed that DMRs were found at the IGF2/H19 locus and that 116 DMRs were located on other chromosomes [14]. Moreover, a GWMA in patients with various imprinting disorders identified patients with multilocus imprinting disturbances (MLID) [15]. Given the potentially shared epigenetic regulation in imprinting disorders, Hara‑Isono et al. recently investigated the methylation signatures associated with the overlapping phenotypes of three imprinting disorders in SRS, temple syndrome (TS14) and PWS. However, no methylation signatures were found to be shared by these three syndromes [16]. We found that these imprinting disorders shared phenotypical similarities concerning growth, development and endocrine and metabolism dysfunctions but showed differences in their neurodevelopmental trajectories. Adult patients with PWS present intellectual disability, social impairment and emotional lability, whereas adult patients with TS14 classically present normal intellectual development and can expect to attend university [17]. We hypothesized that these phenotypical differences would partly explain the results of the authors.
Moreover, although the phenotypical characteristics may differ between syndromes, they may also vary within the same syndrome. Patients with PWS and PWS-related disorders present complete or partial PWS phenotypes depending on the type of mutation in the 15q11–q13 region. For example, a patient described with a SNORD116 microdeletion (MD) [18] displays a complete PWS phenotype and patients with MAGEL2 mutations present with Schaaf–Yang syndrome (SYS), which comprises such PWS features as an early phase of poor feeding, endocrine dysfunction and more severe ASD features.
Considering these data, we aimed to (i) specify the methylation signature in PWS and PWS-related syndromes by considering a group with different mutations of the 15q11–q13 region including SNORD116 MD and MAGEL2 mutation, (ii) associate the signatures with biological pathways and clinical features and (iii) specify the methylation signatures with the two mutations of SNORD116 MD and MAGEL2.
Results
Clinical features
The clinical features, including age and gender, and the genetic data of the patients are presented in Table 1. All patients displayed a complete or partial PWS phenotype. The PWS group included two infants, one child and four adults; two of the patients were female and five were male. The control group was composed of two infants, one female and one male.
Table 1.
Clinical features of the patients; NA: non-applicable
| Group | Age category | Gender | Genotype |
|---|---|---|---|
| PWS1 |
Infant (1 Year) |
Male | Deletion type1 |
| PWS2 |
Child (10 year) |
Male | Deletion type1 |
| PWS3 |
Adult (27 years) |
Female | Deletion type2 |
| PWS4 |
Adult (32 years) |
Male | Deletion type2 |
| PWS5 |
Infant (1 year) |
Male | Uniparental disomy |
| PWS6 |
Adult (32 years) |
Female | SNORD116 microdeletion |
| PWS7 |
Adult (21 years) |
Male | MAGEL2 mutation |
| Control |
Infant (1 year) |
Male | NA |
| Control |
Infant (1 year) |
Female | NA |
Distribution of the DMRs
We performed an analysis of the DMRs by RRBS approach which compared all the patients with PWS with controls. The analysis tested 1,971,050 cytosines and identified 29,234 differentially methylated cytosines, corresponding to 5,308 DMRs. These DMRs matched with 2,280 genes. Table 2 describes the top 50 of the hypomethylated genes and the top 50 of the hypermethylated genes.
Table 2.
Description of the top 100 of the gene with the higher methylation difference (50 hypermethylated genes and 50 hypomethylated genes)
| Genes | Methylation difference (%) | p value | |
|---|---|---|---|
| Top 50 of the hypomethylated genes | GGT6 | − 100 | 0.000514363 |
| GRK5 | − 100 | 0.000,635,702 | |
| KCNJ15 | − 100 | 0.000,170,544 | |
| LOC101927824 | − 100 | 5.8E−23 | |
| MAP1B | − 100 | 0.000,405,688 | |
| PHRF1 | − 100 | 0.000,635,702 | |
| RAB3GAP1 | − 100 | 0.000,621,357 | |
| ZNF592 | − 100 | 0.000,405,688 | |
| ITGA9− AS1 | − 97,44,493,917 | 0.0,000,013 | |
| LYST | − 97,44,493,917 | 0.0,000,013 | |
| APC2 | − 97,3,698,163 | 0.00,109,708 | |
| PSMD12 | − 97,3,698,163 | 0.00,109,708 | |
| LOC728743 | − 96,97,178,877 | 0.000,564,938 | |
| TTC39B | − 96,97,178,877 | 0.000,564,938 | |
| CXADR | − 96,68,781,868 | 0.0,000,176 | |
| TDRD10 | − 96,00,217,622 | 0.0,000,356 | |
| CCER2 | − 95,55,845,589 | 0.00,000,562 | |
| TBC1D22A | − 95,55,845,589 | 0.00,000,562 | |
| ZNF510 | − 92,27,324,274 | 0.000,437,373 | |
| ABCG4 | − 92,05,494,615 | 2.35883E−09 | |
| GJC1 | − 92,05,494,615 | 2.35883E−09 | |
| CYP27A1 | − 91,74,804,204 | 5.85E−08 | |
| WDR78 | − 91,36,703,468 | 0.000,000,003 | |
| LOC102723665 | − 90,51,175,406 | 0.000,568,276 | |
| SH3D19 | − 90,51,175,406 | 0.000,568,276 | |
| ACOT9 | − 89,67,481,584 | 2.12E−49 | |
| LINC00476 | − 89,67,481,584 | 2.12E−49 | |
| CACFD1 | − 87,25,081,098 | 0.00,000,197 | |
| MTCP1 | − 87,25,081,098 | 0.00,000,197 | |
| GOLGA7B | − 87,13,880,766 | 2.60667E−05 | |
| GSDMD | − 86,01,727,055 | 0.000,527,908 | |
| HUS1 | − 86,01,727,055 | 0.000,527,908 | |
| UBXN6 | − 85,98,645,224 | 0.000,102,859 | |
| CERCAM | − 85,61,137,856 | 0.000,358,764 | |
| DOCK11 | − 85,61,137,856 | 0.000,358,764 | |
| C3P1 | − 84,44,523,505 | 1.52E−09 | |
| TBCD | − 84,44,523,505 | 1.52E−09 | |
| C1QTNF9 | − 84,32,653,935 | 0.000,000,467 | |
| IARS1 | − 84,11,532,069 | 2.29667E−06 | |
| BSPRY | − 83,73,678,607 | 0.000,156,053 | |
| TFE3 | − 83,73,678,607 | 0.000,156,053 | |
| SMARCA2 | − 83,57,182,955 | 0.000,004,115 | |
| CCDC62 | − 83,11,684,461 | 9.48193E−09 | |
| FAM117B | − 83,11,684,461 | 9.48193E−09 | |
| DENND2B | − 83,11,642,437 | 0.000,501,791 | |
| ZNF462 | − 82,62,363,647 | 0.000,132,237 | |
| ARID2 | − 81,88,091,076 | 0.000,330,603 | |
| LRRTM1 | − 81,88,091,076 | 0.000,330,603 | |
| SPON1 | − 80,65,841,688 | 0.00,000,314 | |
| VPS4B | − 80,51,171,134 | 0.0,000,114 | |
| Top 50 of the genes hypomethylated | EFCAB2 | 77,73,232,951 | 0.001,120,107 |
| DGKD | 78,91,166,751 | 0.000,352,854 | |
| LOC283683 | 78,91,166,751 | 0.000,352,854 | |
| DLX2 | 81,76,085,867 | 0.000,213,868 | |
| CAMSAP3 | 84,37,216,271 | 0.000,563,998 | |
| SLC25A10 | 84,37,216,271 | 0.000,563,998 | |
| CACTIN-AS1 | 84,99,735,616 | 3.18E−08 | |
| RPS7 | 85,01,990,768 | 0.0,004,312 | |
| CCDC110 | 85,65,159,895 | 0.000,654,831 | |
| TMEM184B | 87,1,437,235 | 0.000,244,851 | |
| CYBC1 | 88,63,637,014 | 0.000,616,386 | |
| MUC16 | 88,63,637,014 | 0.000,616,386 | |
| MLIP | 88,69,043,414 | 0.000,146,656 | |
| DOCK10 | 90,28,688,782 | 0.0,000,496 | |
| MMP17 | 90,28,688,782 | 0.0,000,496 | |
| HIF3A | 90,90,923,198 | 0.0,000,676 | |
| SMOX | 90,90,923,198 | 0.0,000,676 | |
| GMCL1 | 94,26,290,015 | 0.00,000,577 | |
| ITPR2 | 94,26,290,015 | 0.00,000,577 | |
| LINC00205 | 98,14,808,839 | 1.0035E−06 | |
| SPON2 | 98,14,808,839 | 1.0035E−06 | |
| TARBP2 | 98,27,592,303 | 0.000,171,639 | |
| EMD | 99,84,144,366 | 0.000,046 | |
| BCOR | 100 | 0.000,443,279 | |
| CDC42BPA | 100 | 0.0,000,146 | |
| FAF1 | 100 | 0.000,133,428 | |
| FGD5 | 100 | 0.0,000,146 | |
| GALNT8 | 100 | 0.000,358,708 | |
| GSEC | 100 | 0.0,000,634 | |
| HMGXB3 | 100 | 0.000,204,717 | |
| INTS11 | 100 | 0.0,000,194 | |
| KMT2A | 100 | 0.000,063 | |
| MAN2A1 | 100 | 0.0,000,981 | |
| NFKBIL1 | 100 | 0.000,133,428 | |
| POMT2 | 100 | 0.0,000,159 | |
| PPM1B | 100 | 0.000,358,708 | |
| PTPRE | 100 | 0.000,725,654 | |
| RASA2 | 100 | 0.000,533,106 | |
| SDC3 | 100 | 0.0,000,566 | |
| SERPINB9P1 | 100 | 0.0,000,566 | |
| SH2B2 | 100 | 1.12E−19 | |
| SLC27A2 | 100 | 4.92E−21 | |
| TMEM222 | 100 | 1.09E−26 | |
| TMEM242 | 100 | 0.001,111,454 | |
| TMUB2 | 100 | 0.000,063 | |
| TNRC6A | 100 | 0.000,533,106 | |
| TTC23 | 100 | 0.0,000,981 | |
| TTLL6 | 100 | 0.0,000,634 | |
| UNC5A | 100 | 0.0,000,159 | |
| XPR1 | 100 | 0.001,111,454 |
The total distribution for the DMRs indicated that 34% were located in intergenic regions with the following repartition: 58% were located in transcription start sites (TSSs), which are regions that exert a great influence on gene expression regulation [19], and 42% were located in the transposable elements (TEs), which are the highly repetitive DNA sequences that constitute more than 50% of the human genome and contain about 52% of all CpG dinucleotides [20].
Sixty-six percent of the DMRs were located in intragenic regions, with a repartition of 47% in the exons and 53% in the introns. The following analysis in the intragenic regions was conducted in order to identify the associated biological pathways.
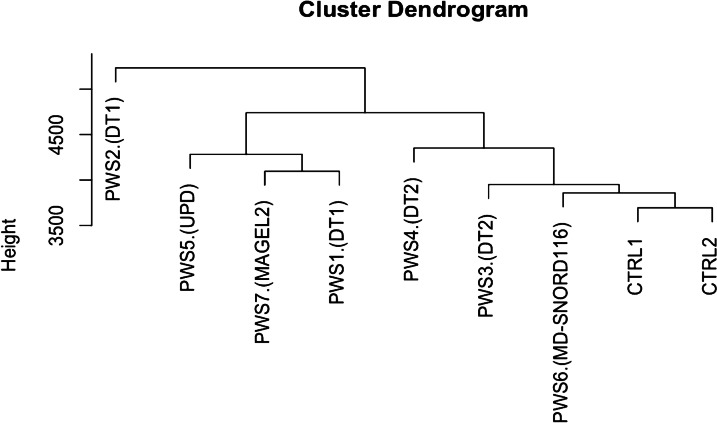
Hierarchical clustering analysis of the DMRs showed that the two controls grouped together. In this analysis, age did not play a notable role in the clustering. Moving from the control cluster, we found the patient with the SNORD116 MD and the two PWS patients with type 2 deletion (short deletion or DT2). Less close were the patients with type 1 deletion (long deletion or DT1), the patient with MAGEL2 mutation and a patient with uniparental disomy (UPD) (Fig. 1). One individual with DT1 (PWS2) was independently clustered.
Fig. 1.
Results of hierarchical clustering including the gender, the age and the DMRs; the dendrogram includes, from right to left: CTRL2: female infant control, CRTL1: male infant control, PWS6 (SNORD116 MD): female adult patient with SNORD116 deletion, PWS3: female adult patient with type 2 deletion, PWS4: male adult patient with type 2 deletion, PWS4: male adult patient with type2 deletion, PWS1: male infant patient with type 1 deletion, PWS7 (MAGEL2): male adult patient with MAGEL2 mutation, PWS5: male infant patient with uniparental disomy (UPD), PWS2: male child patient with type 1 deletion
Functional analyses of genes associated with DMRs and association with neurodevelopmental and nutritional trajectories
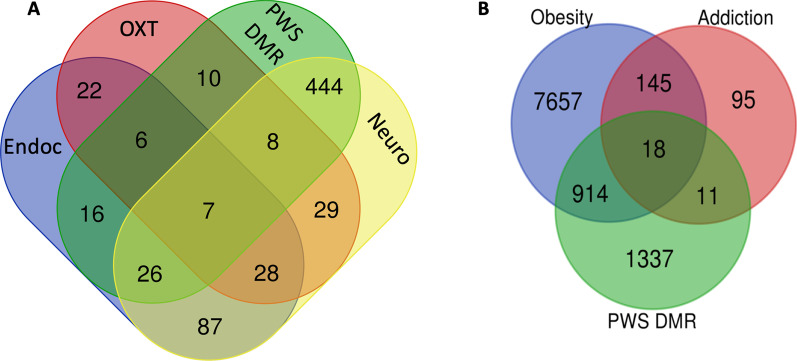
We performed a gene ontology functional pathways analysis that included the 2,280 genes corresponding to the DMRs. The most significant results for the functional pathways associated with the DMRs in PWS included biological processes and pathways related to nervous system development, generation of neurons and neurogenesis, anatomical structure development, synapses, aldosterone synthesis, Cushing syndrome, cortisol synthesis, cholinergic synapse, oxytocin signaling and endocrine resistance (Table 3). Four hundred and eighty-five (21%) of the differentially methylated genes corresponded to nervous system development. In addition, some genes involved in neurodevelopment overlapped with other systems related to the PWS phenotype (endocrine resistance and oxytocin pathway). Figure 2a illustrates this overlap.
Table 3.
Top biological processes and KEEG pathway connected to the PWS DMRs
| Biological process/KEGG pathway | GO/KEGG ID | Adjusted_p_value |
|---|---|---|
| Nervous system development | GO:0007399 | 1.23E−14 |
| Generation of neurons | GO:0048699 | 1.30E−13 |
| Neurogenesis | GO:0022008 | 1.71E−13 |
| Anatomical structure development | GO:0048856 | 1.16E−12 |
| Synapse | GO:0045202 | 1.51E−11 |
| Aldosterone synthesis and secretion | KEGG:04925 | 2.54E−03 |
| Cushing syndrome | KEGG:04934 | 5.01E−03 |
| Cortisol synthesis and secretion | KEGG:04927 | 9.20E−03 |
| Cholinergic synapse | KEGG:04725 | 1.00E−02 |
| Oxytocin signaling pathway | KEGG:04921 | 1.39E−02 |
| Endocrine resistance | KEGG:01522 | 1.54E−02 |
Fig. 2.
a Venn diagram for the PWS DMRs of the oxytocin pathway (OXT), nervous system development (Neuro), the endocrine pathway (Endoc) and the Prader–Willi differentially methylated regions (PWS DMRs). b Venn diagram of the genes implicated in addiction, obesity and the PWS DMRs
Regarding the eating disorders and the specific nutritional trajectory observed in PWS, we explored the connection between PWS DMRs and the genes related to addiction and obesity. The results revealed that 18 of the DMR genes were associated with addiction and obesity (ADCY3; ADCY9; ATF4; CDK5R1; CHRNB2; GABRD; GABRG3; GNB1; GNB3; GRK5; HDAC4; HDAC9; MAP2K1; PDE11A; PDE2A; PDE3A; PPP1CA; SLC6A3). The Venn diagram (Fig. 2b) represents this analysis.
Methylation status
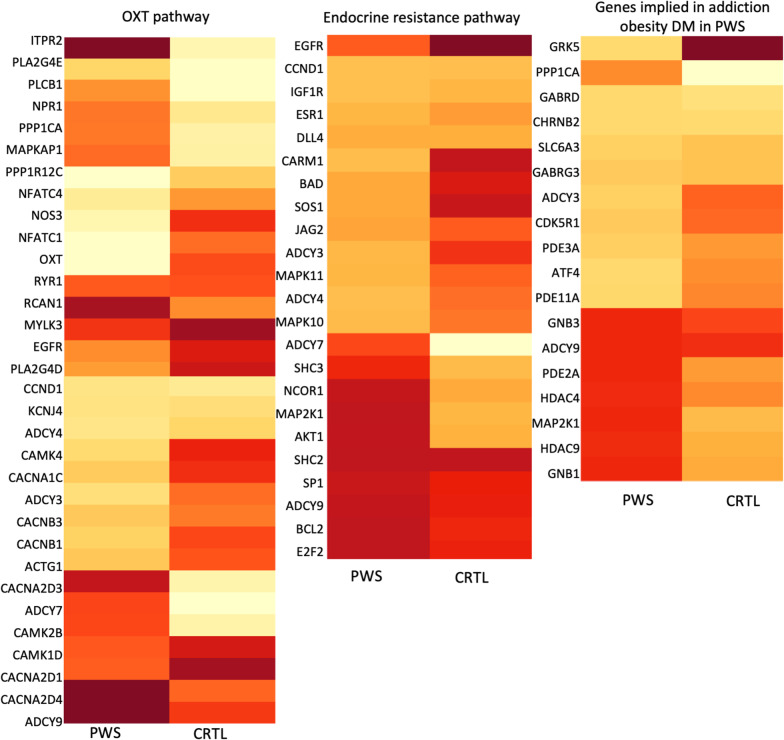
Twelve of the 32 genes involved in the OXT pathway were hypermethylated in PWS versus control, while the OXT gene was hypomethylated. Ten of the 23 genes involved in the endocrine resistance pathway were hypermethylated in PWS versus control. Eight of the 18 genes involved in obesity and addiction were hypermethylated in PWS versus control. Figure 3 details the methylation level for each gene.
Fig. 3.
Representation of the methylation level of the gene of interest. The darkness of the color indicates the level of methylation (pale colors indicate low level of methylation, while dark colors indicate high level of methylation)
SNORD116 and MAGEL2 mutations
The complete and partial PWS phenotypes were associated, respectively, with SNORD116 and MAGEL2 deficits.
The SNORD116 gene corresponds to the so-called minimal critical region that determines the PWS phenotype, and indeed, patients with SNORD116 MD display a phenotype very close to that of PWS [18]. The patient with the SNORD116 MD that we analyzed in this study was the one we previously reported [18]. In order to determine the methylated genes associated with this deletion, we compared the gene methylation in the SNORD116 patient (a young adult female) with the gene methylation of an infant female control patient. Cellular metabolic processes, nervous system development and metabolic processes were associated with the SNORD116 MD.
The patient with the MAGEL2 deletion analyzed in this study was a young adult male. Cellular macromolecule biosynthetic processes, macromolecule biosynthetic processes and organic substance biosynthetic processes were associated with MAGEL2 inactivation. We compared the gene methylation in the MAGEL2 patient (a young adult male) with the gene methylation of an infant male control patient.
The top significant results for MAGEL2 and SNORD116 are presented in Table 4.
Table 4.
Top biological processes and KEEG pathways connected to the MAGEL2 mutation and SNORD116 deletion
| MAGEL2 | Cellular macromolecule biosynthetic process | GO:0034645 | 2.83E−07 |
| Macromolecule biosynthetic process | GO:0009059 | 7.87E−07 | |
| Organic substance biosynthetic process | GO:1901576 | 3.08E−06 | |
| Biosynthetic process | GO:0009058 | 4.21E−06 | |
| Cellular biosynthetic process | GO:0044249 | 9.83E−06 | |
| Regulation of RNA metabolic process | GO:0051252 | 5.03E−05 | |
| Nucleobase-containing compound metabolic process | GO:0006139 | 6.72E−05 | |
| Nucleic acid metabolic process | GO:0090304 | 8.69E−05 | |
| SNORD116 | Cellular metabolic process | GO:0044237 | 8.54E−09 |
| Nervous system development | GO:0007399 | 9.56E−09 | |
| Metabolic process | GO:0008152 | 6.57E−08 | |
| Primary metabolic process | GO:0044238 | 3.31E−07 | |
| Nitrogen compound metabolic process | GO:0006807 | 5.41E−07 | |
| Nucleic acid metabolic process | GO:0090304 | 9.97E−07 | |
| Organic substance metabolic process | GO:0071704 | 1.15E−06 | |
| Central nervous system development | GO:0007417 | 1.34E−06 | |
| Hippo signaling pathway | KEGG:04390 | 6.29E−03 | |
| Chronic myeloid leukemia | KEGG:05220 | 1.25E−02 | |
| Neurotrophin signaling pathway | KEGG:04722 | 4.77E−02 |
Discussion
In this study, we found 5,308 DMRs that matched with 2280 genes in the PWS group and found differences between the analyzed genotypes. These genes are involved in nervous system development, which is relevant to the clinical definition of PWS as a genetic NDD, and are related to the main characteristics and comorbidities of the disease, such as eating disorders with increased fatness and obesity, behavioral disturbances and various comorbidities including endocrine dysfunction, an impaired oxytocin pathway with poor social abilities, addictive behaviors comprising food, smoking and alcohol addiction and dysautonomy. In addition, although poorly documented in the literature, increased sensitivity to drugs has been observed and may be related to these genes [21].
We found DMRs in genes involved in endocrine resistance/sensitivity. Interestingly, patients with PWS display endocrine deficits and are very sensitive to growth hormone (GH) treatment for unknown reasons. We demonstrated in a previous study that children with PWS are more sensitive to GH treatment and need lower doses [22]. Considering our findings, we suggest that this high sensitivity might be partly due to the methylation changes of this pathway. With regard to the endocrine dysfunctions associated with DMRs for the aldosterone pathway, a few cases of unexplained severe hyponatremia were reported in patients with PWS, although the levels of aldosterone and renin were normal [23]. For the cortisol pathway, several reports have hypothesized that PWS patients display central adrenal insufficiency under stressful conditions [24], and small-sized adrenal glands have been documented in autopsies from some cases [25]. However, two recent reports found no cases of central adrenal insufficiency [26, 27], and Ota et al. found that the cortisol response to insulin was normal in patients with PWS, with a peak response that was nevertheless delayed [28].
Notably, we found DMRs in genes involved in the OXT pathway. OXT is a neuropeptide that acts as a neuromodulator in social behavior and a circulating hormone that plays a major role in labor, birth and lactation maintenance. A social deficit has been described in PWS that resembles ASD and can be mild or severe [7]. A dysfunction in the OXT pathway has also been reported, including a hypothalamic OXT prohormone processing deficit and secretion dysfunction [29, 30] and a reduction in the number of hypothalamic OXT neurons [4] with increased plasma levels of OXT [31]. Moreover, we demonstrated in clinical studies that the altered social behaviors and poor sucking are improved in neonates with PWS by OXT treatment administered by the intranasal route [32, 33].
PWS is characterized by a well-described developmental trajectory in terms of nutritional aspects [6, 34]. Indeed, infants with PWS suffer from anorexia in the first so-called nutritional phase [35], with a subsequent shift to hyperphagia with a lack of satiety, leading to early severe obesity [36]. The hyperphagia that most older children and adults display is similar to addictive behavior for food [34]. We found an overlap between PWS DMRs and genes associated with addiction and obesity. Among them, the ADCY3 gene that encodes for adenylate cyclase 3, which plays an essential role in energy metabolism [37], was hypomethylated in patients with PWS. Interestingly, very recent studies linking novel ADCY3 variants to obesity and diabetes have been published [37], and ADCY3 gene mutations with loss of function have been identified in monogenic severe obesity [38]. The melanocortin 4 receptor gene, MC4R, is a key component of the melanocortin system, and its mutation is the most common monogenic cause of severe obesity. Interestingly, MC4R and ADCY3 were specifically colocalized in the primary cilia of a subset of hypothalamic paraventricular nucleus neurons [39]. Moreover, specific inhibition of ADCY3 in the primary cilia resulted in increased food intake and significant weight gain.
We found that SNORD116 MD DMRs are associated with the Hippo signaling pathway, which is associated with the metabolic processes related to chemical reactions and pathways, including anabolism and catabolism and adaptive thermogenesis. A previous study showed that Snord116-deleted mice housed at 22 °C exhibited low body weight, hyperphagia and changes in energy expenditure compared to wild type, and most of these modifications were rescued when the mice were housed at 30 °C [40]. Interestingly, patients with PWS display temperature regulation defects that are most often characterized by episodes of low temperature [40]. Animal models also support a role for the Hippo pathway in regulating adipose cell proliferation, differentiation and adipogenesis [41, 42]. Patients with PWS show unusual body composition and fatness patterns, characterized by reduced lean tissue and increased subcutaneous adiposity [43]. We also found that SNORD116 MD DMRs are associated with the neurotrophin signaling pathway. A lower BDNF level in plasma was found in patients with PWS, as well as lower BDNF transcription in human hypothalamus [44, 45]. Last, we found that SNORD116 MD DMRs were associated with the pathway of chronic myeloid leukemia (CML). One study reported an excess of CML in patients with PWS. In this study of 1160 patients, eight presented leukemia, a prevalence that was 40 times higher than expected. This suggests that the risk of myeloid leukemia may be increased in PWS [46].
Whereas the DMRs associated with SNORD116 MD were found in genes involved in neurodevelopmental and metabolic pathways, the specific inactivation of MAGEL2 mutation showed overlap with the genes involved in macromolecule biosynthesis. The truncating point mutations of the paternally inherited allele of MAGEL2 cause SYS, which has significant phenotypical overlap with PWS, particularly the initial nutritional phase and endocrine dysfunction. However, the developmental trajectory of SYS is clinically distinct, with a particularly high prevalence of ASD (up to 75% of affected individuals) [47], severe intellectual disability and a lower incidence of hyperphagia and obesity.
We acknowledge that this pilot study has several limitations. Notably, we used a small number of samples, especially for the control samples, which carried the risk of high variability between the individuals. The control samples were from infants as two of the PWS samples and this could limit the effects of age and environmental factors. However, the others samples were obtained from children or adults. Moreover, cluster analysis showed that the control patients clustered together and that age did not seem to play a major role in the clustering. Interestingly, the analysis revealed that the patients with DT1, UPD or MAGEL2 mutations were more distant from the controls. The DT1 mutation is associated with more severe clinical symptoms, with these patients scoring lower in adaptive behavior scores and showing poorer reading, math and visual-motor skills [48]. The patient with DT1 (PWS2) clustered apart and displayed not only severe relationship impairment with a need for psychiatric follow-up, but also presented orthopedic problems, with scoliosis and extreme fatigability requiring a wheelchair to move. Patients with UPD more frequently display severe social impairment, including ASD [49]. This suggests that the largest deletions might accentuate the epigenetic modifications observed in the shortest deletions.
Second, our analyses were conducted in blood samples, whereas epigenetic modulation may be tissue- or cell type-specific. Yet, access to specific human tissue, especially brain tissue, is complicated, and most studies focused on DMRs are currently conducted in blood samples.
Third, we chose the analysis of methylation located in the intragenic regions as a first step. However, we acknowledge that methylation occurs in intergenic regions such as the TSS and TE regions and that regions may play a role in epigenetic regulation and phenotype expression.
Last, the DMR analyses accounted for differences in hypermethylation or hypomethylation between patients and controls, therefore suggesting a difference in gene expression. We did not perform RNA sequencing in this study because RNA samples were unavailable. We nevertheless suspect that RNA sequencing would have brought relevant additional information on gene expression.
Despite several limitations, our preliminary results showed that the DMRs we described may be related to the complex phenotype of PWS.
Conclusion
These data suggest that genetic defects of the imprinted chromosomal region 15q11–q13 that lead to PWS are associated with epigenetic methylation signatures. Those epigenetic signatures are associated with pathways involved in brain development, endocrine function and metabolism. The SNORD116 MD and MAGEL2 mutations are also associated with specificities in DMRs that may explain at least partly the complex PWS phenotype. A question of utmost importance arises from these results concerning whether it would be possible to modify the methylation status caused by a lack of expression of SNORD116, MAGEL2 and perhaps other genes in the PWS region [50] with, for example, oxytocin treatment [33] or other drugs and/or social disability rehabilitation.
Methods
The 15q11–q13 deletions enabled the mapping of three main break sites, BP1 (for breakpoint 1) for the most centromeric, BP2, and BP3 for the most telomeric. These three break sites cause two classes of deletions commonly called type 1 when they extend from BP1 to BP3 and type 2 (the more frequent) when they are between BP2 and BP3 (the type 1 deletion, which is more extensive, therefore includes the type 2 deletion). Uniparental disomy (UPD) refers to the situation in which two copies of the 15q11–q13 region come from the mother.
We collected nine blood samples, seven of which were collected from patients with the PWS phenotype. Two patients with PWS carried a type 2 deletion, two patients a type 1 deletion, one patient a maternal UPD, one patient a microdeletion of a region encompassing SNORD116, IPW and SNORD109A as described by Bieth et al. 2015 [18], and one patient displaying SYS showed a de novo frameshift mutation c.2855delC of the paternal MAGEL2 gene. For control, we used DNA samples from infants that had been studied for suspicion of genetic diseases that was not confirmed by the genetic analysis and the clinical follow-up. Two blood samples were collected from control patients: a male infant and a female infant.
Ethics
Before the study for the genetic analysis, in accordance with French law, adult patients gave informed consent if possible or legal guardians in cases of intellectual disabilities, parents gave informed consent for their children. All data were anonymized without any possibility of returning to the patient data. The protocol was submitted to an ethics committee in agreement with the French Jardé law (agreement of the Comité de Protection des Personnes: CPP Sud Ouest et Outremer 1).
DNA extraction
The DNA was extracted from whole blood, and erythrocytes were lysed by a low salt buffer with Tris–HCl, KCl, MgCl2 and EDTA (TKM1 buffer). The samples were digested by proteinase K and precipitated in sodium acetate, then with phenol–chloroform–isoamyl alcohol (PCI), then with chloroform–isoamyl alcohol (CIA) and cold ethanol (99.5%). Last, a DNA pellet was dried in fresh air for 10 min and then suspended in 20 µL of ultra-pure water. DNA concentrations ranged from 70 µg/mL to 100 µg/mL. The samples were conserved in Tris–EDTA pH 8.0 buffer.
Before reduced representation bisulfite sequencing (RRBS) analysis, the 260/230 and 260/280 ratios were measured. In cases of low quality of sample purity, we proceeded to the material purification on AMPure XP beads (Beckman #A63881).
RRBS analysis
The samples were controlled and validated by microfluorometry with the Qubit High-Sensitivity Assay (Life Technologies #Q32851) and agarose gel 0.8% to monitor degradation. The required quality was 200 ng per sample and a minimal concentration of 4 ng/µl. We did not use any heparin in the sample collection as it interferes with bisulfite labeling. The samples were treated with DNAase-free, protease-free RNAse A (Life Technologies #EN0531).
The banks were constructed by enzymatic digestion of DNA by Mspl enzyme (CCGG) enriched in CpG islands. The digested DNA was repaired in extremities and adenylated in 3’ before being treated with bisulfite and amplified by PCR (13 to 15 cycles depending on the pool). The treatment by bisulfite was followed by a quality check. In our experiment, the rate of non-conversion ranged between 0.4% and 1.24%, indicating that the conversion stage was successful.
The clustering and sequencing steps were performed on a NovaSeq 6000 from Illumina using sequencing-by-synthesis (SBS) technology with NovaSeq Reagent Kits (100 cycles). Clusters were generated by denaturation and dilution of the banks. Then, hybridization and clonal expansion were performed on the flow cell (ID AHHMF2DRXX) using a dual indexing method. The thymine was marked with a green fluorophore and the cytosine with a red fluorophore. Image analysis was performed with NovaSeq Control software and base calling with RTA software, both from Illumina. This step serves to correct the intensity and transform it to a nucleotide base in order to obtain sequences.
The quality of the sequences was controlled with FastQC (v0.11.8). This analysis showed good quality with a probability of error of 2/10,000 (NovaSeq score = 37). Base calling reveals that no cycle was affected by base loss (N bases). The contaminant search was performed with the FastQ Screen software with Bowtie2, and this step revealed no contamination.
Bioinformatics analysis
The adaptor sequences were then trimmed with Trim Galore!, and the reads were aligned on the genome of reference [51] using Bismark software [52]. The data were extracted from the BAM files with the Bismark methylation extractor tool.
The statistical analysis was performed with the methylSig R package [53] based on a statistical method, with a beta binomial model then applied to calculate the DMRs encompassing a window of 25pb. The sites that were tested only corresponded to the CpG island. The q-value threshold (p-value after correction for the multiple test of Benjamini–Hochberg [54]) was 5%, and the threshold of methylation percentage difference was 25. The DMRs were annotated according to the nearest gene using the RefGene getnearestgene tool from CisGenome [55].
For the individual analysis, we considered as significant a methylation percentage difference of 25 with the control. This method was used for clustering analysis for the SNORD116 deletion and the MAGEL2 mutation analysis.
Hierarchical clustering and pathway analysis
We performed a hierarchical clustering including the gender, the age and the DMRs.
Gene ontology (GO) analysis was conducted with g:Profiler (http://biit.cs.ut.ee/gprofiler/) by following a g:GOSt–functional enrichment analysis. This currently covers KEGG [56], Reactome [57] and WikiPathways [58]; miRNA targets from miRTarBase [59] and regulatory motif matches from TRANSFAC [60]; tissue specificity based on expression data from the Human Protein Atlas [61]; data on protein complexes from CORUM [62]; and human disease phenotype associations from the Human Phenotype Ontology [63].
The list of genes associated with addiction was constituted by the addition of the list of genes collected from KEGG for cocaine addiction (KEGG:05,030), amphetamine addiction (KEGG:05,031), morphine addiction (KEGG:05,032), nicotine addiction (KEGG:05,033) and alcoholism (KEEG:05,034). The genes associated with obesity were extracted from GeneCards® https://www.genecards.org/Search/Keyword?queryString=obesity.
Acknowledgements
Not applicable.
Abbreviations
- ADHD
Attention deficit hyperactivity disorder
- ASD
Autism spectrum disorder
- ATP
Adenosine triphosphate
- cAMP
Cyclic adenosine monophosphate
- CML
Chronic myeloid leukemia
- CPP
Comité de Protection des Personnes
- DMR
Differentially methylated regions
- GABA
Gamma-aminobutyric acid
- GO
Gene ontology
- GWMA
Genome-wide methylation analysis
- IPSC
Induced pluripotent stem cells
- MLID
Multilocus imprinting disturbances
- NDD
Neurodevelopmental disorder
- OXT
Oxytocin
- PWS
Prader–Willi syndrome
- RRBS
Reduced representation bisulfite sequencing
- TE
Transposable elements
- TSS
Transcription start sites
Authors' contributions
JS analyzed the samples, EB collected the samples, VL and CM collected patient information, and JS, NF, JPS and MT wrote the article. All authors read and approved the final manuscript.
Funding
This work was supported by a grant from the French Association for Prader–Willi Syndrome (grant R15062BB).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
In accordance with French law, all patients gave informed consent before the study for genetic analysis. All data were anonymized without any possibility to return to the patient data. The protocol was submitted to an ethics committee in agreement with French law Jardé (agreement of the Comité de Protection des Personnes (CPP) Sud Ouest et Outremer 1).
Consent for publication.
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Goldstone AP, Holland AJ, Hauffa BP, Hokken-Koelega AC, Tauber M. Speakers contributors at the Second Expert Meeting of the Comprehensive Care of Patients with PWS. Recommendations for the diagnosis and management of Prader-Willi syndrome. J Clin Endocrinol Metab. 2008;93(11):4183–4197. doi: 10.1210/jc.2008-0649. [DOI] [PubMed] [Google Scholar]
- 2.Whittington J, Holland A. Cognition in people with Prader-Willi syndrome: Insights into genetic influences on cognitive and social development. Neurosci Biobehav Rev. 2017;72:153–167. doi: 10.1016/j.neubiorev.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 3.Mantoulan C, Payoux P, Diene G, Glattard M, Rogé B, Molinas C, et al. PET scan perfusion imaging in the Prader-Willi syndrome: new insights into the psychiatric and social disturbances. J Cereb Blood Flow Metab. 2011;31(1):275–282. doi: 10.1038/jcbfm.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swaab DF, Purba JS, Hofman MA. Alterations in the hypothalamic paraventricular nucleus and its oxytocin neurons (putative satiety cells) in Prader-Willi syndrome: a study of five cases. J Clin Endocrinol Metab. 1995;80(2):573–579. doi: 10.1210/jcem.80.2.7852523. [DOI] [PubMed] [Google Scholar]
- 5.Lukoshe A, Hokken-Koelega AC, van der Lugt A, White T. Reduced cortical complexity in children with Prader-Willi Syndrome and its association with cognitive impairment and developmental delay. PLoS ONE. 2014;9(9):e107320. doi: 10.1371/journal.pone.0107320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salles J, Lacassagne E, Benvegnu G, Berthoumieu SÇ, Franchitto N, Tauber M. The RDoC approach for translational psychiatry: Could a genetic disorder with psychiatric symptoms help fill the matrix? the example of Prader-Willi syndrome. Transl Psychiatry. 2020;10(1):274. doi: 10.1038/s41398-020-00964-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bennett JA, Hodgetts S, Mackenzie ML, Haqq AM, Zwaigenbaum L. Investigating autism-related symptoms in children with Prader-Willi syndrome: a case study. Int J Mol Sci. 2017;18:3. doi: 10.3390/ijms18030517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogata H, Ihara H, Murakami N, Gito M, Kido Y, Nagai T. Autism spectrum disorders and hyperactive/impulsive behaviors in Japanese patients with Prader-Willi syndrome: a comparison between maternal uniparental disomy and deletion cases. Am J Med Genet A. 2014;164A(9):2180–2186. doi: 10.1002/ajmg.a.36615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Millan MJ. An epigenetic framework for neurodevelopmental disorders: from pathogenesis to potential therapy. Neuropharmacology. 2013;68:2–82. doi: 10.1016/j.neuropharm.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 10.Plagge A, Gordon E, Dean W, Boiani R, Cinti S, Peters J, et al. The imprinted signaling protein XL alpha s is required for postnatal adaptation to feeding. Nat Genet. 2004;36(8):818–826. doi: 10.1038/ng1397. [DOI] [PubMed] [Google Scholar]
- 11.Garfield AS, Cowley M, Smith FM, Moorwood K, Stewart-Cox JE, Gilroy K, et al. Distinct physiological and behavioural functions for parental alleles of imprinted Grb10. Nature. 2011;469(7331):534–538. doi: 10.1038/nature09651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen M, Wang J, Dickerson KE, Kelleher J, Xie T, Gupta D, et al. Central nervous system imprinting of the G protein G(s)alpha and its role in metabolic regulation. Cell Metab. 2009;9(6):548–555. doi: 10.1016/j.cmet.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferguson-Smith AC. Genomic imprinting: the emergence of an epigenetic paradigm. Nat Rev Genet. 2011;12(8):565–575. doi: 10.1038/nrg3032. [DOI] [PubMed] [Google Scholar]
- 14.Wu D, Gong C, Su C. Genome-wide analysis of differential DNA methylation in Silver-Russell syndrome. Sci China Life Sci. 2017;60(7):692–699. doi: 10.1007/s11427-017-9079-7. [DOI] [PubMed] [Google Scholar]
- 15.Court F, Martin-Trujillo A, Romanelli V, Garin I, Iglesias-Platas I, Salafsky I, et al. Genome-wide allelic methylation analysis reveals disease-specific susceptibility to multiple methylation defects in imprinting syndromes. Hum Mutat. 2013;34(4):595–602. doi: 10.1002/humu.22276. [DOI] [PubMed] [Google Scholar]
- 16.Hara-Isono K, Matsubara K, Fuke T, Yamazawa K, Satou K, Murakami N, et al. Genome-wide methylation analysis in Silver-Russell syndrome, Temple syndrome, and Prader-Willi syndrome. Clin Epigenetics. 2020;12(1):159. doi: 10.1186/s13148-020-00949-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Temple IK, Cockwell A, Hassold T, Pettay D, Jacobs P. Maternal uniparental disomy for chromosome 14. J Med Genet. 1991;28(8):511–514. doi: 10.1136/jmg.28.8.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bieth E, Eddiry S, Gaston V, Lorenzini F, Buffet A, Conte Auriol F, et al. Highly restricted deletion of the SNORD116 region is implicated in Prader-Willi Syndrome. Eur J Hum Genet EJHG. 2015;23(2):252–255. doi: 10.1038/ejhg.2014.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zemach A, Kim MY, Silva P, Rodrigues JA, Dotson B, Brooks MD, et al. Local DNA hypomethylation activates genes in rice endosperm. Proc Natl Acad Sci USA. 2010;107(43):18729–18734. doi: 10.1073/pnas.1009695107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su J, Shao X, Liu H, Liu S, Wu Q, Zhang Y. Genome-wide dynamic changes of DNA methylation of repetitive elements in human embryonic stem cells and fetal fibroblasts. Genomics. 2012;99(1):10–17. doi: 10.1016/j.ygeno.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 21.Bonnot O, Cohen D, Thuilleaux D, Consoli A, Cabal S, Tauber M. Psychotropic treatments in Prader-Willi syndrome: a critical review of published literature. Eur J Pediatr. 2016;175(1):9–18. doi: 10.1007/s00431-015-2670-x. [DOI] [PubMed] [Google Scholar]
- 22.Feigerlová E, Diene G, Conte-Auriol F, Molinas C, Gennero I, Salles J-P, et al. Hyperghrelinemia precedes obesity in Prader-Willi syndrome. J Clin Endocrinol Metab. 2008;93(7):2800–2805. doi: 10.1210/jc.2007-2138. [DOI] [PubMed] [Google Scholar]
- 23.Landau D, Hirsch HJ, Gross-Tsur V. Case report: severe asymptomatic hyponatremia in Prader-Willi Syndrome. BMC Pediatr. 2016;18(16):28. doi: 10.1186/s12887-016-0563-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Lind van Wijngaarden RFA, Otten BJ, Festen DAM, Joosten KFM, de Jong FH, Sweep FCGJ,, et al. High prevalence of central adrenal insufficiency in patients with Prader-Willi syndrome. J Clin Endocrinol Metab. 2008;93(5):1649–1654. doi: 10.1210/jc.2007-2294. [DOI] [PubMed] [Google Scholar]
- 25.Stevenson DA, Anaya TM, Clayton-Smith J, Hall BD, Van Allen MI, Zori RT, Zackai EH, Frank G, Clericuzio CL. Unexpected death and critical illness in Prader-Willi syndrome: report of ten individuals. Am J Med Genet A. 200415;124A(2):158–64. 10.1002/ajmg.a.20370. PMID: 14699614. [DOI] [PubMed]
- 26.Farholt S, Sode-Carlsen R, Christiansen JS, Østergaard JR, Høybye C. Normal cortisol response to high-dose synacthen and insulin tolerance test in children and adults with Prader-Willi syndrome. J Clin Endocrinol Metab. 2011;96(1):E173–180. doi: 10.1210/jc.2010-0782. [DOI] [PubMed] [Google Scholar]
- 27.Nyunt O, Cotterill AM, Archbold SM, Wu JY, Leong GM, Verge CF, et al. Normal cortisol response on low-dose synacthen (1 microg) test in children with Prader Willi syndrome. J Clin Endocrinol Metab. 2010;95(12):E464–467. doi: 10.1210/jc.2010-0647. [DOI] [PubMed] [Google Scholar]
- 28.Oto Y, Matsubara K, Ayabe T, Shiraishi M, Murakami N, Ihara H, et al. Delayed peak response of cortisol to insulin tolerance test in patients with Prader-Willi syndrome. Am J Med Genet A. 2018;176(6):1369–1374. doi: 10.1002/ajmg.a.38713. [DOI] [PubMed] [Google Scholar]
- 29.Schaller F, Watrin F, Sturny R, Massacrier A, Szepetowski P, Muscatelli F. A single postnatal injection of oxytocin rescues the lethal feeding behaviour in mouse newborns deficient for the imprinted Magel2 gene. Hum Mol Genet. 2010;19(24):4895–4905. doi: 10.1093/hmg/ddq424. [DOI] [PubMed] [Google Scholar]
- 30.Ates T, Oncul M, Dilsiz P, Topcu IC, Civas CC, Alp MI, et al. Inactivation of Magel2 suppresses oxytocin neurons through synaptic excitation-inhibition imbalance. Neurobiol Dis. 2019;121:58–64. doi: 10.1016/j.nbd.2018.09.017. [DOI] [PubMed] [Google Scholar]
- 31.Johnson L, Manzardo AM, Miller JL, Driscoll DJ, Butler MG. Elevated plasma oxytocin levels in children with Prader-Willi syndrome compared with healthy unrelated siblings. Am J Med Genet A. 2016;170(3):594–601. doi: 10.1002/ajmg.a.37488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tauber M, Boulanouar K, Diene G, Çabal-Berthoumieu S, Ehlinger V, Fichaux-Bourin P, et al. The use of oxytocin to improve feeding and social skills in infants with Prader-Willi syndrome. Pediatrics. 2017;139:2. doi: 10.1542/peds.2016-2976. [DOI] [PubMed] [Google Scholar]
- 33.Tauber M, Mantoulan C, Copet P, Jauregui J, Demeer G, Diene G, et al. Oxytocin may be useful to increase trust in others and decrease disruptive behaviours in patients with Prader-Willi syndrome: a randomised placebo-controlled trial in 24 patients. Orphanet J Rare Dis. 2011;24(6):47. doi: 10.1186/1750-1172-6-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tauber M, Diene G, Mimoun E, Çabal-Berthoumieu S, Mantoulan C, Molinas C, et al. Prader-Willi syndrome as a model of human hyperphagia. Front Horm Res. 2014;42:93–106. doi: 10.1159/000358317. [DOI] [PubMed] [Google Scholar]
- 35.Beauloye V, Diene G, Kuppens R, Zech F, Winandy C, Molinas C, et al. High unacylated ghrelin levels support the concept of anorexia in infants with prader-willi syndrome. Orphanet J Rare Dis. 2016;11(1):56. doi: 10.1186/s13023-016-0440-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salles J, Lacassagne E, Eddiry S, Franchitto N, Salles J-P, Tauber M. What can we learn from PWS and SNORD116 genes about the pathophysiology of addictive disorders? Mol Psychiatry. 2020;26:51. doi: 10.1038/s41380-020-00917-x. [DOI] [PubMed] [Google Scholar]
- 37.Tian Y, Peng B, Fu X. New ADCY3 variants dance in obesity etiology. Trends Endocrinol Metab TEM. 2018;29(6):361–363. doi: 10.1016/j.tem.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 38.Saeed S, Bonnefond A, Tamanini F, Mirza MU, Manzoor J, Janjua QM, et al. Loss-of-function mutations in ADCY3 cause monogenic severe obesity. Nat Genet. 2018;50(2):175–179. doi: 10.1038/s41588-017-0023-6. [DOI] [PubMed] [Google Scholar]
- 39.Siljee JE, Wang Y, Bernard AA, Ersoy BA, Zhang S, Marley A, et al. Subcellular localization of MC4R with ADCY3 at neuronal primary cilia underlies a common pathway for genetic predisposition to obesity. Nat Genet. 2018;50(2):180–185. doi: 10.1038/s41588-017-0020-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qi Y, Purtell L, Fu M, Sengmany K, Loh K, Zhang L, et al. Ambient temperature modulates the effects of the Prader-Willi syndrome candidate gene Snord116 on energy homeostasis. Neuropeptides. 2017;61:87–93. doi: 10.1016/j.npep.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 41.An Y, Kang Q, Zhao Y, Hu X, Li N. Lats2 modulates adipocyte proliferation and differentiation via hippo signaling. PLoS One. 2013;8(8):e72042. 10.1371/journal.pone.0072042. PMID: 23977200; PMCID: PMC3745423. [DOI] [PMC free article] [PubMed]
- 42.Huang H, Wu W, Zhang L, Liu XY. Drosophila ste-20 family protein kinase, hippo, modulates fat cell proliferation. PLoS One. 2013;8(4):e61740. 10.1371/journal.pone.0061740. PMID: 23637896; PMCID: PMC3630116. [DOI] [PMC free article] [PubMed]
- 43.Theodoro MF, Talebizadeh Z, Butler MG. Body composition and fatness patterns in Prader-Willi syndrome: comparison with simple obesity. Obes Silver Spring Md. 2006;14(10):1685–1690. doi: 10.1038/oby.2006.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han JC, Muehlbauer MJ, Cui HN, Newgard CB, Haqq AM. Lower brain-derived neurotrophic factor in patients with Prader-Willi syndrome compared to obese and lean control subjects. J Clin Endocrinol Metab. 2010;95(7):3532–3536. doi: 10.1210/jc.2010-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bochukova EG, Lawler K, Croizier S, Keogh JM, Patel N, Strohbehn G, et al. A transcriptomic signature of the hypothalamic response to fasting and BDNF deficiency in Prader-Willi syndrome. Cell Rep. 2018;22(13):3401–3408. doi: 10.1016/j.celrep.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davies HD, Leusink GL, McConnell A, Deyell M, Cassidy SB, Fick GH, et al. Myeloid leukemia in Prader-Willi syndrome. J Pediatr. 2003;142(2):174–178. doi: 10.1067/mpd.2003.81. [DOI] [PubMed] [Google Scholar]
- 47.Fountain MD, Schaaf CP. Prader-Willi syndrome and Schaaf-Yang syndrome: neurodevelopmental diseases intersecting at the MAGEL2 gene. Dis Basel Switz. 2016;4:1. doi: 10.3390/diseases4010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Butler MG, Bittel DC, Kibiryeva N, Talebizadeh Z, Thompson T. Behavioral differences among subjects with Prader-Willi syndrome and type I or type II deletion and maternal disomy. Pediatrics. 2004;113(3 Pt 1):565–573. doi: 10.1542/peds.113.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Veltman MWM, Craig EE, Bolton PF. Autism spectrum disorders in Prader-Willi and Angelman syndromes: a systematic review. Psychiatr Genet. 2005;15(4):243–254. doi: 10.1097/00041444-200512000-00006. [DOI] [PubMed] [Google Scholar]
- 50.Jones C, Barrera I, Brothers S, Ring R, Wahlestedt C. Oxytocin and social functioning. Dialogues Clin Neurosci. 2017;19(2):193–201. doi: 10.31887/DCNS.2017.19.2/cjones. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Genome Reference Consortium. Genome Reference Consortium Human Build 37 patch release 13 (GRCh37. p13). NCBI. https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.25/.
- 52.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27(11):1571–2. 10.1093/bioinformatics/btr167. Epub 2011 Apr 14. PMID: 21493656; PMCID: PMC3102221. [DOI] [PMC free article] [PubMed]
- 53.Park Y, Figueroa ME, Rozek LS, Sartor MA. MethylSig: a whole genome DNA methylation analysis pipeline. Bioinformatics. 2014;30(17):2414–22. 10.1093/bioinformatics/btu339. [DOI] [PMC free article] [PubMed]
- 54.Benjamini. Benjamini, Y., & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc: Ser B (Methodol) 1995;57(1):289–300. [Google Scholar]
- 55.Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol. 2008;26(11):1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanehisa M, Sato Y, Furumichi M, Morishima K, Tanabe M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019;47(D1):D590–D595. doi: 10.1093/nar/gky962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2018;46(D1):D649–D655. doi: 10.1093/nar/gkx1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Slenter DN, Kutmon M, Hanspers K, Riutta A, Windsor J, Nunes N, et al. WikiPathways: a multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res. 2018;46(D1):D661–D667. doi: 10.1093/nar/gkx1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chou C-H, Shrestha S, Yang C-D, Chang N-W, Lin Y-L, Liao K-W, et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46 (Database issue):D296–302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34(Database issue):D108–110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 62.Giurgiu M, Reinhard J, Brauner B, Dunger-Kaltenbach I, Fobo G, Frishman G, et al. CORUM: the comprehensive resource of mammalian protein complexes-2019. Nucleic Acids Res. 2019;47(D1):D559–D563. doi: 10.1093/nar/gky973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Köhler S, Carmody L, Vasilevsky N, Jacobsen JOB, Danis D, Gourdine J-P, et al. Expansion of the human phenotype ontology (HPO) knowledge base and resources. Nucleic Acids Res. 2019;47(D1):D1018–D1027. doi: 10.1093/nar/gky1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.