Abstract
Intraspecific competition for limited niches has been recognized as a driving force for adaptive radiation, but results for the role of interspecific competition have been mixed. Here, we report the adaptive diversification of the model bacteria Pseudomonas fluorescens in the presence of different numbers and combinations of four competing bacterial species. Increasing the diversity of competitive community increased the morphological diversity of focal species, which is caused by impeding the domination of a single morphotype. Specifically, this pattern was driven by more diverse communities being more likely to contain key species that occupy the same niche as otherwise competitively superior morphotype, and thus preventing competitive exclusion within the focal species. Our results suggest that sympatric adaptive radiation is driven by the presence or absence of niche-specific competitors.
Keywords: competition, niche, community ecology, adaptive radiation, microbial ecology, evolutionary ecology
Introduction
Adaptive radiation is a key component of biodiversity generation (Dieckmann and Doebeli, 1999; Schluter, 2000; Losos, 2010; Kassen, 2014). Population diversification – the precursor to adaptive radiation – not only depends on genetic characteristics of a population, such as cryptic genetic variation (Zheng et al., 2019) and mutation/recombination rate (Gavrilets and Vose, 2005; Lanfear et al., 2010), but also on abiotic and biotic environmental conditions (Grant, 1986; Rainey and Travisano, 1998; Buckling and Rainey, 2002; Hall and Colegrave, 2007; Meyer and Kassen, 2007; Betts et al., 2018). Intraspecific competition, in particular, is suggested to be a key driver of diversifying selection in a wide range of taxa (Schluter, 2000; MacLean et al., 2005; Svanback and Bolnick, 2007). However, the role of interspecific competition is less clear. Interspecific competitors might contribute to increased diversification by creating new ecological niches or could select for different resource usage to that of coexisting species (Emerson and Kolm, 2005; Svanback and Bolnick, 2007; Erwin, 2008; Calcagno et al., 2017). On the other hand, interspecific competitors may inhibit diversification due to fewer vacant niches or reduced population sizes (Gómez and Buckling, 2013; Ghoul and Mitri, 2016; Schluter and Pennell, 2017; Pontarp and Petchey, 2018; Harvey et al., 2020). Moreover, the neutral theory of community assembly proposes that all species are functionally equivalent for community assembly and maintenance (Hubbell, 2001; Harris et al., 2017), implying a neutral role of interspecific competition in diversification.
Direct experimental tests of the role of interspecific competitors on diversification have typically used the bacterium Pseudomonas fluorescens as the focal species because of its propensity to morphologically diversify over 10s of generations. These studies have generated contrasting results. Gómez and Buckling (2013) demonstrated lower evolved diversity in resource use of an initially isogenic population of P. fluorescens in the presence vs. absence of the natural microbial community in soil; Jousset et al. (2016) reported that increasing the number of competing P. fluorescens isolates resulted in an increased diversifying selection of the focal strain; while Zhang et al. (2012) reported little effect of a competitor (Pseudomonas putida) on P. fluorescens diversification.
We argue that the inconsistent results could be reconciled based on the theoretical framework emerging from biodiversity-ecosystem functioning studies (Schulze and Mooney, 1993; Tilman, 1997; Loreau et al., 2001). Specifically, more diverse competitors may generally reduce the diversification of focal species due to the reduced niche availability and total abundance of a focal species, which is akin to a complementarity effect. Meanwhile, a sampling effect may also take place, as a species-richer competitor community is more likely to contain particular species that can strongly affect the focal species. However, its effect on the diversification within the latter may depend on the specific competitive interactions, either positive (if it competes with a dominant genotype and thus prevent competitive exclusion within the focal species) or negative (if it competes with all genotypes and strongly reduces the total abundance of the focal species). While the Jousset study created a diversity gradient, all the competitors were P. fluorescens isolates, limiting the potential for resident niche occupation (Jousset et al., 2016). A recent study of natural microbial communities from the Earth Microbiome Project found a unimodal relationship between diversity and diversification: community diversity beget diversity in low-diversity biomes and reach plateaus when niches are increasingly filled in high-diversity biomes (Madi et al., 2020).
To further investigate the role of interspecific diversity on diversification, we evolved P. fluorescens SBW25 across a diversity gradient of a synthetic microbial community isolated from potting compost, which consists of four species that can stably coexist in a relatively oligotrophic medium when incubated in the lab (Castledine et al., 2020). Pseudomonas fluorescens can rapidly diversify into three main colony morphotypes when propagated in spatially heterogeneous microcosms (Rainey and Travisano, 1998). These morphotypes can be typically identified as: ancestor-like smooth (SM) that occupies the liquid phase, wrinkly spreader (WS), which arises by spontaneous mutations and forms a self-supporting mat on the air-liquid interface, and fuzzy spreader (FS), which occupies the anaerobic niche (Rainey and Travisano, 1998). The driver of diversification has been shown to be competition for oxygen and other nutrients (Koza et al., 2011; Kassen, 2014). The present study measured the morphological diversification of P. fluorescens in the presence of microbial communities, in which a diversity gradient was set-up using four species, to determine the diversity of competing microbial communities on adaptive diversification.
Materials and Methods
Experimental Evolution
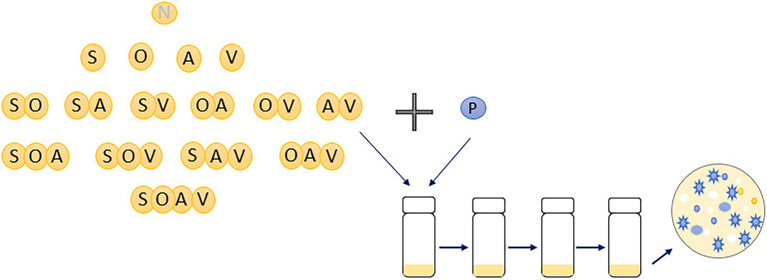
Communities were set up with a SM clone of P. fluorescens SBW25 with isogenic clones of Achromobacter sp. (A), Ochrobactrum sp. (O), Stenotrophomonas sp. (S), and Variovorax sp. (V), which can be distinguished by their unique colony morphologies (Supplementary Figure 1A; Castledine et al., 2020; Padfield et al., 2020 unpublished). Every community combination of the four species with P. fluorescens was set up across all the levels of diversity with six replicates for each community combination. Therefore, a total of 16 different communities were established with total species richness ranging from 1 (only P. fluorescens) to 5 (all five species). Specifically, there were one 1-species community treatment, four 2-species treatments, six 3-species treatments, four 4-species treatments, and one 5-species treatment (Figure 1). Communities were grown in 25 ml glass vials with loosened lids with 6 ml of M9KB media (glycerol 10 g L−1, proteose peptone no.3 20 g L−1, KH2PO4 3 g L−1, NaCl 0.5 g L−1, and NH4Cl 1 g L−1). Prior to experimental set-up, each species was grown for 2 days in M9KB media at 28°C to achieve high-cell densities. Cell densities of each species were diluted into M9 buffer to approximately 104 CFUs uL−1. Each microcosm was inoculated with 20 uL per species (~105 cells) and incubated at 28°C in static, after which 60uL was transferred every 7 days for a total of three transfers. Culture samples were cryogenically frozen at −80°C in 50% glycerol (final concentration: 25%) at each transfer. Species densities within each microcosm at each transfer were determined by plating culture dilutions onto KB agar and counting the number of colony-forming units (CFUs) after 2 days of incubation at 28°C.
Figure 1.
An illustration of experimental design of the present study. Sixteen different microbial communities were set-up with species diversity ranging from 1 (monoculture of Pseudomonas fluorescens) to 5 (with all five species). Six replicates were conducted for each community combination. N represents no other species were inoculated into the microcosms; P, A, O, S, and V represents Pseudomonas fluorescens, Achromobacter, Ochrobactrum, Stenotrophomonas and Variovorax, respectively. Diversity of the focal species, Pseudomonas fluorescens, at the end of the evolution experiment was estimated based on its morphological diversity of 100 randomly chosen colonies (blue ones).
Measurement of Diversity
Community diversity was estimated by species richness (the number of species). The ancestral smooth P. fluorescens diversified into three morphotypes (SM, WS, and FS; Supplementary Figure 1B) at the end point and the sympatric diversity of P. fluorescens populations was obtained by measuring the morphologies of 100 randomly chosen colonies (Buckling et al., 2000) and calculated as Simpson’s index: , where N is the total number of colonies sampled from the focal population and ( is the frequency of the ith morphotype; Simpson, 1949).
Statistical Analysis
All analysis was conducted using R (version 3.5.2; R Core Team, 2018), and all plots were made using the R package “ggplot2” (Wickham, 2016). Simpson’s index was calculated using the package “vegan” (Oksanen et al., 2019). A linear model was used to analyze the relationship between P. fluorescens diversity and the proportion of WS morphotype; the relationship between inoculated density of the competing community and proportion of WS or P. fluorescens diversity; and the relationship between the initial or final density of P. fluorescens and its sympatric diversity. ANOVA was performed to analyze whether the presence or absence of competitors affected P. fluorescens diversity and the proportion of WS. Density data were log-transformed [log10 (1 + CFUs ml−1)].
The effect of community richness on the diversity of P. fluorescens were analyzed by an analysis of variance with sequential sum of squares (type I) using linear models (Schmid et al., 2002; Buzhdygan et al., 2020). To assess whether diversity effects were only due to the presence of single species, we fitted each species (presence/absence) before species richness in separate sequential analyses. If fitting single genotypes before species richness removed the effect of richness indices, the observed diversity effects may have been mainly caused by a sampling effect (the inclusion of a particular genotype in the community; Huston, 1997). To further explain whether species composition matters, we used ANOVA to test the effect of the presence of each species and their interactions on P. fluorescens diversity. The simplest model was obtained by AICc ranking using the R package “MuMIn” (Barton, 2020). The effects of the presence of single species and initial community diversity on the proportion of WS, and on the density of WS were tested with sequential linear models as described above.
Results
Dominant WS Morphotype Inhibits Pseudomonas fluorescens Diversity
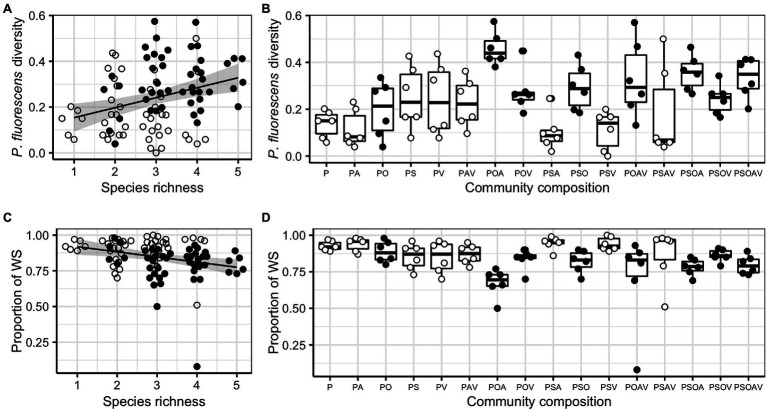
Across all evolution lines, the WS morphotype was found to dominate the diversified P. fluorescens populations (mean ± SD of the proportion of WS in P. fluorescens populations: 0.847 ± 0.128). A negative relationship was found between P. fluorescens diversity and proportion of WS (F1,94 = 369.82, p < 0.001; Figure 2), indicating that P. fluorescens diversity is caused by reduced proportions of WS morphotypes. This implies that factors limiting the dominant WS morphotype would increase the sympatric diversity of P. fluorescens. Although the presence of competing communities was not found to affect P. fluorescens diversity (F1,94 = 3.606, p = 0.061) or the proportion of WS (F1,94 = 2.521, p = 0.116), the density of inoculated competing communities reduced the proportion of WS (F1,88 = 5.346, p = 0.023; Supplementary Figure 2). In addition, a positive relationship was observed between the density of inoculated competing communities and the diversity of focal species (F1,88 = 10.169, p = 0.002; Supplementary Figure 2), suggesting a competing effect of the microbial communities on P. fluorescens.
Figure 2.
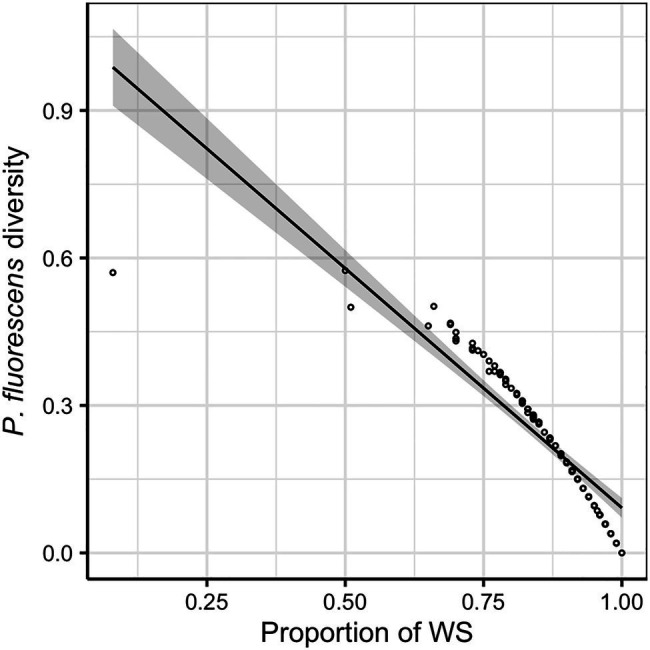
Pseudomonas fluorescens diversity is negatively correlated with the proportion of wrinkly spreader (WS). The regression line represents a significant linear relationship and shaded areas around lines show the 95% confidence intervals: , F1,94 = 369.820, p < 0.001, adjusted R2 = 0.795.
Species-Specific Effects on Pseudomonas fluorescens Morphotype Diversity
Pseudomonas fluorescens diversity was positively affected by increasing species richness (F1,94 = 10.204, p = 0.002). However, the significant effect of species richness was removed when fitted after the presence/absence of the Ochrobactrum species (Figures 3A,B; Table 1), indicating that the observed diversity effect is mainly due to a sampling effect. Further analysis revealed that the diversity of P. fluorescens was also affected by the interaction between Ochrobactrum and Achromobacter species (F1,92 = 9.537, p = 0.003). Qualitatively similar results were obtained using alternative community diversity metric (Simpson’s index; see Supplementary Text).
Figure 3.
The effect of species richness, the presence of Ochrobactrum sp. (O) and community composition on Pseudomonas fluorescens diversity (A,B) and the proportion of wrinkly spreader (WS; C,D). For both responsive variables, regression lines showed significant linear relationships with species richness: , F1,94 = 369.820, p < 0.001, adjusted R2 = 0.088 (A); and , F1,94 = 8.061, p = 0.006, adjusted R2 = 0.069 (B). Shaded areas around lines show the 95% confidence intervals. Treatments containing O (filled dots) and not containing O (open dots) are highlighted to show the sampling effect of O. Tops and bottoms of the bars represent the 75th and 25th percentiles of the data, the middle lines are the medians, and the whiskers extend from their respective hinge to the smallest or largest value no further than 1.5 × interquartile range.
Table 1.
Analysis of variance table of F-values on the effects of presence of specific species, and species richness on the diversity of Pseudomonas fluorescens populations.
| Factor | df | Achromobacter sp. | Ochrobactrum sp. | Stenotrophomonas sp. | Variovorax sp. | ||||
|---|---|---|---|---|---|---|---|---|---|
| F | P | F | P | F | P | F | P | ||
| Species presence | 1 | 2.558 | 0.113 | 31.898 | <0.001 | 0.293 | 0.590 | 0.027 | 0.871 |
| Species richness | 1 | 7.538 | 0.007 | 0.620 | 0.433 | 16.849 | <0.001 | 13.174 | <0.001 |
| Residuals | 93 | ||||||||
The bold values indicate significant effects.
While species Achromobacter (A), Ochrobactrum (O), Stenotrophomonas (S), and Variovorax (V) stably coexisted in a previous study, the presence of P. fluorescens destabilized this community in many combinations (Supplementary Figure 3). V was driven extinct in at least 3/6 replicates in 7/8 species combinations with V going extinct in 2/6 replicates in the PSAV evolution line. Similarly, S went extinct in 4/6 replicates in evolution lines PSO and PSOA, and in 1/6 replicates in evolution lines PSOV and PSV. These extinctions resulted in variable community diversities and the P. fluorescens final densities ranging from 4.40 × 108 to 2.78 × 109. These results indicate that O and A (to a less extent) are the main competitors of P. fluorescens and affect its diversity.
Potential Mechanisms
We first tested if the patterns of diversification could be affected by P. fluorescens density by affecting the supply of mutations and the strength of diversifying selection. Neither inoculated P. fluorescens density nor its final density affected diversification. However, the final population size of P. fluorescens decreased in the presence of competing species (F1,94 = 13.410, p < 0.001), suggesting the interspecific resource competition during the diversification of focus species. As A and O coexisted with P. fluorescens, while S and V went extinct in many combinations (Supplementary Figure 3), they are presumably the main competitors of P. fluorescens.
Given that P. fluorescens diversity is caused by the reduced proportion of WS, the increased diversity of focal species in the presence of O, and O and A together may be a result of competition with WS. Therefore, we tested whether the presence of particular species affected diversity of focal species by affecting the proportion of WS (the sampling effect). Although the proportion of WS was affected by species richness (F1,94 = 8.061, p = 0.006), its effect was removed when fitted after the presence of O (Figures 3C,D; Table 2). In addition, a marginal sampling effect of A was also detected (Table 2). These results supported the competition between WS and O (and to a lesser extent A) in driving the diversification of P. fluorescens. As WS forms mats to occupy the air-liquid interface, we hypothesized that O (and A to a lesser extent) may compete with WS in this niche, reducing invasion of WS. O, A, and WS were found to form mats in media when grown in monoculture while V formed very thin mats and mats were absent for S and SM. The presence of O and A were, indeed, found to affect the density of WS (Supplementary Text; Supplementary Figure 4; and Supplementary Table 1), suggesting the niche competition of O and A with the derived WS morphs of P. fluorescens.
Table 2.
Analysis of variance table of F-values on the effects of presence of specific species, and species richness on the proportion of wrinkly spreader.
| Factor | df | Achromobacter sp. | Ochrobactrum sp. | Stenotrophomonas sp. | Variovorax sp. | ||||
|---|---|---|---|---|---|---|---|---|---|
| F | P | F | P | F | P | F | P | ||
| Species presence | 1 | 3.868 | 0.052 | 18.257 | <0.001 | 0.807 | 0.371 | 0.249 | 0.619 |
| Species richness | 1 | 4.548 | 0.036 | 0.942 | 0.334 | 15.272 | <0.001 | 8.955 | 0.004 |
| Residuals | 93 | ||||||||
The bold values indicate significant effects.
Discussion
Here, we demonstrated that the colony morphological diversity of evolving P. fluorescens populations increased with the number of bacterial taxa it was co-cultured with. This pattern was primarily driven by the presence of a specific species (O) in more diverse communities, although there were also some interactive effects with other species. Pseudomonas fluorescens diversity was negatively correlated with the proportion of evolved WS morphotypes, which had a mean frequency of ~0.8 across all replicates. Most of the remaining P. fluorescens populations were SM morphotypes. Diversity was therefore maximized when the evolved WS less successfully dominating populations.
The presence of O (and to a lesser extent A) impeded the dominance of WS. The other species (S and V) were frequently driven extinct by the end of experiment, and hence would have imposed relatively little competition. We also observed that O and A produced more detectable mats (or biofilms) than the other two species, and mat formation is characteristic of WS [we tried to quantify the biofilms with the resazurin assay, (Peeters et al., 2008) but failed as their mats are not as definable as the biofilms of P. fluorescens]. This suggests that O and A, which are the strongest competitors in the community, are competing more with WS than SM by occupying a similar ecological niche to WS (Kim et al., 2014). Similar constraints on the invasion of evolved WS have been observed when the resident P. fluorescens WS genotypes were present in the populations (Brockhurst et al., 2007; Flohr et al., 2013).
The formation of biofilms and inhibiting the biofilm formation of other microbial populations could be an evolved competitive strategy since these genotypes could gain preferential access to resources and the biofilms could protect cells from environmental hazards (An et al., 2006; Stacy et al., 2016). For example, it has been reported that P. aeruginosa could prevent Agrobacterium from forming its own biofilm by surface blanketing (An et al., 2006), and E. coli can reduce the biofilm formation of Gram-positive and Gram-negative bacteria by producing extracellular polysaccharides or surfactants (Valle et al., 2006; Rendueles et al., 2011).
WS density was highly variable in the presence of O and A, and this may be explained by the advantage of forming biofilms and historical contingences. The diversified WS is a cooperating group that can enable individuals to align with them by overproducing an adhesive polymer and promote the colonization of the air-liquid interface though the overproduction is costly to individuals (Spiers et al., 2002; Rainey and Rainey, 2003). Therefore, if mutations to form WS arose late in a population or P. fluorescens initially grew slowly, WS may lose the chance to spread because the available niche is already partially occupied by competitor species.
It has been previously shown that the Pseudomonas sp. can stably coexist with the other four competing species (Castledine et al., 2020), while S and V were at low densities or went extinct in many combinations by the end of our experiment. Although a different Pseudomonas was involved here, it is not suggested to be a main cause of the instability as family-level bacteria are found to have similar function (Goldford et al., 2018; Louca et al., 2018). Therefore, the instability may be a result of different media used in the present studies that altered community composition (Goldberg and Miller, 1990; Goldford et al., 2018). Specifically, the rich media used here may have increased the likelihood of competitive exclusion by favoring the fast-growing Pseudomonas.
Limitations of the present study are noteworthy. The relative short-term evolution limited the study to further investigate more nuanced processes such as how within-species diversification feedback to affect community structure. Though the three morphotypes of P. fluorescens has been widely used for diversity estimation, less consideration has been given to its genetic diversity (Spiers et al., 2002; Fukami et al., 2007).
Diversity-dependent adaptive radiation theory predicts that more diverse communities are more likely to experience evolutionary diversification (Calcagno et al., 2017). This is not supported by our study despite a greater diversity of focal species in the presence of more diverse competitors was observed. Our study is instead consistent with previous work, suggesting that evolutionary processes are impeded by the presence of competitors (Hall et al., 2018; Scheuerl et al., 2020). In this case, competitors reduced the dominance of the evolved WS genotypes, most likely because of shared niche occupation in spatially structured (static) microcosms. Though the reduced population size of focal species by interspecific competition may affect the adaptive radiation process, P. fluorescens is less affected in our working system as the focal species showed an advantage and dominated the microbial communities after three transfers (Johansson, 2008; Osmond and de Mazancourt, 2012; Zhao et al., 2018). Our results may help to understand the inconsistent effects of interspecific competitors on diversification of resident species across different studies and suggest that the presence of niche-specific competitors is a possible explanation.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://doi.org/10.6084/m9.figshare.14471229.
Author Contributions
All authors contributed to the design of the study. X-LC and MC conducted the experiments. All authors contributed to the data analysis and the writing of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Jesica Soria-Pascual and Elze Hesse for help in the laboratory and Daniel Padfield for helpful advice on statistical analysis.
Glossary
Abbreviations
- A
Achromobacter sp.
- O
Ochrobactrum sp.
- S
Stenotrophomonas sp.
- V
Variovorax sp.
- SM
Smooth morph
- WS
Wrinkly spreaders
- FS
Fuzzy spreader
Footnotes
Funding. The work was funded by NERC (NE/P001130/1). X-LC was funded by the China Scholarship Council. MC was funded by a studentship from the MRC (111846). Q-GZ was supported by the National Natural Science Foundation of China (31725006 and 31670376), and the 111 project (B13008).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articless/10.3389/fmicb.2021.699190/full#supplementary-material
References
- An D., Danhorn T., Fuqua C., Parsek M. R. (2006). Quorum sensing and motility mediate interactions between Pseudomonas aeruginosa and Agrobacterium tumefaciens in biofilm cocultures. Proc. Natl. Acad. Sci. U. S. A. 103, 3828–3833. 10.1073/pnas.0511323103, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton K.. (2020). MuMIn: multi-model inference. Available at: https://cran.r-project.org/web/packages/MuMIn/index.html (Accessed April 15, 2020).
- Betts A., Gray C., Zelek M., MacLean R. C., King K. C. (2018). High parasite diversity accelerates host adaptation and diversification. Science 360, 907–911. 10.1126/science.aam9974, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockhurst M. A., Colegrave N., Hodgson D. J., Buckling A. (2007). Niche occupation limits adaptive radiation in experimental microcosms. PLoS One 2:e193. 10.1371/journal.pone.0000193, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckling A., Kassen R., Bell G., Rainey P. B. (2000). Disturbance and diversity in experimental microcosms. Nature 408, 961–964. 10.1038/35050080, PMID: [DOI] [PubMed] [Google Scholar]
- Buckling A., Rainey P. B. (2002). The role of parasites in sympatric and allopatric host diversification. Nature 420, 496–499. 10.1038/nature01164, PMID: [DOI] [PubMed] [Google Scholar]
- Buzhdygan O. Y., Meyer S. T., Weisser W. W., Eisenhauer N., Ebeling A., Borrett S. R., et al. (2020). Biodiversity increases multitrophic energy use efficiency, flow and storage in grasslands. Nat. Ecol. Evol. 4, 393–405. 10.1038/s41559-020-1123-8, PMID: [DOI] [PubMed] [Google Scholar]
- Calcagno V., Jarne P., Loreau M., Mouquet N., David P. (2017). Diversity spurs diversification in ecological communities. Nat. Commun. 8:15810. 10.1038/ncomms15810, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castledine M., Padfield D., Buckling A. (2020). Experimental (co)evolution in a multi-species microbial community results in local maladaptation. Ecol. Lett. 23, 1673–1681. 10.1111/ele.13599, PMID: [DOI] [PubMed] [Google Scholar]
- Dieckmann U., Doebeli M. (1999). On the origin of species by sympatric speciation. Nature 400, 354–357. 10.1038/22521, PMID: [DOI] [PubMed] [Google Scholar]
- Emerson B. C., Kolm N. (2005). Species diversity can drive speciation. Nature 434, 1015–1017. 10.1038/nature03450, PMID: [DOI] [PubMed] [Google Scholar]
- Erwin D. H. (2008). Macroevolution of ecosystem engineering, niche construction and diversity. Trends Ecol. Evol. 23, 304–310. 10.1016/j.tree.2008.01.013, PMID: [DOI] [PubMed] [Google Scholar]
- Flohr R. C. E., Blom C. J., Rainey P. B., Beaumont H. J. E. (2013). Founder niche constrains evolutionary adaptive radiation. Proc. Natl. Acad. Sci. U. S. A. 110, 20663–20668. 10.1073/pnas.1310310110, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukami T., Beaumont H. J. E., Zhang X. X., Rainey P. B. (2007). Immigration history controls diversification in experimental adaptive radiation. Nature 446, 436–439. 10.1038/nature05629, PMID: [DOI] [PubMed] [Google Scholar]
- Gavrilets S., Vose A. (2005). Dynamic patterns of adaptive radiation. Proc. Natl. Acad. Sci. U. S. A. 102, 18040–18045. 10.1073/pnas.0506330102, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoul M., Mitri S. (2016). The ecology and evolution of microbial competition. Trends Microbiol. 24, 833–845. 10.1016/j.tim.2016.06.011, PMID: [DOI] [PubMed] [Google Scholar]
- Goldberg D. E., Miller T. E. (1990). Effects of different resource additions of species diversity in an annual plant community. Ecology 71, 213–225. 10.2307/1940261 [DOI] [Google Scholar]
- Goldford J. E., Lu N., Bajić D., Estrela S., Tikhonov M., Sanchez-Gorostiaga A., et al. (2018). Emergent simplicity in microbial community assembly. Science 361, 469–474. 10.1126/science.aat1168, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez P., Buckling A. (2013). Real-time microbial adaptive diversification in soil. Ecol. Lett. 16, 650–655. 10.1111/ele.12093, PMID: [DOI] [PubMed] [Google Scholar]
- Grant P. R. (1986). Ecology and Evolution of Darwin’s Finches. Princeton, New Jersey: Princeton University Press. [Google Scholar]
- Hall A. R., Colegrave N. (2007). How does resource supply affect evolutionary diversification? Proc. R. Soc. B 274, 73–78. 10.1098/rspb.2006.3703, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J. P. J., Harrison E., Brockhurst M. A. (2018). Competitive species interactions constrain abiotic adaptation in a bacterial soil community. Evol. Lett. 2, 580–589. 10.1002/evl3.83, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris K., Parsons T. L., Ijaz U. Z., Lahti L., Holmes I., Quince C. (2017). Linking statistical and ecological theory: Hubbell’s unified neutral theory of biodiversity as a hierarchical dirichlet process. Proc. IEEE 105, 516–529. 10.1109/JPROC.2015.2428213 [DOI] [Google Scholar]
- Harvey M. G., Bravo G. A., Claramunt S., Cuervo A. M., Derryberry G. E., Battilana J., et al. (2020). The evolution of a tropical biodiversity hotspot. Science 370, 1343–1348. 10.1126/science.aaz6970, PMID: [DOI] [PubMed] [Google Scholar]
- Hubbell S. P. (2001). The Unified Neutral Theory of Biodiversity and Biogeography. Princeton, New Jersey: Princeton University Press. [Google Scholar]
- Huston M. A. (1997). Hidden treatments in ecological experiments: re-evaluating the ecosystem function of biodiversity. Oecologia 110, 449–460. 10.1007/s004420050180, PMID: [DOI] [PubMed] [Google Scholar]
- Johansson J. (2008). Evolutionary responses to environmental changes: how does competition affect adaptation? Evolution 62, 421–435. 10.1111/j.1558-5646.2007.00301.x, PMID: [DOI] [PubMed] [Google Scholar]
- Jousset A., Eisenhauer N., Merker M., Mouquet N., Scheu S. (2016). High functional diversity stimulates diversification in experimental microbial communities. Sci. Adv. 2:e1600124. 10.1126/sciadv.1600124, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassen R. (2014). Experimental Evolution and the Nature of Biodiversity. Colorado: Roberts and Company Publisher. [Google Scholar]
- Kim W., Racimo F., Schluter J., Levy S. B., Foster K. R. (2014). Importance of positioning for microbial evolution. Proc. Natl. Acad. Sci. U. S. A. 111, E1639–E1647. 10.1073/pnas.1323632111, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koza A., Moshynets O., Otten W., Spiers A. J. (2011). Environmental modification and niche construction: developing O2 gradients drive the evolution of the wrinkly spreader. ISME J. 5, 665–673. 10.1038/ismej.2010.156, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R., Ho S. Y. W., Love D., Bromham L. (2010). Mutation rate is linked to diversification in birds. Proc. Natl. Acad. Sci. U. S. A. 107, 20423–20428. 10.1073/pnas.1007888107, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreau M., Naeem S., Inchausti P., Bengtsson J., Grime J. P., Hector A., et al. (2001). Biodiversity and ecosystem functioning: current knowledge and future challenges. Science 294, 804–808. 10.1126/science.1064088, PMID: [DOI] [PubMed] [Google Scholar]
- Losos J. B. (2010). Adaptive radiation, ecological opportunity, and evolutionary determinism. Am. Nat. 175, 623–639. 10.1086/652433, PMID: [DOI] [PubMed] [Google Scholar]
- Louca S., Polz M. F., Mazel F., Albright M. B. N., Huber J. A., O’Connor M. I., et al. (2018). Function and functional redundancy in microbial systems. Nat. Ecol. Evol. 2, 936–943. 10.1038/s41559-018-0519-1, PMID: [DOI] [PubMed] [Google Scholar]
- MacLean R. C., Dickson A., Bell G. (2005). Resource competition and adaptive radiation in a microbial microcosm. Ecol. Lett. 8, 38–46. 10.1111/j.1461-0248.2004.00689.x [DOI] [Google Scholar]
- Madi N., Vos M., Murall C. L., Legendre P., Shapiro B. J. (2020). Does diversity beget diversity in microbiomes? elife 9:e58999. 10.7554/eLife.58999, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer J. R., Kassen R. (2007). The effects of competition and predation on diversification in a model adaptive radiation. Nature 446, 432–435. 10.1038/nature05599, PMID: [DOI] [PubMed] [Google Scholar]
- Oksanen J., Blanchet F. G., Friendly M., Kindt R., Legendre P., Minchin P. R., et al. (2019). vagan: community ecology package. Available at: https://cran.r-project.org/package=vegan (Accessed November 28, 2020).
- Osmond M. M., de Mazancourt C. (2012). How competition affects evolutionary rescue. Philos. Trans. R. Soc. B Biol. Sci. 368:20120085. 10.1098/rstb.2012.0085, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters E., Nelis H. J., Coenye T. (2008). Comparison of multiple methods for quantification of microbial biofilms grown in microtiter plates. J. Microbiol. Methods 72, 157–165. 10.1016/j.mimet.2007.11.010, PMID: [DOI] [PubMed] [Google Scholar]
- Pontarp M., Petchey O. L. (2018). Ecological opportunity and predator-prey interactions: linking eco-evolutionary processes and diversification in adaptive radiations. Proc. R. Soc. B Biol. Sci. 285:20172550. 10.1098/rspb.2017.2550, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2018). R: A language and environment for statistical computing. Available at: https://www.r-project.org/ (Accessed December 20, 2020).
- Rainey P. B., Rainey K. (2003). Evolution of cooperation and conflict in experimental bacterial populations. Nature 425, 72–74. 10.1038/nature01906, PMID: [DOI] [PubMed] [Google Scholar]
- Rainey P. B., Travisano M. (1998). Adaptive radiation in a heterogeneous environment. Nature 394, 69–72. 10.1038/27900, PMID: [DOI] [PubMed] [Google Scholar]
- Rendueles O., Travier L., Latour-Lambert P., Fontaine T., Magnus J., Denamur E., et al. (2011). Screening of Escherichia coli species biodiversity reveals new biofilm-associated antiadhesion polysaccharides. Mbio 2, e00043–e00011. 10.1128/mBio.00043-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuerl T., Hopkins M., Nowell R. W., Rivett D. W., Barraclough T. G., Bell T. (2020). Bacterial adaptation is constrained in complex communities. Nat. Commun. 11:754. 10.1038/s41467-020-14570-z, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter D. (2000). The Ecology of Adaptive Radiations. New York: Oxford University Press. [Google Scholar]
- Schluter D., Pennell M. W. (2017). Speciation gradients and the distribution of biodiversity. Nature 546, 48–55. 10.1038/nature22897, PMID: [DOI] [PubMed] [Google Scholar]
- Schmid B., Hector A., Huston M. A., Inchasusti P., Nijs I., Leadley P. W. (2002). “The design and analysis of biodiversity experiments,” in Biodiversity and Ecosystem Functioning: Synthesis and Perspectives. eds. Loreau M., Naeem S., Inchausti P. (New York: Oxford University Press; ). [Google Scholar]
- Schulze E. D., Mooney H. A. (1993). Biodiversity and Ecosystem Function. Berlin: Springer-Verlag. [Google Scholar]
- Simpson E. H. (1949). Measurement of diversity. Nature 163, 688–688. 10.1038/163688a0 [DOI] [Google Scholar]
- Spiers A. J., Kahn S. G., Bohannon J., Travisano M., Rainey P. B. (2002). Adaptive divergence in experimental populations of Pseudomonas fluorescens. I. Genetic and phenotypic bases of wrinkly spreader fitness. Genetics 161, 33–46. 10.1093/genetics/161.1.33, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacy A., McNally L., Darch S. E., Brown S. P., Whiteley M. (2016). The biogeography of polymicrobial infection. Nat. Rev. Microbiol. 14, 93–105. 10.1038/nrmicro.2015.8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svanback R., Bolnick D. I. (2007). Intraspecific competition drives increased resource use diversity within a natural population. Proc. R. Soc. B Biol. Sci. 274, 839–844. 10.1098/rspb.2006.0198, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilman D. (1997). Community invasibility, recruitment limitation, and grassland biodiversity. Ecology 78, 81–92. 10.1890/0012-9658(1997)078[0081:CIRLAG]2.0.CO;2, PMID: 16894146 [DOI] [Google Scholar]
- Valle J., Da Re S., Henry M., Fontaine T., Balestrino D., Latour-Lambert P., et al. (2006). Broad-spectrum biofilm inhibition by a secreted bacterial polysaccharide. Proc. Natl. Acad. Sci. U. S. A. 103, 12558–12563. 10.1073/pnas.0605399103, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. (2016). ggplot2: Elegant Graphics for Data Analysis. New York: Springer Press. [Google Scholar]
- Zhang Q. G., Ellis R. J., Godfray H. C. J. (2012). The effect of a competitor on a model adaptive radiation. Evolution 66, 1985–1990. 10.1111/j.1558-5646.2011.01559.x, PMID: [DOI] [PubMed] [Google Scholar]
- Zhao X. F., Buckling A., Zhang Q. G., Hesse E. (2018). Specific adaptation to strong competitors can offset the negative effects of population size reductions. Proc. R. Soc. B Biol. Sci. 285:20180007. 10.1098/rspb.2018.0007, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Payne J. L., Wagner A. (2019). Cryptic genetic variation accelerates evolution by opening access to diverse adaptive peaks. Science 365, 347–353. 10.1126/science.aax1837, PMID: [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://doi.org/10.6084/m9.figshare.14471229.