SUMMARY
Tissue regeneration requires coordinated and dynamic remodeling of stem and progenitor cells and the surrounding niche. Although the plasticity of epithelial cells has been well explored in many tissues, the dynamic changes occurring in niche cells remain elusive. Here, we show that, during lung repair after naphthalene injury, a population of PDGFRα+ cells emerges in the non-cartilaginous conducting airway niche, which is normally populated by airway smooth muscle cells (ASMCs). This cell population, which we term “repair-supportive mesenchymal cells” (RSMCs), is distinct from conventional ASMCs, which have previously been shown to contribute to epithelial repair. Gene expression analysis on sorted lineage-labeled cells shows that RSMCs express low levels of ASMC markers, but high levels of the pro-regenerative marker Fgf10. Organoid co-cultures demonstrate an enhanced ability for RSMCs in supporting club-cell growth. Our study highlights the dynamics of mesenchymal cells in the airway niche and has implications for chronic airway-injury-associated diseases.
Graphical Abstract
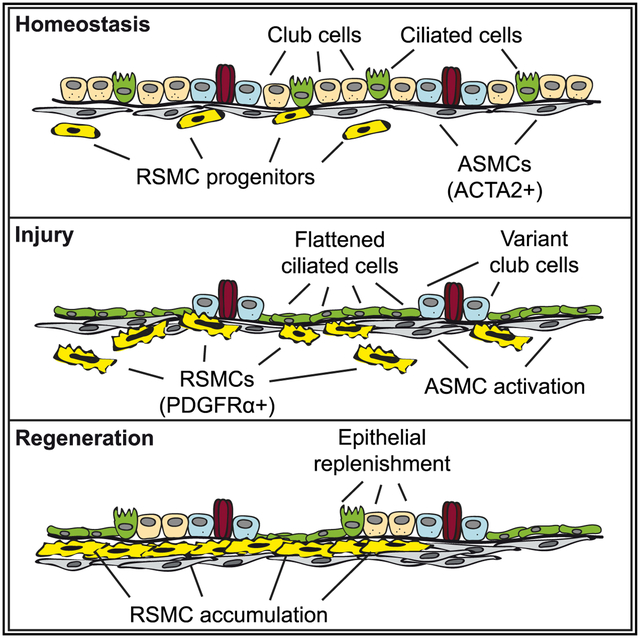
In Brief
Moiseenko et al. explore the dynamics of mesenchymal cells in the peribronchial niche in response to airway injury. They identify a population of mesenchymal cells located in close proximity to airway smooth muscle cells (ASMCs). This population, termed “repair-supportive mesenchymal cells” (RSMCs), is recruited to facilitate airway epithelial regeneration.
INTRODUCTION
The respiratory tract consists of a conducting zone and a respiratory zone. Although the respiratory zone represents the site of gas exchange between inhaled air and the pulmonary circulation, the conducting zone represents the first line of defense and first domain of contact between the respiratory system and the external environment. Club cells, previously known as exocrine bronchiolar cells or Clara cells (Clara, 1937; Winkelmann and Noack, 2010), are dome-shaped, non-ciliated secretory cells that populate the bronchiolar region of the lung epithelium. These cells can be identified by the expression of secretoglobin family 1A member 1 (SCGB1A1). In addition to having a major role in detoxifying the airways, club cells serve as long-term progenitors for ciliated and secretory cells (Rawlins et al., 2009; Stripp et al., 1995). Because of significant expression levels of cytochrome P450 family 2 subfamily f polypeptide 2 (CYP2F2), club cells are selectively targeted by naphthalene (NA); they metabolize it into a toxic derivative and consequently undergo necrosis and depletion (Boyd, 1977; Fanucchi et al., 1997; Mahvi et al., 1977). Interestingly, the surviving variant club cells (CYP2F2-low), located at neuroepithelial bodies (NEBs) or bronchioalveolar duct junctions (BADJs), mediate the repair process (Giangreco et al., 2002; Hong et al., 2001; Reynolds et al., 2000).
During embryonic lung development, the epithelium-derived WNT ligand, WNT7b, acts on adjacent airway and vascular smooth muscle cell (ASMC and VSMC) progenitors to induce platelet-derived growth factor receptor alpha and beta (Pdgfra and Pdgfrb) expression (Cohen et al., 2009). Therefore, the WNT/PDGFR pathway is important for SMC formation in the developing lung. Moreover, early ASMC precursors, located in the distal mesenchyme, express fibroblast growth factor 10 (Fgf10) (El Agha et al., 2014; Kumar et al., 2014; Mailleux et al., 2005), a key developmental gene that is critical for branching morphogenesis and maintenance of epithelial progenitors (Bellusci et al., 1997; Ramasamy et al., 2007; Sekine et al., 1999). FGF10 has been shown to be a potent pro-regenerative growth factor that poses protective and therapeutic effects in the context of various injury models in mice (Gupte et al., 2009; Quantius et al., 2016; Volckaert et al., 2011, 2017). Importantly, decreased FGF10 expression in humans is associated with chronic obstructive pulmonary disease (COPD) (Klar et al., 2011) and bronchopulmonary dysplasia (BPD) (Benjamin et al., 2007).
Previous studies have implicated similar epithelial-mesenchymal interactions in progenitor-cell activation after injury in the adult lung. Following NA injury, most club cells are depleted, and ciliated cells flatten to cover the denuded epithelium and maintain barrier function (Rawlins et al., 2007; Volckaert et al., 2011). As a result, surviving epithelial cells secrete the WNT ligand, WNT7b, which acts on neighboring ASMCs to induce Fgf10 expression (Volckaert et al., 2011). FGF10 then acts on surviving club cells expressing fibroblast growth factor receptor 2-IIIb (Fgfr2b) to induce epithelial regeneration (Volckaert et al., 2011). A similar WNT7b-FGF10 signaling axis has also been shown to maintain basal stem cells in the cartilaginous airways under homeostatic conditions and contributes to the mobilization of those cells in response to NA injury (Volckaert et al., 2017). Another elegant study has shown that leucine-rich repeat-containing G-protein coupled receptor 6 (Lgr6) expression identifies an ASMC subpopulation that promotes epithelial repair after NA injury in a similar WNT-FGF10-mediated fashion (Lee et al., 2017). Finally, a mesenchymal population of WNT-responsive (AXIN2+) cells contributes to de novo ASMC formation after NA injury (Zepp et al., 2017). Therefore, there is a notion that the peribronchiolar region represents a WNT-responsive mesenchymal niche for epithelial stem/progenitor cells and facilitates the repair process after injury by producing FGF10.
In this study, we lineage-traced alpha smooth muscle actin-positive (ACTA2+) cells after NA injury. In addition to bona fide ASMCs, we identified a population of PDGFRα+ cells that we termed “repair-supportive mesenchymal cells” (RSMCs). These cells are a source of FGF10 and display an enhanced ability to support club-cell growth in vitro. Our data highlight the heterogeneity and dynamics of peribronchial mesenchymal cells during airway regeneration.
RESULTS
β-Catenin Signaling and FGF10 Production in ACTA2+ Cells Are Critical for Epithelial Regeneration
To validate that β-catenin signaling in peribronchial ACTA2+ cells is critical for airway repair, Acta2-Cre-ERT2; Ctnnb1flox/flox (experimental) and Ctnnb1flox/flox (control) mice received an intraperitoneal (IP) injection of NA or corn oil and were exposed to tamoxifen after injury (Figure 1A). In line with previous studies (Lee et al., 2017; Volckaert et al., 2011, 2017), experimental mice displayed more severe weight loss, as well as suboptimal recovery in response to injury, compared with controls (Figure 1B). Quantification of SCGB1A1 immunofluorescence showed that, although the expression patterns were comparable at the peak of injury (day 3), experimental mice revealed progressive impairment of club-cell replenishment up to day 14 (Figures 1C and 1D). Gene expression analysis confirmed the histological observations and also revealed decreased expression of the regeneration markers Wnt7b and Fgf10 (Figure 1E). A similar result was obtained using Acta2-Cre-ERT2; Fgf10flox/flox mice in comparison with Fgf10flox/flox controls (Figures 1F–1J) and immunofluorescence showed decreased epithelial proliferation upon loss of Ctnnb1 or Fgf10 (Figure S1). Therefore, β-catenin signaling and FGF10 production in ACTA2+ cells after injury are important for epithelial repair.
Figure 1. Deletion of Ctnnb1 or Fgf10 in ACTA2+ Cells after Naphthalene Injury Impairs Epithelial Regeneration.
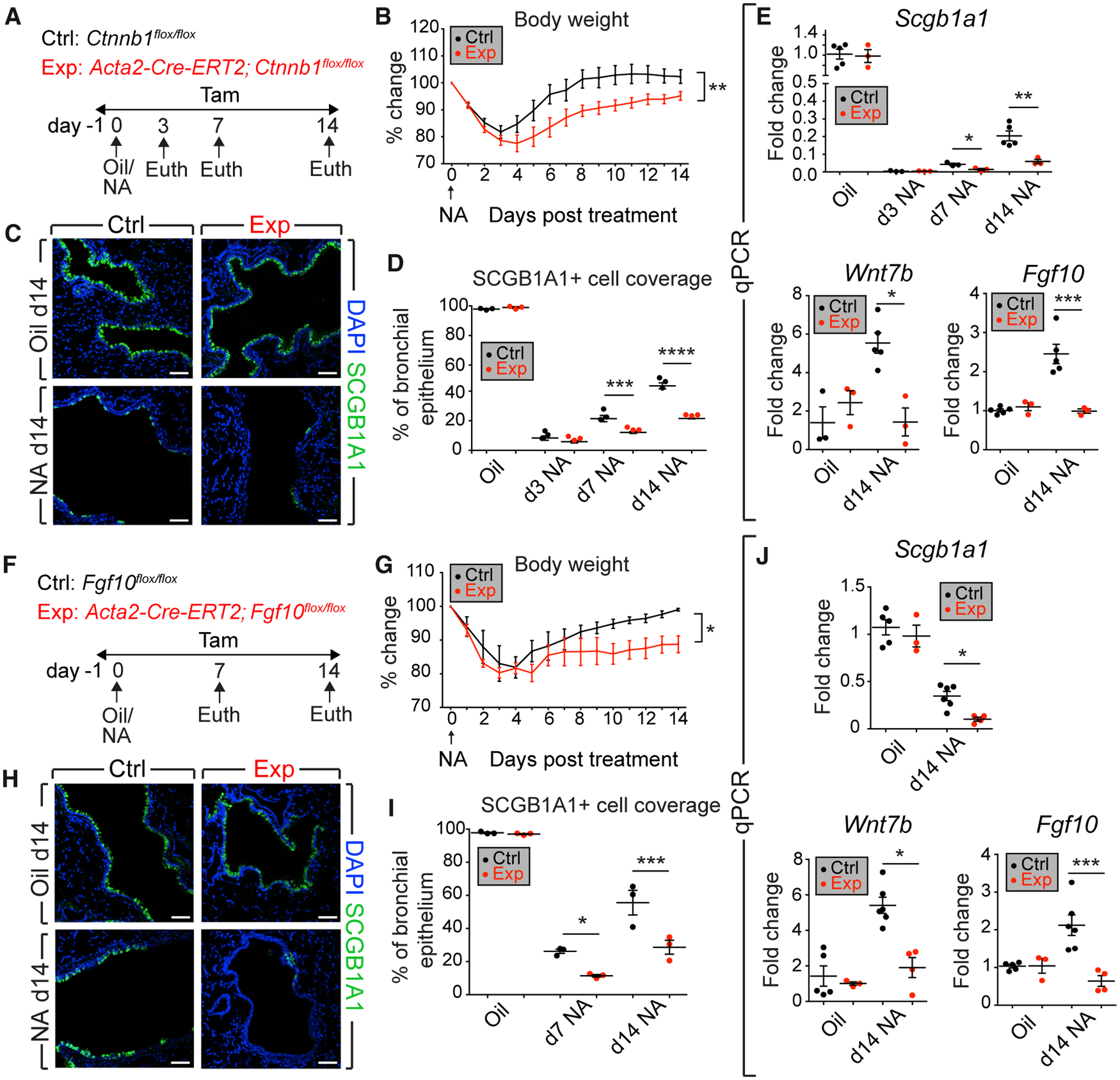
(A and F) Experimental setup and timeline of tamoxifen and NA treatment. Mice were fed tamoxifen-containing food.
(B–D and G–I) Weight-loss curves (B and G), immunofluorescence (C, H), and quantification of immunoreactivity (D, I) are shown.
(E and J) qPCR on lung homogenates.
Ctrl, control mice; Euth, euthanasia; Exp, experimental mice; NA, naphthalene; Tam, Tamoxifen. Scale bars: 50 μm. (B and E) n = 3 per group, except n = 5 for Ctrl-Oil and n = 5 for Ctrl-NA d14; (C, D, H, and I) n = 3 per group; (G and J) n = 5 for Ctrl-Oil, n = 3 for Exp-Oil, n = 6 for Ctrl-NA d14, and n = 4 for Exp-NA d14. Data are represented as means ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. See also Figure S1.
Mesenchymal Cells Expressing Pdgfra Emerge in Response to Epithelial Injury
To visualize peribronchial ACTA2+ cells after injury, Acta2-Cre-ERT2; tdTomatoflox mice received an IP injection of NA or corn oil, followed by five IP injections of tamoxifen (Figures 2A and 2D). Such labeling strategy allows labeling total ACTA2+ cells during repair, including bona fide ASMCs and other potential ACTA2+ cells. As expected, oil-treated lungs showed labeling of bona fide ASMCs around the conducting airways (Figure 2B). Interestingly, NA-treated lungs showed, in addition to ASMCs, a distinct population of lineage-labeled cells (Figure 2E). Those cells partially expressed Acta2 at day 7 (Figure S2A), were present at varying extents along the non-cartilaginous conducting airway, and were mostly observed between the ASMC sheet and the airway epithelium around the proximal lobular airways and at branch points. Because peribronchial PDGFRα+ cells are primed to differentiate along the ASMC lineage during embryonic development (El Agha et al., 2014; Andrae et al., 2014; Cohen et al., 2009; Endale et al., 2017; Gouveia et al., 2017; Ntokou et al., 2015), and based on recent work showing that PDGFRα and ACTA2 mark distinct populations of mesenchymal cells in the remodeled lung (Biasin et al., 2020), we decided to check whether those non-ASMC-lineage-labeled cells express Pdgfra. Indeed, immunofluorescence showed that 74% of those peribronchial non-ASMC-lineage-labeled cells were PDGFRα+ (Figures 2E, 2F, and 2N), whereas the expression of PDGFRα was very weak in oil-treated samples (Figures 2B and 2C). PDGFRα expression was not detected in the ASMC layer in either group (Figures 2C and F). Importantly, we could not detect PDGFRα+ ACTA2+ cells or PDGFRα+ tdTomato+ cells in the alveolar region, indicating that Acta2 is not induced in that region and that our approach does not label alveolar PDGFRα+ cells (Figures S2B and S2C). Because peribronchial PDGFRα+ cells derived from the Acta2 lineage could only be observed in injured lungs, we termed them “repair-supportive mesenchymal cells” (RSMCs). Proliferation could be detected in those cells at day 7 (Figure 2H). Notably, tdTomato+ cells did not express leukocyte, epithelial, or club-cell markers (Figure S3).
Figure 2. A PDGFRα+ Cell Population Emerges in Response to Epithelial Injury.
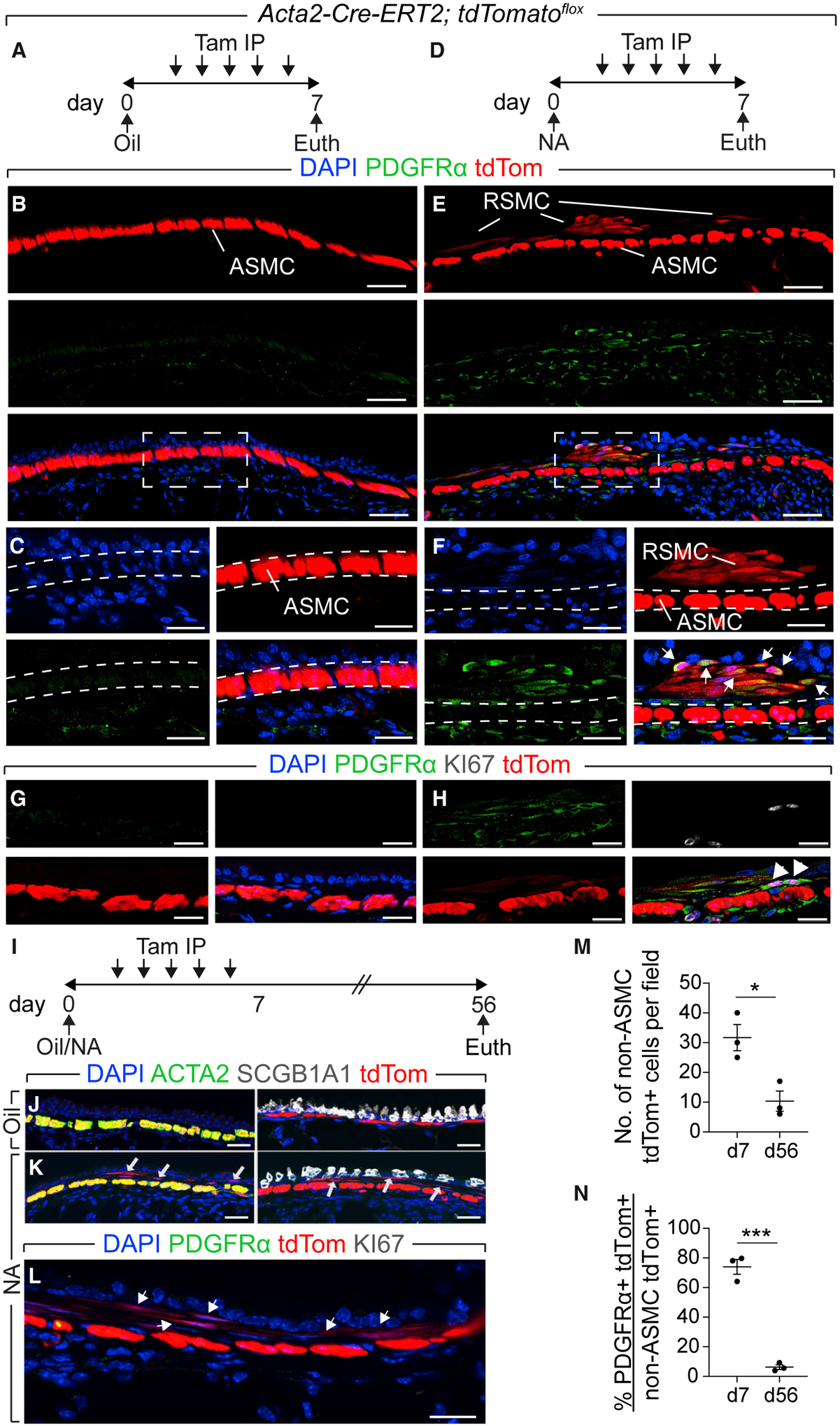
(A, D, and I) Experimental setup and timeline of NA and tamoxifen treatment. Mice received five intraperitoneal injections of tamoxifen.
(B, C, and E–H) Immunofluorescence revealing “repair-supportive mesenchymal cells” (RSMCs) after NA treatment.
(J–L) Immunofluorescence for the indicated markers.
(M and N) Quantification of tdTomato+ cells. IP, intraperitoneal.
Scale bars: (B and E) 50 μm; (C and F) 25 μm; (G, H, and J–L) 20 μm. n = 3 per group. Data are represented as means ± SEM. *p < 0.05, ***p < 0.001. See also Figures S2, S3, and S4.
To investigate the fate of RSMCs when homeostasis is restored, lungs were analyzed 8 weeks after injury (Figure 2I). RSMCs were still detected in NA-treated lungs, albeit at lower numbers compared with day 7 (Figures 2K and 2M), but lost Pdgfra expression (Figures 2L and 2N). When bleomycin injury was carried out (as a second hit) (Figure S4A), fibrosis-associated myofibroblasts (ACTA2+) did not co-express tdTomato and the abundance of RSMC descendants was unchanged (Figures S4B–S4G), indicating that RSMCs are not involved in further remodeling of the lung.
β-Catenin Is Upstream of Pdgfra and Fgf10 in RSMCs
We then decided to analyze the dynamics of the Acta2 lineage after NA injury by flow cytometry (Figures 3A and 3B). The abundance of the PDGFRα+ fraction (RSMC-enriched) out of the total lung suspension steadily increased from day 3 to day 7 and then to day 14 (Figure 3C). The PDGFRα− fraction, which includes ASMCs, also showed an increase in relative abundance from day 3 to days 7 and 14 (Figure 3D) confirming that ASMCs are also amplified in response to NA injury, as previously described (Lee et al., 2017; Volckaert et al., 2011, 2017). As expected, the PDGFRα+ fraction consistently showed greater expression levels for Pdgfra (as an internal control) compared with the PDGFRα− fraction (Figure 3E), whereas the PDGFRα− fraction showed higher expression levels for the SMC markers Acta2 (Figure 3H) and Lgr6 (Lee et al., 2017) (Figure 3I), thus confirming that this fraction is enriched with ASMCs. Interestingly, the PDGFRα+ fraction exhibited higher expression levels for Fgf10 (Figure 3F) and the WNT-responsive gene Axin2 (Figure 3G). compared with the PDGFRα− fraction.
Figure 3. Dynamics of PDGFRα+ tdTomato+ Cells.
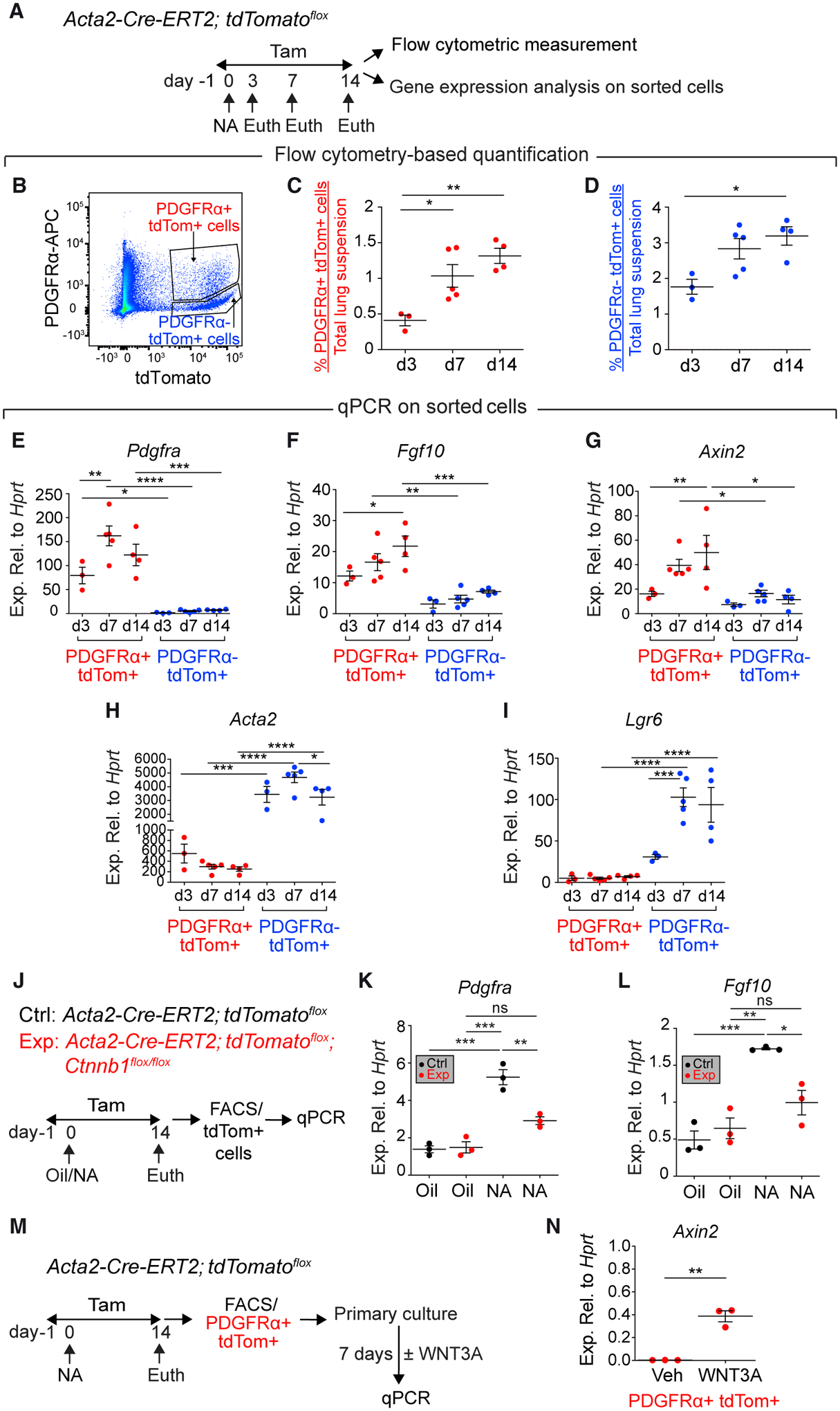
(A, J, and M) Experimental setup and timeline of NA and tamoxifen treatment. Mice were fed tamoxifen-containing food.
(B–D) Gating strategy and quantification of tdTomato+ cells.
(E–I) qPCR on sorted cells.
(K and L) qPCR on sorted tdTomato+ cells.
(N) qPCR on PDGFRα+ tdTomato+ cells.
n = 3 for d3, n = 5 for d7, and n = 4 for d14 (C–I); n = 3 per group (K, L, and N). Data are represented as means ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. See also Figure S2.
Next, Acta2-Cre-ERT2; tdTomatoflox; Ctnnb1flox/flox (experimental) and Acta2-Cre-ERT2; tdTomatoflox (control) mice were subjected to NA injury and exposed to tamoxifen for up to day 14 (Figure 3J). Fluorescence-activated cell sorting (FACS)-sorted tdTomato+ cells, derived from controls, displayed significant upregulation of Pdgfra (Figure 3K) and Fgf10 (Figure 3L). Strikingly, the induction of those markers was significantly attenuated in experimental samples receiving NA (Figures 3K and 3L), thus demonstrating that β-catenin signaling in ACTA2+ cells is critical for inducing Pdgfra and Fgf10 expression. To show that RSMCs respond to WNT ligands, PDGFRα+ tdTomato+ cells were isolated by FACS and cultured for 1 week in the presence or absence of recombinant WNT3A (Figure 3M). Gene expression analysis showed robust upregulation of Axin2 in response to WNT3A treatment (Figure 3N).
RSMCs Support Club-Cell Growth in an In Vitro Co-culture Organoid System
We finally tested whether the PDGFRα+ fraction possesses an enhanced intrinsic capacity to support the growth of club cells in vitro. Club cells were sorted from uninjured Scgb1a1Cre-ERT; tdTomatoflox mice and were co-cultured for 14 days with PDGFRα− tdTomato+ cells or PDGFRα+ tdTomato+ cells derived from NA-treated Acta2-Cre-ERT2; tdTomatoflox lungs (Figure 4A). tdTomato+ cells from oil-treated Acta2-Cre-ERT2; tdTomatoflox lungs were used as controls (Figure 4A). The co-culture conditions yielded predominantly bronchiolospheres (Figures 4B–4G). Strikingly, PDGFRα+ tdTomato+ cells yielded higher numbers of bronchiolospheres (Figure 4H) with larger diameters (Figure 4I) compared with control tdTomato+ cells and PDGFRα− tdTomato+ cells, indicating that the RSMC-enriched population has an enhanced ability to support club-cell growth.
Figure 4. Analysis of the Ability of RSMCs to Support Club-Cell Growth In Vitro.
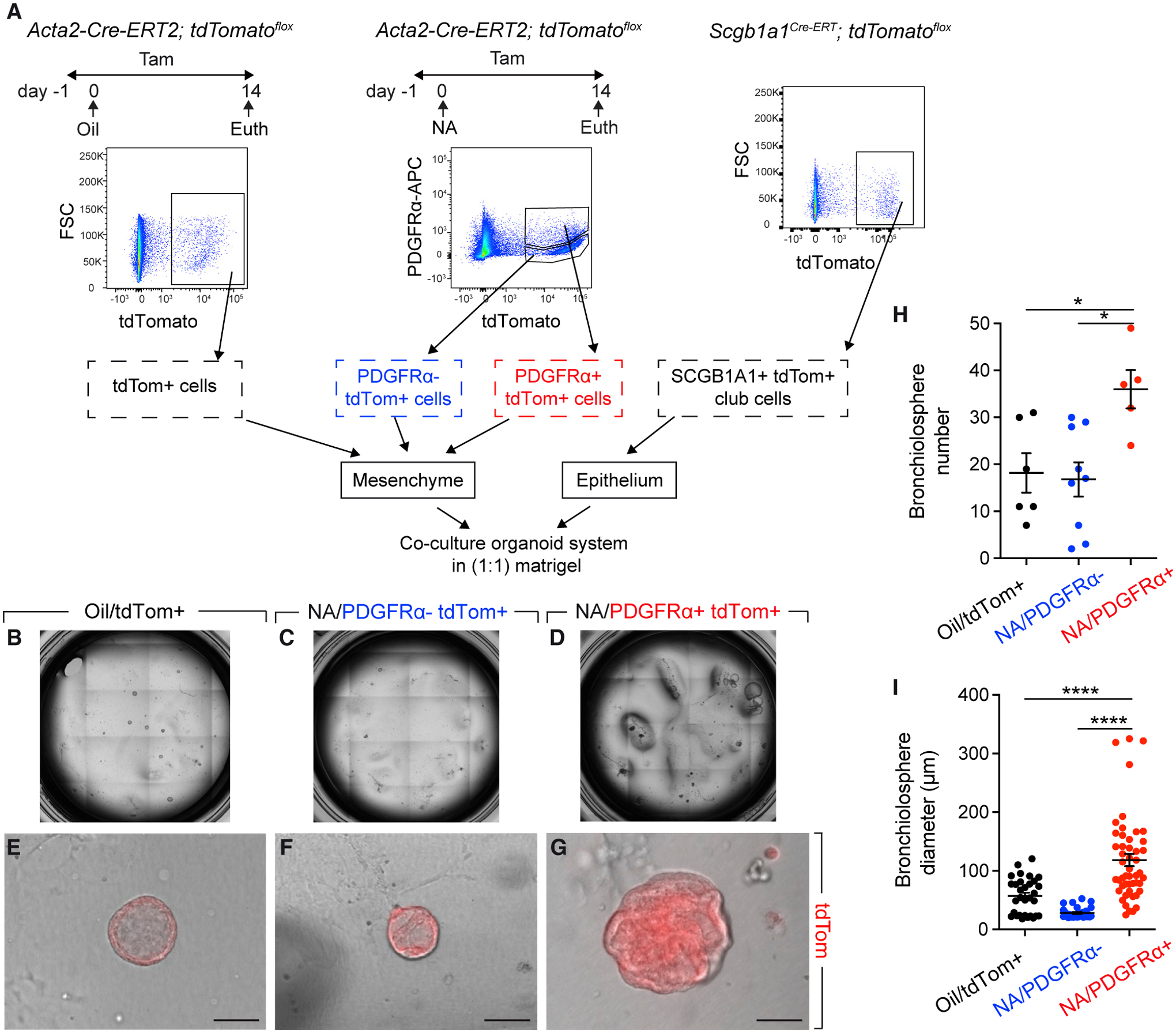
(A) Experimental setup and gating strategy for cell sorting. Indicated mice were fed tamoxifen-containing food.
(B–G) Overview and representative images of bronchiolospheres.
(H and I) Quantification of organoids.
Scale bars: 100 μm. n = 6 for [Oil/tdTomato+] + [SCGB1A1+], n = 9 for [NA/PDGFRα− tdTomato+] + [SCGB1A1+], and n = 5 for [NA/PDGFRα+ tdTomato+] + [SCGB1A1+]. Data are represented as means ± SEM. *p < 0.05, ****p < 0.0001.
DISCUSSION
In this study, we explored the complexity of the peribronchial region of the mature lung and identified RSMCs as a component of the niche required for airway regeneration. RSMCs are distinct from ASMCs in the confined peribronchial space and emerge upon club-cell depletion. A previous study identified a population of mesenchymal alveolar niche cells (MANCs) consisting of PDGFRα+ AXIN2+ cells that are involved in type 2 alveolar epithelial cell (AT2) maintenance in the distal lung, as well as a pool of AXIN2+ myofibrogenic progenitors (AMPs) that are largely PDGFRα− and give rise to ASMCs upon NA injury (Zepp et al., 2017). PDGFRα has also been used to identify lipofibroblasts that support AT2 (Barkauskas et al., 2013). Thus, PDGFRα has, so far, been described as a marker of the (distal) alveolar niche, rather than the (proximal) airway niche. We have recently shown that PDGFRα+ cells are indispensable for generating complex bronchioalveolar lung organoids (containing both proximal and distal structures) when co-cultured with bronchioalveolar stem cells (BASCs) (Vazquez-Armendariz et al., 2020). In the current study, we identify PDGFRα as a marker of RSMCs associated with airway repair. RSMCs highly express Axin2 and respond to WNT ligand stimulation in vitro. Our data are, therefore, in line with the notion that activation of WNT signaling in the peribronchial mesenchyme is important for airway regeneration (Lee et al., 2017; Volckaert et al., 2011; Zepp et al., 2017). Future studies will involve gain of function of β-catenin in these cells to investigate whether such an approach accelerates airway regeneration. In parallel, organoid cultures in the presence or absence of WNT stimulation will help determine whether forced activation of WNT signaling indeed promotes club-cell growth.
Our finding that PDGFRα+ cells derived from the Acta2 lineage possess greater potential to support club-cell growth in vitro compared with their PDGFRα− counterparts suggests that RSMCs are an important constituent of the airway niche. Because AMPs were shown to be largely PDGFRα− (Zepp et al., 2017), it is likely that RSMCs represent a distinct population of peribronchial mesenchymal cells that transiently upregulate Acta2 in response to injury (and are, therefore, captured using the Acta2-Cre-ERT2 driver line when tamoxifen is introduced after injury) and then acquire Axin2 expression. In the Zepp et al. (2017) paper, lineage tracing using Axin2Cre-ERT2 and PdgfraCre-ERT2 lines was performed by administering tamoxifen 1 week before NA injury. That allowed labeling of pre-existing, steady-state AXIN2+ cells and PDGFRα+ cells before injury, and following their fate during repair. In our approach, tamoxifen was administered after injury and this allowed the capture of cells that transit through the Acta2 lineage and upregulate Pdgfra. Timing is, therefore, a critical factor because RSMCs are absent in the uninjured lung and only emerge after epithelial injury. Future studies using other mesenchymal driver lines, such as the Fgf10Cre-ERT2 or Gli1Cre-ERT2, will help uncover the cellular source of RSMCs after NA injury. In that regard, peribronchial GLI1+ cells are a good candidate as they have been shown to be amplified after NA injury (Peng et al., 2015). Another logical follow-up for our work will be the use of dual recombinases (Cre-ERT2 and Dre-ERT2) to exclusively capture the cells of interest and isolate them for single-cell transcriptomics and organoid-based functional assays. Once the cell of origin is identified, the dynamics of the parent cell population as it enters the Acta2 lineage and later acquires Pdgfra expression will be analyzed. That would allow more-comprehensive analysis of the fate of those cells and the proportion of the cells becoming RSMCs versus persisting in an intermediate state (note that not all non-ASMC tdTomato+ cells are PDGFRα+). It will also be interesting to test whether the cells that are maintained in the intermediate state are capable of accelerating recovery after a second NA hit as they are already primed to become RSMCs.
A previous study used the non-inducible smooth muscle myosin heavy chain-Cre (Myh11-Cre) line to elegantly demonstrate that FGF10 produced by ASMCs is important for club-cell replenishment after NA injury (Volckaert et al., 2011). Our data are in line with those findings because we found that ASMCs are indeed amplified after injury (Figure 3D), they express the regeneration markers Fgf10 and Axin2 (Figures 3F and 3G), and are capable of supporting club-cell growth in vitro (Figure 4). However, the RSMC-enriched fraction of lineage-labeled cells showed higher expression of Fgf10 and Axin2, as well as greater potential in supporting bronchiolosphere formation compared with the ASMC-enriched fraction. In the future, it will be imperative to precisely quantify the contribution of RSMCs versus ASMCs to the repair process in the context of the Cre-ERT2/Dre-ERT2 lineage-tracing approach, single-cell transcriptomics, and organoid assays.
Our observation that lineage-traced cells were also found in close proximity to bronchial epithelial cells raised the possibility that our lineage-tracing approach might have labeled surviving epithelial progenitor cells undergoing transient epithelial-to-mesenchymal transition before regeneration. If that scenario were true, tdTomato would have colocalized with SCGB1A1 or CDH1 during the recovery phase when epithelial progenitors re-differentiate into airway epithelial cells. However, no colocalization between tdTomato and epithelial markers was observed at day 14, indicating that labeled cells are indeed of a mesenchymal nature.
When RSMCs were analyzed 8 weeks after NA injury, they had decreased in abundance and lost Pdgfra expression. This indicates that these cells are transiently activated in response to acute injury and are progressively cleared once homeostasis is re-established. When these animals were challenged with bleomycin 8 weeks after NA injury, we found that fibrosis-associated myofibroblasts did not derive from RSMCs. These data provide another piece of evidence that distinct mesenchymal niches are activated or employed depending on the nature and anatomical location of epithelial injury (conducting airways in the case of NA versus distal alveoli in the case of bleomycin).
To sum up, our study demonstrates that RSMCs are an important component of the peribronchial niche and complements the growing knowledge regarding the involvement of mesenchymal cells in lung repair. Our data thus warrant further research on the cellular and molecular mechanisms involved in many life-threatening and chronic respiratory diseases, such as idiopathic pulmonary fibrosis (IPF), asthma, and COPD.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Elie El Agha (elie.el-agha@innere.med.uni-giessen.de).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study did not generate/analyze datasets/code.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice and tamoxifen administration
Mice were housed in a specific-pathogen-free (SPF) environment with free access to food and water. To induce genetic recombination, 8–12-week-old female mice were fed tamoxifen-containing food (0.4 g of tamoxifen per kg of food) (Altromin, Germany) or injected intraperitoneally 5 times with tamoxifen dissolved in corn oil at a dose of 0.1 mg per g of body weight, as indicated. Animal experiments were approved by the Regierungspräsidium Giessen (protocol # G 2/2016), University of Alabama at Birmingham Institutional Animal Care and Use Committee and the Duke University Institutional Animal Care and Use Committee (protocol # A213–16-09). Animals were handled in accordance with the NIH and AAALAC guidelines for humane care and use of laboratory animals.
For naphthalene treatment, 8–12-week-old female mice were used. Naphthalene (Sigma-Aldrich) was dissolved in corn oil (20 mg/mL) and was administered intraperitoneally at a dose of 0.275 mg naphthalene per g of body weight. A subset of these mice was allowed to recover for 6 weeks after naphthalene/corn oil treatment and was then treated with bleomycin. For bleomycin injury, 2.5 U/Kg bleomycin (Hospira) or PBS was administered intranasally and mice were monitored daily. Mice were euthanized 2 weeks after bleomycin injury.
Organoid assay
Sorted mesenchymal cells (PDGFRα+ tdTomato+, PDGFRα− tdTomato+ and tdTomato+) from 8–12-week-old female Acta2-CreERT2; tdTomatoflox mouse lungs and epithelial cells (tdTomato+) from 12–14-week-old female Scgb1a1CreERT; tdTomatoflox mouse lungs were centrifuged and resuspended in Dulbecco’s Modified Eagle Medium (DMEM, Life Technologies). Concentrations were adjusted to 1×103 epithelial cells in 25 μL of medium and 2×104 mesenchymal cells in 25 μL of medium per insert (12 mm cell culture inserts with 0.4 μm membrane (Millipore) were used). Epithelial and mesenchymal cell suspensions were mixed and cold Matrigel® growth factor-reduced (Corning) was added at a 1:1 dilution resulting in 100 μL final volume per insert. Matrigel cell suspensions were added on top of the filter membrane of the insert and incubated at 37°C for 5 min. Then, 350 μL of cell culture medium was added per well. Cells were incubated under air-liquid conditions at 37°C with 5% CO2 for 2 weeks. Media were changed 3 times per week.
METHOD DETAILS
Immunofluorescence
For histological analysis, lungs were perfused with PBS and fixed with 4% paraformaldehyde. Then, tissues were embedded in paraffin and cut into thin sections. Alternatively, lungs were placed in 30% sucrose at 4°C before being incubated in a 1:1 mixture of 30% sucrose (in PBS)/O.C.T. for 1 h at 4°C. Later, they were embedded in O.C.T. and cryosectioned. Immunofluorescence was performed using monoclonal anti-ACTA2 (Sigma-Aldrich, 1:200), monoclonal anti-CC10 (Santa Cruz Biotechnology, 1:200), polyclonal anti-PDGFRα (Abcam, 1:200), polyclonal anti-KI67 (Thermo Fisher Scientific, 1:200), monoclonal anti-CD45 (Novus Biologicals, 1:200) and monoclonal anti-CDH1 (BD Biosciences, 1:200) antibodies. Staining for KI67, CD45 and CDH1 was performed after antigen retrieval with citric buffer and boiling for 15 min. The latter stainings were followed by staining with polyclonal anti-RFP antibodies (Rockland, 1:200) in order to recover the tdTomato signal. Nuclei were counterstained with DAPI (4’,6-diamidino-2-phenylindole) (Life Technologies). Fluorescent images were acquired using Leica DM550 B fluorescence microscope (equipped with Leica DFC360 FX camera) or an upgraded version of a TCS SP5 confocal microscope (Leica Microsystems). For quantitative analysis, multiple images were used (n > 8). For each experiment, sections from at least 3 independent lungs were analyzed. Figures were assembled using Adobe Photoshop and Illustrator.
Flow cytometry analysis and cell sorting
Lungs were isolated in Hank’s balanced salt solution (HBSS, Life Technologies), cut into small pieces with a sharp blade and treated with 0.5% collagenase type IV in HBSS at 37°C for 45 min. Lung homogenates were passed through 18, 21 and 24G needles and then through 70 μm and 40 μm cell strainers (BD Biosciences) to obtain single-cell suspensions. Cells were centrifuged at 4°C at 1000 rpm for 5 min and then resuspended in MACS buffer (7.4% EDTA, 0.5% FCS in PBS, PH 7.2) containing anti-PDGFRα (Biolegend, 1:100) or isotype control for 20 min on ice in the dark. Cells were then washed with MACS buffer. Flow cytometry measurements and cell sorting were done using FACSAria III cell sorter (BD Biosciences) and data were analyzed using FlowJo (FlowJo LLC).
Quantitative real-time PCR
For RNA extraction, accessory lobes were lysed and homogenized using Bullet Blender Blue (Next Advance, USA), and RNA was extracted using RNeasy Mini kit (QIAGEN). Extraction of RNA from sorted cells was performed using RNeasy Micro kit (QIAGEN). Primers were designed using the Universal Probe Library Assay Design center program (Roche). Quantitative real-time PCR (qPCR) was performed using Light Cycler 480 II (Roche). Hypoxanthine guanine phosphoribosyl transferase (Hprt) was used to normalize gene expression. Primers used for qPCR are shown in Table S1.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantitative data were assembled using GraphPad Prism software (GraphPad Software) and presented as average values ± sem. Student’s t test was used to compare 2 groups, one-way ANOVA was used to compare 3 or more groups in the presence of 1 variable (for e.g., treatment, time or genotype), and two-way ANOVA was used to compare multiple groups in the presence of 2 independent variables (for e.g., treatment and genotype or time and genotype). The statistical significance between weight loss curves was calculated using ROC curve analysis. The number of biological replicates is indicated in the corresponding figure legends. Results were considered significant if p < 0.05.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-ACTA2 | Sigma-Aldrich | Cat#F3777; RRID: AB_476977 |
| Mouse monoclonal anti-CC10 | Santa Cruz Biotechnology | Cat#sc-390313 |
| Rabbit polyclonal anti-PDGFRα | Abcam | Cat#ab203491 |
| Rat monoclonal anti-PDGFRα | Biolegend | Cat#135907; RRID: AB_2043969 |
| Rabbit polyclonal anti-KI67 | Thermo Fisher Scientific | Cat#PA5-19462; RRID: AB_10981523 |
| Rat monoclonal anti-CD45 | Novus Biologicals | Cat# NB100-77417; RRID: AB_1083776 |
| Mouse monoclonal anti-CDH1 | BD Biosciences | Cat#610182; RRID: AB_397581 |
| Rabbit polyclonal anti-RFP | Rockland | Cat#600-401-379; RRID: AB_2209751 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Recombinant human WNT3A | R&D Systems | Cat#5036-WN-010 |
| Tamoxifen | Sigma-Aldrich | Cat#T5648 |
| Naphthalene | Sigma-Aldrich | Cat#84679 |
| Bleomycin | Hospira | Cat#NDC 61703-332-18 |
| Critical Commercial Assays | ||
| RNeasy Mini kit | QIAGEN | Cat#74104 |
| RNeasy Micro kit | QIAGEN | Cat#74004 |
| QuantiTect Rev. Transcription kit | QIAGEN | Cat#205311 |
| PowerUp SYBR Green Master Mix | Thermo Fisher Scientific | Cat#A25742 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Tg(Acta2-cre/ERT2)12Pcn | Pierre Chambon (Wendling et al., 2009) | MGI: 3831907 |
| Mouse: B6;129S6-Gt(ROSA)26 Sortm9(CAG-tdTomato)Hze/J | Jackson Laboratory | RRID: IMSR_JAX:007905 |
| Mouse: B6;129-Fgf10tm1.2Sms | Suzanne L. Mansour (Abler et al., 2009; Urness et al., 2010) | RRID: IMSR_JAX:023729 |
| Mouse: B6.129-Ctnnb1tm2Kem/KnwJ | Jackson Laboratory | RRID:IMSR_JAX:004152 |
| Mouse: B6N.129S6(Cg)-Scgb1a1tm1(cre/ERT)Blh/J | Christos Samakovlis | RRID: IMSR_JAX:016225 |
| Mouse: B6.Cg-Gt(ROSA)26 Sortm14(CAG-tdTomato)Hze/J | Christos Samakovlis | RRID: IMSR_JAX:007914 |
| Oligonucleotides | ||
| Primers for qPCR, see Table S1 | Metabion | N/A |
| Software and Algorithms | ||
| GraphPad Prism Software | GraphPad Software | https://www.graphpad.com/scientific-software/prism RRID:SCR_002798 |
| Adobe Photoshop | Adobe | RRID:SCR_014199 |
| Adobe Illustrator | Adobe | RRID:SCR_010279 |
| FlowJo | FlowJo LLC | https://www.flowjo.com/solutions/flowjo RRID: SCR_008520 |
| Universal Probe Library Assay Design center program | Roche | https://lifescience.roche.com/global_en/articles/Universal-ProbeLibrary-System-Assay-Design.html |
Highlights.
The mesenchymal niche is critical for airway epithelial regeneration
β-catenin signaling and FGF10 production are pivotal for proper repair after injury
RSMCs, which are distinct from ASMCs, emerge after club-cell depletion
RSMCs display high potential to support bronchiolosphere formation in vitro
ACKNOWLEDGMENTS
E.E.A. acknowledges the support of the German Research Foundation (DFG; KFO309 P7 and SFB CRC1213-project A04), ILH, CPI (EXC 2026, project ID: 390649896), UKGM, and DZL. S.B. acknowledges the support of the DFG (BE4443/14–1, BE4443/6–1, KFO309 P7, and SFB CRC1213-projects A02 and A04), DZL, the First Affiliated Hospital of Wenzhou Medical University, and CPI. J.-S.Z was funded by a start-up package from Wenzhou Medical University and the National Natural Science Foundation of China (grant number 81472601). C.C. was supported by the Interventional Pulmonary Key Laboratory of Zhejiang Province, the Interventional Pulmonology Key Laboratory of Wenzhou City, the Interventional Pulmonology Innovation Subject of Zhejiang Province, the National Natural Science Foundation of China (81570075 and 81770074), Zhejiang Provincial Natural Science Foundation (LZ15H010001), Zhejiang Provincial Science Technology Department Foundation (2015103253), and the National Key Research and Development Program of China (2016YFC1304000). S.H. was supported by the UKGM (FOKOOPV), DZL, and DFG (KFO309 P2/8, SFB1021 C05, and SFB TR84 B9). We thank Dr. Athanasios Fysikopoulos and Prof. Christos Samakovlis for providing the Scgb1a1Cre-ERT; tdTomatoflox mice.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108549.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Abler LL, Mansour SL, and Sun X (2009). Conditional gene inactivation reveals roles for Fgf10 and Fgfr2 in establishing a normal pattern of epithelial branching in the mouse lung. Dev. Dyn 238, 1999–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrae J, Gouveia L, He L, and Betsholtz C (2014). Characterization of platelet-derived growth factor-A expression in mouse tissues using a lacZ knock-in approach. PLoS ONE 9, e105477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkauskas CE, Cronce MJ, Rackley CR, Bowie EJ, Keene DR, Stripp BR, Randell SH, Noble PW, and Hogan BLM (2013). Type 2 alveolar cells are stem cells in adult lung. J. Clin. Invest 123, 3025–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellusci S, Grindley J, Emoto H, Itoh N, and Hogan BL (1997). Fibroblast growth factor 10 (FGF10) and branching morphogenesis in the embryonic mouse lung. Development 124, 4867–4878. [DOI] [PubMed] [Google Scholar]
- Benjamin JT, Smith RJ, Halloran BA, Day TJ, Kelly DR, and Prince LS (2007). FGF-10 is decreased in bronchopulmonary dysplasia and suppressed by Toll-like receptor activation. Am. J. Physiol. Lung Cell. Mol. Physiol 292, L550–L558. [DOI] [PubMed] [Google Scholar]
- Biasin V, Crnkovic S, Sahu-Osen A, Birnhuber A, El Agha E, Sinn K, Klepetko W, Olschewski A, Bellusci S, Marsh LM, and Kwapiszewska G (2020). PDGFRα and αSMA mark two distinct mesenchymal cell populations involved in parenchymal and vascular remodeling in pulmonary fibrosis. Am. J. Physiol. Lung Cell. Mol. Physiol 318, L684–L697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd MR (1977). Evidence for the Clara cell as a site of cytochrome P450-dependent mixed-function oxidase activity in lung. Nature 269, 713–715. [DOI] [PubMed] [Google Scholar]
- Clara M (1937). Zur histobiologie des bronchalepithels. Z. Mikrosk. Anat. Forsch 41, 321–347. [Google Scholar]
- Cohen ED, Ihida-Stansbury K, Lu MM, Panettieri RA, Jones PL, and Morrisey EE (2009). Wnt signaling regulates smooth muscle precursor development in the mouse lung via a tenascin C/PDGFR pathway. J. Clin. Invest 119, 2538–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Agha E, Herold S, Al Alam D, Quantius J, MacKenzie B, Carraro G, Moiseenko A, Chao C-M, Minoo P, Seeger W, and Bellusci S (2014). Fgf10-positive cells represent a progenitor cell population during lung development and postnatally. Development 141, 296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endale M, Ahlfeld S, Bao E, Chen X, Green J, Bess Z, Weirauch MT, Xu Y, and Perl AK (2017). Temporal, spatial, and phenotypical changes of PDGFRα expressing fibroblasts during late lung development. Dev. Biol 425, 161–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanucchi MV, Murphy ME, Buckpitt AR, Philpot RM, and Plopper CG (1997). Pulmonary cytochrome P450 monooxygenase and Clara cell differentiation in mice. Am. J. Respir. Cell Mol. Biol 17, 302–314. [DOI] [PubMed] [Google Scholar]
- Giangreco A, Reynolds SD, and Stripp BR (2002). Terminal bronchioles harbor a unique airway stem cell population that localizes to the bronchoalveolar duct junction. Am. J. Pathol 161, 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouveia L, Betsholtz C, and Andrae J (2017). Expression analysis of platelet-derived growth factor receptor alpha and its ligands in the developing mouse lung. Physiol. Rep 5, e13092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupte VV, Ramasamy SK, Reddy R, Lee J, Weinreb PH, Violette SM, Guenther A, Warburton D, Driscoll B, Minoo P, and Bellusci S (2009). Overexpression of fibroblast growth factor-10 during both inflammatory and fibrotic phases attenuates bleomycin-induced pulmonary fibrosis in mice. Am. J. Respir. Crit. Care Med 180, 424–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong KU, Reynolds SD, Giangreco A, Hurley CM, and Stripp BR (2001). Clara cell secretory protein-expressing cells of the airway neuroepithelial body microenvironment include a label-retaining subset and are critical for epithelial renewal after progenitor cell depletion. Am. J. Respir. Cell Mol. Biol 24, 671–681. [DOI] [PubMed] [Google Scholar]
- Klar J, Blomstrand P, Brunmark C, Badhai J, Håkansson HF, Brange CS, Bergendal B, and Dahl N (2011). Fibroblast growth factor 10 haploin-sufficiency causes chronic obstructive pulmonary disease. J. Med. Genet 48, 705–709. [DOI] [PubMed] [Google Scholar]
- Kumar ME, Bogard PE, Espinoza FH, Menke DB, Kingsley DM, and Krasnow MA (2014). Mesenchymal cells: defining a mesenchymal progenitor niche at single-cell resolution. Science 346, 1258810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Tammela T, Hofree M, Choi J, Marjanovic ND, Han S, Canner D, Wu K, Paschini M, Bhang DH, et al. (2017). Anatomically and functionally distinct lung mesenchymal populations marked by Lgr5 and Lgr6. Cell 170, 1149–1163.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahvi D, Bank H, and Harley R (1977). Morphology of a naphthalene-induced bronchiolar lesion. Am. J. Pathol 86, 558–572. [PMC free article] [PubMed] [Google Scholar]
- Mailleux AA, Kelly R, Veltmaat JM, De Langhe SP, Zaffran S, Thiery JP, and Bellusci S (2005). Fgf10 expression identifies parabronchial smooth muscle cell progenitors and is required for their entry into the smooth muscle cell lineage. Development 132, 2157–2166. [DOI] [PubMed] [Google Scholar]
- Ntokou A, Klein F, Dontireddy D, Becker S, Bellusci S, Richardson WD, Szibor M, Braun T, Morty RE, Seeger W, et al. (2015). Characterization of the platelet-derived growth factor receptor-α-positive cell lineage during murine late lung development. Am. J. Physiol. Lung Cell. Mol. Physiol 309, L942–L958. [DOI] [PubMed] [Google Scholar]
- Peng T, Frank DB, Kadzik RS, Morley MP, Rathi KS, Wang T, Zhou S, Cheng L, Lu MM, and Morrisey EE (2015). Hedgehog actively maintains adult lung quiescence and regulates repair and regeneration. Nature 526, 578–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quantius J, Schmoldt C, Vazquez-Armendariz AI, Becker C, El Agha E, Wilhelm J, Morty RE, Vadász I, Mayer K, Gattenloehner S, et al. (2016). Influenza Virus Infects Epithelial Stem/Progenitor Cells of the Distal Lung: Impact on Fgfr2b-Driven Epithelial Repair. PLoS Pathog 12, e1005544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy SK, Mailleux AA, Gupte VV, Mata F, Sala FG, Veltmaat JM, Del Moral PM, De Langhe S, Parsa S, Kelly LK, et al. (2007). Fgf10 dosage is critical for the amplification of epithelial cell progenitors and for the formation of multiple mesenchymal lineages during lung development. Dev. Biol 307, 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins EL, Ostrowski LE, Randell SH, and Hogan BL (2007). Lung development and repair: contribution of the ciliated lineage. Proc. Natl. Acad. Sci. USA 104, 410–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins EL, Okubo T, Xue Y, Brass DM, Auten RL, Hasegawa H, Wang F, and Hogan BLM (2009). The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell 4, 525–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds SD, Giangreco A, Power JHT, and Stripp BR (2000). Neuroepithelial bodies of pulmonary airways serve as a reservoir of progenitor cells capable of epithelial regeneration. Am. J. Pathol 156, 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine K, Ohuchi H, Fujiwara M, Yamasaki M, Yoshizawa T, Sato T, Yagishita N, Matsui D, Koga Y, Itoh N, and Kato S (1999). Fgf10 is essential for limb and lung formation. Nat. Genet 21, 138–141. [DOI] [PubMed] [Google Scholar]
- Stripp BR, Maxson K, Mera R, and Singh G (1995). Plasticity of airway cell proliferation and gene expression after acute naphthalene injury. Am. J. Physiol 269, L791–L799. [DOI] [PubMed] [Google Scholar]
- Urness LD, Paxton CN, Wang X, Schoenwolf GC, and Mansour SL (2010). FGF signaling regulates otic placode induction and refinement by controlling both ectodermal target genes and hindbrain Wnt8a. Dev. Biol 340, 595–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Armendariz AI, Heiner M, El Agha E, Salwig I, Hoek A, Hessler MC, Shalashova I, Shrestha A, Carraro G, Mengel JP, et al. (2020). Multilineage murine stem cells generate complex organoids to model distal lung development and disease. EMBO J 39, e103476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volckaert T, Dill E, Campbell A, Tiozzo C, Majka S, Bellusci S, and De Langhe SP (2011). Parabronchial smooth muscle constitutes an airway epithelial stem cell niche in the mouse lung after injury. J. Clin. Invest 121, 4409–4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volckaert T, Yuan T, Chao C-M, Bell H, Sitaula A, Szimmtenings L, El Agha E, Chanda D, Majka S, Bellusci S, et al. (2017). Fgf10-hippo epithelial-mesenchymal crosstalk maintains and recruits lung basal stem cells. Dev. Cell 43, 48–59.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendling O, Bornert J-M, Chambon P, and Metzger D (2009). Efficient temporally-controlled targeted mutagenesis in smooth muscle cells of the adult mouse. Genesis 47, 14–18. [DOI] [PubMed] [Google Scholar]
- Winkelmann A, and Noack T (2010). The Clara cell: a “Third Reich eponym”? Eur. Respir. J 36, 722–727. [DOI] [PubMed] [Google Scholar]
- Zepp JA, Zacharias WJ, Frank DB, Cavanaugh CA, Zhou S, Morley MP, and Morrisey EE (2017). Distinct mesenchymal lineages and niches promote epithelial self-renewal and myofibrogenesis in the lung. Cell 170, 1134–1148.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate/analyze datasets/code.


