Summary
The short half‐life of coagulation factor IX (FIX) for haemophilia B (HB) therapy has been prolonged through fusion with human serum albumin (HSA), which drives the neonatal Fc receptor (FcRn)‐mediated recycling of the chimera. However, patients would greatly benefit from further FIX‐HSA half‐life extension. In the present study, we designed a FIX‐HSA variant through the engineering of both fusion partners. First, we developed a novel cleavable linker combining the two FIX activation sites, which resulted in improved HSA release. Second, insertion of the FIX R338L (Padua) substitution conferred hyperactive features (sevenfold higher specific activity) as for FIX Padua alone. Furthermore, we exploited an engineered HSA (QMP), which conferred enhanced human (h)FcRn binding [dissociation constant (KD) 0·5 nM] over wild‐type FIX‐HSA (KD 164·4 nM). In hFcRn transgenic mice, Padua‐QMP displayed a significantly prolonged half‐life (2·7 days, P < 0·0001) versus FIX‐HSA (1 day). Overall, we developed a novel FIX‐HSA protein with improved activity and extended half‐life. These combined properties may result in a prolonged functional profile above the therapeutic threshold, and thus in a potentially widened therapeutic window able to improve HB therapy. This rational engineering of both partners may pave the way for new fusion strategies for the design of engineered biotherapeutics.
Keywords: albumin fusion proteins, factor IX Padua, FcRn receptor, human serum albumin, protein engineering
Introduction
Haemophilia B (HB) is associated with bleeding symptoms whose severity is related to the degree of coagulation factor IX (FIX) deficiency.1 Prophylaxis, effectively reducing bleeding episodes and preventing joint damage, represents the standard‐of‐care for HB.2 However, it requires frequent administrations, which create vein access problems and interfere with patients’ life, resulting in poor adherence to prophylactic regimens, particularly in young patients.3 Different strategies have been employed to extend the short half‐life of FIX (18–22 h) and to reduce the frequency of intravenous injections. Three extended half‐life products are currently on the market,4 namely Fc‐fused Alprolix® (Bioverativ Therapeutics, Waltham, MA, USA),5, 6 pegylated Refixia® (Novo Nordisk, Bagsværd, Denmark),7, 8 albeit not widely available and not authorised for prophylaxis in children in many countries, and the albumin‐fused Idelvion® (CSL Behring, Marburg, Germany).9, 10 Idelvion is designed with a cleavable linker connecting the two fusion partners that, upon FIX activation, drives the release of albumin and activated FIX.11, 12 Hence, this fusion strategy improves FIX half‐life without compromising its functional properties.9, 10
The fact that albumin is biodegradable, non‐immunogenic and effector negative, which minimises the risk of side effects, makes it attractive in this context. Moreover, as albumin consists of a single polypeptide, it can easily be fused to a protein partner and expressed as a monovalent albumin‐fused product. Indeed, fusion to albumin is being explored to improve the pharmacokinetics of various therapeutic proteins.13, 14 The albumin and immunoglobulin G (IgG)‐Fc, fusion strategies relies on the acquired capacity of these proteins to undergo the neonatal Fc receptor (FcRn)‐mediated recycling pathway.15, 16, 17 Indeed, FcRn is responsible for the average 3‐week long albumin and IgG half‐life.15, 16, 18, 19 FcRn primarily resides in the endosomal compartment of a broad range of cells, including haematopoietic and non‐haematopoietic cells,20, 21 where it can bind albumin and IgG with high affinity. The interactions between FcRn and its ligands are pH‐dependent, and are initiated at slightly acidic (5·5–6·0) but not at neutral (7·4) pH.15, 22, 23, 24, 25 In this way, the receptor can bind fluid‐phase pinocytosed albumin and IgG in low‐pH sorting endosomes, and transport them back via recycling endosomes to the plasma membrane, where they are ultimately released due to the neutral pH.26, 27, 28, 29, 30 Thus, FcRn rescues albumin and IgG from intracellular degradation, which extends their plasma half‐life.
The dissection of the molecular bases underlying albumin‐FcRn interactions have prompted the design of novel albumin variants with improved FcRn binding and extended half‐life.13, 31, 32 Among the three homologous albumin domains,33, 34 the C‐terminal domain III (DIII) holds the principal binding site for FcRn.24, 35 We recently demonstrated that three amino acid substitutions within DIII (E505Q/T527M/K573P, QMP) improve binding of human serum albumin (HSA) to human (h)FcRn by >180‐fold.13 Notably, QMP fusion to recombinant activated coagulation factor VII (rFVIIa) led to a 3·6‐fold extended half‐life (2·9 days) compared to the wild‐type fusion (0·8 days) in hFcRn transgenic mice, without affecting the therapeutic potential of rFVIIa.13
In addition to half‐life extension, the biological properties of coagulation factors may also be improved by increasing their activity. In this scenario, the gain‐of‐function FIX Padua variant (R338L substitution), which has an approximately sevenfold higher specific activity compared to wild‐type FIX,36 may represent an attractive candidate for fusion purposes. Notably, FIX Padua has been explored in HB gene therapy, where it was shown to sustain coagulant levels and decrease the annualised bleeding rate in 10 patients with HB. Importantly, no serious adverse events or development of inhibitory antibodies were reported.37
In the present study, we combined FIX Padua with the QMP albumin variant via a novel linker design, which resulted in a functional product with hyperactive features and extended half‐life in hFcRn transgenic mice.
Materials and methods
Detailed methods are available as Supporting Information.
Plasmids design and protein expression
Chimeric constructs consisting of F9 complementary DNA (cDNA; reference sequences: NM_000133·4, NP_000124·1), including 1·4 kb of intron 1,38 designed cleavable linkers and the cDNA of mature (amino acids 25‐609) HSA (reference sequences: NM_000477·7, NP_000468·1), either wild‐type or QMP, were cloned in the pCDNA3 expression vector. The FIX Padua variant (R338L) was inserted through site‐directed mutagenesis. All constructs were validated by sequencing (primer details in Table SI).
Expression studies in HEK293 cells and stable clone selection were performed as described previously.12, 39
Protein purification and characterisation
Fusion proteins were purified with a HSA affinity matrix followed by size exclusion chromatography and concentrated by centrifugation units.13 The activation profile of FIX and FIX‐HSA proteins was evaluated by incubation with plasma‐derived activated factor XI (pdFXIa) followed by Western blotting as described previously.40
Characterisation of secreted protein levels through polyclonal enzyme‐linked immunosorbent assay (ELISA),40, 41 as well as FIX activity by chromogenic41 or activated partial thromboplastin time (aPTT)‐based42 assays, was as described previously. Specific activity was calculated as the ratio between activity and protein levels, with the value of 1 corresponding to normal specific activity of the reference molecule (FIX or FIX‐HSA).39
FcRn binding studies
The ELISA‐based assays on FIX‐HSA fusions for hFcRn binding were performed as described previously.13, 43
Binding kinetics to hFcRn were measured through surface plasmon resonance (SPR), with FIX‐HSA fusion proteins immobilised on sensor chips and of serial dilutions of soluble monomeric hFcRn‐His injected.13 Binding curves were zero adjusted and the reference flow‐cell value was subtracted. Binding kinetics were determined using the 1:1 Langmuir binding model provided by the Biacore T200 Evaluation Software (version 3·0).
Mouse studies
Studies in mice (male, aged 6–8 weeks, weighing 20–30 g, three to five mice/group) were performed at The Jackson Laboratory, in accordance with guidelines and regulations approved by the local Animal Care and Use Committee. Hemizygous hFcRn transgenic Tg32, homozygous hFcRn transgenic Tg32 albumin knock‐out (KO) and FcRn KO mice received 2 mg/kg fusion proteins, and blood was collected from retro‐orbital sinus at the scheduled time points. The concentration of fusion proteins in plasma was quantified by ELISA.13 The β‐phase half‐life was calculated using the formula: t1/2 = log 0·5/(log Ae/A0) × t, where t1/2 = the half‐life of the variant evaluated, Ae = the concentration remaining, A0 = concentration on day 1 and t = the elapsed time.
Statistical analysis
Statistical differences were analysed by unpaired t‐test, with P < 0·05 considered as statistically significant.
Results
Construction of a FIX‐HSA fusion protein with optimised linker
The efficient release of albumin from the FIX‐HSA chimera is crucial to ensure proper coagulant activity.9, 43 First, FIX was recombinantly fused to the N‐terminal end of mature HSA via a cleavable amino acid linker resembling one of the FIX activation sites, R145‐A146 (linker a; FIX‐a‐HSA), which is used in the clinically approved Idelvion.9 To investigate whether proper activation of the fusion could be further enhanced, two alternative linkers, which included the second FIX activation site, R180‐V181 (linker b; FIX‐b‐HSA), or a head‐to‐tail combination of the two activation sequences (linker ab; FIX‐ab‐HSA), were designed (Fig 1A). Upon transient expression in HEK293 cells, albeit showing protein levels significantly lower (P < 0·0001) than those of wild‐type FIX, the three fusion proteins were efficiently secreted (Fig S1A, left panel), with the FIX‐b‐HSA and FIX‐ab‐HSA being produced at approximately half the levels of FIX‐a‐HSA (P < 0·0001). Importantly, incubation with FXIa, a physiological FIX activator, generated protein fragments of the expected molecular weights (Fig S1B). To assess the functional integrity of fusion proteins, their ability to restore coagulation in FIX‐deficient plasma was evaluated by aPTT‐based assays. The results supported that the three variants were functional, as demonstrated by the shortening of coagulation times upon addition of medium containing any of the three fusion proteins (Fig S1A, right panel). In order to compare the three fusions, specific activity was calculated as the ratio between activity and protein concentration (Fig 1B). The FIX‐ab‐HSA fusion, containing the combination of the two FIX activation sites (linker ab), showed the highest specific activity [mean (SD) 1·20 (0·13)]. This was a significant improvement compared to the Idelvion‐like FIX‐a‐HSA [mean (SD) 1·00 (0·1), P = 0·0046] and the FIX‐b‐HSA [mean (SD) 0·86 (0·12), P = 0·0001] fusions.
Fig 1.
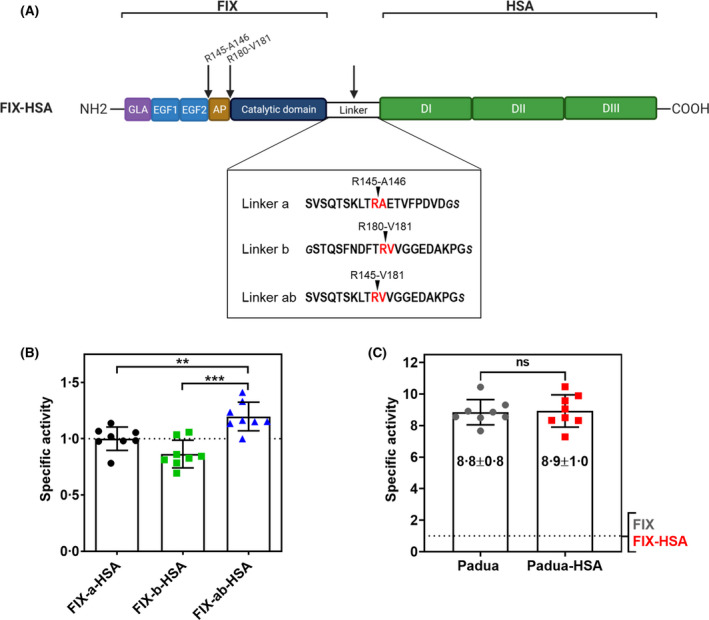
Fusion of FIX to HSA with three different linker sequences. (A) Schematic representation of the FIX‐HSA fusion protein (upper panel) with the amino acid sequences of the three linkers tested (lower panel). The cleavage sites in FIX and the linker sequences are indicated by arrows (upper panel) and highlighted in red (lower panel). According to the Human Gene Variation Society (HGVS) nomenclature,60 whose numbering begins from the first translation initiation methionine‐coding AUG, the corresponding positions for FIX activation sites are R191‐A192 and R226‐R227. Created with Biorender.com. (B) Specific activity of transiently expressed fusion proteins with different cleavable linkers. The dotted line indicates the specific activity (=1) of the reference construct (FIX‐a‐HSA). (C) Specific activity of transiently expressed Padua variants. The dotted line represents the specific activity (=1) of the wild‐type constructs. All results are reported as mean ± standard deviation. Specific activity was calculated as the activity/antigen ratio. The corresponding P values are indicated as not significant (ns, P > 0·05), ** (P ≤ 0·01), *** (P ≤ 0·001), by unpaired t‐test. AP, activation peptide; DI, domain I; DII, domain II; DIII, domain III; EGF1 epidermal growth factor‐like domain 1, EGF2, epidermal growth factor‐like domain 2; FIX, factor IX; GLA, gamma‐carboxyglutamic acid domain; HSA, human serum albumin.
These results demonstrated the favourable features of the designed linker ab, and prompted the selection of the FIX‐ab‐HSA chimera (henceforth referred to as FIX‐HSA) as the scaffold for subsequent engineering steps.
Design of a double‐engineered FIX‐HSA fusion protein
To enhance the therapeutic potential of the FIX‐HSA chimera, we engineered both fusion partners.
First, to improve the procoagulant features, we exploited the gain‐of‐function FIX Padua variant by introducing the R338L substitution in FIX alone (Padua) and in the FIX‐HSA fusion (Padua‐HSA). Upon transient expression, their activity profiles (Fig S1C) demonstrated an approximate eightfold enhanced procoagulant activity of both Padua variants as compared to the corresponding wild‐type versions (Fig 1C). Importantly, no significant difference in activity of the Padua variant was measured as a result of fusion to albumin.
Second, to enhance the pharmacokinetic properties, we fused the engineered QMP albumin13 to the FIX Padua variant (Padua‐QMP) through the optimised linker ab (Fig 2A). The resulting chimera, as well as the wild‐type counterpart, were stably expressed in HEK293 cells and purified. A sodium dodecyl sulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE) analysis revealed pure fractions of FIX‐HSA and of the double engineered fusion, which migrated with the expected molecular weights (~115 kDa) (Fig S2).
Fig 2.
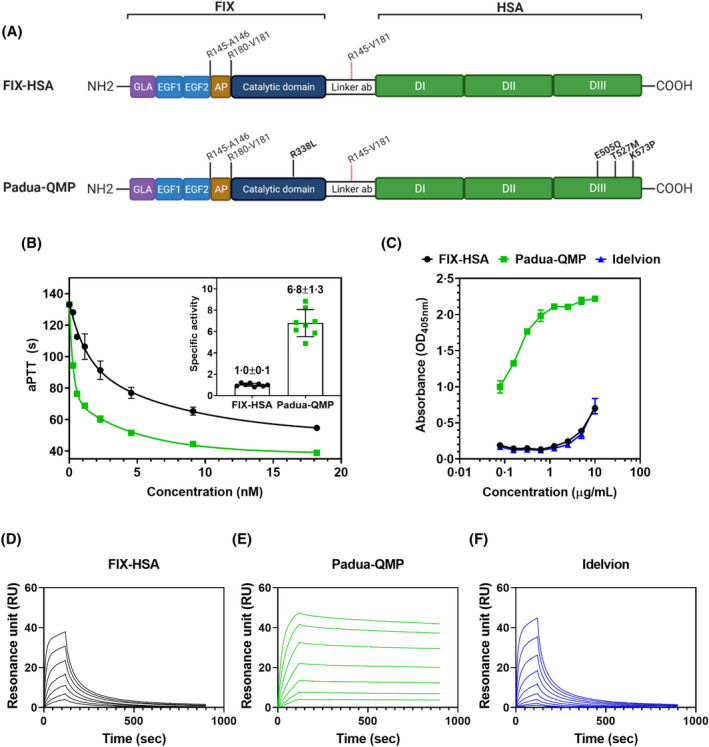
Engineering of the FIX‐HSA fusion improves procoagulant activity and hFcRn binding. (A) Schematic representation of the designed fusion proteins of FIX and HSA connected via the cleavable linker ab. FIX is activated by proteolytic cleavage at the two cleavage sites indicated (R145‐A146 and R180‐V181). Simultaneous detachment of HSA is accomplished as the linker sequence (linker ab) contains an upstream–downstream combination of the two cleavage sites in FIX (R145‐V181). FIX‐HSA consists of wild‐type FIX and wild‐type HSA (upper panel). Padua‐QMP contains point mutations in FIX (R338L, Padua) and HSA (E505Q/T527M/K573P) as indicated in bold. According to Human Gene Variation Society (HGVS) nomenclature,60 the corresponding positions/substitutions are R191‐A192/R226‐R227 (activation sites) and R384L (Padua variant) for FIX, and E529Q/T551M/K597P for the QMP variant. Created with Biorender.com. (B) Representative procoagulant activity of serial dilutions of purified fusion proteins, evaluated by aPTT‐based assays. The resulting specific activity is reported (inset). (C) Binding of titrated amounts of purified fusion proteins to immobilised hFcRn at pH 5·5. Results are reported as mean ± standard deviation of duplicates from one representative experiment. (D–F) Representative sensorgrams showing binding of serial dilutions (0–4 µM) of monomeric hFcRn injected over immobilised (~200 RU) fusion proteins at pH 5·5. AP, activation peptide; aPTT, activated partial thromboplastin time; DI, domain I; DII, domain II; DIII, domain III; EGF1 epidermal growth factor‐like domain 1, EGF2, epidermal growth factor‐like domain 2; FIX, factor IX; GLA, gamma‐carboxyglutamic acid domain; HSA, human serum albumin; hFcRn, human neonatal Fc receptor.
Padua‐QMP shows enhanced procoagulant and hFcRn binding properties
The functional features of the purified FIX‐HSA and Padua‐QMP fusions (Fig 2A) were evaluated in FIX‐deficient human plasma through aPTT‐based assays. Both variants restored coagulation times in a concentration‐dependent manner (Fig 2B), with the specific activity of the Padua‐QMP being sevenfold higher (P < 0·0001) than that of the wild‐type fusion (Fig 2B, inset).
Binding of fusion proteins to hFcRn was evaluated using ELISA, which showed that Padua‐QMP bound more strongly to the receptor than the wild‐type fusion at acidic pH (Fig 2C). As a control, we included the commercial fusion protein Idelvion, which bound hFcRn similarly to the designed wild‐type FIX‐HSA fusion.
Furthermore, to determine the binding kinetics, we performed SPR analysis with the fusion variants immobilised on the sensor chip and monomeric fractions of the receptor injected at pH 5·5. Whereas FIX‐HSA showed a dissociation constant (KD 164·4 nM) comparable to that of Idelvion (KD 150·0 nM), the QMP amino acid substitutions greatly improved the binding affinity, resulting in a KD of 0·5 nM (TableI). Importantly, Padua‐QMP was shown to bind pH dependently, as bound hFcRn was released from the immobilised fusion protein upon injection of a pH 7·4 buffer (Fig 2D–F; Fig S3).
Table I.
Binding kinetics of fusion protein variants to hFcRn at pH 5·5.
| Mean (SD): | |||
|---|---|---|---|
| Protein variant | Ka, × 104 M−1 s−1 | Kd, ×10−3 s−1 | KD, nM |
| FIX‐HSA | 4·5 (0·4) | 7·4 (0·2) | 164·4 |
| Padua‐QMP | 18·2 (0·4) | 0·1 (0·0) | 0·5 |
| Idelvion | 4·2 (0·1) | 6·3 (0·4) | 150·0 |
FIX, factor IX; HSA, human serum albumin; Ka, association constant; Kd, dissociation constant.
The kinetic rate constants were determined by the 1:1 Langmuir bimolecular interaction model. The kinetic values reflect the mean (SD) of three runs for each fusion protein.
Thus, the designed Padua‐QMP displays improved procoagulant activity and favourable pH‐dependent hFcRn binding properties.
Padua‐QMP shows extended half‐life in hFcRn transgenic mice
We have previously demonstrated that HSA, as well as fusions built on HSA, poorly bind to mouse FcRn, which hampers evaluation of such molecules in conventional mouse models.13, 24, 45, 46, 47 Therefore, to study the effect of FIX‐HSA engineering on plasma half‐life we took advantage of transgenic mice genetically modified to express the hFcRn heavy chain and not the mouse counterpart.16, 48, 49 Importantly, these transgenic mice express a chimeric version of the receptor, as the hFcRn heavy chain pairs with the mouse β2 microglobulin, which has been shown to bind mouse and human albumin with comparable affinity.50
First, we determined the half‐life of Idelvion and of our designed fusion proteins in Tg32 hFcRn transgenic mice that express mouse albumin (29 mg/ml).16, 51 Blood samples were collected up to 10 days after injection of fusion proteins. ELISA was performed to quantify plasma levels, which revealed that FIX‐HSA and Idelvion showed a similar half‐life of ~1 day, whereas that of Padua‐QMP was 2·7 days (vs. FIX‐HSA, P = 0·0006; vs. Idelvion, P < 0·0001) (Fig 3A). Thus, the QMP albumin variant extended the half‐life by >2·5‐fold even in the presence of competing endogenous mouse albumin.
Fig 3.
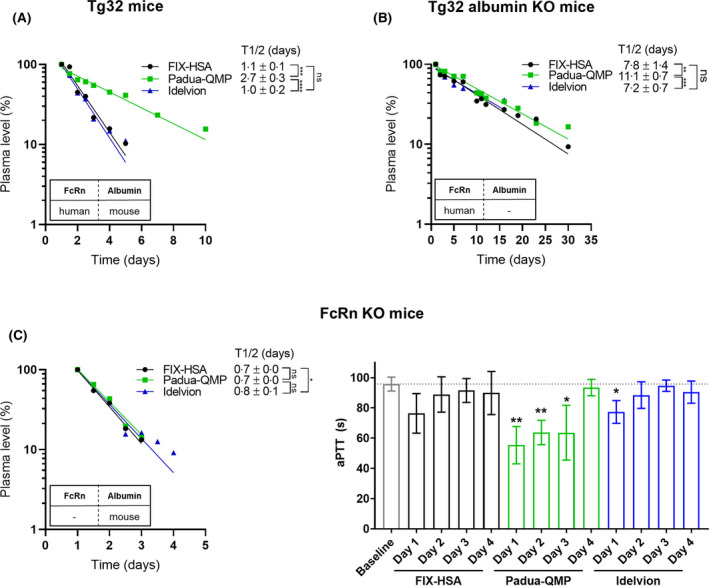
FcRn extends the plasma half‐life of the engineered Padua‐QMP fusion. Clearance curves of FIX‐HSA (black circles), Padua‐QMP (green squares) and Idelvion (blue triangles) in hFcRn transgenic mice (Tg32) (A), hFcRn transgenic mice deficient for mouse albumin (Tg32 albumin KO) (B), and FcRn deficient mice (FcRn KO) (C, left panel). Plasma samples from FcRn KO mice were also evaluated for FIX‐dependent shortening of coagulation times (C, right panel). The mice received 2 mg/kg fusion protein by intravenous injection. Plasma levels are reported as the percentage remaining in the circulation compared to the level measured 24 h after injection (day 1, 100%). The values represent the mean ± standard deviation (SD) of 3–5 mice. Mean β‐phase half‐life (t1/2) ± SD is reported in days and shown in the upper right corner of each figure. The corresponding P values are indicated as not significant (ns, P > 0·05), *(P ≤ 0·05), **(P ≤ 0·01), ***(P ≤ 0·001), ****(P < 0·0001), by unpaired t‐test. P values from unpaired t‐test on in vivo half‐life data are reported in Table SII. FIX, factor IX; HSA, human serum albumin; hFcRn, human neonatal Fc receptor; KO, knock‐out.
The experiment was then performed in hFcRn transgenic mice lacking mouse albumin (Tg32 albumin KO mice).48 From these mice, blood was collected for up to 30 days after injection, as longer half‐lives were expected in the absence of mouse albumin competing for hFcRn binding. Indeed, fusion proteins were detectable in plasma 30 days after administration, with half‐lives of 7·8 days and 11·1 days for FIX‐HSA and Padua‐QMP, respectively (Fig 3B). Similar to the designed wild‐type fusion, a half‐life of 7·2 days was determined for Idelvion. Thus, in the absence of mouse albumin as competitor, a 1·4‐fold longer half‐life was measured for Padua‐QMP in comparison with FIX‐HSA (P < 0·0091) and Idelvion (P < 0·0005) as a result of albumin engineering.
To confirm that improved hFcRn binding was responsible for the extended half‐life, we also injected the fusions in mice lacking expression of FcRn (FcRn KO mice).16 As expected, all three fusion proteins showed equally rapid clearance from the circulation, as their half‐lives dropped to only 17–19 h (Fig 3C, left panel). Prompted by the overlapping clearance curves, we also assessed FIX‐dependent coagulant activity in plasma from FcRn KO mice (Fig 3C, right panel). A slightly significant shortening of coagulation times for reference fusions over the baseline was detectable only for Idelvion until day 1. Noticeably, Padua‐QMP continued to confer a significant increase in FIX activity until day 3 (Fig 3C, right panel).
Altogether, these data demonstrate that the engineered albumin variant extends the half‐life of the Padua‐QMP chimera in an FcRn‐dependent manner and provide in vivo evidence for the hyperactive features of the Padua‐QMP molecule.
Discussion
Fusion to HSA represents an attractive strategy to improve plasma half‐life of therapeutic proteins, and has been applied to increase FIX half‐life (Idelvion) from 22 to 102 h in humans.9, 10, 52 However, the half‐life extension achieved by fusion to wild‐type albumin is limited, and far from that of endogenous albumin, whose high concentration exerts a considerable competitive pressure for hFcRn binding.13 In this view, even a minor increase in receptor binding may favour and improve the ability of hFcRn to rescue a fusion product from intracellular degradation. As such, to provide a more effective product for HB replacement therapy, we designed an engineered FIX‐HSA fusion protein with further extended half‐life and improved functional features.
This was first achieved by optimising the linker design, which represents a pivotal element to ensure proper biological properties.44 To this purpose, two novel cleavable linker sequences were compared with that present in Idelvion.9, 53 We found that a combination of the two FIX activation sequences conferred higher specific activity to the resulting FIX‐HSA chimera, probably due to more efficient release of albumin.
Once the best‐performing fusion strategy was identified, we combined this design with the hyperactive FIX Padua variant. While FIX Padua has been safely explored for gene therapy purposes,37 it has not yet been explored in the context of enhanced half‐life proteins for replacement therapy purposes. When combined with albumin, FIX Padua conferred the expected sevenfold higher specific activity in clotting assays, thus supporting its suitability as a fusion partner to generate a more active product. Moreover, given the high specific activity of FIX Padua, its potential for thrombogenicity has to be carefully addressed. It is worth noting that this variant has been found to be associated with thrombosis in a patient with FIX activity levels of ~770%, but neither in his brother nor his mother displaying FIX levels of ~550% and ~330%, respectively.36 Consistently, in non‐human primates treated with adeno‐associated viral vector serotype 5 (AAV5)‐delivered FIX Padua, increased FIX activity up to 500% was not associated with prothrombotic states, which, together with previous studies, support the FIX Padua safety profile.54, 55, 56 The potential risk for immunogenicity could also be an issue. It is worth noting that, in the above reported studies in mouse and canine HB models, as well as in patients, the viral‐mediated delivery of FIX Padua was not associated with inhibitor development.37, 54, 55 In our present study, no thrombotic events occurred after injecting Padua‐QMP into mice expressing FIX, and the monophasic log‐linear decay up to 30 days of FIX‐HSA plasma levels did not point toward immunogenicity.
Noticeably, the extended plasma half‐life of FIX Padua was achieved by combining it with the recently developed QMP albumin variant,13 which should increase its chance of outcompeting endogenous albumin and thus favour its rescue from intracellular degradation. Due to large cross‐species differences in FcRn binding, transgenic mice expressing the hFcRn heavy chain, instead of the mouse receptor, were used to evaluate FIX‐HSA fusions. Importantly, while mouse albumin binds with higher affinity than HSA to the fully human receptor,32, 45, 46, 47 it binds the chimeric FcRn version expressed by the transgenic mice with an affinity similar to that of HSA.50 This supports that the circulating endogenous mouse albumin represents a suitable competitor for HSA‐based molecules assessed in the Tg32 hFcRn transgenic mice, where the Padua‐QMP gained a >2·5‐fold longer half‐life compared with FIX‐HSA and Idelvion.
It is worth noting that our data on protein levels in Tg32 mice do not take into account the functional contribution of the hyperactive FIX Padua, as these mice express normal FIX levels, not allowing the comparison of FIX‐HSA in vivo efficacy. However, the available HB mice, expressing mouse FcRn, do not either represent a good model to assess the contribution of HSA engineering to the improved HB phenotype correction due to the cross‐species differences. Indeed, HSA binds poorly to mouse FcRn, and although QMP improves binding to the mouse receptor, the kinetic values are far from that measured towards hFcRn.13 Notwithstanding, in FcRn KO mouse plasma, although levels of the three fusion proteins were comparable, only Padua‐QMP demonstrated a significant shortening of FIX‐dependent coagulation times over the baseline until day 3, which highlights its improved functional properties.
In summary, we produced a novel FIX‐HSA fusion protein with enhanced coagulant activity and hFcRn binding properties translating into significantly extended plasma half‐life in hFcRn transgenic mice. These features may allow for a prolonged functional profile above the therapeutic threshold and, consequently, reduce the frequency of injections required in a prophylactic treatment regimen for patients with HB. This, in turn, could allow the widening of the therapeutic window with a consequent lowering of treatment burden.58 As treatment is typically provided in amounts that are just sufficient to limit joint bleeds, a hyper‐functional product, requiring lower doses to achieve the same or even superior functional levels, may favour achieving an optimal functional trough level.57, 58, 59
Overall, our present study demonstrates that combining a natural gain‐of‐function FIX variant with a rationally engineered HSA may result in novel fusion proteins with improved biological properties and bioavailability, which in turn may pave the way for new strategies for the design of engineered biotherapeutics.
Author contributions
Silvia Lombardi designed fusion proteins, performed expression as well as functional studies, analyzed data and wrote the manuscript; Kristin H. Aaen and Jeannette Nilsen purified fusion proteins, performed hFcRn binding studies and ELISA assays on mouse samples, analysed data and wrote the manuscript; Mattia Ferrarese established and validated stable clones and analysed data; Torleif T. Gjølberg performed ELISA on Tg32 albumin KO mouse samples and analysed data; Francesco Bernardi and Mirko Pinotti analysed data and revised the manuscript; Jan T. Andersen and Alessio Branchini conceived the study, designed and performed research, analysed data and wrote the manuscript. All authors approved the final version of the manuscript.
Conflicts of interest
Jan T. Andersen is a co‐inventor of patents, which are entitled ‘Albumin Variants and uses thereof’ and relate to the data described in this paper, for example, EP3063171B1, US10208102 and US10781245. Alessio Branchini received grants and non‐financial support from Grifols. Mirko Pinotti received grants from Novo Nordisk. Alessio Branchini and Mirko Pinotti received grants and personal fees from Pfizer. Francesco Bernardi, received a research grant from Pfizer. The remaining authors have no competing financial interests to declare.
Supporting information
Fig S1. Transient expression of the FIX‐HSA fusion proteins in HEK293 cells. (A) Secreted levels of the fusion proteins and FIX measured by polyclonal anti‐FIX ELISA (left panel), and representative activity profiles of serial dilutions of media in FIX‐deficient plasma evaluated by aPTT‐based assay (right panel). (B) Western blotting analysis of the activation profile of FIX and FIX‐HSA fusions (left panel), in the absence (zym) or in the presence (act) of the physiological activator FXIa. A schematic representation of the protein fragments deriving from FXIa‐mediated cleavage is provided (right panel). (C) Representative activity profile of serial dilutions of transiently expressed fusion proteins, either wild‐type or containing the R384L (Padua) substitution, evaluated by chromogenic assays. The corresponding P values are indicated as not significant (ns, P > 0·05), ****(P < 0·0001), by unpaired t‐test. aPTT, activated partial thromboplastin time; FIX, factor IX; HSA, human serum albumin.
Fig S2. Sodium dodecyl sulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE) analysis of the FIX‐HSA fusion proteins. Purified fractions of Idelvion (lane 2), FIX‐HSA (lane 3) and Padua‐QMP (well 4) were analysed by SDS‐PAGE and Coomassie staining. FIX, factor IX; HSA, human serum albumin.
Fig S3. Complete SPR sensorgrams showing binding of FIX‐HSA fusion proteins to hFcRn. Serial dilutions (0–4 µM) of monomeric hFcRn were injected over immobilised (~200 RU) fusion proteins at pH 5·5. Regeneration was performed by injecting a pH 7·4 buffer at the time point indicated by the arrow. hFcRn, human neonatal Fc receptor; SPR, surface plasmon resonance.
Table SI. Oligonucleotides used to create expression vectors for designed fusion proteins.
Table SII. P values from unpaired t‐test on in vivo half‐life data.
Acknowledgements
This study was financially supported by the Bayer Early Career Investigator Award (Bayer Haemophilia Award Programme) 2018 (Alessio Branchini and Mirko Pinotti). Jeannette Nilsen, Kristin H. Aaen and Jan T. Andersen were supported by the Research Council of Norway (Grant no. 274993; 287927), and Jan T. Andersen by the South‐Eastern Norway Regional Health Authority (Grant no. 2018052; 2019084). T.T.G. was supported by internal funding from Division of Head, Neck and Reconstructive Surgery, Oslo University Hospital. The authors would like to thank Simone Mester for contributing with protein for an in vivo experiment.
Contributor Information
Jan T. Andersen, Email: j.t.andersen@medisin.uio.no.
Alessio Branchini, Email: brnlss@unife.it.
References
- 1.Bolton‐Maggs PH, Pasi KJ. Haemophilias A and B. Lancet. 2003;361:1801–9. [DOI] [PubMed] [Google Scholar]
- 2.Manco‐Johnson MJ, Abshire TC, Shapiro AD, Riske B, Hacker MR, Kilcoyne R, et al. Prophylaxis versus episodic treatment to prevent joint disease in boys with severe hemophilia. N Engl J Med. 2007;357:535–44. [DOI] [PubMed] [Google Scholar]
- 3.Bolton‐Maggs PH. Optimal haemophilia care versus the reality. Br J Haematol. 2006;132:671–82. [DOI] [PubMed] [Google Scholar]
- 4.Ling G, Nathwani AC, Tuddenham EG. Recent advances in developing specific therapies for haemophilia. Br J Haematol. 2018;181:161–72. [DOI] [PubMed] [Google Scholar]
- 5.Peters RT, Low SC, Kamphaus GD, Dumont JA, Amari JV, Lu QI, et al. Prolonged activity of factor IX as a monomeric Fc fusion protein. Blood. 2010;115:2057–64. [DOI] [PubMed] [Google Scholar]
- 6.Shapiro AD, Ragni MV, Valentino LA, Key NS, Josephson NC, Powell JS, et al. Recombinant factor IX‐Fc fusion protein (rFIXFc) demonstrates safety and prolonged activity in a phase 1/2a study in hemophilia B patients. Blood. 2012;119:666–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Østergaard H, Bjelke JR, Hansen L, Petersen LC, Pedersen AA, Elm T, et al. Prolonged half‐life and preserved enzymatic properties of factor IX selectively PEGylated on native N‐glycans in the activation peptide. Blood. 2011;118:2333–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins PW, Young G, Knobe K, Karim FA, Angchaisuksiri P, Banner C, et al. Recombinant long‐acting glycoPEGylated factor IX in hemophilia B: a multinational randomized phase 3 trial. Blood. 2014;124:3880–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Metzner HJ, Weimer T, Kronthaler U, Lang W, Schulte S. Genetic fusion to albumin improves the pharmacokinetic properties of factor IX. Thromb Haemost. 2009;102:634–44. [DOI] [PubMed] [Google Scholar]
- 10.Santagostino E, Negrier C, Klamroth R, Tiede A, Pabinger‐Fasching I, Voigt C, et al. Safety and pharmacokinetics of a novel recombinant fusion protein linking coagulation factor IX with albumin (rIX‐FP) in hemophilia B patients. Blood. 2012;120:2405–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen X, Zaro JL, Shen W‐C. Fusion protein linkers: property, design and functionality. Adv Drug Deliv Rev. 2013;65:1357–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrarese M, Pignani S, Lombardi S, Balestra D, Bernardi F, Pinotti M, et al. The carboxyl‐terminal region of human coagulation factor X as a natural linker for fusion strategies. Thromb Res. 2019;173:4–11. [DOI] [PubMed] [Google Scholar]
- 13.Bern M, Nilsen J, Ferrarese M, Sand KM, Gjølberg TT, Lode HE, et al. An engineered human albumin enhances half‐life and transmucosal delivery when fused to protein‐based biologics. Sci Transl Med. 2020;12:eabb0580. [DOI] [PubMed] [Google Scholar]
- 14.Zaman R, Islam RA, Ibnat N, Othman I, Zaini A, Lee CY, et al. Current strategies in extending half‐lives of therapeutic proteins. J Control Release. 2019;301:176–89. [DOI] [PubMed] [Google Scholar]
- 15.Chaudhury C, Mehnaz S, Robinson JM, Hayton WL, Pearl DK, Roopenian DC, et al. The major histocompatibility complex‐related Fc receptor for IgG (FcRn) binds albumin and prolongs its lifespan. J Exp Med. 2003;197:315–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roopenian DC, Christianson GJ, Sproule TJ, Brown AC, Akilesh S, Jung N, et al. The MHC class I‐like IgG receptor controls perinatal IgG transport, IgG homeostasis, and fate of IgG‐Fc‐coupled drugs. J Immunol. 2003;170:3528–33. [DOI] [PubMed] [Google Scholar]
- 17.Chia J, Louber J, Glauser I, Taylor S, Bass GT, Dower SK, et al. Half‐life‐extended recombinant coagulation factor IX‐albumin fusion protein is recycled via the FcRn‐mediated pathway. J Biol Chem. 2018;293:6363–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peters T. Serum albumin. Adv Protein Chem. 1985;37:161–245. [DOI] [PubMed] [Google Scholar]
- 19.Spiegelberg HL, Fishkin BG. The catabolism of human G immunoglobulins of different heavy chain subclasses. 3. The catabolism of heavy chain disease proteins and of Fc fragments of myeloma proteins. Clin Exp Immunol. 1972;10:599–607. [PMC free article] [PubMed] [Google Scholar]
- 20.Challa DK, Wang X, Montoyo HP, Velmurugan R, Ober RJ, Ward ES. Neonatal Fc receptor expression in macrophages is indispensable for IgG homeostasis. MAbs. 2019;11:848–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montoyo HP, Vaccaro C, Hafner M, Ober RJ, Mueller W, Ward ES. Conditional deletion of the MHC class I‐related receptor FcRn reveals the sites of IgG homeostasis in mice. Proc Natl Acad Sci USA. 2009;106:2788–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raghavan M, Bonagura VR, Morrison SL, Bjorkman PJ. Analysis of the pH dependence of the neonatal Fc receptor/immunoglobulin G interaction using antibody and receptor variants. Biochemistry. 1995;34:14649–57. [DOI] [PubMed] [Google Scholar]
- 23.Andersen JT, Dee Qian J, Sandlie I. The conserved histidine 166 residue of the human neonatal Fc receptor heavy chain is critical for the pH‐dependent binding to albumin. Eur J Immunol. 2006;36:3044–51. [DOI] [PubMed] [Google Scholar]
- 24.Andersen JT, Dalhus B, Cameron J, Daba MB, Plumridge A, Evans L, et al. Structure‐based mutagenesis reveals the albumin‐binding site of the neonatal Fc receptor. Nat Commun. 2012;3:610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oganesyan V, Damschroder MM, Cook KE, Li Q, Gao C, Wu H, et al. Structural insights into neonatal Fc receptor‐based recycling mechanisms. J Biol Chem. 2014;289:7812–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ober RJ, Martinez C, Vaccaro C, Zhou J, Ward ES. Visualizing the site and dynamics of IgG salvage by the MHC class I‐related receptor FcRn . J Immunol. 2004;172:2021–9. [DOI] [PubMed] [Google Scholar]
- 27.Ober RJ, Martinez C, Lai X, Zhou J, Ward ES. Exocytosis of IgG as mediated by the receptor, FcRn: an analysis at the single‐molecule level. Proc Natl Acad Sci USA. 2004;101:11076–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prabhat P, Gan Z, Chao J, Ram S, Vaccaro C, Gibbons S, et al. Elucidation of intracellular recycling pathways leading to exocytosis of the Fc receptor, FcRn, by using multifocal plane microscopy. Proc Natl Acad Sci USA. 2007;104:5889–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidt EG, Hvam ML, Antunes F, Cameron J, Viuff D, Andersen B, et al. Direct demonstration of a neonatal Fc receptor (FcRn)‐driven endosomal sorting pathway for cellular recycling of albumin. J Biol Chem. 2017;292:13312–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grevys A, Nilsen J, Sand KM, Daba MB, Øynebråten I, Bern M, et al. A human endothelial cell‐based recycling assay for screening of FcRn targeted molecules. Nat Commun. 2018;9:621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt M, Townson S, Andreucci A, King B, Schirmer E, Murillo A, et al. Crystal structure of an HSA/FcRn complex reveals recycling by competitive mimicry of HSA ligands at a pH‐dependent hydrophobic interface. Structure. 2013;21:1966–78. [DOI] [PubMed] [Google Scholar]
- 32.Andersen JT, Dalhus B, Viuff D, Ravn BT, Gunnarsen KS, Plumridge A, et al. Extending serum half‐life of albumin by engineering neonatal Fc receptor (FcRn) binding. J Biol Chem. 2014;289:13492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dockal M, Carter DC, Rüker F. The three recombinant domains of human serum albumin. Structural characterization and ligand binding properties. J Biol Chem. 1999;274:29303–10. [DOI] [PubMed] [Google Scholar]
- 34.Guizado TR. Analysis of the structure and dynamics of human serum albumin. J Mol Model. 2014;20:2450. [DOI] [PubMed] [Google Scholar]
- 35.Chaudhury C, Brooks CL, Carter DC, Robinson JM, Anderson CL. Albumin binding to FcRn: distinct from the FcRn−IgG interaction. Biochemistry. 2006;45:4983–90. 10.1021/bi052628y [DOI] [PubMed] [Google Scholar]
- 36.Simioni P, Tormene D, Tognin G, Gavasso S, Bulato C, Iacobelli NP, et al. X‐linked thrombophilia with a mutant factor IX (factor IX Padua). N Engl J Med. 2009;361:1671–5. [DOI] [PubMed] [Google Scholar]
- 37.George LA, Sullivan SK, Giermasz A, Rasko JE, Samelson‐Jones BJ, Ducore J, et al. Hemophilia B Gene Therapy with a High‐Specific‐Activity Factor IX Variant. N Engl J Med. 2017;377:2215–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kurachi S, Hitomi Y, Furukawa M, Kurachi K. Role of intron I in expression of the human factor IX gene. J Biol Chem. 1995;270:5276–81. [DOI] [PubMed] [Google Scholar]
- 39.Pignani S, Todaro A, Ferrarese M, Marchi S, Lombardi S, Balestra D, et al. The chaperone‐like sodium phenylbutyrate improves factor IX intracellular trafficking and activity impaired by the frequent p. R294Q mutation. J Thromb Haemost. 2018;16:2035–43. [DOI] [PubMed] [Google Scholar]
- 40.Ferrarese M, Testa MF, Balestra D, Bernardi F, Pinotti M, Branchini A. Secretion of wild‐type factor IX upon readthrough over F9 pre‐peptide nonsense mutations causing hemophilia B. Hum Mutat. 2018;39:702–8. [DOI] [PubMed] [Google Scholar]
- 41.Branchini A, Ferrarese M, Campioni M, Castaman G, Mari R, Bernardi F, et al. Specific factor IX mRNA and protein features favor drug‐induced readthrough over recurrent nonsense mutations. Blood. 2017;129:2303–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Branchini A, Campioni M, Mazzucconi MG, Biondo F, Mari R, Bicocchi MP, et al. Replacement of the Y450 (c234) phenyl ring in the carboxyl‐terminal region of coagulation factor IX causes pleiotropic effects on secretion and enzyme activity. FEBS Lett. 2013;587:3249–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sand KMK, Bern M, Nilsen J, Dalhus B, Gunnarsen KS, Cameron J, et al. Interaction with both domain I and III of albumin is required for optimal pH‐dependent binding to the neonatal Fc receptor (FcRn). J Biol Chem. 2014;289:34583–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sheffield WP, Mamdani A, Hortelano G, Gataiance S, Eltringham‐Smith L, Begbie ME, et al. Effects of genetic fusion of factor IX to albumin on in vivo clearance in mice and rabbits. Br J Haematol. 2004;126:565–73. [DOI] [PubMed] [Google Scholar]
- 45.Andersen JT, Daba MB, Berntzen G, Michaelsen TE, Sandlie I. Cross‐species binding analyses of mouse and human neonatal Fc receptor show dramatic differences in immunoglobulin G and albumin binding. J Biol Chem. 2010;285:4826–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Andersen JT, Cameron J, Plumridge A, Evans L, Sleep D, Sandlie I. Single‐chain variable fragment albumin fusions bind the neonatal Fc receptor (FcRn) in a species‐dependent manner: implications for in vivo half‐life evaluation of albumin fusion therapeutics. J Biol Chem. 2013;288:24277–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nilsen J, Bern M, Sand KM, Grevys A, Dalhus B, Sandlie I, et al. Human and mouse albumin bind their respective neonatal Fc receptors differently. Sci Rep. 2018;8:14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roopenian DC, Low BE, Christianson GJ, Proetzel G, Sproule TJ, Wiles MV. Albumin‐deficient mouse models for studying metabolism of human albumin and pharmacokinetics of albumin‐based drugs. MAbs. 2015;7:344–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nilsen J, Sandlie I, Roopenian DC, Andersen JT. Animal models for evaluation of albumin‐based therapeutics. Curr Opin Chem Eng. 2017;19:65–76. [Google Scholar]
- 50.Viuff D, Antunes F, Evans L, Cameron J, Dyrnesli H, Thue Ravn B, et al. Generation of a double transgenic humanized neonatal Fc receptor (FcRn)/albumin mouse to study the pharmacokinetics of albumin‐linked drugs. J Control Release. 2016;223:22–30. [DOI] [PubMed] [Google Scholar]
- 51.Stein C, Kling L, Proetzel G, Roopenian DC, de Angelis MH, Wolf E, et al. Clinical chemistry of human FcRn transgenic mice. Mamm Genome. 2012;23:259–69. [DOI] [PubMed] [Google Scholar]
- 52.Santagostino E. Transforming the treatment for hemophilia B patients: update on the clinical development of recombinant fusion protein linking recombinant coagulation factor IX with recombinant albumin (rIX‐FP). Thromb Res. 2016;141(Suppl 3):S5–8. [DOI] [PubMed] [Google Scholar]
- 53.Schulte S. Half‐life extension through albumin fusion technologies. Thromb Res. 2009;124(Suppl 2):S6–8. [DOI] [PubMed] [Google Scholar]
- 54.Finn JD, Nichols TC, Svoronos N, Merricks EP, Bellenger DA, Zhou S, et al. The efficacy and the risk of immunogenicity of FIX Padua (R338L) in hemophilia B dogs treated by AAV muscle gene therapy. Blood. 2012;120:4521–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Crudele JM, Finn JD, Siner JI, Martin NB, Niemeyer GP, Zhou S, et al. AAV liver expression of FIX‐Padua prevents and eradicates FIX inhibitor without increasing thrombogenicity in hemophilia B dogs and mice. Blood. 2015;125:1553–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spronck EA, Liu YP, Lubelski J, Ehlert E, Gielen S, Montenegro‐Miranda P, et al. Enhanced factor IX activity following administration of AAV5‐R338L “Padua” Factor IX versus AAV5 WT Human Factor IX in NHPs. Mol Ther Methods Clin Dev. 2019;15:221–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jiménez‐Yuste V, Auerswald G, Benson G, Lambert T, Morfini M, Remor E, et al. Achieving and maintaining an optimal trough level for prophylaxis in haemophilia: the past, the present and the future. Blood Transfus. 2014;12:314–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lambert T, Benson G, Dolan G, Hermans C, Jiménez‐Yuste V, Ljung R, et al. Practical aspects of extended half‐life products for the treatment of haemophilia. Ther Adv Hematol. 2018;9:295–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Skinner MW, Nugent D, Wilton P, O’Mahony B, Dolan G, O’Hara J, et al. Achieving the unimaginable: Health equity in haemophilia. Haemophilia. 2020;26:17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.den Dunnen JT, Dalgleish R, Maglott DR, Hart RK, Greenblatt MS, McGowan‐Jordan J, et al. HGVS Recommendations for the Description of Sequence Variants: 2016 Update. Hum Mutat. 2016;37:564–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1. Transient expression of the FIX‐HSA fusion proteins in HEK293 cells. (A) Secreted levels of the fusion proteins and FIX measured by polyclonal anti‐FIX ELISA (left panel), and representative activity profiles of serial dilutions of media in FIX‐deficient plasma evaluated by aPTT‐based assay (right panel). (B) Western blotting analysis of the activation profile of FIX and FIX‐HSA fusions (left panel), in the absence (zym) or in the presence (act) of the physiological activator FXIa. A schematic representation of the protein fragments deriving from FXIa‐mediated cleavage is provided (right panel). (C) Representative activity profile of serial dilutions of transiently expressed fusion proteins, either wild‐type or containing the R384L (Padua) substitution, evaluated by chromogenic assays. The corresponding P values are indicated as not significant (ns, P > 0·05), ****(P < 0·0001), by unpaired t‐test. aPTT, activated partial thromboplastin time; FIX, factor IX; HSA, human serum albumin.
Fig S2. Sodium dodecyl sulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE) analysis of the FIX‐HSA fusion proteins. Purified fractions of Idelvion (lane 2), FIX‐HSA (lane 3) and Padua‐QMP (well 4) were analysed by SDS‐PAGE and Coomassie staining. FIX, factor IX; HSA, human serum albumin.
Fig S3. Complete SPR sensorgrams showing binding of FIX‐HSA fusion proteins to hFcRn. Serial dilutions (0–4 µM) of monomeric hFcRn were injected over immobilised (~200 RU) fusion proteins at pH 5·5. Regeneration was performed by injecting a pH 7·4 buffer at the time point indicated by the arrow. hFcRn, human neonatal Fc receptor; SPR, surface plasmon resonance.
Table SI. Oligonucleotides used to create expression vectors for designed fusion proteins.
Table SII. P values from unpaired t‐test on in vivo half‐life data.


