Summary
Plant cellulose is synthesized by a large plasma membrane‐localized cellulose synthase (CesA) complex. However, an overall functional determination of secondary cell wall (SCW) CesAs is still lacking in trees, especially one based on gene knockouts.
Here, the Cas9/gRNA‐induced knockouts of PtrCesA4, 7A, 7B, 8A and 8B genes were produced in Populus trichocarpa. Based on anatomical, immunohistochemical and wood composition evidence, we gained a comprehensive understanding of five SCW PtrCesAs at the genetic level.
Complete loss of PtrCesA4, 7A/B or 8A/B led to similar morphological abnormalities, indicating similar and nonredundant genetic functions. The absence of the gelatinous (G) layer, one‐layer‐walled fibres and a 90% decrease in cellulose in these mutant woods revealed that the three classes of SCW PtrCesAs are essential for multilayered SCW structure and wood G‐fibre. In addition, the mutant primary and secondary phloem fibres lost the n(G + L)‐ and G‐layers and retained the thicker S‐layers (L, lignified; S, secondary). Together with polysaccharide immunolocalization data, these findings suggest differences in the role of SCW PtrCesAs‐synthesized cellulose in wood and phloem fibre wall structures.
Overall, this functional understanding of the SCW PtrCesAs provides further insights into the impact of lacking cellulose biosynthesis on growth, SCW, wood G‐fibre and phloem fibre wall structures in the tree.
Keywords: Cas9, cellulose synthase (CesA), gelatinous layer (G‐layer), gene knockout, gRNA, phloem fibre, Populus trichocarpa , secondary cell wall (SCW), tension wood
Introduction
Plant cellulose microfibrils (CMFs), which are fibrillar crystalline aggregates of β‐1,4‐glucans, are synthesized by the large plasma membrane (PM)‐localized cellulose synthase (CesA) complex (CSC) (Somerville, 2006; McFarlane et al., 2014). In earlier studies, plant CSCs have been visualized as rosette structures by freeze‐fracture studies and each rosette has six particles, each of which is postulated to contain six CesA subunits (Herth, 1983; Kimura et al., 1999; Doblin et␣al., 2002). The CSC is supposed to contain 36 CesAs, which might synthesize 36‐chain CMFs. Other studies have suggested 24‐chain CMFs in celery collenchyma and spruce wood (Fernandes et␣al., 2011; Thomas et␣al., 2013). The data from Nixon et␣al. (2016) and Purushotham et␣al., (2020) support a rosette CSC with 18 CesAs that mediate the synthesis of a fundamental microfibril composed of 18 glucan chains. For higher plant CSCs, the identity and arrangement of CSC CesAs in diverse plant species, especially in trees, remain to be widely investigated at the genetic level.
Higher plants have evolved a gene family including multiple CesAs that are classified into primary cell wall (PCW) and secondary cell wall (SCW) CesA groups. In Arabidopsis, CesA1, 3 and 6 (‐like) synthesize PCW cellulose, whereas CesA4, 7 and 8 synthesize SCW cellulose (Taylor et␣al., 2003; Desprez et␣al., 2007; Persson et␣al., 2007). Mutation of any of the three SCW AtCesAs causes the same irregular xylem (irx) phenotype with collapsed tracheary elements, demonstrating the nonredundant function of each SCW AtCesA (Taylor et␣al., 2004). In cellulose‐rich trees, genetic studies leading to a functional understanding of the SCW CesAs, especially studies using the complete loss‐of‐function, are limited. Populus trichocarpa, as a model tree species with a well‐annotated genome (Tuskan et␣al., 2006; Jansson & Douglas, 2007), has 17 CesA genes and multiple expression data show the involvement of several CesA genes in SCW synthesis (Kalluri & Joshi, 2004; Djerbi et␣al., 2005; Suzuki et␣al., 2006; Dharmawardhana et␣al., 2010). A phylogeny‐based CesA nomenclature presents five Populus CesAs (PtiCesA4, 7A, 7B, 8A and 8B) into the SCW CesA group (Kumar et␣al., 2009). Proteomic data have implied that CesA4, 7A/B and 8A/B, and CesA1A/B, 3C/D and 6E/F assemble into two types of Populus CSCs, which might contribute simultaneously to cellulose biosynthesis in wood SCWs (Song et␣al., 2010; Xi et␣al., 2017). Overexpression of an aspen mutant CesA8 lacking one of the N‐terminal methionines unexpectedly resulted in a silencing of the transgene and revealed the role of CesA8 in vertical tree growth (Joshi et␣al., 2011; Liu et␣al., 2012). Most recently, SCW CesA stoichiometry in developing aspen xylem was suggested by quantitative proteomics to be 3 : 2 : 1 (Zhang et␣al., 2018). To gain insight into the roles of SCW CesAs and their CSC models in trees, it is essential to knock out each of the SCW CesAs.
The wood cell wall is a multilayered structure that includes, from outside to inside, the middle lamella (ML), PCW and SCW. These wall layers have differences in CMF organization and ratios of cellulose to wall matrix components (Plomion et␣al., 2001; Barnett & Bonham, 2004). The SCWs of wood cells comprise three layers (S1–S3), of which S2 is the most important for mechanical support and accounts for 75–85% of the SCW thickness. In response to certain environmental cues (e.g. wind and slope), the stems and branches of trees perceive gravity to determine their orientation and, in response, produce reaction wood to reinforce their position (Du & Yamamoto, 2007; Felten & Sundberg, 2013; Groover, 2016). In angiosperms, the upper sides of these stems and branches create tension wood (TW) to pull them upward. At the ultrastructural level, the TW of poplar stems has shown specific anatomical changes, such as a decrease in vessel density and an increase in fibre length (Jourez et␣al., 2001). In addition, the fibres of the TW develop a thick gelatinous (G) layer in their walls, and so are called G‐fibres, where the G‐layer can replace the S3 or S2 + S3 layer (Wardrop & Dadswell, 1955; Nishikubo et␣al., 2007; Kwon, 2008). The chemical composition of the G‐fibres includes high cellulose content and low lignin, and the polysaccharide composition of the wall matrix in the G‐layers differs from that of the PCWs and SCWs (Mellerowicz & Gorshkova, 2012; Fagerstedt et␣al., 2014) and can produce high glucose yields for biofuel production (Brereton et␣al., 2011, 2012). In phloem fibres, the cell wall structures are classified into three types: S1 + S2, S1 + S2 + G and S1 + S2 + n(G + L), where n indicates the number of repetitions of the G‐layer and the thin lignified layer (L) (Nanko, 1979; Nakagawa et␣al., 2012, 2014). Considering that the G‐layers are developed after the PCWs and SCWs and have a distinct composition, architecture and physical properties (Yamamoto et␣al., 2010; Mellerowicz & Gorshkova, 2012), they should be considered as tertiary cell walls (TCWs; Gorshkova et␣al., 2018). However, little is known about the roles of SCW CesAs in the TCWs of wood and phloem fibres of trees at the genetic level.
Knockout mutants are a crucial genetic tool for uncovering gene functions and biological mechanisms. Genome editing techniques are promising for the production of tree gene knockouts, and Cas9/gRNA genome editing has been demonstrated in poplar, apple and grape to generate null mutations in the first generation (Fan et␣al., 2015; Zhou et␣al., 2015; Osakabe et␣al., 2018). In the present study, we produced multiple knockouts of PtrCesA4, 7A, 7B, 8A and 8B genes in P. trichocarpa through Cas9/gRNA‐targeted mutagenesis. A comprehensive functional analysis revealed that complete loss of PtrCesA4, 7A/B or 8A/B led to similar morphological abnormalities, reduced wood cellulose content by 90% and suggested similar and differential roles in wood and phloem fibre wall structures.
Materials and Methods
Plant material and growth conditions
The Populus trichocarpa genotype Nisqually‐1 was used in this study. Sterile plantlets were propagated for genetic transformation in a growth chamber (25–27°C, 16 h : 8 h, light : dark photoperiod) with a light intensity of 60–80 μmol m−2 s−1. The asexual propagation of the transgenic plants was performed using three methods: apical bud cloning, lateral bud cloning and shoot regeneration. Apical and lateral buds were cut from young trees and water‐cultivated to rooting for three weeks, and shoot regeneration propagation was performed as described previously (Li et␣al., 2017). The plantlets generated were grown for phenotypic analysis in a glasshouse (25–28°C, 16 h : 8 h, light : dark photoperiod) with a light intensity of c. 250 μmol m−2 s−1. In addition, the wild‐type (WT), and ptrcesa4, 7ab, 8a, 8b and 8ab mutants were fixed to sticks to grow straight for 4 months in the glasshouse and TW was induced by inclining the stems to a 45° angle from the vertical direction for 10 d.
gRNA design and vector construction
We used the CRISPRdirect (http://crispr.dbcls.jp/; Naito et␣al., 2015) to acquire efficient gRNA target sites for genes of interest. In addition, the gRNAs were selected as close to the 5′‐ends of the gene coding sequences (CDS) as possible so that the induced frameshift mutations at the target sites would conveniently result in loss‐of‐function alleles. To ensure targeting specificity, the target sequence of each gRNA selected was used in a BlastN search against the P. trichocarpa genome and did not contain single nucleotide polymorphisms (SNPs) or small insertion/deletion polymorphisms (indels).
For cloning of the Cas9/gRNA constructs, we used the method described previously, and pCBC‐DT1T2 and pHSE401 plasmids kindly were provided by Qi‐Jun Chen from China Agricultural University (Xing et␣al., 2014). The PCR fragment was amplified using pCBC‐DT1T2 as a template, and the purified PCR fragment and pHSE401 plasmid were set up for the Golden Gate reaction using BsaI and T4 ligase. The obtained pHSE401‐2gRNA vector, after sequencing, was transferred into Agrobacterium strain GV3101.
Genetic transformation of Nisqually‐1
Agrobacterium‐mediated transformation of Nisqually‐1 was performed according to our protocol (Li et␣al., 2017), and hygromycin selection concentrations for transformants were determined. Agrobacterium carrying the pHSE401‐2gRNA binary vector was incubated to an OD600 of 0.5–0.6, and the pellet after centrifugation was resuspended for transformation. Stem explants from 1‐month‐old sterile plantlets were excised to produce fragments of 1.0–1.2 cm in length and infected for 25 min. The infected explants were co‐cultivated for 2 d, and shoot transformants were induced for 25–30 and 10–15 d in selection media supplemented with 10 and 5 mg l−1 hygromycin, respectively. The hygromycin‐resistant shoots were transferred to rooting medium with 5 mg l−1 hygromycin for 10–15 d, and the rooting shoots were objective transformants.
Identification of mutations and verification of genotype stability
Genomic DNA was extracted from the leaves of WT plants and transformants using a Plant Genomic DNA Extraction Kit (BioTeke Corp., Wuxi, China). The presence of the transgene was determined in the transformants by genomic DNA PCR with zCas (Zea mays‐codon optimized Cas9) and Hyg primers. After the growth of the transgenic plants in the glasshouse for 30 d, their genomic DNA was used for PCR amplification with gene‐specific primers spanning target sites; all primers are shown in Table S1. PCR‐amplified fragments were cloned using the pMD18‐T vector (TaKaRa, Beijing, China), and 25 positive clones for each amplicon were sequenced to identify editing at the target site. To observe the inheritance of the Cas9/gRNA‐induced mutations, the progeny of the ptrcesa mutants were propagated asexually using three methods: apical bud cloning, axillary bud cloning and shoot regeneration. Each target locus was amplified, and the PCR amplicon was sequenced as described above.
Antibody production, protein isolation, and western blot
Two specific peptides for each class of the SCW PtrCesAs (PtrCesA4, 7A/B and 8A/B) were synthesized by Hangzhou HuaBio (http://www.huabio.com) as antigens to raise antibodies in rabbits. Three groups of peptides KDELRPPTRQSATLC/IKHHDHDESNQKNVC, EHKPLKNLDGQVC/CGRGHDDEENSQFP and STMASHLNNSQDVC/CPAQDPAEVYKDAKR represent PtrCesA4, PtrCesA7A/B and PtrCesA8A/B, respectively. Two rabbits were injected for each peptide. The immunoglobulin fraction was purified from rabbit antiserum with protein A‐agarose beads. The obtained antibody each was applied to Western blot with Populus xylem protein as an antigen to examine antibody specificity.
For Western blot analysis, the xylem tissues were ground into powder in liquid nitrogen and homogenized with ultrasonic shaking in extraction buffer (100 mM Tris‐HCl, pH 8.0; 1% SDS) for 30 min at 4°C. The homogenate was boiled in a water bath for 10 min and isolated by centrifugation at 14 000 g for 20 min. Protein extracts were loaded onto an 8% SDS‐PAGE gel and transferred to a PVDF membrane. Each membrane was incubated in blocking solution containing anti‐PtrCesA4 antibody (1 : 1500), anti‐PtrCesA7A/B antibody (1:500), or anti‐PtrCesA8A/B antibody (1 : 500) for 1 h, and then washed five times in wash buffer (1 × TBS with 0.1% Tween 20). Subsequently, the membrane was incubated in blocking solution containing HRP‐conjugated anti‐rabbit secondary antibody (1:5000, ab205718; Abcam, Cambridge, UK). The signals were captured using ECL Western Blotting Substrate (Pierce Biotechnology 32106; Thermo Fisher Scientific, Waltham, MA, USA) by exposure to X‐ray films. The ACTIN was detected as a loading control using an anti‐Actin antibody (ab197345; Abcam).
Immunolocalization of cell wall polysaccharides
Samples were fixed in FAA buffer (50% ethanol, 5% acetic acid and 3.7% formaldehyde) and embedded in paraffin. Transverse sections of 8 μm thickness were cut with a sliding microtome (HM340E; MICROM International GmbH, Walldorf, Germany). Immunolocalization of wall polysaccharides (xylan, β‐(1 → 4)‐galactan, mannan and crystalline cellulose) was conducted in TW and phloem fibres of WTs and mutants. The sections were incubated with carbohydrate‐specific antibody (LM10/5/21, Plant probes, https://plantcellwalls.leeds.ac.uk/plantprobes/) or CBM3a‐6 × His protein (Plant probes) in the dilution buffer (1 : 100). Signals were detected with Alexa Fluor 633 goat anti‐rat IgG (A21094, Invitrogen) in dilution buffer (1 : 100) or anti‐His tag (1 : 500, ab18184, Abcam) as secondary antibodies and then incubated with Alexa Fluor 633 goat anti‐rat IgG (1 : 100). Sections were observed under a Zeiss LSM800 confocal laser‐scanning microscope.
Microscopy analyses
For light microscopy, stem, petiole and root samples were fixed, embedded in paraffin, sectioned and stained with toluidine blue and phloroglucinol‐HCl, as described previously (Liu et␣al., 2015). For scanning electron microscopy (SEM), free‐hand cross‐sections of fresh stem segment samples were coated with gold (Au), transferred to an SEM (S‐4800; Hitachi, Tokyo, Japan) chamber and imaged to analyse the wood wall thickness and cell shapes of xylem fibres, vessels, rays, pith and phloem fibres. For transmission electron microscopy (TEM) observation as described previously (Zhou et␣al., 2009), the stem xylem and bark samples of WT and mutants were cut into 0.5‐mm pieces and fixed. After postfixing in 1% OsO4, the samples were embedded in Epon 812 resin and 80‐nm‐thick ultrathin sections were prepared, which were stained in uranium acetate followed by lead citrate. Section‐mounted grids were observed using a TEM (CM120; Phillips, Eindhoven, the Netherlands) at an acceleration voltage of 80 kV and imaged for analyzing the wood wall thickness and structure.
Wood composition assay
The basal stems from 6‐month‐old mutant and WT trees were peeled, air‐dried and ball‐milled into the fine powder. After the power was washed with 70% aqueous ethanol, chloroform/methanol (1 : 1 v/v) solution and acetone successively, the obtained insoluble residues were prepared into cell wall materials, as previously described (Foster et␣al., 2010b), for crystalline cellulose and lignin content assays. The lignin content was determined using two methods: the acetyl bromide spectrophotometric method (Foster et␣al., 2010a) and Klason lignin and acid‐soluble lignin for the total lignin content, as described previously (Nakano & Meshitsuka, 1992). The crystalline cellulose content was measured according to a method described previously (Foster et␣al., 2010b). The hemicellulose content was determined by the GC‐MS method, as described previously (Zhang et␣al., 2017).
Statistical analysis
Data were analyzed by ANOVA in Spss (17.0). Student’s t‐test was used to determine statistical significance between the mutant and WT samples. Values are the means ± SD, and asterisks indicate statistical significance at different levels (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Accession numbers
The sequences used in this study are available in Phytozome (v.12.0) under the following accessions: PtrCesA4 (Potri.002G257900), PtrCesA7A (Potri.006G181900), PtrCesA7B (Potri.018G103900), PtrCesA8A (Potri.011G069600), and PtrCesA8B (Potri.004G059600).
Results
Cas9/gRNA‐induced knockout of SCW PtrCesAs in P. trichocarpa
In order to produce gene knockouts in P. trichocarpa, we developed the Cas9/gRNA‐targeted mutagenesis system in genotype Nisqually‐1 (Supporting information, Fig. S1). Owing to an observable albino phenotype of chli mutants (Huang & Li, 2009), PtrCHLI genes were tested for the gene editing mutagenesis in Nisqually‐1. For single‐gene editing, 11 biallelic/homozygous (61.1%) and five monoallelic mutation lines showed albino/pale‐green phenotypes, indicating knockout/knockdown of PtrCHLI1 (Fig. S2a–d; Table S2). The Cas9/gRNA‐induced editing efficiency was further tested in PtrCHLI1/2 double genes (Fig. S2e–f; Table S3). Cas9/gCHLI1/2‐a and ‐b caused many homozygous/biallelic mutations in both PtrCHLI1 and PtrCHLI2, suggesting that knockout of double genes is as efficient as that of a single gene. However, Cas9/gCHLI1/2‐c and ‐d produced more mutations in PtrCHLI1 than in PtrCHLI2, suggesting that gRNA specificity is crucial for editing efficiency. Six randomly selected T0 lines with Cas9/gRNA‐induced mutations were analyzed for each off‐target event. No mutation was detected in these potential off‐target sites among the 48 (six lines × eight putative off‐target sites) sequenced samples (Table S4; Methods S1).
In order to investigate the roles of SCW CesAs in trees, the loss‐of‐function mutations in PtrCesA4, 7A, 7B, 8A and 8B were performed using the Cas9/gRNA gene editing method. Two pairs of gRNAs were selected for each class of SCW PtrCesAs to generate multiple mutants (Figs 1a, S3a). After detection of the edited target sites, a total of six, seven, eight, 13, seven, five and eight mutation lines were obtained for the PtrCesA4, 7A, 7B, 7A/B, 8A, 8B and 8A/B genes, respectively (Table S5). In most cases, nucleotide deletions and insertions at target sites by Cas9/gRNA cause frameshift mutations in protein‐coding sequences, which result in the knockout of the target gene. According to the amino acids deduced from the coding sequence, three lines (1#, 4# and 6#) with different mutations showed a putative knockout of PtrCesA4, and multiple knockouts of double genes were generated in PtrCesA7A/B and PtrCesA8A/B, respectively (Fig. 1b). Likewise, knockout of PtrCesA7A, 7B, 8A or 8B gene was shown in a number of the mutant lines (Fig. S3b–c; Table S5). Furthermore, the ptrcesa mutation lines were analyzed for off‐target assays, and no mutation was detected at the potential off‐target sites (Table S6; Methods S1), suggesting the specificity of the selected gRNA for each PtrCesA.
Fig. 1.
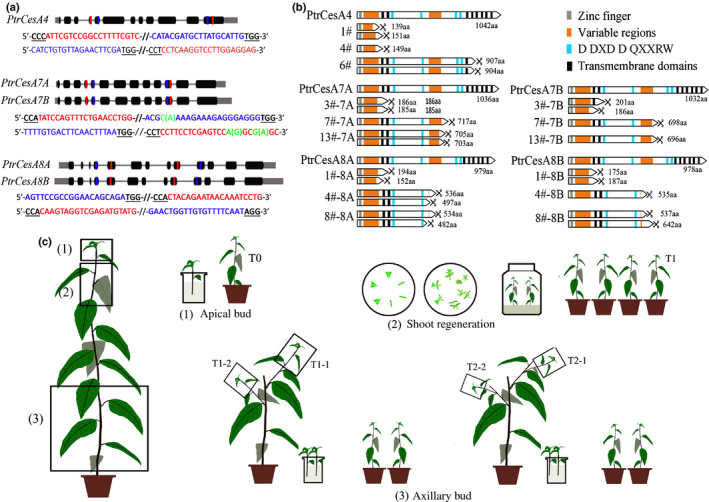
Cas9/gRNA‐induced mutations in PtrCesA4, PtrCesA7A/B, and PtrCesA8A/B genes of Populus trichocarpa (CesA, cellulose synthase). (a) Twelve gRNAs were designed in PtrCesA4, PtrCesA 7A /B and PtrCesA 8A /B genes. Nucleotides in blue and red represent the target sites. (b) The deduced amino acids of protein‐coding regions from the Cas9/gRNA‐edited genes in nine putative ptrcesa knockout mutants (ptrcesa4‐1#, −4# and − 6#, ptrcesa 7a /b‐3#, −7# and − 13#, ptrcesa 8a /b‐1#, −4# and − 8#). The scissors indicate protein‐coding termination. (c) Three asexual propagation methods (apical bud cloning, axillary bud cloning and shoot regeneration) were used to generate progeny from the ptrcesa mutants with the Cas9/gRNA‐induced mutations. The apical buds and axillary buds were rooted hydroponically and planted in soils.
We assessed the inheritance of Cas9/gRNA‐induced mutations in the progeny of ptrcesa mutants. Two T0 lines arbitrarily selected from ptrcesa4, ptrcesa7ab or ptrcesa8ab mutants served as mother plants for generating the progeny by three asexual propagation methods (Fig. 1c; Table S7). Of the 100 sites sequenced, 99 edited sites in PtrCesA4, 7A/B and 8A/B did not change in apical and axillary bud clones, suggesting faithful transmission of the mutations to the next generation (Table S8). A considerable proportion (22 of 60) of new mutations arose in target sites of the progeny propagated by the shoot regeneration method (Table S8). Thus, apical and axillary bud cloning is optimal for asexual propagation of Cas9/gRNA‐induced mutations in P. trichocarpa.
Analysis of the SCW PtrCesA protein concentrations in the Cas9/gRNA‐induced ptrcesa mutants
In order to examine the PtrCesA4, 7A/B and 8A/B protein concentrations in the Cas9/gRNA‐induced ptrcesa mutants, we produced anti‐PtrCesA4, anti‐PtrCesA7A/B and anti‐PtrCesA8A/B antibodies as a tool. Two specific peptides for each class of the SCW PtrCesAs were synthesized as antigens to raise antibodies in rabbits (Fig. S4a,b). As a result, we obtained effective anti‐PtrCesA4, anti‐PtrCesA7A/B and anti‐PtrCesA8A/B polyclonal antibodies, respectively (Fig. S4c). Western blot analysis showed high concentrations of PtrCesA4, 7A/B or 8A/B proteins in WT xylem and their molecular weights of c. 118, 117 and 110 kDa (Fig. 2). However, PtrCesA4, 7A/B and 8A/B proteins were undetectable in putative ptrcesa4, ptrcesa7a/b and ptrcesa8a/b knockout mutants (Fig. 2a), suggesting that they are null mutants. In addition, the abundance of PtrCesA7A/B (or PtrCesA8A/B) proteins decreased significantly in putative ptrcesa7a and ptrcesa7b (or ptrcesa8a and ptrcesa8b) knockout mutants (Fig. 2b).
Fig. 2.
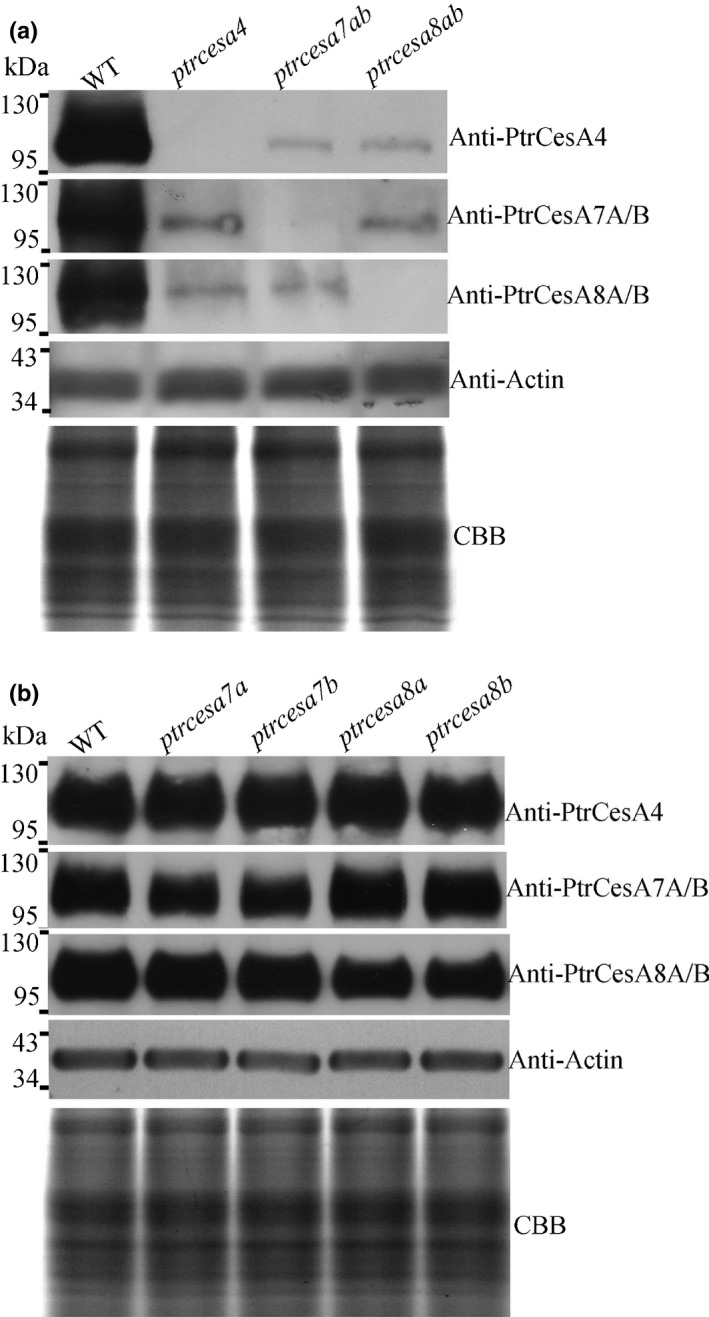
Analysis of the secondary cell wall (SCW) cellulose synthase PtrCesA protein concentrations in the Cas9/gRNA‐induced Populus trichocarpa ptrcesa knockout mutants. (a) Immunoblot analysis of PtrCesA4, PtrCesA7A/B and PtrCesA8A/B protein concentrations in secondary xylem of wild‐type (WT), ptrcesa4, ptrcesa7ab and ptrcesa8ab young trees. (b) Immunoblot analysis of PtrCesA4, PtrCesA7A/B and PtrCesA8A/B protein concentrations in secondary xylem of WT, ptrcesa7a, ptrcesa7b, ptrcesa8a and ptrcesa8b young trees. The ACTIN as control was detected using an anti‐Actin antibody (ab197345, Abcam), indicating equal loading proteins. A replicate Coomassie Brilliant Blue (CBB)‐stained gel is shown to confirm equal loading.
In comparison with the WT, the concentrations of PtrCesA7A/B and 8A/B proteins decreased sharply in ptrcesa4 knockout mutants, and likewise, those of PtrCesA4 and 8A/B (or 7A/B) proteins were significantly reduced in ptrcesa7a/b (or ptrcesa8a/b) knockout mutants (Fig. 2a). These data indicate that deletion of one class of the SCW PtrCesAs (PtrCesA4, 7A/B or 8A/B) diminished the protein concentrations of the other two classes of the SCW PtrCesAs. In addition, PtrCesA7A and PtrCesA7B (or PtrCesA8A and PtrCesA8B) with presumed functional redundancies had no obvious mutual complementation at protein concentrations in the corresponding ptrcesa mutants (Fig. 2b). In addition, RT‐PCR analysis showed that the transcriptional level of each SCW PtrCesA moderately decreased in the corresponding knockout mutant, but those of other SCWPtrCesAs did not change significantly in the mutant (Fig. S5; Methods S2).
Ptrcesa4, 7a/b and 8a/b knockout mutants exhibit similar morphological abnormalities
Knockout of PtrCesA4 caused serious defects in growth and development in transgenic trees (Fig. 3a). The ptrcesa4 mutants showed complete prostrate growth and formed twisted stems. After c. 6 months of growth in the glasshouse, the ptrcesa4 mutants completely lost apical dominance, resulting in weeping woody plants (Fig. S6b). The stems of ptrcesa4 were extremely brittle, revealing major changes in stem mechanical properties (Fig. S6d). In addition, the ptrcesa4 mutants showed significant reductions in stem diameter, internode length, leaf size and root structure, and significantly decreased the sizes of the pavement and guard cells in the leaves (Fig. S7; Methods S3). The ptrcesa7a/b (or ptrcesa8a/b) double mutants presented the same defects as the ptrcesa4 mutants (Figs 3a, S6, S7), implying similar and nonredundant roles of PtrCesA4, 7A/B and 8A/B in P. trichocarpa. Compared with the ptrcesa7a/b double mutant, the knockouts of ptrcesa7a or 7b showed slight growth defects (Fig. S6a,c), indicating redundant roles for PtrCesA7A and 7B. Likewise, the redundant roles of PtrCesA8A and 8B were suggested by comparative phenotypes of single and double mutants.
Fig. 3.
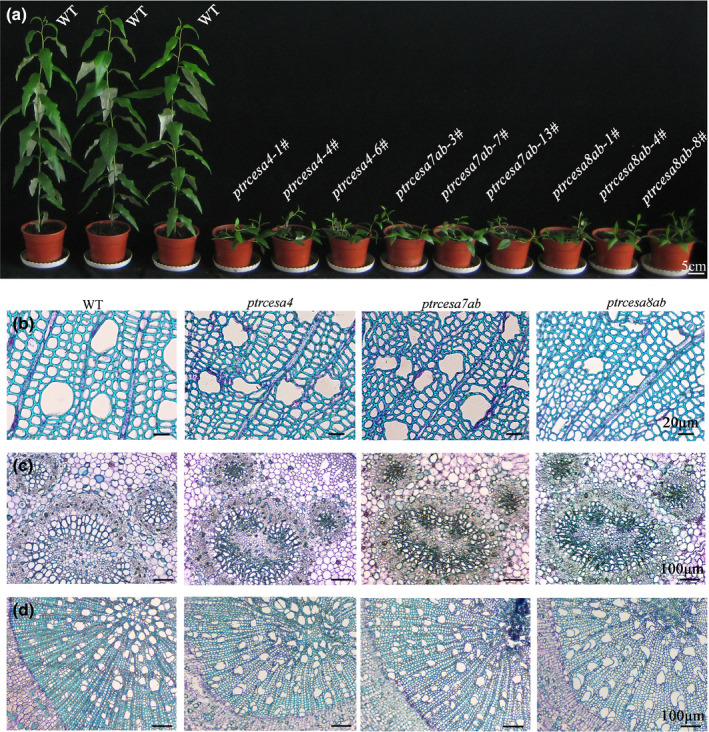
Phenotypes of Populus trichocarpa ptrcesa4, ptrcesa7a/b, and ptrcesa8a/b mutants (CesA, cellulose synthase). (a) Morphology of multiple ptrcesa mutant and wild‐type (WT) trees grown for 3 months in a glasshouse. (b‐d) Light microscopic analysis of stems (b), petioles (c) and roots (d) from 3‐month‐old ptrcesa mutant and WT trees. Sections were stained with toluidine blue. Stem cross‐sections are from the 10th internode; petiole cross‐sections are from the 8th leaf below the apical bud. Bars: (a) 5 cm; (b) 20 μm; (c–d) 100 μm.
Light microscopy analysis showed that the stem xylem was severely collapsed in ptrcesa4, 7a/b and 8a/b mutants but not in ptrcesa7a, 7b, 8a and 8b mutants (Figs 3b, S8, S9). The vessel cells of the 2nd to 4th stem internodes undergoing predominantly primary growth, leaf petioles, and root tissues also displayed collapse in these mutants (Figs 3c,d, S8), which might be the main reason for impaired leaf and root growth. In addition, the inner surfaces of pith parenchyma cell walls of these mutants were undulating (or folded) and had few pits, whereas those of the WT were smooth and had numerous ones (Fig. S10). Approximately 3‐month‐old mutants in the glasshouse showed earlier accumulation of starch granules in pith ray and parenchyma cells, whereas > 6‐month‐old WT trees were capable of accumulating starch granules in these cells (Figs 4, S10), suggesting that the SCW PtrCesAs‐mediated cellulose synthesis is a key pathway that is integrated into carbohydrate metabolism in trees. Thus, the above complete loss‐of‐function phenotypes indicate that PtrCesA4, 7A/B and 8A/B play crucial, similar and nonredundant roles in the growth and development of P. trichocarpa.
Fig. 4.
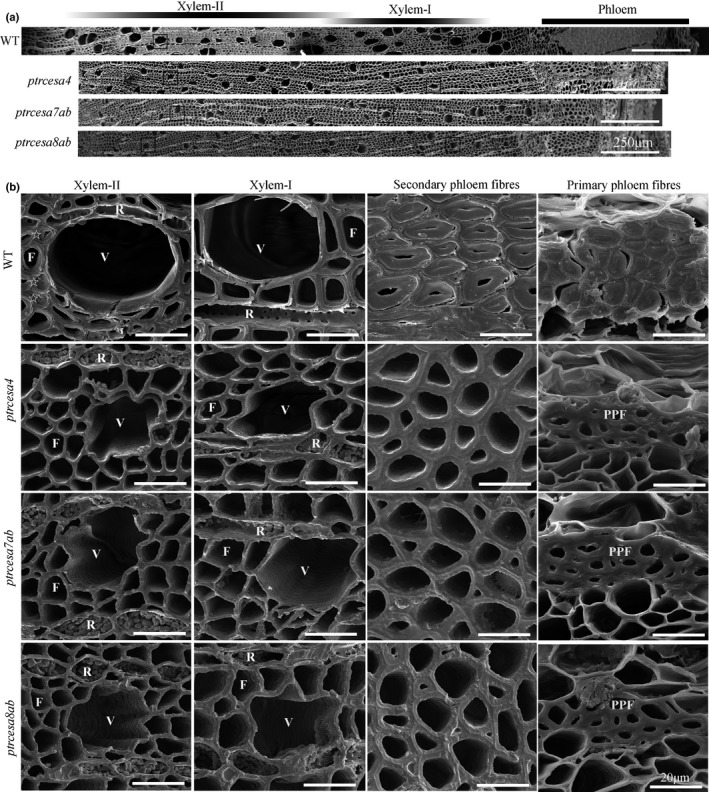
Scanning electron microscopy analysis of stem tissues in Populus trichocarpa ptrcesa4, ptrcesa7a/b and ptrcesa8a/b mutants (CesA, cellulose synthase). (a) Scanning electron microscopy (SEM) images of cross‐sections of basal stems from 6‐month‐old wild‐type (WT) and ptrcesa mutant trees. (b) Xylem fibres, vessels, and ray cells within inset boxes in the (a) and secondary and primary phloem fibres (SPF and PPF) in the WT and these mutants were magnified under SEM. Some mature wood fibres (indicated by pentangles) have developed the thick G‐layers inside the secondary cell walls (SCWs). Xylem‐I, developing xylem; Xylem‐II, mature xylem; V, vessel cell; F, fibre cell; R, ray cell. Bars: (a) 250 μm; (b) 20 μm.
The crucial roles of PtrCesA4, 7A/B and 8A/B in wood cell wall structure
We explored the roles of PtrCesA4, 7A/B and 8A/B in wood wall structure using SEM analysis. As shown in Fig. 4, the wood fibres of WT xylem‐I (developing xylem) developed the SCWs, and some wood fibres of xylem‐II (mature xylem) produced a thick G‐layer (indicated by the pentangle) inside the SCW. However, in addition to the severe collapse of wood fibres and vessels, thin one‐layer‐walled fibres were observed in the xylem‐I and xylem‐II of ptrcesa4, 7a/b and 8a/b mutants. Further TEM analysis clearly showed a multilayered SCW structure in WT wood fibres with visible S1 and S2 layers, whereas these mutants displayed thin one‐layer‐walled fibres in woods (Fig. 5). The thickness of wood fibre walls in these mutants was reduced by approximately half compared with the WT, and the lengths of the mutant wood fibres and vessels were significantly shorter than those of the WT (Figs S11, S12; Methods S4). These data indicate that PtrCesA4, 7A/B and 8A/B are essential for a multilayered SCW structure of wood in Populus.
Fig. 5.
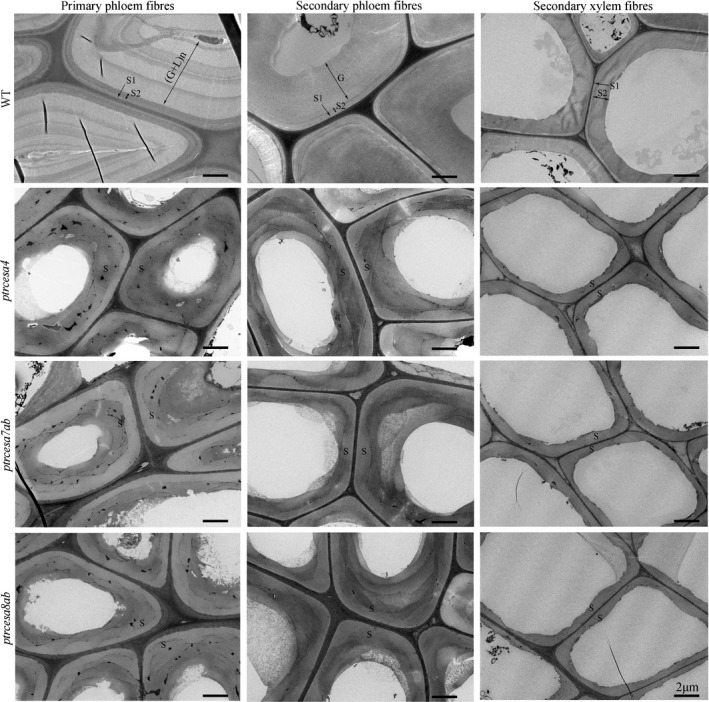
Transmission electron microscopy analysis of the phloem and wood fibre wall structures in Populus trichocarpa ptrcesa4, ptrcesa7a/b and ptrcesa8a/b mutants (CesA, cellulose synthase). Transmission electron microscopy (TEM) images from the basal stem phloem and xylem fibres in 6‐month‐old wild‐type (WT) and ptrcesa mutant trees. The WT primary and secondary phloem fibres showed S1 + S2 + n(G + L) and S1 + S2 + G wall structures, respectively. L, lignified layer; G, gelatinous (G)‐layer; n, number of repetitions of the G and L; S, S‐layer of SCW with S1, S2 and S3. Bars, 2 μm.
Furthermore, we examined the inner surfaces of fibre and vessel cell walls in longitudinal sections of the mutant wood under SEM. From the pith parenchyma outwards spiral and annular vessels were observed in protoxylem (primary xylem) of WT stems, and subsequently, reticulate and pitted vessels were observed in the metaxylem (secondary xylem) (Fig. 6a). Overall, the mutants also developed these types of vessels in protoxylem and metaxylem, whereas the stem xylem was severely collapsed (Fig. 6a). Nevertheless, the inner surfaces of vessel walls in the mutant woods were undulating and uneven, and pit membranes were broken in the mutant wood vessels (Fig. 6b), suggesting that SCW cellulose is an essential component. Likewise, the fibre wall features (wall pits and their patterns) were significantly impaired in the mutant woods (Fig. 6c). Taken together, loss of PtrCesA4, 7A/B or 8A/B leads to serious damage to fibre and vessel wall structure in Populus.
Fig. 6.
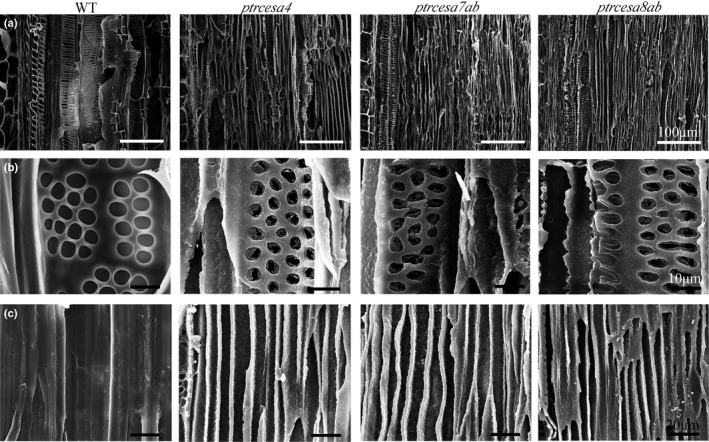
Scanning electron microscopic images of xylem fibres and vessels in longitudinal sections of basal stems from Populus trichocarpa wild‐type (WT) and ptrcesa4, 7a /b and 8a /b mutants (CesA, cellulose synthase). (a‐c) Primary and secondary xylem (a), the pitted pattern vessels of secondary xylem (b) and the fibres of secondary xylem (c). Bars: (a) 100μm; (b) 10 μm; (c) 20 μm.
PtrCesA4, 7A/B, and 8A/B are indispensable for G‐layer formation of TW fibres in Populus
We investigated the roles of the SCW PtrCesAs in G‐layer formation of Populus TW fibres at genetic level. In the leaning stem of young WT trees, the TW side showed eccentric growth, a reduction in the number of vessels and the production of G‐fibres under SEM (Figs 7, S13), which have been considered as the main TW features. Likewise, the first two features were observable in the TW sides of ptrcesa4, ptrcesa7ab and ptrcesa8ab mutants (Fig. S13), indicating a primary response to the incline‐induced gravity stimuli. However, no G‐layer was visible in any of the TW fibres of the ptrcesa4, ptrcesa7ab and ptrcesa8ab mutants (Fig. 7). In addition, the ability of G‐layer formation was slightly impaired in ptrcesa8a or ptrcesa8b mutants, suggesting a redundant role of PtrCesA8A and 8B in this aspect. These data indicate that PtrCesA4, 7A/B and 8A/B are indispensable for the G‐layer formation of TW fibres.
Fig. 7.
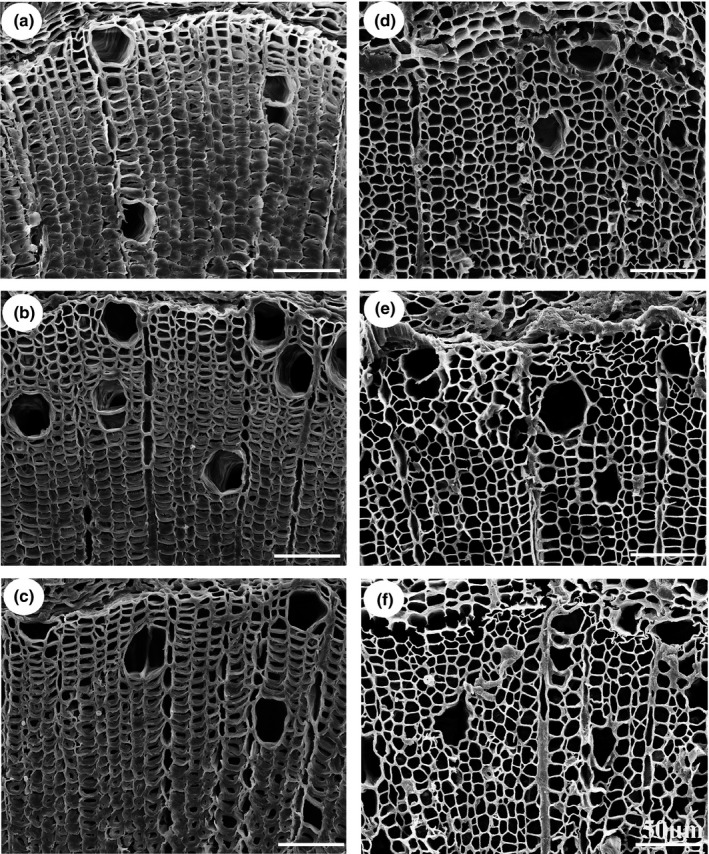
Scanning electron microscopy analysis of tension wood (TW) gelatinous (G)‐fibres in Populus trichocarpa wild‐type (WT) and ptrcesa mutants under gravi‐stimulation (CesA, cellulose synthase). (a–f) The WT (a), and ptrcesa8a (b), ptrcesa8b (c), ptrces8ab (d), ptrces7ab (e) and ptrces4 (f) mutants grown straight in a glasshouse for 4 months were inclined to induce TW by a 45˚ angle from the vertical direction for 10 d. The scanning electron microscopy (SEM) images were taken from freehand cross‐sections of the 16th internode of each sample. Bars, 50 μm.
Furthermore, the distribution of cell wall polymers was examined using the fluorescent immunolabelling technique in the mutant TW and opposite wood (OW). Crystalline cellulose, confirmed by CBM3a immunolabelling, was distributed intensively in the G‐layer of TW fibres and SCWs of OW in the WT, but nearly invisible in TW and OW of these mutants (Figs 8, S14), suggesting that PtrCesA4, 7A/B and 8A/B synthesized complete crystalline cellulose in SCWs of OW and TCWs of TW. Using LM10 to detect xylan (a main noncellulosic polysaccharide of the S‐layer), the fluorescence outlines were narrowed in S‐layers of the mutant OW compared with the WT, but those were likewise thin in S‐layers of TW in the WT and mutants (Figs 8, S15), implying that loss of crystalline cellulose impaired SCW structure in OW, but did not affect SCW thinning during TW. The β‐(1 → 4)‐galactan‐rich G‐layers of TW (Gorshkova et␣al., 2015) were not detected in WT and mutant OW, and conversely, strong fluorescence signals appeared in fibres of mutant TW without TCWs, which was similar to that observed in TW of the WT (Figs 8, S16). In addition, the immunofluorescence for detecting mannan was weak in fibres of the mutant TW but strong in those of the WT (Figs 8, S17), indicating that loss of crystalline cellulose impairs deposition of mannans in TCWs of TW.
Fig. 8.
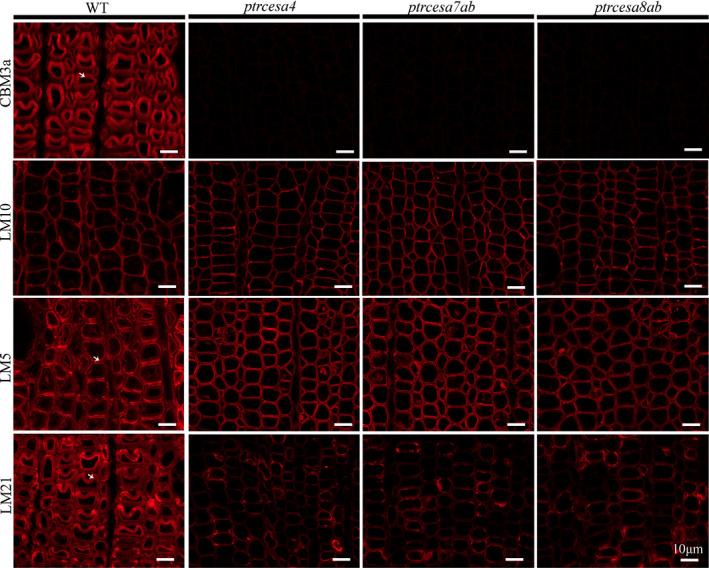
Immunolocalization of crystalline cellulose, xylan, β‐(1 → 4)‐galactan and mannan in the tension wood (TW) side in Populus trichocarpa wild‐type (WT), and ptrcesa4, ptrcesa7ab and ptrcesa8ab mutants (CesA, cellulose synthase). The 8‐μm transverse cross‐sections of the 16th internode from each sample were incubated with carbohydrate‐specific antibody (LM10/5/21) or CBM3a‐6 × His protein and anti‐His antibody. Signals (red) were detected with Alexa Fluor 633 goat anti‐rat IgG. CBM3a, LM10, LM5 and LM21 bind crystalline cellulose, xylan, β‐(1 → 4)‐galactan and mannan in plant cell walla, respectively. Arrowheads indicate TW fibre gelatinous (G)‐layers labelled with immunofluorescence in the WT. Bars, 10 μm.
The roles of PtrCesA4, 7A/B and 8A/B in stem phloem fibre wall structures
In order to investigate the role of the SCW PtrCesAs in phloem fibres, we observed wall structures in ptrcesa4, ptrcesa7ab and ptrcesa8ab mutants under SEM. The WT primary and secondary phloem fibres (PPFs, SPFs) showed extremely thick walls with multilayers and S + G layers, respectively (Fig. 4b). However, the mutant SPF showed a larger lumen, lost the G‐layer, but retained the thick wall with no obvious collapse (Fig. 4b). The mutant PPF also lost the multilayered structure and retained the thicker wall, and cell size was greatly reduced (Fig. 4b). Under TEM, WT SPF showed S1 + S2 + G layers in the wall, and interestingly, the PPF developed an unique wall structure with S1 + S2 + n(G + L) layers, where G and L layers are formed alternately (Fig. 5). By contrast, the mutant SPF and PPF lost S1 + S2 + G and S1 + S2 + n(G + L) wall structures and both remained a thick wall with 3–5 μm thickness, even exceeding that of SCWs in WT wood fibres (Figs 5, S11). The thick walls of the mutant SPF and PPF should be composed of S‐layers, where the boundaries of S1 and S2 layers were indistinguishable under TEM.
We further analyzed the distribution of cell wall polymers in the mutant phloem fibres. The S + G layers of SPF and S + n(G + L) layers of PPF in the WT showed strong CBM3a‐immunolabelled fluorescence (indicating crystalline cellulose), and conversely, the signals in the mutant SPF and PPF were weak but could be detected (Figs 9, S18). LM5‐immunolabelled fluorescence revealed rich β‐(1 → 4)‐galactan in G‐layers of SPF and n(G + L) layers of PPF in the WT, whereas most of the mutant SPFs and PPFs had no significant deposition of β‐(1 → 4)‐galactan in the walls, but only several SPFs (or PPFs) in each phloem fibre region showed clear deposition of β‐(1 → 4)‐galactan (Figs 9, S19). In addition, signals indicating mannans did not appear in the mutant SPF and PPF (Figs 9, S20), whereas they were visible (but weak) in the mutant TW fibres (Fig. 8). Xylan‐immunolabelled signals gathered strongly in S‐layers of WT SPF and PPF and were distributed moderately in n(G + L) layers of PPF. In the mutants, both SPFs and PPFs losing the G/n(G + L) layers showed wider fluorescence outlines (Figs 9, S21), suggesting a thicker S‐layer in the walls. Overall, the findings show that knockout of PtrCesA4, 7A/B or 8A/B leads to similar but differential wall structures in wood and phloem fibres.
Fig. 9.
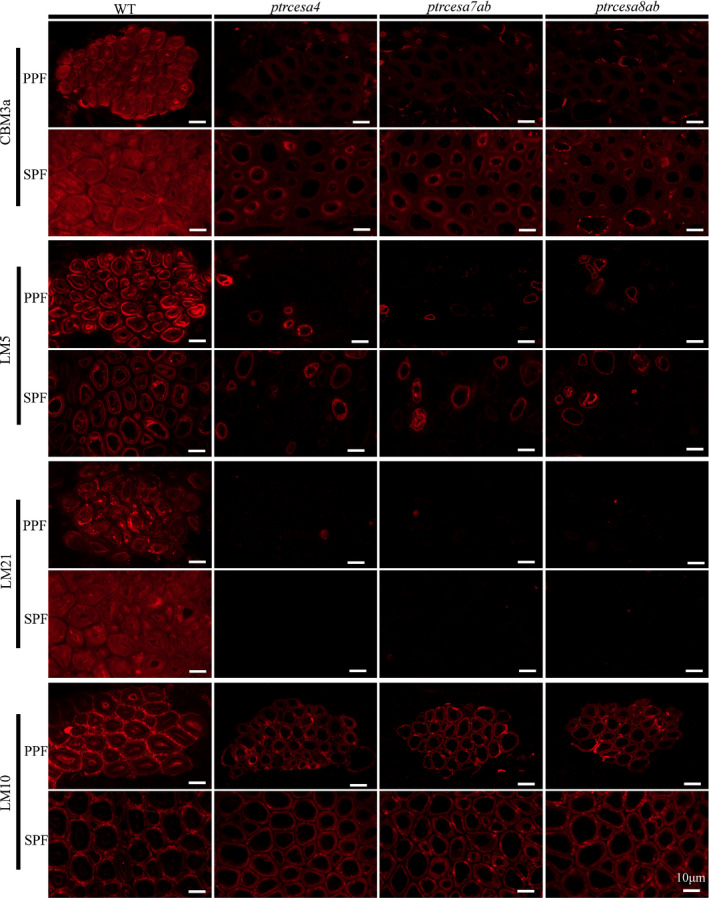
Immunolocalization of crystalline cellulose, xylan, β‐(1 → 4)‐galactan and mannan in phloem fibres in Populus trichocarpa wild‐type (WT), and ptrcesa4, ptrcesa7ab and ptrcesa8ab mutants (CesA, cellulose synthase). Primary and secondary phloem fibers (PPF, SPF) in the 8‐μm transverse cross‐sections of the 20th internode from each sample were incubated with carbohydrate‐specific antibody (LM10/5/21) or CBM3a‐6 × His protein and anti‐His antibody. Signals (red) were detected with Alexa Fluor 633 goat anti‐rat IgG. CBM3a, LM10, LM5 and LM21 bind crystalline cellulose, xylan, β‐(1 → 4)‐galactan and mannan in plant cell walls, respectively. Bars, 10 μm.
Contributions of PtrCesA4, 7A/B and 8A/B to wood chemical composition
We examined the chemical composition of the wood walls in the ptrcesa4, 7a/b and 8a/b mutants (Table 1). The cellulose content of the wood of these mutants, as calculated by the Updegraff method, were c. 4%, one‐tenth of that in WT wood (c. 41%), indicating that PtrCesA4, 7A/B and 8A/B are essential for wood cellulose synthesis. By contrast, wood lignin content as calculated by the Klason or acetyl bromide soluble lignin method were increased nearly two‐fold in these mutants compared with the WT, and the phloroglucinol staining data were in agreement with this result (Fig. S22). Compared with ptrcesa7a/b (or ptrcesa8a/b) double mutants, the decrease in cellulose content and increase in lignin content in wood were not drastic in ptrcesa7a and 7b (or ptrcesa8a and 8b) mutants (Fig. S23), implying the redundant role of PtrCesA7A and 7B (or PtrCesA8A and 8B) in wood cellulose synthesis.
Table 1.
Cellulose, lignin and noncellulosic polysaccharides in dry wood of Populus trichocarpa wild‐type (WT) and ptrcesa mutants (CesA, cellulose synthase).
| Wood composition | WT | ptrcesa4 | ptrcesa7ab | ptrcesa8ab |
|---|---|---|---|---|
| Cellulose (%) | ||||
| Crystalline | 41.36 ± 0.62 | 3.88 ± 0.11*** | 3.89 ± 0.06*** | 4.03 ± 0.15*** |
| Lignin (%) | ||||
| Klason | 20.36 ± 0.41 | 39.02 ± 0.79*** | 39.38 ± 0.51*** | 39.45 ± 0.41*** |
| Acid‐soluble | 2.19 ± 0.06 | 2.12 ± 0.04 | 2.11 ± 0.06 | 2.13 ± 0.05 |
| Total lignin | 22.83 ± 0.36 | 41.14 ± 0.75*** | 41.49 ± 0.45*** | 41.58 ± 0.49*** |
| Acetyl bromide soluble lignin | 22.20 ± 0.55 | 41.62 ± 0.92*** | 42.09 ± 0.88*** | 42.13 ± 0.92*** |
| Polysaccharidea | ||||
| Xylose | 203.29 ± 5.63 | 505.52 ± 10.48*** | 507.39 ± 10.77*** | 504.26 ± 6.10*** |
| Rhamnose | 4.38 ± 0.19 | 8.12 ± 0.15*** | 8.24 ± 0.16*** | 8.44 ± 0.24*** |
| Fucose | 1.02 ± 0.04 | 1.32 ± 0.04*** | 1.33 ± 0.04*** | 1.32 ± 0.03*** |
| Arabinose | 4.15 ± 0.06 | 9.87 ± 0.09*** | 9.93 ± 0.09*** | 9.45 ± 0.09*** |
| Galactose | 8.61 ± 0.27 | 10.95 ± 0.28*** | 10.38 ± 0.29*** | 11.06 ± 0.17*** |
| Mannose | 14.62 ± 0.22 | 5.13 ± 0.11*** | 4.84 ± 0.09*** | 4.73 ± 0.07*** |
| Glucose | 56.15 ± 1.98 | 13.18 ± 0.20*** | 11.33 ± 0.18*** | 13.59 ± 0.40*** |
Values are means ± SD (n = 3). Asterisks denote significant difference from WT by Student’s t‐test (***, P < 0.001). aData are shown as µg mg−1 cell wall residues (AIRs).
The content of xylose, which is a major monosaccharide of hemicelluloses (xylan), increased 2.5‐fold in ptrcesa4, 7a/b and 8a/b mutants, and other low concentrations of monosaccharides (rhamnose, arabinose, fucose and galactose) increased significantly (Table 1), indicating a large amount of hemicellulose accumulation in these mutant woods. However, the mannose and glucose contents of these mutant woods decreased three‐to four‐fold compared with those in the WT (Table 1), which might suggest a significant decrease in mannans and/or glucomannans by the loss of PtrCesA4, 7A/B or 8A/B.
Discussion
Approximately half of the woody biomass synthesized by trees is cellulose. In P. trichocarpa, five cellulose synthases PtrCesAs (PtrCesA4, 7A, 7B, 8A and 8B) are classified into the secondary cell wall (SCW) CesA group (Kumar et␣al., 2009). Here, the defective phenotypes of the Cas9/gRNA‐induced gene knockouts show that the set of SCW PtrCesAs plays a crucial role in growth and development, multilayered wall structure and wood chemical composition. In addition, each class of the SCW PtrCesAs is essential for developing wood gelatinous (G)‐fibres, and our findings suggest differences in the role of SCW PtrCesAs‐synthesized cellulose in the wall architecture of wood and phloem fibres. Overall, this functional understanding of the SCW PtrCesAs provides further insights into the impact of lacking cellulose biosynthesis on growth, SCW, wood G‐fibre and phloem fibre wall structures in the tree.
Genetic cessation of SCW cellulose biosynthesis can impair growth and development in transgenic Populus trees. Here, the ptrcesa4, 7a/b and 8a/b knockout mutants exhibited serious and similar morphological abnormalities. A recent study reported similar and moderate defects in PtrCesA RNAi transgenic plants in P. trichocarpa (Abbas et␣al., 2020). Serious defects of these null mutants exhibited prostrate growth, loss of apical dominance and significant reductions in stem diameters, internode lengths and leaf sizes (Figs 3, S6, S7). Small organs of these mutants suggest the possible roles of PtrCesA4, 7A/B and 8A/B in cell‐size determination, as manifested by the decrease in sizes of wood fibres and vessels, pith parenchyma, and guard and pavement cells (Figs S7g, S10, S12). Although the shapes and sizes of plant cells are defined largely by their surrounding walls (Bögre et␣al., 2008), plant cell expansion should not be undermined by the reduced secondary cellulose. This is because cell expansion generally terminates before SCW deposition. It is very likely that water deficit led to smaller organs in these mutants. Lack of secondary cellulose gave rise to serious collapse in vessel cells of leaf petioles and xylem of stems and roots in these mutants (Figs 3b–d, 6a, S8), which might, in turn, reduce xylem water transport capacity. In Arabidopsis cesa7 mutants, stem xylem is partially collapsed and the small guard cells result from the decreased water supply to the developing leaves (Liang et␣al., 2010). In our mutants (of ptrcesa4, 7a/b and 8a/b), the loss of SCW cellulose also resulted in broken pit membranes in the xylem vessels (Fig. 6b). Pits are essential components in the water transport system and pit membranes account for c. 50% of the hydraulic resistivity of the water transported through xylem conduits (Sperry et␣al., 2006; Choat et␣al., 2008).
The SCW cellulose synthesis could be one of the key pathways integrated into plant carbohydrate metabolism, especially in trees. Starch, as one of the main nonstructural carbohydrates, is stored in the wood parenchyma as long‐term reserves. During spring growth or in adverse environments, starch is mobilized or converted to sucrose for physiological metabolism in trees (Richardson et␣al., 2013; Bellasio et␣al., 2014; Yoshimura et␣al., 2016). Deletion of PtrCesA4, 7A/B or 8A/B caused earlier accumulation of starch granules in the mutant stem pith rays and parenchyma (Figs 4b, S10), implying that the cessation of SCW cellulose synthesis directs the flow of carbohydrates into starch as a sink. The sucrose synthase (SuSy) associated with the CesA complex (CSC) supplies uridine diphosphate (UDP)‐glucose as a substrate for CesAs, and overexpression of the cotton SuSy results in increased cellulose production in poplar (Coleman et␣al., 2009; Fujii et␣al., 2010). It is possible that owing to a lack of conversion of UDP‐glucose into cellulose, excessive UDP‐glucose forms a feedback signal and is redirected to starch synthesis in these mutants. We speculate that perturbation of SCW cellulose synthesis would cause pleiotropic effects in complex carbohydrate metabolism in trees.
The wood fibre SCW contains three layers (S1–S3) with different cellulose microfibril (CMF) orientations. Complete knockout of PtrCesA4, 7A/B or 8A/B disrupted the multilaminar wall structure and formed thin one‐layer‐walled fibres in the wood (Figs 4, 5). Lack of cellulose destroys the multilayer formation of SCWs in mutant wood, presumably due to serious CMF deficiency. It is known that the deficiency of CMFs in SCWs alters the physical properties of wood, as demonstrated by excessive stem brittleness and severe wood vessel and fibre collapse in the mutants (Figs 4, 8, S6). Although Arabidopsis does not produce wood, the AtCesA8, 7 and 4 mutations (irx1, 3 and 5) have shown irregular xylem (irx) phenotypes (Taylor et␣al., 1999, 2000; 2003). The collapsed phenotypes caused by deletion of SCW CesAs in Populus are more severe than those in Arabidopsis. The interfascicular fibres in the irx1, 3 or 5 mutants showed no obvious collapse or distortion, possibly analogous to retention of the S‐layers in phloem fibres of the ptrcesa mutants. Notably, PtrCesA4, 7A/B and 8A/B contribute to complete wood SCW cellulose synthesis because deletion of any of the three classes reduced wood cellulose content to 4% (Table 1), which is inferred to represent plant cell wall (PCW) cellulose.
The formation of wood G‐fibres in trees responds to gravistimulation, and gravitropism and reaction wood are the organizing centres for this biological process (Groover, 2016). The ptrcesa4, 7a/b and 8a/b mutants undergoing gravistimulation showed asymmetrical radial growth, thinned the S‐layers in fibres but lost the G‐layers (Figs 7, 8), implying that they have perceived gravistimulation and produced tension wood (TW). It is inferred that PtrCesA4, 7A/B and 8A/B act as basal components in this pathway, and the CMFs that they synthesize are the component core in TCWs of TW fibres. In addition, rhamnogalacturonan‐I (RG‐I), mannan and xyloglucan are the main polysaccharides in TW fibre TCWs (Nishikubo et␣al., 2007; Mellerowicz & Gorshkova, 2012). RG‐Is with shortened galactan chains are proposed as spacers between the CMFs to prevent their lateral interaction, potentially creating tension in mature G‐fibres (Gorshkova et␣al., 2015). Despite no CMFs of TCWs, the galactans still accumulated in the walls of the mutant TW fibres (Fig. 8), possibly because they interact with the polysaccharides of S‐layer. Additionally, losing CMFs of the TCWs reduced the deposition of mannans in walls of the mutant TW fibres (Fig. 8), seemingly suggesting the association of mannans with the CMFs. In the future, loss‐of‐functions of the genes encoding noncellulosic polysaccharide synthesis enzymes could provide further insights into the architecture of the polymers in TW fibres.
Mature phloem fibres endow structural strength and flexibility to plant stems. Some herbaceous plants normally develop thick G‐layers in mature phloem fibres (Roach et␣al., 2011). In the wild‐type (WT) Populus of the glasshouse, the mature primary and secondary phloem fibres (PPFs and SPFs) developed n(G + L)‐ and G‐layers in the walls, respectively (Figs 4, 5). Differential wall structures in both might be ascribed to different origins, that is, both initiate from the procambium and vascular cambium, respectively. Expression data show that flax phloem fibres recruit both PCW and SCW CesAs during deposition of TCWs (Mokshina et␣al., 2017). Here, both PPFs and SPFs lost individualized wall structures in these SCW ptrcesa mutants and remained similar thick walls without the TCWs. Accordingly, PtrCesA4, 7A/B and 8A/B are essential for developing n(G + L)‐/G‐layers of Populus phloem fibres. The synthesized CMFs are proposed as the skeleton to assemble the wall structures. Additionally, the phenotypes of these mutant phloem and TW fibres (Figs 4, 5, 8, 9) could suggest different roles of the SCW PtrCesAs in the two wall structures. First, the SPF walls accumulated a small amount of crystalline cellulose but TW fibres almost did not. Second, phloem fibres remained thicker walls (than TW fibres), which presumably comprised the S‐layers. Third, phloem fibres displayed weaker deposition of mannan and galactan on the walls (than TW fibres) when the two lost the TCWs in structure. Thus, it is proposed that differential wall deposit mode (or additional CesA‐like proteins) might be involved in SCW biosynthesis of phloem fibres.
In Arabidopsis, CesA4, 7 and 8 are the core components of the SCW CSC (Taylor et␣al., 2000, 2003) and exhibit varying degrees of class specificity (Kumar et␣al., 2017). The ptrcesa4, 7a/b and 8a/b knockout mutants exhibited similar defects in all phenotypes, including growth and development, SCW structure and wood composition (Figs 3, 4, 5, 6; Table 1), indicating that PtrCesA4, 7A/B and 8A/B are not redundant and represent the three classes of SCW CesAs. Deletion of any one class in Populus caused a complete lack of wood SCW cellulose synthesis (Table 1), which implies destructive damage to SCW CSC. This notion is further supported by the evidence that deletion of one class of the SCW PtrCesAs exaggeratedly diminished protein concentrations in the other two classes (Fig. 2a). Similar results have been shown in Arabidopsis cesa4, 7 or 8 mutants, assuming that these mutants do not assemble a functional CSC (Atanassov et␣al., 2009). In this regard, the mode by which the Populus CSC employs the SCW CesA classes is similar to that of Arabidopsis, thus suggesting the conserved core architecture of SCW CSCs in plants, including trees. Different from those of Arabidopsis, Populus CesA7 and CesA8 classes each evolve two members owing to recent whole genome duplication events (Takata & Taniguchi, 2015). The phenotypes of single and double mutants (Figs 3, 7, S6, S9) suggested that PtrCesA7A and 7B (and PtrCesA8A and 8B) are redundant in function. Considering that Populus CesA7 (or CesA8) class has two members, we speculate that Populus SCW CSCs are diversified in the number, identity and arrangement of the PtrCesA units. Whether PtrCesA7A and 7B (or PtrCesA8A and 8B) could heterogeneously constitute a SCW CSC, remains to be identified. In addition, it is a question that a specific CSC is responsible for synthesizing CMFs of the TCWs. A quantitative proteomics study has proposed that the stoichiometries of aspen CesA8A/B‐4‐7A/B are 3 : 2 : 1 and 8 : 3 : 1 in the developing xylem and TW, respectively (Zhang et␣al., 2018). In the present study, no G‐layer of TW fibres in these null mutants (Figs 7, 8) indicates that PtrCesA4, 7A/B and 8A/B are indispensable for synthesizing the CMFs of TCWs. Therefore, it is still unclear how the five SCW CesAs of three classes constitute the CSC in Populus.
Author contributions
WX and YC conceived and designed the research; WX, HC, SZ, JC, HJ, BZ, SC, CW, GT, CZ, LM, YZ and YC performed the experiments and analyzed data; YC provided the funding; WX and YC wrote the manuscript; and YC reviewed and edited the manuscript.
Supporting information
Fig.␣S1 Cas9/gRNA system and stepwise protocol in Nisqually‐1.
Fig.␣S2 Cas9/gRNA‐induced mutations in PtrCHLI1 gene and PtrCHLI1/2 family genes of P. trichocarpa.
Fig.␣S3 Cas9/gRNA‐induced mutations in PtrCesA7A, PtrCesA7B, PtrCesA8A and PtrCesA8B genes of P. trichocarpa.
Fig.␣S4 Production of anti‐PtrCesA4, −7A/B and −8A/B polyclonal antibodies in rabbits.
Fig.␣S5 Transcriptional levels of five SCW PtrCesA genes in ptrcesa mutants using reverse transcription (RT)‐PCR analysis.
Fig.␣S6 Characterization of the ptrcesa4, 7a/b, 8a/b, 7a, 7b, 8a, and 8b mutants.
Fig.␣S7 Observation of stem internodes, mature leaves, and roots of 3‐month‐old ptrcesa4, 7a/b and 8a/b mutants.
Fig.␣S8 Anatomical analysis of different stem internodes from the WT, an ptrcesa4, 7a/b and 8a/b mutants.
Fig.␣S9 Anatomical analysis of different stem internodes from the WT, and ptrcesa7a, 7b, 8a and 8b mutants.
Fig.␣S10 Observation of pith parenchyma in the WT, and ptrcesa4, 7a/b and 8a/b mutants.
Fig.␣S11 Wall thickness of xylem and phloem fibres in the basal stems of 6‐month‐old WT and ptrcesa mutants.
Fig.␣S12 Microscopic analysis of the disaggregated xylem fibres and vessels in the WT, and ptrcesa4, 7a/b and 8a/b mutants.
Fig.␣S13 Induction of TW in the WT and ptrcesa mutants under gravi‐stimulation.
Fig.␣S14 Immunolocalization of crystalline cellulose in TW and OW sides in the WT, and ptrcesa4, 7ab and 8ab mutants.
Fig.␣S15 Immunolocalization of the xylan in TW and OW sides in the WT, and ptrcesa4, 7ab and 8ab mutants.
Fig.␣S16 Immunolocalization of β‐(1 → 4)‐galactan in TW and OW sides in the WT, and ptrcesa4, 7ab and 8ab mutants.
Fig.␣S17 Immunolocalization of the mannan in TW and OW sides in the WT, and ptrcesa4, 7ab and 8ab mutants.
Fig.␣S18 Immunolocalization of crystalline cellulose in phloem fibres of the WT, and ptrcesa4, 7ab and 8ab mutants.
Fig.␣S19 Immunolocalization of β‐(1 → 4)‐galactan in phloem fibres of the WT, ptrcesa4, 7ab and 8ab mutants.
Fig.␣S20 Immunolocalization of the mannan in phloem fibres of the WT, and ptrcesa4, 7ab and 8ab mutants.
Fig.␣S21 Immunolocalization of the xylan in phloem fibres of the WT, and ptrcesa4, 7ab and 8ab mutants.
Fig.␣S22 Lignin phloroglucinol staining in the WT, andptrcesa4, 7ab and 8ab mutants.
Fig.␣S23 Crystalline cellulose and lignin content in wood of the WT, and ptrcesa7a, 7b, 8a and 8b mutants.
MethodsS1 Analysis of putative Cas9/gRNA off‐target sites.
MethodsS2 RNA extraction and RT‐PCR analysis.
MethodsS3 SEM of leaf epidermal cells.
MethodsS4 Wood fibre and vessel cell length analysis.
Table␣S1 Primers used in this study.
Table␣S2 The Cas9/gRNA‐targeted mutations in a single PtrCHLI1 gene of P. trichocarpa.
Table␣S3 The Cas9/gRNA‐targeted mutations in the PtrCHLI1 and 2 genes of P. trichocarpa.
Table␣S4 Analysis of potential off‐target sites among the Cas9/gRNA‐induced ptrch l i1 mutants.
Table␣S5 The Cas9/gRNA‐targeted mutations in PtrCesA4, 7A, 7B, 8A, 8B, 7A /B and 8A /B genes of P. trichocarpa.
Table␣S6 Analysis of potential off‐target sites among the Cas9/gRNA‐induced ptrcesa mutants.
Table␣S7 Inheritance of the Cas9/gRNA‐induced mutations in progeny of ptrcesa4, 7a/b and 8a/b mutants through asexual propagation methods.
Table␣S8 Inheritance of the Cas9/gRNA‐induced mutations in the progeny of the ptrcesa lines.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
We thank Qi‐Jun Chen (China Agricultural University) for providing the pCBC‐DT1T2 and pHSE401 vectors. The research was supported by the National Key Research and Development Program of China (2016YFD0600106), the National Natural Science Foundation of China (31570580 and 31770637), the Fundamental Research Funds for the Central Universities (2572018CL01), the Innovation Project of State Key Laboratory of Tree Genetics and Breeding (2013A02) and the Heilongjiang Touyan Innovation Team Program (Tree Genetics and Breeding Innovation Team). The authors declare no conflict of interest.
Data availability
The data used to support the findings of this study appeared in the submitted article and are available from the corresponding author.
References
- Abbas M, Peszlen I, Shi R, Kim H, Katahira R, Kafle K, Xiang Z, Huang X, Min D, Mohamadamin Met␣al. 2020. Involvement of CesA4, CesA7‐A/B and CesA8‐A/B in secondary wall formation in Populus trichocarpa wood. Tree Physiology 40: 73–89. [DOI] [PubMed] [Google Scholar]
- Atanassov I, Pittman J, Turner S. 2009. Elucidating the mechanisms of assembly and subunit interaction of the cellulose synthase complex of Arabidopsis secondary cell walls. Journal of Biological Chemistry 284: 3833–3841. [DOI] [PubMed] [Google Scholar]
- Barnett JR, Bonham VA. 2004. Cellulose microfibril angle in the cell wall of wood fibres. Biological Reviews 79: 461–472. [DOI] [PubMed] [Google Scholar]
- Bellasio C, Fini A, Ferrini F. 2014. Evaluation of a high throughput starch analysis optimised for wood. PLoS ONE 9: e86645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bögre L, Magyar Z, López‐Juez E. 2008. New clues to organ size control in plants. Genome Biology 9: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brereton NJ, Pitre FE, Ray MJ, Karp A, Murphy RJ. 2011. Investigation of tension wood formation and 2, 6‐dichlorbenzonitrile application in short rotation coppice willow composition and enzymatic saccharification. Biotechnology for Biofuels 4: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brereton NJ, Ray MJ, Shield I, Martin P, Karp A, Murphy RJ. 2012. Reaction wood – a key cause of variation in cell wall recalcitrance in willow. Biotechnology for Biofuels 5: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choat B, Cobb AR, Jansen S. 2008. Structure and function of bordered pits: new discoveries and impacts on whole‐plant hydraulic function. New Phytologist 177: 608–626. [DOI] [PubMed] [Google Scholar]
- Coleman HD, Yan J, Mansfield SD. 2009. Sucrose synthase affects carbon partitioning to increase cellulose production and altered cell wall ultrastructure. Proceedings of the National Academy of Sciences, USA 106: 13118–13123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, Hofte H, Gonneau M, Vernhettes S. 2007. Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 104: 15572–15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmawardhana P, Brunner AM, Strauss SH. 2010. Genome‐wide transcriptome analysis of the transition from primary to secondary stem development in Populus trichocarpa . BMC Genomics 11: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djerbi S, Lindskog M, Arvestad L, Sterky F, Teeri TT. 2005. The genome sequence of black cottonwood (Populus trichocarpa) reveals 18 conserved cellulose synthase (CesA) genes. Planta 221: 739–746. [DOI] [PubMed] [Google Scholar]
- Doblin MS, Kurek I, Jacob‐Wilk D, Delmer DP. 2002. Cellulose biosynthesis in plants: from genes to rosettes. Plant and Cell Physiology 43: 1407–1420. [DOI] [PubMed] [Google Scholar]
- Du S, Yamamoto F. 2007. An overview of the biology of reaction wood formation. Journal of Integrative Plant Biology 49: 131–143. [Google Scholar]
- Fagerstedt K, Mellerowicz E, Gorshkova T, Ruel K, Joseleau J. 2014. Cell wall polymers in reaction wood. In: Gardiner B, Barnett J, Saranpää P, Gril J, eds. The biology of reaction wood. Berlin: Springer, 37–106. [Google Scholar]
- Fan D, Liu T, Li C, Jiao B, Li S, Hou Y, Luo K. 2015. Efficient CRISPR/Cas9 mediated targeted mutagenesis in Populus in the first generation. Scientific Reports 5: 12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felten J, Sundberg B. 2013. Biology, chemistry and structure of tension wood. In: Fromm J, ed. Cellular aspects of wood formation, vol 20. Berlin: Springer, 203–224. [Google Scholar]
- Fernandes AN, Thomas LH, Altaner CM, Callow P, Forsyth VT, Apperley DC, Kennedy CJ, Jarvis MC. 2011. Nanostructure of cellulose microfibrils in spruce wood. Proceedings of the National Academy of Sciences, USA 108: E1195–E1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster CE, Martin TM, Pauly M. 2010a. Comprehensive compositional analysis of plant cell walls (lignocellulosic biomass) part I: Lignin. Journal of Visualized Experiments 37: pii: 1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster CE, Martin TM, Pauly M. 2010b. Comprehensive compositional analysis of plant cell walls (lignocellulosic biomass) part II: carbohydrates. Journal of Visualized Experiments 37: pii: 1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Hayashi T, Mizuno K. 2010. Sucrose synthase is an integral component of the cellulose synthesis machinery. Plant and Cell Physiology 51: 294–301. [DOI] [PubMed] [Google Scholar]
- Gorshkova T, Chernova T, Mokshina N, Ageeva M, Mikshina P. 2018. Plant ‘muscles’: fibers with a tertiary cell wall. New Phytologist 218: 66–72. [DOI] [PubMed] [Google Scholar]
- Gorshkova T, Mokshina N, Chernova T, Ibragimova N, Salnikov V, Mikshina P, Tryfona T, Banasiak A, Immerzeel P, Dupree Pet␣al. 2015. Aspen tension wood fibers contain beta‐(1→4)‐galactans and acidic arabinogalactans retained by cellulose microfibrils in gelatinous walls. Plant Physiology 169: 2048–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groover A. 2016. Gravitropisms and reaction woods of forest trees ‐ evolution, functions and mechanisms. New Phytologist 211: 790–802. [DOI] [PubMed] [Google Scholar]
- Herth W. 1983. Arrays of plasma‐membrane “rosettes” involved in cellulose microfibril formation of Spirogyra . Planta 159: 347–356. [DOI] [PubMed] [Google Scholar]
- Huang YS, Li HM. 2009. Arabidopsis CHLI2 can substitute for CHLI11 . Plant Physiology 150: 636–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson S, Douglas CJ. 2007. Populus: a model system for plant biology. Annual Review of Plant Biology 58: 435–458. [DOI] [PubMed] [Google Scholar]
- Joshi CP, Thammannagowda S, Fujino T, Gou JQ, Avci U, Haigler CH, McDonnell LM, Mansfield SD, Mengesha B, Carpita NCet␣al. 2011. Perturbation of wood cellulose synthesis causes pleiotropic effects in transgenic Aspen. Molecular Plant 4: 331–345. [DOI] [PubMed] [Google Scholar]
- Jourez B, Riboux A, Leclercq A. 2001. Anatomical characteristics of tension wood and opposite wood in young inclined stems of poplar (Populus euramericana cv ‘Ghoy’). IAWA Journal 22: 133–157. [Google Scholar]
- Kalluri UC, Joshi CP. 2004. Differential expression patterns of two cellulose synthase genes are associated with primary and secondary cell wall development in aspen trees. Planta 220: 47–55. [DOI] [PubMed] [Google Scholar]
- Kimura S, Laosinchai W, Itoh T, Cui X, Linder CR, Brown RM Jr. 1999. Immunogold labeling of rosette terminal cellulose‐synthesizing complexes in the vascular plant vigna angularis . The Plant Cell 11: 2075–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Atanassov I, Turner S. 2017. Functional analysis of cellulose synthase (CESA) protein class specificity. Plant Physiology 173: 970–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Thammannagowda S, Bulone V, Chiang V, Han KH, Joshi CP, Mansfield SD, Mellerowicz E, Sundberg B, Teeri Tet␣al. 2009. An update on the nomenclature for the cellulose synthase genes in Populus . Trends in Plant Science 14: 248–254. [DOI] [PubMed] [Google Scholar]
- Kwon M. 2008. Tension wood as a model system to explore the carbon partitioning between lignin and cellulose biosynthesis in woody plants. Journal of Applied Biological Chemistry 51: 83–87. [Google Scholar]
- Li S, Zhen C, Xu W, Wang C, Cheng Y. 2017. Simple, rapid and efficient transformation of genotype Nisqually‐1: a basic tool for the first sequenced model tree. Scientific Reports 7: 2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y‐K, Xie X, Lindsay SE, Wang YB, Masle J, Williamson L, Leyser O, Hetherington AM. 2010. Cell wall composition contributes to the control of transpiration efficiency in Arabidopsis thaliana . The Plant Journal 64: 679–686. [DOI] [PubMed] [Google Scholar]
- Liu J, Hai G, Wang C, Cao S, Xu W, Jia Z, Yang C, Wang JP, Dai S, Cheng Y. 2015. Comparative proteomic analysis of Populus trichocarpa early stem from primary to secondary growild‐typeh. Journal of Proteomics 126: 94–108. [DOI] [PubMed] [Google Scholar]
- Liu Y, Xu F, Gou J, Al‐Haddad J, Telewski FW, Bae HJ, Joshi CP. 2012. Importance of two consecutive methionines at the N‐terminus of a cellulose synthase (PtdCesA8A) for normal wood cellulose synthesis in aspen. Tree Physiology 32: 1403–1412. [DOI] [PubMed] [Google Scholar]
- McFarlane HE, Doring A, Persson S. 2014. The cell biology of cellulose synthesis. Annual Review of Plant Biology 65: 69–94. [DOI] [PubMed] [Google Scholar]
- Mellerowicz EJ, Gorshkova TA. 2012. Tensional stress generation in gelatinous fibres: a review and possible mechanism based on cell wall structure and composition. Journal of Experimental Botany 63: 551–565. [DOI] [PubMed] [Google Scholar]
- Mokshina N, Gorshkov O, Ibragimova N, Chernova T, Gorshkova T. 2017. Cellulosic fibres of flax recruit both primary and secondary cell wall cellulose synthases during deposition of thick tertiary cell walls and in the course of graviresponse. Functional Plant Biology 44: 820–831. [DOI] [PubMed] [Google Scholar]
- Naito Y, Hino K, Bono H, Ui‐Tei K. 2015. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off‐target sites. Bioinformatics 31: 1120–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa K, Yoshinaga A, Takabe K. 2012. Anatomy and lignin distribution in reaction phloem fibres of several Japanese hardwoods. Annals of Botany 110: 897–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa K, Yoshinaga A, Takabe K. 2014. Xylan deposition and lignification in the multi‐layered cell walls of phloem fibres in Mallotus japonicus (Euphorbiaceae). Tree Physiology 34: 1018–1029. [DOI] [PubMed] [Google Scholar]
- Nakano J, Meshitsuka G. 1992. The determination of lignin. In: Lin SY, Dence CW, eds. Methods in lignin chemistry. Berlin, Germany: Springer, 33–61. [Google Scholar]
- Nanko H. 1979. Studies on the development and cell wall structure of sclerenchymatous elements in the secondary phloem of woody dicotyledons and conifers. PhD thesis, Kyoto University, Japan. [Google Scholar]
- Nishikubo N, Awano T, Banasiak A, Bourquin V, Ibatullin F, Funada R, Brumer H, Teeri TT, Hayashi T, Sundberg Bet␣al. 2007. Xyloglucan endo‐transglycosylase (XET) functions in gelatinous layers of tension wood fibers in poplar–a glimpse into the mechanism of the balancing act of trees. Plant Cell Physiology 48: 843–855. [DOI] [PubMed] [Google Scholar]
- Nixon BT, Mansouri K, Singh A, Du J, Davis JK, Lee JG, Slabaugh E, Vandavasi VG, O'Neill H, Roberts EMet␣al. 2016. Comparative structural and computational analysis supports eighteen cellulose synthases in the plant cellulose synthesis complex. Scientific Reports 6: 28696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe Y, Liang Z, Ren C, Nishitani C, Osakabe K, Wada M, Komori S, Malnoy M, Velasco R, Poli Met␣al. 2018. CRISPR‐Cas9‐mediated genome editing in apple and grapevine. Nature Protocols 13: 2844–2863. [DOI] [PubMed] [Google Scholar]
- Persson S, Paredez A, Carroll A, Palsdottir H, Doblin M, Poindexter P, Khitrov N, Auer M, Somerville CR. 2007. Genetic evidence for three unique components in primary cell‐wall cellulose synthase complexes in Arabidopsis . Proceedings of the National Academy of Sciences, USA 104: 15566–15571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plomion C, Leprovost G, Stokes A. 2001. Wood formation in trees. Plant Physiology 127: 1513–1523. [PMC free article] [PubMed] [Google Scholar]
- Purushotham P, Ho R, Zimmer J. 2020. Architecture of a catalytically active homotrimeric plant cellulose synthase complex. Science 369: 1089–1094. [DOI] [PubMed] [Google Scholar]
- Richardson AD, Carbone MS, Keenan TF, Czimczik CI, Hollinger DY, Murakami P, Schaberg PG, Xu X. 2013. Seasonal dynamics and age of stemwood nonstructural carbohydrates in temperate forest trees. New Phytologist 197: 850–861. [DOI] [PubMed] [Google Scholar]
- Roach MJ, Mokshina NY, Badhan A, Snegireva AV, Hobson N, Deyholos MK, Gorshkova TA. 2011. Development of cellulosic secondary walls in flax fibers requires beta‐galactosidase. Plant Physiology 156: 1351–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C. 2006. Cellulose synthesis in higher plants. Annual Review of Cell and Developmental Biology 22: 53–78. [DOI] [PubMed] [Google Scholar]
- Song D, Shen J, Li L. 2010. Characterization of cellulose synthase complexes in Populus xylem differentiation. New Phytologist 187: 777–790. [DOI] [PubMed] [Google Scholar]
- Sperry JS, Hacke UG, Pittermann J. 2006. Size and function in conifer tracheids and angiosperm vessels. American Journal of Botany 93: 1490–1500. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Li L, Sun YH, Chiang VL. 2006. The cellulose synthase gene superfamily and biochemical functions of xylem‐specific cellulose synthase‐like genes in Populus trichocarpa . Plant Physiology 142: 1233–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata N, Taniguchi T. 2015. Expression divergence of cellulose synthase (CesA) genes after a recent whole genome duplication event in Populus . Planta 24: 29–42. [DOI] [PubMed] [Google Scholar]
- Taylor NG, Gardiner JC, Whiteman R, Turner SR. 2004. Cellulose synthesis in the Arabidopsis secondary cell wall. Cellulose 11: 329–338. [Google Scholar]
- Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR. 2003. Interactions among three distinct CesA proteins essential for cellulose synthesis. Proceedings of the National Academy of Sciences, USA 100: 1450–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Laurie S, Turner SR. 2000. Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis . The Plant Cell 12: 2529–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Scheible WR, Cutler S, Somerville CR, Turner SR. 1999. The irregular xylem 3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. The Plant Cell 11: 769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas LH, Forsyth VT, Šturcová A, Kennedy CJ, May RP, Altaner CM, Apperley DC, Wess TJ, Jarvis MC. 2013. Structure of cellulose microfibrils in primary cell walls from collenchyma. Plant Physiology 161: 465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, DiFazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov Aet␣al. 2006. The genome of black cottonwood, Populus trichocarpa (Torr & Gray). Science 313: 1596–1604. [DOI] [PubMed] [Google Scholar]
- Wardrop A, Dadswell H. 1955. The nature of reaction wood. IV. Variations in cell wall organization of tension wood fibres. Australian Journal of Botany 3: 177–189. [Google Scholar]
- Xi W, Song D, Sun J, Shen J, Li L. 2017. Formation of wood secondary cell wall may involve two type cellulose synthase complexes in Populus . Plant Molecular Biology 93: 419–429. [DOI] [PubMed] [Google Scholar]
- Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ. 2014. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biology 14: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Ruelle J, Arakawa Y, Yoshida M, Clair B, Gril J. 2010. Origin of the characteristic hygro‐mechanical properties of the gelatinous layer in tension wood from Kunugi oak (Quercus acutissima). Wood Science and Technology 44: 149–163. [Google Scholar]
- Yoshimura K, Saiki S, Yazaki K, Ogasa M, Shirai M, Nakano T, Yoshimura J, Ishida A. 2016. The dynamics of carbon stored in xylem sapwood to drought‐induced hydraulic stress in mature trees. Scientific Reports 6: 24513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Zhang L, Li F, Zhang D, Liu X, Wang H, Xu Z, Chu C, Zhou Y. 2017. Control of secondary cell wall patterning involves xylan deacetylation by a GDSL esterase. Nature Plants 3: 17017. [DOI] [PubMed] [Google Scholar]
- Zhang X, Dominguez PG, Kumar M, Bygdell J, Miroshnichenko S, Sundberg B, Wingsle G, Niittylä T. 2018. Cellulose synthase stoichiometry in aspen differs from Arabidopsis and Norway spruce. Plant Physiology 177: 1096–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Cheng Y, Yap A, Chateigner‐Boutin AL, Delannoy E, Hammani K, Small I, Huang J. 2009. The Arabidopsis gene YS1 encoding a DYW protein is required for editing of rpoB transcripts and the rapid development of chloroplasts during early growth. The Plant Journal 58: 82–96. [DOI] [PubMed] [Google Scholar]
- Zhou X, Jacobs TB, Xue LJ, Harding SA, Tsai CJ. 2015. Exploiting SNPs for biallelic CRISPR mutations in the outcrossing woody perennial Populus reveals 4‐coumarate: CoA ligase specificity and redundancy. New Phytologist 208: 298–301. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig.␣S1 Cas9/gRNA system and stepwise protocol in Nisqually‐1.
Fig.␣S2 Cas9/gRNA‐induced mutations in PtrCHLI1 gene and PtrCHLI1/2 family genes of P. trichocarpa.
Fig.␣S3 Cas9/gRNA‐induced mutations in PtrCesA7A, PtrCesA7B, PtrCesA8A and PtrCesA8B genes of P. trichocarpa.
Fig.␣S4 Production of anti‐PtrCesA4, −7A/B and −8A/B polyclonal antibodies in rabbits.
Fig.␣S5 Transcriptional levels of five SCW PtrCesA genes in ptrcesa mutants using reverse transcription (RT)‐PCR analysis.
Fig.␣S6 Characterization of the ptrcesa4, 7a/b, 8a/b, 7a, 7b, 8a, and 8b mutants.
Fig.␣S7 Observation of stem internodes, mature leaves, and roots of 3‐month‐old ptrcesa4, 7a/b and 8a/b mutants.
Fig.␣S8 Anatomical analysis of different stem internodes from the WT, an ptrcesa4, 7a/b and 8a/b mutants.
Fig.␣S9 Anatomical analysis of different stem internodes from the WT, and ptrcesa7a, 7b, 8a and 8b mutants.
Fig.␣S10 Observation of pith parenchyma in the WT, and ptrcesa4, 7a/b and 8a/b mutants.
Fig.␣S11 Wall thickness of xylem and phloem fibres in the basal stems of 6‐month‐old WT and ptrcesa mutants.
Fig.␣S12 Microscopic analysis of the disaggregated xylem fibres and vessels in the WT, and ptrcesa4, 7a/b and 8a/b mutants.
Fig.␣S13 Induction of TW in the WT and ptrcesa mutants under gravi‐stimulation.
Fig.␣S14 Immunolocalization of crystalline cellulose in TW and OW sides in the WT, and ptrcesa4, 7ab and 8ab mutants.
Fig.␣S15 Immunolocalization of the xylan in TW and OW sides in the WT, and ptrcesa4, 7ab and 8ab mutants.
Fig.␣S16 Immunolocalization of β‐(1 → 4)‐galactan in TW and OW sides in the WT, and ptrcesa4, 7ab and 8ab mutants.
Fig.␣S17 Immunolocalization of the mannan in TW and OW sides in the WT, and ptrcesa4, 7ab and 8ab mutants.
Fig.␣S18 Immunolocalization of crystalline cellulose in phloem fibres of the WT, and ptrcesa4, 7ab and 8ab mutants.
Fig.␣S19 Immunolocalization of β‐(1 → 4)‐galactan in phloem fibres of the WT, ptrcesa4, 7ab and 8ab mutants.
Fig.␣S20 Immunolocalization of the mannan in phloem fibres of the WT, and ptrcesa4, 7ab and 8ab mutants.
Fig.␣S21 Immunolocalization of the xylan in phloem fibres of the WT, and ptrcesa4, 7ab and 8ab mutants.
Fig.␣S22 Lignin phloroglucinol staining in the WT, andptrcesa4, 7ab and 8ab mutants.
Fig.␣S23 Crystalline cellulose and lignin content in wood of the WT, and ptrcesa7a, 7b, 8a and 8b mutants.
MethodsS1 Analysis of putative Cas9/gRNA off‐target sites.
MethodsS2 RNA extraction and RT‐PCR analysis.
MethodsS3 SEM of leaf epidermal cells.
MethodsS4 Wood fibre and vessel cell length analysis.
Table␣S1 Primers used in this study.
Table␣S2 The Cas9/gRNA‐targeted mutations in a single PtrCHLI1 gene of P. trichocarpa.
Table␣S3 The Cas9/gRNA‐targeted mutations in the PtrCHLI1 and 2 genes of P. trichocarpa.
Table␣S4 Analysis of potential off‐target sites among the Cas9/gRNA‐induced ptrch l i1 mutants.
Table␣S5 The Cas9/gRNA‐targeted mutations in PtrCesA4, 7A, 7B, 8A, 8B, 7A /B and 8A /B genes of P. trichocarpa.
Table␣S6 Analysis of potential off‐target sites among the Cas9/gRNA‐induced ptrcesa mutants.
Table␣S7 Inheritance of the Cas9/gRNA‐induced mutations in progeny of ptrcesa4, 7a/b and 8a/b mutants through asexual propagation methods.
Table␣S8 Inheritance of the Cas9/gRNA‐induced mutations in the progeny of the ptrcesa lines.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Data Availability Statement
The data used to support the findings of this study appeared in the submitted article and are available from the corresponding author.


