ABSTRACT
Objectives
To develop and validate a nomogram based on fetal nuchal translucency thickness (NT) and ultrasonographic facial markers for screening for trisomy 21 in the first trimester of pregnancy.
Methods
This was a retrospective case–control study using stored two‐dimensional midsagittal fetal profile images captured at 11 + 0 to 13 + 6 weeks' gestation in singleton pregnancies. We included images from 302 trisomy‐21 pregnancies and 322 euploid pregnancies. Cases were divided into a training set (200 euploid + 200 with trisomy 21) and a validation set (122 euploid + 102 with trisomy 21) at a ratio of approximately 2:1. For each, the maternal age, gestational age, fetal NT and karyotype were noted, and 12 ultrasonographic fetal facial markers were measured. The least absolute shrinkage and selection operator (LASSO) method and multivariable analysis were used to select automatically the discriminative markers. Logistic regression was used to develop a LASSO model, based on the selected markers, to screen for trisomy 21 in the first trimester of pregnancy. Furthermore, 60 of the 624 images were selected randomly as a retest set to evaluate the model's robustness. The predictive performance of screening for trisomy 21 of a model based on fetal NT and maternal age and of the LASSO model was assessed using the area under the receiver‐operating‐characteristics curve (AUC). A nomogram was developed as an individualized tool to predict patient‐specific probability for trisomy 21, which is a more visual presentation of the LASSO model. The performance of the nomogram was assessed using the C‐index and calibration curve.
Results
Into the LASSO model were incorporated eight markers, including fetal NT, prenasal‐thickness‐to‐nasal‐bone‐length ratio, facial profile line, frontomaxillary facial angle, frontonasal facial angle, mandibulomaxillary facial angle, maxilla‐nasion‐mandible angle and d2 (distance between the anterior edge of the prefrontal skin and the mandibulomaxillary line) (all P < 0.05). The AUCs of the LASSO model for screening for trisomy 21 were 0.983 (95% CI, 0.971–0.994) in the training set and 0.979 (95% CI, 0.966–0.993) in the validation set, and these were higher than the AUCs of all eight individual ultrasonographic markers included in the model. The AUC of the LASSO model in the retest set was 0.997 (95% CI, 0.990–1.000), indicating good robustness of the LASSO model. The AUC of the LASSO model was significantly higher than that of the model based on fetal NT and maternal age in both training and validation sets (P < 0.001 for both). The nomogram of the LASSO model showed good discrimination of trisomy 21, with C‐indices of 0.983 in the training set and 0.981 in the validation set.
Conclusions
We present an individualized nomogram which incorporates fetal NT and a series of ultrasonographic facial profile markers selected by the LASSO method and multivariable analysis. This nomogram can potentially be utilized as a convenient and effective tool in screening for trisomy 21 in the first trimester of pregnancy. © 2020 The Authors. Ultrasound in Obstetrics & Gynecology published by John Wiley & Sons Ltd on behalf of International Society of Ultrasound in Obstetrics and Gynecology.
Keywords: facial profile, first‐trimester screening, machine learning, nomogram, trisomy 21, ultrasonographic markers
CONTRIBUTION —
What are the novel findings of this work?
We present an individualized nomogram, incorporating fetal nuchal translucency thickness (NT) and a series of ultrasonographic facial profile markers, which can potentially be utilized as a convenient and effective tool for screening for trisomy 21 in the first trimester of pregnancy.
What are the clinical implications of this work?
At present, the most widely used ultrasonographic marker for trisomy 21 is fetal NT at 11 to 13 + 6 weeks' gestation. The nomogram developed in this study can improve the ultrasonographic detection rate of trisomy 21 and can be used for screening during the routine prenatal ultrasound examination in early pregnancy.
INTRODUCTION
Trisomy 21 is the most common chromosomal abnormality. Growth delay and intellectual disability are common in children with Down syndrome1. Screening for trisomy 21 in the first trimester of pregnancy is of critical importance, in order to provide women with affected fetuses reproductive choices at the earliest opportunity.
The 2013 ISUOG Practice Guidelines2 state that fetal nuchal translucency thickness (NT) and absence of the nasal bone can be used to screen for chromosomal abnormalities in the first trimester of pregnancy. Furthermore, it has been observed that some trisomy‐21 fetuses have abnormal facial features, such as a flat face and nasal‐bridge collapse. However, the evaluation of these observations is subjective. Considerable effort has been devoted to transforming such abnormal facial features into objective ultrasonographic facial markers. There are essentially three main categories of quantitative fetal facial marker: (1) length and ratio, including prenasal thickness, nasal bone length, prenasal‐thickness‐to‐nasal‐bone‐length ratio and prefrontal space ratio3, 4; (2) angles, including frontomaxillary facial angle, maxilla‐nasion‐mandible angle, frontonasal facial angle and mandibulomaxillary facial angle5, 6, 7, 8; and (3) facial profile line4. Some of these markers, such as prenasal‐thickness‐to‐nasal‐bone‐length ratio, frontomaxillary facial angle, prefrontal space ratio and facial profile line have been assessed individually for the prediction of trisomy 21 in the first trimester3, 4, 5, whilst the others have only been utilized in the second and third trimesters. No study has reported on the performance of screening for trisomy 21 based on a combination of these quantitative markers in the first trimester of pregnancy.
Machine‐learning methodology is a branch of research on artificial intelligence9, 10. Machine‐learning algorithms build a mathematical model based on sample data (known as ‘training data’) to make predictions without being programmed explicitly to perform the task11. In recent years, the machine‐learning method has been utilized widely for the identification of tumors from ultrasonographic images12. This method has yet to be applied in prenatal screening13. The aim, therefore, of this study was to develop and validate a nomogram that incorporates fetal NT and ultrasonographic fetal facial markers for screening for trisomy 21 in the first trimester of pregnancy, in cases in which the fetal face could be visualized adequately.
METHODS
The process of development of the nomogram for screening for trisomy 21 in the first trimester of pregnancy is illustrated in Figure 1.
Figure 1.

Flowchart summarizing development of nomogram for screening for trisomy 21 in first trimester of pregnancy. (a) Marker delineation of euploid fetuses and fetuses with trisomy 21. (b) Marker selection using least absolute shrinkage and selection operator (LASSO) method. (c) Development of LASSO model and nomogram. AUC, area under the receiver‐operating‐characteristics curve; d2, distance between anterior edge of prefrontal skin and mandibulomaxillary line; FMF, frontomaxillary facial angle; FNA, frontonasal facial angle; FPL, facial profile line; MMF, mandibulomaxillary facial angle; MNM, maxilla‐nasion‐mandible angle; NBL, nasal bone length; NT, nuchal translucency thickness; PFSR, prefrontal space ratio; PT, prenasal thickness.
Patients
This was a retrospective case–control study using stored fetal ultrasound images obtained at the Department of Ultrasound at the Beijing Obstetrics & Gynecology Hospital, Maternal & Child Health Centre, Capital Medical University, Beijing, China, between January 2009 and February 2019. We selected from our database two‐dimensional images in the midsagittal plane of the fetal face from singleton pregnancies at 11 to 13 + 6 weeks' gestation. The criteria for selection were as follows: (a) fetal karyotype confirmed by chorionic villus sampling or amniocentesis; (b) fetal nasal bone, frontal bone, skin over the nasal bone and the anterior edges of maxilla and mandible clearly visible on the image.
During the study period, 408 cases of trisomy 21 were confirmed by chorionic villus sampling or amniocentesis. Of these, 83 (20%) cases were excluded because of poor image quality, and a further 23 (6%) were excluded due to absence of the nasal bone, as many of the ultrasonographic markers rely on its presence. Thus, images from 302 trisomy‐21 cases were retrieved for inclusion in the study. These were matched with images from 322 euploid fetuses that were selected randomly using the same criteria as for the trisomy‐21 cases. Images were divided into a training set (200 euploid + 200 with trisomy 21) and a validation set (122 euploid + 102 with trisomy 21) at a ratio of approximately 2:1 (Figure 2).
Figure 2.

Flowchart summarizing development and verification of prediction model (least absolute shrinkage and selection operator (LASSO) model).
Markers
For each of the 624 fetuses, the following data were collected: maternal age, gestational age, fetal karyotype and fetal NT, and the following 12 facial ultrasonographic markers were measured: prenasal thickness, nasal bone length, prenasal‐thickness‐to‐nasal‐bone‐length ratio, d1 (distance between the anterior edge of the frontal bone and the anterior edge of the prefrontal skin), d2 (distance between the anterior edge of the prefrontal skin (at the same point as the d1 measurement) and the mandibulomaxillary line), prefrontal space ratio (calculated as d2/d1), frontomaxillary facial angle, mandibulomaxillary facial angle, maxilla‐nasion‐mandible angle, frontonasal facial angle, facial profile line and the F distance (the largest perpendicular distance between the facial profile line and the outermost part of the frontal bone). These facial markers were measured manually, using retrieved midsagittal images which had been acquired for the measurement of fetal NT, by a single sonographer (Y.S.), using ITK‐snap software (Version 3.6.0, http://www.itksnap.org). The sonographer was blinded to the karyotypic results.
Ultrasound images demonstrating the facial ultrasonographic marker measurements in euploid and trisomy‐21 fetuses are given in Appendix S1. Prenasal thickness was the shortest distance between the anterior edge of the lowest part of the frontal bone and the anterior line of skin. Nasal bone length was measured between the two ends of the ossification line (Figure S1 in Appendix S1)3. For d1 and d2, the mandibulomaxillary line was drawn between the anterior edge of the mandible and the anterior edge of the maxilla, and extended in the direction of the frontal bone. Both d1 and d2 were measured approximately parallel to the inferior edge of the maxilla. d1 was the distance between the anterior edge of the frontal bone and the anterior edge of the prefrontal skin and d2 was the distance between the anterior edge of the prefrontal skin at the same point as the d1 measurement and the mandibulomaxillary line. When the mandibulomaxillary line crossed behind the anterior edge of the prefrontal skin, d1 was measured between the frontal bone and the prefrontal skin, while d2 was measured between the mandibulomaxillary line and the prefrontal skin and multiplied by –1. The prefrontal space ratio was calculated as d2/d1 (Figure S2 in Appendix S1)4. The frontomaxillary facial angle was measured between a line along the upper surface of the palate and a line from the upper‐anterior corner of the maxilla to the external surface of the frontal bone or the external surface of an echogenic line under the skin, representing the metopic suture that remained open (Figure S3 in Appendix S1)5. The mandibulomaxillary facial angle was measured between a line along the upper surface of the palate (as for the frontomaxillary facial angle) and a line from the upper‐anterior corner of the maxilla to the upper anterior corner of the mandible (Figure S3 in Appendix S1)8. The nasion was the apex of the maxilla‐nasion‐mandible angle; this angle was measured between a line from the midpoint of the anterior edge of the maxilla to the midpoint of the nasion and a line from the nasion to the midpoint of the anterior edge of the mandible. The nasion is the intersection of the frontal and nasal bones. When there was no intersection between nasal and frontal bones, the nasion was defined as the intersection between the tangential line of the nasal bone and the tangential line of the lowest part of the frontal bone (Figure S4 in Appendix S1)14. The frontonasal facial angle was measured between a line along the outer profile of the nasal bone and a line traversing the upper corner of the external profile of the nasal bone, extending to the external surface of the forehead (Figure S5 in Appendix S1)15. The facial profile line was drawn from the nasion to the midpoint of the anterior edge of the mandible, and was classified as ‘negative’, ‘zero’ or ‘positive’, which indicated that the facial profile line passed the frontal bone anteriorly, longitudinally or posteriorly, respectively. The F distance (the greatest perpendicular distance between the facial profile line and the outermost part of the frontal bone) was measured when the facial profile line was positive (Figure S6 in Appendix S1)4.
In total, we evaluated 15 markers, including maternal age, fetal NT, fetal NT ≥ 2.5 mm and the 12 facial ultrasonographic markers. We selected the most useful predictive markers for trisomy 21 from these markers using the least absolute shrinkage and selection operator (LASSO) method and multivariable analysis. The LASSO method uses the penalty parameter ln(λ) to shrink automatically regression coefficients of these 15 markers; the greater the value of ln(λ), the more a marker's coefficient will shrink to zero (Figure 3). We excluded any marker whose coefficient was shrunk to approximately zero, and selected for inclusion the remaining markers. Multivariable analysis was used on the markers selected by the LASSO method, and the markers with P < 0.05 were selected for development of the prediction model.
Figure 3.
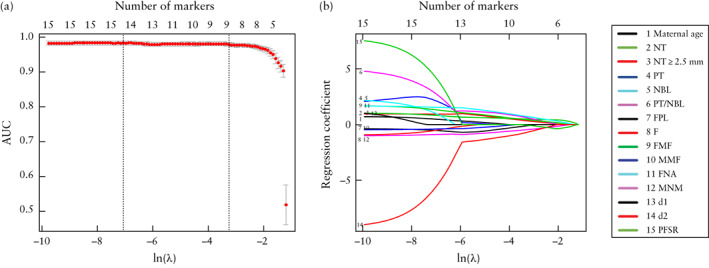
Selection from 15 potential markers of most useful predictive markers for trisomy 21 by least absolute shrinkage and selection operator (LASSO) method. Top x‐axes represent number of selected markers and bottom x‐axes represent penalty parameter ln(λ). (a) Illustration of relationship between area under receiver‐operating‐characteristics curve (AUC) of prediction model (LASSO model) and number of markers in the model. Vertical dotted line on right is at optimal value of ln(λ) (−3.44), at which nine markers were selected. Interval between left and right vertical dotted lines represents the range of good values for ln(λ). (b) Illustration of relationship between regression coefficients of markers and penalty parameter ln(λ). d1, distance between anterior edge of frontal bone and anterior edge of prefrontal skin; d2, distance between anterior edge of prefrontal skin (at same point as d1 measurement) and mandibulomaxillary line; F, largest perpendicular distance between facial profile line and outermost part of frontal bone; FMF, frontomaxillary facial angle; FNA, frontonasal facial angle; FPL, facial profile line; MMF, mandibulomaxillary facial angle; MNM, maxilla‐nasion‐mandible angle; NBL, nasal bone length; NT, nuchal translucency thickness; PFSR, prefrontal space ratio; PT, prenasal thickness; PT/NBL, prenasal‐thickness‐to‐nasal‐bone‐length ratio.
Prediction (LASSO) model
In the training set, a prediction model (LASSO model) for screening for trisomy 21 in the first trimester of pregnancy was developed using logistic regression based on the selected markers. In order to validate the predictive performance of the LASSO model, the area under the receiver‐operating‐characteristics (ROC) curve (AUC) of the LASSO model was calculated for both the training and the validation sets.
In order to examine whether the screening performance of the LASSO model was operator‐dependent, 60 fetuses (30 euploid and 30 with trisomy 21) were selected randomly from our dataset of 624, to act as a retest set. Delineation of the ultrasonographic markers in these 60 retest cases was performed by another sonographer (X.L.), who was blinded to the observations of the first sonographer (Y.S.) and to the karyotype results. The predictive performance of the LASSO model for trisomy 21 in this retest set was also assessed.
For the purpose of comparison, two additional models were developed, using logistic regression: (1) a model based on the combination of fetal NT and maternal age (the standard method of screening in many countries); (2) a model combining all 15 markers considered in this study (maternal age, fetal NT, fetal NT ≥ 2.5 mm, prenasal thickness, nasal bone length, prenasal‐thickness‐to‐nasal‐bone‐length ratio, d1, d2, prefrontal space ratio, frontomaxillary facial angle, maxilla‐nasion‐mandible angle, frontonasal facial angle, mandibulomaxillary facial angle, facial profile line and the F distance). The predictive performance of the LASSO model for trisomy 21 was compared with that of these two models.
Development and validation of nomogram
In order to provide clinicians with an individualized tool to predict patient‐specific probability for trisomy 21, a nomogram was developed based on the LASSO model constructed in the training set. The nomogram provided a visual presentation of the LASSO model. Each marker was assigned a ‘point’ on the nomogram, based on its predictive ability for trisomy 21. The sum of points for a particular fetus was then converted into an overall risk score for trisomy 21.
In order to validate the performance of the nomogram, the concordance index (C‐index) was calculated and a calibration curve for the nomogram was produced.
Statistical analysis
Statistical analysis was performed using R software (Version 3.5.2, http://www.R‐project.org). Differences for numerical and for categorical variables were calculated based on the t‐test, Mann–Whitney U‐test or chi‐square test, as appropriate. The screening performance of each marker and of each of the three models was evaluated by ROC‐curve analysis. Youden's index was used to calculate the optimal threshold (cut‐off) for prediction of trisomy 21. The DeLong test was used to compare ROC curves. The performance of the nomogram was assessed by C‐index and Hosmer‐Lemeshow test (a value > 0.05 shows good performance)16. P < 0.05 was considered statistically significant.
RESULTS
Maternal age and first‐trimester fetal ultrasonographic markers are presented in Table 1. All markers were significantly different between the trisomy‐21 and the euploid fetuses.
Table 1.
Maternal age and first‐trimester fetal ultrasonographic markers in 322 euploid fetuses and 302 fetuses with trisomy 21
| Parameter | Euploid | Trisomy 21 | P |
|---|---|---|---|
| Maternal age (years) | 33 (30–38) | 36 (31–39) | < 0.001 |
| Prenasal thickness (mm) | 1.2 (1.0–1.4) | 1.4 (1.2–1.7) | < 0.001 |
| Nasal bone length (mm) | 2.0 (1.8–2.1) | 1.8 (1.5–2.0) | < 0.001 |
| Prenasal‐thickness‐to‐nasal‐bone‐length ratio | 0.62 (0.48–0.73) | 0.85 (0.71–0.97) | < 0.001 |
| d1 (mm) | 1.2 (1.0–1.4) | 1.4 (1.2–1.7) | < 0.001 |
| d2 (mm) | 1.0 (0.8–1.3) | 0.8 (0.6–1.0) | < 0.001 |
| Prefrontal space ratio (d2/d1) | 0.86 (0.75–1.00) | 0.57 (0.43–0.75) | < 0.001 |
| Mandibulomaxillary facial angle (°) | 93.55 (92.15–96.18) | 93.15 (91.30–94.57) | 0.046 |
| Frontomaxillary facial angle (°) | 83.00 ± 4.88 | 88.84 ± 6.24 | < 0.001 |
| Maxilla‐nasion‐mandible angle (°) | 10.02 ± 1.87 | 7.83 ± 2.41 | < 0.001 |
| Frontonasal facial angle (°) | 115.75 ± 4.81 | 121.35 ± 4.90 | < 0.001 |
| NT (mm) | 1.5 (1.2–1.8) | 3.2 (1.9–4.2) | < 0.001 |
| NT ≥ 2.5 mm | 26 (8.1) | 207 (68.5) | < 0.001 |
| Facial profile line | < 0.001 | ||
| Negative | 0 (0.0) | 37 (12.3) | |
| Zero | 250 (77.6) | 241 (79.8) | |
| Positive | 72 (22.4) | 24 (7.9) | |
| F distance (cm)* | 0.16 (0.15–0.19) | 0.30 (0.26–0.32) | < 0.001 |
Data are given as median (interquartile range), mean ± SD or n (%).
In cases with positive facial profile line.
d1, distance between anterior edge of frontal bone and anterior edge of prefrontal skin; d2, distance between anterior edge of prefrontal skin and mandibulomaxillary line; F distance, largest perpendicular distance between facial profile line and outermost part of frontal bone; NT, nuchal translucency thickness.
In the training set, nine potential markers, including fetal NT, NT ≥ 2.5 mm, prenasal‐thickness‐to‐nasal‐bone‐length ratio, facial profile line, frontomaxillary facial angle, frontonasal facial angle, mandibulomaxillary facial angle, maxilla‐nasion‐mandible angle and d2, were identified by the LASSO method. Multivariable analysis led us to exclude fetal NT ≥ 2.5 mm, with a P‐value of 0.63. Thus, eight markers were used to develop the LASSO model.
LASSO model
The risk scores for trisomy 21 based on the LASSO model are given in Appendix S2 (including Figures S7 and S8). The AUC of the LASSO model was higher than the AUCs of all eight individual ultrasonographic markers included in the model, in both training and validation sets (LASSO model AUC, 0.983 (95% CI, 0.971–0.994) and 0.979 (95% CI, 0.966–0.993), respectively) (Figure 4 and Table 2). The LASSO model showed good discrimination of trisomy 21 in the retest set, with an AUC of 0.997 (95% CI, 0.990–1.000) (Figure 5).
Figure 4.
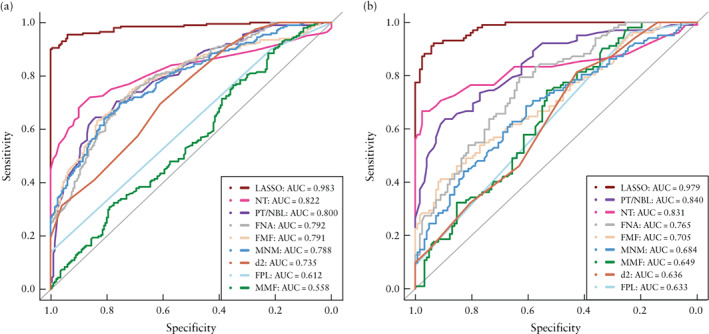
Receiver‐operating‐characteristics curves for prediction of trisomy 21 by least absolute shrinkage and selection operator (LASSO) model and, individually, eight ultrasonographic markers included in LASSO model, in training set (a) and validation set (b). Areas under the curves are given in Table 2. d2, distance between anterior edge of prefrontal skin and mandibulomaxillary line; FMF, frontomaxillary facial angle; FNA, frontonasal facial angle; FPL, facial profile line; MMF, mandibulomaxillary facial angle; MNM, maxilla‐nasion‐mandible angle; NT, nuchal translucency thickness; PT/NBL, prenasal‐thickness‐to‐nasal‐bone‐length ratio.
Table 2.
Areas under receiver‐operating‐characteristics (ROC) curves (AUC) for prediction of trisomy 21 by least absolute shrinkage and selecction operator (LASSO) model and, individually, eight ultrasonographic markers included in LASSO model, in training and validation sets
| Model/marker | Training set | Validation set |
|---|---|---|
| LASSO model | 0.983 (0.971–0.994) | 0.979 (0.966–0.993) |
| Nuchal translucency thickness | 0.822 (0.778–0.865) | 0.831 (0.770–0.892) |
| Prenasal‐thickness‐to‐nasal‐bone‐length ratio | 0.800 (0.758–0.843) | 0.840 (0.788–0.891) |
| Frontonasal facial angle | 0.792 (0.749–0.835) | 0.765 (0.704–0.826) |
| Frontomaxillary facial angle | 0.791 (0.747–0.835) | 0.705 (0.637–0.773) |
| Maxilla‐nasion‐mandible angle | 0.788 (0.744–0.831) | 0.684 (0.614–0.754) |
| d2 | 0.735 (0.688–0.782) | 0.636 (0.565–0.707) |
| Facial profile line | 0.612 (0.574–0.651) | 0.633 (0.587–0.678) |
| Mandibulomaxillary facial angle | 0.558 (0.501–0.614) | 0.649 (0.578–0.720) |
Data are given as AUC (95% CI).
ROC curves are shown in Figure 4.
d2, distance between anterior edge of prefrontal skin and mandibulomaxillary line.
Figure 5.
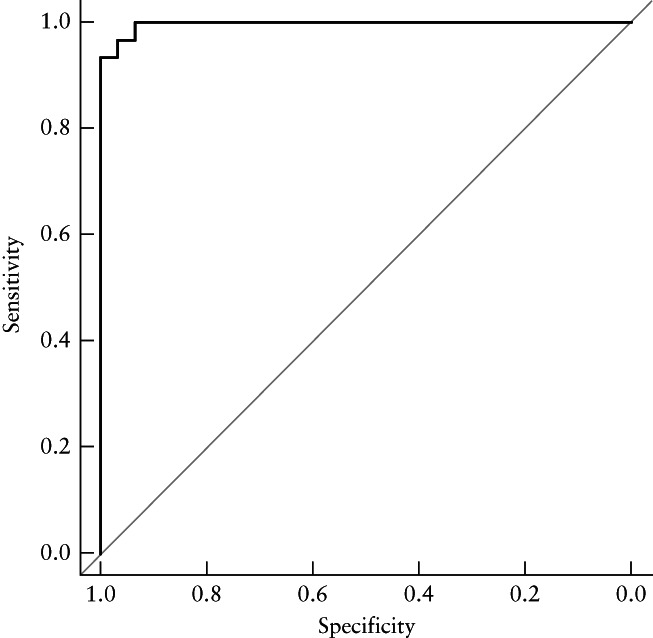
Receiver‐operating‐characteristics curve for prediction of trisomy 21 by least absolute shrinkage and selection operator (LASSO) model in retest set. Area under the curve = 0.997 (95% CI, 0.990–1.000).
The predictive performance in screening for trisomy 21 of the model based on fetal NT and maternal age, the model based on all markers and the LASSO model are presented in Table 3. The corresponding detection rates of the three models, at false‐positive rates (FPRs) of 1%, 3% and 5%, for screening for trisomy 21 are presented in Table 4. The ROC curves of the models are presented in Figure 6. The LASSO model showed significantly higher AUCs in both training (P < 0.001) and validation (P < 0.001) sets when compared with the model based on fetal NT and maternal age (Table 3), and it showed a lower AUC in the training set (P = 0.004), but a higher AUC in the validation set (P = 0.001), when compared with the model based on all markers. The model based on all markers showed significantly higher AUCs in both the training set (P < 0.001) and the validation set (P < 0.001) when compared with the model based on fetal NT and maternal age. For the LASSO model, there was no statistically significant difference in AUCs between the training set and the validation set (P = 0.697). For the model based on all markers, the AUC in the training set was higher than the AUC in the validation set (P < 0.001).
Table 3.
Comparison of predictive performance of three models in screening for trisomy 21
| Model | Detection rate(% (95% CI)) | False‐positive rate(% (95% CI)) | Youden index(cut‐off) | AUC(95% CI) |
|---|---|---|---|---|
| Fetal NT and maternal age | ||||
| Training set | 75.0 (69.0–81.0) | 7.0 (3.5–10.5) | 0.68 (0.561) | 0.883 (0.847–0.918) |
| Validation set | 84.3 (77.3–91.4) | 78.7 (71.4–86.0) | — | 0.797 (0.729–0.866) |
| All‐markers model | ||||
| Training set | 96.0 (93.3–98.7) | 1.0 (0.1–3.9) | 0.95 (0.644) | 0.994 (0.988–1.000) |
| Validation set | 93.1 (88.2–98.0) | 28.7 (26.8–43.7) | — | 0.931 (0.896–0.966) |
| LASSO model | ||||
| Training set | 90.5 (86.4–94.6) | 0.5 (0–1.5) | 0.90 (0.661) | 0.983 (0.971–0.994) |
| Validation set | 92.2 (86.9–97.4) | 7.4 (2.7–12.0) | — | 0.979 (0.966–0.993) |
‘All‐markers’ model included 15 markers: maternal age, fetal nuchal translucency thickness (NT), fetal NT ≥ 2.5 mm, prenasal thickness, nasal bone length, prenasal‐thickness‐to‐nasal‐bone‐length ratio, d1, d2, prefrontal space ratio, frontomaxillary facial angle, mandibulomaxillary facial angle, maxilla‐nasion‐mandible angle, frontonasal facial angle, facial profile line and the F distance.
LASSO model included eight markers: fetal NT, prenasal‐thickness‐to‐nasal‐bone‐length ratio, facial profile line, frontomaxillary facial angle, frontonasal facial angle, mandibulomaxillary facial angle, maxilla‐nasion‐mandible angle and d2.
Receiver‐operating‐characteristics curves are shown in Figure 6.
AUC, area under the receiver‐operating‐characteristics curve; LASSO, least absolute shrinkage and selection operator.
Table 4.
Comparison of detection rates of three models in screening for trisomy 21, at false‐positive rates (FPR) of 1%, 3% and 5%
| Model | Detection rate (% (95% CI)) for: | ||
|---|---|---|---|
| FPR of 1% | FPR of 3% | FPR of 5% | |
| Fetal NT and maternal age | |||
| Training set | 59 (52–66) | 66 (59–73) | 71 (65–77) |
| Validation set | 76 (67–84) | 82 (75–90) | 84 (77–91) |
| All markers | |||
| Training set | 92 (89–96) | 96 (94–99) | 98 (96–100) |
| Validation set | 92 (87–97) | 94 (90–99) | 95 (91–99) |
| LASSO model | |||
| Training set | 90 (86–95) | 91 (87–95) | 93 (90–96) |
| Validation set | 92 (87–97) | 93 (88–98) | 94 (90–99) |
For each model, sensitivity in validation set was calculated according to cut‐off of training set.
LASSO, least absolute shrinkage and selection operator; NT, nuchal translucency thickness.
Figure 6.

Receiver‐operating‐characteristics curves for prediction of trisomy 21 by model based on fetal nuchal translucency thickness and maternal age (a), model based on all 15 markers considered (b) and least absolute shrinkage and selection operator (LASSO) model (c), in training set ( ) and validation set (
) and validation set ( ). Areas under the curves are given in Table 3.
). Areas under the curves are given in Table 3.
Nomogram
The predictive ability of each independent marker included in the LASSO model for trisomy 21 is reflected in the nomogram (Figure 7a); an example, based on a trisomy‐21 case, is illustrated in Figure 7b. The nomogram showed good performance, with C‐indices of 0.983 and 0.981 in the training and validation sets, respectively. The indicators of the Hosmer–Lemeshow test in the training and validation sets were 0.157 and 0.132, respectively. The nomogram also showed a good calibration curve in the training set. However, the calibration curve in the validation set showed that the nomogram had a tendency to overestimate the risk for trisomy 21, in comparison to that in the training set (Figure 8a). The calibration curve of the entire dataset of 624 fetuses is given in Figure 8b.
Figure 7.
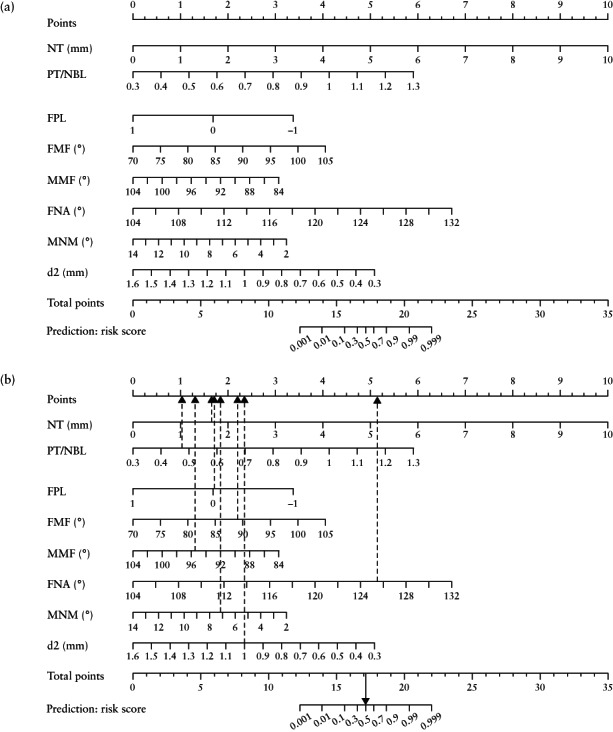
Nomogram for screening for trisomy 21 in first trimester of pregnancy (a), showing example of a case of trisomy 21 (b). Dashed arrows indicate points corresponding to individual markers and solid arrow represents overall risk score, converted from total points. In this case, fetal nuchal translucency thickness (NT) was 1.7 mm, prenasal‐thickness‐to‐nasal‐bone‐length ratio (PT/NBL) was 0.48, facial profile line (FPL) was 0, frontomaxillary facial angle (FMF) was 88.79°, mandibulomaxillary facial angle (MMF) was 95.60°, frontonasal facial angle (FNA) was 125.52°, maxilla‐nasion‐mandible angle (MNM) was 7.32°, distance between anterior edge of prefrontal skin and mandibulomaxillary line (d2) was 1.0 mm, corresponding to points of 1.71 + 1.01 + 1.73 + 2.18 + 1.27 + 5.12 + 1.82 + 2.32 = 17.16, providing a risk score in this case for trisomy 21 of 0.5.
Figure 8.
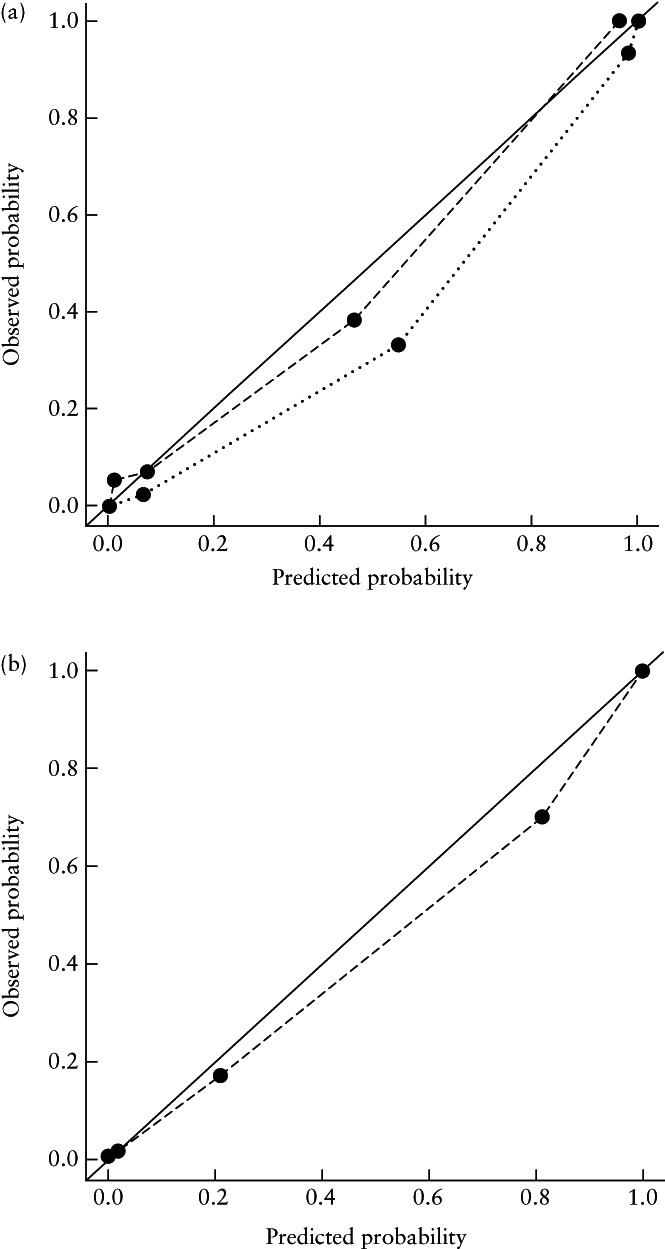
Calibration curves for nomogram for screening for trisomy 21 in first trimester of pregnancy, in training ( ) and validation (
) and validation ( ) sets (a) and for entire dataset of 624 fetal images (
) sets (a) and for entire dataset of 624 fetal images ( ) (b). Solid lines represent ideal performance.
) (b). Solid lines represent ideal performance.
DISCUSSION
Main findings
In this study, we developed and validated a LASSO model‐derived nomogram, which incorporates fetal NT and a series of fetal facial ultrasonographic markers relating to the nasal bone, prenasal thickness and flatness of the face, for the individualized prediction of trisomy 21 in the first trimester of pregnancy. The LASSO model was shown to be superior in the prediction of trisomy 21 to a model based on fetal NT and maternal age, whilst it had a lower AUC in the training set but a higher AUC in the validation set in comparison with a model based on all 15 markers; this may be because the model based on all markers was overfitting due to the use of redundant markers17. The calibration curve of the validation set showed that the nomogram based on the LASSO model has a tendency to overestimate the risk for trisomy 21; this may be related to the limited sample size.
Comparison with previous studies
Other than fetal NT, only four of the ultrasonographic markers selected by the LASSO method had been evaluated previously for their predictive performance for trisomy 21 in the first trimester: prenasal thickness‐to‐nasal bone length ratio, frontomaxillary facial angle, d2 and facial profile line. Our study confirmed that the first‐trimester prenasal thickness‐to‐nasal bone length ratio in trisomy 21 fetuses is greater than that in euploid fetuses3. Borenstein et al.5 demonstrated that adding the frontomaxillary facial angle to first‐trimester combined screening led to an increase in the estimated detection rate of trisomy 21, from 90% to 94% at a FPR of 5% and from 85% to 92% at a FPR of 3%5. We found that the frontomaxillary facial angle was increased in trisomy‐21 fetuses. The same research group also reported that trisomy‐18 fetuses had smaller mandibulomaxillary facial angle compared with euploid fetuses in the first trimester8. Our study proves that the mandibulomaxillary facial angle also has the potential to be an effective marker in the prediction of trisomy 21 in the first trimester. Midfacial hypoplasia is common in trisomy‐21 fetuses. However, while measurement of the frontonasal facial angle is feasible at 11 + 0 to 13 + 6 weeks7, this has not been reported in trisomy 21. We found that the frontonasal facial angle was increased in these fetuses. Our study also found that the prefrontal space ratio and d2 were both lower in the first trimester in trisomy‐21 fetuses compared with euploid fetuses4, confirming the suggestion of Bakker et al.4 that fetuses with trisomy 21 may have increased prenasal thickness, with lagging growth and forward displacement of the maxilla, resulting in a smaller d2 and decreased prefrontal space ratio. Neither prefrontal space ratio nor d1 was included in our LASSO model, while d2 was included. It appears that the prenasal‐thickness‐to‐nasal‐bone‐length ratio is superior to the prefrontal space ratio and d1 in the prediction of trisomy 21 in the first trimester of pregnancy. In the same study, Bakker et al.4 found that the facial profile line showed no clear pattern in relation to trisomy‐21 fetuses. Interestingly, a second‐ and third‐trimester study reported that there were no normal fetuses with a negative facial profile line, which is consistent with our findings18. Weichert et al.19 found the maxilla‐nasion‐mandible angle to be smaller in trisomy‐21 fetuses than in euploid fetuses in the second trimester of pregnancy. We found a similar relationship in our study, suggesting that this angle could be effective in the prediction of trisomy 21 in the first trimester.
The detection rate based on maternal age alone in screening for trisomy 21 is 30% at a 5% FPR20. With the identification of more sensitive and specific fetal markers, one might expect that the use of maternal age will eventually be phased out. This was indeed the case in the present study, with maternal age being excluded from the LASSO model.
Implications for clinical practice
For over a decade, the first‐trimester combined test, including maternal age, fetal NT and maternal serum biochemistry, with a detection rate of 90% at a FPR of 5%, has played a major role in screening for trisomy 21 in the first trimester of pregnancy. In recent years, cell‐free (cf)DNA testing has improved significantly the performance of screening for trisomy 21, with a detection rate of > 99% at a FPR of < 0.1%21, 22, 23, 24. However, the cost of cfDNA testing is high, making it impossible to offer it to all pregnant women. In some countries, the current recommendation is to offer cfDNA testing contingent on the results of the first‐trimester combined test25, 26. In China, the primary screening method for trisomy 21 is a combination of fetal NT and maternal age at 11–13 weeks' gestation. Our study has demonstrated that the LASSO model achieves better performance than does a combination of fetal NT and maternal age. This implies that, if the LASSO model is used as the primary ultrasonographic screening method, cfDNA testing could be offered to < 1% of the screened population.
Limitations
This was a retrospective study. Limitations include the fact that fetuses with ineligible images and those with absence of the nasal bone were excluded. This exclusion may have contributed to bias in our findings. In relation to the latter, in clinical practice it is well established that absence of the nasal bone is the second most important ultrasonographic marker in screening for trisomy 21. We excluded 23 of 325 (7%) cases with absence of the nasal bone. Further research is needed for development of a model that would allow evaluation of facial markers when there is absence of the nasal bone.
Conclusions
In this retrospective, case–control study, we have presented a promising, individualized nomogram that incorporates fetal NT and a series of ultrasonographic facial profile markers selected by the LASSO method and multivariable analysis. The nomogram can potentially be utilized as a convenient and effective tool in screening for trisomy 21 in the first trimester of pregnancy and requires prospective validation in future research.
Supporting information
Appendix S1 Definition and delineation of ultrasonographic markers (including Figures S1–S6)
Appendix S2 Risk scores for trisomy 21 based on the LASSO model (including Figures S7 and S8)
ACKNOWLEDGMENTS
This work was supported by the National Key Research and Development Program of China (2016YFC1000104, 2017YFC1308700, 2017YFA0205200, 2017YFC1309100), National Natural Science Foundation of China (81971619, 91959130, 81971776, 81771924, 81227901, 81671851, 81527805), Beijing Municipal Administration of Hospitals' Ascent Plan (DFL20151302), Beijing Natural Science Foundation (L182061), Bureau of International Cooperation of Chinese Academy of Sciences (173211KYSB20160053), Instrument Developing Project of the Chinese Academy of Sciences (YZ201502) and Youth Innovation Promotion Association CAS (2017175).
Contributor Information
C. Yin, Email: modscn@126.com.
L. C. Poon, Email: liona.poon@cuhk.edu.hk.
J. Tian, Email: jie.tian@ia.ac.cn.
Q. Wu, Email: wuqq2007@163.com.
REFERENCES
- 1.Agarwal GN, Kabra M. Diagnosis and management of Down syndrome. Indian J Pediatr 2014; 81: 560–567. [DOI] [PubMed] [Google Scholar]
- 2.Salomon LJ, Alfirevic Z, Bilardo CM, Chalouhi GE, Ghi T, Kagan KO, Lau TK, Papageorghiou AT, Raine‐Fenning NJ, Stirnemann J, Suresh S, Tabor A, Timor‐Tritsch IE, Toi A, Yeo G. ISUOG Practice Guidelines: performance of first‐trimester fetal ultrasound scan. Ultrasound Obstet Gynecol 2013; 41: 102–113. [DOI] [PubMed] [Google Scholar]
- 3.Manegold‐Brauer G, Bourdil L, Berg C, Schoetzau A, Gembruch U, Geipel A. Prenasal thickness to nasal bone length ratio in normal and trisomy 21 fetuses at 11–14 weeks of gestation. Prenat Diagn 2015; 35: 1079–1084. [DOI] [PubMed] [Google Scholar]
- 4.Bakker M, Pace M, de Jong‐Pleij E, Birnie E, Kagan KO, Bilardo CM. Prenasal Thickness, Prefrontal Space Ratio and Other Facial Profile Markers in First‐Trimester Fetuses with Aneuploidies, Cleft Palate, and Micrognathia. Fetal Diagn Ther 2018; 43: 231–240. [DOI] [PubMed] [Google Scholar]
- 5.Borenstein M, Persico N, Kagan KO, Gazzoni A, Nicolaides KH. Frontomaxillary facial angle in screening for trisomy 21 at 11 + 0 to 13 + 6 weeks. Ultrasound Obstet Gynecol 2008; 32: 5–11. [DOI] [PubMed] [Google Scholar]
- 6.Liu W, Qu S, Wang M, Xu W, Zhang G, Zhang C. Maxilla‐nasion‐mandible (MNM) angle: an indicator to assess fetal facial profile in first‐trimester of pregnancy. Springerplus 2016; 5: 1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vicario R, Pirollo LM, De Angelis C, Narcisi M, Pietropolli A, Piccione E. Frontonasal facial angle in chromosomally normal fetuses at 11 + 0 to 13 + 6 weeks. J Obstet Gynaecol Res 2010; 36: 1179–1184. [DOI] [PubMed] [Google Scholar]
- 8.Borenstein M, Persico N, Strobl I, Sonek J, Nicolaides KH. Frontomaxillary and mandibulomaxillary facial angles at 11 + 0 to 13 + 6 weeks in fetuses with trisomy 18. Ultrasound Obstet Gynecol 2007; 30: 928–933. [DOI] [PubMed] [Google Scholar]
- 9.Dong D, Tang L, Li ZY, Fang MJ, Gao JB, Shan XH, Ying XJ, Sun YS, Fu J, Wang XX, Li LM, Li ZH, Zhang DF, Zhang Y, Li ZM, Shan F, Bu ZD, Tian J, Ji JF. Development and validation of an individualized nomogram to identify occult peritoneal metastasis in patients with advanced gastric cancer. Ann Oncol 2019; 30: 431–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drukker L, Noble JA, Papageorghiou AT. Introduction to artificial intelligence in ultrasound imaging in obstetrics and gynecology. Ultrasound Obstet Gynecol 2020; 56: 498–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bishop CM. Pattern recognition and machine learning. Springer: New York, 2006. [Google Scholar]
- 12.Brattain LJ, Telfer BA, Dhyani M, Grajo JR, Samir AE. Machine learning for medical ultrasound: status, methods, and future opportunities. Abdom Radiol (NY) 2018; 43: 786–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Namburete AI, Stebbing RV, Kemp B, Yaqub M, Papageorghiou AT, Alison NJ. Learning‐based prediction of gestational age from ultrasound images of the fetal brain. Med Image Anal 2015; 21: 72–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Jong‐Pleij EA, Ribbert LS, Manten GT, Tromp E, Bilardo CM. Maxilla‐nasion‐mandible angle: a new method to assess profile anomalies in pregnancy. Ultrasound Obstet Gynecol 2011; 37: 562–569. [DOI] [PubMed] [Google Scholar]
- 15.Ko HS, Lee UY, Choi SK, Park YG, Park IY, Shin JC. Craniofacial inclination at 14 to 39 weeks' gestation in normal Korean fetuses. J Ultrasound Med 2012; 31: 569–576. [DOI] [PubMed] [Google Scholar]
- 16.Kramer AA, Zimmerman JE. Assessing the calibration of mortality benchmarks in critical care: The Hosmer‐Lemeshow test revisited. Crit Care Med 2007; 35: 2052–2056. [DOI] [PubMed] [Google Scholar]
- 17.Van Calster B, McLernon DJ, van Smeden M, Wynants L, Steyerberg EW, Topic Group ‘Evaluating diagnostic tests and prediction models’ of the STRATOS initiative. Calibration: the Achilles heel of predictive analytics. BMC Med 2019; 17: 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Jong‐Pleij EA, Ribbert LS, Pistorius LR, Tromp E, Bilardo CM. The fetal profile line: a proposal for a sonographic reference line to classify forehead and mandible anomalies in the second and third trimester. Prenat Diagn 2012; 32: 797–802. [DOI] [PubMed] [Google Scholar]
- 19.Weichert J, Gembicki M, Ribbat‐Idel J, Hartge DR. Assessment of Midfacial Hypoplasia in Down Syndrome Fetuses ‐ Validity of a Two‐Line Approach and Introduction of a Novel Angle (Maxilla‐Mandible‐Nasion Angle). Ultrasound Int Open 2016; 2: E58–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kagan KO, Wright D, Baker A, Sahota D, Nicolaides KH. Screening for trisomy 21 by maternal age, fetal nuchal translucency thickness, free beta‐human chorionic gonadotropin and pregnancy‐associated plasma protein‐A. Ultrasound Obstet Gynecol 2008; 31: 618–624. [DOI] [PubMed] [Google Scholar]
- 21.Gil MM, Accurti V, Santacruz B, Plana MN, Nicolaides KH. Analysis of cell‐free DNA in maternal blood in screening for aneuploidies: updated meta‐analysis. Ultrasound Obstet Gynecol 2017; 50: 302–314. [DOI] [PubMed] [Google Scholar]
- 22.Quezada MS, Gil MM, Francisco C, Orosz G, Nicolaides KH. Screening for trisomies 21, 18 and 13 by cell‐free DNA analysis of maternal blood at 10–11 weeks' gestation and the combined test at 11–13 weeks. Ultrasound Obstet Gynecol 2015; 45: 36–41. [DOI] [PubMed] [Google Scholar]
- 23.Galeva S, Konstantinidou L, Gil MM, Akolekar R, Nicolaides KH. Routine first‐trimester screening for fetal trisomies in twin pregnancy: cell‐free DNA test contingent on results from combined test. Ultrasound Obstet Gynecol 2019; 53: 208–213. [DOI] [PubMed] [Google Scholar]
- 24.Kagan KO, Maier V, Sonek J, Abele H, Luthgens K, Schmid M, Wagner P, Hoopmann M. False‐Positive Rate in First‐Trimester Screening Based on Ultrasound and Cell‐Free DNA versus First‐Trimester Combined Screening with Additional Ultrasound Markers. Fetal Diagn Ther 2019; 45: 317–324. [DOI] [PubMed] [Google Scholar]
- 25.Nicolaides KH, Syngelaki A, Poon LC, Gil MM, Wright D. First‐trimester contingent screening for trisomies 21, 18 and 13 by biomarkers and maternal blood cell‐free DNA testing. Fetal Diagn Ther 2014; 35: 185–192. [DOI] [PubMed] [Google Scholar]
- 26.Nicolaides KH, Wright D, Poon LC, Syngelaki A, Gil MM. First‐trimester contingent screening for trisomy 21 by biomarkers and maternal blood cell‐free DNA testing. Ultrasound Obstet Gynecol 2013; 42: 41–50. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 Definition and delineation of ultrasonographic markers (including Figures S1–S6)
Appendix S2 Risk scores for trisomy 21 based on the LASSO model (including Figures S7 and S8)


