SUMMARY
Dysregulated glycine metabolism is emerging as a common denominator in cardiometabolic diseases, but its contribution to atherosclerosis remains unclear. Here we demonstrate impaired glycine-oxalate metabolism through alanine-glyoxylate aminotransferase (AGXT) in atherosclerosis. As found in patients with atherosclerosis, glycine/oxalate ratio is decreased in atherosclerotic mice concomitant with suppression of AGXT. Agxt deletion in apolipoprotein E-deficient (Apoe−/−) mice decreases the glycine/oxalate ratio and increases atherosclerosis with induction of hepatic pro-atherogenic pathways, predominantly cytokine/chemokine signaling and dysregulated redox homeostasis. Consistently, circulating and aortic C-C motif chemokine ligand 5 (CCL5) and superoxide in lesional macrophages are increased. Similar findings are observed following dietary oxalate overload in Apoe−/− mice. In macrophages, oxalate induces mitochondrial dysfunction and superoxide accumulation leading to increased CCL5. Conversely, AGXT overexpression in Apoe−/− mice increases the glycine/oxalate ratio, decreases aortic superoxide, CCL5 and atherosclerosis. Our findings uncover dysregulated oxalate metabolism via suppressed AGXT as a driver and therapeutic target in atherosclerosis.
Keywords: AGXT, Amino acids, Atherosclerosis, CCL5, Glycine, Mitochondrial dysfunction, Oxalate
INTRODUCTION
Despite significant advances in diagnosis, drug development and medical treatment, cardiovascular diseases (CVDs) remain a leading cause of death worldwide (Virani et al., 2020). Atherosclerosis, the underlying cause of most CVDs, is a chronic disease of the arteries arising from imbalanced lipid metabolism, maladaptive immune response and dysregulated redox homeostasis (Griendling et al., 2016; Moore et al., 2013). While the association between altered lipid metabolism and CVDs is well-established, recent evidence indicates that dysregulated metabolism of specific amino acids plays an important role in the pathogenesis of atherosclerosis (Grajeda-Iglesias et al., 2018; Nitz et al., 2019; Rom et al., 2018; Zaric et al., 2020). Among all amino acids, lower circulating glycine is emerging as a common denominator in CVDs and related metabolic comorbidities (Rom et al., 2018), including coronary heart disease (Wittemans et al., 2019), myocardial infarction (Ding et al. 2015), obesity (Newgard et al. 2009), type 2 diabetes (T2D), Guasch-Ferré et al., 2016), metabolic syndrome (Li et al., 2018) and non-alcoholic fatty liver disease (NAFLD) (Gaggini et al., 2018, Rom et al., 2020). While numerous studies reported lower circulating glycine in cardiometabolic diseases, the contribution of impaired glycine metabolism to the development of atherosclerosis remains unclear.
Glycine is the simplest, nonessential amino acid and is synthesized primarily in the liver from serine, threonine, alanine and glyoxylate (Wang et al., 2013). Impaired biosynthesis of glycine in atherosclerosis was suggested by evidence of decreased glycine/serine ratio in plasma from patients with unstable atherosclerotic plaques (Hitzel et al., 2018). Furthermore, transcriptomics of human and mouse livers revealed suppression of genes driving glycine biosynthesis from serine, alanine and glyoxylate in NAFLD (Mardinoglu et al., 2014), predominantly alanine-glyoxylate aminotransferase (AGXT) (Asgharpour et al., 2016; Rom et al., 2020; Stepanova et al., 2010). AGXT is expressed primarily in the liver where it catalyzes glycine biosynthesis from alanine and glyoxylate. Functional deficiencies of AGXT lead to accumulation of glyoxylate which is rapidly converted to oxalate (Li et al., 1999; Fodor et al., 2012; Rom et al., 2020; Salido et al., 2006; Wang et al., 2013). Whereas limited literature proposed that dysregulated oxalate metabolism is linked to CVDs (Arafa et al., 2020; Devarajan, 2018), the role of this metabolic pathway in the pathogenesis of atherosclerosis has not been studied yet.
Here, using targeted metabolomics, we identified decreased ratios of glycine to its precursors or related metabolites, serine, threonine and oxalate, in patients with coronary artery disease (CAD). As found in patients with CAD, glycine/oxalate ratio was significantly decreased in atherosclerotic Apoe−/− mice that showed suppression of hepatic AGXT. Utilizing genetic and dietary approaches to manipulate oxalate in Apoe−/− mice combined with studies in isolated macrophages, we demonstrate that increased oxalate exposure drives accelerated atherosclerosis in relation with dysregulated redox homeostasis, increased inflammatory response and enhanced hypercholesterolemia. The therapeutic potential of targeting dysregulated oxalate metabolism in atherosclerosis was studied using adeno-associated virus (AAV)-mediated overexpression of AGXT in Apoe−/− mice that showed lower oxidative stress, inflammation and atherosclerosis. Thus, by studying impaired glycine metabolism in patients and mice with atherosclerosis and using mouse models to manipulate oxalate, we identified dysregulated oxalate metabolism via suppressed AGXT as a driver and therapeutic target in atherosclerosis.
RESULTS
Dysregulated Glycine and Oxalate Metabolism in Patients and Mice with Atherosclerosis
To determine whether glycine metabolism is dysregulated in atherosclerosis, we applied targeted metabolomics focusing on circulating glycine and related metabolites (Figure 1A) in patients with significant CAD (sCAD) compared to those without (No sCAD) (Figure S1A and S1B, Table S1). The patients with sCAD had a higher calcium score (P<0.0001) without significant differences in other atherosclerotic risk factors (Table S1). Metabolomics revealed that the glycine/serine ratio was decreased in patients with sCAD (P=0.0150, Figure 1B), as previously reported (Hitzel et al., 2018), while we additionally found a decrease in the glycine/threonine ratio (P=0.0020, Figure 1C) and a similar trend for the glycine/alanine ratio (P=0.0706, Figure 1D). Interestingly, glycine/oxalate ratio was most significantly decreased in patients with sCAD (by 27%, P=0.0004, Figure 1E). To test whether dysregulated glycine-oxalate metabolism is similarly observed in mice with atherosclerosis, we studied Apoe−/− mice fed a standard diet (SD) or Western diet (WD). First, we confirmed increased atherosclerosis after 12 weeks on WD (Figure 1F). Next, we analyzed the above metabolites and found a similar trend towards decreased glycine/serine ratio in the atherosclerotic mice (P=0.0952, Figure 1G), without significant differences in the glycine/threonine (Figure 1H) or glycine/alanine ratios (Figure 1I). In line with the human data, the glycine/oxalate ratio was most significantly decreased (by 51%, P=0.0162, Figure 1J). Accordingly, Western blot analysis revealed a significant decrease in the protein abundance of AGXT in livers from the atherosclerotic mice (by 75%, P=0.0079, Figure 1K). These findings indicate impaired glycine-oxalate metabolism through suppressed AGXT enzymatic pathway in atherosclerosis.
Figure 1. Dysregulated Glycine and Oxalate Metabolism in Patients and Mice with Atherosclerosis.
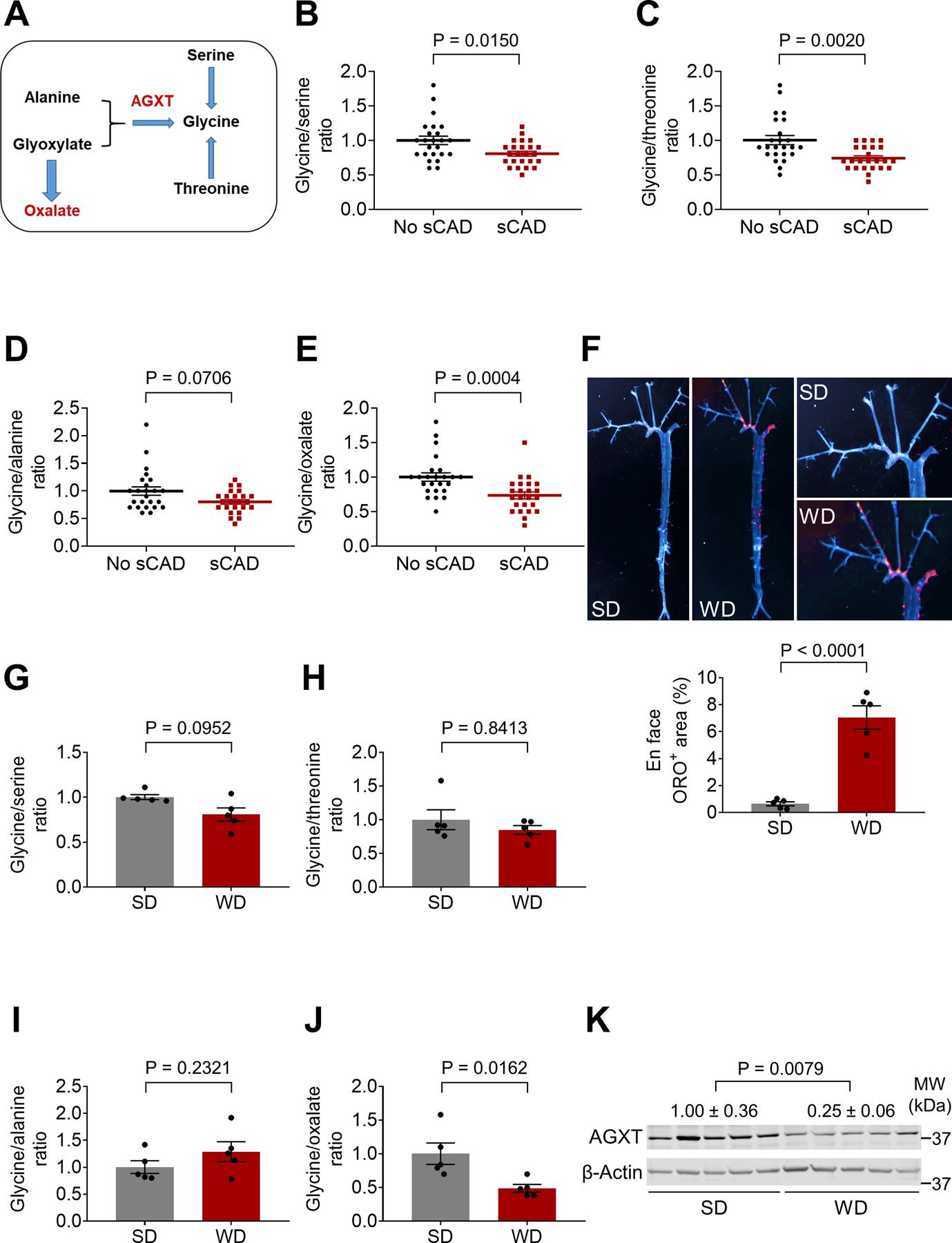
(A) Schematic representation of glycine metabolic pathways.
(B-E) Targeted metabolomics assessing the ratios of glycine to (B) serine, (C) threonine, (D) alanine and (E) oxalate in serum from age- and sex-matched patients with or without sCAD (n=24).
(F) En face analysis of atherosclerotic lesions in male Apoe−/− mice fed a standard diet (SD) or Western diet (WD) for 12 weeks (n=5).
(G-J) Targeted metabolomics assessing the ratios of glycine to (G) serine, (H) threonine, (I) alanine and (J) oxalate in plasma from male Apoe−/− mice fed a SD or WD for 12 weeks (n=5).
(K) Western blot analysis of AGXT protein abundance in livers from Apoe−/− mice fed a SD or WD for 12 weeks (n=5).
Mann-Whitney U test for B-E, G, H and K. Unpaired t test for F, I, J. Data are presented as mean ± SEM. All points and P values are shown.
AGXT Deficiency Exacerbates Atherosclerosis in Male Apoe−/− Mice
AGXT is expressed primarily in the liver and its loss-of-function leads to oxalate accumulation (Li et al. 1999; Rom et al. 2020; Salido et al. 2006). To explore the role of AGXT and oxalate metabolism in atherosclerosis, we generated Agxt−/− mice using CRISPR/Cas9 and crossed them with Apoe−/− mice (Figure S1C and S1D). We first confirmed the loss of AGXT in the liver (Figure 2A) and found a decreased glycine/oxalate ratio in plasma from male Agxt−/−/Apoe−/− mice (Figure 2B). After 12 weeks on WD, no significant differences in body weight (Figure S2A) and fasting blood glucose (Figure S2B) were observed between male Agxt−/−/Apoe−/− mice and Agxt+/+/Apoe−/− littermates. A significant increase in plasma total cholesterol was noted in male Agxt−/−/Apoe−/− mice (by 29.7%, P=0.0062, Figure 2C) with a similar trend for plasma triglycerides (P=0.0743, Figure S2C). We further addressed cholesterol changes in specific lipoprotein particles using fast protein liquid chromatography (FPLC). The FPLC analysis revealed increased cholesterol content in very-low-density lipoprotein (VLDL) and intermediate-density lipoprotein (IDL) particles in Agxt−/−/Apoe−/− mice (Figure S2D). En face analysis revealed a significant increase in atherosclerotic lesions in the whole aortic tree in male Agxt−/−/Apoe−/− mice (by 64.9%, P=0.0015, Figure 2D). We further characterized atherosclerosis in the aortic sinus using H&E and Oil Red O (ORO) staining and immunohistochemistry for macrophage content using Galectin 3 (Mac-2) (Figure 2E–G). We found a significant increase in the atherosclerotic plaque area and ORO positive area (Figure 2E and 2F) in male Agxt−/−/Apoe−/− mice compared to Agxt+/+/Apoe−/− littermates (by 56.2%, P=0.0268 and 50.4%, P=0.0404, respectively). Collectively, these findings indicate that dysregulated oxalate metabolism due to the loss of AGXT promotes the development of atherosclerosis.
Figure 2. AGXT Deficiency Exacerbates Atherosclerosis in Male Apoe−/− Mice.
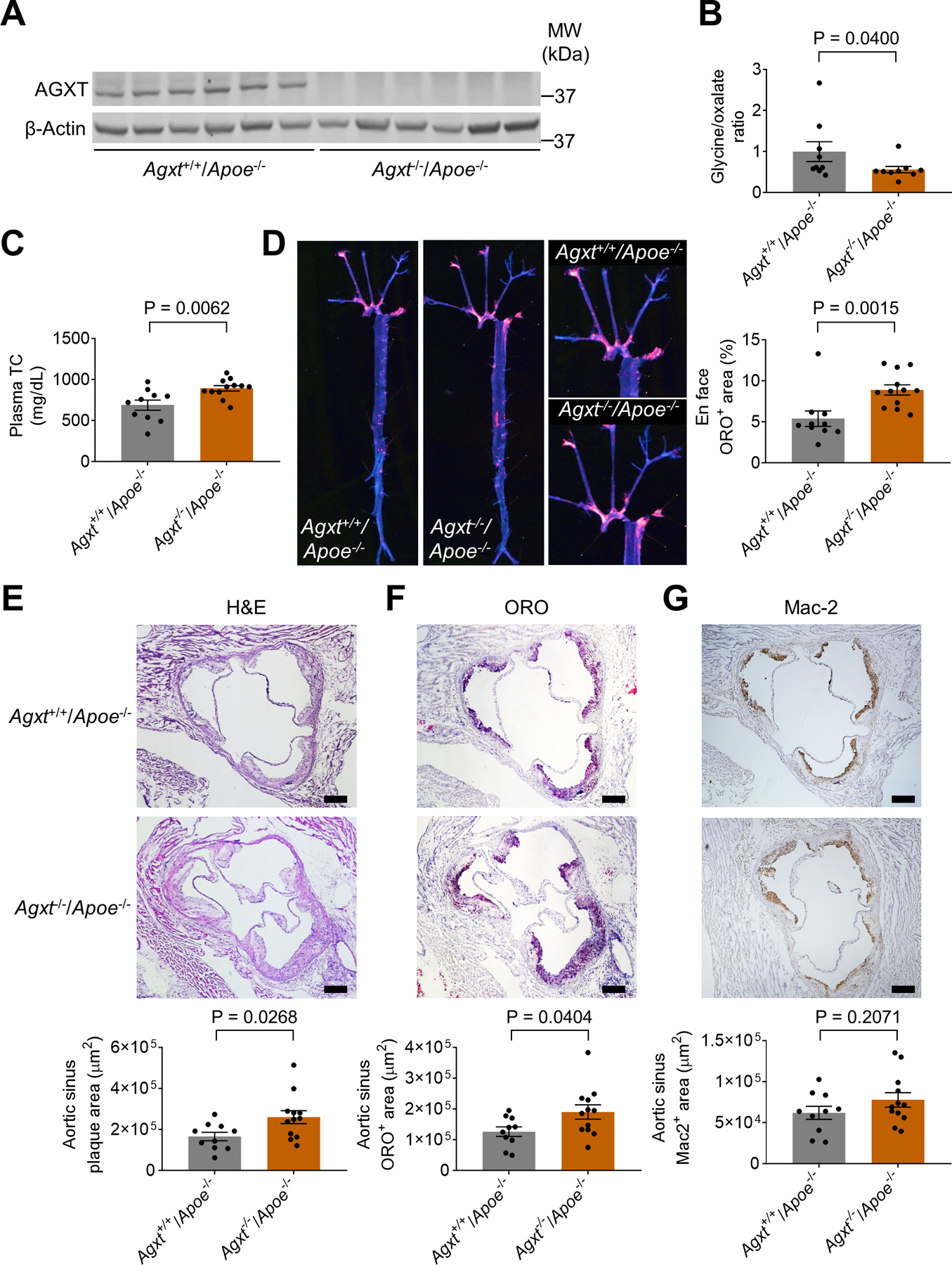
(A) Western blot analysis confirming the loss of AGXT in livers from male Agxt−/−/Apoe−/− mice (n=6).
(B) Glycine to oxalate ratio in plasma from male Agxt−/−/Apoe−/− mice and their Agxt+/+/Apoe−/− littermates (n=9).
(C-G) Male Agxt−/−/Apoe−/− (n=12) and Agxt+/+/Apoe−/− mice (n=10) were fed a WD for 12 weeks: (C) plasma total cholesterol (TC), (D) atherosclerosis in the aortic tree, (E) H&E staining, (F) Oil Red O (ORO) staining, and (G) Mac2 immunohistochemistry of the aortic sinus. (scale bar: 200 μm).
Unpaired t test for B-C and E-G. Mann-Whitney U test for D. Data are presented as mean ± SEM. All points and P values are shown.
Oxalate Homeostasis is Maintained and Atherosclerosis is Unaltered in Female Agxt−/−/Apoe−/− Mice
Previous rodent studies established that oxalate homeostasis is better maintained in females than males. Females present lower hepatic oxalate biosynthesis, urinary oxalate excretion and plasma oxalate concentration (Fan et al., 1999; Liang et al., 2014; Salido et al., 2006; Yoshihara et al., 1999; Zhu et al., 2019). To determine whether enhanced atherosclerosis is caused by loss of AGXT or due to impaired oxalate homeostasis, we next compared female Agxt−/−/Apoe−/− and Agxt+/+/Apoe−/− mice (Figure 3A). In contrast to the males, female Agxt−/−/Apoe−/− mice presented comparable glycine/oxalate ratio to Agxt+/+/Apoe−/− littermates (Figure 3B). As found in the males, no significant differences were observed in body weight and fasting blood glucose (Figure S2E and S2F) between female Agxt−/−/Apoe−/− and Agxt+/+/Apoe−/− mice. Unlike the males, no significant differences in plasma total cholesterol and cholesterol content in VLDL and IDL were noted between female Agxt−/−/Apoe−/− and Agxt+/+/Apoe−/− mice, despite being fed a WD for the same period as the males (Figure 3C, Figure S2G and S2H). Furthermore, atherosclerosis in the whole aortic tree and in the aortic sinus was comparable between female Agxt−/−/Apoe−/− mice and Agxt+/+/Apoe−/− littermates (Figure 3D–G). Taken together, these studies indicate that dysregulated oxalate metabolism, rather than the loss of AGXT itself, promotes atherosclerosis development.
Figure 3. Oxalate homeostasis is maintained and atherosclerosis is unaltered in female Agxt−/−/Apoe−/− mice.
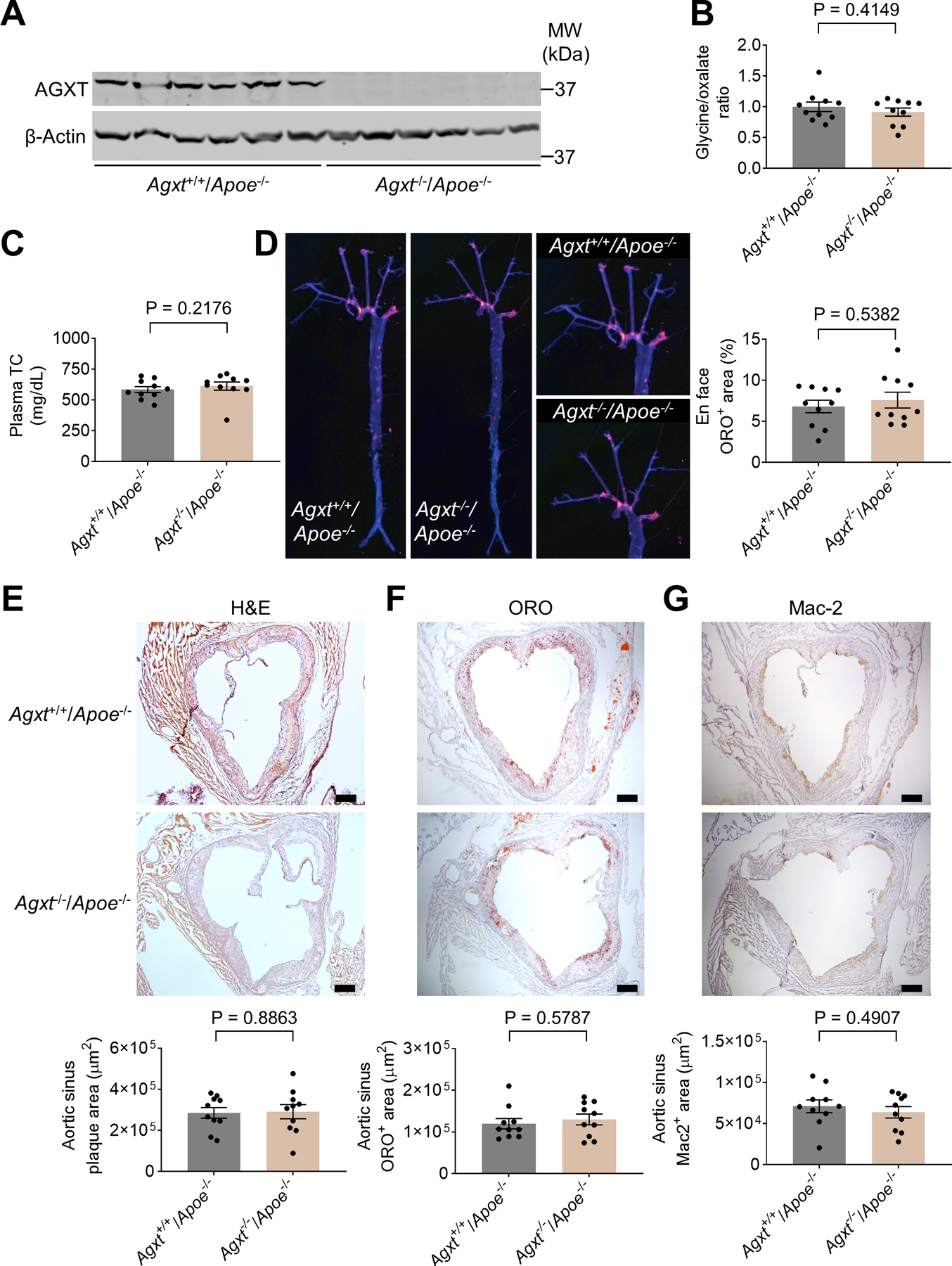
(A) Western blot analysis confirming the loss of AGXT in livers from female Agxt−/−/Apoe−/− mice (n=6).
(B) Glycine to oxalate ratio in plasma from female Agxt−/−/Apoe−/− and Agxt+/+/Apoe−/− mice (n=10).
(C-G) Female Agxt−/−/Apoe−/− and Agxt+/+/Apoe−/− mice were fed a WD for 12 weeks (n=10): (C) plasma total cholesterol (TC), (D) atherosclerosis in the aortic tree, (E) H&E staining, (F) Oil Red O staining, and (G) Mac2 immunohistochemistry of aortic sinus. (scale bar: 200 μm).
Unpaired t test for B, D, E and G. Mann-Whitney U test for C and F. Data are presented as mean ± SEM. All points and P values are shown.
Dysregulated Oxalate Metabolism Increases Pro-Inflammatory Responses, Circulating and Aortic CCL5
To explore potential mechanisms by which dysregulated oxalate metabolism enhances atherosclerosis, we utilized unbiased transcriptomics. Since AGXT is primarily expressed in the liver (Li et al., 1999), we first performed RNA-sequencing of livers from male Agxt−/−/Apoe−/− and Agxt+/+/Apoe−/− mice after 12 weeks on WD. We found 1812 differentially expressed genes (DEGs) with adjusted P value (padj) <0.05. Among those, 164 DEGs were downregulated and 180 DEGs were upregulated with a log2 fold change >1 (Figure 4A). Analysis of the top 100 DEGs revealed that key genes implicated in inflammatory responses (C-C motif chemokine receptor 1, Ccr1, C-X3-C motif ligand 1, Cx3cl1, nitric oxide synthase 2, Nos2, and triggering receptor expressed on myeloid cells 2, Trem2), redox homeostasis (glutathione S-transferase, alpha 2, Gsta2 and Gsta4) and fibrosis (collagen, type I, alpha 1, Col1a1, Col1a2 and Col3a1) were overexpressed in livers from Agxt−/−/Apoe−/− mice (Figure S3A). Pathway analysis revealed a most significant enrichment of pathways regulating cytokine-cytokine receptor interactions (padj=3.6×10−4) in Agxt−/−/Apoe−/− mice, followed by pathways regulating glutathione metabolism (padj=2.7×10−3), fluid shear stress and atherosclerosis (padj=1.1×10−2) and cell adhesion molecules (padj=2.1×10−2). The only pathway that was significantly downregulated (padj=2.9×10−2) was steroid hormone biosynthesis (Figure 4B). Analysis of DEGs linking the above pathways to atherosclerosis further underscored that genes promoting pro-inflammatory and pro-fibrotic responses and genes regulating redox homeostasis were overexpressed in Agxt−/−/Apoe−/− mice, while genes regulating sterol/steroid metabolism were suppressed (Figure 4C).
Figure 4. Dysregulated Oxalate Metabolism Induces a Pro-inflammatory Response and CCL5 Release.
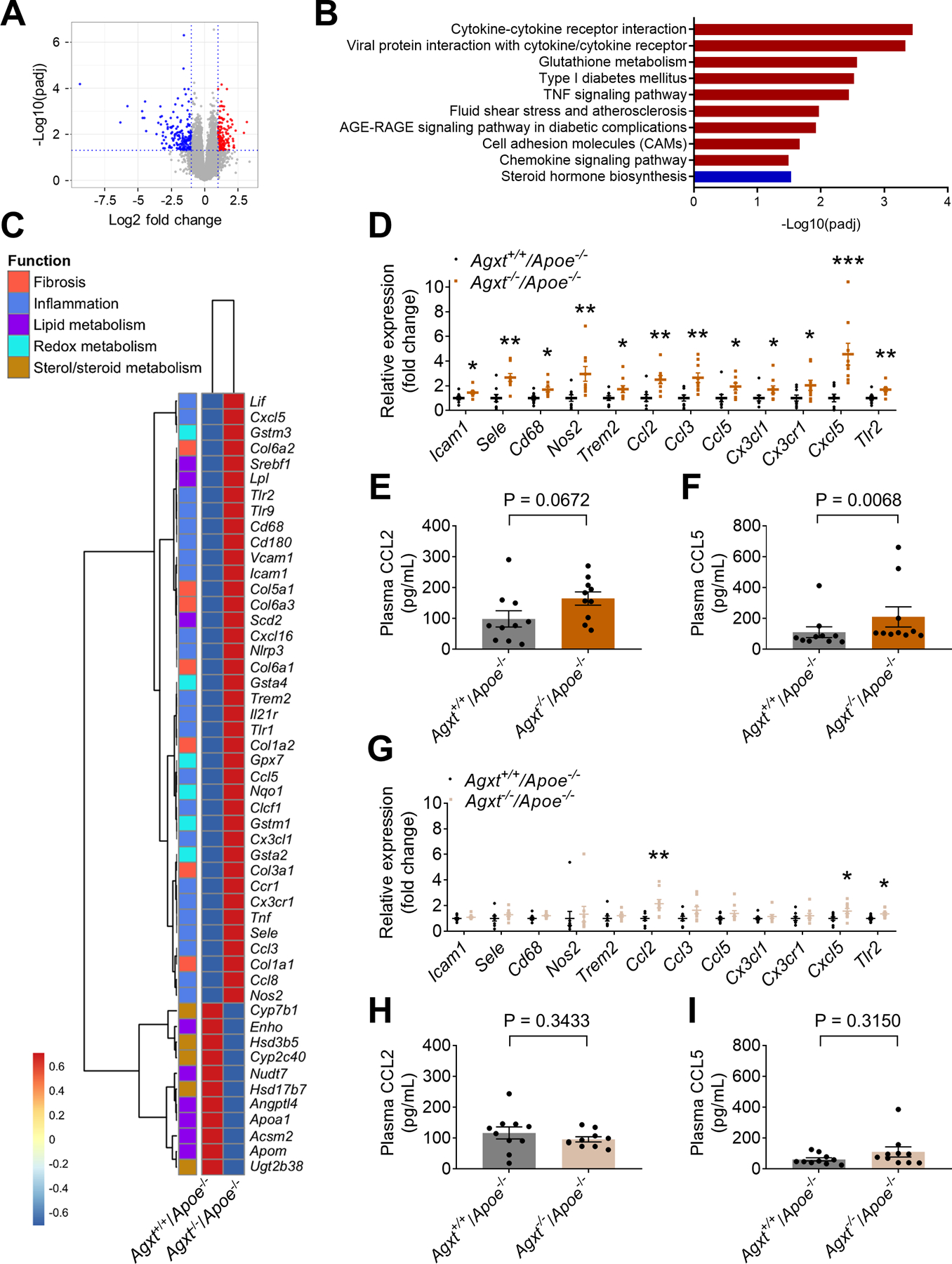
(A-C) RNA-sequencing of livers collected from male Agxt−/−/Apoe−/− and Agxt+/+/Apoe−/− mice fed a WD for 12 weeks (n=5): (A) Volcano plot of DEGs (padj <0.05, log2 fold change >1) in male Agxt−/−/Apoe−/− vs. Agxt+/+/Apoe−/ mice (Blue: downregulated; Red: upregulated). (B) Kyoto Encyclopedia of Genes and Genomes (KEGG)-based pathway analysis. The significance of the enrichment was determined by right-tailed Fisher’s exact test followed by Benjamini-Hochberg multiple testing adjustment. (C) Heatmap-based representation of 50 DEGs linking inflammation, fibrogenesis and the metabolism of lipids and sterols/steroids with atherosclerosis.
(D) qPCR analyses using independent samples of DEGs regulating inflammatory responses in livers from male Agxt−/−/Apoe−/− vs. Agxt+/+/Apoe−/ mice (n=9). Gene expression levels were normalized to 18S.
(E) CCL2 concentrations in plasma from male Agxt−/−/Apoe−/− vs. Agxt+/+/Apoe−/ mice (n=10).
(F) CCL5 concentrations in plasma from male Agxt−/−/Apoe−/− vs. Agxt+/+/Apoe−/ mice (n=10).
(G) qPCR analyses of genes regulating inflammatory responses in livers from female Agxt−/−/Apoe−/− vs. Agxt+/+/Apoe−/ mice (n=9). Gene expression levels were normalized to 18S.
(E) CCL2 concentrations in plasma from female Agxt−/−/Apoe−/− vs. Agxt+/+/Apoe−/ mice (n=10).
(F) CCL5 concentrations in plasma from female Agxt−/−/Apoe−/− vs. Agxt+/+/Apoe−/ mice (n=10).
Statistical differences in gene expression (D and G) were tested using unpaired t test or Mann-Whitney U test, depending on normality tests. *P<0.05, **P<0.01, ***P<0.001 vs. Agxt+/+/Apoe−/− mice. Unpaired t test for E and H. Mann-Whitney U test for F and I. Data are presented as mean ± SEM.
To determine whether the observed pro-inflammatory response is caused by the absence of AGXT or due to impaired oxalate homeostasis, we performed qPCR analyses both in male and female Agxt−/−/Apoe−/− mice and determined the circulating levels of pro-atherogenic chemokines (Noels et al., 2019). The RNA-sequencing findings were confirmed in male Agxt−/−/Apoe−/− mice using independent qPCR analyses. Genes encoding cell adhesion molecules (intercellular adhesion molecule 1, Icam1, and selectin E, Sele), key regulators of the inflammatory response and inflammatory markers (Cd68, Nos2, Trem2 and toll-like receptor 2, Tlr2) as well as chemokines and their receptors (Ccl2, Ccl3, Ccl5, Cx3cl1, Cx3cr1 and C-X-C motif chemokine 5, Cxcl5) were all significantly upregulated in livers from male Agxt−/−/Apoe−/− mice (Figure 4D). Accordingly, a trend towards increased plasma CCL2 was noted (Figure 4E), and plasma CCL5 was significantly increased in male Agxt−/−/Apoe−/− mice (by 90.0%, P=0.0068, Figure 4F). We next applied immunofluorescence to study changes in CCL5 within the atherosclerotic plaque. In line with increased hepatic expression and circulating levels, CCL5 was significantly increased in the aortic sinus of male Agxt−/−/Apoe−/− compared to Agxt+/+/Apoe−/− mice (by 79.5%, P=0.0487, Figure S3B). In contrast to the males, only Ccl2, Cxcl5 and Tlr2 were up-regulated in livers from female Agxt−/−/Apoe−/− mice (Figure 4G). Furthermore, plasma CCL2 and CCL5 (Figure 4H and 4I) were comparable between female Agxt−/−/Apoe−/− and Agxt+/+/Apoe−/− mice. Accordingly, CCL5 immunofluorescence in the aortic sinus was also comparable between female Agxt−/−/Apoe−/− and Agxt+/+/Apoe−/− mice (Figure S3C). Altogether, these studies indicate that dysregulated oxalate metabolism, not the absence of AGXT, induces a pro-inflammatory response including increased hepatic expression, circulating and aortic levels of CCL5.
Dysregulated Oxalate Metabolism Alters Cholesterol-Bile Acid Homeostasis
The RNA-sequencing revealed a suppression of key genes regulating sterol/steroid metabolism (cytochrome P450, family 7, subfamily b, polypeptide 1, Cyp7b1) and lipoprotein metabolism (Apoa1) and upregulation of major genes driving lipogenesis (sterol regulatory element binding transcription factor 1, Srebf1, and stearoyl-Coenzyme A desaturase 2, Scd2) and fibrogenesis in livers from male Agxt−/−/Apoe−/− mice. Using independent qPCR analyses, we confirmed the upregulation of pro-fibrotic genes (Col1a1, Col1a2, Col3a1 and Col5a1) and lipogenic genes (Srebf1 and Scd2) in livers from male Agxt−/−/Apoe−/− mice (Figure S4A and S4B). We next studied genes regulating sterol/steroid and lipoprotein metabolism to address the observed cholesterol differences in male Agxt−/−/Apoe−/− and Agxt+/+/Apoe−/− mice. We confirmed the suppression of Cyp7b1, but not Apoa1, in livers from male Agxt−/−/Apoe−/− mice (Figure S4C). Western blot analysis revealed that the protein abundance of CYP7B1 was decreased in livers from male Agxt−/−/Apoe−/− mice (by 52%, P=0.0068, Figure S4D). CYP7B1 plays a major role in the acidic pathway of cholesterol metabolism to bile acids (BAs) in the liver (Pandak and Kakiyama. 2019). In line, total BAs were significantly decreased in livers from male Agxt−/−/Apoe−/− mice (Figure S4E). Unlike the males, no significant changes in the hepatic expression of genes regulating fibrogenesis and lipid metabolism were found in female Agxt−/−/Apoe−/− mice except for Scd2 (Figure 4SF–4SH). Notably, both the mRNA and protein levels (Figure S4H and S4I) of CYP7B1 were comparable between female Agxt−/−/Apoe−/− and Agxt+/+/Apoe−/− mice. Altogether, these findings indicate that dysregulated oxalate metabolism suppresses cholesterol metabolism to BAs that may account for the increased hypercholesterolemia found in male Agxt−/−/Apoe−/− mice.
Oxalate Induces Mitochondrial Dysfunction and Overproduction of Superoxide Leading to CCL5 Release in Macrophages
The RNA-sequencing showed an overexpression of genes regulating redox homeostasis in livers from male Agxt−/−/Apoe−/− mice. In independent qPCR analyses, we confirmed that genes regulating glutathione metabolism (glutathione peroxidase 7, Gpx7, glutathione S-transferase, mu 1, Gstm1, Gstm3, Gsta2 and Gsta4) and antioxidant responses (NAD(P)H dehydrogenase, quinone, Nqo1) were significantly upregulated in livers from male Agxt−/−/Apoe−/− mice (Figure 5A), indicating a response to a pro-oxidant insult. To explore the link between dysregulated oxalate metabolism, redox homeostasis and atherosclerosis, we next evaluated reactive oxygen species (ROS) in the aortic sinuses from male Agxt+/+/Apoe−/− and Agxt−/−/Apoe−/− mice. Superoxide is one of the major ROS and its generation by macrophages is known to promote atherosclerosis (Vendrov et al., 2007). Dihydroethidium (DHE) fluorescence which is commonly used to detect superoxide in cardiovascular systems (Griendling et al., 2016; Vendrov et al., 2007), was significantly increased in the atherosclerotic lesions of male Agxt−/−/Apoe−/− mice (by 43.5%, P=0.0291). Using Mac-2 immunofluorescence, increased colocalization of the DHE signal in macrophages was observed in the atherosclerotic lesions of male Agxt−/−/Apoe−/− mice (Figure 5B). In contrast to the males, only Gstm1 was upregulated in the livers from female Agxt−/−/Apoe−/− mice (Figure 5C). Furthermore, no significant differences in DHE fluorescence were noted in the atherosclerotic lesions of female Agxt−/−/Apoe−/− and Agxt+/+/Apoe−/− mice (Figure 5D). Altogether, the transcriptome dysregulation, increased CCL5 and superoxide in the aortic sinuses of male, but not female Agxt−/−/Apoe−/− mice, indicate that dysregulated redox homeostasis and enhanced inflammatory responses are mediators likely to be associated with accelerated atherosclerosis driven by impaired oxalate metabolism.
Figure 5. Oxalate Overload Induces Mitochondrial Dysfunction and Overproduction of Superoxide Leading to CCL5 Release in Macrophages.
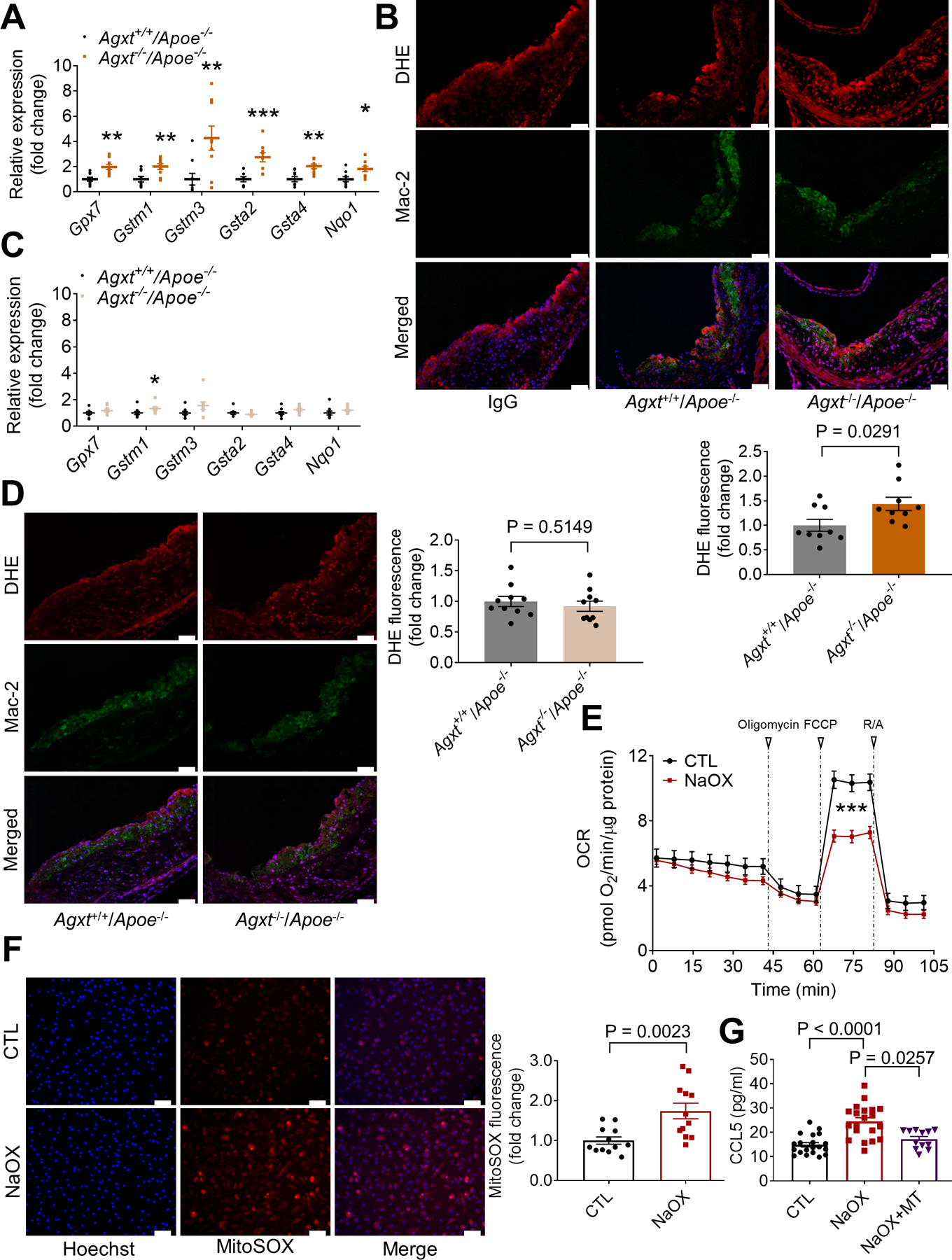
(A) qPCR analyses of genes regulating redox homeostasis in livers from male Agxt−/−/Apoe−/− vs. Agxt+/+/Apoe−/ mice (n=9). Gene expression levels were normalized to 18S.
(B) DHE fluorescence and Mac-2 immunofluorescence in the aortic sinuses of male Agxt−/−/Apoe−/− vs. Agxt+/+/Apoe−/ mice (scale bar: 50 μm. n=9).
(C) qPCR analyses of genes regulating redox homeostasis in livers from female Agxt−/−/Apoe−/− vs. Agxt+/+/Apoe−/ mice (n=9). Gene expression levels were normalized to 18S.
(D) DHE fluorescence and Mac-2 immunofluorescence in the aortic sinuses of female Agxt−/−/Apoe−/− vs. Agxt+/+/Apoe−/ mice (scale bar: 50 μm. n=10).
(E) Oxygen consumption rate (OCR) measured in BMDM treated with 0.75 mM NaOX for 30 min and in control cells (CTL) using Seahorse extracellular flux analyzer (n=16). Oligomycin, FCCP and rotenone+antimycin A (R/A) were used at final concentrations of 1.5 μM, 1 μM and 0.5 μM, respectively.
(F) Mitochondrial superoxide measured in BMDM treated with 0.75 mM NaOX for 30 min using the MitoSOX Red probe (n=12, scale bar: 50 μm).
(G) Medium concentration of CCL5 in BMDM treated with 0.75 mM NaOX for 18 h in the absence or presence of mitoTEMPO (MT, 10 μM, n=12–20).
Statistical differences in gene expression (A and C) were tested using unpaired t test or Mann-Whitney U test, depending on normality tests. *P<0.05, **P<0.01, ***P<0.001 vs. Agxt+/+/Apoe−/− mice. Unpaired t test for B, D and F. Mann-Whitney U test for E. Kruskal-Wallis test followed by Dunn’s post-hoc test for G. Data are presented as mean ± SEM. All points are shown.
To explore the link between oxalate, macrophage redox homeostasis and inflammation, we next performed experiments using bone marrow derived macrophages (BMDM). Characterized by respiratory chain dysfunction and increased ROS production, mitochondrial dysfunction in macrophages is known to promote inflammation and atherosclerosis (Madamanchi and Runge, 2007; Wang et al., 2014; Vendrov et al., 2007). We measured the oxygen consumption rate (OCR), mitochondrial superoxide production and mitochondrial mass in BMDM treated with non-cytotoxic levels of sodium oxalate (NaOX). Based on cell viability (Figure S5A) and previous studies (Patel et al., 2018), a concentration of 0.75 mM NaOX was chosen. NaOX caused mitochondrial dysfunction as evidenced by a significant decrease in maximal respiratory capacity (Figure 5E), increased mitochondrial superoxide production (Figure 5F) and decreased mitochondrial DNA (Figure S5B). Genes regulating redox homeostasis that were overexpressed in livers from male Agxt−/−/Apoe−/− mice (Gpx7, Gstm1, Gstm3 and Nqo1) were significantly upregulated in BMDM treated with NaOX (Figure S5C). Similarly, regulators of the inflammatory response, inflammatory markers, cytokines and chemokines, including Ccl5 (5.3-fold, P<0.0001), that were overexpressed in vivo were also significantly upregulated in BMDM treated with NaOX (Figure S5D). Accordingly, the release of CCL5 by BMDM treated with NaOX was significantly increased (by 64.6%, P<0.0001) and attenuated by pre-treatment with MitoTEMPO, a mitochondria-specific superoxide scavenger (Figure 5G). Taken together, these results indicate that oxalate induces mitochondrial dysfunction and overproduction of superoxide in macrophages leading to increased release of the pro-atherogenic chemokine, CCL5.
Exogenous Oxalate Overload Modulates Redox Hemostasis, Inflammation and Cholesterol Metabolism Leading to Enhanced Atherosclerosis
We next studied a direct effect of oxalate on redox homeostasis, inflammation and cholesterol metabolism in atherosclerosis by applying a dietary model of oxalate overload in Apoe−/− mice. The non-proteinogenic amino acid hydroxy-L-proline (HLP) is a precursor of oxalate. Supplementation of mice with HLP at 5% for 4 weeks is commonly used to study oxalate overload but was reported to cause lethargy in mice (Khan et al. 2006; Khan and Glenton. 2010; Tzou et al. 2016; Bilbault and Haymann. 2016). Therefore, we adopted a milder approach and developed a WD enriched with 3% HLP (WD+HLP). We fed Apoe−/− mice a standard WD or WD+HLP for 4 weeks. As expected, feeding the WD+HLP increased plasma oxalate (by 54.5%, P=0.003, Figure S5E). Genes regulating redox homeostasis that were overexpressed in livers from male Agxt−/−/Apoe−/− mice or in BMDM treated with NaOX (Gpx7, Gstm1, Gstm3, Gsta2 and Gsta4) were significantly upregulated in livers from Apoe−/− mice fed the WD+HLP (Figure S5F). In line, Apoe−/− mice fed the WD+HLP showed increased levels of superoxide and CCL5 (Figure 5SG and S5H) in the atherosclerotic plaque (by 67.7%, P=0.0385 and 75.1%, P=0.0207, respectively). To address a direct role of oxalate in cholesterol metabolism, we analyzed the hepatic expression of genes regulating sterol/steroid metabolism that were suppressed in livers from Agxt−/−/Apoe−/− mice. In line, Cyp7b1 was significantly downregulated in livers from Apoe−/− mice fed the WD+HLP together with a significant decrease in CYP7B1 protein abundance (Figure S6A and S6B). Accordingly, plasma total cholesterol and cholesterol content in VLDL and IDL were increased in Apoe−/− mice fed the WD+HLP (Figure S6C–E). Furthermore, atherosclerosis in the whole aortic tree and in the aortic sinus was significantly enhanced in Apoe−/− mice fed the WD+HLP (Figure S6F–I). En face analysis revealed a 2.8-fold increase in atherosclerotic lesions (P<0.0001, Figure S6F), and histological analysis of the aortic sinus showed increased atherosclerotic plaque area, ORO and Mac-2 positive areas in Apoe−/− mice fed the WD+HLP (by 70.5%, P=0.0252, 2-fold, P=0.0159, and 82.0%, P=0.0133, respectively). Collectively, by studying both endogenous dysregulation of oxalate and exogenous oxalate overload, we show that dysregulated oxalate metabolism modulates redox homeostasis, inflammation and cholesterol metabolism leading to accelerated atherosclerosis.
AAV-mediated Overexpression of AGXT Reduces Oxidative Stress and Inflammation
To determine whether impaired oxalate metabolism could be targeted for the treatment of atherosclerosis, we tested AAV-mediated overexpression of AGXT in male Apoe−/− mice. The mice were injected with AAV-AGXT or AAV-GFP and fed a WD for 12 weeks. We first confirmed the overexpression of AGXT in the liver (Figure 6A) and found a consistent increase in the glycine/oxalate ratio in plasma from mice treated with AAV-AGXT (by 61.8%, P=0.0057, Figure 6B). Next, we studied the hepatic expression of genes regulating redox homeostasis and inflammatory responses. In sharp contrast to male Agxt−/−/Apoe−/− mice, genes regulating glutathione metabolism (Gpx7, Gstm1, Gstm3, Gsta2 and Gsta4) were significantly downregulated in Apoe−/− mice treated with AAV-AGXT (Figure 6C). Similarly, genes encoding cell adhesion molecules (Icam1 and Sele), regulators of the inflammatory response and inflammatory markers (Cd68, Nos2, Trem2 and Tlr2), cytokines and their receptors (Ccl2, Ccl3, Ccl5, Cx3cl1 and Cxcl5) were all significantly downregulated in livers from mice overexpressing AGXT (Figure 6D). In contrast to the male Agxt−/−/Apoe−/− mice, plasma concentrations of the pro-atherogenic chemokines, CCL2 and CCL5, were significantly reduced in Apoe−/− mice treated with AAV-AGXT (by 49.5%, P=0.0065, and 40.2%, P=0.0070, respectively, Figure 6E and 6F). Consistently, analysis of the atherosclerotic plaque revealed lower CCL5 (Figure S7A) and superoxide (Figure 6G) in Apoe−/− mice treated with AAV-AGXT (by 34.7%, P=0.0280, and 72.4%, P=0.0379). Altogether, the AGXT loss- and gain-of-function studies in Apoe−/− mice indicate that an oxidant insult and a pro-inflammatory response are major mediators linking dysregulated oxalate metabolism to enhanced atherosclerosis.
Figure 6: AAV-mediated Overexpression of AGXT Reduces Oxidative Stress and Inflammation.
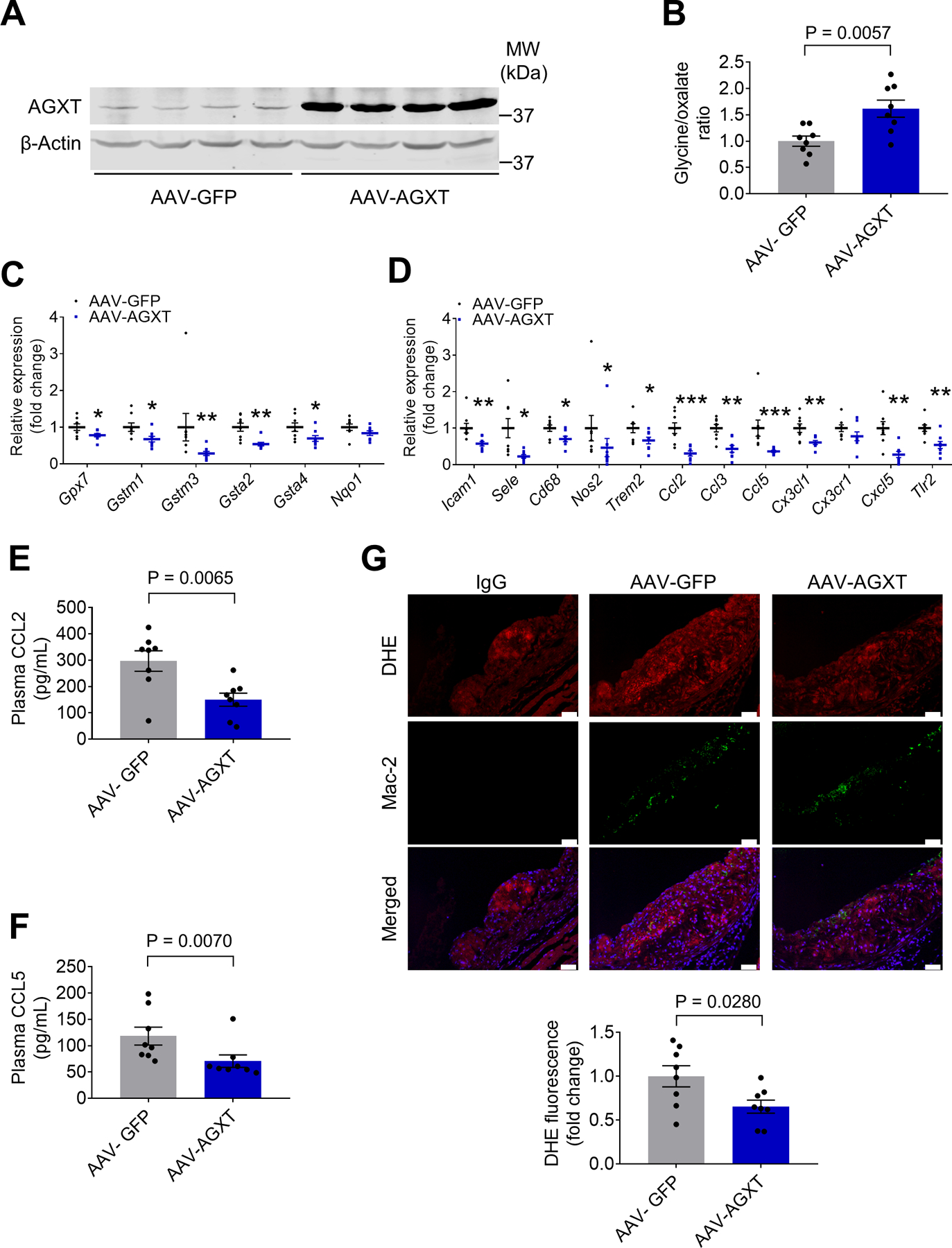
AAV-AGXT or AAV-GFP were injected into male Apoe−/− mice and the mice were fed a WD for 12 weeks (n=8).
(A) Western blot analysis confirming the overexpression of AGXT in livers from mice treated with AAV-AGXT.
(B) Plasma glycine/oxalate ratio.
(C-D) qPCR analyses of genes regulating (C) redox homeostasis and (D) inflammatory responses. Gene expression levels were normalized to 18S.
(E-F) Plasma concentrations of (E) CCL2 and (F) CCL5.
(G) DHE fluorescence and Mac-2 immunofluorescence in the aortic sinuses (scale bar: 50 μm).
Data are presented as mean ± SEM. All points are shown. *P<0.05, **P<0.01, ***P<0.001 vs. AAV-GFP.
AAV-mediated Overexpression of AGXT Reduces Atherosclerosis
Apoe−/− mice treated with AAV-AGXT and those treated with AAV-GFP showed comparable body weight and blood glucose after 12 weeks on a WD (Figure S7B and S7C). qPCR analyses of genes regulating fibrogenesis and lipid metabolism in the liver revealed some opposing effects to those found in male Agxt−/−/Apoe−/− mice including suppression of pro-fibrotic genes and upregulation of Cyp7b1 (Figure S7D–S7F). Nevertheless, the protein abundance of CYP7B1 and total BAs in the liver (Figure S7G and S7H) were comparable between Apoe−/− mice treated with AAV-AGXT and AAV-GFP. Furthermore, no significant differences in lipid profile were found (Figure 7A, Figure S7I and S7J). En face analysis revealed that Apoe−/− mice overexpressing AGXT had significantly less atherosclerotic lesions (by 36.9%, P=0.0001, Figure 7B). Accordingly, histological analysis of the aortic sinuses (Figure 7C–E) revealed a significant decrease in the atherosclerotic plaque area and Mac-2 positive area in Apoe−/− mice treated with AAV-AGXT (by 25.4%, P=0.0314, and 25.5%, P=0.0340, respectively). Lastly, we addressed the relationship between atherosclerosis and dysregulated oxalate metabolism versus hypercholesterolemia in the loss- and gain-of-function studies. As expected, a positive and significant correlation was found between the atherosclerotic lesion area in the whole aortic tree and plasma total cholesterol with a similar trend for the lesional macrophage content. Interestingly, a negative and significant correlation was found between the lesion area and plasma glycine/oxalate ratio (Figure S7K). These findings indicate dysregulated oxalate metabolism via suppressed AGXT as a driver in atherosclerosis and AGXT overexpression as a therapeutic approach.
Figure 7: AAV-mediated Overexpression of AGXT Reduces Atherosclerosis Independent of Circulating Cholesterol.
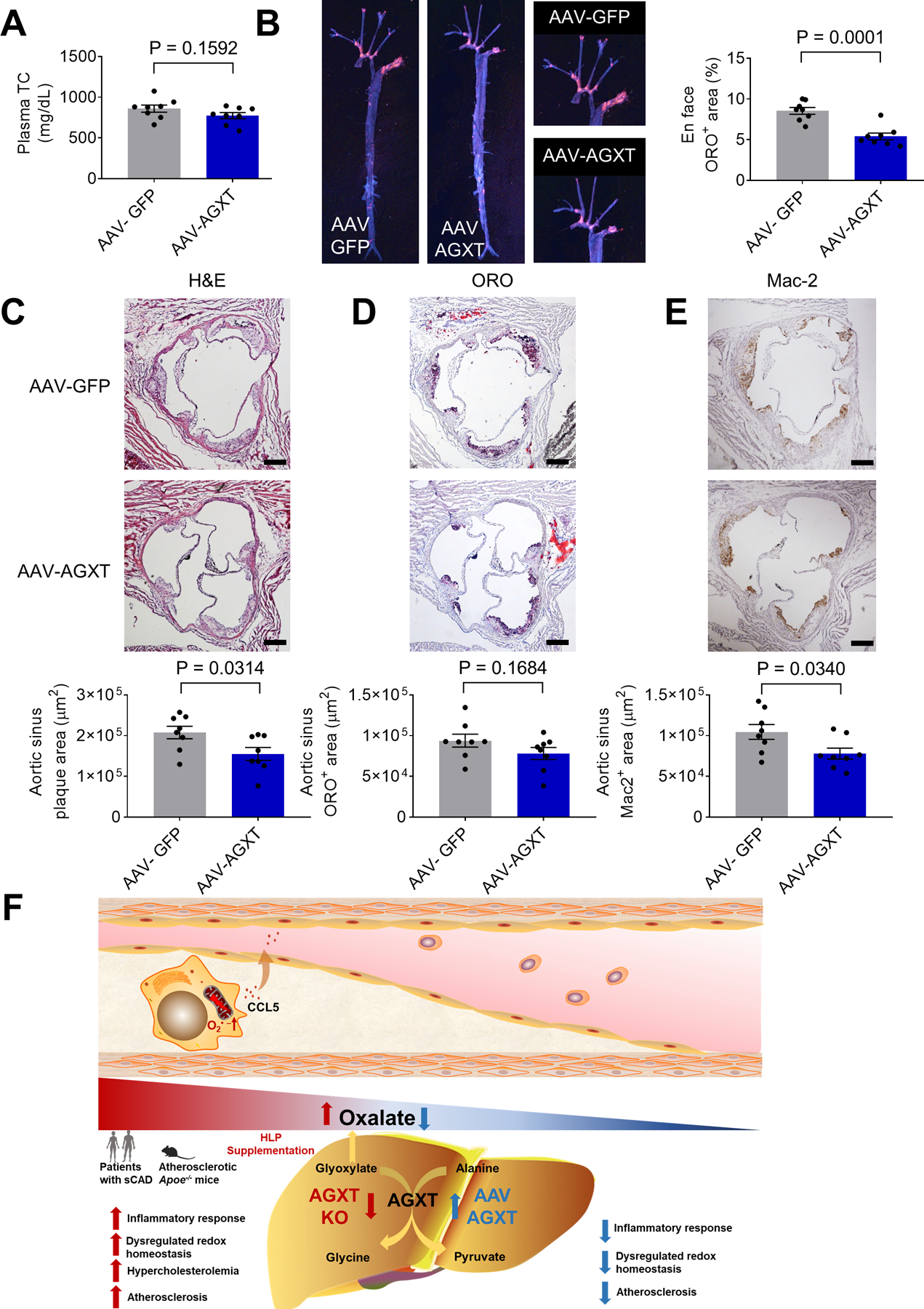
AAV-AGXT or AAV-GFP were injected into male Apoe−/− mice and the mice were fed a WD for 12 weeks (n=8).
(A) Plasma total cholesterol (TC).
(B) Atherosclerosis in the aortic tree.
(C) H&E staining, (D) Oil Red O staining, (E) Mac2 immunohistochemistry of aortic sinus. (scale bar: 200 μm).
(F) Proposed model of dysregulated oxalate metabolism as a driver and therapeutic target in atherosclerosis.
Unpaired t test for A-E. Data are presented as mean ± SEM. All points and P values are shown.
DISCUSSION
Atherosclerosis, the main driver of most CVDs, develops in response to the biologic effects of underlying risk factors including dyslipidemia, obesity, T2D and NAFLD (Lonardo et al., 2018; Virani et al., 2020). Dysregulated lipid metabolism is a hallmark feature in the pathogenesis of atherosclerosis (Moore et al., 2013). Nevertheless, despite remarkable advances in lipid-lowering therapies, CVDs remain a leading cause of death likely due to lack of influence or detrimental effects on other risk factors beyond dyslipidemia (de Carvalho et al., 2018; Swerdlow et al., 2015). Thus, identification of metabolic pathways that contribute to the pathogenesis of atherosclerosis, beyond altered lipid metabolism, may lead to the development of novel therapeutics. Recent evidence indicates that dysregulated amino acid metabolism plays a role in atherosclerosis (Grajeda-Iglesias et al., 2018; Nitz et al., 2019; Rom et al., 2018; Zaric et al., 2020). Specifically, lower circulating glycine has been consistently reported in various cardiometabolic diseases including coronary heart disease (Wittemans et al., 2019), myocardial infarction (Ding et al., 2015), T2D (Guasch-Ferré et al., 2016), metabolic syndrome (Li et al., 2018), obesity (Newgard et al., 2009) and NAFLD (Gaggini et al., 2018, Rom et al., 2020).
Here, targeted metabolomics revealed impaired glycine biosynthesis in patients with sCAD indicated by decreased ratios of glycine to its precursors, serine and threonine. This is in line with a recent report of decreased glycine/serine ratio in patients with unstable atherosclerotic plaques (Hitzel et al., 2018), further supporting the notion of impaired glycine biosynthesis in CVDs. Importantly, we report a significant decrease in the glycine/oxalate ratio in patients with sCAD that was recapitulated in atherosclerotic mice. In line, AGXT which catalyzes glycine biosynthesis from alanine and glyoxylate was significantly suppressed in livers from atherosclerotic mice. Using a combination of genetic and dietary approaches to manipulate oxalate exposure in vivo and in macrophages, we found that oxalate causes mitochondrial dysfunction, oxidative stress and pro-inflammatory responses associated with enhanced atherosclerosis. The therapeutic potential of targeting dysregulated oxalate metabolism was demonstrated in Apoe−/− mice overexpressing AGXT that showed lower oxidative stress, inflammation and atherosclerosis (Figure 7F).
Previous transcriptomics studies in humans and mice revealed a suppression of genes driving glycine biosynthesis from serine, alanine and glyoxylate in NAFLD (Mardinoglu et al., 2014), with a most significant downregulation of AGXT (Asgharpour et al., 2016; Rom et al., 2020; Stepanova et al., 2010). Analyzing samples from liver transplantation donors, we reported that AGXT expression is inversely correlated with hepatic fat (Rom et al., 2020). We further found that loss of AGXT accelerates diet-induced NAFLD (Rom et al., 2020). In the current investigation, we studied Apoe−/− mice lacking AGXT and found exacerbated atherosclerosis in male Agxt−/−/Apoe−/− mice. Our previous studies (Rom et al., 2020), together with the current findings, demonstrate an important role of AGXT both in NAFLD and in atherosclerosis. Considering the challenges of simultaneously treating CVDs and NAFLD (Nielsen et al., 2020), these findings indicate AGXT as a potential therapeutic target for the cotreatment of these diseases.
AGXT is expressed primarily in the liver (Li et al., 1999), and its loss-of-function causes accumulation of glyoxylate which is rapidly converted to oxalate and cannot be further metabolized (Fodor et al., 2012). Functional deficiencies of AGXT cause primary hyperoxaluria type 1 (PH1) characterized by hepatic overproduction of oxalate (Fodor et al., 2012), that is recapitulated in Agxt−/− mice (Salido et al., 2006). Although hypothesized to increase the risk of CVDs and atherosclerosis (Arafa et al., 2020; Devarajan, 2018), evidence linking oxalate to increased cardiovascular risk are limited to a few case reports (Jorge et al., 2013; Palka et al., 2001). Here, by studying male and female Agxt−/−/Apoe−/− mice, we provide evidence that dysregulated oxalate metabolism promotes atherosclerosis development. Studies utilizing endogenous or exogenous models of increased oxalate exposure established that oxalate homeostasis is better maintained in females than males (Fan et al., 1999; Liang et al., 2014; Salido et al., 2006; Yoshihara et al., 1999; Zhu et al., 2019). Androgens are known to increase, while estrogens decrease hepatic oxalate biosynthesis, urinary oxalate excretion and plasma oxalate concentration (Fan et al., 1999; Liang et al., 2014; Yoshihara et al., 1999; Zhu et al., 2019). These sex differences allowed us to determine whether the increased atherosclerosis was caused by loss of AGXT or impaired oxalate metabolism. The comparable glycine/oxalate ratio and atherosclerosis in female Agxt−/−/Apoe−/− and Agxt+/+/Apoe−/− mice together with enhanced atherosclerosis in Apoe−/− mice on WD+HLP indicate a causative role of oxalate in atherosclerosis.
Through unbiased transcriptomics, we identified pathways involved in pro-oxidant/inflammatory responses linking dysregulated oxalate metabolism to enhanced atherosclerosis. A significant upregulation of genes involved in the response to pro-oxidant insult and genes encoding key regulators of the inflammatory response was evident in livers from male Agxt−/−/Apoe−/− mice. In line, male Agxt−/−/Apoe−/− mice showed increased plasma CCL5, a known pro-atherogenic chemokine (Noels et al., 2019). CCL5 was also increased in the atherosclerotic plaque together with enhanced superoxide in lesional macrophages. Importantly, these effects were not observed in female Agxt−/−/Apoe−/− mice but were evident in Apoe−/− mice fed WD+HLP, indicating a direct contribution of oxalate to pro-oxidant/inflammatory responses in atherosclerosis. Interestingly, Apoe−/− mice fed WD+HLP for 4 weeks showed increased lesional macrophages, which was not significant in male Agxt−/−/Apoe−/− mice fed WD for 12 weeks, suggesting a different response to exogenous or endogenous oxalate at different stages of atherosclerosis development. Nevertheless, Apoe−/− mice overexpressing AGXT showed a significant decrease in lesional macrophages in line with marked reductions in circulating CCL2 and CCL5.
Mitochondrial dysfunction and oxidative stress in macrophages are known to promote the inflammatory response and atherosclerosis (Madamanchi and Runge, 2007; Wang et al., 2014; Vendrov et al., 2007). Mechanistically, we showed that oxalate induces mitochondrial dysfunction and superoxide accumulation leading to increased CCL5 release in macrophages. The latter was attenuated by scavenging mitochondrial superoxide using MitoTEMPO. In support, oxalate exposure was previously reported to cause mitochondrial dysfunction, oxidative stress and inflammation in other models. Early studies showed that oxalate inhibits mitochondrial respiration in renal cells (McMartin and Wallace, 2005), and decreases membrane potential leading to ROS accumulation (Cao et al., 2004). Oxalate was also shown to induce mitochondrial dysfunction and disrupt redox homeostasis in the THP-1 monocyte cell line (Patel et al., 2018). In endothelial cells, oxalate was reported to increase intracellular calcium and proposed to exert pro-atherogenic effects by preventing re-endothelialization (Recht et al. 2004). Utilizing different in vivo and in vitro models, oxalate was shown to induce renal inflammation through NLR family, pyrin domain-containing 3 (NLRP3)-mediated interleukin 1β secretion (Knauf et al., 2013; Mulay et al., 2013). Whereas previous studies demonstrated that oxalate promotes pro-oxidant/inflammatory responses, we provide here evidence linking these effects to enhanced atherosclerosis.
Previously, proteomics revealed significant alterations in pathways regulating glucose and lipid metabolism in livers from Agxt−/− mice (Hernández-Fernaud and Salido, 2010). We reported accelerated NAFLD in male Agxt−/− mice in association with enhanced hepatic inflammation, fibrogenesis and hyperlipidemia (Rom et al., 2020). Here, enhanced hypercholesterolemia, higher cholesterol contents in VLDL and IDL, and altered expression of genes regulating lipid metabolism and fibrogenesis were evident in male, but not in female Agxt−/−/Apoe−/− mice, and in Apoe−/− mice fed WD+HLP, indicating the contribution of dysregulated oxalate homeostasis to dyslipidemia, atherosclerosis and NAFLD. Through transcriptomics and follow-up studies on significantly altered pathways, we addressed the cholesterol differences in the different study cohorts. Genes regulating sterol/steroid metabolism were significantly suppressed in livers from Agxt−/−/Apoe−/− mice including Cyp7b1. In the liver, cholesterol is metabolized to BAs through two main pathways: the classical/neutral pathway and the alternative/acidic pathway. CYP7B1 plays a main role in the alternative/acidic pathway which can become the predominant pathway for BA synthesis during liver insult and disease (Pandak and Kakiyama. 2019). In line with the transcriptomics, the protein abundance of CYP7B1 was significantly decreased in livers from male Agxt−/−/Apoe−/− mice, suggesting impaired metabolism of cholesterol to BAs. Accordingly, we found a significant decrease in total BAs in livers from male Agxt−/−/Apoe−/− mice. Whereas CYP7B1 expression was unalerted in female Agxt−/−/Apoe−/− mice, it was significantly suppressed in livers from mice fed WD+HLP that showed enhanced hypercholesterolemia. These studies, combining a sex-based comparison in Agxt−/−/Apoe−/− mice and exogenous oxalate overload, indicate that cholesterol differences may contribute to accelerated atherosclerosis due to dysregulated oxalate metabolism. Nevertheless, we did not observe significant differences in CYP7B1 protein levels, total BAs and hypercholesterolemia in Apoe−/− mice overexpressing AGXT.
To date, there is no pharmaceutical therapy to reduce hepatic oxalate production and gene transfer has been proposed as an attractive therapeutic option for PH1 (Rumsby and Cochat, 2013; Salido et al., 2011). Treatment of Agxt−/− mice with AAV-AGXT was shown to restore the hepatic enzymatic activity of AGXT and to reduce hyperoxaluria (Salido et al., 2011). In the current study, we sought to determine whether treatment of Apoe−/− mice (that have intact hepatic AGXT) with AAV-AGXT could lower atherosclerosis. We found that the pro-oxidant/inflammatory responses that were enhanced in the male Agxt−/−/Apoe−/− mice were attenuated in Apoe−/− mice overexpressing AGXT. These included suppression of hepatic genes implicated in pro-oxidant/inflammatory responses, reduction of plasma and aortic CCL5 and decreased superoxide in lesional macrophages. Importantly, atherosclerosis was significantly reduced in Apoe−/− mice overexpressing AGXT. Thus, beyond the therapeutic potential in PH1 (Rumsby and Cochat, 2013; Salido et al., 2011), our studies indicate that AGXT overexpression to lower oxalate can reduce atherosclerosis by attenuating oxidative stress and inflammation.
In conclusion, combining data from patients and mice with atherosclerosis and complementary genetic and dietary approaches to manipulate oxalate exposure in vivo and in vitro, the current study uncovered dysregulated oxalate metabolism as a driver of atherosclerosis due to dysregulated redox homeostasis, enhanced inflammatory response and altered cholesterol metabolism. Furthermore, this study demonstrated the therapeutic potential of AGXT and targeting dysregulated oxalate metabolism to reduce atherosclerosis.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Oren Rom (oren.rom@lsuhs.edu)
Materials Availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
RNA-sequencing data have been deposited at NCBI’s SRA and are publicly available as of the date of publication. Accession number is listed in the key resources table. All raw data have been deposited at Mendeley and are publicly available as of the date of publication (doi: 10.17632/n8nkdgd258.1). This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-mouse β-Actin antibody | Cell Signaling Technology | Cat#4967; RRID: AB_330288 |
| Mouse anti-mouse AGXT antibody | Santa Cruz Biotechnology | Cat# SC-517388 |
| Rabbit anti-mouse CYP7B1 antibody | Proteintech | CAT# 24889–1-AP RRID: AB_2879780 |
| Mouse anti-mouse B-ACTIN antibody | Cell Signaling Technology | CAT#3700 RRID: AB_2242334 |
| Rat anti-Galectin 3 antibody | Thermo Fisher Scientific | Cat#13-5301-81; RRID: AB_837112 |
| CCL5 (RANTES) Recombinant Polyclonal Antibody | ThermoFisher Scientific | Cat# 710001, RRID: AB_2532515 |
| 800CW Donkey anti-mouse IRDye | LI-COR Biosciences | Cat# 926–32212; RRID: AB_621847 |
| 680RD Donkey anti-rabbit IRDye | LI-COR Biosciences | Cat# 926–68073; RRID: AB_10954442 |
| 800CW Donkey anti-rabbit IRDye | LI-COR Biotechnology | CAT# 926–32213 RRID: AB_621848 |
| 680RD Donkey anti-mouse IRDye | LI-COR Biotechnology | CAT# 926–68072 RRID: AB_10953628 |
| Goat anti-Rat IgG (H+L) Secondary Antibody, HRP | ThermoFisher Scientific | Cat# 31470, RRID: AB_228356 |
| Alexa Fluor 647 donkey anti-rat | Jackson ImmunoResearch Laboratories | Cat# 712-605-153; RRID: AB_2340694 |
| Alexa Fluor 594 donkey anti-rabbit | Jackson ImmunoResearch Laboratories | Cat#711-585-152, RRID: AB_2340621 |
| Bacterial and Virus Strains | ||
| Stable Competent E. coli | New England Biolabs | Cat # C3040H |
| AAV-TBG-GFP | This paper | N/A |
| AAV-TBG-AGXT | This paper | N/A |
| Biological Samples | ||
| Serum from patients with or without sCAD | This paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Polyethylenimine | Sigma-Aldrich | Cat# 408727 |
| Dihydroethidium | Sigma-Aldrich | Cat# D7008 |
| MitoTEMPO | Sigma-Aldrich | Cat# SML0737 |
| Sodium oxalate | Sigma-Aldrich | Cat# 223433 |
| Oil red O | Sigma-Aldrich | Cat# O0625 |
| Sodium azide | Sigma-Aldrich | CAT#S2002 |
| Proteinase K | Sigma-Aldrich | CAT# 1245680500 |
| Trans-4-Hydroxy-L-proline | ACROS Organics | CAT#121785000 |
| UltraPure™ Phenol:Chloroform:Isoamyl Alcohol | ThermoFisher Scientific | CAT#15593–031 |
| Poloxamer 188 | Sigma-Aldrich | Cat# P5556 |
| Harris hematoxylin | ThermoFisher Scientific | Cat# 842 |
| Eosin Y | ThermoFisher Scientific | Cat# 832 |
| Clarifier Bluing reagent | ThermoFisher Scientific | Cat# 7401 |
| Bluing reagent | ThermoFisher Scientific | Cat# 7301 |
| MitoSOX Red | ThermoFisher Scientific | Cat# M36008 |
| Hoechst | ThermoFisher Scientific | Cat# H3570 |
| Trizol reagent | ThermoFisher Scientific | Cat# 5596026 |
| Canonical amino acid mix unlabeled standard | Cambridge Isotopes Laboratory | Cat# MSK-CAA-US-1 |
| Canonical amino acid mix | Cambridge Isotopes Laboratory | Cat# MSK-CAA-1 |
| O.C.T. Compound | Tissue-Tek | Cat# 4583 |
| iQ SYBR Green Supermix | Bio-Rad | Cat# 1708880 |
| Critical Commercial Assays | ||
| Wako Cholesterol E | FUJIFILM medical system | Cat# 999–02601 |
| RNeasy Maxi kit | QIAGEN | Cat# 75162 |
| EndoFree Plasmid Maxi Kit | QIAGEN | Cat# 12362 |
| Oxalate assay kit | Abcam | Cat# ab196990 |
| Mouse CCL2/JE/MCP-1 Quantikine ELISA Kit | R&D Systems | Cat# MJE00B |
| Mouse/Rat CCL5/RANTES Quantikine ELISA Kit | R&D Systems | Cat# MMR00 |
| Glucose test strips | Contour Next | Cat# NDC: 0193-7308-50 |
| Seahorse XFp Cell Mito Stress Test Kit | Agilent Technologies | Cat# 103010–100 |
| Gibson assembly kit | New England Biolabs | Cat# E5510 |
| NEBNext Ultra II Directional RNA Library Prep Kit for Illumina | New England Biolabs | CAT# E7760L |
| Poly(A) mRNA Magnetic Isolation Module | New England Biolabs | Cat# E7490L |
| NEBNext Multiplex Oligos for Illumina Unique dual | New England Biolabs | Cat# E6440L |
| Protease inhibitor | Thermo Fisher Scientific | Cat# A32965 |
| Phosphatase inhibitor | Roche Diagnostics | Cat# 04906845001 |
| SuperScript™ III First-Strand Synthesis System | ThermoFisher Scientific | Cat# 18080051 |
| CCK8 assay kit | Sigma-Aldrich | Cat# 96992 |
| SignalStain® DAB Substrate Kit | Cell Signaling Techonology | CAT#8059 |
| Total Bile Acid Assays Kit | Cell Biolabs, INC | CAT#STA-631 |
| Cholesterol fluorometric assay kit | Cayman Chemical | CAT#10007640 |
| LabAssay™ Triglyceride kit | FUJIFILM medical system | CAT# 290–63701 |
| Deposited Data | ||
| RNA-sequencing | This paper | NCBI’s SRA (accession number: PRJNA671451) |
| Raw data | This paper | Mendeley Data (doi:10.17632/n8nkdgd258.1) |
| Experimental Models: Cell Lines | ||
| HEK293T | ATCC | CRL-3216; RRID: CVCL_0063 |
| L929 cells | ATCC | CRL-1; RRID: CVCL_0462 |
| Bone marrow derived-macrophages | This paper | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | JAX: 000664; RRID: IMSR_JAX:000664 |
| Mouse: Apoe−/− | The Jackson Laboratory | JAX: 002052; RRID: IMSR_JAX:002052 |
| Mouse: Agxt−/− | Rom et al., 2020 | N/A |
| Mouse: Agxt+/+/Apoe−/− | This paper | N/A |
| Mouse: Agxt−/−/Apoe−/− | This paper | N/A |
| Oligonucleotides | ||
| Guide-RNA targeting the mouse Agxt exon1: 5’-GGGTCCGGGGCCCTCCAACC-3’ | Integrated DNA Technologies | N/A |
| Primer sequences for Agxt genotyping: forward: 5’-ACACCTCCACTGTCCTGTCC-3’ | Integrated DNA Technologies | N/A |
| Primer sequences for Agxt genotyping: reverse: 5’-GGTCAGATCTGCCTGCTACC-3’ | Integrated DNA Technologies | N/A |
| Primer for Sanger sequencing: 5’-GCAGAGCTAGCTGGGAAATG-3’ | Integrated DNA Technologies | N/A |
| Primer sequences for Apoe genotyping: forward: 5′-GCCTAGCCGAGGGAGAGCCG-3′ | Integrated DNA Technologies | N/A |
| Primer sequences for Apoe genotyping: reverse 1: 5′-TGTGACTTGGGAGCTCTGCAGC-3′ | Integrated DNA Technologies | N/A |
| Primer sequences for Apoe genotyping: reverse 2: 5′-GCCGCCCCGACTGCATCT-3’ | Integrated DNA Technologies | N/A |
| Primer sequences for qPCR, see Table S2 | Integrated DNA Technologies | N/A |
| Recombinant DNA | ||
| pAdDeltaF6 | Jiandie Lin’s lab, University of Michigan; Guo et al., 2017 | N/A |
| pAAV2/8 | Jiandie Lin’s lab, University of Michigan; Guo et al., 2017 | N/A |
| pAAV-TBG-GFP | Jiandie Lin’s lab, University of Michigan; Guo et al., 2017 | N/A |
| pAAV-TBG-MCS | Jiandie Lin’s lab, University of Michigan; Guo et al., 2017 | N/A |
| AGXT Human Tagged ORF Clone | Origene | CAT# RG212899 |
| Software and Algorithms | ||
| ImageJ | National Institutes of Health | RRID:SCR_003070 https://imagej.nih.gov/ij/download.html |
| GraphPad Prism | GraphPad Software | Version 8.0 RRID:SCR_002798 https://www.graphpad.com/scientific-software/prism/ |
| R statistics | Hosted by Vienna, University of Economics and Business | http://www.R-project.org/. |
| RStudio | RStudio Team (2020). | http://www.rstudio.com/.RRID:SCR_000432 |
| IBM SPSS software | IBM Corporation | Version 23.0; RRID:SCR_019096 https://www.ibm.com/analytics/spss-statistics-software |
| FastQC | Babraham Bioinformatics | Version 0.11.8 RRID:SCR_014583 https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| Trimmomatic | Bolger et al., 2014 | Version 0.35 RRID:SCR_011848 http://www.usadellab.org/cms/?page=trimmomatic |
| HISAT2 | Kim et al., 2015 | Version 2.1.0.13; RRID:SCR_015530 http://daehwankimlab.github.io/hisat2/download/ |
| HTSeq | Anders et al., 2015 | Version 0.6.0; RRID:SCR_005514 https://htseq.readthedocs.io/en/master/install.html |
| Analyst software | Applied Biosystems | Version 1.6.2; RRID:SCR_015785 https://sciex.com/products/software/analyst-software |
| LI-COR Image studio | LI-COR Biosciences | Version 2.1; RRID:SCR_015795 https://www.licor.com/bio/image-studio-lite/download |
| SPOT Basic software | SPOT imaging | RRID:SCR_014313 https://www.spotimaging.com/software/spot-basic/ |
| Other | ||
| Mouse standard diet | LabDiet | Cat# 5L0D |
| Mouse Western diet | Envigo | Cat# TD.88137 |
| Mouse Western diet supplemented with hydroxy-L-proline (HLP) | Envigo | CAT# TD.190842 |
| Dulbecco’s Modified Eagle Medium | Gibco | Cat# 11965092 |
| Iscove’s Modified Dulbecco’s Media | Gibco | Cat# 12440053 |
| Agilent Poroshell 120 EC-C18 column | Agilent Technologies | Cat# 699775–902 |
| Superose 6 Increase 10/300 GL columns | Sigma-Aldrich | CAT# GE29-0915-96 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animal studies
Animal procedures were approved by the Institutional Animal Care & Use Committee of the University of Michigan (PRO00008239) and performed in accordance with the institutional guidelines. Agxt−/− mice on the C57BL/6J background (The Jackson Laboratory, JAX: 000664) were generated using CRISPR/Cas9, with guide-RNA targeting exon 1 of Agxt: 5’-GGGTCCGGGGCCCTCCAACC-3’ (Rom et al., 2020). Genotyping was performed using the following primers: forward: 5’-ACACCTCCACTGTCCTGTCC-3’, reverse: 5’-GGTCAGATCTGCCTGCTACC-3’. Sanger sequencing using the following primer: 5’-GCAGAGCTAGCTGGGAAATG-3’ confirmed an A deletion 3 bases from the protospacer adjacent motif (PAM) (Figure S2A). The frame-shift mutation after amino acid 53 of the ORF, introduced a premature stop-codon, and the absence of AGXT in the liver was confirmed by Western blot. No CRISPR off-target effects were detected as assessed using CRISPOR (Haeussler et al., 2016). Apoe−/− mice (B6.129P2-Apoetm1Unc/J, Stock: JAX 002052) were purchased from the Jackson Laboratory. Agxt−/− mice were cross-bred with Apoe−/− mice to generate Agxt−/−/Apoe−/− mice. Agxt+/+/Apoe−/− littermates served as controls. For Apoe genotyping, the wild-type allele (155 bp) was detected using the following primers: forward: 5′-GCCTAGCCGAGGGAGAGCCG-3′, reverse: 5′-TGTGACTTGGGAGCTCTGCAGC-3′. The knockout allele (245 bp) was detected using the following primers: forward: 5′-GCCTAGCCGAGGGAGAGCCG-3′, reverse: 5′-GCCGCCCCGACTGCATCT-3’ (Figure S2B). AAV8-AGXT expressing human AGXT and AAV8-GFP (control) driven by the liver specific thyroxine binding globulin (TBG) promoter were produced in HEK293T cells. Purified AAV was injected intraperitoneally into 7-week-old male Apoe−/− mice at a dosage of 2×1011 viral genomes per mouse. Hepatic AAV overexpression was confirmed by Western blot. Male Apoe−/− mice, male and female Agxt+/+/Apoe−/− and Agxt−/−/Apoe−/− mice and male Apoe−/− mice overexpressing AAV or GFP were maintained on a 12/12 h light/dark cycle. Starting from 8 weeks of age, the mice were fed ad libitum either a standard diet (SD, LabDiet 5L0D, 13% fat) or a high-fat and high-cholesterol Western diet (WD, Envigo TD.88137, 42% fat). For the dietary model of oxalate overload, 8-week-old male Apoe−/− mice were fed ad libitum either the standard WD (TD.88137) or a WD supplemented with 3% (w/w) hydroxy-L-proline (HLP) (WD+HLP, Envigo TD.190842) for 4 weeks.
Human study
The study was approved by the local Institutional Review Board at Rambam Health Care Campus, Haifa, Israel (protocol 0373-18-RMB, Clinical Trial Registration: NCT03646019, https://www.clinicaltrials.gov). All participants gave informed consent. The study included patients admitted to the chest pain unit in the Department of Internal Medicine E after presented with acute chest pain suspected to be of cardiac origin. Exclusion criteria included a prior diagnosis of CAD, allergy to iodine contrast agents, contraindication for radiations, asthma exacerbation, current use of steroids or other immunomodulating drugs, renal insufficiency (creatinine level ≥ 1.5 mg/dl), fever during the last 48 h prior to admission, concomitant inflammatory diseases (infections, autoimmune disorders, kidney and liver diseases, and recent major surgical procedure), valvular, myocardial or pericardial diseases and poor quality of computed tomography (CT) image due to motion artifacts or inappropriate contrast delivery resulting in non-diagnostic image quality. Patients that met the above criteria underwent coronary computed tomography angiography (CCTA) to assess atherosclerosis in their coronary arteries. Information on patient age/gender can be found in Table S1. The patients were divided in groups with significant CAD (sCAD, coronary artery stenosis >49%) or with no sCAD (coronary artery stenosis ≤49% Figure S1A and S1B). Twenty-four patients were diagnosed with sCAD. Using case-control matching and randomization (IBM SPSS software version 23.0), 24 age- and sex-matched patients that were found to have no sCAD were randomly assigned as controls. Blood samples were obtained following a 10 h overnight fast. Serum biochemical analyses and targeted metabolomics were performed by technicians blinded to the groups. There were no significant differences in demographics, drug use and biochemical parameters between patients with or without sCAD (Table S1).
Bone marrow derived macrophages (BMDM)
BMDM were obtained as previously described (Villacorta et al., 2018), with minor modifications. Briefly, bone marrow was flushed from the femur and tibia of 6-10-week-old male C57BL/6J mice. Bone marrow cells were cultured on 15-cm petri dishes with Iscove’s Modified Dulbecco’s Media (IMDM, Gibco) supplemented with 10% heat-inactivated FBS (Gibco), 1% sodium pyruvate (Gibco), 1% non-essential amino acids (Gibco), 1% antibiotics (pen/strep, Gibco) and 30% L929 supernatant as a source of macrophage-stimulating factor. The L-929 cells were grown in DMEM (Gibco) supplemented with 10% FBS and 1% antibiotics. After 10 days, the culture medium was removed, and filtered (0.22 μm filter). After 6–7 days, adherent macrophages were scraped and seeded in cell culture plates for following experiments. Differentiated BMDM were cultured in IMDM supplemented with 10% heat-inactivated FBS and 1% antibiotics. Following sodium oxalate (NaOX) treatment, cell viability was determined using the CCK8 assay (Sigma-Aldrich).
METHOD DETAILS
Targeted metabolomics and plasma analyses
Analyses were performed by investigators blinded to the study groups. Liquid chromatography (LC)-mass spectrometry (MS)/MS was used to determine circulating amino acids in human and mouse samples. For amino acid analysis, a mixture of the 20 canonical amino acids (MSK-CAA-US-1, Cambridge Isotopes Laboratory) was used for calibration. A mixture of the corresponding 20 stable isotope-labelled amino acids (MSK-CAA-1, Cambridge Isotopes Laboratory) was used as internal standard. The mixture was reconstituted with 1 mL methanol-water (40:60, v/v) containing 0.1M HCl to prepare a stock solution. The concentration of each amino acid in the stock solutions was 2.5 mM (with the exception for L-cystine at 1.25 mM). Working solutions were prepared by serial dilutions with water at concentrations of 0.1, 0.25, 0.5, 1.0, 2.5, 5.0, 10.0, 25.0, and 50.0 μM (0.05, 0.125, 0.25, 0.5, 1.25, 2.50, 5.00, 12.5, and 25.0 μM for L-Cystine). Internal standard stock solution was diluted with methanol to prepare the internal standard working solution at the concentration of 0.5 μM for each of the isotope-labeled amino acids (0.25 μM for isotope-labeled cysteine). Internal standard solutions were diluted with methanol to a concentration of 0.5 μM of the isotope-labelled amino acid. The LC-MS/MS analysis was performed using a Shimadzu LC-20AD HPLC system (Shimadzu Corporation) coupled in-line to an AB Sciex QTrap 5500 system (Applied Biosystems) equipped with an electrospray ionization source. Chromatographic separation was conducted on an Agilent Poroshell 120 EC-C18 column (50 mm × 2.1 mm I.D., 2.7 μm; Agilent) at a flow rate of 0.4 mL/min. The mobile phase consisted of water (A) and acetonitrile containing 0.1% (v/v) formic acid (B). The linear gradient program was utilized as follows: 0 min, 2% B; 2 min, 2% B; 4 min, 95% B; 5 min, 99% B; 5.1 min, 2% B; and 9 min, 2% B. The autosampler temperature was set at 4°C. The mass spectrometer was operated in positive ionization mode with multiple reaction monitoring (MRM). The source parameters were as follows: curtain gas, 30 psi; nebulizer gas (GS1), 40 psi; turbo gas (GS2), 50 psi; ion spray voltage, 1500 V; source temperature, 500°C; and dwell time, 10 ms. Analyst version 1.6.2 software (Applied Biosystems) was used for data acquisition and analysis. Twenty microliters of the standard working solution was added to a 200 μL aliquot of internal standard working solution. The mixture was vortex-mixed for 1 min and centrifuged at 11,000 × g for 5 min at 4°C, and then a 2 μL aliquot of the resulting solution was injected into the LC-MS/MS system for analysis. Samples were diluted 30-fold with water for the determination of lysine, glutamine, and histidine, and diluted 5-fold with water for the determination of the other 17 amino acids. Twenty microliters of the diluted samples were added to a 200 μL aliquot of internal standard working solution. The mixture was vortex-mixed for 1 min and centrifuged at 11,000 × g for 5 min at 4°C, and then a 2 μL aliquot of the resulting solution was injected into the LC–MS/MS system for analysis. The calibration curves were generated by plotting the peak area ratios (analyte/internal standard) versus analyte concentrations using a weighted (1/x2) least-squares linear regression. Oxalate levels were measured using the Abcam oxalate assay kit in accordance with the manufacturer’s instructions. Blood glucose was measured using glucometer and test strips (Contour Next). Plasma samples were analyzed for total cholesterol using the Wako Diagnostics kit (FUJIFILM medical system). Plasma CCL2 and CCL5 were measured using the Mouse CCL2/JE/MCP-1 Quantikine ELISA Kit and the Mouse/Rat CCL5/RANTES Quantikine ELISA Kit (R&D Systems) in accordance with the manufacturer’s instructions.
Analysis of atherosclerosis in mice
All analyses were conducted in a blinded fashion and in accordance with the American Heart Association recommendations for experimental atherosclerosis studies (Daugherty et al., 2017). For en face analysis, the entire length of the aorta from the heart to the iliac bifurcation was removed. After clearing the adventitial tissue, aortas were stained with Oil Red O (ORO), opened longitudinally and pinned flat onto a dark wax plate. The percentage of ORO positive area was calculated using ImageJ (National Institutes of Health, NIH) (Xiong et al., 2018). For analysis of atherosclerosis in the aortic sinus, histological procedures were performed by technicians at the In-Vivo Animal Core histology laboratory within the Unit for Laboratory Animal Medicine, Michigan Medicine. Formalin-fixed samples were cryoprotected in 20% sucrose at 4°C overnight, blotted, then liquid nitrogen-snap frozen in O.C.T. Compound (Tissue-Tek) and stored at −80°C until cryo-sectioning. Prior to sectioning, frozen blocks were brought up to approximately −20°C, then sectioned at 5 microns on a Leica Cryostat (Leica #CM3050S). Slides were stored at −80°C until stained. Prior to staining, the slides were thawed to room temperature for 30 min. For H&E staining, slides were stained with Harris hematoxylin (ThermoFisher Scientific), rinsed in tap water and treated with Clarifier (ThermoFisher Scientific), rinsed in tap water and the nuclei were counter stained with bluing reagent (ThermoFisher Scientific), rinsed in 70% Ethanol, stained with eosin Y, alcoholic (ThermoFisher Scientific), then dehydrated and cleared through graded alcohols and xylene and mounted with Micromount (Leica cat# 3801731) using a Leica CV5030 automatic coverslipper. For ORO staining, slides were rinsed with water and with 60% isopropanol for 20 sec before staining with an ORO working solution for 30 minutes (100 mg ORO, #O0625 Sigma, in 35 ml methanol and 10 ml 1M sodium hydroxide, filtered 3 times with 0.22 μM filter). Slides were then rinsed with 60% isopropanol for 10 sec and proceed with standard hematoxylin staining procedures. For Galectin 3 (Mac-2) immunohistochemistry, slides were rinsed for 2 min with PBS (x3) and then blocked with 5% BSA for 30 min at room temperature. Slides were then incubated overnight with an anti-Galectin 3 (Mac-2) Rat monoclonal antibody (eBioM3/38 (M3/38), eBioscience, working dilution 1:150) at 4°C. Slides were then rinsed for 2 min with PBS (x3) and incubated with a HRP-conjugated secondary antibody (# 31470, ThermoFisher) at room temperature for 1 h in the dark. After washing with PBS (x3), slides were incubated with diaminobenzidine (DAB) substrate (Cat#8059, Cell Signaling Technology) for 1 min, rinsed with PBS (x3) and then proceed to standard hematoxylin procedures. For Mac-2 and dihydroethidium (DHE) immunofluorescence, slides were rinsed for 2 min with PBS (x3) and then blocked with 5% BSA for 30 min at room temperature. Slides were then incubated overnight with an anti-Galectin 3 (Mac-2) monoclonal antibody (eBioscience, working dilution 1:150) at 4°C. Slides were then rinsed for 2 min with PBS (x3) and incubated with a secondary antibody (Alexa Fluor 647 donkey anti-rat, #712-605-153, Jackson ImmunoResearch Laboratories, working dilution 1:500) together with 5 μM DHE (Sigma-Aldrich) at 37°C for 30 min in the dark. After washing with PBS (x3), the slides were mounted with ProLong™ Gold Antifade Mountant with DAPI (Invitrogen, P36935), and images were captured using an Olympus DP73 microscope. For CCL5 and Mac-2 immunofluorescence, slides were rinsed for 2 min with PBS (x3) and then blocked with 5% BSA for 30 min at room temperature. Slides were then incubated overnight with an Rat monoclonal antibody (eBioM3/38 (M3/38), eBioscience, working dilution 1:150) and an anti-CCL5 polyclonal antibody (Rabbit Polyclonal, #710001, ThermoFisher, working concentration: 10 μg/ml) at 4°C. Slides were then rinsed for 2 min with PBS (x3) and incubated with secondary antibodies (Alexa Fluor 647 donkey anti-rat, #712-605-153, Alexa Fluor 594 donkey anti-rabbit, # 711-585-152, Jackson ImmunoResearch Laboratories, working dilution 1:500) at room temperature for 1 hour in the dark. After washing with PBS (x3), the slides were mounted with ProLong™ Gold Antifade Mountant with DAPI (Invitrogen, P36935), and images were captured using an Olympus DP73 microscope. The atherosclerotic plaque area and DHE fluorescence at the 3 valve leaflets were quantified using ImageJ software (NIH).
Lipoprotein analysis
Cholesterol content in specific lipoprotein particles was analyzed using fast protein liquid chromatography (FPLC) as previously described (Rigotti et al. 1997) with minor modifications. Pooled plasma (50 μl total from five mice) was subjected to FPLC using Superose 6 Increase 10/300 GL columns (Sigma, CAT# GE29-0915-96) with elution buffer (154 mM NaCl, 0.02% sodium azide). Proteins were eluted at 0.6 ml/min. Forty fractions were collected, eluted and total cholesterol in each fraction was determined using Cholesterol fluorometric assay kit (Cayman Chemical, CAT#10007640).
Cloning and production of AAV
Plasmids for AAV8 package (pAdDeltaF6, pAAV2/8, pAAV-TBG-GFP, pAAV-TBG-MCS) were gifts from Dr. Jiandie Lin’s lab at the University of Michigan (Guo et al., 2017). AAV-AGXT expressing human AGXT and AAV-GFP (control) were produced in HEK293T cells in accordance with Addgene’s instructions (https://www.addgene.org/protocols/aav-production-hek293-cells/). We cloned the human AGXT sequence from plasmid #RG212899 (Origene) into the AAV backbone plasmid pAAV-TBG-MCS using NEB Gibson assembly kit (New England Biolabs). The correct sequence and proper insertion of human AGXT were confirmed with Sanger sequencing. The pAAV-TBG-GFP with the same backbone but expressing GFP was used as a control. To produce AAV8-GFP and AAV8-AGXT, 70 μg of AAV shuttle vector, 200 μg Delta F6 helper plasmid and 70 μg AAV2/8- Rep/Cap plasmid were prepared with EndoFree Plasmid Maxi Kit (QIAGEN) and transfected into ten 15-cm plates of HEK293T cells using PEI transfection reagent (Sigma-Aldrich). Approximately 96 hours after transfection, cells were scraped off the plates, washed once with PBS and resuspended in lysis buffer (20 mM Tris, pH 8.0, 150 mM NaCl). After three freeze-thaw cycles between liquid nitrogen and 37°C water bath, 1 M MgCl2 and 25 KU/ml Benzonase were added accordingly for final concentrations of 1 mM and 25 U/ml, respectively. Cell lysates were incubated in 37°C for 15 min and then centrifuged at 4000 rpm at 4°C for 30 min. AAV in the supernatant was purified in a density gradient iodixanol solution by ultracentrifugation (Thermo scientific WX Ultra 80) with a T865 rotor for 160 min at 60,000 rpm at 14°C. AAV was concentrated in PBS with 0.01% Poloxamer 188 (Sigma-Aldrich) using a 100 kDa filter tube (Millipore, Cat# 910096) and the titer was quantified by qPCR. Purified AAV was injected into 7-week old male Apoe−/− mice intraperitoneally in a dosage of 2×1011 viral genomes per mouse and a final volume of 200 μl diluted with saline.
Protein extraction and Western blot
Mouse liver samples were lysed in RIPA buffer supplemented with a protease inhibitor (Thermo Fisher Scientific) and a phosphatase inhibitor (Roche Diagnostics). Proteins were resolved in 10% SDS-PAGE and transferred to nitrocellulose membranes (Bio-Rad). The membranes were incubated with the following primary antibodies at 4°C overnight: mouse monoclonal anti-AGXT (Santa Cruz Biotechnology, working dilution 1:500), rabbit polyclonal anti-CYP7B1(Proteintech # 24889-1-AP, working dilution 1:1000), rabbit polyclonal anti-β-actin antibody (Cell signaling, working dilution 1:2000) and mouse monoclonal anti-β-actin antibody (Cell signaling, #7300, working dilution 1:2000). After TBST washing, membranes were incubated with secondary antibodies (LI-COR Biotechnology, working dilution 1:10000; donkey anti-mouse IRDye 926–68072 and donkey anti-rabbit IRDye 926–32213, working dilution 1:10000) for 1 hour at room temperature. After TBST washing, bands were visualized and quantified using an Odyssey Infrared Imaging System (LI-COR Biosciences, version 2.1).
Total bile acid analysis
The total bile acid in the liver were measured using the Cell Biolabs Total Bile Acid Assay Kit (Cell Biolabs, #STA-631) in accordance with the manufacturer’s instructions. In brief, liver samples were homogenized in cold PBS and centrifuged at 10,000 × g for 10 minutes at 4°C. The levels of total bile acids were measured in the supernatant and normalized to sample weight.
RNA-sequencing and data analysis
RNA-sequencing and data analysis were performed as previously described (Rom et al., 2019; Rom et al., 2020). Total RNA from liver samples of male Agxt+/+/Apoe−/− and Agxt−/−/Apoe−/− was extracted using QIAGEN’s RNeasy kit (QIAGEN). Library preparation and sequencing were performed by the University of Michigan DNA Sequencing Core. RNA was assessed for quality using the TapeStation (Agilent). All samples had RNA integrity numbers (RINs) >8. Samples were prepared using the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (New England Biolabs) with Poly(A) mRNA Magnetic Isolation Module (New England Biolabs) and NEBNext Multiplex Oligos for Illumina Unique dual (New England Biolabs), where 10 ng to 1 μg of total RNA were subjected to mRNA polyA purification. The mRNA was then fragmented and copied into first strand cDNA using reverse transcriptase and dUTP mix. Samples underwent end repair and dA-Tailing step followed by ligation of NEBNext adapters. The products were purified and enriched by PCR to create the final cDNA library. Final libraries were checked for quality and quantity by TapeStation (Agilent) and qPCR using Kapa’s library quantification kit for Illumina Sequencing platforms (Kapa Biosystems, KK4835). Libraries were paired-end sequenced on a NovaSeq 6000 Sequencing System (Illumina). The quality of the raw FASTQ files was checked through FastQC v0.11.8 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Trimmomatic v.0.35 was used to trim the low-quality reads with the parameters: SLIDINGWINDOW:4:20 MINLEN:25 (Bolger et al., 2014). The resulted high-quality reads were then mapped to the mouse reference genome (GRCm38.90) using HISAT2 v.2.1.0.13 (Kim et al., 2015). Gene level quantification was performed using HTSeq-counts v0.6.0 based on the GRCm38.90 genome annotations (Anders et al. 2015). The R package DESeq2 was then used to identify significant differentially expressed genes (DEGs) (Love et al., 2014). Genes with adjusted P value <0.05 were considered as significant DEGs. The up- and down-regulated DEGs were analyzed for significantly enriched KEGG pathways using the clusterProfiler package (Yu et al., 2012). The significance of the enrichment was determined by right-tailed Fisher’s exact test followed by Benjamini-Hochberg multiple testing adjustment. RNA-sequencing data have been deposited in NCBI’s SRA (accession number: PRJNA671451)
RNA preparation and RT-qPCR analysis
Total RNA from mouse liver samples was extracted using Trizol reagent (ThermoFisher Scientific) and purified with QIAGEN’s RNeasy kit (QIAGEN). Total RNA from mouse bone marrow derived macrophages (BMDM) was extracted using QIAGEN’s RNeasy kit (QIAGEN). RNA was reverse-transcribed into cDNA with SuperScript III and random primers (Thermo Fisher Scientific). Specific transcript levels were assessed by a real-time PCR system (Bio-Rad) using iQ SYBR Green Supermix (Bio-Rad) and the ΔΔCt threshold cycle method of normalization. Gene expression levels were normalized to 18S. Primer pairs used for qPCR were obtained from Integrated DNA Technologies and are listed in Supplemental Table S2.
Seahorse extracellular flux analysis
Oxygen consumption rate (OCR) was assessed using a Seahorse XFe96 Analyzer (Agilent). BMDM cells were seeded at 3×104 cells/well in XF96 cell culture microplates (Agilent). The next day, cells were treated with or without 0.75 mM NaOX for 30 min. XFe96 sensor cartridges were hydrated in accordance with the manufacturer’s protocol. Oligomycin, FCCP and rotenone+antimycin A (R/A) were used at final concentrations of 1.5 μM, 1 μM and 0.5 μM, respectively (Agilent, Seahorse XFp Cell Mito Stress Test Kit).
Mitochondrial superoxide analysis
BMDM were seeded at 3×104 cells/well in 96-well black wall and clear bottom plates. The next day, BMDM were treated with or without 0.75 mM NaOX for 30 min. A stock of MitoSOX Red (ThermoFisher Scientific) was prepared in DMSO in accordance with the manufacturer’s instructions. BMDM were stained with 2 μM MitoSOX Red at 37°C for 10 min. After washing with PBS (x2), nuclei were stained with 10 μg/ml Hoechst (ThermoFisher Scientific). Finally, BMDM were visualized using a fluorescent microscope (Olympus, IX71). MitoSOX and Hoechst images were merged and quantified using ImageJ software (NIH).
Mitochondrial DNA copy number
BMDM were seeded at 5×105 cells/well in 6-well plates. The next day, BMDM were treated with 0.75 mM NaOX for 18h. BMDM were lysed using cell lysis buffer (5 mM EDTA, 200 mM NaCl, 100 mM Tris-HCl and 0.2% sodium dodecyl sulfate) with proteinase K and DNA was extracted with phenol chloroform. The mitochondrial DNA (mtDNA) and nuclear DNA (nDNA) copy numbers were quantified by qPCR as previously described (Malik et al 2016) using the following primers: mtDNA sequences: forward: 5’-CTAGAAACCCCGAAACCAAA-3’; reverse: 5’-CCAGCTATCACCAAGCTCGT-3’ targeting Mus musculus mitochondrion, complete genome. nDNA sequences: forward: 5’-ATGGGAAGCCGAACATACTG-3’; reverse: 5’-CAGTCTCAGTGGGGGTGAAT-3’ targeting Mus musculus beta-2 microglobulin.
CCL5 release from BMDM
BMDM were seeded at 5×105 cells/well in 6-well plates. The next day, BMDM were treated with 0.75 mM NaOX for 18h in the absence or presence of MitoTEMPO (10 μM, Sigma-Aldrich). CCL5 released into the culture media was measured using the Mouse/Rat CCL5/RANTES Quantikine ELISA Kit (R&D Systems) in accordance with the manufacturer’s instructions.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data are presented as mean ± SEM throughout the manuscript. Statistical analyses were performed using GraphPad Prism version 8.0. Case-control matching and randomization was performed using IBM SPSS software version 23.0. All data were tested for normality using the Shapiro-Wilk test. If passed, Student’s t test was used to compare two groups or one-way ANOVA followed by Bonferroni post-hoc test for comparisons among >2 groups. Otherwise, nonparametric tests (Mann-Whitney U test or Kruskal-Wallis test followed by Dunn’s post-hoc test) were used. Differences between categorical variables were tested using Fisher’s exact test. Statistical details can be found in the figure and table legends. P<0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
This study was partially supported by the National Institutes of Health grants HL150233 (O.R.), HL137214, HL109946, HL147527, and HL134569 (Y.E.C.), HL138139 (J.Z.), DK106540 (W-Q.L.), HL145176 (Y.F.), HL098435, HL133497, and HL141155 (A.W.O.), the American Heart Association Postdoctoral Fellowship 19POST34380224 (O.R.), and the Michigan-Israel Partnership for Research and Education (O.R., Y.E.C., T.H., and M.A.). We utilized services from the University of Michigan In-Vivo Animal Core supported by grant U2CDK110768.
Footnotes
Declaration of Interests
The authors declare no competing interests.
REFERENCES
- Anders S, Pyl PT, and Huber W (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arafa A, Eshak ES, and Iso H (2020). Oxalates, urinary stones and risk of cardiovascular diseases. Med. Hypotheses 137:109570. [DOI] [PubMed] [Google Scholar]
- Asgharpour A, Cazanave SC, Pacana T, Seneshaw M, Vincent R, Banini BA, Kumar DP, Daita K, Min HK, Mirshahi F, et al. (2016). A diet-induced animal model of non-alcoholic fatty liver disease and hepatocellular cancer. J. Hepatol 65:579–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilbault H, and Haymann JP (2016). Experimental models of renal calcium stones in rodents. World J. Nephrol 5:189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao LC, Honeyman TW, Cooney R, Kennington L, Scheid CR, and Jonassen JA (2004). Mitochondrial dysfunction is a primary event in renal cell oxalate toxicity. Kidney Int 66:1890–1900. [DOI] [PubMed] [Google Scholar]
- Daugherty A, Tall AR, Daemen MJAP, Falk E, Fisher EA, García-Cardeña G, Lusis AJ, Owens AP 3rd, Rosenfeld ME, Virmani R; American Heart Association Council on Arteriosclerosis, Thrombosis and Vascular Biology; and Council on Basic Cardiovascular Sciences. (2017). Recommendation on Design, Execution, and Reporting of Animal Atherosclerosis Studies: A Scientific Statement From the American Heart Association. Circ. Res 121:e53–e79. [DOI] [PubMed] [Google Scholar]
- de Carvalho LSF, Campos AM, and Sposito AC (2018). Proprotein Convertase Subtilisin/Kexin Type 9 (PCSK9) Inhibitors and Incident Type 2 Diabetes: A Systematic Review and Meta-analysis With Over 96,000 Patient-Years. Diabetes Care 41:364–367. [DOI] [PubMed] [Google Scholar]
- Devarajan A (2018). Cross-talk between renal lithogenesis and atherosclerosis: an unveiled link between kidney stone formation and cardiovascular diseases. Clin. Sci. (Lond) 132:615–626. [DOI] [PubMed] [Google Scholar]
- Ding Y, Svingen GF, Pedersen ER, Gregory JF, Ueland PM, Tell GS, and Nygård OK (2015). Plasma Glycine and Risk of Acute Myocardial Infarction in Patients With Suspected Stable Angina Pectoris. J. Am. Heart. Assoc 5:e002621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Chandhoke PS, and Grampsas SA (1999). Role of sex hormones in experimental calcium oxalate nephrolithiasis. J. Am. Soc. Nephrol 10:S376–380. [PubMed] [Google Scholar]
- Fodor K, Wolf J, Erdmann R, Schliebs W, and Wilmanns M (2012). Molecular requirements for peroxisomal targeting of alanine-glyoxylate aminotransferase as an essential determinant in primary hyperoxaluria type 1. PLoS Biol 10:e1001309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaggini M, Carli F, Rosso C, Buzzigoli E, Marietti M, Della Latta V, Ciociaro D, Abate ML, Gambino R, Cassader M, et al. (2018). Altered amino acid concentrations in NAFLD: Impact of obesity and insulin resistance. Hepatology 67:145–158. [DOI] [PubMed] [Google Scholar]
- Grajeda-Iglesias C, Rom O, and Aviram M (2018). Branched-chain amino acids and atherosclerosis: friends or foes? Curr. Opin. Lipidol 29:166–169. [DOI] [PubMed] [Google Scholar]
- Griendling KK, Touyz RM, Zweier JL, Dikalov S, Chilian W, Chen YR, Harrison DG, and Bhatnagar A; American Heart Association Council on Basic Cardiovascular Sciences. (2016). Measurement of Reactive Oxygen Species, Reactive Nitrogen Species, and Redox-Dependent Signaling in the Cardiovascular System: A Scientific Statement From the American Heart Association. Circ. Res 119:e39–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guasch-Ferré M, Hruby A, Toledo E, Clish CB, Martínez-González MA, Salas-Salvadó J, and Hu FB (2016). Metabolomics in Prediabetes and Diabetes: A Systematic Review and Meta-analysis. Diabetes Care 39:833–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Zhang P, Chen Z, Xia H, Li S, Zhang Y, Kobberup S, Zou W, and Lin JD (2017). Hepatic neuregulin 4 signaling defines an endocrine checkpoint for steatosis-to-NASH progression. J. Clin. Invest 127:4449–4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeussler M, Schönig K, Eckert H, Eschstruth A, Mianné J, Renaud JB, Schneider-Maunoury S, Shkumatava A, Teboul L, Kent J, et al. (2016). Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol 17:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Fernaud JR, and Salido E (2010). Differential expression of liver and kidney proteins in a mouse model for primary hyperoxaluria type I. FEBS J 277:4766–4774. [DOI] [PubMed] [Google Scholar]
- Hitzel J, Lee E, Zhang Y, Bibli SI, Li X, Zukunft S, Pflüger B, Hu J, Schürmann C, Vasconez AE, et al. (2018). Oxidized phospholipids regulate amino acid metabolism through MTHFD2 to facilitate nucleotide release in endothelial cells. Nat. Commun 9:2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorge P, García González MJ, Rebollo SG, García S, Bosa F, Laynez I, and De la Rosa A (2013). Myocardial infiltration by oxalate: a rare case of cardiomyopathy by accumulation of oxalate in a 53-year-old woman. J. Am. Coll. Cardiol 62:e525. [DOI] [PubMed] [Google Scholar]
- Khan SR, Glenton PA, and Byer KJ (2006). Modeling of hyperoxaluric calcium oxalate nephrolithiasis: experimental induction of hyperoxaluria by hydroxy-L-proline. Kidney Int 70:914–923. [DOI] [PubMed] [Google Scholar]
- Khan SR, and Glenton PA (2010). Experimental induction of calcium oxalate nephrolithiasis in mice. J. Urol 184:1189–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, and Salzberg SL (2015). HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauf F, Asplin JR, Granja I, Schmidt IM, Moeckel GW, David RJ, Flavell RA, and Aronson PS (2013). NALP3-mediated inflammation is a principal cause of progressive renal failure in oxalate nephropathy. Kidney Int 84:895–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Sun L, Zhang W, Li H, Wang S, Mu H, Zhou Q, Zhang Y, Tang Y, Wang Y, et al. (2018). Association of serum glycine levels with metabolic syndrome in an elderly Chinese population. Nutr. Metab. (Lond) 15:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XM, Salido EC, and Shapiro LJ (1999). The mouse alanine:glyoxylate aminotransferase gene (Agxt1): cloning, expression, and mapping to chromosome 1. Somat. Cell. Mol. Genet 25:67–77. [DOI] [PubMed] [Google Scholar]
- Liang L, Li L, Tian J, Lee SO, Dang Q, Huang CK, Yeh S, Erturk E, Bushinsky D, Chang LS, et al. (2014). Androgen receptor enhances kidney stone-CaOx crystal formation via modulation of oxalate biosynthesis & oxidative stress. Mol. Endocrinol 28:1291–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonardo A, Nascimbeni F, Mantovani A, and Targher G (2018). Hypertension, diabetes, atherosclerosis and NASH: Cause or consequence? J. Hepatol 68:335–352. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madamanchi NR, and Runge MS (2007). Mitochondrial dysfunction in atherosclerosis. Circ. Res 100:460–473. [DOI] [PubMed] [Google Scholar]
- Malik AN, Czajka A, and Cunningham P (2016). Accurate quantification of mouse mitochondrial DNA without co-amplification of nuclear mitochondrial insertion sequences. Mitochondrion 29:59–64. [DOI] [PubMed] [Google Scholar]
- Mardinoglu A, Agren R, Kampf C, Asplund A, Uhlen M, and Nielsen J (2014). Genome-scale metabolic modelling of hepatocytes reveals serine deficiency in patients with non-alcoholic fatty liver disease. Nat. Commun 5:3083. [DOI] [PubMed] [Google Scholar]
- McMartin KE, and Wallace KB (2005). Calcium oxalate monohydrate, a metabolite of ethylene glycol, is toxic for rat renal mitochondrial function. Toxicol. Sci 84:195–200. [DOI] [PubMed] [Google Scholar]
- Moore KJ, Sheedy FJ, and Fisher EA (2013). Macrophages in atherosclerosis: a dynamic balance. Nat. Rev. Immunol 13:709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulay SR, Kulkarni OP, Rupanagudi KV, Migliorini A, Darisipudi MN, Vilaysane A, Muruve D, Shi Y, Munro F, Liapis H, et al. (2013). Calcium oxalate crystals induce renal inflammation by NLRP3-mediated IL-1β secretion. J. Clin. Invest 123:236–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, Haqq AM, Shah SH, Arlotto M, Slentz CA, et al. (2009). A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab 9:311–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen JB, Rom O, Surakka I, Graham SE, Zhou W, Roychowdhury T, Fritsche LG, Gagliano Taliun SA, Sidore C, Liu Y, et al. (2020). Loss-of-function genomic variants highlight potential therapeutic targets for cardiovascular disease. Nat. Commun 11:6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitz K, Lacy M, and Atzler D (2019). Amino Acids and Their Metabolism in Atherosclerosis. Arterioscler. Thromb. Vasc. Biol 39:319–330. [DOI] [PubMed] [Google Scholar]
- Noels H, Weber C, and Koenen RR (2019). Chemokines as Therapeutic Targets in Cardiovascular Disease. Arterioscler. Thromb. Vasc. Biol 39:583–592. [DOI] [PubMed] [Google Scholar]
- Palka P, Duhig E, Carey L, and Galbraith A (2001). Primary oxalosis with cardiac involvement: echocardiographic features of an unusual form of cardiomyopathy. Circulation 103:E122–123. [DOI] [PubMed] [Google Scholar]
- Pandak WM, and Kakiyama G (2019) The acidic pathway of bile acid synthesis: Not just an alternative pathway. Liver Res 3:88–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M, Yarlagadda V, Adedoyin O, Saini V, Assimos DG, Holmes RP, and Mitchell T (2018). Oxalate induces mitochondrial dysfunction and disrupts redox homeostasis in a human monocyte derived cell line. Redox Biol 15:207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recht PA, Tepedino GJ, Siecke NW, Buckley MT, Mandeville JT, Maxfield FR, and Levin RI (2004) Oxalic acid alters intracellular calcium in endothelial cells. Atherosclerosis 173:321–328. [DOI] [PubMed] [Google Scholar]
- Rigotti A, Trigatti BL, Penman M, Rayburn H, Herz J, and Krieger M (1997). A targeted mutation in the murine gene encoding the high density lipoprotein (HDL) receptor scavenger receptor class B type I reveals its key role in HDL metabolism. Proc. Natl. Acad. Sci. U. S. A 94:12610–12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rom O, Xu G, Guo Y, Zhu Y, Wang H, Zhang J, Fan Y, Liang W, Lu H, Liu Y, et al. (2019). Nitro-fatty acids protect against steatosis and fibrosis during development of nonalcoholic fatty liver disease in mice. EBioMedicine 41:62–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rom O, Liu Y, Liu Z, Zhao Y, Wu J, Ghrayeb A, Villacorta L, Fan Y, Chang L, Wang L, et al. (2020). Glycine-based treatment ameliorates NAFLD by modulating fatty acid oxidation, glutathione synthesis, and the gut microbiome. Sci. Transl. Med 12:eaaz2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rom O, Villacorta L, Zhang J, Chen YE, and Aviram M (2018). Emerging therapeutic potential of glycine in cardiometabolic diseases: dual benefits in lipid and glucose metabolism. Curr. Opin. Lipidol 29:428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumsby G and Cochat P (2013). Primary hyperoxaluria. N. Engl. J. Med 369:2163. [DOI] [PubMed] [Google Scholar]
- Salido EC, Li XM, Lu Y, Wang X, Santana A, Roy-Chowdhury N, Torres A, Shapiro LJ, and Roy-Chowdhury J (2006). Alanine-glyoxylate aminotransferase-deficient mice, a model for primary hyperoxaluria that responds to adenoviral gene transfer. Proc. Natl. Acad. Sci. USA 103:18249–18254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salido E, Rodriguez-Pena M, Santana A, Beattie SG, Petry H, and Torres A (2011). Phenotypic correction of a mouse model for primary hyperoxaluria with adeno-associated virus gene transfer. Mol. Ther 19:870–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova M, Hossain N, Afendy A, Perry K, Goodman ZD, Baranova A, and Younossi Z (2010). Hepatic gene expression of Caucasian and African-American patients with obesity-related non-alcoholic fatty liver disease. Obes. Surg 20:640–650. [DOI] [PubMed] [Google Scholar]
- Swerdlow DI, Preiss D, Kuchenbaecker KB, Holmes MV, Engmann JE, Shah T, Sofat R, Stender S, Johnson PC, Scott RA, et al. (2015). HMG-coenzyme A reductase inhibition, type 2 diabetes, and bodyweight: evidence from genetic analysis and randomised trials. Lancet 385:351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzou DT, Taguchi K, Chi T, and Stoller ML (2016). Animal models of urinary stone disease. Int. J. Surg 36:596–606. [DOI] [PubMed] [Google Scholar]
- Vendrov AE, Hakim ZS, Madamanchi NR, Rojas M, Madamanchi C, and Runge MS (2007). Atherosclerosis is attenuated by limiting superoxide generation in both macrophages and vessel wall cells. Arterioscler. Thromb. Vasc. Biol 27:2714–2721. [DOI] [PubMed] [Google Scholar]
- Villacorta L, Minarrieta L, Salvatore SR, Khoo NK, Rom O, Gao Z, Berman RC, Jobbagy S, Li L, Woodcock SR, et al. (2018). In situ generation, metabolism and immunomodulatory signaling actions of nitro-conjugated linoleic acid in a murine model of inflammation. Redox Biol 15:522–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virani SS, Alonso A, Benjamin EJ, Bittencourt MS, Callaway CW, Carson AP, Chamberlain AM, Chang AR, Cheng S, Delling FN, et al. (2020). American Heart Association Council on Epidemiology and Prevention Statistics Committee and Stroke Statistics Subcommittee. Circulation 141:e139–e596. [DOI] [PubMed] [Google Scholar]
- Wang W, Wu Z, Dai Z, Yang Y, Wang J, and Wu G (2013). Glycine metabolism in animals and humans: implications for nutrition and health. Amino Acids 45:463–77. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang GZ, Rabinovitch PS, and Tabas I (2014). Macrophage mitochondrial oxidative stress promotes atherosclerosis and nuclear factor-κB-mediated inflammation in macrophages. Circ. Res 114:421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittemans LBL, Lotta LA, Oliver-Williams C, Stewart ID, Surendran P, Karthikeyan S, Day FR, Koulman A, Imamura F, Zeng L, et al. (2019). Assessing the causal association of glycine with risk of cardio-metabolic diseases. Nat. Commun 10:1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W, Zhao X, Villacorta L, Rom O, Garcia-Barrio MT, Guo Y, Fan Y, Zhu T, Zhang J, Zeng R, et al. (2018). Brown Adipocyte-Specific PPARγ (Peroxisome Proliferator-Activated Receptor γ) Deletion Impairs Perivascular Adipose Tissue Development and Enhances Atherosclerosis in Mice. Arterioscler. Thromb. Vasc. Biol 38:1738–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara H, Yamaguchi S, and Yachiku S (1999). Effect of sex hormones on oxalate-synthesizing enzymes in male and female rat livers. J. Urol 161:668–673. [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, and He QY (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16:284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaric BL, Radovanovic JN, Gluvic Z, Stewart AJ, Essack M, Motwalli O, Gojobori T, and Isenovic ER (2020). Atherosclerosis Linked to Aberrant Amino Acid Metabolism and Immunosuppressive Amino Acid Catabolizing Enzymes. Front. Immunol 11:551758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Zhao Z, Chou FJ, Zuo L, Liu T, Bushinsky D, Chang C, Zeng G, and Yeh S (2019). The Protective Roles of Estrogen Receptor β in Renal Calcium Oxalate Crystal Formation via Reducing the Liver Oxalate Biosynthesis and Renal Oxidative Stress-Mediated Cell Injury. Oxid. Med. Cell. Longev 5305014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-sequencing data have been deposited at NCBI’s SRA and are publicly available as of the date of publication. Accession number is listed in the key resources table. All raw data have been deposited at Mendeley and are publicly available as of the date of publication (doi: 10.17632/n8nkdgd258.1). This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-mouse β-Actin antibody | Cell Signaling Technology | Cat#4967; RRID: AB_330288 |
| Mouse anti-mouse AGXT antibody | Santa Cruz Biotechnology | Cat# SC-517388 |
| Rabbit anti-mouse CYP7B1 antibody | Proteintech | CAT# 24889–1-AP RRID: AB_2879780 |
| Mouse anti-mouse B-ACTIN antibody | Cell Signaling Technology | CAT#3700 RRID: AB_2242334 |
| Rat anti-Galectin 3 antibody | Thermo Fisher Scientific | Cat#13-5301-81; RRID: AB_837112 |
| CCL5 (RANTES) Recombinant Polyclonal Antibody | ThermoFisher Scientific | Cat# 710001, RRID: AB_2532515 |
| 800CW Donkey anti-mouse IRDye | LI-COR Biosciences | Cat# 926–32212; RRID: AB_621847 |
| 680RD Donkey anti-rabbit IRDye | LI-COR Biosciences | Cat# 926–68073; RRID: AB_10954442 |
| 800CW Donkey anti-rabbit IRDye | LI-COR Biotechnology | CAT# 926–32213 RRID: AB_621848 |
| 680RD Donkey anti-mouse IRDye | LI-COR Biotechnology | CAT# 926–68072 RRID: AB_10953628 |
| Goat anti-Rat IgG (H+L) Secondary Antibody, HRP | ThermoFisher Scientific | Cat# 31470, RRID: AB_228356 |
| Alexa Fluor 647 donkey anti-rat | Jackson ImmunoResearch Laboratories | Cat# 712-605-153; RRID: AB_2340694 |
| Alexa Fluor 594 donkey anti-rabbit | Jackson ImmunoResearch Laboratories | Cat#711-585-152, RRID: AB_2340621 |
| Bacterial and Virus Strains | ||
| Stable Competent E. coli | New England Biolabs | Cat # C3040H |
| AAV-TBG-GFP | This paper | N/A |
| AAV-TBG-AGXT | This paper | N/A |
| Biological Samples | ||
| Serum from patients with or without sCAD | This paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Polyethylenimine | Sigma-Aldrich | Cat# 408727 |
| Dihydroethidium | Sigma-Aldrich | Cat# D7008 |
| MitoTEMPO | Sigma-Aldrich | Cat# SML0737 |
| Sodium oxalate | Sigma-Aldrich | Cat# 223433 |
| Oil red O | Sigma-Aldrich | Cat# O0625 |
| Sodium azide | Sigma-Aldrich | CAT#S2002 |
| Proteinase K | Sigma-Aldrich | CAT# 1245680500 |
| Trans-4-Hydroxy-L-proline | ACROS Organics | CAT#121785000 |
| UltraPure™ Phenol:Chloroform:Isoamyl Alcohol | ThermoFisher Scientific | CAT#15593–031 |
| Poloxamer 188 | Sigma-Aldrich | Cat# P5556 |
| Harris hematoxylin | ThermoFisher Scientific | Cat# 842 |
| Eosin Y | ThermoFisher Scientific | Cat# 832 |
| Clarifier Bluing reagent | ThermoFisher Scientific | Cat# 7401 |
| Bluing reagent | ThermoFisher Scientific | Cat# 7301 |
| MitoSOX Red | ThermoFisher Scientific | Cat# M36008 |
| Hoechst | ThermoFisher Scientific | Cat# H3570 |
| Trizol reagent | ThermoFisher Scientific | Cat# 5596026 |
| Canonical amino acid mix unlabeled standard | Cambridge Isotopes Laboratory | Cat# MSK-CAA-US-1 |
| Canonical amino acid mix | Cambridge Isotopes Laboratory | Cat# MSK-CAA-1 |
| O.C.T. Compound | Tissue-Tek | Cat# 4583 |
| iQ SYBR Green Supermix | Bio-Rad | Cat# 1708880 |
| Critical Commercial Assays | ||
| Wako Cholesterol E | FUJIFILM medical system | Cat# 999–02601 |
| RNeasy Maxi kit | QIAGEN | Cat# 75162 |
| EndoFree Plasmid Maxi Kit | QIAGEN | Cat# 12362 |
| Oxalate assay kit | Abcam | Cat# ab196990 |
| Mouse CCL2/JE/MCP-1 Quantikine ELISA Kit | R&D Systems | Cat# MJE00B |
| Mouse/Rat CCL5/RANTES Quantikine ELISA Kit | R&D Systems | Cat# MMR00 |
| Glucose test strips | Contour Next | Cat# NDC: 0193-7308-50 |
| Seahorse XFp Cell Mito Stress Test Kit | Agilent Technologies | Cat# 103010–100 |
| Gibson assembly kit | New England Biolabs | Cat# E5510 |
| NEBNext Ultra II Directional RNA Library Prep Kit for Illumina | New England Biolabs | CAT# E7760L |
| Poly(A) mRNA Magnetic Isolation Module | New England Biolabs | Cat# E7490L |
| NEBNext Multiplex Oligos for Illumina Unique dual | New England Biolabs | Cat# E6440L |
| Protease inhibitor | Thermo Fisher Scientific | Cat# A32965 |
| Phosphatase inhibitor | Roche Diagnostics | Cat# 04906845001 |
| SuperScript™ III First-Strand Synthesis System | ThermoFisher Scientific | Cat# 18080051 |
| CCK8 assay kit | Sigma-Aldrich | Cat# 96992 |
| SignalStain® DAB Substrate Kit | Cell Signaling Techonology | CAT#8059 |
| Total Bile Acid Assays Kit | Cell Biolabs, INC | CAT#STA-631 |
| Cholesterol fluorometric assay kit | Cayman Chemical | CAT#10007640 |
| LabAssay™ Triglyceride kit | FUJIFILM medical system | CAT# 290–63701 |
| Deposited Data | ||
| RNA-sequencing | This paper | NCBI’s SRA (accession number: PRJNA671451) |
| Raw data | This paper | Mendeley Data (doi:10.17632/n8nkdgd258.1) |
| Experimental Models: Cell Lines | ||
| HEK293T | ATCC | CRL-3216; RRID: CVCL_0063 |
| L929 cells | ATCC | CRL-1; RRID: CVCL_0462 |
| Bone marrow derived-macrophages | This paper | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | JAX: 000664; RRID: IMSR_JAX:000664 |
| Mouse: Apoe−/− | The Jackson Laboratory | JAX: 002052; RRID: IMSR_JAX:002052 |
| Mouse: Agxt−/− | Rom et al., 2020 | N/A |
| Mouse: Agxt+/+/Apoe−/− | This paper | N/A |
| Mouse: Agxt−/−/Apoe−/− | This paper | N/A |
| Oligonucleotides | ||
| Guide-RNA targeting the mouse Agxt exon1: 5’-GGGTCCGGGGCCCTCCAACC-3’ | Integrated DNA Technologies | N/A |
| Primer sequences for Agxt genotyping: forward: 5’-ACACCTCCACTGTCCTGTCC-3’ | Integrated DNA Technologies | N/A |
| Primer sequences for Agxt genotyping: reverse: 5’-GGTCAGATCTGCCTGCTACC-3’ | Integrated DNA Technologies | N/A |
| Primer for Sanger sequencing: 5’-GCAGAGCTAGCTGGGAAATG-3’ | Integrated DNA Technologies | N/A |
| Primer sequences for Apoe genotyping: forward: 5′-GCCTAGCCGAGGGAGAGCCG-3′ | Integrated DNA Technologies | N/A |
| Primer sequences for Apoe genotyping: reverse 1: 5′-TGTGACTTGGGAGCTCTGCAGC-3′ | Integrated DNA Technologies | N/A |
| Primer sequences for Apoe genotyping: reverse 2: 5′-GCCGCCCCGACTGCATCT-3’ | Integrated DNA Technologies | N/A |
| Primer sequences for qPCR, see Table S2 | Integrated DNA Technologies | N/A |
| Recombinant DNA | ||
| pAdDeltaF6 | Jiandie Lin’s lab, University of Michigan; Guo et al., 2017 | N/A |
| pAAV2/8 | Jiandie Lin’s lab, University of Michigan; Guo et al., 2017 | N/A |
| pAAV-TBG-GFP | Jiandie Lin’s lab, University of Michigan; Guo et al., 2017 | N/A |
| pAAV-TBG-MCS | Jiandie Lin’s lab, University of Michigan; Guo et al., 2017 | N/A |
| AGXT Human Tagged ORF Clone | Origene | CAT# RG212899 |
| Software and Algorithms | ||
| ImageJ | National Institutes of Health | RRID:SCR_003070 https://imagej.nih.gov/ij/download.html |
| GraphPad Prism | GraphPad Software | Version 8.0 RRID:SCR_002798 https://www.graphpad.com/scientific-software/prism/ |
| R statistics | Hosted by Vienna, University of Economics and Business | http://www.R-project.org/. |
| RStudio | RStudio Team (2020). | http://www.rstudio.com/.RRID:SCR_000432 |
| IBM SPSS software | IBM Corporation | Version 23.0; RRID:SCR_019096 https://www.ibm.com/analytics/spss-statistics-software |
| FastQC | Babraham Bioinformatics | Version 0.11.8 RRID:SCR_014583 https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| Trimmomatic | Bolger et al., 2014 | Version 0.35 RRID:SCR_011848 http://www.usadellab.org/cms/?page=trimmomatic |
| HISAT2 | Kim et al., 2015 | Version 2.1.0.13; RRID:SCR_015530 http://daehwankimlab.github.io/hisat2/download/ |
| HTSeq | Anders et al., 2015 | Version 0.6.0; RRID:SCR_005514 https://htseq.readthedocs.io/en/master/install.html |
| Analyst software | Applied Biosystems | Version 1.6.2; RRID:SCR_015785 https://sciex.com/products/software/analyst-software |
| LI-COR Image studio | LI-COR Biosciences | Version 2.1; RRID:SCR_015795 https://www.licor.com/bio/image-studio-lite/download |
| SPOT Basic software | SPOT imaging | RRID:SCR_014313 https://www.spotimaging.com/software/spot-basic/ |
| Other | ||
| Mouse standard diet | LabDiet | Cat# 5L0D |
| Mouse Western diet | Envigo | Cat# TD.88137 |
| Mouse Western diet supplemented with hydroxy-L-proline (HLP) | Envigo | CAT# TD.190842 |
| Dulbecco’s Modified Eagle Medium | Gibco | Cat# 11965092 |
| Iscove’s Modified Dulbecco’s Media | Gibco | Cat# 12440053 |
| Agilent Poroshell 120 EC-C18 column | Agilent Technologies | Cat# 699775–902 |
| Superose 6 Increase 10/300 GL columns | Sigma-Aldrich | CAT# GE29-0915-96 |


