Abstract
Fibroblasts have recently attracted attention as a key stromal component that controls the immune responses in lymphoid tissues. The thymus has a unique microenvironment comprised of a variety of stromal cells, including fibroblasts and thymic epithelial cells (TECs), the latter of which is known to be important for T cell development because of their ability to express self‐antigens. Thymic fibroblasts contribute to thymus organogenesis during embryogenesis and form the capsule and medullary reticular network in the adult thymus. However, the immunological significance of thymic fibroblasts has thus far only been poorly elucidated. In this review, we will summarize the current views on the development and functions of thymic fibroblasts as revealed by new technologies such as multicolor flow cytometry and single cell–based transcriptome profiling. Furthermore, the recently discovered role of medullary fibroblasts in the establishment of T cell tolerance by producing a unique set of self‐antigens will be highlighted.
Keywords: Thymus, fibroblast, T cell, capsule, medulla
1. INTRODUCTION
The thymus is an organ in which T cells develop and their antigen recognition repertoire is established.1 In the three‐dimensional microenvironment composed of thymic stromal cells, immature T cells (called thymocytes) undergo stepwise developmental processes, including differentiation, proliferation, and cell fate determination in order to give rise to mature T cells expressing a diverse T cell receptor (TCR) repertoire.2
The thymus parenchyma is subdivided into two regions, the cortex and medulla, wherein distinct subsets of thymic epithelial cells (TECs) form a reticular meshwork that houses developing thymocytes.3, 4 The cortex is the outer region with cortical TECs (cTECs) and thymocytes of immature stages, while the medulla is the inner region and is characterized by medullary TECs (mTECs) and mature thymocytes (Figure 1A). TECs play an essential role in T cell development, providing various signals in support of the survival, proliferation, migration, differentiation, and repertoire selection of thymocytes.
FIGURE 1.
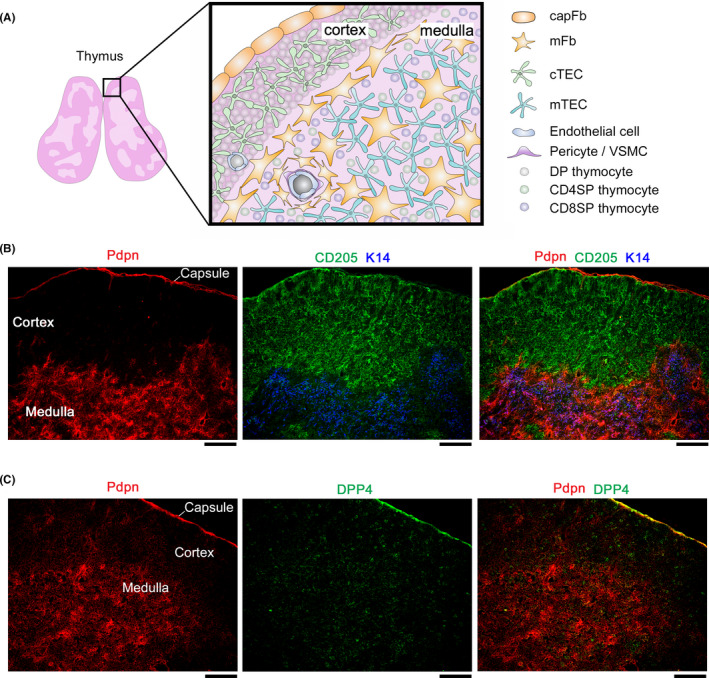
Thymic architecture and stromal cell localization. (A) Schematic depicting the thymic structure and the localization of stromal cells and thymocytes. (B and C) Thymus sections from 5‐week‐old C57BL/6 mice were stained for the indicated markers; Pdpn (fibroblasts), CD205 (cTECs), keratin 14 (K14) (mTECs), and DPP4 (capFbs). The scale bars indicate 100 µm. (B) Thymic fibroblasts expressing Pdpn are localized in the medulla as well as the capsule of the thymus. (C) DPP4 expression segregates the Pdpn+ thymic fibroblasts into capFbs and mFbs
Early T‐cell progenitors (ETPs) from the fetal liver or adult bone marrow differentiate into CD4−CD8− (double negative, DN) thymocytes in the thymic cortex. Guided by cTECs, DN thymocytes are committed to the T‐cell lineage and undergo rearrangements of the genes encoding the TCR.3, 4, 5 In the adult thymus, ETPs arrive at the cortico‐medullary junction (CMJ) where blood vessels are enriched, and developing DN thymocytes migrate through the cortex toward the subcapsular region and differentiate into CD4+CD8+ (double positive, DP) thymocytes in the outer cortex. DP thymocytes that have completed gene rearrangement express the rearranged TCR on the cell surface. Upon interaction between the TCR and self‐peptide/MHC complexes, the cells with functional TCR are induced to differentiate into CD4+CD8− (CD4 single positive, CD4SP) or CD4−CD8+ (CD8SP) thymocytes (positive selection), while cells expressing self‐reactive TCR are deleted (negative selection).6 The positive selection of such a diverse TCR repertoire depends on the ability of cTECs to produce and present a unique set of self‐peptides via MHC molecules.7, 8, 9, 10, 11, 12
Positively selected SP thymocytes migrate from the cortex to the medulla, attracted by chemokines produced by mTECs.13, 14, 15, 16 In the medulla, mTECs express a large number of highly diverse antigens called tissue‐restricted antigens (TRAs) that represent almost all of the tissues in the entire body.17, 18 The coordination of the mTEC expression of TRAs and relocation of SP thymocytes ensures the negative selection and/or regulatory T cell (Treg) conversion of SP thymocytes that recognize such TRAs, thus establishing self‐tolerance in T cells.15, 16, 19, 20, 21, 22, 23, 24
In addition to TECs, which control T cell differentiation and selection, a variety of non‐TEC stromal cells support the thymic microenvironment. The blood vasculature is an important parenchymal component of the thymus that supplies oxygen and nutrients and provides entry and exit points for T cells as well as other immune cells. The cortex contains a network of capillaries, while the CMJ and medulla are enriched with arterioles and postcapillary venules.25, 26 These are made up of functionally distinct endothelial cells that control the influx of bloodborne molecules as well as ETP ingress and mature T cell egress.27, 28, 29
The thymus also contains mesenchymal cells that originate from neural crest (NC) cells. These NC‐derived mesenchymal cells are important for the differentiation and expansion of TECs during embryogenesis. In the postnatal thymus, the mesenchymal cells are predominantly found as fibroblasts in the capsule and medulla, and also as vascular mural cells (Figure 1A). However, despite their abundance in the thymus, the immunological significance of thymic fibroblasts in the postnatal thymus has been both less explored and understood than that of TECs.
Fibroblasts have been generally considered to be ordinary cells without specific features, distributed in tissues throughout the body. However, recent studies have revealed the functional heterogeneity of fibroblasts under various physiological and pathological conditions,30, 31, 32 including immune responses in secondary lymphoid organs33, 34 or upon viral infection.35 This review will provide a current state‐of‐the‐art overview of thymic fibroblasts, focusing on historical studies and recently reported findings on their nature and immunological functions.
2. FIBROBLASTS IN THE THYMUS
2.1. Overview of thymic mesenchymal cells
In the adult thymus, NC‐derived mesenchymal cells are predominantly found in the capsule and medulla.36, 37 The capsule of the mouse thymus comprises a monolayer of fibroblasts (capsular fibroblasts, capFbs) that covers the surface of the thymic parenchyma.25 The human thymus is covered by a capsule from which interlobular septa arise and divide the parenchyma into lobes.38, 39 There are some sparsely distributed fibroblasts in the cortex, but their structural features are not presently known. In the medulla, NC‐derived cells are found as medullary fibroblasts (mFbs) and vascular mural cells. mFbs form the reticular network40 as well as the blood vessel adventitial layer.41 Mural cells are subdivided into pericytes and vascular smooth muscle cells (VSMCs), both of which are embedded in the basement membrane and ensheath the endothelial tubes. Pericytes and VSMCs are distinguished by the absence or the presence of contractility.42, 43 Although the definition of these cells has been different or even deemed controversial in different studies, in this review we define pericytes as non‐contractile cells and VSMCs as contractile cells expressing α‐smooth muscle actin (α‐SMA).
Traditionally, monoclonal antibodies such as ER‐TR7 and MTS‐15 have been used for the detection of thymic fibroblasts. ER‐TR7 reacts with an unidentified intracellular epitope of fibroblasts,44 while MTS‐15 binds glycosphingolipid on the fibroblast surface.45, 46, 47 It was shown that in flow cytometry approximately one‐half of PDGFRα+ thymic fibroblasts are MTS‐15+.47 Although these antibodies recognize not only fibroblasts but also endothelial cells and mural cells, studies using these antibodies have led to the discovery of more specific molecular markers of thymic fibroblasts.
As summarized in Table 1, several proteins have been reported as thymic stromal cell subset markers, including fibroblasts. PDGFRα and PDGFRβ are markers widely used for detecting thymic fibroblasts.48, 49, 50 PDGFRα is highly expressed in capFbs and mFbs, while PDGFRβ is prominent in pericytes and VSMCs. capFbs and mFbs also express podoplanin (Pdpn, also called gp38) and CD34 at high levels.40, 41 Pericytes and VSMCs can be distinguished from fibroblasts by their high expression of Mcam (CD146) and integrin α7 (Itga7).41, 51 These markers allow the detection of thymic fibroblast subsets by immunohistochemistry (Figure 1B,C) and flow cytometry (Figure 2), as described below (Section 2.3).
TABLE 1.
Molecular markers of mouse thymic stromal cells
| Protein | Gene | Expression pattern | References | ||||||
|---|---|---|---|---|---|---|---|---|---|
| capFb | mFb | PC | VSMC | EC | cTEC | mTEC | |||
| PDGFRα (CD140a) | Pdgfra | ++ | ++ | + | + | +/− | − | − | 36, 48, 49, 50, 71, 73 |
| PDGFRβ (CD140b) | Pdgfrb | + | + | ++ | ++ | +/− | − | − | 36, 49, 50, 73 |
| Podoplanin (gp38) | Pdpn | ++ | ++ | − | − | − | +/− | +/− | 40, 41, 72, 73, 133 |
| FSP1 (S100A4) | S100a4 | + | + | ++ | ++ | + | − | + | 132 |
| CD34 | Cd34 | ++ | ++ | ‐ | − | ++ | ‐ | − | 41, 73 |
| DPP4 (CD26) | Dpp4 | ++ | − | − | − | − | − | − | 72 |
| Endosialin (CD248) | Cd248 | ++ | + | + | + | − | − | − | 97 |
| PECAM‐1 (CD31) | Pecam1 | − | − | − | − | ++ | − | − | 37, 45, 69, 71 |
| Mcam (CD146) | Mcam | − | +/− | ++ | ++ | + | − | − | 72, 73 |
| Integrin α7 | Itga7 | − | − | ++ | ++ | − | − | − | 41 |
| α‐SMA | Acta2 | − | − | − | ++ | − | − | − | 36, 37, 41, 50 |
| Ly51 (CD249) | Enpep | − | − | + | + | − | ++ | − | 45, 46, 71 |
| EpCAM (CD326) | Epcam | − | − | − | − | − | ++ | ++ | 45, 46, 71 |
| MHC‐II | H2‐Aa,H2‐Ab1 | − | − | − | − | − | ++ | ++ | 45, 70, 71 |
| H2‐Ea,H2‐Eb1 | |||||||||
| CD80 | Cd80 | − | − | − | − | − | − | ++ | 71 |
Abbreviations: capFb, capsular fibroblast; cTEC, cortical thymic epithelial cell; EC, endothelial cell; mFb, medullary fibroblast; mTEC, medullary thymic epithelial cell; PC, pericyte; VSMC, vascular smooth muscle cell.
−, negative; +, positive; ++, strongly positive; +/−, partially positive.
FSP1 and α‐SMA are intracellular proteins, and the others are cell surface proteins.
FIGURE 2.
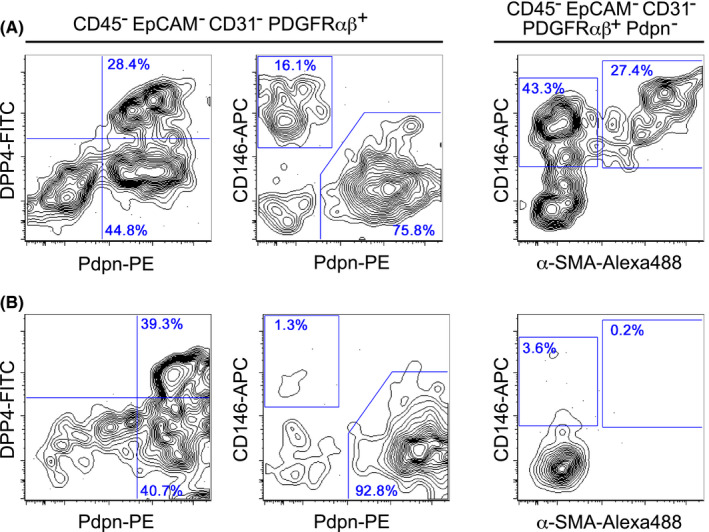
Flow cytometry detection of thymic fibroblast subsets. Thymic stromal cells were prepared using 0.01% Liberase TM (Roche) (A) or 0.125% collagenase D (Roche) (B) from 5‐week‐old C57BL/6 mice, as described previously.71, 72 Representative flow cytometry profiles of gated stromal cell populations are shown. (A) In the Liberase TM‐dissociated, CD45− EpCAM− CD31− PDGFRαβ+ cell population, Pdpn+ DPP4+ cells (capFbs) and Pdpn+ DPP4− cells (mFbs) were detected. The Pdpn− CD146+ cells contain α‐SMA− pericytes and α‐SMA+ VSMCs. (B) In the collagenase D‐dissociated cell suspension, the Pdpn− CD146+ α‐SMA− pericytes and Pdpn− CD146+ α‐SMA+ VSMCs were found at a very low frequency or were almost undetectable, while capFbs and mFbs were readily detectable
Most of our understanding of thymic fibroblasts has come from studies using the thymus of animals such as mice and rats. Even though human thymus samples can be obtained from aborted fetuses or neonatal cardiac surgery, these specimens are generally not readily available in many countries. Therefore, previous studies on human thymic fibroblasts have been limited mainly to histological observations using fixed thymus specimens. Recently, however, some have attempted to clarify the functional classification and age‐related changes of human thymic fibroblasts using new technologies such as single‐cell transcriptomics. These studies will be discussed in detail later (Section 2.5).
In order to understand how these thymic fibroblasts develop and are localized within the thymus, it is necessary to have a close look at the organogenesis of the thymus.
2.2. Mesenchymal cells in organogenesis of the thymus
The thymus originates from the 3rd pharyngeal pouch, a temporary embryonic structure composed of evaginated endodermal epithelial cells.52 The epithelial cells are surrounded by NC‐derived mesenchymal cells, which support pouch patterning, organogenesis of the thymus as well as parathyroid grand, and differentiation of the epithelial cells into TECs. Along with the proliferation of TECs and organization of epithelial parenchyma, the surrounding mesenchymal cells form the capsule that covers the surface, while a fraction of these cells invaginate into the thymus across the epithelial layers to establish an intrathymic network of fibroblasts.49, 53 Along with this migration, mesoderm‐derived progenitor cells enter into the thymus and differentiate into blood vessel endothelial cells in order to form a vascular network.54 Thus, the thymic epithelial, mesenchymal, and endothelial cells spatially and functionally interact in a coordinated manner in order to organize the thymic microenvironment.
NC‐derived mesenchymal cells are required for the differentiation and proliferation of TECs, thus maximizing the thymic capacity for T cell production.48, 49, 53, 55, 56 The production of extracellular matrixes secreted by mesenchymal cells may be important for incorporating immature TECs into a three‐dimensional microenvironment and presenting cytokines to developing thymocytes.57, 58 Mesenchymal cell‐derived signaling proteins that control fetal TEC differentiation and expansion have been reported, including insulin‐like growth factor‐1 (IGF1), IGF2, fibroblast growth factor‐7 (FGF7), FGF10, bone morphogenic protein‐4 (BMP4), and the Wnt ligands.48, 56, 59, 60, 61, 62, 63, 64 Mesenchymal cells produce the vitamin A metabolite retinoic acid, which inhibits TEC proliferation in embryonic thymus.65 Thus, thymic mesenchymal cells may also exert a negative regulatory function on TECs.
Kernfeld et al performed single‐cell RNA sequencing (RNA‐seq) of whole cell types from embryonic thymus, including mesenchymal cells and TECs.66 Figure 3 shows the uniform manifold approximation and projection (UMAP) clustering of their data (GSE107910). Igf1, Fgf7, Fgf10, and Aldh1a2 (a gene encoding an enzyme for retinoic acid biosynthesis) were specifically expressed in mesenchymal cells (cluster 2), suggesting the non‐redundant role of mesenchymal cells as a source of these key factors. Bmp4 is expressed in both mesenchymal cells and TECs (cluster 1), consistent with a previous report that the deletion of Bmp4 in both NC‐derived cells and endoderm‐derived cells (but not either one alone) resulted in defects in thymus organogenesis.62 The Wnt ligand Wnt4 is reported to induce the expression of FoxN1 in TECs63 and is highly expressed in TECs, but only slightly in mesenchymal cells, suggesting a role for the Wnt pathway in fetal TEC differentiation, mainly in an autocrine manner. This single‐cell study also revealed that the thymic mesenchyme strongly expresses Delta‐like non‐canonical Notch ligand 1 (Dlk1, also called Pref1), which is reported to support thymocyte cellularity in organ culture.67
FIGURE 3.
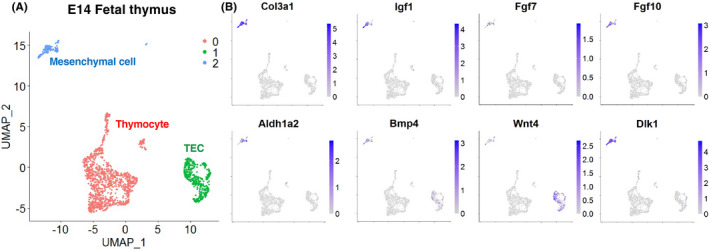
Single‐cell transcriptome of fetal thymic cells. (A) Two‐dimensional representation of E14 fetal thymic cells. Data from the Gene Expression Omnibus (GEO) database under accession no. GSE107910 were used for UMAP clustering.66 Each dot represents a single cell. The full source code for analysis is available in GitHub (https://github.com/nittatakeshi/ImmunolRev_Fig3). (B) Expression profiles of the genes which are known to control fetal TEC differentiation and expansion. Col3a1 was used as a marker of mesenchymal cells
Fetal thymic mesenchymal cells are thought to be a heterogeneous mixture of cells with different characteristics rather than a homogeneous population, and are remotely located in the outer and inner sites of the thymus. However, the mechanism underlying their heterogeneity has yet to be elucidated.
In the following Sections 2.3 to 2.5, recent advances in the identification and characterization of fibroblast subsets as well as other mesenchymal cells in the adult thymus by flow cytometry and transcriptome analyses will be reviewed.
2.3. Flow cytometry of the thymic fibroblast subsets
Since several different markers are co‐expressed in different cell types (Table 1), thymic stromal cell subsets have been difficult to evaluate by histological studies alone. An early study by Izon et al applied flow cytometry analysis to characterize thymic stromal cells.68 Later studies improved on this method, establishing protocols to efficiently dissociate cells from thymus tissue and to distinguish different stromal cell populations using multicolor cytometry.45, 46, 69, 70 To date, the collagenase extracted from Clostridium histolyticum has been widely used for dissociating thymic stromal cells. Liberase research grade enzymes, a blend of purified collagenase and other proteases, are able to dissociate thymic epithelial cells in higher yield than using crude collagenase products,71 and are now most widely used as the standard protocol. Although they cleave certain cell surface epitopes and thereby weaken the staining intensity, Liberase enzymes are also useful for preparing non‐epithelial thymic stromal cells including fibroblasts, endothelial cells, and vascular mural cells in high yield and quality. Figure 2A shows the flow cytometry profiles of thymic mesenchymal stromal cells (Ter119− CD45− EpCAM− CD31− PDGFRαβ+) dissociated from the mouse thymus with Liberase TM. Among the thymic mesenchymal cells, a dominant population is the Pdpn+ CD146− fibroblasts, which are characterized by a high expression of PDGFRα and CD34.41 A minor population, Pdpn− CD146+ cells co‐expressing PDGFRβ, Ly51, and integrin α7,41 contains α‐SMA− pericytes and α‐SMA+ VSMCs.
Recently, we developed a gradual method of digestion using the Liberase enzyme that allows for the fractionation of thymic cells based on their location within the thymus.72 This location‐based fractionation method allows the physical separation of remotely localized thymic fibroblast subsets, capFbs, and mFbs. We identified a cell‐surface protein, dipeptidyl peptidase‐4 (DPP4, also called CD26), which is highly expressed in capFbs but not in mFbs, and consequently established a method to separate capFbs (DPP4+ Pdpn+) and mFbs (DPP4− Pdpn+) by flow cytometry (Figure 2A) as well as histological staining (Figure 1C).
Among the thymic stromal cells, mural cells are relatively difficult to dissociate. When dissociated with collagenase D, a crude collagenase preparation, the yield of Pdpn− CD146+ mural cells, including pericytes and VSMCs is very low or even undetectable compared to the yield when Liberase TM is used (Figure 2B). This might be due to the excessive cellular damage caused by contaminating components such as endotoxin in the crude enzyme preparations, and possibly explains the reason why these cells have not been readily detected in previous studies (see Section 2.5). Even with Liberase, it is still possible that some unnoticed stromal cell types are lost during enzymatic digestion.
Collectively, however, by using cell dissociation with Liberase and multicolor flow cytometry, it has now been made possible to determine and isolate almost all of the types of stromal cells that compose thymic microenvironment.
2.4. Population‐based transcriptome profiling of thymic fibroblasts
In order to characterize the nature and function of thymic fibroblasts, many studies have sought to reveal their gene expression profiles. However, it has been difficult to delineate a unified gene expression pattern for thymic fibroblasts, because the markers used to isolate the cell subsets vary from study to study.
Patenaude and Perreault performed whole transcriptome analysis by RNA‐seq of thymic mesenchymal cells (Lineage− EpCAM− CD31− Sca1+).73 Their results revealed that Sca1+ thymic mesenchymal cells exhibit a higher expression of genes involved in epithelial interaction, apoptotic cell clearance, and T‐cell progenitor expansion, compared to their bone or skin counterparts, suggesting a pivotal role for these cells in the thymic microenvironment. However, since the Sca1+ mesenchymal cell population is a mixture of the fibroblasts, pericytes, and VSMCs, the cell subsets which express each of the key genes remain to be determined.
In a study by Sitnik et al, thymic mesenchymal cells (Lineage− EpCAM− CD31− PDGFRβ+) were divided into two subsets, Pdpn+ Ly51− fibroblasts and Pdpn− Ly51+ mural cells, and then, their entire transcriptome was analyzed by microarray.41 The Pdpn+ Ly51− cells expressed genes regulating vascular and epithelial cell growth (Vegfc, Vegfd, Igf1, Igf2, Fgf2, Fgf7, and Fgf10), suggesting that thymic fibroblasts play a role in maintaining vascular and epithelial niches. It is likely that the Pdpn− Ly51+ cells contained VSMCs, as indicated by the high expression of the Acta2 gene (α‐SMA).
Our recent results have determined the whole transcriptome of the isolated thymic fibroblast subsets by RNA‐seq.72 Both capFbs and mFbs highly express certain fibroblast‐associated genes such as collagens (Col1a1, Col1a2, Col3a1, and Col6a1), extracellular matrix proteins (Dcn, Lum, Mgp, and Sparc), extracellular proteases (Htra1, Htra3, Mmp2, Mmp3, and Mmp14), and protease inhibitors (Serping1 and Serpinh1). These gene expression signatures are similar to those of fibroblastic cells in secondary lymphoid organs reported in previous studies.41, 73 A set of genes was found to be differentially expressed in capFbs and mFbs (see Sections 3 and 4).
We also analyzed the transcriptome of Pdpn− CD146+ mural cells. These cells displayed a gene expression pattern reminiscent of a certain type of fibroblast (Col3a1, Col4a1, and Col6a1) and a potent expression of genes associated with pericytes (Cspg4) or muscle cells (Acta2, Myl9, and Myh11), indicating that the Pdpn− CD146+ cell population is a mixture of pericytes and VSMCs. This is consistent with the results of flow cytometry that indicate Pdpn− CD146+ cells comprise α‐SMA− pericytes and α‐SMA+ VSMCs (Figure 2A).
These population‐based transcriptome datasets of thymic fibroblast subsets should provide a powerful tool for understanding the development and function of the thymic microenvironment, especially in combination with the recently advanced single cell–based transcriptome datasets described below.
2.5. Single cell–based transcriptome profiling of thymic fibroblasts
Bornstein et al reported single‐cell RNA‐seq analysis of mouse thymic stromal cells.74 Figure 4 shows the UMAP clustering of their data of whole thymic stromal cells (GSE103967). The TEC subpopulations, endothelial cells, and contaminating lymphocytes and myeloid cells were clustered according to the gene expression signatures specific for each cell types (Figure 4A,B). Clusters 2, 4, 7, and 8 exhibited a high level of expression of Col3a1, Pdgfra, and Pdgfrb, indicating that these clusters comprise thymic fibroblasts. Cluster 7 expressed Dpp4, Pi16, and Mfap5, corresponding to capFb (Figure 4C,D) based on the results of the population‐based transcriptome.72 Both clusters 2 and 4 exhibited a high expression of Serpine2 and Apod and no expression of Dpp4, corresponding to mFb. Pericytes and VSMCs were not clearly clustered at this resolution, most likely because most of these cells were lost during the collagenase D digestion performed for cell isolation.74
FIGURE 4.
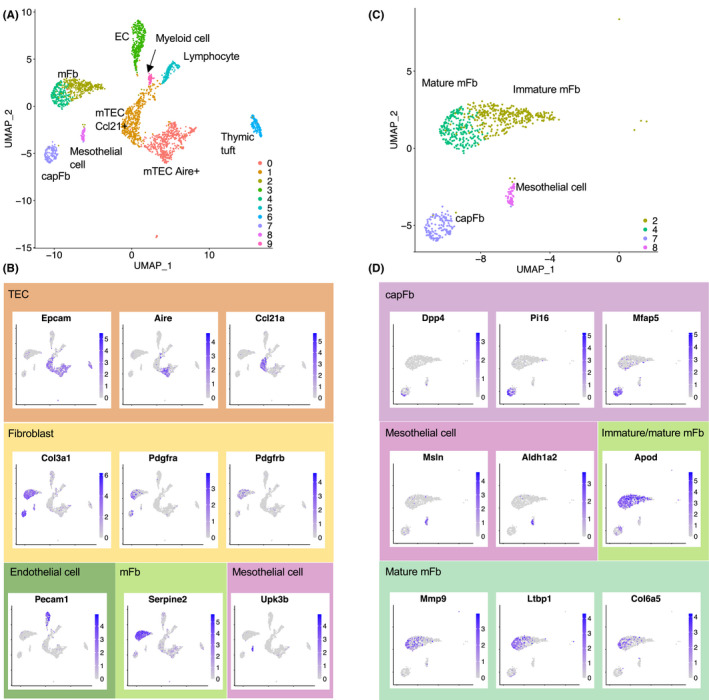
Single‐cell transcriptome of adult thymic stromal cells. Single‐cell RNA‐seq data of mouse thymic stromal cells (GEO accession no. GSE103967, Experiment ID thymus_stroma_WT) were used for UMAP clustering.74 The full source code for analysis is available in GitHub (https://github.com/nittatakeshi/ImmunolRev_Fig4). (A) Two‐dimensional representation of cells by UMAP. Each dot represents one cell. (B) Projection of representative genes. Clusters 0, 1, and 6 are TEC subsets defined by the expression of Epcam as well as key genes such as Aire or Ccl21a. Cluster 3 represents endothelial cells (ECs) expressing Pecam1. Clusters 2, 4, 7, and 8 represent thymic fibroblasts characterized by the expression of Col3a1, Pdgfra, and Pdgfrb. Contaminating lymphocytes (cluster 5) and myeloid cells (cluster 9) are also included. (C) Thymic fibroblast subsets (clusters 2, 4, 7, and 8) are highlighted. (D) Cluster 7 represents capFbs expressing Dpp4, Pi16, and Mfap5. Cluster 8 represents mesothelial cells defined by a high expression of Msln. mFbs (clusters 2 and 4) can be subdivided into immature and mature mFbs, and the latter express Mmp9, Ltbp1, and Col6a5
Cluster 8 is a relatively minor cell population that expresses the Dpp4 and Pdpn that are similar to cluster 7 (capFb), but also exhibits a high expression of Msln and Upk3b with only negligible expression of Pi16 and Mfap5, which is markedly different from cluster 7. It is likely that cluster 8 comprises mesothelial cells that can be defined by a high expression of mesothelin (Msln).75, 76 These mesothelial cells are specifically detectable in the outermost fractions of the thymus (unpublished data), suggesting that these cells compose the thymic capsule together with capFbs, or that they are derived from the visceral pleura in contact with the thymus. Thus, the DPP4+ Pdpn+ cells that we called capFbs (Section 2.2 and Figure 2A) contain two subpopulations, a major population expressing Pi16 and a minor population corresponding to mesothelial cells. In this review, we refer to the former as capFb.
In a recent study, Park et al performed single‐cell RNA‐seq to create a comprehensive atlas of human thymic cells including stromal cells.77 In their dataset, thymic mesenchymal cells were classified into fibroblast type 1 (Fb1), fibroblast type 2 (Fb2), cycling fibroblasts, and VSMCs.78 Fb1 and Fb2, respectively, correspond to mesothelial cells and capFbs in the mouse (Figure 4C,D). These clustering results did not contain clusters that correspond to mouse mFbs, probably due to the large variability in human data, which includes a variety of samples in a range from the fetus to the adult.
The same group also published a single‐cell RNA‐seq dataset of the mouse thymus78 generated with previously reported data from the fetal and postnatal thymus,66, 74 in which thymic mesenchymal cells were classified into four groups, Fb_Aldh1a2 (corresponding to mesothelial cells), Fb_Pi16 (corresponding to capFb), Fb_Postn (corresponding to mFb), and VSMCs (containing pericytes and VSMCs). These clusters are all consistent with the cell populations revealed by flow cytometry and the population‐based transcriptome.
In the following sections, we will focus on the thymic fibroblast subsets capFb and mFb, summarizing how they develop and regulate T cell development, referring to the studies with transcriptome data as well as genetically modified mouse models.
3. THE CAPSULAR FIBROBLAST (CAPFB)
3.1. Development of capFbs
The surface of the thymus is covered by a monolayer of fibroblasts that contacts the epithelial parenchyma across the basement membrane. capFbs are derived from the NC‐derived mesenchymal cells that surround the embryonic thymus primordium and remain outside. In the mouse, at around E13, thymic mesenchymal cells diverge into two populations, a perithymic cell population remaining outside the organ that forms the thymic capsule, and another population that migrates into the thymus across the epithelial layers to give rise to mFbs, pericytes, and VSMCs. At E15, DPP4 begins to be expressed in the capsular populations, which allows the two populations capFb and mFb to be distinguished by flow cytometry analysis.72 It was also shown that in human thymus the fibroblasts expressing DPP4 and PI16 (likely capFbs) increase during fetal development.77 The mechanisms that induce the expression of capFb‐associated genes, including Dpp4, are still unclear.
DPP4 is a useful marker for the detection and isolation of capFbs. DPP4 is reportedly expressed by activated fibroblasts in fibrotic tissues such as the skin of patients with systemic sclerosis as well as in cases of liver fibrosis79, 80, 81, 82 or the breast implant capsule in patients with capsular contraction.83, 84 Genetic ablation or pharmacological inhibition of DPP4 ameliorates fibrosis in mice, indicating that DPP4 activity is important for fibroblast activation and tissue fibrosis.82, 85 Whether DPP4 is involved in the function of the thymic capsule remains to be elucidated.
3.2. Control of TEC development by capFbs
Figure 5A shows the KEGG pathway enrichment analysis of the transcriptome data of the thymic fibroblast subsets.72 The Wnt signaling pathway was found to be significantly enriched in capFbs compared with mFbs. Indeed, capFbs express many Wnt family ligands and regulators (Wnt2, Wnt5a, Wnt9a, Wnt11, Sfrp2, and Sfrp4) at higher levels than mFbs as well as other thymic stromal cells (Figure 5B). Wnt signals reportedly critically control the differentiation of TECs and thymocytes,63, 86, 87, 88, 89 suggesting a role for capFb‐derived Wnt signals in the thymus. Although a previous study demonstrated that TECs themselves act as a source of Wnt ligands for maintaining TEC cellularity and thymus size,90 the contribution of capFb‐derived Wnt signals to the regulation of TEC and T cell development remains to be determined in future study.
FIGURE 5.
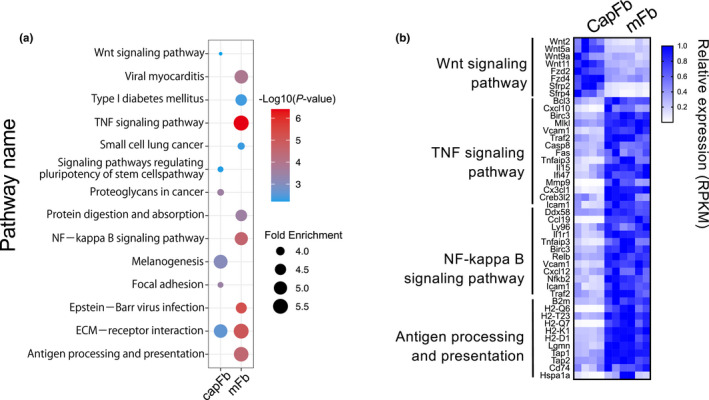
Gene expression profiling of capFbs and mFbs by whole transcriptome analysis. (A) KEGG pathway analysis of genes differentially expressed in capFbs and mFbs. Genes highly expressed in capFbs (capFbs/mFbs > 2, mean RPKM of capFbs > 10, P <.05) or in mFbs (mFb/capFb > 2, mean RPKM of mFb > 10, P <.05) were collected from bulk RNA‐seq data (GEO accession no. GSE147357)72 and subjected to KEGG pathway enrichment analysis using DAVID 6.8. (B) Heat map showing the relative expression of genes categorized as being in the Wnt signaling pathway, TNF signaling pathway, and NF‐κB signaling pathway, as well as antigen processing and presentation
Thymic mesenchymal cells (Pdpn+ Ly51−) are a major thymic source of retinoic acid, which exerts an inhibitory effect on TEC proliferation.65 The transcriptome data show that the genes encoding retinoic acid‐producing enzymes Aldh1a1, Aldh1a2, and Aldh1a3 are strongly expressed in capFbs and/or mesothelial cells: Aldh1a1 in both, Aldh1a2 in mesothelial cells, and Aldh1a3 in capFbs (Figure 4D and data not shown). Mice with TECs unable to respond to retinoic acid display an aberrant cTEC phenotype, including increased proliferation and the accumulation of an immature population, with a subsequent reduction in thymic cellularity.91 Thus, the retinoic acid produced in the outermost layer of the thymus acts as a regulator of TECs and is important for normal T cell development.
3.3. Control of T cell development by capFbs
The genes uniquely expressed in capFbs in the thymus include the extracellular protease DPP4 (Dpp4), peptidase inhibitor‐16 (Pi16), Wnt ligands, and semaphorin ligands (Sema3c and Sema3d). The semaphorin ligands expressed in capFbs may control the migration of developing thymocytes, since it is reported that the semaphorin receptor Plexin D1 is strongly expressed by DP thymocytes and the absence of Plexin D1 disturbs the medulla localization of newly generated SP thymocytes.92
CD248 (Endosialin) is known to be a marker of mesenchymal cells.93, 94, 95 Immunohistochemical analysis indicated that CD248 expression is prominently detected in the perithymic mesenchyme in the mouse embryo, then downregulated postnatally.96, 97 From transcriptome analysis of human and mouse thymus, Cd248 mRNA is expressed at the highest level in capFbs and mesothelial cells,72, 77, 78 suggesting that CD248 may exert effects in the outermost niches of the thymus. CD248‐deficient mice display age‐dependent decline of thymus size and thymocyte cellularity, and, in particular, a marked reduction of DN3 thymocytes.97 The proliferation of DN3 thymocytes that occurs in the subcapsular zone may be regulated by the CD248 expressed in capFbs. It was also shown that CD248‐deficient mice exhibit delayed recovery of thymus size and vascularization following infection‐induced atrophy. Although the mechanism remains unclear, CD248 may promote re‐vascularization and thymocyte growth during postinfection regeneration.
How capFbs and mesothelial cells control the outermost barrier of the thymus and thymus integrity needs to be clarified in future. The interplay between capFbs and subcapsular cTECs may also be important for supporting T cell development in the subcapsular zone and outer cortex, but determining its physiological significance and molecular basis still remains a challenge.
4. THE MEDULLARY FIBROBLAST (MFB)
4.1. Development of mFbs
On histological analysis, mFbs are detected as a reticular structure interwoven with but also clearly separated from the network of mTECs.40, 72 A population of mFbs expressing CD34 forms adventitial layers that surround mural cells and endothelial cells, and thus referred to as adventitial cells.41 In flow cytometry analysis combined with the location‐based fractionation method, mFbs are found to be enriched in the medullary fraction together with mTECs, consistent with the histological findings.72
Studies using a fate‐mapping strategy with various Cre lines specific to NC (Wnt1a‐Cre, Sox10‐Cre, or Twist2‐Cre) or thymic epithelium (Foxn1‐Cre) have demonstrated that mFbs are derived from the NC‐derived mesenchymal cells that surround the embryonic thymus primordium, not from TECs36, 37, 72 (this is also discussed in Section 5). mFbs in the fetal thymus express low levels of marker proteins such as Pdpn and ICAM1, and their expression increases in the course of postnatal development,41 indicating that the maturation of mFbs is developmentally regulated during ontogeny (see Section 4.3). It has also been shown that Pdpnhi ICAM1hi mFbs are capable of generating Pdpn− Esam1+ mural cells or lymph node stroma‐like cells when reaggregated under appropriate conditions.41 Hence, the mFbs at the population level contain progenitor cells for multiple lymphoid tissue stromal cells, and their differentiation capacity is dependent on the particular environmental context.
4.2. Gene expression in mFbs
As a result of KEGG pathway analysis, the genes for TNF signaling and NF‐κB signaling as well as antigen processing and presentation were found to be significantly enriched in mFbs compared with capFbs (Figure 5A). These findings suggest that TNF signaling and NF‐κB activation pathways play important roles in development and/or function of mFbs and that mFbs are differentiated such that they have a higher antigen presentation capacity than capFbs. Also, a set of genes, including certain collagens (Col6a5, Col6a6), matrix metalloprotease‐9 (Mmp9), metabolic enzymes (Hmgcs2, Ltc4s, and Qprt), and TGFβ‐binding proteins (Ltbp1 and Ltbp2) are predominantly expressed in mFbs among all of the thymic stromal cell types.
mFbs form a conduit‐like structure that resembles the one formed by fibroblastic reticular cells (FRCs) in the lymph nodes.40, 98 To determine the functional cue for mFbs, the transcriptome was compared between mFbs and lymph node FRCs.72 Gene ontology (GO) term analysis revealed that mFbs are significantly enriched for genes associated with extracellular matrix organization and cell adhesion but have a lower association with genes for angiogenesis, the inflammatory response, and the immune response, suggesting functional differences between these two morphologically similar cell types (Figure 6A,B).
FIGURE 6.
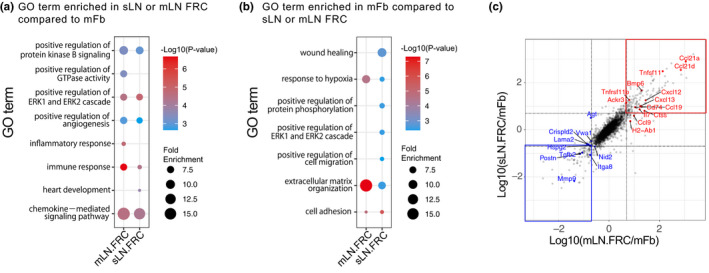
Comparison of the whole transcriptome between thymic mFbs and lymph node FRCs. RNA‐seq data of mFbs, mesenteric lymph node (mLN) FRCs, and skin‐draining lymph node (sLN) FRCs (GEO accession no. GSE147357) were used.72 (A, B) GO term enrichment analysis of genes preferentially expressed in lymph node (LN) FRCs or mFbs. Genes preferentially expressed in LN FRCs compared to mFbs (mLN FRCs: mLN FRCs/mFbs > 5, sLN FRCs: sLN FRCs/mFbs > 5) (A) or in mFbs compared to LN FRCs (mLN.FRCs: mFbs /mLN FRCs > 5, sLN.FRCs: sLN FRCs/mFbs > 5) (RPKM > 10 in any of the groups) were subjected to GO term enrichment analysis using DAVID 6.8. (C) Scatter plot of the gene expression ratio between mLN FRCs/mFbs and sLN FRCs/mFbs. The genes associated with the immune response and the chemokine‐mediated signaling pathway in (A) are highlighted in red. The genes associated with extracellular matrix organization in (B) are highlighted in blue
Lymph node FRCs can be divided into several subtypes expressing different sets of key cytokines and chemokines, such as T‐cell zone reticular cells (TRCs) expressing Ccl19, Ccl21a and Il7, follicular dendritic cells (FDCs) expressing Cxcl13, marginal reticular cells (MRCs) expressing Tnfsf11, and medullary reticular cells (medRCs) expressing Cxcl12, Il6, Tnfsf13, and Tnfsf13b.33, 34, 99 Most of these FRC‐associated cytokines and chemokines are not or only just barely expressed in mFbs (Figure 6C),72 but are predominantly expressed in TECs or thymocytes,100, 101, 102 suggesting that the roles played by FRCs in the lymph nodes are replaced by TECs and thymocytes in the thymus. mFbs highly express other sets of chemokine genes, such as Cx3cl1 and Cxcl14, that are barely expressed in lymph node FRCs, possibly contributing to the regulation of cell migration in the thymic medulla.72 Thus, mFbs comprise a thymic‐specific subset of fibroblasts that is functionally distinct from lymph node FRCs.
It has been reported that patients with autoimmune diseases such as myasthenia gravis or autoimmune‐prone mice exhibit an abnormal accumulation of B cells in the thymus.103, 104, 105 In lupus‐prone BWF1 mice, thymic B cells proliferate within the perivascular space and cluster in structures that resemble ectopic germinal centers, where B cells differentiate to secrete autoantibodies.106 Other early studies also reported that the thymus from lupus‐prone NZB mice or diabetes‐prone NOD mice contained giant perivascular spaces, which are filled with mature T cells, B cells, and fibroblast‐associated extracellular matrix proteins.107, 108, 109, 110 Whether these autoimmune‐associated, “germinal center‐like” structures involve any subset(s) of thymic fibroblasts, like as the formation of the canonical germinal centers in the lymph nodes requires FDCs, remains an open and interesting question.
4.3. LTβR‐dependent maturation of mFbs
Single‐cell RNA‐seq analysis of mouse thymic stromal cells demonstrated that the genes highly expressed in mFbs, such as Serpine2 and Apod, are prominently detected in clusters 2 and 4 (Figure 4C,D). Certain mFb‐associated genes, including Mmp9, Ltbp1, and Col6a5, are detectable in cluster 4 but not cluster 2. These cluster 4‐specific genes are expressed in mFbs from adult but not neonatal mice,72 suggesting that clusters 4 and 2 represent mature and immature mFbs, respectively. In addition, most of these mature mFb‐associated genes are expressed under the control of the lymphotoxin signal.
The TNF superfamily ligand lymphotoxin (LTα1β2) is predominantly expressed by developing SP thymocytes in the thymus and binds to the lymphotoxin β receptor (LTβR) expressed in thymic stromal cells to induce intracellular signal transduction. The LTβR is expressed at the highest level in mFbs among the thymic stromal cells.72 In mFbs from LTβR‐deficient mice, the expression of a large fraction of mFb‐associated genes was diminished. Indeed, LTβR‐deficient mFbs displayed a reduced expression of mFb‐associated proteins such as Pdpn, ICAM‐1, and VCAM‐1.41, 111 Thus, the LTβR signal critically controls the functional maturation of mFbs. It is known that the LTβR signal is required for the maturation of lymph node FRCs,112 offering an analogy that shows that these distinct fibroblast subsets share common signaling pathways for maturation.
4.4. Self‐antigen expression by mFbs for the induction of immune tolerance
Early studies showed that fibroblasts are capable of presenting self‐antigens to induce the positive selection of thymocytes, suggesting that the ability to mediate positive selection is not limited to the thymic epithelium.113, 114 However, a subsequent series of studies revealed that positive selection requires proteasomes and lysosomal proteases that are uniquely expressed in cTECs.7, 8, 9, 10, 11, 115, 116, 117 Also, the major stromal cells that interact with preselected DP thymocytes are cTECs,118 and fibroblasts are scarce in the thymic cortex. Therefore, it has been thought that fibroblasts are not important for positive selection. On the other hand, mFbs are localized in the medulla, where the negative selection of TRA‐reactive thymocytes occurs. Transcriptome analysis indicates that mFbs are highly associated with genes for antigen presentation, suggesting a contribution to negative selection (Figure 5A).
Insights into the roles of thymic fibroblasts in T cell selection have come from studies of the LTβR. Pioneering studies by Boehm and colleagues reported that the LTβR expressed in thymic stroma is important for the induction of T cell tolerance,119, 120 and later studies demonstrated the requirement of LTβR in optimum mTEC differentiation and gene expression.121, 122, 123, 124 However, mice lacking LTβR specifically in TECs do not exhibit signs of autoimmunity, while mice systemically lacking LTβR do, indicating that the key target of lymphotoxin signaling in the context of tolerance induction must be non‐TEC stromal cells.123, 124 Recently, we found that fibroblast‐specific LTβR‐deficient mice displayed signs of autoimmunity against peripheral tissues, similar to systemic LTβR‐deficient mice.72 TCR repertoire analysis revealed that certain TCR clones escape negative selection in fibroblast‐specific LTβR‐deficient mice. The LTβR in mFbs controls the expression of a set of mFb‐specific genes. Such LTβR‐dependent genes expressed in mFbs include certain TRAs that have been defined based on mathematical methods for evaluating tissue‐specific gene expression (Figure 7). Mice specifically lacking the LTβR in fibroblasts exhibit a marked production of autoantibodies against these TRAs. Collectively, these findings indicate that mFbs act as a primary source of certain self‐antigens for the induction of T cell tolerance, and the lymphotoxin signal is a key mediator of this process (Figure 8).
FIGURE 7.
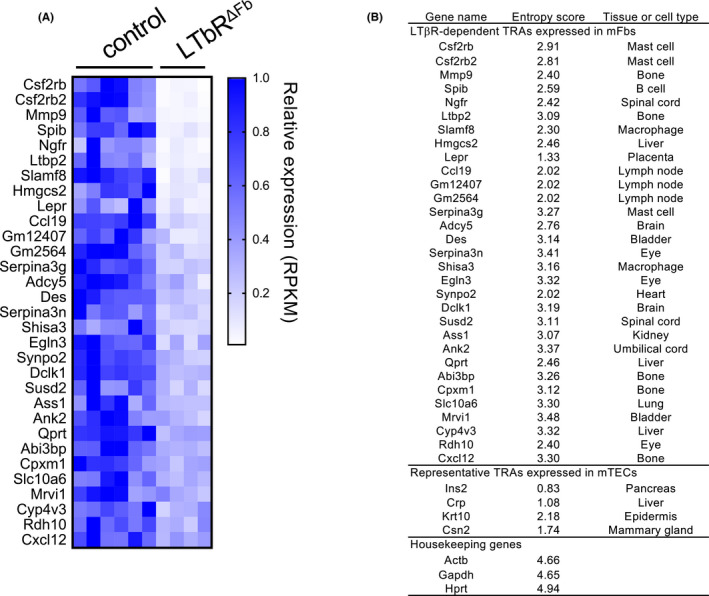
LTβR‐dependent genes in mFbs include TRAs. To define the TRAs, the Shannon entropy score was calculated using the gene expression profiles (GEO accession no. GSE10246).147 The full source code for analysis is available in GitHub (https://github.com/nittatakeshi/ImmunolRev_Fig7). Genes with an entropy score of less than 3.5 are defined as TRA genes. Although these TRA genes were extracted from comprehensive transcriptome data by unbiased mathematical calculation, they may also contain genes that encode functional proteins in mFbs and fibroblast lineage‐specific proteins. (A) The gene expression data on the mFbs from LtbrΔFb mice (Twist2‐Cre Ltbrflox/flox (n = 4)) compared to those from control mice (C57BL/6 (n = 2) and Ltbrflox/flox (n = 4)) are from an RNA‐seq dataset (GEO accession no. GSE147357).72 TRA genes with a mean RPKM >10 in the control mFbs and the ratio of RPKM (LtbrΔFb /control) > 0.5 and significant (P <.05) are shown. (B) LTβR‐dependent TRA genes expressed in mFbs, representative TRA genes expressed in mTECs, and representative housekeeping genes are listed. Expression specificity was determined by computationally extracting the tissues or cell types that showed the highest mRNA expression values
FIGURE 8.
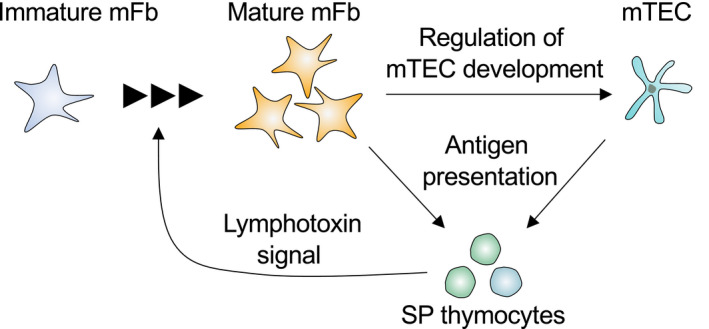
Induction of T cell tolerance by mFbs and mTECs. The immature mFbs give rise to mature mFbs upon interaction with SP thymocytes expressing lymphotoxin (LTα1β2). The mature mFbs promote mTEC development. Both mTECs and mFbs express and present self‐antigens, thus contributing to the deletion of self‐reactive SP thymocytes and the establishment of central tolerance
The expression of TRAs in mFbs is induced by the non‐canonical NF‐κB pathway downstream of the LTβR.72 This is apparently different from the regulatory mechanisms for TRA expression in mTECs, where hundreds to thousands TRAs are expressed by virtue of chromatin modification and transcription regulators such as Aire and Fezf2.18, 19, 125 Considering the large number of self‐antigens encoded by genomes, the induction of central tolerance likely does not rely only on the mTEC expression of TRAs, but instead is achieved by expression in the thymic medulla of genes representing cell types that exist across multiple tissues and organs. The number of TRAs expressed in mFbs may be less than that of mTEC‐expressed TRAs, but mFbs express a set of fibroblast‐specific antigens for developing T cells to delete self‐reactive clones and thereby induce self‐tolerance. It is likely that the thymic medulla needs to be populated with various cell lineages to ensure T cell tolerance to self‐antigens, as indeed mTECs also express antigens specific for various cell lineages by differentiating themselves into peripheral epithelial cells such as keratinocyte‐like cells or tuft cells.74, 126 These findings have led us to propose that the thymic medulla needs to contain different cell types, each of which express cell type‐restricted antigens (CRAs) to maximize the variety of self‐antigens available for T cell selection.72
The expression and presentation of self‐antigens have also been observed in fibroblastic stromal cells in the lymph nodes.127 Lymph node stromal cells present antigens to T cells directly, or indirectly through interaction with dendritic cells (DCs).128, 129 Similarly, in the thymus, mFb‐specific antigens might also be transferred to and presented by thymic DCs so as to induce T cell tolerance, since a substantial portion (about half) of mTEC‐derived self‐antigens are indirectly presented by thymic DCs.130, 131 Indeed, it was demonstrated that the cytoplasmic proteins produced in thymic fibroblasts can be transferred to thymic DCs.72 This mechanism explains how LTβR deficiency in fibroblasts results in the production of autoantibodies against mFb‐specific antigens.
4.5. Regulation of mTEC development by mFbs
It is also possible that mFbs indirectly promote T cell tolerance by controlling mTECs, since the fibroblast‐specific deletion of the LTβR causes a reduction in the number of mTECs.72 Consistent with this, it was shown that the LTβR signal influences the localization of mFbs and their interaction with mTECs.50 In contrast, the loss of mTECs has no influence on mFb cellularity, indicating that mFbs lie upstream of mTECs in the hierarchy of stromal interactions within the medullary microenvironment.72 LTβR‐dependent genes such as cell adhesion molecules (ICAM‐1 and VCAM‐1), extracellular proteases (MMP9), and extracellular matrix proteins (collagens and related proteins) might play key roles in controlling the development and/or maintenance of mTECs.
LTβR‐independent mechanisms in fibroblasts also reportedly induce TEC differentiation. IGF1 is predominantly produced by capFbs and mFbs in the thymus, and the administration of IGF1 by continuous infusion induces an expansion of both cTECs and mTECs,60 although the physiological significance of this induction has not been clarified by loss‐of‐function studies. FGF7 and FGF10, both reported to be involved in TEC proliferation during embryogenesis, are also expressed in adult thymic fibroblasts. FGF7 is expressed in capFbs, while FGF10 is expressed in mFbs, suggesting different roles for these factors in the regulation of postnatal TECs.
Sun et al reported the role played by fibroblast‐specific protein 1 (FSP1, also called S100a4).132 FSP1 is a cytoplasmic and secreted protein expressed in mFbs at higher levels than in capFbs. FSP1 promoter‐driven, inducible cell ablation in mice resulted in a marked reduction of mTECs. Furthermore, FSP1 itself may function as a direct regulator of mTECs, as FSP1‐deficient mice exhibited a smaller sized thymus and reduced number of mTECs, and the addition of purified FSP1 protein increased the mTEC number in organ culture. Collectively, this study demonstrates the pivotal role of FSP1‐expressing fibroblasts in controlling the mTEC number. However, as FSP1 expression (at least at the mRNA level) is not limited to fibroblasts but is also detectable in other thymic stromal cells, including mural cells and mTECs themselves,72 so whether FSP1 is a molecule representative of the function of thymic fibroblasts is in need of further clarification. More specific markers or reporters as well as cell ablation systems need to be developed to better investigate the physiological functions of the thymic fibroblast subsets.
4.6. Regulation of T cell migration by mFbs
The LTβR signal induces the expression of Pdpn in mFbs. Pdpn is a mucin type glycoprotein expressed in various types of stromal cells and in particular is highly expressed in FRCs in the lymph nodes. In the thymus, Pdpn is expressed in capFbs and mFbs as well as a fraction of TECs (Table 1).133, 134, 135 The extracellular domain of Pdpn binds to various proteins that are secreted by or displayed on other cells.136 Pdpn+ mFbs form conduit‐like structures in the medulla and bind the chemokine CCL21 produced by mTECs.40 In the absence of Pdpn, CCL21 fails to efficiently localize in the medulla, a failure which is accompanied by both inefficient migration and generation of Tregs in the medulla. A similar phenotype is observed in mice lacking CCL21 or CCR7, the receptor for CCL21, suggesting a role for Pdpn‐immobilized CCL21 on mFbs in thymic Treg generation.
Two very recent reports demonstrated that CCL21 is displayed on the surface of mFbs and pericytes around blood vessels.137, 138 Cell‐surface binding of CCL21 is mediated by the heparan sulfate strongly expressed by these cells, and consistent with this, the EXT family genes, Ext1 and Ext2, that encode the glycosyl transferases for heparan sulfate biosynthesis are highly expressed in mFbs and pericytes.138 James et al reported that CCL21 captured by the blood‐vessel surrounding fibroblasts and pericytes promotes T cell emigration from the neonatal mouse thymus, suggesting a synergy between TEC‐dependent production and mesenchymal cell‐dependent immobilization of chemokines for controlling T cell migration.137 A study by Hsu et al demonstrated that elimination of heparan sulfate in the thymus by genetic deletion of Ext1 resulted in a marked decrease in the number of thymic fibroblasts and TECs as well as thymocytes.138 Thus, heparan sulfate produced by mesenchymal cells may also be important for thymic stromal cell homeostasis via the immobilization of secreted proteins including chemokines, although the mechanism has not yet been elucidated.
As shown by transcriptome analyses, mFbs themselves express certain chemokine genes, such as Cxcl14 and Cx3cl1, but not Ccl21.72 Whether and how these mFb‐specific chemokines contribute to cell migration in the thymus and exert immunological functions are still presently unknown.
5. FIBROBLASTS IN AGE‐RELATED THYMIC INVOLUTION AND ADIPOSIS
The thymus undergoes an age‐related progressive atrophy called involution that is characterized by qualitative and quantitative changes in stromal cells as well as their replacement with adipocytes.3, 139, 140 In particular, mTECs exhibit a marked decrease in cellularity and an alteration in gene expression patterns with aging.46, 141 In contrast, the frequency of thymic fibroblasts increases with aging, so the ratio of fibroblasts to TECs is markedly increased in aged mice.46 It was shown that TECs in aged mice can give rise to fibroblasts and further into adipocytes, by a process called epithelial‐to‐mesenchymal transition (EMT).142, 143 This suggests the possibility that a fraction of thymic fibroblasts may be of TEC origin. However, fate‐mapping studies using TEC‐specific Cre lines (Psmb11‐Cre or FoxN1‐Cre) demonstrated that only a small percentage of fibroblasts (up to approximately 10%) may be derived from the TEC lineage, and these cells do not increase with age.5, 72 Therefore, the majority of thymic fibroblasts in the adult thymus is indeed NC‐derived, so the contribution of EMT to the development of thymic fibroblasts, if there is any, is limited.
Ucar et al reported that the thymic stroma in adult mice contains cells that under low‐attachment culture conditions form spheres (termed thymospheres),144 and Sheridan et al subsequently demonstrated that those cells do not belong to the TEC lineage but rather, contain mesenchymal stem cells.145 These thymosphere‐forming cells were shown to be capable of giving rise to fibroblasts and adipocytes under appropriate culture conditions. It was also shown in an early study that mesenchymal stromal cells isolated from human thymus are able to differentiate in vitro into adipocytes.146 Nevertheless, at present, there is no conclusive evidence as to which TECs or thymic fibroblasts are responsible for the age‐related adiposis of the thymus. This is an important issue for understanding the contribution of the entire repertoire of thymic stromal cells, including fibroblasts, to age‐related thymic atrophy, as well as for exploring the possible technologies that would allow thymic regeneration.
6. CONCLUDING REMARKS
With the recent advance of large‐scale datasets of stromal cells across multiple organs, we now stand at a new beginning for a comprehensive understanding of cellular characteristics and interactions in the immune system. Such bioinformatics approaches, along with certain long‐sought results in histology and embryology, have unveiled the versatile range of functions of thymic fibroblasts in supporting thymus organogenesis and T cell development. Efforts to understand thymic fibroblast function are now being applied to the studies of human thymus, which may shed light on the role of thymic fibroblasts in human health and disease. In particular, medullary fibroblasts are an emerging subset of thymic stromal cells that is essential for the self‐antigen expression to induce immune tolerance. This finding also suggests an intrinsic need for the thymic medulla to embrace a variety of cell types, each of which expresses cell type‐restricted antigens in order to produce the diverse array of self‐antigens required to accomplish T cell selection. A major issue that remains to be addressed in future is the cellular and molecular basis for fibroblast cooperation with other stromal cells in age‐related thymic atrophy as well as autoimmunity. Elucidating the lineage relationships and cell‐cell interactions of stromal cells as well as their significance in TCR repertoire formation in the degenerating thymus will open up possibilities to better understand and control the thymic microenvironment in future therapeutic applications.
CONFLICT OF INTEREST
There is no conflict of interest to declare.
ACKNOWLEDGMENTS
We thank Sachiko Nitta for continuous assistance and encouragement. This research was supported by the Japan Society for Promotion of Science (JSPS) (KAKENHI 15H05703 to HT, 16H05202, 17H05788, 19H03485, and 19H04802 to TN, and 19J00248 and 19K16602 to RM), the CREST program of the Japan Agency for Medical Research and Development (AMED) (21gm1210008 to HT), and the Sasakawa Scientific Research Grant from the Japan Science Society (2020‐4057 to TI). A. Ota is supported by the MD Scientist Training Program of the University of Tokyo. R. Muro is supported by a JSPS Research Fellowship for Young Scientists.
Nitta T, Ota A, Iguchi T, Muro R, Takayanagi H. The fibroblast: An emerging key player in thymic T cell selection. Immunol Rev. 2021;302:68–85. 10.1111/imr.12985
This article is part of a series of reviews covering Immunological functions of fibroblasts in human health and disease appearing in Volume 302 of Immunological Reviews.
REFERENCES
- 1.Miller JFAP. The function of the thymus and its impact on modern medicine. Science. 2020;369(6503):eaba2429. [DOI] [PubMed] [Google Scholar]
- 2.Han J, Zúñiga‐Pflücker JC. A 2020 view of thymus stromal cells in T cell development. J Immunol. 2021;206:249‐256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abramson J, Anderson G. Thymic epithelial cells. Annu Rev Immunol. 2017;35:85‐118. [DOI] [PubMed] [Google Scholar]
- 4.Kadouri N, Nevo S, Goldfarb Y, Abramson J. Thymic epithelial cell heterogeneity: TEC by TEC. Nat Rev Immunol. 2020;20:239‐253. [DOI] [PubMed] [Google Scholar]
- 5.Ohigashi I, Kozai M, Takahama Y. Development and developmental potential of cortical thymic epithelial cells. Immunol Rev. 2016;271:10‐22. [DOI] [PubMed] [Google Scholar]
- 6.Klein L, Kyewski B, Allen PM, Hogquist KA. Positive and negative selection of the T cell repertoire: what thymocytes see (and don't see). Nat Rev Immunol. 2014;14:377‐391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murata S, Sasaki K, Kishimoto T, et al. Regulation of CD8+ T cell development by thymus‐specific proteasomes. Science. 2007;316:1349‐1353. [DOI] [PubMed] [Google Scholar]
- 8.Nitta T, Murata S, Sasaki K, et al. Thymoproteasome shapes immunocompetent repertoire of CD8+ T cells. Immunity. 2010;32:29‐40. [DOI] [PubMed] [Google Scholar]
- 9.Honey K, Nakagawa T, Peters C, Rudensky A. Cathepsin L regulates CD4+ T cell selection independently of its effect on invariant chain: a role in the generation of positively selecting peptide ligands. J Exp Med. 2002;195:1349‐1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gommeaux J, Gregoire C, Nguessan P, et al. Thymus‐specific serine protease regulates positive selection of a subset of CD4+ thymocytes. Eur J Immunol. 2009;39:956‐964. [DOI] [PubMed] [Google Scholar]
- 11.Viret C, Lamare C, Guiraud M, et al. Thymus‐specific serine protease contributes to the diversification of the functional endogenous CD4 T cell receptor repertoire. J Exp Med. 2011;208:3‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murata S, Takahama Y, Kasahara M, Tanaka K. The immunoproteasome and thymoproteasome: functions, evolution and human disease. Nat Immunol. 2018;19:923‐931. [DOI] [PubMed] [Google Scholar]
- 13.Ueno T, Saito F, Gray DHD, et al. CCR7 signals are essential for cortex‐medulla migration of developing thymocytes. J Exp Med. 2004;200:493‐505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurobe H, Liu C, Ueno T, et al. CCR7‐dependent cortex‐to‐medulla migration of positively selected thymocytes is essential for establishing central tolerance. Immunity. 2006;24:165‐177. [DOI] [PubMed] [Google Scholar]
- 15.Nitta T, Nitta S, Lei Y, Lipp M, Takahama Y. CCR7‐mediated migration of developing thymocytes to the medulla is essential for negative selection to tissue‐restricted antigens. Proc Natl Acad Sci USA. 2009;106:17129‐17133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozai M, Kubo Y, Katakai T, et al. Essential role of CCL21 in establishment of central self‐tolerance in T cells. J Exp Med. 2017;214:1925‐1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derbinski J, Schulte A, Kyewski B, Klein L. Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat Immunol. 2001;2:1032‐1039. [DOI] [PubMed] [Google Scholar]
- 18.Derbinski J, Gabler J, Brors B, et al. Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels. J Exp Med. 2005;202:33‐45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson MS, Venanzi ES, Klein L, et al. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298:1395‐1401. [DOI] [PubMed] [Google Scholar]
- 20.Liston A, Lesage S, Wilson J, Peltonen L, Goodnow CC. Aire regulates negative selection of organ‐specific T cells. Nat Immunol. 2003;4:350‐354. [DOI] [PubMed] [Google Scholar]
- 21.Anderson MS, Venanzi ES, Chen ZB, Berzins SP, Benoist C, Mathis D. The cellular mechanism of Aire control of T cell tolerance. Immunity. 2005;23:227‐239. [DOI] [PubMed] [Google Scholar]
- 22.Aschenbrenner K, D'Cruz LM, Vollmann EH, et al. Selection of Foxp3+ regulatory T cells specific for self antigen expressed and presented by Aire+ medullary thymic epithelial cells. Nat Immunol. 2007;8:351‐358. [DOI] [PubMed] [Google Scholar]
- 23.Legoux FP, Lim JB, Cauley AW, et al. CD4+ T cell tolerance to tissue‐restricted self antigens is mediated by antigen‐specific regulatory T cells rather than deletion. Immunity. 2015;43:896‐908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malhotra D, Linehan JL, Dileepan T, et al. Tolerance is established in polyclonal CD4+ T cells by distinct mechanisms, according to self‐peptide expression patterns. Nat Immunol. 2016;17:187‐195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petrie HT. Role of thymic organ structure and stromal composition in steady‐state postnatal T‐cell production. Immunol Rev. 2002;189:8‐19. [DOI] [PubMed] [Google Scholar]
- 26.Kato S. Thymic microvascular system. Microsc Res Tech. 1997;38:287‐299. [DOI] [PubMed] [Google Scholar]
- 27.Buono M, Facchini R, Matsuoka S, et al. A dynamic niche provides Kit ligand in a stage‐specific manner to the earliest thymocyte progenitors. Nat Cell Biol. 2016;18:157‐167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buono M, Thezenas ML, Ceroni A, Fischer R, Nerlov C. Bi‐directional signaling by membrane‐bound KitL induces proliferation and coordinates thymic endothelial cell and thymocyte expansion. Nat Commun. 2018;9:4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagatake T, Zhao YC, Ito T, et al. Selective expression of claudin‐5 in thymic endothelial cells regulates the blood‐thymus barrier and T‐cell export. Int Immunol. 2020;33:171‐182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lynch MD, Watt FM. Fibroblast heterogeneity: implications for human disease. J Clin Invest. 2018;128:26‐35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guerrero‐Juarez CF, Dedhia PH, Jin S, et al. Single‐cell analysis reveals fibroblast heterogeneity and myeloid‐derived adipocyte progenitors in murine skin wounds. Nat Commun. 2019;10:650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mizoguchi F, Slowikowski K, Wei K, et al. Functionally distinct disease‐associated fibroblast subsets in rheumatoid arthritis. Nat Commun. 2018;9:789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perez‐Shibayama C, Gil‐Cruz C, Ludewig B. Fibroblastic reticular cells at the nexus of innate and adaptive immune responses. Immunol Rev. 2019;289:31‐41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fletcher AL, Baker AT, Lukacs‐Kornek V, Knoblich K. The fibroblastic T cell niche in lymphoid tissues. Curr Opin Immunol. 2020;64:110‐116. [DOI] [PubMed] [Google Scholar]
- 35.Krausgruber T, Fortelny N, Fife‐Gernedl V, et al. Structural cells are key regulators of organ‐specific immune responses. Nature. 2020;583:296‐302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Foster K, Sheridan J, Veiga‐Fernandes H, et al. Contribution of neural crest‐derived cells in the embryonic and adult thymus. J Immunol. 2008;180:3183‐3189. [DOI] [PubMed] [Google Scholar]
- 37.Muller SM, Stolt CC, Terszowski G, et al. Neural crest origin of perivascular mesenchyme in the adult thymus. J Immunol. 2008;180:5344‐5351. [DOI] [PubMed] [Google Scholar]
- 38.Haynes BF. The human thymic microenvironment. Adv Immunol. 1984;36:87‐142. [DOI] [PubMed] [Google Scholar]
- 39.Berrih S, Savino W, Cohen S. Extracellular‐matrix of the human thymus ‐ immunofluorescence studies on frozen‐sections and cultured epithelial‐cells. J Histochem Cytochem. 1985;33:655‐664. [DOI] [PubMed] [Google Scholar]
- 40.Fuertbauer E, Zaujec J, Uhrin P, et al. Thymic medullar conduits‐associated podoplanin promotes natural regulatory T cells. Immunol Lett. 2013;154:31‐41. [DOI] [PubMed] [Google Scholar]
- 41.Sitnik KM, Wendland K, Weishaupt H, et al. Context‐dependent development of lymphoid stroma from adult CD34+ adventitial progenitors. Cell Rep. 2016;14:2375‐2388. [DOI] [PubMed] [Google Scholar]
- 42.Armulik A, Genove G, Betsholtz C. Pericytes: developmental, physiological, and pathological perspectives, problems, and promises. Dev Cell. 2011;21:193‐215. [DOI] [PubMed] [Google Scholar]
- 43.Holm A, Heumann T, Augustin HG. Microvascular mural cell organotypic heterogeneity and functional plasticity. Trends Cell Biol. 2018;28:302‐316. [DOI] [PubMed] [Google Scholar]
- 44.Van Vliet E, Melis M, Van Ewijk W. Monoclonal antibodies to stromal cell types of the mouse thymus. Eur J Immunol. 1984;14:524‐529. [DOI] [PubMed] [Google Scholar]
- 45.Gray DHD, Chidgey AP, Boyd RL. Analysis of thymic stromal cell populations using flow cytometry. J Immunol Methods. 2002;260:15‐28. [DOI] [PubMed] [Google Scholar]
- 46.Gray DHD, Seach N, Ueno T, et al. Developmental kinetics, turnover, and stimulatory capacity of thymic epithelial cells. Blood. 2006;108:3777‐3785. [DOI] [PubMed] [Google Scholar]
- 47.Gray DHD, Tull D, Ueno T, et al. A unique thymic fibroblast population revealed by the monoclonal antibody MTS‐15. J Immunol. 2007;178:4956‐4965. [DOI] [PubMed] [Google Scholar]
- 48.Jenkinson WE, Rossi SW, Parnell SM, Jenkinson EJ, Anderson G. PDGFRα‐expressing mesenchyme regulates thymus growth and the availability of intrathymic niches. Blood. 2007;109:954‐960. [DOI] [PubMed] [Google Scholar]
- 49.Itoi M, Tsukamoto N, Yoshida H, Amagai T. Mesenchymal cells are required for functional development of thymic epithelial cells. Int Immunol. 2007;19:953‐964. [DOI] [PubMed] [Google Scholar]
- 50.Odaka C. Localization of mesenchymal cells in adult mouse thymus: their abnormal distribution in mice with disorganization of thymic medullary epithelium. J Histochem Cytochem. 2009;57:373‐382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seftalioglu A, Karakoc L. Expression of CD146 adhesion molecules (MUC18 or MCAM) in the thymic microenvironment. Acta Histochem. 2000;102:69‐83. [DOI] [PubMed] [Google Scholar]
- 52.Gordon J, Manley NR. Mechanisms of thymus organogenesis and morphogenesis. Development. 2011;138:3865‐3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suniara RK, Jenkinson EJ, Owen JJ. An essential role for thymic mesenchyme in early T cell development. J Exp Med. 2000;191:1051‐1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bryson JL, Griffith AV, Hughes B, et al. Cell‐autonomous defects in thymic epithelial cells disrupt endothelial‐perivascular cell interactions in the mouse thymus. PLoS One. 2013;8:e65196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anderson G, Jenkinson EJ, Moore NC, Owen JJT. MHC class‐II‐positive epithelium and mesenchyme cells are both required for T‐cell development in the thymus. Nature. 1993;362:70‐73. [DOI] [PubMed] [Google Scholar]
- 56.Jenkinson WE, Jenkinson EJ, Anderson G. Differential requirement for mesenchyme in the proliferation and maturation of thymic epithelial progenitors. J Exp Med. 2003;198:325‐332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anderson G, Anderson KL, Tchilian EZ, Owen JJT, Jenkinson EJ. Fibroblast dependency during early thymocyte development maps to the CD25+ CD44+ stage and involves interactions with fibroblast matrix molecules. Eur J Immunol. 1997;27:1200‐1206. [DOI] [PubMed] [Google Scholar]
- 58.Banwell CM, Partington KM, Jenkinson EJ, Anderson G. Studies on the role of IL‐7 presentation by mesenchymal fibroblasts during early thymocyte development. Eur J Immunol. 2000;30:2125‐2129. [DOI] [PubMed] [Google Scholar]
- 59.Erickson M, Morkowski S, Lehar S, et al. Regulation of thymic epithelium by keratinocyte growth factor. Blood. 2002;100:3269‐3278. [DOI] [PubMed] [Google Scholar]
- 60.Chu YW, Schmitz S, Choudhury B, et al. Exogenous insulin‐like growth factor 1 enhances thymopoiesis predominantly through thymic epithelial cell expansion. Blood. 2008;112:2836‐2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bleul CC, Boehm T. BMP signaling is required for normal thymus development. J Immunol. 2005;175:5213‐5221. [DOI] [PubMed] [Google Scholar]
- 62.Gordon J, Patel SR, Mishina Y, Manley NR. Evidence for an early role for BMP4 signaling in thymus and parathyroid morphogenesis. Dev Biol. 2010;339:141‐154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Balciunaite G, Keller MP, Balciunaite E, et al. Wnt glycoproteins regulate the expression of FoxN1, the gene defective in nude mice. Nat Immunol. 2002;3:1102‐1108. [DOI] [PubMed] [Google Scholar]
- 64.Sultana DA, Tomita S, Hamada M, et al. Gene expression profile of the third pharyngeal pouch reveals role of mesenchymal MafB in embryonic thymus development. Blood. 2009;113:2976‐2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sitnik KM, Kotarsky K, White AJ, Jenkinson WE, Anderson G, Agace WW. Mesenchymal cells regulate retinoic acid receptor‐dependent cortical thymic epithelial cell homeostasis. J Immunol. 2012;188:4801‐4809. [DOI] [PubMed] [Google Scholar]
- 66.Kernfeld EM, Genga RMJ, Neherin K, Magaletta ME, Xu P, Maehr R. A single‐cell transcriptomic atlas of thymus organogenesis resolves cell types and developmental maturation. Immunity. 2018;48:1258‐1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaneta M, Osawa M, Sudo K, Nakauchi H, Farr AG, Takahama Y. A role for Pref‐1 and HES‐1 in thymocyte development. J Immunol. 2000;164:256‐264. [DOI] [PubMed] [Google Scholar]
- 68.Izon DJ, Nieland JD, Godfrey DI, Boyd RL, Kruisbeek AM. Flow cytometric analysis reveals unexpected shared antigens between histologically defined populations of thymic stromal cells. Int Immunol. 1994;6:31‐39. [DOI] [PubMed] [Google Scholar]
- 69.Gray DHD, Fletcher AL, Hammett M, et al. Unbiased analysis, enrichment and purification of thymic stromal cells. J Immunol Methods. 2008;329:56‐66. [DOI] [PubMed] [Google Scholar]
- 70.McLelland BT, Gravano D, Castillo J, Montoy S, Manilay JO. Enhanced isolation of adult thymic epithelial cell subsets for multiparameter flow cytometry and gene expression analysis. J Immunol Methods. 2011;367:85‐94. [DOI] [PubMed] [Google Scholar]
- 71.Seach N, Wong K, Hammett M, Boyd RL, Chidgey AP. Purified enzymes improve isolation and characterization of the adult thymic epithelium. J Immunol Methods. 2012;385:23‐34. [DOI] [PubMed] [Google Scholar]
- 72.Nitta T, Tsutsumi M, Nitta S, et al. Fibroblasts as a source of self‐antigens for central immune tolerance. Nat Immunol. 2020;21:1172‐1180. [DOI] [PubMed] [Google Scholar]
- 73.Patenaude J, Perreault C. Thymic mesenchymal cells have a distinct transcriptomic profile. J Immunol. 2016;196:4760‐4770. [DOI] [PubMed] [Google Scholar]
- 74.Bornstein C, Nevo S, Giladi A, et al. Single‐cell mapping of the thymic stroma identifies IL‐25‐producing tuft epithelial cells. Nature. 2018;559:622‐626. [DOI] [PubMed] [Google Scholar]
- 75.Chang K, Pai LH, Batra JK, Pastan I, Willingham MC. Characterization of the antigen (Cak1) recognized by monoclonal antibody K1 present on ovarian cancers and normal mesothelium. Cancer Res. 1992;52:181‐186. [PubMed] [Google Scholar]
- 76.Chang K, Pastan I. Molecular cloning of mesothelin, a differentiation antigen present on mesothelium, mesotheliomas, and ovarian cancers. Proc Natl Acad Sci USA. 1996;93(1):136‐140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Park JE, Botting RA, Conde CD, et al. A cell atlas of human thymic development defines T cell repertoire formation. Science. 2020;367:eaay3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. https://developmentcellatlas.ncl.ac.uk/datasets/HCA_thymus/
- 79.Yazbeck R, Jaenisch SE, Abbott CA. Potential disease biomarkers: dipeptidyl peptidase 4 and fibroblast activation protein. Protoplasma. 2018;255:375‐386. [DOI] [PubMed] [Google Scholar]
- 80.Lay AJ, Zhang HE, McCaughan GW, Gorrell MD. Fibroblast activation protein in liver fibrosis. Front Biosci‐Landmrk. 2019;24:1‐17. [DOI] [PubMed] [Google Scholar]
- 81.Desjardins‐Park HE, Chinta MS, Foster DS, et al. Fibroblast heterogeneity in and its implications for plastic and reconstructive surgery: a basic science review. Plast Glob Open. 2020;8:e2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Soare A, Györfi HA, Matei AE, et al. Dipeptidylpeptidase 4 as a marker of activated fibroblasts and a potential target for the treatment of fibrosis in systemic sclerosis. Arthritis Rheumatol. 2020;72:137‐149. [DOI] [PubMed] [Google Scholar]
- 83.Borrelli MR, Patel RA, Nguyen D, Momeni A, Longaker MT, Wan DC. CD26+ fibroblasts increase in abundance in breast capsule tissue surrounding irradiated breasts. J Am Coll Surgeons. 2019;229:S220. [Google Scholar]
- 84.Borrelli MR, Irizzary D, Patel RA, et al. Pro‐fibrotic CD26‐positive fibroblasts are present in greater abundance in breast capsule tissue of irradiated breasts. Aesthet Surg J. 2020;40:369‐379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kawakubo M, Tanaka M, Ochi K, et al. Dipeptidyl peptidase‐4 inhibition prevents nonalcoholic steatohepatitis‐associated liver fibrosis and tumor development in mice independently of its anti‐diabetic effects. Sci Rep. 2020;10:983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pongracz J, Hare K, Harman B, Anderson G, Jenkinson EJ. Thymic epithelial cells provide Wnt signals to developing thymocytes. Eur J Immunol. 2003;33:1949‐1956. [DOI] [PubMed] [Google Scholar]
- 87.Weerkamp F, Baert MRM, Naber BAE, et al. Wnt signaling in the thymus is regulated by differential expression of intracellular signaling molecules. Proc Natl Acad Sci USA. 2006;103(9):3322‐3326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zuklys S, Gill J, Keller MP, et al. Stabilized β‐catenin in thymic epithelial cells blocks thymus development and function. J Immunol. 2009;182:2997‐3007. [DOI] [PubMed] [Google Scholar]
- 89.Kvell K, Fejes AV, Parnell SM, Pongracz JE. Active Wnt/β‐catenin signaling is required for embryonic thymic epithelial development and functionality ex vivo. Immunobiology. 2014;219:644‐652. [DOI] [PubMed] [Google Scholar]
- 90.Brunk F, Augustin I, Meister M, Boutros M, Kyewski B. Thymic epithelial cells are a nonredundant source of Wnt ligands for thymus development. J Immunol. 2015;195:5261‐5271. [DOI] [PubMed] [Google Scholar]
- 91.Wendland K, Niss K, Kotarsky K, et al. Retinoic acid signaling in thymic epithelial cells regulates thymopoiesis. J Immunol. 2018;201:524‐532. [DOI] [PubMed] [Google Scholar]
- 92.Choi YI, Duke‐Cohan JS, Ahmed WB, et al. PlexinD1 Glycoprotein controls migration of positively selected thymocytes into the medulla. Immunity. 2008;29:888‐898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.MacFadyen JR, Haworth O, Roberston D, et al. Endosialin (TEM1, CD248) is a marker of stromal fibroblasts and is not selectively expressed on tumour endothelium. FEBS Lett. 2005;579:2569‐2575. [DOI] [PubMed] [Google Scholar]
- 94.MacFadyen J, Savage K, Wienke D, Isacke CM. Endosialin is expressed on stromal fibroblasts and CNS pericytes in mouse embryos and is downregulated during development. Gene Expr Patterns. 2007;7:363‐369. [DOI] [PubMed] [Google Scholar]
- 95.Simonavicius N, Robertson D, Bax DA, Jones C, Huijbers IJ, Isacke CM. Endosialin (CD248) is a marker of tumor‐associated pericytes in high‐grade glioma. Modern Pathol. 2008;21:308‐315. [DOI] [PubMed] [Google Scholar]
- 96.Lax S, Hou TZ, Jenkinson E, et al. CD248/endosialin is dynamically expressed on a subset of stromal cells during lymphoid tissue development, splenic remodeling and repair. FEBS Lett. 2007;581:3550‐3556. [DOI] [PubMed] [Google Scholar]
- 97.Lax S, Ross EA, White A, et al. CD248 expression on mesenchymal stromal cells is required for post‐natal and infection‐dependent thymus remodelling and regeneration. FEBS Open Bio. 2012;2:187‐190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Drumea‐Mirancea M, Wessels JT, Muller CA, et al. Characterization of a conduit system containing laminin‐5 in the human thymus: a potential transport system for small molecules. J Cell Sci. 2006;119:1396‐1405. [DOI] [PubMed] [Google Scholar]
- 99.Rodda LB, Lu E, Bennett ML, et al. Single‐cell RNA sequencing of lymph node stromal cells reveals niche‐associated heterogeneity. Immunity. 2018;48:1014‐1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hikosaka Y, Nitta T, Ohigashi I, et al. The cytokine RANKL produced by positively selected thymocytes Fosters medullary thymic epithelial cells that express autoimmune regulator. Immunity. 2008;29:438‐450. [DOI] [PubMed] [Google Scholar]
- 101.Ueno T, Hara K, Willis MS, et al. Role for CCR7 Ligands in the emigration of newly generated T lymphocytes from the neonatal thymus. Immunity. 2002;16:205‐218. [DOI] [PubMed] [Google Scholar]
- 102.Hara T, Shitara S, Imai K, et al. Identification of IL‐7‐producing cells in primary and secondary lymphoid organs using IL‐7‐GFP knock‐in mice. J Immunol. 2012;189:1577‐1584. [DOI] [PubMed] [Google Scholar]
- 103.Berrih‐Aknin S, Ragheb S, Le Panse R, Lisak RP. Ectopic germinal centers, BAFF and anti‐B‐cell therapy in myasthenia gravis. Autoimmun Rev. 2013;12:885‐893. [DOI] [PubMed] [Google Scholar]
- 104.Ishikawa S, Sato T, Abe M, et al. Aberrant high expression of B lymphocyte chemokine (BLC/CXCL13) by C11b+CD11c+ dendritic cells in murine lupus and preferential chemotaxis of B1 cells towards BLC. J Exp Med. 2001;193:1393‐1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sato T, Ishikawa S, Akadegawa K, et al. Aberrant B1 cell migration into the thymus results in activation of CD4 T cells through its potent antigen‐presenting activity in the development of murine lupus. Eur J Immunol. 2004;34:3346‐3358. [DOI] [PubMed] [Google Scholar]
- 106.Hidalgo Y, Nunez S, Fuenzalidal MJ, et al. Thymic B cells promote germinal center‐like structures and the expansion of follicular helper T cells in lupus‐prone mice. Front Immunol. 2020;11:696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fletcher AL, Seach N, Reiseger JJ, et al. Reduced thymic Aire expression and abnormal NF‐kappa B2 signaling in a model of systemic autoimmunity. J Immunol. 2009;182:2690‐2699. [DOI] [PubMed] [Google Scholar]
- 108.Savino W, Carnaud C, Luan JJ, Bach JF, Dardenne M. Characterization of the extracellular matrix‐containing giant perivascular spaces in the NOD mouse thymus. Diabetes. 1993;42:134‐140. [DOI] [PubMed] [Google Scholar]
- 109.Colomb E, Savino W, Wicker L, Peterson L, Dardenne M, Carnaud C. Genetic control of giant perivascular space formation in the thymus of NOD mice. Diabetes. 1996;45:1535‐1540. [DOI] [PubMed] [Google Scholar]
- 110.Mendes‐da‐Cruz DA, Lemos JP, Passos GA, Savino W. Abnormal T‐cell development in the thymus of non‐obese diabetic mice: possible relationship with the pathogenesis of type 1 autoimmune diabetes. Front Endocrinol. 2018;9:381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.James KD, Cosway EJ, Lucas B, et al. Endothelial cells act as gatekeepers for LTβR‐dependent thymocyte emigration. J Exp Med. 2018;215:2984‐2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chai Q, Onder L, Scandella E, et al. Maturation of lymph node fibroblastic reticular cells from myofibroblastic precursors is critical for antiviral immunity. Immunity. 2013;38:1013‐1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pawlowski T, Elliott JD, Loh DY, Staerz UD. Positive selection of T‐lymphocytes on fibroblasts. Nature. 1993;364:642‐645. [DOI] [PubMed] [Google Scholar]
- 114.Hugo P, Kappler JW, McCormack JE, Marrack P. Fibroblasts can induce thymocyte positive selection in vivo. Proc Natl Acad Sci USA. 1993;90(21):10335‐10339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ripen AM, Nitta T, Murata S, Tanaka K, Takahama Y. Ontogeny of thymic cortical epithelial cells expressing the thymoproteasome subunit β5t. Eur J Immunol. 2011;41:1278‐1287. [DOI] [PubMed] [Google Scholar]
- 116.Xing Y, Jameson SC, Hogquist KA. Thymoproteasome subunit‐β5T generates peptide‐MHC complexes specialized for positive selection. Proc Natl Acad Sci USA. 2013;110:6979‐6984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tomaru U, Konno S, Miyajima S, et al. Restricted expression of the thymoproteasome is required for thymic selection and peripheral homeostasis of CD8+ T cells. Cell Rep. 2019;26:639‐651. [DOI] [PubMed] [Google Scholar]
- 118.Nakagawa Y, Ohigashi I, Nitta T, et al. Thymic nurse cells provide microenvironment for secondary T cell receptor α rearrangement in cortical thymocytes. Proc Natl Acad Sci USA. 2012;109:20572‐20577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Boehm T, Scheu S, Pfeffer K, Bleul CC. Thymic medullary epithelial cell differentiation, thymocyte emigration, and the control of autoimmunity require lympho‐epithelial cross talk via LTβR. J Exp Med. 2003;198:757‐769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Martins VC, Boehm T, Bleul CC. LTβR signaling does not regulate Aire‐dependent transcripts in medullary thymic epithelial cells. J Immunol. 2008;181:400‐407. [DOI] [PubMed] [Google Scholar]
- 121.Chin RK, Zhu M, Christiansen PA, et al. Lymphotoxin pathway‐directed, autoimmune regulator‐independent central tolerance to arthritogenic collagen. J Immunol. 2006;177:290‐297. [DOI] [PubMed] [Google Scholar]
- 122.Seach N, Ueno T, Fletcher AL, et al. The lymphotoxin pathway regulates Aire‐independent expression of ectopic genes and chemokines in thymic stromal cells. J Immunol. 2008;180:5384‐5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cosway EJ, Lucas B, James KD, et al. Redefining thymus medulla specialization for central tolerance. J Exp Med. 2017;214:3183‐3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wu W, Shi Y, Xia H, et al. Epithelial LTβR signaling controls the population size of the progenitors of medullary thymic epithelial cells in neonatal mice. Sci Rep. 2017;7:44481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Takaba H, Morishita Y, Tomofuji Y, et al. Fezf2 orchestrates a thymic program of self‐antigen expression for immune tolerance. Cell. 2015;163:975‐987. [DOI] [PubMed] [Google Scholar]
- 126.Miller CN, Proekt I, von Moltke J, et al. Thymic tuft cells promote an IL‐4‐enriched medulla and shape thymocyte development. Nature. 2018;559:627‐631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fletcher AL, Lukacs‐Kornek V, Reynoso ED, et al. Lymph node fibroblastic reticular cells directly present peripheral tissue antigen under steady‐state and inflammatory conditions. J Exp Med. 2010;207:689‐697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dubrot J, Duraes FV, Potin L, et al. Lymph node stromal cells acquire peptide‐MHCII complexes from dendritic cells and induce antigen‐specific CD4+ T cell tolerance. J Exp Med. 2014;211:1153‐1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kedl RM, Lindsay RS, Finlon JM, Lucas ED, Friedman RS, Tamburini BAJ. Migratory dendritic cells acquire and present lymphatic endothelial cell‐archived antigens during lymph node contraction. Nat Commun. 2017;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Perry JSA, Lio CWJ, Kau AL, et al. Distinct contributions of Aire and antigen‐presenting‐cell subsets to the generation of self‐tolerance in the thymus. Immunity. 2014;41:414‐426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Perry JSA, Russler‐Germain EV, Zhou YW, et al. CD36 mediates cell‐surface antigens to promote thymic development of the regulatory T cell receptor repertoire and allo‐tolerance. Immunity. 2018;48:923‐936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sun L, Sun C, Liang Z, et al. FSP1+ fibroblast subpopulation is essential for the maintenance and regeneration of medullary thymic epithelial cells. Sci Rep. 2015;5:14871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Farr A, Nelson A, Hosier S. Characterization of an antigenic determinant preferentially expressed by type‐I epithelial‐cells in the murine thymus. J Histochem Cytochem. 1992;40:651‐664. [DOI] [PubMed] [Google Scholar]
- 134.Farr AG, Berry ML, Kim A, Nelson AJ, Welch MP, Aruffo A. Characterization and cloning of a novel glycoprotein expressed by stromal cells in T‐dependent areas of peripheral lymphoid‐tissues. J Exp Med. 1992;176:1477‐1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Onder L, Mair F, Ledermann B, Becher B, Waisman A, Ludewig B. Alternative NF‐κB signaling regulates mTEC differentiation from podoplanin‐expressing precursors in the cortico‐medullary junction. Eur J Immunol. 2015;45:2218‐2231. [DOI] [PubMed] [Google Scholar]
- 136.Ugorski M, Dziegiel P, Suchanski J. Podoplanin ‐ a small glycoprotein with many faces. Am J Cancer Res. 2016;6:370‐386. [PMC free article] [PubMed] [Google Scholar]
- 137.James KD, Legler DF, Purvanov V, et al. Medullary stromal cells synergize their production and capture of CCL21 for T‐cell emigration from neonatal mouse thymus. Blood Adv. 2021;5:99‐112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hsu HP, Chen YT, Chen YY, et al. Heparan sulfate is essential for thymus growth. J Biol Chem. 2021;296:100419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Taub DD, Longo DL. Insights into thymic aging and regeneration. Immunol Rev. 2005;205:72‐93. [DOI] [PubMed] [Google Scholar]
- 140.Chinn IK, Blackburn CC, Manley NR, Sempowski GD. Changes in primary lymphoid organs with aging. Semin Immunol. 2012;24:309‐320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Baran‐Gale J, Morgan MD, Maio S, et al. Ageing compromises mouse thymus function and remodels epithelial cell differentiation. Elife. 2020;9.e56221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yang HW, Youm YH, Dixit VD. Inhibition of thymic adipogenesis by caloric restriction is coupled with reduction in age‐related thymic involution. J Immunol. 2009;183:3040‐3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Youm YH, Yang HW, Sun YX, et al. Deficient Ghrelin receptor‐mediated signaling compromises thymic stromal cell microenvironment by accelerating thymic adiposity. J Biol Chem. 2009;284:7068‐7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ucar A, Ucar O, Klug P, et al. Adult thymus contains FoxN1− epithelial stem cells that are bipotent for medullary and cortical thymic epithelial lineages. Immunity. 2014;41:257‐269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sheridan JM, Keown A, Policheni A, et al. Thymospheres are formed by mesenchymal cells with the potential to generate adipocytes, but not epithelial cells. Cell Rep. 2017;21:934‐942. [DOI] [PubMed] [Google Scholar]
- 146.Mouiseddine M, Mathieu N, Stefani J, Demarquay C, Bertho JM. Characterization and histological localization of multipotent mesenchymal stromal cells in the human postnatal thymus. Stem Cells Dev. 2008;17:1165‐1174. [DOI] [PubMed] [Google Scholar]
- 147.Schug J, Schuller WP, Kappen C, Salbaum JM, Bucan M, Stoeckert CJ Jr. Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol. 2005;6:R33. [DOI] [PMC free article] [PubMed] [Google Scholar]


