Abstract
Vancomycin-resistant Enterococcus faecium (VRE) is a major cause of nosocomial infections. A new study by McKenney et al. (Cell Host Microbe 2019;25:695-705.e5) reports that VRE undergo a morphotype switch in response to lithocholic acid (LCA) to facilitate gastrointestinal (GI) tract colonization. This metabolic cue is a potential target to decrease VRE colonization and subsequent transmission of antibiotic resistance.
Vancomycin-resistant Enterococcus faecium (VRE) are serious nosocomial pathogens with few therapeutic options. Patients colonized by VRE in the gastrointestinal (GI) tract appear to have an increased risk of bloodstream infections and subsequently worse clinical outcomes [1]. Controlling VRE colonization is a target priority to reduce serious invasive VRE infections and potentially prevent the spread of antibiotic resistance.
Bile Acids Affect the Ability of VRE to Colonize the Host GI Tract
Understanding how pathogens establish GI tract colonization is an important step in targeting the cycle of transmission. McKenney and colleagues have observed a morphological switch in enterococci (from a diplococcal form to long chains) when exposed to certain bile acids [2]. They postulate that this change may contribute to better adherence to the mucosal surface, favoring colonization [3]. The ‘chaining’ morphology has been proposed to promote recognition by the host innate immune system in Streptococcus pneumoniae [4], suggesting that the morphological switch must occur only when inside the intestinal lumen. McKenney et al. show that chaining is associated with the induction of biofilm formation, and occurs upon exposure to the secondary bile acid, lithocholic acid (LCA, a compound found almost exclusively in the GI tract [5]. Thus, LCA (and bile acids in general) may serve as an important physiological cue that allows VRE to colonize the host environment.
The authors also observed an increase in enterococcal chain length upon treatment with both LCA-supplemented liquid media and in an ex vivo model involving exposure of VRE to mouse cecal extracts [2]. Morphologically, LCA-treated cells had typical septa with appropriate localization of peptidoglycan synthesis. However, bacteria exposed to LCA had increased uptake of fluorescently labeled peptidoglycan precursors at sites of eventual cell separation, and were noted to be deficient in spontaneous autolysis, suggesting alterations in cell wall metabolism. Both chaining and biofilm formation were shown to be dependent on the autolysin AtlA and were modulated by cation concentration in the microenvironment, even in the presence of LCA. Using fluorescence in situ hybridization in a mouse GI tract colonization model, this phenotype was found to persist in vivo, with aggregation of VRE in the cecum of LCA-fed mice [2]. These results link VRE exposure to LCA to altered cell division, biofilm formation, and host colonization.
Morphotype Changes and Antimicrobial Resistance
The cell morphology observed with the VRE morphotype switch is reminiscent of that seen in enterococci resistant to daptomycin, a lipopeptide antibiotic similar to cationic antimicrobial peptides (CAMPs) of the innate immune system. Daptomycin relies on divalent cations, specifically calcium, to insert into the bacterial membrane at the anionic phospholipid-rich division septa, displacing membrane-associated proteins and leading to cell death. Resistance to this antibiotic has been associated with alterations in membrane architecture and cell surface charge modulated via two primary stress response pathways, the LiaFSR and YycFG systems [6].
To determine the genetic pathways behind the morphotype switch, McKenney and colleagues used a modified serial passage experiment. By removing supernatant from the top of a static culture treated with LCA, the authors enriched for bacteria that had lost the ability to form long chains (which would sink to the bottom of the culture tube). They isolated strains locked in the diplococcal morphotype and used whole-genome sequencing to identify mutations shared across the strains. Interestingly, mutations in yycG and liaR were common, and these strains displayed an increased susceptibility to daptomycin [2].
LiaFSR is a three-component cell envelope stress response system that mediates resistance to membrane stressors, including daptomycin, in enterococci [6]. Inactivation of the system by deletion of liaR, encoding the response regulator, results in hypersusceptibility to daptomycin and CAMPs [6]. Substitutions in LiaR and LiaS (the histidine kinase) observed in clinical enterococcal strains are associated with activation of LiaFSR and daptomycin resistance [6]. Furthermore, activation of the LiaFSR system appears to have an important role in protecting enterococci from the innate immune response [7]. The increased susceptibility to daptomycin associated with mutations observed in this study suggests that the diplococcal morphotype leads to downregulation of the LiaFSR system.
YycFG (also known as WalKR) is an essential system conserved in Firmicutes, involved in mediating the balance between peptidoglycan synthesis and cell wall autolysis [8]. McKenney and colleagues identified amino acid substitutions A88R and R262P in the histidine kinase YycG, implicating this system in the chaining phenotype and biofilm formation affecting GI tract colonization [2]. Notably, substitutions in YycG (S333L) have been also found in daptomycin-resistant E. faecium and changes in the Yyc system in conjunction with changes in phospholipid metabolism proteins are sufficient for daptomycin resistance in enterococci [9].
While the authors identified common genetic changes across several morphotype classes, it is unclear precisely what role each has in this complex phenotype. The increased susceptibility to daptomycin suggests that the observed mutations in liaR result in impaired function of the LiaFSR system. However, further studies including allelic exchange would be necessary to confirm this hypothesis. In addition, bile acids are one of many stimuli that enterococci would be likely to encounter in the intestinal environment. Thus, the regulation of a morphotype switch is likely to be a nuanced response incorporating many inputs to establish host colonization. Whether this change impacts susceptibility to additional classes of antibiotic or host antimicrobial peptide remains to be determined.
Implications for VRE Treatment
McKenney and colleagues also investigated the adaptability of enterococci in maintaining a survival advantage in the host GI tract and drew a link between the membrane stress response, changes in cell morphology, biofilm formation, and antimicrobial resistance (Figure 1) [2]. This malleability underlies the clinical challenge of treating serious VRE infections. While daptomycin remains a preferred agent for these infections, the findings that the colonization phenotype promotes expression of pathways linked to daptomycin resistance is concerning. However, leveraging an understanding of the molecular basis of this transition may lead to a personalized approach to the treatment of VRE.
Figure 1. Recognition of Lithocholic Acid (LCA) in the Gastrointestinal (GI) Tract Triggers Morphotype Switch in Vancomycin-Resistant Enterococcus faecium (VRE).
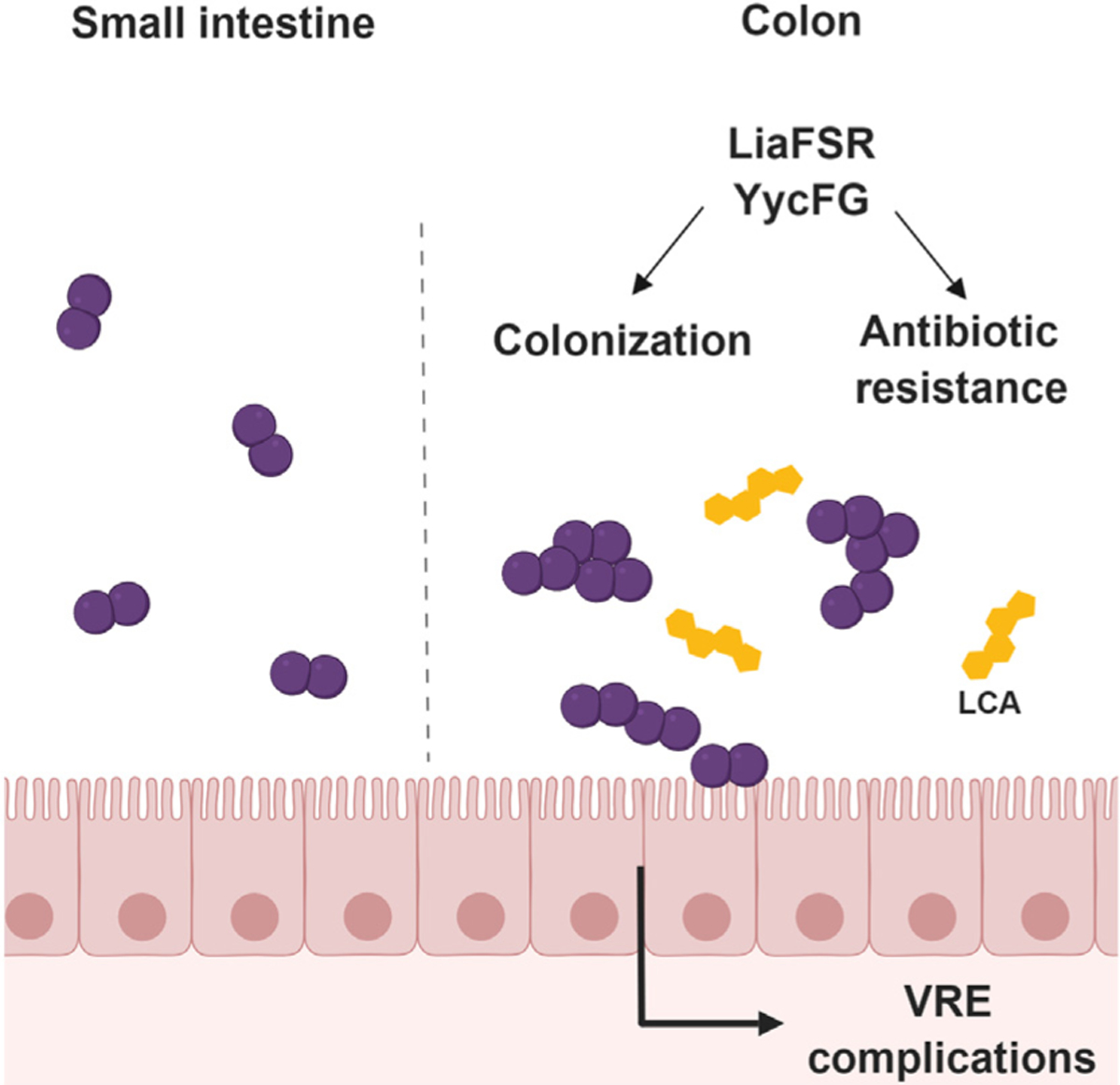
LCA is a secondary bile acid present in the large intestines of mice and humans. Exposure of VRE to LCA causes a morphological switch from a diplococcal form to long chains to aid colonization. This process is associated with changes in the LiaFSR and YycFG systems, resulting in not only increased biofilm formation, but potentially also alterations in antibiotic susceptibility.
Mutations were found in genes encoding proteins involved in both the YycFGH and LiaFSR cell envelope stress systems under LCA selective pressure [2]. This suggests that these regulatory systems help modulate colonization of the GI tract, opening the door to a potential therapeutic target. By disarming the bacterial stress response, it may be possible to decrease the burden of colonization, restore the activity of antibiotics, and potentiate the effectiveness of the host innate immune response. Fecal microbiota transplant (FMT) has been shown to be effective for clearing VRE colonization from mouse GI tracts [10]. McKenney and colleagues demonstrated that VRE with an impaired morphotype switch were more quickly cleared by FMT compared with parental strains that retained an ability to form chains [2]. Additionally, part of the innate immunity in the GI tract involves secretion of CAMPs from epithelial cells. For example, RegIIIγ secreted in response to commensal Gram-negative bacteria is able to inhibit colonization of VRE [11]. Thus, an antiadaptation agent would provide a two-fold benefit: impairing the chaining phenotype involved in adhering to host tissue and potentiating CAMPs and the host immune response. Whether this strategy would provide benefits in invasive infections, such as infective endocarditis, deserves further study.
Acknowledgments
C.A.A. is supported by National Institutes of Health (NIH)/National Institutes of Allergy and Infectious Diseases (NIAID) grants K24 AI121296, R01 AI134637, and R21 AI143229, and University of Texas System STARS Award and UTHealth Presidential Award. W.R.M. is supported by a K08 AI135093 award from NIH/NIAID.
Disclaimer Statement
W.R.M. has received grants from Merck and Entasis Therapeutics, and honoraria from Achaogen and Shionogi. C.A.A. has received grants from Merck, MeMed Diagnostics, and Entasis Therapeutics.
References
- 1.Jung E et al. (2014) Vancomycin-resistant Enterococcus colonization in the intensive care unit : clinical outcomes and attributable costs of hospitalization. Am. J. Infect. Control 42, 1062–1066 [DOI] [PubMed] [Google Scholar]
- 2.McKenney PT et al. (2019) Intestinal bile acids induce a morphotype switch in vancomycin-resistant Enterococcus that facilitates intestinal colonization. Cell Host Microbe 25, 695–705.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodriguez JL et al. (2012) Increased chain length promotes pneumococcal adherence and colonization. Infect. Immun 80, 3454–3459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dalia AB and Weiser JN (2011) Minimization of bacterial size allows for complement evasion and is overcome by the agglutinating effect of antibody. Cell Host Microbe 10, 486–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hofmann AF et al. (2010) Bile salts of vertebrates: structural variation and possible evolutionary significance. J. Lipid Res 51, 226–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tran TT et al. (2015) Mechanisms of drug resistance: daptomycin resistance. Ann. N. Y. Acad. Sci 1354, 32–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rincon S et al. (2019) Disrupting membrane adaptation restores in vivo efficacy of antibiotics against multidrug-resistant enterococci and potentiates killing by human neutrophils. J. Infect. Dis Published online April 2, 2019. 10.1093/infdis/jiz131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Türck M and Bierbaum G (2012) Purification and activity testing of the full-length YycFGHI proteins of Staphylococcus aureus. PLoS One 7, e30403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tran TT et al. (2013) Whole-genome analysis of a daptomycin-susceptible Enterococcus faecium strain and its daptomycin-resistant variant arising during therapy. Antimicrob. Agents Chemother 57, 261–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ubeda C et al. (2013) Intestinal microbiota containing Barnesiella species cures vancomycin-resistant Enterococcus faecium colonization. Infect. Immun 81, 965–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubin K and Pamer EG (2014) Enterococci and their interactions with the intestinal microbiome. Microbiol. Spectr Published online November 17, 2014. 10.1128/microbiolspec.BAD-0014-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]


