Abstract
Objective
To undertake a genome-wide association study (GWAS) to identify genetic variants for stroke in an Indian population.
Methods
In a hospital-based case-control study, 8 teaching hospitals in India recruited 4,088 participants, including 1,609 stroke cases. Imputed genetic variants were tested for association with stroke subtypes using both single-marker and gene-based tests. Association with vascular risk factors was performed with logistic regression. Various databases were searched for replication, functional annotation, and association with related traits. Status of candidate genes previously reported in the Indian population was also checked.
Results
Associations of vascular risk factors with stroke were similar to previous reports and show modifiable risk factors such as hypertension, smoking, and alcohol consumption as having the highest effect. Single-marker–based association revealed 2 loci for cardioembolic stroke (1p21 and 16q24), 2 for small vessel disease stroke (3p26 and 16p13), and 4 for hemorrhagic stroke (3q24, 5q33, 6q13, and 19q13) at p < 5 × 10−8. The index single nucleotide polymorphism of 1p21 is an expression quantitative trait locus (plowest = 1.74 × 10−58) for RWDD3 involved in SUMOylation and is associated with platelet distribution width (1.15 × 10−9) and 18-carbon fatty acid metabolism (p = 7.36 × 10−12). In gene-based analysis, we identified 3 genes (SLC17A2, FAM73A, and OR52L1) at p < 2.7 × 10−6. Eleven of 32 candidate gene loci studied in an Indian population replicated (p < 0.05), and 21 of 32 loci identified through previous GWAS replicated according to directionality of effect.
Conclusions
This GWAS of stroke in an Indian population identified novel loci and replicated previously known loci. Genetic variants in the SUMOylation pathway, which has been implicated in brain ischemia, were identified for association with stroke.
The burden of stroke as the second leading cause of death and the leading cause of long-term disability is being felt all over the world, especially in developing countries such as India, which will have 80% of the world stroke cases by 2030.1 The etiology of stroke is multifactorial and includes both genetic and environmental factors.2 Studies of Indian immigrants in the United Kingdom and United States have suggested a higher vulnerability of Indian individuals to stroke, probably due to genetic factors.3 To date, genome-wide association studies (GWAS) are the most effective way for gene discovery when it comes to complex disorders such as stroke.4 A large number of GWAS and their meta-analysis have been conducted for stroke and revealed ≈40 loci.4 However, except for a few studies, the majority of the participants in stroke GWAS are of European origin. It has been suggested that such scenarios could lead to health disparities later because many of the findings based on Europeans are not transferable to populations of other ethnicities.5 GWAS of stroke from India are completely lacking.6 The Indian population of 1.35 billion people with a rich diversity of ethnic groups and ancestral populations with large founder effects and distinct genetic architecture in terms of allele frequency and linkage disequilibrium (LD) blocks offers tremendous opportunities for genetic studies.7-10
In this large-scale GWAS of stroke from India, we also study association with vascular risk factors and check the status of previous stroke GWAS loci and candidate genes studied in Indian individuals.
Methods
Participant Recruitment
This study involved a multicentric hospital-based case-control design. Eight teaching hospitals from different parts of India recruited phenotypically well-characterized stroke cases and ethnically matched controls (figure e-1 and appendix e-1 at doi.org/10.5061/dryad.qv9s4mwcv). Stroke was diagnosed from guidelines set by the World Health Organization by trained neurologists and was primarily of vascular origin. Eligibility criteria included age of 18 to 85 years, both sexes with a history of the first-ever stroke in life, Indian ancestry, and absence of any major neurologic disorders (epilepsy, Parkinson disorder, Alzheimer disease, multiple sclerosis, brain tumor, etc). Ischemic strokes were classified according to subtype classification in Trial of Org 10172 in Acute Stroke Treatment (TOAST).11 In total 4,088 participants were recruited, which included 1,609 stroke cases and 2,479 controls. Stroke-free status was assessed by a well-validated questionnaire.6 Ethnically matched individuals diagnosed as stroke-free with this questionnaire and no history of any serious neurologic disorder were recruited as controls. Controls were selected from spouses, who serve as better controls because they would have similar environmental exposure, hence limiting bias due to the effect of environmental exposures. When spouses were unavailable, then unrelated blood donors fulfilling inclusion and exclusion criteria were included as controls. Each study site recruited its controls from the same site after ethnicity matching. The controls thus selected approximated the distribution of exposure to that in the population from which cases arose.
Inclusion criteria for controls were as follows: no prior stroke by the Questionnaire for Stroke-Free Status, spouse or unrelated blood donors, age of 18 to 85 years (both sexes), willingness to provide written informed consent by self or legal representative, and no evidence of any serious brain disorders. Exclusion criteria for controls were unwillingness to provide written informed consent (by self or legal representative), pregnancy, or any serious brain disorder.
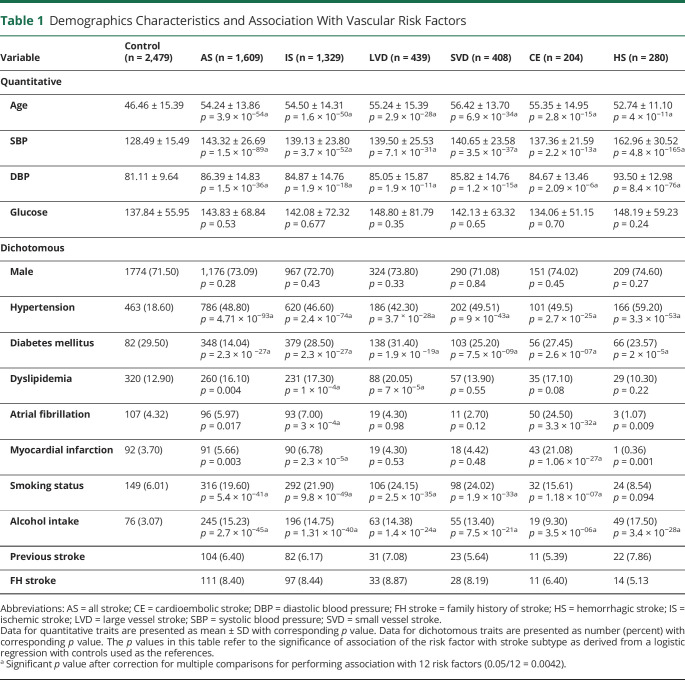
Detailed demographic characteristics of the cases and controls are presented in table 1 and table e-1 (doi.org/10.5061/dryad.qv9s4mwcv).
Table 1.
Demographics Characteristics and Association With Vascular Risk Factors
Standard Protocol Approvals, Registrations, and Patient Consents
The study protocol was approved by the ethics committees of the respective participating institutions, and signed informed consent was obtained from the participants before enrolling them into the study.
Power of the Study
We estimated the power of the study at GWAS significance levels of p = 5 × 10−8, assuming a population risk of 10% to detect an odds ratio (OR) in the range of 1.1 to 2.0 and allele frequency ranges of 0.01 to 0.50 using Quanto version 1.2.4 (figure e-2, doi.org/10.5061/dryad.qv9s4mwcv).
Genome-Wide Genotyping
Genome-wide genotyping was performed on an Illumina platform using the genome screen array version 2.0 (with additional multidisease content), which comprises ≈750,000 markers. Intensity data files were imported into Illumina's GenomeStudio software for intensity normalization, scaling, and offset estimation, followed by single nucleotide polymorphism (SNP) clustering, genotype assignment, and calling. Initial quality control was performed on the basis of Illumina-designed quality control probes (sample independent and sample dependent) to assess experiment and sample quality. For association analysis, cluster files generated from GenomeStudio were used to convert intensity data files to variant call format (VCF) files with Illumina's GTCtoVCF tool. Individual VCF files were merged using bcftools to create a single project–level VCF file. Emphasis was on getting a VCF file rather than the usual PLINK format .ped and .map file, which is the default option in GenomeStudio, because allele coding in VCF files is as per the reference genome on the positive strand and allele coding is not omitted for variants for which the alternate allele was not observed. With the availability of multiple tools designed to interrogate large VCF files being generated through large sequencing projects in a very short time, VCF format has become a popular choice.
Genotype and Sample Quality Checks
All marker and sample quality checks were carried out mainly with bcftools and plink2 unless mentioned otherwise. We implemented a stringent marker quality check and included only variants with genotype call rate >95%, deviation from Hardy-Weinberg equilibrium p > 1 × 10−5), minor allele frequency (MAF) >0.01, biallelic markers, and autosomal and X-chromosome markers. Both SNPs and short insertion and deletion were included for association analysis. After this first round of marker quality check, we were left with 399,541 markers. The numbers of markers failing quality checks in the various categories are given in table e-2 (doi.org/10.5061/dryad.qv9s4mwcv). Major loss of markers was due to the presence of nonpolymorphic and very low-frequency markers (34.71%) in this Indian population due to the nature of the chip design to capture variants that could be specific to populations across the world (table e-2).
During sample quality check, we excluded samples with a call rate <95%, heterozygosity beyond ±3 SDs, and genotype and pedigree sex mismatch. Markers were pruned with the indep-pairwise 50 5 0.2 function of plink to retain only markers with values of r2 < 0.2 in a window size of 50 markers and slide of 5 markers per window. Pruned markers were used to ascertain cryptic relatedness using the genome function in plink, and among the pairs of duplicated samples with PI_HAT values >0.6, only 1 of the samples with the highest call rate was retained. These samples with linkage disequilibrium pruned markers were used to perform principal components analysis (PCA) using FastPCA function implemented in EIGENSOFT version 7.2.1. The samples were projected on the samples of the 1,000 Genomes project using the first 2 PCAs, and PCA outliers were excluded. The sample quality check was blinded to the disease status of the participant. The number of samples excluded at the various stages is listed in table e-3 (doi.org/10.5061/dryad.qv9s4mwcv). After sample quality checks, the marker quality check was repeated (round 2) due to a change in the number of participants. Statistics such as call rate, MAF, and Hardy-Weinberg equilibrium p value were recalculated, which led to exclusion of 4,286 markers, leaving us in total 395,255 good-quality genotyped markers (table e-2).
Imputation
Imputation was carried out with the TOPMed reference panel (version R2 on GRC38). Whole-genome sequencing data of 97,256 individuals were available in this version of the reference panel. Genotypes were on the positive strand, and allele checks were made along with other quality control as suggested (topmedimpute.readthedocs.io/en/latest/) before submission of genotypes for imputation. Phasing and imputation were carried out on the TOPMedimputationserver.12 Analysis of the imputed genotypes was restricted to variants with MAF >0.01 and imputation quality >0.7.
Association Analysis and Functional Annotation
Association analyses with stroke and its subtypes were performed using the fastGWA function in GCTA tool, which uses the generalized mixed model. The use of generalized mixed models has been shown to be useful to account for relatedness and residual population stratifications using principal components. We used the first 10 principal components as covariates to remove any residual population stratification. Age and sex were also used as covariates in association analysis. We calculated effective allele count as 2 × MAF cases × number of cases × imputation quality and reported association analysis of only variants with an effective allele count >10. Such an effective allele count restricts association analysis to only situations in which at least 10 counts of good imputation quality minor allele are observed in cases, thereby ruling out false positives due to rare variants. We performed gene-based tests using the MAGMA tool as implemented in FUMA.13 For biological interpretation and better understating, annotation was performed with ANNOVAR, Online Mendelian Inheritance in Man, Kyoto Encyclopedia of Genes and Genomes, and Gene Ontology databases. We looked up previously published GWAS data of stroke and related phenotypes to check for replications and better biological insights using the following resources: Cerebrovascular Disease Knowledge Portal,14 Type 2 Diabetes Knowledge Portal,15 and Gene ATLAS.16 The GTEx17 and Phenoscanner18 resources were accessed to identify expression quantitative trait loci (eQTLs) for the top associated SNPs.
Replication of Top Hits in UK Biobank Samples of South Asian Origin
A total of 8,235 participants of South Asian origin were included for replication analysis based on Data-Field 21,000 of the UK Biobank dataset. Only individuals self-identifying as of Indian, Pakistani, or Bangladeshi origin were included, while any individual with mixed ethnicity was excluded. These included 211 all stroke cases, 98 ischemic stroke cases, and 25 hemorrhagic stroke cases. Classification into stroke cases was obtained from Data-Fields 42,007, 42,009, and 42,011. Association analysis was performed with the glm function of PLINK2 after accounting for age, sex, and first 10 principal components as covariates in the logistic regression model.
Replication of Previously Reported GWAS Loci of Stroke and Subtypes
We extracted the summary statistics of all previously reported loci based on GWAS studies of stroke (appendix e-1, doi.org/10.5061/dryad.qv9s4mwcv). This included GWAS studies on European and non-European individuals and their meta-analysis. The first replication was tested according to only directionality of effect because the current study is not powered enough to replicate associations performed in large studies like those of MEGASTROKE19 (>500,000 participants). Later, we also tested association on the basis of both directionality of effect and p < 0.05. Finally, we also report whether any loci stood the multiple testing threshold after correcting for the number of independent loci being queried.
Candidate Gene Studies on Indian Population
Details of the search strategy to identify candidate gene studies that investigated the association with stroke and its subtypes in the Indian population are presented in appendix e-1 (doi.org/10.5061/dryad.qv9s4mwcv). Briefly, we searched PubMed for studies investigating the association of genetic variants with stroke in the Indian population and extracted the association statistics from the publications. Later, we extracted results of the same genetic variants in the current study and checked whether they replicated at p < 0.05 and if the direction of effect was the same. Multiple-testing corrections were implemented after correction for the number of independent loci.
Data Availability
Summary statistics of the association of variants for all stroke subtypes at p < 1 × 10−5 are provided in the supplementary tables (doi.org/10.5061/dryad.qv9s4mwcv). Summary statistics of all variants in the GWAS of various stroke subtypes will be provided on request to the corresponding author.
Results
Association With Vascular Risk Factors
A total of 1,255 cases and 2,154 controls met the quality control criteria for the GWAS study and were considered for the association study. The study flow diagram adopted for this study is presented in figure 1. The stroke cases in this study were on an average 6 to 10 years older than the controls, and the majority (71%–74%) were men, but there was no significant sex difference between cases and controls (table 1). Hypertension status and measures of blood pressure were strongly associated with stroke, with the strongest association being for hemorrhagic stroke (mean systolic blood pressure 162.00 mm Hg, mean diastolic blood pressure 93.50 mm Hg, hypertension status 59.2%). Tobacco smoking and alcohol consumption were also strongly associated with stroke. Among the risk factors, dyslipidemia and fasting glucose levels had mild to moderate effect except for large artery stroke, which showed a strong association with dyslipidemia (p = 7 × 10−5). Atrial fibrillation was associated mainly with cardioembolic stroke (p = 3.3 × 10−32). Disease comorbid conditions such as diabetes and myocardial infarction were also more prevalent in stroke cases than controls.
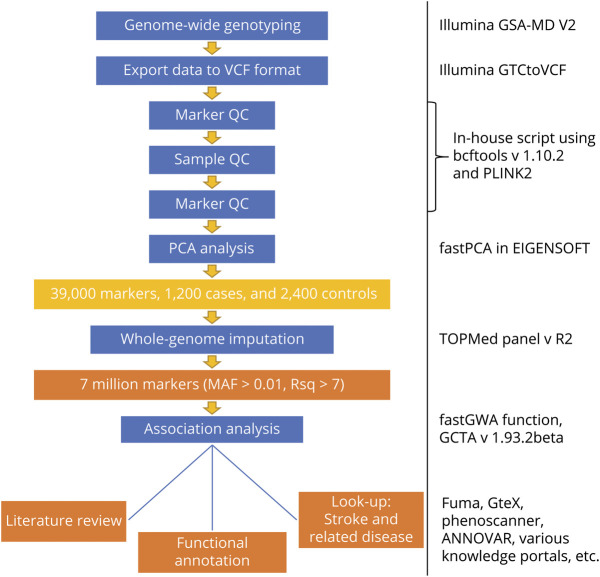
Figure 1. Study Design for the Indian Stroke Genome-Wide Association Study.
MAF = minor allele frequency; PCA = principal component = components analysis; QC = quality control; Rsq = r2; VCF = variant call format.
Association With Genetic Risk Factors
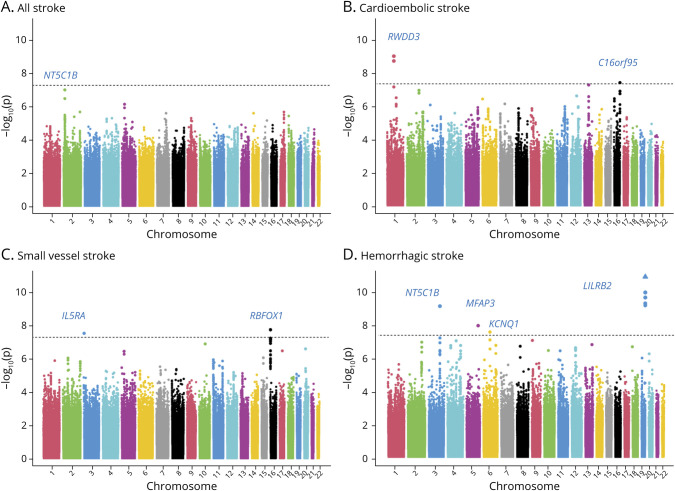
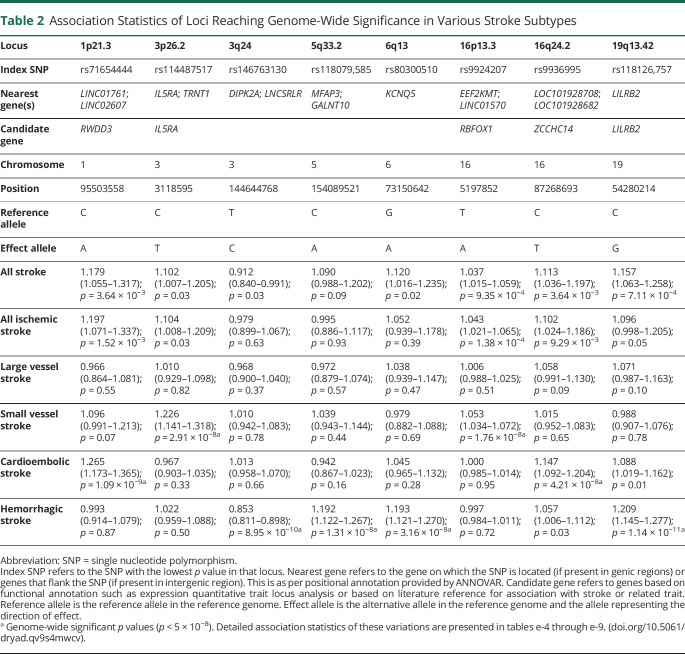
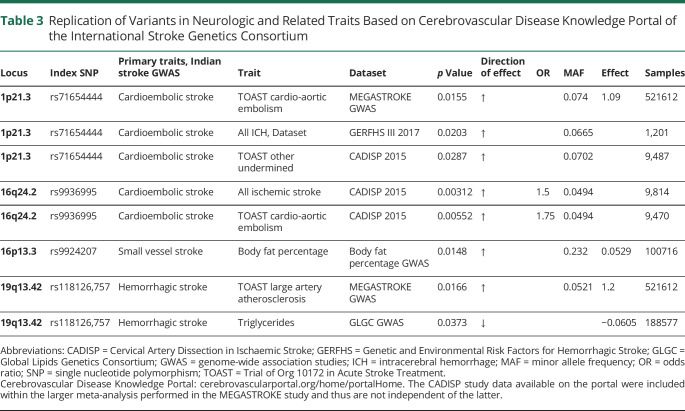
Using 395,255 good-quality genotyped markers, we were able to impute 7,827,597 variations using the TOPMed reference panel with MAF >0.01 and imputation quality >0.7. After excluding variants with effective allele count <10, we observed that 8 independent loci reached genome-wide significance threshold (p < 5 × 10−8) in 3 stroke subtypes (figure 2 and figure e-3, doi.org/10.5061/dryad.qv9s4mwcv). No major inflation was observed in the test statistics as detected on the quantile-quantile plot and median of χ2 value (λ value) being close to 1 (figure e-4). Two loci at 1p21 (OR 1.265, p = 1.09 × 10−9) and 16q24 (OR 1.147, p = 4.21 × 10−8) were associated with cardioembolic stroke (figure 2 and table 2). Two loci were identified for small vessel disease stroke at 3p26 (OR 1.226, p = 2.91 × 10−8) and 16p13 (OR 1.053, p = 1.76 × 10−8) (figure 2 and table 2). The rest of the 4 newly identified loci were for hemorrhagic stroke at 3q24 (OR 0.853, p = 8.95 × 10−10), 5q33 (OR 1.192, p = 1.31 × 10−8), 6q13 (OR 1.193, p = 3.16 × 10−8), and 19q13 (OR 1.209; p = 1.14 × 10−11) (figure 2 and table 2). The nearest genes for these loci as annotated with ANNOVAR are presented in table 2, along with the candidate gene in the region likely affecting the risk of stroke based on eQTL analysis and literature review. Lists of variants that were significant at p < 1 × 10−5 in various stroke subtypes considered in the study are presented in tables e-4 through e-9 and e-19. In the gene-based analysis, we identified 1 gene for large vessel disease (SLC17A2, p = 1.26 × 10−6) and 2 for small vessel disease (FAM73A, p = 2.60 × 10−6 and OR52L1, p = 1.38 × 10−6) that were significant after correction for multiple testing (p <2.701 × 10−6) (figure e-5 and table e-10). Apart from rs9924207 (16p13.3) associated with small vessel disease stroke, the rest of the 7 variants are in the low-frequency range (table 2). The regional plots of the top 8 loci and the variant showing the most significant association with all stroke are presented in figures e-6 through e-9. Data for 4 of 8 of the top loci were available in MEGASTROKE, and we observed that 3 of them (1p21.3, 16p13.3, 19q13.42) were associated with 1 of the stroke subtypes at p < 0.05 (table e-11). The loci 1p21.3 and 16q24.2 also show a nominal association with stroke subtypes in other smaller studies like Cervical Artery Dissection in Ischaemic Stroke and Genetic and Environmental Risk Factors for Hemorrhagic Stroke III according to the Cerebrovascular Knowledge Portal hosted by International Stroke Genetics Consortium (table 3). Replication in the UK Biobank samples of South Asian origin suggested that there was consistency in direction of effects with similar risk alleles, although the associations did not reach nominal significance of p < 0.05 (table e-18). TOAST subtyping data were not available in the UK Biobank, restricting head to head comparison of subtype specific associations.
Figure 2. Manhattan Plots for (A–D) Association With Stroke Subtypes.
Table 2.
Association Statistics of Loci Reaching Genome-Wide Significance in Various Stroke Subtypes
Table 3.
Replication of Variants in Neurologic and Related Traits Based on Cerebrovascular Disease Knowledge Portal of the International Stroke Genetics Consortium
Functional Annotation and Association With Related Traits
Functional annotation revealed that the index SNP of 1p21 (rs71654444) is an eQTL for the gene RWDD3(plowest = 1.74 × 10−58) (table e-12, doi.org/10.5061/dryad.qv9s4mwcv). Data from multiple tissues suggested that rs71654444 is an eQTL of RWDD3 (table e-12). The variant rs118126,757 was identified as an eQTL for LILRA3(plowest = 1.74 × 10−58). Similarly, we identified rs9936995 as an eQTL for the gene KLHDC4 (table e-12). A search for association with related traits revealed that the variant rs71654444-RWDD3 was associated with platelet distribution width (p = 1.15 × 10−9) in the UK Biobank study population (figure e-10 and table e-13) and also affected the metabolism of steric acid (p = 7.36 × 10−12) and palmitic acid (p = 4.44 × 10−7) (table e-14). To check the association of these top 8 loci with related risk factors, we also searched the Diabetes Knowledge Portal, and the results are presented in table e-15. The most significant association of rs71654444-RWDD3 was extreme chronic kidney disease (p = 0.000669).
Replication of Loci Identified by Previous GWAS
The status of previously reported 32 genome-wide significant loci that replicated in the MEGASTROKE dataset is shown in table e-16 (doi.org/10.5061/dryad.qv9s4mwcv). Using similar directionality of effect, we were able to replicate 19 to 21 of these loci, depending on replication in any stroke subtype (table e-16). Checking for both directionality of effect and nominal significance at p < 0.05, we observed association of 6 loci (KCNK3, LOC100505841, Chr9p21, LINC01492, ZFHX3, SMARCA4–LDLR) with at least 1 of the stroke subtypes (table e-16). However, if stroke subtype–specific replication is considered at p < 0.05 according to original subtype in which it was discovered, then replication is observed for only ZFHX3 and Chr9p21 (table e-16). The strongest association was observed for the locus ZFHX3 with cardioembolic stroke in the current study (p = 0.0004), which was also associated primarily with cardioembolic stroke in MEGASTROKE analysis. Association of ZFHX3 remained significant after correction for multiple testing (0.05/32 = 0.0016).
Status of Candidate Gene Studies Performed in Indian Population
In total, 32 independent loci have been investigated for association with stroke in 50 publications (appendix e-1 and table e-17, doi.org/10.5061/dryad.qv9s4mwcv). We observed that 11 loci were associated with at least 1 stroke subtype in this study (p < 0.05) and that the strongest association was observed for the variant rs1800610 of TNF (p = 0.0032) with small vessel disease stroke. However, none of the associations of the candidate genes previously studied in the Indian population were significant after correction for multiple testing (0.05/32 = 0.0016).
Discussion
This first GWAS of stroke in an Indian population identified 11 novel loci (8 based on a single variant and 3 on gene-based test) and replicated previously known loci. Using functional annotation and association with stroke-related risk factors, variants in genes for novel pathways not implicated previously by genetic studies for stroke were also identified.
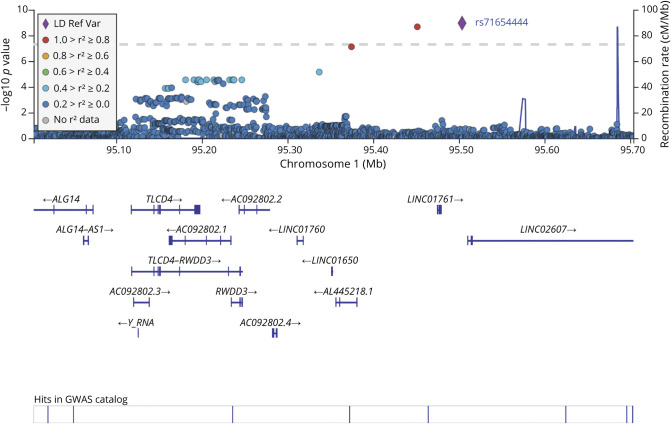
The locus 1p21.3 with the index SNP rs71654444 affecting the expression of the gene RWDD3 was our most important finding. This locus, which was associated with cardioembolic stroke in this study at the genome significance level, was also found to replicate in MEGASTROKE, the largest genetic study of stroke to date with cardioembolic stroke. Strong association with platelet distribution width and 18-carbon fatty acid metabolism also provides evidence that this locus might be affecting known biological mechanisms that affect cardioembolic stroke. Platelet distribution width is an established marker for embolism, and long carbon fatty acid metabolism is specifically implicated in cardioembolic stroke, not with other stroke subtypes.20,21 Considered together, these findings strengthen the evidence of a stroke subtype–specific association, suggesting the potential influence of the 1p21.3 variant with the risk of cardioembolic stroke. However, this locus was not associated with risk of atrial fibrillation in public databases, which may suggest that this locus affects cardioembolic stroke by mechanism other than atrial fibrillation. It has been observed in an Indian population that the risk of atrial fibrillation among cardiac embolic cases is ≈18%, whereas the same is >50% in European individuals.22,23 Factors such as rheumatic heart diseases may play a greater role in the occurrence of cardioembolic stroke in the Indian population, but it has been almost irradicated in populations of European origin.24 From gene expression data obtained on multiple tissues (GTEx portal), it was observed that the index SNP rs71654444 of 1p21 is an eQTL for RWDD3. The gene RWDD3 is also known as RSUME and has a conserved sequence of 110 amino acids containing repeats (R) of tryptophan (W) and aspartic acid (D). This RWD domain–containing protein helps in SUMOylation and posttranslational modifications similar to ubiquitination but involves the addition of a small ubiquitin-like modifier (SUMO) instead of ubiquitin.25,26 Several studies have provided evidence that activation of SUMOylation pathway protects against brain ischemia.27–31
Another variant, rs9936995, located on 16q24 also reached the GWAS significance level for association with cardioembolic stroke. This locus also showed evidence of nominal association with cardio-aortic embolism (p = 0.0052) and all ischemic strokes (p = 0.0031) in the smaller CADISP 2015 dataset (Cerebrovascular Knowledge Portal). This locus is located near the gene ZCCHC14, which has previously been implicated in all stroke (MEGASTROKE) and small vessel disease stroke, including white matter hyperintensities.19,32 However, the index SNP at this locus in the current study did not show any correlation with SNPs previously reported to be associated with stroke at this region (r2 = 0.001 between rs9936995 and rs12445022).
The index SNP rs118126757 of the 19q13.42 locus was found to affect the expression of its nearest gene, LILRB2, and a nearby gene, TSEN34. The LILRB2 gene has been implicated in a rare vascular disorder, Takayasu arteritis, which leads to inflammation of the aorta and its branches, and its orthologs have been implicated in platelet activation.33,34 However, more evidence is required to associate this locus with stroke. The associations of loci 16q24 (cardioembolic stroke) and 19q13.42 (hemorrhagic stroke) need further validation in the specific stroke subtypes in independent samples to confirm the findings.
Among the 3 genes identified through gene-based studies, SLC17A2 has been shown to be associated with carotid intima-media thickness and ankle-brachial index, both of which in turn are associated with the risk of stroke.35,36
While checking for replication of established loci for stroke identified through GWAS, we were able to replicate many of them, 22 in total, if we were to consider only directionality of effects. The sample size in this study is very small compared to some of the large studies like MEGASTROKE; hence, we do not expect to replicate most of the loci if we were to consider association based on p values. Despite this, we observed a strong association of ZFHX3 with a cardioembolic stroke, which remained significant even after correction for multiple testing. However, it is to be noted that PITX2, which is the strongest known signal for cardioembolic stroke, did not show association in the current study. This could be due to differences in LD pattern, but only larger studies including cardioembolic subtyping can resolve these ambiguities. We also note that we did not observe association with large artery or atherothrombotic stroke, which has been observed to have high heritability. One explanation for the lack of association may be biological differences in ethnicity: intracranial atherosclerosis/stenosis is more prevalent among South Asian people; in contrast, extracranial atherosclerosis/stenosis is more prevalent in European/American individuals.37 Alternatively, lack of association may be function of relatively small sample size, leading to type II error. There is a clear need to conduct an adequately powered study with a large sample size to determine the association between genetic risk factor and large vessel disease subtype of stroke in the Indian population.
A large number of studies from India have investigated the association of polymorphisms in candidate genes and stroke. Our search revealed at least 50 such studies that had investigated 38 independent loci. We were interested in checking the status of these candidate genes in our study because very few of such loci (like NOS2) have been confidently implicated in stroke, especially since previous GWAS have had little evidence of their support. This could have been due to allele frequency and LD differences between Indian genetic makeup and other populations in which GWAS was performed. However, we were able to replicate only a few of them at nominal p values, and none were significant after correction for multiple testing. This could reflect false-negative results due to small sizes or false-positive results due to population stratification, etc, of the candidate gene studies. Even the current study is small in sample size considering the ischemic and hemorrhagic stroke subtypes. Hence, larger studies in the Indian population are required to better resolve these issues.
We also investigated the nongenetic factors such as the vascular risk factors because this is one of the largest studies on stroke from India. We observed an association for risk of stroke very similar to that previously reported in many studies investigating different ethnicities. The modifiable risk factors such as hypertension, smoking, and alcohol intake appeared to have high effect estimates across stroke subtypes.
The big limitations of the study are the small number of samples for ischemic stroke subtypes and the lack of subtyping for hemorrhagic stroke, which can limit the interpretation of low-frequency variants. Another limitation was the lack of replication in an Indian sample set from India. Although the South Asian samples from UK Biobank fulfilled this gap, the study still lacked TOAST-based subtyping, limiting direct comparison of subtype-specific associations. The difference in reginal diet and cultural practices among the 3 centers also could affect the results to some extent, but due to a lack of data to account for these differences, it is difficult to comment on these important gene-environment interactions. However, this first GWAS of stroke in an Indian population was nonetheless able to identify robustly associated loci, especially the 1q21 locus (figure 3), of cardioembolic stroke given the strong evidence for association with traits that are associated with stroke and nominal replication in independent datasets. This also led to reporting of variants in genes of the SUMOylation pathway, which is actively being investigated as a therapeutic option for protection against brain ischemia. Future population genetic studies involving larger datasets of Indian origin followed by functional studies in animal models guided by RNA sequencing and gene network analysis are required for further interpretation of these findings.
Figure 3. Regional Plot of Locus 1q21 Associated With Cardioembolic Stroke.
GWAS = genome-wide association study; LD = linkage disequilibrium; Ref Var = reference variant.
Acknowledgment
The authors thank the All India Institute of Medical Sciences, New Delhi, India, for providing support and resources for successful completion of this study. They acknowledge the UK-India Education Research Initiative for providing technical support and funding for the collection of samples from East and South India. Finally, they acknowledge the support of study participants for their participation in the present study. Ganesh Chauhan is supported by DBT-Ramalingaswami Re-Entry Fellowship. The UK Biobank datasets used in this study were obtained under application No. 37489.
Glossary
- CADISP
Cervical Artery Dissection in Ischaemic Stroke
- eQTL
expression quantitative trait locus
- GWAS
genome-wide association studies
- LD
linkage disequilibrium
- MAF
minor allele frequency
- OR
odds ratio
- PCA
principal components analysis
- RWD
repeats of tryptophan and aspartic acid
- SNP
single nucleotide polymorphism
- SUMO
small ubiquitin-like modifier
- TOAST
Trial of Org 10172 in Acute Stroke Treatment
- VCF
variant call format
Appendix. Authors
Footnotes
Editorial, page 159
Study Funding
The present study was funded by the Department of Biotechnology, Ministry of Science and Technology, Government of India (grant BT/01/COE/06/02/09-II). The funding support for sample collection from South India and East India was provided by the UK-India Education Research Initiative.
Disclosure
The authors report no disclosures. Go to Neurology.org/N for full disclosures.
References
- 1.GBD 2016 Stroke Collaborators. Global, regional, and national burden of stroke, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019;18(5):439-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bevan S, Traylor M, Adib-Samii P, et al. . Genetic heritability of ischemic stroke and the contribution of previously reported candidate gene and genomewide associations. Stroke. 2012;43(12):3161-3167. [DOI] [PubMed] [Google Scholar]
- 3.Gunarathne A, Patel JV, Gammon B, Gill PS, Hughes EA, Lip GYH. Ischemic stroke in South Asians: a review of the epidemiology, pathophysiology, and ethnicity-related clinical features. Stroke. 2009;40(6):e415-e423. [DOI] [PubMed] [Google Scholar]
- 4.Chauhan G, Debette S. Genetic risk factors for ischemic and hemorrhagic stroke. Curr Cardiol Rep. 2016;18(12):124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet. 2019;51(4):584-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar A, Kumar P, Kathuria P, et al. . Genetics of ischemic stroke: an Indian scenario. Neurol India. 2016;64(1):29-37. [DOI] [PubMed] [Google Scholar]
- 7.Indian Genome Variation Consortium. The Indian Genome Variation database (IGVdb): a project overview. Hum Genet. 2005;118(1):1-11. [DOI] [PubMed] [Google Scholar]
- 8.Indian Genome Variation Consortium. Genetic landscape of the people of India: a canvas for disease gene exploration. J Genet. 2008;87(1):3-20. [DOI] [PubMed] [Google Scholar]
- 9.Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461(7263):489-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakatsuka N, Moorjani P, Rai N, et al. . The promise of discovering population-specific disease-associated genes in South Asia. Nat Genet. 2017;49(9):1403-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adams HP, Bendixen BH, Kappelle LJ, et al. . Classification of subtype of acute ischemic stroke: definitions for use in a multicenter clinical trial. TOAST: Trial of Org 10172 in Acute Stroke Treatment. Stroke. 1993;24(1):35-41. [DOI] [PubMed] [Google Scholar]
- 12.Kowalski MH, Qian H, Hou Z, et al. , TOPMed Imputation server. PLoS Genet; 2019. Accessed April 17, 2020. imputation.biodatacatalyst.nhlbi.nih.gov/index.html#! [Google Scholar]
- 13.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional Mapping and Annotation of Genetic Associations With FUMA. Nat Commun. 2017;8(1):1826. Accessed May 22, 2020. fuma.ctglab.nl/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crawford KM, Gallego-Fabrega C, Kourkoulis C, et al. , Cerebrovascular Disease Knowledge Portal: An Open-Access Data Resource to Accelerate Genomic Discoveries in Stroke. International Stroke Genetics Consortium Stroke. 2018;49(2):470-475. Accessed July 2, 2020. cd.hugeamp.org/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Type 2 Diabetes Knowledge Portal. T2D-GENES Consortium, DIAGRAM Consortium, Broad Institute, the European Bioinformatics Institute, the University of California at San Diego and the University of Michigan. Accessed July 2, 2020. type2diabetesgenetics.org. [Google Scholar]
- 16.Canela-Xandri O, Rawlik K, Tenesa A. Gene ATLAS. Nat Genet. 2018; 50(11):1593-1599. Accessed July 2, 2020. geneatlas.roslin.ed.ac.uk/phewas/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013; 45(6):580-5. Accessed July 2, 2020. gtexportal.org/home/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamat MA, Blackshaw JA, Young R, et al. , PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics. 2019;35(22):4851-4853. Accessed July 2, 2020. phenoscanner.medschl.cam.ac.uk/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malik R, Chauhan G, Traylor M, et al. . Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet. 2018;50(4):524-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vagdatli E, Gounari E, Lazaridou E, Katsibourlia E, Tsikopoulou F, Labrianou I. Platelet distribution width: a simple, practical and specific marker of activation of coagulation. Hippokratia. 2010;14(1):28-32. [PMC free article] [PubMed] [Google Scholar]
- 21.Sun D, Tiedt S, Yu B, et al. . A prospective study of serum metabolites and risk of ischemic stroke. Neurology. 2019;92(16):e1890-e1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manorenj S, Barla S, Jawalker S. Prevalence, risk factors and clinical profile of patients with cardioembolic stroke in South India: a five-year prospective study: 2020th-06-26th. Int J Community Med Public Health. 2020(7): 2708-2714. [Google Scholar]
- 23.Díaz Guzmán J. Cardioembolic stroke: epidemiology [in Spanish]. Neurol Barc Spain. 2012;27(suppl 1):4-9. [DOI] [PubMed] [Google Scholar]
- 24.Wang D, Liu M, Lin S, et al. . Stroke and rheumatic heart disease: a systematic review of observational studies. Clin Neurol Neurosurg. 2013;115(9):1575-1582. [DOI] [PubMed] [Google Scholar]
- 25.Alontaga AY, Ambaye ND, Li Y-J, et al. . RWD domain as an E2 (Ubc9)-interaction module. J Biol Chem. 2015;290(27):16550-16559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cel Biol. 2007;8(12):947-956. [DOI] [PubMed] [Google Scholar]
- 27.Anderson DB, Zanella CA, Henley JM, Cimarosti H. Sumoylation: implications for neurodegenerative diseases. Adv Exp Med Biol. 2017;963:261-281. [DOI] [PubMed] [Google Scholar]
- 28.Peters M, Wielsch B, Boltze J. The role of SUMOylation in cerebral hypoxia and ischemia. Neurochem Int. 2017;107:66-77. [DOI] [PubMed] [Google Scholar]
- 29.Zhang H, Huang D, Zhou J, Yue Y, Wang X. SUMOylation participates in induction of ischemic tolerance in mice. Brain Res Bull. 2019;147:159-164. [DOI] [PubMed] [Google Scholar]
- 30.Lee Y, Hallenbeck JM. SUMO and ischemic tolerance. Neuromolecular Med. 2013;15(4):771-781. [DOI] [PubMed] [Google Scholar]
- 31.Yang W, Sheng H, Thompson JW, et al. . Small ubiquitin-like modifier 3-modified proteome regulated by brain ischemia in novel small ubiquitin-like modifier transgenic mice: putative protective proteins/pathways. Stroke. 2014;45(4):1115-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Traylor M, Malik R, Nalls MA, et al. . Genetic variation at 16q24.2 is associated with small vessel stroke. Ann Neurol. 2017;81(3):383-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Terao C, Yoshifuji H, Matsumura T, et al. . Genetic determinants and an epistasis of LILRA3 and HLA-B*52 in Takayasu arteritis. Proc Natl Acad Sci USA. 2018;115(51):13045-13050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan X, Shi P, Dai J, et al. . Paired immunoglobulin-like receptor B regulates platelet activation. Blood. 2014;124(15):2421-2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arya R, Escalante A, Farook VS, et al. . A genetic association study of carotid intima-media thickness (CIMT) and plaque in Mexican Americans and European Americans with rheumatoid arthritis. Atherosclerosis. 2018;271:92-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kardia SL, Greene MT, Boerwinkle E, Turner ST, Kullo IJ. Investigating the complex genetic architecture of ankle-brachial index, a measure of peripheral arterial disease, in non-Hispanic whites. BMC Med Genomics. 2008;1:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moussouttas M, Aguilar L, Fuentes K, et al. . Cerebrovascular disease among patients from the Indian subcontinent. Neurology. 2006;67(5):894-896. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Summary statistics of the association of variants for all stroke subtypes at p < 1 × 10−5 are provided in the supplementary tables (doi.org/10.5061/dryad.qv9s4mwcv). Summary statistics of all variants in the GWAS of various stroke subtypes will be provided on request to the corresponding author.