Abstract
The catastrophe of the coronavirus continues from one part of the world to another, and hardly a country is left without its devastations. Millions of people were infected and several hundred thousand died of the COVID-19 pandemic across the world. There is no clear targeted drug therapy available for the treatment of the patients. The discovery of vaccines is not enough to curtail its spread and disastrous implications. An instantly qualifying approach is needed to utilize the current drugs and isolated compounds. The purpose of this work is to determine potent inhibitors against the target proteins of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). For this purpose, molecular docking study of pathogenic spike glycoproteins (S), nucleocapsid phosphoprotein (N), an envelope protein (E), two drugs i.e., cefixime, etoposide, and a previously isolated compound nebrodenside A is performed. Promising results were obtained via complimentary analysis of molecular dynamics (MD) simulations performed for the complexes of three proteins with etoposide drug. Minimum values were recorded for the docking scores and binding energies of the complexes. These results were further supported by the RMSD, RMSF data for the stability of proteins and ligands. Additionally, ligand properties and ligand-protein contacts were also explained with histograms of every simulation trajectory. The computational studies confirmed that cefixime, etoposide, and nebrodenoside A can be used as potent inhibitors of COVID-19. Nevertheless, additional experimental investigations and validation of the selected candidates are mandatory to confirm their applicability for clinical trials.
Keywords: Coronavirus, Pathogenic protein, Etoposide, Cefixime
Graphical Abstract
1. Introduction
Coronavirus disease 2019 (COVID-19) is a contagious disease triggered by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). It was discovered in Wuhan, China, and has since outspread around the world like a pandemic, causing more than 150.6 million infections and about 32 hundred thousand deaths as of May 8, 2021 [1,2]. The World Health Organization called the disorder COVID-19 (coronavirus disease 2019). Genome sequencing reveals high similarity between SARS-CoV-2 and bat coronaviruses (CoV) especially beta-CoV and RaTG13 viruses that cause human diseases such as the Middle East respiratory syndrome (MERS) and the Severe acute respiratory syndrome (SARS) and to a smaller extent HCoV-OC43 and HCoV-HKU1 [3,4]. The nucleocapsid protein, spike (S) protein, envelope (E) protein, membrane (M) protein, and coronavirus key protease participate actively in gene formulation and break polyproteins into replication-associated proteins. They are all encoded in the genome of the new coronavirus (SARS-COV-2) [5]. The SARS-CoV-2 genome, like that of other coronaviruses, encrypt spike (S) glycoproteins, which project from the exterior of grown-up virions. The S glycoprotein is required for virus adhesion, incorporation, and infection of the host cell. The S glycoprotein's external spot makes it a straightforward target for the host immune feedbacks, making it a principal target for compensating the antibodies. The S protein is the subject of several vaccination campaigns and treatments due to its critical role in viral infection and innate immunity [6,7]. The nucleocapsid phosphoprotein (N) is a multifunctional and immunogenic virulence and pathogenicity determinant whose primary roles are oligomerization and packing of the single-stranded RNA (ssRNA) viral genome [8]. The envelope (E) protein, a vital protein of the SARS-CoV-2 virus, creates a homopentameric cation channel that is essential for virus pathogenesis. E is a 75-residual viroporin that makes a cation-selective network through the ERGIC coating. E facilitates the budding and activation of progeny viruses in SARS-CoV-1, and triggers the host inflammasome. Amantadine (AMT) and Hexamethylene amiloride (HMA) inhibit E's channel activity; the latter also inhibits the HIV-1 and influenza A virus viroporins. In certain coronaviruses, deletion of the E gene results in attenuated viruses. Virus pathogenicity is decreased by E mutations that disable channel function. As a result, E may be a target for antiviral drugs and a vaccine candidate for SARS-CoV-2 [9].
Many of the existing drugs and the isolated compounds are being investigated as potent inhibitors of the viral proteins of COVID-19. Considerable efforts are made to discover potent inhibitors for structural and non-structural proteins of SARS-CoV-2 through in silico methods such as computer-aided drug design (CADD), molecular docking, and molecular dynamic simulations [10]. Abu -saleh et al carried out a ligand-based virtual screening followed by a docking screening for testing approved drugs and bioactive compounds as potent inhibitors of SARS-CoV-2 main protease. The study produced useful results for additional optimizations and designs of potent SARS-CoV-2 main protease inhibitors [11]. Ngo and coworkers carried out molecular docking, fast pulling of ligand, and free energy perturbation calculations to discover potential inhibitors of SARS-CoV-2 main protease. The results revealed that the study might contribute to the discovery and development of potent drug candidates to suppress SARS-CoV-2 [12]. Accelerated free energy perturbation-based virtual screening of the existing drugs was performed to discover potential inhibitors of SARS-CoV-2 main protease. Out of 25 drugs, 15 were observed to be potent inhibitors [13]. Molecular docking and molecular dynamics simulations were performed to discover potential inhibitors versus the main protease of SARS-CoV-2 from previously known SARS-3CL protease inhibitors. The in-silico results indicated that some selected N-substituted isatin and pyrazolone derivatives could be utilized as potent inhibitors and might also possess an anti-viral effect versus SARS-CoV-2 [14]. Virtual screening, pharmacophore modeling, docking, and molecular dynamics were performed to discover novel potent inhibitors of the N-terminal domain of nucleocapsid protein of SARS-CoV-2. The study revealed that three molecules may serve as a potential lead for the discovery of potent inhibitors against SARS-CoV-2 [15]. Abdellatiif et al suggested a computational in silico bond among FDA-approved drug candidates and COVID-19-associated proteins and receptors. The compound with the best docking score was expected to be a potential therapeutic inhibitor of the COVID-19 [16]. Molecular docking, pharmacological and toxicological property predictions (ADMET), efficiency calculations of ligands, and molecular dynamics simulations in conjunction with MM-GBSA binding free energy predictions of 26 coumarin and quinoline derivatives were carried out to discover potential inhibitors of SARS-CoV-2 main protease. The investigations proved valuable for the discovery and development of novel anti-COVID-19 therapeutic candidates in the future [17]. Abel et al performed computational prediction of approved drugs, inhibited drugs, and phytochemicals to discover potent inhibitors of SARS-CoV-2 main protease. The in-silico data attained in this work might also contribute to the discovery of potent inhibitors of the SARS-CoV-2 main protease [18]. In this work, two drugs cefixime, etoposide, and a previously isolated compound nebrodenoside A are selected for the computational study [19]. It is noteworthy that this study is purely theoretical and speculative. Subsequently, in vitro and in vivo investigations would be needed to validate the inhibitory effects of the selected candidates against the target proteins of COVID-19.
2. Methodology
Protein formulation for docking
The x-rays crystal structures of three pathogenic proteins were recovered from the protein data bank of high resolutions (http://www.rcsb.org/pdb): SARS-CoV-2 spike glycoprotein (PDB ID: 6VXX: Resolution 2.80 Å) [20], nucleocapsid phosphoprotein (N) (PDB ID: (6YUN) [8], and envelope protein E (PDB ID: 7K3G) [9] were downloaded. For molecular docking, three binding pockets containing active sites were selected [21], and three independent docking investigations were executed through Molegro Virtual Docker (MVD) software in a computer cluster system provided by EXPER, model-FQC-01266 running Intel Core i3–2100 CPU @3.10GHz Processor, 64 BİT, 4 GB RAM, 1TB hard disk, and NVIDIA GeForce GT 630 Graphic card. MolDock Score was chosen at the scoring function and the search algorithm. After docking, energy minimization and H-bond optimizations were executed. The docking simulation was reproduced for each ligand 20 times. The top binding scores were applied for additional study. Likewise, Discovery Studio Visualizer 2020 was employed for the comprehensive study of docking results.
Molecular Dynamics Study (MDS)
For each protein, the structures of the leading-docked complex were chosen for 100 ns molecular dynamics simulation (MDS). The MDS with CHARMM 36 force field was conducted using NAMD software [22]. The complexes for the MDS are prepared using VMD [23]. The equilibration of complexes was done through the CHARMM GUI web server followed by a production run for 100 ns on a supercomputer [24]. Following conditions were applied for equilibration; protein-small molecule was solvated in the TIP3P water model and 0.154 M NaCl solution at 310 K temperature and pH 7 [25]. For trajectories analysis, VMD was used while cluster analysis was done through Chimera software of UCSF [26]. After trajectory clustering, the five most populous clusters were characterized by a conformation and were evaluated for their binding to the protein. AutoDock Vina software was employed in the binding energy computation utilizing 40 Å × 40 Å × 40 Å box dimensions [27,28].
3. Results and discussions
Binding energies
The docking scores of cefixime, etoposide, and nebrodenside A are presented in Table 1 for three crystal structures. The results indicate that etoposide expressed the lowest binding energies with all three proteins, followed by nebrodenside A and cefixime. The complexes with the lowest binding energies were selected for molecular dynamics simulations to validate the docking results.
Table 1.
Binding energy of cefixime, etoposide, and nebrodenside A.
| Cefixime Binding Energy ∆G | Etoposide Binding Energy ∆G | Nebrodenside A Binding Energy ∆G | |
|---|---|---|---|
| 6VXX | -6.1 | -7.5 | 6.5 |
| 6YUN | -6.4 | -8.4 | -6.6 |
| 7K3G | -4.5 | -5.7 | -4.7 |
Interaction analysis of protein 6VXX
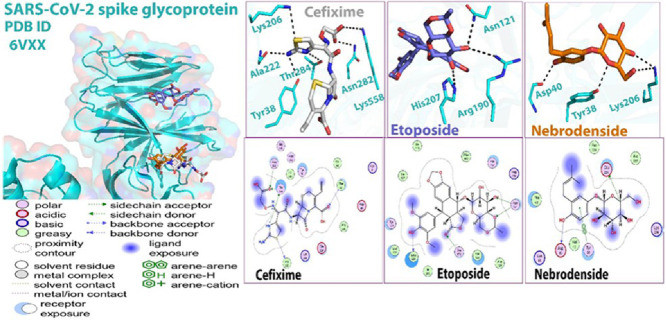
The results indicated that cefixime expressed five hydrogen bonding interactions with residues Lys 206, Ala 222, Thr 284, Asn 282, and Phe 43. In the case of etoposide, three hydrogen-bonding interactions were observed with residues Asn 121, Ser 172, and Leu 226, while nebrodenside A established two hydrogen-bonding interactions with residues Glu 224, and Asp 40 in addition to one arene-arene interaction with residue Tyr 38 (Fig. 1 ).
Fig. 1.
Molecular docking (2D and 3D) images of protein 6VXX with drugs.
The details of interactions are presented in Table 2 which show that etoposide expressed good values in terms of binding energies.
Table 2.
Interactions of ligands with protein 6VXX.
| 6vxx/ Cefixime | ||||
|---|---|---|---|---|
| Ligand | Receptor | Interaction | Distance | E (kcal/mol) |
| N1 9 | OG1 THR 284 (A) | H-donor | 3.48 | -1.2 |
| S1 12 | O ALA 222 (A) | H-donor | 3.90 | -1.6 |
| O3 20 | ND2 ASN 282 (A) | H-acceptor | 3.25 | -2.3 |
| O3 20 | OG1 THR 284 (A) | H-acceptor | 3.06 | -1.1 |
| O6 35 | CA PHE 43 (A) | H-acceptor | 3.31 | -0.7 |
| 6vxx / Etoposide | ||||
|---|---|---|---|---|
| Ligand | Receptor | Interaction | Distance | E (kcal/mol) |
| O6 42 | O LEU 226 (A) | H-donor | 2.89 | -2.7 |
| O4 17 | OG SER 172 (A) | H-acceptor | 2.96 | -1.0 |
| O11 25 | ND2 ASN 121 (A) | H-acceptor | 3.16 | -1.9 |
| O12 30 | ND2 ASN 121 (A) | H-acceptor | 2.96 | -1.0 |
| 6vxxa / Nebrodenside A | ||||
|---|---|---|---|---|
| Ligand | Receptor | Interaction | Distance | E (kcal/mol) |
| C13 3 | OE1 GLU 224 (A) | H-donor | 3.17 | -0.7 |
| O1 19 | O ASP 40 (A) | H-donor | 2.88 | -0.9 |
| 6-ring | 6-ring TYR 38 (A) | pi-pi | 3.77 | 0.0 |
Interaction analysis of protein 6YUN
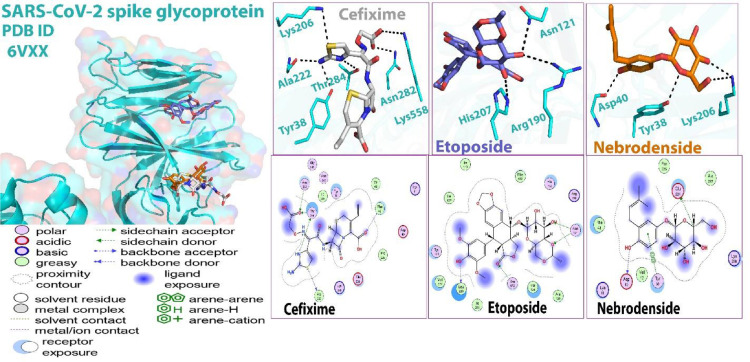
Cefixime formed three hydrogen bonding interactions with residues Tyr 87, Phe 28, and Arg 31, whereas etoposide established two interactions; one hydrogen bonding with residue Arg 31 and one arene-arene interaction with residue Phe 40. Nebrodenside A established only one arene-arene interaction with residue Phe 28 (Fig. 2 ).
Fig. 2.
Molecular docking (2D and 3D) images of protein 6YUN with drugs.
The details of interactions are presented in Table 3 . The data show that E (Kcal/mol) of etoposide is the lowest which is an indication of its favorable interactions.
Table 3.
Interactions of all ligands with protein 6YUN.
| 6yun / Cefixime | ||||
|---|---|---|---|---|
| Ligand | Receptor | Interaction | Distance | E (kcal/mol) |
| S1 12 | O THR 36 (A) | H-donor | 3.70 | -0.3 |
| 6yun / Etoposide | ||||
|---|---|---|---|---|
| Ligand | Receptor | Interaction | Distance | E (kcal/mol) |
| O12 30 | NH2 ARG 31 | (A) H-acceptor | 3.39 | -1.5 |
| 6-ring 6-ring PHE 68 | (A) pi-pi | 3.93 | 0.0 | |
| 6yun / Nebrodenside A | ||||
|---|---|---|---|---|
| Ligand | Receptor | Interaction | Distance | E (kcal/mol) |
| 6-ring | 6-ring PHE 28 (A) | pi-pi | 3.78 | 0.0 |
Interaction analysis of 7K3G
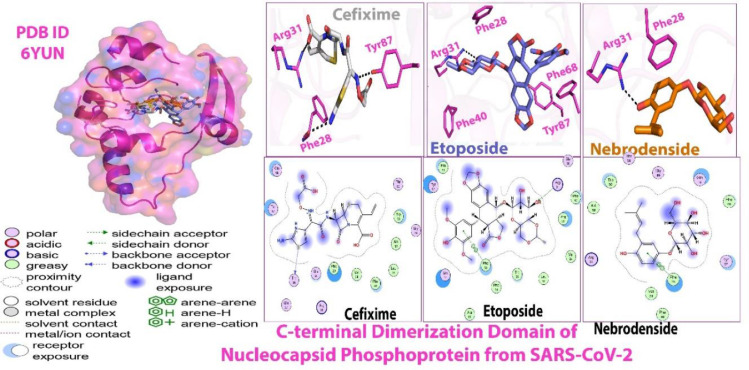
In the case of protein 7K3G, none of the drugs expressed any interaction (Fig. 3 ).
Fig. 3.
Molecular docking (2D and 3D) images of protein 7K3G with drugs.
Molecular Dynamics Simulations (MDS)
Simulation study deals with the physical movement of molecules and atoms. MDS analysis was performed for the three complexes of the etoposide drug because it exhibited the best results compared to the other two drugs. The RMSD and RMSF parameters were used to explain the stability of the protein during simulation.
Ligand properties were explained through four RMSD parameters; the radius of gyration (rGyr), molecular surface area (molSA), solvent-accessible surface area (SASA), and polar surface area (PSA).
The protein-ligand and contact histograms were used to determine the protein-ligand interactions which were classified into four categories i.e. hydrogen bonding, hydrophobic, ionic, and water bridges.
Hydrogen bonding plays a key role in ligand-receptor complex formation and is imperative in drug design because it has a noticeable impact on drug specificity, metabolism, and adsorption.
Ionic or polar interactions are between two oppositely charged atoms that are at 3.7 Å of each other. Moreover, these interactions do not contain hydrogen bonding.
Water bridges are hydrogen-bonded protein-ligand interactions facilitated by a water molecule.
The hydrogen-bond configuration is somewhat eased off from the standard hydrogen bond description. The present geometric standards for a protein-water or water-ligand hydrogen bonds are; a distance of 2.8 Å between the donor and acceptor atoms (D—H…A); a donor angle of ≥ 110o between the donor-hydrogen-acceptor atoms (D—H….A); and an acceptor angle of ≥ 90o between the hydrogen-acceptor-bonded atom (H…A—X).
Stability of protein (6VXX) and ligand
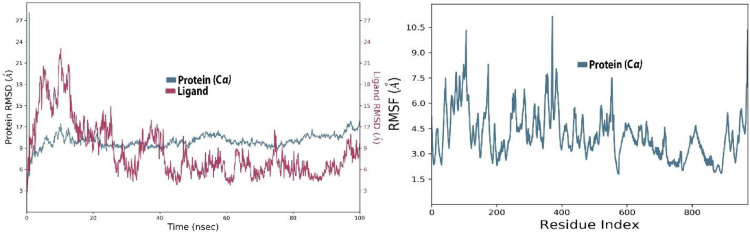
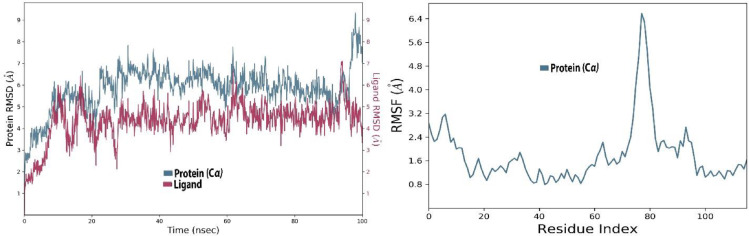
The RMSD value for both ligand and protein was calculated from the graph and its value was noted at the beginning of the simulation for protein. However, the value expressed massive fluctuations till 10 ns followed by the equilibrium state at 10 Å during simulation time. For the ligand, the RMSD value was noted to be 3-21 Å after half time of the simulation, whereas the least fluctuations were exhibited from 5 Å to 6 Å during the rest of the time.
The RMSF values exhibited exclusive fluctuation in various ranges, for example from the first residue to 200 the RMSF value is in the range of 2-10.4 Å, from 201 to 400 the value is noted in the range of 4.5-10.7 Å, from 401 to 600 the value least fluctuated and stayed in the range of 4.5-4.7 Å. Similarly, the rest of the residues show the least fluctuations in the 4.5 Å range during simulation (Fig. 4 ).
Fig. 4.
The protein-ligand (etoposide-6VXX) RMSD of the complex and RMSF trajectory during the 100 ns.
Ligand properties
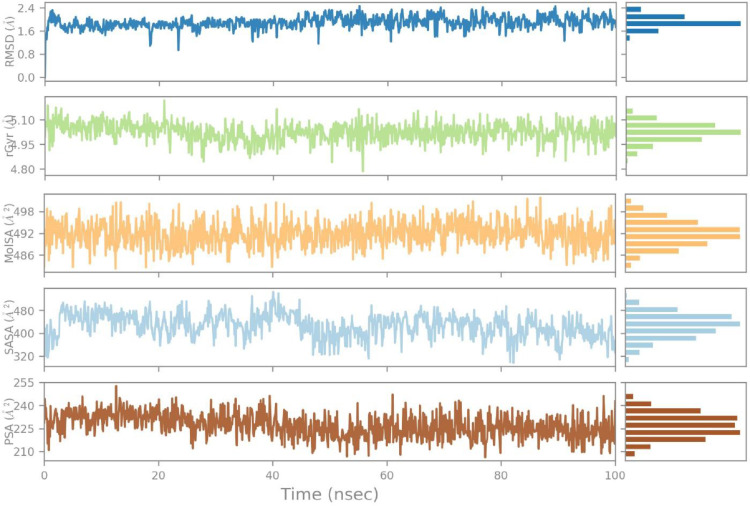
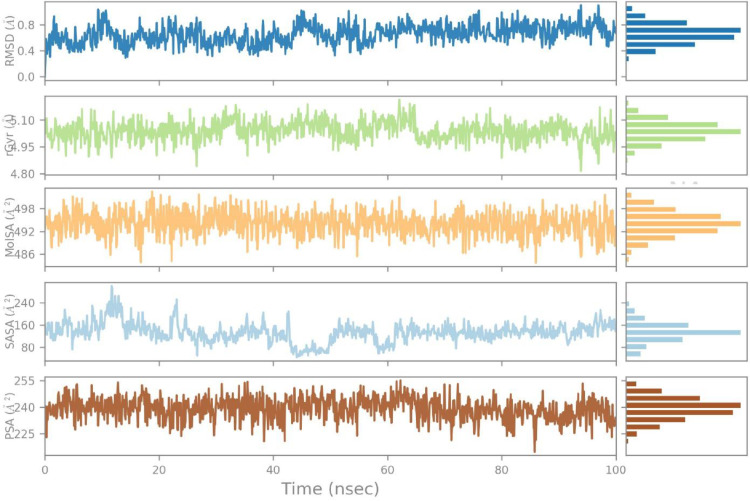
The RMSD value of the ligand stabilizes in a range of 1.6-1.8 Å throughout the simulation time, the radius of gyration value is found in a range of 4.95-4.10 Å with minor fluctuations, and the molecular surface area value is noted in a range of 492-498 Å2 during most of the simulation time.
Additionally, the solvent accessible area expressed fluctuations starting from 320 Å2 and ending at 480 Å2 during 40 ns but was then found constant at 400 Å2 throughout 100 ns with the least fluctuations.
Similarly, the polar surface area value fluctuated between 225 Å2 to 240 Å2 for 40 ns but afterward reached the equilibrium state of 225 Å2 for 100 ns (Fig. 5 ).
Fig. 5.
The ligand property trajectory of the (Etopside-6VXX) complex through 100 ns simulation.
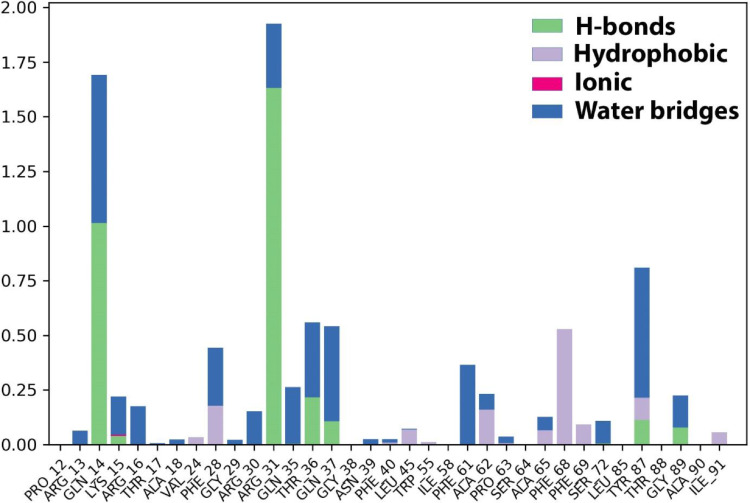
Protein-ligand contacts
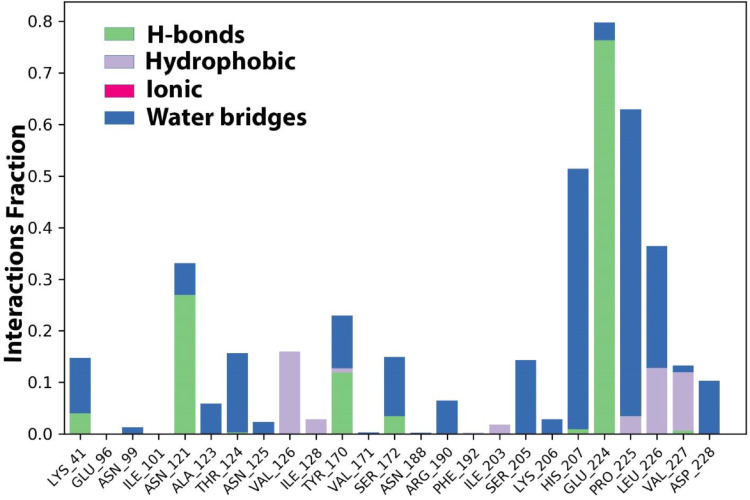
The protein-ligand contacts involved hydrogen bonding with the following six residues; Glu224, His 207, Ser172, Tyr170, Asn121, Lys 41. The hydrophobic contacts are developed by the residues Pro225, Leu226, Val 227, Val126, Ile128, and Ile203. Additionally, water bridges exist in a maximum of residues along with hydrogen bonding and hydrophobic interactions (Fig. 6 ).
Fig. 6.
The histogram of protein-ligand (Etopside-6VXX) contact throughout the trajectory.
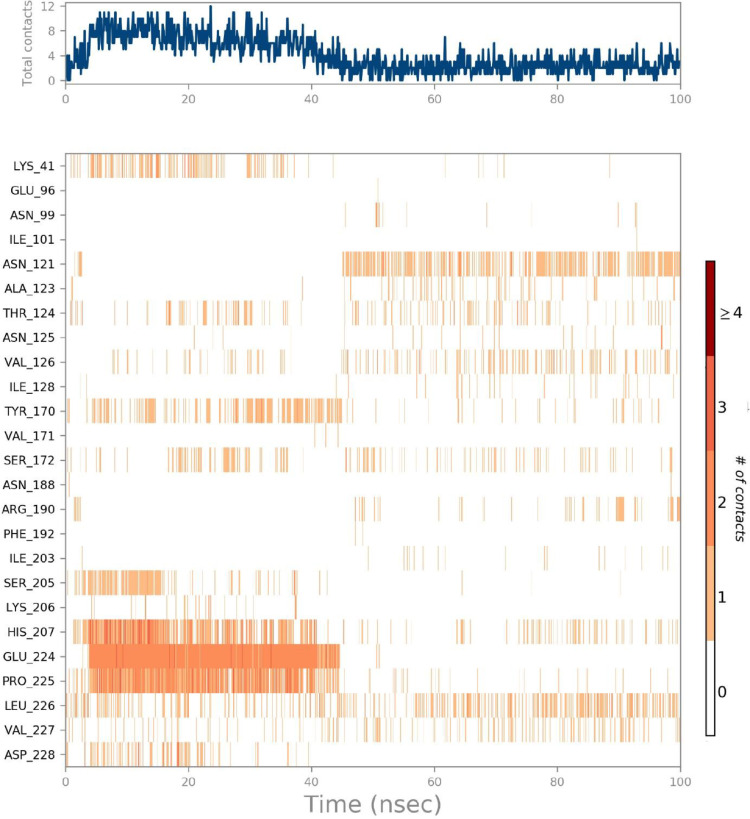
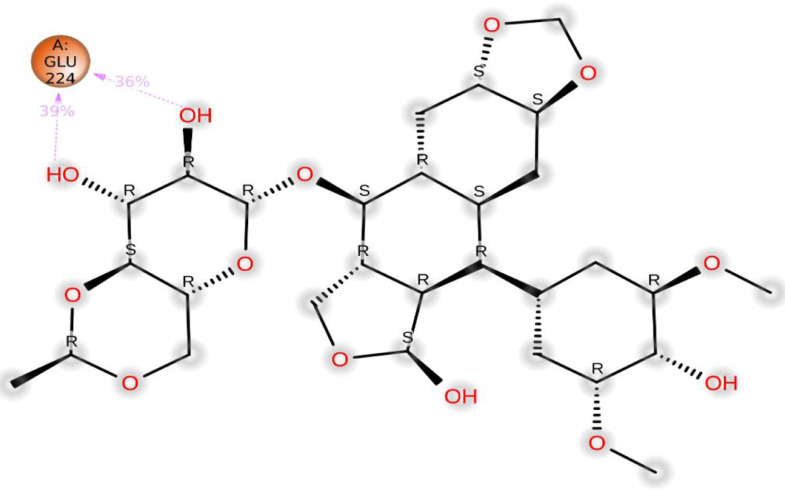
The ligand-receptor interactions histogram consists of two panels which explain the overall interactions between the ligand and protein, the top panel expressed the total explicit contacts between the ligand and protein for every trajectory frame and the bottom panel represent each residue contribution in contacts and is shown by the dark orange line at residues Pro225.Glu224.His 207, Asn121, and Lys41, expressing their interactions (Fig. 7 a).
Fig. 7.
(a) The total number of contacts/interactions in each trajectory framework of (Etopside-6VXX) complex. (b) Interaction is represented by the effective site of amino acids in each trajectory framework of the (Etopside-6VXX) complex. (b). Hydrogen bonding interaction (% age) of residue Glu224 during the simulation.
Additionally, the residue Glu224 established 39% and 36% hydrogen bonding interactions during the simulation (Fig. 7b).
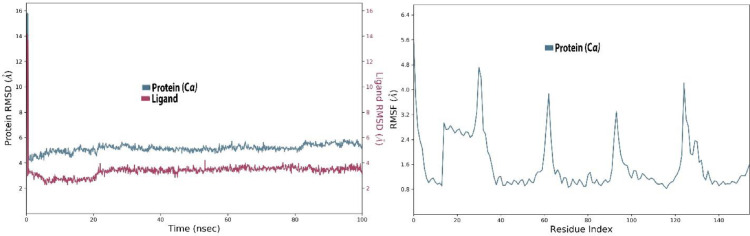
Stability of protein (6YUN) and ligand
RMSD factor is used to find out the stability of protein and ligand. In this case, both protein and the ligand express high fluctuations, the protein shows fluctuations in the range 2.5-7.8 Å throughout 30 ns which then equilibrated in a range of 6-7 Å during 90 ns, however, a high fluctuation was observed again during 100 ns. Whereas, the ligand also expressed fluctuations in a range of 1.5-6 Å through 25 ns but then attained an equilibrium state in the range of 4.5-5 Å throughout the simulation period with minor fluctuations (Fig. 8 ).
Fig. 8.
The protein-ligand (etoposide-6YUN) RMSD of the complex and RMSF trajectory during 100 ns.
The RMSF value shows the least fluctuations and for most of the residues, it stayed in a range of 1.6-2.0 Å throughout the simulation period except that a spike in RMSF value was observed between residues 60 and 80 only. The exhibition of the least fluctuations confirmed the strong attachment of ligand with protein (Fig. 8).
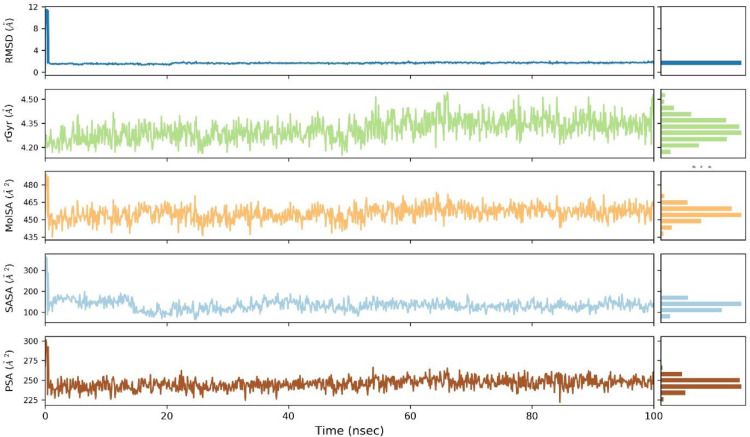
Ligand properties
The RMSD value for ligand attained equilibrium at 0.7 Å after an initial fluctuation from 0.4 Å to 0.6 Å for most of the simulation time, the radius of gyration was found in the range of 4.97-4.10 Å during much of the simulation time with the least fluctuations. The molecular surface area expressed fluctuations above 4.98 Å2 through 40 ns but subsequently stabilized to 4.94 Å2 during the complete course of the simulation period. The solvent-accessible surface area value fluctuated during the simulation period from 80 Å2 to 160 Å2 and touched both high as well as low peaks, overall expressing unstable conditions. The polar surface area also expressed variations but got to a constant value of 240 Å2 for a long period of the simulation (Fig. 9 ).
Fig. 9.
The ligand property trajectory of the (Etopside-6YUN) complex through 100 ns simulation.
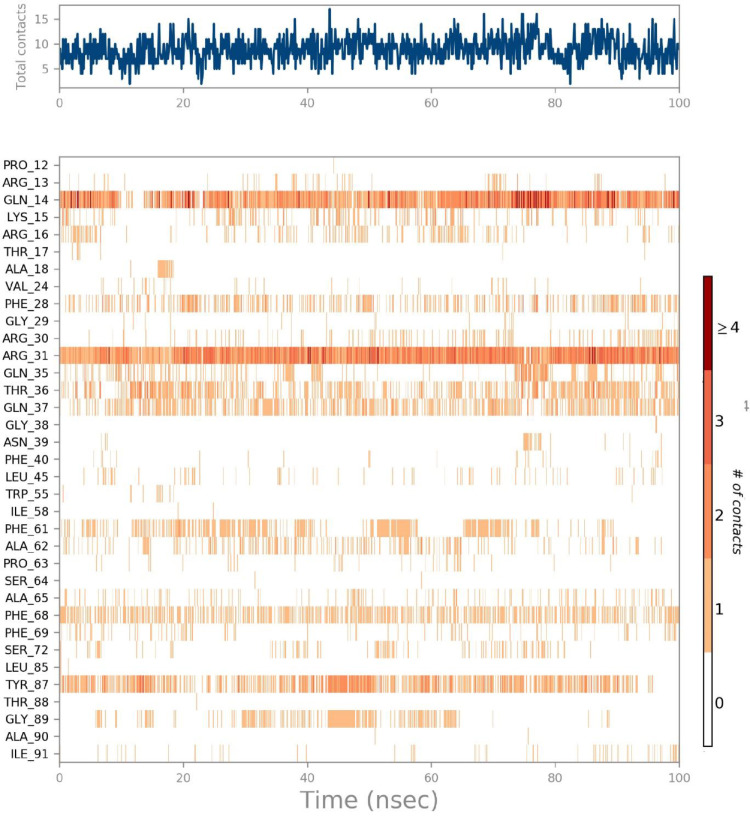
Protein-ligand contacts
Protein-ligand interactions play a key role in drug discovery. Such interactions are expressed through four types of contacts; hydrogen bonding, hydrophobic, ionic, and water bridges. In hydrogen bonding, the subsequent five residues are involved; Gln 14, Lys15, Arg 31, Thr 37, and Gln37, while hydrophobic interactions are exhibited by three residues namely, Val 24, Phe 28, and Phe 68, however, a single residue Lys 15 is ionic contact. Still, numerous residues expressed water bridges in the protein-ligand contacts (Fig. 10 ).
Fig. 10.
The histogram of protein-ligand (Etopside-6YUN) contact throughout the trajectory.
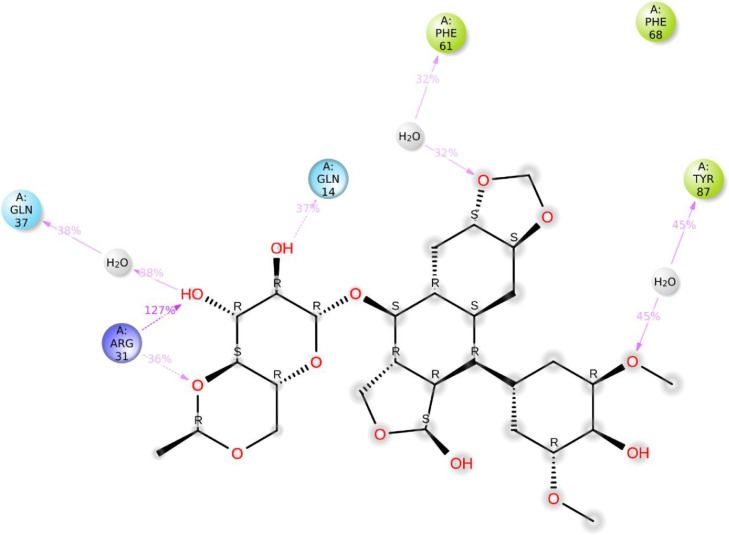
To sum up the whole process, a ligand-receptor interactions timeline histogram was used. The top panel shows specific contacts for every trajectory frame, while the lower panel identifies the amino acids interacting with the ligand. The dark orange shades are shown by the succeeding four residues; Gln 14, Arg31, Tyr87, and Phe 68 (Fig. 11 a).
Fig. 11.
(a) The total number of contacts/interactions in each trajectory framework of the (Etopside-6YUN) complex. (b) Interaction is represented by the effective site of amino acids in every trajectory framework of the (Etopside-6YUN) complex. b: Hydrogen bonding interaction (% age) of Gln14, Arg31, Tyr87, Phe68, and Gln37 during the simulation time.
The residues Gln14 constituted 37% of the hydrogen-bonding interactions during the simulation time involving water molecules. Whereas, Arg31, Tyr87, Phe68, and Gln37 possessed 127%, 36%, 45%, and 32% interactions, respectively during the simulation time (Fig. 11b).
Stability of protein (7K3G) and ligand
The protein attained 4 Å RMSD at the beginning of the simulation and stayed at equilibrium till the end of simulation during 100 ns, while the ligand expressed deviation from 4 Å with a small fluctuation near 20 ns but later attained equilibrium through 100 ns (Fig. 12 ). The RMSF of the protein shows the stability of protein per residue during the simulation. Since there is no hydrogen bonding in this protein, therefore, residual fluctuations were observed for most of the protein, and the RMSF value reached 4.8 Å for 20-40 residues, whereas the fluctuation touched 4 Å for 60 residues. Similarly, the RMSF value increased to 3.2 Å for 90-100 residues and reached again to 4 Å for 120 residues (Fig. 12).
Fig. 12.
The protein-ligand (etoposide-7K3G) RMSD of the complex and RMSF trajectory during the 100 ns.
Ligand properties
The RMSD value for ligand stabilized at 2 Å throughout the simulation time of 100 ns and expressed stability. For ligand, the radius of gyration values ranged 4.20-4.35Å during 50 ns followed by a fluctuation between 4.35 Å and 4.50 Å during 100 ns with an average value of 4.30 Å.
MolSA represents surface calculation with a 1.4 Å probe radius. This value is equivalent to a van der Waals surface area. For ligand, its value is constant in the range of 450-465 Å with minimum fluctuations throughout the simulation period.
The solvent-accessible surface area for the ligand is observed in the range of 100-150 Å2 with minor fluctuations throughout the simulation period. The polar solvent accessible area in the molecule is furnished solely by nitrogen and oxygen atoms. For ligand, its range is found constant from 230 Å2 to 250 Å2 with minor fluctuations in the simulation period (Fig. 13 ). The majority of the ligand properties expressed the least fluctuation only at the beginning and middle of the simulation but overall equilibrium states were found in most of the simulation period which indicated the strength of ligand inside the dynamic pocket of protein (Fig. 13).
Fig. 13.
The ligand property trajectory of the (Etopside-7K3G) complex throughout 100 ns simulation.
Protein-ligand contacts
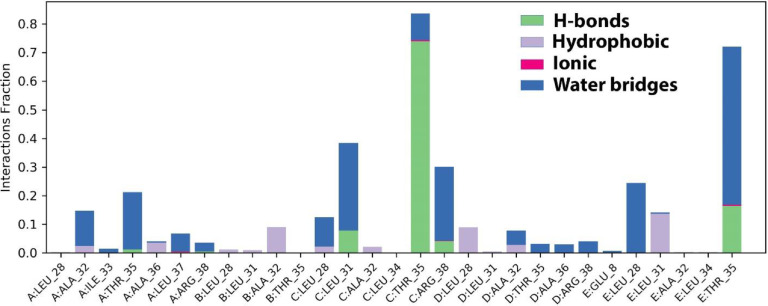
The histogram shows that amino acid residues namely Thr35, and Arg34 are involved in hydrogen bonding interactions, while Leu 31, 28 Ala 28, 33, and 36 established hydrophobic interactions. In this case, only one residue namely Leu37 formed an ionic interaction (Fig. 14 ).
Fig. 14.
The histogram of protein-ligand (Etopside-7K3G) contact throughout the trajectory.
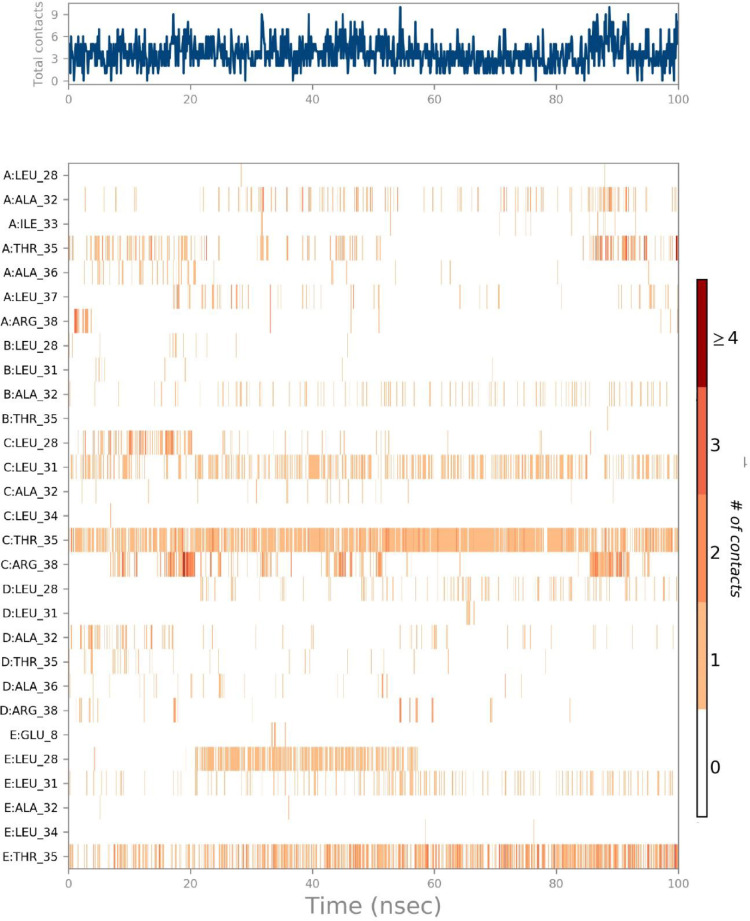
The interactions and contacts (Hydrogen bonds, Hydrophobic, Ionic, and Water bridges) timeline representation is outlined in the ligand-receptor interaction (histogram) which consists of two panels (Figs. 15 a,b). The top panel shows all specific contacts of the proteins with the ligand for every trajectory frame. From zero to nine, the contact numbers vary throughout the trajectory. (Fig. 15a). Whereas, the bottom panel is used to analyze the amino acid contribution in making interaction with ligand (Fig. 15b). It is used to identify the amino acid involved in interaction with ligand in each trajectory frame.
Fig. 15.
(a) The total number of contacts/interactions in each trajectory framework of the (Etopside-7K3G) complex. (b) Interaction is represented by the effective site of amino acids in every trajectory framework of the (Etopside-7K3G) complex. b: Hydrogen bonding interaction (% age) of Thr35 during the simulation.
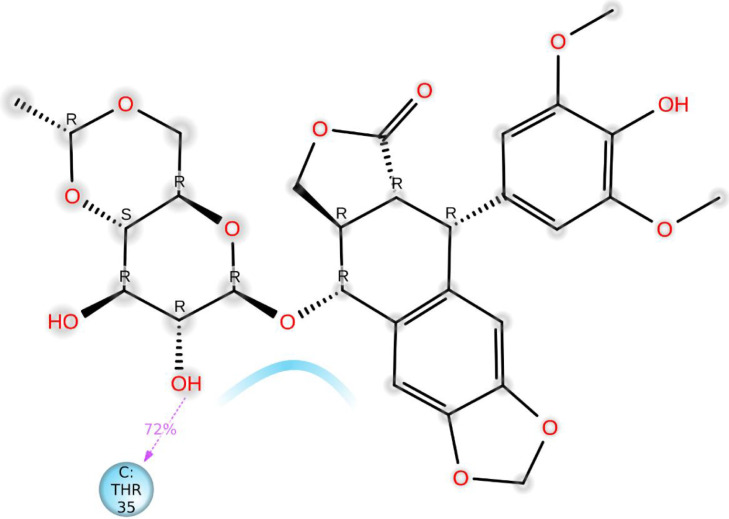
The darker orange shade represents some amino acid residues involved in more than one precise contact with the ligand in a specific trajectory framework according to the scale given below in the plot. In the complex, there is a single deep orange band with residue Thr 35, which explains that this specific residue is in strong interaction with the ligand in all probable alignments (geometry). The results are analogous to the histogram data (Fig. 15a,b). The residue Thr35 formed 72% of hydrogen bonding interaction during the simulation time (Fig. 15b).
4. Conclusions
Identification of already existing and approved drugs as a potent inhibitor of COVID-19 is a remarkable step toward the early discovery of drugs, vaccines alone are not enough to halt or minimize its devastations. The two drugs namely cefixime, etoposide, and a previously isolated compound nebrodenside A showed strong binding interactions in the active pockets of target proteins of SARS-CoV-2 with the least binding energies. Etoposide showed more promising results through docking and simulation analyses as compared to cefixime and nebrodenside A. Furthermore, etoposide and three protein complexes were selected for the molecular dynamics simulations to validate the ligand-receptors contacts. The results were found to perfectly match the physical parameters. RMSD and RMSF were used to explain the stability of proteins and ligand. Moreover, specific parameters such as the radius of gyration (rGyr), solvent-accessible area (SASA), molecular surface area (MolSA), and polar surface area (PolSA) determined the properties of ligand for each trajectory during simulation. The ligand-receptor contacts including hydrogen bonding, hydrophobic interactions, ionic and water bridges were presented through a histogram. Additionally, specific amino acids involved in the aforementioned interactions were identified through a timeline histogram for every trajectory, and the in-silico results of the two techniques supported each other. Nevertheless, in vivo and in vitro studies are needed to remodel and establish solid experimental evidence of the three inhibitors versus COVID-19.
CRediT authorship contribution statement
Haroon ur Rashid: Formal analysis, Reviewing & editing the original draft, Literature survey, Preparing the revision. Nasir Ahmad: Writing the mauscript, Conceptulization and supervision. Mohnad Abdalla: Writing the manuscript, Conceptulization, Data analysis. Khalid Khan: Methodology, Data curation. Marco Antonio Utrera Martines: Methodology, Technical input. Samah Shabana: Methodolgy, editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The authors are thankful to the Universidade Federal de Mato Grosso do Sul. This work was funded in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) – Finance Code 001, and CAPES-PrInt, Finance Code 88881.311799/2018-01. The authors are also grateful to the Conselho Nacional de Desenvolvimento Científico e Tecnológico— Brasil (CNPq)—Finance Code 420852/2018-2 and Fundação de Apoio ao Desenvolvimento do Ensino, Ciência e Tecnologia do Estado de Mato Grosso do Sul—Brasil (FUNDECT-MS)—grants 036/2017 PPSUS-MS (59/300.074/2017) providing funding to execute this project. The authors also acknowledge the financial support of Shandong University and the Chinese Scholarship Council to execute this project.
References
- 1.Mogi R., Spijker J. The influence of social and economic ties to the spread of COVID-19 in Europe. J. Pop. Res. 2021 doi: 10.1007/s12546-021-09257-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization . 2021. Coronavirus.https://www.who.int/emergencies/diseases/novel-coronavirus-2019 Accessed on May 8. [Google Scholar]
- 3.Lu R., Zhao X., Li J., Niu P., Yang B., et al. Genomic characterization and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graham R.L., Sparks J.S., Eckerle L.D., Sims A.C., Denison M.R. SARS coronavirus replicase proteins in pathogenesis. Virus Res. 2008;133(1):88–100. doi: 10.1016/j.virusres.2007.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prasad A., Prasad M. SARS-CoV-2: the emergence of a viral pathogen causing havoc on human existence. J. Genet. 2020;99(37):1–4. doi: 10.1007/s12041-020-01205-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duan L., Zheng Q., Zhang H., Niu Y., Lou Y., Wang H. The SARS-CoV-2 Spike Glycoprotein Biosynthesis, Structure, Function, and Antigenicity: Implications for the Design of Spike-Based Vaccine Immunogens. Front. Immunol. 2020;11:1–12. doi: 10.3389/fimmu.2020.576622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zinzula L., Basquin J., Bohn S., Beck F., Klumpe S., Pfeifer G., Nagy I., Bracher A., Hartl F.U., Baumeister W. High-resolution structure and biophysical characterization of the nucleocapsid phosphoprotein dimerization domain from the Covid-19 severe acute respiratory syndrome coronavirus 2. Biochem. Biophys. Res. Commun. 2021;538:54–62. doi: 10.1016/j.bbrc.2020.09.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mandala V.S., McKay M.J., Shcherbakov A.A., Dregni A.J., Kolocouris A., Hong M. Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers. Nat. Struct. Mol. Biol. 2020;27:1202–1208. doi: 10.1038/s41594-020-00536-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaliamurthi S., Selvaraj G., Selvaraj C., Singh S.K., Wei D.Q., Peslherbe G.H. Structure-Based Virtual Screening Reveals Ibrutinib and Zanubrutinib as Potential Repurposed Drugs against COVID-19. Int J Mol Sci. 2021;22(13):7071. doi: 10.3390/ijms22137071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abu-Saleh A.A., Awad I.E., Yadav A., Poirier R.A. Discovery of potent inhibitors for SARS-CoV-2′s main protease by ligand-based/structure-based virtual screening, MD simulations, and binding energy calculations†. Phys. Chem. Chem. Phys. 2020;22:23099–23106. doi: 10.1039/d0cp04326e. [DOI] [PubMed] [Google Scholar]
- 12.Ngo S.T., Pham N.Q.A., Le L., Pham D.-H., Vu V. Computational determination of potential inhibitors of SARS-CoV‑2 main protease. J. Chem. Inf. Model. 2020;60(12):5771–5780. doi: 10.1021/acs.jcim.0c00491. [DOI] [PubMed] [Google Scholar]
- 13.Li Z., Li X., Huang Y-Y., Wu Y., Liu R., Zhou L., Lin Y., Wu D., Zhang L., Liu H., Xu X., Yu K., Zhang Y., Cui J., Zhan C-G., Wang X., Luo H-B. Identify potent SARS-CoV-2 main protease inhibitors via accelerated free energy perturbation-based virtual screening of existing drugs. Proc. Natl. Acad. Sci. 2020;117(44):27381–27387. doi: 10.1073/pnas.2010470117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Motiwale M., Yadav N.S., Kumar S., Kushwaha T., Choudhir G., Sharma S., Singour P.K. Finding potent inhibitors for COVID-19 main protease (Mpro): an in silico approach using SARS-CoV-3CL protease inhibitors for combating CORONA. J. Biomol. Struct. Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1829501. [DOI] [PubMed] [Google Scholar]
- 15.Dhankhar P., Dalal V., Singh V., Tomar S., Kumar P. Computational guided identification of novel potent inhibitors of N-terminal domain of nucleocapsid protein of severe acute respiratory syndrome coronavirus 2. J. Biomol. Struct. Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1852968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdellatiif M.H., Ali Am., Ali Ab., Hussien M.A. Computational studies by molecular docking of some antiviral drugs with COVID-19 receptors are an approach to medication for COVID-19. Open Chem. 2021;19:245–264. [Google Scholar]
- 17.Osvaldo Y., Isaías O.M., Eugenio U., Carlos A., William T., Pérez-Donoso J.M., García-Beltrán O., González-Nilo F. In silico study of coumarins and quinolines derivatives as potent inhibitors of SARS-CoV-2 Main Protease. Front. Chem. 2021;8:1–15. doi: 10.3389/fchem.2020.595097. 595097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abel R., María P.R., Qiaofeng C., Pérez-Sánchez H., Coluzzi F., Rocco M., Marchetti P., Mura C., Simmaco M., Bourne P.E., Preissner R., Banerjee P. Computational Prediction of Potential Inhibitors of the Main Protease of SARS-CoV-2. Front. Chem. 2020;8:1–19. doi: 10.3389/fchem.2020.590263. 590263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khan K., Rasool S., Khan K., Badshah S.L., Ahmad N., Jan M.T., Hizbullah S.M., Khan I., Ullah A., Muhammad A. Computational evaluation and anti-inflammatory and analgesic activities of nebrodenside a isolated from Dodonaea viscosa. Nat. Prod. Commun. 2019;14(5):1–5. [Google Scholar]
- 20.Walls A.C., Park Y.J., Tortorici M.A., Wall A. 2020. Structure of the SARS-CoV-2 spike glycoprotein (closed state), Seattle Structural Genomics Center for Infectious Disease (SSGCID) McGuire AT, Veesler D. [DOI] [Google Scholar]
- 21.Cavasotto C.N., Lamas M.S., Maggini J. Functional and druggability analysis of the SARS-CoV-2 proteome. Eur. J. Pharmacol. 2021;890 doi: 10.1016/j.ejphar.2020.173705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang J., MacKerell A.D., Jr. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 2013;34(25):2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Humphrey W., Dalke A., Schulten K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 24.Jo S., Kim T., Iyer V.G, Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29(11):1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 25.Mark P., Nilsson L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. A. 2001;105(43):9954–9960. [Google Scholar]
- 26.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 27.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]