Abstract
Patient‐derived organoids (PDOs) represent promising preclinical models in various tumor types. In the context of prostate cancer (PCa), however, their establishment has been hampered by poor success rates, which impedes their broad use for translational research applications. Along with the necessity to improve culture conditions, there is a need to identify factors influencing outcomes and to determine how to assess success versus failure in organoid generation. In the present study, we report our unbiased efforts to generate PDOs from a cohort of 81 PCa specimens with diverse pathological and clinical features. We comprehensively analyzed histological features of each enrolled sample (Gleason score, tumor content, proliferation index) and correlated them with organoid growth patterns. We identified improved culture conditions favoring the generation of PCa organoids, yet no specific intrinsic tumor feature was broadly associated with sustained organoid growth. In addition, we performed phenotypic and molecular characterization of tumor–organoid pairs using immunohistochemistry, immunofluorescence, fluorescence in situ hybridization, and targeted sequencing. Morphological and immunohistochemical profiles of whole organoids altogether provided a fast readout to identify the most promising ones. Notably, primary samples were associated with an initial take‐rate of 83% (n = 60/72) in culture, with maintenance of cancer cells displaying common PCa alterations, such as PTEN loss and ERG overexpression. These cancer organoids were, however, progressively overgrown by organoids with a benign‐like phenotype. Finally, out of nine metastasis samples, we generated a novel organoid model derived from a hormone‐naïve lung metastasis, which displays alterations in the PI3K/Akt and Wnt/β‐catenin pathways and responds to androgen deprivation. Taken together, our comprehensive study explores determinants of outcome and highlights the opportunities and challenges associated with the establishment of stable tumor organoid lines derived from PCa patients. © 2021 The Authors. The Journal of Pathology published by John Wiley & Sons, Ltd. on behalf of The Pathological Society of Great Britain and Ireland.
Keywords: prostate cancer, patient‐derived organoids, disease modeling, personalized medicine, phenotypic and molecular features
Introduction
In the last decade, novel opportunities for in vitro modeling have emerged with the establishment of three‐dimensional (3D) models derived from patients' material and referred to as patient‐derived organoids (PDOs) [1]. In the prostate, seminal studies have reported the successful establishment of organoid lines derived from benign tissue and from patients with advanced prostate cancer (PCa) [2, 3, 4, 5, 6]. In particular, organoids derived from advanced PCa recapitulated molecular and phenotypic traits commonly observed in PCa patients, as well as rare aggressive phenotypes such as neuroendocrine prostate cancer (NEPC) [3, 5, 6]. With about 16% for highly metastatic samples, these studies have, however, highlighted poor success rates associated with the generation of organoids derived from advanced PCa that limit their broad use for personalized medicine applications; in contrast, other cancer types such as bladder, breast, and colorectal cancers have reported success rates ranging from 70% to 90% [7, 8, 9].
In addition, the establishment of PCa organoid lines derived from primary tumors has not been successful so far, which hinders their use for studies aiming at uncovering the functional relevance of specific factors in the early‐stage setting or at deciphering mechanisms underlying castration resistance [2, 3]. Challenges associated with the generation of such organoids may be attributed to the frequent multifocal nature of primary PCa and the potential selection and expansion in culture of minor cell subsets, which do not well reflect the main features of the original tissue [10]. In particular, investigators have reported the frequent overgrowth of benign epithelial cells concomitant with PCa clinical specimens, highlighting the necessity to thoroughly characterize organoid cultures [2, 3, 4]. This may be inferred by analysis of markers associated with basal and luminal cells, which predominantly compose the prostate epithelium in the normal setting; in contrast, the majority of prostate cancers are characterized by expansion of malignant cells with luminal‐like features and the absence of basal epithelial cells [11, 12].
Here, we sought to explore determinants of organoid generation by attempting to establish organoid cultures derived from a large and diverse cohort of PCa samples and comprehensively characterizing the outcomes in terms of growth efficiency and morphological, histological, phenotypic, and genomic features. Along with these analyses, we highlight improved culture conditions for organoid generation and report the successful generation of novel short‐term and long‐term organoid cultures.
Materials and methods
Patient samples and organoid culture
All PCa samples were obtained under approval by the Ethics Committee of Northwestern and Central Switzerland (EKBB 37/13) from patients operated at the University Hospital of Basel (USB) and the St Claraspital Basel (SCB), Switzerland. Detailed clinical and pathological characteristics of the patients and associated samples are described in Supplementary materials and methods; Table 1; and supplementary material, Table S1. Tissue pieces were finely cut with scissors in Advanced Dulbecco's Modified Eagle Medium/Ham's F‐12 (Ad‐DMEM/F‐12) and processed to generate a cell suspension according to published protocols [4]. Alternatively, PCa tissues were mechanically cut to generate small tissue fragments referred to as ‘aggregates’. Aggregates or cell suspensions were seeded in growth factor‐reduced Matrigel (Corning, Corning, NY, USA) domes at an approximate 25%:75% tissue:Matrigel ratio and cultured according to published protocols [4] (see also Supplementary materials and methods and supplementary material, Table S2).
Table 1.
Characteristics of patients enrolled in organoid experiments (n = 75).
| Gleason score | n (%) |
| 6 | 1 (1.3%) |
| 3 + 4 | 20 (26.7%) |
| 4 + 3 | 18 (24.0%) |
| 8 | 5 (6.7%) |
| 9 | 23 (30.7%) |
| 10 | 3 (4.0%) |
| NEPC | 1 (1.3%) |
| Not available | 4 (5.3%) |
| Type | n (%) |
| Hormone‐naïve (HN) | 57 (76.0%) |
| Castration‐resistant (CR) | 18 (24.0%) |
| Age (years) | |
| Median (inter‐quartile range) | 67 (62–73) |
| Last PSA (μg/l) | |
| Median (inter‐quartile range) | 152 (10–459) |
Imaging of organoids and assessment of growth
Images of organoids were captured once to twice a week with an inverted cell culture microscope (Olympus CKX53, Shinjuku, Tokyo, Japan), at 4×, 10×, and 20× magnification. To define organoid growth, two 4× images were analyzed at around 14 days of culture (before passaging); three growth groups were defined: group ‘–’: no significant organoid growth; group ‘+’: between one and five organoids per well; and group ‘++’: more than five organoids per well. Short‐term and long‐term organoid cultures were defined as organoids that were expanded for less than five passages (i.e. either failed to grow or were voluntarily stopped) and for more than five passages, respectively.
Histology, immunohistochemistry, and immunofluorescence
Histological analysis was performed by standard hematoxylin and eosin (H&E) staining. Immunohistochemistry (IHC) was conducted as previously described [13]. For all samples, the Gleason score (GS) was defined and percentages of tumor cells and of Ki67‐positive tumor cells (proliferation index) were scored by a pathologist (LB). Analyses were blinded to the initial pathological assessment of the resection specimen and the outcomes in terms of organoid generation. Immunofluorescence staining was performed on 4‐μm‐thick sections, following antigen retrieval with boiling citrate acid‐based antigen masking solution (Vector Labs, Burlingame, CA, USA), with a protocol adapted from [14]. Immunofluorescence images were captured using a Nikon Ti2 microscope (DBM Imaging Facility). Details of all antibodies and dilutions are provided in supplementary material, Table S3.
Statistical analysis and data representation
Statistical analyses were conducted using a two‐tailed Welch t‐test, one‐ and two‐way ANOVA, or Kolmogorov–Smirnov tests as appropriate. P values less than 0.05 were considered significant. Prism software (version 8.0; GraphPad Inc, San Diego, CA, USA) was used for statistical analyses and data representation.
Further details of Patients and clinical samples, Tissue processing and organoid culture, Imaging and quantification of organoid number and size for comparative analyses, Fluorescence in situ hybridization (FISH) analyses, DNA/RNA extraction and targeted sequencing, and Drug treatments and cell viability assays are provided in Supplementary materials and methods.
Results
Generation of organoid cultures derived from primary and advanced PCa
To improve culture conditions, we reasoned that it may be crucial to preserve cellular interactions which are critical for the maintenance of key properties characterizing tumor cells [15]. To test this approach, PCa tissues were mechanically disrupted to generate tissue aggregates (‘Ag’ method) or mechanically and enzymatically digested to generate cell suspensions (‘Cs’ method), and further processed similarly to the method of Drost et al [4] (Figure 1A). Using these methods, we attempted to generate organoids derived from 81 PCa samples, which mainly included radical prostatectomy (RP, n = 56), transurethral resection of the prostate (TUR‐P, n = 14), and metastasis samples (MET, n = 9) with distinct clinical and pathological features (Figure 1B and Table 1). To determine the rate of initial organoid growth, we assessed patterns of growth at passage 0 after a minimum of 14 days in culture according to specific criteria (++: n > 5; +: 1 < n < 5; or −: n = 0 organoids per well). Notably, the majority of RP‐derived organoids grew with high efficiency, while TUR‐P‐derived organoids exhibited the opposite trend (1/56 RP versus 10/14 TUR‐P with no growth, Figure 1C). Metastasis samples exhibited variability, with 4/9 samples displaying organoid formation at passage 0 (Figure 1C). Thus, organoids derived from PCa patients initially grow with variable efficiency, which may depend on the source and the intrinsic characteristics of the samples.
Figure 1.
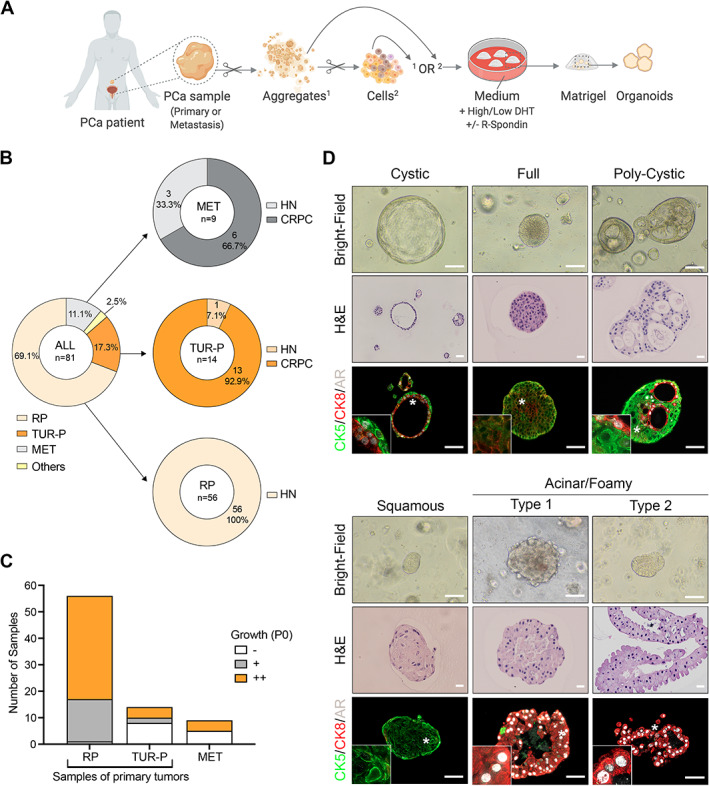
Generation of organoids derived from PCa patients. (A) Schematic diagram depicting the experimental strategy and the distinct conditions used to establish organoids derived from primary PCa or PCa metastasis. Aggregates (1) or cell suspensions (2) are seeded in Matrigel domes immersed in organoid medium. Scissors symbols indicate mechanical or enzymatic digestion. Created with BioRender.com. (B) Summary of sample types used to establish organoid cultures (n = 81). RP, radical prostatectomy; TUR‐P, transurethral resection of the prostate; MET, metastasis. ‘Others’ corresponds to a primary PCa resection obtained from an autopsy of a man with metastatic CRPC and one local progression of primary PCa obtained from a bladder resection. HN, hormone‐naïve; CRPC, castration‐resistant PCa. (C) Growth efficiency for each sample category at passage 0 (P0). Group ‘–’: no significant organoid growth; group ‘+’: between one and five organoids per well; group ‘++’: more than five organoids per well. (D) Bright‐field images, corresponding H&E staining, and immunofluorescence analysis for the indicated antibodies in selected organoids representative of five major morpho‐histological subtypes. Insets show higher magnification of a representative region indicated by a white asterisk on the original image. Scale bars: 100 μm for bright‐field images; 20 μm for H&E images; 50 μm for immunofluorescence images.
In comparison with the Cs method, the Ag method overall resulted in organoids more accurately recapitulating the phenotype of the original tumor and was associated with better maintenance of organoids in terms of passages (n = 47 samples; supplementary material, Figure S1). To understand the potential advantage of growth in Ag‐ versus Cs‐derived organoids, we investigated the presence of stromal cells, which have been shown to promote the viability and growth of prostate‐derived organoids [16]. In Ag and Cs conditions, organoids were composed of pure epithelial cell populations, as suggested by positivity for epithelial‐specific markers (i.e. E‐cadherin, cytokeratin 5/8) and negativity for stromal markers [alpha‐smooth muscle actin (aSMA), vimentin (supplementary material, Figure S2)]. In specific cases of Ag‐derived cultures, however, we observed aSMA and/or vimentin‐positive cells within aggregates adjacent to the organoids, indicating the presence of stromal cells in proximity to organoids. In contrast to Ag versus Cs, other variations of the culture conditions, such as in the concentration of R‐spondin‐1 or dihydrotestosterone (DHT), had little effect on the initial efficiency of organoid formation but may affect their maintenance at long term (supplementary material, Figure S3).
PCa organoids display a wide range of morphological and histological profiles
From the various generated organoid cultures, we identified five major subtypes that were associated with distinct histological and morphological features (Figure 1D and supplementary material, Figure S4A). To elucidate whether these subtypes correlated with cellular phenotypes, we analyzed the expression of CK5 as a basal marker, CK8 as a luminal marker, and androgen receptor (AR) in 18 selected organoid cultures (n = 141 analyzed organoids from n = 16 patients). The ‘cystic’ morphology was characterized by a spherical mono‐ or bi‐layer of cells surrounding a large lumen, while ‘full’ organoids displayed a regular spherical shape filled with numerous packed cells. In contrast, the ‘polycystic’ morphology exhibited glandular‐like structures with several lumina containing secretions. Organoids with a cystic, full, or polycystic morphology typically displayed a mix of CK5‐ and CK8‐positive cells (Figure 1D and supplementary material, Figure S4A,B). Within the same organoids, CK8‐positive luminal cells were characterized by higher levels of AR compared with CK5‐positive basal cells, similar to what is observed in prostate tissue [11]. Based on their histological and phenotypic features, these three subtypes appeared to be more reminiscent of benign prostate glands than PCa.
In addition, the ‘squamous’‐like morphology was often observed in PCa‐derived organoids and was mainly characterized by large cells with abundant and dense cytoplasm consistent with squamous differentiation (Figure 1D and supplementary material, Figure S4A). Squamous‐like organoids nearly exclusively consisted of CK5‐positive basal‐like cells and expressed very low or undetectable levels of AR, consistent with a benign squamous phenotype (Figure 1D and supplementary material, Figure S4A,B). Finally, organoids with an irregular ‘foamy/acinar’ morphology displayed a delicate and finely vacuolar cytoplasm and histological features more typical of PCa cells (Figure 1D and supplementary material, Figure S4A). Consistent with their histological features reminiscent of PCa tissues, this subtype was enriched and often exclusively composed of CK8‐positive cells expressing high levels of AR (Figure 1D and supplementary material, Figure S4A,B). Notably, this morphology was more frequently observed in Ag‐derived than in Cs‐derived organoid cultures (supplementary material, Figure S4C).
Taken together, these analyses suggest that morpho‐histological features of PCa‐derived organoids may inform on their cellular phenotypes.
Histological features of PCa primary samples do not dictate organoid growth
Since primary PCa and PCa metastases are clinically and molecularly distinct entities, we considered them in an independent manner in our detailed analyses of organoid generation. We first focused on primary PCa samples and assessed whether specific intrinsic pathological features of the tumor tissues may determine the growth of their derived organoids. We assigned a GS/ISUP grade group and scored the percentage of tumor cells and of Ki67‐positive cells in each clinical sample enrolled in the organoid experiments, independently of whether their derived organoids grew or not (detailed in supplementary material, Table S4). Out of 71 tissues obtained from primary samples (i.e. RP and TUR‐P), 12 were found to be tumor‐free and were therefore excluded from subsequent analyses (n = 7 RP and 5 TUR‐P, Figure 2A). Among the remaining 59 samples, the majority were represented by GS 3 + 4 (ISUP grade 2, 26/59, 44.1%) and GS 4 + 3 (ISUP grade 3, 11/59, 18.6%) samples, followed by GS 8 (ISUP grade 4, 9/59), and GS 9–10 (ISUP grade 5, 6/59). The only case of NEPC was represented by the CRPC primary resection collected at autopsy, which did not grow in organoid culture.
Figure 2.
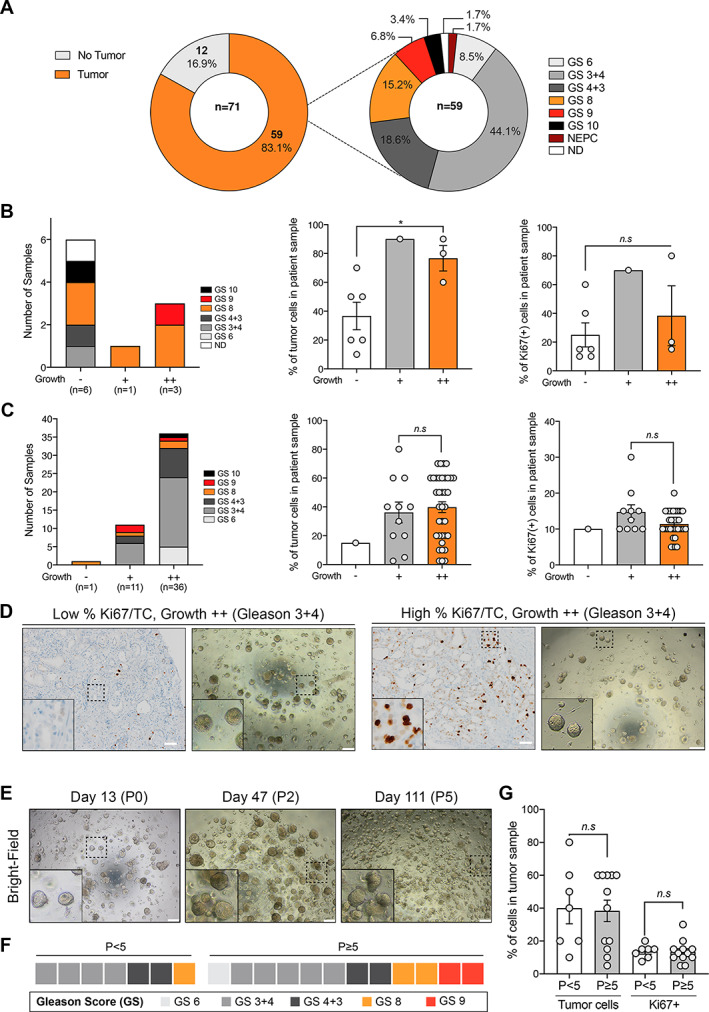
Histological determinants of organoid growth for primary PCa samples. (A) Summary of pathological features associated with primary PCa samples enrolled in organoid studies (n = 71). GS, Gleason score. (B) Gleason score (GS, left), percentage of tumor cells (TCs, middle), and percentage of Ki67‐positive cells (right) according to growth efficiency in transurethral resection (TUR‐P) primary samples and (C) radical prostatectomy (RP) samples. Analyses were performed using a two‐tailed Welch t‐test (*p < 0.05; n.s., not significant). Means with SEM error bars. Group ‘–’: no significant organoid growth; group ‘+’: between one and five organoids per well; group ‘++’: more than five organoids per well. Whenever two methods were tested, the most efficient method was taken into account. (D) Representative bright‐field images and matched Ki67 staining for two RP samples (P20‐15 and P19‐27) with distinct proliferative index and tumor content (i.e. low versus high percentage Ki67/TC). Note that both samples are associated with GS 3 + 4 and grow with high efficiency despite very different % Ki67/TC. Insets show greater magnification of a representative region indicated by a dashed rectangle in the original image. Scale bars represent 200 μm for bright‐field images and 50 μm for IHC images. (E) Example of the P19‐15 organoid line maintained over five passages and time (P: passage). Insets show higher magnification of a representative region indicated by a dashed rectangle in the original image. (F) Heatmap depicting the GS associated with RP samples maintained for at least five passages (n = 12) compared with those that stopped growing before passage 5 (n = 7). (G) Percentage of tumor cells and of Ki67+ tumor cells in RP samples maintained for at least five passages (n = 12, grey bars) compared with samples that stopped growing before passage 5 (n = 7, white bars). Analyses were performed using a two‐tailed Welch t‐test (n.s., not significant). Means with SEM.
The percentage of tumor cells (TCs) and Ki67‐positive cells was highly variable in TUR‐P (n = 48) and RP patients' specimens (n = 10) but TUR‐P displayed a significantly higher proliferative index and a trend towards higher tumor content compared with RP samples (p = 0.03, p = 0.14, respectively; two‐tailed Welch t‐test; supplementary material, Figure S5). For both TUR‐P and RP specimens, well‐growing samples (++) were not associated with specific Gleason patterns (Figure 2A,B, left panels). While no difference was found in terms of Ki67‐positive cells (p = 0.60), the percentage of TCs was significantly higher in TUR‐P samples that grew than in those that did not grow (− versus ++, p = 0.02, two‐tailed Welch t‐test, Figure 2B). Out of three TUR‐P samples which initially grew well in organoid culture (n = 3/10), two were further passaged but could not be maintained after passage 2 (supplementary material, Figure S6A).
In contrast to TUR‐P samples, organoid growth was observed for almost all RP samples (n = 47/48) but could be distinguished by high (++) versus low (+) growth (n = 36 and n = 11, respectively). We did not observe any significant difference in terms of the percentage of TCs or Ki67‐positive cells in RP samples that grew very well and the ones that grew less well (+ versus ++, p = 0.66 and p = 0.13, respectively; two‐tailed Welch t‐test, Figure 2C,D). In contrast to TUR‐P samples, RP specimens were passaged with high efficiency and a subset of them was maintained after passage 5 (n = 12/19, Figure 2E and supplementary material, Figure S6B). Samples that ‘failed’ before passage 5 (n = 7/19) were not associated with specific histological features (i.e. GS, TC, Ki67+), compared with samples that were maintained for at least five passages (Figure 2F,G). Thus, histological parameters such as GS, percentage of TCs, and proliferative index do not seem to predict how organoids will grow in the RP cohort.
Primary prostate cancer cells are progressively lost in organoid culture
It is critical to determine whether organoids derived from tumor specimens truly comprise malignant cells, which may be greatly facilitated by analysis of cancer‐specific markers. ERG and PTEN alterations represent the most frequent genomic events in primary PCa (n = 333, cBioPortal [17, 18, 19], Figure 3A). At the protein level, overexpression of ERG and loss of PTEN expression are robust surrogate markers for TMPRSS2–ERG fusion and homozygous PTEN deletion, respectively [20, 21, 22]. Therefore we assessed ERG and PTEN expression status by IHC in a large subset of our primary PCa cohort. Compared with the TCGA cohort, we observed a higher incidence of samples presumably harboring genomic alterations in either one gene or both, as evident by either PTEN loss or ERG overexpression or both (n = 23/30, 76.7% in the organoid cohort versus 49.7% in TCGA, Figure 3A). Out of these 30 tested samples, 23 were associated with high organoid growth and were almost equally distributed in the four molecular subtypes (22% of PTENpos ERGneg samples, 26% for all other categories, Figure 3B).
Figure 3.
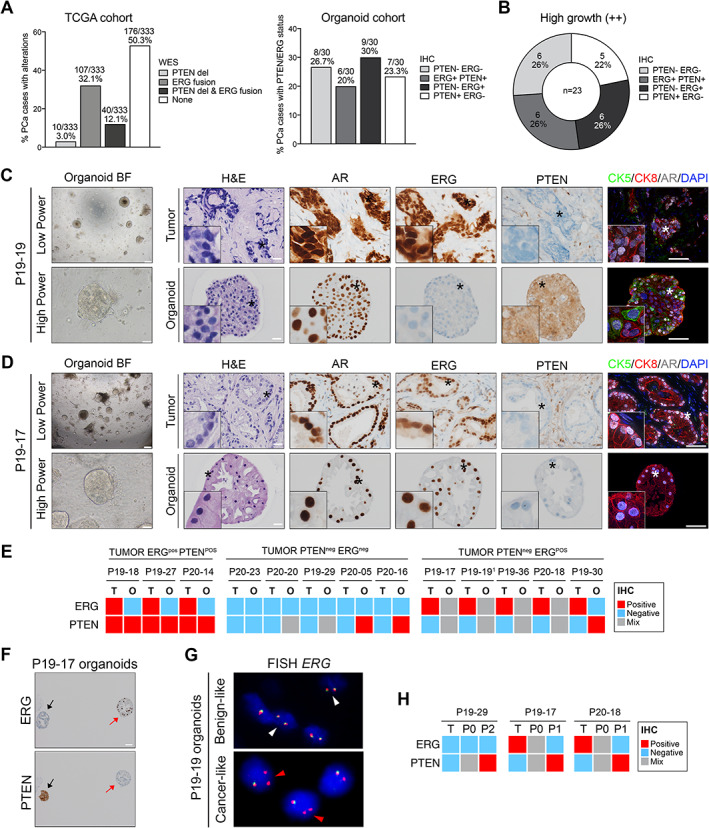
Prostate cancer cells are progressively lost in organoid cultures derived from primary PCa specimens. (A) Left: prevalence of PTEN deep deletions (del) and ERG fusions as analyzed by whole‐exome sequencing (WES) in a published cohort of primary PCa samples (TCGA, n = 333). Right: prevalence of PTEN and ERG positivity/negativity as analyzed by immunohistochemistry (IHC) in a subset of primary PCa samples used to generate organoids (n = 30). (B) Prevalence of samples with distinct PTEN/ERG profiles which grew well in organoid culture at passage 0 (n = 23). (C, D) Morphological and phenotypical analyses of tumor samples and their matched organoid lines. Representative examples of organoids which dissemble (C, P19‐19, day 13) or resemble (D, P19‐17, day 14) the tumor samples they are derived from are shown. Note that the two cultures exhibit a mixed phenotype, while both patient's samples predominantly show PTEN loss and ERG positivity. Bright‐field (BF) and H&E images, as well as immunostaining for the indicated antibodies are presented. Insets show greater magnification of a representative region indicated by an asterisk in the original image. Scale bars: 200 μm and 50 μm for bright‐field images (low and high power); 20 μm for IHC; 50 μm for IF images. (E) Heatmap depicting ERG and PTEN status (IHC) for primary PCa samples (T) and their matched organoid lines at passage 0 (O). (F) Example of a line with a ‘mixed’ phenotype comprising ERGneg PTENpos (black arrow) and ERGpos PTENneg (red arrow) organoids. (G) FISH analysis with ERG dual color break‐apart probe. In cells without ERG rearrangement (‘benign‐like’), two orange/green fusion signals per nucleus are seen (white arrowheads). In contrast, interstitial deletion of the chromosomal region 21q22.2 resulting in the TMPRSS2–ERG fusion leads to a loss of one green signal, as indicated by red arrowheads (‘cancer‐like’). (H) Heatmap comparing ERG and PTEN status (IHC) for tumor samples (T), selected organoids at passage 0, and later passage (P1 or P2).
To easily track down cancer cells in culture, we focused particularly on analyzing organoids derived from samples harboring at least one alteration (i.e. PTEN, ERG, or both). We compared the ERG and PTEN status of tumors and their matched organoids at an early time‐point in IHC (passage 0, 30 days in culture maximum, Figure 3C–E). Notably, none of the four organoid cultures derived from samples with ERG overexpression (ERGpos PTENpos subtype) exhibited positivity for ERG (Figure 3E). Out of six organoid cultures derived from samples with loss of PTEN expression (PTENneg ERGneg subtype), one displayed similar PTEN negativity, three were mixed (i.e. comprising both PTEN‐negative and PTEN‐positive organoids), and two showed intact PTEN expression (Figure 3E). Finally, out of five cultures derived from samples with both PTEN loss and ERG overexpression (PTENneg ERGpos subtype), four displayed a hybrid phenotype with a mix of organoids either displaying molecular features similar to their original tumor (PTENneg ERGpos) or the opposite pattern (PTENpos ERGneg) (Figure 3C–E). One line was exclusively composed of PTENpos ERGneg organoids.
To better characterize the nature of PTENpos ERGneg organoids which did not exhibit clear cancer hallmarks and may therefore represent benign components, we stained selected tumor/organoid pairs for AMACR, a marker associated with PCa, and for GSTP1, a marker whose expression is frequently lost in PCa [23, 24]. Notably, hybrid cultures comprised a mix of organoids displaying either positivity or negativity for both markers, further suggesting the presence of both benign‐ and cancer‐like organoids (supplementary material, Figures S7–S9). In addition, expression of the prostate‐specific marker PSA was observed in a subset of PCa‐derived organoids, demonstrating the presence of cells with a fully differentiated phenotype (supplementary material, Figures S7A–S9). Thus, although to different extents, the presence of cells with a ‘benign‐like’ phenotype can be observed alongside tumor cells in most organoid cultures (12/13) derived from primary tumor specimens.
In agreement with their presumably benign phenotype, PTENpos ERGneg organoids were composed of both CK5+ basal and CK8+ luminal cells, while ‘tumor‐like’ PTENneg ERGpos organoids were exclusively composed of CK8+ luminal cells (Figure 3C,D). Notably, in organoid cultures with hybrid phenotypes, we observed inter‐ and intra‐organoid cellular heterogeneity for PCa markers, mutual exclusivity for ERG and PTEN expression, and heterogeneity for ERG genomic rearrangement (Figure 3F,G and supplementary material, Figure S8). To elucidate whether cells with cancer‐like characteristics are maintained over time, we analyzed the phenotype of mixed organoid lines at later passage (passage 1–2), whenever possible. In four tested lines, cancer‐like characteristics (i.e. ERG and/or AMACR expression, PTEN and/or GSTP1 loss) were no longer observed in the following passages (Figure 3H and supplementary material, Figure S9). For three additional lines with a mixed phenotype, organoids stopped growing at later passage and analyses could therefore not be performed (supplementary material, Table S4). To understand whether culture conditions may favor the maintenance and expansion of normal versus cancer cells, we cultured in parallel cancer and matched benign‐like tissue obtained from the same patients after RP (n = 4). For three out of four patients, we observed lower organoid growth in cancer versus matched benign samples, as evident by the percentage of organoid formation efficiency, the diameter of organoids, and the maintenance of the lines over time (p < 0.05, two‐way ANOVA, supplementary material, Figure S10).
Taken together, these analyses suggest that organoids displaying clear PCa hallmarks are initially present but are progressively lost in culture. Moreover, benign epithelial cells may generally have a growth advantage in these specific culture conditions.
PCa metastasis‐derived organoids are established with poor efficiency and low stability
In addition to primary prostate specimens, we received metastasis samples from diverse tissue sites including lymph nodes (n = 4), bone (n = 2), brain (n = 1), liver (n = 1), and lung (n = 1), as well as one local progression of primary PCa resected from the bladder (Figure 4A and supplementary material, Table S1). Of these ten samples, half did not grow at all (5/10, ‘No growth’ group) and four initially grew but stopped at early passage (‘P ≤ 2’ group, Figure 4B). In the latter group, organoids typically exhibited an irregular shape reminiscent of the acinar/foamy morpho‐histological subtype (Figure 1D) but they either stopped growing or switched to a mesenchymal‐like phenotype upon passaging (Figure 4C). As an example, the P20‐24‐LN line was derived from a cervical lymph node of a CRPC patient and exhibited clear PCa hallmarks, as evident by ERG and PSMA expression, loss of PTEN expression, and AR amplification (Figure 4D). Furthermore, matched tumor and organoids shared similar mutation profiles, with one mutation in TP53 (p.Thr125Met) and one in the beta‐catenin gene CTNNB1 (p.Asp32Asn) at similar allelic frequency in both sample types (Figure 4E). Despite these aggressive features, P20‐24‐LN organoids failed to grow following the second passage. In contrast, one hormone‐naïve lung metastasis resection (P20‐11‐Lg) was cultured for more than 7 months and underwent freezing and thawing cycles (expanded for eight passages and voluntarily stopped). P20‐11‐Lg organoids were quickly formed and maintained the same morphological appearance in culture over passages, despite slower growth and a smaller size (Figure 4F).
Figure 4.
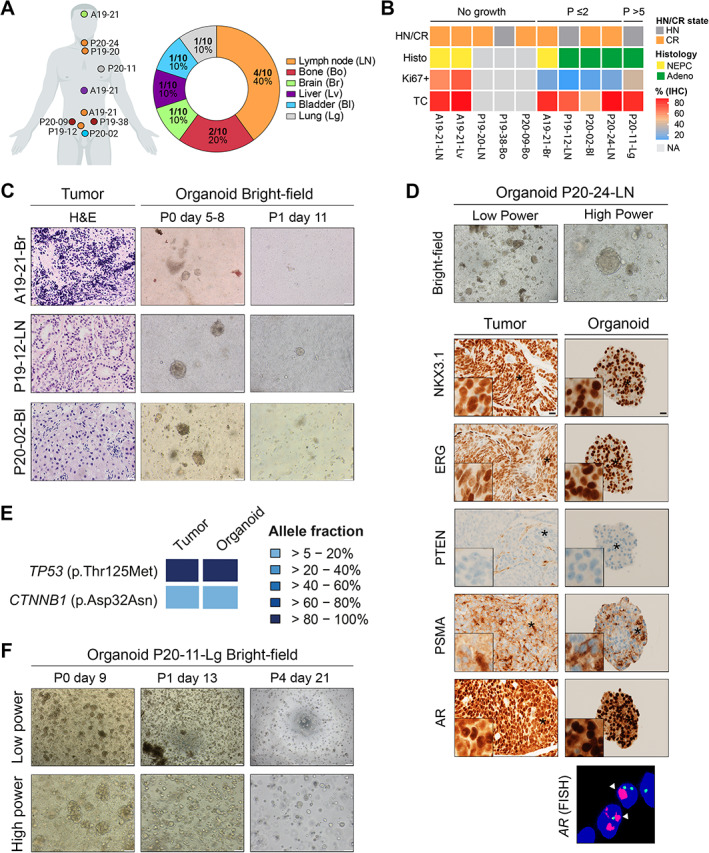
Organoids derived from metastasis specimens are established with poor efficiency and low stability. (A) Summary of tissue sites for advanced samples used to generate organoids (n = 9 metastasis and 1 local progression). Each patient code and its corresponding tissue site are indicated on the left schematic. Created in part with BioRender.com. (B) Heatmap depicting characteristics of samples separated into three groups of organoid growth: ‘No growth’; ‘P ≤ 2’: less than two passages; and ‘P > 5’: more than five passages. HN, hormone‐naïve; CR, castration‐resistant; NEPC, neuroendocrine prostate cancer; Adeno, adenocarcinoma; TC, tumor cells; NA, not available. (C) Examples of samples which initially grew in organoid culture but stopped after passaging. Representative H&E images of tumor samples and bright‐field images of their derived organoids at passage 0 (P0) and passage 1 (P1) are shown. Scale bars: 20 μm for H&E images, 100 μm for bright field images. (D) Morphological and molecular analyses of a lymph node metastasis sample (P20‐24‐LN) and its matched organoid line which stopped growing at passage 2. Bright‐field (BF) images and immunostaining for the indicated antibodies are presented (day 9). Insets show higher magnification of a representative region indicated by an asterisk. Right panel: FISH analysis using AR (red) and CEPX (green) probes. White arrowheads indicate AR amplification. Scale bars represent 200 and 100 μm for bright‐field images and 20 μm for IHC images. (E) Targeted sequencing identifies specific TP53 and CTNNB1 mutations in the P20‐24‐LN tumor sample and its derived organoid line. The heatmap indicates the variant allele fractions of the somatic mutations (blue, see color key). (F) Representative bright‐field images of the organoid line derived from a lung metastasis (P20‐11‐Lg) at passage 0 (P0) and later passages (P1: passage 1; P4: passage 4). Scale bars: 200 μm for low‐power images; 100 μm for high‐power images.
From the three distinct growth groups, there was no obvious enrichment of specific characteristics in terms of hormone status, TC content, or proliferation index (Figure 4B). Thus, no tested intrinsic tumor feature appears to be broadly associated with the growth of organoids derived from PCa metastases.
P20‐11‐Lg organoids recapitulate phenotypic and molecular features of the parental tumor and display sensitivity to androgen levels
At passage 0, P20‐11‐Lg organoids displayed an irregular morphology and phenotypic features similar to their original tumor (Figure 5A–D). In particular, they exhibited typical characteristics of PCa cells such as highly prominent nucleoli and small acinar architecture, expressed high levels of AR and of the prostate identity marker NKX3.1 [14], and displayed a CK5−CK8+ luminal phenotype (Figure 5A–C). Furthermore, homogeneous PTEN loss, ERG overexpression, and ERG genomic rearrangement were observed in both tumor and organoids (Figure 5D). DNA/RNA targeted sequencing confirmed the TMPRSS2–ERG fusion and highlighted a high concordance in the mutation profiles of the tumor sample and its matched organoid line, with two mutations in PTEN (pThr319fs and c.209+2T>A) and one in CTNNB1 (p‐Ser45Pro) with similar allelic frequency (Figure 5E). Notably, ERG overexpression, PTEN loss, and a similar mutation profile were confirmed at later passage in P20‐11‐Lg organoids (supplementary material, Figure S11A,B).
Figure 5.
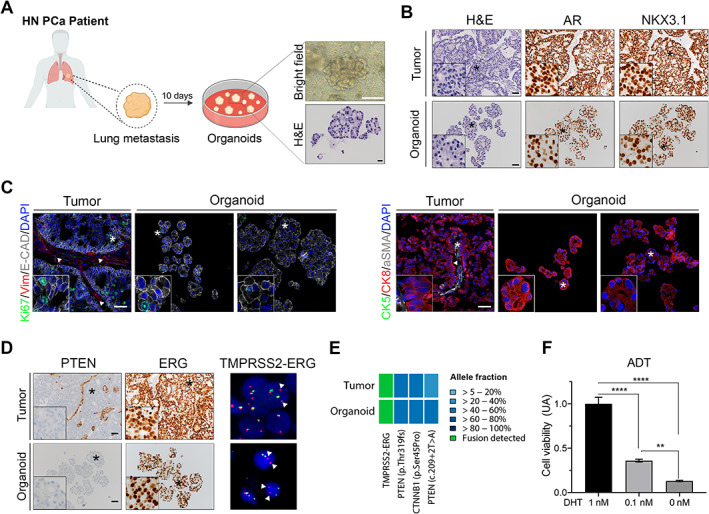
P20‐11‐Lg organoids recapitulate phenotypic and molecular characteristics of their parental tumor sample and display sensitivity to androgen levels. (A) Organoids derived from a hormone‐naïve lung metastasis sample (P20‐11‐Lg) were established after 10 days of culture. High‐power bright‐field image and corresponding H&E of a representative organoid are shown. Scale bars: 100 μm for the bright‐field image; 20 μm for the H&E image. Created in part with BioRender.com. (B) Phenotypical analyses of the P20‐11‐Lg tumor sample (top) and its derived organoid line (bottom). Representative H&E images and IHC staining for the indicated antibodies are shown. Insets show higher magnification of a representative region indicated with an asterisk. Scale bars: 50 μm. (C) Immunofluorescence analyses of P20‐11‐Lg tumor and organoids. Representative images for the indicated antibodies are shown. Vim, vimentin; E‐cad, E‐cadherin. White arrowheads indicate the Vim+ αSMA+ stromal component of the tumor tissue. Insets show greater magnification of a representative region indicated with an asterisk. Scale bars represent 50 μm. (D) Cancer‐associated features of P20‐11‐Lg tumor sample (top) and its derived organoid line (bottom). Left and middle panels: representative IHC staining for PTEN and ERG. Insets show greater magnification of a representative region indicated with an asterisk. Scale bars: 50 μm. Right panels: FISH analysis with ERG dual‐color break apart probe. Interstitial deletion of the chromosomal region 21q22.2 resulting in the TMPRSS2–ERG fusion leads to a loss of one green signal, as indicated by white arrowheads. (E) Heatmap showing the presence of the TMPRSS2–ERG fusion (green) and the identification of specific PTEN and CTNNB1 shared mutations (variant allele fractions, blue) in both tumor sample and its derived organoid line. (F) Culture of P20‐11‐Lg organoids with low (0.1 nm) or no DHT (0 nm) results in a significant decrease in organoid cell viability. Analyses were performed using a two‐way ANOVA test (**p < 0.01, ****p < 0.0001). Data are represented as means relative to the 1 nm DHT control (AU: arbitrary unit) and error bars show SD. Two independent biological replicates were performed, and one representative replicate is shown.
Given that P20‐11‐Lg was obtained from a patient with metastatic hormone‐naïve disease, we hypothesized that its derived organoids may depend on androgen signaling for their growth. We observed a significant reduction of viability in low androgen conditions (0.1 nm DHT) compared with organoids cultured with high DHT levels (1 nm DHT), which was further enhanced in androgen‐deprived (no DHT) conditions (one‐way ANOVA, p < 0.0001 and p < 0.01, respectively; Figure 5F). Treatment of androgen‐deprived P20‐11‐Lg organoids with 0.1 and 1 μm enzalutamide or abiraterone did not significantly affect their viability (one‐way ANOVA, not significant; supplementary material, Figure S11C).
P20‐11‐Lg therefore represents a new organoid model which recapitulates phenotypic and molecular traits commonly observed in PCa and responds to androgen deprivation.
Discussion
Factors that may influence and dictate the success of PCa organoid establishment are undetermined, which impedes their use for personalized medicine applications. In addition, whether organoid cultures derived from primary PCa samples and truly composed of cancer cells can be established reliably is unclear. Here, we provide a transparent report of our efforts to establish and characterize organoid cultures derived from 81 diverse primary and metastasis PCa samples. Among primary samples, RP‐derived organoids initially grew with high efficiency and 63% of them were maintained long term; in contrast, TUR‐P specimens initially grew with moderate success and could not be maintained over long periods of time (Figure 2 and supplementary material, Figure S6). For these samples, we observed a significant correlation between the percentage of TCs in the parental sample and organoid growth, which implies that tumor purity may represent a predictive factor for organoid growth if confirmed in larger sample cohorts. In contrast, other histological features associated with PCa aggressiveness (i.e. high GS and proliferation index [22]) were not enriched in samples growing with high efficiency, suggesting that they do not dictate organoid growth.
As the overgrowth of normal cellular components in PCa organoids is commonly observed [3], it is crucial to demonstrate that they comprise true cancer cells. Yet genomic characterization is often limited by the low frequency of mutations characterizing primary PCa, as well as low material input available at early stage of culture [17]. To overcome these limitations, we thoroughly evaluated PCa‐associated hallmarks in organoid cultures at early passage. First, we identified the ‘acinar/foamy’ morpho‐histological subtype, which was associated with histological and luminal features reminiscent of PCa tissues (Figure 1 and supplementary material, Figure S4) [11]. Furthermore, routine IHC analyses provided easy and fast readouts for the presence of cells with PTEN and/or ERG alterations in culture. Thus, morphological IHC analyses altogether represent a quick readout to determine cancer‐associated features in culture and may help to identify the most relevant organoid lines early in time.
Using this strategy, we demonstrated the presence of cancer cells in at least seven short‐term organoid cultures derived from primary PCa clinical specimens (Figure 3). These organoids typically displayed an acinar/foamy morphology and comprised cells characterized by ERG overexpression and/or PTEN loss of expression. To the best of our knowledge, this is the first study to clearly demonstrate the maintenance of PCa primary cells exhibiting these frequent genomic alterations after a few weeks of culture in patient‐derived 3D models [4, 25, 26]. Furthermore, we show that cells with cancer‐associated features are often accompanied by others which exhibit a benign‐like phenotype within the same organoids and/or the same organoid lines (supplementary material, Figure S8). This intra‐ and inter‐organoid heterogeneity is likely attributable to the selection of a minor subset of benign cells, which may have an advantage of growth in these culture conditions [2, 4]. Solidifying this hypothesis, we show that organoids derived from benign areas generally grow with a significantly higher efficiency compared with those derived from matched cancer areas (supplementary material, Figure S10). Notably, frequent overgrowth by normal cells has also been observed in organoids derived from lung cancer and compounds were used to select genomically‐distinct tumor cells in culture [27]. As we currently do not have reliable strategies to select for cancer versus normal prostate cells, this approach may not be easily amenable to primary PCa samples. Alternative strategies may include the establishment of organoids from single cells, but this method may be associated with very low organoid yields.
In addition to primary specimens, we generated three short‐term and one long‐term organoid lines derived from metastasis specimens (Figure 4). Our success rate (1/9) is in line with previous studies reporting the establishment of organoid lines derived from PCa metastases [3, 5]. The newly generated organoid model displays alterations in the PI3K/Akt and Wnt/β‐catenin pathways, which are commonly altered in advanced PCa [28, 29] . Our data do not point to any specific genomic or phenotypic factor associated with sustained organoid growth, which may be dictated by alternative unidentified genetic, epigenetic, and/or transcriptomic drivers.
Altogether, our study highlights the crucial need to define novel culture conditions that may be more adapted to the lineage specificity and the unique characteristics of PCa cells. In this framework, we show that using tissue aggregates instead of cell suspensions may improve the establishment of PCa PDOs (supplementary material, Figures S1 and S2). Multiple factors, such as cell‐to‐cell contacts or signals from neighboring stromal cells, may promote this potential advantage and will have to be investigated in future studies [15, 16]. Beshiri et al recently reported refined conditions that improve the establishment of organoids from a heterogeneous cohort of patient‐derived xenografts (PDXs) [26]. Similar to our findings in patient samples, there were no obvious genomic or phenotypic correlates of PDXs that grew long term and those that did not. Culture conditions may have to be further personalized to ensure the reliable establishment of organoids derived from specific PCa molecular subtypes, such as recently done for NEPC samples [30]. Finally, it is worth noticing that organoids established from PDXs generally grow with a higher efficiency than those directly derived from patients' clinical material [26, 31]; to achieve higher success rates without prior grafting in mice, future strategies should aim at identifying molecular factors promoting these growth advantages.
Author contributions statement
CL supervised the study. RS and CL planned and designed the study. HS, CAR, LB and CL acquired financial support for the project. TV, HP, DM, TZ, AJT, HS, CAR and LB collected samples and clinicopathologic data. LB and TV performed the pathological assessment. RS, MG, MB, AG and SM carried out experiments and analyzed data. SP and MAR supervised some of the analyses. RS and CL wrote the original draft manuscript. TV, DCM, AJT, SP, MAR, CAR and LB critically reviewed and edited the manuscript and all the authors approved its final version.
Supporting information
Supplementary materials and methods
Figure S1. Comparisons between the aggregate (Ag) method and the cell suspension (Cs) method
Figure S2. Expression of stromal‐ and epithelial‐associated markers in Ag‐ and Cs‐derived organoids
Figure S3. Comparisons of distinct organoid conditions
Figure S4. Five morpho‐histological subtypes in PCa organoids
Figure S5. Tumor content and proliferation index in RP and TUR‐P primary PCa specimens
Figure S6. Passaging and take‐rates of organoids derived from primary PCa samples
Figure S7. Expression of AMACR, GSTP1, and PSA in tumor/organoid pairs
Figure S8. Inter‐ and intra‐organoid phenotypic heterogeneity
Figure S9. Histological and phenotypic changes in organoids before and after passaging
Figure S10. Organoid growth in benign versus cancer areas derived from the same primary samples
Figure S11. Additional analyses of P20‐11‐Lg organoids
Table S1. Metastasis and local progression samples enrolled in organoid experiments
Table S2. Organoid medium composition
Table S3. List of antibodies used in this study
Table S4. Tumor‐organoid master file
Acknowledgements
We thank all patients who consented to participate in the study and all clinicians, pathologists, and study nurses involved in the acquisition of samples. We are grateful to Nicola Aceto, Zoi Diamantopoulo, Viola Paradiso, Ilaria Aborelli, and Loréline Genschik for experimental help. We acknowledge the IHC/FISH facility of the Institute of Pathology, and the microscopy facility of the DBM for technical assistance. Financial support was provided by funding from the University of Basel (3MS1025 to CL), Fond'Action contre le Cancer (YIA 2020 to CL), Krebsliga beider Basel (KLbB‐4480‐03‐2018 to CL), the Swiss Cancer League (KFS‐4983‐02‐2020 to CL and KFS‐4988‐02‐2020‐R to SP), the Department of Surgery of the University Hospital Basel, and the Theron Foundation, Vaduz (SP). Components of several figures were created with BioRender.com.
No conflicts of interest were declared.
References
References 32–35 are cited only in the supplementary material.
- 1.Fatehullah A, Tan SH, Barker N. Organoids as an in vitro model of human development and disease. Nat Cell Biol 2016; 18: 246–254. [DOI] [PubMed] [Google Scholar]
- 2.Karthaus WR, Iaquinta PJ, Drost J, et al. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell 2014; 159: 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao D, Vela I, Sboner A, et al. Organoid cultures derived from patients with advanced prostate cancer. Cell 2014; 159: 176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drost J, Karthaus WR, Gao D, et al. Organoid culture systems for prostate epithelial and cancer tissue. Nat Protoc 2016; 11: 347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Puca L, Bareja R, Prandi D, et al. Patient derived organoids to model rare prostate cancer phenotypes. Nat Commun 2018; 9: 2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pauli C, Hopkins BD, Prandi D, et al. Personalized in vitro and in vivo cancer models to guide precision medicine. Cancer Discov 2017; 7: 462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sachs N, de Ligt J, Kopper O, et al. A living biobank of breast cancer organoids captures disease heterogeneity. Cell 2018; 172: 373–386.e10. [DOI] [PubMed] [Google Scholar]
- 8.van de Wetering M, Francies HE, Francis JM, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 2015; 161: 933–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee SH, Hu W, Matulay JT, et al. Tumor evolution and drug response in patient‐derived organoid models of bladder cancer. Cell 2018; 173: 515–528.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haffner MC, Zwart W, Roudier MP, et al. Genomic and phenotypic heterogeneity in prostate cancer. Nat Rev Urol 2021; 18: 79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen MM, Abate‐Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev 2010; 24: 1967–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang G, Zhao D, Spring DJ, et al. Genetics and biology of prostate cancer. Genes Dev 2018; 32: 1105–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Federer‐Gsponer JR, Müller DC, Zellweger T, et al. Patterns of stemness‐associated markers in the development of castration‐resistant prostate cancer. Prostate 2020; 80: 1108–1117. [DOI] [PubMed] [Google Scholar]
- 14.Dutta A, Le Magnen C, Mitrofanova A, et al. Identification of an NKX3.1‐G9a‐UTY transcriptional regulatory network that controls prostate differentiation. Science 2016; 352: 1576–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.West J, Newton PK. Cellular interactions constrain tumor growth. Proc Natl Acad Sci U S A 2019; 116: 1918–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richards Z, McCray T, Marsili J, et al. Prostate stroma increases the viability and maintains the branching phenotype of human prostate organoids. iScience 2019; 12: 304–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The Cancer Genome Atlas Research Network . The molecular taxonomy of primary prostate cancer. Cell 2015; 163: 1011–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cerami E, Gao J, Dogrusoz U, et al. The cBio Cancer Genomics Portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012; 2: 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013; 6: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Falzarano SM, Zhou M, Carver P, et al. ERG gene rearrangement status in prostate cancer detected by immunohistochemistry. Virchows Arch 2011; 459: 441–447. [DOI] [PubMed] [Google Scholar]
- 21.Jamaspishvili T, Berman DM, Ross AE, et al. Clinical implications of PTEN loss in prostate cancer. Nat Rev Urol 2018; 15: 222–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vlajnic T, Bubendorf L. Molecular pathology of prostate cancer: a practical approach. Pathology 2021; 53: 36–43. [DOI] [PubMed] [Google Scholar]
- 23.Rubin MA, Zhou M, Dhanasekaran SM, et al. α‐Methylacyl coenzyme A racemase as a tissue biomarker for prostate cancer. JAMA 2002; 287: 1662–1670. [DOI] [PubMed] [Google Scholar]
- 24.Martignano F, Gurioli G, Salvi S, et al. GSTP1 methylation and protein expression in prostate cancer: diagnostic implications. Dis Markers 2016; 2016: 4358292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garraway IP, Sun W, Tran CP, et al. Human prostate sphere‐forming cells represent a subset of basal epithelial cells capable of glandular regeneration in vivo . Prostate 2010; 70: 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beshiri ML, Tice CM, Tran C, et al. A PDX/organoid biobank of advanced prostate cancers captures genomic and phenotypic heterogeneity for disease modeling and therapeutic screening. Clin Cancer Res 2018; 24: 4332–4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dijkstra KK, Monkhorst K, Schipper LJ, et al. Challenges in establishing pure lung cancer organoids limit their utility for personalized medicine. Cell Rep 2020; 31: 107588. [DOI] [PubMed] [Google Scholar]
- 28.Robinson D, Van Allen EM, Wu YM, et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015; 162: 454. [DOI] [PubMed] [Google Scholar]
- 29.Abida W, Cyrta J, Heller G, et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci U S A 2019; 116: 11428–11436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mosquera JM, Bareja R, Bernheim J, et al. Extracellular microenvironment in patient‐derived hydrogel organoids of prostate cancer regulates therapeutic response. medRxiv 2020; 10.1101/2020.05.17.20104349. [Not peer reviewed]. [DOI]
- 31.Karkampouna S, La Manna F, Benjak A, et al. Patient‐derived xenografts and organoids model therapy response in prostate cancer. Nat Commun 2021; 12: 1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salami SS, Hovelson DH, Kaplan JB, et al. Transcriptomic heterogeneity in multifocal prostate cancer. JCI Insight 2018; 3: e123468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garofoli A, Paradiso V, Montazeri H, et al. PipeIT: a singularity container for molecular diagnostic somatic variant calling on the ion torrent next‐generation sequencing platform. J Mol Diagn 2019; 21: 884–894. [DOI] [PubMed] [Google Scholar]
- 34.Chang MT, Asthana S, Gao SP, et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol 2016; 34: 155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao J, Chang MT, Johnsen HC, et al. 3D clusters of somatic mutations in cancer reveal numerous rare mutations as functional targets. Genome Med 2017; 9: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials and methods
Figure S1. Comparisons between the aggregate (Ag) method and the cell suspension (Cs) method
Figure S2. Expression of stromal‐ and epithelial‐associated markers in Ag‐ and Cs‐derived organoids
Figure S3. Comparisons of distinct organoid conditions
Figure S4. Five morpho‐histological subtypes in PCa organoids
Figure S5. Tumor content and proliferation index in RP and TUR‐P primary PCa specimens
Figure S6. Passaging and take‐rates of organoids derived from primary PCa samples
Figure S7. Expression of AMACR, GSTP1, and PSA in tumor/organoid pairs
Figure S8. Inter‐ and intra‐organoid phenotypic heterogeneity
Figure S9. Histological and phenotypic changes in organoids before and after passaging
Figure S10. Organoid growth in benign versus cancer areas derived from the same primary samples
Figure S11. Additional analyses of P20‐11‐Lg organoids
Table S1. Metastasis and local progression samples enrolled in organoid experiments
Table S2. Organoid medium composition
Table S3. List of antibodies used in this study
Table S4. Tumor‐organoid master file


