Abstract
NGLY1 deficiency is a rare disorder caused by mutations in the NGLY1 gene which codes for the highly conserved N-glycanase1 (NGLY1). This enzyme functions in cytosolic deglycosylation of N-linked glycoproteins. An induced pluripotent stem cell (iPSC) line was generated from the dermal fibroblasts of a 2-year-old patient carrying compound heterozygous mutations, p.R390P and p.L318P in the NGLY1 gene. This cell-based iPSC disease model provides a resource to study disease pathophysiology and to develop a cell-based disease model for drug development for NGLY1 patients.
1. Resource utility
Currently, there are no effective treatments for NGLY1 deficiency. This patient-specific hiPSC line provides a unique approach for cell-based disease modeling to further investigate the pathogenesis and pathophysiology of the NGLY1 disease.
2. Resource details
N-glycanase 1 (NGLY1) deficiency is a rare autosomal recessive congenital disorder of deglycosylation (CDDG) caused by mutations in the NGLY1 gene. This conserved deglycosylation enzyme is responsible for the cleavage of N-linked glycans in glycoproteins including misfolded glycoproteins in the ERAD pathway [1,2]. Mutations in this gene that cause a loss of NGLY1 function leads to inactivation of Nrf1 and proteasomal dysfunction [1,3]. In this particular compound heterozygote, the missense mutations (L318P and R390P) most likely lead to a misfolding and degradation of NGLY1 as little to no protein can be detected in these cells (data not shown). Patients with this disorder have severe clinical symptoms such as developmental delay, movement disorder, hypotonia and alacrimia. New disease models can aid in the development of novel therapeutics for patients [3].
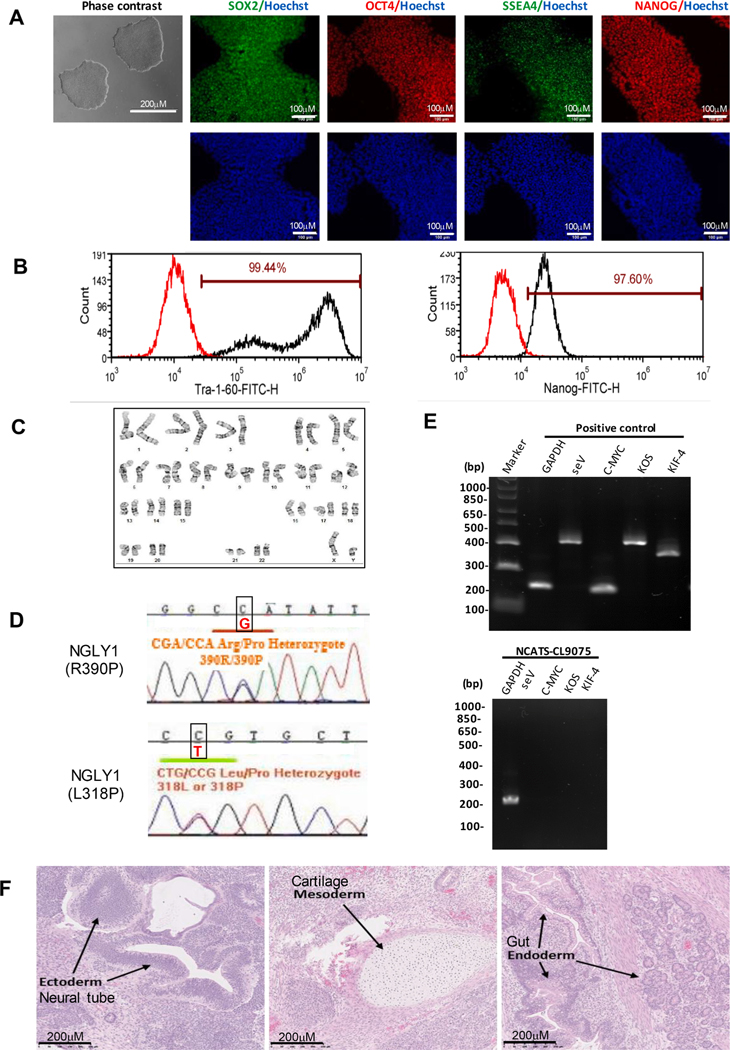
In this study, a patient-specific iPS cell line (NCATS-CL9075) was derived from dermal fibroblasts of a 2-year-old male patient (GM26602, Coriell Institute) carrying heterozygous mutations of p.L318P and p. R390P in the NGLY1 gene. Sendai virus (SeV) vector containing four essential transcription factors OCT3/4, KLF4, SOX2 and c-MYC were used to reprogram the fibroblasts. The iPSC’s were expanded and then validated using cell morphology and pluripotential markers SOX2, OCT4, NANOG, SSEA4 and TRA-1–60 (Fig. 1A). Similarly, the quantitative analysis revealed expression levels of 99.44% and 97.60% for TRA-1–60 and Nanog, respectively (Fig. 1B). G-banded karyotype showed diploid 46, XY without any unusual abnormalities (Fig. 1C). Furthermore, the presence of two mutations, p.L318P and p.R390P, on the NGLY1 gene was confirmed using Sanger sequencing, consistent with the original description on the Coriell Cell Repository website (Fig. 1D). After passage 15, both the reprograming factors and the vector were cleared from NCATS-CL9075 iPSCs (Fig. 1E). Routine mycoplasma checks showed no presence of contamination (Supplemental Fig. S1). The experiment of in vivo teratoma formation validated the pluripotency of the iPSC line through differentiation into three germ layers of ectoderm, mesoderm and endoderm (Fig. 1F). Last, the 18 loci STR DNA profiling analysis of NCATS-CL9075 revealed a match with the parental fibroblasts (data not shown) (see Table 1).
Fig. 1.
Characterization of NCATS-CL9075 iPSC line. A) Left: Phase contrast imaging of NCATS-CL9075 colonies on Matrigel coated plates at passage 12. Right: Immunofluorescent montage of iPSCs marker expression: SOX2, OCT4, TRA-1–60, NANOG, and SSEA4 with Hoechst 33,342 labelled nucleus (in blue). B) Flow cytometry analysis of pluripotency protein markers: TRA-1–60 and NANOG respectively. C) G-banding analysis showed a normal karyotype (46, XY). D) Detection of heterozygous gene mutations p.L318P p.R390P in the NGLY1 gene. E) RT-PCR verification of SeV and factors clearance from reprogrammed cells using sendai virus vector transduced fibroblasts as a positive control. F) Histopathological analysis of teratoma displaying normal ectodermal, mesodermal and endodermal differentiation.
Table 1.
Characterization and validation.
| Classification | Test | Result | Data |
|---|---|---|---|
| Morphology | Photography | Normal | Fig. 1 Panel A |
| Phenotype | Immunocytochemistry | SOX2, OCT4, NANOG, SSEA-4, TRA-1-60 | Fig. 1 Panel A |
| Flow cytometry | TRA-1-60 (99.44%); NANOG (97.60%) |
Fig. 1 Panel B | |
| Genotype | Karyotype (G-banding) and resolution | 46XY Resolution: 450-550 |
Fig. 1 Panel C |
| Identity | Microsatellite PCR (mPCR) | Not performed | N/A |
| STR analysis | 18 Loci tested, all sites matched | Supplemental Fig. S1 | |
| Mutation analysis (IF APPLICABLE) | Sequencing | Heterozygous mutations LEU318PRO (L318P), ARG390PRO (R390P) |
Fig. 1 Panel D |
| Southern Blot OR WGS | N/A | N/A | |
| Microbiology and virology | Mycoplasma | Mycoplasma testing by luminescence. Negative result | Supplemental Fig. S1 |
| Differentiation potential | Teratoma formation | Three germ layer formation: ectoderm (neural tube), mesoderm (cartilage) and endoderm (gut) | Fig. 1 Panel F |
| Donor screening (OPTIONAL) | HIV 1 + 2 Hepatitis B, Hepatitis C | N/A | N/A |
| Genotype additional info (OPTIONAL) | Blood group genotyping | N/A | N/A |
| HLA tissue typing | N/A | N/A |
3. Materials and methods
3.1. Cell culture and reprogramming
The original patient dermal fibroblasts (GM22602) were acquired from Coriell Cell Repositories. The cells were cultured in DMEM medium (Thermo Fisher) supplemented with 10% fetal bovine serum, 100 ug/ml streptomycin, 100 units/ml penicillin,and incubated at 37 °C with 5% CO2 and 5% O2. After a few passages, fibroblasts were reprogrammed into iPSCs using the integration-free CytoTune Sendai viral vector kit (A16517, Thermo Fisher Scientific) following a previous method [4]. The generated iPSCs were cultured on Matrigel (Corning)-coated six-well plate in Stemflex medium (Stem cell technologies) at 5% CO2 and 5% O2. The iPSCs were cultured at 70% confluency using EZ-Lift (Sigma-Aldrich) and then frozen using CryoStor® CS10 (StemCell Technologies) freezing medium.
4. NGLY1 mutation analysis
The genome analysis for the NGLY1 mutations was conducted by Genecopoeia (Rockville, MD, USA). Genomic DNA was extracted from the iPSCs. Primers (Table 2) were designed specifically for the amplification of the two target sites. Sanger sequencing analysis was used to confirm the p.L318p and p.R390P mutations in the patient iPSC line.
Table 2.
Reagents details.
| Antibodies for immunocytochemistry/flow-cytometry | |||
|---|---|---|---|
| Antibody | Dilution | Company Cat # and RRID | |
| Pluripotency Primary Marker |
Mouse anti-SOX2 | 1:50 | R & D systems, Cat# MAB2018, RRID: AB_358009 |
| Pluripotency Primary Marker |
Rabbit anti-NANOG | 1:400 | Cell signaling, Cat# 4903, RRID: AB_10559205 |
| Pluripotency Primary Marker |
Rabbit anti-OCT4 | 1:400 | Thermo Fisher, Cat# A13998, RRID: AB_2534182 |
| Pluripotency Primary Marker |
Mouse anti-SSEA4 | 1:1000 | Cell signaling, Cat# 4755, RRID: AB_1264259 |
| Secondary Antibodies | Donkey anti-Mouse IgG (Alexa Fluor 488) | 1:400 | Thermo Fischer, Cat# A21202, RRID: AB_141607 |
| Secondary Antibodies | Donkey anti-Rabbit IgG (Alexa Fluor 594) | 1:400 | Thermo Fischer, Cat# A21207, RRID: AB_141637 |
| Flow Cytometry Antibodies |
Anti-Tra-1-60-DyLight 488 | 1:50 | Thermo Fischer, Cat# MA1-023-D488X, RRID: AB_2536700 |
| Flow Cytometry Antibodies |
Anti-Nanog-Alexa Fluor 488 | 1:50 | Millipore, Cat# FCABS352A4, RRID: AB_10807973 |
| Flow Cytometry Antibodies |
Mouse-IgM-DyLight 488 | 1:50 | Thermo Fischer, Cat# MA1-194-D488, RRID: AB_2536969 |
| Flow Cytometry Antibodies |
Rabbit IgG-Alexa Fluor 488 | 1:50 | Cell Signaling, Cat# 4340S, RRID: AB_10694568 |
| Primers | |||
| Target | Forward/Reverse primer (5′−3′) | ||
| SeV specific primers (RT-PCR) | SeV /181 bp | F: GGATCACTAGGTGATATCGAGC R: ACCAGACAAGAGTTTAAGAGATATGTATC |
|
| SeV specific primers (RT-PCR) | KOS/528 bp | F: ATGCACCGCTACGACGTGAGCGC R: ACCTTGACAATCCTGATGTGG | |
| SeV specific primers (RT-PCR) | Klf4/410 bp | F: TTCCTGCATGCCAGAGGAGCCC R: AATGTATCGAAGGTGCTCAA | |
| SeV specific primers (RT-PCR) | C-Myc/523 bp | F: TAACTGACTAGCAGGCTTGTCG R: TCCACATACAGTCCTGGATGATGATG |
|
| House-Keeping gene (RT-PCR) | GAPDH/197 bp | F: GGAGCGAGATCCCTCCAAAAT R: GGCTGTTGTCATACTTCTCATGG |
|
| Targeted mutation analysis (PCR) | NGLY1L318P /467 bp | F: GCACCTGTAGTCACAGATACTCTGGAGG R: GGTCAGACTGACAAGGCCAAAAAGTAAC |
|
| Targeted mutation analysis (PCR) | NGLY1R390P/571 bp | F: TATAGTCCCAGCTACTCAGGAGGCTG R: CTTTGAAATGAGACAGTTTAATCCAAAATAACTC |
|
5. Immunocytochemistry
The iPSCs were fixed in 96-well plates with 4% paraformaldehyde for 30 mins at room temperature (RT), permeabilized with 0.3% Triton X-100 in Dulbecco’s phosphate-buffered saline (DPBS) for 15 mins, and washed with DPBS. Next, the cells were blocked using Image-iT™ FX signal enhancer (Thermo Fisher Scientific) for 30 mins at RT. After the removal of the blocking buffer, cells were incubated overnight at 4 °C with primary antibodies (Table 2) diluted in the blocking buffer. Following, the cells were washed with DPBS and incubated with corresponding secondary antibodies conjugated with Alexa Fluor 488 or Alexa Fluor 594 (Table 2) for 1 h at RT. Finally, the nuclei were stained with Hoechst 33,342 for 15 mins and imaged using an INCell Analyzer 2500 imaging system (Cytiva, Marlborough, MA) with 20x objective lens and Texas Red, FITC and DAPI filter sets.
6. G-banding karyotyping and short tandem repeat (STR) analysis
The G-banding karyotype analysis was performed at WiCell Research Institute (Madison, WI, USA) using standard cytogenetic protocols. A total of 20 cells at metaphase were examined and analyzed to check for potential clonal abnormalities.
The STR analysis for both the fibroblasts and iPSCs were conducted at the Johns Hopkins University Genetic Resources Core Facility using the Promega PowerPlex 18D kit. The PCR product was electrophoresed on an ABI Prism® 3730xl Genetic Analyzer and analyzed using GeneMapper® v 4.0 software (Applied Biosystems).
7. Flow cytometry analysis
The iPSCs were detached using TrypLE Express (Thermo Fisher Scientific) and fixed with 4% paraformaldehyde for 10 min at RT. Following, the cells were washed with DPBS and permeabilized with 0.2% Tween-20 in DPBS for 10 min at RT. They were then stained with fluorophore-conjugated antibodies (Table 2) for 1 h at 4 °C on a shaker. After incubation, cells were analyzed on a BD Accuri™ C6 Flow Cytometry system (BD Biosciences).
8. Sendai virus detection
RNA extraction, cDNA synthesis and amplification were performed on the iPSCs following a previously published protocol [5]. Human fibroblasts (GM05659, Coriell Institute) transfected with Sendai virus served as the positive control. The final PCR products were loaded on E-Gel® 1.2% with SYBR Safe™ gel, and imaged by G: Box Chemi-XX6 gel doc system (Syngene).
9. Teratoma formation assay
Patient iPSCs cultured in 6-well plates were dissociated using EZ-Lift (Sigma-Aldrich). Approximately 1 × 107 dissociated cells were resuspended in 400 μl culture medium supplemented with 25 mM HEPES (pH 7.4) and chilled on ice. Next, 200 μl of cold Matrigel (Corning, 354277) was added to the cells. This mixture was injected subcutaneously into NSG mice (JAX No. 005557) at 150 μl per injection site. Visible tumors were removed 6–8 weeks post injection, immediately fixed in 10% neutral buffered formalin and embedded in paraffin. Finally, the tissue was stained with hematoxylin and eosin for visualization of teratoma formation.
10. Mycoplasma detection
Mycoplasma levels were detected and analyzed using the Lonza MycoAlert Kit following manufacturer’s instruction (Ratio B/A > 1.2 mycoplasma positive; 0.9–1.2 ambiguous result; <0.9 mycoplasma negative).
Supplementary Material
Resource table
| Unique stem cell line identifier | NCATS-CL39075 |
| Alternative name(s) of stem cell line | HT594C |
| Institution | National Institutes of Health National Center for Advancing Translational Sciences Bethesda, Maryland, USA |
| Contact information of distributor | Dr. Wei Zheng Wei.Zheng@nih.gov |
| Type of cell line | iPSC |
| Origin | Human |
| Additional origin info | Age: 2-year-old Sex: Male Ethnicity: Caucasian |
| Cell Source | Skin fibroblasts |
| Clonality | Clonal |
| Method of reprogramming | Integration-free Sendai viral vectors |
| Genetic Modification | No |
| Type of Modification | Hereditary |
| Associated disease | NGLY1 Deficiency |
| Gene/Locus | NGLY1 gene, located at 3p24.2, mutations L318P and R390P |
| Method of modification | N/A |
| Name of transgene or resistance | N/A |
| Inducible/constitutive system | N/A |
| Date archived/stock date | 4/20/2020 |
| Cell line repository/bank | N/A |
| Ethical approval | NIGMS Informed Consent Form was obtained from patient at time of sample submission. Confidentiality Certificate: CC-GM-15-004 |
Acknowledgment
This work was supported by the Intramural Research Programs of the National Center for Advancing Translational Sciences, National Institutes of Health, and was a CRADA collaboration between NCATS, CDG CARE (cdgcare.org), and Travere Therapeutics.
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scr.2021.102400.
References
- [1].Tambe MA, Ng BG, Freeze HH, 2019. N-Glycanase 1 Transcriptionally Regulates Aquaporins Independent of Its Enzymatic Activity. Cell Reports 29 (13), 4620–4631.e4. 10.1016/j.celrep.2019.11.097. [DOI] [PubMed] [Google Scholar]
- [2].Han SY, Pandey A, Moore T, Galeone A, Duraine L, Cowan TM, Jafar-Nejad H, Hietakangas V, 2020. A conserved role for AMP-activated protein kinase in NGLY1 deficiency. PLOS Genetics 16 (12), e1009258. 10.1371/journal.pgen.1009258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Tomlin FM, Gerling-Driessen UIM, Liu Y-C, Flynn RA, Vangala JR, Lentz CS, … Bertozzi CR (2017). Inhibition of NGLY1 Inactivates the Transcription Factor Nrf1 and Potentiates Proteasome Inhibitor Cytotoxicity. ACS Central Science, 3(11), 1143–1155. 10.1021/acscentsci.7b00224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Beers J, Linask KL, Chen JA, Siniscalchi LI, Lin Y, Zheng W, Rao M, Chen G, 2015. A cost-effective and efficient reprogramming platform for large-scale production of integration-free human induced pluripotent stem cells in chemically defined culture. Sci. Rep 5 (1) 10.1038/srep11319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Li R, Pradhan M, Xu M, Baskfield A, Farkhondeh A, Cheng Y-S, Zheng W, 2019. Generation of an induced pluripotent stem cell line (TRNDi002-B) from a patient carrying compound heterozygous p. Q208X and p.G310G mutations in the NGLY1 gene. Stem Cell Research 34, 101362. 10.1016/j.scr.2018.101362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.