SUMMARY
Phytophthora infestans is a pathogenic oomycete that causes the infamous potato late blight disease. Resistance (R) genes from diverse Solanum species encode intracellular receptors that trigger effective defense responses upon the recognition of cognate RXLR avirulence (Avr) effector proteins. To deploy these R genes in a durable fashion in agriculture, we need to understand the mechanism of effector recognition and the way the pathogen evades recognition. In this study, we cloned 16 allelic variants of the Rpi‐chc1 gene from Solanum chacoense and other Solanum species, and identified the cognate P. infestans RXLR effectors. These tools were used to study effector recognition and co‐evolution. Functional and non‐functional alleles of Rpi‐chc1 encode coiled‐coil nucleotide‐binding leucine‐rich repeat (CNL) proteins, being the first described representatives of the CNL16 family. These alleles have distinct patterns of RXLR effector recognition. While Rpi‐chc1.1 recognized multiple PexRD12 (Avrchc1.1) proteins, Rpi‐chc1.2 recognized multiple PexRD31 (Avrchc1.2) proteins, both belonging to the PexRD12/31 effector superfamily. Domain swaps between Rpi‐chc1.1 and Rpi‐chc1.2 revealed that overlapping subdomains in the leucine‐rich repeat (LRR) domain are responsible for the difference in effector recognition. This study showed that Rpi‐chc1.1 and Rpi‐chc1.2 evolved to recognize distinct members of the same PexRD12/31 effector family via the LRR domain. The biased distribution of polymorphisms suggests that exchange of LRRs during host–pathogen co‐evolution can lead to novel recognition specificities. These insights will guide future strategies to breed durable resistant varieties.
Keywords: NLR cluster, leucine‐rich repeat, Phytophthora infestans, late blight resistance gene, RXLR effector, Solanum species, potato
Significance statement
In this manuscript the cloning of novel late blight resistance genes and their cognate effectors is described. The mechanism of recognition, host pathogen co‐evolution, and implications for deployment of the knowledge in agriculture are discussed.
INTRODUCTION
Potato (Solanum tuberosum) is the third largest food crop in terms of human consumption in the world, after rice (Oryza sativa) and wheat (Triticum aestivum), with more than 370 million tonnes produced in 2019 (FAO, 2020; Devaux et al., 2020). Potato late blight, caused by the oomycete Phytophthora infestans, is one of the most infamous potato diseases. During the mid‐1840s, this pathogen caused the Great Irish Famine, from which around one million people died (Callaway, 2013). Nowadays, losses from late blight are estimated to reach 16% of the world production. The main disease management is based on biocide applications. Including yield losses and crop protection measures, late blight causes a global economic loss of €5.2 billion per year (Haverkort et al., 2016).
Phytophthora infestans is an oomycete with sexual and asexual life cycles, which exhibits a hemibiotrophic lifestyle on potato. Together with its large and fast evolving genome (estimated to be 240 Mb), its population diversity leads to the regular emergence of new aggressive and virulent strains (Haas et al., 2009). The infection starts when a spore lands on the plant surface, germinates, and forms a penetration structure called appressorium. Alternatively, spores can also enter through natural openings such as stomata. After passing the epidermis, hyphae spread intercellularly projecting haustorium structures into the mesophyll cells. These haustoria are specialized infection structures that secrete both apoplastic and cytoplasmic effectors to create an intimate association with the host cell and facilitate nutrient uptake (Fry, 2008). Effectors are pathogen molecules that interact with different host targets to suppress the host defense response and enable colonization. The publication of the P. infestans T30‐4 genome revealed the presence of 563 effector genes encoding the conserved Arg–any amino acid–Leu–Arg (RXLR) peptide motif (Haas et al., 2009). These effectors rapidly evolve by gaining and losing repeat‐rich domains through recombination with different paralogs, transposon movement, and point mutations (Goss et al., 2013). During co‐evolution, potato has evolved receptors to recognize some of these effectors and trigger an immune response.
Wild Solanum species are the main source of resistance (R) genes to P. infestans (Rpi). To date, over 20 Rpi genes have been characterized in different Solanum species, e.g., R1, R2, R3a, R3b, R8, and R9a from Solanum demissum, Rpi‐blb1 and Rpi‐blb12 from Solanum bulbocastanum, Rpi‐vnt1 from Solanum venturii, and Rpi‐amr1 from Solanum americanum (Ballvora et al., 2002; Huang et al., 2005; Jo et al., 2015; Li et al., 2011; Lokossou et al., 2009; Pel et al., 2009; van der Vossen et al., 2003, 2005; Vossen et al., 2016; Witek et al., 2021). All these receptors belong to the nucleotide‐binding (NB)–leucine‐rich repeat (NLR) type of receptors and contain a coiled‐coil (CC) domain in their N‐termini, referred to as CC‐NB‐LRR (CNL). The recognition of a specific effector or avirulence factor (Avr) leads to the activation of effector‐triggered immunity (ETI) and the restriction of the pathogen growth (Jones et al., 2016). ETI is mostly monogenic and therefore well suited and commonly deployed for resistance breeding and crop protection strategies. To keep up with the fast evolution of effectors, NLR genes are also very diverse and rapidly evolving. Gene duplications, recombinations, unequal crossing‐over, and transpositions have been proposed to provide the basis for the evolution of the NLR recognition spectrum (Leister, 2004; Mcdowell and Simon, 2006). This fast evolution can lead to the independent development of new receptors in different geographical locations that recognize the same effector. For instance, the effector Avr2 from P. infestans is recognized by the unrelated R2 and Rpi‐mcq1 CNLs (Aguilera‐Galvez et al., 2018). R2 is located on chromosome IV in the Mexican species S. demissum, while Rpi‐mcq1 is located on chromosome IX from a Peruvian accession of Solanum mochiquense (Foster et al., 2009; Smilde et al., 2005). More recently, Rpi‐amr1 alleles have been described to cause differential recognition of Avramr1 homologs from several P. infestans isolates (Witek et al., 2021). When the doubled‐monoploid DM1‐3 519 R44 potato genome was published, 755 NLR genes were identified (Jupe et al., 2013). Many of them were found in clusters together with closely related paralogs. All of these clusters were formed in ancestral species and had sequence homology to syntenic genomic regions from other Solanum species harboring late blight resistance genes. Thus, alleles from functional Rpi genes that do not provide resistance (rpi) can be found in all studied Solanum genomes.
Here, we studied Solanum chacoense, a diploid wild potato relative from South America considered a source of resistance to P. infestans (Karki et al., 2021; Vossen et al., 2009). We identified two functionally distinct receptors, Rpi‐chc1.1 and Rpi‐chc1.2, which are allelic variants that recognize distinct P. infestans effectors from the same PexRD12/31 effector superfamily. Remarkably, only Rpi‐chc1.1 is able to provide resistance against several P. infestans isolates. The expression and recognition of PexRD12 effectors was associated with Rpi‐chc1.1‐mediated resistance and, therefore, they were designated as Avrchc1.1 effectors. PexRD31 effectors were still expressed in several P. infestans isolates, but were rapidly downregulated during the interaction with potato. This potentially explains the inability of Rpi‐chc1.2 to provide late blight resistance. We postulate that Rpi‐chc1.2 is a ubiquitous ancient R gene that was recently overcome and PexRD31 may have functioned as Avrchc1.2. An allele‐mining strategy revealed Rpi‐chc1 orthologs in different wild Solanum accessions and potato cultivars that could be classified by their sequence and recognition spectrum of Avrchc1.1 or Avrchc1.2 or non‐functionality. Finally, using domain swaps, we found that the LRR domain harbors the recognition specificity of both Avrchc1.1 and Avrchc1.2. The specificities resided in overlapping LRR subdomains and could not be combined into one active protein using domain exchanges.
RESULTS
Cloning and characterization of Rpi‐chc1.1
The S. chacoense accession CHC543 from Bolivia is a previously described wild potato relative harboring resistance to P. infestans (Vleeshouwers et al., 2011a). To identify the genetic locus of resistance, the resistant seedling CHC543‐5 was crossed with the susceptible seedling CHC544‐5 to generate the F1 population 7650, consisting initially of 212 individuals. This population was challenged with P. infestans isolate 90128 in a detached leaf assay (DLA). A clear 1:1 segregation was observed, indicating the presence of a single dominant resistance gene which will henceforth be referred to as Rpi‐chc1. Cleaved amplified polymorphic sequence markers from chromosome 10 were tested as this chromosome was known to harbor Rpi‐ber from the related species Solanum berthaultii (Vossen et al., 2013). The marker TG63 in chromosome 10 was indeed linked to the Rpi‐chc1 resistance. Successive fine‐mapping in a recombinant population representing 2357 individuals was performed using markers derived from RH89‐39‐16 BAC clones from chromosome 10 (PGSC) (Sharma et al., 2013; The Potato Genome Sequencing Consortium, 2011). A narrow genetic window between markers RH106G03‐T and RH97D21_C21‐4 was identified to contain Rpi‐chc1 (Figure 1a). To generate a physical map of the mapping interval, two bacterial artificial chromosome (BAC) clones, B1 and B2, were selected from a BAC library that was derived from CHC543‐5 genomic DNA. After sequencing the BAC clones, two and six NLR genes were identified in clones B1 and B2, respectively. Further fine‐mapping revealed that only the last six were located within the mapping interval and only three (B2‐1, B2‐2, and B2‐3) encoded complete NLR proteins (Figure 1b). The three candidates were subcloned including their native 5′ and 3′ regulatory elements, and complementation analyses were performed in Nicotiana benthamiana. After 2 days, the agroinfiltrated area was challenged with P. infestans 90128. Rpi‐blb1, which was shown to provide resistance to P. infestans, was used as a positive control. The leaves agroinfiltrated with candidate B2‐3 and Rpi‐blb1 showed severely compromised pathogen growth, while leaves with candidates B2‐1 and B2‐2 were completely susceptible to P. infestans 90128 (Figure 1c). This result suggested that B2‐3 is the gene in CHC543‐5 that provides resistance to P. infestans. To verify this result, the three candidates B2‐1, B2‐2, and B2‐3 were stably transformed into the susceptible S. tuberosum cv. ‘Desiree’. Indeed, only the events containing candidate B2‐3 showed resistance to P. infestans (Figure 1d). Furthermore, three different single guide RNAs (sgRNAs) were designed to specifically target the LRR domain of candidate B2‐3 (Figure 2a). The resistant CHC543‐5 genotype was stably transformed with CRISPR‐Cas9 and these sgRNAs. The transformation events were challenged with P. infestans 90128 and IPO‐C isolates, and 48% of the transformants had become susceptible to both isolates (Table S1; Figure 2b), which suggested that the active late blight resistance gene was specifically and successfully mutated. Four transformants were genotyped and frameshift mutations were found only in the three susceptible transformants (Figure 2c). Therefore, we concluded that B2‐3 is the gene from CHC543‐5 that is causal for late blight resistance. Henceforth, we refer to gene B2‐3 as Rpi‐chc1.1 as it is the first Rpi‐chc1 allele that is identified in S. chacoense.
Figure 1.
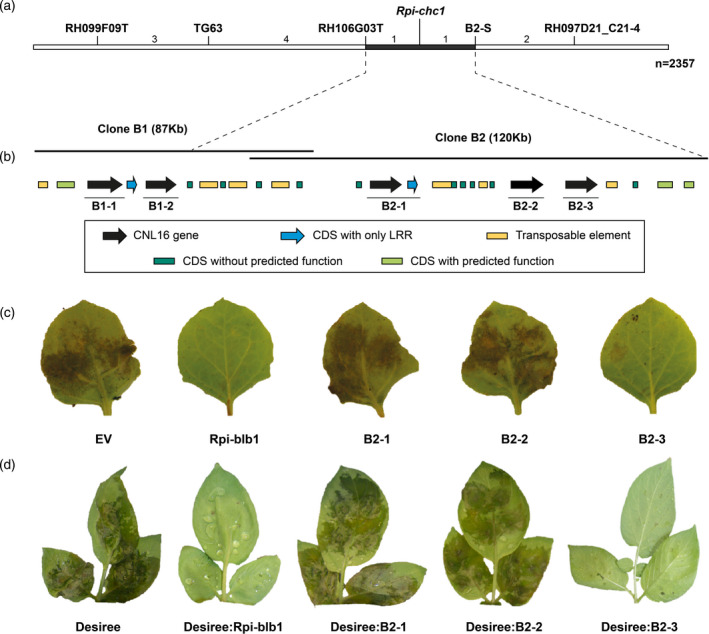
Map‐based cloning of Rpi‐chc1.1.
(a) Genetic map of P. infestans (isolate 90128) resistance from CHC543‐5. The number between the markers represents the number of recombinants found in a population derived from 2357 seedlings. Markers starting with RH were derived from BAC end sequences generated by PGSC. Marker B2‐S represents the BAC end marker from clone 2. The black horizontal line represents the interval of Rpi‐chc1.1. (b) Two BAC clones were isolated to generate the physical map. Annotation revealed the presence of NB‐LRR genes, genes with or without predicted function, and transposable elements. Three complete NB‐LRR (B2‐1, B2‐2, and B2‐3) genes between flanking markers RH106G03T and B2‐S were selected as candidates. (c) The three candidates were expressed through agroinfiltration in N. benthamiana leaves. An empty vector (EV) and Rpi‐blb1 were used as negative and positive controls, respectively. Only candidate B2‐3 was able to compromise the growth of P. infestans isolate 90128. (d) The three candidates were stably transformed into the potato variety Desiree. After inoculation with isolate 90128, only the candidate B2‐3 was able to provide resistance. Untransformed Desiree and Desiree plants stably transformed with Rpi‐blb1 were used as negative and positive controls, respectively.
Figure 2.
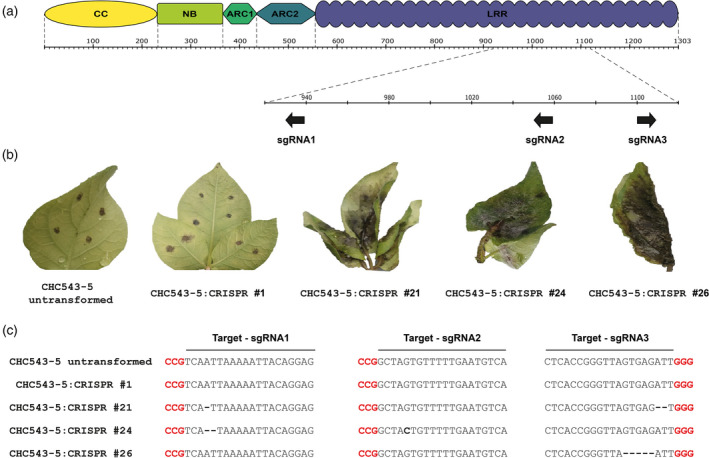
CRISPR‐Cas9‐induced frameshifts in candidate B2‐3 (Rpi‐chc1.1) lead to Phytophthora susceptibility.
(a) Three different sgRNAs were designed to target the LRR domain of candidate B2‐3. (b) A construct with Cas9 and all three sgRNAs was stably transformed into the resistant genotype CHC543‐5. Transformation events were inoculated with P. infestans 90128 and IPO‐C isolates. 48% of the transformation events became susceptible to both isolates (Table S1). (c) One resistant (#1) and three susceptible events (#21, #24, #26) were genotyped together with the untransformed plant CHC543‐5. The three susceptible events contained frameshift mutations, while the tested resistant event had no mutations in the B2‐3 candidate.
Rpi‐chc1.1 encodes a CNL protein that belongs to the CNL16 (Witek et al., 2016) immune receptor family (Figure 3a). Rpi‐chc1.1 has one uninterrupted open reading frame of 3909 bp, which is predicted to be translated into 1303 amino acids (Figure S2). No introns were predicted. The CC domain contains the N‐terminal MADA motif, four predicted α‐helices, and the typical hhGRExE, but the distinctive EDVID motif was less conserved (Rairdan et al., 2008). The NB domain contains the characteristic Kinase 1a VYND, Kinase 2, and Kinase 3a motifs (Campbell, 2003; Leipe et al., 2004; Pal et al., 2007; Saraste et al., 1990; Wendler et al., 2012). The ARC1 domain contains the RNBS‐C motif, Motif 3, and the GLPL motif. The ARC2 domain contains Motif 2, the RNBS‐D motif, and two copies of the MHDL motif (Danot et al., 2009; Reubold et al., 2011; Sukarta et al., 2016). The LRR domain consists of 29 imperfect repeats. Both LRR3 and LRR4 contain a central VLDL motif which is conserved in the third LRR of most functional NLRs (Bendahmane et al., 2002; Warren et al., 1998).
Figure 3.
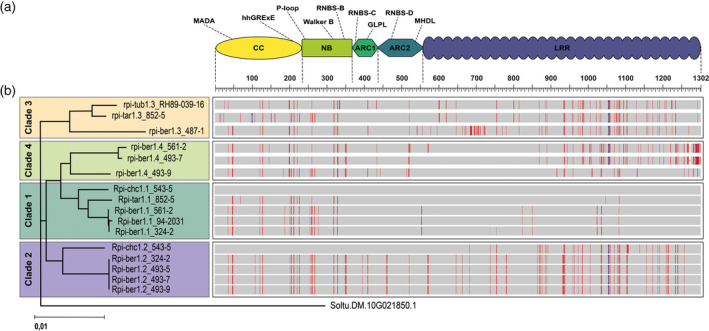
Rpi‐chc1 allele mining.
(a) The Rpi‐chc1.1 allele belongs to the immune receptor family. Different motifs were found in the different CNL receptor domains. The LRR domain consists of 29 imperfect repeats. (b) Sixteen Rpi‐chc1.1‐like sequences were cloned from eleven different diploid Solanum accessions. The phylogenetic analysis of the DNA sequences led to the identification of four clades. The branch lengths represent the percentage of phylogenetic distance. In the protein alignment mismatches are highlighted in red, gaps are indicated by dashes (–), and insertions relative to Rpi‐chc1.1 (anchor) are indicated by a blue bracket.
Identification of Rpi‐chc1.1 allelic variants
In order to identify different Rpi‐chc1.1 allelic variants, we pursued an allele‐mining approach in closely related resistant and susceptible S. chacoense, S. berthaultii, Solanum tarijense, and S. tuberosum accessions. Homologous sequences were amplified using primers overlapping with the start and stop codons of Rpi‐chc1.1. The PCR fragments of the expected 3.9 kb size were cloned and sequenced, resulting in the identification of 15 Rpi‐chc1.1‐like sequences. The mined Rpi‐chc1.1 variants contained between 1296 and 1303 amino acids (Figure 3b; Figure S2). From the selected diploid accessions one or two sequence variants were identified, suggesting that indeed Rpi‐chc1 alleles were mined rather than paralogs. In case only one variant was mined from an accession, this suggested that the second allele has significant sequence polymorphisms at (one of) the primer annealing sites. Phylogenetic analysis of the sequences showed strong sequence similarities among the alleles (94.6–100% identity). Even within this high identity range, the presence of four main clades was revealed (Figure 3b). In clade 1, the Rpi‐chc1.1 allele was found, together with three sequences from S. berthaultii that were nearly identical to each other and a sequence from S. tarijense. From clade 1, together with Rpi‐chc1.1, we selected one sequence from S. berthaultii (94‐2031) and one from S. tarijense (TAR852‐5) for complementation analysis. Transformation of the corresponding genes to susceptible Desiree plants showed that they provide resistance to P. infestans isolates 90128 and IPO‐C, like Rpi‐chc1.1 (Table S2). We therefore concluded that clade 1 contains functional homologs of Rpi‐chc1.1. The S. tarijense allele will be referred to as Rpi‐tar1.1. The S. berthaultii allele will be referred to as Rpi‐ber1.1, which matches to the previously described Rpi‐ber and Rpi‐ber1 genes that were derived from the same accession (PI473331) at similar genetic positions (Rauscher et al., 2006; Tan et al., 2010; Vossen et al., 2013).
The allele mining in accession CHC543‐5 resulted not only in the re‐identification of the active Rpi‐chc1.1, but also in the identification of a presumed allelic variant, which we will refer to as Rpi‐chc1.2. To test if Rpi‐chc1.1 and Rpi‐chc1.2 were indeed alleles of the same gene, we tested Rpi‐chc1.2‐specific markers in the recombinant population 6750 (CHC543‐5 × CHC544‐5). We found a perfect repulsion between Rpi‐chc1.2 and Rpi‐chc1.1, strongly suggesting that both genes are allelic variants (Table S3). Additionally, this analysis proved that Rpi‐chc1.2 does not cause resistance against P. infestans 90128, even though Rpi‐chc1.2 is expressed during infection (Table S2; Figure S1f). The Rpi‐chc1.2 protein sequence clusters in clade 2 together with four identical sequences from S. berthaultii. Close to clade 2, we can observe clade 3, which consisted of an S. berthaultii, an S. tarijense, and an S. tuberosum allele from RH89‐039‐16, a diploid clone previously characterized as susceptible to P. infestans (Vleeshouwers et al., 2011a). The clade 3 allele from S. tarijense contained an in‐frame stop codon, making it unlikely that this allele produces an active resistance protein. Clade 4 contained only S. berthaultii alleles. The allelic variants were numbered according to the clade in which they were found (i.e., Rpi‐ber1.1 from clade 1, Rpi‐ber1.2 from clade 2, etc.), followed by an extension to indicate the genotype from which the allele was derived.
Rpi‐chc1.1 recognizes the RXLR PexRD12 effector family from P. infestans
To understand the resistance mechanism of the S. chacoense CHC543‐5 accession, we searched for the effector recognized by Rpi‐chc1.1. A collection of 90 P. infestans extracellular (Pex) proteins in a PVX agroinfectious vector, of which 54 contained the RXLR‐DEER motif (PexRD), was screened. Individual clones from the Pex collection were co‐agroinfiltrated with Rpi‐chc1.1 in N. benthamiana leaves. As a positive control, we used a mix of the R3a/Avr3a R gene effector pair, which is known to trigger a strong hypersensitive response (HR) in N. benthamiana leaves. Only two effectors from the Pex collection, PexRD12‐1 and PexRD12‐2 (PITG_16233 and PITG_16240, respectively), were able to trigger an Rpi‐chc1.1‐dependent HR (Figure 4a). Neither the inactive paralogs B2‐1 and B2‐2 nor R3a produced an HR upon co‐agroinfiltration with PexRD12. These results showed that PexRD12 is specifically recognized by Rpi‐chc1.1. We could further confirm this finding using transgenic Desiree potato plants that were transformed with Rpi‐chc1.1. About half of this transgenic population showed late blight resistance, while the other half was susceptible, probably due to impaired transgene expression. Interestingly, the plants that showed late blight resistance also showed PexRD12 recognition, while the susceptible transgenic plants did not show any response upon PexRD12 agroinfiltration (Table S4). We sought for further evidence that PexRD12 was indeed causing avirulence on Rpi‐chc1.1‐expressing plants. In a field trial with natural infection, we found isolates that were virulent on plants containing Rpi‐chc1.1. The infected material was collected and used for gene expression analysis via quantitative reverse transcription‐PCR (RT‐qPCR). PexRD12 expression was not detected in the Rpi‐chc1.1 resistance‐breaking isolates, while other effector genes such as Avrsto1 were normally expressed (Vleeshouwers et al., 2008). Reciprocally, we found that Rpi‐sto1 breaking isolates still expressed PexRD12 normally (Figure 4b). Altogether, these results suggest that PexRD12 corresponds to Avrchc1.1.
Figure 4.
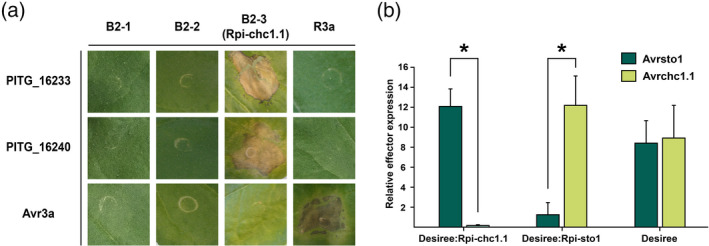
The RXLR effector PexRD12 corresponds to Avrchc1.1.
(a) The three Rpi‐chc1.1 candidates were co‐agroinfiltrated with the Pex effector collection in N. benthamiana leaves to screen for Avrchc1.1. Rpi‐chc1.1 induces cell death when co‐expressed with both PITG_16233 and PITG_16240, from the PexRD12 family. R3a and Avr3a were used as negative controls, and a mix of R3a and Avr3a was used as a positive control. (b) Relative Avrchc1.1 and Avrsto1 effector expression was measured in infected plant material from a field trial with natural P. infestans infection (2013, Wageningen). Untransformed Desiree plants and Desiree plants transformed with Rpi‐chc1.1 (Desiree:Rpi‐chc1.1) or Rpi‐sto1 (Desiree:Rpi‐sto1) were used in this study. Relative expression data were obtained by dividing the relative effector gene expression by the expression of the P. infestans elongation factor 2 (ef2) gene. Three independent samples were included in the RT‐qPCR experiment. Stars represent a statistically significant difference (two‐sample t‐test, P < 0.008).
The PexRD12/31 superfamily is a complex P. infestans RXLR effector family
Using Blast analyses of the T30‐4 proteome, we found nine homologs of PexRD12 in the P. infestans T30‐4 genome. Additionally, we found that PexRD12 proteins had strong homology with nine members of the PexRD31 family and two additional, more distantly related sequences (Table S5). These 20 effectors are encoded by clusters of paralogs mainly in three supercontigs (Figure S3) and will henceforth be referred to as the PexRD12/31 superfamily (see also Petre et al., 2020). All PexRD12/31 effectors are small proteins that include a signal peptide in the N‐terminus, an effector domain in the C‐terminus, and the conserved RXLR and EER motifs in the center, except for PITG_16243 and PITG_09577, which contain RXXR‐EER and RXXLR‐EER motifs, respectively (Figure 5a).
Figure 5.
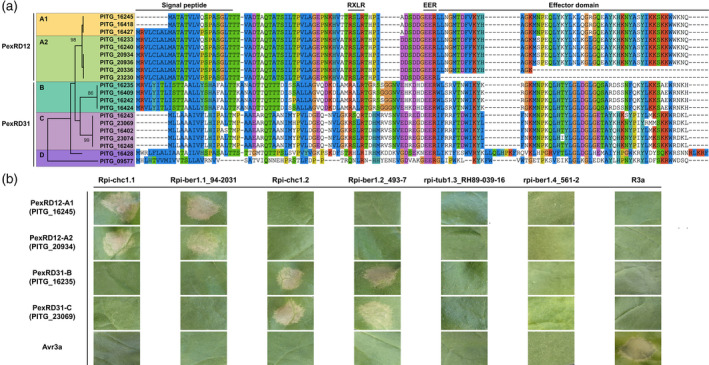
Rpi‐chc1 alleles show non‐overlapping recognition of the PexRD12/31 effector superfamily.
(a) Twenty members of the PexRD12/31 superfamily were found in P. infestans isolate T30‐4. In the amino acid sequence, we can distinguish a signal peptide in the N‐terminus, the conserved RXLR‐EER motifs in the center, and the effector domain in the C‐terminus. Some PexRD12/31 family members differed at the nucleotide level but were identical at the protein level (PITG_16245 = PITG_16418; PITG_16233 = PITG_16240; PITG_20934 = PITG_20936; PITG_16409 = PITG_16424). The phylogenetic analysis of the complete protein sequences led to the identification of five clades. This analysis was performed in mega x by using the maximum likelihood method based on the JTT matrix‐based model. The tree with the highest log‐likelihood (−766) is shown. The bootstrapping values, which indicate the percentage of trees that had the particular branch, are shown in each branch. In the protein alignment: blue, hydrophobic residues (A, I, L, M, F, W, V, and C); red, positively charged residues (K and R); magenta, negatively charged residues (E and D); green, polar residues (N, Q, S, and T); pink, cysteine residues (C); orange, glycine residues (G); yellow, proline residues (P); cyan, aromatic residues (H and Y); and white, unconserved residues or gaps. (b) Different Rpi‐chc1 allelic variants were co‐agroinfiltrated in N. benthamiana with a member from each PexRD12/31 clade. While variants from clade 1 recognize both PexRD12‐A1 and ‐A2 clades, Rpi‐chc1 variants from clade 2 recognize PexRD31‐B and ‐C. Receptors from clades 3 and 4 do not recognize any PexRD12/31 effector. A mix of R3a and Avr3a was used as a positive control.
The alignment of the protein sequences and the phylogenetic analysis of the PexRD12/31 superfamily members resulted in five main clades (Figure 5a). Two highly homologous clades, PexRD12‐A1 and PexRD12‐A2, can be distinguished to form the PexRD12 family. Clade PexRD12‐A2 also includes truncated versions which partly or completely miss the effector domain. In addition, two related clades, PexRD31‐B and PexRD31‐C, constitute the PexRD31 family. Additionally, PITG_16428 and PITG_09577 were much less related and are together referred to as PexRD12/31 group D.
To determine the degree to which PexRD12/31 members are expressed in planta, as observed for other Avr effectors of P. infestans (Rietman et al., 2012; Vleeshouwers et al., 2011b), we tested their expression during infection with qPCR on cDNA using clade A‐, B‐, and C‐specific primers. The relative expression was calculated and normalized for the relative amount of P. infestans. Three different P. infestans isolates were evaluated at different time points after inoculation of different susceptible potato genotypes (Figure S1a–d). In all the tested genotypes, PexRD12 showed the highest relative expression. In two isolates maximum expression was found from 4 to 24 h after inoculation and expression remained high until after 48 h in all four isolates. The PexRD31‐B effectors were expressed in two isolates but were rapidly downregulated in the first hours after inoculation with hardly any expression left. PexRD31‐C expression was mostly undetectable along the inoculation time course. Similar results were observed when PexRD12/31 expression was analyzed during infection of the four different P. infestans isolates EU_13_A2, Ec1, EU_6_A1, and US23 (Figure S4), using the data from the PenSeq dataset (Lin et al., 2020).
Rpi‐chc1.2 recognizes the RXLR PexRD31 effector family from P. infestans
In order to describe the spectrum of effector recognition by different Rpi‐chc1 alleles, several representatives from each clade were selected and co‐agroinfiltrated with different PexRD12/31 members in N. benthamiana. Rpi‐chc1.1_543‐5 and Rpi‐ber1.1_94‐2031‐01 from clade 1, Rpi‐chc1.2_543‐5 and Rpi‐ber1.2_493‐7 from clade 2, rpi‐tub1‐RH89‐039‐16 from clade 3, and rpi‐ber1.4_561‐2 from clade 4 were selected. As a representation from each of the clades of the PexRD12/31 effector superfamily, we selected PITG_16245 (PexRD12‐A1), PITG_20934 (PexRD12‐A2), PITG_16235 (PexRD31‐B), and PITG_23069 (PexRD31‐C). The different Rpi‐chc1 alleles were co‐agroinfiltrated with the PexRD12/31 effectors in N. benthamiana leaves. Three days after agroinfiltration, we observed that the members from clade 1, Rpi‐chc1.1 and Rpi‐ber1.1, specifically recognized both PexRD12‐A1 and PexRD12‐A2 effectors (Figure 5b). This result showed that Rpi‐chc1.1 and Rpi‐ber1.1 recognize multiple members of the PexRD12 family, suggesting that Avrchc1.1 is encoded by multiple redundant paralogs. On the other hand, Rpi‐chc1.2 and Rpi‐ber1.2 from clade 2 specifically recognized both PexRD31‐B and PexRD31‐C effectors (Figure 5b). This suggests that multiple PexRD31 paralogs correspond to Avrchc1.2. The selected alleles from clades 3 and 4, rpi‐tub1.3_RH89‐039‐16 and rpi‐ber1.4_561‐2, were not able to recognize any of the PexRD12/31 members (Figure 5b), showing that clades 3 and 4 encode more functionally distant receptors, in agreement with the known susceptibility of RH89‐039‐16 to P. infestans (Vleeshouwers et al., 2011a).
The LRR domain of the Rpi‐chc1 variants determines the PexRD12/31 effector recognition specificity
Since the allelic variants of Rpi‐chc1 could be divided into three activity groups (recognition of Avrchc1.1, Avrchc1.2, or none), while having an amino acid identity up to 96%, they provide ideal tools to investigate the Rpi‐chc1 mechanism of recognition. Therefore, we performed progressive exchanges of the different receptor domains. The chimeric receptors were co‐agroinfiltrated with the PexRD12/31 effectors in N. benthamiana leaves to evaluate their recognition specificity. First, we selected Rpi‐chc1.2 and rpi‐tub1.3_RH89‐039‐16 as representatives of clades B and C, respectively. When aligning the protein sequences, 54 single amino acid polymorphisms (SAPs) were found and most of them were located in the LRR domain (Figure 6a). As previously mentioned, Rpi‐chc1.2 specifically recognizes PexRD31‐B and PexRD31‐C, while rpi‐tub1.3_RH89‐039‐16 does not recognize any of the PexRD12/31 effectors. When the complete rpi‐tub1.3_RH89‐039‐16 LRR domain was exchanged for the Rpi‐chc1.2 LRR, the chimeric receptor RH::C2_2‐29 was able to recognize both PexRD31‐B and PexRD31‐C. Reciprocally, the exchange of the Rpi‐chc1.2 LRR for the rpi‐tub1.3_RH89‐039‐16 in C2::RH_2‐29 led to the inability to recognize any of the PexRD12/31 effectors. This result demonstrates the importance of the LRR domain during Avrchc1.2 recognition. Additional domain exchanges were performed in order to identify the essential LRRs for effector recognition. The LRRs required for Avrchc1.2 recognition could be narrowed down with the construct RH::C2_14‐19 to nine amino acid polymorphisms (Figure 6a). Due to the absence of polymorphisms in LRRs 14 and 15, we can conclude that the exchange of the amino acid polymorphisms present in LRRs 16 to 19 activates rpi‐tub1.3_RH89‐039‐16 to recognize Avrchc1.2. Interestingly, the majority of these nine amino acid polymorphisms are particularly situated in the solvent‐exposed domain (xxLxLxxxx) of every LRR. The exchange of the tested solvent‐exposed residues led to the partial or complete loss of effector recognition, suggesting the contribution of an effector‐binding surface (Figure 6b).
Figure 6.
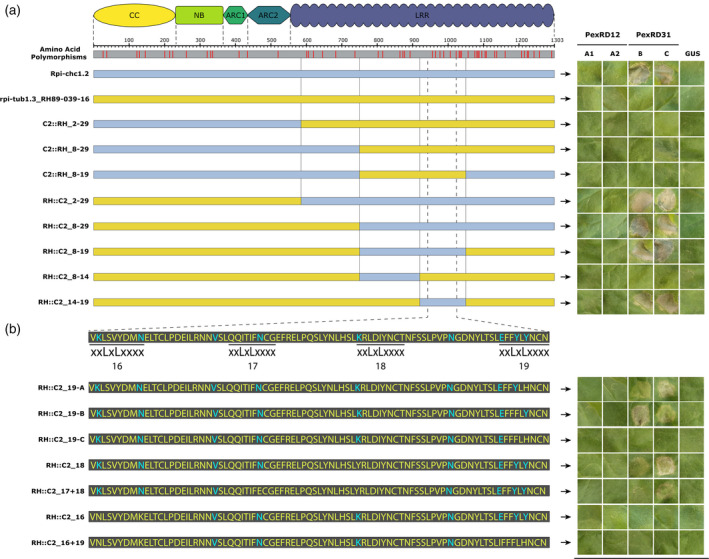
Domain exchanges between Rpi‐chc1.2 and rpi‐tub1.3_RH89‐039‐16.
(a) The positions of SAPs and the corresponding protein domains are indicated on top. Rpi‐chc1.2 and rpi‐tub1.3_RH89‐039‐16 are represented as light blue and yellow bars, respectively. Below, the domain exchanges are shown. The chimeric constructs were co‐agroinfiltrated with the PexRD12/31 effectors in N. benthamiana leaves. After 4 days, the HR was visible and recorded. Experiments were repeated three times with 12 inoculation sites each time. A representative leaf for the response of each chimeric construct is shown in the right panel. GUS was used as a negative control. It is concluded that the exchange of the complete LRR domain led to recognition of PexRD31. With the final construct, RH::C2_14‐19, the exchange of only nine amino acids led to the activation of the rpi‐tub1.3_RH89‐036‐16 protein. (b) Seven new modified receptors were derived from RH::C2_14‐19 in order to pinpoint the amino acids involved in the Rpi‐chc1.2 recognition specificity. SAPs present in Rpi‐chc1.2 are highlighted in blue font. Most of the SAPs are located in the solvent‐exposed xxLxLxxxx motifs of LRRs 16–19. The chimeric constructs were co‐agroinfiltrated with the PexRD12/31 effectors in N. benthamiana leaves. A representative leaf for the response of each chimeric construct is shown in the right panel. Experiments were repeated three times with 12 inoculation sites each time. GUS was used as a negative control. The modification of the tested residues of the Rpi‐chc1.2 solvent‐exposed specific amino acids (blue) for the corresponding amino acid present in rpi‐tub1.3_RH89‐039‐16 (yellow) led to the partial or complete loss of effector recognition.
To understand the difference in effector recognition specificity between Rpi‐chc1.1 and Rpi‐chc1.2 and to explore the possibility to combine both recognitions in one receptor, we performed a similar progressive domain exchange approach between Rpi‐chc1.1 and Rpi‐chc1.2 (Figure 7). The exchange of the LRR domain in the chimeric receptors C1::C2_8‐29 and C2::C1_8‐29 led to a shift in effector recognition, from Avrchc1.1 to Avrchc1.2. Further exchanges revealed that LRRs 14 to 23 from Rpi‐chc1.2 led to an opposite effector recognition pattern as the chimeric receptor C1::C2_14‐23 was only able to recognize Avrchc1.2. With reciprocal domain exchanges of Rpi‐chc1.1 into Rpi‐chc1.2, we found that LRRs 8 to 29 led to Avrchc1.1 recognition. In an attempt to further reduce the length of the exchanged sequence, the recognition of Avrchc1.1 resulted in partial (C2::C1_8‐25 and C2::C1_8‐23) or complete (C2::C1_14‐25 and C2::C1_14‐23) loss of recognition. Especially, when comparing the receptors C2::C1_8‐29 and C2::C1_8‐25, already the modification of the last five SAPs led to the reduced recognition of Avrchc1.1. But, apparently not only the last LRRs are involved in effector recognition; also the first LRRs, from 8 to 14, are important for Avrchc1.1 recognition as C2::C1_8‐25 was able to partially recognize Avrchc1.1, while C2::C1_14‐25 did not trigger any HR. We conclude that LRRs 8 to 29 in Rpi‐chc1.1 are important for Avrchc1.1 recognition, which overlap with LRRs 16–19 from Rpi‐chc1.2, which are required for Avrchc1.2 recognition.
Figure 7.
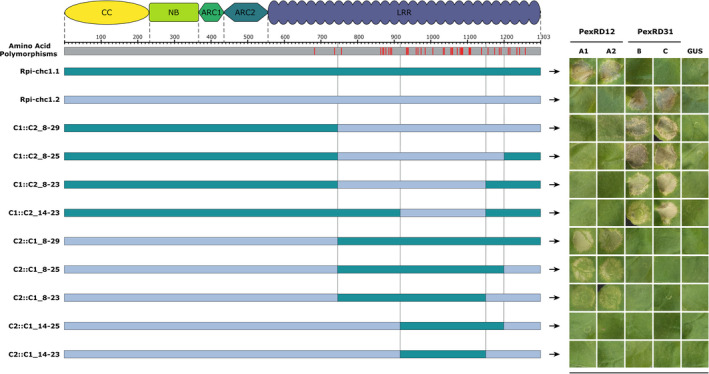
The effector recognition specificity could be exchanged between Rpi‐chc1.1 and Rpi‐chc1.2.
The alignment of Rpi‐chc1.1 and Rpi‐chc1.2 shows that all the 41 amino acid polymorphisms (red bars) are located in the LRR domain. The chimeric constructs were co‐agroinfiltrated with the PexRD12/31 effectors in N. benthamiana leaves. A representative leaf for the response of each chimeric construct is shown in the right panel. Experiments were repeated three times with 12 inoculation sites each time. GUS was used as a negative control. In construct C1::C2_14‐23, LRRs 16–19 appear again, determining PexRD31 recognition. The required domain exchanges of the Rpi‐chc1.1 LRR are more complex and encompass almost the complete LRR.
DISCUSSION
In this study, we identified Rpi‐chc1.1 and 15 additional allelic variants from S. chacoense, S. berthaultii, S. tarijense, and S. tuberosum. Phylogenetic analysis of the encoded protein sequences revealed four clades. These four clades were not only supported by sequence similarity but also by differences in effector and P. infestans recognition. Clade 1 genes encode receptors that recognize PexRD12 effectors and includes the active orthologs Rpi‐chc1.1, Rpi‐ber1.1, and Rpi‐tar1.1 (Figures 3 and 5). Clade 2 receptors could be distinguished by the recognition of the PexRD31 effectors (Figure 5). Receptors encoded by clades 3 and 4 do not recognize PexRD12/31 effectors and no other activity has been found. Interestingly, clade 3 alleles are also present in domesticated potato clones that are susceptible to late blight, e.g., RH89‐039‐16 (Figure 3) and the varieties Colomba and Altus (to be published elsewhere), implying that the encoded receptors are not able to effectively provide resistance against P. infestans.
Rpi‐ber1.1_94‐2031‐01 was derived from the same accession as the previously described Rpi‐ber (Rauscher et al., 2006; Tan et al., 2010; Vossen et al., 2009) and Rpi‐ber1 genes (Park et al., 2009). In these four studies, Rpi‐ber and Rpi‐ber1 mapped close to marker TG63 but slightly different genetic positions were reported. The population from Park et al. was quite small and a single recombination event may have caused the deviating genetic distance. In the case of Tan et al., a single mis‐phenotyping could explain the mapping of Rpi‐ber distal to TG63. We therefore assume that Rpi‐ber and Rpi‐ber1 are the same genes and adopt the Rpi‐ber1 naming as it is more consistent with current nomenclature for late blight resistance genes. Rpi‐ber2, as described by Park et al., was derived from the same accession that was used in our allele‐mining studies (BER493). We could not find a clade 1 Rpi‐chc1 allele from the BER493 accessions, which supports the idea that a more distantly related CNL16 member may be present that lacks sufficient match to the primer sequences, explaining the Rpi‐ber2 map position distal from TG63.
The presence of Rpi‐chc1 alleles in S. tarijense and S. berthaultii suggests a functional common ancestor existed before their speciation. However, it must be noted that the geographic locations where the accessions were found are close to each other in Bolivia. Since S. chacoense, S. tarijense, and S. berthaultii are closely related, the presence of functional Rpi‐chc1 alleles in the three species might be a result from a recent species intercrossing.
Sequence similarity among the studied allelic variants correlated with their functionality, deduced by their ability to provide late blight resistance and P. infestans effector recognition (Figures 4 and 5). This is not the first described case of R gene allelic variants across Solanum species. Rpi‐blb1, Rpi‐sto1, and Rpi‐pta1, from the Mexican species S. bulbocastanum, Solanum stoloniferum, and Solanum papita are allelic variants that recognize the same IpiO or PexRD6 P. infestans effector (Vleeshouwers et al., 2008). Among allelic variants of late blight resistance genes (i.e., Rpi‐blb3 and Rpi‐hjt1 that recognize Avr2 effectors), overlapping recognition specificities have been previously described (Champouret, 2010) but also more recently (Witek et al., 2021). Moreover, highly similar, but non‐allelic R genes from the same CNL cluster had different recognition specificities, i.e., Rpi‐vnt1, Rpi‐mcq1, R9a, Ph‐3 (Foster et al., 2009; Jo et al., 2015; Smilde et al., 2005; Zhang et al., 2014). In the current report, we describe for the first time that allelic variants of a late blight resistance gene show non‐overlapping effector recognition specificities in the combinations that were tested. Remarkably, the recognized effectors belong to the same effector family, which is a further refinement of our insight in host–pathogen co‐evolution.
When studying the Rpi‐chc1 protein domain structure, we identified most of the conserved CNL motifs. Remarkably, the MADA motif (Adachi et al., 2019) was not located downstream of the starting methionine, but downstream of the second methionine in position 46 of the Rpi‐chc1 protein. Further research is needed to show if either or both methionines are used as translational start codons. Interestingly, we recently cloned the functional late blight resistance gene from the late blight‐resistant variety Carolus (Rpi‐Carolus gene, to be published elsewhere). Rpi‐Carolus differs only at seven amino acid positions from Rpi‐ber1, but its N‐terminus is shorter as a stop codon is present between the first two methionine codons. This strongly suggests that translation in Rpi‐Carolus starts from the second methionine while retaining biological activity.
In contrast to the relatively conserved N‐termini of the proteins encoded by the Rpi‐chc1 alleles, most interallelic sequence variation localized to the LRR regions. Through domain interchange between the rpi‐tub1.3_RH89‐039‐16 and Rpi‐chc1.2 alleles and between Rpi‐chc1.1 and Rpi‐chc1.2, we discovered that the LRR domain defines recognition specificity (Figures 6 and 7). Polymorphisms in the LRRs of some NLR receptors were previously shown to determine the effector recognition specificity (Catanzariti et al., 2010; Dodds et al., 2001; Krasileva et al., 2010; Ravensdale et al., 2012; Shen et al., 2003). In one example, a domain exchange between Rx1 and Gpa2 converted the virus resistance into nematode resistance, and vice versa (Slootweg et al., 2017). The recognition of both nematode and virus could not be combined into one chimeric receptor, as we also observed with Rpi‐chc1.1 and Rpi‐chc1.2. The reason for this is the overlap between the LRRs involved in recognition.
Most of the amino acids in Rpi‐chc1.2 that are required for Avrchc1.2 recognition are located in the LRR solvent‐exposed motif (xxLxLxxxx), and modification of the tested solvent‐exposed amino acids led to the partial or complete loss of PexRD31 recognition (Figure 6b). The co‐requirement of these solvent‐exposed amino acids suggests that they are involved in recognition of a particular epitope. This observation, combined with the observation of unequal distribution of SAPs, allow us to hypothesize that Rpi‐chc1 alleles evolved through insertion of a stretch of DNA into the LRR domain rather than through accumulation of independent mutations. A similar model of evolution was recently proposed for allelic variants of Rpi‐amr1 (Witek et al., 2021). Such insertions may happen through unequal crossing‐over with paralog sequences or through retro‐transposition. Interestingly, the evolution of integrated domains in R genes has been postulated to be caused by an unknown recombination‐ or transposon‐independent translocation mechanism (Bailey et al., 2018). The same mechanism may be active in LRR exchange to evolve recognition of non‐integrated domains like guardees or decoys (Kourelis and van der Hoorn, 2018) or direct effector recognition.
Interestingly, some of the PexRD31 family members have been previously identified as one of the most rapidly diversifying and fast evolving RXLR effectors in the T30‐4 genome, with ⍵ values higher than 1.55 (Haas et al., 2009). Additionally, several members of the PexRD12/31 superfamily have recently been characterized to target the host vesicle trafficking machinery by interacting with the vesicle‐associated membrane protein 72 family (Petre et al., 2020). Even though both PexRD12 and PexRD31 have the same or functionally similar host targets, they are differentially expressed during P. infestans infection. While PexRD12 is highly expressed in all the tested isolates, PexRD31 is expressed at low levels after contact with potato (Figures S1 and S4). This would also explain why Rpi‐chc1.2 is not able to provide resistance against P. infestans, since most of the isolates have low or undetectable expression levels of Avrchc1.2 (Figure S4). Consequently, clade A (PexRD12) may have evolved to avoid detection by Rpi‐chc1.2 while retaining its targeting of the vesicle trafficking machinery.
Another step in the co‐evolution between Rpi‐chc1.1 and the PexRD12/31 family was found by analyzing the effector expression in plants expressing Rpi‐chc1.1. The isolates that overcome the Rpi‐chc1.1 resistance no longer express PexRD12, while the expression in untransformed Desiree plants was normal and comparable to the expression of Avrsto1 (Figure 4b). Similarly, evasion of recognition through transcriptional suppression was previously observed in plants expressing Rpi‐vnt1 infected with P. infestans (Pel, 2010). Once more, we confirmed the plasticity of the P. infestans effector secretion and the fast evolution capacity of some aggressive isolates to break down single Rpi resistances.
The introgression of single R genes drives P. infestans to evolve and evade recognition. In order to durably deploy late blight resistance in agriculture, we need novel strategies informed by knowledge of disease resistance genes in varieties, their recognition specificities, and the presence of the cognate effectors in the pathogen populations. Virulence information from the field must be rapidly translated to decision support systems (DSSs) for the risk prediction and calculation of biocide spraying intervals. Additionally, DSSs can be used to determine the R gene composition of (novel) varieties to be deployed in the next season. To meet these requirements, novel breeding strategies are needed to rapidly tailor the R gene contents of the potato varieties to the pathogen populations. In current breeding schemes, it takes 10–15 years to select a late blight‐resistant potato variety. Moreover, susceptible varieties with dominant market shares will not be easy to replace. A system of varieties with flexible R gene content is needed. In other crops this has been accomplished through F1 hybrid varieties. In potato, this route has only recently been opened (Su et al., 2020) and no hybrid potato varieties have reached the market yet. A proof of principle for flexible late blight resistance varieties produced through cisgenesis was provided several years ago (Haverkort et al., 2016). Unfortunately, the EU legislation does not distinguish between cisgenic and transgenic products, making it impossible to market cisgenic varieties. In other parts of the world similar improvement technologies are being deployed to obtain late blight resistance (Ghislain et al., 2019; Habig et al., 2018). It is promising that many different national authorities now apply or consider separate oversight regulations for events that have been enriched with ‘cisgenes’ or ‘genes with a history of safe use’. Knowledge as obtained in this study is essential to pursue such enrichment strategies, which can be achieved through transformation of cisgenes or though novel gene editing tools. We now know how inactive resistance genes from susceptible varieties could be repaired by replacing minimal fragments with the corresponding fragments of alleles from wild relatives. This would provide an unprecedented accuracy and speed which is not present in introgression breeding.
EXPERIMENTAL PROCEDURES
Plant materials and growth conditions
The wild Solanum species used in this study are listed in Table S6 (Tan et al., 2010; Vleeshouwers et al., 2011a). The potato plants were maintained in vitro on MS20 at 24°C under a 16/8‐h day/night regime (Domazakis et al., 2017). The 7650 F1 population was generated by crossing S. chacoense (CHC543‐5) × S. chacoense (CHC544‐5). Solanum tuberosum cv. ‘Desiree’ was used for stable transformations of the different Rpi‐chc1.1 candidates. Four‐week‐old N. benthamiana leaves were used for agroinfiltration. The agroinfiltrated plants were kept in climate‐regulated greenhouse compartments of Unifarm (Wageningen University & Research) at 20–25°C and under a 16/8‐h day/night regime.
BAC clone isolation and sequencing
The procedure has been described in patent US9551007B2. Briefly, two different BAC libraries were produced using partial digestion of CHC543‐5 genomic DNA with HindIII. Fragments larger than 100 kb were ligated into pBeloBAC or pCC1BAC arms (Epicenter). The BAC clones were collected and stored as bacterial pools of approximately 700–1000 white colonies. BAC pools were screened with selected markers and individual clones were identified using colony PCR. The ends of positive individual BACs were sequenced for the purpose of fine‐mapping RH106G03T and RH137D14_C37‐7‐4. The complete inserts were sequenced using shotgun sequencing of 2‐kb library fragments generated by partial digestion with EcoRI by Macrogen Inc. (Seoul, South Korea). Assembly of the sequences resulted in contigs as indicated in Figure 1 (GenBank accession number MW383255).
Cloning of Rpi‐chc1 allelic variants and chimeric constructs
The Rpi‐chc1 allelic variants were amplified using genomic DNA from the different wild Solanum species using PCR primers as described in Table S7 and DNA polymerase with proofreading activity. The fragments were cloned into pGEM‐T easy vector (Promega, Leiden, the Netherlands) for sequencing. GenBank submission numbers are provided in Table S6. The Rpi‐ber1.1 and Rpi‐tar1.1 genes were amplified using primers in the promotor and terminator. The resulting PCR fragments were cloned into pBINPLUS‐PASSA (Jo et al., 2016) and were expressed in transgenic Desiree plants under the control of their native regulatory elements. For transient expression analyses, the coding sequences of the allelic variants were cloned under the Rpi‐chc1.1 regulatory elements (900‐bp promotor and 400‐bp terminator) into pDEST using a multisite gateway protocol. Escherichia coli strain DH10ß was transformed with the gateway reaction products and clones with the correct insert were selected. Agrobacterium tumefaciens AGL1+VirG was used for transient and stable transformations of N. benthamiana leaves and S. tuberosum cv. ‘Desiree’.
The chimeric constructs were cloned using the Golden Gate modular cloning principle. As acceptor vector, we used a Golden Gate‐compatible version of pBINPLUS (McBride and Summerfelt, 1990), PBINPLUS‐GG (van de Vossenberg et al., 20192019). The final acceptor vector was constructed to contain 800‐bp Rpi‐chc1.1 promoter::CDS::1000‐bp Rpi‐ber terminator (Figure S5). The different PCR fragments were amplified using the Phusion High‐Fidelity PCR Kit (Thermo Scientific, Ochten, the Netherlands) and primers with BsaI sites as overhang (Table S7) and purified using the DNA Clean&Concentrator Kit (Zymo Research, Freiburg im Breisgau, Germany). PCR fragments and the acceptor vector were incubated in Buffer G (Thermo Scientific) with 1 mm ATP for 30 cycles of 37°C for 5 min and 16°C for 5 min. We included an additional step at 37°C for 10 min to digest the wrongly assembled plasmids and a final step at 65°C for 20 min to heat‐inactivate the BsaI enzyme.
Hypersensitive cell death assays
Transient expression of the different receptors and PexRD12/31 effectors was performed in 4‐week‐old N. benthamiana leaves. R3a/Avr3a was used as a positive control. All the constructs were agroinfiltrated at an OD600 of 0.5. Each construct was agroinfiltrated twice on two leaves of four plants in at least two independent experiments. Cell death responses were observed at 3–4 days post‐inoculation.
Phylogenetic analysis of Rpi‐chc1.1 homologs and the PexRD12/31 superfamily
The sequences of the PexRD12/31 effectors were retrieved from the P. infestans T30‐4 genome (Haas et al., 2009). Twenty family members were found to form the PexRD12/31 superfamily. The coding sequences of the Rpi‐chc1 variants as obtained in this study were aligned using muscle and a neighbor‐joining tree was constructed using Megalign from the DNAstar package. The closest homolog of Rpi‐chc1.1 from the DM reference genome (SoltuDM10G021850.1) was used as an outgroup.
The protein sequences of PexRD12/31 effectors were aligned using Clustal omega and manually edited in mega x (Kumar et al., 2018; Sievers et al., 2011). The phylogenetic relationship was inferred using the maximum likelihood method based on the JTT matrix‐based model in mega x with 1000 bootstraps (Jones et al., 1992). The tree with the highest log‐likelihood is shown. The two more distant effectors PITG_16428 and PITG_09577 served as an outgroup.
Phytophthora infestans isolates and the detached leaf assay
The P. infestans isolates used in this study (90128, IPO‐C, and NL08645) were retrieved from our in‐house collection. Isolates were grown at 15°C on solid rye medium in the dark (Caten and Jinks, 1968). After 2 weeks, sporulating mycelium was flooded with 20 ml of ice‐cold water, adjusted to 70 zoospores/μl, and incubated at 4°C for 2–3 h. After the incubation, the detached leaves were inoculated with 10 μl of the zoospore suspension on the abaxial side of the leaves. Detached leaves were inserted into wet floral foam. For each biological replicate the three leaflets from four leaves from two independent plants were used. Twelve spots on each leaf were inoculated with the zoospore suspension and closed in a plastic bag to maintain high humidity. The leaves were kept in a climate cell at 18°C for 5 days. Disease resistance was scored on a scale from 1 to 10 for each leaflet, where 10 = no symptoms; 9 = HR no larger than the inoculum droplet; 8 = HR lesion of up to 0.5 cm diameter; 7 = diffuse lesions up to 1 cm diameter, no sporulation, no water soaking; 5 = lesions larger than 1 cm sometimes with water soaking, no sporulation; 4 = large water‐soaked lesions with sporulation only visible through binoculars; 2 = large lesions with macroscopically visible sporulation on one side of the leaflet; and 1 = large lesions with macroscopically visible sporulation on both sides of the leaflet.
Relative effector and R gene expression
The P. infestans effectors used in this study are listed in Table S5. The different genotypes were inoculated with the different P. infestans isolates and samples were collected after 0, 3, 8, 24, 48, 72, 96, and 120 h. Infected plant material with the different P. infestans isolates was collected and RNA was isolated using an RNA Purification Kit (Qiagen, Venlo, the Netherlands). The isolated RNA was converted into cDNA using the QuantiTect Reverse Transcription Kit (Qiagen). The primers used in this study are listed in Table S7. The expression of the different effectors in the infected material was evaluated using RT‐qPCR SYBR Green (Bio‐Rad, Hercules, CA, USA). The samples were heated to 95°C for 2 min, followed by 40 cycles of 15 s at 95°C, 30 s at 60°C, and 30 s at 72°C. Fluorescence was measured after each cycle. After the final amplification cycle a melting curve was calculated. Relative gene expression was calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001). Gene expression was normalized by dividing the relative gene expression by the relative expression of the P. infestans elongation factor 2 (ef2) gene.
SgRNA and CRISPR‐Cas9 construct design
The CRISPOR web tool (http://crispor.org) was used to design the sgRNAs with lower off‐target and higher on‐target potentials (Concordet and Haeussler, 2018).
A modular cloning system based on the Golden Gate cloning technology was used to assemble the different sgRNAs and binary vectors as previously described for tomato (Solanum lycopersicum) mutagenesis (Engler et al., 2008; Weber et al., 2011). Briefly, each sgRNA was fused to the Arabidopsis thaliana U6‐26 promoter as AtU6‐26::gRNA. The Level 1 constructs pICH47732‐pNOS::NPTII::tOCS and pICH47742‐p2x35S::hCas9::tNOS and the linker pICH41780 were used to build the Level 2 vector pICSL4723 (Werner et al., 2012). The primers used for cloning the gRNAs are listed in Table S7.
AUTHOR CONTRIBUTIONS
JV planned and designed the research; DML, MN, and LK performed the majority of the experiments; SK provided the Pex‐RD set; DML and JV wrote the manuscript; RV proofread the manuscript and provided the essential research environment; SA, HS, and KS contributed by mapping, cloning, and characterizing Rpi‐chc1 allelic variants; RS, AL, and AAH contributed by identifying Avrchc1 and their differential recognition specificities by Rpi‐chc1 allelic variants.
CONFLICT OF INTEREST
Some results presented in this manuscript have been included in a patent application.
Supporting information
Figure S1. Effector and R gene expression in potato leaves inoculated with P. infestans.
Figure S2. Rpi‐chc1.1 protein domain organization.
Figure S3. Localization of PexRD12/31 effectors in the P. infestans T30‐4 contigs.
Figure S4. PexRD12/31 effector expression in potato leaves infected with P. infestans from the PenSeq dataset.
Figure S5. pBINPLUS‐PASSA‐GG vector map.
Table S1. CRISPR‐Cas9 targeting of the Rpi‐chc1.1 allele in S. chacoense resistant accession CHC543‐5.
Table S2. Late blight resistance assessment of different Rpi‐chc1 alleles.
Table S3. Segregation of markers and late blight resistance of Rpi‐chc1.1 and Rpi‐chc1.2.
Table S4. Functional expression of Rpi‐chc1.1 in Desiree transgenic events correlates with responsiveness to PexRD12.
Table S5. Phytophthora infestans effectors used in this study.
Table S6. Accession numbers of Solanum genotypes and Rpi‐chc1 sequences.
Table S7. Primers used in this study.
ACKNOWLEDGMENTS
The first part of this research was performed in the DuRPh program, funded by the Ministry of Agriculture, Nature, and Food Quality in the Netherlands. The second part of the research was funded by the Ministry of Infrastructure and Water Management in the Netherlands through the NWO‐TTW program Biotechnology and Safety (project number 15815). We thank Jan de Boer and Adillah Tan for advice in selecting BAC clones and marker sequences from chr10. Evert Jacobsen and Clemens van der Wiel are thanked for their advice about regulation and safety aspects of plant biotechnology. Sidrat Abdullah is thanked for testing late blight resistance of transgenic potato plants harboring Rpi‐chc1 allelic variants. Last but not least, we thank Marjan Bergervoet, Gert van Arkel, Koen Pelgrom, Dirk Jan Huigen, and Isolde Pereira for plant transformations, molecular biology assistance, and plant maintenance.
DATA AVAILABILITY STATEMENT
Supplementary figures and tables are available as supporting information from the TPJ website. All described sequences have been submitted to GenBank. Accession numbers are listed in Table S6.
References
- Adachi, H., Contreras, M.P., Harant, A., Wu, C.‐H., Derevnina, L., Sakai, T.et al. (2019) An N‐terminal motif in NLR immune receptors is functionally conserved across distantly related plant species. eLife, 8, e49956. 10.7554/eLife.49956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera‐Galvez, C., Champouret, N., Rietman, H., Lin, X., Wouters, D., Chu, Z.et al. (2018) Two different R gene loci co‐evolved with Avr2 of Phytophthora infestans and confer distinct resistance specificities in potato. Studies in Mycology, 89, 105–115. 10.1016/j.simyco.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, P.C., Schudoma, C., Jackson, W., Baggs, E., Dagdas, G., Haerty, W.et al. (2018) Dominant integration locus drives continuous diversification of plant immune receptors with exogenous domain fusions. Genome Biology, 19(1), 23. 10.1186/s13059-018-1392-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballvora, A., Ercolano, M.R., Weiss, J., Meksem, K., Bormann, C.A., Oberhagemann, P.et al. (2002) The R1 gene for potato resistance to late blight (Phytophthora infestans) belongs to the leucine zipper/NBS/LRR class of plant resistance genes. The Plant Journal, 30(3), 361–371. 10.1046/j.1365-313X.2001.01292.x. [DOI] [PubMed] [Google Scholar]
- Bendahmane, A., Farnham, G., Moffett, P. & Baulcombe, D.C. (2002) Constitutive gain‐of‐function mutants in a nucleotide binding site‐leucine rich repeat protein encoded at the Rx locus of potato. The Plant Journal, 32(2), 195–204. 10.1046/j.1365-313X.2002.01413.x. [DOI] [PubMed] [Google Scholar]
- Callaway, E. (2013) Pathogen genome tracks Irish potato famine back to its roots. Nature, 10.1038/nature.2013.13021. [DOI] [Google Scholar]
- Campbell, T.A. (2003) Investigation of variations in NBS motifs in alfalfa (Medicago sativa), M. edgeworthii, and M. ruthenica . Canadian Journal of Plant Science, 83(2), 371–376. 10.4141/P02-002. [DOI] [Google Scholar]
- Catanzariti, A.‐M., Dodds, P.N., Ve, T., Kobe, B., Ellis, J.G. & Staskawicz, B.J. (2010) The AvrM effector from flax rust has a structured C‐terminal domain and interacts directly with the M resistance protein. Molecular Plant‐Microbe Interactions®, 23(1), 49–57. 10.1094/MPMI-23-1-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caten, C.E. & Jinks, J.L. (1968) Spontaneous variability of single isolates of Phytophthora infestans. I. Cultural variation. Canadian Journal of Botany, 46(4), 329–348. 10.1139/b68-055. [DOI] [Google Scholar]
- Champouret, N. (2010) Functional genomics of Phytophthora infestans effectors and Solanum resistance genes. PhD Dissertation, Wageningen University. https://edepot.wur.nl/138174 [Google Scholar]
- Concordet, J.‐P. & Haeussler, M. (2018) CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Research, 46(W1), W242–W245. 10.1093/nar/gky354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danot, O., Marquenet, E., Vidal‐Ingigliardi, D. & Richet, E. (2009) Wheel of life, wheel of death: a mechanistic insight into signaling by STAND proteins. Structure, 17(2), 172–182. 10.1016/j.str.2009.01.001. [DOI] [PubMed] [Google Scholar]
- Devaux, A., Goffart, J.‐P., Petsakos, A., Kromann, P., Gatto, M., Okello, J., Suarez, V. & Hareau, G. (2020) Global food security, contributions from sustainable potato agri‐food systems. In: Campos, H. & Ortiz, O. (Eds.) The potato crop. Cham: Springer International Publishing, pp. 3–35. 10.1007/978-3-030-28683-5_1. [DOI] [Google Scholar]
- Dodds, P.N., Lawrence, G.J. & Ellis, J.G. (2001) Six amino acid changes confined to the leucine‐rich repeat β‐strand/β‐turn motif determine the difference between the P and P2 rust resistance specificities in flax. The Plant Cell, 13(1), 163–178. 10.1105/tpc.13.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domazakis, E., Lin, X., Aguilera‐Galvez, C., Wouters, D., Bijsterbosch, G., Wolters, P.J. & Vleeshouwers, V.G.A.A. (2017) Effectoromics‐based identification of cell surface receptors in potato. In: Shan, L. & He, P. (Eds.) Plant Pattern Recognition Receptors. New York, NY: Springer; New York (Methods in Molecular Biology), pp. 337–353. 10.1007/978-1-4939-6859-6_29. [DOI] [PubMed] [Google Scholar]
- Engler, C., Kandzia, R. & Marillonnet, S. (2008) A one pot, one step, precision cloning method with high throughput capability. PLoS One, 3(11), e3647– 10.1371/journal.pone.0003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Food and Agriculture Organization of the United Nations (FAO) (2020) fao.org/faostat/en/#data/QC
- Foster, S.J., Park, T.‐H., Pel, M., Brigneti, G., Śliwka, J., Jagger, L.et al. (2009) Rpi‐vnt1.1, a Tm‐2 Homolog from Solanum venturii, confers resistance to potato late blight. Molecular Plant‐Microbe Interactions®, 22(5), 589–600. 10.1094/MPMI-22-5-0589 [DOI] [PubMed] [Google Scholar]
- Fry, W. (2008) Phytophthora infestans: the plant (and R gene) destroyer. Molecular Plant Pathology, 9(3), 385–402. 10.1111/j.1364-3703.2007.00465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghislain, M., Byarugaba, A.A., Magembe, E., Njoroge, A., Rivera, C., Román, M.L.et al. (2019) Stacking three late blight resistance genes from wild species directly into African highland potato varieties confers complete field resistance to local blight races. Plant Biotechnology Journal, 17(6), 1119–1129. 10.1111/pbi.13042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss, E.M., Press, C.M. & Grünwald, N.J. (2013) Evolution of RXLR‐class effectors in the oomycete plant pathogen Phytophthora ramorum . PLoS One, 8(11), e79347– 10.1371/journal.pone.0079347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, B.J., Kamoun, S., Zody, M.C., Jiang, R.H.Y., Handsaker, R.E., Cano, L.M.et al. (2009) Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans . Nature, 461(7262), 393–398. 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- Habig, J.W., Rowland, A., Pence, M.G. & Zhong, C.X. (2018) Food safety evaluation for R‐proteins introduced by biotechnology: a case study of VNT1 in late blight protected potatoes. Regulatory Toxicology and Pharmacology, 95, 66–74. 10.1016/j.yrtph.2018.03.008. [DOI] [PubMed] [Google Scholar]
- Haverkort, A.J., Boonekamp, P.M., Hutten, R., Jacobsen, E., Lotz, L.A.P., Kessel, G.J.T.et al. (2016) Durable late blight resistance in potato through dynamic varieties obtained by cisgenesis: scientific and societal advances in the DuRPh project. Potato Research, 59(1), 35–66. 10.1007/s11540-015-9312-6. [DOI] [Google Scholar]
- Huang, S., Van Der Vossen, E.A.G., Kuang, H., Vleeshouwers, V.G.A.A., Zhang, N., Borm, T.J.A.et al. (2005) Comparative genomics enabled the isolation of the R3a late blight resistance gene in potato: cloning the potato late blight R3a gene by synteny. The Plant Journal, 42(2), 251–261. 10.1111/j.1365-313X.2005.02365.x. [DOI] [PubMed] [Google Scholar]
- Jo, K.‐R., Zhu, S., Bai, Y., Hutten, R.C.B., Kessel, G.J.T., Vleeshouwers, V.G.A.A.et al. (2016) Problematic crops: 1. Potatoes: towards sustainable potato late blight resistance by cisgenic R gene pyramiding. In: Collinge, D.B. (Ed.) Plant pathogen resistance biotechnology. Hoboken, NJ: John Wiley & Sons Inc, pp. 171–191. 10.1002/9781118867716.ch9 [DOI] [Google Scholar]
- Jo, K.‐R., Visser, R.G.F., Jacobsen, E. & Vossen, J.H. (2015) Characterisation of the late blight resistance in potato differential MaR9 reveals a qualitative resistance gene, R9a, residing in a cluster of Tm‐2 2 homologs on chromosome IX. Theoretical and Applied Genetics, 128(5), 931–941. 10.1007/s00122-015-2480-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, D.T., Taylor, W.R. & Thornton, J.M. (1992) The rapid generation of mutation data matrices from protein sequences. Bioinformatics, 8(3), 275–282. 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Jones, J.D.G., Vance, R.E. & Dangl, J.L. (2016) Intracellular innate immune surveillance devices in plants and animals. Science, 354(6316), aaf6395. 10.1126/science.aaf6395. [DOI] [PubMed] [Google Scholar]
- Jupe, F., Witek, K., Verweij, W., Śliwka, J., Pritchard, L., Etherington, G.J.et al. (2013) Resistance gene enrichment sequencing (RenSeq) enables reannotation of the NB‐LRR gene family from sequenced plant genomes and rapid mapping of resistance loci in segregating populations. The Plant Journal, 76(3), 530–544. 10.1111/tpj.12307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki, H.S., Jansky, S.H. & Halterman, D.A. (2021) Screening of wild potatoes identifies new sources of late blight resistance. Plant Disease, 105(2), 368–376. 10.1094/PDIS-06-20-1367-RE. [DOI] [PubMed] [Google Scholar]
- Kourelis, J. & van der Hoorn, R.A.L. (2018) Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. The Plant Cell, 30(2), 285–299. 10.1105/tpc.17.00579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasileva, K.V., Dahlbeck, D. & Staskawicz, B.J. (2010) Activation of an arabidopsis resistance protein is specified by the in planta association of its leucine‐rich repeat domain with the cognate oomycete effector. The Plant Cell, 22(7), 2444–2458. 10.1105/tpc.110.075358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. (2018) ‘MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35(6), 1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipe, D.D., Koonin, E.V. & Aravind, L. (2004) STAND, a class of P‐Loop NTPases including animal and plant regulators of programmed cell death: multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. Journal of Molecular Biology, 343(1), 1–28. 10.1016/j.jmb.2004.08.023. [DOI] [PubMed] [Google Scholar]
- Leister, D. (2004) Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance genes. Trends in Genetics, 20(3), 116–122. 10.1016/j.tig.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Li, G., Huang, S., Guo, X., Li, Y., Yang, Y., Guo, Z.et al. (2011) Cloning and Characterization of R3b; members of the R3 superfamily of late blight resistance genes show sequence and functional divergence. Molecular Plant‐Microbe Interactions®, 24(10), 1132–1142. 10.1094/MPMI-11-10-0276. [DOI] [PubMed] [Google Scholar]
- Lin, X., Song, T., Fairhead, S., Witek, K., Jouet, A., Jupe, F.et al. (2020) Identification of Avramr1 from Phytophthora infestans using long read and cDNA pathogen‐enrichment sequencing (PenSeq). Molecular Plant Pathology, 21(11), 1502–1512. 10.1111/mpp.12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J. & Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2−ΔΔCT method. Methods, 25(4), 402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lokossou, A.A., Park, T.‐H., van Arkel, G., Arens, M., Ruyter‐Spira, C., Morales, J.et al. (2009) Exploiting knowledge of R/Avr genes to rapidly clone a new LZ‐NBS‐LRR family of late blight resistance genes from potato linkage group IV. Molecular Plant‐Microbe Interactions®, 22(6), 630–641. 10.1094/MPMI-22-6-0630. [DOI] [PubMed] [Google Scholar]
- McBride, K.E. & Summerfelt, K.R. (1990) Improved binary vectors for Agrobacterium‐mediated plant transformation. Plant Molecular Biology, 14(2), 269–276. 10.1007/BF00018567. [DOI] [PubMed] [Google Scholar]
- Mcdowell, J.M. & Simon, S.A. (2006) Recent insights into R gene evolution. Molecular Plant Pathology, 7(5), 437–448. 10.1111/j.1364-3703.2006.00342.x. [DOI] [PubMed] [Google Scholar]
- Pal, A., Chakrabarti, A. & Basak, J. (2007) New motifs within the NB‐ARC domain of R proteins: probable mechanisms of integration of geminiviral signatures within the host species of Fabaceae family and implications in conferring disease resistance. Journal of Theoretical Biology, 246(3), 564–573. 10.1016/j.jtbi.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Park, T.‐H., Foster, S., Brigneti, G. & Jones, J.D.G (2009) Two distinct potato late blight resistance genes from Solanum berthaultii are located on chromosome 10. Euphytica, 165(2), 269–278. 10.1007/s10681-008-9784-4. [DOI] [Google Scholar]
- Pel, M.A., Foster, S.J., Park, T.‐H., Rietman, H., van Arkel, G., Jones, J.D.G.et al. (2009) Mapping and cloning of late blight resistance genes from Solanum venturii using an interspecific candidate gene approach. Molecular Plant‐Microbe Interactions®, 22(5), 601–615. 10.1094/MPMI-22-5-0601. [DOI] [PubMed] [Google Scholar]
- Pel, M.A. (2010) Mapping, isolation and characterization of genes responsible for late blight resistance in potato. Wageningen: Wageningen University. [Google Scholar]
- Petre, B., Contreras, M.P., Bozkurt, T.O., Schattat, M.H., Sklenar, J., Schornack, S.et al. (2020) Host‐interactor screens of Phytophthora infestans RXLR proteins reveal vesicle trafficking as a major effector‐targeted process. preprint. Plant Biology. 10.1101/2020.09.24.308585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rairdan, G.J., Collier, S.M., Sacco, M.A., Baldwin, T.T., Boettrich, T. & Moffett, P. (2008) The coiled‐coil and nucleotide binding domains of the potato Rx disease resistance protein function in pathogen recognition and signaling. The Plant Cell, 20(3), 739–751. 10.1105/tpc.107.056036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauscher, G.M., Smart, C.D., Simko, I., Bonierbale, M., Mayton, H., Greenland, A.et al. (2006) Characterization and mapping of Rpi‐ber, a novel potato late blight resistance gene from Solanum berthaultii . Theoretical and Applied Genetics, 112(4), 674–687. 10.1007/s00122-005-0171-4. [DOI] [PubMed] [Google Scholar]
- Ravensdale, M., Bernoux, M., Ve, T., Kobe, B., Thrall, P.H., Ellis, J.G.et al. (2012) Intramolecular interaction influences binding of the flax L5 and L6 resistance proteins to their AvrL567 ligands. PLoS Path, 8(11), e1003004– 10.1371/journal.ppat.1003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reubold, T.F., Wohlgemuth, S. & Eschenburg, S. (2011) Crystal structure of full‐length Apaf‐1: how the death signal is relayed in the mitochondrial pathway of apoptosis. Structure, 19(8), 1074–1083. 10.1016/j.str.2011.05.013. [DOI] [PubMed] [Google Scholar]
- Rietman, H., Bijsterbosch, G., Cano, L.M., Lee, H.‐R., Vossen, J.H., Jacobsen, E.et al. (2012) Qualitative and quantitative late blight resistance in the potato cultivar Sarpo Mira is determined by the perception of five distinct RXLR effectors. Molecular Plant‐Microbe Interactions®, 25(7), 910–919. 10.1094/MPMI-01-12-0010-R. [DOI] [PubMed] [Google Scholar]
- Saraste, M., Sibbald, P.R. & Wittinghofer, A. (1990) The P‐loop — a common motif in ATP‐ and GTP‐binding proteins. Trends in Biochemical Sciences, 15(11), 430–434. 10.1016/0968-0004(90)90281-F. [DOI] [PubMed] [Google Scholar]
- Sharma, S.K., Bolser, D., de Boer, J., Sønderkær, M., Amoros, W., Carboni, M.F.et al. (2013) Construction of reference chromosome‐scale pseudomolecules for potato: integrating the potato genome with genetic and physical maps. G3: Genes|Genomes|Genetics, 3(11), 2031–2047. 10.1534/g3.113.007153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Q.‐H., Zhou, F., Bieri, S., Haizel, T., Shirasu, K. & Schulze‐Lefert, P. (2003) Recognition specificity and RAR1/SGT1 dependence in barley Mla disease resistance genes to the powdery mildew fungus. The Plant Cell, 15(3), 732–744. 10.1105/tpc.009258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers, F., Wilm, A., Dineen, D., Gibson, T.J., Karplus, K., Li, W.et al. (2011) Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Molecular Systems Biology, 7(1), 539. 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slootweg, E., Koropacka, K., Roosien, J., Dees, R., Overmars, H., Lankhorst, R.K.et al. (2017) Sequence exchange between homologous NB‐LRR genes converts virus resistance into nematode resistance, and vice versa. Plant Physiology, 175(1), 498–510. 10.1104/pp.17.00485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smilde, W.D., Brigneti, G., Jagger, L., Perkins, S. & Jones, J.D.G. (2005) Solanum mochiquense chromosome IX carries a novel late blight resistance gene Rpi‐moc1. Theoretical and Applied Genetics, 110(2), 252–258. 10.1007/s00122-004-1820-8. [DOI] [PubMed] [Google Scholar]
- Su, Y., Viquez‐Zamora, M., den Uil, D., Sinnige, J., Kruyt, H., Vossen, J.H.et al. (2020) Introgression of genes for resistance against Phytophthora infestans in diploid potato. American Journal of Potato Research, 97(1), 33–42. 10.1007/s12230-019-09741-8. [DOI] [Google Scholar]
- Sukarta, O.C.A., Slootweg, E.J. & Goverse, A. (2016) Structure‐informed insights for NLR functioning in plant immunity. Seminars in Cell & Developmental Biology, 56, 134–149. 10.1016/j.semcdb.2016.05.012. [DOI] [PubMed] [Google Scholar]
- Tan, M.Y.A., Hutten, R.C.B., Visser, R.G.F. & van Eck, H.J. (2010) The effect of pyramiding Phytophthora infestans resistance genes R Pi‐mcd1 and R Pi‐ber in potato. Theoretical and Applied Genetics, 121(1), 117–125. 10.1007/s00122-010-1295-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Potato Genome Sequencing Consortium (2011) Genome sequence and analysis of the tuber crop potato. Nature, 475(7355), 189–195. 10.1038/nature10158. [DOI] [PubMed] [Google Scholar]
- van der Vossen, E., Sikkema, A., te Lintel Hekkert, B., Gros, J., Stevens, P., Muskens, M., Wouters, D., Pereira, A., Stiekema, W.et al. (2003) An ancient R gene from the wild potato species Solanum bulbocastanum confers broad‐spectrum resistance to Phytophthora infestans in cultivated potato and tomato. The Plant Journal, 36(6), 867–882. 10.1046/j.1365-313X.2003.01934.x [DOI] [PubMed] [Google Scholar]
- van de Vossenberg, B.T.L.H., Prodhomme, C., van Arkel, G., van Gent‐Pelzer, M.P.E., Bergervoet, M., Brankovics, B.et al. (2019) The Synchytrium endobioticum AvrSen1 Triggers a Hypersensitive Response in Sen1 Potatoes While Natural Variants Evade Detection. Molecular Plant‐Microbe Interactions, 32, 1536–1546. 10.1094/MPMI-05-19-0138- [DOI] [PubMed] [Google Scholar]
- Vleeshouwers, V.G.A.A., Finkers, R., Budding, D., Visser, M., Jacobs, M.M.J., van Berloo, R.et al. (2011a) SolRgene: an online database to explore disease resistance genes in tuber‐bearing Solanum species. BMC Plant Biology, 11(1), 116. 10.1186/1471-2229-11-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vleeshouwers, V.G.A.A., Raffaele, S., Vossen, J.H., Champouret, N., Oliva, R., Segretin, M.E.et al. (2011b) Understanding and exploiting late blight resistance in the age of effectors. Annual Review of Phytopathology, 49(1), 507–531. 10.1146/annurev-phyto-072910-095326. [DOI] [PubMed] [Google Scholar]
- Vleeshouwers, V.G.A.A., Rietman, H., Krenek, P., Champouret, N., Young, C., Oh, S.‐K.et al. (2008) Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. PLoS One, 3(8), e2875. 10.1371/journal.pone.0002875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vossen, E.A.G., Gros, J., Sikkema, A., Muskens, M., Wouters, D., Wolters, P.et al. (2005) The Rpi‐blb2 gene from Solanum bulbocastanum is an Mi‐1 gene homolog conferring broad‐spectrum late blight resistance in potato: Isolation of the late blight resistance gene Rpi‐blb2. The Plant Journal, 44(2), 208–222. 10.1111/j.1365-313X.2005.02527.x. [DOI] [PubMed] [Google Scholar]
- Vossen, J.H.,Nijenhuis M., Arens M, van der Vossen E.A.G., Jacobsen E., & Visser R.G.F. (2009) Cloning and exploitation of a functional R gene from Solanum chacoense . Patent application WO2011034433 A1. Published by the world intellectual property organization 18 September 2009
- Vossen, J.H., Dezhsetan, S., Esselink, D., Arens, M., Sanz, M.J., Verweij, W.et al. (2013) Novel applications of motif‐directed profiling to identify disease resistance genes in plants. Plant Methods, 9(1), 37. 10.1186/1746-4811-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vossen, J.H., van Arkel, G., Bergervoet, M., Jo, K.‐R., Jacobsen, E. & Visser, R.G.F. (2016) The Solanum demissum R8 late blight resistance gene is an Sw‐5 homologue that has been deployed worldwide in late blight resistant varieties. Theoretical and Applied Genetics, 129(9), 1785–1796. 10.1007/s00122-016-2740-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren, R.F., Henk, A., Mowery, P., Holub, E. & Innes, R.W. (1998) A mutation within the leucine‐rich repeat domain of the arabidopsis disease resistance gene RPS5 partially suppresses multiple bacterial and downy mildew resistance genes. The Plant Cell, 10(9), 1439–1452. 10.1105/tpc.10.9.1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, E., Engler, C., Gruetzner, R., Werner, S. & Marillonnet, S. (2011) A modular cloning system for standardized assembly of multigene constructs. PLoS One, 6(2), e16765– 10.1371/journal.pone.0016765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendler, P., Ciniawsky, S., Kock, M. & Kube, S. (2012) Structure and function of the AAA+ nucleotide binding pocket. Biochimica et Biophysica Acta (BBA) ‐ Molecular Cell Research, 1823(1), 2–14. 10.1016/j.bbamcr.2011.06.014. [DOI] [PubMed] [Google Scholar]
- Werner, S., Engler, C., Weber, E., Gruetzner, R. & Marillonnet, S. (2012) Fast track assembly of multigene constructs using Golden Gate cloning and the MoClo system. Bioengineered, 3(1), 38–43. 10.4161/bbug.3.1.18223. [DOI] [PubMed] [Google Scholar]
- Witek, K., Jupe, F., Witek, A.I., Baker, D., Clark, M.D. & Jones, J.D.G. (2016) Accelerated cloning of a potato late blight–resistance gene using RenSeq and SMRT sequencing. Nature Biotechnology, 34(6), 656–660. 10.1038/nbt.3540. [DOI] [PubMed] [Google Scholar]
- Witek, K., Lin, X., Karki, H.S., Jupe, F., Witek, A.I., Steuernagel, B.et al. (2021) A complex resistance locus in Solanum americanum recognizes a conserved Phytophthora effector. Nature Plants, 7(2), 198–208. 10.1038/s41477-021-00854-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C., Liu, L., Wang, X., Vossen, J., Li, G., Li, T.et al. (2014) The Ph‐3 gene from Solanum pimpinellifolium encodes CC‐NBS‐LRR protein conferring resistance to Phytophthora infestans . Theoretical and Applied Genetics, 127(6), 1353–1364. 10.1007/s00122-014-2303-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Effector and R gene expression in potato leaves inoculated with P. infestans.
Figure S2. Rpi‐chc1.1 protein domain organization.
Figure S3. Localization of PexRD12/31 effectors in the P. infestans T30‐4 contigs.
Figure S4. PexRD12/31 effector expression in potato leaves infected with P. infestans from the PenSeq dataset.
Figure S5. pBINPLUS‐PASSA‐GG vector map.
Table S1. CRISPR‐Cas9 targeting of the Rpi‐chc1.1 allele in S. chacoense resistant accession CHC543‐5.
Table S2. Late blight resistance assessment of different Rpi‐chc1 alleles.
Table S3. Segregation of markers and late blight resistance of Rpi‐chc1.1 and Rpi‐chc1.2.
Table S4. Functional expression of Rpi‐chc1.1 in Desiree transgenic events correlates with responsiveness to PexRD12.
Table S5. Phytophthora infestans effectors used in this study.
Table S6. Accession numbers of Solanum genotypes and Rpi‐chc1 sequences.
Table S7. Primers used in this study.
Data Availability Statement
Supplementary figures and tables are available as supporting information from the TPJ website. All described sequences have been submitted to GenBank. Accession numbers are listed in Table S6.


