Abstract
Obesity is a major public health burden worldwide and is characterized by chronic low-grade inflammation driven by the cooperation of the innate immune system and dysregulated metabolism in adipose tissue and other metabolic organs. Receptor-interacting serine/threonine-protein kinase 1 (RIPK1) is a central regulator of inflammatory cell function that coordinates inflammation, apoptosis and necroptosis in response to inflammatory stimuli. Here we show that genetic polymorphisms near the human RIPK1 locus associate with increased RIPK1 gene expression and obesity. We show that one of these single nucleotide polymorphisms is within a binding site for E4BP4 and increases RIPK1 promoter activity and RIPK1 gene expression in adipose tissue. Therapeutic silencing of RIPK1 in vivo in a mouse model of diet-induced obesity dramatically reduces fat mass, total body weight and improves insulin sensitivity, while simultaneously reducing macrophage and promoting invariant natural killer T cell accumulation in adipose tissue. These findings demonstrate that RIPK1 is genetically associated with obesity, and reducing RIPK1 expression is a potential therapeutic approach to target obesity and related diseases.
Obesity is characterized by the interplay of dysregulated energy balance and non-resolving inflammation. Subcutaneous and visceral adipose tissue depots expand over time by the differentiation of stromal stem cells to mature adipocytes and the esterification of free fatty acids to glycerol for storage in lipid droplets. As these adipose depots expand, the synergistic conversion of tissue-resident macrophages to an M1-like phenotype and expression of cytokines and adipokines by adipocytes promote a proinflammatory state1,2. This metabolically triggered systemic inflammation, particularly in the visceral adipose depots, increases the risk of developing other diseases, including atherosclerosis, type 2 diabetes and cancer. Accordingly, large clinical trials are underway to target innate immune signals to treat type 2 diabetes and reduce cardiovascular disease.
Despite the strong environmental drivers that lead to excess fat accumulation, twin studies clearly demonstrate the heritable nature of body weight3. Both monogenic and polygenic drivers of obesity are commonly associated with the leptin/melanocortin pathways, which act as a conduit between the central nervous system and peripheral adipose and muscle tissue. Large genome-wide association studies (GWAS) have revealed common variants in the FTO locus, which contains the coding region for the α-ketoglutarate-dependent dioxygenase enzyme, that associate with the risk of obesity4–7. Individuals harbouring the minor/risk allele of this locus have increased caloric intake8, and mouse studies suggest that this distal enhancer regulates mitochondrial metabolism in adipose tissue independently of the expression of the main FTO gene product α-ketoglutarate-dependent dioxygenase9. Similarly, variants near MC4R (encoding the melanocortin-4 receptor) associate with early-onset obesity due to defects in the appetite-control pathway and signals of satiety in the brain10,11. We have previously examined single nucleotide polymorphisms (SNPs) across the human genome and revealed that SNPs in inflammatory genes account for 28% of the heritability of obesity12. However, to date, there have been no specific inflammatory genes implicated in the genetic risk of developing obesity in humans.
We have previously investigated the RIP kinase family in vascular disease in mice and humans and found that therapeutic inhibition of RIPK1 activity in a chronic mouse model of atherosclerosis reduces lesion complexity and stabilizes vulnerable plaques13. RIPK1 serves as a central node of inflammatory signalling and can orchestrate activation of the transcription factor NFκB and cell survival or promote cell death via programmed apoptosis or necroptosis14. The role of RIPK1 in driving inflammation in inflammatory states such as psoriasis, arthritis and infection are known15; however, whether RIPK1 also promotes inflammation and/or cell death during metabolic disease has never been tested. We show herein that genetic variants in the RIPK1 locus associate with higher expression levels of RIPK1 in human adipose tissue, and we identify E4BP4 as a new transcriptional regulator of RIPK1 expression. Accordingly, blockade of Ripk1 in mice fed a high-fat diet (HFD) reduces adiposity, adipose tissue inflammation and improves insulin resistance. Together, these data place RIPK1 at the nexus of metabolic inflammation and indicate that targeting of RIPK1 may be an effective way to combat inflammation in the setting of obesity and metabolic syndrome.
Results
SNPs in human RIPK1 associate with obesity.
We set out to test the hypothesis that elevated RIPK1, which is known to drive other cardiometabolic diseases15, including atherosclerosis13, may also play a role in driving obesity. To begin, we examined the RIPK1 locus for genetic variants and their relationship to obesity in humans. In the Finnish Metabolic Syndrome In Men (METSIM) cohort of over 700 individuals, we found strong expression quantitative trait loci (eQTL) for RIPK1 mRNA in subcutaneous adipose tissue (P = 1.2 × 10−25; Fig. 1a and Extended Data Fig. 1a)16,17. These RIPK1 eQTL were also found in the Genotype-Tissue Expression (GTEx) database, where individuals carrying the minor allele of the SNP (7–10% of the population) have increased RIPK1 expression in subcutaneous and visceral adipose tissue (P value from 10−5 to 10−14, Extended Data Fig. 1b–d)18. In a cohort of individuals with extreme obesity19, carriers of the minor alleles for SNPs in RIPK1 in linkage disequilibrium (LD) had a 1.6–1.8-fold risk of being obese compared to individuals with the protective alleles (Extended Data Fig. 1e). To further probe the causal relationship between RIPK1 eQTL and obesity, we performed Mendelian randomization (MR) to estimate the casual association between RIPK1 expression and obesity by using genotypic variants at the RIPK1 locus (with an r2 < 0.2). Using the GSMR algorithm, which compares the effect sizes of the SNPs on the exposure (RIPK1 expression) with the outcome (obesity), we estimated the causal effect of elevated RIPK1 expression on obesity using generalized least-squares regression20–23. Using this MR approach and its associated assumptions, we found evidence supporting an association between RIPK1 expression and obesity across multiple independent cohorts and were able to use genetic variation to estimate the likely causal effect (Fig. 1b,c, Table 1 and Extended Data Fig. 1f–i). These estimates suggest that RIPK1 expression that is one standard deviation (s.d.) above the mean (β = 0.27) causally increases obesity (odds ratio = 1.3; Table 1). Together, these data suggest that participants who are genetically susceptible to higher expression of RIPK1 have a higher risk of obesity.
Fig. 1 |. Association of human SNPs with RIPK1 in obesity.
a, eQTL analysis of SNP rs2064310 in the RIPK1 locus and levels of RIPK1 mRNA in adipose tissue from the METSIM cohort. b,c, MR analysis of SNPs that are independently (r2 < 0.2) and significantly (P < 10−4) associated with expression of RIPK1 and obesity. The beta and standard error (s.e.) values of the effect sizes of RIPK1 expression (eQTL; x axis) and the effect sizes on obesity (y axis) are plotted. Data in b were derived from Riveros-McKay et al.77 and GTEx18 and data in c were derived from Westra et al.23 and the obese versus lean (OBLE) cohort19. d, RIPK1 mRNA expression in adipose tissue from humans categorized by a BMI of less than or greater than 25, and the correlation of relative RIPK1 mRNA expression and BMI. Data represent the mean ± s.e.m. and were analysed using a two-tailed Student’s t-test; *P < 0.05 (P = 0.025) and linear regression (n = 4 participants, BMI < 25; n = 6 participants, BMI > 25). e, Ripk1 mRNA expression in adipose tissue in mice from the HMDP, and the correlation with fat mass measured by NMR and visceral fat mass in male mice.
Table 1 |.
MR demonstrating a positive association between RIPK1 expression and obesity risk by using data from GWAS and eQTL sources
| RIPK1 eQTL study (exposure) | Obesity study (outcome) | Tissue | Beta (MR) | s.e. (MR) | P value (MR) | Number of SNPs (MR) |
|---|---|---|---|---|---|---|
| METSIM | OBLE19 | Adipose (subcutaneous) | 0.26941 | 0.13983 | 0.054018 | 3 |
| GTEx18 | Riveros-McKay et al.77 | Adipose (visceral omentum) | 0.666114 | 0.282809 | 0.0185633 | 3 |
| GTEx18 | OBLE19 | Adipose (subcutaneous) | 0.742815 | 0.242662 | 0.0022052 | 8 |
| METSIM | Riveros-McKay et al.77 | Adipose (subcutaneous) | 0.163246 | 0.065646 | 0.0128912 | 9 |
| GTEx18 | UK-Biobank (impedance of whole body) | Adipose (subcutaneous) | −0.00985636 | 0.00457773 | 0.0313101 | 23 |
| GTEx18 | UK-Biobank (impedance of whole body) | Adipose (subcutaneous) | −0.841754 | 0.398739 | 0.0347689 | 23 |
| Lloyd-Jones et al.78 | OBLE19 | Blood | 0.293619 | 0.133475 | 0.027821 | 29 |
| Westra et al.23 | Riveros-McKay et al.77 | Blood | 0.349417 | 0.14985 | 0.019712 | 6 |
| Westra et al.23 | OBLE19 | Blood | 1.41146 | 0.350033 | 5.522 × 10−5 | 5 |
| Lloyd-Jones et al.78 | UK Biobank (impedance of whole body) | Blood | −0.00624607 | 0.00313365 | 0.0462366 | 18 |
| Lloyd-Jones et al.78 | UK Biobank (impedance of whole body) | Blood | −0.551498 | 0.273545 | 0.0437882 | 18 |
To better understand the expression of RIPK1 in adipose tissue, we analysed single-nuclei sequencing data from human adipose tissue. We found that RIPK1 was expressed broadly across cell types, with the most abundant expression in adipocyte, endothelial and perivascular clusters but also across clusters of immune cells (dendritic, macrophage and T cells; Extended Data Fig. 2a). In a cohort of participants categorized as overweight (body mass index (BMI) > 25), RIPK1 mRNA in adipose tissue was significantly elevated compared to levels in normal-weight participants (BMI < 25; P < 0.001; Fig. 1d) and correlated positively with BMI (R2 = 0.70; P < 0.01). We further examined the expression of Ripk1 mRNA in the Hybrid Mouse Diversity Panel (HMDP), which contains transcriptome data from approximately 100 inbred strains of mice24. The expression of Ripk1 in adipose tissue from HFD-fed mice correlated strongly with total fat mass assessed by nuclear magnetic resonance (NMR) and visceral fat mass in both male and female mice (Fig. 1e and Extended Data Fig. 2b). Moreover, adipose Ripk1 expression significantly correlated with other metabolic traits including circulating insulin levels and HOMA-IR, a marker of insulin resistance, in HFD-fed male mice (Extended Data Fig. 2b). Together, these data demonstrate that obesity-associated polymorphisms in the RIPK1 gene functionally result in elevated RIPK1 expression in adipose tissue in humans, and elevated Ripk1 expression in mice strongly associates with adiposity and metabolic dysfunction.
The RIPK1 polymorphism rs5873855 disrupts a transcriptional repressor binding site.
We postulated that one or more of the SNPs within the RIPK1 locus might disrupt a gene regulatory element(s), which subsequently alters the transcription of RIPK1. We cloned regions spanning the tightly linked SNPs in the RIPK1 locus that are linked to RIPK1 expression and tested whether the major or minor allele differed in their ability to activate the RIPK1 promoter upstream of a luciferase reporter. The minor allele for rs5873855 increased RIPK1 promoter activation compared to the major/protective allele (Fig. 2a). None of the other SNPs showed differences in promoter activation between the major or minor alleles (Extended Data Fig. 2c). We next queried HaploReg25 for a potential functional impact of rs5873855 on the RIPK1 locus, and found that this SNP located within a binding motif for a well-characterized transcription factor, E4 promoter-binding protein 4 (E4BP4, also known as NFIL3)26. E4BP4 is a repressive transcription factor that has been shown to control invariant natural killer T (iNKT) cell function27 and is induced during M1 and M2 macrophage polarization28. The deletion at rs5873855 in the common allele is predicted to allow E4BP4 binding and repression, whereas the obesity-associated TA insertion is rare and is predicted to disrupt the E4BP4 binding site to derepress RIPK1 transcription and expression (Fig. 2b). Our data from these promoter assays are consistent with the prediction that this variant disrupts E4BP4 binding and derepresses RIPK1 promoter activation. We next assessed whether endogenous E4BP4 is capable of binding the intronic sequence containing the obesity-associated SNPs. Chromatin immunoprecipitation assays in human SW872 cells confirmed that E4BP4 binding was enriched at the RIPK1 intronic region containing obesity-associated SNPs, similarly to what has been shown with binding to other E4BP4 target genes, such as peroxisome proliferator-activated receptor gamma (PPARG; Fig. 2c)29. In adipose tissue from patients who are either homozygous (T/T) or heterozygous (T/TA) for the rs5873855 major allele, we observed an enrichment of E4BP4 binding to this locus in homozygous patients, as compared with IgG controls, which was not observed in heterozygous patients (Fig. 2d). Further, silencing of E4BP4 with small interfering RNA (siRNA) led to the activation of RIPK1 mRNA expression under tumour-necrosis factor-α (TNF-α)-stimulated conditions (Fig. 2e). Together, these data demonstrate the role of E4BP4 in the regulation of RIPK1 expression.
Fig. 2 |. Relationship between E4BP4 and RIPK1 in adipose tissue and regulation by the polymorphism rs5873855.
a, Human SW872 cells were transfected with a plasmid containing the rs5873855 sequence (− or TA) and the human RIPK1 promoter upstream of the firefly luciferase gene and a plasmid encoding human E4BP4. Luciferase activity was measured 24 h after transfection. Data are shown as the mean ± s.e.m. of n = 4 independent experiments. Significance was assessed using a two-tailed Student’s t-test; *P < 0.05 (P = 0.003). b, Schematic model depicting how a candidate SNP regulates E4BP4 to affect RIPK1 expression. c, Chromatin immunoprecipitation assay using antibodies against E4BP4 (or IgG as control) showing enrichment of the RIPK1 locus in SW872 cells. PPARG, a known E4BP4 target gene, serves as a positive control. Data are shown as the mean ± s.d. of n = 3 independent experiments performed in technical duplicate and analysed using two-way ANOVA and Sidak’s multiple-comparison test; **P ≤ 0.01 (P = 0.003) and ***P ≤ 0.001 (P = 0.0009) versus IgG control. d, Chromatin immunoprecipitation assay as described in c but using omental adipose tissue biopsies from patients with obesity either homozygous (T/T; n = 5) or heterozygous (T/TA; n = 2) for the reference allele. Data are shown as the mean ± s.d. of independent experiments performed in technical duplicate or triplicate and analysed using a one-way ANOVA with Tukey’s multiple-comparison test; *P < 0.05 (P = 0.0435 versus IgG). e, siRNA knockdown of E4BP4 in SW872 liposarcoma cells and levels of RIPK1 mRNA expression in untreated cells or cells treated with TNF-α (20 ng ml−1) for 24 h. Data are shown as the mean ± s.d. of n = 3 independent experiments performed in technical duplicate and analysed using two-way ANOVA with Holm–Sidak’s multiple-comparison test; *P < 0.05 (P = 0.009). f, Analysis of E4bp4 and Ripk1 mRNA in NKT cells from adipose tissue or the spleen of mice. Expression is depicted as a ratio of gene expression in spleen:adipose. E4bp4 levels are high and Ripk1 levels are low in adipose NKT cells, whereas spleen NKTs have low expression of E4bp4 and high expression of Ripk1. Data were analysed from the dataset GSE63358. g, mRNA expression data from WAT of mice stimulated with vehicle or αGalCer. Data were analysed from the dataset GSE36032 and expressed as log fold change versus vehicle control (n = 3 vehicle, n = 4 αGalCer; adjusted P value corrected for multiple comparisons). h,i, The percentage of iNKT cells expressing RIPK1 and E4BP4 in omental adipose tissue (h) or liver (i) from patients with obesity (n = 9). iNKT cells were identified as a CD19−CD3+ αGalCer-loaded CD1d tetramer+ population based on negative control (isotype IgG) staining. The gating strategy for flow cytometry analyses is available in Supplementary Fig. 1. Significance was assessed using a paired Student’s t-test; ***P ≤ 0.001.
E4BP4 has an important role in regulating iNKT cell function in adipose tissue. iNKT cells in the adipose tissue protect against diet-induced obesity30, and E4BP4 is required for the production of interleukin (IL)-10 (ref. 27) to drive M2 macrophage polarization31. Given that E4BP4 serves as a transcriptional repressor of RIPK1, we postulated that the levels of RIPK1 and E4BP4 might be inversely correlated in adipose tissue iNKT cells. Previous studies in mice profiling the gene expression in iNKT cells from adipose tissue and other depots (spleen and liver) enabled us to examine the relationship between Ripk1 and E4bp4 expression using publicly available transcriptome data. E4bp4 expression was higher in adipose tissue iNKT cells compared with iNKT cells from the spleen, whereas Ripk1 expression was low in adipose tissue and high in splenic iNKT cells (Fig. 2f; from dataset GSE63358). Analysis of immune cell clusters by single-cell RNA sequencing (scRNA-seq) over the course of HFD feeding in mice, when dramatic shifts in the immune landscape of the adipose tissue occur and iNKT cells are lost, revealed decreased E4bp4 expression in the NK/NKT cluster at the point where Ripk1 expression peaks (Extended Data Fig. 2d,e; from dataset GSE128518 (ref. 32)). We next analysed available data from the adipose tissue of obese mice treated with or without αGalCer, a lipid antigen specific for the activation of iNKT cells31. This activation increases E4bp4 expression and concomitantly reduces Ripk1 expression (Fig. 2g; from dataset GSE36032; wild-type (WT) vehicle versus WTαGalCer). To understand whether this inverse relationship is consistent in humans, we analysed iNKT cells from adipose and liver tissue from patients with obesity and found that iNKT cells in the adipose tissue show an inverse relationship between RIPK1 and E4BP4 levels, where the number RIPK1+ cells and RIPK1 expression is high, and the number of E4BP4+ cells and E4BP4 expression is low (Fig. 2h and Extended Data Fig. 3a). This reciprocal relationship was not found in liver iNKT cells from the same individuals (Fig. 2i and Extended Data Fig. 3a). Consistently, when we examined RIPK1 expression across CD45+ immune cells in human adipose by scRNA-seq, we found that, while widely expressed, clusters with higher RIPK1+ cells tended to have fewer E4BP4+ cells (Extended Data Fig. 3b). In contrast, NKT cell clusters in human liver showed fewer E4BP4+ and RIPK1+ cells overall (Extended Data Fig. 4)33. This tissue difference is also reflected by the differences in IL-10 expression in the liver compared to the adipose tissue: in liver, IL-10+ cells are largely myeloid derived, whereas in the adipose tissue, IL-10 expression is also found in NKT cells. These observations are in keeping with the pro- versus anti-inflammatory roles of NKT cells in liver and adipose tissue, respectively30,34. Together, these data suggest that RIPK1 and E4BP4 are inversely regulated in adipose tissue NKT cells, and that this relationship may be unique to adipose tissue.
Inhibition of Ripk1 reduces diet-induced obesity and improves insulin resistance in mice.
To determine whether RIPK1 expression drives obesity, we tested whether knockdown of Ripk1 by using antisense oligonucleotides (ASOs) could rescue the adverse metabolic consequences associated with diet-induced obesity. Male C57BL/6J mice were fed a HFD (60% kcal) for 24 weeks and received weekly injections of control ASOs or one of two ASOs targeting Ripk1 (ASO-A or ASO-B). Ripk1 ASO therapy reduced whole body weight over the course of HFD feeding (Fig. 3a). In mice fed a normal chow diet, Ripk1 inhibition also limited weight gain, but differences in body weight were observed only after 16 weeks of treatment (Extended Data Fig. 5a). In mice fed a HFD, the difference in body weight upon Ripk1 inhibition was the result of a substantial reduction in total fat mass, with no effects on lean body mass, as assessed by NMR (15.3 ± 3 g in control versus 5.1 ± 1 g or 7.0 ± 4 g in Ripk1 ASO-A or ASO-B groups, respectively; P < 0.001; Fig. 3b), which was particularly evident in the perigonadal fat pads of these mice (Fig. 3c). We confirmed Ripk1 ASO delivery to the adipose tissue by immunostaining (Fig. 3d) where, accordingly, RIPK1 protein and mRNA expression were markedly reduced (Fig. 3e and Extended Data Fig. 5b). Ripk1 ASO treatment also improved glucose homeostasis, including fasted blood glucose (Fig. 3f), plasma insulin (Fig. 3g) and response to both glucose and insulin challenge (fasting glucose tolerance test (GTT), Fig. 3h; or insulin tolerance test (ITT), Fig. 3i). To further examine whole-body metabolic function, we performed open-circuit indirect calorimetry and found little difference in the overall metabolic rates of these mice. Mice treated with Ripk1 ASO-A had a slight decrease in VO2 and a corresponding small increase in respiratory exchange ratio compared to those in both controls and Ripk1 ASO-B, consistent with improved carbohydrate metabolism in these mice (Extended Data Fig. 5c,d). Despite the reduction in body fat, there was no significant difference in physical activity (Extended Data Fig. 5e) or food consumption (Extended Data Fig. 5f) in mice treated with either of the Ripk1 ASOs compared to controls. There was a slight increase in serum alanine aminotransferase (ALT) in mice receiving Ripk1 ASO-A compared to other groups (Extended Data Fig. 5g), although these levels were still within the normal range for obese mice35. We next ascertained the role of RIPK1 kinase activity on the obesogenic function of RIPK1. We evaluated the response to HFD feeding in mice with a mutation of the RIPK1 catalytic lysine K45 (RIPK1K45A)36 as compared to WT littermates. After 20 weeks on a HFD, RIPK1K45A mice gained equivalent weight to their WT counterparts and had similar fat mass in the perigonadal white adipose tissue (WAT; Extended Data Fig. 6a,b). When challenged with a bolus of either glucose or insulin, mice with defective RIPK1 K45 activity responded in a similar manner to their WT littermates (Extended Data Fig. 6c,d). Together, these data demonstrate that silencing Ripk1 reduces body weight and fat mass, with no effects on lean mass and is accompanied by improved glucose tolerance and insulin sensitivity, but this is independent of the kinase activity of RIPK1.
Fig. 3 |. Therapeutic knockdown of Ripk1 in a diet-induced obesity mouse model.
a–i, Male WT C57BL/6J mice (n = 8 per group) were fed a HFD for 24 weeks with simultaneous administration of 50 mg kg−1 control or Ripk1-A or Ripk1-B ASOs weekly. a, Total body weights. b, Fat, lean and total body mass measured using NMR (EchoMRI). c, Liver and epididymal adipose tissue weights at termination of study. d, Immunohistochemistry staining for the 2′F methoxyethyl (MOE) moiety on the ASO (left), RIPK1 (right) in epididymal WAT after 24 weeks of treatment. Negative control staining is shown in Supplementary Fig. 5. Scale bar: 0.02 mm. e, Ripk1 mRNA expression in subcutaneous WAT, epididymal WAT and liver in mice treated with control and Ripk1 ASO after 24 weeks as determined using NanoString (n = 3 replicates of n = 8 pooled mice; *P < 0.05, two-tailed Student’s t-test). f,g, Fasting blood glucose (f) and fasting blood insulin (g) after 18 weeks of treatment. Data are representative of the mean ± s.e.m. of n = 8 mice per group and were analysed using one-way ANOVA and Dunnet’s multiple-comparison test; *P < 0.05 and **P < 0.01. h,i, Fasting GTT (h) or insulin tolerance test (ITT; i) after 14 or 18 weeks of treatment; AUC, area under the curve. Data are representative of the mean ± s.e.m. of n = 8 mice per group and were analysed using two-way ANOVA and Tukey’s multiple-comparison test; *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001; NS, not significant.
Blocking Ripk1 reduces adipose tissue inflammation.
RIPK1 is an activator of multiple inflammatory pathways, including nuclear factor kappa B (NFκB). To better understand the inflammatory status of mice upon Ripk1 knockdown, we examined inflammatory gene activation in liver and adipose tissue of Ripk1 ASO-treated mice. We observed a substantial reduction of key inflammatory genes known to be downstream of RIPK1 (Fig. 4a). Importantly, there was a dramatic decrease in lipid staining as detected by Oil Red O staining in the Ripk1 ASO-treated mice (Fig. 4b), demonstrating that Ripk1 ASO therapy reduces overall liver inflammation and lipid accumulation—both of which are known to contribute to obesity and associated complications. Strikingly, we observed a decrease in both adipocyte size (Fig. 4c) and macrophage accumulation (Fig. 4d) in Ripk1 ASO-treated mice. RIPK1 governs activation of NFκB and elicits a proinflammatory response; therefore, we tested downstream activation of inflammatory genes in Ripk1 ASO-treated mice. Expression analysis of epididymal adipose tissue revealed a downregulation of proinflammatory genes in the adipose tissue of Ripk1 ASO- versus control ASO-treated mice, including Cd68 and Il1a (Fig. 4e), whereas genes associated with M2 macrophage polarization, Fizz1, Stat3 and Il10, were elevated when Ripk1 was knocked down (Fig. 4f). Of note, there were no detectable differences in circulating cytokines in Ripk1 ASO-treated mice compared to controls, indicating that the changes in inflammatory gene activation were localized to the adipose tissue and did not represent a global defect in inflammatory signalling (Extended Data Fig. 7). These data indicate that Ripk1 inhibition reduces activation of inflammatory pathways known to be downstream of RIPK1 in the liver and adipose tissue of obese mice.
Fig. 4 |. Knockdown of Ripk1 decreases inflammation in liver and adipose tissue.
a, Hepatic mRNA expression of inflammatory genes. Bar graphs depict mean ± s.e.m. of an average of n = 8 samples per group. Significance was assessed using a two-tailed Student’s t-test; *P < 0.05 (P = 0.01), **P < 0.01 (P = 0.005) and ****P < 0.0001 (P = 0.00008). b, Liver Oil Red O staining for neutral lipids. c, Adipocyte area in control ASO- and Ripk1 ASO-treated mice analysed from H&E stained sections. Bar graphs depict the mean ± s.e.m. of an average of five sections per mouse (n = 8 mice per group). *P < 0.05 (P = 0.009); two-tailed Student’s t-test. d, Images are representative of Mac2 staining of epididymal WAT (left) and quantification of Mac2+ area (right). Bar graphs depict the mean ± s.e.m. of an average of five sections per mouse (n = 8 mice per group). *P < 0.05 (P = 0.024); two-tailed Student’s t-test. Scale bar: 0.02 mm. e,f, Gene expression in epididymal WAT for proinflammatory genes Cd68 and Il1a (e) and M2 macrophage markers Fizz1, Stat3 and Il10 (f) analysed by quantitative PCR (qPCR; Cd68, Il1a and Il10 normalized to housekeeping genes) and NanoString (Fizz1 and Stat3 normalized to housekeeping genes and expressed as counts). Bar graphs depict the mean ± s.e.m. of an average of three samples per group of pooled RNA from n = 8 mice per group. *P < 0.05, **P < 0.01 and ****P < 0.0001; two-tailed Student’s t-test. All samples were analysed after 24 weeks of treatment.
Ripk1 inhibition augments iNKT cells in obese mice.
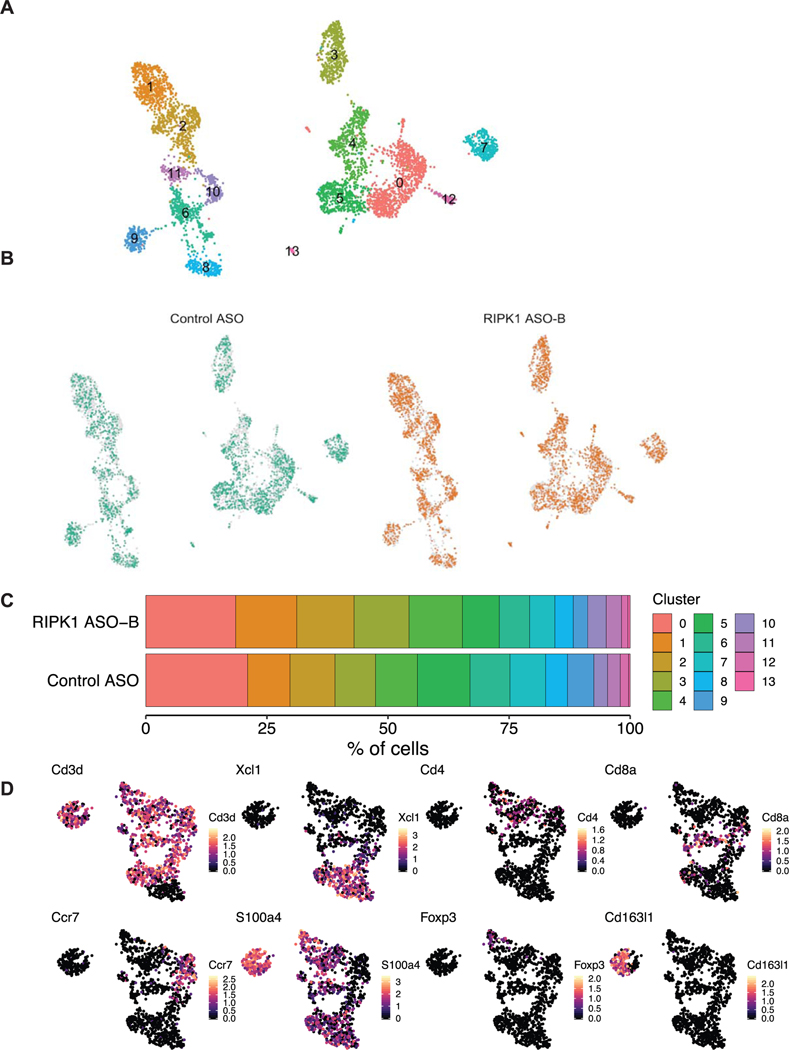
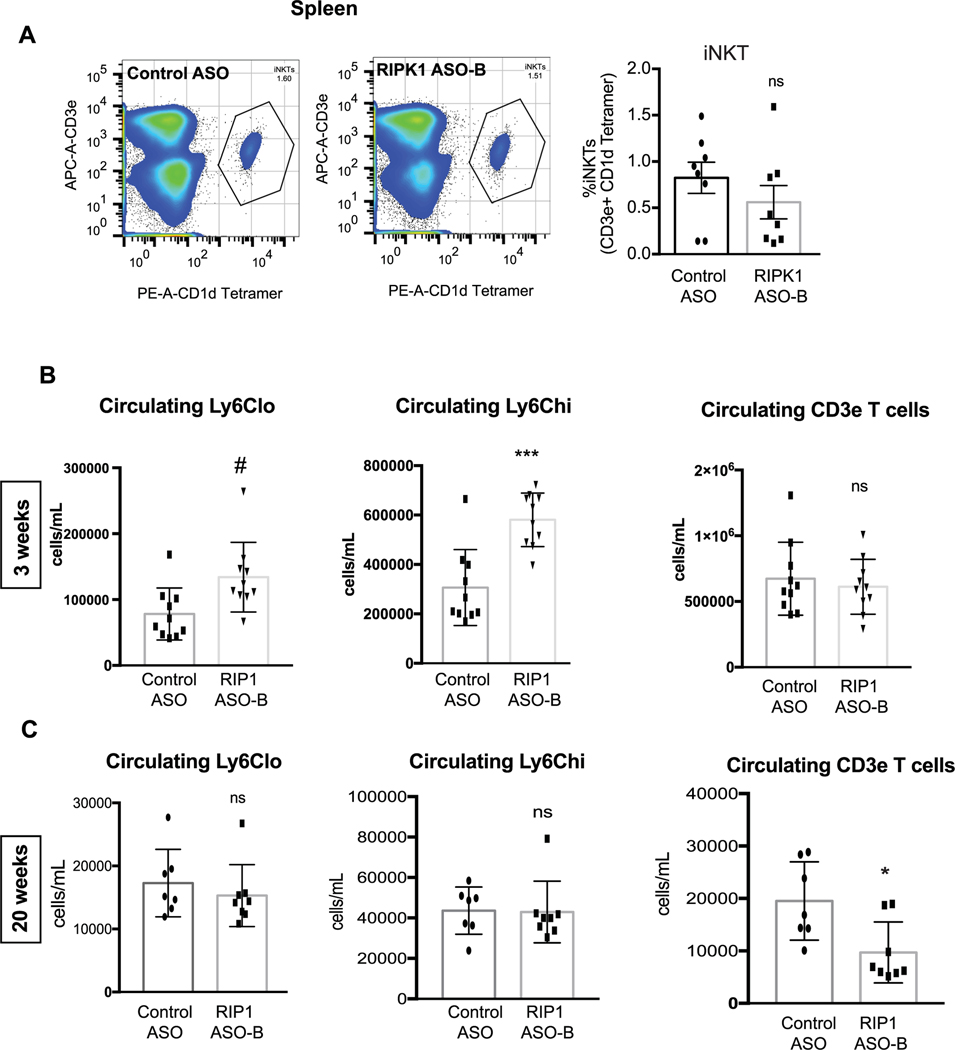
Given that E4BP4 represses Ripk1 expression, and we observed a strong inverse correlation between RIPK1 and E4BP4 levels in iNKT cells, we hypothesized that Ripk1 inhibition might affect iNKT cells in adipose tissue. To begin, we used scRNA-seq to broadly characterize immune cell populations in the adipose tissue upon short-term HFD feeding and Ripk1 knockdown. According to UMAP cluster analysis, there was an increase in a cluster identified as putative NKT cells (based on the expression marker Xcl1; ref. 37) in Ripk1 ASO-treated mice compared to controls (Fig. 5a,b and Extended Data Fig. 8). Using an αGalCer-loaded phycoerythrin (PE)-conjugated CD1d tetramer to allow flow cytometric detection of iNKT cells, we found an increase in adipose tissue iNKT cells after short-term HFD feeding following Ripk1 silencing compared to that in controls (allophycocyanin (APC)-conjugated CD3e+CD1d tetramer+; Fig. 5c,d). These differences were unique to the adipose tissue, as the proportion of splenic iNKT cells was not different between groups (Extended Data Fig. 9a). The increased proportion of iNKT cells in Ripk1 ASO-treated mice persisted after long-term HFD feeding (Fig. 5e). There were no changes in circulating T cells in mice treated with Ripk1 ASOs compared to control ASOs after 3 weeks and, in fact, circulating CD3+ lymphoid cells decreased after 22 weeks of HFD feeding (Extended Data Fig. 9b,c), suggesting that these effects on iNKT cells were not a result of a global increase in T cell populations. Because iNKT cells are a source of IL-10, we tested whether this increase in the proportion of iNKT cells in the adipose tissue was accompanied by an increase in IL-10 expression in these cells and, indeed, there were more IL-10+ iNKT cells in the Ripk1 ASO-treated mice (Fig. 5f). Other T cell clusters were either unchanged or decreased upon Ripk1 inhibition, as compared with controls, including Foxp3+ (regulatory T cells) and CD163l1+ (gamma delta T cells; Fig. 5b and Extended Data Figs. 8 and 10). These data suggest that Ripk1 inhibition may protect against diet-induced obesity by maintaining the anti-obesogenic population of iNKT cells and the expression of IL-10 uniquely within the adipose tissue.
Fig. 5 |. Adipose tissue invariant NKT cells are increased in mice treated with Ripk1 ASO.
a,b, scRNA-seq analysis of CD45+ cells from the adipose tissue of mice treated with control ASO or Ripk1 ASO-B for 5 weeks, including 3 weeks of HFD feeding. a, Clustered UMAP embedding of scRNA-seq data from T cells of mouse adipose tissue. The specific subsets of T cells were assigned to clusters based on the top gene expression markers of each cluster (Extended Data Fig. 8). b, Relative frequency of each type in control ASO and Ripk1 ASO-B samples. c–e, Flow cytometry analysis of the isolated stromal vascular fraction of adipose tissue from mice treated with control ASO or Ripk1 ASO-B for 5 weeks, including 3 weeks of HFD feeding. c, Flow cytometry plots are representative of adipose NKT cells identified as αGalCer-loaded CD1d tetramer+CD3e+ (outlined area). d, The percentage of NKT (expressed as percentage of F4/80− cells that were αGalCer-loaded CD1d tetramer+CD3e+) in adipose tissue from mice treated for 5 weeks, including 3 weeks of HFD feeding, with control or Ripk1 ASO (n = 8 per group). Significance was assessed using a two-tailed Student’s t-test; *P < 0.05 (P = 0.044). e, NKT percentage of T cells from adipose tissue of control or Ripk1 ASO-treated mice after 22 weeks of HFD feeding. *P < 0.05 P = 0.03); two-tailed Student’s t-test. f, The number of iNKT cells expressing IL-10 was measured as in b (n = 7–8 per group, pooled for flow cytometry analysis). *P < 0.05 (P = 0.04); two-tailed Student’s t-test. The gating strategy for flow cytometry analyses is available in Supplementary Figs 2–4.
Discussion
Inflammation is implicated in the pathophysiology of obesity, but whether this represents a cause or a consequence of disease remains unclear. Herein, we identify a genetic link between RIPK1 and obesity in humans. RIPK1 is a central regulator of immune signalling that receives receptor–ligand signals, particularly via TNF-α, to coordinate the activation of NFκB and cell survival15. We analysed human genetic variants and found a causal association between higher expression of RIPK1 and risk of obesity. In individuals with obesity, the expression level of RIPK1 is elevated in adipose tissue, and silencing Ripk1 in mice dramatically reduces adiposity and promotes improved metabolic function. Our work suggests that RIPK1 directly induces activation of proinflammatory signalling in adipose tissue, which promotes accumulation of macrophages and drives metabolic inflammation.
While an environment of excess caloric intake and decreased energy expenditure promotes adipose tissue expansion, there remains a strong genetic component in obesity. Thus far, GWAS have identified over 300 significant (P < 10−8) loci for adiposity related traits, the majority of which implicate genes involved in appetite control (for example, LEP (encoding leptin) and MCR4) and resting energy expenditure (for example, FTO)38. By this approach, few variants within inflammatory pathways are found to associate with obesity risk at the genome-wide level. Others have used annotation of genome-wide significant variants with tissue-specific regulatory elements and found enrichment of B-cell-specific signatures, further suggesting a role for immune cells in the genetic risk of obesity39. We previously used a holistic approach considering all SNPs across the genome and found that SNPs within inflammatory genes explain 28% of heritability of BMI12. While this analysis suggests that inflammation is linked to the genetic risk of obesity, the effect sizes of individual inflammatory gene SNPs are small and, as such, few reach genome-wide significance. Moreover, given that inflammation is a downstream consequence of obesity, it is important to determine whether these loci contribute to obesity directly or indirectly by using other methodologies. Under these circumstances, alternative methods such as MR can be used to investigate potential functional and causal gene–disease relationships. MR analysis is a useful approach for the detection of the collective contribution of SNPs that have small effect sizes on a given outcome. To date, the studies that have used MR to investigate inflammation and obesity have considered only a few SNPs in a small number of inflammatory genes and, as such, suffer heavily from weak instrument bias40. In contrast, we performed MR analysis using multiple genome-wide studies of obesity and eQTL datasets to test specifically whether RIPK1 was causally associated with obesity, as SNPs near the RIPK1 locus do not appear in any GWAS of obesity. Using this approach, we found that participants who were genetically susceptible to higher expression of RIPK1 tended to have higher risk of obesity. The data in HFD-fed mice further support a causal role for RIPK1 in obesity, in which blocking RIPK1 resulted in significant protection from weight gain and glucose intolerance. There is the possibility that variants within the RIPK1 locus are linked to obesity by other mechanisms independent of inflammation (for example, due to RIPK1’s role as a scaffolding protein or in regulating cell death). Additionally, other loci that are identified as ‘inflammatory’ that contribute to the heritability of BMI may also drive adiposity by non-inflammatory mechanisms. Furthermore, although we used robust analytical methods to remove pleiotropy across SNPs, no single method can account for all of the potential pleiotropy, and thus it is possible that RIPK1 SNPs could influence other traits. While we accounted for reverse causation in our MR methodology, one cannot discount the possibility that RIPK1 or other inflammatory loci act downstream of obesity. A more in-depth analysis of RIPK1 functions during obesity in humans will help to resolve these unknowns. Regardless of its mechanism of action, our study demonstrates that hyperactivation of RIPK1 is a heritable and therapeutically targetable contributor to obesity and its associated complications.
The regulatory mechanisms governing RIPK1 transcriptional activation are relatively unknown. We identified the promoter of human RIPK1 located approximately 5 kb upstream of the previously annotated transcriptional start site and a putative regulatory element in the RIPK1 gene that harbours SNPs associated with increased expression of RIPK1. One particular variant, rs5873855, with a frequency of 0.07, contains the insertion TTA in the minor allele in a region predicted to be bound by the transcription factor E4BP4. We found E4BP4 bound to this site, and the presence of the TTA insertion enhanced the activation of the RIPK1 promoter by E4BP4, suggesting E4BP4 binds to and represses this region. Although rs5873855 represented an interesting RIPK1 functional candidate, we cannot rule out effects of the other linked SNPs as regulators of RIPK1 expression, in particular across different cell types that may be less abundant in adipose tissue. Other SNPs may also affect the activation of E4BP4 or may alter RIPK1 expression by another mechanism. However, the majority of the other obesity-associated RIPK1 SNPs did not alter the promoter activity of RIPK1. In addition to adipose tissue, RIPK1 SNPs are eQTL for RIPK1 expression in other tissues, including the brain, thyroid and whole blood, suggesting that alterations in RIPK1 gene expression may drive inflammation or other processes and pathways in other tissues. SNPs in the RIPK1 locus are also associated with behavioural phenotypes such as preschool-age internalizing problems and depressive symptoms. Whether these represent the known proinflammatory roles for RIPK1 in other tissues or whether these other traits associated with RIPK1 also contribute to obesity remains to be investigated. Additionally, a small number of variants in the RIPK1 locus are also eQTL for other transcripts, for example, RP11–506K6.4, a noncoding antisense transcript in an intron of the SCL22A23 gene, and TUBB2B (tubulin beta 2B class IIb). Although these genes have not been investigated in obesity, they could also play a role in the adipose tissue environment.
RIPK1 is a member of a family of kinases that includes RIPK3, which have divergent functions in inflammation. RIPK1 is modified by its reciprocal ubiquitination and phosphorylation status41. Ubiquitinated RIPK1 promotes NFκB activation, whereas phosphorylated RIPK1 acts to activate RIPK3 to promote programmed necrosis14,42,43. Complete genetic ablation of Ripk1 results in early postnatal lethality, as RIPK1 appears to perform a scaffolding function during development44. When the kinase domain of RIPK1 is mutated, mice have reduced TNF-α-dependent necroptosis in vitro and in vivo, owing to the dependence of RIPK1 kinase activity in the execution of programmed necrosis36. We found that the kinase activity of RIPK1 (K45) is dispensable for RIPK1 in promoting obesity. Our data suggest that it is the NFκB activation by RIPK1 that mediates the proinflammatory and pro-obesogenic function in the adipose tissue; however, further mechanistic studies are needed to define this. This is further supported by contrasting the current study with genetic loss of Ripk3, which has been shown to exacerbate diet-induced obesity and insulin resistance45. This is attributed to the expansion of adipocytes in the absence of RIPK3-dependent adipocyte apoptosis and turnover. With antisense knockdown of Ripk1 in the present study, we see a decrease in adipocyte size and decreased macrophage accumulation in the adipose tissue of Ripk1 ASO-treated mice, arguing against excessive cell death in this model. Instead, silencing Ripk1 reduces proinflammatory NFκB gene expression specifically within the adipose tissue environment. These data suggest that the roles for RIPK1 and RIPK3 in controlling adipocyte cell death are distinct. Notably, loss of RIPK1 in intestinal epithelial cells and keratinocytes can promote both necroptosis and apoptosis, exacerbating inflammation and a loss of barrier function46. This was attributed to the dominant role of RIPK1 as a scaffold for pro-survival proteins to inhibit cell death in barrier tissues. In our mice treated with Ripk1 ASOs, we did not observe evidence of skin lesions or intestinal abnormalities, and nutrient absorption was similar in both control and RIPK1 groups (data not shown). These data suggest that while ASOs do indeed target epithelial layers like the small intestine, they may allow sufficient expression of RIPK1 for barrier tissue function to remain intact in these mice.
We found a role for iNKT cells in modulating the effects of RIPK1 in obesity. Upon activation, iNKT cells play a unique immunomodulatory function by production of either TH1-like or TH2-like cytokines47. In adipose tissue, resident iNKT cells recognize lipid antigens presented by the major histocompatibility complex (MHC) class I-like CD1d on adipocytes to promote IL-10, IL-13 and IL-4 production to maintain an anti-inflammatory state48,49, and the majority of studies show iNKT cells protect against obesity30,50,51. In humans, iNKT cells are enriched in adipose tissue but decline sharply during obesity30,52. The pro- or anti-inflammatory role of iNKT cells is highly tissue dependent, as shown by the comparison of gene expression signatures among splenic, liver and adipose iNKT cells in mice27,53 and humans34. Indeed, in mice we observed high expression of Ripk1 in splenic iNKT cells in comparison to levels found in adipose iNKT cells, in alignment with the proinflammatory nature of the spleen iNKT cell reservoir. In contrast, in adipose iNKT cells activated with the lipid antigen αGalCer, expression of E4bp4 increases while Ripk1 expression decreases, also in agreement with their anti-inflammatory state. In individuals with obesity, we found that RIPK1 is broadly expressed in immune and non-immune cells, and similarly found an inverse relationship between RIPK1 and E4BP4 expression in adipose iNKT cells, where RIPK1 expression is comparatively higher than E4BP4 in the proinflammatory obese environment. Interestingly, this relationship is not observed in iNKT cells in the liver. We also found that in human liver, IL-10+ cells are largely restricted to the monocyte cluster, whereas adipose tissue expresses IL10 in both monocytes and NKT cells34, further confirming a depot-specific iNKT cell gene signature. We observed a significant increase in iNKT cell number in the adipose tissue of mice treated with RIPK1 inhibitors compared to iNKT cell numbers in control mice, which was evident as early as 3 weeks after HFD feeding and persisted to the end of the study. Conversely, other T cell populations in the adipose tissue remained either unchanged or decreased upon RIPK1 inhibition. These data suggest that RIPK1 functions differently within immune cell subsets, potentially by directing downstream pathways (for example, apoptosis, necroptosis and inflammatory cytokines) in the adipose tissue. Together, our data indicate that elevated RIPK1 can drive adipose tissue inflammation via changes in iNKT cells and could explain the anti-inflammatory milieu of the Ripk1 ASO-treated adipose tissue.
Drug development increasingly relies on human genetics to prioritize therapeutic targets54,55, and drug targets supported by genetic evidence are more likely to be successful in the clinic56,57. These approaches have led to the identification of proteins whose expression drives disease and as such can be targeted to reduce protein expression or activity. The use of ASOs is one way to directly reduce protein expression and is increasingly used in the clinic. Examples of this gene-guided therapeutic strategy include the identification of driver mutations in APOC3 and PCSK9 in hyperlipidaemia and the subsequent development of the antisense drugs volanesorsen58,59 and inclisiran60, respectively. Our study uses similar antisense chemistry to block Ripk1 expression, which is used clinically for these and other chronic diseases, for example, mipomersen (antisense to APOB for hypercholesterolaemia61) and patisiran (antisense to TRR (encoding transthyretin) for amyloidosis62). Antisense therapy is emerging as a safe and effective approach to treat chronic diseases63,64, and this has the potential to enable rapid translation of RIPK1 antisense therapies into the clinic to treat obesity.
In conclusion, our study identifies RIPK1 as an important genetic driver of obesity in mice and humans. In humans, we reveal a causal association between higher expression of RIPK1 and risk of obesity. In mice, RIPK1 expression in adipose tissue correlates with adiposity and measures of glucose intolerance and, as such, RIPK1 inhibition dramatically reduces body weight and adipose tissue accumulation, and improves insulin sensitivity, suggesting that elevated RIPK1 might be a central driver of obesity. Because therapeutically blocking RIPK1 in mice prevents the activation of local macrophage inflammatory pathways, and this improves the local inflammatory milieu to dampen metabolic dysfunction associated with a HFD, this may be valuable for the treatment of the expanding epidemic of obesity and its associated comorbidities.
Methods
Mendelian randomization.
To investigate whether the expression of RIPK1 drives obesity, we performed forward MR analysis. Namely, we obtained summary association statistics (beta and s.e. values) for SNPs that were independently (r2 < 0.2) associated with a RIPK1 expression probe (P < 10−4) and used these as an instrument to investigate a causal effect. This meant that, for SNPs in the instrument, we also obtained their summary association statistics (beta and s.e. values) with obesity and compared the effect sizes of the SNPs on RIPK1 expression (exposure) with the effect sizes of the SNPs on obesity (outcome) to estimate the causal effect of RIPK1 expression on obesity. In this context, a significant positive association indicates that individuals who are genetically susceptible have higher probe values (RIPK1 expression) and are at greater risk of obesity. MR analysis was performed using the GSMR algorithm implemented in GCTA software (version 1.92)22. As compared to other methods for two-sample MR analysis, GSMR automatically detects and removes SNPs that have pleiotropic effects on both exposure and outcome using the HEIDI test; in addition, GSMR accounts for the sampling variance in beta estimates and the LD among SNPs, and as such, it is statistically more powerful than other MR approaches22.
The GSMR algorithm also requires access to individual-level genotype data (an LD reference panel) to estimate the LD between SNPs. The LD reference panel must match the exposure and outcome data with regard to ethnicity. Our exposure and outcome summary statistics were obtained from studies carried out in European individuals. As such, we used genotype data from the INTERHEART study, which is a sample of 854 participants of European ancestry, that we previously used in 1000 Genomes-based meta-analysis of CAD GWAS results65. We did not use the genotype data from 1000 Genomes because of the small number of European participants in 1000 Genomes with available genotype data (n = 364).
GSMR then takes the following steps to make a causal inference:
Finds SNPs that are significantly associated with the exposure.
Keeps only those SNPs that have summary statistics in both the exposure and outcome datasets.
Removes SNPs that have a difference of >0.2 between their allele frequency in the exposure/outcome datasets and the LD reference panel.
At each SNP, aligns summary statistics in both exposure and outcome datasets to the same reference allele at the LD reference panel and excludes SNPs that do not match.
Performs the HEIDI test (test of pleiotropy) at each SNP and excludes the pleiotropic SNPs (HEIDI P value < 0.01) from the instrument.
Keeps only SNPs that are in linkage equilibrium.
Contrasts the effect sizes of the SNPs on the exposure with the effect sizes of the SNPs on the outcome, and estimates the causal effect of the exposure on the outcome using generalized least-squares regression.
Finally, to exclude associations due to reverse causation (obesity → RIPK1 expression), we then carried out reverse-MR analysis, where we identified independent SNPs (r2 < 0.2) that are associated with obesity (P < 5 × 10−8) and contrasted their effect size on obesity with their effect size on a RIPK1 expression and excluded any obesity–probe associations that showed significant evidence of reverse causation (P < 0.05). In summary, the purpose of forward MR analysis was to find out whether RIPK1 expression drives obesity, whereas reverse MR investigates whether obesity drives RIPK1 expression.
eQTL analysis of METSIM.
RIPK1 eQTL in the METSIM cohort were calculated as described previously16. We applied the probabilistic estimation of expression residuals (PEER) method to infer and account for complex nongenetic factors affecting gene expression levels. This method is designed to detect the maximum number of cis eQTL. To optimize the discovery of trans eQTL within the same analysis, we performed PEER analysis by examining 10–50 inferred factors (Nk) at increments of five factors. We then used Matrix-eQTL to assess the genetic association with inverse normal-transformed PEER-processed residuals from robust multi-array average (RMA)-normalized expression data. We examined variants to determine the number of trans eQTL target genes by using various numbers of PEER factors. We selected Nk = 35 as a single analysis to maximize the number of target genes at this locus; this threshold captured 94.8% of cis eQTL identified with 50 PEER factors. For downstream eQTL mapping, we used the inverse normal-transformed PEER-processed residuals after accounting for 35 factors. Correlations were calculated with the biweight midcorrelation method (bicor) as implemented in the WGCNA R package.
Human tissue collection.
Human tissue was collected after informed consent was obtained from all participants and according to approved institutional review board (IRB) protocol at New York University Medical Center (for gene expression analysis), the University of Helsinki (for single-nuclei RNA sequencing) or approved research ethics board (REB) protocol by the Ottawa Health Science Network (for scRNA-seq, flow cytometry analysis and chromatin immunoprecipitation assay and OBLE study). For gene expression analyses, adipose tissue samples were homogenized in TRIzol reagent (Ambion, Life Technologies) and RNA was purified using the Direct-zol RNA MiniPrep kit (Zymo Research) as described previously66. Extracted mRNA was then reverse transcribed using the iScript cDNA synthesis kit (Bio-Rad). Real-time qPCR was performed with KAPA SYBR FAST qPCR kits (KAPA Biosystems) on a QuantStudio 3 (Applied Biosystems) in triplicates. Results were expressed as normalized fold change calculated by the comparative cycle method (2−ΔΔCt).
Primer sequences used were as follows:
28S: F TGGGAATGCAGCCCAAG, R CCTTACGGTACTTGTTGACTATCG
RIPK1: F AGACTGCTCGTCAAGTGTGG, R TTGAGCTGTAGCCTGA ACCTTA
RIPK1 promoter and dual-luciferase assays.
We first inserted the RIPK1 promoter sequence into the multiple cloning site of the pGL3-Basic vector (Promega) by digesting pGL3-Basic with MluI and HindIII. The RIPK1 genomic promoter sequence was PCR amplified using primers with MluI and HindIII restriction sites (Table 2). The amplified promoter sequence was ligated with digested pGL3-Basic using T4 DNA ligase (New England Biolabs). Next, a 309-bp intronic sequence containing the rs5873855 SNP was inserted downstream of the luciferase gene in pGL3-Basic using Gibson assembly (New England Biolabs). pGL3-Basic RIPK1 promoter was digested with the SalI to linearize the plasmid, run on an agarose gel and gel purified with the GeneAid GenepHlow kit (Frogga Biosciences). The rs5873855 genomic sequence and linearized pGL3-Basic RIPK1 promoter were assembled using the Gibson Assembly cloning kit (New England Biolabs).
Table 2 |.
List of primer sequences for cloning
| Region | Primer direction | Sequence (5′–3′) |
|---|---|---|
| RIPK1 promoter | Forward | GATTACGCGTTGCAGTTCCAACTTTGCTC |
| Reverse | GATTAAGCTTTGGCTTCTTCAAGTGTAAG | |
| rs5873855 element | Forward | gtaaaatcgataaggatccgGGCAGTAAGTACAGTCCC |
| Reverse | aaggctctcaagggcatcggTCAAAGCCACCTAAGTGC | |
| rs5873855 SDM | Forward | taTATATAAGTGGAATCATACAGTATTTGTCC |
| Reverse | AGGCACCCAGTGCAGTTA |
SDM, site-directed mutagenesis.
SW872 cells were grown in 24-well plates until they reached 40–50% confluence. Each well was transfected with 250 ng of firefly luciferase reporter DNA (containing the intronic region of RIPK1, as described above), 10 ng of pRL-TK Renilla reporter (Promega) and co-transfected with 250 ng E4BP4 expression plasmid (OriGene, in pCMV6-XL5 backbone). All transfections were carried out using Lipofectamine 3000 reagent (Thermo Fisher Scientific). SW872 cells were harvested with 100 μl of 1× passive lysis buffer per well 24 h after transfection. Dual-luciferase assays were conducted using 20 μl of lysate, the dual-luciferase reporter assay kit (Promega) and the GloMax luminometer (Promega) per the manufacturer’s instructions.
Chromatin immunoprecipitation.
SW872 cell genomic DNA was first genotyped for the rs5873855 variant (homozygous for no insertion) via Sanger sequencing. SW872 cells were seeded in 10-cm dishes and grown to confluence before harvesting. Cells were fixed with 1% paraformaldehyde for 10 min before being quenched with glycine. Cells from two 10-cm dishes were washed twice with cold 1× PBS, pooled after scraping and spun for 10 min at 1,000g at 4 °C, and pellets were lysed with 500 μl of nuclear lysis buffer (50 mM Tris-HCl (pH 8), 150 mM NaCl, 1 mM EDTA (pH 8), 0.1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS and protease inhibitors). Cross-linked chromatin was sheared into fragments of approximately 100–500 bp using the BioRuptor sonicator (Diagenode) and clarified using centrifugation. PureProteome Protein A/G magnetic beads (Millipore) were preblocked with sonicated salmon sperm DNA and BSA for 30 min at 4 °C. Before the immunoprecipitation, the 500 μl of total chromatin was incubated with resuspended preblocked beads to further reduce background. Each chromatin immunoprecipitation reaction used 200 μg of SW872 chromatin and 10 μg of either NFIL3 antibody cocktail (5 μg sc-74415X and 5 μg sc-374451X, Santa Cruz Biotechnology) or 10 μg of nonspecific mouse IgG (Abcam ab37355). SW872 chromatin was first incubated with antibody for 1 h at 4 °C with rotation (chromatin samples were diluted to 1 ml). Next, 10 μl of resuspended preblocked beads were added to each tube and protein–antibody–bead complexes were incubated overnight at 4 °C with rotation. Complexes were washed four times with ChIP wash buffer, once with high salt wash buffer and eluted from the beads with elution buffer containing 1% SDS and 100 mM sodium bicarbonate and heating for 15 min at 65 °C. Cross-links were reversed by the addition of NaCl to a final concentration of 200 mM along with RNase A and shaking at 65 °C for 5 h. Eluted DNA was purified using the GenepHlow gel/PCR kit (GeneAid) and qPCR was performed using SYBR Green I Master (Roche) and the LightCycler 480 II machine (Roche). The primer sequences specific for the rs5873855 DNA sequence were: forward: 5′-GTCCCATAGTACATTCACTTTCC-3′ and reverse: 5′-AAGCCAGACACAAAAGGACAA-3′. For human adipose tissue biopsies, the same protocol as above was followed, except tissue was fixed in paraformaldehyde, minced into ~2 mm3 pieces and sonicated four times with of Bioruptor (5 min each; 30 s on, 30 s off). Supernatant was then cleared by centrifugation and processed as above.
Animals.
All animal use was approved by the University of Ottawa Animal Care Committee according to the Canada Council on Animal Care. Eight-week old male C57BL/6J mice were purchased from Charles River, and RIPK1K45A and their corresponding WT littermates were a generous gift from P. Gough (GlaxoSmithKline). Male mice (eight mice per group) aged 8–10 weeks were fed a HFD containing 60% kcal from fat (D12492, Research Diets) and received weekly subcutaneous injections of saline or control ASOs or two independently targeting RIP1 MOE gapmer ASOs (Ionis Pharmaceuticals) for 24 weeks, or 2 weeks for short-term studies, before HFD feeding for another 3 weeks. Body weight was measured weekly throughout the study, and food consumption was measured weekly for 2 weeks. Mice were fasted for 6 h and anaesthetized with isoflurane before cardiac puncture, exsanguination and perfusion with PBS as previously described67. Tissues were collected, snap frozen and stored at −80 °C or processed immediately for flow cytometry.
Glucose and insulin tolerance tests.
As described previously67, mice were fasted for 6 h and then received intraperitoneal injections of 1 g kg−1 of D-glucose (Fisher Scientific) or 0.75 units per kg of insulin (HI0213, Eli Lilly). Blood glucose was measured from the tail vein at the depicted time intervals with an Accu-Check Aviva Nano glucose meter (Roche). Blood glucose levels in each group were averaged and plotted for each time point, and the AUC was determined for each individual mouse. The mean ± s.e.m. for each group is represented within the GTT/ITT graph.
Metabolic outputs and energy expenditure.
Mice were placed individually in a customized 12-chamber Oxymax open-circuit indirect calorimeter equipped with laser beam sets (Columbus Instruments) and allowed to acclimatize for 48 h, and data were collected for another 48-h period. The 2.5-l plexiglass chambers were supplied with air at 0.5 l min−1 and were maintained at 28 °C throughout a 12 h light/dark cycle. In each chamber, concentrations of O2 and CO2 in dry air were measured for 60 s every 8–12 min. Whole-body O2 consumption (VO2) corrected for lean body mass (EchoMRI) was collected and respiratory exchange ratios were calculated as previously described67.
RNA isolation, real-time qPCR and NanoString analysis.
Tissues were placed in Trizol reagent (Invitrogen) and homogenized using the Bullet Blender homogenizer (Next Advance). Total RNA was isolated and cDNA was synthesized using iScript Reverse Transcription kit (Bio-Rad) according to the manufacturer’s instructions. Real-time qPCR was performed in triplicate using either EvaGreen Express qPCR MasterMix (ABM) or Taqman gene expression assays per the manufacturer’s instructions, and mRNA level of target genes were normalized to an average of at least two housekeeping genes (B2m, Hprt and Sdha). For some genes (that is, Fizz1, Stat3 and Ripk1), NanoString nCounter Mouse Inflammatory CodeSet was used and average counts were compared using nSolver analysis software after normalization to housekeeping genes and internal controls according to the manufacturer’s instructions.
Immunohistochemistry and Oil Red O staining.
Formalin-fixed frozen liver and adipose tissue sections (10-mm thickness) were stained with primary antibodies (anti-ASO, gift from Ionis Pharmaceuticals, 1:10,000; anti-RIP1, Life Techonologies PA5–20811, 1:40; and anti-Mac2, Cedarlane CL8942AP, 1:500) at 4° C overnight or Oil Red O at 60 °C for 12 min as previously described11. Slides were then counterstained with Harris haematoxylin (SH30, Fisher Scientific) and mounted in Aquatex (108562, Merck). Quantification of staining was performed using ImageJ software. Negative control staining is shown in Supplementary Fig. 5.
Flow cytometry analysis.
All flow cytometry was analysed using FlowJo (version 10).
Human iNKT analysis from adipose and liver.
Adipose tissue and liver biopsies were directly collected in 10% FBS-DMEM. The tissue was immediately processed to obtain a single-cell suspension. Tissue was cut into small pieces and incubated with 0.2 mg ml−1 type II collagenase (adipose) or 0.5 mg ml−1 of type IV collagenase (liver; Worthington) for 30 min at 37 °C on a rotary shaker. Cells were washed and passed through a 300-μm cell strainer (adipose, pluriSelect) or 100-μm cell strainer (liver, vWR) to remove adipocyte or hepatocytes, respectively. The suspension was centrifuged at 500g for 10 min, and the pellet was lysed with HcK lysis buffer to remove red blood cells. The cell suspension was passed through a 40-μm nylon cell strainer (vWR), and cells were then counted and fixed with 1% PFA. Fixed cells were incubated with Foxp3 permeabilization buffer (BD Biosciences) for 20 min at 4 °C and then centrifuged at 900g for 5 min. The staining on 10 × 106 cells (equal number from adipose and liver) was performed in three steps: (1) intracellular staining was performed in 10% FBS-PBS 1× with antibodies for human RIPK1 (Thermo Fisher, PA5–20811; 10 μg) and antibodies for human E4BP4 (Santa Cruz, C-6, sc-374451X; 20 μg) or isotype control (rabbit IgG ab37415–5 or mouse IgG ab37355, both 10 μg; Abcam) for 30 min, and cells were washed with 10% FBS-PBS and centrifuged at 900g for 5 min; (2) secondary antibodies (Alexa Fluor 594 goat anti-rabbit, A11037; and Alexa Fluor 488 goat anti-mouse, A11029; both 4 μg ml−1; Invitrogen) were incubated with cells in 10% FBS-PBS for 30 min, then cells were washed with 10% FBS-PBS and centrifuged at 900g for 5 min; (3) the last staining step was performed with fluorescent directly coupled antibodies against CD3-APC (BD Biosciences), 557597; 1:50), CD19−BV711 (BD Biosciences), clone SJ25C1; 1:100), PE-loaded tetramer or PE-unloaded tetramer (the National Institutes of Health (NIH) Tetramer Core Facility; 1:100) for 30 min. Cells were then washed, centrifuged at 900g for 5 min before analysis by flow cytometer (BD Biosciences, FACSAria IIIu). The strategies used to detect iNKT cells were based on the CD3+CD19−tetramer+ population and gating according to isotype control (Supplementary Fig. 1). The comparison between E4BP4 and RIPK1 staining in adipose or liver iNKT cells was analysed using mean fluorescence intensity (MFI).
iNKT cells and macrophages from mouse adipose tissue and spleen.
iNKT cells were isolated as previously described50. Briefly, cells were digested from adipose tissue using collagenase type IV or dissociated from spleen using two glass slides and the single-cell suspensions were passed through a 40-μm mesh filter. The cell suspensions were first blocked with FBS, unlabelled streptavidin, Fc receptor blocking antibody (CD16/CD32) and BV711-labelled CD8a and MHC class II (BD Biosciences, all 1:100) and then incubated with PE-labelled PBS57 CD1d tetramers (NIH Tetramer Core Facility). Unloaded CD1d tetramers were used as negative controls (Supplementary Fig. 2). Cells were also labelled with APC-Cy7-labelled F4/80 (BioLegend; 1:100), APC-labelled CD3e and FITC-labelled CD45 (BD Biosciences; 1:100) and BV421-labelled IL-10 (BD Biosciences; 1:100). Gating strategies used to detect iNKT cell and F4/80 populations are shown in Supplementary Fig. 3).
Circulating blood leukocytes in mice.
Mouse blood was collected in heparin-coated tubes and red blood cells were lysed in lysis buffer (150 mM NH4Cl, 10 mM KHCO3 and 0.1 mM Na2EDTA (pH 7.4)) for 5 min on ice. Leukocytes were isolated via centrifugation at 340g for 10 min, and cells were then divided into two and stained with two antibody panels (all 1:100): (1) BV421-CD45, FITC-CD11b, PerCP5.5-Ly6C, APC-Cy7-Ly6G, APC-CD115, PE-NK.1.1 and Pe-Cy7-CD3e; and (2) BV421-CD45, FITC-CD4, Alexa Fluor 700-CD8a, PE-CD3e, Pe-Cy7-B220 and APC-Cy7-CD11b. Cells were washed and analysed for MFI using FACSAria III (BD Biosciences). Gating strategies are shown in Supplementary Fig. 4.
scRNA-seq of mouse and human transcripts.
Single-cell suspensions from mouse or human omental adipose tissue were prepared as follows: mouse CD45+ cells were sorted using FACSAria III (n = 3 mice per group pooled; BD Biosciences); human CD45+ cells (from n = 3 bariatric surgery patients, as above) were isolated using the Miltenyi Human CD45 MicroBead MS selection kit (Miltenyi, 130–045-801) according to the manufacturer’s instructions. Cells were processed using 10x Genomics Chromium v2 Single Cell 3′ RNA-seq (10x Genomics) and an Illumina NextSeq 500 Sequencer by the Ottawa StemCore Laboratories (mouse) or the University of California, Los Angeles (UCLA) Neuroscience Genomics Core (UNGC) Sequencing Core (human). Input cells were quantified and checked for viability using the Countess II (Thermo Fisher). Suspended cells were loaded on a Chromium controller single-cell instrument to generate barcoded single-cell Gel beads-in-EMulsion (GEMs). GEMs were broken and the barcoded cDNA were amplified using the C1000 Touch Thermal cycler with 96-well reaction module. The amplified barcoded cDNA was then fragmented, A-tailed and ligated with adaptors. Finally, PCR amplification was performed to enable sample indexing and enrichment of the 3′ RNA-seq libraries. The final libraries were quantified using Qubit dsDNA HS Assay kit (Thermo Fisher; Q32854), and the fragment-size distribution of the libraries was determined using the High Sensitivity NGS fragment analysis kit (AATI; DNF-486–0500). Pooled libraries were then sequenced using Illumina Nextseq. All samples were sequenced to an average of >50,000 reads per cell, per 10x Genomics recommendations.
Sequencing reads were aligned to the mm10 (mouse) or GRCh38 (human) genomes using Cell Ranger 2.2.0 from 10x Genomics with default parameters. The resultant expression matrices were imported into R and processed with Seurat v3 (3.0.0.9)68. Cells containing fewer than 200 detectable genes and those with high proportions of mitochondria-associated transcripts were removed. The remaining data was normalized using SCTransform69, which also performed feature selection for downstream dimensionality reduction. Principal-component analysis (PCA) of the top 2,000 variable genes was used for initial dimensionality reduction of the filtered expression matrix. Embeddings on the first 30 PCs were used for further dimensionality reduction using UMAP70,71. To cluster the cells and identify distinct cell types in the data, these PC embeddings were also used to construct a nearest neighbour graph using Seurat’s FindNeighbors function, which was then clustered using the Louvain algorithm for modularity optimization implemented in the FindClusters function.
Integration of human datasets.
Due to sample availability, human samples were processed on different dates, resulting in observable batch effects in the data (large shifts in PCA/UMAP embeddings). This batch effect was removed using the integration method implemented in Seurat v3 (ref. 68). Integration was performed directly on the Pearson residuals produced from SCTransform rather than the log-transformed gene counts.
Single-nucleus RNA sequencing of human subcutaneous adipose tissue.
Abdominal subcutaneous adipose tissue was collected from six participants (as part of the Finnish Twin Study) and frozen immediately and processed separately for each of the six samples. Tissue was minced over dry ice and transferred into ice-cold lysis buffer consisting of 0.1% IGEPAL, 10 mM Tris-HCl, 10 mM NaCl and 3 mM MgCl2. After a 10-min incubation period, the lysate was gently homogenized using a dounce and filtered through a 70-μm MACS smart strainer (Miltenyi Biotec; 130–098-462) to remove debris. Nuclei were centrifuged at 500g for 5 min at 4 °C and resuspended in wash buffer consisting of 1× PBS, 1% BSA and 0.2 U μl−1 RNase inhibitor. We further filtered nuclei using a 40-μm Flowmi cell strainer (Sigma-Aldrich; BAH136800040) and centrifuged at 500g for 5 min at 4 °C. Pelleted nuclei were resuspended in wash buffer and immediately processed with the 10x Chromium platform following the Single Cell 3′ v2 protocol. After library generation with the 10x platform, libraries were sequenced on an Illumina NovaSeq S2 at a sequencing depth of 50,000 reads per cell.
Reads were aligned to the GRCh38 human genome reference and Gencode v26 (ref. 72) gene annotations using the 10x Cell Ranger 2.1.1 pipeline. A custom pre-mRNA reference was generated to account for unspliced mRNA by merging all introns and exons of a gene into a single meta-exon. The gene-barcode matrix was processed with Seurat v3.0.0 (ref. 73). We removed droplets with less than 200 genes detected and the percentage of mitochondrial reads over 5%. Furthermore, we noticed that downstream clusters showed higher expression of mitochondrial genes, which is indicative of background RNA contamination; therefore, we removed cells belonging to clusters characterized by high levels of mitochondrial expression. To remove doublets, we also removed barcodes with unique molecular index counts that were two standard deviations above the mean in log space. Expression data was normalized using the SCTransform69 method implemented in Seurat. The normalized expression data from six samples were merged together, with no observable batch effects. PCA was performed on the top 2,000 variable genes to extract PCs for clustering. Then, the Louvain algorithm was used on the nearest neighbour graph of the top 30 PCs using a default resolution parameter of 0.8. We noticed there were adipocyte subclusters sharing a majority of marker genes, so we merged them into one single adipocyte cluster. Cell type identities were assigned based on known gene markers.
Bioinformatic analysis of publicly available data.
Affymetrix MoGene 1.1ST CEL files were downloaded from the Gene Expression Omnibus (GEO) under accession numbers GSE36032 and GSE63358 (mouse) and GSE115469 (ref. 33; human liver) and RMA normalized using the R Bioconductor ‘oligo’ library74. Fold-change analysis was performed using the ‘limma’ library75 with false discovery rate (FDR)-adjusted P values. Gene symbol annotations were assigned to transcript cluster identifiers using the ‘mogene11sttranscriptcluster.db’ library76. Data from GTEx V6 (dbGaP accession no. phs000424.v6.p1 derived from GENCODE v19) was accessed in January–June 2017 for eQTL analyses and November 2019–May 2020 for MR.
Statistics and reproducibility.
Comparisons between two groups were performed using a two-tailed Student’s t-test, and multiple groups were compared using a one-way ANOVA for single parameters or two-way ANOVA for multiple parameters with multiple test correction (GraphPad Prism v8.2). For analysis of gene expression using NanoString, the nSolver Analysis software was used and data were expressed as the mean ± s.d. All data are depicted from independent samples or replicates as indicated and graphs show the mean ± s.e.m. unless otherwise indicated.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Human and mouse scRNA-seq data have been deposited in the GEO database (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE151889. Other publicly available data used in this study are available at the GTEx database, or under GEO accession numbers GSE36032 and GSE63358 (mouse) and GSE115469 (human liver). All other data that support the findings of this study are available from the corresponding author upon request. Source data are provided with this paper.
Extended Data
Extended Data Fig. 1 |. SNPs in the RiPK1 locus associate with obesity in humans.
a, eQTL analysis of SNPs rs2272990 and rs2064310 in the RIPK1 locus and levels of RIPK1 mRNA in adipose tissue from the METSIM cohort. b, Regional association plots of eQTLs for RIPK1 in subcutaneous adipose from GTEX (probe ENSG00000137275.9). SNPs are coloured based on their linkage disequilibrium (r2) with the labeled top eQTL (rs2064310), which has the smallest P value in the region. Linkage disequilibrium and recombination rate calculations are based on the European population from 1000 Genomes reference panel (Phase I; release 3). c, d, Individual SNPs from the GTEx database and their association with RIPK1 expression in visceral (c) and subcutaneous (d) adipose tissue. Number of subjects homozygous or heterozygous for the minor (Ref) or major (Alt) alleles are indicated. p-values as calculated by the GTEx Consortium detailed at: https://www.gtexportal.org. Box plot shows ranked normalized gene expression in median, 1st and 3rd quartiles, 1.5 interquartile range (IQR) of 1st and 3rd quartiles. e, Odds ratio for the index SNP rs6907943 and LD SNPs in the Ottawa OBLE cohort and comparison to rs9939609 and LD SNPs in the FTO locus. f–i, Mendelian Randomization analysis of SNPs that are independently (linkage disequilibrium r2<0.2) associated with expression of RIPK1 and significantly (P<0.05) associated with obesity metrics. The beta and standard error of the effect sizes of RIPK1 expression (eQTL, x-axis) and the effect sizes on obesity metrics (y-axis) are plotted. f- UKHLS& GTEX (Adipose tissue) g- UKHLS & METSIM (Adipose tissue) h- UK Biobank & GTEX (Adipose tissue), i- UK Biobank & Llyod-Jones, 2017 (Whole Blood).
Extended Data Fig. 2 |. Single nuclei sequencing of human adipose tissue and HMDP data from mice.
a, Single-nuclei RNA sequencing on human adipose tissue: UMAP visual representation of 10 cell types derived from single-nucleus RNA-seq of human abdominal subcutaneous adipose tissue. Clusters are identified using the Seurat pipeline and cell types are assigned by biological function of marker genes. Vertical bar plot of average cell type-specific expression of RIPK1 across the 10 cell types identified in human abdominal subcutaneous adipose tissue. Expression was normalized using SCTransform. b, Data from the Hybrid Mouse Diversity Panel (HMDP) showing correlations of Ripk1 mRNA expression and NMR, visceral fat mass at sacrifice, serum insulin and HOMA-IR index in males from approximately 100 inbred strains of mice19. c, Luciferase reporter activity containing the variant sequence plus the human RIPK1 promoter upstream of the firefly luciferase gene (as in Fig. 2b). Data is shown as mean ± SEM of n=4 independent experiments, d–f. Single cell RNA sequencing of mouse CD45+ immune cells (from GSE128518) after 6, 12 and 18 weeks high fat diet feeding. d, Horizontal bar plot of cell clusters in normal chow (NC) and high fat diet (HFD). e, UMAP plot of Xcl1 cluster (cluster 9, representing NK/NKT cells) and (f) the corresponding expression of Nfil3 and Ripk1 in cluster 9.
Extended Data Fig. 3 |. Analysis of flow cytometry and single cell RNA sequencing of human adipose.
a, Mean fluorescence intensity (MFI) of RIPK1 and E4BP4 in iNKT cells from human omental adipose tissue (left) and liver (right) from obese patients (n= 12 patients total). iNKT were identified as CD19−CD3+αGalCer-loaded CD1d-tetramer+ population and based on negative control (isotype IgG) staining. Outliers were removed according to ROUT method with a specified FDR of 1% (adipose, two E4BP4 samples; liver one E4BP4 and one RIPK1 sample). Data is mean ± SD, ***p≤0.001 by paired two-tailed Student’s t-test. b, UMAP embeddings of scRNA-seq data from human omentum samples. The colour maps represent the log2-based gene expression values of various T/NK cell markers (CD3D, XCL1, NKG7, CD8A) and myeloid markers (LYZ, CD14). The bottom panels show RIPK1 and NFIL3 expression across these populations. Data deposited in the GEO database (accession number GSE151889).
Extended Data Fig. 4 |. Analysis of single cell RNA sequencing of human adipose and liver.
Single-cell RNA sequencing comparison of human adipose tissue (from Supplementary Fig. 3) and human liver (from GSE115469), showing expression of major cluster IDs XCL1 (NK/NKT), CD3D (T-cells), CD14 (monocytes) and genes of interest IL10, RIPK1 and NFIL3.
Extended Data Fig. 5 |. Metabolic analyses of RiPK1 ASO and cont ASO treated mice.
a, C57BL6 mice were treated with 50mg/kg control ASO, RIPK1 ASO-A or ASO-B over 20 weeks while fed a normal chow diet, and body weight was measured weekly. Data depicts mean ± SEM of n=3 mice per group. Two-way ANOVA using Holm-Sidak’s multiple comparison test. *p<0.05 control ASO vs. RIPK1 ASO-A and ASO-B. b, Western blot of RIPK1 (top) and GAPDH (bottom) from adipose tissue of ASO treated mice after 5 weeks of treatment. Quantification of RIPK1 band density normalized to GAPDH band density was performed using Image J. Data representative of mean ± SD of n=7–8 mice/group, p=0.09 RIPK1 ASO-A vs cont ASO, p=0.02 RIPK1 ASO-B vs. cont ASO by two-tailed Student’s t-test. One outlier was removed according to ROUT method with a specified FDR of 1%. c–f, Mice were caged in metabolic cages with indirect calorimetric and activity monitoring. VO2 (c) and respiratory exchange rations (RER) (d) and beam breaks were recorded as a measure of physical activity (e). Total food consumption (f) was measured over a 48h period. Data representative of mean ± SEM of n=8 mice/group, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, one-way ANOVA using Tukey’s multiple comparison test. g, Plasma levels of alanine transaminase (ALT) and aspartate transaminase (AST) were determined after 20 weeks of ASO treatment. Data representative of mean ± SEM of n=8 mice/group analyzed by one-way ANOVA using Tukey’s multiple comparison test. *p<0.05 vs. control ASO and RIPK1 ASO-B.
Extended Data Fig. 6 |. Analysis of diet-induced obesity in RiPK1 kinase dead mice.
WT or RIP1K45A mice (male, n=8/group) were fed a high fat diet for 20 weeks. a, Body weight. b, Epididymal adipose tissue, liver and spleen weights at termination of study. c, d, Fasting glucose tolerance test (GTT, c) or insulin tolerance test (ITT, d) and area under the curve. Data representative of mean ± SEM and analyzed by Two-way ANOVA (body weight, GTT and ITT) or two-tailed Student’s t-test (tissues weights).
Extended Data Fig. 7 |. Serum cytokine analysis of cont ASO and RiPK1 ASO treated mice.
Serum cytokines from mice treated with cont ASO or RIPK1 ASO-B for 24 weeks. Cytokine analysis was done using the Bioplex Pro Cytokine 23-plex assay. No significant differences between cytokines IL-1α, TNFα, IL-4 or IL-10 were observed. Data representative of mean ± SEM of n=8 mice/group and was analysed by one-way ANOVA using Dunnett’s multiple comparison test.
Extended Data Fig. 8 |. Analysis of single cell RNA sequencing of mouse adipose tissue.
a, Clustered UMAP embedding of scRNA-seq data from mouse adipose tissue. The colours and labels represent each of the 13 distinct clusters. b, The same embeddings as in (a), but separately coloring the cells derived from either Control ASO or RIPK1 ASO-B samples. c, A graph showing the percentage of cells from each sample that correspond to each cell type. d, UMAP embedding of scRNA-seq data from T cell populations of mouse adipose tissue. T cells were defined as CD3+ clusters. Plots show the log-transformed gene expression of various markers defining NKT (Xcl1), CD4+ T (Cd4), CD8+ T (Cd8a), Memory CD4+ T (Ccr7), Naive CD4+ T (S100a4), Regulatory T (Foxp3), and Gamma Delta T (Cd163l1). Data deposited in the GEO database (accession number GSE151889).
Extended Data Fig. 9 |. Flow cytometry of adipose and circulating immune cells from cont ASo and RiPK1 ASo treated mice.
a, Representative flow cytometry plots and NKT % (expressed as % of F4/80− cells that are αGalCer-loaded CD1d-tetramer+CD3e+) in spleen from mice treated for 5 weeks with cont ASO or RIPK1 ASO (n=8/group). Data representative of mean ± SEM of n=8 mice/group, analyzed by two-tailed Student’s t-test. b, c, Flow cytometry analysis of circulating leukocytes from mice treated with control ASO or RIP1 ASO-B at 5 weeks: Ly6Clow monocytes *p<0.05 (p=0.0452) Ly6Chigh monocytes *p<0.05 (p=0.0108), and CD3e+ T cells. (c) Flow cytometry analysis of circulating leukocytes from mice treated with control ASO or RIP1 ASO-B at 22 weeks: Ly6Clow monocytes, Ly6Chigh monocytes and CD3e+ T cells (p=0.0132). Data in b, c representative of mean ± SEM of n=8 mice/group, analyzed by two-tailed Student’s t-test.
Extended Data Fig. 10 |. Flow cytometry of adipose immune cells from cont ASo and RiPK1 ASo treated mice.
a, Regulatory T-cell (CD25+Foxp3+) and NK (NK1.1+) cells analysis by flow cytometry of isolated SVF of adipose tissue from mice treated with control ASO or RIP1 ASO-B for 5 weeks while fed a high fat diet (n=7–8/group). b, Gating strategy for selecting CD45+, CD3e+, CD4+ cells then those that were positive for CD25 and Foxp3 (to represent T regulatory cells) or NK1.1 (to represent NK cells). Data depicts mean ± SEM of n=8 mice/group.
Supplementary Material
Acknowledgements
The authors thank X. Zhao, K. Sheikheleslamy and G. Palidwor for their technical and statistical support. We thank J. Kaprio and A. Rissanen for their contributions and the individuals who participated in the Finnish Twin study. We also thank the University of Ottawa StemCore Facility and the UNGC Sequencing Core at UCLA for performing RNA sequencing. This study was supported with funding from the following organizations: Canadian Institutes of Health Research (K.J.R., R.M., M.E.H., B.C.V. and D.P.C.), NIH (R01 HL119047 to K.J.R.; DK117850 and HL147883 to A.J.L., HL095056, HL28481 and U01 DK105561 to P.P.), Foundation Leduc (A.J.L.), National Psoriasis Foundation (D.K.), Academy of Finland (M.L.; 272376, 266286, 314383 and 315035 to K.H.P.), European Union (M.L.) and Sigrid Juselius Foundation (M.L. and K.H.P.); Finnish Medical Foundation (K.H.P.) and the Finnish Diabetes Research Foundation, Novo Nordisk Foundation, Gyllenberg Foundation, Helsinki University Hospital Research Funds and University of Helsinki (K.H.P.); University of Ottawa Heart Institute Cardiac Endowment Fellowship (A.C.D.); and HHMI Gilliam Fellowship (M.A.). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the article.
Footnotes
Competing interests
R.L. is an employee of Ionis Pharmaceuticals, S.B.B. is an employee of Janssen Pharmaceuticals, P.J.G. is an employee of Inzen Therapeutics and J.B. is an employee of GlaxoSmithKline. The other authors declare no competing interests.
Additional information
Extended data is available for this paper at https://doi.org/10.1038/s42255-020-00279-2.
Supplementary information is available for this paper at https://doi.org/10.1038/s42255-020-00279-2.
Peer review information Primary Handling Editor: Pooja Jha.
References
- 1.Berg AH & Scherer PE Adipose tissue, inflammation and cardiovascular disease. Circ. Res 96, 939–949 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Weisberg SP et al. Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Invest 112, 1796–1808 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stunkard AJ, Foch TT & Hrubec Z. A twin study of human obesity. JAMA 256, 51–54 (1986). [PubMed] [Google Scholar]
- 4.Frayling TM et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 316, 889–894 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerken T. et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science 318, 1469–1472 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scuteri A. et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 3, e115 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dina C. et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat. Genet 39, 724–726 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Speakman JR, Rance KA & Johnstone AM Polymorphisms of the FTO gene are associated with variation in energy intake, but not energy expenditure. Obesity (Silver Spring) 16, 1961–1965 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Smemo S. et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature 507, 371–375 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loos RJ et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat. Genet 40, 768–775 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thorleifsson G. et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat. Genet 41, 18–24 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Nikpay M, Turner AW & McPherson R. Partitioning the pleiotropy between coronary artery disease and body mass index reveals the importance of low frequency variants and central nervous system-specific functional elements. Circ. Genom. Precis. Med 11, e002050 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Karunakaran D. et al. Targeting macrophage necroptosis for therapeutic and diagnostic interventions in atherosclerosis. Sci. Adv 2, e1600224 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silke J, Rickard JA & Gerlic M. The diverse role of RIP kinases in necroptosis and inflammation. Nat. Immunol 16, 689–697 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Kondylis V, Kumari S, Vlantis K. & Pasparakis M. The interplay of IKK, NFκB and RIPK1 signaling in the regulation of cell death, tissue homeostasis and inflammation. Immunol. Rev 277, 113–127 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Civelek M. et al. Genetic regulation of adipose gene expression and cardio-metabolic traits. Am. J. Hum. Genet 100, 428–443 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laakso M. et al. The Metabolic Syndrome in Men study: a resource for studies of metabolic and cardiovascular diseases. J. Lipid. Res 58, 481–493 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davies RW et al. A 680-kb duplication at the FTO locus in a kindred with obesity and a distinct body fat distribution. Eur. J. Hum. Genet 21, 1417–1422 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Rheenen W, Peyrot WJ, Schork AJ, Lee SH & Wray NR Genetic correlations of polygenic disease traits: from theory to practice. Nat. Rev. Genet 20, 567–581 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Pingault JB et al. Using genetic data to strengthen causal inference in observational research. Nat. Rev. Genet 19, 566–580 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Zhu Z. et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat. Commun 9, 224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westra HJ et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet 45, 1238–1243 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Nas A. et al. The systems genetics resource: a web application to mine global data for complex disease traits. Front. Genet 4, 84 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ward LD & Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40, D930–D934 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cowell IG E4BP4/NFIL3, a PAR-related bZIP factor with many roles. Bioessays 24, 1023–1029 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Lynch L. et al. Regulatory iNKT cells lack expression of the transcription factor PLZF and control the homeostasis of Treg cells and macrophages in adipose tissue. Nat. Immunol 16, 85–95 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roy S. et al. Redefining the transcriptional regulatory dynamics of classically and alternatively activated macrophages by deepCAGE transcriptomics. Nucleic Acids Res. 43, 6969–6982 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takahashi S. et al. A promoter in the novel exon of hPPARγ directs the circadian expression of PPARγ. J. Atheroscler. Thromb 17, 73–83 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Lynch L. et al. Adipose tissue iNKT cells protect against diet-induced obesity and metabolic disorder through regulatory cytokine production. Immunity 37, 574–587 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ji Y. et al. Activation of natural killer T cells promotes M2 macrophage polarization in adipose tissue and improves systemic glucose tolerance via IL-4/STAT6 protein signaling axis in obesity. J. Biol. Chem 287, 13561–13571 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaitin DA et al. Lipid-associated macrophages control metabolic homeostasis in a Trem2-dependent manner. Cell 178, 686–698(2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacParland SA et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat. Commun 9, 4383 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kenna T. et al. NKT cells from normal and tumor-bearing human livers are phenotypically and functionally distinct from murine NKT cells. J. Immunol 171, 1775–1779 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Fengler VH et al. Susceptibility of different mouse wild-type strains to develop diet-induced NAFLD/AFLD-associated liver disease. PLoS ONE 11, e0155163 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berger SB et al. Cutting edge: RIP1 kinase activity is dispensable for normal development but is a key regulator of inflammation in SHARPIN-deficient mice. J. Immunol 192, 5476–5480 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woo YDC, Kim HY & Chung DH Invariant NKT cells recruit CD103+ dendritic cells via XCL1–XCR1 axis, thereby promoting allergic airway resistance. J. Immunol 198, 1 (2017). [Google Scholar]
- 38.van der Klaauw AA & Farooqi IS The hunger genes: pathways to obesity. Cell 161, 119–132 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Akiyama M. et al. Genome-wide association study identifies 112 new loci for body mass index in the Japanese population. Nat. Genet 49, 1458–1467 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Welsh P. et al. Unraveling the directional link between adiposity and inflammation: a bidirectional Mendelian randomization approach. J. Clin. Endocrinol. Metab 95, 93–99 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vucic D, Dixit VM & Wertz IE Ubiquitylation in apoptosis: a post-translational modification at the edge of life and death. Nat. Rev. Mol. Cell. Biol 12, 439–452 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Chan FK, Luz NF & Moriwaki K. Programmed necrosis in the cross talk of cell death and inflammation. Annu. Rev. Immunol 33, 79–106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peltzer N, Darding M. & Walczak H. Holding RIPK1 on the ubiquitin leash in TNFR1 signaling. Trends Cell Biol. 26, 445–461 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Roderick JE et al. Hematopoietic RIPK1 deficiency results in bone marrow failure caused by apoptosis and RIPK3-mediated necroptosis. Proc. Natl Acad. Sci. USA 111, 14436–14441 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gautheron J. et al. The necroptosis-inducing kinase RIPK3 dampens adipose tissue inflammation and glucose intolerance. Nat. Commun 7, 11869 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dannappel M. et al. RIPK1 maintains epithelial homeostasis by inhibiting apoptosis and necroptosis. Nature 513, 90–94 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berzins SP, Smyth MJ & Baxter AG Presumed guilty: natural killer T cell defects and human disease. Nat. Rev. Immunol 11, 131–142 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Lynch L. Adipose invariant natural killer T cells. Immunology 142, 337–346 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huh JY et al. Deletion of CD1d in adipocytes aggravates adipose tissue inflammation and insulin resistance in obesity. Diabetes 66, 835–847 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Vieth JA et al. TCRα–TCRβ pairing controls recognition of CD1d and directs the development of adipose NKT cells. Nat. Immunol 18, 36–44 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Mookerjee-Basu J. & Kappes DJ iNKT cells do a fat lot of good. Nat. Immunol 18, 10–12 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Lynch L. et al. Invariant NKT cells and CD1d+ cells amass in human omentum and are depleted in patients with cancer and obesity. Eur. J. Immunol 39, 1893–1901 (2009). [DOI] [PubMed] [Google Scholar]
- 53.Engel I. et al. Innate-like functions of natural killer T cell subsets result from highly divergent gene programs. Nat. Immunol 17, 728–739 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Plenge RM Priority index for human genetics and drug discovery. Nat. Genet 51, 1073–1075 (2019). [DOI] [PubMed] [Google Scholar]
- 55.Fang H. et al. A genetics-led approach defines the drug target landscape of 30 immune-related traits. Nat. Genet 51, 1082–1091 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nelson MR et al. The support of human genetic evidence for approved drug indications. Nat. Genet 47, 856–860 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Plenge RM, Scolnick EM & Altshuler D. Validating therapeutic targets through human genetics. Nat. Rev. Drug Discov 12, 581–594 (2013). [DOI] [PubMed] [Google Scholar]
- 58.Paik J. & Duggan S. Volanesorsen: first global approval. Drugs 79, 1349–1354 (2019). [DOI] [PubMed] [Google Scholar]
- 59.Witztum JL et al. Volanesorsen and triglyceride levels in familial chylomicronemia syndrome. N. Engl. J. Med 381, 531–542 (2019). [DOI] [PubMed] [Google Scholar]
- 60.Fitzgerald K. et al. A highly durable RNAi therapeutic inhibitor of PCSK9. N. Engl. J. Med 376, 41–51 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Duell PB et al. Long-term mipomersen treatment is associated with a reduction in cardiovascular events in patients with familial hypercholesterolemia. J. Clin. Lipidol 10, 1011–1021 (2016). [DOI] [PubMed] [Google Scholar]
- 62.Adams D. et al. Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N. Engl. J. Med 379, 11–21 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Stein CA & Castanotto D. FDA-approved oligonucleotide therapies in 2017. Mol. Ther 25, 1069–1075 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Levin AA Treating disease at the RNA level with oligonucleotides. N. Engl. J. Med 380, 57–70 (2019). [DOI] [PubMed] [Google Scholar]
- 65.Nikpay M. et al. A comprehensive 1000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet 47, 1121–1130 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ramkhelawon B. et al. Netrin-1 promotes adipose tissue macrophage retention and insulin resistance in obesity. Nat. Med 20, 377–384 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Karunakaran D. et al. Therapeutic inhibition of miR-33 promotes fatty acid oxidation but does not ameliorate metabolic dysfunction in diet-induced obesity. Arterioscler. Thromb. Vasc. Biol 35, 2536–2543 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Butler A, Hoffman P, Smibert P, Papalexi E. & Satija R. Integrating single-cell transcriptomic data across different conditions, technologies and species. Nat. Biotechnol 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hafemeister C. & Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Becht E. et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol 37, 38–44 (2018). [DOI] [PubMed] [Google Scholar]
- 71.McInnes L, Healy J, Saul N. & Großberger L. UMAP: uniform manifold approximation and projection. J. Open Source Softw 3, 861 (2018). [Google Scholar]
- 72.Frankish A. et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 47, D766–D773 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stuart T. et al. Comprehensive integration of single cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Carvalho BS & Irizarry RA A framework for oligonucleotide microarray preprocessing. Bioinformatics 26, 2363–2367 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ritchie ME et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.MacDonald JW mogene11sttranscriptcluster.db: Affymetrix mogene11 annotation data (chip mogene11sttranscriptcluster). R package version 8.7.0. (2017). [Google Scholar]
- 77.Riveros-McKay F. et al. Genetic architecture of human thinness compared to severe obesity. PLoS Genet. 15, e1007603 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lloyd-Jones LR et al. The genetic architecture of gene expression in peripheral blood. Am. J. Hum. Genet 100, 228–237 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Human and mouse scRNA-seq data have been deposited in the GEO database (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE151889. Other publicly available data used in this study are available at the GTEx database, or under GEO accession numbers GSE36032 and GSE63358 (mouse) and GSE115469 (human liver). All other data that support the findings of this study are available from the corresponding author upon request. Source data are provided with this paper.