Abstract
Accurately mapping changes in cellular membrane potential across large groups of neurons is crucial for understanding the organization and maintenance of neural circuits. Measuring cellular voltage changes by optical means allows greater spatial resolution than traditional electrophysiology methods and is adaptable to high-throughput imaging experiments. VoltageFluors, a class of voltage-sensitive dyes, have recently been used to optically study the spontaneous activity of many neurons simultaneously in dissociated culture. VoltageFluors are particularly useful for experiments investigating differences in excitability and connectivity between neurons at different stages of development and in different disease models. The protocols in this article describe general procedures for preparing dissociated cultures, imaging spontaneous activity in dissociated cultures with VoltageFluors, and analyzing optical spontaneous activity data.
Basic Protocol 1:
Preparation of dissociated rat hippocampal or cortical cultures
Alternate Protocol:
Preparation of microisland dissociated cultures
Basic Protocol 2:
Imaging of spontaneous activity in dissociated cultures using voltage-sensitive dyes
Basic Protocol 3:
Analysis of spontaneous activity imaging data
Keywords: dissociated culture, imaging, SpikeConnect, spontaneous activity, VoltageFluor
INTRODUCTION
This article describes a general procedure for imaging spontaneous activity in dissociated culture using a voltage-sensitive dye. Rapid changes in membrane potential underlie signaling and cell physiology in excitable cells such as neurons and cardiomyocytes. Many electrical methods have been developed to directly measure membrane potential changes as a means to interrogate neuronal signaling. This pioneering work allowed researchers to work out how individual neurons change the electrochemical gradient across their membranes in response to stimuli and how changes in membrane potential lead to the release of neurotransmitters that propagate the signal on to other neurons. As the field of neuroscience has advanced, focus has shifted away from individual neurons to neural circuits and larger systems. However, the electrical tools developed for individual neurons run into limitations when larger numbers of neurons are considered. The invasive nature of direct measurement of membrane potential with an electrode complicates the simultaneous measurement of membrane potential changes from all neurons in a neural circuit.
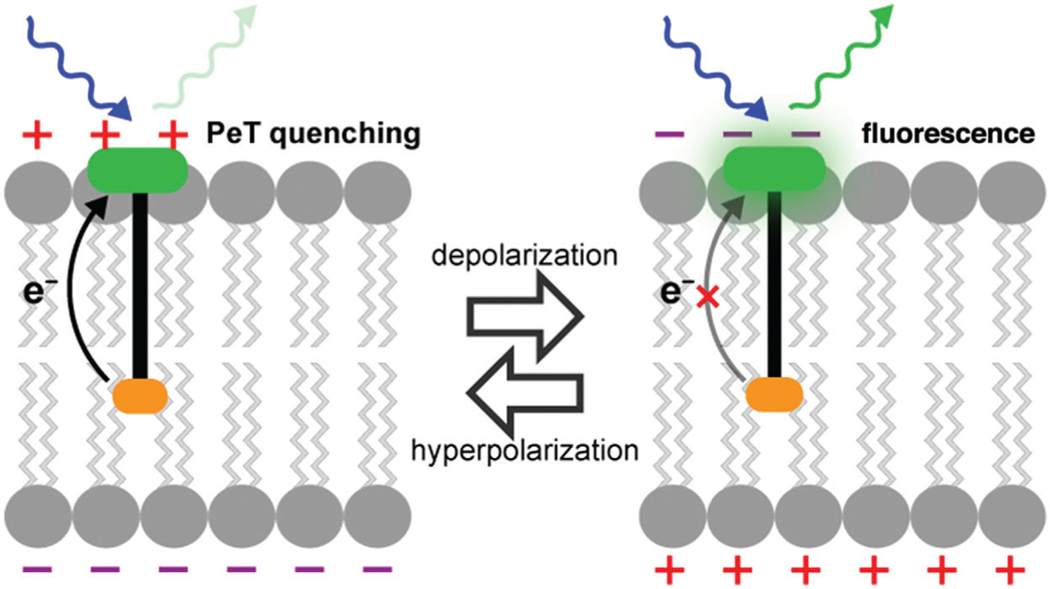
In response to these challenges, voltage-sensitive dyes have been developed whose optical properties change in response to changes in membrane potential. Measuring voltage optically is less invasive than traditional electrophysiology methods and facilitates the simultaneous measurement of membrane potential changes from many distinct neurons. This protocol focuses on one class of voltage-sensitive dyes, VoltageFluors, which insert into neuronal membranes and change their fluorescence quantum yields in response to changes in membrane potential (Miller, 2016). VoltageFluors consist of a fluorescent dye attached to a donor group through a lipophilic molecular wire region. Under resting membrane potential conditions, the fluorescent dye is largely quenched by the donor group through a photo-induced electron transfer (PET) process. However, when the membrane potential depolarizes, the rate of PET is slowed, resulting in an increase in the fluorescence quantum yield (see Fig. 1).
Figure 1.
Schematic depicting the mechanism of action of VoltageFluor dyes. Adapted from Miller (2016).
This protocol focuses on one VoltageFluor derivative, Berkeley Red Sensor of Transmembrane Potential (BeRST 1; Huang, Walker, & Miller, 2015). BeRST 1’s optical properties reduce background signal, while the dye scaffold retains excellent voltage sensitivity. Thus, BeRST 1 displays high signal-to-noise ratios when measuring spontaneous voltage changes in dissociated culture. BeRST 1’s properties enabled the imaging of spontaneous activity from up to 25 neurons simultaneously and the investigation of excitability and connectivity in bulk dissociated culture (Walker, Raliski, & Karbasi, et al., 2020) and microisland cultures (Walker, Raliski, & Nguyen, et al., 2020). However, one challenge that arose during these studies was the difficulty of processing large amounts of spontaneous activity imaging data. Therefore, along with developing BeRST 1 imaging experiments, our lab also developed a MATLAB program, SpikeConnect, to facilitate analysis of spontaneous activity imaging data (Walker, Raliski, & Nguyen, et al., 2020).
This protocol outlines the complete process of imaging spontaneous neuronal activity with BeRST 1 and analyzing the resulting data. First, you prepare dissociated culture either in bulk (Basic Protocol 1) or arranged in microislands (Alternate Protocol). Then, you stain your chosen culture with BeRST 1 and record movies of spontaneous voltage changes on a fluorescence microscope (Basic Protocol 2). Finally, you process the fluorescence movies using SpikeConnect (Basic Protocol 3) to analyze neuronal excitability and connectivity.
STRATEGIC PLANNING
The success of any spontaneous activity imaging experiment depends on having a well-developed experimental plan. The first choice you must make is whether you want to perform experiments on bulk culture (Basic Protocol 1) or on microislands (Alternate Protocol). Microislands are created by placing a 4-polydimethylsiloxane (PDMS) stencil on coverslips before plating the dissociated cultures. This creates “islands” of neurons that are electrically isolated from each other and have a size defined by the dimensions of the PDMS stencil. Using a stencil with holes the same size as the typical imaging window on a selected microscope makes it possible to image an entire closed neural circuit simultaneously. Therefore, microislands are particularly useful for studies of neuronal connectivity. However, fabricating the silicon wafer mold and casting the PDMS stencils can be quite labor intensive and require access to microfabrication equipment. If your research program is more focused on neuronal excitability, then bulk culture is the more straightforward choice. Another experimental factor to consider is whether you plan to perform immunohistochemistry on the neurons after the imaging experiments are completed. This can be useful as a means to identify the cell type of every neuron imaged and investigate differential effects on the excitability of specific cell types. If cell type information is desired, use of gridded coverslips is recommended to facilitate the re-identification of areas of neurons after imaging.
Once an appropriate cell culture system has been chosen, imaging settings that yield optical traces which faithfully reproduce electrical traces must be determined. This can be accomplished by imaging cells stained with dye (Basic Protocol 2) while simultaneously recording electrical traces from the same cells. In this way, a signal-to-noise threshold can be set (Basic Protocol 3) for the optical traces to delineate the point at which a spike in the optical trace is labeled as an action potential. For our fluorescence movies, we typically collect images (2048 × 400 px2, pixel size 0.325 × 0.325 μm2) continuously for 10 s at a sampling rate of 500 Hz, with 4 × 4 binning, using a 1.0-NA, 20× water-immersion objective. These parameters allow us to observe up to 25 neurons at a time and record quickly enough to measure on the same timescale as the timescale of action potentials. To excite BeRST 1, we use a 633-nm LED with an intensity of 13 mW/mm2. This allows us to record for 10 s continuously without any appreciable photobleaching. Good agreement between the optical and electrophysiology traces can be obtained by using a signal-to-noise threshold of 16 for the optical trace recorded by BeRST 1 under these imaging settings (Walker, Raliski, & Nguyen, et al., 2020). However, if a desired imaging experiment requires different imaging parameters, the agreement between the optical and electrophysiology traces will need to be reassessed. Also, the excitation light and movie length must be adjusted to avoid appreciable dye photobleaching, as otherwise analysis by SpikeConnect (Basic Protocol 3) will be confounded by a changing fluorescence baseline.
Finally, to facilitate analysis of the fluorescence movies by SpikeConnect (see Basic Protocol 3), your imaging data should be organized in as follows: Each coverslip imaged should be its own individual folder that contains separate folders for each area imaged on that coverslip; and each area folder should contain a brightfield image of the area and the fluorescence movies recorded from that area (see Basic Protocol 2). In this way, SpikeConnect can associate brightfield images with the corresponding movies (see Basic Protocol 3). This is important because regions of interest are drawn based on brightfield images and applied to the fluorescence movies during analysis.
BASIC PROTOCOL 1
PREPARATION OF DISSOCIATED RAT HIPPOCAMPAL OR CORTICAL CULTURES
This protocol outlines the preparation of bulk dissociated cultures for spontaneous activity imaging experiments. When starting this research program, coordinate with the animal facility at your institution to ensure that your laboratory is following the proper protocols and animal handling guidelines. If you plan to image spontaneous activity over many weeks, it is good practice to establish a regular rotation of personnel who prepare the dissociated cultures at the same time each week. Upon following this protocol, you should obtain bulk dissociated rat hippocampal or cortical cultures at your desired density for planned imaging experiments. This protocol describes the optimal reagent amounts for plating dissociated rat hippocampal or cortical neurons on 12-mm glass coverslips in plastic 24-well cell culture plates. If the use of other coverslips, such as gridded coverslips for correlative experiments, is desired (see Strategic Planning, above), adjust the reagent amounts according to the guidelines of your chosen well plates and coverslips.
Materials
1 M HCl (CAS 7647-01-0)
Ethanol (CAS 64-17-5)
Poly-D-lysine (Sigma-Aldrich P7280-5 mg)
Sodium borate buffer (see recipe)
Dulbecco’s Phosphate-Buffered Saline (DPBS; Gibco 14200-075)
MEM++++ culture medium (see recipe)
Sprague Dawley rats (Charles River Laboratories)
Dissection medium: HBSS, Ca2+- and Mg2+-free, with phenol red (Invitrogen 14170-161), ice cold
2.5% trypsin (Invitrogen 15090-046)
Neurobasal (NB++) medium (see recipe)
12-mm round German glass coverslips for rat neurons (VWR 100499-634)
Glass Petri dishes (VWR 75845-542)
Synergy Water Purification System (Millipore Sigma Synergy W-R), for preparing double-distilled water
General-purpose heating and drying oven (Fisher Scientific 15-103-0503)
24-well cell culture plates (VWR 62406-183)
Incubator (ThermoFisher Scientific Heracell VIOS 160i)
50-ml 0.2-μm-pore-size sterile filter (VWR 82030-938)
Tissue culture hood (Baker SterilGARD e3)
Dumont no. 5 fine-tip forceps (Fine Science Tools 11251-20), for rat dissection
Narrow-pattern forceps (Fine Science Tools 11002-12), for rat dissection
Curved scissors (VWR 25608-225), for rat dissection
6 1/2-inch sharp-tip dissecting scissors (VWR 82027-592), for rat dissection
Positive-action tweezers, Style 5 (Electron Microscopy Services 72706-01), for rat dissection
50- and 15-ml Corning centrifuge tubes (VWR 21008-725 and VWR 21008-673)
Dissection microscope (Olympus SZ40 Stereo Zoom)
2-ml aspirator pipets (VWR 53106-450)
1-, 5-, and 10-ml serological pipets (VWR 29443-041, VWR 29443-045, and VWR 29443-047)
Pasteur pipets (FisherScientific 13-678-20C)
Hemocytometer for phase scope (VWR 15170-089)
Mouse dissection
Before starting this protocol, coordinate with your animal facility or animal supplier to produce one timed pregnant (embryonic day 17–19 [E17-E19]) Sprague Dawley rat for each week you intend to perform this protocol.
Two weeks before dissection, place 12-mm glass coverslips in 1 M HCl in a clean glass petri dish and shake 3–5 hr at 90 rpm. Remove the acid solution and wash with ethanol while shaking at 100 rpm overnight. Repeat this procedure twice more, for a total of three overnight washes. Wash with double-distilled water three times overnight while shaking at 100 rpm. Remove the water and place the petri dish into a glassware oven (150°C) for 2–5 hr or until completely dry.
Once the coverslips cool, you can store them at room temperature indefinitely and use as needed.
To maintain sterility, we have found it best to leave the petri dish sealed with Parafilm or tape to prevent accidental contamination.
3. One day before the dissection, dissolve solid poly-D-lysine (PDL) in sodium borate buffer at 1 mg/ml and dilute this solution 1:9 in DPBS. Then add the final solution to well plates of desired size containing pre-sterilized coverslips (see step 1).
The final PDL concentration is 0.1 mg/ml. For a 24-well plate, we use 12-mm glass coverslips and 250 μl PDL solution per well. We use fresh PDL solution every time.
4. Allow the plates to sit overnight in a humidified 37°C incubator.
This step can be done for a shorter time, but we find that incubating overnight yields the best results.
5. On the day of the dissection, remove the PDL solution by aspiration and wash the coverslips twice with sterile Milli-Q water and then twice with DPBS.
6. Sterile filter the MEM++++ culture medium and add half the total plating volume of medium to the coverslips. Return the plates to the humidified incubator to allow the medium to equilibrate with the CO2 in the incubator atmosphere.
For 24-well plates, the total volume per well is 750 μl. We add 400 μl MEM++++ to equilibrate and add the remaining 350 μl when plating the neurons.
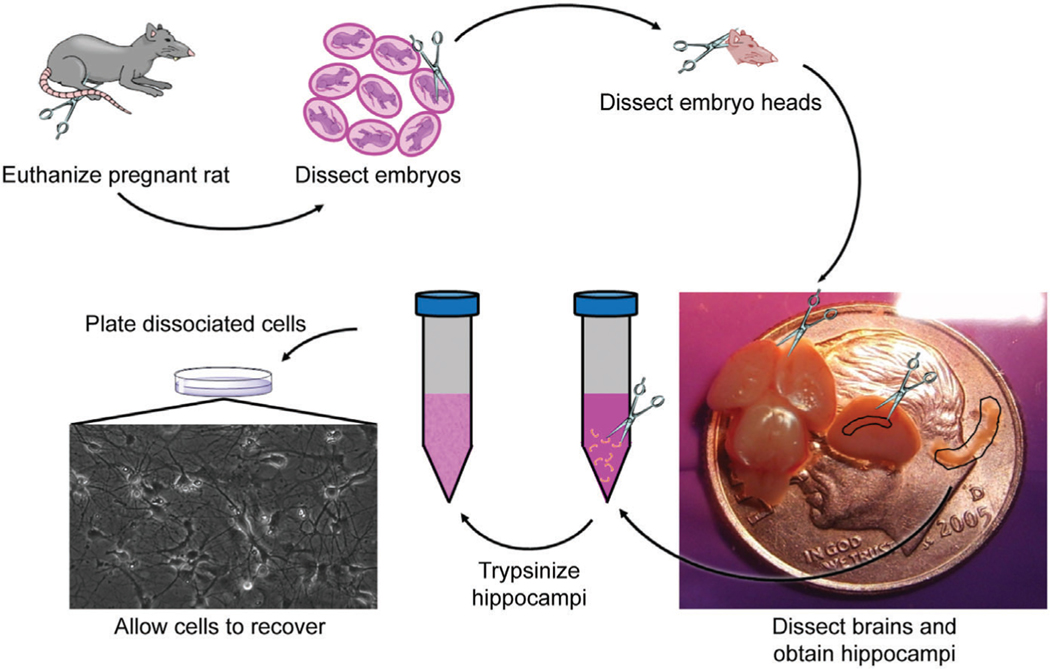
7. Euthanize a timed pregnant (E17-E19) Sprague Dawley rat with CO2 in accordance with regulations. In a tissue culture hood, remove embryos and decapitate, placing the heads into a 50-ml tube filled with ice-cold dissection medium. Discard all carcasses, blood, and tissue as medical waste in accordance with regulations.
Figure 2 shows these dissection steps in more detail.
Figure 2.
Workflow for preparing dissociated rat hippocampal cultures, with an example of a healthy culture plated at a density of 30,000/well.
Preparation of dissociated hippocampal or cortical cells
8. Working on a dissection microscope in a tissue culture hood, puncture the optic area of each head with tweezers to hold the head steady. With another pair of tweezers, remove the skin from the skull. Then, cut with tweezers along the longitudinal fissure of the skull from the posterior to the anterior and peal the skull away from the brain. Remove the brain from the remainder of the head. Cut the cortices away from the midbrain. For a hippocampal culture, remove the meninges from the cortices and dissect out the hippocampus from each cortex. Place the hippocampi in a 15-ml tube filled with dissection medium. For a cortical culture, remove the meninges from the cortices and dissect the tissue above the hippocampus. Place the cortical tissue in a 15-ml tube filled with dissection medium.
For more details see Figure 2 and Audesirk, Audesirk, & Ferguson (2001).
9. Transfer the hippocampi or cortical tissue to 1 ml 2.5% trypsin and incubate the solution for 15 min in a water bath at 37°C.
We aliquot our 2.5% trypsin solution as 1-ml aliquots and store them at –20°C indefinitely.
10. Use a pipet to remove the trypsin solution from above the hippocampi or cortical tissue. Wash the hippocampi or cortical tissue three times with dissection medium, and remove the medium from the last wash.
Be very careful to not suck the tissue samples up into the pipet during these washing steps
11. Add 1 ml MEM++++ to the tissue samples. Triturate the hippocampi or cortical tissue three times using three progressively smaller flame-polished Pasteur pipets until the solution appears homogeneous.
We flame-polish three different sizes of Pasteur pipets and autoclave them before use in this procedure. Triturating too much causes excessive debris and unhealthy culture.
12. Dilute the eluate to 3 ml with MEM++++ and measure the cell density using a hemocytometer.
We typically measure 1–2 million cells/ml at this step.
Culture of dissociated neurons
13. Make appropriate dilutions in MEM++++ and plate the neurons at the desired density.
We typically plate dissociated rat hippocampal or cortical neurons at a density of 30,000 per well for a 24-well plate. See Figure 2 for an example of dissociated hippocampal neurons plated at this density.
14. At 1 day in vitro (DIV), replace 50% of the culture medium with Neurobasal medium (NB++).
We usually take out 350 μl of culture medium and add 400 μl of fresh NB++.
15. Top up the medium with NB++ at 7 DIV or when it is low.
At 7 DIV, we add 250 μl NB++ to each well for a 24-well plate.
ALTERNATE PROTOCOL
PREPARATION OF MICROISLAND DISSOCIATED CULTURES
This protocol outlines the preparation of microisland dissociated cultures for spontaneous voltage imaging experiments. Microisland cultures are created by plating cells onto a glass coverslip which is partially covered by a stencil. Once the stencil is removed, cells that grew on the stencil are also removed and the only cells that remain are those that grew on the glass coverslip in the holes of the stencil. This technique creates electrically isolated “islands” of neurons. By designing stencils to create microislands smaller than a set imaging window, one can image spontaneous voltage changes simultaneously from each neuron in an entire closed neural circuit. Thus, microislands are particularly useful for studies of neuronal connectivity. This protocol describes the preparation of stencils and the addition of stencils to glass coverslips. Once the glass coverslips have been prepared with stencils, the culture preparation proceeds identically to Basic Protocol 1.
Materials
Trimethylchlorosilane (CAS 75-77-4)
SYLGARD 184 Silicone Elastomer Kit (Fisher Scientific NC9285739)
70% (v/v) ethanol (CAS 64-17-5)
Dulbecco’s phosphate-buffered saline (DPBS; Gibco 14200-075)
100-mm silicon wafer (University Wafer 452)
0.6-ml clear microcentrifuge tubes (Microtubes MCT-060-C)
Vacuum desiccator (Thermo Scientific 53100250)
Overhead projector acetate transparency film (Hygloss Products 75905)
Glass plates
Bulldog clips
General-purpose heating and drying oven (Fisher Scientific 15-103-0503)
Razor blade
Duckbill point tweezers (Fisher Scientific 17-456-104)
Ruler
50-ml Corning centrifuge tubes (VWR 21008-725)
Forceps
12-mm round German glass coverslips for rat neurons (VWR 100499-634)
24-well cell culture plates (VWR 62406-183)
20-ml glass liquid scintillation vials (Wheaton V7520-6)
Reactive-ion etching (RIE) system (Plasma Etch PE-100)
Tissue culture hood (Baker SterilGARD e3)
Preparation of stencils
Fabricate a silicon wafer in the microfabrication center at your institution with pillars of defined size for the stencils you wish to make.
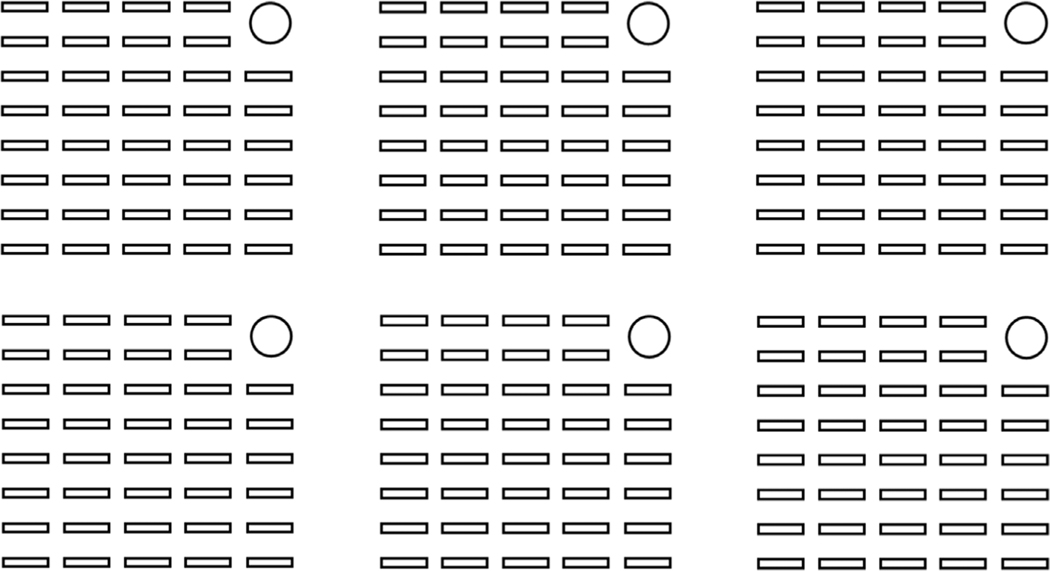
We designed our wafer so that our stencils would have holes slightly smaller than the size of our typical imaging window (650 μm × 120 μm). A schematic of the final wafers is shown in Figure 3.
Figure 3.
Schematic of patterned microislands. Dimensions of each microisland (small rectangles) are ~120 μm × 650 μm, and rectangles are spaced ~220 μm apart horizontally and 370 μm apart vertically. The circle in the upper right allows the user to identify microislands for post-hoc immunostaining after functional imaging.
2. Pipet 100 μl trimethylchlorosilane into a microcentrifuge tube, and place the tube into a vacuum desiccator along with the silicon wafer. Turn on the vacuum and leave for at least 2 hr.
We re-silanize our wafer after every four PDMS coatings to prevent the PDMS from bonding to the wafer.
3. Prepare 2.5 ml PDMS mixture (SYLGARD 184; 10:1 monomer:catalyst) in a plastic cup and degas for 1 hr in a vacuum desiccator.
4. Slowly pour the PDMS mixture onto the silanized wafer, and place a transparency film on top of the PDMS. Gently smooth out the PDMS mixture with your hands, ensuring that the entire wafer is covered. Check that the transparency is in contact with the wafer pillars.
5. Position a second transparency under the wafer, and place the wafer and transparencies between two square glass plates. Secure the entire assembly with bulldog clips. Cure the PDMS for 2 hr in a 65°C oven.
6. Disassemble the transparency and glass assembly. Remove the PDMS stencil sheet from the wafer by first running a razor blade around the wafer edge and then peeling the PDMS away from the wafer with tweezers.
Working across the long edge of the wafer pillars is the best way to avoid tearing the PDMS. PDMS stencil sheets can be rolled up and stored indefinitely in 50-ml tubes in 50% (v/v) isopropyl alcohol or 70% (v/v) ethanol.
7. Use a razor blade and a ruler to cut stencils from a PDMS sheet. For 12-mm coverslips, we use stencils containing 4 × 8 islands. Larger stencils can be cut if needed for larger coverslips
8. Discard any stencil in which fewer than 75% of the stencils are punched through, and store the stencils indefinitely in a scintillation vial in 70% ethanol.
Preparation of stencil-covered coverslips
9. Arrange 12-mm coverslips in the lid of a 24-well plate. Use forceps to remove the stencils from the ethanol solution and place them on one finger of your gloved hand. Use the forceps to remove a single stencil from the stack and dip it in 70% ethanol briefly before placing it on the center of one coverslip. When all coverslips have stencils, allow them to dry for at least 2 hr. Partially cover the lid to protect the coverslips from dust.
10. Use a reactive-ion etching (RIE) system in your institution’s microfabrication center to etch the coverslips with oxygen plasma according to the following details.
Briefly, turn on the O2 tank and radiofrequency (RF) generator. Next, vent the reaction chamber, and then open the lid and wipe it clean. Add the coverslips, close the chamber, close the vent, and start the vacuum pump. When the pressure is stable, open the O2 to 20%, apply RF to 150 W, and treat with O2 plasma for 2 min. After O2 plasma treatment, turn off the RF generator, O2 tank, and vacuum pump. Finally, vent the reaction chamber and remove the coverslips.
Coverslips can be stored in a 24-well plate in ethanol at 4°C until needed.
Preparation of microisland dissociated cultures
11. In a tissue culture hood, add the coverslips to the wells of a fresh 24-well plate and wash them with ethanol followed by DPBS. Perform steps 1–12 from Basic Protocol 1.
12. At 4–7 DIV, remove stencils from the coverslips using ethanol-sterilized forceps.
Removing stencils at this stage caused no detrimental effects. Removal of stencils should always be done at least 2 days before imaging.
13. Perform step 13 from Basic Protocol 1.
BASIC PROTOCOL 2
IMAGING OF SPONTANEOUS ACTIVITY IN DISSOCIATED RAT HIPPOCAMPAL OR CORTICAL CULTURES USING VOLTAGE-SENSITIVE DYES
This protocol outlines the imaging of spontaneous activity in dissociated cultures using the VoltageFluor BeRST 1. Once dissociated cultures have been prepared (see Basic Protocol 1 or Alternate Protocol) and imaging settings have been optimized (see Strategic Planning), imaging spontaneous activity with a voltage-sensitive dye is straightforward. After staining the cultures with dye, you then record a brightfield image and fluorescence movies from the areas of neurons you wish to study. This raw imaging data can then be analyzed with SpikeConnect (see Basic Protocol 3).
Materials
14- to 17-DIV dissociated cultures (from Basic Protocol 1 or Alternate Protocol)
250 μM BeRST 1 in DMSO (available from the corresponding author upon request)
HBSS with calcium and magnesium but without phenol red (Gibco 14025-092)
Dulbecco.s phosphate-buffered saline (DPBS; Gibco 14200-075), proceeding to immunohistochemistry
Incubator (ThermoFisher Scientific Heracell VIOS 160i)
Tissue culture hood (Baker SterilGARD e3)
Attofluor cell chamber, for microscopy (ThermoFisher Scientific A7816)
Microscope (Zeiss Axio Observer.Z1)
20 min before beginning imaging, remove the cell culture plate containing the cells you wish to image from the incubator and place it in a tissue culture hood. Aspirate the medium from one coverslip and replace it with 500 μl of a 500 nM solution of BeRST 1 in HBSS (diluted from 250 μM BeRST stock in DMSO). Place the well plate back into the incubator and let the dye load onto the cells for 20 min.
We typically image mature 14- to 17-DIV cultures. If you are looking at changes in spontaneous activity across different developmental timepoints, you can image younger cultures.
2. After 20 min, take the cell culture plate back out of the incubator, remove the coverslip from the dye solution, and transfer the slide to a cell chamber that fits into the stage on your microscope and contains enough HBSS to completely cover the coverslip.
3. Take the cell chamber to your microscope and take a brightfield image of the area of neurons you wish to observe. Next, record movies of spontaneous activity using the imaging parameters worked out for your specific microscope, ensuring that your optical recordings are reliably reproducing electrical recordings (see Strategic Planning). Repeat this process for a few areas on each coverslip.
As we image at ambient temperature and atmosphere, we commonly do not image coverslips for any longer than 10 min.
4. After imaging, if you wish to determine the cell types of your imaged neurons (see Strategic Planning), fix the coverslips by covering them in 4% paraformaldehyde (PFA) for 20 min. After fixing the coverslips, wash them three times with PBS and leave them in PBS at 4°C for up to 1 week. When ready, follow your chosen immunohistochemistry protocol to determine the cell types of your imaged neurons.
Using gridded coverslips or microislands are both good strategies to facilitate the correlation of imaging data and immunohistochemistry data.
BASIC PROTOCOL 3
ANALYSIS OF SPONTANEOUS ACTVITY IMAGING DATA
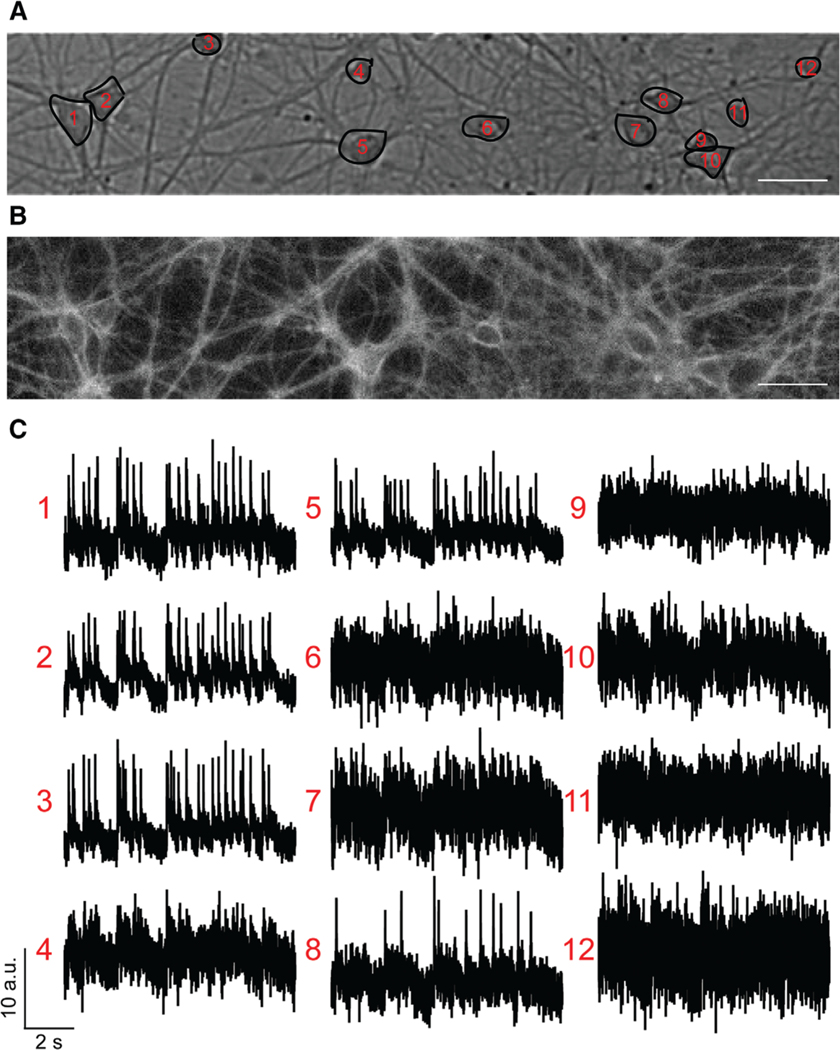
This protocol outlines the analysis of spontaneous activity data measured by voltage-sensitive dyes by SpikeConnect. Imaging spontaneous activity in neurons as described above can generate a tremendous amount of data. VoltageFluor dyes, which respond to voltage changes on a submillisecond timescale and are relatively photostable, enable the routine recording of 10-s movies at a sampling rate of 500 Hz (see Strategic Planning). Furthermore, each imaging movie typically contains 10–20 neurons, and processing the data for each region of interest can be a very tedious and labor-intensive process. In response to these challenges, our lab developed a MATLAB program, SpikeConnect, to partially automate our analysis of spontaneous activity data (Walker, Raliski, & Nguyen, et al., 2020). The following protocol describes the basic components and workflow of SpikeConnect (see Internet Resources for how to obtain SpikeConnect from GitHub). After following this protocol, you will be able to detect action potential spikes in your imaging data (Fig. 4).
Figure 4.
Voltage imaging with BeRST 1. (A) Example brightfield image with selected regions of interest (ROIs). (B) Example fluorescence image from the same field of view (FOV). (C) Raw fluorescence traces from the selected ROIs. These traces will be analyzed by SpikeConnect in Basic Protocol 3. Scale bar, 10 μm.
Materials
Personal computer
External hard drive
MATLAB (MathWorks, any version newer than 2017a)
SpikeConnect (see Internet Resources)
Imaging data, organized for SpikeConnect (see Basic Protocol 2 and Strategic Planning)
Drawing Pad (Wacom Intuos S)
Set up MATLAB on your personal computer. Download the SpikeConnect folder from GitHub (see Internet Resources) and add the folder to your MATLAB path.
It is helpful to have an external hard drive to handle the large file sizes associated with this type of data.
2. Ensure that your imaging data are organized in the manner described in Strategic Planning, and begin processing your first area of imaging by running “selectroi_gui”. Select the brightfield image (as a .tiff file) associated with the movies (also as .tiff files) you wish to analyze (see Basic Protocol 2) and import the movie files. Ensure the frame rate parameter matches the recording frame rate. Click on the graphic user interface and press “C” to begin drawing regions of interest (ROIs) around each neuron you wish to analyze (you must press “C” before each ROI). Once you are done, click on “Save ROIs and choose background”. This will bring up the first frame of the movie .tiff stack. Draw a ROI around an area of background and save it. Finally, repeat this process for every area that you would like to analyze in one batch before moving on to the next step. The output from this step is a “roi-*.mat” file where “*” is the name of the brightfield image.
A drawing pad can be helpful for drawing large numbers of ROIs when analyzing large datasets. Also, be sure to draw a full ROI for the background. Clicking on a point of background and saving it will cause the next step in the analysis to fail. Finally, this program can only analyze up to 25 ROIs from each brightfield image.
3. After drawing all the ROIs you wish to analyze, run “batchkmeans_gui”. You can choose whether you want to analyze the raw traces, the background-corrected traces, or the background-subtracted traces. We always analyze the background-corrected traces. Next, select the folder containing all the “roi-*.mat” files you wish to analyze. This step of the analysis will compute a fluorescence trace for each ROI in each recording and use k-means clustering to identify possible action potentials (spikes), subthreshold events, and baseline.
Once you select a folder, the k-means clustering algorithm will begin running, so you must choose your background option before choosing the folder. For more information on this step of analysis, see Walker, Raliski, & Nguyen, et al. (2020).
4. Next, run “thresholding_gui”. You must choose the same folder that you just analyzed with the k-means clustering algorithm. A histogram of signal-to-noise ratios (SNRs) for the traces contained in that folder is displayed. Set an SNR threshold, re-arm factor, and press “Set Threshold”. The SNR threshold is the minimum SNR above which a signal will be labeled as a spike. The re-arm factor sets the minimum number of frames between spikes to prevent over-counting. Press “Preview” to preview the spikes that have been identified with these thresholds to ensure that they match up well with the fluorescence traces. Adjust the thresholds as needed and press “Save Spikes to file” to save the spikes as a “spikes-*.mat” file, where “*” is the name of the movie.
When starting this research program, it is good practice to simultaneously image neurons while obtaining electrical recordings (see Strategic Planning). In this way, a reasonable SNR threshold for your microscope setup can be determined upon comparison of the optical and electrical traces. We typically use an SNR threshold of 16 and re-arm factor of 3.
5. If you have cell type information available from correlative immunostaining (see Strategic Planning), SpikeConnect can label hippocampal neuron cell types to facilitate later analysis. To label neurons, run “labelroi” and select a folder that contains a brightfield image with labeled ROIs from step 2. Click through each ROI and label it as a DGC neuron, inhibitory neuron, CA1 neuron, CA3 neuron, or unknown.
Currently, SpikeConnect only lists hippocampal neuron cell types as labels.
6. Export the spiking frequencies for each ROI by running “freqexport_gui”. You will select the folders to be exported and a destination for the excel file generated. The Excel file generated contains average frequency (Hz), instantaneous frequency (Hz), and interspike interval (ms) data for each ROI. The Excel file also contains summary columns for each of these parameters; see Figure 5 (example frequency comparison).
Figure 5.
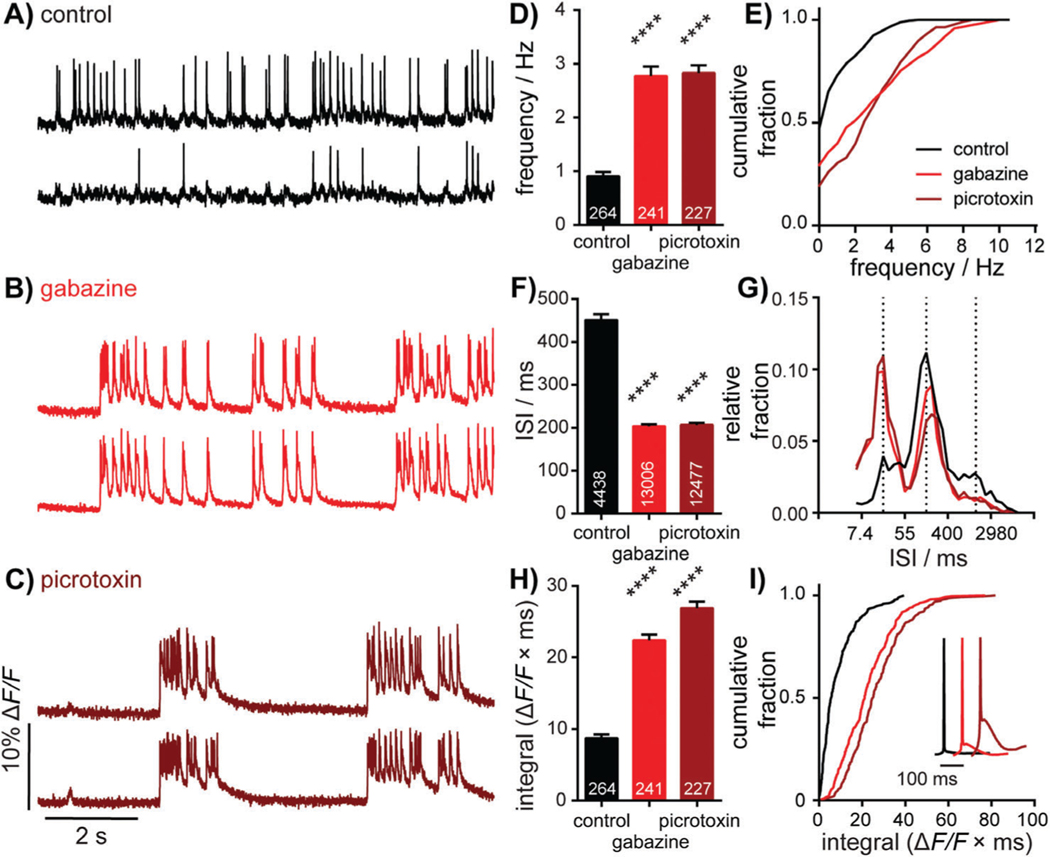
Characterization of neuronal responses to pharmacological manipulation. Representative ΔF/F voltage imaging traces of spontaneous spiking activity measured by BeRST 1 in hippocampal neurons under control conditions (A) or following acute administration of 10 μM gabazine (B) or 50 μM picrotoxin (C).Traces are of two neurons from the same acquisition. Summarized data show plots of frequency (D and E), inter-spike interval (ISI; F and G), and integrated area (H and I) for spontaneously active neurons following acute treatment with gabazine or picrotoxin compared to sister control neurons. Data are represented as bar plots (D, F, and H), cumulative frequency plots (E and I), or relative frequency distribution of ISI data (G). Insets in I show mean traces scaled for amplitude. Biological n is 3 for gabazine and picrotoxin treatments. Values indicated on bar graphs in D and H the indicate number of individual neurons used to determine frequency (D) and integrated area (H). Values on bar graph in F indicate the number of pairs of consecutive action potentials used to determine ISI. Statistical tests are Kruskal-Wallis ANOVAs with multiple-comparisons tests to control data. ****p < 0.0001. Figure adapted from Walker, Raliski, & Karbasi, et al. (2020).
REAGENTS AND SOLUTIONS
Use Milli-Q purified water in all recipes and protocol steps.
MEM++++ culture medium
10 ml B27 (Invitrogen 17504-044)
5 ml GlutaMAX (Invitrogen 35050-061)
25 ml fetal bovine serum (VWR 89510-186)
10 ml 1 M dextrose (FisherScientific D16-500; sterile filtered)
500 ml MEM medium for rat dissection (Invitrogen 11090-081)
Combine all components, sterile filter, and transfer aliquots into 50-ml tubes. Store at 4°C indefinitely.
Neurobasal (NB++) culture medium
10 ml B27 (Invitrogen 17504-044)
5 ml GlutaMAX (Invitrogen 35050-061)
500 ml Neurobasal medium (Invitrogen 21103-049)
Combine all components, sterile filter, and transfer aliquots into 50-ml tubes. Store at 4°C indefinitely.
Sodium borate buffer
1.55 g boric acid
4.50 g sodium tetraborate decahydrate
500 ml Milli-Q water
Combine all components, bring the pH to 8.5, and sterile filter. Store at 4°C indefinitely.
COMMENTARY
Background Information
The collective firing of neural circuits underlies our entire human experience. Historically, however, most understanding of the electrical behavior of neurons comes from single-cell recordings. Thus, numerous techniques have been developed in the past few decades to obtain a more complete picture of voltage changes in neural circuits. Multielectrode arrays (MEAs) can contain thousands of microelectrodes that record electrical signals across an entire dissociated culture. However, without imaging, MEAs still offer poor spatial resolution as they cannot connect specific neurons with specific field potentials. Measuring voltage changes optically presents an attractive alternative to electrical techniques by removing the requirement for physical contact with the neurons being analyzed and offering much higher spatial resolution. Thus, over the past five decades many small-molecule voltage-sensitive dyes and genetically encoded voltage indicators (GEVIs) have been developed in attempts to turn voltage signals into robust optical signals (Kulkarni & Miller, 2017).
Early GEVIs were constructed from the combinations of voltage-sensing domains (VSDs) and fluorescent proteins (FPs; Kulkarni & Miller, 2017). Though sensitive to voltage changes, these early indicators experienced severe trafficking defects that limited the amount of indicator in the cell membrane (Siegel & Isacoff, 1997). Over the past two decades, much work has been done to improve the trafficking, sensitivity, and kinetics of these VSD-FP fusion constructs (Xu, Zou, & Cohen, 2017). However, the slow kinetics and capacitive load of such constructs still limit their use in certain applications (Kulkarni & Miller, 2017). The other main class of GEVIs that has been developed more recently is based on light-gated ion pumps or channels that are then run in reverse to display a change in fluorescent properties in response to voltage changes (Kralj, Douglass, Hochbaum, MacLaurin, & Cohen, 2012). Constructs based on these opsin proteins show faster kinetics and better membrane targeting but have complex photocycles, can generate photocurrents, and are generally dim. Fluorescent proteins or small-molecule dyes can be combined with opsin proteins to improve brightness, but these reporters show decreases in fluorescence in response to voltage changes and still have complex photocycles (Abdelfattah et al., 2019).
Similar to GEVIs, there are also many types of voltage-sensitive small molecules. Historically, electrochromic dyes and oxonol dyes were the most widely used voltage indicators. Electrochromic dyes display slight shifts their absorbance and emission properties in response to voltage changes (Miller, 2016). As a result, these compounds respond quickly enough to voltage changes to sense individual action potentials but display low voltage sensitivity (Fluhler, Burnham, & Loew, 1985). Oxonol dyes are charged lipophilic molecules that sense voltage changes by redistributing within the plasma membrane (González & Tsien, 1995). They therefore respond too slowly to sense individual action potentials, but they have higher voltage sensitivities than electrochromic dyes. Recently, dyes that use a photo-induced electron transfer (PeT) process to sense membrane voltage changes were developed as a way to achieve both fast responses and high voltage sensitivity (Miller et al., 2012). These voltage-sensitive fluorophores, or VoltageFluors, consist of a fluorescent dye head, a lipophilic molecular wire that inserts into cell membranes, and an electron-rich donor group that quenches the fluorescent dye by photoinduced electron transfer (PeT). The voltage across a cell membrane affects the rate of PeT, which is observed as a change in the quantum yield of the dye (Miller, 2016). As PeT is an electron-mediated process, VoltageFluors can respond to voltage changes fast enough to sense individual action potentials and display high voltage sensitivities. Our lab has developed a number of VoltageFluor derivatives (Liu & Miller, 2020). To date, BeRST 1 displays the highest signal-to-noise ratios for action potential detection in dissociated culture (Huang et al., 2015). To handle the large amount of data that is generated in a spontaneous voltage imaging experiment, our lab has also developed a MATLAB program for semiautomated spike detection, termed SpikeConnect (Walker, Raliski, & Nguyen, et al., 2020). Thus, this protocol serves as guide to the spontaneous voltage imaging experiments that are becoming routine in our lab and in academic labs across the globe.
Critical Parameters
Cell health
When beginning spontaneous activity imaging experiments, it is critical that the dissociated cultures not vary widely in cell health or density from week to week. Cells grown at different densities or culture environments can have different baseline firing rates, which will complicate analysis of data obtained over multiple weeks of imaging. As stated in the introduction to Basic Protocol 1, having a regular rotation of personnel preparing the cultures can help to ensure reproducible cell health and density.
Imaging parameters
The most critical imaging parameter is imaging speed. The microscope must sample at 500 Hz or faster to reliably detect action potentials. As a result, the imaging window is confined by the hardware limitations of a specific microscope: If the microscope is unable to sample the whole field of view at 500 Hz, your imaging size will need to be reduced. Furthermore, appreciable photobleaching over the timescale of one imaging movie will complicate analysis. If photobleaching is observed, the excitation light power or the length of imaging will need to be reduced. Another good practice is to reduce the excitation light power when adjusting the focus in the fluorescence channel to reduce the amount of excessive light the neurons experience.
Drawing regions of interest
The main human input into the SpikeConnect program is the drawing of regions of interest around neurons. However, dissociated cultures also contain glial cells and cell debris in addition to neurons. Therefore, mislabeling of glial cells or cell debris as neurons can be a large source of error in analyzing spontaneous activity imaging data. When starting analysis, it is good practice to have personnel with experience in neuroanatomy train other researchers who will be carrying out the analysis. Then, both experienced and inexperienced personnel should label regions of interest on the same dataset so that errors can be identified and corrected.
Time Considerations
Basic Protocol 1 and Alternate Protocol should both take ~1 month to complete. This is because the rat must be at least 17 days pregnant before performing the dissection and cell plating. During this time, other supplies can be obtained or fabricated such as the microisland stencils. After the cells are plated, the cultures must be maintained for 2 weeks before they are mature and ready for imaging experiments. Basic Protocol 2 should only take 1 day to complete. However, if data from multiple weeks and cultures are desired, this step will last as long as dictated by the experimental plan. Basic Protocol 3 should only take 1 day to complete. Even if multiple weeks of imaging are planned, it is good practice to analyze the data for each week immediately after completing imaging.
Acknowledgments
The authors thank the National Institute of Neurological Disorders and Stroke (NINDS, NS098088) and the National Science Foundation (NeuroNex, 1707350) for support.
Footnotes
Internet Resources
https://github.com/evanwmiller/spikeconnect.git
GitHub repository for the MATLAB code for SpikeConnect.
Literature Cited
- Abdelfattah AS, Kawashima T, Singh A, Novak O, Liu H, Shuai Y, … Schreiter ER (2019). Bright and photostable chemigenetic indicators for extended in vivo voltage imaging. Science, 365, 699–704. doi: 10.1126/science.aav6416. [DOI] [PubMed] [Google Scholar]
- Audesirk G, Audesirk T, & Ferguson C. (2001). Culturing rat hippocampal neurons. Current Protocols in Toxicology, 1–17. doi: 10.1002/0471140856.tx1203s04. [DOI] [PubMed] [Google Scholar]
- Fluhler E, Burnham VG, & Loew LM (1985). Spectra, membrane binding, and potentiometric responses of new charge shift probes. Biochemistry, 24(21), 5749–5755. doi: 10.1021/bi00342a010. [DOI] [PubMed] [Google Scholar]
- González JE, & Tsien RY (1995). Voltage sensing by fluorescence resonance energy transfer in single cells. Biophysical Journal, 69(4), 1272–1280. doi: 10.1016/S0006-3495(95)80029-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YL, Walker AS, & Miller EW (2015). A photostable silicon rhodamine platform for optical voltage sensing. Journal of the American Chemical Society, 137, 10767–10776. doi: 10.1021/jacs.5b06644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kralj JM, Douglass AD, Hochbaum DR, MacLaurin D, & Cohen AE (2012). Optical recording of action potentials in mammalian neurons using a microbial rhodopsin. Nature Methods, 9, 90–95. doi: 10.1038/nmeth.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni RU & Miller EW (2017). Voltage imaging: Pitfalls and potential. Biochemistry, 56, 5171–5177. doi: 10.1021/acs.biochem.7b00490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, & Miller EW (2020). Electrophysiology, unplugged: Imaging membrane potential with fluorescent indicators. Accounts of Chemical Research, 53(1), 11–19. doi: 10.1021/acs.accounts.9b00514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EW (2016). Small molecule fluorescent voltage indicators for studying membrane potential. Current Opinion in Chemical Biology, 33, 74–80. doi: 10.1016/j.cbpa.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EW, Lin JY, Frady EP, Steinbach PA, Kristan WB, & Tsien RY (2012). Optically monitoring voltage in neurons by photoinduced electron transfer through molecular wires. Proceedings of the National Academy of Sciences of the United States of America, 109(6), 2114–2119. doi: 10.1073/pnas.1120694109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel MS, & Isacoff EY (1997). A genetically encoded optical probe of membrane voltage. Neuron, 19(4), 735–741. doi: 10.1016/S0896-6273(00)80955-1. [DOI] [PubMed] [Google Scholar]
- Walker AS, Raliski BK, Karbasi K, Zhang P, Sanders K, & Miller EW (2021). Optical spike detection and connectivity analysis with a far-red voltage-sensitive fluorophore reveals changes to network connectivity in development and disease. Frontiers in Neuroscience, 15, 643859. doi: 10.3389/fnins.2021.643859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AS, Raliski BK, Nguyen DV, Zhang P, Sanders K, Karbasi K, & Miller EW (2021). Imaging voltage in complete neuronal networks within patterned microislands reveals preferential wiring of excitatory hippocampal neurons. Frontiers in Neuroscience, 15, 43868. doi: 10.3389/fnins.2021.643868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zou P, & Cohen AE (2017). Voltage imaging with genetically encoded indicators. Current Opinion in Chemical Biology, 39, 1–10. doi: 10.1016/j.cbpa.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]