Abstract
Aspergillus niger, one of the most common and important fungal species, is ubiquitous in various environments. A. niger isolates possess a large number of cryptic biosynthetic gene clusters (BGCs) and produce various biomolecules as secondary metabolites with a broad spectrum of application fields covering agriculture, food, and pharmaceutical industry. By extensive literature search, this review with a comprehensive summary on biological and chemical aspects of A. niger strains including their sources, BGCs, and secondary metabolites as well as biological properties and biosynthetic pathways is presented. Future perspectives on the discovery of more A. niger-derived functional biomolecules are also provided in this review.
Keywords: Aspergillus niger, secondary metabolite, bioactivity, biosynthesis, application
Introduction
Aspergillus, one sizeable genus belonging to Aspergillaceae family, comprises as many as 492 species registered on the database of the National Center for Biotechnology Information (NCBI) to date. Its section Nigri is an important group of species, and the A. niger aggregate represents its most complicated taxonomic subgroup with eight morphologically indistinguishable taxa (Perrone et al., 2011). Owing to superior adaptability and survivability, A. niger is ubiquitous in nature, including in terrestrial soil (Xie et al., 2006), ocean (Li et al., 2016; Uchoa et al., 2017), the Arctic (Singh et al., 2011), and space. It also occupies a wide spectrum of habitats in plants and animals such as herb (Shreelalitha and Sridhar, 2015; Manganyi et al., 2018), shrub (Kaur et al., 2015; Liu et al., 2016), tree (Soltani and Moghaddam, 2014; Wang et al., 2019), lichen (Elissawy et al., 2019), shrimp (Liu et al., 2013; Fang et al., 2016), and marine sponge (Takano et al., 2001; Hiort et al., 2004). A. niger strain grows well in various media with different carbon sources, including glucose, bran, maltose, xylan, xylose, sorbitol, and lactose (Toghueo et al., 2018). However, its metabolism is remarkably affected by culture conditions, such as medium composition and fermentation mode.
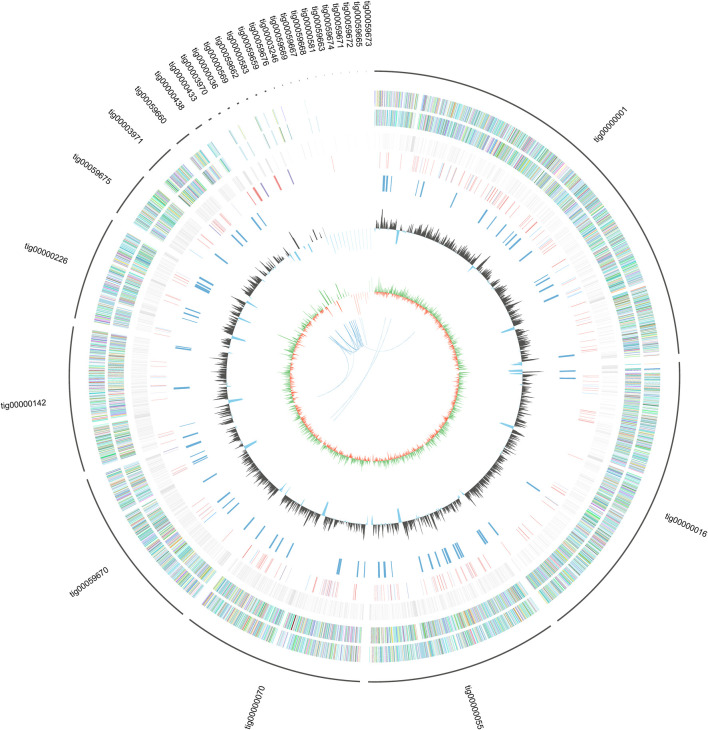
The genome features of strain L14 are summarized in a polycyclic graph (Figure 1), which consists of in-paralog pair, GC skew, widely, SM biosynthetic gene cluster (BGC), ncRNA, repeat, strand coding sequence (CDS) annotation, and scaffold. There are some in-paralog pairs between different scaffolds, and SM BGCs and CDS distributed widely in genome. As shown in Table 1, genome sizes of WT A. niger strains range from 33.8 to 36.1 Mb. Their G + C% and gene numbers are closely similar, while the numbers of scaffolds are different owing to various sequencing and assembling manners. The antibiotics and Secondary Metabolite Analysis Shell (antiSMASH) results indicated that each WT A. niger strain harbors at least 20 cryptic SM BGCs, including PKS, NRPS, NRPS-like, and their hybrids (Figure 2 and Supplementary Table S1) (Blin et al., 2019). These BGCs involving in indole and terpene biosynthesis are ubiquitous and have great potential to synthesize therapeutical agents and pesticides, such as AbT1, azanigerone A, fusarin, ferrichrome, nidulanin A, melanin, TAN-1612, yanuthone D, and aflavarin (Supplementary Table S2).
FIGURE 1.
General genome features of marine strain Aspergillus niger L14 (From the inside out: In-paralog pairs; GC skew, the green part represent positive value while the orange part represent negative value; G + C%; SMs gene clusters; ncRNA; repeat; minus strand coding sequence (CDS) annotation; plus strand CDS annotation; scaffolds).
TABLE 1.
General genomic features of 12 Aspergillus niger strains from NCBI database.
| Strain | Genome size (Mb) | G + C% | Scaffold | Gene | tRNA | Protein-coding genes | Isolation source | Assembly ID |
|---|---|---|---|---|---|---|---|---|
| ATCC 1015 | 34.8 | 50.3 | 24 | 10947 | – | 10950 | – | GCA_000230395.2 |
| CBS 513.88 | 34.0 | 50.4 | 20 | 10828 | 263 | 14165 | – | GCA_000002855.2 |
| SH-2 | 34.6 | 50.3 | 349 | – | – | – | Soil | GCA_000633045.1 |
| ATCC 13496 | 35.7 | 49.5 | 133 | 12468 | 273 | 12194 | – | GCA_003344705.1 |
| An76 | 34.9 | 49.4 | 669 | 10373 | – | 10373 | Soil | GCA_001515345.1 |
| JSC-093350089 | 36.1 | 49.5 | 223 | – | – | – | International space station environmental surface | GCA_001931795.1 |
| H915-1 | 36.0 | 49.2 | 30 | – | – | – | Soil | GCA_001741905.1 |
| L2 | 36.4 | 49.2 | 30 | – | – | – | Soil | GCA_001741915.1 |
| A1 | 34.6 | 50.1 | 319 | – | – | – | Soil | GCA_001741885.1 |
| MOD1-FUNGI2 | 33.8 | 50.4 | 3199 | – | – | – | Red seedless grapes | GCA_004634315.1 |
| RAF 106 | 35.1 | 49.1 | 10 | – | – | – | Pu-er tea | GCA_011316255.1 |
| L14 | 36.1 | 49.3 | 30 | 11524 | 296 | – | Marine sponge | JADEYF000000000 |
FIGURE 2.
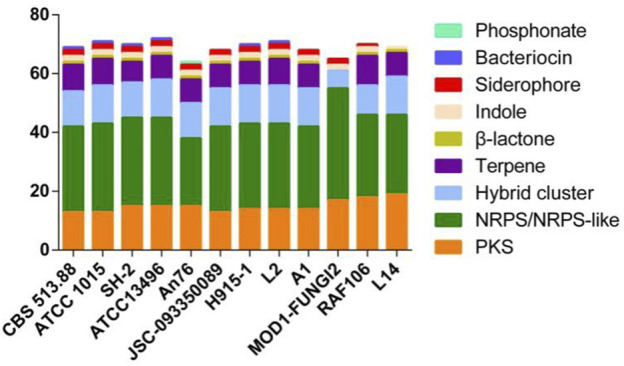
Biosynthetic gene clusters of secondary metabolites of 12 A. niger strains.
It is a matter of controversy that some A. niger isolates are renowned for biosynthesis of valuable natural products of nutritional, agrochemical, and pharmaceutical interest, while others are reputed to cause the “black mold” disease (Hayden et al., 1994; Ozer and Koycu, 2006) and produce a plethora of mycotoxins (Sanchez et al., 2012). A. niger possesses a bulk warehouse of prolific genes, which involve in regulation of primary and secondary metabolisms (Pel et al., 2007). A genome-scale metabolic network for A. niger has been established on account of its high efficiency in rational metabolic design and systems biology studies, such as strain improvement and process optimization (Sun et al., 2007; Lu et al., 2017). Numerous A. niger strains have been applied in many fields for a long time. For instance, citric acid as one of incredible organic acids in food industry had been produced on a large scale by A. niger 100 years ago (Cairns et al., 2018; Li et al., 2020). It is important that A. niger is one of the excellent producers of valuable proteases, which had been widely used as detergents and food ingredients and additives, such as acetylesterase, amylase, fucosidase, glucose oxidase, glucosidase, mannanase, phospholipase, phytase, prolyl endopeptidase, triacylglycerol lipase, trehalase, and xylanase. In addition, numerous chemical studies have indicated that A. niger is one of the rich sources of bioactive SMs, with great potential application in agriculture and medicine. Moreover, endoxylanase isozymes of A. niger have great potential transforming lignocellulose in pulp and paper industry as industrial bleaching aids (Duarte and Costaferreira, 1994). Furthermore, A. niger is also able to deal with the phenolic contaminants in waste water of fermentation broth from industry (Duarte and Costaferreira, 1994). Since genetic engineering is inefficient for fully exploiting in the filamentous fungi industry, a CRISPR (clustered regularly interspaced short palindromic repeats)–Cas9 system had been developed (Nødvig et al., 2015; Nødvig et al., 2018). Based on these genome-editing toolbox, gene inactivation and knockout, gene insertion, base editing, promoter replacement, and regulation of gene expression in A. niger have come true. In the future, more importance may be focused on traceless gene editing, multiple gene editing and fine regulation of gene expression in A. niger.
Secondary Metabolites From Aspergillus niger
By extensive search on the database of Dictionary of Natural Products (DNP), as many as 166 A. niger–derived secondary metabolites (1–166) were detected till 2020. On the basis of chemical structures, these chemicals are grouped into five types: pyranone, alkaloid, cyclopentapeptide, polyketide, and sterol and, respectively, introduced as follows. (More detailed information about these substances is provided in the Supplementary Materials (Supplementary Table S3).)
Pyranones
γ-Naphthylpyradone Monomers
Pyranone derivatives are the most isolated SMs from A. niger, including γ-naphthylpyradones (1–31), α-pyranones (32–56), and γ-pyranones (57–60). A. niger–derived naphthylpyradones are sorted into two classifications: monomers and dimers, with linear and angular naphtho-γ-pyrone. Fonsecin (1) is one of the most frequently isolated γ-naphthylpyradone produced by several A. niger strains from various sources, including terrestrial soil (Sakurai et al., 2002), marine (Leutou et al., 2016; Zhou et al., 2016), and plants (Bouras et al., 2005; Fernand et al., 2017; Akinfala et al., 2020). Biological tests suggested that compound 1 possesses dose-dependent inhibitory effect on the interleukin-4 (IL-4) signal transduction and stronger radical scavenging activity against 2,2-diphenyl-1-picrylhydrazyl (DPPH) than ascorbic acid (Sakurai et al., 2002; Leutou et al., 2016). Two analogs TMC-256A1 (3) and TMC-256C1 (8) also effectively inhibited the IL-4 driven luciferase (Sakurai et al., 2002). However, fonsecin B (2) and nigerasperone A (4) exhibited weak bioactivity against luciferase and DPPH (Sakurai et al., 2002; Zhang et al., 2007b). One new cytotoxic and antimicrobial rubrofusarin B (5) was purified from strain IFB-E003 endophytic on Cynodon dactylon Linn. (Song et al., 2004). When cultivated in NaBr or CaBr2-containing medium, one marine-derived strain MSA773 was found to secrete a new brominated derivative 6,9-dibromoflavasperone (9) with potent radical scavenging activity (Leutou et al., 2016).
γ-Naphthylpyradone Dimers
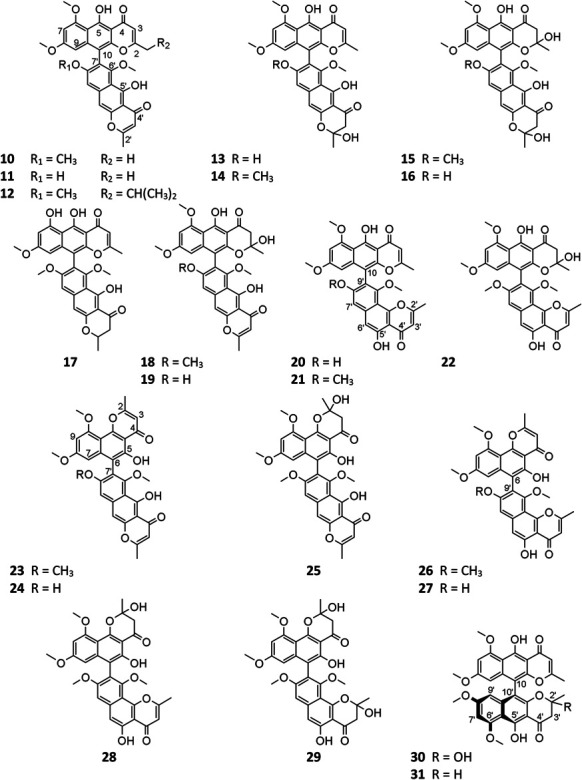
A. niger–derived dimeric naphthylpyradones (10–31) consist of two monomers with linear and/or angular structure(s). It is interesting that most of these bis-naphtho-γ-pyrones were produced by symbiotic A. niger strains. Chemical investigation of eight A. niger strains led to isolation of the same SM aurasperone A (10) (Tanaka et al., 1966; Tanaka et al., 1972; Akiyama et al., 2003; Zhang et al., 2007b; Fang et al., 2016; Li et al., 2016; Wang et al., 2018; Padhi et al., 2020), which possessed a broad spectrum of bioactivities including moderate cytotoxicity (Fang et al., 2016; Padhi et al., 2020), strong antimicrobial effect (Lu et al., 2014; Padhi et al., 2020), and xanthine oxidase (XO) inhibitory and anti-hyperuricosuric activity (Song et al., 2004). Aurasperone B (15) had potent radical scavenging activity against DPPH with an IC50 value of 0.01 μM (Leutou et al., 2016). Marine strain SCSIO Jcsw6F30 was a prolific producer of asperpyrone-type bis-naphtho-γ-pyrones (BNPs) 10, 13–16, 18, 20–22, 24, and 27, among which compounds 13, 16, and 20 exhibited remarkable inhibitory effects on COX-2 (Fang et al., 2016). In addition to nigerasperone A (4), two dimeric naphthylpyradones nigerasperones B (29) and C (19) were obtained from strain EN-13 and shown to exhibit a moderate radical scavenging effect on DPPH (Zhang et al., 2007b). Bioassay-guided fractionation of the crude extract of strain AKRN associated with Entandrophragma congoënse afforded a new antibacterial naphtho-γ-pyrone dimer 2-hydroxydihydronigerone (30) (Happi et al., 2015).
One possible pathway for biosynthesis of γ-naphthylpyradone derivatives had been first proposed by Obermaier and Muller (2019). As shown in Figure 3, one acetyl-CoA and six malonyl-CoA clusters were used as substrates for the biosynthesis of compounds 1–3 and 8 by successive catalytic reactions in a nonreducing PKS (nrPKS) system. Two of these monomers further dimerized at various carbon positions (C-6, C-7, C-9, or C-10) and resulted in the formation of dimers 16, 21, 27, and 28. Lately, one nrPKS gene D8.t287 responsible for the biosynthesis of the initial precursor heptaketone was identified and characterized by target gene knockout experiment and UPLC-MS analysis (Hua et al., 2020). However, the role of the gene AunB or BfoB is not confirmed so far.
FIGURE 3.
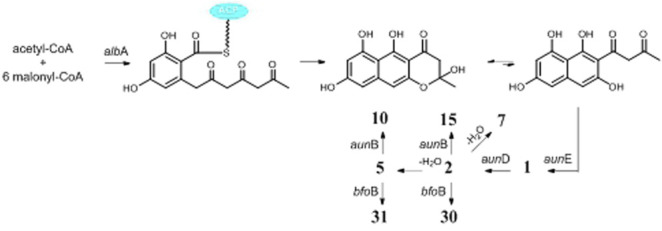
Proposed biosynthetic pathway of γ-naphthylpyridones.
α-Pyranones
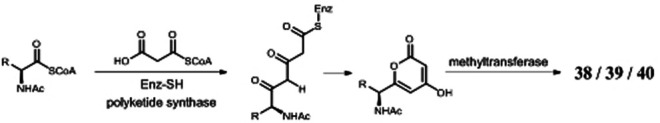
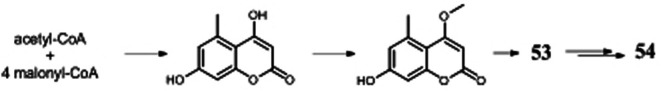
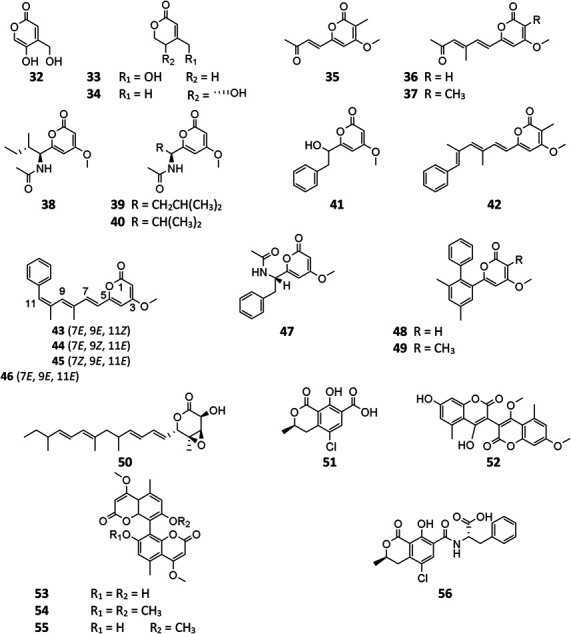
A. niger–derived α-pyranones contain 14 monocyclic compounds (32–40, 50), 7 dicyclics (41–47, 51), three tricyclics (48, 49, and 56), and four tetracyclics (52–55). Chemical analysis of an endophytic A. niger strain colonizing in liverwort Heteroscyphus tener (Steph.) Schiffn resulted in isolation of three new amide campyrones A–C (38–40) together with compounds 33 and 34 (Talontsi et al., 2013; Li et al., 2015). One possible biosynthetic pathway proposed by Reber and Burdge (2018) suggested that compounds 38–40 were, respectively, formed by one malonyl-CoA and three N-acetyl aliphatic amino acids including L-valine, L-leucine, and L-isoleucine (Figure 4), along with two congeners asnipyrones A (42) and B (46) and nigerapyrones A-H (35–37, 43–45, 48–49) were first discovered from a mangrove plant–derived strain MA-132 (Liu et al., 2011). Unfortunately, none of these compounds showed potent cytotoxic or antimicrobial activities. Nafuredin (50) and bicoumanigrin (52) were new α-pyranone analogs produced by marine sponge-derived A. niger strains; the former exhibited a powerful and selective inhibitory effect on NFRD (NADH-fumarate reductase) (Takano et al., 2001; Ui et al., 2001) and the latter 3,3′-bicoumarin had moderate cytotoxicity against leukemia and carcinoma cell lines (Hiort et al., 2004). Three 8,8′-bicoumarins, orlandin (53), kotanin (54), and 7-desmethyl-kotanin (55) were produced by a number of A. niger strains from various sources, and 53 showed potent inhibitory activity against wheat coleoptile growth at 1 mM but not toxic to day-old cockerels (Cutler et al., 1979; Ovenden et al., 2004; Sorensen et al., 2009; Jomori et al., 2020). Biosynthetically, one acetyl-CoA and four malonyl-CoAs comprised one coumarin through several successive reactions catalyzed by PKSs, followed by formation of compounds 52–55 through dimerization (Figure 5) (Huttel et al., 2003; Huttel and Muller, 2007; Girol et al., 2012). In this pathway, PKS gene ktnS was responsible for origination of dimeric coumarins 52–55, gene ktnB encode O-methyltransferase, and gene ktnC encode CYP450 monooxygenase, manipulating the dimerization of 52–55.
FIGURE 4.
Proposed biosynthetic pathway of campyrones.
FIGURE 5.
Proposed biosynthetic pathway of orlandin (53) and kotanin (54).
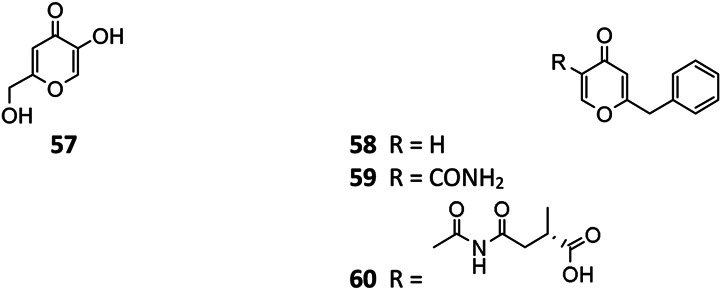
γ-Pyranones
To the best of our knowledge, only four γ-pyranone derivatives (57–60) had been detected in SM of A. niger. Among these substances, kojic acid (57) is the most common product with weak antimicrobial property (Liu et al., 2011; Happi et al., 2015; Padhi et al., 2020). In addition to carbonarone A (59) and tensidol B (60), one new benzyl γ-pyranone nigerpyrone (58) was discovered from a mutant strain FGSC A1279 ΔgcnE and was found to have potent and selective activity against Candida parapsilosis (Wang et al., 2018; Padhi et al., 2020).
Alkaloids
Pyrroles
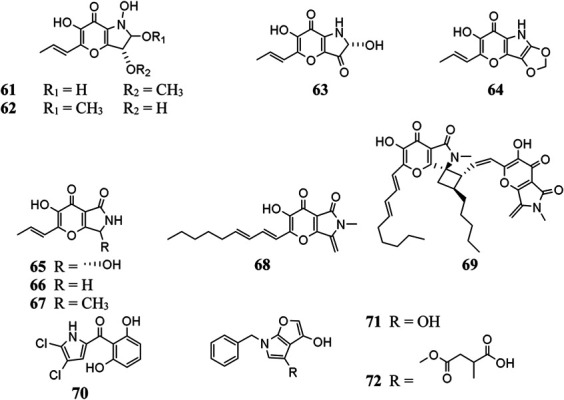
Pyranonigrin derivatives (61–69) are a family characterized by pyrano [2,3-b] pyrrole skeleton, and their biosynthesis are manipulated by the pyn gene cluster in A. niger (Riko et al., 2014; Yamamoto et al., 2015). Chemical investigation of one marine sponge–derived strain afforded four pyranonigrins B-D (61, 62, 64) and Ab (63), which 63 showed a strong inhibitory effect on the growth of neonate larvae of the plant pest insect Spodoptera littoralis (Hiort et al., 2004). Pyranonigrins A (65), S (66), and E (67) were important agents with potent radical scavenging activity toward DPPH and superoxide (Miyake et al., 2007; Riko et al., 2014). One possible biosynthetic pathway of pyranonigrin E (67) had been first proposed by Yamamoto et al. (2015) and coworker in 2015, in which the start units contained one acetyl-CoA, six malonyl-CoAs, and one L-Ser (Figure 6), under the action of gene pynA (PKS-NRPS hybrid synthase), pynI (encode thioesterase), pynC (encode methyltransferase), pynG (encode flavin-dependent oxidase), pynD (encode CYP450), and pynH (encode aspartyl protease). After non-enzymatic reaction, two pyranonigrin E (67) units could be dimerized to form pyranonigrin F (69). One soil-derived A. niger strain was found to produce a new dichlorinated pyrrole pyoluteorin (70), which obviously induced cell cycle arrest and apoptosis in human triple-negative breast cancer cells MDA-MB-231 (Ding et al., 2020). Two benzyl furopyrrols tensidols A (71) and B (72) from strain FKI-2342 were potentiators of antifungal miconazole activity (Fukuda et al., 2006) and lately corrected as compounds 59 and 60 (Henrikson et al., 2011).
FIGURE 6.
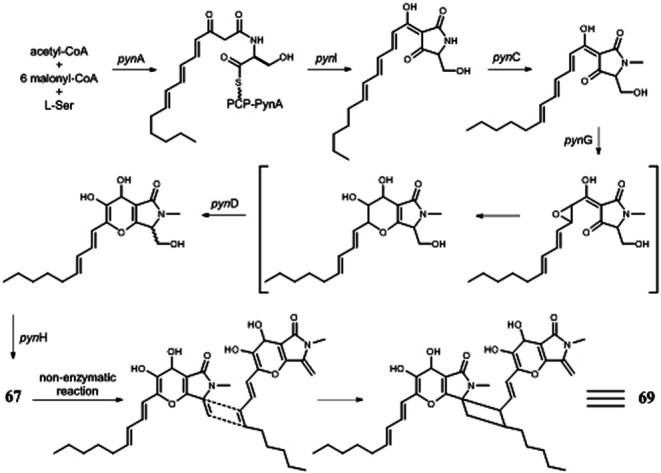
Proposed biosynthetic pathway of pyranonigrin E (67) and pyranonigrin F (69).
Pyridones
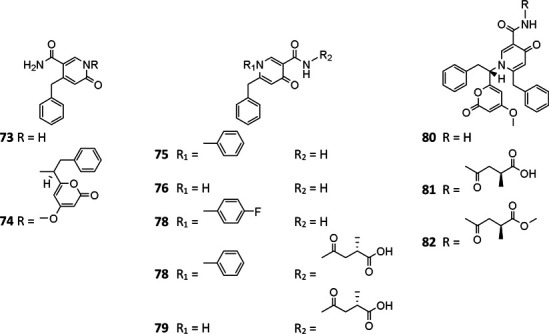
A. niger–derived pyridone derivatives (73–82) have one benzyl group and possess antimicrobial and cytotoxic properties (chatr 4). Two new α-pyridones aspernigrins A (73) and Bb (74) were isolated from one A. niger strain of marine sponge Axinella damicornis and showed moderate cytotoxicity and a potent neuroprotective effect, respectively, (Hiort et al., 2004). When cultivated in fermentation medium containing suberoylanilide hydroxamic acid (SAHA) and p-fluoro SAHA, strain ATCC 1015 was discovered to produce three antifungal γ-pyridones, nygerones A (78), B (75), and p-fluoro nygerone B (77) (Henrikson et al., 2009; Henrikson et al., 2011). In addition to three γ-naphthylpyradones (1, 3, and 5) and one cyclic peptide (111), three 2-benzyl-γ-pyridones aspernigrins B-D (80–82) were obtained from the marine strain SCSIO Jcsw6F30, and 81 was found to have potent inhibitory activity toward HIV-1 SF162-infected TZM-bl cells (Zhou et al., 2016).
Other Alkaloids
Three fatty amines fumonisins B2 (83), B1 (84), and B4 (85) from stains FGSC A1279 and IBT 28144 were carcinogenic (Nielsen et al., 2009; Sorensen et al., 2009; Li et al., 2019). The aza gene cluster in strain ATC C1015 was found be responsible for biosynthesis of azanigerone D (86) (Zabala et al., 2012). In addition to pyoluteorin (70), phenazine-1-carboxylic acid (87) was produced by the soil A. niger strain (Ding et al., 2020). Two new piperazines nigragillin (88) and nigerazine B (89) were purified from strain ATCC 11414, and their biosynthesis were regulated by the naphthopyrone precursor BGC alb gene cluster (Chiang et al., 2011). Endophytic strain IFB-E003–derived aspernigerin (90) displayed a potent effect on the tumor cell lines nasopharyngeal epidermoid KB, cervical carcinoma Hela, and colorectal carcinoma SW1116 (Shen et al., 2006).
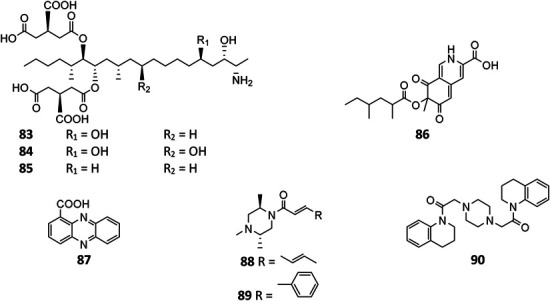
Amides
Till the end of 2020, only six amides (91–96) had been isolated and characterized from A. niger strains. Fractionation of crude extract of marine strain BRF-074A afforded one furan ester derivative (91), one cerebroside chrysogeside D (93), and two spiro amides pseurotins A (95) and D (96), among which 91 exerted a cytotoxic effect on HCT-116 cell line (Uchoa et al., 2017). When cultivated on wheat bran, strains CFR-W-105 and MTCC-5166 were discovered to produce nigerloxin (92) with free radical DPPH scavenging activity and inhibitory effect on lipoxygenase-I (LOX-1) and rat lens aldose reductase (RLAR) (Rao et al., 2002; Chakradhar et al., 2009). Ergosterimide (94) was a new natural Diels–Alder adduct of ergosteroid and maleimide produced by the strain EN-13 from marine alga (Zhang et al., 2007a).
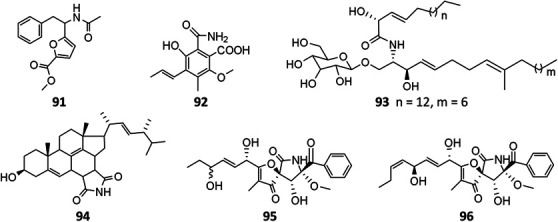
Cyclopeptides
All peptides of A. niger are cyclic and consist of ten dipeptides (97–106), eight pentapeptides (107–114), and three bis(dipeptide)s (115–117). In addition to α-pyranones 32–34, 38, and 40, four diketopiperazines (97, 99, 115, and 116) were isolated from an endophytic strain of liverwort Heteroscyphus tener (Steph.). Schiffn, and compounds 115 and 116 showed weak activity against the human ovarian carcinoma cancer cell line A2780 (Li et al., 2015). However, 115 exhibited significant selective cytotoxicity to human leukemia murine colon 38 and human colon H116 and CX1 cell lines (Varoglu et al., 1997; Varoglu and Crews, 2000). One strain BRF-074A from Northeast Brazilian coast was a prolific producer of cyclopeptides (101–107, 114) (Uchoa et al., 2017). Phytochemcial analysis of an uncoded marine strain afforded a new diketopiperazine dimer (117) and nine monomers (98–106) (Ovenden et al., 2004; Zhang et al., 2010; Uchoa et al., 2017). Compounds 98 and 99 had been reported to regulate plant growth (Kimura et al., 1996; Kimura et al., 2005), and 101 had selectively potential cytotoxicity (Graz et al., 2000). Eight malformin analogs (107–114) were a group of SMs containing structural skeleton of cyclo-D-cysteinyl-D-cysteinyl-L-amino acid-D-amino acid-L-amino acid (Kim et al., 1993). Malformin A (107) demonstrated antibacterial (Suda and Curtis, 1966; Liu et al., 2013) and anticancer activities (Wang et al., 2015), while malformin C (114) exhibited a broad spectrum of biological properties including anti-HIV-1 (Zhou et al., 2016), cytotoxic (Jomori et al., 2020), anticancer (Wang et al., 2015), and antibacterial (Suda and Curtis, 1966; Liu et al., 2013).
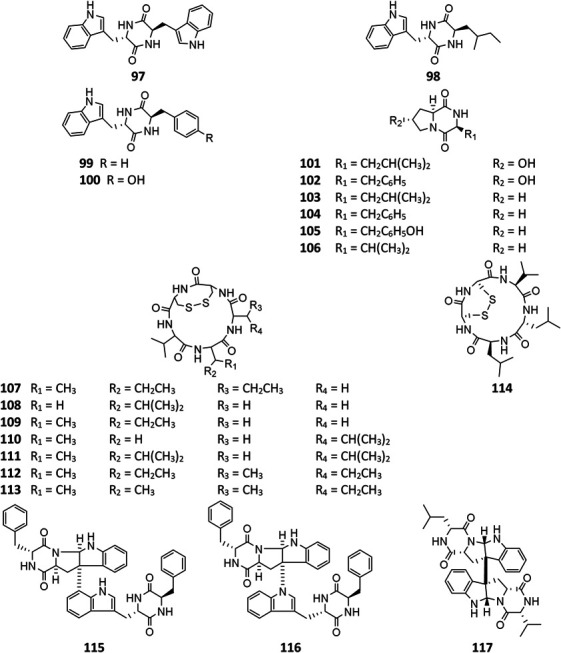
Polyketides
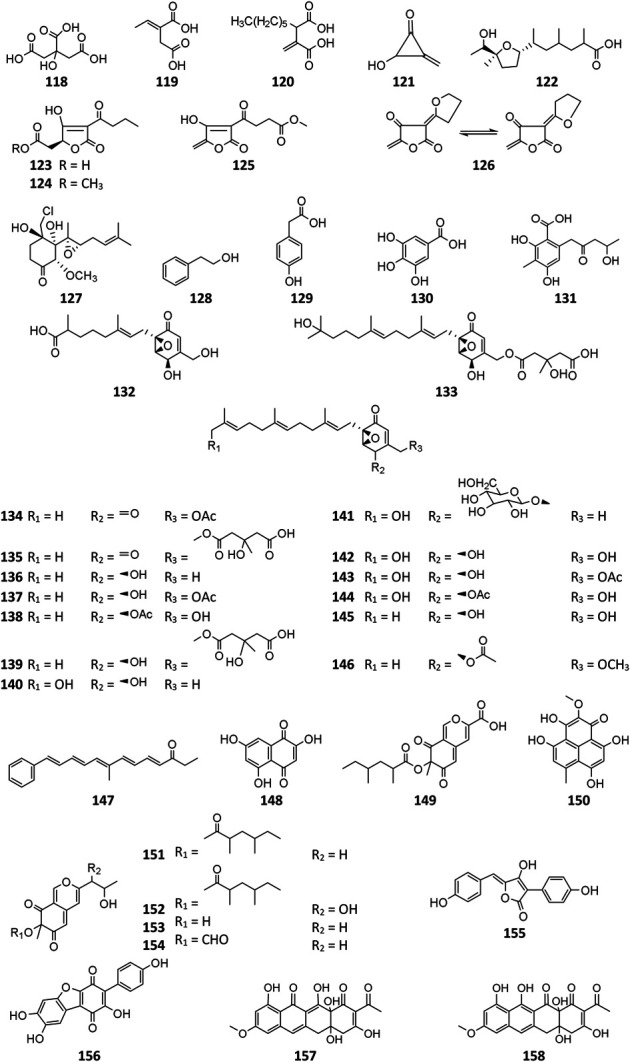
Polyketides (118–155) are the largest group of SMs produced by A. niger. Citric acid (118) and itaconic acid (119) have been large-scale products in food and pharmaceutical industry for decades (Andersen et al., 2011; Li et al., 2012). Some other valuable chemicals with low molecular weight are also produced by A. niger, such as 2-phenylethanol (128) (Etschmann et al., 2015), p-hydroxyphenylacetic acid (129) (Happi et al., 2015), gallic acid (130) (Saeed et al., 2020), benzoic acid derivative (131) (Zabala et al., 2012), and asperyellone (147) (Jefferson, 1967; Chidananda et al., 2008). In comparison with 119, the biological activity of hexylitaconic acid (120) dramatically attenuated (Varoglu et al., 1997; Varoglu and Crews, 2000). By overexpression of transcriptional regulator pBARAGA-CaaR of BGC caa in glucose minimal medium, strain ATC C1015 successfully produced three acyltetronic acid derivatives carlosic acid (123), carlosic acid methyl ester (124), and agglomerin F (125) (Yang et al., 2014). Chemical analysis of two strains KB1001 and F97S11 afforded fifteen meroterpenoid derivatives (132–146), in which biosynthesis was deduced to be manipulated by the yan gene cluster in strain KB1001 (Figure 7) (Bugni et al., 2000; Holm et al., 2014). Furthermore, yan gene cluster consisted of gene yanA [encode 6-methylsalicylic acid synthase (6-MSAS)] together with eight additional genes yanB (encode decarboxylase), yanC (encode CYP450), yanD (encode dehydrogenase), yanE (unknown), yanF (encode oxidase), yanI (encode O-mevalon tiransferase), yanH (encode CYP450), and yanG (encode prenyl transferase).
FIGURE 7.
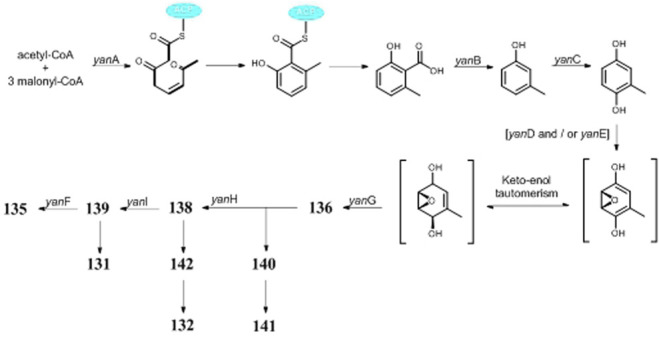
Proposed biosynthetic pathway of yanuthones.
Asperyellone (147) was the common product of strains NRRL-3 and CFTRI 1105 (Jefferson, 1967; Chidananda et al., 2008) and exhibited inhibitory effect on lipoxygenase and human platelet aggregation (Rao et al., 2002), UVB protection (Santhakumaran et al., 2019), and antifungal activity (Ayer et al., 1996). In addition to γ-pyridone (86), five highly oxygenated pyranoquinones (86, 149, and 151–154) were detected in SMs of strain T1 by activation of the aza gene cluster (Zabala et al., 2012) (Figure 8). In biosynthesis of pyranoquinones, genes azaE (encode ketoreductase), azaF (encode acyl:CoA ligase), azaG (encode FAD-dependent oxygenase), azaH (encode salicylate monooxygenase), azaI (encode CYP450), azaJ (encode dehydrogenase), and azaL (encode FAD-dependent oxygenase) play important roles. Funalenone (150), one phenalene derivative, was obtained from strain ATCC 11414 whether the albA gene was auxotrophic or not (Chiang et al., 2011). Meanwhile, funalenone (150) was also found in A. niger mutant ΔgcnE (strain FGSC A1279 lacking epigenetic regulatorgcnE) (Wang et al., 2018). Two tetracycline analogs BMS-192548 (157) and TAN-1612 (158) were, respectively, obtained from strains WB2346 and ATC C1015 and shown to be acyclic binding inhibitors of neuropeptide Y receptors (Kodukula et al., 1995; Shu et al., 1995; Li et al., 2011).
FIGURE 8.
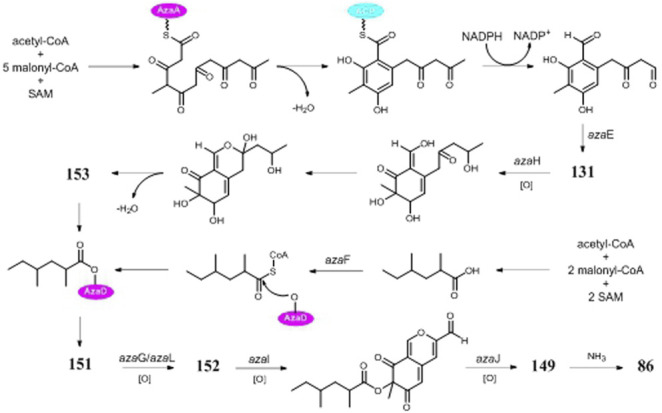
Proposed biosynthetic pathway of pyranoquinones in strain ATCC 1015.
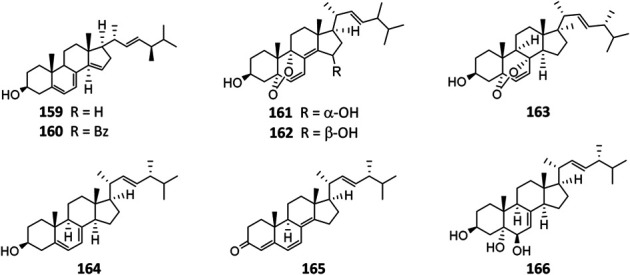
Sterols
As the by-product of manufacture of citric acid, 14-dehydroergosterol (159) and its benzoate (160) were the first steroids isolated from A. niger (Barton and Bruun, 1951) and possessed anti-inflammatory and cytotoxic properties (Ano et al., 2017). Strain MA-132–derived nigerasterols A (161) and B (162) had potent antiproliferative activity against human promyelocytic leukemia (HL60) and human lung carcinoma (A549), with IC50 values of 0.11 and 0.43 μM, respectively, (Liu et al., 2013). In addition to ergosterimide (94), four steroid derivatives (163–166) were discovered from the endophytic strain EN-13 associated with marine brown alga (Zhang et al., 2007a).
Conclusion and Future Prospects
A. niger strains are ubiquitous in nature and occupy a wide spectrum of habitats in animal and plant environments, and they are economically important both as harmful or beneficial microorganisms. Numerous chemical studies suggest that A. niger is one of the prolific sources of functional biomolecules, including organic acids, vitamins, pesticides, valuable proteases, and therapeutic agents, which have potential application in various fields including agriculture, food industry, and medicine. However, the number of new bioactive compounds from A. niger has been decreasing for the past 5 years. This deteriorating trend will result in a negative impact on discovery and development of new A. niger–derived valuable substances, such as new drug leads. Therefore, more efforts should be made to explore more sources for isolation of new A. niger strains and to awaken their silent BGCs to manufacture novel functional biomolecules using new strategies, such as one strain many compounds (OSMAC) approach (Hemphill et al., 2017; Pan et al., 2019) and genetic mining combined with metabolic engineering (Zhang et al., 2019; Li et al., 2020; Wei et al., 2021). Moreover, functional genomics should allow for an in-depth understanding of the underlying biosynthetic logic of A. niger–derived SMs (He et al., 2018). In order to accelerate development of valuable products from A. niger, construction and breeding of robust strains as well as optimization of their cultivation and fermentation processes should be intensively conducted at various levels (Zou et al., 2015; Xu et al., 2019).
Author Contributions
RY made a draft of this review; JL and YW searched and collected all references; HW helped in critical assessing this manuscript; and HZ conceived and revised this review.
Funding
Financial supports from the National Key R&D Program of China (2018YFC0311004), the National Natural Science Foundation of China (41776139) and the Fundamental Research Fund for the Provincial Universities of Zhejiang (RF-C2019002) were greatly appreciated.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2021.701022/full#supplementary-material
References
- Akinfala T. O., Houbraken J., Sulyok M., Adedeji A. R., Odebode A. C., Krska R., et al. (2020). Moulds and Their Secondary Metabolites Associated with the Fermentation and Storage of Two cocoa Bean Hybrids in Nigeria. Int. J. Food Microbiol. 316, 108490. 10.1016/j.ijfoodmicro.2019.108490 [DOI] [PubMed] [Google Scholar]
- Akiyama K., Teraguchi S., Hamasaki Y., Mori M., Tatsumi K., Ohnishi K., et al. (2003). New Dimeric Naphthopyrones from Aspergillus niger . J. Nat. Prod. 66 (1), 136–139. 10.1021/np020174p [DOI] [PubMed] [Google Scholar]
- Andersen M. R., Salazar M. P., Schaap P. J., van de Vondervoort P. J. I., Culley D., Thykaer J., et al. (2011). Comparative Genomics of Citric-Acid-Producing Aspergillus niger ATCC 1015 versus Enzyme-Producing CBS 513.88. Genome Res. 21 (6), 885–897. 10.1101/gr.112169.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ano Y., Ikado K., Shindo K., Koizumi H., Fujiwara D. (2017). Identification of 14-dehydroergosterol as a Novel Anti-inflammatory Compound Inducing Tolerogenic Dendritic Cells. Sci. Rep. 7. 10.1038/s41598-017-14446-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayer W. A., Muir D. J., Chakravarty P. (1996). Phenolic and Other Metabolites of Phellinus Pini, a Fungus Pathogenic to pine. Phytochemistry 42 (5), 1321–1324. 10.1016/0031-9422(96)00125-2 [DOI] [Google Scholar]
- Barton D. H. R., Bruun T. (1951). 607. A New Sterol from a Strain of Aspergillus niger . J. Chem. Soc., 2728. 10.1039/jr9510002728 [DOI] [Google Scholar]
- Blin K., Shaw S., Steinke K., Villebro R., Ziemert N., Lee S. Y., et al. (2019). antiSMASH 5.0: Updates to the Secondary Metabolite Genome Mining Pipeline. Nucleic Acids Res. 47 (W1), W81–W87. 10.1093/nar/gkz310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouras N., Mathieu F., Coppel Y., Lebrihi A. (2005). Aurasperone F - a New Member of the Naphtho-Gamma-Pyrone Class Isolated from a Cultured Microfungus,Aspergillus nigerC-433. Nat. Product. Res. 19 (7), 653–659. 10.1080/14786410412331286955 [DOI] [PubMed] [Google Scholar]
- Bugni T. S., Abbanat D., Bernan V. S., Maiese W. M., Greenstein M., Van Wagoner R. M., et al. (2000). Yanuthones: Novel Metabolites from a Marine Isolate ofAspergillusniger. J. Org. Chem. 65 (21), 7195–7200. 10.1021/jo0006831 [DOI] [PubMed] [Google Scholar]
- Cairns T. C., Nai C., Meyer V. (2018). How a Fungus Shapes Biotechnology: 100 Years of Aspergillus niger Research. Fungal Biol. Biotechnol. 5, 13. 10.1186/s40694-018-0054-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakradhar D., Javeed S., Sattur A. P. (2009). Studies on the Production of Nigerloxin Using Agro-Industrial Residues by Solid-State Fermentation. J. Ind. Microbiol. Biotechnol. 36 (9), 1179–1187. 10.1007/s10295-009-0599-7 [DOI] [PubMed] [Google Scholar]
- Chiang Y.-M., Meyer K. M., Praseuth M., Baker S. E., Bruno K. S., Wang C. C. C. (2011). Characterization of a Polyketide Synthase in Aspergillus niger Whose Product Is a Precursor for Both Dihydroxynaphthalene (DHN) Melanin and Naphtho-γ-Pyrone. Fungal Genet. Biol. 48 (4), 430–437. 10.1016/j.fgb.2010.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chidananda C., Kumar C. M., Sattur A. P. (2008). Strain Improvement of Aspergillus niger for the Enhanced Production of Asperenone. Indian J. Microbiol. 48 (2), 274–278. 10.1007/s12088-008-0026-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler H. G., Crumley F. G., Cox R. H., Hernandez O., Cole R. J., Dorner J. W. (1979). Orlandin: a Nontoxic Fungal Metabolite with Plant Growth Inhibiting Properties. J. Agric. Food Chem. 27 (3), 592–595. 10.1021/jf60223a043 [DOI] [PubMed] [Google Scholar]
- Ding T., Yang L.-J., Zhang W.-D., Shen Y.-H. (2020). Pyoluteorin Induces Cell Cycle Arrest and Apoptosis in Human Triple-Negative Breast Cancer Cells MDA-MB-231. J. Pharm. Pharmacol. 72 (7), 969–978. 10.1111/jphp.13262 [DOI] [PubMed] [Google Scholar]
- Duarte J. C., Costaferreira M. (1994). Aspergilli and Lignocellulosics - Enzymology and Biotechnological Applications. FEMS Microbiol. Rev. 13 (2-3), 377–386. 10.1111/j.1574-6976.1994.tb00038.x [DOI] [PubMed] [Google Scholar]
- Elissawy A. M., Ebada S. S., Ashour M. L., El-Neketi M., Ebrahim W., Singab A. B. (2019). New Secondary Metabolites from the Mangrove-Derived Fungus Aspergillus Sp. AV-2. Phytochemistry Lett. 29, 1–5. 10.1016/j.phytol.2018.10.014 [DOI] [Google Scholar]
- Etschmann M. M. W., Huth I., Walisko R., Schuster J., Krull R., Holtmann D., et al. (2014). Improving 2-phenylethanol and 6-Pentyl-α-Pyrone Production with Fungi by Microparticle-Enhanced Cultivation (MPEC). Yeast 32 (1), a–n. 10.1002/yea.3022 [DOI] [PubMed] [Google Scholar]
- Fang W., Lin X., Wang J., Liu Y., Tao H., Zhou X. (2016). Asperpyrone-Type Bis-Naphtho-γ-Pyrones with COX-2-Inhibitory Activities from Marine-Derived Fungus Aspergillus niger. Molecules 21 (7), 941. 10.3390/molecules21070941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernand M. G., Roullier C., Guitton Y., Lalande J., Lacoste S., Dupont J., et al. (2017). Fungi Isolated from Madagascar Shrimps - Investigation of the Aspergillus niger Metabolism by Combined LC-MS and NMR Metabolomics Studies. Aquaculture 479, 750–758. 10.1016/j.aquaculture.2017.07.015 [DOI] [Google Scholar]
- Fukuda T., Hasegawa Y., Hagimori K., Yamaguchi Y., Masuma R., Tomoda H., et al. (2006). Tensidols, New Potentiators of Antifungal Miconazole Activity, Produced by Aspergillus niger FKI-2342. J. Antibiot. 59 (8), 480–485. 10.1038/ja.2006.67 [DOI] [PubMed] [Google Scholar]
- Girol C. G., Fisch K. M., Heinekamp T., Guenther S., Huettel W., Piel J., et al. (2012). Regio- and Stereoselective Oxidative Phenol Coupling in Aspergillus niger . Angew. Chemie-International Edition 51 (39), 9788–9791. 10.1002/anie.201203603 [DOI] [PubMed] [Google Scholar]
- Graz C. J. M., Grant G. D., Brauns S. C., Hunt A., Jamie H., Milne P. J. (2010). Cyclic Dipeptides in the Induction of Maturation for Cancer Therapy. J. Pharm. Pharmacol. 52 (1), 75–82. 10.1211/0022357001773535 [DOI] [PubMed] [Google Scholar]
- Happi G. M., Kouam S. F., Talontsi F. M., Nkenfou C. N., Longo F., Zühlke S., et al. (2015). A New Dimeric Naphtho-γ-Pyrone from an Endophytic Fungus Aspergillus niger AKRN Associated with the Roots of Entandrophragma Congoënse Collected in Cameroon. Z. Naturforschung Section B-a J. Chem. Sci. 70 (9), 625–630. 10.1515/znb-2015-0036 [DOI] [Google Scholar]
- Hayden N. J., Maude R. B., Proctor F. J. (1994). Studies on the Biology of Black Mould (Aspergillus niger) on Temperate and Tropical Onions. 1. A Comparison of Sources of the Disease in Temperate and Tropical Field Crops. Plant Pathol. 43 (3), 562–569. 10.1111/j.1365-3059.1994.tb01591.x [DOI] [Google Scholar]
- He Y., Wang B., Chen W., Cox R. J., He J., Chen F. (2018). Recent Advances in Reconstructing Microbial Secondary Metabolites Biosynthesis in Aspergillus Spp . Biotechnol. Adv. 36 (3), 739–783. 10.1016/j.biotechadv.2018.02.001 [DOI] [PubMed] [Google Scholar]
- Hemphill C. F. P., Sureechatchaiyan P., Kassack M. U., Orfali R. S., Lin W., Daletos G., et al. (2017). OSMAC Approach Leads to New Fusarielin Metabolites from Fusarium Tricinctum . J. Antibiot. 70 (6), 726–732. 10.1038/ja.2017.21 [DOI] [PubMed] [Google Scholar]
- Henrikson J. C., Ellis T. K., King J. B., Cichewicz R. H. (2011). Reappraising the Structures and Distribution of Metabolites from Black Aspergilli Containing Uncommon 2-Benzyl-4h-Pyran-4-One and 2-Benzylpyridin-4(1h)-One Systems. J. Nat. Prod. 74 (9), 1959–1964. 10.1021/np200454z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrikson J. C., Hoover A. R., Joyner P. M., Cichewicz R. H. (2009). A Chemical Epigenetics Approach for Engineering the in Situbiosynthesis of a Cryptic Natural Product from Aspergillus niger. Org. Biomol. Chem. 7 (3), 435–438. 10.1039/b819208a [DOI] [PubMed] [Google Scholar]
- Hiort J., Maksimenka K., Reichert M., Perović-Ottstadt S., Lin W. H., Wray V., et al. (2004). New Natural Products from the Sponge-Derived FungusAspergillus niger. J. Nat. Prod. 67 (9), 1532–1543. 10.1021/np030551d [DOI] [PubMed] [Google Scholar]
- Holm D. K., Petersen L. M., Klitgaard A., Knudsen P. B., Jarczynska Z. D., Nielsen K. F., et al. (2014). Molecular and Chemical Characterization of the Biosynthesis of the 6-MSA-Derived Meroterpenoid Yanuthone D in Aspergillus niger . Chem. Biol. 21 (4), 519–529. 10.1016/j.chembiol.2014.01.013 [DOI] [PubMed] [Google Scholar]
- Hua Y., Pan R., Bai X., Wei B., Chen J., Wang H., et al. (2020). Aromatic Polyketides from a Symbiotic Strain Aspergillus fumigatus D and Characterization of Their Biosynthetic Gene D8.T287 . Mar. Drugs 18 (6), 324. 10.3390/md18060324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttel W., Müller M. (2007). Regio- and Stereoselective Intermolecular Oxidative Phenol Coupling in Kotanin Biosynthesis byAspergillus Niger. Chembiochem 8 (5), 521–529. 10.1002/cbic.200600434 [DOI] [PubMed] [Google Scholar]
- Huttel W., Nieger M., Muller M. (2003). A Short and Efficient Total Synthesis of the Naturally Occurring Coumarins Siderin, Kotanin, Isokotanin A and Desertorin C. Synthesis-Stuttgart (12), 1803–1808. 10.1055/s-2003-41027 [DOI] [Google Scholar]
- Jefferson W. E., Jr. (1967). The Isolation and Characterization of Asperenone, a New Phenylpolyene from Aspergillus niger*. Biochemistry 6 (11), 3479–3484. 10.1021/bi00863a019 [DOI] [PubMed] [Google Scholar]
- Jomori T., Hara Y., Sasaoka M., Harada K., Setiawan A., Hirata K., et al. (2020). Mycobacterium Smegmatis Alters the Production of Secondary Metabolites by marine-derived Aspergillus niger . J. Nat. Med. 74 (1), 76–82. 10.1007/s11418-019-01345-0 [DOI] [PubMed] [Google Scholar]
- Kaur T., Singh B., Kaur A., Kaur S. (2015). Endophyte-mediated Interactions between Cauliflower, the Herbivore Spodoptera Litura, and the Ectoparasitoid Bracon Hebetor. Oecologia 179 (2), 487–494. 10.1007/s00442-015-3358-7 [DOI] [PubMed] [Google Scholar]
- Kim K.-W., Sugawara F., Yoshida S., Murofushi N., Takahashi N., Curtis R. W. (1993). Structure of Malformin B, a Phytotoxic Metabolite Produced byAspergillus niger. Biosci. Biotechnol. Biochem. 57 (5), 787–791. 10.1271/bbb.57.787 [DOI] [PubMed] [Google Scholar]
- Kimura Y., Sawada A., Kuramata M., Kusano M., Fujioka S., Kawano T., et al. (2005). Brevicompanine C, Cyclo-(d-Ile-L-Trp), and Cyclo-(d-Leu-L-Trp), Plant Growth Regulators from Penicillium Brevi-Compactum. J. Nat. Prod. 68 (2), 237–239. 10.1021/np040178p [DOI] [PubMed] [Google Scholar]
- Kimura Y., Tani K., Kojima A., Sotoma G., Okada K., Shimada A. (1996). Cyclo-(L-tryptophyl-L-phenylalanyl), a Plant Growth Regulator Produced by the Fungus Penicillium Sp . Phytochemistry 41 (3), 665–669. 10.1016/0031-9422(95)00693-1 [DOI] [Google Scholar]
- Kodukula K., Arcuri M., Cutrone J. Q., Hugill R. M., Lowe S. E., Pirnik D. M., et al. (1995). BMS-192548, a Tetracyclic Binding Inhibitor of Neuropeptide Y Receptors, from Aspergillus niger WB2346. I. Taxonomy, Fermentation, Isolation and Biological Activity. J. Antibiot. 48 (10), 1055–1059. 10.7164/antibiotics.48.1055 [DOI] [PubMed] [Google Scholar]
- Leutou A. S., Yun K., Son B. W. (2016). Induced Production of 6,9-dibromoflavasperone, a New Radical Scavenging Naphthopyranone in the marine-mudflat-derived Fungus Aspergillus niger . Arch. Pharm. Res. 39 (6), 806–810. 10.1007/s12272-016-0764-2 [DOI] [PubMed] [Google Scholar]
- Li A., Pfelzer N., Zuijderwijk R., Punt P. (2012). Enhanced Itaconic Acid Production in Aspergillus niger Using Genetic Modification and Medium Optimization. BMC Biotechnol. 12. 10.1186/1472-6750-12-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Zhou J., Du G., Chen J., Takahashi S., Liu S. (2020). Developing Aspergillus niger as a Cell Factory for Food Enzyme Production. Biotechnol. Adv. 44, 107630. 10.1016/j.biotechadv.2020.107630 [DOI] [PubMed] [Google Scholar]
- Li D.-H., Han T., Guan L.-P., Bai J., Zhao N., Li Z.-L., et al. (2016). New Naphthopyrones from marine-derived Fungus Aspergillus niger 2HL-M-8 and Their In Vitro Antiproliferative Activity. Nat. Product. Res. 30 (10), 1116–1122. 10.1080/14786419.2015.1043553 [DOI] [PubMed] [Google Scholar]
- Li M., Yu R., Bai X., Wang H., Zhang H. (2020). Fusarium: a Treasure Trove of Bioactive Secondary Metabolites. Nat. Prod. Rep. 37, 1568–1588. 10.1039/d0np00038h [DOI] [PubMed] [Google Scholar]
- Li X.-B., Li Y.-L., Zhou J.-C., Yuan H.-Q., Wang X.-N., Lou H.-X. (2015). A New Diketopiperazine Heterodimer from an Endophytic Fungus Aspergillus niger. J. Asian Nat. Prod. Res. 17 (2), 182–187. 10.1080/10286020.2014.959939 [DOI] [PubMed] [Google Scholar]
- Li X., Pan L., Wang B., Pan L. (2019). The Histone Deacetylases HosA and HdaA Affect the Phenotype and Transcriptomic and Metabolic Profiles of Aspergillus niger . Toxins 11 (9), 520. 10.3390/toxins11090520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Chooi Y.-H., Sheng Y., Valentine J. S., Tang Y. (2011). Comparative Characterization of Fungal Anthracenone and Naphthacenedione Biosynthetic Pathways Reveals an α-Hydroxylation-Dependent Claisen-like Cyclization Catalyzed by a Dimanganese Thioesterase. J. Am. Chem. Soc. 133 (39), 15773–15785. 10.1021/ja206906d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Li X.-M., Li C.-S., Wang B.-G. (2013). Nigerasterols A and B, Antiproliferative Sterols from the Mangrove-Derived Endophytic FungusAspergillus nigerMA-132. Hca 96 (6), 1055–1061. 10.1002/hlca.201200332 [DOI] [Google Scholar]
- Liu D., Li X.-M., Meng L., Li C.-S., Gao S.-S., Shang Z., et al. (2011). Nigerapyrones A-H, α-Pyrone Derivatives from the Marine Mangrove-Derived Endophytic FungusAspergillus nigerMA-132. J. Nat. Prod. 74 (8), 1787–1791. 10.1021/np200381u [DOI] [PubMed] [Google Scholar]
- Liu Y., Nan L., Liu J., Yan H., Zhang D., Han X. (2016). Isolation and Identification of Resveratrol-Producing Endophytes from Wine Grape Cabernet Sauvignon . Springerplus 5. 10.1186/s40064-016-2571-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Cao W., Ouyang L., Xia J., Huang M., Chu J., et al. (2017). Comprehensive Reconstruction and In Silico Analysis ofAspergillus Nigergenome-Scale Metabolic Network Model that Accounts for 1210 ORFs. Biotechnol. Bioeng. 114 (3), 685–695. 10.1002/bit.26195 [DOI] [PubMed] [Google Scholar]
- Lu S., Tian J., Sun W., Meng J., Wang X., Fu X., et al. (2014). Bis-naphtho-γ-pyrones from Fungi and Their Bioactivities. Molecules 19 (6), 7169–7188. 10.3390/molecules19067169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manganyi M. C., Regnier T., Kumar A., Bezuidenhout C. C., Ateba C. N. (2018). Biodiversity and Antibacterial Screening of Endophytic Fungi Isolated from Pelargonium Sidoides . South Afr. J. Bot. 116, 192–199. 10.1016/j.sajb.2018.03.016 [DOI] [Google Scholar]
- Miyake Y., Ito C., Itoigawa M., Osawa T. (2007). Isolation of the Antioxidant Pyranonigrin-A from rice Mold Starters Used in the Manufacturing Process of Fermented Foods. Biosci. Biotechnol. Biochem. 71 (10), 2515–2521. 10.1271/bbb.70310 [DOI] [PubMed] [Google Scholar]
- Nielsen K. F., Mogensen J. M., Johansen M., Larsen T. O., Frisvad J. C. (2009). Review of Secondary Metabolites and Mycotoxins from the Aspergillus niger Group. Anal. Bioanal. Chem. 395 (5), 1225–1242. 10.1007/s00216-009-3081-5 [DOI] [PubMed] [Google Scholar]
- Nødvig C. S., Hoof J. B., Kogle M. E., Jarczynska Z. D., Lehmbeck J., Klitgaard D. K., et al. (2018). Efficient Oligo Nucleotide Mediated CRISPR-Cas9 Gene Editing in Aspergilli. Fungal Genet. Biol. 115, 78–89. 10.1016/j.fgb.2018.01.004 [DOI] [PubMed] [Google Scholar]
- Nødvig C. S., Nielsen J. B., Kogle M. E., Mortensen U. H. (2015). A CRISPR-Cas9 System for Genetic Engineering of Filamentous Fungi. PLoS One 10 (7), e0133085. 10.1371/journal.pone.0133085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermaier S., Müller M. (2019). Biaryl-Forming Enzymes from Aspergilli Exhibit Substrate-dependent Stereoselectivity. Biochemistry 58 (22), 2589–2593. 10.1021/acs.biochem.9b00291 [DOI] [PubMed] [Google Scholar]
- Ovenden S. P. B., Sberna G., Tait R. M., Wildman H. G., Patel R., Li B., et al. (2004). A Diketopiperazine Dimer from a Marine-Derived Isolate ofAspergillusniger. J. Nat. Prod. 67 (12), 2093–2095. 10.1021/np0497494 [DOI] [PubMed] [Google Scholar]
- Özer N., Köycü N. D. (2006). The Ability of Plant Compost Leachates to Control Black Mold (Aspergillus niger) and to Induce the Accumulation of Antifungal Compounds in Onion Following Seed Treatment. Biocontrol 51 (2), 229–243. 10.1007/s10526-005-1035-1 [DOI] [Google Scholar]
- Padhi S., Masi M., Panda S. K., Luyten W., Cimmino A., Tayung K., et al. (2020). Antimicrobial Secondary Metabolites of an Endolichenic Aspergillus niger Isolated from Lichen Thallus of Parmotrema Ravum . Nat. Product. Res. 34 (18), 2573–2580. 10.1080/14786419.2018.1544982 [DOI] [PubMed] [Google Scholar]
- Pan R., Bai X., Chen J., Zhang H., Wang H. (2019). Exploring Structural Diversity of Microbe Secondary Metabolites Using OSMAC Strategy: a Literature Review. Front. Microbiol. 10. 10.3389/fmicb.2019.00294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pel H. J., de Winde J. H., Archer D. B., Dyer P. S., Hofmann G., Schaap P. J., et al. (2007). Genome Sequencing and Analysis of the Versatile Cell Factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 25 (2), 221–231. 10.1038/nbt1282 [DOI] [PubMed] [Google Scholar]
- Perrone G., Stea G., Epifani F., Varga J., Frisvad J. C., Samson R. A. (2011). Aspergillus niger Contains the Cryptic Phylogenetic Species A. Awamori. Fungal Biol. 115 (11), 1138–1150. 10.1016/j.funbio.2011.07.008 [DOI] [PubMed] [Google Scholar]
- Rao K. C. S., Divakar S., Babu K. N., Rao A. G. A., Karanth N. G., Sattur A. P. (2002). Nigerloxin, a Novel Inhibitor of Aldose Reductase and Lipoxygenase with Free Radical Scavenging Activity from Aspergillus niger CFR-W-105. J. Antibiot. 55 (9), 789–793. 10.7164/antibiotics.55.789 [DOI] [PubMed] [Google Scholar]
- Reber K. P., Burdge H. E. (2018). Total Synthesis of Pyrophen and Campyrones A-C. J. Nat. Prod. 81 (2), 292–297. 10.1021/acs.jnatprod.7b00720 [DOI] [PubMed] [Google Scholar]
- Riko R., Nakamura H., Shindo K. (2014). Studies on Pyranonigrins-Isolation of Pyranonigrin E and Biosynthetic Studies on Pyranonigrin A. J. Antibiot. 67 (2), 179–181. 10.1038/ja.2013.91 [DOI] [PubMed] [Google Scholar]
- Saeed S., Aslam S., Mehmood T., Naseer R., Nawaz S., Mujahid H., et al. (2020). Production of Gallic Acid under Solid-State Fermentation by Utilizing Waste from Food Processing Industries. Waste Biomass Valor. 12 (1), 155–163. 10.1007/s12649-020-00980-z [DOI] [Google Scholar]
- Sakurai M., Kohno J., Yamamoto K., Okuda T., Nishio M., Kawano K., et al. (2002). TMC-256A1 and C1, New Inhibitors of IL-4 Signal Transduction Produced by Aspergillus niger Var niger TC 1629. J. Antibiot. 55 (8), 685–692. 10.7164/antibiotics.55.685 [DOI] [PubMed] [Google Scholar]
- Sanchez J. F., Somoza A. D., Keller N. P., Wang C. C. C. (2012). Advances in Aspergillus Secondary Metabolite Research in the post-genomic Era. Nat. Prod. Rep. 29 (3), 351–371. 10.1039/c2np00084a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santhakumaran I., Kesavan S. S., Arumugam G. (2019). Asperyellone Pretreatment Protects HaCaT Cells from UVB Irradiation Induced Oxidative Damages: Assessment under In Vitro and In Vivo Conditions and at Molecular Level. J. Cel Biochem 120 (6), 10715–10725. 10.1002/jcb.28363 [DOI] [PubMed] [Google Scholar]
- Shen L., Ye Y.-H., Wang X.-T., Zhu H.-L., Xu C., Song Y.-C., et al. (2006). Structure and Total Synthesis of Aspernigerin: A Novel Cytotoxic Endophyte Metabolite. Chem. Eur. J. 12 (16), 4393–4396. 10.1002/chem.200501423 [DOI] [PubMed] [Google Scholar]
- Shreelalitha S. J., Sridhar K. R. (2015). Endophytic Fungi of Wild Legume Sesbania Bispinosa in Coastal Sand Dunes and Mangroves of the Southwest Coast of India. J. For. Res. 26 (4), 1003–1011. 10.1007/s11676-015-0103-3 [DOI] [Google Scholar]
- Shu Y.-Z., Cutrone J. Q., Klohr S. E., Huang S. (1995). BMS-192548, a Tetracyclic Binding Inhibitor of Neuropeptide Y Receptors, from Aspergillus niger WB2346. II. Physico-Chemical Properties and Structural Characterization. J. Antibiot. 48 (10), 1060–1065. 10.7164/antibiotics.48.1060 [DOI] [PubMed] [Google Scholar]
- Singh S. M., Yadav L. S., Singh S. K., Singh P., Singh P. N., Ravindra R. (2011). Phosphate Solubilizing Ability of Two Arctic Aspergillus niger Strains. Polar Res. 30. 10.3402/polar.v30i0.7283 [DOI] [Google Scholar]
- Soltani J., Moghaddam M. S. H. (2014). Diverse and Bioactive Endophytic Aspergilli Inhabit Cupressaceae Plant Family. Arch. Microbiol. 196 (9), 635–644. 10.1007/s00203-014-0997-8 [DOI] [PubMed] [Google Scholar]
- Song Y. C., Li H., Ye Y. H., Shan C. Y., Yang Y. M., Tan R. X. (2004). Endophytic Naphthopyrone Metabolites Are Co-inhibitors of Xanthine Oxidase, SW1116 Cell and Some Microbial Growths. FEMS Microbiol. Lett. 241 (1), 67–72. 10.1016/j.femsle.2004.10.005 [DOI] [PubMed] [Google Scholar]
- Sørensen L. M., Lametsch R., Andersen M. R., Nielsen P. V., Frisvad J. C. (2009). Proteome Analysis of Aspergillus niger: Lactate Added in Starch-Containing Medium Can Increase Production of the Mycotoxin Fumonisin B2 by Modifying Acetyl-CoA Metabolism. BMC Microbiol. 9, 255. 10.1186/1471-2180-9-255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda S., Curtis R. W. (1966). Antibiotic Properties of Malformin. Appl. Microbiol. 14 (3), 475–476. 10.1128/aem.14.3.475-476.1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Lu X., Rinas U., Zeng A. (2007). Metabolic Peculiarities of Aspergillus niger Disclosed by Comparative Metabolic Genomics. Genome Biol. 8 (9), R182. 10.1186/gb-2007-8-9-r182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano D., Nagamitsu T., Ui H., Shiomi K., Yamaguchi Y., Masuma R., et al. (2001). Absolute Configuration of Nafuredin, a New Specific NADH-Fumarate Reductase Inhibitor. Tetrahedron Lett. 42 (16), 3017–3020. 10.1016/s0040-4039(01)00355-0 [DOI] [Google Scholar]
- Talontsi F. M., Tatong M. D. K., Dittrich B., Douanla-Meli C., Laatsch H. (2013). Structures and Absolute Configuration of Three Alpha-Pyrones from an Endophytic Fungus Aspergillus niger . Tetrahedron 69 (34), 7147–7151. 10.1016/j.tet.2013.05.098 [DOI] [Google Scholar]
- Tanaka H., Pie-Lang W., Namiki M. (1972). Structure of Aurasperone C. Agric. Biol. Chem. 36 (13), 2511–2517. 10.1080/00021369.1972.10860562 [DOI] [Google Scholar]
- Tanaka H., Wang P.-L., Yamada Late O., Tamura T. (1966). Yellow Pigments ofAspergillus nigerandAsp. Awamori. Agric. Biol. Chem. 30 (2), 107–113. 10.1080/00021369.1966.10858561 [DOI] [Google Scholar]
- Toghueo R. M. K., Sahal D., Zabalgogeazcoa Í., Baker B., Boyom F. F. (2018). Conditioned media and Organic Elicitors Underpin the Production of Potent Antiplasmodial Metabolites by Endophytic Fungi from Cameroonian Medicinal Plants. Parasitol. Res. 117 (8), 2473–2485. 10.1007/s00436-018-5936-1 [DOI] [PubMed] [Google Scholar]
- Uchoa P. K. S., Pimenta A. T. A., Braz-Filho R., de Oliveira M. d. C. F., Saraiva N. N., Rodrigues B. S. F., et al. (2017). New Cytotoxic Furan from the marine Sediment-Derived Fungi Aspergillus niger . Nat. Product. Res. 31 (22), 2599–2603. 10.1080/14786419.2017.1283499 [DOI] [PubMed] [Google Scholar]
- Ui H., Shiomi K., Yamaguchi Y., Masuma R., Nagamitsu T., Takano D., et al. (2001). Nafuredin, a Novel Inhibitor of NADH-Fumarate Reductase, Produced by Aspergillus niger FT-0554. J. Antibiot. 54 (3), 234–238. 10.7164/antibiotics.54.234 [DOI] [PubMed] [Google Scholar]
- Varoglu M., Corbett T. H., Valeriote F. A., Crews P. (1997). Asperazine, a Selective Cytotoxic Alkaloid from a Sponge-Derived Culture ofAspergillus niger. J. Org. Chem. 62 (21), 7078–7079. 10.1021/jo970568z [DOI] [PubMed] [Google Scholar]
- Varoglu M., Crews P. (2000). Biosynthetically Diverse Compounds from a Saltwater Culture of Sponge-DerivedAspergillus niger. J. Nat. Prod. 63 (1), 41–43. 10.1021/np9902892 [DOI] [PubMed] [Google Scholar]
- Wang B., Li X., Yu D., Chen X., Tabudravu J., Deng H., et al. (2018). Deletion of the Epigenetic Regulator GcnE in Aspergillus niger FGSC A1279 Activates the Production of Multiple Polyketide Metabolites. Microbiol. Res. 217, 101–107. 10.1016/j.micres.2018.10.004 [DOI] [PubMed] [Google Scholar]
- Wang J., Jiang Z., Lam W., Gullen E. A., Yu Z., Wei Y., et al. (2015). Study of Malformin C, a Fungal Source Cyclic Pentapeptide, as an Anti-cancer Drug. PLoS One 10 (11), e0140069. 10.1371/journal.pone.0140069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.-X., Ren L.-L., Liu X.-B., Shi J., Wang J.-Z., Luo Y.-Q. (2019). Effects of Endophytic Fungi in Mongolian pine on the Selection Behavior of Woodwasp (Sirex noctilio) and the Growth of its Fungal Symbiont. Pest Manag. Sci. 75 (2), 492–505. 10.1002/ps.5146 [DOI] [PubMed] [Google Scholar]
- Wei Q., Bai J., Yan D., Bao X., Li W., Liu B., et al. (2021). Genome Mining Combined Metabolic Shunting and OSMAC Strategy of an Endophytic Fungus Leads to the Production of Diverse Natural Products. Acta Pharmaceutica Sinica B 11 (2), 572–587. 10.1016/j.apsb.2020.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H., Zhuang X., Bai Z., Qi H., Zhang H. (2006). Isolation of Levoglucosan-Assimilating Microorganisms from Soil and an Investigation of Their Levoglucosan Kinases. World J. Microbiol. Biotechnol. 22 (9), 887–892. 10.1007/s11274-006-9133-5 [DOI] [Google Scholar]
- Xu Y., Shan L., Zhou Y., Xie Z., Ball A. S., Cao W., et al. (2019). Development of a Cre-loxP-Based Genetic System in Aspergillus niger ATCC1015 and its Application to Construction of Efficient Organic Acid-Producing Cell Factories. Appl. Microbiol. Biotechnol. 103 (19), 8105–8114. 10.1007/s00253-019-10054-3 [DOI] [PubMed] [Google Scholar]
- Yamamoto T., Tsunematsu Y., Noguchi H., Hotta K., Watanabe K. (2015). Elucidation of Pyranonigrin Biosynthetic Pathway Reveals a Mode of Tetramic Acid, Fused γ-Pyrone, and Exo-Methylene Formation. Org. Lett. 17 (20), 4992–4995. 10.1021/acs.orglett.5b02435 [DOI] [PubMed] [Google Scholar]
- Yang X.-L., Awakawa T., Wakimoto T., Abe I. (2014). Three Acyltetronic Acid Derivatives: Noncanonical Cryptic Polyketides fromAspergillus nigerIdentified by Genome Mining. Chembiochem 15 (11), 1578–1583. 10.1002/cbic.201402172 [DOI] [PubMed] [Google Scholar]
- Zabala A. O., Xu W., Chooi Y.-H., Tang Y. (2012). Characterization of a Silent Azaphilone Gene Cluster from Aspergillus niger ATCC 1015 Reveals a Hydroxylation-Mediated Pyran-Ring Formation. Chem. Biol. 19 (8), 1049–1059. 10.1016/j.chembiol.2012.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Li X.-M., Feng Y., Wang B.-G. (2010). Phenethyl-α-pyrone Derivatives and Cyclodipeptides from a marine Algous Endophytic Fungus Aspergillus niger EN-13. Nat. Product. Res. 24 (11), 1036–1043. 10.1080/14786410902940875 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Li X.-M., Proksch P., Wang B.-G. (2007a). Ergosterimide, a New Natural Diels-Alder Adduct of a Steroid and Maleimide in the Fungus Aspergillus niger . Steroids 72 (9-10), 723–727. 10.1016/j.steroids.2007.05.009 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Li X.-M., Wang B.-G. (2007b). Nigerasperones A∼C, New Monomeric and Dimeric Naphtho-γ-Pyrones from a Marine Alga-Derived Endophytic Fungus Aspergillus niger EN-13. J. Antibiot. 60 (3), 204–210. 10.1038/ja.2007.24 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhang X., Yu Q., Yuan Z., Chen X. (2019). Mutant Breeding of Furfural Tolerant Aspergillus terreus for Itaconic Acid Production. j biobased mat bioenergy 13 (6), 799–805. 10.1166/jbmb.2019.1909 [DOI] [Google Scholar]
- Zhou X., Fang W., Tan S., Lin X., Xun T., Yang B., et al. (2016). Aspernigrins with Anti-HIV-1 Activities from the marine-derived Fungus Aspergillus niger SCSIO Jcsw6F30. Bioorg. Med. Chem. Lett. 26 (2), 361–365. 10.1016/j.bmcl.2015.12.005 [DOI] [PubMed] [Google Scholar]
- Zou S.-P., Zhong W., Xia C.-J., Gu Y.-N., Niu K., Zheng Y.-G., et al. (2015). Mutagenesis Breeding of High Echinocandin B Producing Strain and Further Titer Improvement with Culture Medium Optimization. Bioproc. Biosyst Eng 38 (10), 1845–1854. 10.1007/s00449-015-1425-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.