Abstract
As the most common malignancy, lung cancer is characterised by high rates of occurrence and mortality. Although circular RNAs (circRNAs) are known to act as important regulators in cancer, their role in lung cancer remains poorly understood. In this study, circ_GRHPR expression was found to be significantly upregulated in the serum of five patients with non‐small cell lung cancer (NSCLC), compared to that in healthy controls. It is expressed at high levels in NSCLC cell lines, as revealed by qRT‐PCR analysis. Functionally, we demonstrated that circ_GRHPR promotes NSCLC proliferation and invasion in vitro and in vivo by cell proliferation, transwell, cell cycle, and tumour‐forming assays. Mechanistically, RNA pull‐down and RNA immunoprecipitation assays showed that circ_GRHPR interacts with the RNA‐binding protein poly(rC)‐binding protein 2 (PCBP2) and regulates its subcellular localisation by forming the circ_GRHPR/PCBP2 complex, localizing PCBP2 mainly in the cytoplasm and reducing the proportion found in the nucleus. Furthermore, we demonstrated that four‐and‐a‐half LIM‐only protein 3 (FHL3) is a tumour‐stimulating factor in NSCLC that interacts with and is influenced by PCBP2. Circ_GRHPR increased FHL3 expression in the nucleus of NSCLC cells by decreasing PCBP2 expression therein and promoting the proliferation and invasion of NSCLC cells. Therefore, our study identified that circ_GRHPR promotes NSCLC proliferation and invasion, providing a possible explanation for its mechanism of action.
Keywords: circular RNA, FHL3, non‐small cell lung cancer, PCBP2
1. INTRODUCTION
Lung cancer is the most common malignancy, with an estimated 2.2 million new cancer cases and 1.8 million deaths worldwide. It was the second most commonly diagnosed cancer and the major cause of cancer‐related death in 2020, representing approximately one in ten (11.4%) of cancers diagnosed and one in five (18.0%) of cancer‐related deaths.1 Over 85% of all new lung cancer cases are non‐small cell lung cancers (NSCLCs). Due to insufficient early screening and delayed clinical symptoms, most patients are diagnosed with advanced disease; thus, the prognosis is poor.2 Therefore, understanding the mechanisms underlying NSCLC progression and identifying the possible molecular markers are extremely important.
The structure of circRNAs makes them highly conserved and widely expressed. Compared with linear RNAs, circRNAs have a longer half‐life and greater resistance to digestion by RNase R.3 Because of these advantages, they are potential candidates for diagnostic markers and therapeutic targets. Many studies have discussed their unique expression characteristics and important biological roles in different kinds of cancers. For example, CircLIFR synergises with MSH2 to attenuate chemoresistance via the MutSα/ATM‐p73 axis in bladder cancer;4 CircPTPRA blocks the recognition of RNA N 6‐methyladenosine by interacting with IGF2BP1 to suppress bladder cancer progression.5 However, the biological functions of circRNAs in NSCLC remain unclear and need further studies.
In this study, we found a circular RNA with significant difference in expression in NSCLC—circ_GRHPR, and further explore its function and mechanism in the occurrence and development of NSCLC, ultimately providing novel insights into the pathogenesis of NSCLC.
2. RESULTS
2.1. Validation and stability analysis of circ_GRHPR in NSCLC
Hierarchical clustering analysis revealed the circRNA expression profiles in the serum of lung cancer patients and healthy controls (Figure 1A). We found 201 circRNAs that were differentially expressed between the sera of lung cancer patients and healthy controls. The characteristics of the circRNA profile in NSCLC are shown in Figure S1. Of these, hsa_circ_0001861—also known as hsa_circ_GRHPR (circ_GRHPR)—was significantly different between lung cancer patients and normal controls. Circ_GRHPR is derived from exons 2, 3 and 4 of the glyoxylate and hydroxypyruvate reductase (GRHPR) gene on chr9 (37424841‐37426651), with a length of 321 bp. To confirm its structure, Sanger sequencing was performed on the PCR products of circ_GRHPR, confirming that the PCR products had the expected size and predicted splicing sites (Figure 1B). We know that head‐to‐tail splicing is caused not only from the reverse splicing of cDNA but also from genomic rearrangements; thus, we designed convergent and divergent primers for circ_GRHPR to perform PCR using cDNA or genomic DNA from H1299 cells. Our results showed that circ_GRHPR could be amplified by divergent primers from cDNA, but not from gDNA (Figure 1C). To confirm the stability of circ_GRHPR, we used RNase R to treat RNA from H1299 cells; quantitative reverse transcriptase PCR (qRT‐PCR) demonstrated that linear GRHPR expression decreased greatly under RNase R treatment but circ_GRHPR expression did not (Figure 1D). Consequently, we identified the circular characteristics of circ_GRHPR. The function of circRNA is usually related to its localisation in NSCLC cells; we detected the subcellular localisation of circ_GRHPR in NSCLC cells by fluorescence in situ hybridisation (FISH) and nuclear‐cytoplasmic fractionation and found that it was mainly present in the cytoplasm of NSCLC cells, with only a small proportion in the nucleus (Figure 1E,F).
FIGURE 1.
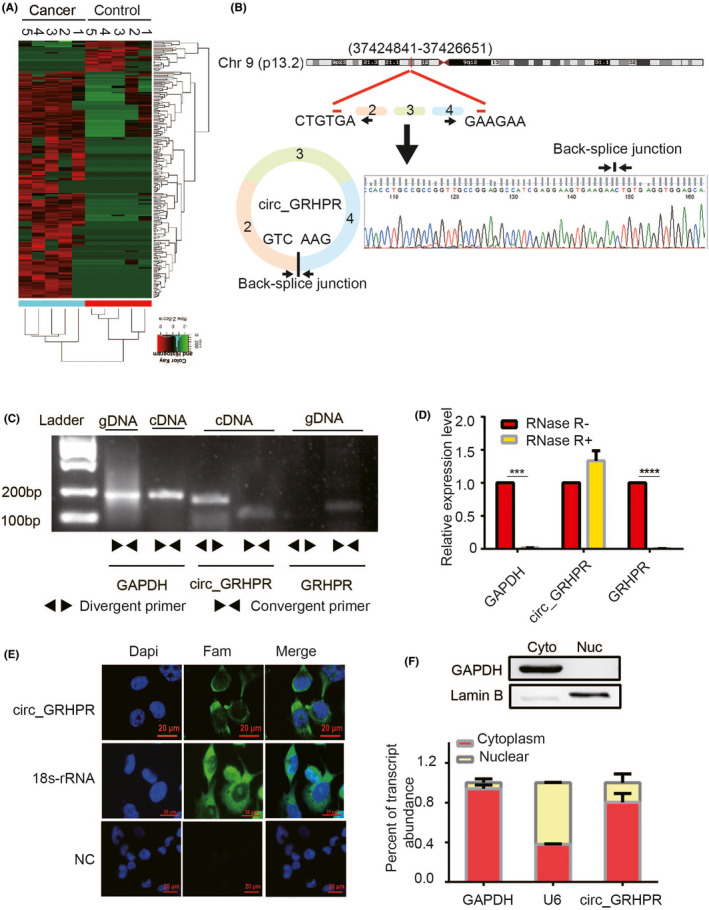
Validation and stability analysis of circ_GRHPR in non‐small cell lung cancer (NSCLC). (A) Heat maps of differential expression of circular RNAs in lung cancer patients (n = 5) and controls (n = 5) are shown (red indicates upregulated expression; green indicates downregulated expression). (B) The schematic diagram shows the structure of circ_GRHPR via the circularisation of exons 2, 3, and 4 from GRHPR in Chr9; Sanger sequencing of the RT‐PCR products of circ_GRHPR is shown on the right. (C) circ_GRHPR and GRHPR were amplified by cDNA and gDNA of H1299 cells with divergent primers and convergent primers, respectively. (D) The expression of circ_GRHPR and GRHPR in H1299 cells were detected by qRT‐PCR in the presence or absence of RNase R. GAPDH was used as an internal control. (E) Subcellular localisation of circ_GRHPR was detected by FISH, 18S‐rRNA was used as the positive control, and a negative control probe was used as the negative control in NSCLC cells. (F) Location of circ_GRHPR in NSCLC cells. (Left) GAPDH and U6 were applied as positive controls in the cytoplasm and nucleus, respectively. Western blotting was used to detect the partitioning between the nucleus and cytoplasm. Data are shown as the mean ± SD, p < 0.05 was considered statistically significant (*p < 0.05/**p < 0.01/***p < 0.001)
2.2. Circ_GRHPR promotes the proliferation and invasion of NSCLC cells
To investigate the biological role of circ_GRHPR in NSCLC, we assessed circ_GRHPR expression levels in different NSCLC cells (Figure 2A). The results showed that the expression levels of circ_GRHPR in NSCLC cell lines were significantly higher than those in BEAS‐2B cells. Among them, circ_GRHPR had the highest expression level in H1299 cells and the lowest in PC‐9 cells. Therefore, we constructed circ_GRHPR overexpression plasmids (PLC5‐circ_GRHPR‐OE) and control plasmids (pLC5‐circ_GRHPR‐NC) and transfected them into PC‐9 cells. To further determine whether circ_GRHPR knockdown had opposite effects, two siRNAs were designed to specifically target the junction site of circ_GRHPR and were then transfected into H1299 cells. Figure 2B shows the transfection efficiency by qRT‐PCR. circ_GRHPR expression was higher in circ_GRHPR‐overexpressing PC‐9 cells and lower in circ_GRHPR knockdown H1299 cells, whereas the expression of the corresponding parental gene GRHPR showed no significant changes. Functionally, the real‐time cellular analysis (RTCA) assay revealed that circ_GRHPR stimulated NSCLC cell proliferation (Figure 2C). Moreover, the transwell assay showed that circ_GRHPR significantly promoted the invasion ability of NSCLC cells (Figure 3D). Cell‐cycle experiments (Figure S2A) showed that the G1 phase cell ratio of si‐circ_GRHPR was approximately 5% higher than that in H1299 cells. Similarly, PC‐9 circ_GRHPR‐OE decreased by approximately 5% in the G1 phase compared to that in the control. This result indicated that circ_GRHPR delayed the NSCLC cell‐cycle process. To test whether circ_GRHPR had a similar function in vivo, we conducted a subcutaneous tumorigenesis experiment in nude mice. The experimental results showed that the tumour growth in the high expression group was much faster than that in the control group and the weight of the solid tumour in the high expression group was much higher than that in the control group (Figure 2E). Taken together, these results indicated that overexpression of circ‐GRHPR effectively promotes the proliferation and invasion of NSCLC in vitro and in vivo.
FIGURE 2.
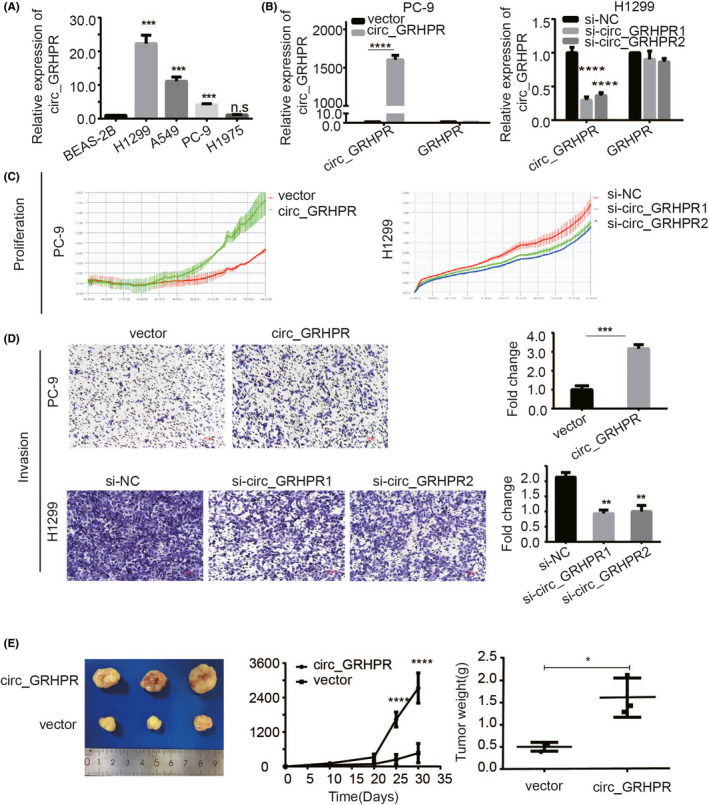
Circ_GRHPR promotes the proliferation and invasion of non‐small cell lung cancer (NSCLC) cells. (A) qRT‐PCR analysis of circ_GRHPR expression in normal lung epithelial BEAS‐2B cells and NSCLC cells. (B) The transfection efficacy of circ_GRHPR and GRHPR expression in PC‐9 and H1299 cells was detected using qRT‐PCR analysis. (C) The proliferation status of PC‐9 and H1299 cells was determined by RTCA assay. (D) The invasion status of PC‐9 and H1299 cells was determined by transwell assay. (Right) average number of cells in five fields under a 10× microscope. (E) Circ_GRHPR promoted the proliferation of NSCLC cells in nude mice. The three pictures represent solid tumour volume, tumour growth curve, and tumour weight. Data are shown as the mean ± SD, p < 0.05 was considered statistically significant (*p < 0.05/**p < 0.01/***p < 0.001)
FIGURE 3.
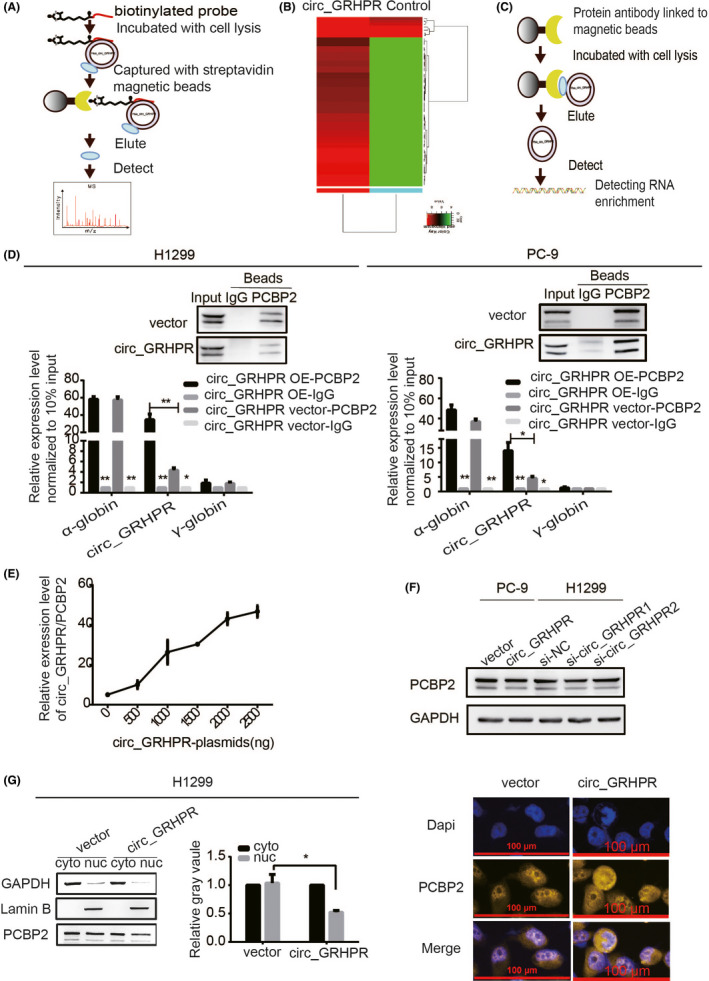
Circ_GRHPR interacts with the RNA‐binding protein PCBP2 and regulates its subcellular localisation. (A) The schematic diagram provides a summary of the RNA pull‐down procedure. (B) The cluster analysis showed 58 candidate proteins that might interact with circ_GRHPR. (Red and green represent up‐ and downregulation, respectively). (C) The schematic diagram shows the RIP procedure. (D) circ_GRHPR molecules enriched by PCBP2 protein and IgG protein were detected by RIP assay in H1299 and PC‐9 cells. α‐globin is a positive control that has been shown to interact with PCBP2; γ‐globin was the negative control. Western blotting shows the reliability of the RIP experimental system data. (E) The relationship between the amount of circ_GRHPR/PCBP2 complex and circ_GRHPR was observed by the RIP experimental system. With an increase in the circ_GRHPR plasmid, the expression levels of circ_GRHPR increased and then the number of circ_GRHPR enriched by PCBP2 increased (F) PCBP2 expression in PC‐9 and H1299 cells was detected by western blotting after transfecting the circ_GRHPR overexpression plasmid and siRNA, respectively. (G) PCBP2 expression levels in the nucleus and cytoplasm of circ_GRHPR‐OE and NC cells were detected in H1299 cells. The grey value of their protein bands is shown on the right. (H) The location of PCBP2 protein was detected in circ_GRHPR‐OE and NC cells of H1299 by FISH. Data are presented as the mean ± SD, p < 0.05 was considered statistically significant (*p < 0.05/**p < 0.01/***p < 0.001)
2.3. Circ_GRHPR interacts with the RNA‐binding protein PCBP2 and regulates its subcellular localisation
To further investigate the mechanism of circ_GRHPR function, H1299 lysates were incubated with a biotinylated circ_GRHPR probe and subjected to an RNA pull‐down assay and mass spectrometry (Figure 3A). Figure 3B shows the hierarchical clustering of 58 candidate proteins that may interact with circ_GRHPR. The top ten proteins are listed in Table S1. We chose the RNA‐binding protein poly(rC)‐binding protein 2 (PCBP2) as the research target. We use RBPmap6 database to predict circ_ GRHPR has at least eight conserved binding sites to PCBP2 (Figure S3A). To further verify whether circ_GRHPR interacts with PCBP2, we designed an RNA immunoprecipitation (RIP) experiment (Figure 3C). The results showed that the complex precipitated with anti‐PCBP2 antibody contained a high level of circ_GRHPR compared to the control with anti‐IgG antibody. α‐globin is a positive control that has been shown to interact with PCBP2; γ‐globin is a negative control7 (Figure 3D). This phenomenon may reflect the specificity of circ_GRHPR and PCBP2 binding. For further verification, we transfected H1299 cells with different concentrations of circ_GRHPR overexpression plasmids. We found that more circ_GRHPR was overexpressed and more circ_GRHPR/PCBP2 complexes were bound, reflecting the specificity of circ_GRHPR binding with PCBP2 (Figure 3E). However, as shown in Figure 3F, circ_GRHPR did not affect PCBP2 expression but regulated the subcellular localisation of PCBP2 and increased the cytoplasm/nucleus ratio of PCBP2 in H1299 (Figure 3G) and PC‐9 cells (Figure S3B). While circ_GRHPR was highly expressed, PCBP2 was concentrated in the cytoplasm, even though the level of PCBP2 in the nucleus decreased. Immunofluorescence staining also showed that circ_GRHPR mainly retained PCBP2 in the cytoplasm (Figure 3H). In summary, circ_GRHPR interacts with PCBP2 and the complex regulates the subcellular localisation of the latter by increasing its cytoplasm/nucleus ratio.
2.4. Circ_GRHPR promotes the nuclear expression of FHL3 through PCBP2
Next, we explored how changes in the nucleocytoplasmic localisation of PCBP2 cause downstream changes, ultimately altering the proliferation and invasion ability of NSCLC. Studies have shown that PCBP2 belongs to a class of proteins that bind to poly(rC) fragments of RNA and DNA.8 PCBP2 has been shown to have many functions, with the main one being the maintenance of mRNA stability and translation regulation.8 Wei Han9 studied glioma and showed that 35 types of mRNA can be enriched by RIP chip analysis of PCBP2 binding protein that can bind directly to PCBP2. Thus, we analysed this finding from the perspective of mRNA regulation by the RNA binding protein PCBP2, which regulates NSCLC cell proliferation and invasion. We detected nine of the most interesting mRNAs from these 35 mRNAs through RIP experiment in H1299 cell line, of which FHL3 was finally chosen as our target mRNA as the research object (Figure S4A). To verify the binding of PCBP2 to FHL3, an RIP assay was performed in the PC‐9 cell line too. Compared with IgG, PCBP2 protein aggregated a significant number of FHL3 mRNAs in NSCLC cells (Figure 4A). To further verify the relationship between PCBP2 and FHL3, we constructed low‐expression cell lines of PCBP2 in PC‐9 cells and overexpression cell lines of PCBP2 in H1299 cells and detected the protein and mRNA expression levels of FHL3. Our results showed that the expression of FHL3 increased when PCBP2 was downregulated in PC‐9 cells and decreased when PCBP2 was overexpressed in H1299 cells (Figure 4B). To test our hypothesis, we detected FHL3 expression in the nucleus in the circ_GRHPR‐OE and circ_GRHPR‐NC cells. Figure 4C shows the results of the nucleocytoplasmic separation. At both the protein and mRNA levels, FHL3 expression was higher in the nucleus of circ_GRHPR‐OE cells than in the nucleus of circ_GRHPR‐NC cells, confirming our hypothesis. As shown in Figure 3G, we verified that overexpression of circ_GRHPR decreased the expression of PCBP2 in the nucleus and PCBP2 inhibited FHL3 expression in NSCLC. When circ_GRHPR was overexpressed, nuclear FHL3 expression increased compared to that in the control (Figure 4C). Therefore, we propose that overexpression of circ‐GRHPR and re‐localisation of PCBP2 in the cell reduces PCBP2 expression and changes its function, which results in an increase in FHL3 expression.
FIGURE 4.
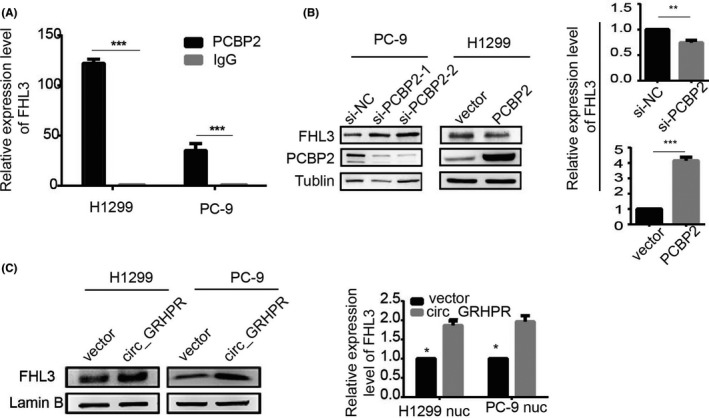
Circ_GRHPR promotes the expression of FHL3 in the nucleus through PCBP2. (A) The RIP assay was performed on the PC‐9 and H1299 cell lines to verify the binding of PCBP2 to FHL3. Compared with IgG, PCBP2 protein aggregated a significant number of FHL3 mRNAs in non‐small cell lung cancer (NSCLC) cells. (B) PCBP2 could degrade FHL3 protein and mRNA levels in NSCLC cells. (C) circ_GRHPR promoted protein expression of FHL3 in the nucleus on the left. Circ_GRHPR promoted the expression of FHL3 mRNA in the nucleus on the right. Data are presented as the mean ± SD, p < 0.05 was considered statistically significant (*p < 0.05/**p < 0.01/***p < 0.001)
2.5. FHL3 promotes proliferation and invasion of NSCLC
To verify the FHL3 phenotype, we studied 483 patients with lung adenocarcinoma (LUAD) with 59 normal controls, and 486 patients with lung squamous cell carcinoma (LUSC) with 50 normal controls from The Cancer Genome Atlas using Gene Expression Profiling Interactive Analysis (GEPIA2) (http://gepia2.cancer‐pku.cn/). We also analysed the correlation between overall survival rates and FHL3 expression in NSCLC. A boxplot using disease state as a variable was plotted to calculate the differential FHL3 expression (Figure 5A). A clear trend was observed with significantly increased FHL3 expression in NSCLC tumour tissues. In LUAD patients, higher FHL3 expression was correlated with significantly poorer survival (p = 0.0085). Following a similar pattern, FHL3 was associated with slightly poorer survival by quartile expression (p = 0.15) in LUSC tissues, indicating that FHL3 is a cancer‐promoting factor in NSCLC. Through cell‐cycle experiments, we also verified that FHL3 reduces the number of cells in the G1 phase, allowing more cells to enter the G2 phase, thus promoting tumour growth (Figure S5A). To further verify the relationship between circ_GRHPR and FHL3, we transfected circ_GRHPR and FHL3 into H1299 and PC‐9 cells simultaneously. The results showed that FHL3 promotes NSCLC cell proliferation and invasion; its function is synergetic with that of circ_GRHPR. (Figure 5B,C).
FIGURE 5.
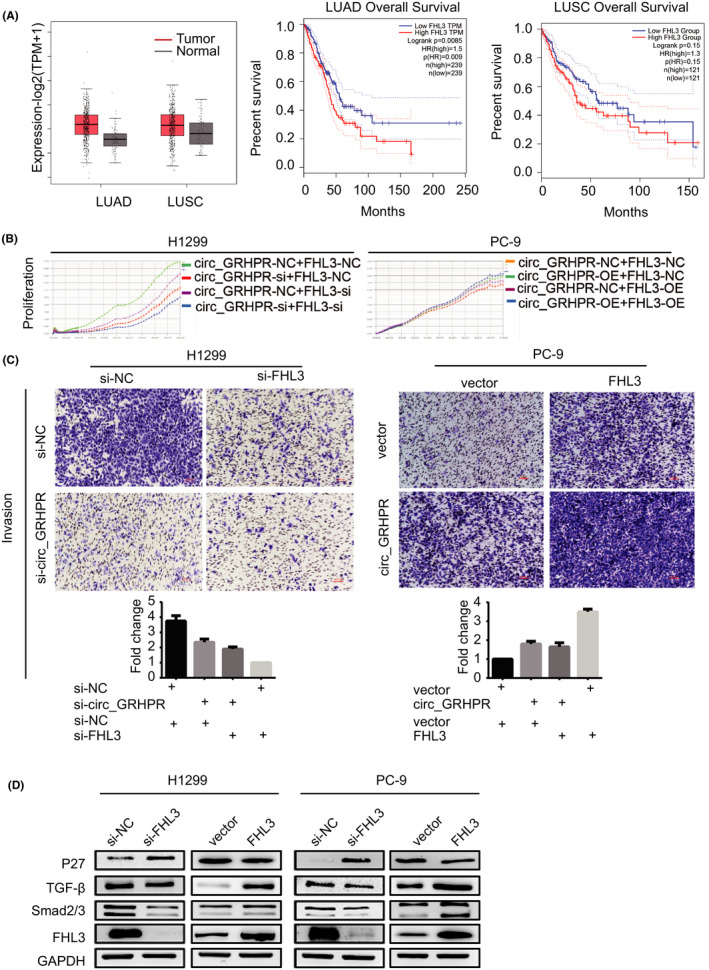
FHL3 promotes proliferation and invasion of non‐small cell lung cancer (NSCLC) cells. (A) Expression levels of FHL3 and overall survival of the FHL3 genes were validated in LUAD, LUSC, and normal tissues with GEPIA2. |log2FC| > 1 and p‐value <0.01 were considered statistically significant. Tumour and normal tissues are shown in red and grey, respectively. (B) The proliferation status of FHL3 was determined by the RTCA assay in H1299 and PC‐9 cells. (C) The invasion status of FHL3 in H1299 and PC‐9 cells was determined by a transwell assay. The number of cells was quantified in five fields under a 10× microscope. (D) P27, TGF‐β, and Smad2/3 were downstream target genes of FHL3 in NSCLC cells. Data are presented as the mean ± SD, p < 0.05 was considered statistically significant (*p < 0.05/**p < 0.01/***p < 0.001)
According to reports by Niu10 and Mao,11 genes such as P27, TGF‐β, and Smad2/3 are the downstream targets of FHL3. We examined expression of these genes in NSCLC cells and found that FHL3 overexpression inhibited P27 expression, disturbing the cell cycle and promoting the expression of TGF‐β and Smad2/3—all of which promote cell invasion (Figure 5D).
3. DISCUSSION
CircRNAs are novel non‐coding RNA molecules that perform various biological functions. To date, circRNAs have four known mechanisms: (a) circRNA adsorbs microRNA to prevent target mRNA from being degraded by miRNA;12, 13, 14, 15 (b) circRNAs regulate transcription and splicing by recruiting epigenetic regulators or transcription factors;16, 17 (c) circRNAs translate and encode proteins or polypeptides;18, 19 and (d) circRNAs interact with RNA‐binding proteins (RBPs).20 Among these mechanisms, its best‐known function is that of a miRNA sponge; however, the mechanism by which circRNAs affect the development of tumours via RBPs is poorly understood. RBPs play a significant role in the post‐transcriptional regulation of RNAs, participating in mRNA stabilisation, translation, and localisation, as well as protein binding. Therefore, there must be a strong relationship between RBPs and circRNAs; however, the mechanism underlying this relationship is largely unclear. Our research sheds light on the role of circ_GRHPR in promoting the development of NSCLC by affecting the subcellular localisation of PCBP2. In our article, circ_GRHPR could bind PCBP2 directly and the binding is specific. Since circ_GRHPR is mainly expressed in the cytoplasm, with the increase of circ_GRHPR expression in NSCLC, similar with sponge absorption, more PCBP2 were delayed in the cytoplasm. Of course, several other proteins may also bind to circ_GRHPR, we cannot exclude the possibility that some of these proteins may also play roles in mediating the role of circ_GRHPR in NSCLC. This will be studied in the future.
PCBP2 is one of the most abundant RBPs in mammalian cells. At the current stage, PCBP2 has been proved to play an important role in RNA stabilization, translation, and enhancement.21, 22 As Wei Han9 studied, PCBP2 may be negatively regulating FHL3 mRNA translation in gliomas, and the major binding determinant of PCBP2 resides in the FHL3‐3′A fragments. In our article, we also demonstrated that PCBP2 may negatively regulate the expression of FHL3 in NSCLC. We also found that PCBP2 had other downstream targets that might be regulated by other novel mechanisms and they might also regulate the occurrence and development of NSCLC. The detailed mechanisms need to be further explored.
FHL3 is a member of the four‐and‐a‐half LIM domain (FHL) family. FHL3 interacts with transcription factors and multiple cell‐signalling molecules in glioma, breast cancer, and liver cancer.9, 10, 23 Although most studies mentioned that FHL3 is a tumour suppressor, we identified that FHL3 promotes NSCLC cell proliferation and invasion, potentially affecting the biological behaviour of NSCLC cells by disturbing the cell cycle via P27 and invasion by the TGF‐β signalling pathway, as reported in other studies.10, 11 Meanwhile, the tumour‐promoting effect of FHL3 is closely correlated with the survival rate and prognosis. Taken together, these results demonstrate that FHL3 acts as a tumour‐promoting factor in NSCLC.
In conclusion, we revealed the characteristics of circRNA profiles in NSCLC from human serum samples. We demonstrated that circ_GRHPR promotes NSCLC cell proliferation and invasion and interacts with PCBP2, changing PCBP2's subcellular localisation. This translocation regulates FHL3 expression in the nucleus, promoting the proliferation and invasion ability of NSCLC cells. Overall, we demonstrated the role of circ_GRHPR in the occurrence and development of NSCLC and provided clear evidence for the possible mechanism underlying its action.
4. METHODS
4.1. CircRNAs array analysis
Serum samples of five lung cancer patients and healthy controls were obtained from Ruijin Hospital Affiliated to Medical College of Shanghai Jiaotong University (Shanghai, China). High throughput sequencing was performed by CloudSeq Biotech (Shanghai, China). Methods are described in the previous literature.24
4.2. Cell culture
Immortalised human bronchial epithelial cells (BEAS‐2B cells) were purchased from the Cell Bank of the Chinese Academy of Sciences, and human NSCLC cell lines (A549, H1975, H1299, PC‐9) were gifted by the Shanghai Chest Hospital Affiliated to Shanghai Jiaotong University. The cells were maintained in RPMI 1640, DMEM, and IMDM supplemented with 10% foetal bovine serum and 1% antibiotics.
4.3. RNase R treatment
For RNase R digestion, 2 μg RNA was diluted in a buffer containing 0.2 μL of RNase R (20 U/μL) and 0.6 μL 10× RNase R reaction buffer; 0.2 μL DEPC‐treated water was used instead of RNase R for the control. The samples were cultured at 37°C for 30 min. GAPDH was used as an internal control.
4.4. RNA extraction and qRT‐PCR
Total RNA was extracted from cell lines using Trizol Reagent (Invitrogen/Thermo Fisher Scientific). The RNA from nuclear and cytoplasmic fractions was separated using the PARIS Kit (Invitrogen). qRT‐PCR was performed using Evo M‐MLV RT Kit with gDNA Clean for qPCR (Accurate Bio‐Medical Technology). All primers are listed in Table S2.
4.5. RNA interference and overexpression
All siRNA pairs and PCBP2 and FHL3 overexpression plasmids were constructed and purified by Shanghai GenePharma Co., Ltd.. Circ_GRHPR overexpression (pLC5‐circ_GRHPR) and control plasmids (pLC5‐circ) were synthesised by Geneseed. Transfection was performed using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen). We collected total RNA and protein 48 h after transfection.
4.6. Protein extraction and western blot analysis
We lysed cells in lysis buffer (1% sodium dodecyl sulphate [SDS], 1% phenylmethylsulphonyl fluoride [PMSF], and 1% cocktail), incubated on ice for 30 min, and then, centrifuged for 10 min (12 000 g, 4°C). The supernatant was collected, and protein concentration was calculated using a Pierce BCA protein assay kit (Thermo Fisher Scientific). All antibodies are listed in Table S3.
4.7. Cell proliferation assay
RTCA is a real‐time marker‐free cell function analyser. After digestion, 5 × 103 cells were seeded on the electrode plate and cultured for 72 h. Cell proliferation was observed.
4.8. Invasion assay
Cell invasion was detected using chambers (pore size 8 μm, 24‐well; Becton, Dickinson and Company) with Matrigel according to the manufacturer's instructions (Corning Life Sciences). The lower chamber was supplemented with 600 μL medium containing 20% foetal bovine serum; 5 × 104 cells were added to the upper chamber.
4.9. Cell cycle
For the cell‐cycle assay, H1299 and PC‐9 cells were harvested, fixed in ice‐cold ethanol, and stored at 4°C overnight. Next, the fixed cells were resuspended in PBS and stained with propidium iodide (PI). The cell‐cycle phase was evaluated using a FACSCanto II with a FACS Canto II (BD Biosciences); the results were analysed using ModFit LT 2.0 (Verity Software House).
4.10. Xenograft tumourigenesis model
Sixteen 4‐week‐old male nude mice were obtained from the Ruijin Hospital Experimental Animal Centre (Shanghai, China) and divided into two equal groups for circ_GRHPR‐NC and circ_GRHPR‐overexpression treatment; 5 × 106 stable H1299 cells were injected subcutaneously into the armpit. Tumour volumes were calculated using the length (a) and width (b) as follows: volume (mm3) = ab2/2. Five weeks after injection, the animals were killed.
4.11. Fluorescent in situ hybridisation
Cy3‐labeled locked nucleic acid circ_GRHPR probes were designed and synthesised by GenePharma Co., Ltd. The probe signals were detected using a FISH kit according to the manufacturer's instructions (RiboBio Co., Ltd). The probes are listed in Table S3.
4.12. RNA pull‐down assays
Circ_GRHPR biotinylated‐probe and NC biotinylated‐probe were designed and synthesised by CloudSeq. Briefly, H1299 cells were harvested, lysed, and sonicated. To generate probe‐coated beads, the circ_GRHPR probe and NC probe were incubated with magnetic beads. After 2 h of incubation, cell lysates were incubated with the probes overnight. The bound RNAs were then washed and purified for analysis.
4.13. RNA‐binding protein immunoprecipitation
RNA‐binding protein immunoprecipitation experiments were performed using the Magna RIP RNA‐Binding Protein Immunoprecipitation Kit according to the manufacturer's instructions (Millipore).
4.14. Immunofluorescence
Non‐small cell lung cancer cells were grown on a confocal dish (Corning) for 24 h and then incubated with primary antibody at 4°C overnight. Then, they were incubated with secondary antibodies for 30 min at 37°C, followed by DAPI staining.
4.15. Gene Expression Profiling Interactive Analysis
The expression of FHL3 and survival analysis in NSCLC and normal tissue were analysed using the GEPIA2 online analysis program (http://gepia2.cancer‐pku.cn/). To identify genes that affect overall survival in FHL3, patient populations were split into two groups by median or quartile expression (high vs. low expression).
4.16. Statistical analysis
SPSS Version 19.0 (IBM Corp.) was used for statistical analysis, and images were plotted using GraphPad Prism 6.02 (GraphPad Software). A t‐test was used for comparison between two groups and one‐way ANOVA for more than two groups. Statistical significance was set at p < 0.05.
CONFLICT OF INTEREST
None of the authors have any financial conflicts of interest that might have influenced the results or interpretation of the article.
PEER REVIEW
The authors, reviewers and editors affirm that in accordance to the policies set by the journal of Clinical and Experimental Pharmacology and Physiology, this manuscript presents an accurate and transparent account of the study being reported and that all critical details describing the methods and results are present.
Supporting information
Figure S1
Figure S2
Figure S3
Figure S4
Figure S5
Table S1
Table S2
Table S3
ACKNOWLEDGEMENTS
We thank Editage for English editing of the manuscript. This work was supported by the Project of Shanghai Municipal Commission of Health and Family Planning (NO. 201740055) and Shanghai Sailing Program (20YF1426300 to Q.L.).
Yanyan Hou, Jiafei Lin and Danyang Wang contributed equally to this study.
Contributor Information
Xiangfan Liu, Email: liuxiangfanbxnll@163.com.
Peihua Ni, Email: nipeihua000@163.com.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1.Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209‐249. [DOI] [PubMed] [Google Scholar]
- 2.Gridelli C, Rossi A, Carbone DP, et al. Non‐small‐cell lung cancer. Nat Rev Dis Primers. 2015;1:10059. [DOI] [PubMed] [Google Scholar]
- 3.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453‐461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang H, Xiao X, Wei W, et al. CircLIFR synergizes with MSH2 to attenuate chemoresistance via MutSα/ATM‐p73 axis in bladder cancer. Mol Cancer. 2021;20:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie F, Huang C, Liu F, et al. CircPTPRA blocks the recognition of RNA N6‐methyladenosine through interacting with IGF2BP1 to suppress bladder cancer progression. Mol Cancer. 2021;20(1):68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paz I, Kosti I, Ares M Jr, Cline M, Mandel‐Gutfreund Y. RBPmap: a web server for mapping binding sites of RNA‐binding proteins. Nucleic Acids Res. 2014;42:W361‐W367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xin Z, Han W, Zhao Z, et al. PCBP2 enhances the antiviral activity of IFN‐α against HCV by stabilizing the mRNA of STAT1 and STAT2. PLoS One. 2011;6:e25419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Makeyev AV, Liebhaber SA. The poly(C)‐binding proteins: a multiplicity of functions and a search for mechanisms. RNA. 2002;8:265‐278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han W, Xin Z, Zhao Z, et al. RNA‐binding protein PCBP2 modulates glioma growth by regulating FHL3. J Clin Invest. 2013;123:2103‐2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niu C, Yan Z, Cheng L, et al. Downregulation and antiproliferative role of FHL3 in breast cancer. IUBMB Life. 2011;63:764‐771. [DOI] [PubMed] [Google Scholar]
- 11.Mao J, Sun Z, Cui Y, et al. PCBP2 promotes the development of glioma by regulating FHL3/TGF‐β/Smad signaling pathway. J Cell Physiol. 2020;235:3280‐3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen B, Ji F, Wen X, Jin Z. Circular RNA circ_ASAP2 promotes cell viability, migration, and invasion of gastric cancer cells by regulating the miR‐770‐5p/CDK6 axis. Int J Clin Exp Pathol. 2020;13:2806‐2819. [PMC free article] [PubMed] [Google Scholar]
- 13.Guo X, Zhou Q, Su D, et al. Circular RNA circBFAR promotes the progression of pancreatic ductal adenocarcinoma via the miR‐34b‐5p/MET/Akt axis. Mol Cancer. 2020;19:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jian X, He H, Zhu J, et al. Hsa_circ_001680 affects the proliferation and migration of CRC and mediates its chemoresistance by regulating BMI1 through miR‐340. Mol Cancer. 2020;19:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsiao KY, Lin YC, Gupta SK, et al. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77:2339‐2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z, Huang C, Bao C, et al. Exon‐intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256‐264. [DOI] [PubMed] [Google Scholar]
- 17.Tsai MC, Manor O, Wan Y, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689‐693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conn VM, Hugouvieux V, Nayak A, et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R‐loop formation. Nature Plants. 2017;3:17053. [DOI] [PubMed] [Google Scholar]
- 19.Chen L, Kong R, Wu C, et al. Circ‐MALAT1 functions as both an mRNA translation brake and a microRNA sponge to promote self‐renewal of hepatocellular cancer stem cells. Adv Sci (Weinh). 2020;7(4):1900949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu Y‐J, Zheng B, Luo G‐J, et al. Circular RNAs negatively regulate cancer stem cells by physically binding FMRP against CCAR1 complex in hepatocellular carcinoma. Theranostics. 2019;9:3526‐3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wan C, Gong C, Zhang H, et al. β2‐adrenergic receptor signaling promotes pancreatic ductal adenocarcinoma (PDAC) progression through facilitating PCBP2‐dependent c‐myc expression. Cancer Lett. 2016;373:67‐76. [DOI] [PubMed] [Google Scholar]
- 22.López‐Manríquez E, Vashist S, Ureña L, et al. Norovirus genome circularization and efficient replication are facilitated by binding of PCBP2 and hnRNP A1. J Virol. 2013;87:11371‐11387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding L, Wang Z, Yan J, et al. Human four‐and‐a‐half LIM family members suppress tumor cell growth through a TGF‐beta‐like signaling pathway. J Clin Invest. 2009;119:349‐361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luan J, Jiao C, Kong W, et al. circHLA‐C plays an important role in lupus nephritis by sponging miR‐150. Mol Ther Nucleic Acids. 2018;10:245‐253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Figure S2
Figure S3
Figure S4
Figure S5
Table S1
Table S2
Table S3
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


