Abstract
Acetic acid stress represents a frequent challenge to counteract for yeast cells under several environmental conditions and industrial bioprocesses. The molecular mechanisms underlying its response have been mostly elucidated in the budding yeast Saccharomyces cerevisiae, where acetic acid can be either a physiological substrate or a stressor. This review will focus on acetic acid stress and its response in the context of cellular transport, pH homeostasis, metabolism and stress‐signalling pathways. This information has been integrated with the results obtained by multi‐omics, synthetic biology and metabolic engineering approaches aimed to identify major cellular players involved in acetic acid tolerance. In the production of biofuels and renewable chemicals from lignocellulosic biomass, the improvement of acetic acid tolerance is a key factor. In this view, how this knowledge could be used to contribute to the development and competitiveness of yeast cell factories for sustainable applications will be also discussed.
Keywords: acetic acid stress, cell factory, industrial biotechnology, Saccharomyces cerevisiae , signalling, yeast
Take Away
Acetic acid stress is a frequent challenge for budding yeast.
Signalling pathways dissection and system‐wide approaches reveal a complex picture.
Cell fitness and adaptation under acid stress conditions is environment dependent.
Tolerance to acetic acid is a key factor in yeast‐based industrial biotechnology.
There is no ‘magic bullet’: An integrated approach is advantageous to develop performing yeast cell factories.
Multiple factors influenced acetic acid stress and tolerance in the budding yeast Saccharomyces cerevisiae.
1. INTRODUCTION
Acetic acid stress represents a frequent challenge to counteract for yeast cells under several environmental conditions and industrial bioprocesses. In the budding yeast Saccharomyces cerevisiae, acetic acid is a normal co‐product of alcoholic fermentation; thus, cells under physiological conditions do not normally sense this compound as toxic and can use acetate as a regular carbon source by channelling it into respiratory metabolism. On the other hand, extracellular acetic acid can be an environmental challenge and trigger adaptive responses at sublethal concentrations or alternatively be toxic and leading to cell death. In a physiological scenario where acetic acid derives from cellular metabolism, its accumulation occurs in parallel with glucose consumption and became maximal in stationary‐phase yeast cells. In a stress‐promoting context, the final toxicity for the cell is a combined function of extracellular acetic acid concentration, extracellular pH and consequent intracellular accumulation due to its uptake (Casal et al., 1996). The mechanisms underlying acetic acid stress response have been mostly clarified in laboratory yeast strains. Results may differ depending on experimental settings, including strains used, growth conditions (pH of the medium and composition), concentration of acetic acid and time of exposure. This review will highlight the knowledge concerning acetic acid stress response in budding yeast focusing on its molecular mechanisms involving cellular transport, pH homeostasis, metabolism and stress‐signalling pathways. This knowledge represents a precious resource for industrial biotechnology, where the improvement of acetic acid tolerance affected by other inhibitors like furfural and hydroxymethylfurfural is a key factor for fermentation processes using lignocellulosic raw materials as substrates. In this view, how this knowledge could be used to contribute to the development of cell factories for sustainable applications will be also discussed.
2. MOLECULAR MECHANISMS OF ACETIC ACID STRESS RESPONSE
Cellular transport, pH homeostasis, metabolism and stress‐signalling pathways represent overall the main factors affecting yeast response to acetic acid stress: how much extracellular acetic acid is transported into the cell, how much acetic acid will enter the metabolism and which is the threshold level of acetic acid tolerance? Answers to these questions will provide the final concentration of acetic acid in the cell and explain the activation of tolerance or toxicity mechanisms.
2.1. Cellular transport
Transport of acetic acid inside the cells has to be considered within the environmental context, in terms of extracellular pH and specific nutrient‐related conditions, such as the carbon source and medium composition, minimal or rich. In addition, acetic acid transport processes are also dependent on cellular growth phase. In glucose‐repressed cells, when the extracellular pH is below acetic acid pKa (4.76), it exists mainly in the undissociated form, which can freely diffuse through the plasma membrane with a rate that depends on the membrane solubility of the molecule (Casal et al., 1996). A passive diffusion facilitated by the aquaglyceroporin Fps1 has also been proposed for acetic acid by Mollapour and Piper, although this might be condition dependent (Mollapour & Piper, 2007). When the extracellular pH is higher than 4.76, acetic acid is present mainly as acetate anions, entering the cells through the two main electroneutral proton symporter, Jen1 and Ady2 (Casal et al., 2016). Both Jen1 and Ady2 transporters have also been suggested to be involved in the export of monocarboxylates, yet it is unclear how these efflux processes are regulated (Alves et al., 2020, and references therein).
Biophysical and chemical properties of the plasma membrane can also affect cell permeability to acetic acid, which itself can cause modifications in the plasma membrane composition. The amount of acetic acid entering the cells and the rate of its diffusion are partially related either to the structure of the plasma membrane, particularly to lipid composition and cell wall assembly (Berterame et al., 2016; Lindberg et al., 2013; Mira, Palma, et al., 2010; Palma et al., 2018) or to the partitioning of selected compounds, such as ethanol and n‐butanol, into the plasma membrane (Lindhal et al., 2017). At this regard, the ABC transporter Pdr18 has been reported to play a role in ergosterol homeostasis contributing to counteract acetic acid‐induced changes in lipid composition and permeability (Godinho et al., 2018).
Once inside the cells, acetic acid in the more alkaline environment of the cytoplasm dissociates into acetate ions and protons, causing acetate accumulation and intracellular acidification. This can lead to both inhibition of growth and metabolic activity, such as glucose consumption, in addition to other deleterious effects, including oxidative damage and energy depletion, depending on acetic acid concentration, intracellular acidification and glucose availability (Mira, Palma, et al., 2010; Pampulha & Loureiro‐Dias, 1989). A detailed model for the dynamic dependence of biomass yield and growth rate on pH conditions, acetic acid and extracellular glucose concentrations has been provided by Kitanovic et al., indicating that intracellular acidification due to accumulation of dissociated acetic acid in the cytosol is required for acetic acid toxicity, which creates a state of energy deficiency and nutrient starvation (Kitanovic et al., 2012). This study points the attention on the strict relationship between acetic acid tolerance, environmental context and cell growth phase. Increased acetic acid resistance has been observed in exponential cells treated with the same concentration of acetic acid but grown in glucose limiting or de‐repressing conditions (Guaragnella et al., 2013). Accordingly, yeast cells in diauxic shift after glucose exhaustion or in stationary‐phase, are much more tolerant to acetic acid at low pH compared to exponential cells (Ludovico et al., 2002).
2.2. pH homeostasis
Intracellular acidification due to accumulation of dissociated acetic acid in the cytosol can be counteracted by yeast cells through the activation of the plasma membrane H+‐ATPase (Pma1), which pumps protons out of the cell, and the vacuolar H+‐ATPase, which pumps protons into the lumen of the vacuole (Carmelo et al., 1997; Martínez‐Muñoz & Kane, 2008). Proton pumping is a high energy‐demanding process, consuming up to 20% of the cellular ATP produced in actively growing cells in the presence of glucose (Morsomme et al., 2000). Intracellular pH recovery due to Pma1p activity is a key determinant of acetic acid resistance, for which intracellular acidification is the major cause of growth inhibition (Ullah et al., 2012). This is also confirmed by the enhanced tolerance to acetic acid in PMA1‐overexpressing strains (Lee et al., 2017). Differently from other weak organic acids, acetate accumulation is not very toxic even at high concentrations and PDR12, responsible for acetate anions efflux, is not involved in acetic acid resistance (Ullah et al., 2012). This tolerance to anion accumulation can be explained by yeast capacity to produce and metabolise acetate at relatively high concentrations (Gancedo 1992). Also, VMA3, one of the major V‐ATPase assembly genes, contributes to acetic acid stress resistance by counteracting intracellular acidification and activating PKA signalling and glycolysis in the presence of glucose (Konarzewska et al., 2017).
2.3. Metabolism and stress‐signalling pathways
The threshold between tolerance and toxicity defines cell capacity to adapt or succumb to an environmental stress. At this regard, it is important to consider both mechanisms of tolerance (physiological state) and mechanisms of toxicity (pathological state) for a complete overview on acetic acid stress response. For acetic acid tolerance, a comprehensive picture focused on omics approaches is described in the work by Geng et al. (2017, and references therein). Omics studies revealed that acetic acid tolerance is controlled by multiple genes; the network of interaction among such genes is highly complex, and its elucidation is challenging even in a relatively simple model organism such as S. cerevisiae. Transcriptomic analysis identified Haa1p as the main player controlling yeast tolerance to acetic acid (Mira, Becker, & Sá‐Correia, 2010). On the other hand, the involvement in acetic acid tolerance of many genes related to carbohydrate metabolism, protein folding, lipid metabolism, cell wall function and transport has been elucidated by functional genomics screening and genome wide analysis (Mira, Palma, et al., 2010). Omics approaches could provide the theoretical basis and define genes for genetic modification in yeast, but the combination of methods able to capture complexity is necessary to obtain a clear picture of the cell‐wide acetic acid response in yeast. Therefore, high‐throughput techniques and systems metabolic analysis and advance genome scale modelling, also including regulation at certain levels, are needed for a deeper understanding of regulatory networks and, above all, their successful harnessing for cell factory development.
Most of the knowledge on the intracellular signalling of acetic acid toxicity comes from studies performed under extreme conditions affecting cell viability in different growth phases. Cell treatment of dividing cells with increasing concentration of acetic acid in the presence of glucose as sole carbon source at pH 3.0 is associated to forms of regulated cell death sharing apoptosis‐like (80–120 mM) or necrotic‐like (160–200 mM) features, respectively (Ludovico et al., 2001). Acetic acid concentrations higher than 120 mM are required to induce regulated cell death in stationary phase cells (Ludovico et al., 2002). Moreover, acetic acid deriving from ethanol under respiratory metabolism can mediate cell death through Sch9p and RAS/PKA pathway activation negatively affecting longevity (Burtner et al., 2009).
Overall, these evidences links acetic stress sensitivity to the environmental context, metabolic and energetic cellular profile. A strict relationship between carbon source and acetic acid tolerance is suggested by full resistance to acetic acid of dividing yeast cells grown on raffinose, but not on glucose, indicating that either intracellular metabolism or stress response pathways are activated under derepressing conditions (Guaragnella et al., 2013). In the detailed picture of cell components and mechanisms of yeast acetic acid‐induced regulated cell death (AA‐RCD), the pivotal role played by reactive oxygen species (ROS) and mitochondria emerge (Eisenberg et al., 2007; Guaragnella et al., 2012; Pereira et al., 2008). In particular, hydrogen peroxide appears to be a second messenger in the AA‐RCD cascade of events, as also shown by the inhibitory effect of the ROS scavenger N‐acetyl cysteine on AA‐RCD process (Guaragnella et al., 2010). Mitochondria play a dual role in AA‐RCD as for the release of apoptotic proteins (pro‐death), such as cytochrome c, as for the energy and metabolic supply (pro‐life). In a late phase of AA‐RCD, cytochrome c is degraded and mitochondrial dysfunction occurred with a decrease of the respiratory control index (RCI), a collapse of the mitochondrial membrane potential, a reduction in cytochrome c oxidase (COX) activity and in cytochromes a–a3 levels (Giannattasio et al., 2008; Ludovico et al., 2002). Thus, genetic or metabolic interventions to activate the antioxidant system or certain mitochondrial stress‐signalling pathways can counteract acetic acid toxicity. In this regard, low‐pH pre‐conditioning before acetic acid exposure or the overexpression of the cytosolic isoform of catalase, CTT1, also considered a general stress marker enzyme, has been shown to completely rescue AA‐RCD (Giannattasio et al., 2005; Guaragnella et al., 2008). The retrograde (RTG) response is a mitochondria‐to‐nucleus communication pathway for adaptation to mitochondrial dysfunction (Liu & Butow, 2006). Experimental evidences have shown that AA‐RCD resistance in raffinose grown exponential cells is partially due to the RTG pathway activation (Guaragnella et al., 2013; Ždralević et al., 2012). Interestingly, reduced oxidative stress and cyt c release together with simultaneous SNF1‐dependent relief of carbon catabolite repression have been observed in these cells, suggesting possible links between the central carbon metabolism reprogramming, mitochondria‐dependent AA‐RCD and the RTG pathways (Laera et al., 2016). This indicates that crosstalk between cell death and adaptation mechanisms are activated by acetic acid stress (Giannattasio et al., 2013; Sousa et al., 2013). The master regulator of transcription High‐osmolarity glycerol 1 (HOG1) is the main responsible of AA‐RCD resistance after low‐pH pre‐conditioning (Giannattasio et al., 2005; Guaragnella et al., 2019). Under these conditions, the RTG pathway is not activated but still involved with one of its positive regulator, Rtg2, acting as a general stress sensor and required for Hog1 activation (Guaragnella et al., 2019). Hog1 has also been reported to contribute to acetic acid resistance by inhibiting the acid entry through post‐translational regulation of the acetate channel Fps1 (Mollapour & Piper, 2007). Other pathways linked to RTG, TOR and Ras–cAMP–PKA, regulating nutrient sensing, metabolism, stress resistance and cell growth, are causally involved in the signalling of yeast AA‐RCD (Almeida et al., 2009; Giannattasio et al., 2013).
Several studies based on omics approaches, such as transcriptomics, proteomics and metabolomics, have been carried out to gain insight into the molecular determinants of acetic acid stress response. However, the different experimental settings, such as medium composition and pH, acetic acid concentration and duration of the treatment or cellular growth phase, do not always allow proper comparisons to obtain a homogenous picture. In addition, a strain‐dependent tolerance to acetic acid has to be also considered (Table 1).
TABLE 1.
Acetic acid concentrations, pH and the resulting undissociated acetic acid concentrations in the cited studies
| Reference | Strain background | Acetic acid (mM) | Acetic acid (g/l) | pH |
|---|---|---|---|---|
| Almeida et al. (2009) | BY4742 | 140–200 | 8.4–12.0a | 3.0 |
| Ask et al. (2013) | CEN.PK 113‐7D | 55a | 3.3 | 5.0 |
| Burtner et al. (2009) | BY4742 | 10 | 0.6a | 2.5 |
| Casal et al. (1996) | IGC4072 | 87b | 5.2b | 5.0 |
| Y. Chen et al. (2016) | D452‐e2 | 182a | 10.9 | 4.0 |
| C. Chen et al. (2016) | BY4741 | 92a | 5.5 | 4.0, 4.8 |
| Cunha et al. (2018) | PE‐2 | 73a | 4.4 | 4.0 |
| Ding et al. (2015) | S288C | 140 | 8.4a | 4.8 |
| Dong et al. (2017) | W303 | 30–150 | 1.8–9.0a | 3.0 |
| Giannattasio et al. (2008) | W303 | 20–80 | 1.2–4.8a | 3.0 |
| Giannattasio et al. (2005) | W303 | 20–200 | 1.2–12.0a | 3.0 |
| Godinho et al. (2018) | BY4741 | 50–100 | 3.0–6.0a | 4.0 |
| Guaragnella et al. (2008) | W303 | 80 | 4.8a | 3.0 |
| Guaragnella et al. (2010) | W303 | 20–200 | 1.2–12a | 3.0 |
| Guaragnella et al. (2006) | W303 | 80 | 4.8a | 3.0 |
| Guaragnella et al. (2019) | W303 | 80 | 4.8a | 3.0 |
| Guaragnella et al. (2013) | W303 | 80 | 4.8a | 3.0 |
| Gurdo et al. (2018) | BAFC 3084 | 83a | 5.0 | 6.0 |
| Hu et al. (2019) | BY4742 | 150 | 9.0a | 3.0 |
| Kim et al. (2019) | BY4741 | 30 | 1.8a | 7.4 |
| Kitanovic et al. (2012) | FF 18984 | 30–120 | 1.8–7.2a | 3.0–7.0 |
| Konarzewska et al. (2017) | BY4741 | 25, 50 | 1.5, 3a | N/A |
| Laera et al. (2016) | W303 | 80 | 4.8a | 3.0 |
| Lee et al. (2017) | BY4741 | 105b | 6.3b | 4.5 |
| Lindberg et al. (2013) | CEN.PK 113‐7D | 30–200a | 1.8–12.0 | 5.0 |
| Longo et al. (2015) | W303 | 80 | 4.8a | 3.0 |
| Ludovico et al. (2001) | W303 | 20–200 | 1.2–12.0a | 3.0 |
| Ludovico et al. (2002) | W303 | 120–240 | 7.2–14.4a | 3.0 |
| Meijnen et al. (2016) | Ethanol Red, JT22689 | 110–167b | 6.6–10.0b | 4.0 |
| Mira, Palma, et al. (2010) | BY4741 | 60 | 3.6a | 4.0 |
| Mira, Becker, and Sá‐Correia (2010) | BY4741 | 70–110 | 4.2–6.6a | 4.5 |
| Mollapour and Piper (2007) | BY4741 | 100 | 6.0a | 4.5 |
| Oh et al. (2019) | CEN.PK2‐1D | 83a | 5.0 | 4.0 |
| Pampulha and Loureiro‐Dias (1989) | IGC 3507 III | 40–200 | 2.4–12a | 3.5–5.5 |
| Raghavendran et al. (2020) | CEN.PK 113‐7D | 55a | 3.3 | 5.0 |
| Sousa et al. (2013) | BY4741 | 400 | 24a | 3.0 |
| Swinnen et al. (2017) | BY4741 | 200 | 12a | 4.5 |
| Tanaka et al. (2012) | S288C | 167a | 10.0 | 4.2 |
| Ullah et al. (2012) | BY4741 | 42 | 2.5a | 5.0 |
| Wu et al. (2016) | BY4741 | 75a | 4.5 | 4.0 |
| Zhang et al. (2017) | BY4741 | 60–167a | 3.6–10 | 4.5 |
| Zhang et al. (2015) | BY4741 | 7a | 4.3 | 4.8 |
Note: Values separated by ‘–’designate a range, and values separated by commas designate the specific values used.
Abbreviation: N/A, not available.
Recalculated from grams per litre or millimolar. When values in millimolar were given, the corresponding concentration in grams per litre has been recalculated and vice versa.
Recalculated from % v/v.
In AA‐RCD regulation, the yeast metacaspase Yca1 has been shown to play an extensive role causing significant alterations in carbohydrate catabolism, lipid metabolism, proteolysis and stress‐response as judged by proteome and metabolome profiling of YCA1‐knock out cells during AA‐RCD (Guaragnella et al., 2006; Longo et al., 2015). Particularly, a shift from the main glycolytic pathway to the pentose phosphate pathway and a proteolytic mechanism to cope with oxidative stress characterise AA‐RCD in the presence of YCA1, while AA‐RCD occurs through the activation of ceramide metabolism in its absence (Longo et al., 2015). It is of note that metacaspases are ancestors of mammalian caspases and their extensive biological functions reflects the involvement in a regulated death process occurring in a context of a failing response to internal or external mild stress. In this regard, obvious differences exist between mammalian apoptosis and yeast regulated cell death in terms of specific morphologic and biochemical features, effectors and key molecular players (Carmona‐Gutierrez et al., 2018; Kulkarni et al., 2019).
For the modulation of AA‐RCD in yeast, the relevance of certain genes, especially involved in mitochondrial function, glucose repression and oxidative stress response, has been confirmed at genome‐wide scale (Sousa et al., 2013). The work by Almeida et al. (2009) identifies TOR pathway as an important regulatory node during a late phase of AA‐RCD and indicates that the proteomic alterations are directly or indirectly linked to a TOR‐dependent regulation and include amino acid uptake, transportation and synthesis (Almeida et al., 2009). That acetic acid affects amino acids transport, particularly between cytosol and vacuole, has been recently confirmed by a study on knock out yeast cells lacking the vacuolar autophagy‐related protein ATG22 (Hu et al., 2019).
A transcriptomic and metabolomic analysis show temporal‐ and spatial‐specific expression in acetic acid treated cells, implying the upregulation of genes involved in transcription and protein synthesis in an early‐phase; protein fate, cell cycle and DNA processing in a middle stage and regulation of metabolism and protein function in a late phase. Meanwhile, genes involved in cellular transport, transport facilities and transport routes were reduced in both early and middle stage (Dong et al., 2017).
3. STRATEGIES TO IMPROVE ACETIC ACID TOLERANCE
Acetic acid is an important source of stress and a potent inhibitor during yeast‐based industrial fermentation processes, such as wine making and production of fuels and chemicals from renewable carbohydrate feedstocks, including lignocellulosic biomass. This latter procedure involves preliminary pretreatment and hydrolysis steps to convert polysaccharides into sugar monomers, which are then converted to produce desired products. Acetic acid is the main by‐product of both lignocellulose pretreatment and fermentation process, inhibition affects productivity and growth of S. cerevisiae and the capacity of fermenting alternative yet abundant carbon sources, such as xylose in recombinant strains. For this reason, implementing efficient and economical strategies to obtain robust cell factories and increase acetic acid resistance represent a critical challenge for commercial production of cellulosic fuels and chemicals. Numerous studies and different approaches, mostly developed in laboratory S. cerevisiae strains, allowed to identify important factors, components and regulatory networks for constructing more robust industrial yeast strains to be used in the field of ethanologenic fermentation. A list of genes involved in acetic acid tolerance and identified through different methodological approaches has been reported in Table 2.
TABLE 2.
Genes involved in acetic acid tolerance
| Gene | Function | Acetic acid tolerance | Methodology | Strain background | Reference |
|---|---|---|---|---|---|
| FPS1 | Aquaglyceroporin | + | Deletion | BY4741 | Mollapour and Piper (2007) |
| ADY2 | Acetate transporter | + | Deletion | BY4741 | Zhang et al. (2017) |
| PDR18 | Transporter of ABC family | − | Deletion | BY4741 | Godinho et al. (2018) |
| PMA1 | Plasma Membrane H+‐ATPase | + | Overexpression | BY4741 | Lee et al. (2017) |
| VMA3 | Vacuolar H+‐ATPase | − | Deletion | BY4741 | Konarzewska et al. (2017) |
| HAA1 | Transcriptional activator for weak acid stress | + |
Deletion/Transcriptomics Overexpression |
BY4741 S288C |
Mira, Becker, and Sá‐Correia (2010) and Tanaka et al. (2012) |
| RTG2 | Sensor of mitochondrial dysfunction | − | Deletion | W303 | Guaragnella et al. (2013) |
| HOG1 | Mitogen‐activated protein kinase | − | Deletion | W303 | Guaragnella et al. (2019) |
| RCK1 | Protein kinase | + | Overexpression | D452‐2 | Oh et al. (2019) |
| WHI2 | Protein phosphatase activator | +/− |
Overexpression Deletion |
D452‐2 | Chen et al. (2016) |
| SET5 | Methyltransferase | + | Overexpression | BY4741 | Zhang et al. (2015) |
| PPR1 | Zinc finger transcription factor | + | Overexpression | BY4741 | Zhang et al. (2015) |
| CTT1 | Cytosolic catalase | + | Overexpression | W303 | Guaragnella et al. (2008) |
| JJJ1 | ATPase activator | + | Deletion | BY4741 | Wu et al. (2016) |
| RTT109 | Histone acetyltransferase | + | Deletion | BY4741 | Cheng et al. (2016) |
| ACS1 | AcetylCoA‐synthetase 1 | + | Overexpression | XM19/XM20 | Ding et al. (2015) |
| ACS2 | AcetylCoA‐synthetase 2 | + | Overexpression | S288C | Qin et al. (2020) |
To enhance the competitiveness of industrial lignocellulosic fuels and chemicals, robust enzymes and cell factories are vital. Lignocellulose‐derived streams contain a cocktail of inhibitors, including acetic acid, that drain the cell of ATP and redox potential, causing a multi‐level cell response. Also in this case, results from applied research for cell factory development point to the fact that there is no ‘magic bullet’ and targeting a single gene, without taking into consideration a cell‐wide response, often results in unforeseen cell response, counteracting or neutralising the expected potential beneficial effects.
3.1. Overexpression or deletion of individual genes
Although studies on the effects of overexpression or deletion of single genes on acetic tolerance differ for strains and experimental settings, they mostly converge on common defence mechanisms, involving general stress response, particularly oxidative stress, metabolic and energetic aspects. Overexpression of RCK1, coding for a protein kinase, has been recently reported to confer higher resistance to acetic acid by reducing oxidative stress (Oh et al., 2019). This is in accordance with the proposed mechanism for Whi2, whose overexpression could activate the transcription factors Msn2/Msn4 and consequently the expression of stress response genes related to acetic acid tolerance (Chen et al., 2016). The strict link between oxidative stress and acetic acid resistance is also confirmed by the overexpression of SET5 and PPR1 exerting their functions in the presence of acetic acid through both global gene transcription and metabolic regulation via ROS detoxification upon a pH of 3.5 (Zhang et al., 2015). At this regard, a specific role for the antioxidant enzyme catalase has been related to increased acetic acid tolerance either in CTT1 overexpressing cells or in JJJ1 and RTT109 knockout cells (Cheng et al., 2016; Guaragnella et al., 2008; Wu et al., 2016). These evidences reinforce the concept that the maintenance of intracellular redox balance and particularly detoxification of H2O2 and not O2˙ is a key condition for multiple stress tolerance, including acetic acid, in S. cerevisiae (Gurdo et al., 2018). Also, the deletion of the plasma membrane acetate transporter ADY2 improves growth and fermentation under acetic acid stress, by reducing ROS accumulation and increasing cell membrane integrity (Zhang et al., 2017). Increasing cell capacity to consume acetic acid through the overexpression of acetyl‐coenzyme A synthetases 1 or 2, ACS1 or ACS2, is another possible strategy for improving acetic acid tolerance by accelerating carbon and energy metabolism (Ding et al., 2015; Qin et al., 2020).
Interestingly, a beneficial effects of flocculin genes and zinc supplementation on growth and fermentation capacity has been revealed in the development of acetic acid tolerant S. cerevisiae strains by metabolic engineering (Cheng et al., 2017, and references therein). Upon zinc supplementation and acetic acid stress, a protective effect has been observed after overexpression of ADE genes enhancing cell growth, improving ethanol productivity, controlling purine and amino acid biosynthesis (Zhang et al., 2017).
3.2. Manipulation of Haa1‐Regulon
Another possibility to improve strain robustness against acetic acid is acting on the Haa1‐regulon. The major role played by this transcription factor in acetic acid stress has been largely demonstrated at genomic, genetic and molecular level (Kim et al., 2019; Meijnen et al., 2016; Mira, Becker, & Sá‐Correia, 2010). Haa1 overexpression itself or in combination with the overexpression of the phosphoribosyl pyrophosphate synthetase encoded by PRS3 in a recombinant industrial S. cerevisiae strain boosts yeast tolerance towards acetic acid (Cunha et al., 2018; Tanaka et al., 2012). The known mechanisms of Haa1‐mediated acetic acid stress response involve the cell wall integrity pathway and the activation of plasma membrane multidrug transporters to reduce intracellular acetate accumulation (Cunha et al., 2018; Swinnen et al., 2017). Also, the phosphorylation state of Haa1 can affect its transcriptional activity and consequently stress response to acetic acid (Collins et al., 2017).
3.3. Synthetic biology and metabolic engineering approaches
The ‘omics’ era has offered a new opportunity and perspective in the comprehension of biological processes and allowed the development of synthetic biology and metabolic engineering approaches. Also, the knowledge on acetic acid tolerance has been extended to a genomic point of view and important achievements based on the manipulation of genomes or metabolic fluxes have been reached to counteract acetic acid toxicity (Palma et al., 2018). An industrial S. cerevisiae strain tolerant to high concentrations of acetic acid and with improved performance during 2G ethanol production was obtained through the evolutionary adaptation strategy of metabolic engineering conferring physiological or genotype variations, specifically higher oxidative stress resistance (Gurdo et al., 2018). Evolutionary engineering combined with genome shuffling via mating allowed to generate recombinants S. cerevisiae strains with improved tolerance to key inhibitory by‐products present in hydrolysates of lignocellulosic biomass, including acetic acid and to identify the specific mutations responsible for the improved fitness by whole‐genome resequencing (Cheng et al., 2015). Transcriptome profile of cells exposed to acetic acid under various conditions and different times confirmed that enhanced acetic acid tolerance is attributed to multiple factors. At this regard, an engineered S. cerevisiae strain with improved xylose utilisation and ethanol production has been obtained by creating a feedback regulation system through the simultaneous overexpression of selected genes regulated by stress‐driven promoters and strengthening the GSH (glutathione) biosynthesis and acetate degradation pathways (Qin et al., 2020). This is consistent with other observations, demonstrating the higher overall tolerance to lignocellulosic hydrolysates of GSH‐accumulating strains (Ask et al., 2013).
Notwithstanding the positive effect on enhanced GSH accumulation, such multiple overexpression system may in themselves become the cause of cell stress, causing metabolic burden and unforeseen cells responses, as shown by recent results obtained with recombinant strains accumulating high levels of GSH (Raghavendran et al., 2020).
4. CONCLUSIONS
Reflecting on the molecular mechanisms of acetic acid stress response in budding yeast gives the opportunity to get an integrated view connecting aspects related to cellular transport, pH homeostasis, metabolism and stress‐signalling pathways (Figure 1). All this knowledge supports the relevance of an environment‐dependent cell fitness and adaptation under acid stress conditions. Integrating this information with the results obtained by synthetic biology and metabolic engineering approaches in the ‘omics’ era is a valuable resource in industrial biotechnology, where the improvement of acetic acid tolerance is a key factor during yeast‐based fermentation processes. In this view, the production of biofuels and renewable chemicals from lignocellulosic biomass represents one of the most promising goals to promote a sustainable circular and biobased economy. In order to develop the performing cell factories required for the competitiveness of this sector, the knowledge on the single molecular signalling pathways that has accumulated over the years, as well as the current capacity of developing system‐wide approaches for modelling and engineering cell pathways have to come into play.
FIGURE 1.
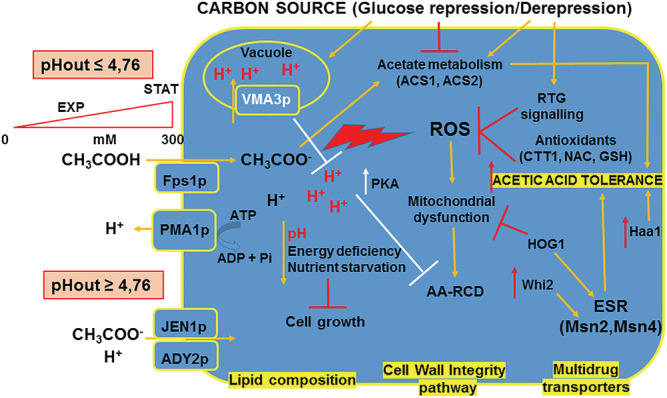
Major factors and cell components involved in acetic acid stress and its response in budding yeast. Extracellular and intracellular pH, cellular growth phase and environmental growth conditions, plasma membrane composition and cell wall assembly, activity of plasma membrane/vacuolar proton pumps and monocarboxylate transporters, metabolic and antioxidant genes, antioxidant molecules, transcription factors and stress‐signalling pathways [Colour figure can be viewed at wileyonlinelibrary.com]
ACKNOWLEDGEMENTS
We thank the Swedish Energy Agency, the Swedish Research Council Energy‐oriented basic research (project number P43524‐1. Title: “Cell membrane engineering as a novel target for the improvement of microbial strains for biofuels production: investigating the relationship between membrane constituents and membrane permeability”) and the Chalmers University of Technology Area of Advance Energy for supporting this work.
Guaragnella N, Bettiga M. Acetic acid stress in budding yeast: From molecular mechanisms to applications. Yeast. 2021;38:391–400. 10.1002/yea.3651
Funding information Swedish Energy Agency, Grant/Award Number: P43524‐1
REFERENCES
- Almeida, B., Ohlmeier, S., Almeida, A. J., Madeo, F., Leão, C., Rodrigues, F., & Ludovico, P. (2009). Yeast protein expression profile during acetic acid‐induced apoptosis indicates causal involvement of the TOR pathway. Proteomics, 9(3), 720–732. 10.1002/pmic.200700816 [DOI] [PubMed] [Google Scholar]
- Alves, R., Sousa‐Silva, M., Vieira, D., Soares, P., Chebaro, Y., Lorenz, M. C., Casal, M., Soares‐Silva, I., & Paiva, S. (2020). Carboxylic acid transporters in Candida pathogenesis. MBio, 11(3), e00156‐20. 10.1128/mBio.00156-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ask, M., Mapelli, V., Höck, H., Olsson, L., & Bettiga, M. (2013). Engineering glutathione biosynthesis of Saccharomyces cerevisiae increases robustness to inhibitors in pretreated lignocellulosic materials. Microbial Cell Factories, 12, 87. 10.1186/1475-2859-12-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berterame, N. M., Porro, D., Ami, D., & Branduardi, P. (2016). Protein aggregation and membrane lipid modifications under lactic acid stress in wild type and OPI1 deleted Saccharomyces cerevisiae strains. Microbial Cell Factories, 15, 39. 10.1186/s12934-016-0438-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtner, C. R., Murakami, C. J., Kennedy, B. K., & Kaeberlein, M. (2009). A molecular mechanism of chronological aging in yeast. Cell Cycle, 8, 1256–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmelo, V., Bogaerts, P., & Sá‐Correia, I. (1997). Activity of plasma membrane H+‐ATPase and expression of PMA1 and PMA2 genes in Saccharomyces cerevisiae cells grown at optimal and low pH. Archives of Microbiology, 166(5), 315–320. [DOI] [PubMed] [Google Scholar]
- Carmona‐Gutierrez D., Bauer M. A., Zimmermann A., Aguilera A., Austriaco N., Ayscough K., Balzan R., Bar‐Nun S., Barrientos A., Belenky P., Blondel M., Braun R. J., Breitenbach M., Burhans W.C., Buettner S., Cavalieri D., Chang M., Cooper K. F., Côrte‐Real M., Costa V., Cullin C., Dawes I., Dengjel J., Dickman M. B., Eisenberg T., Fahrenkrog B., Fasel N., Froehlich K.U., Gargouri A., Giannattasio S., Goffrini P., Gourlay C. W., Grant C. M., Greenwood M. T., Guaragnella N., Heger T., Heinisch J., Herker E., Herrmann J. M., Hofer S., Jiménez‐Ruiz A., Jungwirth H., Kainz K., Kontoyiannis D. P., Ludovico P., Manon S., Martegani E., Mazzoni C., Megeney L. A., Meisinger C., Nielsen J., Nystroem T., Osiewacz H. D., Outeiro T. F., Park H. O., Pendl T., Petranovic D., Picot S., Polčic P., Powers T., Ramsdale M., Rinnerthaler M., Rockenfeller P., Ruckenstuhl C., Schaffrath R., Segovia M., Severin F. F., Sharon A., Sigrist S. J., Sommer‐Ruck C., Sousa M. J., Thevelein J. M., Thevissen K., Titorenko V., Toledano M. B., Tuite M., Voegtle F. N., Westermann B., Winderickx J., Wissing S., Woelfl S., Zhang Z. J., Zhao R. Y., Zhou B., Galluzzi L., Kroemer G., Madeo F. (2018) Guidelines and recommendations on yeast cell death nomenclature. Microbial Cell, 5(1), 4–31. 10.15698/mic2018.01.607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal, M., Cardoso, H., & Leão, C. (1996). Mechanisms regulating the transport of acetic acid in Saccharomyces cerevisiae . MicrobiologyReading, 142(Pt 6), 1385–1390. 10.1099/13500872-142-6-1385 [DOI] [PubMed] [Google Scholar]
- Casal, M., Queirós, O., Talaia, G., Ribas, D., & Paiva, S. (2016). Carboxylic acids plasma membrane transporters in Saccharomyces cerevisiae . Advances in Experimental Medicine and Biology, 892, 229–251. 10.1007/978-3-319-25304-6_9 [DOI] [PubMed] [Google Scholar]
- Chen, Y., Stabryla, L., & Wei, N. (2016). Improved acetic acid resistance in Saccharomyces cerevisiae by overexpression of the WHI2 gene identified through inverse metabolic engineering. Applied and Environmental Microbiology, 82(7), 2156–2166. 10.1128/AEM.03718-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, C., Almario, M. P., & Kao, K. C. (2015). Genome shuffling to generate recombinant yeasts for tolerance to inhibitors present in lignocellulosic hydrolysates. Biotechnology Letters, 37(11), 2193–2200. 10.1007/s10529-015-1895-0 [DOI] [PubMed] [Google Scholar]
- Cheng, C., Zhang, M., Xue, C., Bai, F., & Zhao, X. (2017). Development of stress tolerant Saccharomyces cerevisiae strains by metabolic engineering: New aspects from cell flocculation and zinc supplementation. Journal of Bioscience and Bioengineering, 123, 141e146. [DOI] [PubMed] [Google Scholar]
- Cheng, C., Zhao, X., Mingming Zhang, M., & Bai, F. (2016). Absence of Rtt109p, a fungal‐specific histone acetyltransferase, results in improved acetic acid tolerance of Saccharomyces cerevisiae . FEMS Yeast Research, 16(2), 1–9, fow010. 10.1093/femsyr/fow010 [DOI] [PubMed] [Google Scholar]
- Collins, M. E., Black, J. J., & Liu, Z. (2017). Casein kinase I isoform Hrr25 is a negative regulator of haa1 in the weak acid stress response pathway in Saccharomyces cerevisiae . Applied and Environmental Microbiology, 83(13), e00672–e00617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha, J. T., Costa, C. E., Ferraz, L., Romaní, A., Johansson, B., Sá‐Correia, I., & Domingues, L. (2018). HAA1 and PRS3 overexpression boosts yeast tolerance towards acetic acid improving xylose or glucose consumption: unravelling the underlying mechanisms. Applied Microbiology and Biotechnology, 102(10), 4589–4600. 10.1007/s00253-018-8955-z [DOI] [PubMed] [Google Scholar]
- Ding, J., Holzwarth, G., Penner, M. H., Patton‐Vogt, J., & Bakalinsky, A. T. (2015). Overexpression of acetyl‐CoA synthetase in Saccharomyces cerevisiae increases acetic acid tolerance. FEMS Microbiology Letters, 362(3), 1–7. 10.1093/femsle/fnu042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, Y., Hu, J., Fan, L., & Chen, Q. (2017). RNA‐Seq‐based transcriptomic and metabolomic analysis reveal stress responses and programmed cell death induced by acetic acid in Saccharomyces cerevisiae . Scientific Reports, 7, 42659. 10.1038/srep42659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg, T., Büttner, S., Kroemer, G., & Madeo, F. (2007). The mitochondrial pathway in yeast apoptosis. Apoptosis, 12(5), 1011–1023. [DOI] [PubMed] [Google Scholar]
- Gancedo, J. M. (1992). Carbon catabolite repression in yeast. European Journal of Biochemistry, 206, 297–313. [DOI] [PubMed] [Google Scholar]
- Gancedo, J. M., & Gancedo, C. (1986). Catabolite repression mutants of yeast. FEMS Microbiology Reviews, 32, 179–187. [Google Scholar]
- Geng, P., Zhang, L., & Shi, G. Y. (2017). Omics analysis of acetic acid tolerance in Saccharomyces cerevisiae . World Journal of Microbiology and Biotechnology, 33(5), 94. 10.1007/s11274-017-2259-9 [DOI] [PubMed] [Google Scholar]
- Giannattasio, S., Atlante, A., Antonacci, L., Guaragnella, N., Lattanzio, P., Passarella, S., & Marra, E. (2008). Cytochrome c is released from coupled mitochondria of yeast en route to acetic acid‐induced programmed cell death and can work as an electron donor and a ROS scavenger. FEBS Letters, 582(10), 1519–1525. 10.1016/j.febslet.2008.03.048 [DOI] [PubMed] [Google Scholar]
- Giannattasio, S., Guaragnella, N., Corte‐Real, M., Passarella, S., & Marra, E. (2005). Acid stress adaptation protects Saccharomyces cerevisiae from acetic acid‐induced programmed cell death. Gene, 354, 93–98. 10.1016/j.gene.2005.03.030 [DOI] [PubMed] [Google Scholar]
- Giannattasio, S., Guaragnella, N., Zdralević, M., & Marra, E. (2013). Molecular mechanisms of Saccharomyces cerevisiae stress adaptation and programmed cell death in response to acetic acid. Frontiers in Microbiology, 4, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho, C. P., Prata, C. S., Pinto, S. N., Cardoso, C., Bandarra, N. M., Fernandes, F., & Sá‐Correia, I. (2018). Pdr18 is involved in yeast response to acetic acid stress counteracting the decrease of plasma membrane ergosterol content and order. Scientific Reports, 8(1), 7860. 10.1038/s41598-018-26128-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guaragnella, N., Antonacci, L., Giannattasio, S., Marra, E., & Passarella, S. (2008). Catalase T and Cu,Zn‐superoxide dismutase in the acetic acid‐induced programmed cell death in Saccharomyces cerevisiae . FEBS Letters, 582(2), 210–214. 10.1016/j.febslet.2007.12.007 [DOI] [PubMed] [Google Scholar]
- Guaragnella, N., Passarella, S., Marra, E., & Giannattasio, S. (2010). Knock‐out of metacaspase and/or cytochrome c results in the activation of a ROS‐independent acetic acid‐induced programmed cell death pathway in yeast. FEBS Letters, 584(16), 3655–3660. 10.1016/j.febslet.2010.07.044 [DOI] [PubMed] [Google Scholar]
- Guaragnella, N., Pereira, C., Sousa, M. J., Antonacci, L., Passarella, S., Côrte‐Real, M., Marra, E., & Giannattasio, S. (2006). YCA1 participates in the acetic acid induced yeast programmed cell death also in a manner unrelated to its caspase‐like activity. FEBS Letters, 580(30), 6880–6884. [DOI] [PubMed] [Google Scholar]
- Guaragnella, N., Stirpe, M., Marzulli, D., Mazzoni, C., & Giannattasio, S. (2019). Acid stress triggers resistance to acetic acid‐induced regulated cell death through Hog1 activation which requires RTG2 in yeast. Oxidative Medicine and Cellular Longevity, 4651062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guaragnella, N., Zdralević, M., Antonacci, L., Passarella, S., Marra, E., & Giannattasio, S. (2012). The role of mitochondria in yeast programmed cell death. Frontiers in Oncology, 2, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guaragnella, N., Ždralević, M., Lattanzio, P., Marzulli, D., Pracheil, T., Liu, Z., Passarella, S., Marra, E., & Giannattasio, S. (2013). Yeast growth in raffinose results in resistance to acetic‐acid induced programmed cell death mostly due to the activation of the mitochondrial retrograde pathway. Biochimica et Biophysica Acta, 1833(12), 2765–2774. 10.1016/j.bbamcr.2013.07.017 [DOI] [PubMed] [Google Scholar]
- Gurdo, N., Novelli Poisson, G. F., Juárez, A. B., Rios de Molina, M. C., & Galvagno, M. A. (2018). Improved robustness of an ethanologenic yeast strain through adaptive evolution in acetic acid is associated with its enzymatic antioxidant ability. Journal of Applied Microbiology, 125(3), 766–776. 10.1111/jam.13917 [DOI] [PubMed] [Google Scholar]
- Hu, J., Dong, Y., Wang, W., Zhang, W., Lou, H., & Chen, Q. (2019). Deletion of Atg22 gene contributes to reduce programmed cell death induced by acetic acid stress in Saccharomyces cerevisiae . Biotechnology for Biofuels, 12, 298. 10.1186/s13068-019-1638-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M. S., Cho, K. H., Park, K. H., Jang, J., & Hahn, J. S. (2019). Activation of Haa1 and War1 transcription factors by differential binding of weak acid anions in Saccharomyces cerevisiae . Nucleic Acids Research, 47(3), 1211–1224. 10.1093/nar/gky1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitanovic, A., Bonowski, F., Heigwer, F., Ruoff, P., Kitanovic, I., Ungewiss, C., & Wölfl, S. (2012). Acetic acid treatment in S. cerevisiae creates significant energy deficiency and nutrient starvation that is dependent on the activity of the mitochondrial transcriptional complex Hap2‐3‐4‐5. Front. Oncologia, 2, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarzewska, P., Sherr, G. L., Ahmed, S., Ursomanno, B., & Shen, C. H. (2017). Vma3p protects cells from programmed cell death through the regulation of Hxk2p expression. Biochemical and Biophysical Research Communications, 493(1), 233–239. 10.1016/j.bbrc.2017.09.041 [DOI] [PubMed] [Google Scholar]
- Kulkarni, M., Stolp, Z. D., & Hardwick, J. M. (2019). Targeting intrinsic cell death pathways to control fungal pathogens. Biochemical Pharmacology, 162, 71–78. 10.1016/j.bcp.2019.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laera, L., Guaragnella, N., Ždralević, M., Marzulli, D., Liu, Z., & Giannattasio, S. (2016). The transcription factors ADR1 or CAT8 are required for RTG pathway activation and evasion from yeast acetic acid‐induced programmed cell death in raffinose. Microbial Cell, 3(12), 621–631. 10.15698/mic2016.12.549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y., Nasution, O., Lee, Y. M., Kim, E., Choi, W., & Kim, W. (2017). Overexpression of PMA1 enhances tolerance to various types of stress and constitutively activates the SAPK pathways in Saccharomyces cerevisiae . Applied Microbiology and Biotechnology, 101, 229–239. 10.1007/s00253-016-7898-5 [DOI] [PubMed] [Google Scholar]
- Lindahl, L., Genheden, S., Faria‐Oliveira, F., Allard, S., Eriksson, L. A., Olsson, L., & Bettiga, M. (2017). Alcohols enhance the rate of acetic acid diffusion in S. cerevisiae: Biophysical mechanisms and implications for acetic acid tolerance. Microbial Cell, 5(1), 42–55. 10.15698/mic2018.01.609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg, L., Santos, A. X., Riezman, H., Olsson, L., & Bettiga, M. (2013). Lipidomic profiling of Saccharomyces cerevisiae and Zygosaccharomyces bailii reveals critical changes in lipid composition in response to acetic acid stress. PLoS ONE, 8(9), e73936. 10.1371/journal.pone.0073936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z., & Butow, R. A. (2006). Mitochondrial retrograde signalling. Annual Review of Genetics, 40, 159–185. 10.1146/annurev.genet.40.110405.090613 [DOI] [PubMed] [Google Scholar]
- Longo, V., Ždralević, M., Guaragnella, N., Giannattasio, S., Zolla, L., & Timperio, A. M. (2015). Proteome and metabolome profiling of wild‐type and YCA1‐knock‐out yeast cells during acetic acid‐induced programmed cell death. Journal of Proteomics, 128, 173–188. 10.1016/j.jprot.2015.08.003 [DOI] [PubMed] [Google Scholar]
- Ludovico, P., Rodrigues, F., Almeida, A., Silva, M. T., Barrientos, A., & Côrte‐Real, M. (2002). Cytochrome c release and mitochondria involvement in programmed cell death induced by acetic acid in Saccharomyces cerevisiae . Molecular Biology of the Cell, 13(8), 2598–2606. 10.1091/mbc.e01-12-0161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludovico, P., Sousa, M. J., Silva, M. T., Leão, C. L., & Côrte‐Real, M. (2001). Saccharomyces cerevisiae commits to a programmed cell death process in response to acetic acid. Microbiology, 147(Pt 9), 2409–2415. 10.1099/00221287-147-9-2409 [DOI] [PubMed] [Google Scholar]
- Martínez‐Muñoz, G. A., & Kane, P. (2008). Vacuolar and plasma membrane proton pumps collaborate to achieve cytosolic pH homeostasis in yeast. The Journal of Biological Chemistry, 283(29), 20309–20319. 10.1074/jbc.M710470200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijnen, J. P., Randazzo, P., Foulquié‐Moreno, M. R., van den Brink, J., Vandecruys, P., Stojiljkovic, M., Dumortier, F., Zalar, P., Boekhout, T., Gunde‐Cimerman, N., Kokošar, J., Štajdohar, M., Curk, T., Petrovič, U., & Thevelein, J. M. (2016). Polygenic analysis and targeted improvement of the complex trait of high acetic acid tolerance in the yeast Saccharomyces cerevisiae . Biotechnology for Biofuels, 9(5), 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira, N. P., Becker, J. D., & Sá‐Correia, I. (2010). Genomic expression program involving the Haa1p‐regulon in Saccharomyces cerevisiae response to acetic acid. OMICS, 14(5), 587–601. 10.1089/omi.2010.0048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira, N. P., Palma, M., Guerreiro, J. F., & Sá‐Correia, I. (2010). Genome‐wide identification of Saccharomyces cerevisiae genes required for tolerance to acetic acid. Microbial Cell, 9, 79. 10.1186/1475-2859-9-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollapour, M., & Piper, P. W. (2007). Hog1 mitogen‐activated protein kinase phosphorylation targets the yeast Fps1 aquaglyceroporin for endocytosis, thereby rendering cells resistant to acetic acid. Molecular and Cellular Biology, 27(18), 6446–6456. 10.1128/MCB.02205-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morsomme, P., Slayman, C. W., & Goffeau, A. (2000). Mutagenic study of the structure, function and biogenesis of the yeast plasma membrane H+‐ATPase. Biochimica et Biophysica Acta, 1469, 133–157. 10.1016/S0304-4157(00)00015-0 [DOI] [PubMed] [Google Scholar]
- Oh, E. J., Wei, N., Kwak, S., Kim, H., & Jin, Y. S. (2019). Overexpression of RCK1 improves acetic acid tolerance in Saccharomyces cerevisiae . Journal of Biotechnology, 292, 1–4. 10.1016/j.jbiotec.2018.12.013 [DOI] [PubMed] [Google Scholar]
- Palma, M., Guerreiro, J. F., & Sa‐Correia, I. (2018). Adaptive response and tolerance to acetic acid in Saccharomyces cerevisiae and Zygosaccharomyces bailii: A physiological genomics perspective. Frontiers in Microbiology, 9, 274. 10.3389/fmicb.2018.00274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pampulha, M. E., & Loureiro‐Dias, M. C. (1989). Combined effect of acetic acid, pH and ethanol on intracellular pH of fermenting yeast. Applied Microbiology and Biotechnology, 31, 547–550. [Google Scholar]
- Pereira, C., Silva, R. D., Saraiva, L., Johansson, B., Sousa, M. J., & Côrte‐Real, M. (2008). Mitochondria‐dependent apoptosis in yeast. Biochimica et Biophysica Acta, 1783(7), 1286–1302. 10.1016/j.bbamcr.2008.03.010 [DOI] [PubMed] [Google Scholar]
- Qin, L., Dong, S., Yu, J., Ning, X., Xu, K., Zhang, S. J., Xu, L., Li, B. Z., Li, J., Yuan, Y. J., & Li, C. (2020). Stress‐driven dynamic regulation of multiple tolerance genes improves robustness and productive capacity of Saccharomyces cerevisiae in industrial lignocellulose fermentation. Metabolic Engineering, 61, 160–170. 10.1016/j.ymben.2020.06.003 [DOI] [PubMed] [Google Scholar]
- Raghavendran, V., Marx, C., Olsson, L., & Bettiga, M. (2020). The protective role of intracellular glutathione in Saccharomyces cerevisiae during lignocellulosic ethanol production. AMB Express, 10(1), 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa, M. J., Duarte, A. M., Fernandes, T. R., Chaves, S. R., Pacheco, A., Leão, C., Côrte‐Real, M., & Sousa, M. J. (2013). Genome‐wide identification of genes involved in the positive and negative regulation of acetic acid‐induced programmed cell death in Saccharomyces cerevisiae . BMC Genomics, 14(1), 838. 10.1186/1471-2164-14-838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinnen, S., Henriques, S. F., Shrestha, R., Ho, P., Sá‐Correia, I., & Nevoigt, E. (2017). Improvement of yeast tolerance to acetic acid through Haa1 transcription factor engineering: Towards the underlying mechanisms. Microbial Cell Factories, 16(1), 7. 10.1186/s12934-016-0621-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, K., Ishii, Y., Ogawa, J., & Shima, J. (2012). Enhancement of acetic acid tolerance in Saccharomyces cerevisiae by overexpression of the HAA1 gene, encoding a transcriptional activator. Applied and Environmental Microbiology, 78, 8161–8163. 10.1128/AEM.02356-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah, A., Orij, R., Brul, S., & Smits, G. J. (2012). Quantitative analysis of the modes of growth inhibition by weak organic acids in Saccharomyces cerevisiae . Applied and Environmental Microbiology, 78(23), 8377–8387. 10.1128/AEM.02126-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X., Zhang, L., Jin, X., Fang, Y., Zhang, K., Qi, L., & Zheng, D. (2016). Deletion of JJJ1 improves acetic acid tolerance and bioethanol fermentation performance of Saccharomyces cerevisiae strains. Biotechnology Letters, 38, 1097–1106. 10.1007/s10529-016-2085-4 [DOI] [PubMed] [Google Scholar]
- Ždralević, M., Guaragnella, N., Antonacci, L., Marra, E., & Giannattasio, S. (2012). Yeast as a tool to study signalling pathways in mitochondrial stress response and cytoprotection. The Scientific World Journal, 912147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M., Zhang, K., Mehmood, M. A., Zhao, Z. K., Bai, F., & Zhao, X. (2017). Deletion of acetate transporter gene ADY2 improved tolerance of Saccharomyces cerevisiae against multiple stresses and enhanced ethanol production in the presence of acetic acid. Bioresource Technology, 245, 1461–1468. 10.1016/j.biortech.2017.05.191 [DOI] [PubMed] [Google Scholar]
- Zhang, M. M., Zhao, X. Q., Cheng, C., & Bai, F. W. (2015). Improved growth and ethanol fermentation of Saccharomyces cerevisiae in the presence of acetic acid by overexpression of SET5 and PPR1. Biotechnology Journal, 10(12), 1903–1911. 10.1002/biot.201500508 [DOI] [PubMed] [Google Scholar]


