Abstract
Background
Antiretroviral therapy (ART) can reduce opportunistic infections and mortality rates among individuals infected with human immunodeficiency virus (HIV); however, some HIV-infected individuals exhibit poor immune recovery after ART. Hence, we explored the association between metabolome profiles and immune recovery in HIV-infected individuals following ART.
Methods
An untargeted metabolomics approach was used to analyze plasma samples from 18 HIV-negative individuals and 20 HIV-infected individuals, including 10 immunological non-responders (INR, CD4+ T cell rise < 100 cells/μl) and 10 immunological responders (IR, CD4+ T cell rise > 300 cells/μl) after 2 years of ART. These individuals were followed for the next 6 years and viral loads and CD4+ T cell count were measured regularly. Orthogonal projection on latent structures discriminant analysis (OPLS-DA), ANOVA, correlation, receiver operating characteristic (ROC), and survival analyses were used for selection of discriminant metabolites.
Results
Eighteen lipid metabolites were identified which could distinguish among control, INR, and IR groups. Among them, myristoylcarnitine (MC), palmitoylcarnitine (PC), stearoylcarnitine (SC), and oleoylcarnitine (OC) were significantly elevated in INR plasma samples compared with those from the IR and control groups and were negatively associated with CD4+ T cell count. Additionally, ROC analysis using a combination of MC, PC, SC, and OC had high sensitivity and specificity for differentiating INR from IR (AUC = 0.94). Finally, survival analysis for the combination of MC, PC, SC, and OC demonstrated that it could predict CD4+ T cell count in patients undergoing long-term ART.
Conclusions
High levels of lipid metabolites, MC, PC, SC, and OC are associated with poor immune recovery in patients receiving ART and these data provide potential new insights into immune recovery mechanisms.
Keywords: HIV, ART, Immune recovery, Metabolomics, Biomarkers
Background
Antiretroviral therapy (ART) can suppress plasma viral RNA to undetectable levels, promote immune recovery, and efficiently reduce opportunistic infections and mortality rates among individuals infected with human immunodeficiency virus (HIV) [1–4]; however, some HIV-infected individuals, referred to as “immunological non-responders” (INR) have poor immune recovery after ART, and are at increased risk of rapid disease progression and death [5]. Poor immune recovery has been associated with sex, baseline CD4+ and CD8+ T cell count, and HIV RNA levels, as well as related to alterations in T cell phenotype, function, and PD-1 expression. Further, recent reports have demonstrated that various metabolites, such as serine and polysaccharides, can affect the proliferation and immune function of CD4+ T cells in healthy people [6–9].
Metabolomics is a powerful discovery tool for exploration of biomarkers and potential disease mechanisms, in conditions such as liver disease, cardiovascular disease, prostate cancer, obesity, and diabetes. A recent study measured lipoproteins by nuclear magnetic resonance and found that HIV-infected patients with high levels of large HDL particles, HDL cholesterol, and larger sized LDL particles had superior immunological recovery after treatment [10]. In addition, a Spanish research team investigated the metabolic characteristics of gut bacteria in immunological responders (IR) and INR, demonstrating that the gut microbiome interacts with immune recovery [11]. Nevertheless, limited information is available regarding the association of plasma metabolite profiles with immune recovery in HIV-infected individuals following ART. Here, we attempted to discover associations between metabolites and immune recovery, and to identify biomarkers which could predict CD4+ T cell count during continuous treatment and after ART in HIV-infected individuals, using a metabolomic approach.
In this study, we describe the plasma metabolite profiles of controls, IR, and INR, detected by ultra-high-performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS). We demonstrate associations of myristoylcarnitine (MC), palmitoylcarnitine (PC), stearoylcarnitine (SC), and oleoylcarnitine (OC) with poor immune recovery in individuals receiving ART and found that levels of these lipids could predict CD4+ T cell count in later treatment.
Methods
Study subjects
All subjects enrolled were men who have sex with men recruited from the First Affiliated Hospital of China Medical University and provided written informed consent. The study protocol was approved by the Research and Ethics Committee of The First Affiliated Hospital of China Medical University, in compliance with the Declaration of Helsinki. Twenty ART-treated HIV-infected individuals, including 10 INR and 10 IR, where INR and IR were defined by a CD4+ T cell count rise after 2 years of < 100 or > 300 cells/μl of ART (viral loads < 20 copies/ml) were collected from the Red Ribbon Clinic. Treatment regimens comprised nucleoside reverse transcriptase inhibitors (NRTI) plus non-NRTI (TDF/AZT + 3TC + EFV/NVP). Subjects selected for inclusion did not have any laboratory abnormalities related to glucose, or kidney or liver function measurements, and were not co-infected with hepatitis, tuberculosis, syphilis, or other infectious diseases. In addition, 18 HIV-negative individuals were used as healthy controls. Age and body mass index (BMI) did not differ significantly among three groups. Participant characteristics are presented in Table 1.
Table 1.
Characteristics of subjects enrolled in this study
| Characteristics | Controls | INR | IR | p-value (IR vs INR) |
|---|---|---|---|---|
| Number of subjects | 18 | 10 | 10 | |
| Male (%) | 100 | 100 | 100 | |
| Age (years) | 37 (35, 38) | 33 (29, 49) | 39 (30, 43) | > 0.05 |
| Body mass index (kg/m2) | 23 (20.3, 23.0) | 21.7 (20.6, 22.8) | 21.5 (20.9, 21.9) | > 0.05 |
| Glucose (mmol/L) | NA | 5.3 (4.5, 5.6) | 5.0 (4.9, 5.4) | > 0.05 |
| AST (U/L) | NA | 26.5 (23.8, 30.2) | 24.6 (23.0, 37.0) | > 0.05 |
| ALT (U/L) | NA | 26.5 (20.3, 35.0) | 31.0 (19.0, 50.0) | > 0.05 |
| Triglyceride (mmol/L) | NA | 1.23 (1.00, 1.33) | 1.28 (1.09, 1.79) | > 0.05 |
| Total cholesterol (mmol/L) | NA | 3.44 (3.36, 3.90) | 4.06 (3.56, 4.28) | > 0.05 |
| Viral load (copies/mL) | NA | < 20 | < 20 | |
| CD4+ T cell count (cells/μL) | ||||
| Baseline | NA | 162 (95, 179) | 97 (79, 154) | > 0.05 |
| At 2 years of ART | NA | 219 (163, 234) | 456 (383, 510) | < 0.05 |
| Drug regimen | ||||
| EFV + 3TC + AZT | NA | 2 | 1 | > 0.05 |
| EFV + 3TC + TDF | NA | 1 | 1 | |
| NVP + 3TC + AZT | NA | 7 | 8 | |
Data are expressed as the median (interquartile range), unless otherwise stated. NA not available, INR immunological non-responders, IR immunological responders, AST aspartate aminotransferase, ALT alanine aminotransferase
HIV viral load test
Plasma samples were prepared and analyzed using the COBAS® TaqMan® system with COBAS® AmpliPrep/COBAS® TaqMan® HIV-1 Test Kits v2.0 (Roche Molecular Systems, USA). The upper and lower detection limits of the kit were 107 copies/mL and 20 copies/mL, respectively.
Detection of CD4+ T cell count
Fresh anticoagulant blood was stained with TriTEST anti-CD4-FITC/CD8-PE/CD3-PerCP reagent in Trucount tubes (BD Biosciences, San Jose, CA, USA). Then, hemolysin was added to lyse the red blood cells. CD4+ T cells were detected and enumerated using a FACS Calibur flow cytometer (BD Biosciences, San Jose, CA, USA).
Metabolomic profiling
Venous blood samples were drawn after an overnight fasting. Plasma samples were obtained and assigned a unique identifier using a laboratory information management system, and kept at -80℃ until processed. An automated MicroLab STAR® system (Hamilton Company, Reno, NV, U.S.A) was used to prepare samples, as described previously [12, 13]. Briefly, several recovery standards were added before extraction for quality control purposes; proteins were precipitated in methanol by vigorous shaking, followed by centrifugation to remove proteins; the resulting extracts were divided into five parts, of which four were analyzed using different methods and one was retained as a backup. Samples were placed momently on a TurboVap® (Zymark, Westborough, MA, USA) to deplete the organic solvent, then stored overnight in nitrogen before analysis.
Detailed descriptions of non-targeted metabolomics analysis can be found in published work [13]. In summary, sample extracts were dried and resuspended in specific solvents for each of four methods: one aliquot was analyzed by hydrophilic interaction liquid chromatography/UPLC-MS/MS, with negative ion mode electrospray ionization (ESI); two aliquots were analyzed using two separate reverse phases (RP)/UPLC-MS/MS methods, with positive ion mode ESI; and the final aliquot was analyzed by RP/UPLC-MS/MS with negative ion mode ESI. All methods were conducted using a Waters ACQUITY UPLC and a Thermo Scientific Q-Exactive high resolution/accurate mass spectrometer, which were coupled with a heated electrospray ionization (HESI-II) source and an Orbitrap mass analyzer, with a working mass resolution of 35,000. The scanning range varied slightly between different methods but covered 70–1000 m/z. Raw data files were extracted and archived.
Data extraction and compound identification procedures were as previously described [14]. Briefly, Metabolon software and hardware were used to extract the original data, identify peak values, and carry out quality control processes, while proprietary visualization and interpretation software were used to confirm peak identification.
Data processing, bioinformatics, and statistical analysis
For metabolomics data, variables with > 20% missing values were excluded [15]. For the remaining variables, missing values were replaced with the lower limit of detection. Metabolite data detected across multiple days were normalized by setting the medians to equal one and then normalizing each data point proportionately. The multivariate statistical methods, orthogonal partial least square discriminant analysis (OPLS-DA) was conducted using SIMCA 14.1 (Umetrics, Sweden). One-way ANOVA with Fisher’s LSD test was performed for comparisons of variables among the three groups, and the Benjamini and Hochberg false discovery rate (FDR) method was used for corrections, with a cut-off level of 5%. A heatmap of the 30 metabolites that differed significantly among the three groups was produced using MultiExperiment Viewer 4.9.0 [16]. Fold-change values of A/B were calculated as the ratios of mean levels of metabolites in group A to those in group B. Metabolite Set Enrichment Analysis were performed using MetaboAnalyst 4.0 [17].Correlation analysis was performed using the Spearman correlation test. Receiver operating characteristic (ROC) analysis was used to assess ability to predict immune recovery. For ROC analysis of the four-metabolite combination, predicted probability was calculated for inclusion in the ROC analysis by binary logistic regression. Then, the best diagnostic thresholds for metabolites, with the highest sensitivity and specificity values, were selected and used to regroup the 20 HIV-infected subjects, followed by Kaplan–Meier survival analysis plus a log-rank test to evaluate the influence of potential biomarkers on CD4+ T cell count. Statistical analyses were conducted using R (http://cran.r-project.org/) and GraphPad Prism 8.0.2. Two-sided test p values < 0.05 were considered significant.
Results
Overall metabolic signatures of plasma samples
An untargeted metabolomics assay was performed to detect plasma metabolites in controls, IR and INR after 2 years of ART. A total of 330 known compounds were identified. To reduce noise in the analysis, variables with > 20% missing values and xenobiotics were excluded. The remaining 241 metabolites included 125 lipids (52%), 68 amino acids (28%), 7 peptides (3%), 14 carbohydrates (6%), 12 cofactors and vitamins (5%), 9 nucleotides (4%), and 6 energy metabolites (2%).
OPLS-DA, a supervised clustering method, was applied to determine which metabolites significantly contributed to the observed separation among groups. All metabolites were used to build OPLS-DA models and model parameters are presented in Table 2. The score plot showed good separation among the three groups (INR, IR, and controls), with R2Y = 0.782, and Q2 (cum) = 0.184, indicating a good capacity for fitting and prediction (Fig. 1a). Since OPLS-DA may lead to over-fitted models, a permutation test (999 iterations) was performed and the Q2 intercept value was -0.294, indicating that the model was not overfitted.
Table 2.
Parameters for OPLS-DA models
| Model | OPLS-DA models | Permutation | ||||
|---|---|---|---|---|---|---|
| Components# | R2X (cum) | R2Y (cum) | Q2 (cum) | R2 intercept | Q2 intercept | |
| Controls vs IR vs INR | 2P + 1O | 0.250 | 0.782 | 0.184 | 0.513 | − 0.294 |
| Controls vs INR | 1P + 4O | 0.408 | 0.999 | 0.763 | 0.994 | − 0.249 |
| Controls vs IR | 1P + 5O | 0.421 | 1.000 | 0.462 | 0.999 | − 0.142 |
| IR vs INR | 1P + 1O | 0.134 | 0.601 | 0.076 | 0.936 | − 0.017 |
#P predictive, O orthogonal
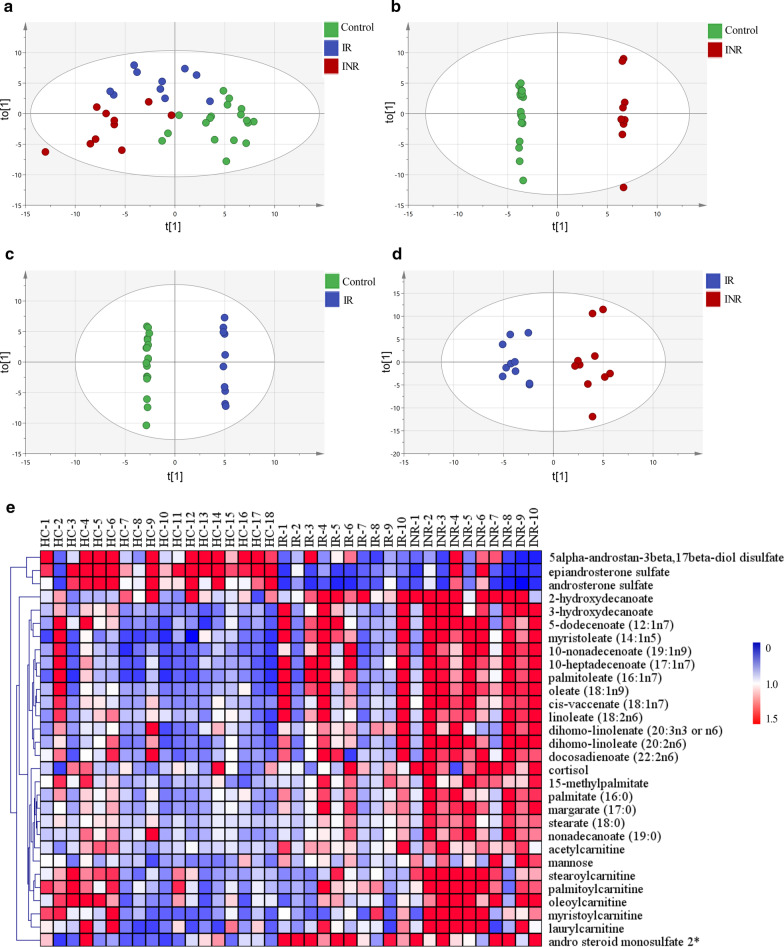
Fig. 1.
Metabolite profiles in the control, INR, and IR groups. a–d OPLS-DA score plots derived from untargeted metabolism profiles of plasma samples. e Hierarchical cluster analysis and heat map visualization of the top 30 variables ranked by ANOVA
Furthermore, OPLS-DA models for binary group comparisons were generated and variable influence in projection (VIP) values were calculated. For control and INR samples, the model had 1 predictive and 4 orthogonal components, and its validation parameters were R2Y = 0.999, and Q2 (cum) = 0.763 (Fig. 1b). For control and IR samples, the model had 1 predictive and 5 orthogonal components, and its validation parameters were R2Y = 1.000, and Q2 (cum) = 0.462 (Fig. 1c). For IR and INR samples, the model had 1 predictive and 1 orthogonal component, and its validation parameters were R2Y = 0.601, and Q2 (cum) = 0.076 (Fig. 1d). In all these models, permutation tests (999 iterations) were conducted, and all resulting Q2 intercept values were negative, indicating that the models were not overfitted. VIP scores > 2 were considered to indicate variables contributing to the separation of groups.
Identification of metabolites which could distinguish IR from INR metabolomics profiles
To select potential biomarkers of immune recovery, metabolites were identified which differed significantly among the three groups and between IR and INR with a p value < 0.05 (FDR < 5%) based on the results of ANOVA. We found that the levels of 30 metabolites differed significantly among the three groups: most were lipids, all differed between INR and control samples, 14 differed between IR and control samples, and 16 between IR and INR. The heatmap depicts levels of these 30 metabolites and shows two main variable clusters (Fig. 1e). The levels of some steroids were decreased in IR and INR compared with control samples, while high levels of fatty acids and acylcarnitines were detected in INR samples compared with control and IR samples.
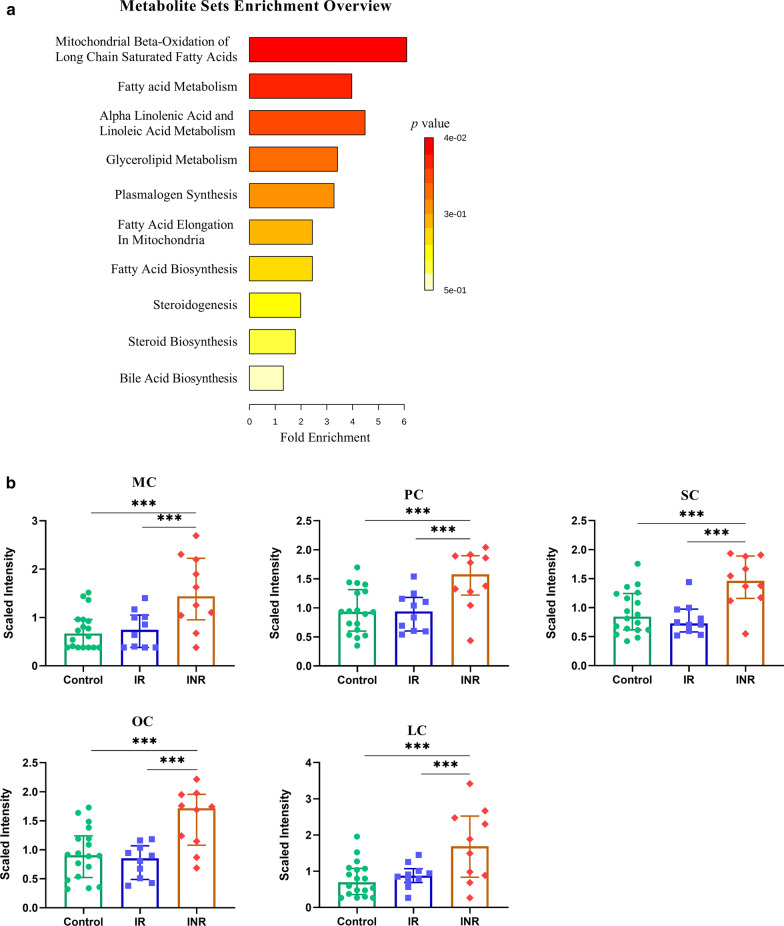
Furthermore, fold-change values (calculated as the ratio of the means between two groups) > 1.5 were taken into consideration. Using these screening criteria, 18 metabolites were identified; all were lipids and were elevated in INR relative to IR and control samples (Table 3). Acylcarnitines, including MC, LC, OC, SC, and PC had both high fold change and VIP values which could distinguish INR from IR and control samples. In addition, various fatty acids including polyunsaturated fatty acids and long chain fatty acids, also made contributions to distinguishing INR from controls. Metabolite set enrichment analysis showed that mitochondrial beta-oxidation of long chain saturated fatty acids and fatty acid metabolism were the main impaired pathways, which also suggest that acylcarnitines were the main differentiators between IR and INR (Fig. 2a). The levels of SC, MC, OC, PC and LC were not significantly different between control and IR samples, which suggest the mitochondrial beta-oxidation of IR samples were similar to control samples but were seriously impaired in INR samples (Fig. 2b).
Table 3.
Metabolites with differing plasma levels among groups
| Name | Pathway | Sub pathway | IR/control | INR/control | INR/IR | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| VIP | p value | FC | VIP | p value | FC | VIP | p value | FC | |||
| Myristoylcarnitine (MC) | Lipid | Fatty Acid Metabolism (Acyl Carnitine) | 0.11 | 9.13E−01 | 1.06 | 1.81 | 4.12E−04 | 2.11 | 2.13 | 1.97E−03 | 1.99 |
| Laurylcarnitine (LC) | Lipid | Fatty Acid Metabolism (Acyl Carnitine) | 0.46 | 6.93E−01 | 1.2 | 1.69 | 7.29E−04 | 2.32 | 2.27 | 6.16E−03 | 1.94 |
| Oleoylcarnitine (OC) | Lipid | Fatty Acid Metabolism (Acyl Carnitine) | 0.68 | 4.34E−01 | 0.91 | 1.64 | 1.29E−03 | 1.74 | 2.86 | 5.79E−04 | 1.9 |
| Stearoylcarnitine (SC) | Lipid | Fatty Acid Metabolism (Acyl Carnitine) | 0.71 | 4.14E−01 | 0.9 | 1.71 | 8.77E−04 | 1.63 | 2.80 | 3.73E−04 | 1.81 |
| Palmitoylcarnitine (PC) | Lipid | Fatty Acid Metabolism (Acyl Carnitine) | 0.09 | 9.21E−01 | 1.03 | 1.64 | 1.83E−03 | 1.65 | 2.54 | 4.21E−03 | 1.6 |
| 2-hydroxydecanoate | Lipid | Fatty Acid, Monohydroxy | 2.07 | 8.42E−03 | 0.54 | 1.62 | 5.35E−03 | 0.52 | 0.48 | 8.77E−01 | 0.95 |
| 10-nonadecenoate (19:1n9) | Lipid | Long Chain Fatty Acid | 1.92 | 1.53E−02 | 1.88 | 2.33 | 1.00E−05 | 2.70 | 1.55 | 2.76E−02 | 1.44 |
| Cis-vaccenate (18:1n7) | Lipid | Long Chain Fatty Acid | 1.64 | 4.05E−02 | 1.36 | 2.34 | 4.47E−06 | 1.90 | 1.95 | 6.33E−03 | 1.4 |
| Oleate (18:1n9) | Lipid | Long Chain Fatty Acid | 1.71 | 2.08E−02 | 1.56 | 2.37 | 1.80E−05 | 2.13 | 1.63 | 3.15E−02 | 1.37 |
| Margarate (17:0) | Lipid | Long Chain Fatty Acid | 2.03 | 1.24E−02 | 1.49 | 2.32 | 3.87E−06 | 1.92 | 1.55 | 1.73E−02 | 1.28 |
| Palmitate (16:0) | Lipid | Long Chain Fatty Acid | 1.61 | 5.35E−02 | 1.38 | 2.07 | 5.20E−05 | 1.76 | 1.48 | 2.74E−02 | 1.27 |
| Stearate (18:0) | Lipid | Long Chain Fatty Acid | 1.44 | 1.14E−01 | 1.23 | 2.19 | 8.39E−06 | 1.51 | 1.96 | 3.19E−03 | 1.23 |
| 10-heptadecenoate (17:1n7) | Lipid | Long Chain Fatty Acid | 2.18 | 3.42E−03 | 2.16 | 2.21 | 7.44E−05 | 2.62 | 0.95 | 2.43E−01 | 1.21 |
| Palmitoleate (16:1n7) | Lipid | Long Chain Fatty Acid | 2.09 | 2.64E−03 | 2.35 | 2.11 | 1.61E−04 | 2.73 | 0.80 | 3.88E−01 | 1.16 |
| Dihomo-linoleate (20:2n6) | Lipid | Polyunsaturated Fatty Acid (n3 and n6) | 1.68 | 1.06E−01 | 1.53 | 2.11 | 2.27E−05 | 2.45 | 1.63 | 7.34E−03 | 1.6 |
| Epiandrosterone sulfate | Lipid | Steroid | 2.81 | 2.58E−05 | 0.28 | 1.80 | 2.07E−04 | 0.40 | 0.77 | 5.41E−01 | 1.41 |
| Androsterone sulfate | Lipid | Steroid | 2.36 | 4.13E−04 | 0.27 | 1.58 | 1.41E−03 | 0.36 | 0.65 | 7.03E−01 | 1.35 |
| Andro steroid monosulfate | Lipid | Steroid | 2.52 | 1.26E−03 | 2.84 | 1.94 | 1.02E−03 | 2.88 | 0.12 | 9.48E−01 | 1.01 |
Features that meet all following conditions are highlighted using bold text: variable important in projection (VIP) values > 2, p values < 0.05 (FDR < 5%), or fold-change (FC) values > 1.5 or < 0.67
Fig. 2.
Acylcarnitines were identified as the metabolites exhibiting the main differences between IR and INR. a Metabolite Set Enrichment Analysis using 18 metabolites which could distinguish controls, IR, and INR. b Box-and-whisker plots of the metabolite levels in control (n = 18), IR (n = 10), and INR (n = 10) samples. ANOVA with Fisher’s LSD test (FDR < 5%) was used to compare the three groups. *p < 0.05, **p < 0.01, ***p < 0.001
Acylcarnitine levels were negatively correlated with CD4+ T cell count
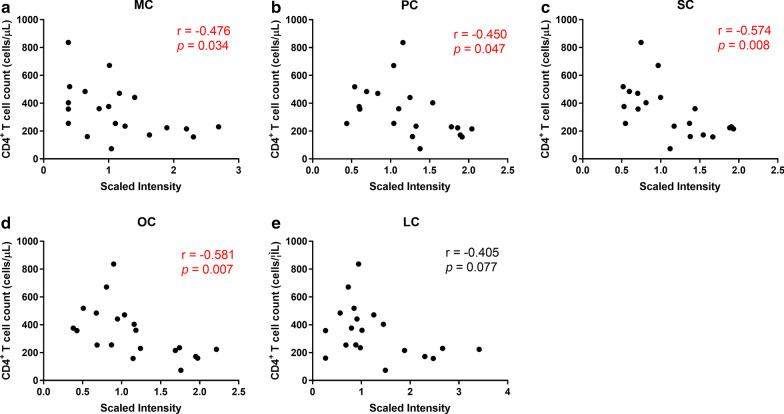
Based on our finding that acylcarnitines were the main differentiators between INR and IR, we further explored the associations of acylcarnitines with immune recovery. We found that CD4+ T cell count in HIV-infected subjects were negatively correlated with MC (p = 0.034, r = -0.476; Fig. 3a), PC (p = 0.047, r = -0.45; Fig. 3b), SC (p = 0.008, r = -0.574; Fig. 3c), and OC (p = 0.007, r = -0.581; Fig. 3d), while LC was not significantly associated with CD4+ T cell count (p = 0.077, r = -0.406; Fig. 3e). These results indicate that elevated levels of MC, PC, SC, and OC in INR may be associated with decreased CD4+ T cell count.
Fig. 3.
Correlations between metabolite levels and CD4+ T cell count. a–e The Spearman rank test was used to evaluate the correlation between acylcarnitines and CD4+ T cell count
Levels of MC, PC, SC, and OC could be used to evaluate immune recovery
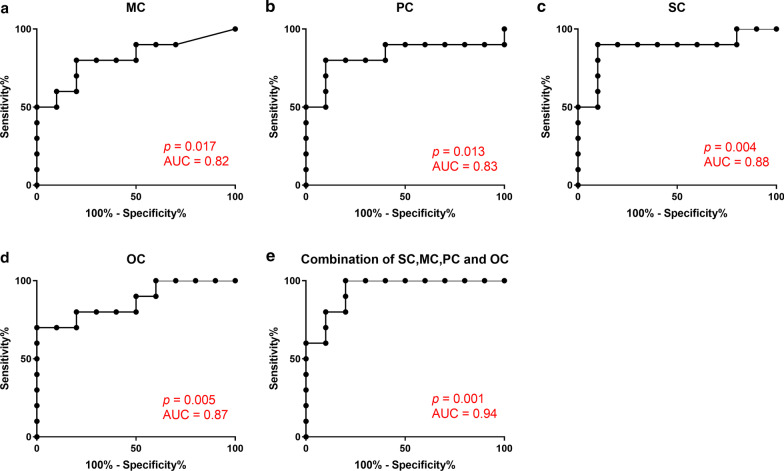
Given the negative correlations between levels of acylcarnitines and CD4+ T cell count, we considered that these acylcarnitines were potential biomarkers for evaluation of immune recovery; therefore, we used ROC curves to evaluate the clinical value of these metabolites. The area under the curve (AUC) values for MC (p = 0.017; Fig. 4a), PC (p = 0.013; Fig. 4b), SC (p = 0.004; Fig. 4c), and OC (p = 0.005; Fig. 4d) were 0.82, 0.83, 0.88, and 0.87, respectively, indicating that these metabolites may be useful for evaluation of immune recovery. Next, we conducted ROC analysis of the four metabolites combined and the resulting AUC value was 0.94, suggesting that this combination of metabolites may be more meaningful for evaluation of immune recovery (p = 0.001; Fig. 4e).
Fig. 4.
ROC analysis of acylcarnitine levels to distinguish INR and IR groups. a–e ROC analysis of the levels of MC, PC, SC, OC, or the combination of MC, PC, SC, and OC
Levels of MC, PC, SC, and OC could predict CD4+ T cell count during long term ART
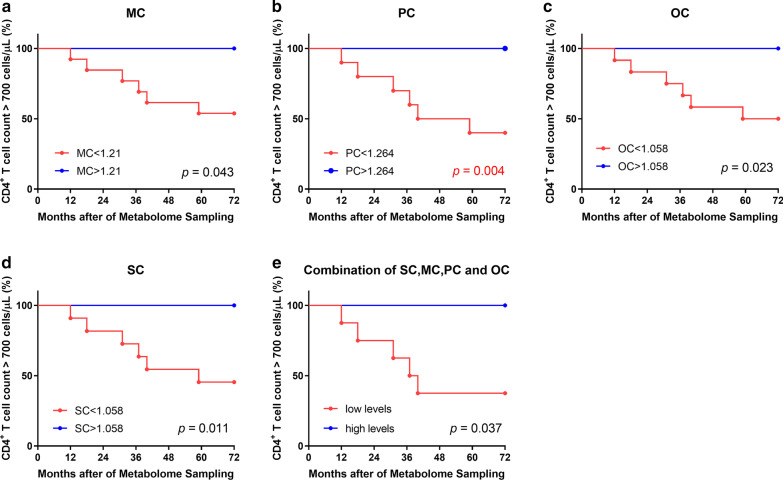
To further validate these findings, we determined the cut-off point that generated from the largest sum of sensitivity plus specificity values in the ROC analysis, to regroup the 20 HIV-infected individuals, based on CD4+ T cell count data collected within 6 years after metabolome sampling (at 2 years after ART), and set CD4+ T cell count reaching 700 cells/μL as the outcome to conduct survival analysis. The results showed that MC (p = 0.043; Fig. 5a), PC (p = 0.004; Fig. 5b), OC (p = 0.023; Fig. 5c), and SC (p = 0.011; Fig. 5d) could help to predict CD4+ T cell count within 6 years. Moreover, we regrouped the samples according to levels of MC, PC, SC, and OC: samples with all metabolites at high levels were classified into the ‘high levels’ group, while samples with all metabolites at low levels were classified into the ‘low levels’ group. The results showed that analysis combining MC, PC, SC, and OC could predict CD4+ T cell count during long term ART (p = 0.037; Fig. 5e).
Fig. 5.
Analysis of the ability of acylcarnitine levels to predict CD4+ T cell count. a–e Survival analysis of levels of MC, PC, OC, SC, or the combination of MC, PC, SC, and OC. Subjects were divided into high-level and low-level groups, according to the cutoff points with the largest sum of sensitivity plus specificity in ROC curves. CD4+ T cell count of > 700 cells/μL were considered the end point. Kaplan–Meier survival curves and the log-rank test were used to assess predictive ability
Discussion
In this investigation, we analyzed metabolite profiles in plasma samples from controls, IR, and INR after 2 years of ART. We identified 18 metabolites that could distinguish the control, INR, and IR groups. The acylcarnitines, MC, PC, OC, and SC, were identified as the metabolites with the main profile differences between IR and INR and were associated with poor immune recovery of HIV-infected individuals receiving ART.
Our results indicate that lipid metabolites are the main differentiators that distinguish control, IR, and INR groups. HIV can disturb lipid and amino acid metabolism [18], while our data show that levels of lipid metabolites in HIV-infected patients are still higher than that of HIV- negative individuals. In addition, some ART drugs can affect lipid metabolism, probably by inhibiting the degradation of adipogenic transcription factors, such as SREBPs and PPAR-c [19–21]. All HIV-infected subjects in our study received NRTI combined with non-NRTI therapy for two years, and their viral loads were under the detection limit; therefore, the effect of the virus and drugs can be discounted, and the main cause of differences in lipid profiles can be attributed to immune recovery, in this study.
Further, we found that the fatty acid metabolites, acylcarnitines, were clearly elevated in INR. Acylcarnitines are synthesized by the combination of carnitine and acyl-CoA (activated fatty acid), and transported into the mitochondria by carnitine-acylcarnitine translocase [22]. Acylcarnitines are reconverted to acyl-CoAs with the help of carnitine-palmitoyltransferase-2 (CPT-2), and acyl-CoAs undergo β-oxidation in the mitochondrial matrix [23]. Increased levels of acylcarnitines are associated with reduced CPT-2 activity [24, 25], hence, the increased levels of acylcarnitines identified in the INR group in our study may indicate mitochondrial dysfunctional; for example, impaired translocase activity, which could result in the accumulation of acylcarnitines.
The association of high levels of acylcarnitines with poor immune recovery detected in this study may have been caused by CD4+ T cell apoptosis. PC can stimulate the activity of caspase-3/7 and caspase-8 to induce apoptosis of murine CD4+CD25+ T cells and the Jurkat cell line [26, 27]. Further, the combination of PC and carnitine can induce apoptosis of a colon cancer cell line, with O2− generation during β-oxidation in the mitochondria [28]. Therefore, we speculate that PC may induce apoptosis of CD4+ T cells, by being absorbed by cells and then transported into mitochondria, where the increase in substrate results in O2− accumulation, potentially causing apoptosis. In addition, there is evidence that MC or PC can activate JNK and ERK in proinflammation and stress signaling pathways in some murine cell lines [29, 30]. Acylcarnitines can also disrupt membrane barriers to solutes, leading to membrane solubilization [31, 32]. Hence, we hypothesize that long chain acylcarnitines may induce apoptosis of CD4+ T cells by promoting β-oxidation, leading to elevated oxidative stress, or membrane disruption; however, the specific mechanism underlying the induction of CD4+ T cell apoptosis by acylcarnitines remains to be explored.
It was reported that the accumulation of long-chain acylcarnitines in plasma were associated with liver or renal fibrosis in non-HIV infection[33–35], while the fibrosis was contributed to the loss of CD4+ T cells in HIV infection [36, 37]. Therefore, we postulated that acylcarnitines might associated with lymph node fibrosis and lead to poor immune recovery.
Our ROC analysis results demonstrate that MC, PC, SC, and OC can distinguish INR from IR, and the combination of these four metabolites exhibited higher sensitivity and specificity than any of them individually. Survival analysis also indicated that the combination of these four metabolites could be biomarkers for prediction of CD4+ T cell count to determine immune recovery in patients undergoing long term ART. Hence MC, PC, SC, and OC are important biomarkers of immune recovery and potential targets for immune intervention and treatments.
This study has some limitations. Although the age and BMI of the control, IR, and INR groups were matched, the data of lifestyle factors, diets and dietary supplements unavailable may have affected the results. We identified four metabolites which could be used to clearly distinguish INR from IR and predict CD4+ T cell count in patients undergoing long term ART; however, the results require verification in a future study with a larger sample size.
Conclusion
In conclusion, we compared the metabolite profiles in plasma samples among control, IR, and INR groups in a Chinese cohort and found that MC, PC, SC, and OC were negatively correlated with CD4+ T cell count. The combined analysis of MC, PC, SC, and OC could be useful for indicating the immune status, and predicting immune recovery, during long term ART, potentially guiding clinical treatment decisions.
Acknowledgements
The authors wish to acknowledge all the study participants who contributed to this work, as well as the clinical research staff of the participating institutions, who made this research possible.
Abbreviations
- HIV
Human immunodeficiency virus
- ART
Antiretroviral therapy
- INR
Immunological non-responders
- IR
Immunological responders
- NRTI
Nucleoside reverse transcriptase inhibitors
- UPLC-MS/MS
Ultra-high-performance liquid chromatography-tandem mass spectrometry
- FC
Fold change
- MC
Myristoylcarnitine
- PC
Palmitoylcarnitine
- SC
Stearoylcarnitine
- OC
Oleoylcarnitine
- ROC
Receiver operating characteristic
- FDR
False discovery rate
- OPLS-DA
Orthogonal partial least square discriminant analysis
- VIP
Variable influence in projection
- AUC
Area under curve
- BMI
Body mass index
- ESI
Electrospray ionization
- RP
Reverse phase
Authors' contributions
SQ, XC, TW and YS collected the samples, performed the experiments and analyzed the data. SQ, XC, and YJJ wrote and revised the manuscript. YJJ designed the experiments. ZNZ, JJX, XXH, HBD and YJF enrolled patients from the hospital and provided the clinical information. All authors read and approved the final manuscript.
Funding
The authors would like to acknowledge support from the Mega-Projects of National Science Research for the 12th Five-Year Plan (2012ZX10001-006) and the Mega-Projects of National Science Research for the 13th Five-Year Plan (2017ZX10201101). None of the funders of this research was involved in the design of the study; collection, analysis, and interpretation of data; or writing the manuscript.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
All enrolled subjects provided written informed consent and the protocol of this study was approved by the Research and Ethics Committee of The First Affiliated Hospital of China Medical University, in compliance with the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Shi Qian, Xi Chen and Tong Wu contributed equally to this work
References
- 1.Davenport MP, Khoury DS, Cromer D, Lewin SR, Kelleher AD, Kent SJ. Functional cure of HIV: the scale of the challenge. Nat Rev Immunol. 2019;19(1):45–54. doi: 10.1038/s41577-018-0085-4. [DOI] [PubMed] [Google Scholar]
- 2.Sun Y, Fu Y, Zhang Z, Tang T, Liu J, Ding H, Han X, Xu J, Chu Z, Shang H, et al. The investigation of CD4+T-cell functions in primary HIV infection with antiretroviral therapy. Medicine (Baltimore) 2017;96(28):e7430. doi: 10.1097/MD.0000000000007430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mirani G, Williams PL, Chernoff M, Abzug MJ, Levin MJ, Seage GR, 3rd, Oleske JM, Purswani MU, Hazra R, Traite S, et al. Changing trends in complications and mortality rates among US youth and young adults with HIV infection in the era of combination antiretroviral therapy. Clin Infect Dis. 2015;61(12):1850–1861. doi: 10.1093/cid/civ687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palella FJ, Delaney KM, Moorman AC, Loveless MO, Fuhrer J, Satten GA, Aschman DJ, Holmberg SD. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N Engl J Med. 1998;338(13):853–860. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- 5.Massanella M, Negredo E, Clotet B, Blanco J. Immunodiscordant responses to HAART–mechanisms and consequences. Expert Rev Clin Immunol. 2013;9(11):1135–1149. doi: 10.1586/1744666X.2013.842897. [DOI] [PubMed] [Google Scholar]
- 6.Boatman JA, Baker JV, Emery S, Furrer H, Mushatt DM, Sedlacek D, Lundgren JD, Neaton JD, Group ISS Risk factors for low CD4+ count recovery despite viral suppression among participants initiating antiretroviral treatment with CD4+ Counts > 500 cells/mm3: findings from the strategic timing of antiretroviral treatment (START) trial. J Acquir Immune Defic Syndr. 2019 doi: 10.1097/QAI.0000000000001967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muenchhoff M, Adland E, Roider J, Kloverpris H, Leslie A, Boehm S, Keppler OT, Ndung'u T, Goulder PJR. Differential Pathogen-Specific Immune Reconstitution in Antiretroviral Therapy-Treated Human Immunodeficiency Virus-Infected Children. J Infect Dis. 2019 doi: 10.1093/infdis/jiy668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma EH, Bantug G, Griss T, Condotta S, Johnson RM, Samborska B, Mainolfi N, Suri V, Guak H, Balmer ML, et al. Serine is an essential metabolite for effector T cell expansion. Cell Metab. 2017;25(2):345–357. doi: 10.1016/j.cmet.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 9.Postler TS, Ghosh S. Understanding the holobiont: how microbial metabolites affect human health and shape the immune system. Cell Metab. 2017;26(1):110–130. doi: 10.1016/j.cmet.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodriguez-Gallego E, Gomez J, Pacheco YM, Peraire J, Vilades C, Beltran-Debon R, Mallol R, Lopez-Dupla M, Veloso S, Alba V, et al. A baseline metabolomic signature is associated with immunological CD4+ T-cell recovery after 36 months of antiretroviral therapy in HIV-infected patients. AIDS. 2018;32(5):565–573. doi: 10.1097/QAD.0000000000001730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serrano-Villar S, Rojo D, Martinez-Martinez M, Deusch S, Vazquez-Castellanos JF, Bargiela R, Sainz T, Vera M, Moreno S, Estrada V, et al. Gut bacteria metabolism impacts immune recovery in HIV-infected individuals. EBioMedicine. 2016;8:203–216. doi: 10.1016/j.ebiom.2016.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem. 2009;81(16):6656–6667. doi: 10.1021/ac901536h. [DOI] [PubMed] [Google Scholar]
- 13.Dehaven CD, Evans AM, Dai H, Lawton KA. Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J Cheminform. 2010;2(1):9. doi: 10.1186/1758-2946-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halama A, Guerrouahen BS, Pasquier J, Diboun I, Karoly ED, Suhre K, Rafii A. Metabolic signatures differentiate ovarian from colon cancer cell lines. J Transl Med. 2015;13:223. doi: 10.1186/s12967-015-0576-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bijlsma S, Bobeldijk I, Verheij ER, Ramaker R, Kochhar S, Macdonald IA, van Ommen B, Smilde AK. Large-scale human metabolomics studies: a strategy for data (pre-) processing and validation. Anal Chem. 2006;78(2):567–574. doi: 10.1021/ac051495j. [DOI] [PubMed] [Google Scholar]
- 16.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34(2):374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 17.Chong J, Soufan O, Li C, Caraus I, Li S, Bourque G, Wishart DS, Xia J. MetaboAnalyst 40: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46(W1):W486–W494. doi: 10.1093/nar/gky310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X, Wu T, Jiang Y, Zhang Z, Han X, Geng W, Ding H, Kang J, Wang Q, Shang H. Plasma metabolic changes in Chinese HIV-infected patients receiving lopinavir/ritonavir based treatment: Implications for HIV precision therapy. Cytokine. 2018;110:204–212. doi: 10.1016/j.cyto.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 19.Bastard JP, Caron M, Vidal H, Jan V, Auclair M, Vigouroux C, Luboinski J, Laville M, Maachi M, Girard PM, et al. Association between altered expression of adipogenic factor SREBP1 in lipoatrophic adipose tissue from HIV-1-infected patients and abnormal adipocyte differentiation and insulin resistance. Lancet. 2002;359(9311):1026–1031. doi: 10.1016/S0140-6736(02)08094-7. [DOI] [PubMed] [Google Scholar]
- 20.Parker RA, Flint OP, Mulvey R, Elosua C, Wang F, Fenderson W, Wang S, Yang WP, Noor MA. Endoplasmic reticulum stress links dyslipidemia to inhibition of proteasome activity and glucose transport by HIV protease inhibitors. Mol Pharmacol. 2005;67(6):1909–1919. doi: 10.1124/mol.104.010165. [DOI] [PubMed] [Google Scholar]
- 21.Minami R, Yamamoto M, Takahama S, Ando H, Miyamura T, Suematsu E. Comparison of the influence of four classes of HIV antiretrovirals on adipogenic differentiation: the minimal effect of raltegravir and atazanavir. J Infect Chemother. 2011;17(2):183–188. doi: 10.1007/s10156-010-0101-5. [DOI] [PubMed] [Google Scholar]
- 22.Schooneman MG, Vaz FM, Houten SM, Soeters MR. Acylcarnitines: reflecting or inflicting insulin resistance? Diabetes. 2013;62(1):1–8. doi: 10.2337/db12-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melone MAB, Valentino A, Margarucci S, Galderisi U, Giordano A, Peluso G. The carnitine system and cancer metabolic plasticity. Cell Death Dis. 2018;9(2):228. doi: 10.1038/s41419-018-0313-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scholte HR, Jennekens FG, Bouvy JJ. Carnitine palmitoyltransferase II deficiency with normal carnitine palmitoyltransferase I in skeletal muscle and leucocytes. J Neurol Sci. 1979;40(1):39–51. doi: 10.1016/0022-510X(79)90007-8. [DOI] [PubMed] [Google Scholar]
- 25.Scholte HR, Luyt-Houwen IE, Vaandrager-Verduin MH. The role of the carnitine system in myocardial fatty acid oxidation: carnitine deficiency, failing mitochondria and cardiomyopathy. Basic Res Cardiol. 1987;82(Suppl 1):63–73. doi: 10.1007/978-3-662-08390-1_8. [DOI] [PubMed] [Google Scholar]
- 26.Mutomba MC, Yuan H, Konyavko M, Adachi S, Yokoyama CB, Esser V, McGarry JD, Babior BM, Gottlieb RA. Regulation of the activity of caspases by L-carnitine and palmitoylcarnitine. FEBS Lett. 2000;478(1–2):19–25. doi: 10.1016/S0014-5793(00)01817-2. [DOI] [PubMed] [Google Scholar]
- 27.Hulme HE, Meikle LM, Wessel H, Strittmatter N, Swales J, Thomson C, Nilsson A, Nibbs RJB, Milling S, Andren PE, et al. Mass spectrometry imaging identifies palmitoylcarnitine as an immunological mediator during Salmonella Typhimurium infection. Sci Rep. 2017;7(1):2786. doi: 10.1038/s41598-017-03100-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wenzel U, Nickel A, Daniel H. Increased carnitine-dependent fatty acid uptake into mitochondria of human colon cancer cells induces apoptosis. J Nutr. 2005;135(6):1510–1514. doi: 10.1093/jn/135.6.1510. [DOI] [PubMed] [Google Scholar]
- 29.Rutkowsky JM, Knotts TA, Ono-Moore KD, McCoin CS, Huang S, Schneider D, Singh S, Adams SH, Hwang DH. Acylcarnitines activate proinflammatory signaling pathways. Am J Physiol Endocrinol Metab. 2014;306(12):E1378–1387. doi: 10.1152/ajpendo.00656.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCoin CS, Knotts TA, Ono-Moore KD, Oort PJ, Adams SH. Long-chain acylcarnitines activate cell stress and myokine release in C2C12 myotubes: calcium-dependent and -independent effects. Am J Physiol Endocrinol Metab. 2015;308(11):E990–E1000. doi: 10.1152/ajpendo.00602.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Requero MA, Goni FM, Alonso A. The membrane-perturbing properties of palmitoyl-coenzyme A and palmitoylcarnitine A comparative study. Biochemistry. 1995;34(33):10400–10405. doi: 10.1021/bi00033a011. [DOI] [PubMed] [Google Scholar]
- 32.Goni FM, Requero MA, Alonso A. Palmitoylcarnitine, a surface-active metabolite. FEBS Lett. 1996;390(1):1–5. doi: 10.1016/0014-5793(96)00603-5. [DOI] [PubMed] [Google Scholar]
- 33.Miyaaki H, Kobayashi H, Miuma S, Fukusima M, Sasaki R, Haraguchi M, Nakao K. Blood carnitine profiling on tandem mass spectrometry in liver cirrhotic patients. BMC Gastroenterol. 2020;20(1):41. doi: 10.1186/s12876-020-01190-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Enooku K, Nakagawa H, Fujiwara N, Kondo M, Minami T, Hoshida Y, Shibahara J, Tateishi R, Koike K. Altered serum acylcarnitine profile is associated with the status of nonalcoholic fatty liver disease (NAFLD) and NAFLD-related hepatocellular carcinoma. Sci Rep. 2019;9(1):10663. doi: 10.1038/s41598-019-47216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J, Muise ES, Han S, Kutchukian PS, Costet P, Zhu Y, Kan Y, Zhou H, Shah V, Huang Y, et al. Molecular Profiling Reveals a Common Metabolic Signature of Tissue Fibrosis. Cell Rep Med. 2020;1(4):100056. doi: 10.1016/j.xcrm.2020.100056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Estes JD, Haase AT, Schacker TW. The role of collagen deposition in depleting CD4+ T cells and limiting reconstitution in HIV-1 and SIV infections through damage to the secondary lymphoid organ niche. Semin Immunol. 2008;20(3):181–186. doi: 10.1016/j.smim.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeng M, Smith AJ, Wietgrefe SW, Southern PJ, Schacker TW, Reilly CS, Estes JD, Burton GF, Silvestri G, Lifson JD, et al. Cumulative mechanisms of lymphoid tissue fibrosis and T cell depletion in HIV-1 and SIV infections. J Clin Invest. 2011;121(3):998–1008. doi: 10.1172/JCI45157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.