Abstract
The interferon-regulated antiviral responses are essential for the induction of both innate and adaptive immunity in mammals. Production of virus-derived small-interfering RNAs (vsiRNAs) to restrict virus infection by RNA interference (RNAi) is a recently identified mammalian immune response to several RNA viruses, which cause important human diseases such as influenza and Zika virus. However, little is known about Dicer processing of viral double-stranded RNA replicative intermediates (dsRNA-vRIs) in mammalian somatic cells. Here we show that infected somatic cells produced more influenza vsiRNAs than cellular microRNAs when both were produced by human Dicer expressed de novo, indicating that dsRNA-vRIs are not poor Dicer substrates as previously proposed according to in vitro Dicer processing of synthetic long dsRNA. We report the first evidence both for canonical vsiRNA production during wild-type Nodamura virus infection and direct vsiRNA sequestration by its RNAi suppressor protein B2 in two strains of suckling mice. Moreover, Sindbis virus (SINV) accumulation in vivo was decreased by prior production of SINV-targeting vsiRNAs triggered by infection and increased by heterologous expression of B2 in cis from SINV genome, indicating an antiviral function for the induced RNAi response. These findings reveal that unlike artificial long dsRNA, dsRNA-vRIs made during authentic infection of mature somatic cells are efficiently processed by Dicer into vsiRNAs to direct antiviral RNAi. Interestingly, Dicer processing of dsRNA-vRIs into vsiRNAs was inhibited by LGP2 (laboratory of genetics and physiology 2), which was encoded by an interferon-stimulated gene (ISG) shown recently to inhibit Dicer processing of artificial long dsRNA in cell culture. Our work thus further suggests negative modulation of antiviral RNAi by a known ISG from the interferon response.
Author summary
The function and mechanism of the interferon-regulated antiviral responses have been extensively characterized. Recent studies have demonstrated induction of antiviral RNA interference (RNAi) in somatic cells against several mammalian RNA viruses rendered incapable of RNAi suppression. However, little is known about Dicer-mediated production of virus-derived small-interfering RNAs (vsiRNAs) in these cells active in the type I interferon response. Here we show that the dsRNA precursors of influenza vsiRNAs were processed more efficiently than cellular precursor microRNA hairpins by wild-type human Dicer expressed de novo in Dicer-knockout somatic cells. We found that infection of two strains of suckling mice with wild-type Nodamura virus (NoV) was associated with production of silencing-active vsiRNAs and direct sequestration of duplex vsiRNAs by its RNAi suppressor protein B2. Our findings from in vivo infection with Sindbis virus recombinants expressing NoV B2 or carrying a vsiRNA-targeted insert provide evidence for an antiviral function of the induced RNAi response. Interestingly, NoV infection induces expression of RIG-I-like receptor LGP2 to inhibit vsiRNA biogenesis and promote virulent infection in suckling mice. Our findings together reveal efficient Dicer processing of vsiRNA precursors in interferon-competent somatic cells and suckling mice in contrast to synthetic long dsRNA examined previously by in vitro dicing.
Introduction
Dicer enzymes in the RNase III family mediate the biogenesis of microRNAs (miRNAs) and small interfering RNAs (siRNAs) in plants and animals [1,2]. Mature miRNAs are produced by Dicer from hairpin precursor miRNAs (pre-miRNAs) to repress the translation of the target mRNAs [3,4]. However, siRNAs are processed from long double-stranded RNA (dsRNA) to initiate RNA interference (RNAi), defined as specific slicing of complementary RNAs by an Argonaute protein (AGO) in RNA-induced silencing complex (RISC) [1,5]. The RNAi pathway functions as a potent antiviral immunity in plants and invertebrates because these hosts produce highly abundant virus-derived siRNAs (vsiRNAs) by Dicer from viral dsRNA precursors to guide RISC-dependent clearance of virus RNAs [6]. In counter defense, plant and insect viruses have evolved viral suppressors of RNAi (VSRs) to block various steps in the antiviral RNAi pathway [6–8].
In vertebrate animals, the type I interferon (IFN) response is a major first line of defense against virus infection before the activation of adaptive immunity [9,10]. The IFN antiviral response is frequently initiated by cytoplasmic sensing of viral RNA ligands by retinoic acid-inducible gene I (RIG-I) or melanoma differentiation factor 5 (MDA5). Upon RNA binding, these RIG-I-like receptors (RLRs) interact with mitochondrial antiviral-signaling protein (MAVS, also known as VISA, IPS-1 or Cardif) to activate RLR signal transduction, leading to transcriptional induction of the genes encoding type I IFN and other genes in the nucleus [11]. Binding of the type I IFN by IFN-α/β receptor (IFNAR) on cell surface then activates an intracellular signaling cascade to drive the expression of hundreds of IFN-stimulated genes (ISGs). Some ISGs encode virus restriction factors whereas other ISG factors such as 2’-5’ oligoadenylate synthetases and dsRNA-dependent protein kinase R (PKR) inhibit cell growth by inducing global RNA degradation and protein translation shutdown, respectively [11]. LGP2 (laboratory of genetics and physiology 2) is the third member of RLRs, but lacks the amino-terminal domains conserved in RIG-I and MDA5 necessary for independent signal-transducing activity. Although not essential for the induction of the IFN response, LGP2 can modulate antiviral defense by promoting MDA5-mediated responses or acting as an inhibitor of RIG-I signaling [12–19].
Recent studies have provided evidence for the induction of the antiviral RNAi response in mammals [20–28]. These studies, including ours, have demonstrated production of abundant vsiRNAs predominantly 22 nucleotides (nt) long during the infection of undifferentiated and differentiated cells as well as mice with positive- or negative-strand RNA viruses [23–30]. Notably, Nodamura virus (NoV), influenza A virus (IAV), human enterovirus 71 (HEV71), and dengue virus-2 (DNV2) from 4 distinct RNA virus families encode structurally unrelated VSRs necessary for infection and active to suppress Dicer processing of the cognate viral dsRNA replicative intermediates (dsRNA-vRIs) into vsiRNAs [23–25,27,29].
Plants and most invertebrate species encode two or more Dicer genes with at least one member being dispensable for miRNA biogenesis and dedicated instead to processing long dsRNA into siRNA and antiviral RNAi [6,31–33]. In contrast, mammals encode a single Dicer [1], which recognizes pre-miRNAs as more efficient substrates than long dsRNA in in vitro dicing assays [34,35]. Moreover, because of the induction of cell growth inhibition and cell death in IFN-competent differentiated cells, artificial long dsRNA has been shown to trigger RNAi only in pluripotent embryonic stem cells (ESCs) and embryonic carcinoma cells defective in canonical IFN signaling [36–40]. Recent genetic studies also support an antagonistic role of the IFN response to RNAi since long dsRNA induces RNAi in differentiated cells after PKR knockout or inactivation of the type I IFN response by removal of MAVS or IFNAR [41,42]. Consistently, LGP2 has recently been identified as an ISG factor to block long dsRNA-induced RNAi in mouse embryonic fibroblasts (MEFs) by inhibiting Dicer processing of long dsRNA into siRNAs [43].
As shown in synthetic RNAi induced by long dsRNA or short hairpin RNA, mammalian vsiRNAs are processed from dsRNA-vRIs by Dicer to direct AGO2-dependent antiviral RNAi [23–25,27,29,44]. In contrast to long dsRNA, however, abundant vsiRNAs are processed from dsRNA-vRIs made by viral RNA-dependent RNA polymerase (RdRP) during infection in several commonly used lines of differentiated cells and/or mice with mutant viruses, including NoV, IAV, HEV71 and DNV2, rendered defective in the expression or the activity of the cognate VSR, but remained competent in the induction of the IFN response [23,25,27,29,30]. All of the 4 validated VSRs are dsRNA-binding proteins and suppress Dicer processing of long dsRNA into siRNAs in vitro. However, a dominant population of vsiRNAs becomes undetectable after infection with any of these viruses expressing a functional VSR, which is thus similar to previous deep sequencing profiling of total small RNAs in mature cells infected with a range of wild type RNA viruses [45–50]. Although two wild type RNA viruse induce production of abundant vsiRNAs in undifferentiated cells [24,26], production of vsiRNAs in the IFN-defective mouse ESCs induced by NoV infection is potently suppressed by the cognate VSR protein B2 [24], suggesting widespread suppression of vsiRNA biogenesis during mammalian virus infection [6,20]. Moreover, MAVS knockout in MEFs activates synthetic RNAi induced by artificial long dsRNA without enhancing RNAi-mediated antiviral defense shown to be active in wildtype MEFs [42,44]. A receent study also indicates that antiviral RNAi requires AGO4, which is dispensable for RNA slicing by synthetic siRNAs [51].
Several key questions remain unresolved in mammalian antiviral RNAi induced in differentiated cells and in vivo. First, it is unclear whether vsiRNA precursors are inherently poor substrates of human Dicer in differentiated cells as it is widely known for long dsRNA dicing in vitro [34,35]. Second, it is also unclear whether canonical vsiRNAs are produced during infection of differentiated cells or in vivo by wild type viruses with or without a validated VSR although it is known that natural infection of MEFs by wild type NoV and IAV is inhibited by RNAi [29,44]. Third, recent studies have provided evidence for vsiRNA-dependent RNA degradation [25,44]. However, it remains unknown whether the vsiRNAs can mediate specific inhibition of virus accumulation in vivo. In this work, we designed and performed experiments to address these unresolved questions in mammalian antiviral RNAi. Moreover, we investigated whether the recently identified inhibitor of artificial long dsRNA Dicer processing, LGP2, is also active against vsiRNA production in vitro and in vivo. Our findings reveal key aspects of antiviral RNAi induced by RNA virus infection that are different or similar to the induction of RNAi by artificial long dsRNA in mammalian somatic cells.
Results
Human Dicer recognized and processed more viral dsRNA replication products than cellular pre-miRNA hairpins in somatic cells
We first developed an experimental system to compare the accumulation of vsiRNAs and cellular miRNAs processed from their precursors in differentiated human cells. We and others have previously demonstrated Dicer-mediated biogenesis of the influenza vsiRNAs from dsRNA-vRI precursors in human 293T cells infected with PR8/delNS1, a VSR-NS1 (non-structural protein 1) deletion mutant of IAV-strain PR8 [29,30]. In this work, we infected the Dicer-deficient (NoDice) human 293T cells [52] with PR8/delNS1 6 hours post-transfection with a human Dicer (hDcr)-expressing plasmid and sequenced the total small RNAs in the infected cells 24 hours post-infection. NoDice cells accumulate little miRNAs [52]. Thus, it became possible in our experimental system to compare the relative accumulation levels of vsiRNAs and cellular miRNAs processed from their respective precursors by the Dicer enzyme expressed de novo in the same sequenced small RNA library.
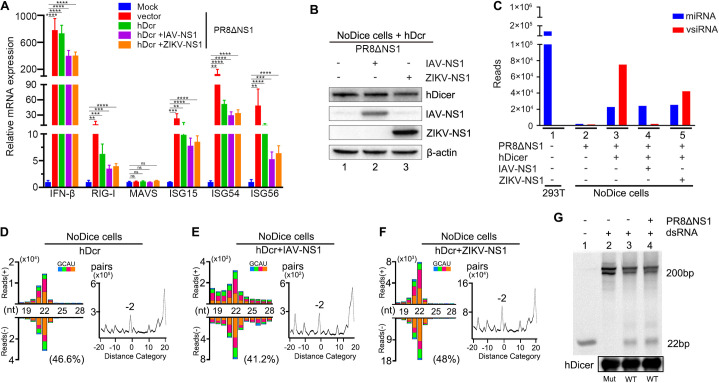
We found that infection with the VSR-deficient IAV induced expression of ISGs, including IFNβ and RIG-I, in NoDice cells with or without the ectopic expression of hDcr (Fig 1A), indicating induction of the type I IFN response by PR8/delNS1 infection as expected [53]. Ectopic expression of hDcr was confirmed by Western blotting (Fig 1B). Deep sequencing verified rescue of the severely impaired miRNA biogenesis in NoDice cells by the ectopically expressed hDcr (Fig 1C and S1 Table) as previously reported [52]. Also as described previously [29], we detected hDcr-dependent production of a typical population of predominantly 22-nt influenza vsiRNAs in NoDice cells (Fig 1D–1F). By comparison, however, the influenza vsiRNAs made during the one-day infection were 229.5% more abundant than the total human miRNAs accumulated in NoDice cells (Fig 1C, lane 3 and S1 Table). These influenza vsiRNAs had an approximately equal ratio of positive and negative strands, and contained a dominant population of 22-nt vsiRNA duplexes with 2-nt 3’ overhangs, indicating that they were Dicer products processed from dsRNA-vRIs of the 8 negative-strand genomic RNAs (Figs 1D–1F and S1). Thus, the influenza dsRNA-vRIs synthesized by the viral RdRP were recognized as the main substrate of hDcr in the infected human cells with an active type I IFN response.
Fig 1. Efficient Dicer processing of IAV derived-dsRNA in human 293T cells active in type I IFN response.
A. Induction of type I IFN response in NoDice cells. NoDice cells were mock-inoculated or infected with PR8/delNS1 at 6 hours post-transfection with the plasmid vector, an hDcr-expressing plasmid alone or together another plasmid to express IAV-NS1 or ZIKV-NS1. The accumulation levels of IFN-β, RIG-I, MAVS, ISG15, ISG54, and ISG56 mRNAs were determined by RT-qPCR at 24 hours post-infection. Each experiment was repeated at least three times independently and error bars indicate the standard deviation. The mRNA level of mock-inoculated cells without plasmid transfection was set as 1. ** indicates p<0.01, *** indicates p<0.001, **** indicates p<0.0001 (Student’s t-test). ns indicates no significance. B. Western blotting detection of the ectopically expressed human Dicer and/or IAV-NS1 /hDcr+ZIKV-NS1 as described in (A). Endogenous β-actin was detected as a loading control. C. Relative abundance of cellular miRNAs and IAV-derived 21- to 23-nt small RNAs per million total 18- to 28-nt reads in the individual small RNA libraries prepared from the NoDice cells at 24 hours post-infection as described in (A). D-F. Size distribution of IAV-derived 18- to 28-nt small RNAs (left) and duplex pattern of 22-nt IAV vsiRNAs (right) in the 3 libraries (no. 3, 4 and 5) presented in (C) from PR8/delNS1-infected NoDice cells ectopically expressing hDcr alone (D) or with IAV-NS1 (E) or ZIKV-NS1 (F). Data information: Reads were shown as per million total 18- to 28-nt reads. The 5’ terminal nucleotide of 18- to 28-nt viral small RNAs was indicated by color and 1U % of 21- to 23-nt vsiRNA given in parenthesis. The “-2” peak corresponded to the pair of canonical 22-nt vsiRNAs with a 20-nt duplex region plus 2-nt 3’ overhangs, calculated by an algorithm described previously counting pairs of complementary 22-nt vsiRNAs in each distance category (in nucleotides) between 5’ and 3’ ends of each pair. Length (nt) indicates x-axis for size distribution. Distance category indicates x-axis for duplex pattern. G. In vitro dicing of a synthetic 200 bp dsRNA by FLAG-tagged wild-type (lanes 3 & 4) or mutant (lane 2) human Dicer immune-precipitated from NoDice cells with or without PR8/delNS1. The Dicer substrate and product RNAs were fractionated by 15% PAGE and detected by GelRed staining. Lane 1: 22bp marker. Each experiment was repeated at least three times independently with one represented image shown.
We further sequenced the total small RNAs in PR8/delNS1-infected NoDice cells that co-expressed hDcr with the non-structural protein 1 (NS1) encoded by either IAV or Zika virus (ZIKV). The two viral NS1 proteins antagonize the IFN response by distinct mechanisms [53,54] and IAV NS1 exhibits an additional activity to suppress vsiRNA biogenesis [29,55]. The expression of both NS1 proteins was confirmed by Western blotting (Fig 1B). We found that expression of neither IFN antagonistic protein had an obvious effect on the abundance of the total human miRNAs made by hDcr in the infected NoDice cells (Fig 1C, lanes 3, 4 and 5, left; S1 Table). A typical population of the influenza vsiRNAs was produced by hDcr in the infected NoDice cells expressing NS1 of either IAV or ZIKV (Fig 1E and 1F). We noted that the virus genome distribution patterns of vsiRNAs were similar in all three libraries (S1 Fig). However, expression of both viral NS1 proteins reduced the abundance of the influenza vsiRNAs with the lowest abundant vsiRNAs detected in cells expressing IAV NS1 (Fig 1C, lanes 3, 4 and 5, right), consistent with its known VSR activity [29,55]. Interestingly, we found that the influenza vsiRNAs were 66.2% more abundant than host miRNAs in the NoDice cells expressing NS1 of ZIKV (Fig 1C, lane 5, right and S1 Table). Compared to cellular pre-miRNA hairpins, therefore, the influenza dsRNA-vRIs remained as the dominant substrates of human Dicer in the infected cells when type-I IFN production is inhibited by ZIKV NS1 shown previously at the step of TBK1 complex formation [54].
To verify the dicing activity of the ectopically expressed hDcr, we assayed in vitro processing of 200-nt dsRNA by FLAG-tagged hDcr co-immunoprecipitated from NoDice cells with or without induction of the IFN response by PR8/delNS1 infection. As shown in Fig 1G, we detected efficient processing of the dsRNA into 22-nt siRNAs by wild-type hDcr, but not a Dicer mutant in which the catalytic sites were mutated (Fig 1G, lanes 2 and 3). Importantly, we observed no obvious differences in the processing of the long dsRNA by hDcr purified from either the mock- or PR8/delNS1-infected cells (Fig 1C, lanes 3 and 4), indicating that hDcr remained active after the induction of the IFN response. Together, our results demonstrate that when the biogenesis of both vsiRNAs and cellular miRNAs was mediated by the Dicer enzyme expressed de novo, viral dsRNA replicative intermediates served as the more dominant substrates than pre-miRNAs in the IFN-competent human cells.
Production and direct VSR sequestration of canonical duplex vsiRNAs in two strains of mice infected with a wild type RNA virus
We next examined the hypothesis that VSR-expressing wild type virus infection in vivo also triggers production of canonical vsiRNAs, which, however, are not readily visible from deep sequencing of total small RNAs due to the presence of abundant viral RNA degradation products. It is known that vsiRNAs are selectively loaded into RISC and the VSR-B2 protein encoded by NoV and the closely related Flock house virus exhibits duplex siRNA-binding activity in vitro [6,56–59]. Thus, we initially searched for the presence of in vivo vsiRNAs by sequencing total small RNAs both before and after co-immunoprecipitation (IP) with B2-specific antibodies or a pan-Argonaute antibody from NoV-infected BALB/c suckling mice at 3 days post-inoculation (dpi).
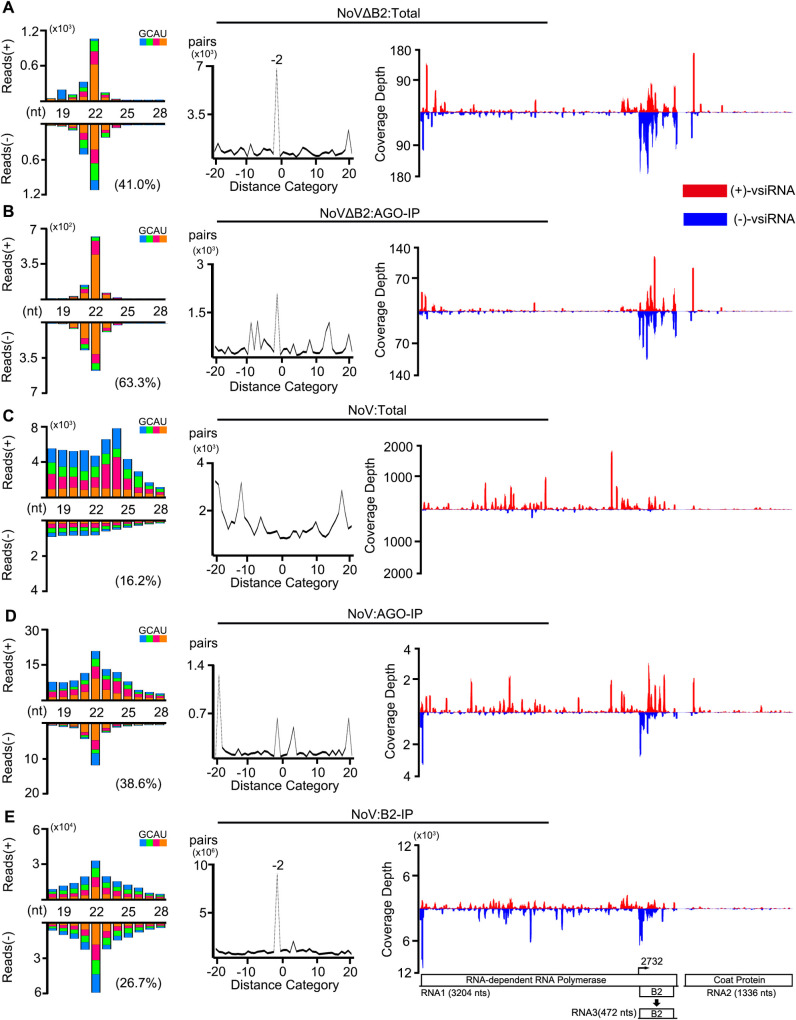
We detected abundant vsiRNAs in the Argonaute precipitants from NoVΔB2-infected suckling mice in the control experiments. Similar to the total vsiRNAs, the most dominant size class in Argonaute-bound vsiRNAs of both polarities was 22-nt (Fig 2A and 2B, left panels). The distribution patterns of vsiRNA hot spots on the two positive-strand genomic RNAs of NoV were similar between the total and Argonaute-bound populations (Fig 2A and 2B, right panels), suggesting that the vsiRNAs produced by mice in response to infection are loaded proportionally into the Argonaute complexes. However, Argonaute-bound vsiRNAs exhibited an increased enrichment for vsiRNAs in the size range of 21- to 23-nt (92.8%) and for 1U vsiRNAs (63.3%), which were 85.9% and 41.0% for the total vsiRNAs, respectively (Fig 2A and 2B and S1 Table). Similar to our recent findings from the mutant mice defective in adaptive immunity [44], these findings indicate that the vsiRNAs produced in IFN-competent wild type mice are in vivo loaded into Argonaute complexes with strong selection for 21- to 23-nt 1U vsiRNAs. As a further indicative measure of selective loading [45], the population of 22-nt duplex vsiRNAs with 2-nt overhangs (shown by the -2 peak in Fig 2A and 2B, middle panel) was less dominant for Argonaute-bound vsiRNAs than the total vsiRNAs.
Fig 2. NoV infection in vivo induces production of abundant canonical duplex vsiRNAs sequestered in VSR-B2 protein complex.
A-E. Size distribution of 18- to 28-nt virus-derived small RNAs (left), duplex pattern of the 22-nt vsiRNAs (middle) and the distribution of 21- to 23-nt vsiRNAs along the viral genomic RNAs 1 and 2 sequenced from BALB/c suckling mice infected with NoVΔB2 (A, B) or NoV (C, D, E) at 3 dpi, either with or without co-immunoprecipitation (co-IP) by antibodies specific to mouse AGOs (B, D) or the viral B2 protein (E). Data information: Same as in Fig 1.
As described previously [23], the total vsRNAs sequenced from NoV-infected mice were highly abundant, but showed no preference in the size range of Dicer products and were mostly positive strands (Fig 2C). Notably, B2 immunoprecipitants from the same NoV-infected mice contained highly abundant vsRNAs with approximately equal ratios of positive and negative strands and the 22-nt as the most dominant species for both polarities. Moreover, B2-bound 22-nt vsRNAs were overwhelmingly enriched for 22-nt canonical siRNA duplexes with 2-nt 3′ overhangs without 1U preference (Fig 2E). Argonaute-bound vsiRNAs were also detectable in NoV-infected mice, but were approximately 25-fold less abundant in NoV-infected mice than those in NoVΔB2-infected mice (Fig 2B and 2D and S1 Table). These findings reveal Dicer-mediated production of vsiRNAs from viral dsRNA replicative intermediates in NoV-infected mice, which were sequestered by B2 before Argonaute loading.
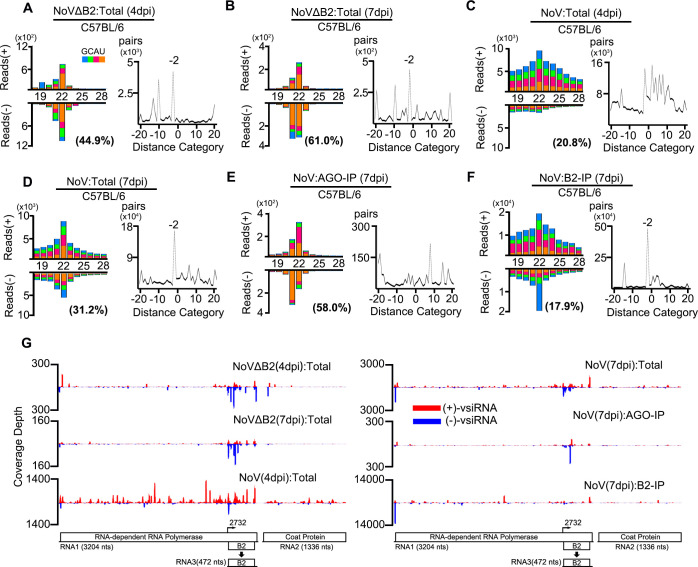
C57BL/6 suckling mice developed a delayed lethal disease after NoV infection compared to BALB/c mice (see below), which allowed us to sequence total and co-immunoprecipitated small RNAs from suckling mice at both 4 and 7 dpi (S2 Fig). As in BALB/c mice (Fig 2), we observed vsiRNA production and the interference of vsiRNA biogenesis in C57BL/6 mice 4 days after infection with NoVΔB2 and NoV, respectively (Fig 3). The vsiRNAs remained abundant in NoVΔB2-infected mice at 7 dpi (Fig 3B). In contrast to the earlier time points in BALB/c and C57BL/6 (Figs 2C and 3C), however, NoV-infected mice at 7 dpi accumulated highly abundant vsRNAs exhibiting clearly visible properties of vsiRNAs, including a dominant peak at 22 nt for both strands and strong enrichment for 22-nt vsiRNA duplexes with 2-nt 3’ overhangs (Fig 3D). The percentage of 21- to 23-nt vsiRNAs relative to total mature miRNAs from NoV-infected C57BL/6 mice was able to reach 7.1% (S1 Table). The 22-nt canonical vsiRNA duplexes accumulated to high levels in B2 complexes from NoV-infected C57BL/6 mice (Fig 3F), providing further evidence for in vivo B2 sequestration of duplex vsiRNAs. Nevertheless, the Argonaute immunoprecipitants from NoV-infected C57BL/6 mice at 7 dpi contained much more abundant 1U vsiRNAs than those from NoV-infected BALB/c mice at 3 dpi (Figs 2D and 3E). Together, our findings provide the first evidence for the in vivo production and sequestration of canonical duplex vsiRNAs by VSR-B2 in response to the infection with a VSR-expressing wild type virus.
Fig 3. Abundant vsiRNAs and their sequestration by VSR-B2 in C57BL/6 mice infected with wild-type NoV.
A-F. Size distribution of 18- to 28-nt virus-derived small RNAs (left) and duplex pattern of the 22-nt vsiRNAs (right) sequenced from NoVΔB2 and NoV-infected C57BL/6 suckling mice at 4 (A, C) or 7 dpi (B, D), either with or without co-IP by antibodies specific to mouse AGOs proteins (E) or the viral B2 protein (F). G. The distribution of 21- to 23-nt vsiRNAs along the viral genomic RNAs 1 and 2 from the six libraries presented from (A) to (F). Reads are shown as per million total 18- to 28-nt reads. Data information: Same as in Fig 1.
In cis expression of NoV VSR-B2 from Sindbis virus interfered with vsiRNA biogenesis and enhanced virus accumulation in vivo
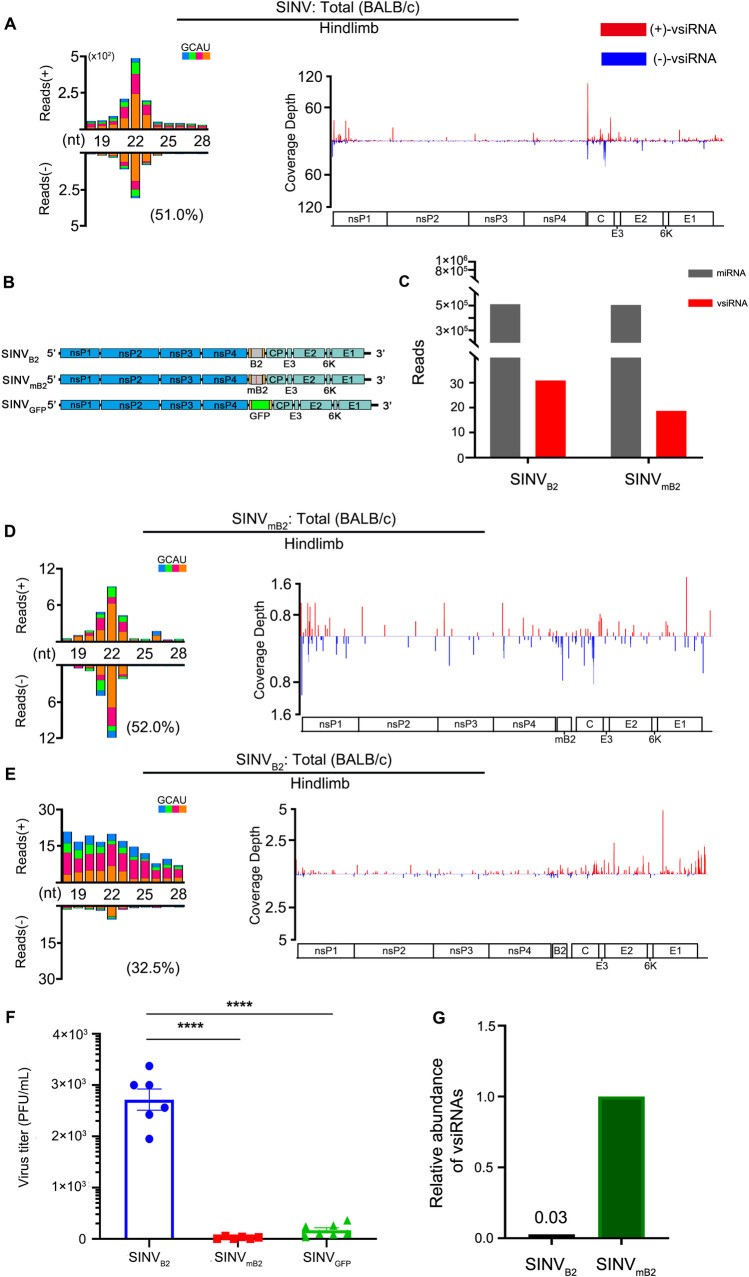
We further determined whether the B2 protein of NoV acts as a functional VSR when expressed from a heterologous positive-strand RNA virus, Sindbis virus (SINV), recently documented to trigger production of vsiRNAs in infected brain tissues of suckling mice after intracranial inoculation [28]. We first cloned and sequenced the total small RNAs from hind limb muscle tissue of BALB/c suckling mice inoculated by intraperitoneal injection. The profile of SINV-derived small RNAs exhibited the “hallmark” of vsiRNAs, with the 22-nt vsiRNAs as the most abundant population (Fig 4A), suggesting lack of strong viral suppression of vsiRNA biogenesis in wild-type SINV-infected mice as has been shown in mosquitoes [60].
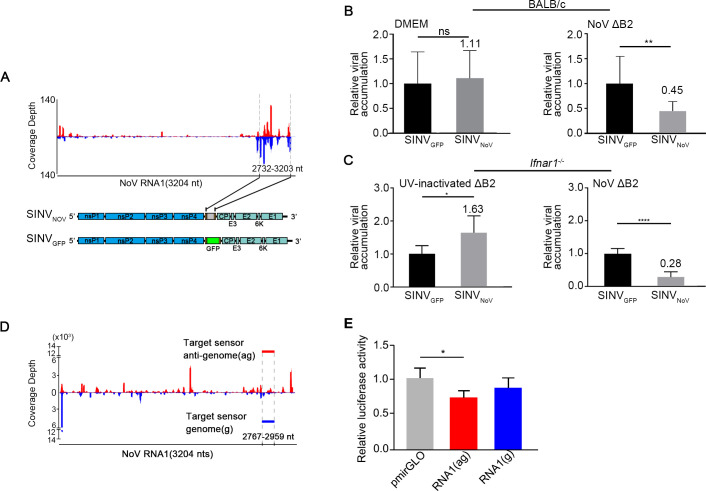
Fig 4. In vivo function of a heterologous VSR expressed in cis from Sindbis virus genome.
A, D, and E: Virus-derived small RNAs produced by BALB/c suckling mice in response to the infection with wild-type (A) or recombinant SINV expressing wild-type (E) or mutant (D) VSR-B2 of NoV by intraperitoneal injection at 3 dpi. Size distribution of 18- to 28-nt virus-derived small RNAs (left) and the distribution of 21- to 23-nt vsiRNAs along the viral genomic RNA (right) were shown as per million total mature miRNAs. B. The genomic structure of SINVB2, SINVmB2, and SINVGFP. C. Relative abundance of mature miRNAs and 21- to 23-nt vsiRNAs sequenced from BALB/c suckling mice infected with SINVB2 or SINVmB2 at 3 dpi. Reads counts were shown as per million total 18- to 28-nt reads. F. Viral titer (PFU/ml) in the hindlimb of 6 to 7 individual BALB/c suckling mice infected with recombinant SINVB2, SINVmB2, or SINVGFP at 3dpi was measured by a standard plaque assay and normalized by tissue mass. **** indicates p<0.0001, Student’s t-test. G. Relative abundance of cellular miRNAs and 21- to 23-nt vsiRNAs sequenced from BALB/c suckling mice infected with SINVB2 or SINVmB2 at 3 dpi. Reads counts were normalized by both per million total 18- to 28-nt reads and virus accumulation levels determined by RT-qPCR.
We next constructed three SINV recombinants for mouse infection (Fig 4B). We found that the hind limb muscle tissue of BALB/c mice also accumulated a typical population of vsiRNAs after intraperitoneal injection with SINVmB2, a recombinant SINV engineered to express a non-functional B2 mutant (mB2) with a single amino acid substitution (Fig 4D). In contrast to infection with SINV or SINVmB2, deep sequencing of small RNAs from mice infected with SINVB2 engineered to express wild-type B2, revealed a population of mostly positive-strand vsRNAs without the size preference of vsiRNAs (Fig 4E), indicating that in cis expression of VSR-B2 interfered with the biogenesis of vsiRNAs during SINVB2 infection. Furthermore, viral plaque assays showed that SINVB2 replicated to significantly enhanced levels in the infected mice than either SINVmB2 or SINVGFP, engineered to express GFP from the same genomic position of SINV as B2 (Figs 4F and S3). Although total 21–23 nt vsRNA reads in mice infected with SINVB2 were slightly more abundant than those in SINVmB2-infected mice (Fig 4C), they were much less abundant in SINVB2-infected mice than SINVmB -infected mice after normalization by viral accumulation (Fig 4G). These results indicate that in cis expression of a functional VSR-B2 from SINV genome interfered with the biogenesis of vsiRNAs and enhanced the accumulation of the heterologous virus in the infected suckling mice, suggesting in vivo inhibition of SINV infection by antiviral RNAi.
Biological activity of mammalian vsiRNAs produced in the response to NoV infection in vivo
We next investigated whether in vivo production of vsiRNAs mediates specific inhibition of virus accumulation. Production of vsiRNAs confers resistance in plants against heterologous viruses engineered to contain a segment from the vaccinating virus [61]. Sequence specific degradation of chimeric reporter mRNAs has been observed in cultured cells induced to produce complementary vsiRNAs [25]. We recently took a similar approach to determine the homology-dependent viral RNA degradation in vivo since this strategy avoids the unintended consequences of the genetic manipulation of Dicer or Argonaute-2 on endogenous miRNAs and development [28,62]. In this work, we investigated whether production of SINV-targeting vsiRNAs in mice prior to SINV inoculation could enhance the inhibition of SINV infection.
We constructed a recombinant Sindbis virus, SINVNoV, which contained an insert corresponding to a region of NoV genomic RNA1 targeted by high densities of vsiRNAs in NoVΔB2-infected mice (Fig 5A). It is predicted that SINVNoV, but not SINVGFP used as a control, will be specifically targeted for RNAi by the vsiRNAs produced in the mice after infection with NoVΔB2. Indeed, we found that SINVNoV accumulated to significantly lower levels than SINVGFP in mice pre-inoculated with NoVΔB2, but not with buffer DMEM (Fig 5B). Infection with live NoVΔB2 also induced significant inhibition on the accumulation of SINVNoV compared to that of SINVGFP in type I IFN receptor knockout mice (Ifnar1-/-, Fig 5C), which are defective in the signaling by type I IFNs [63]. It appeared that NoVΔB2-induced suppression of SINVNoV accumulation was more effective in Ifnar1-/- mice than BALB/c mice (Fig 5B and 5C). For unknown reason, however, SINVNoV replicated to higher levels than SINVGFP in Ifnar1-/- mice immunized with UV-inactivated NoVΔB2 (Fig 5C). We propose that prior production of SINVNoV-targeting vsiRNAs induced by NoVΔB2 immunization in BALB/c and Ifnar1-/- suckling mice directed suppression of virus infection by RNAi in a mechanism similar to the sequence-specific protection from incoming RNA viruses in Ifnar1-/- MEFs induced by transfection with long dsRNA [42].
Fig 5. The viral siRNAs of NoV inducing homology-dependent viral RNA degradation.
A. The genomic structure of a recombinant SINV (SINVNoV) carrying a segment of NoV RNA1 known to be targeted by high densities of vsiRNAs in NoVΔB2-infected mice shown at the top. Also shown below was the genomic structure of SINVGFP introduced above. B. In vivo genomic RNA levels of SINVNoV and SINVGFP one day after challenge inoculation of NoVΔB2-infected (right) or DMEM mock-infected (left) BALB/c suckling mice (n = 7 per DMEM group, n = 10 per NoVΔB2 group). The viral genomic RNA accumulation level was determined by RT-qPCR amplification of the viral nsP2 coding region and normalized by endogenous actin mRNA with SINVGFP level set as 1. Error bars represent SD. ** indicates p<0.01 (Student’s t-test). C. In vivo genomic RNA levels of SINVNoV and SINVGFP measured as in (B) one day after challenge inoculation of Ifnar1-/- suckling mice (n = 5~7 per group) inoculated two days earlier with live (right) or UV-inactivated (left) NoVΔB2. Error bars represent SD. * indicates <0.05, **** indicates p<0.0001 (Student’s t-test). The viral RNA accumulation of SINVGFP was set as 1. D. Diagram showing the genomic position of the 200-nt NoV sequence in the genome (g, blue) or anti-genome (ag, red) sense inserted into the 3′ untranslated region (UTR) of firefly luciferase reporter mRNA, to be targeted respectively by antisense and sense vsiRNAs extracted from VSR-B2 immuno-precipitants of NoV-infected C57BL/6 suckling mice. E. Relative luciferase activity of the control and chimeric reporter constructs with a 3’-UTR containing a NoV sequence targeted by the positive- or negative-strand vsiRNAs sequestered in VSR-B2 complex. Error bars indicate standard deviation of three replicates. Error bars represent SD. * indicates p<0.05 (Student’s t-test).
We also investigated the silencing activity of the vsiRNAs produced in NoV-infected mice using an mRNA reporter approach similar to that described by Qiu et al (2017). A vsiRNA-targeted fragment of NoV RNA1 in sense or antisense orientation was inserted into the 3′ UTR of a dual luciferase reporter plasmid (Fig 5D). Human 293T cells were transfected with one of the reporter constructs together with the NoV-specific vsiRNAs extracted from B2 immunoprecipitants obtained from NoV-infected C57BL/6 suckling mice as described above (Fig 3F). Although we observed suppression of the luciferase reporter containing the NoV fragment in either orientation compared to the control 24 hours after transfection, the difference was statistically significant only for the luciferase reporter targeted by antisense vsiRNAs (Fig 5E), indicating gene silencing activity for the vsiRNAs made by C57BL/6 mice in response to NoV infection and sequestered by VSR B2.
LGP2 inhibits Dicer-mediated biogenesis of vsiRNAs from dsRNA-vRIs
Finally, we examined the role of LGP2 encoded by DHX58 gene in NoV infection in vivo, which has recently been shown to block artificial long dsRNA-induced RNAi in IFNAR knockout MEFs by inhibiting Dicer processing of long dsRNA into siRNAs [43]. Both C57BL/6 suckling mice and Dhx58-/- suckling mice on a pure C57BL/6 background [12] were infected with the same dose of NoV and NoVΔB2 by intraperitoneal injection. We monitored mouse survival and virus accumulation in the limb muscular tissues of the infected mice (Fig 6). NoVΔB2 induced no signs of disease in both genotypes of mice; however, Dhx58-/- mice survived approximately 1 or 2 days longer than C57BL/6 mice after NoV infection and the difference in survival was statistically significant (Fig 6A). Northern and Western blotting as well as quantitative RT-PCR revealed that NoV accumulated to lower levels in Dhx58-/- mice than wild-type mice at 4 dpi (Fig 6B–6D). Moreover, although NoVΔB2 was cleared in both genotypes of mice, Dhx58-/- mice also supported lower levels of NoVΔB2 accumulation than C57BL/6 mice at 4 dpi (Fig 6B–6D). These results suggest that LGP2 expression enhanced the in vivo accumulation of both NoV and NoVΔB2.
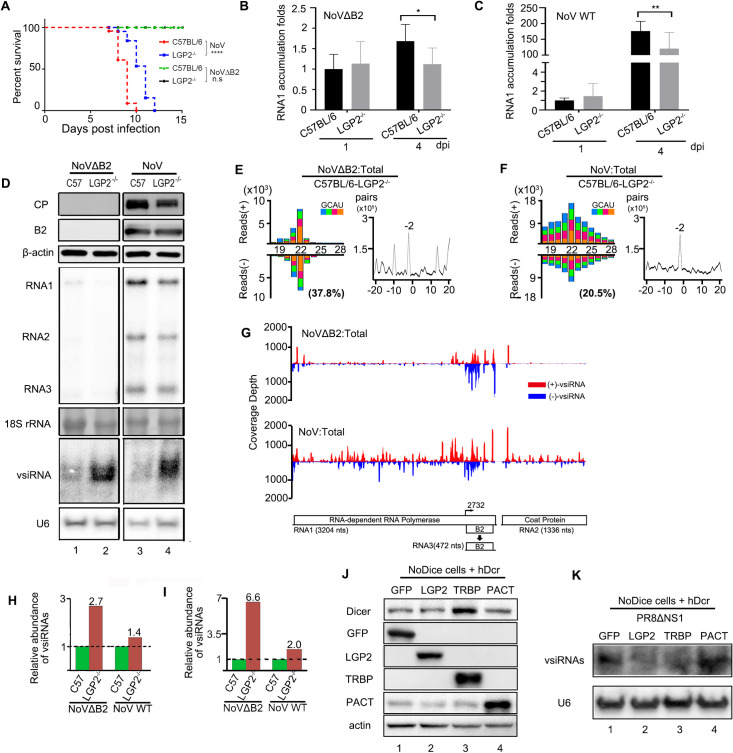
Fig 6. Enhanced production of vsiRNAs and reduced virulence of NoV in Dhx58-/- suckling mice compared to parental C57BL / 6 mice.
A. Survival curve of C57BL/6 and Dhx58-/- suckling mice inoculated with WT NoV. Data analysis was performed using GraphPad Prism. n = 12~14. Log-rank (Mantel-Cox) test. **** indicates p<0.0001. B, C RNA1 accumulation level of NoVΔB2 (B) or WT NoV (C) measured by RT-qPCR from hindlimb of C57BL/6 or Dhx58-/-suckling mice at 1 or 4 dpi. The RNA1 level of NoVΔB2 infected C57BL/6 suckling mice at 1 dpi was set as 1. Error bars indicate standard deviation of three replicates. * indicates p<0.05, ** indicates p<0.01 (Student’s t-test). D. Western blotting detection (upper 3 panels) of the viral coat protein (CP) and B2 protein in C57BL/6 or Dhx58-/- suckling mice infected with NoVΔB2 or NoV at 4 dpi. Detection of the endogenous β-actin was used as a loading control. Northern blotting detection (lower 4 panels) of viral genomic RNAs and vsiRNAs in C57BL/6 and Dhx58-/- suckling mice inoculated with NoVΔB2 or NoV at 4 dpi. Detection of 18S rRNAs and U6 were used as loading controls. E, F. Size distribution 18- to 28-nt virus-derived small RNAs (left) and duplex pattern of the 22-nt vsiRNAs (right) from NoVΔB2 (E) or WT NoV (F) infected Dhx58-/- suckling mice at 4 dpi. Reads shown as per million total mature miRNAs. The 5’ terminal nucleotide and 1U % of 21- to 23-nt vsiRNAs were indicated. G. The distribution of 21- to 23-nt vsiRNAs along the viral genomic RNAs 1 and 2 from the libraries of (E) and (F). H, I. Relative abundance of 21- to 23-nt vsiRNAs sequenced from C57BL/6 and Dhx58-/- suckling mice inoculated with NoVΔB2 or NoV at 4dpi. Reads were normalized either by mature miRNAs only (H) or by both mature miRNAs and virus titer determined by RT-qPCR (I). J, K. Western blotting detection of GFP, LGP2, TRBP, and PACT proteins (J) and Northern blotting of vsiRNAs (K) in PR8/delNS1-infected NoDice cells ectopically co-expressing hDcr with GFP, LGP2, TRBP or PACT. Detection of the endogenous β-actin protein and U6 rRNA were used as loading controls. Each experiment was repeated at least three times independently.
The induction of IFNβ, DHX58 and several other ISGs were verified in suckling mice after infection with either NoV or NoVΔB2 (S4–S6 Figs). Deep sequencing of small RNAs showed that whereas both the strand ratio and the size preference of the vsiRNAs were similar, the relative abundance of vsiRNAs triggered by NoVΔB2 infection was higher in Dhx58-/- mice (2.7%) than in wild-type mice (1.0%) (Figs 3A and 6E and S1 Table). Northern blotting of small RNAs further confirmed that NoVΔB2 infection triggered an increased accumulation of vsiRNAs in Dhx58-/- mice compared to wild-type mice even though NoVΔB2 replicated to lower levels in Dhx58-/- mice (Fig 6D). Furthermore, several properties of vsiRNAs were readily visible for the total vsRNAs sequenced from NoV-infected Dhx58-/- mice at 4 dpi (Fig 6F), unlike those from NoV-infected wild-type C57BL/6 mice at 4 dpi (Fig 3C). These included a dominant 22-nt peak for both the positive and negative strands, an increased abundance of the negative strands (37.4% vs 14.3% in wild-type mice), an increased preference in the size range of 21- to 23-nt (46.1% vs 39.1%), and strong enrichment of a single dominant population of 22-nt canonical vsiRNA duplexes with 2-nt 3’ overhangs (Figs 3C and 6F and S1 Table). Stronger signals in the size range of vsiRNAs were detectable by Northern blotting for RNA samples extracted from NoV-infected Dhx58-/- mice than wild-type mice (Fig 6D). vsiRNAs were more abundant in Dhx58-/- mice than C57BL/6 mice infected with either NoVΔB2 or WT NoV (Fig 6H), particularly after normalization by virus titer (Fig 6I). These results suggest that LGP2 inhibited Dicer processing of the dsRNA-vRIs into vsiRNAs during the in vivo infection with both NoV and NoVΔB2.
We further compared the production of the influenza vsiRNAs triggered by PR8/delNS1 infection in NoDice 293T cells expressing hDcr together with GFP or LGP2 (Fig 6J and 6K). Northern blotting revealed strong inhibition of the influenza vsiRNA accumulation by LGP2 compared to the control (Fig 6K). In this assay (Fig 6K), the biogenesis of the influenza vsiRNAs appeared to be suppressed by TAR-RNA binding protein (TRBP), but not the protein activator of PKR (PACT), both of which are dsRNA-binding proteins involved in Dicer-mediated miRNA biogenesis and formation of the RISC complex. Our findings indicate that LGP2 inhibits Dicer-mediated biogenesis of vsiRNAs from dsRNA-vRIs, revealing another similarity between RNAi induced by dsRNA-vRIs and artificial long dsRNA [43].
Discussion
Results from this work reveal several new insights into the induction and suppression of Dicer-mediated production of mammalian vsiRNAs during infection of mammalian somatic cells. We found that influenza vsiRNA precursors became the more dominant Dicer substrates than pre-miRNA hairpins in mature human cells when both vsiRNAs and cellular miRNAs were processed by human Dicer expressed de novo. Unlike synthetic long dsRNA identified as less efficient Dicer substrates than pre-miRNA hairpins from previous in vitro biochemical studies [34,35], therefore, our findings show that the dsRNA-vRIs made by viral RdRP are not inherently poor substrates of human Dicer in the infected cells.
Our results provide the first evidence that canonical vsiRNAs processed from dsRNA-vRIs were produced in two strains of IFN-competent suckling mice after wild-type virus infection. Similar to previous studies on cell culture infections with a range of wild-type viruses [20–28,45–50], we were not able to find canonical vsiRNAs from NoV-infected mice at 3 or 4 dpi by deep sequencing of total small RNAs. However, the hallmarks of vsiRNAs were clearly visible when the same samples were enriched by VSR-B2 co-immunoprecipitation before sequencing. These VSR-sequestered vsiRNAs made in vivo directed gene silencing of a luciferase reporter in human cells, providing the first example for in vivo sequestration of functional vsiRNAs by a mammalian VSR. Interestingly, canonical vsiRNAs became readily detectable by deep sequencing without prior VSR-B2 co-immunoprecipitation in NoV-infected wild-type C57BL/6 mice by 7 dpi. In these infected mice, abundant vsiRNAs were loaded into RISC and the Argonaute-bound vsiRNAs exhibited strong selection for 1U vsiRNAs of 21 to 23 nucleotides in length, similarly to those found in NoVΔB2-infected wild-type suckling mice, cultured cells [29] or mutant adult mice defective in the adaptive immunity reported recently by Han et al [44]when this manuscript was under preparation. These findings support our earlier conclusion that NoV-infected mice produce RNA slicing-competent vsiRNAs-RISC [44]. Our results suggest that examining in vivo infection at different time points or removing non-specific small RNAs by Argonaute or VSR co-immunoprecipitation may facilitate detection of vsiRNAs by deep sequencing.
Our characterization of suckling mouse infection with SINV recombinants expressing VSR-B2 (SINVB2) or a segment of NoV genomic RNA 1 (SINVNoV) indicated an in vivo function of the vsiRNA response induced by dsRNA-vRIs. Similar to the infection of mosquitoes active in antiviral RNAi [60], we found that SINVB2 accumulated to significantly higher levels in BALB/c mice than both SINVGFP and SINVmB2, which expressed GFP or a mutant B2 protein unable to interfere with the biogenesis of the vsiRNAs. By contrast, SINVNoV replicated to significantly lower levels than SINVGFP in the suckling mice accumulating abundant vsiRNAs to target the inserted NoV sequence due to prior infection/immunization with NoVΔB2. Together, our findings suggest an in vivo antiviral activity of the vsiRNAs triggered by infection with SINV recombinants or NoVΔB2.
Finally, we found that over-expression of the ISG factor LGP2 in human 293T cells potently inhbited Dicer processing of the influenza dsRNA-vRIs into vsiRNAs. Thus, LGP2 inhibits Dicer processing of both dsRNA-vRIs in 293T cells shown in this work and artificial long dsRNA reported previously in IFN-deficient MEFs [43]. As expected, infection of suckling mice with either NoV or NoVΔB2 induced expression of the LGP2 gene Dhx58. Notably, both viruses replicated to lower levels, but triggered production of vsiRNAs at higher levels in Dhx58-/- mice than wild-type mice. These findings suggest negative modulation of antiviral RNAi by LGP2, which may explain why immunization with NoVΔB2 induced weaker suppression of SINVNoV accumulation in wild-type mice than Ifnar1-/- mice defective in the IFN-mediated induction of Dhx58. As noted previously [43], our findings also support the functional similarity of LGP2 with one of the three RLRs encoded by the single-Dicer nematode Caenorhabditis elegans, Dicer-related helicase 2 (DRH-2), which suppresses antiviral RNAi and lacks the N-terminal domain conserved in DRH-1 and DRH-3 [64,65]. Together, our findings further highlight the differences and similarities in the induction of RNAi by the dsRNA-vRIs made by viral RdRP and artificial long dsRNA in mammalian somatic cells.
Materials and methods
Ethics statement
All animal experiments in China were performed in Fudan University of China and under the guidelines of the Institutional Animal Care and Use Committee. Animals in USA were housed in the Animal Resources Facility according to the guidelines described under the federal Animal Welfare Regulations Act and the procedures were approved by the Institutional Animal Care and Use Committee at the University of California, Riverside.
Viruses and cell culture
Wildtype Nodamura virus (NoV) and mutant NoVΔB2 strains used in this study were described previously [23]. Influenza A virus (IAV), PR8/delNS1 (NS1 deletion mutant) was gift from Dr. A. Garcıa-Sastre and Dr. P. Palese. Sindbis virus (SINV) was rescued from the plasmids as previously described [28]. The NoDice Human embryonic kidney (293T) cell line and its parental 293T cell line were gifts from from Dr. B. Cullen. African green monkey kidney epithelial cells (Vero) were purchased from the American Type Culture Collection (ATCC). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum at 37°C with 5% CO2.
Animals
Experimental animals, including BALB/c, C57BL/6 and Ifnar1-/-, were purchased from Jackson labs (Bar Harbor, ME), Shanghai SLAC laboratory Animal Co., Ltd., and Cyagen Biosciences. Dhx58-/- mice was gift from Dr. Michael Gale, Jr., University of Washington.
Plasmids
The sequences encoding NS1 of IAV-WSN or ZIKV were amplified by reverse transcription-polymerase chain reaction (RT-PCR) using specific primers containing the restriction enzyme sites and cloned into pcDNA3.1(-) vector to generate plasmids expressing NS1 protein of IAV and ZIKV. The expression plasmids for human Dicer (hDcr) were purchased from Addgene (cat. # 19873, #41584 and #41585). The plasmids expressing EGFP and human LGP2, TRBP, PACT were constructed into pEF-BOS entry vector using standard molecular cloning technique. The pTE/5’2J/GFP and pTE/5’2J were gifts from Dr. C.M. Rice. To obtain recombination SINV virus expressing B2 protein of NoV and sequence of RNA1, we constructed the plasmids of pTE/5’2J/B2, pTE/5’2J/mB2 and pTE/5’2J/NoV by ligating PCR products of NoV B2, mB2 (mutant B2, R59Q) and NoV RNA1 (2732–3203 nt) flanked by Xba I and Apa I into the MCS of pTE/5’2J, which was used to rescue recombinant SINV virus of SINVB2, SINVmB2, SINVNOV as previously described [28]. The pmirGLO Dual-Luciferase miRNA Target Expression Vector was purchased from promega, which encode the firefly luciferase and Renilla luciferase genes. The pmirGLO-RNA1(ag)/RNA1(g) reporters were designed by cloning part of the viral genome sequence NoV (anti-genome and genome of 2767–2959 nt respectively) into pmirGLO plasmid using Xba I sites and ClonExpress II One Step Cloning Kit (Vazyme). Primers used are listed in S2 Table. All plasmids were confirmed by sequencing.
Western and Northern blotting analyses
Western and Northern blotting analysis was performed as described previously [29,66]. Antibodies to IAV-NS1, NoV B2 protein and coat protein (Capsid) were described previously [23,66], antibodies to Dicer, LGP2, GFP, TRBP and PACT (Santa Cruz Biotechnology, sc 136979, sc 134668, sc 9996, sc 100909, sc 81569), ZIKV-NS1 (GeneTex) are sourced from commercial suppliers. Probes used for Northern blotting to detect miRNA are listed in S2 Table and to measure vsRNAs were shown previously [23,29].
Cell culture transfection and infection
NoDice 293T cells (2.5 × 106 per 6 cm plate) were seeded one day before transfection. Cells were transfected with different recombination plasmids, including 8 μg plasmid encoding hDicer and mock or 4 μg following plasmids, pcDNA-IAV-NS1, pcDNA-ZIKV-NS1, pEF-BOS-GFP, pEF-BOS-LGP2, pEF-BOS-TRBP and pEF-BOS-PACT using Lipofectamine 2000 (Thermo) according to the supplier’s recommended protocol and then infected by PR8/delNS1 (MOI = 1) at six hours after transfection. The infected cells were harvested for the extraction of total protein and RNA using TRIzol 24 hours after infection.
Preparation of long dsRNA
The dsRNAs used for in vitro assay of Dicer activity were prepared as described before [42]. Briefly, to generate (+)-sense RNA GFP and (-)-sense RNA GFP, two PCR fragments were amplified from pcDNA3.1-GFP plasmid (Promega) corresponding to the first 200 nt of GFP. The PCR products were purified using QIAquick PCR purification kit (Qiagen), and in vitro transcription (IVT) with T7 RNA polymerase (T7 MEGAscript kit, Ambion) was performed according to the manufacturer’s instructions. The dsRNAs were generated by mixing an equal volume of (+)-sense IVT RNA and (-)-sense IVT RNA, incubation at 95°C for 10 min followed by cool down to RT. The dsRNAs were purified and separated on a 2% agarose gel to verify size and integrity.
In vitro assay of Dicer activity
Human Flag-Dicer and Flag-mutant-Dicer in pCAGGS (Addgene #41584 and #41585) were transfected into NoDice 293T cells using Lipofectamine 2000. Mutant Dicer construct? NoDice 293T cells were seeded in a 6-well plate at a density of 5 × 105 per well one day before transfection. NoDice 293T cells were transfected with 3 μg plasmids. 48 hours after transfection, hDcr-KO 293T cells were infected by PR8/delNS1 (MOI = 1) or DMEM and the infected cells were lysed in cell lysis buffer (CST) 24 hours after infection. The Flag-Dicer was retrieved using FLAG beads (GenScript) according to the manufacturer’s instructions. For dicing assays, Flag-Dicer was incubated with 50 nM dsRNA in dicing buffer [30 mM Tris pH 6.8, 50 mM NaCl, 3 mM MgCl2, 5% glycerol, 1 mM DTT, RNAsin (Promega)] for 1 h at 37°C followed by Trizol purification. The RNA was resuspended in formamide sample buffer without xylene blue (47.5% formamide, 0.01% SDS, 0.01% bromophenol blue, 0.5 mM EDTA), loaded onto a 15% TBE-Urea gel and visualised by GelRed staining.
NoV and SINV infection
NoV or NoVΔB2 preparations shown to contain 7x106 copies of genomic RNA1 from the titrated set of stocks was inoculated to each of suckling mice of 6 to 8 days old after birth by intraperitoneal injection (i.p.) as described previously [23]. 50 PFU of SINV was inoculated to each of suckling mice of 6 to 8 days old after birth by intraperitoneal injection. Total RNAs and proteins were purified from the hind limb muscle tissues of infected suckling mice at different time.
Co-immunoprecipitation
Hind limb muscle tissue of suckling mice infection with NoV or NoVΔB2 were co-immunoprecipitated (co-IP) by anti-pan Argonaute (Ago) antibody (Millipore, Billerica, MA) and by B2 antibodies as described. Briefly, 3 μg of rabbit or mouse IgG and 15 μl of protein A/G PLUS-Agarose beads (Santa Cruz Biotechnology) were incubated with 100 μg of muscle tissue lysates in 1 ml RIPA. After pre-cleared, 3 μg of anti-pan Ago, B2 antibodies, or controls IgG antibody immobilized to protein A/G PLUS-Agarose beads were added for 2 hours at 4°C. The total RNAs obtained from precipitated complexes were used for the construction of small RNA libraries as described [23]. Due to the extremely low RNA content in the controls IgG antibody immunoprecipitants, these samples were not included in subsequent library construction and sequencing.
Viral plaque assays
In brief, hindlimb muscles were isolated from infected or mock infected suckling mice. After homogenization, 1mL DMEM were added to per 50mg tissue. The tissue homogenates were used for plague assay. Vero cells were plated at a density of 5.0 × 105 cells/ well in 3 ml DMEM, 10% FBS, on 6-well plates and incubated at 37°C, 5% CO2 atmosphere. On the next day, 1ml of 10-fold gradient dilutions of supernatants were added to each well. After incubation of 1 h, supernatants were discarded and the cells were overlaid with 5mL 1×MEM containing 0.5% agarose in each well. After incubation of 2 days, the agarose gels were removed, and cell layer was stained with commassie brilliant blue. Virus content of the supernatants was calculated as plaque forming units (PFU)/ml.
In vivo recombinant SINV reporter experiments
For BALB/c suckling mice, seven-day-old mice were inoculated by i.p. with NoVΔB2 (shown to contain 7x106 copies of genomic RNA1) or with the same volume of DMEM (mock). Two days after inoculation, the mice were infected by i.p. with SINVGFP or SINVNoV viruses of 500 PFU. Each group of suckling mice were euthanized one day after SINV infection to determine virus titers in the hind limb tissue by RT-qPCR. For Ifnar1-/- mice, seven-day-old mice were inoculated by i.p. with NoVΔB2 (shown to contain 7x106 copies of genomic RNA1) or with UV-inactivation NoVΔB2. Two days after inoculation, Ifnar1-/- mice were infected by i.p. with SINVGFP or SINVNoV viruses of 500 PFU and euthanized one day after SINV infection to determine virus titers in the hind limb tissue by RT-qPCR.
Luciferase reporter assay
293T cells were seeded in a 12-well plate at a density of 5 × 105 per plate one day before transfection. Cells were transfected with 1 μg of pmirGLO-Control vector or pmirGLO-RNA1(ag)/RNA1(g) vector, together with 0.3 μg of total small RNA extracted from B2 immunoprecipitants described above, with TransIT-TKO Transfection Reagent (Mirus). Luciferase activity was analyzed using the Dual-Luciferase Reporter Assay System (Cat. #E1910, Promega) and measured on Fluoreskan Ascent FL (Thermo Scientific) after 24 hours transfection, and the firefly luciferase values were divided by Renilla luciferase values to normalize for transfection efficiency.
RT-qPCR
Total RNA was extracted using TRIzol reagent following the manufacturer’s instructions (Thermo Fisher Scientific). 1 μg of RNA was reverse transcribed using PrimeScript RT Reagent Kit with gDNA Eraser (Takara), and qPCR was performed using iQ SYBR Green Supermix (Bio-rad). Reactions were carried out using CFX-Connect Real-Time System (Bio-rad). Relative expression values were calculated using the ΔΔCt method normalized by β-actin housekeeping gene. All primers used for RT–qPCR were listed in S2 Table.
Statistical analysis
All statistical analysis of RT-qPCR data were performed by GraphPad Prism 8 using unpaired Student’s t test. All experiments were repeated independently at least three times. A P value of <0.05 was considered statistically significant.
RNA-seq and data analysis
Total RNA was extracted from infected and mock hind limb muscle tissue using TRIzol reagent. The integrity of the purified RNA was analyzed by the Agilent 2200 Electrophoresis Bioanalyzer System (Agilent Technologies). Enrichment of poly (A)-RNA preparation for RNA Sequencing was performed using NEBNext Poly (A) mRNA Magnetic Isolation Module (NEB) kit. The cDNA libraries were constructed for each pooled RNA sample using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina according to the manufacturer’s instructions. The products were purified and enriched by PCR to create the final cDNA libraries and quantified by Agilent2200. The tagged cDNA libraries were pooled in equal ratio and used for 150 bp paired-end sequencing in a single lane of the Illumina HiSeqXTen. Clean reads were obtained from the raw reads by removing the adaptor sequences, reads with > 5% ambiguous bases (noted as N) and low-quality reads containing more than 20 percent of bases with qualities of < 20. The clean reads were then aligned to mouse genome (version: GRCh38 NCBI) using the hisat2 [67]. HTSeq software was used to count the number of reads mapped to each gene [68]. Differential expression analysis of any two groups was performed using the DESeq package [69] and the differentially expressed genes between samples were identified after the significant analysis and false discovery rate (FDR) analysis under the following criteria [70]: (1) fold change > 2 or < 0.5; (2) FDR< 0.05. Gene Ontology (GO) analysis was applied to analyze the primary functions of the differentially expressed genes according to the GO [71]. Fisher’s exact test was applied to identify the significant GO categories and FDR was used to correct the p-values.
Construction of small RNA libraries
RNA preparations in this study were used for the construction of small RNA libraries by the method that depends on the 5’ monophosphate of small RNAs as described previously with the TruSeq Small RNA Sample Preparation Kit of Illumina (San Diego, CA) [29].
Deep sequencing and bioinformatic analysis of small RNAs
Libraries of small RNAs were cloned from the RNA samples (mice n = 3; cell samples, repeat once) and sequenced by Illumina HiSeq 2000/2500. 19 libraries in total were sequenced from this work (S1 Table). Mapping was done by Bowtie 1.1.2 with perfect match. Subsequent bioinformatics analysis of virus-derived small RNAs was carried out using in-house Perl scripts as described previously [29]. Pairs of complementary 22-nt vsiRNAs in each library with different base-pairing lengths were computed using a previously described algorithm [23]. The reference sequences used in this study are either identical with those described previously or as listed below:
NoV RNAs 1 and 2: AF174533.1 and AF174534.1
NoVΔB2 RNAs 1 and 2: the same as NoV except for 3 substitutions in RNA1: U2745C, U2754C, C2757G.
PR8/delNS1: Obtained from A/Puerto Rico/8/34 (H1N1) (PR8-WT) by deleting nucleotides 57 to 528 in the NS segment. The sequence of PR8-WT: AF389115.1, AF389116.1, AF389117.1, AF389118.1, AF389119.1, AF389120.1, AF389121.1 and AF389122.1.
SINV: j02363.1
Mature miRNAs and miRNA precursors: miRBase 21 (http://www.mirbase.org/).
Supporting information
A-C. PR8/delNS1-infected NoDice 293T cells ectopically expressing hDcr (A), hDcr+IAV-NS1 (B) and hDcr+ZIKV-NS1 (C). Genomic coverage depth of each nucleotide position by 21- to 23-nt vsiRNAs sequenced from RNA above. Reads are shown as per million total 18- to 28-nt reads.
(TIF)
A. RNA1 accumulation level of WT NoV or NoVΔB2 measured by RT-qPCR from hind limb of C57BL/6 suckling mice at 7 dpi (n = 5 per group). The RNA1 level of NoVΔB2 infected C57BL/6 suckling mice was set as 1. B. Expression level of NoV capsid and B2 protein measured by Western blotting. Actin were used as a loading control.
(TIF)
The genomic structure of SINVB2, SINVmB2 or SINVGFP (right) and relative viral accumulation determined by RT-qPCR from hindlimb of BALB/c suckling mice infected with SINVB2, SINVmB2 or SINVGFP at 3dpi. n = 6~7 per group. Error bars represent SD. * indicates p<0.05 (Student’s t-test). The viral RNA accumulation of SINVGFP was set as 1.
(TIF)
A, B. Expression levels of IFN (A) and RNAi (B) related genes in C57BL/6 suckling mice with NoV infection at 7dpi. All data were measured as the mean ± SD of three independent experiments. Asterisks indicate a significant difference level compared to control (Student’s t-test, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001).
(TIF)
A, B. Volcano plots showing demonstrate false discovery rate (FDR) and fold-change (FC) of gene expression levels determined by RNAseq comparing BALB/c suckling mice inoculated by NoVΔB2 vs Mock (A), WT NoV vs Mock (B). Genes up-regulated (fold change > 2 and FDR< 0.05) are indicated in red and those down-regulated (fold change < 0.5 and FDR< 0.05) are indicated in blue. C. Differential expression of IFN pathway related genes from mRNA-seq data of NoV or NoVΔB2 inoculated BALB/c suckling mice at 3 dpi. Fold changes (FC) of 2 or 0.5 (|log2FC| = 1) are indicated by dotted lines. The log2FC was taken as 20 when the ratio of experimental group and mock group tended to be infinite due to a small denominator.
(TIF)
A, B. Volcano plots showing demonstrate false discovery rate (FDR) and fold-change (FC) of gene expression levels determined by RNAseq comparing C57BL/6 suckling mice inoculated by NoVΔB2 vs Mock (A), WT NoV vs Mock (B). Genes up-regulated (fold change > 2 and FDR< 0.05) are indicated in red and those down-regulated (fold change < 0.5 and FDR< 0.05) are indicated in blue. C. Differential expression of IFN pathway related genes from mRNA-seq data of NoV or NoVΔB2 inoculated C57BL/6 suckling mice at 3 dpi. Fold changes (FC) of 2 or 0.5 (|log2FC| = 1) are indicated by dotted lines. The log2FC was taken as 20 when the ratio of experimental group and mock group tended to be infinite due to a small denominator.
(TIF)
(DOCX)
(DOCX)
Acknowledgments
The authors thank Dr. C. Rice, M. McDonald, B.R. Cullen, A. Garcia-Sastre, P. Palese and M. Gale, Jr. for providing materials.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.The RNA sequencing data have been deposited to the NCBI GEO database under the accession number GSE155146.
Funding Statement
This work was supported by the National Natural Science Foundation of China (91640111 and 31770179) and Innovation Program of Shanghai Municipal Education Commission (2017-01-07-00-07-E00015) to Y.L., NIH grants AI52447 and AI110579 and College of Natural and Agricultural Sciences, University of California, Riverside (to S.-W.D.). W-X.L. and S.-W.D. were supported by NIH grant AI141887 (to S.-W.D.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10(2):126–39. Epub 2009/01/24. doi: 10.1038/nrm2632 . [DOI] [PubMed] [Google Scholar]
- 2.Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Bio. 2019;20(1):5–20. WOS:000456596800006. [DOI] [PubMed] [Google Scholar]
- 3.Gebert LFR, MacRae IJ. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol. 2019;20(1):21–37. Epub 2018/08/16. doi: 10.1038/s41580-018-0045-7 ; PubMed Central PMCID: PMC6546304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15(8):509–24. Epub 2014/07/17. doi: 10.1038/nrm3838 . [DOI] [PubMed] [Google Scholar]
- 5.Baulcombe D. RNA silencing in plants. Nature. 2004;431(7006):356–63. Epub 2004/09/17. doi: 10.1038/nature02874 . [DOI] [PubMed] [Google Scholar]
- 6.Guo Z, Li Y, Ding SW. Small RNA-based antimicrobial immunity. Nat Rev Immunol. 2019;19(1):31–44. Epub 2018/10/12. doi: 10.1038/s41577-018-0071-x . [DOI] [PubMed] [Google Scholar]
- 7.Csorba T, Burgyán J. Antiviral Silencing and Suppression of Gene Silencing in Plants: Current Research Topics in Plant Virology; 2016. [Google Scholar]
- 8.Ding SW. RNA-based antiviral immunity. Nat Rev Immunol. 2010;10(9):632–44. Epub 2010/08/14. doi: 10.1038/nri2824 . [DOI] [PubMed] [Google Scholar]
- 9.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14(1):36–49. Epub 2013/12/24. doi: 10.1038/nri3581 ; PubMed Central PMCID: PMC4084561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu MM, Shu HB. Cytoplasmic Mechanisms of Recognition and Defense of Microbial Nucleic Acids. Annu Rev Cell Dev Biol. 2018;34:357–79. Epub 2018/08/11. doi: 10.1146/annurev-cellbio-100617-062903 . [DOI] [PubMed] [Google Scholar]
- 11.Rehwinkel J, Gack MU. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat Rev Immunol. 2020;20(9):537–51. Epub 2020/03/24. doi: 10.1038/s41577-020-0288-3 ; PubMed Central PMCID: PMC7094958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suthar MS, Ramos HJ, Brassil MM, Netland J, Chappell CP, Blahnik G, et al. The RIG-I-like receptor LGP2 controls CD8(+) T cell survival and fitness. Immunity. 2012;37(2):235–48. Epub 2012/07/31. doi: 10.1016/j.immuni.2012.07.004 ; PubMed Central PMCID: PMC3910444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esser-Nobis K, Hatfield LD, Gale M Jr. Spatiotemporal dynamics of innate immune signaling via RIG-I-like receptors. Proc Natl Acad Sci U S A. 2020;117(27):15778–88. Epub 2020/06/24. doi: 10.1073/pnas.1921861117 ; PubMed Central PMCID: PMC7354926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rothenfusser S, Goutagny N, DiPerna G, Gong M, Monks BG, Schoenemeyer A, et al. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J Immunol. 2005;175(8):5260–8. Epub 2005/10/08. doi: 10.4049/jimmunol.175.8.5260 . [DOI] [PubMed] [Google Scholar]
- 15.Venkataraman T, Valdes M, Elsby R, Kakuta S, Caceres G, Saijo S, et al. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J Immunol. 2007;178(10):6444–55. Epub 2007/05/04. doi: 10.4049/jimmunol.178.10.6444 . [DOI] [PubMed] [Google Scholar]
- 16.Uchikawa E, Lethier M, Malet H, Brunel J, Gerlier D, Cusack S. Structural Analysis of dsRNA Binding to Anti-viral Pattern Recognition Receptors LGP2 and MDA5. Mol Cell. 2016;62(4):586–602. Epub 2016/05/21. doi: 10.1016/j.molcel.2016.04.021 ; PubMed Central PMCID: PMC4885022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quicke KM, Kim KY, Horvath CM, Suthar MS. RNA Helicase LGP2 Negatively Regulates RIG-I Signaling by Preventing TRIM25-Mediated Caspase Activation and Recruitment Domain Ubiquitination. J Interferon Cytokine Res. 2019;39(11):669–83. Epub 2019/06/27. doi: 10.1089/jir.2019.0059 ; PubMed Central PMCID: PMC6820871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bruns AM, Leser GP, Lamb RA, Horvath CM. The innate immune sensor LGP2 activates antiviral signaling by regulating MDA5-RNA interaction and filament assembly. Mol Cell. 2014;55(5):771–81. Epub 2014/08/16. doi: 10.1016/j.molcel.2014.07.003 ; PubMed Central PMCID: PMC4156907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanchez David RY, Combredet C, Najburg V, Millot GA, Beauclair G, Schwikowski B, et al. LGP2 binds to PACT to regulate RIG-I- and MDA5-mediated antiviral responses. Science signaling. 2019;12(601). Epub 2019/10/03. doi: 10.1126/scisignal.aar3993. [DOI] [PubMed] [Google Scholar]
- 20.Ding SW, Han Q, Wang J, Li WX. Antiviral RNA interference in mammals. Curr Opin Immunol. 2018;54:109–14. Epub 2018/07/18. doi: 10.1016/j.coi.2018.06.010 ; PubMed Central PMCID: PMC6196099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maillard PV, van der Veen AG, Poirier EZ, Reis e Sousa C. Slicing and dicing viruses: antiviral RNA interference in mammals. EMBO J. 2019;38(8). Epub 2019/03/16. doi: 10.15252/embj.2018100941; PubMed Central PMCID: PMC6463209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berkhout B. RNAi-mediated antiviral immunity in mammals. Curr Opin Virol. 2018;32:9–14. Epub 2018/07/18. doi: 10.1016/j.coviro.2018.07.008 . [DOI] [PubMed] [Google Scholar]
- 23.Li Y, Lu J, Han Y, Fan X, Ding SW. RNA interference functions as an antiviral immunity mechanism in mammals. Science. 2013;342(6155):231–4. Epub 2013/10/12. doi: 10.1126/science.1241911 ; PubMed Central PMCID: PMC3875315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maillard PV, Ciaudo C, Marchais A, Li Y, Jay F, Ding SW, et al. Antiviral RNA interference in mammalian cells. Science. 2013;342(6155):235–8. Epub 2013/10/12. doi: 10.1126/science.1241930 ; PubMed Central PMCID: PMC3853215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiu Y, Xu Y, Zhang Y, Zhou H, Deng YQ, Li XF, et al. Human Virus-Derived Small RNAs Can Confer Antiviral Immunity in Mammals. Immunity. 2017;46(6):992–1004 e5. Epub 2017/06/22. doi: 10.1016/j.immuni.2017.05.006 . [DOI] [PubMed] [Google Scholar]
- 26.Xu YP, Qiu Y, Zhang B, Chen G, Chen Q, Wang M, et al. Zika virus infection induces RNAi-mediated antiviral immunity in human neural progenitors and brain organoids. Cell Res. 2019;29(4):265–73. Epub 2019/03/01. doi: 10.1038/s41422-019-0152-9 ; PubMed Central PMCID: PMC6461993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiu Y, Xu YP, Wang M, Miao M, Zhou H, Xu JY, et al. Flavivirus induces and antagonizes antiviral RNA interference in both mammals and mosquitoes. Science Advances. 2020;6(6):eaax7989. ARTN eaax7989. doi: 10.1126/sciadv.aax7989 WOS:000512902300013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Li Z, Ye Z, Xu Y, Wang B, Wang C, et al. The activation of antiviral RNA interference not only exists in neural progenitor cells but also in somatic cells in mammals. Emerging Microbes & Infections. 2020;9(1):1580–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Basavappa M, Lu J, Dong S, Cronkite DA, Prior JT, et al. Induction and suppression of antiviral RNA interference by influenza A virus in mammalian cells. Nat Microbiol. 2016;2(3):16250. Epub 2016/12/06. doi: 10.1038/nmicrobiol.2016.250; PubMed Central PMCID: PMC5488270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsai K, Courtney DG, Kennedy EM, Cullen BR. Influenza A virus-derived siRNAs increase in the absence of NS1 yet fail to inhibit virus replication. Rna. 2018;24(9):1172–82. Epub 2018/06/16. doi: 10.1261/rna.066332.118 ; PubMed Central PMCID: PMC6097656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat Immunol. 2006;7(6):590–7. Epub 2006/03/24. doi: 10.1038/ni1335 . [DOI] [PubMed] [Google Scholar]
- 32.Li H, Li WX, Ding SW. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296(5571):1319–21. Epub 2002/05/23. doi: 10.1126/science.1070948 . [DOI] [PubMed] [Google Scholar]
- 33.Wang XH, Aliyari R, Li WX, Li HW, Kim K, Carthew R, et al. RNA interference directs innate immunity against viruses in adult Drosophila. Science. 2006;312(5772):452–4. Epub 2006/03/25. doi: 10.1126/science.1125694 ; PubMed Central PMCID: PMC1509097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chakravarthy S, Sternberg SH, Kellenberger CA, Doudna JA. Substrate-specific kinetics of Dicer-catalyzed RNA processing. J Mol Biol. 2010;404(3):392–402. Epub 2010/10/12. doi: 10.1016/j.jmb.2010.09.030 ; PubMed Central PMCID: PMC3005596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma E, MacRae IJ, Kirsch JF, Doudna JA. Autoinhibition of human dicer by its internal helicase domain. J Mol Biol. 2008;380(1):237–43. Epub 2008/05/30. doi: 10.1016/j.jmb.2008.05.005 ; PubMed Central PMCID: PMC2927216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Billy E, Brondani V, Zhang H, Müller U, Filipowicz W. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc Natl Acad Sci U S A. 2001;98(25):14428–33. Epub 2001/11/29. doi: 10.1073/pnas.261562698 ; PubMed Central PMCID: PMC64698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16(8):948–58. Epub 2002/04/18. doi: 10.1101/gad.981002 ; PubMed Central PMCID: PMC152352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang S, Tutton S, Pierce E, Yoon K. Specific double-stranded RNA interference in undifferentiated mouse embryonic stem cells. Mol Cell Biol. 2001;21(22):7807–16. Epub 2001/10/18. doi: 10.1128/MCB.21.22.7807-7816.2001 ; PubMed Central PMCID: PMC99950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu X, Dao Thi VL, Huang Y, Billerbeck E, Saha D, Hoffmann HH, et al. Intrinsic Immunity Shapes Viral Resistance of Stem Cells. Cell. 2018;172(3):423–38.e25. Epub 2017/12/19. doi: 10.1016/j.cell.2017.11.018 ; PubMed Central PMCID: PMC5786493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu X, Kwong AC, Rice CM. Antiviral resistance of stem cells. Curr Opin Immunol. 2019;56:50–9. Epub 2018/10/24. doi: 10.1016/j.coi.2018.10.004 ; PubMed Central PMCID: PMC6462420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kennedy EM, Whisnant AW, Kornepati AV, Marshall JB, Bogerd HP, Cullen BR. Production of functional small interfering RNAs by an amino-terminal deletion mutant of human Dicer. Proc Natl Acad Sci U S A. 2015;112(50):E6945–54. Epub 2015/12/02. doi: 10.1073/pnas.1513421112 ; PubMed Central PMCID: PMC4687602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maillard PV, Van der Veen AG, Deddouche-Grass S, Rogers NC, Merits A, Reis e Sousa C. Inactivation of the type I interferon pathway reveals long double-stranded RNA-mediated RNA interference in mammalian cells. EMBO J. 2016;35(23):2505–18. Epub 2016/11/07. doi: 10.15252/embj.201695086 ; PubMed Central PMCID: PMC5167344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Veen AG, Maillard PV, Schmidt JM, Lee SA, Deddouche-Grass S, Borg A, et al. The RIG-I-like receptor LGP2 inhibits Dicer-dependent processing of long double-stranded RNA and blocks RNA interference in mammalian cells. EMBO J. 2018;37(4). Epub 2018/01/21. doi: 10.15252/embj.201797479; PubMed Central PMCID: PMC5813259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han Q, Chen G, Wang J, Jee D, Li WX, Lai EC, et al. Mechanism and Function of Antiviral RNA Interference in Mice. mBio. 2020;11(4). Epub 2020/08/06. doi: 10.1128/mBio.03278-19; PubMed Central PMCID: PMC7407090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parameswaran P, Sklan E, Wilkins C, Burgon T, Samuel MA, Lu R, et al. Six RNA viruses and forty-one hosts: viral small RNAs and modulation of small RNA repertoires in vertebrate and invertebrate systems. PLoS Pathog. 2010;6(2):e1000764. Epub 2010/02/20. doi: 10.1371/journal.ppat.1000764; PubMed Central PMCID: PMC2820531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Umbach JL, Yen HL, Poon LL, Cullen BR. Influenza A virus expresses high levels of an unusual class of small viral leader RNAs in infected cells. mBio. 2010;1(4). Epub 2010/09/16. doi: 10.1128/mBio.00204-10; PubMed Central PMCID: PMC2934610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perez JT, Varble A, Sachidanandam R, Zlatev I, Manoharan M, García-Sastre A, et al. Influenza A virus-generated small RNAs regulate the switch from transcription to replication. Proc Natl Acad Sci U S A. 2010;107(25):11525–30. Epub 2010/06/11. doi: 10.1073/pnas.1001984107 ; PubMed Central PMCID: PMC2895093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Girardi E, Chane-Woon-Ming B, Messmer M, Kaukinen P, Pfeffer S. Identification of RNase L-dependent, 3’-end-modified, viral small RNAs in Sindbis virus-infected mammalian cells. mBio. 2013;4(6):e00698–13. Epub 2013/11/21. doi: 10.1128/mBio.00698-13 ; PubMed Central PMCID: PMC3870239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bogerd HP, Skalsky RL, Kennedy EM, Furuse Y, Whisnant AW, Flores O, et al. Replication of many human viruses is refractory to inhibition by endogenous cellular microRNAs. J Virol. 2014;88(14):8065–76. Epub 2014/05/09. doi: 10.1128/JVI.00985-14 ; PubMed Central PMCID: PMC4097787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Otsuka M, Jing Q, Georgel P, New L, Chen JM, Mols J, et al. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity. 2007;27(1):123–34. doi: 10.1016/j.immuni.2007.05.014 WOS:000248398100014. [DOI] [PubMed] [Google Scholar]
- 51.Adiliaghdam F, Basavappa M, Saunders TL, Harjanto D, Prior JT, Cronkite DA, et al. A Requirement for Argonaute 4 in Mammalian Antiviral Defense. Cell Rep. 2020;30(6):1690–701 e4. Epub 2020/02/13. doi: 10.1016/j.celrep.2020.01.021 ; PubMed Central PMCID: PMC7039342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bogerd HP, Whisnant AW, Kennedy EM, Flores O, Cullen BR. Derivation and characterization of Dicer- and microRNA-deficient human cells. Rna. 2014;20(6):923–37. Epub 2014/04/24. doi: 10.1261/rna.044545.114 ; PubMed Central PMCID: PMC4024645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.García-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, et al. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252(2):324–30. Epub 1999/01/08. doi: 10.1006/viro.1998.9508 . [DOI] [PubMed] [Google Scholar]
- 54.Xia H, Luo H, Shan C, Muruato AE, Nunes BTD, Medeiros DBA, et al. An evolutionary NS1 mutation enhances Zika virus evasion of host interferon induction. Nat Commun. 2018;9(1):414. Epub 2018/01/31. doi: 10.1038/s41467-017-02816-2; PubMed Central PMCID: PMC5788864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li WX, Li H, Lu R, Li F, Dus M, Atkinson P, et al. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc Natl Acad Sci U S A. 2004;101(5):1350–5. Epub 2004/01/28. doi: 10.1073/pnas.0308308100 ; PubMed Central PMCID: PMC337056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu R, Maduro M, Li F, Li HW, Broitman-Maduro G, Li WX, et al. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature. 2005;436(7053):1040–3. Epub 2005/08/19. doi: 10.1038/nature03870 ; PubMed Central PMCID: PMC1388260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sullivan CS, Ganem D. A virus-encoded inhibitor that blocks RNA interference in mammalian cells. J Virol. 2005;79(12):7371–9. Epub 2005/05/28. doi: 10.1128/JVI.79.12.7371-7379.2005 ; PubMed Central PMCID: PMC1143619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chao JA, Lee JH, Chapados BR, Debler EW, Schneemann A, Williamson JR. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat Struct Mol Biol. 2005;12(11):952–7. Epub 2005/10/18. doi: 10.1038/nsmb1005 . [DOI] [PubMed] [Google Scholar]
- 59.Aliyari R, Wu Q, Li HW, Wang XH, Li F, Green LD, et al. Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila. Cell Host Microbe. 2008;4(4):387–97. Epub 2008/10/16. doi: 10.1016/j.chom.2008.09.001 ; PubMed Central PMCID: PMC2584229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Myles KM, Wiley MR, Morazzani EM, Adelman ZN. Alphavirus-derived small RNAs modulate pathogenesis in disease vector mosquitoes. Proc Natl Acad Sci U S A. 2008;105(50):19938–43. Epub 2008/12/03. doi: 10.1073/pnas.0803408105 ; PubMed Central PMCID: PMC2604946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ratcliff FG, MacFarlane SA, Baulcombe DC. Gene silencing without DNA: RNA-mediated cross-protection between viruses. Plant Cell. 1999;11(7):1207–15. doi: 10.1105/tpc.11.7.1207 WOS:000081674300003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Otsuka M, Zheng M, Hayashi M, Lee JD, Yoshino O, Lin S, et al. Impaired microRNA processing causes corpus luteum insufficiency and infertility in mice. J Clin Invest. 2008;118(5):1944–54. Epub 2008/04/10. doi: 10.1172/JCI33680 ; PubMed Central PMCID: PMC2289794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lazear HM, Govero J, Smith AM, Platt DJ, Fernandez E, Miner JJ, et al. A Mouse Model of Zika Virus Pathogenesis. Cell Host Microbe. 2016;19(5):720–30. Epub 2016/04/14. doi: 10.1016/j.chom.2016.03.010 ; PubMed Central PMCID: PMC4866885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lu R, Yigit E, Li WX, Ding SW. An RIG-I-Like RNA helicase mediates antiviral RNAi downstream of viral siRNA biogenesis in Caenorhabditis elegans. PLoS Pathog. 2009;5(2):e1000286. Epub 2009/02/07. doi: 10.1371/journal.ppat.1000286; PubMed Central PMCID: PMC2629121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guo X, Zhang R, Wang J, Ding SW, Lu R. Homologous RIG-I-like helicase proteins direct RNAi-mediated antiviral immunity in C. elegans by distinct mechanisms. Proc Natl Acad Sci U S A. 2013;110(40):16085–90. Epub 2013/09/18. doi: 10.1073/pnas.1307453110 ; PubMed Central PMCID: PMC3791698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Y, Anderson DH, Liu Q, Zhou Y. Mechanism of influenza A virus NS1 protein interaction with the p85beta, but not the p85alpha, subunit of phosphatidylinositol 3-kinase (PI3K) and up-regulation of PI3K activity. J Biol Chem. 2008;283(34):23397–409. Epub 2008/06/07. doi: 10.1074/jbc.M802737200 . [DOI] [PubMed] [Google Scholar]
- 67.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–60. Epub 2015/03/10. doi: 10.1038/nmeth.3317 ; PubMed Central PMCID: PMC4655817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–9. Epub 2014/09/28. doi: 10.1093/bioinformatics/btu638 ; PubMed Central PMCID: PMC4287950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. Epub 2010/10/29. doi: 10.1186/gb-2010-11-10-r106; PubMed Central PMCID: PMC3218662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125(1–2):279–84. Epub 2001/10/30. doi: 10.1016/s0166-4328(01)00297-2 . [DOI] [PubMed] [Google Scholar]
- 71.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–9. Epub 2000/05/10. doi: 10.1038/75556 ; PubMed Central PMCID: PMC3037419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A-C. PR8/delNS1-infected NoDice 293T cells ectopically expressing hDcr (A), hDcr+IAV-NS1 (B) and hDcr+ZIKV-NS1 (C). Genomic coverage depth of each nucleotide position by 21- to 23-nt vsiRNAs sequenced from RNA above. Reads are shown as per million total 18- to 28-nt reads.
(TIF)
A. RNA1 accumulation level of WT NoV or NoVΔB2 measured by RT-qPCR from hind limb of C57BL/6 suckling mice at 7 dpi (n = 5 per group). The RNA1 level of NoVΔB2 infected C57BL/6 suckling mice was set as 1. B. Expression level of NoV capsid and B2 protein measured by Western blotting. Actin were used as a loading control.
(TIF)
The genomic structure of SINVB2, SINVmB2 or SINVGFP (right) and relative viral accumulation determined by RT-qPCR from hindlimb of BALB/c suckling mice infected with SINVB2, SINVmB2 or SINVGFP at 3dpi. n = 6~7 per group. Error bars represent SD. * indicates p<0.05 (Student’s t-test). The viral RNA accumulation of SINVGFP was set as 1.
(TIF)
A, B. Expression levels of IFN (A) and RNAi (B) related genes in C57BL/6 suckling mice with NoV infection at 7dpi. All data were measured as the mean ± SD of three independent experiments. Asterisks indicate a significant difference level compared to control (Student’s t-test, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001).
(TIF)
A, B. Volcano plots showing demonstrate false discovery rate (FDR) and fold-change (FC) of gene expression levels determined by RNAseq comparing BALB/c suckling mice inoculated by NoVΔB2 vs Mock (A), WT NoV vs Mock (B). Genes up-regulated (fold change > 2 and FDR< 0.05) are indicated in red and those down-regulated (fold change < 0.5 and FDR< 0.05) are indicated in blue. C. Differential expression of IFN pathway related genes from mRNA-seq data of NoV or NoVΔB2 inoculated BALB/c suckling mice at 3 dpi. Fold changes (FC) of 2 or 0.5 (|log2FC| = 1) are indicated by dotted lines. The log2FC was taken as 20 when the ratio of experimental group and mock group tended to be infinite due to a small denominator.
(TIF)
A, B. Volcano plots showing demonstrate false discovery rate (FDR) and fold-change (FC) of gene expression levels determined by RNAseq comparing C57BL/6 suckling mice inoculated by NoVΔB2 vs Mock (A), WT NoV vs Mock (B). Genes up-regulated (fold change > 2 and FDR< 0.05) are indicated in red and those down-regulated (fold change < 0.5 and FDR< 0.05) are indicated in blue. C. Differential expression of IFN pathway related genes from mRNA-seq data of NoV or NoVΔB2 inoculated C57BL/6 suckling mice at 3 dpi. Fold changes (FC) of 2 or 0.5 (|log2FC| = 1) are indicated by dotted lines. The log2FC was taken as 20 when the ratio of experimental group and mock group tended to be infinite due to a small denominator.
(TIF)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.The RNA sequencing data have been deposited to the NCBI GEO database under the accession number GSE155146.