Abstract
Objective
To compare immune cell phenotype and function in psoriatic arthritis (PsA) versus psoriasis in order to better understand the pathogenesis of PsA.
Methods
In‐depth immunophenotyping of different T cell and dendritic cell subsets was performed in patients with PsA, psoriasis, or axial spondyloarthritis and healthy controls. Subsequently, we analyzed cells from peripheral blood, synovial fluid (SF), and skin biopsy specimens using flow cytometry, along with high‐throughput transcriptome analyses and functional assays on the specific cell populations that appeared to differentiate PsA from psoriasis.
Results
Compared to healthy controls, the peripheral blood of patients with PsA was characterized by an increase in regulatory CD4+ T cells and interleukin‐17A (IL‐17A) and IL‐22 coproducing CD8+ T cells. One population specifically differentiated PsA from psoriasis: i.e., CD8+CCR10+ T cells were enriched in PsA. CD8+CCR10+ T cells expressed high levels of DNAX accessory molecule 1 and were effector memory cells that coexpressed skin‐homing receptors CCR4 and cutaneous lymphocyte antigen. CD8+CCR10+ T cells were detected under inflammatory and homeostatic conditions in skin, but were not enriched in SF. Gene profiling further revealed that CD8+CCR10+ T cells expressed GATA3, FOXP3, and core transcriptional signature of tissue‐resident memory T cells, including CD103. Specific genes, including RORC, IFNAR1, and ERAP1, were up‐regulated in PsA compared to psoriasis. CD8+CCR10+ T cells were endowed with a Tc2/22‐like cytokine profile, lacked cytotoxic potential, and displayed overall regulatory function.
Conclusion
Tissue‐resident memory CD8+ T cells derived from the skin are enhanced in the circulation of patients with PsA compared to patients with psoriasis alone. This may indicate that aberrances in cutaneous tissue homeostasis contribute to arthritis development.
INTRODUCTION
New critical insights into the pathogenesis of psoriasis and psoriatic arthritis (PsA) have been made in recent years, including the role of the interleukin‐23 (IL‐23)/IL‐17 axis (1, 2). This finding further propelled the development of novel drugs with potential to vastly improve psoriasis, but their strength at halting arthritis is less impressive. A clearer understanding of the pathogenesis of PsA could guide the development of therapeutics capable of resolving arthritis.
The prevalence of PsA in patients with psoriasis is ~20% (2), and in its simplest form, PsA presents with arthritis in a patient with a history of psoriasis. However, the relationship between skin and joint manifestations encompasses a spectrum: patients can have severe psoriasis without musculoskeletal symptoms, while others have minimal psoriasis and severe arthritis.
This raised the question of whether these diseases are part of a single spectrum or are separate entities (3, 4). More specifically, it is currently unknown whether immunologic processes in the skin and the joint are directly related. One possibility is that immunologic processes in the skin and the joint occur in parallel, but independently from each other. Some arguments for this hypothesis are the aforementioned clinical disease heterogeneity of PsA and the finding that response to specific treatment targets varies by tissue site (2). Also, observations that local tissue damage (e.g., the Koebner phenomenon) can trigger local inflammation (5) fit this concept.
Another possibility is that the pathophysiologic processes are directly linked and occur sequentially. Supporting this hypothesis is the fact that psoriasis itself is a strong risk factor for the development of arthritis and that psoriasis typically precedes the onset of arthritis by several years. In this scenario, immune cells, cytokines, and/or other mediators induced by skin inflammation could trigger a second hit at the joint. Indeed, soluble factors have been shown to be capable of inducing models of spondyloarthritis (SpA) (6), potentially sparking responses of resident, innate lymphocytes at musculoskeletal sites (7).
While the role of different immune cells in the skin and joints has been described (1), there have been few studies explicitly comparing immune cells between psoriasis and PsA (8, 9, 10, 11). In this study, we first performed in‐depth immunophenotyping in patients with psoriasis and patients with PsA who had not been treated with disease‐modifying antirheumatic drugs (DMARDs) and were matched with the psoriasis patients for skin disease activity as measured by Psoriasis Area and Severity Index (PASI) (12). This was followed by phenotypic, transcriptomic, and functional investigations to determine the specific CD8+ T cell subset that best distinguished PsA from psoriasis.
PATIENTS AND METHODS
Study cohort and samples
The study was conducted at the Department of Rheumatology and Clinical Immunology, University Medical Centre Utrecht (UMCU), in accordance with the Declaration of Helsinki and with approval from the institutional review board. Written informed consent was obtained from all patients before participation. Patients with psoriasis had a dermatologist‐confirmed diagnosis. All patients classified as having psoriasis alone underwent clinical evaluation to exclude concomitant PsA. Patients with PsA fulfilled the Classification of Psoriatic Arthritis (CASPAR) Study Group criteria (13). Patients with axial SpA met the Assessment of SpondyloArthritis international Society classification criteria (14) and did not have concomitant psoriasis.
In the first phase of the study, the frequency of T cells and dendritic cell (DC) subsets in peripheral blood mononuclear cells (PBMCs) from patients with PsA (n = 21) was compared to that in healthy controls (n = 20), patients with psoriasis (n = 21), and patients with axial SpA (n = 16). Patients with psoriasis and patients with PsA were matched for key clinical parameters, including PASI (Supplementary Table 1, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41652/abstract). In the second phase, we specifically investigated the properties of CD8+CCR10+ T cells, for which additional samples were collected: PBMCs (from 32 healthy controls, 17 patients with psoriasis, and 18 patients with PsA), synovial fluid (SF) (from 8 patients with PsA), and skin biopsy samples (from 6 patients with PsA and 8 patients with psoriasis). With the exception of SF samples, all samples were obtained from patients who were not being treated with DMARDs at the time of participation.
Sample collection
PBMCs were isolated by density centrifugation using Ficoll‐Paque Plus (GE Healthcare) from lithium‐heparinized venous blood and first stored in liquid nitrogen. Four‐millimeter punch biopsy sections from lesional psoriatic skin sites (donor‐dependent lesional sites) and nonlesional skin sites (always dorsal thorax) were obtained and placed in phosphate buffered saline on ice before further processing. The skin biopsy samples were subjected to mechanical and tissue digestion according to the manufacturer’s protocol (Whole Skin Dissociation Kit, human; Miltenyi Biotec), after which flow cytometry was performed on the freshly digested skin biopsy samples. SF mononuclear cells were isolated by density centrifugation using a Ficoll‐Paque Plus gradient procedure and first stored in liquid nitrogen.
Flow cytometry
Four different flow cytometry panels were used to identify and enumerate in thawed PBMCs the relative frequency of a broad range of T cell and DC subsets (Supplementary Figures 1–4, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41652/abstract), using a standardized flow‐cytometry protocol, as previously described (15). Antibodies used are listed in Supplementary Table 2 (http://onlinelibrary.wiley.com/doi/10.1002/art.41652/abstract). Fixation and permeabilization solution was used for intracellular antibody staining according to the manufacturer’s instructions (eBioscience). Fluorescence minus one was used as negative control for determining manual gating strategy. Flow cytometry data were acquired using a BD LSRFortessa Cell analyzer, and flow cytometric cell sorting was performed using a BD FACSAria III cell sorter (BD Bioscience). Different subsets of CD8+ T cells were flow sorted based on expression of CCR10 and/or CCR4 (Supplementary Figure 5, http://onlinelibrary.wiley.com/doi/10.1002/art.41652/abstract) for proliferation suppression assays and RNA sequencing.
Functional assays
For quantification of intracellular cytokine production by flow cytometry, PBMCs were restimulated for 4 hours in culture medium (RPMI 1640 with 10% fetal calf serum), with phorbol 12‐myristate 13‐acetate (PMA), ionomycin calcium salt, and BD GolgiPlug (BD Biosciences) at 37°C.
Proliferation suppression assays were performed in accordance with established protocols for Treg cells (16). For this assay, “regulatory‐type” cells and “target cells” were derived from fresh PBMCs isolated from 6 healthy controls. The “regulatory‐type” cells were different flow‐sorted CD8+ T cell subsets (Supplementary Figure 5, http://onlinelibrary.wiley.com/doi/10.1002/art.41652/abstract). The target cells were CellTrace Violet (Thermo Fisher) labeled autologous PBMCs. The “regulatory‐type” cells and target cells were resuspended in culture medium (RPMI 1640 with 10% fetal calf serum) and cocultured at a ratio of 1:2 cells, respectively, in anti‐CD3–coated plates for 4 days. The readout was the percentage of proliferated target cells. As reference for the readout, regulatory CD4+ T cells (CD3+CD4+CD25high CD127−) and effector CD4+ T cells (CD3+CD4+CD25−CD127high) were flow sorted from the same donors in each experiment, which indicated that the suppression assay worked (results not shown).
RNA sequencing
Distinct CD8+ T cell subsets were flow sorted from thawed PBMCs (8 healthy controls, 6 patients with psoriasis, and 6 patients with PsA) (Supplementary Figure 5, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41652/abstract). Cells were lysed using Buffer RLT Plus in the presence of β‐mercaptoethanol (final concentration 1%), and RNA was isolated according to the manufacturer’s protocol (Qiagen Universal Kit). In total, 57 samples were used for analysis, and all passed internal quality control checks. RNA sequencing was performed using an Illumina HiSeq 4000 sequencer (paired‐end, 150 bp) at GenomeScan in Leiden, The Netherlands using standard manufacturer’s protocols. FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used to check the quality of the raw reads obtained from RNA‐Seq. STAR aligner was used to align the reads to the human genome (GRCh38 build 79) (17, 18). HTSeq was used to obtain read counts for each annotated gene (19). Differentially expressed genes (DEGs) were identified using Bioconductor/R package DESeq2 (20). Wald’s test was used to identify differential gene expressions between conditions (healthy controls, patients with psoriasis, and patients with PsA) and cell subset pairs (Supplementary Figure 5, http://onlinelibrary.wiley.com/doi/10.1002/art.41652/abstract). Variance stabilizing transformation was applied to the raw read count data to obtain normalized gene counts (variance‐stabilized data), which were used for subsequent plotting. The heatmap, principal components analysis, and violin plots were plotted using R.
Statistical analysis
Categorical variables were compared using chi‐square tests, and group differences were compared using the Mann‐Whitney U test or independent‐samples t‐test (based on normality distribution). Group differences were compared using Wilcoxon’s signed rank test for paired samples. Spearman’s rank correlation was used to test the association between clinical parameters and flow cytometry results.
Flow cytometry data were analyzed using FlowJo software (TreeStar). Statistical analysis and visual representation of the data were performed using SPSS version 25 and GraphPad Prism software, version 7.0. The heatmap of flow cytometry results was made with MetaboAnalyst 4.0, using Ward’s clustering algorithm and autoscaling of features (21). Venn diagrams were made using Venny 2.1 (22). P values less than 0.05 were considered significant.
RESULTS
Higher frequency of circulating CD8+CD45RO+CCR10+ T cells in patients with PsA compared to patients with psoriasis
We compared the frequency of T cell and DC subsets in peripheral blood from patients with PsA versus healthy controls, patients with psoriasis, and axial SpA using our standardized immunophenotyping panels (Supplementary Table 1 and Supplementary Figures 1–4, http://onlinelibrary.wiley.com/doi/10.1002/art.41652/abstract). The results of immunophenotyping indicate that PBMC subsets from patients with psoriasis and PsA are generally similar (Figure 1A). The CD8+CD45RO+CCR10+ subset was the only cell population that was significantly different between patients with PsA and those with psoriasis, with higher levels found in patients with PsA (P < 0.05) (Figure 1B). Compared to healthy controls, patients with PsA had increased frequencies of CD8+CCR10+ T cells, CD8+IL‐17A+IL‐22+ T cells, and regulatory CD4+ T cells. Patients with PsA also had reduced frequencies of plasmacytoid DCs and CD8+CD161+CCR6+ T cells (mucosal‐associated invariant T–like cells) compared to healthy controls (Figures 1B–F). Considering that there are few studies that have examined CCR10 expression on CD8+ T cells (23, 24, 25), we subsequently set out to further characterize the phenotype, origin, and function of CCR10 expression on CD8+ T cells in general and in relation to PsA.
Figure 1.

Phenotype and function of circulating immune cell subsets in psoriatic arthritis (PsA), and frequencies of T cell and dendritic cell subsets in peripheral blood mononuclear cells. A, Heatmap showing the top 20 flow cytometry features that best distinguished the different groups studied (healthy controls [HCs] and patients with axial spondyloarthritis [AxSpA], psoriasis [Pso], or psoriatic arthritis [PsA]), as assessed by analysis of variance. B, CCR10+ cells within CD8+CD45RO+ T cells. C, IL‐17A+IL‐22+ cells within CD8+ T cells. D, FoxP3+CD25+CD45RO+ cells within CD4+ T cells. E, CD161+CCR6+ cells within CD8+CD45RO+ T cells. F, CD303+ cells within DR+CD14−CD16−cells. Symbols represent individual subjects; bars show the median. * = P < 0.05. IL‐17A = interleukin‐17A.
CCR10 expression on CD8+ T cells marks a memory, DNAM‐1high phenotype
We first reexamined the phenotype of CD8+ T cells that expressed CCR10 (not using CD45RO+ T cells as a prerequisite). As expected, when directly gating on CD8+CCR10+ T cells, we found that the majority were classified as either central memory T (Tcm) cells or effector memory T (Tem) cells (Supplementary Figures 6A and B, http://onlinelibrary.wiley.com/doi/10.1002/art.41652/abstract). CCR10 expression was significantly elevated in both Tcm and Tem subsets in patients with PsA (Supplementary Figures 6C and D). CD8+CCR10+ T cells were also enriched for CCR6 coexpression. The expression of CD45RO, CD27, CCR6, and CXCR3 with respect to CD8+CCR10+ T cells was similar across patient groups (data not shown).
We broadly screened additional CD8 T cell markers and found that CD8+CCR10+ T cells coexpressed DNAX accessory molecule 1 (DNAM‐1) (Figure 2A). DNAM‐1 is an activating receptor, and T cell immunoreceptor with Ig and immunoreceptor tyrosine‐based inhibition motif domains (TIGIT) is an inhibitory receptor on T cells, that compete for binding to CD155, an immunoglobulin‐like adhesion molecule. Based on DNAM‐1 and TIGIT, distinct coexpression patterns were detected on CD8+ T cells (Figure 2B). Overall, CD8+CCR10+ T cells were typically DNAM‐1high (Figure 2C), but had less TIGIT coexpression in patients with PsA (Figure 2D).
Figure 2.
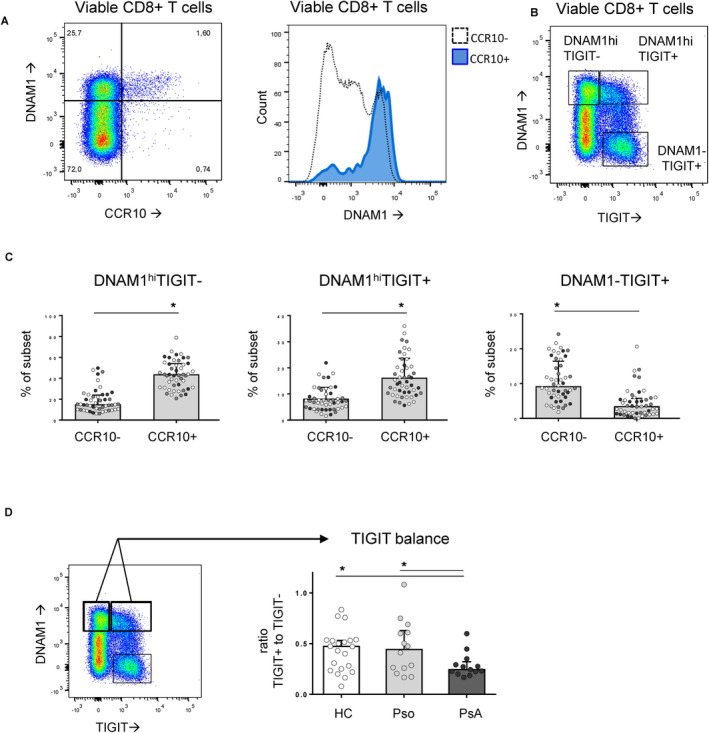
CD8+CCR10+ T cells are prototypically DNAM‐1high. A, CD8+CCR10+ T cells coexpressed high levels of DNAX accessory molecule 1 (DNAM‐1). B, Distinct populations of CD8+ T cells were distinguishable based on expression of DNAM‐1 and T cell immunoreceptor with Ig and immunoreceptor tyrosine‐based inhibition motif domains (TIGIT), including DNAM‐1highTIGIT−, DNAM‐1highTIGIT+, and DNAM‐1−TIGIT+. C, The majority of CD8+CCR10+ T cells were either DNAM‐1highTIGIT− or DNAM‐1highTIGIT+. D, Within CD8+CCR10+ T cells with high expression of DNAM‐1, TIGIT coexpression was reduced in patients with psoriatic arthritis (PsA). In C and D, symbols represent individual subjects (healthy controls [HCs] [white symbols], patients with psoriasis [Pso] [gray symbols], and patients with PsA [black symbols]); bars show the median and interquartile range. * = P < 0.05. Color figure can be viewed in the online issue, which is available at http://onlinelibrary.wiley.com/doi/10.1002/art.41652/abstract.
Enrichment of CD8+CCR10+ T cells in the skin, but not the joint
To investigate their tissue origin, we enumerated the frequency of CD8+CCR10+ T cells in skin biopsy samples and in SF mononuclear cells (SFMCs). The frequency of CD8+CCR10+ T cells was significantly higher in the skin compared to paired PBMCs. In contrast, there was no enrichment of CD8+CCR10+ T cells in SFMC samples compared to non‐paired PBMCs (Figure 3A and Supplementary Figure 7, http://onlinelibrary.wiley.com/doi/10.1002/art.41652/abstract). Previous studies have shown that CCR10 is a chemokine receptor found on skin‐tropic T cells. We confirmed that the majority of circulating CD8+CCR10+ T cells coexpressed the skin‐homing markers CCR4 and cutaneous lymphocyte antigen (CLA) (Figures 3B and C). Conversely, CD8+CCR10+ T cells did not coexpress β7 integrin, a marker associated with gut‐homing properties (Figure 3D).
Figure 3.
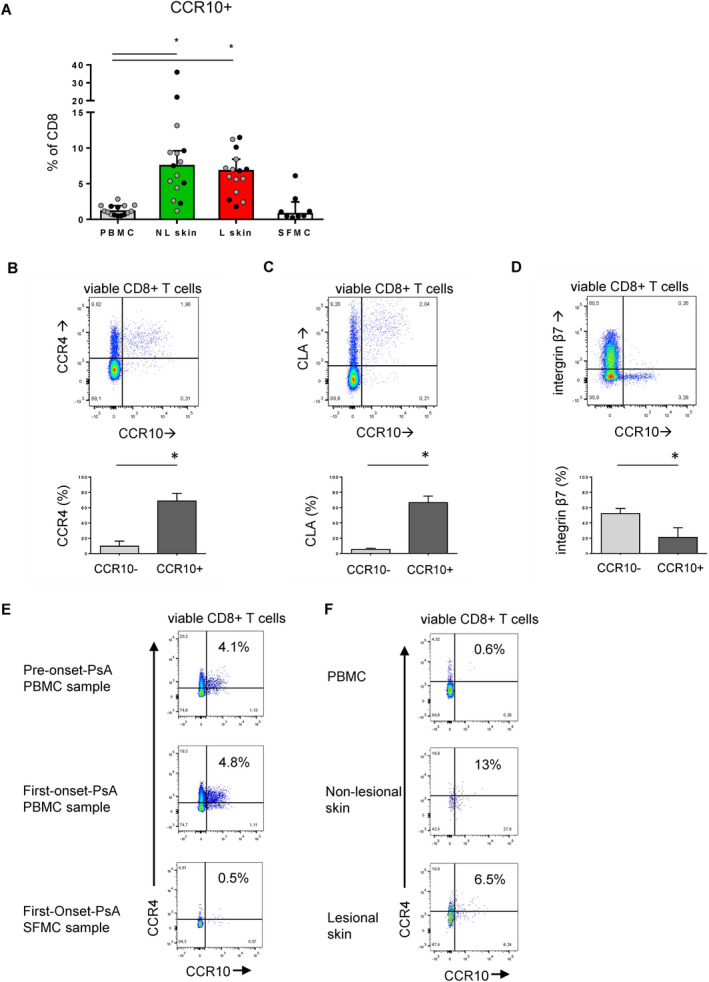
Enrichment of CD8+CCR10+ T cells in skin, but not in synovial fluid (SF). A, CD8+CCR10+ T cell frequency in paired nonlesional (NL) skin, lesional (L) skin, and peripheral blood mononuclear cells (PBMCs), and in nonpaired SF mononuclear cells (SFMCs). B–D, Expression of CCR4 (B), cutaneous lymphocyte antigen (CLA) (C), and β7 integrin (D) in CCR10+ versus CD8+CCR10− T cells. E, CD8+CCR10+ T cell frequency in PBMCs from a patient with psoriasis before psoriatic arthritis (PsA) onset and in PBMCs and SFMCs from the same patient at initial PsA onset. F, Frequency of CD8+CCR10+ T cells in paired nonlesional skin, lesional skin, and PBMCs from a patient with PsA. Symbols in A represent individual subjects (patients with psoriasis [Pso] [gray symbols] and patients with PsA (black symbols); bars in A–D show the median and interquartile range. * = P < 0.05. Color figure can be viewed in the online issue, which is available at http://onlinelibrary.wiley.com/doi/10.1002/art.41652/abstract.
We then analyzed whether CD8+CCR10+ T cell frequency was related to measures of disease activity. The frequency of CD8+CCR10+ T cells in PBMCs was not related to tender or swollen joint count (data not shown). In addition, we detected a stable frequency of CD8+CCR10+ T cells in PBMCs obtained from a patient with psoriasis before PsA onset and after PsA onset. The frequency of CD8+CCR10+ T cells was not higher in SFMCs from this patient at disease onset (Figure 3E). These results indicate that the joint compartment is an unlikely source of CD8+CCR10+ T cells.
As expected, we found that lesional skin contained a much larger absolute number of CD8+ T cells than nonlesional skin (Supplementary Figure 7, http://onlinelibrary.wiley.com/doi/10.1002/art.41652/abstract). However, the fraction of CD8+ T cells expressing CCR10 was either similar or lower in lesional skin compared to nonlesional skin (Figures 3A and F). The frequency of CD8+CCR10+ T cells in PBMCs was not related to PASI score (data not shown). CD103 expression, a marker of tissue retention, was significantly elevated in CD8+CCR10+ T cells from both PBMCs and skin (Supplementary Figure 8, http://onlinelibrary.wiley.com/doi/10.1002/art.41652/abstract). These results indicate that CCR10 is a marker for skin‐tropic CD8+ T cells in circulation and is enriched in both lesional and nonlesional psoriatic skin, but not in SF.
GATA3 and FOXP3 expression and Trm cell profile in CD8+CCR10+ T cells
We next performed transcriptome analyses of circulating CD8+ T cells from healthy controls, patients with psoriasis, and patients with PsA. Three populations of viable CD8+ T cells were sorted: CCR10+, CCR4+, and CCR10−CCR4− (Figure 4A and gating exhibited in Supplementary Figure 5, http://onlinelibrary.wiley.com/doi/10.1002/art.41652/abstract). We determined that the CCR10+ subset and CCR4+ subset were largely overlapping, while both were very distinct from the CCR10−CCR4− fraction (Figures 4B and C). Compared to the CCR10−CCR4− fraction, the CCR10+ subset was different with respect to numerous well‐characterized genes, including the up‐regulation of GATA3, CCR8, IL‐4R, and CD44 (Supplementary Figure 9, http://onlinelibrary.wiley.com/doi/10.1002/art.41652/abstract). In addition, the CCR10+ subset exhibited high expression of FOXP3 and lacked expression of genes associated with cytotoxic potential, e.g., GZMB and PRF1 (Figure 4C). Moreover, CD8+CCR10+ T cells displayed a prototypical gene expression pattern resembling Trm: high expression of ITGAE (CD103), CD69, CCR8, and CD44, and low expression of KLRG1 and CX3CR1 (Figure 4D).
Figure 4.
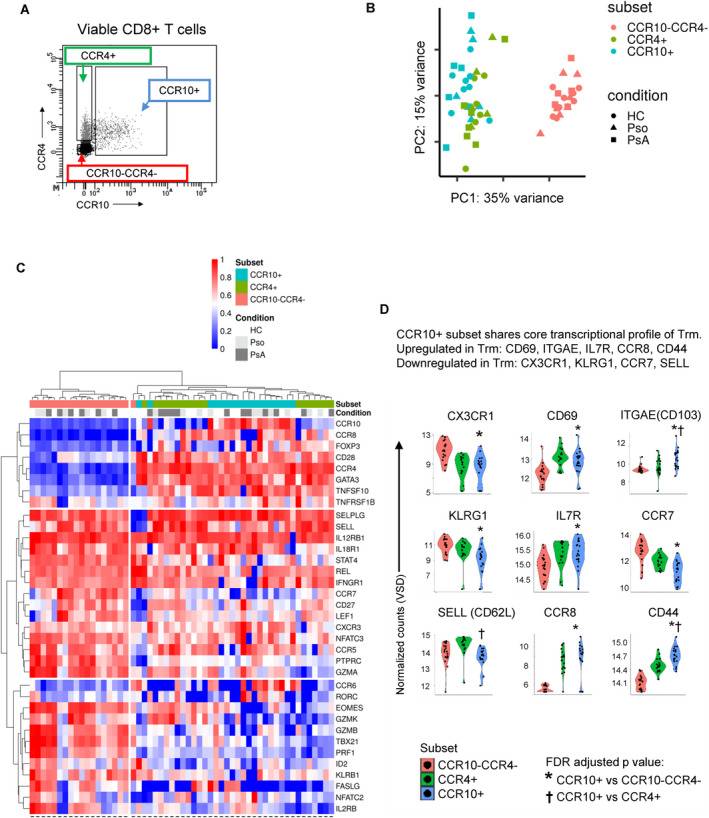
CD8+CCR10+ T cells express GATA3 and FOXP3 and exhibit a tissue‐resident memory (Trm) cell profile. A, Three different subsets of CD8+ T cells were flow sorted based on the presence/absence of CCR10 and CCR4. Detailed data regarding the full gating strategy are shown in Supplementary Figure 5 (http://onlinelibrary.wiley.com/doi/10.1002/art.41652/abstract). B, Principal components (PC) analysis was performed based on preselection of 5,268 genes that were differentially expressed in any of the cell subsets (nominal P < 0.05). C, Heatmap shows expression levels, on CD8+ cell subsets from healthy controls (HCs) and patients with psoriasis (Pso) or psoriatic arthritis (PsA), of genes previously reported as being critical for CD8+ T cells. D, Violin plots indicate the main transcriptional features attributed to Trm cells. Expression on the CCR10+ subset was compared to expression on the CCR10−CCR4− subset and the CCR4+ subset. VSD = variance‐stabilized data; FDR = false discovery rate.
As expected, the dominant factors determining the overall transcriptomic profile were the cell subsets rather than patient/health status. Exploratory analysis on the CD8+CCR10+ subset in patients with PsA identified 536 DEGs unique to patients with PsA compared to those with psoriasis and healthy controls (nominal P < 0.05), including up‐regulation of RORC, MYD88, and IFNAR1 (Supplementary Figure 10, http://onlinelibrary.wiley.com/doi/10.1002/art.41652/abstract). In summary, the results indicate that CD8+CCR10+ T cells are characterized by high expression of GATA3, FOXP3, and core transcripts defining Trm cells.
CD8+CCR10+ T cells exhibit a Tc2/Tc22 cytokine profile with net regulatory function
Consistent with the transcriptomic profile, CD8+CCR10+ T cells produced significantly more IL‐17A and IL‐22 compared to bulk CD8+ T cells on ex vivo restimulation. The production of IL‐4, IL‐13, and IL‐10 was also enriched in CD8+CCR10+ T cells. In contrast, CD8+CCR10+ T cells had reduced overall capacity to produce interferon γ and lacked markers (granzyme B and perforin) associated with cytotoxic capacity of CD8+ T cells (Figures 5A–F, and Supplementary Figure 11, http://onlinelibrary.wiley.com/doi/10.1002/art.41652/abstract).
Figure 5.
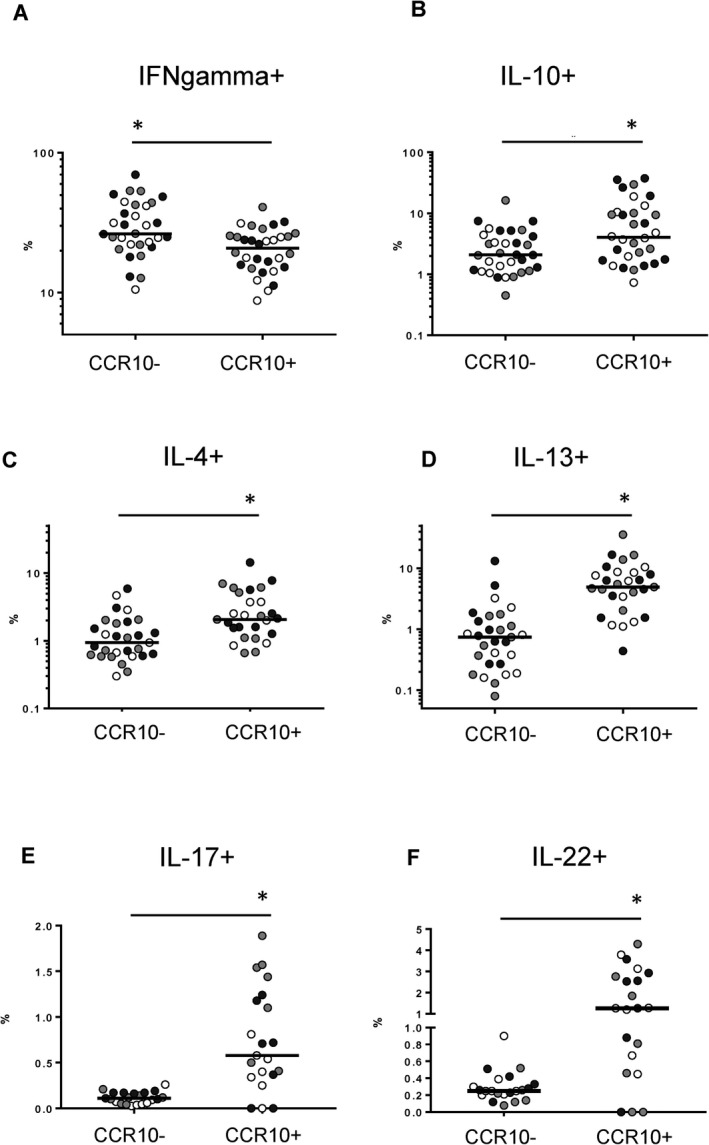
CD8+CCR10+ T cells exhibit a Tc2/Tc22‐like cytokine profile. Peripheral blood mononuclear cells were restimulated with phorbol 12‐myristate 13‐acetate and ionomycin calcium salt. The frequency of intracellular production of interferon‐γ (IFNγ) (A), interleukin‐10 (IL‐10) (B), IL‐4 (C), IL‐13 (D), IL‐17 (E), and IL‐22 (F) was compared between CD8+ T cells based on positivity or negativity for CCR10. Symbols represent individual subjects (healthy controls [white symbols], patients with psoriasis [gray symbols], and patients with psoriatic arthritis [black symbols]); bars show the median. * = P < 0.05.
CD8+CCR10+ T cells coexpressed FoxP3, and the frequency of CCR10+ CD8+ T cells in PBMCs was strongly correlated with the frequency of CD8+ T cells that expressed a “regulatory” phenotype (CD25+FoxP3+ and CD25+CD127−) (Figures 6A and B and Supplementary Figure 11, http://onlinelibrary.wiley.com/doi/10.1002/art.41652/abstract). Considering that CD8+CCR10+ T cells exhibited a pleiotropic cytokine‐producing profile (Tc2/22‐like), we next determined whether these cells have an overall immunoregulatory function. To this end, we performed immunosuppression assays using sorted CD8+ T cell subsets (selected based on the markers CCR10 and CCR4) and cocultured them with autologous, CellTrace Violet–labeled T cells (Figure 6C). Compared to coculture with bulk CD8+ T cells, the coculture with CD8+CCR10+ T cells significantly reduced the proliferation of both CD4+ T cells and CD8+ T cells (Figure 6D). Our functional assays thus confirmed the transcriptome profile data presented in Figure 4, indicating that CD8+CCR10+ T cells are Tc2/22‐like cells with an overall regulatory function.
Figure 6.
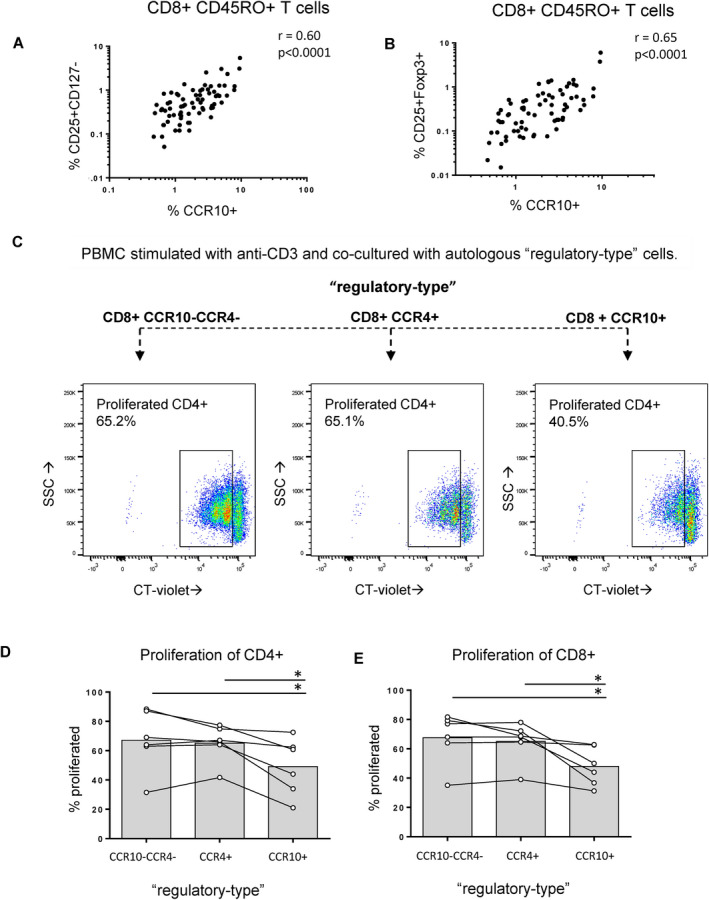
CD8+CCR10+ T cells exhibit regulatory properties. A and B, In peripheral blood mononuclear cells (PBMCs), the ex vivo fraction of memory CD8+ T cells that expressed CCR10 strongly correlated with the ex vivo fraction of CD8+ T cells that were CD25+CD127− (A) and CD25+FoxP3+ (B). Circles indicate pooled data from healthy controls. Additional data are shown in Supplementary Table 1 (http://onlinelibrary.wiley.com/doi/10.1002/art.41652/abstract). C, Examples of suppression assays are shown. Fresh PBMCs from 5 healthy controls were incubated with CellTrace Violet (CT‐violet) and cocultured with different CD8+ T cell subsets. Data regarding the gating strategy used for flow sorting are shown in Supplementary Figure 5, http://onlinelibrary.wiley.com/doi/10.1002/art.41652/abstract). D and E, The suppressive effect on CD4+ (D) and CD8+ (E) T cell proliferation was determined on day 4. Symbols represent individual subjects; bars show the median.* = P < 0.05. Color figure can be viewed in the online issue, which is available at http://onlinelibrary.wiley.com/doi/10.1002/art.41652/abstract.
DISCUSSION
In this study, we discovered and investigated in detail an increase in CD8+CCR10+ T cells in the peripheral blood of patients with PsA compared to patients with psoriasis. CD8+CCR10+ T cells are Tem cells with Tc2/22‐like cytokine profile and regulatory function. This CD8+ T cell subset was further endowed with a transcriptomic profile comparable to that observed in Trm cells, which originated in skin, but not the joint.
To our knowledge, this is the broadest immunophenotyping study thus far, as performed in a PASI‐matched cohort of patients with PsA and patients with psoriasis who were not being treated with immunomodulatory drugs. Overall, the results underscore the role of memory CD8+ T cells in the pathogenesis of PsA, which is consistent with findings in previous immunophenotyping studies and genetic association studies (1, 11, 26, 27, 28). Specifically, we have identified a novel role of CD8+CCR10+ T cells, which may be important in the pathogenesis of PsA.
The facts that skin‐homing markers on T cells in SF/tissue have previously been described (28, 29, 30), and that therapeutics blocking specific integrins can induce arthritis (31, 32), have raised the question of whether skin‐tropic T cells could be redirected into the joint. However, our results do not indicate that CD8+CCR10+ T cells are derived from SF, nor do they appear to preferentially migrate to this site. Instead, we discovered a typical pattern of coexpression with the skin‐tropic markers CLA and CCR4, consistent with previous studies indicating that CCR10 guides trafficking of T cells towards the skin (23, 33). The CD8+CCR10+ T cells are most likely found in the epidermis (34). Notably, we found that both lesional and nonlesional skin harbors CD8+CCR10+ T cells. There was a trend toward fewer regulatory CD8+CCR10+ T cells in lesional skin, which may contribute to pathology. We suspect that CD8+CCR10+ T cells from nonlesional skin contributes to the fraction detected in PBMCs for several reasons, including: 1) patients with psoriasis and patients with PsA had similar PASI scores, 2) there was no relationship between the frequency of these cells and PASI scores, and 3) even in healthy individuals, these cells are detected in the circulation (35). Larger studies are needed to compare the quantity of CD8+CCR10+ T cells in skin from patients with psoriasis and patients with PsA, particularly to examine whether there are subtle differences between these groups with respect to both lesional and nonlesional skin sites.
Phenotypically, CD8+CCR10+ T cells were mostly Tem cells based on classic nomenclature, but we also detected a strong transcriptional overlap with skin‐derived Trm cells (36). In human skin, CD8+ T cells can be classified as those that pass through the tissue and those that remain in the tissue, the latter being termed Trm cells. These cells can be divided into CD69+CD103− or CD69+CD103+, the latter being more prevalent in the epidermis and exerting potent effector functions (37, 38). Our transcriptomic analysis of CD8+CCR10+ T cells revealed a striking resemblance to Trm cells: high expression of IL‐7R (CD127), CD69, and ITGAE (CD103), and low expression of KLRG1 and CX3CR1 (33, 37, 38). Furthermore, transcriptomic analysis revealed high expression of CCR8, which has recently been linked to a skin‐resident CD8+ T cell population (33). In a recent study using a murine model memory precursor CD8+ T cell clones from circulation were tracked, and it was found that high expression of CCR10 was present in CD8+ T cells committed to a skin Trm fate (39). Strictly speaking, the CD8+CCR10+ T cells we characterized in PBMCs should not be termed Trm cells, since Trm cells are, by definition, noncirculating (40). However, this dichotomy may be too simplistic, at least in regard to CD4+ T cells. CLA+CD4 Trm cells can exit the skin, reenter the circulation, and occupy distant skin sites (35). Our data fit well into this recent concept of circulating Trm cells, and specifically adds strength to the notion that there are circulating human CD8+ Trm cells (41, 42).
Regarding function, CCR10 has classically been attributed to Th2 and Th22 subsets of CD4+ T cells (43). The CD8+CCR10+ T cells we analyzed indeed exhibited Tc2/22‐like function, but are distinct from Tc17 that was recently described in PsA SF (28), including the fact that the Tc17 expressed high levels of granzymes. CCR10 has also been described as a common marker on regulatory CD4+ T cells (44). Loss of CCR10 in murine models results in loss of Treg cells in skin, which enhances IL‐17A and tumor necrosis factor production at the cost of IL‐10 production (25, 45, 46). This indicates that CCR10 is important for the regulatory function of tissue‐resident T cells in noninflamed murine skin (25, 46). Overall, the transcriptomic profile and functional assays performed in our study indicate that these cells could have an important regulatory function in human skin.
There are different proposed mechanisms by which CD8+ T cells can induce a suppressive function (47). These exact mechanisms are beyond the scope of the present study. The potential role of immune checkpoints, including the DNAM‐1/TIGIT axis, warrants further investigation, since we noted phenotypic disturbances in the PsA group. The balance of regulation versus inflammation could also be secondary to environmental cues that skew cell plasticity and the cytokine profile, as shown for CD4+ T cells and innate lymphoid cells (48, 49). Consistent with the latter concept is our finding that RORC and IFNAR1, among other genes, differentiated CD8+CCR10+ T cells from patients with PsA compared to patients with psoriasis and healthy controls. While RORC is critical to the pathogenic effects of innate immune cells in SpA (50), our transcriptomic analysis within the PsA group remains exploratory and requires confirmation in a larger group of patients.
We suggest that the potential novel role of CD8+ Trm cells in PsA should be viewed in the context of the more established role of CD8+ Trm cells in psoriasis (34, 41, 51, 52). Clinically, unaffected skin from patients with psoriasis has perturbations in keratinocytes and Trm cell populations, which has been suggested to poise the skin for an excessive inflammatory response (52). Also, after psoriasis lesions have clinically resolved, there is long‐term persistence of epidermal CD8+ Trm cells with IL‐17A–producing capacities (41, 51). Of further interest is the fact that the effector function of CD8+ Trm cells in nonlesional skin is related to disease duration (34).
Our study has certain limitations, which includes the fact that it was cross‐sectional in design and that patients with inflammatory rheumatic diseases other than SpA (e.g., rheumatoid arthritis or gout) were not included. Although we adhered to a standardized protocol for flow cytometry (15), viability staining was only performed in the functional and transcriptomic analysis. Also, upon identification of CCR10 as a marker that discriminated between PsA and psoriasis within CD8+CD45RO+ T cells, for further experiments we made the practical choice to exclude CD45RO as a marker in the gating, which may have resulted in the inclusion of some naive CD8+ T cells. Importantly, it will be necessary for these results to be validated in an independent cohort of patients, regardless of any subtle modifications to the flow‐cytometric gating strategy used.
One question that remains unanswered is whether CD8+CCR10+ T cells play a role in the pathogenesis of PsA (e.g., contributing to the production of soluble factors affecting distant musculoskeletal sites) or if these cells should instead be seen as the flag of disturbed cutaneous homeostasis that is principally driven by other cells, such as keratinocytes or other stromal cells.
Taken together, our findings show that PsA is marked by alterations in circulating, skin‐derived, regulatory CD8+ Trm cells. These data support the notion that events occurring in the skin may drive the development of arthritis in patients with psoriasis.
AUTHOR CONTRIBUTIONS
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Dr. Leijten had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design
Leijten, van Kempen, Nordkamp, Pouw, Balak, Verhagen, Hartgring, Pandit, Radstake, Boes.
Acquisition of data
Leijten, van Kempen, Nordkamp, Pouw, Kleinrensink, Vincken, Mertens, Balak, Verhagen, Hartgring, Pandit, Radstake, Boes.
Analysis and interpretation of data
Leijten, van Kempen, Nordkamp, Pouw, Verhagen, Hartgring, Lubberts, Tekstra, Pandit, Radstake, Boes.
Supporting information
Supplementary Material
Acknowledgments
We thank the patients for their participation in the study, the clinical study team (Anne Karien Marijnissen, Anneloes van Loo, Karin Schrijvers, and Joke Nijdeken), and the flow core facility of the Center for Translational Immunology. We also thank Jonas Kuipers for his scientific input.
Drs. Radstake and Boes contributed equally to this work.
Dr. Balak has received consulting fees from Janssen (less than $10,000). Dr. Radstake has received consulting fees from Janssen (less than $10,000) and owns stock or stock options in AbbVie. No other disclosures relevant to this article were reported.
References
- 1.Veale DJ, Fearon U. The pathogenesis of psoriatic arthritis. Lancet 2018;391:2273–84. [DOI] [PubMed] [Google Scholar]
- 2.Ritchlin CT, Colbert RA, Gladman DD. Psoriatic arthritis. N Engl J Med 2017;376:957–70. [DOI] [PubMed] [Google Scholar]
- 3.Sakkas LI, Bogdanos DP. Are psoriasis and psoriatic arthritis the same disease? The IL‐23/IL‐17 axis data [review]. Autoimmun Rev 2017;16:10–5. [DOI] [PubMed] [Google Scholar]
- 4.Boehncke WH. Psoriasis and psoriatic arthritis: flip sides of the coin? Acta Derm Venereol 2016;96:436–41. [DOI] [PubMed] [Google Scholar]
- 5.McGonagle D, Aydin SZ, Gül A, Mahr A, Direskeneli H. ’MHC‐I‐opathy’‐unified concept for spondyloarthritis and Behçet disease [review]. Nat Rev Rheumatol 2015;11:731–40. [DOI] [PubMed] [Google Scholar]
- 6.Sherlock JP, Joyce‐Shaikh B, Turner SP, Chao CC, Sathe M, Grein J, et al. IL‐23 induces spondyloarthropathy by acting on ROR‐γt+ CD3+CD4‐CD8‐ entheseal resident T cells. Nat Med 2012;18:1069–76. [DOI] [PubMed] [Google Scholar]
- 7.Reinhardt A, Prinz I. Whodunit? The contribution of interleukin (IL)‐17/IL‐22‐producing γδ T cells, αβ T cells, and innate lymphoid cells to the pathogenesis of spondyloarthritis [review]. Front Immunol 2018;9:885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abji F, Pollock RA, Liang K, Chandran V, Gladman DD. CXCL10 is a possible biomarker for the development of psoriatic arthritis among patients with psoriasis. Arthritis Rheumatol 2016;68:2911–6. [DOI] [PubMed] [Google Scholar]
- 9.Abji F, Pollock RA, Liang K, Chandran V, Gladman DD. Th17 gene expression in psoriatic arthritis synovial fluid and peripheral blood compared to osteoarthritis and cutaneous psoriasis. Clin Exp Rheumatol 36:486–9. [PubMed] [Google Scholar]
- 10.Pollock RA, Abji F, Liang K, Chandran V, Pellett FJ, Virtanen C, et al. Gene expression differences between psoriasis patients with and without inflammatory arthritis [letter]. J Invest Dermatol 2015;135:620–3. [DOI] [PubMed] [Google Scholar]
- 11.Benham H, Norris P, Goodall J, Wechalekar MD, Fitzgerald O, Szentpetery A, et al. Th17 and Th22 cells in psoriatic arthritis and psoriasis. Arthritis Res Ther 2013;15:R136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fredriksson T, Pettersson U. Severe psoriasis—oral therapy with a new retinoid. Dermatologica 1978;157:238–44. [DOI] [PubMed] [Google Scholar]
- 13.Taylor W, Gladman D, Helliwell P, Marchesoni A, Mease P, Mielants H, et al. Classification criteria for psoriatic arthritis: development of new criteria from a large international study. Arthritis Rheum 2006;54:2665–73. [DOI] [PubMed] [Google Scholar]
- 14.Rudwaleit M, van der Heijde D, Landewe R, Listing J, Akkoc N, Brandt J, et al. The development of Assessment of SpondyloArthritis international Society classification criteria for axial spondyloarthritis (part II): validation and final selection. Ann Rheum Dis 2009;68:777–83. [DOI] [PubMed] [Google Scholar]
- 15.Verhagen FH, Hiddingh S, Rijken R, Pandit A, Leijten E, Nordkamp MO, et al. High‐dimensional profiling reveals heterogeneity of the Th17 subset and its association with systemic immunomodulatory treatment in non‐infectious uveitis. Front Immunol 2018;9:2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wehrens EJ, Mijnheer G, Duurland CL, Klein M, Meerding J, van Loosdregt J, et al. Functional human regulatory T cells fail to control autoimmune inflammation due to PKB/c‐akt hyperactivation in effector cells. Blood 2011;118:3538–48. [DOI] [PubMed] [Google Scholar]
- 17.Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, et al. Ensembl 2015. Nucleic Acids Res 2015;43:D662–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA‐seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anders S, Pyl PT, Huber W. HTSeq: a Python framework to work with high‐throughput sequencing data. Bioinformatics 2015;31:166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia J, Wishart DS. Using MetaboAnalyst 3.0 for comprehensive metabolomics data analysis. Curr Protoc Bioinforma 2016;55:14. [DOI] [PubMed] [Google Scholar]
- 22.Oliveros JC. Venny 2.1. URL: https://bioinfogp.cnb.csic.es/tools/venny/index.html.
- 23.Sigmundsdottir H, Pan J, Debes GF, Alt C, Habtezion A, Soler D, et al. DCs metabolize sunlight‐induced vitamin D3 to “program” T cell attraction to the epidermal chemokine CCL27. Nat Immunol 2007;8:285–93. [DOI] [PubMed] [Google Scholar]
- 24.Homey B, Alenius H, Müller A, Soto H, Bowman EP, Yuan W, et al. CCL27‐CCR10 interactions regulate T cell‐mediated skin inflammation. Nat Med 2002;8:157–65. [DOI] [PubMed] [Google Scholar]
- 25.Xia M, Hu S, Fu Y, Jin W, Yi Q, Matsui Y, et al. CCR10 regulates balanced maintenance and function of resident regulatory and effector T cells to promote immune homeostasis in the skin. J Allergy Clin Immunol 2014;134:634–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menon B, Gullick NJ, Walter GJ, Rajasekhar M, Garrood T, Evans HG, et al. Interleukin‐17+CD8+ T cells are enriched in the joints of patients with psoriatic arthritis and correlate with disease activity and joint damage progression. Arthritis Rheumatol 2014;66:1272–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bowes J, Budu‐Aggrey A, Huffmeier U, Uebe S, Steel K, Hebert HL, et al. Dense genotyping of immune‐related susceptibility loci reveals new insights into the genetics of psoriatic arthritis. Nat Commun 2015;6:6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steel KJ, Srenathan U, Ridley M, Durham LE, Wu SY, Ryan SE, et al. Polyfunctional, proinflammatory, tissue‐resident memory phenotype and function of synovial interleukin‐17A+CD8+ T cells in psoriatic arthritis. Arthritis Rheumatol 2020;72:435–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones SM, Dixey J, Hall ND, McHugh NJ. Expression of the cutaneous lymphocyte antigen and its counter‐receptor E‐selectin in the skin and joints of patients with psoriatic arthritis. Br J Rheumatol 1997;36:748–57. [DOI] [PubMed] [Google Scholar]
- 30.Pitzalis C, Cauli A, Pipitone N, Smith C, Barker J, Marchesoni A, et al. Cutaneous lymphocyte antigen–positive T lymphocytes preferentially migrate to the skin but not to the joint in psoriatic arthritis. Arthritis Rheum 1996;39:137–45. [DOI] [PubMed] [Google Scholar]
- 31.Viguier M, Richette P, Aubin F, Beylot‐Barry M, Lahfa M, Bedane C, et al. Onset of psoriatic arthritis in patients treated with efalizumab for moderate to severe psoriasis. Arthritis Rheum 2008;58:1796–802. [DOI] [PubMed] [Google Scholar]
- 32.Dubash S, Marianayagam T, Tinazzi I, Al‐Araimi T, Pagnoux C, Weizman AV, et al. Emergence of severe spondyloarthropathy‐related entheseal pathology following successful vedolizumab therapy for inflammatory bowel disease. Rheumatology (Oxford) 2019;58:963–8. [DOI] [PubMed] [Google Scholar]
- 33.McCully ML, Ladell K, Andrews R, Jones RE, Miners KL, Roger L, et al. CCR8 expression defines tissue‐resident memory T cells in human skin. J Immunol 2018;200:1639–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vo S, Watanabe R, Koguchi‐Yoshioka H, Matsumura Y, Ishitsuka Y, Nakamura Y, et al. CD8 resident memory T cells with interleukin 17A‐producing potential are accumulated in disease‐naïve nonlesional sites of psoriasis possibly in correlation with disease duration [letter]. Br J Dermatol 2019;181:410–2. [DOI] [PubMed] [Google Scholar]
- 35.Klicznik MM, Morawski PA, Höllbacher B, Varkhande SR, Motley SJ, Kuri‐Cervantes L, et al. Human CD4 + CD103 + cutaneous resident memory T cells are found in the circulation of healthy individuals. Sci Immunol 2019;4:eaav8995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Romero P, Zippelius A, Kurth I, Pittet MJ, Touvrey C, Iancu EM, et al. Four functionally distinct populations of human effector‐memory CD8+ T lymphocytes. J Immunol 2007;178:4112–9. [DOI] [PubMed] [Google Scholar]
- 37.Watanabe R, Gehad A, Yang C, Scott LL, Teague JE, Schlapbach C, et al. Human skin is protected by four functionally and phenotypically discrete populations of resident and recirculating memory T cells. Sci Transl Med 2015;7:279ra39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mackay LK, Rahimpour A, Ma JZ, Collins N, Stock AT, Hafon ML, et al. The developmental pathway for CD103(+)CD8+ tissue‐resident memory T cells of skin. Nat Immunol 2013;14:1294–301. [DOI] [PubMed] [Google Scholar]
- 39.Kok L, Dijkgraaf FE, Urbanus J, Bresser K, Vredevoogd DW, Cardoso RF, et al. A committed tissue‐resident memory T cell precursor within the circulating CD8+ effector T cell pool. J Exp Med 2020;217:e20191711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morris SE, Farber DL, Yates AJ. Tissue‐resident memory T cells in mice and humans: towards a quantitative ecology. J Immunol 2019;203:2561–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheuk S, Schlums H, Sérézal IG, Martini E, Chiang SC, Marquardt N, et al. CD49a expression defines tissue‐resident CD8 + T cells poised for cytotoxic function in human skin. Immunity 2017;46:287–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khalil S, Bardawil T, Kurban M, Abbas O. Tissue‐resident memory T cells in the skin. Inflamm Res 2020;69:245–54. [DOI] [PubMed] [Google Scholar]
- 43.Trifari S, Kaplan CD, Tran EH, Crellin NK, Spits H. Identification of a human helper T cell population that has abundant production of interleukin 22 and is distinct from T(H)‐17, T(H)1 and T(H)2 cells. Nat Immunol 2009;10:864–71. [DOI] [PubMed] [Google Scholar]
- 44.Eksteen B, Miles A, Curbishley SM, Tselepis C, Grant AJ, Walker LS, et al. Epithelial inflammation is associated with CCL28 production and the recruitment of regulatory T cells expressing CCR10. J Immunol 2006;177:593–603. [DOI] [PubMed] [Google Scholar]
- 45.Fu Y, Yang J, Xiong N. Cutting edge: skin CCR10+ CD8+ T cells support resident regulatory T cells through the B7.2/receptor axis to regulate local immune homeostasis and response. J Immunol 2016;196:4859–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li C, Xu M, Coyne J, Wang WB, Devila M, Wang Y, et al. Psoriasis‐associated impairment of CCL27/CCR10‐derived regulation leads to IL‐17A/IL‐22‐producing skin T cell over‐activation. J Allergy Clin Immunol 2021;147:759–63.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu Z, Ho S, Chang CC, Zhang QY, Vasilescu ER, Vlad G, et al. Molecular and cellular characterization of human CD8 T suppressor cells [review]. Front Immunol 2016;7:549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bernink JH, Ohne Y, Teunissen MB, Wang J, Wu J, Krabbendam L, et al. c‐Kit‐positive ILC2s exhibit an ILC3‐like signature that may contribute to IL‐17‐mediated pathologies. Nat Immunol 2019;20:992–1003. [DOI] [PubMed] [Google Scholar]
- 49.Zielinski CE, Mele F, Aschenbrenner D, Jarrossay D, Ronchi F, Gattorno M, et al. Pathogen‐induced human TH17 cells produce IFN‐γ or IL‐10 and are regulated by IL‐1β. Nature 2012;484:514–8. [DOI] [PubMed] [Google Scholar]
- 50.Venken K, Jacques P, Mortier C, Labadia ME, Decruy T, Coudenys J, et al. RORγt inhibition selectively targets IL‐17 producing iNKT and γδ‐T cells enriched in Spondyloarthritis patients. Nat Commun 2019;10:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheuk S, Wikén M, Blomqvist L, Nylén S, Talme T, Ståhle M, et al. Epidermal Th22 and Tc17 cells form a localized disease memory in clinically healed psoriasis. J Immunol 2014;192:3111–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sérézal IG, Hoffer E, Ignatov B, Martini E, Zitti B, Ehrström M, et al. A skewed pool of resident T cells triggers psoriasis‐associated tissue responses in never‐lesional skin from patients with psoriasis. J Allergy Clin Immunol 2019;143:1444–54. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


