Abstract
Mutations in the X-linked cell adhesion protein PCDH19 lead to seizures, cognitive impairment, and other behavioral comorbidities when present in a mosaic pattern. Neither the molecular mechanisms underpinning this disorder nor the function of PCDH19 itself are well understood. By combining RNA in situ hybridization with immunohistochemistry and analyzing single-cell RNA sequencing datasets, we reveal Pcdh19 expression in cortical interneurons and provide a first account of the subtypes of neurons expressing Pcdh19/PCDH19, both in the mouse and the human cortex. Our quantitative analysis of the Pcdh19 mutant mouse exposes subtle changes in cortical layer composition, with no major alterations of the main axonal tracts. In addition, Pcdh19 mutant animals, particularly females, display preweaning behavioral changes, including reduced anxiety and increased exploratory behavior. Importantly, our experiments also reveal an effect of the social environment on the behavior of wild-type littermates of Pcdh19 mutant mice, which show alterations when compared with wild-type animals not housed with mutants.
Keywords: cortical lamination, impact of mutant littermates, neuronal subtypes, open field, single-cell RNAseq
Significance Statement
PCDH19 mutations cause epileptic encephalopathy in humans, but the underlying pathophysiology is not completely understood. Here, we provide the first quantitative analysis of the cortical neuronal types expressing Pcdh19 in the mouse and human neocortex, and of cortical layer composition in Pcdh19 mutant animals, revealing the expression of Pcdh19 in interneurons and the presence of small, but significant, changes in neuronal distribution. The findings of our behavioral analysis indicate not only reduced anxiety and increased exploratory behavior, but also an impact of the mutant genotype on the behavior of wild-type animals when housed in the same cage. This finding underscores the importance of selecting appropriate control cohorts to avoid missing relevant behavioral changes in mutant animals.
Introduction
PCDH19 is one of several genes located on the X chromosome known to impact neurodevelopment and behavior. Mutations in this gene were identified in patients with EIEE9 (Epileptic Encephalopathy, Early Infantile, 9; #300088, OMIM), also known as Girls Clustering Epilepsy (GCE), over a decade ago (Dibbens et al., 2008). Since then, over 140 mutations have been described (Kolc et al., 2019), consolidating PCDH19 as the second most relevant gene in epilepsy after SCNA1 (Depienne and Leguern, 2012; Duszyc et al., 2015). The pathogenicity of PCDH19 mutations is dependent on cellular mosaicism, and therefore the disorder follows an unusual inheritance, manifesting in heterozygous (HET) females and in males with somatic mutations (Depienne et al., 2009; Terracciano et al., 2016). Symptoms develop in affected patients during early infancy, often within the first year of life, and display clustered seizures, varying degrees of cognitive impairment, and other comorbidities, including autism spectrum disorder (ASD), attention deficits, and obsessive-compulsive features (Kolc et al., 2020).
PCDH19 codes for Protocadherin 19, a calcium-dependent cell–cell adhesion molecule of the cadherin superfamily. This δ2-protocadherin has six extracellular cadherin repeats, a single transmembrane domain, and a cytoplasmic tail with two conserved motives of unknown function (CM1 and CM2; Wolverton and Lalande, 2001). In addition, a WRC (WAVE regulatory complex) interacting receptor sequence (WIRS) downstream of CM2 allows PCDH19 to interact with the WAVE (WASP family Verprolin homolog) regulatory complex, enhancing its Rac1-mediated activation (Chen et al., 2014). PCDH19 is involved in different processes, ranging from neurulation and organization of the optic tectum in zebrafish (Emond et al., 2009; Cooper et al., 2015) to neurogenesis and regulation of GABAergic transmission in mammals (Fujitani et al., 2017; Bassani et al., 2018; Homan et al., 2018; Lv et al., 2019; Serratto et al., 2020). In addition, PCDH19 is involved in gene expression regulation with estrogen receptor α (Pham et al., 2017), and mutations in PCDH19 lead to a deficiency of the neurosteroid allopregnanolone and of other neuroactive steroids (Tan et al., 2015; Trivisano et al., 2017). Two very recent publications have also addressed the role of PCDH19 in synapse formation in hippocampal cells (Hoshina et al., 2021; Mincheva-Tasheva et al., 2021).
To date, three different Pcdh19 knock-out (KO) mouse models have been developed to explore the function of PCDH19. The first, produced by Taconic Biosciences, has the first three exons of the gene replaced by a β-galactosidase and neomycin (LacZ-neo) resistance cassette (Pederick et al., 2016). The second model retains exons 2 and 3, with a LacZ-neo selection cassette replacing exon 1, which encodes the entire extracellular and transmembrane domains (Hayashi et al., 2017). The third was created by CRISPR-Cas9-mediated deletion of exon 1 (Hoshina et al., 2021). Lack of Pcdh19 mRNA and protein was confirmed for two of the models (Pederick et al., 2016; Hoshina et al., 2021), and no major anatomic defects were reported in any of the three mutant animal lines. However, increased neuronal migration has been described (Pederick et al., 2016), as well as behavioral alterations (Hayashi et al., 2017; Lim et al., 2019; Hoshina et al., 2021). In addition, heterozygous females display a striking segregation of Pcdh19-expressing and nonexpressing progenitors in the developing cortex and altered electrocorticogram traces (Pederick et al., 2018), as well as presynaptic defects in the hippocampal mossy fiber synapse that lead to long-term potentiation abolishment (Hoshina et al., 2021).
Although no major abnormalities in cortical architecture have been reported in either KO mouse model, no detailed quantitative analysis has been conducted yet. Similarly, while RNA in situ hybridization (ISH) revealed the strongest Pcdh19 expression in layers II/III and V(a) in mice (Pederick et al., 2016; Hayashi et al., 2017), the neuronal subtypes expressing Pcdh19 have not been characterized, possibly because of the difficulty of labeling PCDH19-expressing cells with current antibodies. Here we report on the identity of Pcdh19-expressing excitatory and inhibitory neurons in the mouse and human cortex, focusing mainly on somatosensory areas. We also uncover alterations in cortical neuronal distribution in the somatosensory cortex of the Taconic Biosciences Pcdh19 mutant animals, as well as robust differences in the behavior of heterozygous females, including preweaning alterations and an impact of mutant animals on the behavior of their wild-type (WT) littermates.
Materials and Methods
Experimental animals
Animals were housed under a 12 h light/dark cycle with ad libitum access to water and food, and controlled temperature and humidity. All experiments using mice were approved by the local ethical boards and conducted following the directions of the UK Animal Scientific Procedures Act (update 1986).
C57BL6/J WT animals were purchased from Charles River Laboratories, and the Pcdh19 KO line (TF2108) was acquired from Taconic Biosciences.
Experimental matings for anatomic and cellular characterization, as well as for behavioral analysis were set up using wild-type males and Pcdh19 HET females to produce litters with WT males and females, KO males, and HET females.
Analysis of single-cell RNA sequencing datasets
Gene expression matrices and metadata were downloaded from https://portal.brain-map.org/atlases-and-data/rnaseq. Analysis and visualization were conducted using R version 3.6.3, assisted by RStudio version 1.2.1335. Raw counts were normalized to account for library size (total sum of counts per cell) and transformed to counts per million (CPM) using R package scater version 1.16.2. Violin plots were generated with R packages gridExtra version 2.3 and ggplot2 version 3.3.1. River plots were made with R packages gridExtra version 2.3, ggplot2 version 3.3.1, and ggforce version 0.3.2.
Tissue processing
Animals were perfused with PBS followed by 4% paraformaldehyde (PFA) in PBS. After perfusion, brains were extracted and postfixed in PFA 4% overnight at 4°C. For RNA ISH, brains were then cryoprotected in 30% sucrose in PBS before embedding in OCT compound (Tissue-Tek) before freezing. Samples were stored at −80°C until sectioning. Sections of 12 or 20 μm were cut with a cryostat (model CM3050, Leica Systems) and stored at −80°C until use. For immunostaining, fixed brains were briefly washed in PBS and embedded in 4% low-melting point agarose. Sections of 50 μm were cut with a vibrating microtome (model VT1000S, Leica Systems) and stored in PBS with 0.05% sodium azide at 4°C until use.
RNA in situ hybridization and immunohistochemistry
The probe to detect Pcdh19 has been described previously (Gaitan and Bouchard, 2006). Its sequence was amplified using primers Pcdh19e1-F, 5′-CACCAAGCAGAAGATTGACCGAG-3′, and Pcdh19e1-R, 5′-GCCTCCCATCCACAAGAATAGTG-3′, and cloned into pCRII-Blunt-TOPO (Thermo Fisher Scientific). This plasmid was then used to generate digoxigenin (DIG)-labeled sense and antisense probes.
Thawed sections were postfixed in 4% PFA, endogenous peroxidases were quenched with 3% hydrogen peroxidase, and slices were then acetylated in a 0.25% acetic anhydride solution. Prehybridization took place in prewarmed hybridization buffer (50% formamide, 0.1% Tween-20, 0.25% CHAPS, 250 μg/ml yeast tRNA, 500 μg/ml herring sperm, 5× Denhardt’s solution, 5× SSC, 50 μg/ml heparin, 2.5 mm EDTA) for 1 h at 65°C. Slices were hybridized with the denatured sense or antisense probes overnight at 65°C in a humidified chamber. The next day, slides were washed with 0.2× SSC (Thermo Fisher Scientific) and PBST, and then blocked in ISH blocking solution (10% Denhardt’s solution and 0.1% Triton X-100 in PBS) for 20 min at room temperature (RT). After blocking, brain slices were incubated in primary antibody for 1 h at RT, washed in PBST, and incubated in secondary antibody for 1 h at RT. The antibodies used are described below. Slides were then washed in PBST, equilibrated in TN buffer (150 mm NaCl and 100 mm Tris, pH 7.5 in water), and incubated for 30 min in 1:2000 HRP-coupled anti-DIG antibody (catalog #11207733910, Sigma-Aldrich). Following the incubation, tissue was rinsed in TNT (TN+ 0.5% Tween) and immersed in Cy3-Tyramide (TSATM Plus Cy3 Fluorescence Kit; catalog #NEL744001KT, Perkin-Elmer) in a 1:50 dilution dissolved in the amplification diluent. Slides were then washed, counterstained with DAPI, and mounted with DAKO Mounting Medium.
Immunohistochemistry
Antigen retrieval was performed for staining with antibodies against RORB (RAR Related Orphan Receptor B), SATB2 (Special AT-Rich Sequence-Binding Protein 2), parvalbumin (Pvalb), and calretinin (CR), with the tissue either immersed in a 10 mm citrate buffer, pH 6, at 95°C for 5 min (RORB and SATB2) or 10 min (Pvalb, CR) before blocking. Coronal sections of 50 μm were blocked (4% BSA, 3% donkey serum, 0.1% Triton X-100 in PBS) at RT for 1 h. The tissue was then incubated in primary antibody diluted in blocking solution overnight at 4°C. Primary antibodies used for immunostaining were as follows: anti-CUX1 (Cut Like Homeobox 1) rabbit polyclonal (1:200; catalog #11733, Proteintech, or catalog #sc-13 024, Santa Cruz Biotechnology); anti-CTIP2 (COUP-TF-interacting protein 2) rat monoclonal (1:250; catalog #ab18465, Abcam); anti-SATB2 mouse monoclonal (1:400; catalog #ab51502, Abcam); anti-RORB rabbit polyclonal (1:200; catalog #17635-1AP, Proteintech); anti-TBR1 (T-Box Brain Transcription) rabbit polyclonal (1:350; catalog #ab31940, Abcam); anti-Pvalb rabbit polyclonal (1:10,000 or 1:500 for ISH; catalog #PV27, Swant); anti-CB rabbit polyclonal (1:5000; catalog #CB38, Swant); anti-CR mouse polyclonal (1:1000; catalog #AB5054, Merck); anti-SST (somatostatin) rat monoclonal (1:200; catalog #MAB354, Merck); anti-L1CAM (L1 Cell Adhesion Molecule) rat monoclonal (1:500; catalog #MAB5272, Merck); and anti-Neuropilin1 goat polyclonal (1:300; catalog #AF566, R&D Systems).
Slices were then rinsed in PBS and incubated with secondary antibodies coupled to fluorochromes (Alexa Fluor range, Thermo Fisher Scientific) for 1 h at RT. Nuclei were counterstained with DAPI for 10 min, washed again in PBS, and mounted with DAKO Mounting Medium.
Image acquisition and analysis
Images were acquired using a confocal laser scanning microscope (Model LSM 780, Carl Zeiss) and ZEN Black software (version 2.0; Carl Zeiss). Image analysis was conducted with ImageJ Fiji software (Schindelin et al., 2012). For quantification, the cortical wall was divided into 10 horizontal bins of equal width. The number of marker-positive cells in each bin was quantified and is shown as the mean (±SEM) percentage relative to the total number of cells in all 10 bins.
Behavioral analysis
Behavioral tests were conducted at postnatal day 21 (P21; preweaning) and in young adults (P60 and over). Two different WT controls were tested: WT littermates of the mutant animals [mixed-genotype housed (MGH) mice] and animals from pure WT litters [single-genotype housed (SGH) mice]. The WT parents of the SGH animals were derived from the Pcdh19 colony. Mice were habituated to the new environment by taking them to the behavioral room 30 min before the tests. Mice were handled with open hands to reduce anxiety levels and a maximum of one behavioral test was performed per day.
Open field
Open field behavioral analysis was performed on 2 consecutive days, using the first day to habituate the mice to the new environment. Mice were allowed to explore freely, in the dark, for 20 min, in an open field arena (40 × 40 cm). Spontaneous locomotion was recorded using a computer-linked video camera (The Imaging Source) located above the arena and an infrared illumination box (Tracksys) located underneath the arena. The EthoVision XT software (Noldus) was used to analyze the total distance traveled, the distance traveled in intervals of 5 min, and the time spent in the center of the arena. The center of the arena was defined as the area separated from the wall by ≥5 cm.
Elevated plus maze
Each mouse was left to explore freely for 5 min in a maze consisting of the following four perpendicular arms (40 × 7 cm): two open arms (1 cm high) and two closed arms (16 cm high), in a well lit room. Behavior was recorded using a computer-linked video camera (The Imaging Source) located above the maze. The total time spent in the open arms was measured using EthoVision XT software (Noldus).
Social interaction
At P21, test pups were habituated to the arena for 3 min. Subsequently, WT females in estrus, unfamiliar to the pups, were added to the cage, and both mice were allowed to interact with each other for another 3 min in a well lit room. The interaction between the pups and the females was recorded using a computer-linked video camera (The Imaging Source) located above the arena. Videos were manually scored, and interaction was recorded when both mice were within 2 cm of each other, not including tail–tail interactions.
At P60, only female mice were tested for social interaction. In this case the unknown WT females were not required to be in estrus.
To determine which females were in estrus, vaginal smears were stained with Giemsa solution (Polysciences; Caligioni, 2009) before the experiment.
Twenty-four hour activity
P60 experimental mice were placed in individual clear boxes (40 × 24 × 18 cm) and allowed to roam free for 24 h with ad libitum access to food and water and their normal 12 h light/dark cycle. Three infrared beams traversed each cage at the bottom. Data were analyzed using the MED-PC IV software suite and extracted using the MPC2XL program. The number of beam breaks in 24 h and in 1 h slots, as well as the total number of beam breaks during the light and dark periods were analyzed.
Experimental design and statistical analysis
For all experiments, individual animals were considered the experimental unit and the data obtained from each animal were averaged if more than one quantification was performed (e.g., when analyzing several brain slices from the same animal). Experimenters were blind to the genotype of the animals until all quantification or scoring was completed. Statistical analysis was performed using GraphPad Prism version 9 (cortical lamination analysis) or R version 3.6.2 (behavior; R Foundation for Statistical Computing). Normality of the data was tested using the Shapiro–Wilk test, and homogeneity of variance was assessed with Levene’s test. If either assumption was violated, an appropriate nonparametric test was used. Comparisons between two groups were performed using a two-tailed independent-samples t test for normal data, or a Mann–Whitney test if data distribution did not meet normality criteria. If the variance of the two groups differed, a Welch correction was applied. For comparison of more than two groups, ANOVA was used for normal data and a Kruskal–Wallis test if the assumption of normality was not met. If only the assumption of homogeneity of variance was not met, a Welch’s ANOVA was used. The post hoc test following ANOVA was adjusted according to Tukey’s test HSD or, in the case of the social interaction analysis, Dunnett’s test. A Kruskal–Wallis test was followed by Dunn’s correction, and Welch’s ANOVA was followed by Games–Howell correction. Statistical data are presented as the mean ± SEM for formal tests. To carry out estimation statistics for the behavioral experiments, data were introduced into the form available at www.estimationstats.com, in the section for multiple two-group arrangements to obtain the mean differences between groups and their corresponding 95% confidence intervals (CIs). The y-axis limits were set for optimal display of the raw data, and the graphs obtained were directly used in the figures of the article. Calculation of the unbiased Cohen’s d for each comparison, as well as its 95% CI, was conducted using the esci module on jamovi version 1.6 (The jamovi Project; https://www.jamovi.org).
Results
Pcdh19 is expressed by different subtypes of cortical projection neurons and interneurons
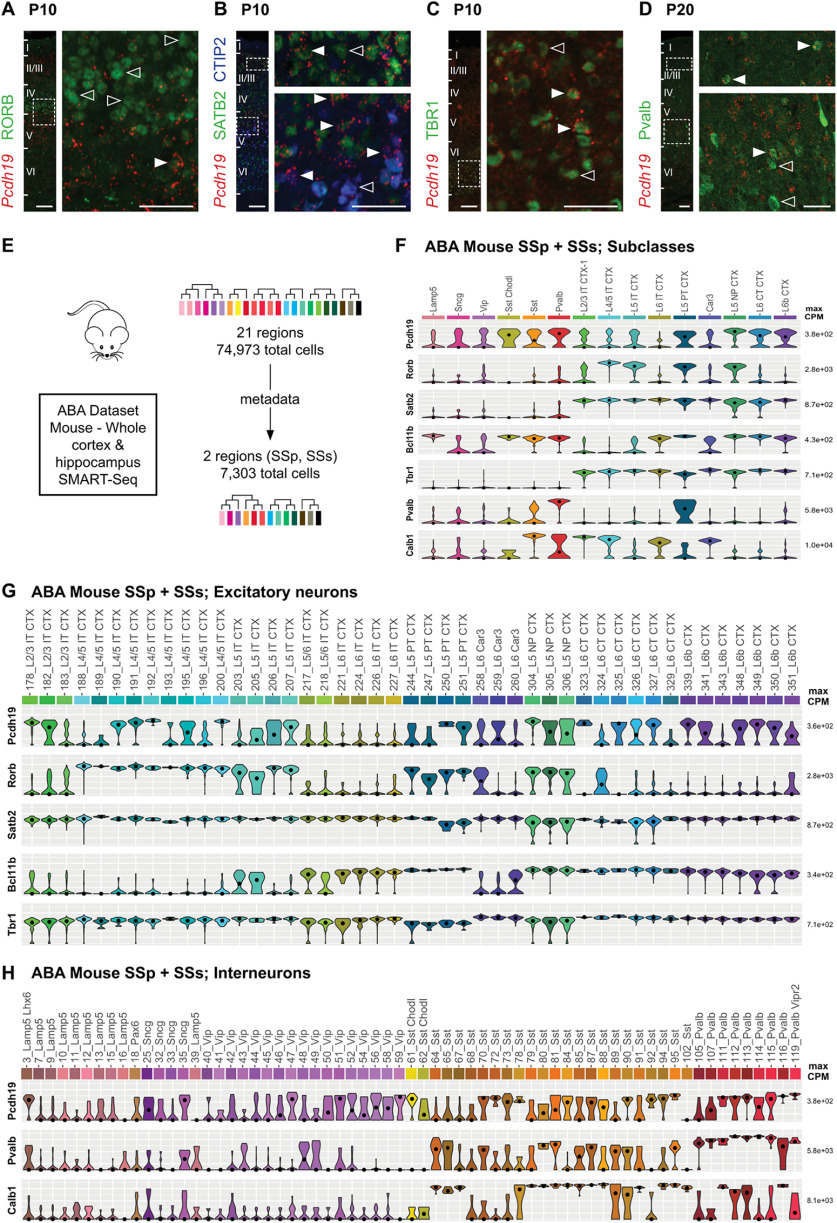
Previous RNA ISH studies have shown two main areas of Pcdh19 expression in the adult cortex, corresponding to the upper regions of layer V (layer Va) and II/III (Hertel and Redies, 2011; Pederick et al., 2016). However, a detailed analysis of the cortical neuronal subtypes expressing Pcdh19, an important consideration given the cellular diversity of the cortex, is still lacking. To address this question, ISH against Pcdh19 was combined with immunohistochemistry (IHC) against several cortical markers for principal neurons and interneurons in the somatosensory cortex at P10 and P20, respectively (Fig. 1A–D). At P10, Pcdh19+ cells were found to coexpress markers for layer IV neurons (RORB; Fig. 1A), callosal projection neurons (SATB2; Fig. 1B), corticospinal neurons (CTIP2; Fig. 1B), and corticothalamic neurons (TBR1; Fig. 1C). The strongest coexpression was seen in SATB2+ neurons, whereas RORB+ cells showed weaker expression and in a smaller proportion of cells. CTIP2+ neurons with strong Pcdh19 expression tended to be located in the upper half of layer V, whereas TBR1+ cells coexpressing Pcdh19 were generally located at lower levels. At P20, we identified interneurons coexpressing Pcdh19 with parvalbumin in layers II/III and V (Fig. 1D), as well as double-positive cells for calbindin and Pcdh19 (data not shown). These data suggest that in juvenile animals Pcdh19 is expressed in both intratelencephalic and corticofugal projection neurons and reveal a previously unreported expression in subpopulations of cortical interneurons.
Figure 1.
Pcdh19 is expressed by excitatory and inhibitory neurons in the mouse cortex. A–D, Confocal micrographs of P10 (A–C) and P20 (D) cortical slices hybridized with an RNA probe against Pcdh19 (red) and antibodies against RORB (green; A), SATB2 and CTIP2 (green and blue, respectively; B), TBR1 (green; C), and parvalbumin (Pvalb, green; D). The left panel shows the entire cortical wall, with boxes indicating the regions enlarged in the right panels. White arrowheads point to double-positive cells, empty arrowheads point to single-positive cells (Pcdh19 negative). Scale bars: left panels, 100 μm; right panels, 50 μm. E, Strategy of the analysis of the mouse whole cortex and hippocampus dataset. F, Violin plots representing gene expression and distribution for Pcdh19 and the markers used in A–D in the 15 subclasses that the SSp and SSs neurons analyzed belong to. Four extra subclasses with five or fewer cells are not included in the figure. G, H, Violin plots representing gene expression and distribution for Pcdh19 and the markers used in A–D in the different excitatory (G) and interneuronal (H) clusters defined in the study by Yao et al. (2020); Allen Brain Atlas, Whole Cortex & Hippocampus - SMART-SEQ (2019) with 10× Smart-Seq Taxonomy (2020). Dots indicate the median value of the cluster in CPM. CPM values are displayed on a log10 scale. For simplicity, clusters belonging to the four subclasses not included in F and any cluster with less than three neurons also are not represented in this figure. Gene expression and distribution of Pcdh19 in cortical excitatory and inhibitory neurons of the Allen Brain Atlas Mouse Whole Cortex & Hippocampus dataset, both globally and by specific brain region, can be found in Extended Data Figures 1-1 and 1-2, respectively.
Gene expression and distribution of Pcdh19 in cortical excitatory projection neurons of the Allen Brain Atlas Mouse Whole Cortex & Hippocampus - SMART-SEQ (2019) with 10x Smart-Seq Taxonomy (2020), represented by violin plots. The first row shows the overall expression of Pcdh19 in the combined dataset excluding hippocampal regions for simplicity. Subsequent rows show expression by cortical region. Dots indicate the median value of the population. Absence of a violin plot in a row indicates that none or fewer than three cells from that particular cortical region were mapped to the corresponding neuronal cluster. Black and red lines indicate consistent low and high expression of Pcdh19 across areas, respectively; asterisks highlight clusters with marked variation in Pcdh19 expression across cortical regions. ACA, Anterior cingulate area; AI, agranular insular area; AUD, auditory areas; GU, gustatory areas; MOp, primary motor area; MOs-FRP, secondary motor area-frontal pole, cerebral cortex; ORB, orbital area; PL-ILA, prelimbic - infralimbic areas; PTLp, posterior parietal association areas; RSP, retrosplenial area; TEa-PERI-ECT, temporal association areas-perirhinal area-ectorhinal area; VIS, visual areas; VISp, primary visual area. Download Figure 1-1, TIF file (12.1MB, tif) .
Gene expression and distribution of Pcdh19 in cortical inhibitory neurons of the Allen Brain Atlas Mouse Whole Cortex & Hippocampus - SMART-SEQ (2019) with 10x Smart-Seq Taxonomy (2020), represented by violin plots. The first row shows the overall expression of Pcdh19 in the combined dataset excluding hippocampal regions, for simplicity. Subsequent rows show expression by cortical region. Dots indicate the median value of the population. Absence of a violin plot in a row indicates that none or fewer than three cells from that particular cortical region were mapped to the corresponding neuronal cluster. Black and red lines indicate consistent low and high expression of Pcdh19 across areas, respectively; asterisks highlight clusters with marked variation in Pcdh19 expression across cortical regions. ACA, Anterior cingulate area; AI, agranular insular area; AUD, auditory areas; GU, gustatory areas; MOp, primary motor area; MOs-FRP, secondary motor area-frontal pole, cerebral cortex; ORB, orbital area; PL-ILA, prelimbic-infralimbic areas; PTLp, posterior parietal association areas; RSP, retrosplenial area; TEa-PERI-ECT, temporal association areas-perirhinal area-ectorhinal area; VIS, visual areas; VISp, primary visual area. Download Figure 1-2, TIF file (13.1MB, tif) .
The previous approach does not allow the identification of distinct molecular subtypes of excitatory and inhibitory neurons populating the neocortex. We thus turned to publicly available datasets of cortical single-cell RNA expression to ascribe molecular identities to Pcdh19-expressing neurons in the mouse adult somatosensory cortex. We chose the “Whole Cortex & Hippocampus - SMART-SEQ (2019) with 10×-Smart-Seq Taxonomy (2020)” dataset from the Allen Brain Atlas (available at https://portal.brain-map.org/atlases-and-data/rnaseq) that includes 76,307 single-cell transcriptomes with cluster-assigned identity isolated from a total of 21 adult cortical and hippocampal regions, including primary and secondary somatosensory cortex. The 74,973 cells for which metadata are available in this dataset are classified into 379 cell types, of which 236 are glutamatergic, 119 GABAergic, and 24 non-neuronal (Yao et al., 2020). We filtered for neurons originating from the primary somatosensory (SSp) and supplemental somatosensory (SSs) cortices using the dataset metadata, which yielded a total of 7303 neurons (Fig. 1E). Those neurons are assigned to 19 subclasses (Fig. 1F), although 4 of them contain <10 cells [Meis2 (5 cells), L2 IT RHP (4 cells), L5 IT TPE-ENT (3 cells), and L2/3 IT CTX-2 (2 cells)] and have not been included in Figure 1. Our analysis shows that, in agreement with our P10 and P20 results, Pcdh19 expression is maintained in both excitatory and inhibitory populations in the adult somatosensory cortex that coexpress the markers of our ISH analysis (Fig. 1E–H).
In excitatory neurons of the adult somatosensory cortex, Pcdh19 expression is lowest in the L6 IT CTX and L6 Car subclasses, where all clusters show consistent low median expression. However, in the remaining subclasses there is always at least one cluster that shows higher expression, indicating that there are Pcdh19-expressing neuronal populations in layers II/III and V, but also in layers VI and VIb, and possibly in layer IV, matching the results of our ISH analysis (Fig. 1G). The neurons expressing Pcdh19 and SATB2 in layers II/III that we identified at P10 (Fig. 1B) could potentially represent clusters 178 and 182 of L2/3 intratelencephalically (IT) projecting neurons. In layer V, neurons expressing Pcdh19 and CTIP2 may correspond to clusters 250 and 251, representing layer V neurons that project outside the cortex (PT), and/or clusters 304–306 of near-projecting neurons, whereas those expressing Pcdh19 and SATB2, but not CTIP2, would be layer V IT neurons, matching those in clusters 190–192, 200, and 207. We also identified neurons expressing Pcdh19 and TBR1 in layer VI (Fig. 1C) that could be corticothalamic neurons (clusters 323, 325, and 327) or layer VIb neurons (clusters 339 and 348–350).
A comparison between different brain regions (Extended Data Fig. 1-1) shows that, although expression levels in the different clusters are generally conserved across brain regions, there are also marked variations in several clusters that tend to manifest in just one or two specific regions.
As in the case of projection neurons, Pcdh19 expression in interneurons of the adult somatosensory cortex is strongly cluster dependent. More specifically, the strongest average expression is found in the Sst-Chodl and Pvalb subclasses (Fig. 1F); however, there is considerable variation and several Sncg, Vip, and Sst clusters also express Pcdh19 widely (Fig. 1H). To assign more meaningful identities to the interneuronal clusters expressing Pcdh19, we made use of the correlation provided between the GABAergic clusters generated from this dataset and the previous taxonomy from Tasic et al. (2018; Yao et al., 2020). Sncg neurons are Vip+, Cck+ multipolar or basket cells located mainly in upper layers, and two of their four subtypes have consistent Pcdh19 expression. Three clusters of Vip interneurons also show relevant Pcdh19 expression (Vip clusters 47, 51, and 59), with at least one of them corresponding to bipolar or multipolar cells (47_Vip). Within the Pvalb subclass, Pcdh19 is expressed by chandelier cells (119_Pvalb Vipr2) and several subtypes of basket cells (Pvalb clusters 112–116). Finally, within the Sst subclass, Pcdh19 expression is strongest in some subtypes of upper layer basket and Martinotti cells (Sst clusters 94 and 95), and in the long-range projecting population (61_Sst-Chodl). Again, variations in the level of Pcdh19 expression within GABAergic clusters can be seen between brain regions (Extended Data Fig. 1-2), but, as was the case for excitatory neurons, differences tend to be limited to a few regions per cluster.
In summary, our analysis demonstrates that mouse Pcdh19 expression is cluster specific in all glutamatergic and GABAergic subclasses in the somatosensory cortex and other cortical areas, being expressed by a heterogeneous neuronal population that includes discrete subtypes of cortical projection neurons and interneurons, with some variation between brain areas. Expression in non-neuronal cells is very low (data not shown).
Human PCDH19 is also expressed in excitatory and inhibitory neurons
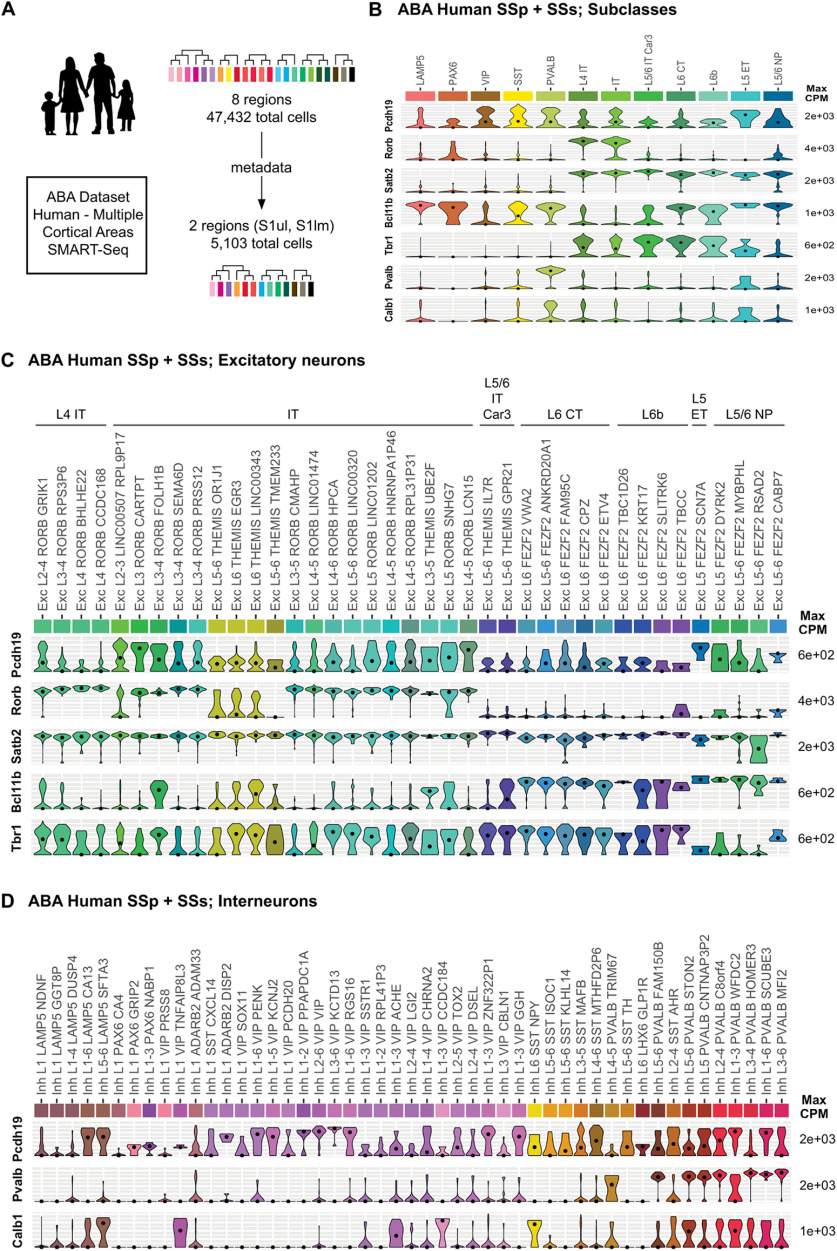
Mutations in PCDH19 cause severe impairments in brain function, yet the expression profile in human cortical neurons is unclear. We therefore extended our analysis to a publicly available human dataset from the Allen Brain Atlas (Human – Multiple Cortical Areas – SMART-seq; available at https://portal.brain-map.org/atlases-and-data/rnaseq), obtained from several brain areas (middle temporal gyrus, anterior cingulate gyrus, primary visual cortex, primary motor cortex, primary somatosensory cortex, and primary auditory cortex). This dataset comprises 49,417 cell nuclei (metadata available for 47,432 cell nuclei) and has allowed the definition of 56 excitatory and 54 inhibitory subtypes. We applied the same strategy as with the mouse dataset, filtering for those neurons originating in the somatosensory cortex, which reduced the dataset to 5103 neurons ascribed to 12 subclasses (Fig. 2A,B). Analysis of PCDH19 expression in this restricted dataset revealed that, within glutamatergic neurons, PCDH19 is primarily expressed in several excitatory neuronal subtypes, particularly Exc L5 FEZF2 SCN7A, which contains layer V neurons that project outside the cortex, and a series of clusters of intracortically projecting neurons spanning layers II–V, such as Exc L3 RORB CARTPT, Exc L3-4 RORB FOLH1B, Exc L5 RORB SNHG7, and Exc L4-5 RORB LCN15 (Fig. 2C). Low expression is evident in many other excitatory neurons of layers III–VI, although several layer IV and VI clusters tend to express much lower levels of PCDH19. A comparison between different brain regions beyond the somatosensory cortex (sSC) shows good correlation between the levels of PCDH19 expression within clusters, with only a few exceptions (Extended Data Fig. 2-1). Regarding interneurons, PCDH19 expression is highest in the L3-6 VIP KCTD13 subtype, with strong expression in most cells. In addition, PCDH19 is also relatively highly expressed in several other VIP, LAMP5, SST, and PVALB subpopulations (Fig. 2D). A comparison between different brain regions reveals that, in general, PCDH19 is expressed in each cluster at similar levels across areas. However, there are some exceptions, like L1 VIP PCDH20 interneurons, which show much higher PCDH19 expression in the primary visual cortex (V1C) than in somatosensory areas [primary somatosensory cortex lower limb region (S1lm) and primary somatosensory cortex upper limb region (S1ul)] or L1-2 VIP RPL41P3, with higher PCDH19 expression in motor areas (Extended Data Fig. 2-2).
Figure 2.
PCDH19 is expressed by excitatory and inhibitory neurons in the human cortex. A, Strategy of the analysis of the Human – Multiple Cortical Areas SMART Seq dataset. B, Violin plots representing gene expression and distribution for Pcdh19 and the markers used in A–D in the 12 subclasses that the sSC neurons analyzed belong to. C, D, Gene expression and distribution of PCDH19 in the glutamatergic (C) and GABAergic (D) cell clusters of the human sSC, represented by violin plots. For the excitatory clusters, the corresponding subclasses are indicated at the top. Dots indicate the median value of the cluster in CPM. CPM values are displayed on a log10 scale. For simplicity, any cluster with less than three neurons is not represented in this figure. Gene expression and distribution of PCDH19 in cortical excitatory and inhibitory neurons of the Allen Brain Atlas Human Multiple Cortical Areas dataset, both globally and by specific brain region, can be found in Extended Data Figures 2-1 and 2-2, respectively. For the strategy to indirectly correlate human and mouse clusters, the specific mouse and human neuronal GABAergic subtypes assigned to the different homology clusters and the correspondence between the nuclei from the MTG and the Multiple Cortical Areas datasets please see Extended Data Figure 2-3. IT (intratelencephalic), CT (corticothalamic), ET (extratelencephalic).
Gene expression and distribution of PCDH19 in cortical excitatory projection neurons of the Allen Brain Atlas Human Multiple Cortical Areas dataset, represented by violin plots. The first row shows overall expression of PCDH19 in the combined dataset; subsequent rows show expression by brain region. Dots indicate the median value of the population. Absence of a violin plot in a row indicates that none or fewer than three cells from that particular brain region were mapped to the corresponding neuronal subtype. Black and red lines indicate consistent low and high expression of PCDH19 across areas, respectively; asterisks highlight clusters with marked variation in PCDH19 expression across cortical regions. CgG, Anterior cingulate gyrus; M1lm, primary motor cortex, lower limb region; M1ul primary motor cortex, upper limb region, A1C, primary auditory cortex. Download Figure 2-1, TIF file (11.2MB, tif) .
Gene expression and distribution of PCDH19 in cortical inhibitory neurons of the Allen Brain Atlas Human Multiple Cortical Areas dataset, represented by violin plots. The first row shows the overall expression of PCDH19 in the combined dataset; subsequent rows show expression by brain region. Dots indicate the median value of the population. Absence of a violin plot in a row indicates that none or fewer than three cells from that particular brain region were mapped to the corresponding neuronal subtype. Black and red lines indicate consistent low and high expression of PCDH19 across areas, respectively; asterisks highlight clusters with marked variation in PCDH19 expression across cortical regions. CgG, Anterior cingulate gyrus; M1lm, primary motor cortex, lower limb region; M1ul primary motor cortex, upper limb region, A1C, primary auditory cortex. Download Figure 2-2, TIF file (12MB, tif) .
A, Diagram indicating the existing correlations between ABA mouse and human cortical datasets. B, Diagram showing the homology clusters defined by Hodge et al. (2019) and the corresponding mouse and human neuronal subtypes assigned to each cluster for GABAergic neuronal clusters. C, River plot showing the mapping of the nuclei from the MTG dataset to the subtypes defined by the Multiple Cortical Areas dataset, for inhibitory neurons. Download Figure 2-3, TIF file (16.5MB, tif) .
Having determined the levels of Pcdh19/PCDH19 expression in the different clusters of excitatory and inhibitory neurons in mouse and human sSC, we set out to evaluate whether expression levels are correlated between clusters in the two species, a relevant issue when using the mouse to investigate a human disorder. No direct equivalents have been described for the clusters of these two datasets, so we took an indirect route, using additional information from the metadata of the Mouse V1 & ALM - SMART-SEQ (2018) and Human MTG - SMART-SEQ (2018) datasets (both available at https://portal.brain-map.org/atlases-and-data/rnaseq; Extended Data Fig. 2-3A). This analysis was only possible for GABAergic neurons, as their clusters (but not the glutamatergic ones) have been correlated between the Whole Cortex & Hippocampus - SMART-SEQ (2019) with 10×-Smart-Seq Taxonomy (2020) and the Mouse V1 & ALM - SMART-SEQ (2018) datasets (Yao et al., 2020). We first determined the composition of the homologous cell types described for these additional mouse and human datasets (Hodge et al., 2019; Extended Data Fig. 2-3B), and then determined the correlation between the human middle temporal gyrus (MTG) and Multiple Brain Areas clusters (Extended Data Fig. 2-3C). This allowed us to establish an indirect comparison between the clusters with highest Pcdh19/PCDH19 expression in mouse and human sSC (Table 1). In general, there is a relatively good correlation between the clusters with highest Pcdh19 expression, particularly for the 3_Lamp Lhx6 cluster, which seems to correspond to chandelier cells in layers V/VI (chandelier type 2 cells; Paul et al., 2017; Tasic et al., 2018), and most (but not all) of the Vip clusters and several Pvalb clusters, including the chandelier cells of 110_Pvalb Vipr2. Correlation in the Sst-Chodl subclass is lower, with mouse long-projecting interneurons expressing higher levels of Pcdh19 than their human counterparts. Levels of expression in clusters of the Sst subclass also tend to show higher variability between the two species.
Table 1.
Comparison of GABAergic clusters with high Pcdh19 expression in mouse and human sSC
| Mouse Whole Cortex & Hippocampus - SMART-SEQ (2019) with 10×-Smart-Seq Taxonomy (2020) |
Mouse V1 & ALM - SMART- SEQ (2018) |
Homologous cell type taxonomy (Hodge et al., 2019) |
Human MTG - SMART-SEQ (2018) |
Human MULTIPLE CORTICAL AREAS - SMART-SEQ (2019) |
|---|---|---|---|---|
| 3_Lamp5 Lhx6 (H) | Lamp5 Lhx6 | Lamp5 Lhx6 | Inh L2-5 LAMP5 CA1 | Inh L1-6 LAMP5 CA13 (H) Inh L5-6 LAMP5 SFTA3 (H) |
| 25_Sncg (M-H) 35_Sncg (H) |
Sncg Vip Nptx2 Sncg Gpr50 Sncg Vip Itih5 |
Vip Sncg | Inh L1-2 VIP TSPAN12 | Inh L1 VIP PRSS8 (L) |
| 40-41_Vip (L) 44-47_Vip (L) |
Serpinf Aqp5 Vip Vip Pygm C1ql1 Vip Chat Htr1f |
Vip 3 | Inh L1-2 VIP PCDH20 | Inh L1-2 VIP PPAPDC1A (H) |
| 47_Vip (H) | Vip Rspo4 Rxfp1 Chat Vip Rspo1 Itga4 |
Vip 4 | Inh L2-4 VIP CBLN1 Inh L1-3 VIP CCDC184 Inh L1-3 VIP GGH Inh L1-3 VIP CHRM2 |
Inh L3 VIP CBLN1 (L) Inh L1-3 VIP ACHE (M) Inh L1-3 VIP GGH (H) Inh L1-2 VIP ZNF322P1 (H) |
| 51_Vip (H) | Vip Gpc3 Slc18a3 | Vip 2 | Inh L2-6 VIP QPCT Inh L3-6 VIP HS3ST3A1 |
Inh L1-6 VIP RGS16 (H) Inh L2-6 VIP VIP (H) Inh L3-6 VIP KCTD13 (H) |
| 59_Vip (H) | Vip Igfbp6 Car10 | Vip 1 | Inh L1-4 VIP PENK Inh L1-3 VIP ADAMTSL1 Inh L1-2 SST BAGE2 |
Inh L1-6 VIP PENK (H) Inh L1-5 VIP KCNJ2 (H) Inh L1 VIP CXCL14 (L) Inh L1 ADARB2 DISP2 (H) |
| 61_Sst Chodl (H) | Sst Chodl | Sst Chodl | Inh L3-6 SST NPY | Inh L6 SST NPY (M) |
| 64_Sst (L) 66_Sst (N.P.) 67_Sst (L) 79_Sst (L) 80-82_Sst (M) |
Sst Myh8 Fibin Sst Chrna2 Glra3 Sst Myh8 Etv1 Sst Nr2f2 Necab1 Sst Chrna2 Ptgdr |
Sst 1 | Inh L3-6 SST HPGD Inh L4-6 SST B3GAT2 |
Inh L4-6 SST MTHFD2P6 (M) |
| 70_Sst (H) 72_Sst (H) 73_Sst (H) 78_Sst (H) |
Sst Tac2 Tacstd2 Sst Rxfp1 Eya1 Sst Rxfp1 Prdm8 |
Sst 3 | Inh L4-6 SST GXYLT2 Inh L5-6 SST NPM1P10 |
Inh L5-6 SST KLHL14 (L) Inh L5-6 SST ISOC1 (L) |
| 84_Sst (H) | Sst Esm1 | Sst 2 | Inh L5-6 SST KLHDC8A (only 3 cells) |
no equivalent |
| 90_Sst (H) 92_Sst (H) 94_Sst (H) 95_Sst (H) |
Sst Calb2 Pdlim5 Sst Tac1 Tacr3 Sst Calb2 Necab1 Sst Tac1 Htr1d |
Sst 5 | Inh L1-3 SST CALB1 | Inh L3-5 SST MAFB (M) |
| 111_Pvalb (H) | Pvalb Akr1c18 Ntf3 | Pvalb 1 | Inh L5-6 PVALB LGR5 Inh L5-6 SST TH Inh L4-5 PVALB MEPE Inh L5-6 SST MIR548F2 |
Inh L5-6 PVALB FAM150B (M) Inh L5-6 SST TH (M) Inh L5 PVALB CNTNAP3P2 (M) Inh L5-6 PVALB STON2 (M) |
| Pvalb Sema3e Kank4 Palb Calb1 Sst |
Pvalb 2 | Inh L2-4 PVALB WFDC2 Inh L4-6 PVALB SULF1 |
Inh L2-4 PVALB C8ORF4 (M) Inh L5 PVALB CNTNAP3P2 (M) Inh L1-3 PVALB WFDC2 (H) Inh L3-4 PVALB HOMER3 (L) |
|
| 112_Pvalb (H) | Pvalb Gpr149 Islr | Pvalb 1 | Inh L5-6 PVALB LGR5 Inh L5-6 SST TH Inh L4-5 PVALB MEPE Inh L5-6 SST MIR548F2 |
Inh L5-6 PVALB FAM150B (M) Inh L5-6 SST TH (M) Inh L5 PVALB CNTNAP3P2 (M) Inh L5-6 PVALB STON2 (M) |
| 113_Pvalb (H) 114_Pvalb (M) 115_Pvalb (H) |
Pvalb Tpbg Pvalb Reln Tac1 Pvalb Reln Itm2a |
Pvalb 2 | Inh L2-4 PVALB WFDC2 Inh L4-6 PVALB SULF1 |
Inh L2-4 PVALB C8ORF4 (M) Inh L5 PVALB CNTNAP3P2 (M) Inh L1-3 PVALB WFDC2 (H) Inh L3-4 PVALB HOMER3 (L) |
| 116_Pvalb (H) | Sst Tac1 Tacr3 Sst Tac1 Htr1d |
Sst 5 | Inh L1-3 SST CALB1 | Inh L3-5 SST MAFB (M) |
| Palb Calb1 Sst Pvalb Tpbg |
Pvalb 2 | Inh L2-4 PVALB WFDC2 Inh L4-6 PVALB SULF1 |
Inh L2-4 PVALB C8ORF4 (M) Inh L5 PVALB CNTNAP3P2 (M) Inh L1-3 PVALB WFDC2 (H) Inh L3-4 PVALB HOMER3 (L) |
|
| 119_Pvalb (H) | Pvalb Vipr2 | Chandelier | Inh L2-5 PVALB SCUBE3 | Inh L1-6 PVALB SCUBE3 (H) |
GABAergic clusters with high Pcdh19 expression in the sSC from either the mouse “Whole Cortex & Hippocampus - SMART-SEQ (2019) with 10×-Smart-Seq taxonomy (2020)” dataset or the human “Multiple Cortical Areas – SMART-SEQ (2019)” dataset are listed in the left and right columns of the table, respectively. The middle columns list the clusters and homologous cell type taxonomy groups that have allowed the indirect correlation between them. H, High expression; M, medium expression; L, low expression; N.P., cluster is not present in the sSC.
Subtle changes in layer composition in Pcdh19 mutant animals
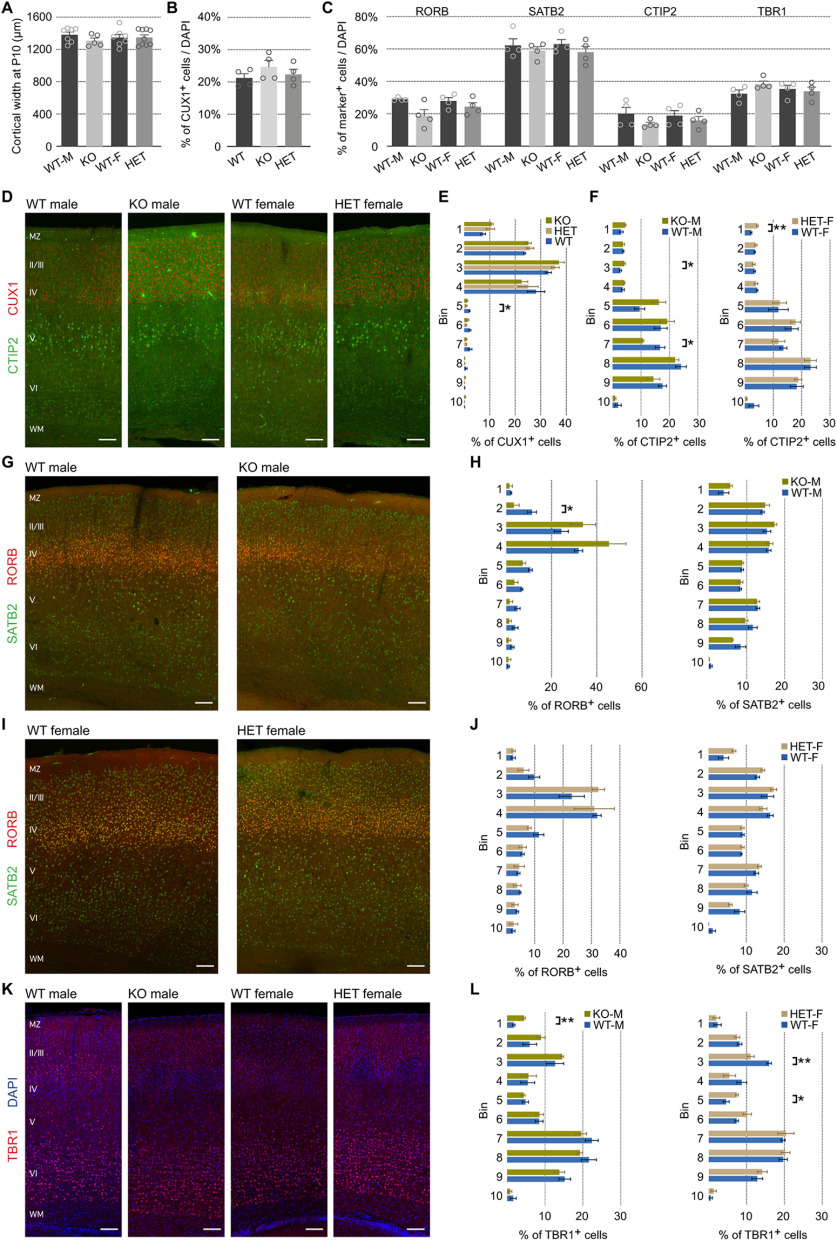
Although no major morphologic defects have been described in Pcdh19 mutant brains (Pederick et al., 2016; Hayashi et al., 2017), a detailed, quantitative study of cortical lamination has not been performed so far. Given that Pcdh19 is expressed in projection neurons and interneurons, we performed an analysis with markers for both neuronal populations in the somatosensory cortex. We first selected cortical markers for projection neurons of deep and upper layers (CUX1, SATB2, RORB, CTIP2, and TBR1) and performed immunohistochemistry at P10, once radial migration is completed. For each marker, we determined the proportion of positive cells, as well as their distribution within 10 bins covering the whole width of the cortical plate. We analyzed males and females separately, using WT male (WT-M) controls for the KO males and WT female (WT-F) controls for the HET animals (except for CUX1, where this was not possible for technical reasons).
In accordance with previous reports (Pederick et al., 2016), we found no differences in cortical width between genotypes (WT-M average = 1381.47 ± 33.72 μm, KO = 1309.10 ± 32.07 μm, WT-F = 1346.85 ± 39.67 μm, HET = 1348.47 ± 32.46 μm; Fig. 3A, Table 2, a). The proportion of positive neurons for all five examined markers was also unaltered (Fig. 3B,C, Table 2, b–f). CUX1+ cells made up approximately one-fifth of all DAPI+ cells (WT = 21.24 ± 1.32%, HET = 22.34 ± 1.64%, KO = 24.66 ± 2.05%), and SATB2+ cells represented more than one-half of all cells (WT-M = 62.20 ± 4.09%, KO = 58.95 ± 2.45%, WT-F = 63.01 ± 2.78%, HET = 57.96 ± 3.64%). The proportion of RORB+ cells seemed lower in KO brains compared with WT-M brains (WT-M = 28.96 ± 0.50%, KO = 18.86 ± 3.74%, WT-F = 27.86 ± 2.15%, HET = 24.37 ± 2.49%), but statistical analysis revealed that this difference was not significant (Mann–Whitney test, U = 3, p = 0.2). CTIP2+ cells were also equally abundant among the four groups (WT-M = 19.97 ± 3.94%, KO = 13.58 ± 1.15%, WT-F = 18.81 ± 3.16%, HET = 15.89 ± 2.46%), and TBR1+ cells added up to approximately one-third of all cells (WT-M = 32.40 ± 2.26%, KO = 38.43 ± 1.80%, WT-F = 35.21 ± 2.40%, HET = 33.85 ± 2.64%).
Figure 3.
Subtle, but significant, changes in the distribution of cortical excitatory neurons in Pcdh19 mutant animals. A, Quantification of cortical width at P10 in Pcdh19 WT and mutant animals, separated by sex. B, Relative percentage of CUX1+ cells examined with respect to total DAPI+ cells in Pcdh19 WT, HET, and KO animals. C, Relative percentages of the different cortical markers examined with respect to total DAPI+ cells. Analysis performed separately for males and females. D, Representative confocal micrographs of immunohistochemistry with anti-CUX1 (red) and anti-CTIP2 (green) antibodies on WT male, KO male, WT female, and HET female tissue. E, Quantification of the percentage of CUX+ cells in each of 10 equal bins spanning the cortical wall. F, Distribution of CTIP2+ cells in each of 10 equal bins spanning the cortical wall, shown as a percentage, for males (left) and females (right). G, I, Representative confocal micrographs of immunohistochemistry with anti-RORB (red) and anti-SATB2 (green) antibodies on WT and KO male tissue (G) and WT and HET female tissue (I). H, J, Quantification of the percentage of RORB+ (left) and SATB2+ (right) cells in each of 10 equal bins spanning the cortical wall. K, Representative confocal micrographs of immunohistochemistry with anti-TBR1 (red) antibodies in WT male, KO male, WT female, and HET female tissue. Nuclei are counterstained with DAPI (blue). L, Distribution of TBR1+ cells in each of 10 equal bins spanning the cortical wall, shown as a percentage for males (left) and females (right). All results are indicated as the mean ± SEM. A minimum of three images per brain, obtained from four animals originating from three different litters were analyzed for each condition. *p < 0.05; **p < 0.01. Scale bars, 200 μm.
Table 2.
Statistical analysis of cortical width and marker composition at P10
| Data | Comparison (n) | Data structure (normality?) | Equal variance? | Test | Results |
|---|---|---|---|---|---|
| Cortical width (a) | WT-M (7) vs KO-M (5) | Yes | Yes | Unpaired t test |
t(2,10) = 1.495 p = 0.1658 |
| WT-F (7) vs HET-F (9) | No | Yes | Mann–Whitney |
U = 31 p > 0.9999 |
|
| WT-M (7) vs WT-F (7) | Yes | Yes | Unpaired t test |
t(2,12) = 0.6648 p = 0.5187 |
|
| % CUX1 over DAPI (b) | WT (4) vs KO (4) vs HET (4) | Yes | Yes | one-way ANOVA |
F(2,9) = 1.065 p = 0.3846 |
| % RORB over DAPI (c) | WT-M (4) vs KO-M (4) | No | Yes | Mann–Whitney |
U = 3 p = 0.2 |
| WT-F (4) vs HET-F (4) | Yes | Yes | Unpaired t test |
t(2,6) = 1.060 p = 0.3301 |
|
| WT-M (4) vs WT-F (4) | No | Yes | Mann–Whitney |
U = 7 p = 0.8857 |
|
| % SATB2 over DAPI (d) | WT-M (4) vs KO-M (4) | Yes | Yes | Unpaired t test |
t(2,6) = 0.6827 p = 0.5203 |
| WT-F (4) vs HET-F (4) | Yes | Yes | Unpaired t test |
t(2,6) = 1.105 p = 0.3113 |
|
| WT-M (4) vs WT-F (4) | Yes | Yes | Unpaired t test |
t(2,6) = 0.1644 p = 0.8749 |
|
| % CTIP2 over DAPI (e) | WT-M (4) vs KO-M (4) | Yes | Yes | Unpaired t test |
t(2,6) = 1.557 p = 0.1704 |
| WT-F (4) vs HET-F (4) | Yes | Yes | Unpaired t test |
t(2,6) = 0.7295 p = 0.4932 |
|
| WT-M (4) vs WT-F (4) | Yes | Yes | Unpaired t test |
t(2,6) = 0.2306 p = 0.8253 |
|
| % TBR1 over DAPI (f) | WT-M (4) vs KO-M (4) | No | Yes | Mann–Whitney |
U = 1 p = 0.0571 |
| WT-F (4) vs HET-F (4) | Yes | Yes | Unpaired t test |
t(2,6) = 0.3816 p = 0.7159 |
|
| WT-M (4) vs WT-F (4) | Yes | Yes | Unpaired t test |
t(2,6) = 0.8509 p = 0.4275 |
The table lists the data analyzed and the groups that have been compared, including the number of independent samples. Normality of the data and equality of variance for the groups compared are indicated, as well as the statistical test performed and the obtained results. The details of the tests performed for the layer distribution of individual markers have not been included, for simplicity.
The distribution of SATB2+ neurons between the 10 bins was unchanged for males and females (Fig. 3G–J). However, we detected some deviations in the distribution of CUX1+, CTIP2+, RORB+, and TBR1+ neurons (Fig. 3D–L). Regarding CUX1, the difference was apparent in bin 5 (Fig. 3E). Pcdh19-HET animals showed a significant 2.4-fold reduction in the percentage of CUX1+ neurons in this bin compared with wild types (WT = 2.08 ± 0.18%, HET = 0.86 ± 0.27%, KO = 1.14 ± 0.32%; one-way ANOVA, F(2,9) = 5.81, p = 0.0239; Tukey’s test: q(1,9) = 4.60, p = 0.0245 HET vs WT). For CTIP2, we found differences in bins 3 (1.7-fold increase) and 7 (1.6-fold reduction) in KO males, suggesting a redistribution of CTIP2+ neurons to higher positions in layer V (bin 3: WT-M = 2.76 ± 0.37%, KO-M = 4.17 ± 0.34%; independent t test, t(2,6) = 2.787, p = 0.0317; bin 7: WT-M = 16.74 ± 1.67%, KO-M = 10.68 ± 0.34%; independent t test with Welch correction for unequal variance, t = 3.556, p = 0.0333). HET females showed double the percentage of cells in bin 1 than their WT siblings (WT-F = 2.20 ± 0.29%, HET-F = 4.42 ± 0.29%; independent t test, t(2,6) = 5.391, p = 0.0017; Fig. 3D,F). Differences in RORB+ distribution were only present in males, specifically in bin 2, with a 3.4-fold reduction (WT-M = 11.38 ± 2.00%, KO-M = 3.36 ± 2.37%; independent t test, t(2,6) = 2.585, p = 0.0415; Fig. 3G,H). However, the graphs for KO and HET animals suggest that the distribution of RORB+ cells tended to be more condensed in those animals. Finally, KO males showed a 2.4-fold increase in the percentage of TBR1+ cells in bin 1 compared with their WT counterparts (WT-M = 1.77 ± 0.33%, KO-M = 4.50 ± 0.33%; independent t test, t(2,6) = 5.818, p = 0.0011), and HET females had a 1.4-fold reduction in the percentage of TBR1+ cells in bin 3 (WT-F = 15.98 ± 0.58%, HET-F = 11.10 ± 0.92%; independent t test, t(2,6) = 4.473, p = 0.0042) and a 1.6-fold increase in bin 5 (WT-F = 4.62 ± 0.79%, HET-F = 7.46 ± 0.35%; independent t test, t(2,6) = 3.268, p = 0.0171; Fig. 3K,L). A comparison between WT males and females did not reveal any differences in the distribution of the four markers analyzed for excitatory neurons (data not shown).
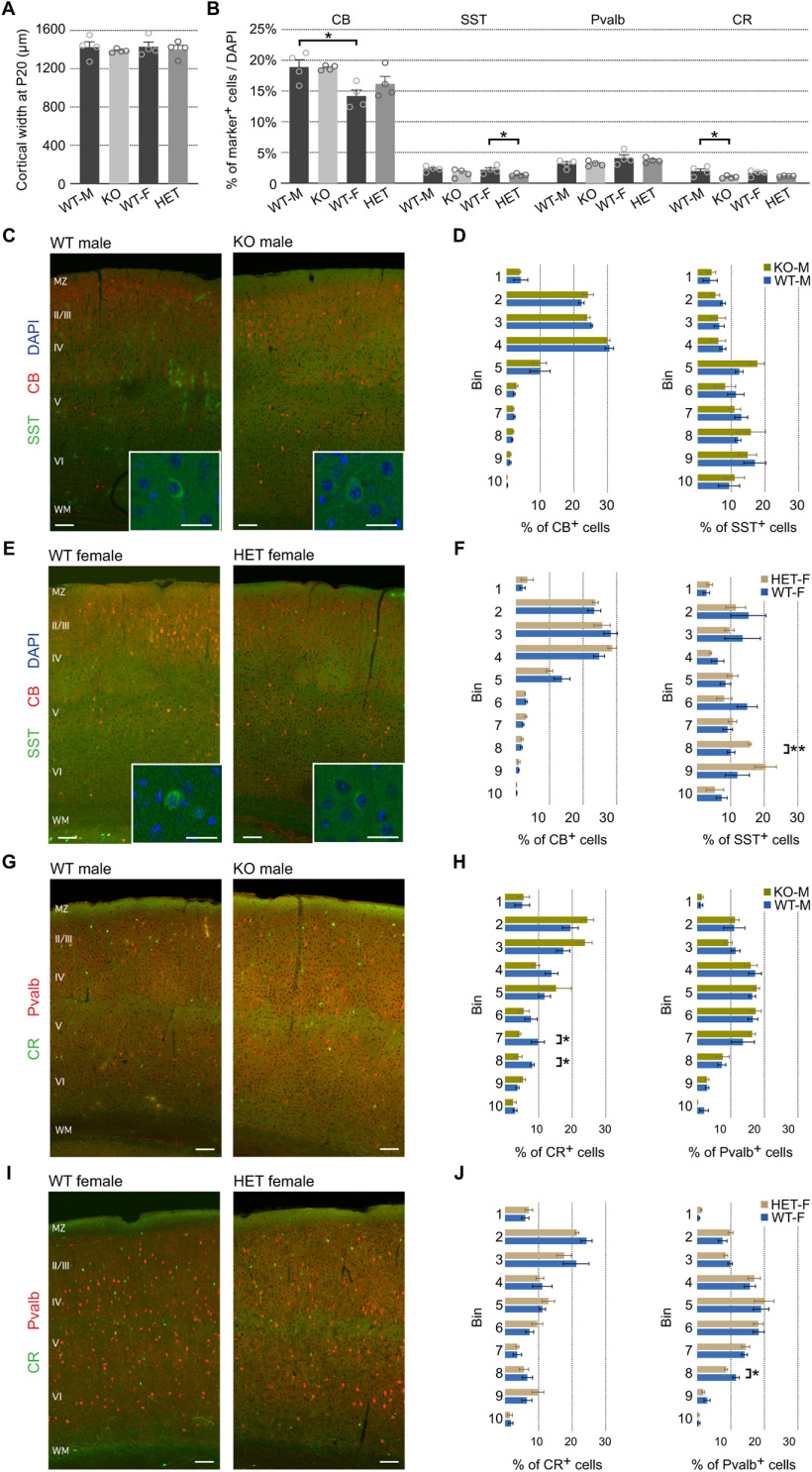
To complete our analysis on cortical composition and lamination, we stained the sSC with four different interneuronal markers (SST, PVALB, CB, and CR) in P20 brains. As before, cortical thickness showed no difference between genotypes of matched sex (WT-M average = 1424.49 ± 57.19 μm, KO = 1387.02 ± 9.88 μm, WT-F = 1429.61 ± 48.84 μm, HET = 1402.97 ± 42.92 μm; Fig. 4A, Table 3, a). However, in this case, some differences were apparent in the overall proportion of three types of interneurons, which may be due in part to the smaller number of cells that test positive for these markers (Fig. 4B, Table 3, b–e). The most abundant type was CB+ cells (WT-M = 18.91 ± 1.20%, KO = 18.77 ± 0.20%, WT-F = 14.19 ± 0.98%, HET = 16.20 ± 1.21%), which, despite no changes between genotypes within males or females, displayed a significantly lower proportion in WT females than in WT males (unpaired t test, t(2,6) = 3.054, p = 0.0224). PVALB+, SST+, and CR+ accounted for <5% of DAPI+ cells each (Fig. 4B). The proportion of PVALB+ interneurons was very similar across the four groups (WT-M = 3.16 ± 0.33%, KO = 3.15 ± 0.21%, WT-F = 4.06 ± 0.55%, HET = 3.70 ± 0.20%), but HET females showed a slight decrease in SST+ cells (WT-M = 2.31 ± 0.23%, KO = 1.61 ± 0.33%, WT-F = 2.19 ± 0.31%, HET = 1.34 ± 0.11%; unpaired t test, WT-F vs HET: t(2,6) = 2.578, p = 0.0419) and KO males a similarly small decrease in CR+ interneurons (WT-M = 1.98 ± 0.39%, KO = 0.98 ± 0.10%, WT-F = 1.63 ± 0.24%, HET = 1.14 ± 0.04%; unpaired t test, WT-M vs KO, t(2,6) = 2.509, p = 0.0459).
Figure 4.
Subtle changes in the distribution of inhibitory neurons in the cortex of Pcdh19 mutant animals. A, Quantification of cortical width at P20 in Pcdh19 WT and mutant animals, separated by sex. B, Relative percentages of the different cortical markers examined with respect to total DAPI+ cells in the somatosensory cortex. Analysis performed separately for males and females. C, E, Representative confocal micrographs of immunohistochemistry with anti-calbindin (CB; red) and anti-SST (green) antibodies on WT and KO male tissue (C), and WT and HET female tissue (E). Insets, High-magnification image of SST+ cells. Nuclei were counterstained with DAPI (blue). D, F, Quantification of the percentage of CB+ (left) and SST+ (right) cells in each of 10 equal bins spanning the cortical wall for males (D) and females (F). G, I, Representative confocal micrographs of immunohistochemistry with anti-parvalbumin (Pvalb; red) and anti-CR (green) antibodies on WT and KO male tissue (G), and WT and HET female tissue (I). H, J, Distribution of CR+ (left) and Pvalb+ (right) cells in each of 10 equal bins spanning the cortical wall, shown as a percentage; male data are shown in H, and female data are shown in J. All results are indicated as the mean ± SEM. A minimum of three images per brain, obtained from four animals originating from three different litters were analyzed for each condition. *p < 0.05, **p < 0.01. Scale bars: 200 μm; insets, 50 μm.
Table 3.
Statistical analysis of cortical width and marker composition at P20
| Data | Comparison (n) | Data structure (normality?) | Equal variance? | Test | Results |
|---|---|---|---|---|---|
| Cortical width (a) | WT-M (4) vs KO-M (4) | Yes | No | Welch’s t test |
t(2,3.179) = 0.6456 p = 0.1658 |
| WT-F (4) vs HET-F (4) | Yes | Yes | Unpaired t test |
t(2,6) = 0.4098 p = 0.6962 |
|
| WT-M (4) vs WT-F (4) | Yes | Yes | Unpaired t test |
t(2,6) = 0.06806 p = 0.9480 |
|
| % CB over DAPI (b) | WT-M (4) vs KO-M (4) | Yes | No | Welch's t test |
t(2,3.168) = 0.1169 p = 0.9140 |
| WT-F (4) vs HET-F (4) | Yes | Yes | Unpaired t test |
t(2,6) = 1.291 p = 0.2443 |
|
| WT-M (4) vs WT-F (4) | Yes | Yes | Unpaired t test |
t(2,6) = 3.054 p = 0.0224 |
|
| % SST over DAPI (c) | WT-M (4) vs KO-M (4) | Yes | Yes | Unpaired t test |
t(2,6) = 1.733 p = 0.1339 |
| WT-F (4) vs HET-F (4) | Yes | Yes | Unpaired t test |
t(2,6) = 2.578 p = 0.0419 |
|
| WT-M (4) vs WT-F (4) | Yes | Yes | Unpaired t test |
t(2,6) = 0.3061 p = 0.7698 |
|
| % PVALB over DAPI (d) | WT-M (4) vs KO-M (4) | Yes | Yes | Unpaired t test |
t(2,6) = 0.01984 p = 0.9848 |
| WT-F (4) vs HET-F (4) | Yes | Yes | Unpaired t test |
t(2,6) = 0.6266 p = 0.5540 |
|
| WT-M (4) vs WT-F (4) | Yes | Yes | Unpaired t test |
t(2,6) = 1.421 p = 0.2051 |
|
| % CR over DAPI (e) | WT-M (4) vs KO-M (4) | Yes | Yes | Unpaired t test |
t(2,6) = 0.0459 p = 2.509 |
| WT-F (4) vs HET-F (4) | Yes | No | Welch's t test |
t(2,3.172) = 2.026 p = 0.1308 |
|
| WT-M (4) vs WT-F (4) | Yes | Yes | Unpaired t test |
t(2,6) = 0.7616 p = 0.4752 |
The table includes the data analyzed and the comparisons made, listing the number of independent samples. Normality of the data and equality of variance for the groups compared are included, as well as the statistical test performed and the obtained results. The details of the tests performed for the layer distribution of individual markers have not been included, for simplicity.
Regarding cellular distribution in the sSC, no differences were apparent for CB+ cells in KO males or HET females (Fig. 4C–F). However, we detected changes in the distribution of SST+ (HET females), CR+ (KO males), and PVALB+ (HET females) interneurons (Fig. 4C–J). HET brains displayed a 1.6-fold increase in the percentage of SST+ cells in bin 8 when compared with gender matched WT brains (WT-F = 10.13 ± 1.15%, HET-F = 15.79 ± 0.4%; independent t test, t(2,6) = 4.647, p = 0.0035; Fig. 4E,F). Although not significant because of higher variability, bin 9 also reflects an increase in SST+ interneurons in HET brains, whereas bins 2 and 3 seem to have reduced numbers, suggesting a potential redistribution of SST+ cells toward deeper layers in HET females. Changes in CR+ cell distribution were found in bin 8 of KO brains, which displayed an approximately twofold reduction over WT male brains (bin 8: WT-M = 8.16 ± 0.57%, KO-M = 4.06 ± 1.05%; Mann–Whitney test, p = 0.0286; Fig. 4G,H). This change, combined with another decrease in bin 7 and concomitant increases in bins 2 and 3 that did not reach statistical significance, might indicate a tendency of CR+ interneurons to occupy higher positions within the cortex in KO animals. As for PVALB+ cells, HET brains showed a reduced percentage in bin 8 (WT-F = 11.54 ± 0.96%, HET-F = 8.61 ± 0.44%; independent t test, t(2,6) = 2.777, p = 0.0321; Fig. 4I,J). In this case, some differences were found in the distribution of CB+ (bin 4), CR+ (bin 8), and PVALB+ (bin 7) interneurons between WT males and females (data not shown, but see Discussion).
In summary, despite relative neuronal proportions and distributions being mostly normal in the sSC of Pcdh19 mutant animals, subtle but significant differences in distribution are apparent for many of the analyzed neuronal markers.
No obvious defects in axonal tracts in Pcdh19 mutant animals
Our results indicate that Pcdh19 is expressed in cortical projection neurons that project through the corpus callosum (layer II–III and some layer V neurons), as well as in neurons projecting outside the cortex, mainly through the pyramidal tract (layer V PT neurons). Although several members of the cadherin superfamily, including δ-protocadherins 7, 10, 17, and 18, have been shown to play a role in axonal outgrowth (Uemura et al., 2007; Piper et al., 2008; Hayashi et al., 2014), fasciculation (Williams et al., 2011; Hayashi et al., 2014), and arborization (Biswas et al., 2014), it is not known whether mutations in Pcdh19 have an impact on any of these processes. We therefore conducted a general characterization of axonal tracts in Taconic Biosciences Pcdh19 male and female WT, male KO, and female HET animals by immunostaining against the cell adhesion molecule L1CAM (Fig. 5A). No differences were apparent for males or females between genotypes in the major axonal tracts, including the internal capsule, stria terminalis, fimbria, or corpus callosum. Next, we analyzed the corpus callosum in more detail by labeling dorsally located axons with Neuropilin-1, which allows the analysis of topographical organization at the midline. Again, the dorsoventral extension of the corpus callosum and the dorsal restriction of Neuropilin-1-expressing axons was similar between genotypes for both male and female animals (Fig. 5B–D, Table 4, a and b). Thus, our results revealed no major abnormalities in the main axonal tracts, although they do not preclude the existence of more subtle defects that would require a more detailed analysis to be revealed.
Figure 5.
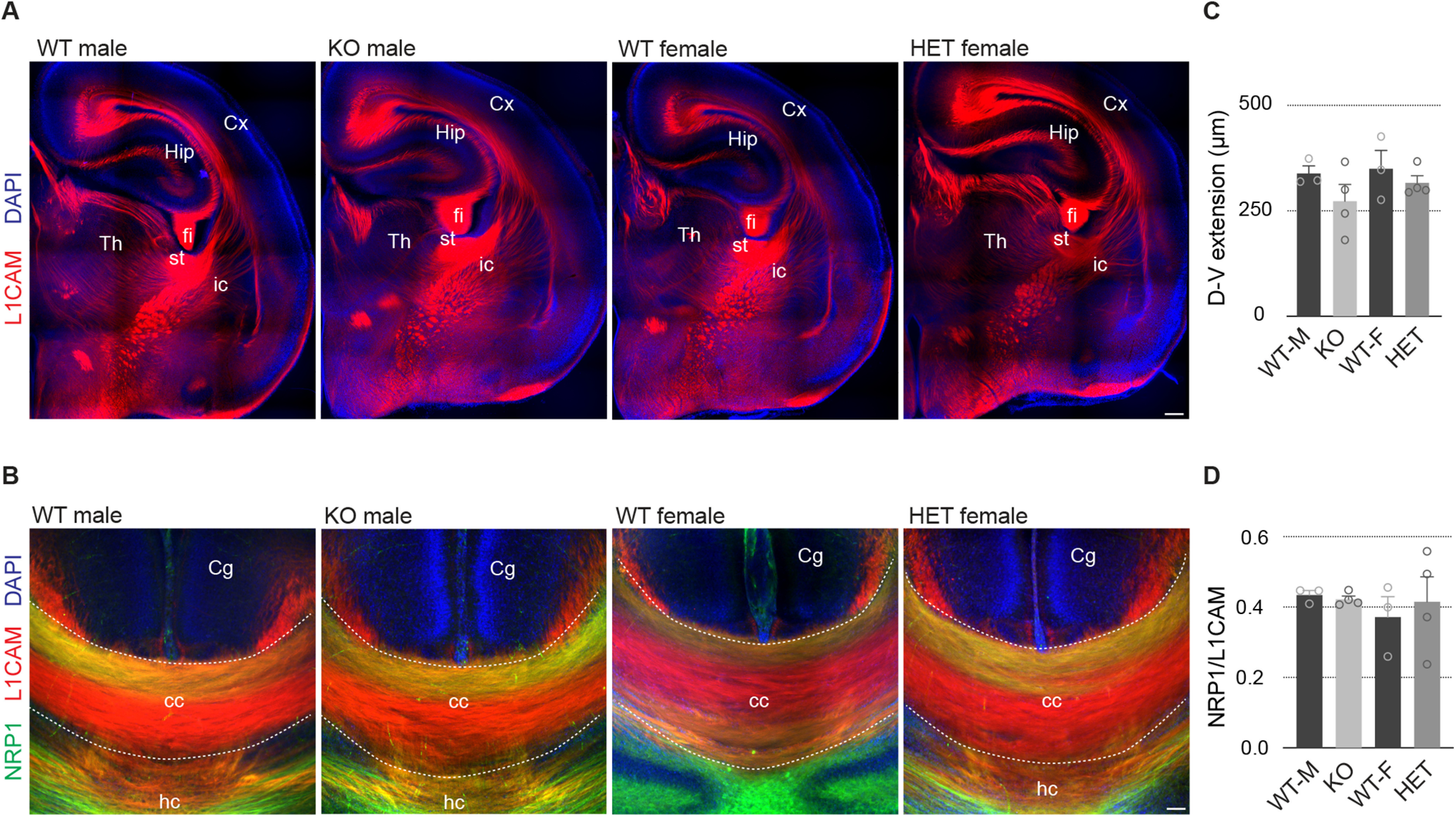
No major anomalies in the main axonal tracts in Pcdh19 mouse mutants. A, Confocal micrographs of P0–P1 mouse hemispheres stained with anti-L1CAM (red). Nuclei were counterstained with DAPI (blue). B, Confocal micrographs of the corpus callosum of P0–P1 mice stained with anti-L1CAM (red) and anti-Neuropilin-1 (green), and counterstained with DAPI (blue). C, Quantification of the dorsoventral extension of the corpus callosum in WT and mutant animals, separated by sex. D, Quantification of the dorsal restriction of Neuropilin-1+ axons in WT and mutant animals, separated by sex. All results are indicated as the mean ± SEM. Two images per brain, obtained from four animals originating from three different litters, were analyzed for each condition. Cx, Cortex; Hip, hippocampus; Th, thalamus, fi, fimbria; st, striatum; ic, internal capsule; Cg, cingulate cortex; cc, corpus callosum; hc, hippocampal commissure. Scale bars: A, 200 μm; B, 50 μm.
Table 4.
Statistical analysis of dorsoventral extension and NRP1/L1CAM ratio in the corpus callosum of wild-type and Pcdh19 mutant pups
| Data | Comparison (n) | Data structure (normality?) | Equal variance? | Test | Results |
|---|---|---|---|---|---|
| D-V extension (a) | WT-M (3) vs KO-M (4) | Yes | Yes | Unpaired t test |
t(2,5) = 1.338 p = 0.2385 |
| WT-F (3) vs HET-F (4) | No | Yes | Mann–Whitney |
U = 5 p = 0.8571 |
|
| WT-M (3) vs WT-F (3) | Yes | Yes | Unpaired t test |
t(2,4) = 0.2420 p = 0.8206 |
|
| NRP1/L1CAM ratio (b) | WT-M (3) vs KO-M (4) | No | Yes | Mann–Whitney |
U = 5 p = 0.8571 |
| WT-F (3) vs HET-F (4) | Yes | Yes | Unpaired t test |
t(2,5) = 0.4525 p = 0.6699 |
|
| WT-M (3) vs WT-F (3) | No | Yes | Mann–Whitney |
U = 3 p = 0.7000 |
The table lists the data analyzed and the groups that have been compared, including the number of independent samples. Normality of the data and equality of variance for the groups compared are indicated, as well as the statistical test performed and the obtained results.
Altered behavior in Pcdh19 mutant animals and their littermates
While there are no major lamination defects in the cortex and in the main axonal tracts of the brain of Pcdh19 mutant animals, the changes in the distribution of specific neuronal subtypes revealed by our quantitative analysis could lead to local connectivity defects that could become apparent at the behavioral level. Indeed, synaptic defects have recently been described between Pcdh19 WT and KO neurons (Hoshina et al., 2021; Mincheva-Tasheva et al., 2021). Thus, we also conducted a series of tests to determine whether these animals present any behavioral alterations. The paradigms included the open field test to evaluate general locomotor activity, anxiety, and exploratory behavior; elevated plus maze (EPM) test to measure anxiety; and a social interaction test. We assessed animals at preweaning age and as adults, to account for any developmental effects. In addition to the WT littermates that Pcdh19 mutant animals were housed with, we included a further control of SGH WT animals (WTSGH; Fig. 6A). Indeed, we note that a previous study on the X-linked ASD-related gene Nlgn3, also a membrane protein expressed in the developing cerebral cortex, revealed that housing conditions impact the behavior of wild-type animals when housed together with mutant animals (Kalbassi et al., 2017). The parents of the animals used to analyze behavior in the single-genotype housed WT condition originated from our Pcdh19 colony, and behavior was analyzed separately for male and female mice. For the behavioral analysis, we have added estimation statistics with CIs to the more common statistical inference analysis (one-way ANOVA or Kruskal–Wallis test among the three groups) to improve the interpretation of results. Because estimation statistics compare the means of only two groups, we provide the average mean difference (Mdiff) and unbiased Cohen’s d of the particular comparison with their corresponding 95% confidence intervals, followed by the results of the overall comparison with ANOVA or Kruskal–Wallis test and the relevant post hoc analysis. When the means of the three groups were not deemed different by any of the two methods, we only present the common statistical inference analysis for brevity.
Figure 6.
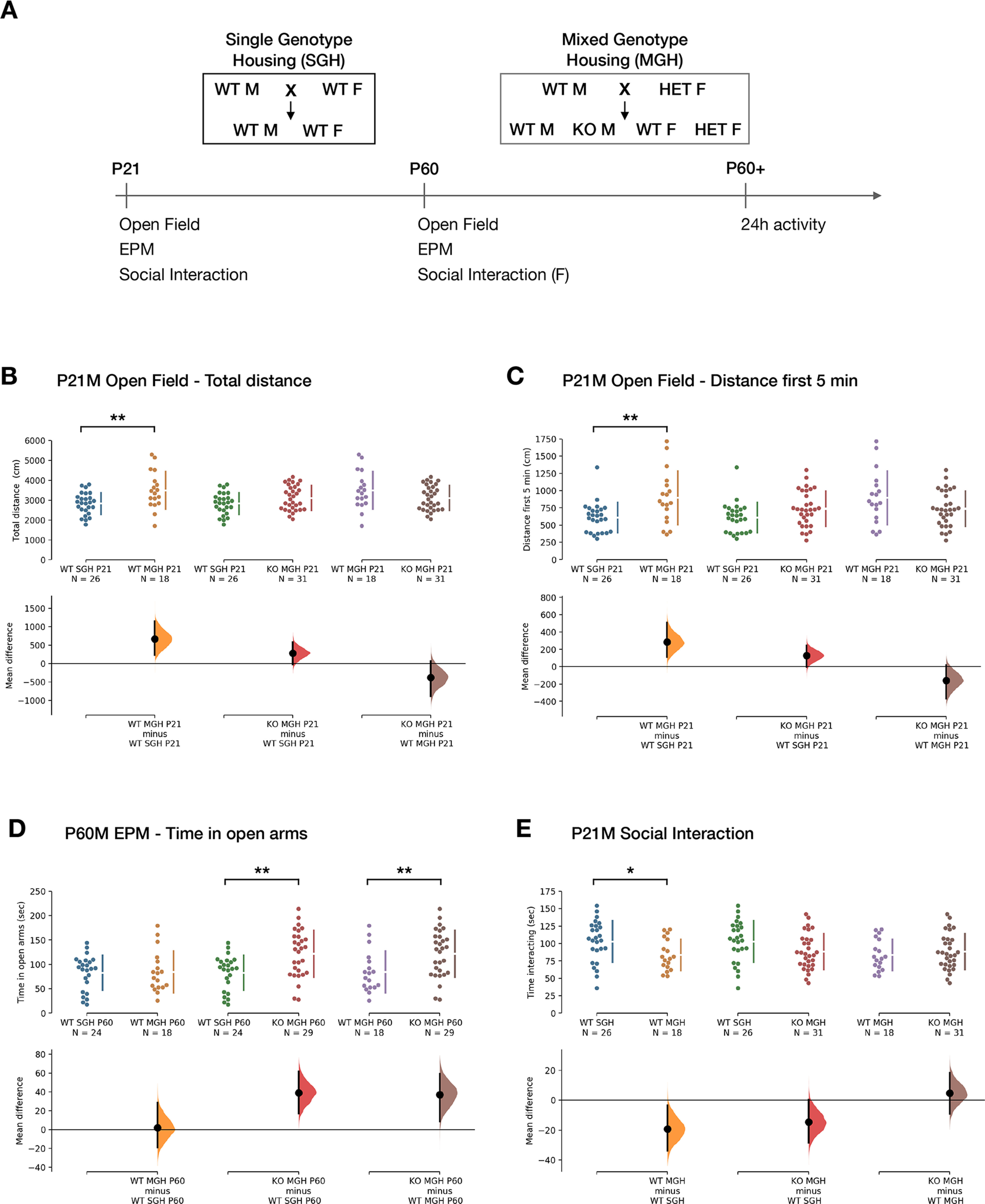
Behavioral alterations in Pcdh19 KO males and their WT littermates. A, Schematic of the behavioral experiments conducted. B, Total distance traveled by males during the 20 min of the open field test at P21. C, Distance traveled in the open field test by P21 males in the first 5 min interval of the open field test. Open field test results correspond to the second day of testing in A and B. D, Total time spent by males in the open arms of the elevated plus maze during the 5 min test at P60. E, Time spent by P21 males interacting with a nonfamiliar female in estrus. The total duration of the test was 5 min. B–D, The top axis shows the raw data points for each group. To their right, the gap in the line indicates the mean, and the lines extending vertically represent the SD. The group and group sizes are indicated at the bottom. Note that each group appears twice in every graph, but with two different colors. The mean difference for each comparison is plotted in the lower axis as a bootstrap sampling distribution. The black dot represents the mean, and the vertical bar its 95% confidence interval. At the top of each graph the significance scores of the one-way ANOVA or Kruskal–Wallis test and their post hoc test are indicated. *p < 0.05; **p < 0.01. Test results with male animals that did not reach significance are presented in Extended Data Figure 6-1.
A, Total distance travelled in the open field test by P60 males. B, Distance travelled in the open field test by P21 males, split into 5 min intervals. The data for the first 5 min are shown in Figure 6. C, Distance travelled in the open field test by P60 males, split into 5 min intervals. D, Time spent by males in the center of the arena during the 20 min open field test at P21 and P60. E, Time spent in the open arms of the elevated plus maze by P21 males. F, Number of beam breaks during the 24 h activity test for the males of the different conditions. Light, light phase; dark, dark phase. G, Number of beam breaks by hour in the 24 h activity test for the males of the different conditions. The time of the day is shown on the x-axis, and the gray square indicates the hours of the dark period. The numbers of tested animals were as follows: at P21: WTSGH, 26; WTMGH, 18; KO, 31; at P60: WTSGH, 24; WTMGH, 18; KO, 29. For the 24 h activity test, the numbers were as follows: WTSGH, 17; WTMGH, 10; KO, 10. Results are indicated as the mean ± SEM. *p < 0.05. Download Figure 6-1, TIF file (5.5MB, tif) .
Differences in male behavior were evident at P21 (Fig. 6B–E, Table 5). MGH WT (WTMGH) males traveled on average 23% more distance during the 20 min open field paradigm than WTSGH males. The unpaired Mdiff was 667.54 cm (95% CI, 233.04, 1150.34; Fig. 6B) and the unbiased Cohen’s d for this comparison was 0.89 (95% CI, 0.29, 1.59), indicating a strong effect of housing (one-way ANOVA, F(2,72) = 5.02, p = 0.0091; post hoc Tukey’s test, WTMGH vs WTSGH: q(1,72) = 4.48, p = 0.0063). In this experiment, KO animals also traveled a higher distance than WTSGH (Mdiff = 281.06 cm, 95% CI, −25.36, 576.08), but an effect of genotype cannot be confirmed with these data. An analysis by 5 min slots showed that the increased distance traveled by WTMGH males compared with WTSGH males was mainly because of a 47% increase in activity during the first 5 min [Mdiff = 285.95 cm; 95% CI, 112.15, 510.92; unbiased Cohen’s d = 0.94 (95% CI, 0.34, 1.65); Kruskal–Wallis test: H(2) = 9.35, p = 0.0093; post hoc Dunn’s test, WTMGH vs WTSGH: z = 3.01, p = 0.0079; Fig. 6C]. Although KO males showed a 21% increase in activity during this period when compared with WTSGH males [Mdiff = 127.75 cm; 95% CI, −4.02, 243.94; unbiased Cohen’s d = 0.53 (95% CI, 0, 1.09)], this difference again does not seem to reflect a real change in behavior, suggesting that increased activity might be an effect of housing in males, rather than of genotype (Kruskal–Wallis test: H(2) = 9.35, p = 0.0093; post hoc Dunn’s test, KO vs WTSGH: z = 3.01, p = 0.1711; Fig. 6C). The increased activity of WTMGH males over WTSGH males disappeared after the first 5 min and also when animals were tested again at ≥P60 [total distance Mdiff = 351.78 cm (95% CI, −197.54, 934.76); unbiased Cohen’s d = 0.36 (95% CI, −0.26, 1.02); one-way ANOVA: F(2,68) = 1.13, p = 0.329; first 5 min: Mdiff = 84.50 cm (95% CI, −96.61, 256.31); unbiased Cohen’s d = 0.27 (95% CI, −0.35, 0.92); one-way ANOVA: F(2,68) = 1.31, p = 0.2759; Extended Data Fig. 6-1A–C]. In accordance with these results, spontaneous activity (number of beam breaks) over a 24 h period in adult male mice did not differ significantly between conditions (Extended Data Fig. 6-1F,G), when analyzed in total (one-way ANOVA: F(2,34) = 0.48, p = 0.621), in the light (one-way ANOVA: F(2,34) = 3.03, p = 0.0615), or in the dark period (one-way ANOVA: F(2,34) = 0.31, p = 0.733). Isolated differences at individual time points (7:00 P.M.: Kruskal–Wallis test, H(2) = 16.08, p = 0.0003; post hoc Dunn’s test, KO vs WTMGH: z = 4.01, p = 0.0002; 8:00 P.M.: one-way ANOVA, F(2,34) = 5.18, p = 0.0109; post hoc Tukey’s test, HET vs KO vs WTSGH: q(1,34) = 4.42, p = 0.0099; 10:00 A.M.: Kruskal–Wallis test: H(2) = 10.78, p = 0.0046; post hoc Dunn’s test, KO vs WTMGH: z = 3.11, p = 0.0056; KO vs WTSGH: z = 2.62, p = 0.0267; 8:00 A.M.: Kruskal–Wallis test, H(2) = 7.17, p = 0.0277; post hoc Dunn’s test, WTMGH vs KO: z = 2.51, p = 0.0361; Extended Data Fig. 6-1G) do not seem to point to an overall activity defect and might be because of a smaller number of animals being tested.
Table 5.
Statistical analysis of the behavioral experiments in P21 and adult males
| Behavioral test | Sex | Age | Normal data? | Equal variance? | Test | Results |
|---|---|---|---|---|---|---|
| Open field test: total distance day 2 | M | P21 | Yes | Yes | One-way ANOVA |
F(2,72) = 5.017; p = 0.0091 Post hoc Tukey’s test: WTSGH vs WTMGH; p = 0.0063 WTSGH vs KO; p = 0.2796 WTMGH vs KO; p = 0.1468 |
| Open field test: total distance day 2 | M | P60 | Yes | Yes | One-way ANOVA | F(2,68) = 1.13; p = 0.329 |
| Open field test: intervals day 2: 0–5 min | M | P21 | No | Yes | Kruskal–Wallis |
H(2) = 9.354; p = 0.0093 Post hoc Dunn’s test: WTSGH vs WTMGH; p = 0.0079 WTSGH vs KO; p = 0.1711 WTMGH vs KO; p = 0.4797 |
| Open field test: intervals day 2: 5–10 min | M | P21 | Yes | Yes | One-way ANOVA | F(2,72) = 0.719; p = 0.491 |
| Open field test: intervals day 2: 10–15 min | M | P21 | Yes | Yes | One-way ANOVA | F(2,72) = 0.976; p = 0.382 |
| Open field test: intervals day 2: 15–20 min | M | P21 | Yes | Yes | One-way ANOVA | F(2,72) = 2.184; p = 0.12 |
| Open field test: intervals day 2: 0–5 min | M | P60 | Yes | Yes | One-way ANOVA | F(2,68) = 1.312; p = 0.276 |
| Open field test: intervals day 2: 5–10 min | M | P60 | Yes | Yes | One-way ANOVA | F(2,68) = 1.292; p = 0.2813 |
| Open field test: intervals day 2: 10–15 min | M | P60 | Yes | Yes | One-way ANOVA | F(2,68) = 0.13; p = 0.879 |
| Open field test: intervals day 2: 15–20 min | M | P60 | No | Yes | Kruskal Wallis | H(2) = 1.56; p = 0.4584 |
| Open field test: time in left day 2 | M | P21 | No | Yes | Kruskal Wallis | H(2) = 2.7579; p = 0.2518 |
| Open field test: time in left day 2 | M | P60 | No | Yes | Kruskal Wallis | H(2) = 3.2761; p = 0.1671 |
| 24 h activity: total | M | >P60 | Yes | Yes | One-way ANOVA | F(2,34) = 0.4831; p = 0.6210 |
| 24 h activity: light period | M | >P60 | Yes | Yes | One-way ANOVA | F(2,34) = 3.031; p = 0.0615 |
| 24 h activity: dark period | M | >P60 | Yes | Yes | One-way ANOVA | F(2,34) = 0.3135; p = 0.7330 |
| Elevated plus maze | M | P21 | Yes | Yes | One-way ANOVA | F(2,72) = 1.994; p = 0.144 |
| Elevated plus maze | M | P60 | Yes | Yes | One-way ANOVA |
F(2,68) = 6.879; p = 0.0019 Post hoc Tukey’s test: WTSGH vs WTMGH; p = 0.9893 WTSGH vs KO; p = 0.0042 WTMGH vs KO; p = 0.0138 |
| Social interaction | M | P21 | Yes | Yes | One-way ANOVA |
F(2,72) = 2.911; p = 0.039 Post hoc Dunnett’s test (all vs WT SGH): WTSGH vs WTMGH; p = 0.0382 WTSGH vs KO; p = 0.0771 |
The table includes the behavioral test analyzed, sex and age of the animals, data and variance distribution, statistical test used, and results obtained. M, Male.
To investigate whether the increased distance traveled by preweaned mixed-genotype housed WT animals in the first 5 min of the open field test could be because of increased anxiety, we analyzed the time spent in the center of the arena. No differences were found among the three conditions, either at P21 (Kruskal–Wallis test: H(2) = 2.76, p = 0.2518) or at P60 (Kruskal–Wallis test: H(2) = 3.58, p = 0.1671; Extended Data Fig. 6-1D). The results of the elevated plus maze confirmed the lack of differences at P21 (Kruskal–Wallis test: H(2) = 4.57, p = 0.1016; Extended Data Fig. 6-1E). However, this was not the case for adult animals, as adult KO males spent >40% more time in the open arms than their WTMGH littermates and WTSGH controls, pointing to an effect of genotype in reducing anxiety [KO vs WTMGH: Mdiff = 37.21 s; 95% CI, 8.57, 59.45; unbiased Cohen’s d = 0.80 (95% CI, 0.21, 1.46); KO vs WTSGH: Mdiff = 39.07 s (95% CI, 16.96, 61.53); unbiased Cohen’s d = 0.90 (95% CI, 0.35, 1.52); one-way ANOVA: F(2,68) = 6.88; p = 0.0019; Tukey’s test, KO vs WTMGH: q(1,68) = 4.10, p = 0.0138; KO vs WTSGH: q(1,68) = 4.68, p = 0.0042; Fig. 6D].
Interestingly, we also detected a subtle difference in social behavior at P21. In this case, WTMGH males spent 19% less time interacting with an unfamiliar female in estrus than single-genotype housed WT males [Mdiff = −19.26 s; 95% CI, −33.73, −3.32; unbiased Cohen’s d = −0.70 (95% CI, −1.38, −0.10); one-way ANOVA: F(2,72) = 3.39; p = 0.039; Dunnett’s test, WTMGH vs WTSGH: q(1,72) = 2.37, p = 0.0382; Fig. 6E]. Although KO males also showed a trend toward reduced interaction, with a 14% decrease [Mdiff = −14.59 s (95% CI, −28.54, 0.18); unbiased Cohen’s d = −0.52 (95% CI, −1.08, 0.00)], this difference is even smaller than for WTMGH males and is unlikely to reflect a real change in behavior (Dunnett’s test, KO vs WTSGH: q(1,72) = 2.07, p = 0.0771). This result again points to an effect of housing on the social behavior of WTMGH males.
In summary, adult KO males displayed a robust phenotype of reduced anxiety in the elevated plus maze test, and WTMGH males showed altered behavior at P21, with increased activity during the first 5 min of the open field test and reduced social interaction.
Changes in behavior were more pronounced in female mice than in their male counterparts (Table 6). We found again differences in the total distance traveled during the open field test at P21, with HET and WTMGH females displaying increases of 35% and 19%, respectively, when compared with single-genotype housed controls [HET vs WTSGH: Mdiff = 913.74 cm (95% CI, 494.07, 1314.30); unbiased Cohen’s d = 1.29 (95% CI, 0.68, 2.04); WTMGH vs WTSGH: Mdiff = 486.76 cm (95% CI, 108.12, 853.27); unbiased Cohen’s d = 0.69 (95% CI, 0.14, 1.31); one-way ANOVA: F(2,69) = 9.54; p = 0.0002; Tukey’s test, HET vs WTSGH: q(1,69) = 6.17; p = 0.0001; for WTMGH vs WTSGH: q(1,69) = 3.55; p = 0.0382; Fig. 7A]. Unlike in males, this effect was maintained at P60, but only in HET females, which traveled on average 19% greater distance than WTSGH animals [Mdiff = 682.77 cm (95% CI, 189.66, 1149.25); unbiased Cohen’s d = 0.83 (95% CI, 0.23, 1.51); one-way ANOVA: F(2,69) = 3.99; p = 0.0229; Tukey’s test, HET vs WTSGH: q(1,69) = 3.87; p = 0.0214; Fig. 7B].
Table 6.
Statistical analysis of the behavioral experiments in P21 and adult females
| Behavioral test | Sex | Age | Normal data? | Equal variance? | Test | Results |
|---|---|---|---|---|---|---|
| Open field test: total distance day 2 | F | P21 | Yes | Yes | One-way ANOVA |
F(2,69) = 9.539; p = 0.0002 Post hoc Tukey’s test: WTSGH vs WTMGH; p = 0.0382 WTSGH vs HET; p = 0.0001 WTMGH vs HET; p = 0.0837 |
| Open field test: total distance day 2 | F | P60 | Yes | Yes | One-way ANOVA |
F(2,69) = 3.990; p = 0.0229 Post hoc Tukey’s test: WTSGH vs WTMGH; p = 0.1094 WTSGH vs HET; p = 0.0214 WTMGH vs HET; p = 0.6459 |
| Open field test: intervals day 2: 0–5 min | F | P21 | No | Yes | Kruskal–Wallis |
H(2) = 21.86; p < 0.0001 Post hoc Dunn’s test: WTSGH vs WTMGH; p = 0.0055 WTSGH vs HET; p < 0.0001 WTMGH vs HET; p = 0.2018 |
| Open field test: intervals day 2: 5–10 min | F | P21 | Yes | Yes | One-way ANOVA |
F(2,69) = 3.290; p = 0.0432 Post hoc Tukey’s test: WTSGH vs WTMGH; p = 0.5888 WTSGH vs HET; p = 0.0359 WTMGH vs HET; p = 0.2036 |
| Open field test: intervals day 2: 10–15 min | F | P21 | Yes | Yes | One-way ANOVA | F(2,69) = 2.102; p = 0.13 |
| Open field test: intervals day 2: 15–20 min | F | P21 | Yes | Yes | One-way ANOVA | F(2,69) = 1.038; p = 0.36 |
| Open field test: intervals day 2: 0–5 min | F | P60 | Yes | Yes | One-way ANOVA |
F(2,69) = 17.95; p < 0.0001 Post hoc Tukey’s test: WTSGH vs WTMGH; p < 0.0001 WTSGH vs HET; p < 0.0001 WTMGH vs HET; p = 0.9276 |
| Open field test: intervals day 2: 5–10 min | F | P60 | Yes | Yes | One-way ANOVA | F(2,69) = 0.228; p = 0.797 |
| Open field test: intervals day 2: 10–15 min | F | P60 | Yes | Yes | One-way ANOVA | F(2,69) = 1.068; p = 0.349 |
| Open field test: intervals day 2: 15–20 min | F | P60 | No | Yes | Kruska–Wallis | H(2) = 3.2334; p = 0.1986 |
| Open field test: time in left day 2 | F | P21 | No | Yes | Kruska–Wallis | H(2) = 4.6819; p = 0.0962 |
| Open field test: time in left day 2 | F | P60 | No | Yes | Kruska–Wallis | H(2) = 4.0863; p = 0.1296 |
| 24 h activity: total | F | >P60 | Yes | Yes | One-way ANOVA | F(2,36) = 1.077; p = 0.3512 |
| 24 h activity: light period | F | >P60 | Yes | Yes | One-way ANOVA | F(2,36) = 2.290; p = 0.1159 |
| 24 h activity: dark period | F | >P60 | Yes | Yes | One-way ANOVA | F(2,36) = 1.103; p = 0.3429 |
| Elevated plus maze | F | P21 | No | Yes | Kruskal–Wallis |
H(2) = 20.943; p < 0.001 Post hoc Dunn’s test: WTSGH vs WTMGH; p = 0.8101 WTSGH vs HET; p = 0.0042 WTMGH vs HET; p < 0.0001 |
| Elevated plus maze | F | P60 | Yes | Yes | One-way ANOVA |
F(2,69) = 5.085; p = 0.0041 Post hoc Tukey’s test: WTSGH vs WTMGH; p = 0.5689 WTSGH vs HET; p = 0.0043 WTMGH vs HET; p = 0.0401 |
| Social interaction | F | P21 | Yes | Yes | One-way ANOVA | F(2,69) = 1.297; p = 0.2425 |
| Social interaction | F | P60 | Yes | Yes | One-way ANOVA |
F(2,69) = 3.536; p = 0.0398 Post hoc Dunnett’s test (all vs WT SGH): WTSGH vs WTMGH; p = 0.0432 WTMGH vs HET; p = 0.9654 |
The table includes the behavioral test analyzed, sex and age of the animals, data and variance distribution, statistical test used, and results obtained. F, female.
Figure 7.
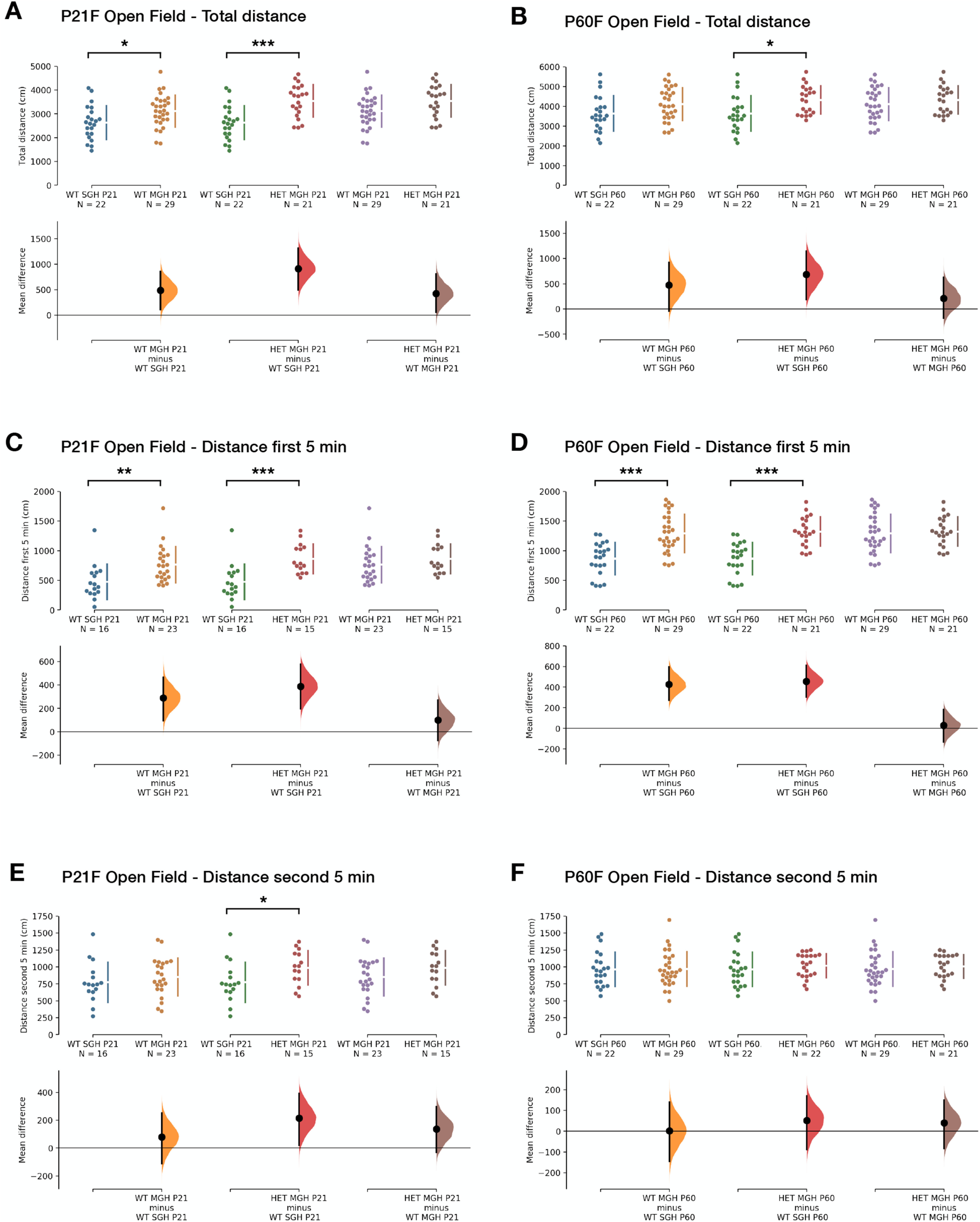
Behavioral alterations in the open field test in Pcdh19 HET females and their WT littermates. A, B, Total distance traveled by females during the 20 min of the open field test at P21 (A) and P60 (B). C, D, Distance traveled by females during the first 5 min of the open field test at P21 (C) and P60 (D). E, F, Distance traveled by females during the second 5 min of the open field test at P21 (E) and P60 (F). Results correspond to the second day of testing at each age. For all panels, the top axis shows the raw data points for each group. To their right, the gap in the line indicates the mean, and the lines extending vertically represent the SD. The group and group sizes are indicated at the bottom. Note that each group appears twice in every graph, but with two different colors. The mean difference for each comparison is plotted in the lower axis as a bootstrap sampling distribution. The black dot represents the mean, and the vertical bar represents the 95% confidence interval. At the top of each graph the significance scores of the one-way ANOVA or Kruskal–Wallis test and their post hoc test are indicated. *p < 0.05; **p < 0.01; ***p < 0.001. Test results with female animals for the open field and 24 h activity that did not reach significance are presented in Extended Data Figure 7-1.
A, B, Distance travelled in the last two 5 min intervals of the open field test by P21 (A) and P60 (B) females. C, Number of beam breaks during the 24 h activity test for the females of the different conditions. Light, Light phase; dark, dark phase. D, Number of beam breaks by hour in the 24 h activity test for the females of the different conditions. The time of the day is shown on the x-axis, and the gray square indicates the hours of the dark period. E, F, Time spent by females in the center of the arena during the 20 min open field test at P21 (E) and P60 (F). In these two panels, raw data are depicted in the top axis, with the mean (gap) and SD (vertical bars) to their right. Group and group sizes are indicated at the bottom. Note that each group appears twice in every graph, but in two different colors. The mean difference for each comparison is plotted in the lower axis as a bootstrap sampling distribution. The mean is indicated by the black dot, and the 95% CI by the vertical bars. The numbers of tested animals were as follows: at P21 and P60: WTSGH, 22; WTMGH, 29; HET, 21. For the 24 h activity test, numbers were as follows: WTSGH, 18; WTMGH, 11; HET, 10. Results are indicated as the mean ± SEM. *p < 0.05. Download Figure 7-1, TIF file (18.5MB, tif) .
Analysis by 5 min intervals showed that the increase in total distance was mainly because of increased activity during the first 5 min in the open field arena both at preweaning age and in adults (Fig. 7C,D). This effect was strong at both ages for HET females and their WT siblings when compared with single-genotype housed females, with increases of 95% (HET) and 54% (WTMGH) at P21, and 53% (HET) and 49% (WTMGH) in adult animals. At P21, the Mdiff between HET and WTSGH was 388.61 cm (95% CI, 195.54, 576.41) with an unbiased Cohen’s d = 1.49 (95% CI, 0.87, 2.27). Between WTMGH and WTSGH, Mdiff = 289.11 cm (95% CI, 94.48, 465.99) with an unbiased Cohen’s d = 0.96 (95% CI, 0.40, 1.61; Kruskal–Wallis test: H(2) = 21.86; p < 0.0001; Dunn’s test, HET vs WTSGH: z = 4.61; p < 0.0001; WTMGH vs WTSGH: z = 3.12; p = 0.0055; Fig. 7C). Despite smaller percentage increases, the Mdiff values between HET and WTSGH, and WTMGH and WTSGH at P60 rose to 456.75 cm (95% CI, 304.66, 609.57) and 426.36 cm (95% CI, 271.11, 595.22), respectively. The unbiased Cohen’s d values for those comparisons were 1.73 (95% CI, 1.09, 2.55) and 1.39 (95% CI, 0.82, 2.09; one-way ANOVA: F(2,69) = 17.95; p < 0.0001; Tukey’s test, HET vs WTSGH: q(1,69) = 7.38; p < 0.0001; WTMGH vs WTSGH: q(1,69) = 7.43; p < 0.0001; Fig. 7D). HET females also traveled a 25% longer distance than WTSGH females during the second 5 min interval at P21 [Mdiff = 215.18 cm (95% CI, 20.06, 391.89); unbiased Cohen’s d = 0.85 (95% CI, 0.25, 1.54); one-way ANOVA: F(2,69) = 3.29; p = 0.0432; Tukey’s test, HET vs WTSGH: q(1,69) = 3.58; p = 0.0359; Fig. 7E], suggesting a potential effect of genotype in addition to the housing effect. By P60, though, there was no average change between the distance run in the second 5 min by any of the groups (Fig. 7F), and no other differences were apparent during the rest of the testing period (Extended Data Fig. 7-1A,B). Similar to male mice, the spontaneous activity over 24 h, measured as the number of beam breaks, was not altered for any of the three experimental groups in the light (one-way ANOVA, main effect of genotype: F(2,36) = 2.29; p = 0.1159), dark (one-way ANOVA, main effect of genotype: F(2,36) = 1.10; p = 0.3429), or total periods (one-way ANOVA, main effect of genotype: F(2,36) = 1.08; p = 0.3512; Extended Data Fig. 7-1C,D). Again, isolated differences were evident at two time points during the dark phase (10:00 P.M.: one-way ANOVA, main effect of genotype: F(2,36) = 3.84; p = 0.0309; Tukey’s test, WTMGH vs WTSGH, q(1,69) = 3.65; p = 0.0364; 4:00 A.M.: Welch’s ANOVA, W(2,23.61) = 8.52; p = 0.0016; Dunnett’s T3, WTMGH vs HET: t(2,18.32) = 3.83; p = 0.0036; Dunnett’s T3, WTMHG vs WTSGH: t(2,23.41) = 2.65; p = 0.0417; Extended Data Fig. 7-1D), but no overall changes in activity were apparent in this test.
Since the increase in distance traveled during the first 5 min in the open field test does not seem to be caused by the overall hyperactivity of HET animals and their WT siblings, we again analyzed anxiety-related behaviors in these animals. There were no differences in the time spent in the center of the open field arena for any of the conditions at P21 (Kruskal–Wallis test: H(2) = 4.68; p = 0.0962) or P60 (Kruskal–Wallis test: H(2) = 4.09; p = 0.1296; Extended Data Fig. 7-1E,F), but, similar to the results obtained with male animals, HET females spent considerably more time in the open arms of the elevated plus maze than any of the WT females at P21 and P60 (Fig. 8A,B). The increases against WTSGH and WTMGH animals amounted to 76% and 103% at preweaning age [HET vs WTSGH: Mdiff = 50.69 s (95% CI, 28.24, 78.00); unbiased Cohen’s d = 1.20 (95% CI, 0.59, 1.94); HET vs WTMGH: Mdiff = 59.71 s (95% CI, 34.28, 84.71); unbiased Cohen’s d = 1.32 (95% CI, 0.74, 2.02); Kruskal–Wallis test: H(2) = 20.94, p < 0.0001; Dunn’s test, HET vs WTSGH: z = 3.19, p = 0.042; WTMGH vs WTSGH: z = 4.49, p < 0.0001]. In adults, the increase was down to 60% and 39% [HET vs WTSGH: Mdiff = 42.40 s (95% CI, 15.09, 69.81); unbiased Cohen’s d = 0.90 (95% CI, 0.30, 1.60); HET vs WTMGH: Mdiff = 31.69 s (95% CI, 7.28, 59.92); unbiased Cohen’s d = 0.71 (95% CI, 0.15, 1.34); one-way ANOVA: F(2,69) = 5.95; p = 0.0041; Tukey’s test, HET vs WTSGH: q(1,69) = 4.67; p = 0.0043; HET vs WTMGH: q(1,69) = 3.72; p = 0.0281]. These results indicate a strong effect of genotype on reducing anxiety, as also seen for adult male KO animals.
Figure 8.
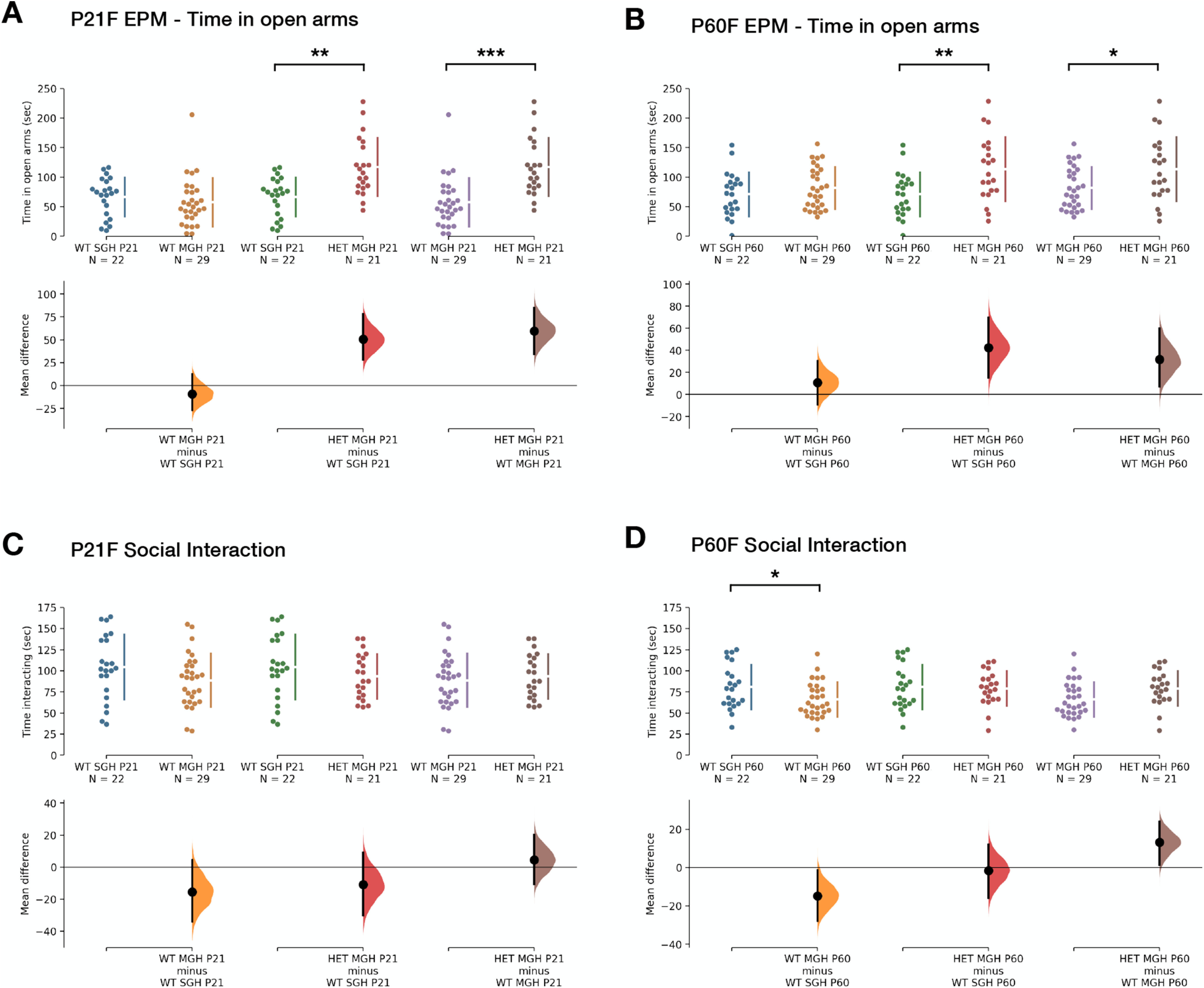
Behavioral alterations in the EPM and social interaction tests in Pcdh19 HET females and their WT littermates. A, B, Total time spent by females in the open arms of the elevated plus maze during the 5 min test at P21 (A) and P60 (B). C, D, Time spent by females interacting with a nonfamiliar female at P21 (C) and P60 (D). The total duration of the test was 5 min. For all panels, the top axis shows the raw data points for each group. To their right, the gap in the line indicates the mean, and the lines extending vertically represent the SD. The group and group sizes are indicated at the bottom. Note that each group appears twice in every graph, but with two different colors. The mean difference for each comparison is plotted in the lower axis as a bootstrap sampling distribution. The black dot represents the mean, and the vertical bar represents the 95% confidence interval. At the top of each graph the significance scores of the one-way ANOVA or Kruskal–Wallis test and their post hoc test are indicated. *p < 0.05; **p < 0.01; ***p < 0.001.
As in the case of male mice, the social interaction test revealed differences between single-genotype and mixed-genotype housed WT females (Fig. 8C,D). However, this effect was present only in adult animals, with WTMGH females spending 15% less time interacting with an unfamiliar female in estrus [Mdiff = −14.69 s (95% CI, −27.79, −1.29); unbiased Cohen’s d = −0.62 (95% CI, −1.24, −0.07); one-way ANOVA: F(2,69) = 3.38; p = 0.0398; Dunnett’s test, WTMHG vs WTSGH: q(1,69) = 2.32, p = 0.0432].
Overall, we found significant behavioral differences between wild-type and mutant animals that were generally more pronounced in HET females than in KO males. HET females displayed consistent hyperactivity during the first 5 min of the open field test and, similar to the mutant males, a robust phenotype of decreased anxiety in the elevated plus maze, in this case both at preweaning and at adult stages. Importantly, we also uncovered an effect of housing on the behavior of WT animals, with WTMGH males and females presenting significant differences in the open field and social interaction tests when compared with single-genotype housed WT animals.
Discussion
Recent studies have shed light on the different functions of PCDH19 (Pederick et al., 2016, 2018; Hayashi et al., 2017; Pham et al., 2017; Bassani et al., 2018; Homan et al., 2018; Serratto et al., 2020; Hoshina et al., 2021; Mincheva-Tasheva et al., 2021; for review, see Gerosa et al., 2019; Gecz and Thomas, 2020), but we still have limited knowledge about the neuronal types expressing PCDH19 and the consequences of Pcdh19 mutations on fine cortical composition, despite the relevance of these factors to understand the pathologic mechanisms underpinning EIEE9. Here we present a detailed analysis of neuronal subtypes expressing Pcdh19 in the mouse somatosensory cortex and a comparison with human data. Our study reveals that Pcdh19/PCDH19 is not only expressed in pyramidal neurons, but also in different types of interneurons, and that, in general, higher expression is limited to specific subpopulations in both cases. Our analysis also rules out major anomalies in the main axonal tracts and provides a quantitative assessment of cortical composition and lamination. Despite the lack of major architectural defects, our data reveal subtle defects in layer composition that could contribute to the pathophysiology of EIEE9. Indeed, mutant animals display behavioral alterations in the open field test (females) and elevated plus maze test (males and females). Importantly, and as previously revealed with the analysis of Nlgn3 mutants (Kalbassi et al., 2017), the Pcdh19 mutation affects the behavior of wild-type littermates when housed in the same cage.
Hitherto, the characterization of the neuronal populations expressing PCDH19 has been hindered by the lack of specific antibodies that perform satisfactorily in immunohistochemistry analyses. In addition, as PCDH19 is likely distributed in both axons and dendrites (Pederick et al., 2016; Hayashi et al., 2017; Bassani et al., 2018), the unambiguous identification of cell bodies expressing PCDH19 is a challenging objective, as is the case for most membrane proteins in the cortex. To overcome this difficulty, we focused on the expression of Pcdh19 mRNA, which is detected in the cell soma and allows a better assessment of coexpression with other neuronal markers, which tend to be either nuclear or cytoplasmic. Although mRNA and protein expression are not necessarily correlated, available data show a good match between the regions with the strongest mRNA and protein signals (Hayashi et al., 2017; Pederick et al., 2018). Our ISH/IHC combination approach provides experimental evidence for the expression of Pcdh19 by different neuronal types across cortical layers, including interneurons. We chose the somatosensory cortex to carry out the analysis because it is a very well characterized area with a good definition of cortical layers. We then confirmed the results obtained in the postnatal sSC by choosing single-cell RNA sequencing (scRNAseq) datasets that include neurons from various cortical regions (including sSC) from adult brain, which allowed us to obtain a global view of Pcdh19/PCDH19 expression across cortical areas in mouse and human.
Our analysis of a mouse dataset of whole cortex and hippocampus confirmed that Pcdh19 is expressed by excitatory neurons in layer V, projecting both intracortically and extracortically, as well as by certain subtypes of layer II/III projection neurons, in agreement with the ISH data. Expression in layer IV is harder to judge from the scRNAseq results, as there are no clusters representing neurons from layer IV exclusively, but several clusters in layers VI and VIb also show high Pcdh19 expression. In interneurons, the expression is widespread in the Pvalb subclass; cluster specific in the Sncg, Vip, and Sst subclasses; and very low in the Lamp5 and Pax6 clusters, except for Lamp5 Lhx6, which shows high expression. These results demonstrate that while Pcdh19 is expressed by a variety of excitatory and inhibitory neurons, expression remains specific for particular clusters. This cluster specificity would suggest a role for PCDH19 in the establishment of neuronal circuits as a potential neuronal recognition molecule.
Human PCDH19 follows a similar pattern, with expression in both excitatory and inhibitory neuronal types. Expression in human excitatory neurons of the sSC is more graded, with many more subtypes showing intermediate expression levels than in mouse, likely reflecting an averaging effect because of the smaller number of human clusters defined for that dataset. In any case, highest expression corresponds to clusters in layers III and V, in line with RNA ISH results in mice. Regarding interneurons, high PCDH19 expression can be found in subtypes of LAMP5, VIP, SST, and PVALB interneurons, which generally show a good correlation with their murine counterparts. This is a relevant finding that supports the use of mouse models to investigate some aspects of PCDH19 GCE. However, it is important to note that there are some differences as well, like the comparatively lower expression of PCDH19 in long-range projecting interneurons in humans (Inh L6 SST NPY in human, Sst_Chodl in mouse). The functional relevance of Pcdh19/PCDH19 expression in particular neuronal subtypes will need to be established experimentally, but our results provide a framework to support those functional studies in the future, not least because of regional differences in the expression of this gene within neuronal subtypes.
To date, no detailed quantitative characterization of cortical composition and lamination has been performed in the three existing Pcdh19 KO models (Pederick et al., 2016; Hayashi et al., 2017; Hoshina et al., 2021). We have quantified five excitatory and four inhibitory markers, looking at overall abundance, as well as distribution throughout the cortical plate in the somatosensory cortex. Our analysis, which was conducted separately in males and females, reveals no differences in the abundance of the different excitatory neuronal types analyzed, but points to small decreases in somatostatin-expressing interneurons in HET females and calretinin+ cells in KO males. We confirm the lack of major lamination defects (Pederick et al., 2016; Hayashi et al., 2017; Hoshina et al., 2021); however, our quantitative approach exposes more subtle changes in the distribution of certain neuronal types, indicating altered composition of specific layers or sublayers. Although some changes might represent false-positive findings, such as the ones for HET Pvalb bin 8 and KO CR bin 7, which might be explained by abnormal distributions that were apparent in the comparison between WT males and females, it is worth noting that changes between genotypes were more frequent and, in many cases, more significant than between WT animals of opposite sex. Indeed, we did not find a single difference between WT males and females at P10, suggesting that, although subtle, changes in layer composition cannot be ruled out in Pcdh19 mutants. Given the degree of neuronal diversity revealed by recent scRNAseq studies, our results also support the possibility of more widespread differences affecting other neuronal subtypes not covered by the antibodies used in our analysis. The origin of these differences is unknown, but one possibility is that they could arise as a consequence of altered neurogenesis, since PCDH19 has been shown to play a role in this process (Fujitani et al., 2017; Homan et al., 2018; Lv et al., 2019). It is also important to consider that we conducted our analysis mainly in the sSC, but, given that Pcdh19 expression varies between cortical regions, it is possible that different areas might be affected in different ways by a total or partial loss of PCDH19. Reports of focal cortical dysplasia and limbic abnormalities in EIEE9 patients (Kurian et al., 2018; Pederick et al., 2018; Lenge et al., 2020) and focal areas of disorganization in ASD patients (Stoner et al., 2014) seem to support this possibility.
Despite the involvement of other δ-protocadherins in the development of axonal tracts (Uemura et al., 2007; Piper et al., 2008; Biswas et al., 2014; Hayashi et al., 2014), our data do not support a major role of PCDH19 in this process. We did not detect any alterations in the main axonal tracts in the brain after staining for the axonal protein L1CAM, and a more detailed analysis of the corpus callosum also showed no differences in its dorsoventral extension or the dorsal restriction of Neuropilin-1-expressing axons. This is in agreement with the lack of defects found by Hayashi et al. (2017) in the projection of axons through this particular tract. More subtle defects in specific tracts would require much deeper analyses to be revealed, as the defects in cortical axonal arborization recently described in Pcdh19 HET animals (Mincheva-Tasheva et al., 2021).
Regardless of any anatomic alterations, investigating behavior allows a relevant functional assessment of the consequences of Pcdh19 loss. Our analysis differed from those in previous studies (Hayashi et al., 2017; Lim et al., 2019) in two main ways. First, in addition to adult animals, we also tested animals at a much younger age (preweaning, P21), as EIEE9 is a developmental disorder and therefore it is relevant to determine when any behavioral changes begin. Second, we added a second cohort of control animals: wild-type single-genotype housed mice, which have only been exposed to other WT animals during their life. An effect of WT littermates on the behavior of mutant animals was shown by Yang et al. (2011), when they demonstrated that raising less sociable BTBR T+tf/J mice with highly sociable C57BL6/J animals improved BTBR T+tf/J sociability. However, the impact of social environment on the behavior of WT littermates has only recently been demonstrated in a study with mice mutant for Nlgn3, an X-linked cell adhesion protein that has been implicated in ASD (Kalbassi et al., 2017). Therefore, this is further evidence to suggest that mutant mice can alter the behavior of their WT littermates and to support the addition of single-genotype housed WT controls.
In line with a previous mouse study (Hayashi et al., 2017) and with the findings in human patients, changes in behavior were more apparent in HET females than in their KO male siblings. Pcdh19 KO males only showed increased time spent in the open arms of the EPM, indicating reduced anxiety, when tested as adults. This same behavior was displayed by young Pcdh19 HET females (P21), which maintained it into adulthood. However, HET females also exhibited increased exploratory behavior, or maybe hypersensitivity to new environments, from a young age, as demonstrated by their consistently higher distance traveled during the first 5 min in the open field test at P21 and P60. It is important to consider that animals were placed into the open field arena four times in total, as they were tested on 2 consecutive days at both ages. Although habituation to the environment would be expected in this situation, the increased distance traveled during the first 5 min was apparent in all four trials, indicating a robust behavioral response. These results also suggest that behavioral changes in Pcdh19 heterozygous animals start early in life, validating them as a good model for a developmental condition.
Open field and EPM tests were also performed in the study by Hayashi et al. (2017). They found no differences in the EPM test, but this could be because of differences in experimental design or in the mouse model used for the test. Regarding the open field test, Hayashi et al. (2017) found no differences in total distance or time in the center when the test was conducted at 11–12 weeks of age. However, when they repeated the test 23 weeks later, Pcdh19 HET females spent significantly more time in the center of the open field arena, suggesting reduced anxiety. Although our animals did not display such behavior, they were tested at approximately P60, which would be in agreement with the data from their first open field test. In addition, the results of our EPM test also indicate reduced anxiety in our animals, which could therefore represent a behavioral characteristic of Pcdh19 mutant animals. Because no specific analysis of the first 5 min was conducted in that study, it is difficult to assess whether their animals exhibited increased activity during that period. Nevertheless, the fact that WT females display the same behavioral phenotype as their HET siblings indicates an effect of the social environment that can only be detected through the inclusion of single-genotype housed WT animals. Interestingly, this effect was also present in young males, with WTMGH animals traveling a higher distance in the first 5 min of the open field test than KO or WTSGH males. However, unlike in the female population, this behavior disappeared in adulthood. Because adult male and female animals are housed separately, it is tempting to speculate that this effect of the social environment is somehow mediated by the HET females, although other causes, like a maternal effect, cannot be ruled out based on our experiments.
One of the comorbidities of EIEE9 patients is ASD (Kolc et al., 2020), and changes in PCDH19 have also been linked to ASD cases (Piton et al., 2011; van Harssel et al., 2013). Indeed, a recent behavioral study with the Taconic Biosciences Pcdh19 KO mouse model has revealed social interaction deficits in the three-chamber test in KO males and HET females, as well as increased repetitive behavior in males (females were not tested; Lim et al., 2019). In our analysis, we also found differences in social behavior, but, interestingly, only in WTMGH animals. Both males and females spent less time interacting with a stranger female at P21 and P60, respectively, than with WTSGH animals, in what appears to be another example of the effect of the environment on mouse behavior. Since males were not tested at P60, because at that age it becomes a measure of courtship behavior rather than simple social interaction and as such is not comparable to the P21 behavior, we do not know whether this phenotype would be maintained into adulthood or whether, similar to the results of the open field test, it would revert to normal with age. The fact that HET and KO animals did not differ in their behavior from their WT littermates is in contradiction with the results from the study by Lim et al. (2019), although different tests were conducted in the two studies, making a direct comparison difficult. In summary, our behavioral characterization of the Pcdh19 Taconic Biosciences mouse model reveals a stronger effect of Pcdh19 mutation in HET females than in KO males and a significant effect of the social environment on the behavior of WT littermates, as previously described for Nlgn3 mutant animals (Kalbassi et al., 2017). This is a relevant finding, and this effect should be taken into consideration for the design of future behavioral experiments, as the failure to do so could result in the misinterpretation of data and missed behavioral phenotypes. It is important to note that, despite the subtle differences found in cortical composition in the sSC, we believe that a correlation between those changes and the observed behavioral alterations cannot be made at this point. Different cortical and brain regions are involved in the control of the behavioral paradigms that we have analyzed, so isolated cellular results of one cortical area, however widespread they might be, cannot be linked to any specific aspects of behavior. Such a correlation would require functional assays of neuronal function to go beyond mere speculation.
Finally, an important question is why the mutation of Pcdh19 in mice leads to much milder defects than in humans, with the absence of seizures as the most striking difference. It is worth noting that similar results have been described for other neurodevelopmental disorders that present with epilepsy, such as CDKL5 deficiency disorder or fragile X syndrome (FXS). Mice carrying either a null allele for Cdkl5 or the disease-causing mutation R59X do not display behavioral seizures, but they exhibit network hyperexcitability that manifests as decreased threshold to pharmacologically induced seizures (Wang et al., 2012; Amendola et al., 2014). In the case of FXS, in which epilepsy develops in ∼20% of patients (Musumeci et al., 1999; Sabaratnam et al., 2001), none of the KO mouse models presents spontaneous seizures. However, they are susceptible to audiogenic seizures and display alterations in cortical EEG frequency (Musumeci et al., 2000; Lovelace et al., 2018; Goswami et al., 2019). Similarly, cortical network activity is altered in Pcdh19 heterozygous animals (Pederick et al., 2018), indicating that mutations in those genes in mice do alter cortical connectivity, but not enough to trigger seizures. The smaller size and reduced complexity of the mouse brain probably account, at least partially, for these discrepancies, maybe by conferring a generally lower susceptibility to seizures in mice. Therefore, considering recent progress in the use of brain organoids for the study of neuronal connectivity (Quadrato et al., 2017), this emerging model might be needed in the future to dissect the effects of PCDH19 mutations on human connectivity.
Acknowledgments
Acknowledgements: We thank all members of the Martinez-Garay laboratory, as well as Y. Barde for insightful comments on this manuscript.
Synthesis
Reviewing Editor: Alfonso Represa, INSERM
Decisions are customarily a result of the Reviewing Editor and the peer reviewers coming together and discussing their recommendations until a consensus is reached. When revisions are invited, a fact-based synthesis statement explaining their decision and outlining what is needed to prepare a revision will be listed below. The following reviewer(s) agreed to reveal their identity: Shinji Hirano.
Dear Authors,
With reviewer we appreciate the changes that were performed in the manuscript that solved some of the concerns evoked in the previous review. It was evoked there that 'histological” and “behavioral” data, were not necessarily correlated but we understand that little can be done to clarify this issue. However, we agree that cellular data will be of interest for readers in the field and that behavioral data, particularly the differences related to housing conditions, will be important for a wider audience.
The reorganization of data focusing mainly now on somatosensory cortex, contributes to clarify the first part of the manuscript.
In response to point 4, gender data provided indicates some differences. Though the phenotype is subtle to make clear conclusions, we think they highlight the interest for taking gender issues into consideration.
Concerning point 5, we understand the technical difficulties in the analysis of arborization of the cortical neurons and withdraw our previous comment on this.
References
- Amendola E, Zhan Y, Mattucci C, Castroflorio E, Calcagno E, Fuchs C, Lonetti G, Silingardi D, Vyssotski AL, Farley D, Ciani E, Pizzorusso T, Giustetto M, Gross CT (2014) Mapping pathological phenotypes in a mouse model of CDKL5 disorder. PLoS One 9:e91613. 10.1371/journal.pone.0091613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassani S, Cwetsch AW, Gerosa L, Serratto GM, Folci A, Hall IF, Mazzanti M, Cancedda L, Passafaro M (2018) The female epilepsy protein PCDH19 is a new GABAAR binding partner that regulates GABAergic transmission as well as migration and morphological maturation of hippocampal neurons. Hum Mol Genet 27:1027–1038. 10.1093/hmg/ddy019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S, Emond MR, Duy PQ, Hao LT, Beattie CE, Jontes JD (2014) Protocadherin-18b interacts with Nap1 to control motor axon growth and arborization in zebrafish. Mol Biol Cell 25:633–642. 10.1091/mbc.E13-08-0475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caligioni CS (2009) Assessing reproductive status/stages in mice. Curr Protoc Neurosci Appendix 4:Appendix4I–Appendix4A.4I.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Brinkmann K, Chen Z, Pak CW, Liao Y, Shi S, Henry L, Grishin NV, Bogdan S, Rosen MK (2014) The WAVE regulatory complex links diverse receptors to the actin cytoskeleton. Cell 156:195–207. 10.1016/j.cell.2013.11.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper SR, Emond MR, Duy PQ, Liebau BG, Wolman MA, Jontes JD (2015) Protocadherins control the modular assembly of neuronal columns in the zebrafish optic tectum. J Cell Biol 211:807–814. 10.1083/jcb.201507108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depienne C, Leguern E (2012) PCDH19‐related infantile epileptic encephalopathy: an unusual X‐linked inheritance disorder. Hum Mutat 33:627–634. 10.1002/humu.22029 [DOI] [PubMed] [Google Scholar]
- Depienne C, Bouteiller D, Keren B, Cheuret E, Poirier K, Trouillard O, Benyahia B, Quelin C, Carpentier W, Julia S, Afenjar A, Gautier A, Rivier F, Meyer S, Berquin P, Hélias M, Py I, Rivera S, Bahi-Buisson N, Gourfinkel-An I, et al. (2009) Sporadic infantile epileptic encephalopathy caused by mutations in PCDH19 resembles Dravet syndrome but mainly affects females. PLoS Genet 5:e1000381. 10.1371/journal.pgen.1000381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibbens LM, Tarpey PS, Hynes K, Bayly MA, Scheffer IE, Smith R, Bomar J, Sutton E, Vandeleur L, Shoubridge C, Edkins S, Turner SJ, Stevens C, O'Meara S, Tofts C, Barthorpe S, Buck G, Cole J, Halliday K, Jones D, et al. (2008) X-linked protocadherin 19 mutations cause female-limited epilepsy and cognitive impairment. Nat Genet 40:776–781. 10.1038/ng.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duszyc K, Terczynska I, Hoffman-Zacharska D (2015) Epilepsy and mental retardation restricted to females: x-linked epileptic infantile encephalopathy of unusual inheritance. J Appl Genet 56:49–56. 10.1007/s13353-014-0243-8 [DOI] [PubMed] [Google Scholar]
- Emond MR, Biswas S, Jontes JD (2009) Protocadherin-19 is essential for early steps in brain morphogenesis. Dev Biol 334:72–83. 10.1016/j.ydbio.2009.07.008 [DOI] [PubMed] [Google Scholar]
- Fujitani M, Zhang S, Fujiki R, Fujihara Y, Yamashita T (2017) A chromosome 16p13.11 microduplication causes hyperactivity through dysregulation of miR-484/protocadherin-19 signaling. Mol Psychiatry 22:364–374. 10.1038/mp.2016.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaitan Y, Bouchard M (2006) Expression of the δ-protocadherin gene Pcdh19 in the developing mouse embryo. Gene Expr Patterns 6:893–899. 10.1016/j.modgep.2006.03.001 [DOI] [PubMed] [Google Scholar]
- Gerosa L, Francolini M, Bassani S, Passafaro M (2019) The Role of protocadherin 19 (PCDH19) in neurodevelopment and in the pathophysiology of early infantile epileptic encephalopathy‐9 (EIEE9). Dev Neurobiol 79:75–84. 10.1002/dneu.22654 [DOI] [PubMed] [Google Scholar]
- Gecz J, Thomas PQ (2020) Disentangling the paradox of the PCDH19 clustering epilepsy, a disorder of cellular mosaics. Curr Opin Genet Dev 65:169–175. 10.1016/j.gde.2020.06.012 [DOI] [PubMed] [Google Scholar]
- Goswami S, Cavalier S, Sridhar V, Huber KM, Gibson JR (2019) Local cortical circuit correlates of altered EEG in the mouse model of fragile X syndrome. Neurobiol Dis 124:563–572. 10.1016/j.nbd.2019.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, Inoue Y, Kiyonari H, Abe T, Misaki K, Moriguchi H, Tanaka Y, Takeichi M (2014) Protocadherin-17 mediates collective axon extension by recruiting actin regulator complexes to interaxonal contacts. Dev Cell 30:673–687. 10.1016/j.devcel.2014.07.015 [DOI] [PubMed] [Google Scholar]
- Hayashi S, Inoue Y, Hattori S, Kaneko M, Shioi G, Miyakawa T, Takeichi M (2017) Loss of X-linked Protocadherin-19 differentially affects the behavior of heterozygous female and hemizygous male mice. Sci Rep 7:5801. 10.1038/s41598-017-06374-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel N, Redies C (2011) Absence of layer-specific cadherin expression profiles in the neocortex of the reeler mutant mouse. Cereb Cortex 21:1105–1117. 10.1093/cercor/bhq183 [DOI] [PubMed] [Google Scholar]
- Hodge RD, Bakken TE, Miller JA, Smith KA, Barkan ER, Graybuck LT, Close JL, Long B, Johansen N, Penn O, Yao Z, Eggermont J, Höllt T, Levi BP, Shehata SI, Aevermann B, Beller A, Bertagnolli D, Brouner K, Casper T, et al. (2019) Conserved cell types with divergent features in human versus mouse cortex. Nature 573:61–68. 10.1038/s41586-019-1506-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homan CC, Pederson S, To T-H, Tan C, Piltz S, Corbett MA, Wolvetang E, Thomas PQ, Jolly LA, Gecz J (2018) PCDH19 regulation of neural progenitor cell differentiation suggests asynchrony of neurogenesis as a mechanism contributing to PCDH19 girls clustering epilepsy. Neurobiol Dis 116:106–119. 10.1016/j.nbd.2018.05.004 [DOI] [PubMed] [Google Scholar]
- Hoshina N, Johnson-Venkatesh EM, Hoshina M, Umemori H (2021) Female-specific synaptic dysfunction and cognitive impairment in a mouse model of PCDH19 disorder. Science 372:eaaz3893. 10.1126/science.aaz3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalbassi S, Bachmann SO, Cross E, Roberton VH, Baudouin SJ (2017) Male and female mice lacking neuroligin-3 modify the behavior of their wild-type littermates. eNeuro 4:ENEURO.0145-17.2017. 10.1523/ENEURO.0145-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolc KL, Sadleir LG, Scheffer IE, Ivancevic A, Roberts R, Pham DH, Gecz J (2019) A systematic review and meta-analysis of 271 PCDH19-variant individuals identifies psychiatric comorbidities, and association of seizure onset and disease severity. Mol Psychiatry 24:241–251. 10.1038/s41380-018-0066-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolc KL, Sadleir LG, Depienne C, Marini C, Scheffer IE, Møller RS, Trivisano M, Specchio N, Pham D, Kumar R, Roberts R, Gecz J (2020) A standardized patient-centered characterization of the phenotypic spectrum of PCDH19 girls clustering epilepsy. Transl Psychiatry 10:127. 10.1038/s41398-020-0803-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurian M, Korff CM, Ranza E, Bernasconi A, Lübbig A, Nangia S, Ramelli GP, Wohlrab G, Nordli DR, Bast T (2018) Focal cortical malformations in children with early infantile epilepsy and PCDH19 mutations: case report. Dev Med Child Neurol 60:100–105. 10.1111/dmcn.13595 [DOI] [PubMed] [Google Scholar]
- Lenge M, Marini C, Canale E, Napolitano A, De Masi S, Trivisano M, Mei D, Longo D, Espagnet R, Lucenteforte MC, Barba C, Specchio N, Guerrini R (2020) Quantitative MRI-based analysis identifies developmental limbic abnormalities in PCDH19 encephalopathy. Cereb Cortex 12:526. [DOI] [PubMed] [Google Scholar]
- Lim J, Ryu J, Kang S, Noh HJ, Kim CH (2019) Autism-like behaviors in male mice with a Pcdh19 deletion. Mol Brain 12:95. 10.1186/s13041-019-0519-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovelace JW, Ethell IM, Binder DK, Razak KA (2018) Translation-relevant EEG phenotypes in a mouse model of fragile X syndrome. Neurobiol Dis 115:39–48. 10.1016/j.nbd.2018.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv X, Ren S-Q, Zhang X-J, Shen Z, Ghosh T, Xianyu A, Gao P, Li Z, Lin S, Yu Y, Zhang Q, Groszer M, Shi S-H (2019) TBR2 coordinates neurogenesis expansion and precise microcircuit organization via Protocadherin 19 in the mammalian cortex. Nat Commun 10:3946. 10.1038/s41467-019-11854-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mincheva-Tasheva S, Guil AFN, Homan CC, Gecz J, Thomas PQ (2021) Disrupted excitatory synaptic contacts and altered neuronal network activity underpins the neurological phenotype in PCDH19-clustering epilepsy (PCDH19-CE). Mol Neurobiol 58:2005–2018. 10.1007/s12035-020-02242-4 [DOI] [PubMed] [Google Scholar]
- Musumeci SA, Hagerman RJ, Ferri R, Bosco P, Bernardina BD, Tassinari CA, De Sarro GB, Elia M (1999) Epilepsy and EEG findings in males with fragile X syndrome. Epilepsia 40:1092–1099. 10.1111/j.1528-1157.1999.tb00824.x [DOI] [PubMed] [Google Scholar]
- Musumeci SA, Bosco P, Calabrese G, Bakker C, De Sarro GB, Elia M, Ferri R, Oostra BA (2000) Audiogenic seizures susceptibility in transgenic mice with fragile X syndrome. Epilepsia 41:19–23. 10.1111/j.1528-1157.2000.tb01499.x [DOI] [PubMed] [Google Scholar]
- Paul A, Crow M, Raudales R, He M, Gillis J, Huang ZJ (2017) Transcriptional architecture of synaptic communication delineates GABAergic neuron identity. Cell 171:522–539.e20. 10.1016/j.cell.2017.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederick DT, Homan CC, Jaehne EJ, Piltz SG, Haines BP, Baune BT, Jolly LA, Hughes JN, Gecz J, Thomas PQ (2016) Pcdh19 loss-of-function increases neuronal migration in vitro but is dispensable for brain development in mice. Sci Rep 6:26765. 10.1038/srep26765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederick DT, Richards KL, Piltz SG, Kumar R, Mincheva-Tasheva S, Mandelstam SA, Dale RC, Scheffer IE, Gecz J, Petrou S, Hughes JN, Thomas PQ (2018) Abnormal cell sorting underlies the unique X-linked inheritance of PCDH19 epilepsy. Neuron 97:59–66.e5. 10.1016/j.neuron.2017.12.005 [DOI] [PubMed] [Google Scholar]
- Pham DH, Tan CC, Homan CC, Kolc KL, Corbett MA, McAninch D, Fox AH, Thomas PQ, Kumar R, Gécz J (2017) Protocadherin 19 (PCDH19) interacts with paraspeckle protein NONO to co-regulate gene expression with estrogen receptor alpha (ERα). Hum Mol Genet 26:2042–2052. 10.1093/hmg/ddx094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper M, Dwivedy A, Leung L, Bradley RS, Holt CE (2008) NF-protocadherin and TAF1 regulate retinal axon initiation and elongation in vivo. J Neurosci 28:100–105. 10.1523/JNEUROSCI.4490-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piton A, Gauthier J, Hamdan FF, Lafrenière RG, Yang Y, Henrion E, Laurent S, Noreau A, Thibodeau P, Karemera L, Spiegelman D, Kuku F, Duguay J, Destroismaisons L, Jolivet P, Côté M, Lachapelle K, Diallo O, Raymond A, Marineau C, et al. (2011) Systematic resequencing of X-chromosome synaptic genes in autism spectrum disorder and schizophrenia. Mol Psychiatry 16:867–880. 10.1038/mp.2010.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrato G, Nguyen T, Macosko EZ, Sherwood JL, Min Yang S, Berger DR, Maria N, Scholvin J, Goldman M, Kinney JP, Boyden ES, Lichtman JW, Williams ZM, McCarroll SA, Arlotta P (2017) Cell diversity and network dynamics in photosensitive human brain organoids. Nature 545:48–53. 10.1038/nature22047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabaratnam M, Vroegop PG, Gangadharan SK (2001) Epilepsy and EEG findings in 18 males with fragile X syndrome. Seizure 10:60–63. 10.1053/seiz.2000.0492 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serratto GM, Pizzi E, Murru L, Mazzoleni S, Pelucchi S, Marcello E, Mazzanti M, Passafaro M, Bassani S (2020) The epilepsy-related protein PCDH19 regulates tonic inhibition, GABAAR kinetics, and the intrinsic excitability of hippocampal neurons. Mol Neurobiol 57:5336–5351. 10.1007/s12035-020-02099-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoner R, Chow ML, Boyle MP, Sunkin SM, Mouton PR, Roy S, Wynshaw-Boris A, Colamarino SA, Lein ES, Courchesne E (2014) Patches of disorganization in the neocortex of children with autism. N Engl J Med 370:1209–1219. 10.1056/NEJMoa1307491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C, Shard C, Ranieri E, Hynes K, Pham DH, Leach D, Buchanan G, Corbett M, Shoubridge C, Kumar R, Douglas E, Nguyen LS, Mcmahon J, Sadleir L, Specchio N, Marini C, Guerrini R, Moller RS, Depienne C, Haan E, et al. (2015) Mutations of protocadherin 19 in female epilepsy (PCDH19-FE) lead to allopregnanolone deficiency. Hum Mol Genet 24:5250–5259. 10.1093/hmg/ddv245 [DOI] [PubMed] [Google Scholar]
- Tasic B, Yao Z, Graybuck LT, Smith KA, Nguyen TN, Bertagnolli D, Goldy J, Garren E, Economo MN, Viswanathan S, Penn O, Bakken T, Menon V, Miller J, Fong O, Hirokawa KE, Lathia K, Rimorin C, Tieu M, Larsen R, et al. (2018) Shared and distinct transcriptomic cell types across neocortical areas. Nature 563:72–78. 10.1038/s41586-018-0654-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terracciano A, Trivisano M, Cusmai R, De Palma L, Fusco L, Compagnucci C, Bertini E, Vigevano F, Specchio N (2016) PCDH19-related epilepsy in two mosaic male patients. Epilepsia 57:e51–e55. 10.1111/epi.13295 [DOI] [PubMed] [Google Scholar]
- Trivisano M, Lucchi C, Rustichelli C, Terracciano A, Cusmai R, Ubertini GM, Giannone G, Bertini ES, Vigevano F, Gécz J, Biagini G, Specchio N (2017) Reduced steroidogenesis in patients with PCDH19‐female limited epilepsy. Epilepsia 58:e91–e95. 10.1111/epi.13772 [DOI] [PubMed] [Google Scholar]
- Uemura M, Nakao S, Suzuki ST, Takeichi M, Hirano S (2007) OL-protocadherin is essential for growth of striatal axons and thalamocortical projections. Nat Neurosci 10:1151–1159. 10.1038/nn1960 [DOI] [PubMed] [Google Scholar]
- van Harssel JJT, Weckhuysen S, van Kempen MJA, Hardies K, Verbeek NE, de Kovel CGF, Gunning WB, van Daalen E, de Jonge MV, Jansen AC, Vermeulen RJ, Arts WFM, Verhelst H, Fogarasi A, de Rijk-van Andel JF, Kelemen A, Lindhout D, De Jonghe P, Koeleman BPC, Suls A, et al. (2013) Clinical and genetic aspects of PCDH19-related epilepsy syndromes and the possible role of PCDH19 mutations in males with autism spectrum disorders. Neurogenetics 14:23–34. 10.1007/s10048-013-0353-1 [DOI] [PubMed] [Google Scholar]
- Wang I-TJ, Allen M, Goffin D, Zhu X, Fairless AH, Brodkin ES, Siegel SJ, Marsh ED, Blendy JA, Zhou Z (2012) Loss of CDKL5 disrupts kinome profile and event-related potentials leading to autistic-like phenotypes in mice. Proc Natl Acad Sci U S A 109:21516–21521. 10.1073/pnas.1216988110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams EO, Sickles HM, Dooley AL, Palumbos S, Bisogni AJ, Lin DM (2011) Delta protocadherin 10 is regulated by activity in the mouse main olfactory system. Front Neural Circuits 5:9. 10.3389/fncir.2011.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolverton T, Lalande M (2001) Identification and characterization of three members of a novel subclass of protocadherins. Genomics 76:66–72. 10.1006/geno.2001.6592 [DOI] [PubMed] [Google Scholar]
- Yang M, Perry K, Weber MD, Katz AM, Crawley JN (2011) Social peers rescue autism‐relevant sociability deficits in adolescent mice. Autism Res 4:17–27. 10.1002/aur.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, van Velthoven CTJ, Nguyen TN, Goldy J, Sedeno-Cortes AE, Baftizadeh F, Bertagnolli D, Casper T, Chiang M, Crichton K, Ding S-L, Fong O, Garren E, Glandon A, Gouwens NW, Gray J, Graybuck LT, Hawrylycz MG, Hirschstein D, Matthew K, et al. (2020) A taxonomy of transcriptomic cell types across the isocortex and hippocampal formation. Cell 184:3222-3241.e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene expression and distribution of Pcdh19 in cortical excitatory projection neurons of the Allen Brain Atlas Mouse Whole Cortex & Hippocampus - SMART-SEQ (2019) with 10x Smart-Seq Taxonomy (2020), represented by violin plots. The first row shows the overall expression of Pcdh19 in the combined dataset excluding hippocampal regions for simplicity. Subsequent rows show expression by cortical region. Dots indicate the median value of the population. Absence of a violin plot in a row indicates that none or fewer than three cells from that particular cortical region were mapped to the corresponding neuronal cluster. Black and red lines indicate consistent low and high expression of Pcdh19 across areas, respectively; asterisks highlight clusters with marked variation in Pcdh19 expression across cortical regions. ACA, Anterior cingulate area; AI, agranular insular area; AUD, auditory areas; GU, gustatory areas; MOp, primary motor area; MOs-FRP, secondary motor area-frontal pole, cerebral cortex; ORB, orbital area; PL-ILA, prelimbic - infralimbic areas; PTLp, posterior parietal association areas; RSP, retrosplenial area; TEa-PERI-ECT, temporal association areas-perirhinal area-ectorhinal area; VIS, visual areas; VISp, primary visual area. Download Figure 1-1, TIF file (12.1MB, tif) .
Gene expression and distribution of Pcdh19 in cortical inhibitory neurons of the Allen Brain Atlas Mouse Whole Cortex & Hippocampus - SMART-SEQ (2019) with 10x Smart-Seq Taxonomy (2020), represented by violin plots. The first row shows the overall expression of Pcdh19 in the combined dataset excluding hippocampal regions, for simplicity. Subsequent rows show expression by cortical region. Dots indicate the median value of the population. Absence of a violin plot in a row indicates that none or fewer than three cells from that particular cortical region were mapped to the corresponding neuronal cluster. Black and red lines indicate consistent low and high expression of Pcdh19 across areas, respectively; asterisks highlight clusters with marked variation in Pcdh19 expression across cortical regions. ACA, Anterior cingulate area; AI, agranular insular area; AUD, auditory areas; GU, gustatory areas; MOp, primary motor area; MOs-FRP, secondary motor area-frontal pole, cerebral cortex; ORB, orbital area; PL-ILA, prelimbic-infralimbic areas; PTLp, posterior parietal association areas; RSP, retrosplenial area; TEa-PERI-ECT, temporal association areas-perirhinal area-ectorhinal area; VIS, visual areas; VISp, primary visual area. Download Figure 1-2, TIF file (13.1MB, tif) .
Gene expression and distribution of PCDH19 in cortical excitatory projection neurons of the Allen Brain Atlas Human Multiple Cortical Areas dataset, represented by violin plots. The first row shows overall expression of PCDH19 in the combined dataset; subsequent rows show expression by brain region. Dots indicate the median value of the population. Absence of a violin plot in a row indicates that none or fewer than three cells from that particular brain region were mapped to the corresponding neuronal subtype. Black and red lines indicate consistent low and high expression of PCDH19 across areas, respectively; asterisks highlight clusters with marked variation in PCDH19 expression across cortical regions. CgG, Anterior cingulate gyrus; M1lm, primary motor cortex, lower limb region; M1ul primary motor cortex, upper limb region, A1C, primary auditory cortex. Download Figure 2-1, TIF file (11.2MB, tif) .
Gene expression and distribution of PCDH19 in cortical inhibitory neurons of the Allen Brain Atlas Human Multiple Cortical Areas dataset, represented by violin plots. The first row shows the overall expression of PCDH19 in the combined dataset; subsequent rows show expression by brain region. Dots indicate the median value of the population. Absence of a violin plot in a row indicates that none or fewer than three cells from that particular brain region were mapped to the corresponding neuronal subtype. Black and red lines indicate consistent low and high expression of PCDH19 across areas, respectively; asterisks highlight clusters with marked variation in PCDH19 expression across cortical regions. CgG, Anterior cingulate gyrus; M1lm, primary motor cortex, lower limb region; M1ul primary motor cortex, upper limb region, A1C, primary auditory cortex. Download Figure 2-2, TIF file (12MB, tif) .
A, Diagram indicating the existing correlations between ABA mouse and human cortical datasets. B, Diagram showing the homology clusters defined by Hodge et al. (2019) and the corresponding mouse and human neuronal subtypes assigned to each cluster for GABAergic neuronal clusters. C, River plot showing the mapping of the nuclei from the MTG dataset to the subtypes defined by the Multiple Cortical Areas dataset, for inhibitory neurons. Download Figure 2-3, TIF file (16.5MB, tif) .
A, Total distance travelled in the open field test by P60 males. B, Distance travelled in the open field test by P21 males, split into 5 min intervals. The data for the first 5 min are shown in Figure 6. C, Distance travelled in the open field test by P60 males, split into 5 min intervals. D, Time spent by males in the center of the arena during the 20 min open field test at P21 and P60. E, Time spent in the open arms of the elevated plus maze by P21 males. F, Number of beam breaks during the 24 h activity test for the males of the different conditions. Light, light phase; dark, dark phase. G, Number of beam breaks by hour in the 24 h activity test for the males of the different conditions. The time of the day is shown on the x-axis, and the gray square indicates the hours of the dark period. The numbers of tested animals were as follows: at P21: WTSGH, 26; WTMGH, 18; KO, 31; at P60: WTSGH, 24; WTMGH, 18; KO, 29. For the 24 h activity test, the numbers were as follows: WTSGH, 17; WTMGH, 10; KO, 10. Results are indicated as the mean ± SEM. *p < 0.05. Download Figure 6-1, TIF file (5.5MB, tif) .
A, B, Distance travelled in the last two 5 min intervals of the open field test by P21 (A) and P60 (B) females. C, Number of beam breaks during the 24 h activity test for the females of the different conditions. Light, Light phase; dark, dark phase. D, Number of beam breaks by hour in the 24 h activity test for the females of the different conditions. The time of the day is shown on the x-axis, and the gray square indicates the hours of the dark period. E, F, Time spent by females in the center of the arena during the 20 min open field test at P21 (E) and P60 (F). In these two panels, raw data are depicted in the top axis, with the mean (gap) and SD (vertical bars) to their right. Group and group sizes are indicated at the bottom. Note that each group appears twice in every graph, but in two different colors. The mean difference for each comparison is plotted in the lower axis as a bootstrap sampling distribution. The mean is indicated by the black dot, and the 95% CI by the vertical bars. The numbers of tested animals were as follows: at P21 and P60: WTSGH, 22; WTMGH, 29; HET, 21. For the 24 h activity test, numbers were as follows: WTSGH, 18; WTMGH, 11; HET, 10. Results are indicated as the mean ± SEM. *p < 0.05. Download Figure 7-1, TIF file (18.5MB, tif) .